Abstract
Monkeypox virus (MPV) causes a human disease which resembles smallpox but with a lower person-to-person transmission rate. To determine the genetic relationship between the orthopoxviruses causing these two diseases, we sequenced the 197-kb genome of MPV isolated from a patient during a large human monkeypox outbreak in Zaire in 1996. The nucleotide sequence within the central region of the MPV genome, which encodes essential enzymes and structural proteins, was 96.3% identical with that of variola (smallpox) virus (VAR). In contrast, there were considerable differences between MPV and VAR in the regions encoding virulence and host-range factors near the ends of the genome. Our data indicate that MPV is not the direct ancestor of VAR and is unlikely to naturally acquire all properties of VAR.
Keywords: Monkeypox virus, Smallpox virus, Genome, Virulence factor, Ankyrin-like protein
Abbreviations: MPV, monkeypox virus; MPV-ZAI, MPV-CNG, monkeypox virus strains Zaire-96-I-16 and Congo-8; VAR, variola virus; VAR-IND, VAR-BSH, VAR-GAR, variola virus strains India-1967, Bangladesh-1975, Garcia-1966; VAC, vaccinia virus; VAC-COP, vaccinia virus strain Copenhagen; CPV, cowpox virus; CPV-GRI, cowpox virus strain GRI-90; PKR, double-stranded RNA-dependent protein kinase; ORF, open reading frame; IFN, interferon; IL, interleukin; SPI, serine protease inhibitor
1. Introduction
Smallpox, an epidemic disease of humans with a high mortality rate caused by variola virus (VAR), a member of the genus Orthopoxvirus of the family Poxviridae, is the only example of an infectious disease that was eradicated by the international community under the aegis of the World Health Organization [1], [2]. Two factors contributed to the success of this unprecedented campaign: the exclusively human host range of VAR and the availability of a highly effective and inexpensive live vaccine derived from the closely related vaccinia virus (VAC). Mass vaccination of populations together with a thorough epidemiological surveillance led to the global eradication of smallpox by 1977 [1]. Following smallpox eradication, vaccination was terminated throughout the world, resulting in a growing population that is completely unprotected from VAR as well as related orthopoxviruses. This situation provides an opportunity for zoonotic orthopoxviruses, such as monkeypox (MPV), cowpox (CPV), and buffalopox, to spread in the human population and possibly undergo adaptive mutations. MPV is of the highest medical concern because it causes a human disease similar to smallpox in its clinical manifestations and mortality rate [3]. The first case of human monkeypox was discovered in Congo in 1970 [4]. Investigations from 1981 to 1986 demonstrated that human monkeypox is a sporadic disease caused by MPV transmission from animals to humans in the tropical rainforest regions of Central and Western Africa. Secondary human-to-human spread accounted for about 28% of the cases; tertiary and quaternary chains of transmission were rare [3]. From 1970 to 1995, 388 of the 418 recorded cases of human monkeypox occurred in Zaire (now the Democratic Republic of the Congo, DRC) [5]. The number of suspected cases of human monkeypox in DRC exceeded 500 in 1996 and 1997, and the DRC health authorities reported several hundred new cases in January 1999 [5], [6]. Although most of these cases have not been confirmed by laboratory testing, there is concern that human monkeypox is increasing [7]. In addition, the clinical similarities of human monkeypox and smallpox raised questions regarding the genetic relationships of the viruses and whether MPV could evolve into a VAR-like virus with a high frequency of human-to-human transmission [5], [7], [8]. Although some preliminary genetic information indicating differences between VAR, MPV and other orthopoxviruses was obtained by comparative analysis of restriction endonuclease site maps and short DNA sequences [6], [9], [10], [11], [12], less than 7% of the MPV genome sequence was in GenBank prior to this study. To provide a definitive comparison of MPV and VAR, the DNA of MPV strain Zaire-96-I-16 (MPV-ZAI), isolated during the 1996 outbreak of human monkeypox in Zaire [6] was sequenced. Comparative analysis has shown that the central genomic regions of MPV and VAR, which encode essential enzymes and structural proteins, are nearly identical whereas the terminal genomic regions, which encode virulence and host-range factors, are substantially different. Mutations of two interferon (IFN) resistance genes as well as the presence of an interleukin-1β (IL-1β) inhibitor in MPV could contribute to the different properties of the two viruses and may also limit the usefulness of monkeypox as a model for smallpox. While the extensive genetic differences are reassuring and establish that MPV was not a direct ancestor of VAR, they do not rule out future adaptation of MPV to humans, making continued monitoring of human monkeypox of great importance.
2. Materials and methods
2.1. MPV isolation and DNA sequencing
MPV-ZAI was isolated in rhesus monkey kidney cell culture LLC-MK2 from a scab sample of a monkeypox patient residing in the Sankuru subregion, Kasdai Oriental, Zaire during a human monkeypox outbreak in 1996. Viral DNA, after the second passage in LLC-MK2 cells, was extracted from the cytoplasm [13]. The fragments of MPV-ZAI DNA were cloned in plasmids pMGC20 and pZEro-2.1. The viral genome was sequenced using the Maxam-Gilbert technique as described earlier [14] and by primer walking with an automatic sequencer (Applied Biosystems, model ABI PRISM 310 DNA Analyzer). Each base was determined one or more times from each strand, with no discrepancies. The genome sequence of MPV-ZAI was deposited with GenBank under accession number AF380138. In addition, the open reading frames (ORFs) that differed most significantly from those of VAR and other orthopoxviruses [15], [16], [17], [18], [19] were also sequenced using the MPV strain Congo-8 (MPV-CNG) [4] DNA to determine whether they were species- or isolate-specific.
2.2. Analysis of the sequence data
Sequences were analyzed using software developed at the State Research Center of Virology and Biotechnology ‘Vector’, Koltsovo, Russia [20]. Protein homology searches were done with BLAST analyses [21] using NCBI internet resources. Sequences alignments were prepared using Clustal W [22]. Phylogenetic analyses were performed with the neighbor-joining (NJ) method [23] implemented in MEGA software [24]. The reliability of the phylogenetic relationship was statistically evaluated from 1000 bootstrap replicates.
3. Results and discussion
The 196 858 bp of MPV-ZAI DNA, comprising the entire genome contains 190 largely non-overlapping ORFs of ≥60 amino acid residues as well as structural features and a GC content of 31.1% similar to other orthopoxviruses. The sequence of MPV genome was compared to that of the India-1967 (VAR-IND) [15] and Bangladesh-1975 (VAR-BSH) [25] strains of VAR major, which caused severe disease and with genomic sequence of the variola minor alastrim Garcia-1966 (VAR-GAR) strain, which caused relatively mild disease [18]. For simplicity, as the genomes of VAR-IND and VAR-BSH are very similar [16], most of our references are to VAR-IND. The coding sequence of MPV-ZAI DNA, limited by the utmost left and utmost right ORFs, is 195 118 bp long, whereas those of VAR-IND and VAR-GAR are 184 151 and 185 846 bp, respectively. The greater length of the MPV DNA results mainly from duplication of the four left terminal ORFs on the right side of the genome as a part of terminal inverted repeat (TIR), whereas the VAR genomes lack the repetition of these ORFs and display very short gene-free TIRs (Fig. 1 ). The degrees of identity between the overlapping genomic nucleotide sequences of MPV-ZAI with VAR-IND and VAR-GAR are 84.6 and 84.5%, respectively.
Fig. 1.
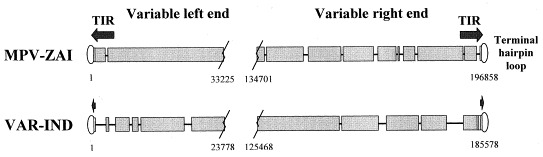
Schematic representation of the terminal species-specific variable genomic regions of MPV-ZAI and VAR-IND. TIRs are designated with arrows, and regions of short tandem terminal repeats are designated with rectangles. Coinciding sequences are shown by wide black blocks; deletions in one genome relative to others are shown as lines. Borders of the variable genomic regions are marked by nucleotide numbers corresponding to their positions in the genomes.
The central genomic regions of orthopoxviruses contain mostly highly conserved essential genes [16], [18]. This region of MPV-ZAI DNA, which is delimited by ORFs C10L and A25R, comprises 101 466 bp with an overall 96.3% identity to the corresponding part of the VAR-IND genome. The virion proteins encoded in this region of MPV-ZAI are 91.7–99.2% identical in amino acid sequences to those of VAR-IND. For comparison, the corresponding ORFs of VAR-GAR are 98.7–100% identical to the isologs of VAR-IND. In contrast to the central region, the two terminal areas of the genomes of MPV-ZAI and VAR-IND exhibit considerable variation caused by deletions (Fig. 1) and ORF truncations (Table 1, Table 2 ) in one DNA relative to the other. The variable terminal regions of orthopoxvirus genomes contain the majority of the virulence and host-range genes that were identified earlier for VAC and CPV [17], [19]. The amino acid sequences of the putative virulence and immunomodulatory factors common to MPV-ZAI and VAR-IND were 83.5–93.6% identical, compared to a 97.3–100% identity between the corresponding ORFs of VAR-GAR and VAR-IND (Table 1). Notably, MPV-ZAI and MPV-CNG have mutations that affect translation of two IFN resistance genes encoding intracellular proteins that are intact in VAR and other orthopoxviruses. One of these (C3L in VAR-IND, Table 1), a homolog of eukaryotic translation initiation factor 2α (eIF-2α), inhibits the antiviral activity of double-stranded RNA-dependent protein kinase (PKR) by acting as a decoy [26]. Unlike VAR, the MPV (strains ZAI and CNG) genome does not code this protein due to multiple mutations in the corresponding gene. It has been shown that a mutant VAC lacking this gene exhibited IFN sensitivity, and virus yields were reduced approximately 100-fold compared to the parental virus [27]. The other IFN resistance gene (E3L in VAR-IND, Table 1), present in VAR and other orthopoxviruses, is expressed from the first or second methionine as a long or short form, respectively [28] (Fig. 2A ). The long form contains an N-terminal domain that in VAC mediates binding to Z-DNA, nuclear localization, and PKR interaction and is required for virulence [29]. Both the short and long forms contain the C-terminal domain, which binds double-stranded RNA, inhibits activation of IFN-induced PKR and 2-5A-synthetase and is required for IFN resistance and host range of VAC [30], [31]. Interestingly, the N-terminal Z-DNA binding motif is also present in the cellular dsRNA-binding protein adenosine deaminase [32]. MPV strains ZAI and CNG have an apparent mutation in the first translation initiation codon and downstream non-sense mutations so that only the short form can be translated (Fig. 2A). It would seem likely that both mutations affecting IFN resistance would be attenuating in vivo, possibly contributing to the less efficient person-to-person spread of MPV compared to VAR.
Table 1.
Comparison of virulence factors of MPV-ZAI, VAR-IND, and VAR-GAR
| Function | MPV-ZAI |
VAR-IND |
VAR-GAR |
AA identityc |
||||
| ORFa | AAb | ORFa | AAb | ORFa | AAb | ZAI/INDd | IND/GARe | |
| Chemokine binding | J3R | 246 | G3R | 253 | G3R | 253 | 83.5 | 99.3 |
| Viral growth factor | D3R | 142 | D2R | 140 | B3R | 140 | 83.6 | 97.1 |
| Tumor necrosis factor binding | J2R | 348 | G2R | 349 | G2R | 349 | 85.1 | 98.9 |
| IFN resistance, eIF-2α homolog | – | – | C3L | 88 | P3L | 88 | –f | 100 |
| IFN resistance, dsRNA binding | F3L | 153 | E3L | 190 | C3L | 192 | 85.6 | 97.3 |
| IFN-α/β binding | B16R | 352 | B20R | 354 | D9R | 355 | 85.6 | 99.2 |
| IFN-γ binding | B9R | 267 | B9R | 266 | H9R | 266 | 90.9 | 98.5 |
| Complement binding | D15L | 216 | D12L | 263 | B18L | 263 | 91.7 | 99.2 |
| IL-18 binding | D6L | 126 | D5L | 126 | B6L | 126 | 92.1 | 99.2 |
| Serine protease inhibitor (SPI) homolog, apoptosis inhibitor | B19R | 357 | B25R | 357 | D14R | 357 | 93.0 | 99.5 |
| SPI homolog, apoptosis inhibitor | B12R | 344 | B13R | 344 | D2R | 344 | 93.6 | 99.1 |
| IL-1β binding | B14R | 326 | B15R | 63 | D4R | 63 | –f | 98.4 |
| 3-β-Hydroxy-δ-5-steroid dehydrogenase | A45L | 346 | A50L | 61 | A54L | 61 | –f | 98.4 |
ORF, open reading frame.
AA, number of amino acids constituting the protein encoded by the ORF.
AA identity, percent identity of deduced amino acid sequences of proteins were calculated by FASTA analysis.
ZAI/IND, comparison of corresponding ORFs of MPV-ZAI and VAR-IND.
e IND/GAR, comparison of corresponding ORFs of VAR-IND and VAR-GAR.
Indicates a deletion in the coding sequence of one virus relative to the other.
Table 2.
Comparison of ankyrin-like proteins of MPV-ZAI, VAR-IND, and VAR-GAR
| MPV-ZAI |
VAR-IND |
VAR-GAR |
AA identityc |
||||
| ORFa | AAb | ORFa | AAb | ORFa | AAb | ZAI/INDd | IND/GARe |
| D1L | 437 | – | – | – | – | –f | – |
| D7L | 660 | D6L | 452 | B8L | 355 | 92.2 | 99.1 |
| D9L | 630 | D7L | 153 | B12L | 132 | 70.3 | 99.2 |
| O1L | 442 | O1L | 446 | Q1L | 449 | 95.9 | 99.8 |
| C1L | 284 | C1L | 66 | P1L | 66 | 89.4 | 100 |
| B5R | 561 | B6R | 558 | H6R | 558 | 91.8 | 99.6 |
| – | – | B19R | 574 | D8R | 574 | –f | 99.5 |
| B17R | 793 | B21R | 787 | D10R | 787 | 87.9 | 99.1 |
| J1R | 587 | G1R | 585 | G1R | 585 | 89.4 | 98.8 |
| C1L | 284 | O3L | 70 | Q3L | 70 | –f | 100 |
ORF, open reading frame.
AA, number of amino acids constituting the protein encoded by the ORF.
AA identity, percent identity of deduced amino acid sequences of proteins were calculated by FASTA analysis.
ZAI/IND, comparison of corresponding ORFs of MPV-ZAI and VAR-IND.
IND/GAR, comparison of corresponding ORFs of VAR-IND and VAR-GAR.
Indicates a deletion in the coding sequence of one virus relative to the other.
Fig. 2.
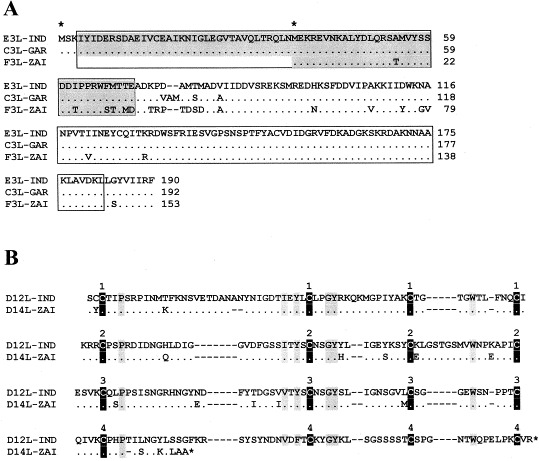
A: Alignment of amino acid sequences of orthopoxviral E3L IFN resistance factor encoded by the corresponding genes of VAR-IND, VAR-GAR, and MPV-ZAI. Blocks containing the N-terminal adenosine deaminase Z-α domain [32] (marked gray) and C-terminal double-stranded RNA binding motif [30] are indicated. Amino acid residues that are identical to those in VAR-IND are marked with dots; amino acid deletions are marked with dashes. The first and second methionine residues from which begins a long or short form of the protein are marked by asterisks above the sequence. B: Alignment of amino acid sequences of the orthopoxviral complement-binding proteins. ORFs of VAR-IND and MPV-ZAI are shown. The conserved cysteine residues are marked with black vertical blocks, other conserved residues are marked with gray vertical blocks. The numbers above the blocks indicate four repeating domains typical of the complement-control proteins [33].
MPV also encodes a form of the complement-binding protein with only three short consensus repeats instead of four found in other orthopoxviruses [17], [33] (Fig. 2B). Although, the MPV protein retains some complement-inhibitory activity in vitro, the effect of one less repeat has not been quantified [34]. On the other hand, MPV encodes a secreted IL-1β-binding protein and 3-β-hydroxy-δ-5-steroid dehydrogenase, discovered originally in VAC [35], [36], whereas the VAR strains do not have intact versions of these ORFs (Table 1) [18], [37], [38]. Notably, deletion of the VAC gene encoding the IL-1β-binding protein has been correlated with fever and pathogenicity [39]. Thus, in this case the presence of the IL-1β-binding protein in MPV may contribute to its lower ability to cause disease than VAR.
ORFs encoding proteins with ankyrin repeats, some of which have host-range functions (orthopoxviral isologs of the MPV-ZAI D7L and C1L, Table 2), comprise the largest orthopoxvirus gene family [17], [40]. Of the 10 genes belonging to this family (Table 2), the one corresponding to B19R of VAR-IND is deleted in the MPV-ZAI genome and the one corresponding to MPV-ZAI gene D1L is deleted in both VAR strains. In addition, four VAR genes in this family (D6L, D7L, C1L, and O3L in VAR-IND) are truncated relative to their MPV homologs. At this time, we can only speculate as to how these differences might affect host range or virulence.
The presence of DNA in VAR that is absent from MPV and vice versa indicated that neither virus is the direct ancestor of the other. To better understand genetic relationships, a phylogenetic analysis of the terminal variable genomic regions of four orthopoxvirus species pathogenic for humans was made based on 117 600 bp of aligned DNA (Fig. 3 ). The major and minor subspecies of VAR are closely related to each other and MPV seems slightly more distant from VAR than VAC. Camelpox and taterapox viruses appear closer to VAR based on a comparison of short DNA sequences [41]. CPV may be nearest to the progenitor orthopoxvirus, as it contains all genes collectively present in the other members of this genus [17].
Fig. 3.
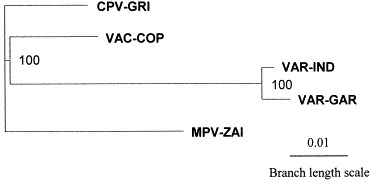
Unrooted phylogenetic tree derived from alignments of the nucleotide sequences in the terminal variable genomic regions of MPV-ZAI, VAR-GAR [18], VAR-IND [15], VAC-COP (VAC strain Copenhagen) [19], and CPV-GRI (CPV strain GRI-90) [17]. The total alignment length was 117 600 bp. NJ analysis of aligned sequences provided bootstrap confidence intervals (values in bold) after 1000 heuristic search replicates. ClustralX software version 1.81 was used.
In summary, a comparative analysis of MPV and VAR genomes indicated that MPV is a discrete species exhibiting multiple differences in virulence genes from both major and minor strains of VAR. MPV and VAR most likely evolved independently from an orthopoxvirus ancestor resembling CPV that contained a more complete set of genes. Although concerns regarding the use of VAR for bioterrorism have prompted surrogate studies of MPV in monkeys, the genetic differences between VAR and MPV raise considerable doubts regarding the validity of this model. Nevertheless, the severe disease caused by MPV makes this virus important in its own right and we need to closely monitor infection rates in Africa to ensure that it does not undergo human adaptation by spontaneous or recombinational means in an unvaccinated population with a high frequency of AIDS.
Acknowledgements
We thank T. Senkevich, J. Yewdell, and E. Berger for comments on the manuscript. This work was supported by Grants 884-97 and 884-2p of ISTC and NAS, Grant RN2-413 of CRDF and NIH, and Grant 00-04-49558 of the Russian Foundation of Basic Research.
References
- 1.Fenner, F., Henderson, D.A., Arita, I., Jezek, Z. and Ladnyi, I.D. (1988) Smallpox and its Eradication, World Health Organization, Geneva.
- 2.Marennikova, S.S. and Shchelkunov, S.N. (1998) Orthopoxviruses Pathogenic for Humans, KMK Press, Moscow.
- 3.Jezek Z., Fenner F. Monogr. Virol. 1988;17:1–140. [Google Scholar]
- 4.Marennikova S.S., Shelukhina E.M., Maltseva N.N., Cimiskjan K.L., Macevic G.R. Bull. WHO. 1972;46:599–611. [PMC free article] [PubMed] [Google Scholar]
- 5.Breman J.G. Emerg. Infect. 2000;4:45–67. [Google Scholar]
- 6.Mukinda V.B.K., Mwema G., Kilundu M., Heymann D.L., Khan A.S., Esposito J.J. Lancet. 1997;349:1448–1450. doi: 10.1016/S0140-6736(05)63725-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen J. Science. 1997;277:312–313. doi: 10.1126/science.277.5324.312. [DOI] [PubMed] [Google Scholar]
- 8.Fenner F. Prog. Med. Virol. 1977;23:1–21. [PubMed] [Google Scholar]
- 9.Mackett M., Archard L.C. J. Gen. Virol. 1979;45:683–701. doi: 10.1099/0022-1317-45-3-683. [DOI] [PubMed] [Google Scholar]
- 10.Esposito J.J., Knight J.C. Virology. 1984;135:561–567. doi: 10.1016/0042-6822(84)90212-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esposito J.J., Knight J.C. Virology. 1985;143:230–251. doi: 10.1016/0042-6822(85)90111-4. [DOI] [PubMed] [Google Scholar]
- 12.Douglass N., Dumbell K. J. Virol. 1992;66:7565–7567. doi: 10.1128/jvi.66.12.7565-7567.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esposito J., Condit R., Obijeski J. J. Virol. Methods. 1981;2:175–179. doi: 10.1016/0166-0934(81)90036-7. [DOI] [PubMed] [Google Scholar]
- 14.Shchelkunov S.N., Blinov V.M., Totmenin A.V., Marennikova S.S., Kolykhalov A.A., Frolov I.V., Chizhikov V.E., Gutorov V.V., Gashnikov P.V., Belanov E.F., Belavin P.A., Resenchuk S.M., Andzhaparidze O.G., Sandakhchiev L.S. Virus Res. 1993;27:25–35. doi: 10.1016/0168-1702(93)90110-9. [DOI] [PubMed] [Google Scholar]
- 15.Shchelkunov S.N., Resenchuk S.M., Totmenin A.V., Blinov V.M., Marennikova S.S., Sandakhchiev L.S. FEBS Lett. 1993;327:321–324. doi: 10.1016/0014-5793(93)81013-p. [DOI] [PubMed] [Google Scholar]
- 16.Shchelkunov S.N., Massung R.F., Esposito J.J. Virus Res. 1995;36:107–118. doi: 10.1016/0168-1702(94)00113-q. [DOI] [PubMed] [Google Scholar]
- 17.Shchelkunov S.N., Safronov P.F., Totmenin A.V., Petrov N.A., Ryazankina O.I., Gutorov V.V., Kotwal G.J. Virology. 1998;243:432–460. doi: 10.1006/viro.1998.9039. [DOI] [PubMed] [Google Scholar]
- 18.Shchelkunov S.N., Totmenin A.V., Loparev V.N., Safronov P.F., Gutorov V.V., Chizhikov V.E., Knight J.C., Parsons J.M., Massung R.F., Esposito J.J. Virology. 2000;266:361–386. doi: 10.1006/viro.1999.0086. [DOI] [PubMed] [Google Scholar]
- 19.Goebel S.J., Johnson G.P., Perkus M.E., Davis S.W., Winslow J.P., Paoletti E. Virology. 1990;179:247–266. doi: 10.1016/0042-6822(90)90294-2. [DOI] [PubMed] [Google Scholar]
- 20.Resenchuk S.M., Blinov V.M. Comput. Appl. Biosci. 1995;11:7–11. doi: 10.1093/bioinformatics/11.1.7. [DOI] [PubMed] [Google Scholar]
- 21.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson J.D., Higgins D.G., Gibson T.J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saitou N., Nei M. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 24.Kumar, S., Tamura, K. and Nei, M. (1993) MEGA: Molecular Evolutionary Genetics Analysis, The Pennsylvania State University, University Park, PA.
- 25.Massung R.F., Liu L.-I., Qi J., Knight J.C., Yuran T.E., Kerlavage A.R., Parsons J.M., Venter J.C., Esposito J.J. Virology. 1994;201:215–240. doi: 10.1006/viro.1994.1288. [DOI] [PubMed] [Google Scholar]
- 26.Davies M.V., Elroy-Stein O., Jagus R., Moss B., Kaufman R.J. J. Virol. 1992;66:1943–1950. doi: 10.1128/jvi.66.4.1943-1950.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beattie E., Tartaglia J., Paoletti E. Virology. 1991;183:419–422. doi: 10.1016/0042-6822(91)90158-8. [DOI] [PubMed] [Google Scholar]
- 28.Yuwen H., Cox J.H., Yewdell J.W., Bennink J.R., Moss B. Virology. 1993;195:732–744. doi: 10.1006/viro.1993.1424. [DOI] [PubMed] [Google Scholar]
- 29.Brandt T.A., Jacobs B.L. J. Virol. 2001;75:850–856. doi: 10.1128/JVI.75.2.850-856.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang H.-W., Jacobs B.L. Virology. 1993;194:537–547. doi: 10.1006/viro.1993.1292. [DOI] [PubMed] [Google Scholar]
- 31.Rivas C., Gil J., Melkova Z., Esteban M., Diaz-Guerra M. Virology. 1998;243:406–414. doi: 10.1006/viro.1998.9072. [DOI] [PubMed] [Google Scholar]
- 32.Liu, Y., Herbert, A., Rich, A. and Samuel, C.E. (1998) Methods: a Companion to Methods in Enzymology, vol. 15, pp. 199–205
- 33.Kotwal G.J., Moss B. Nature. 1988;335:176–178. doi: 10.1038/335176a0. [DOI] [PubMed] [Google Scholar]
- 34.Smith S.A., Mullin N.P., Parkinson J., Shchelkunov S.N., Totmenin A.V., Loparev V.N., Srisatjaluk R., Reynolds D.N., Keeling K.L., Justus D.E., Barlow P.N., Kotwal G.J. J. Virol. 2000;74:5659–5666. doi: 10.1128/jvi.74.12.5659-5666.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spriggs M.K., Hruby D.E., Maliszewski C.R., Pickup D.J., Sims J.E., Buller R.M.L., Van Slyke J. Cell. 1992;71:145–152. doi: 10.1016/0092-8674(92)90273-f. [DOI] [PubMed] [Google Scholar]
- 36.Moore J.B., Smith G.L. EMBO J. 1992;11:1973–1980. doi: 10.1002/j.1460-2075.1992.tb05251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shchelkunov S.N., Blinov V.M., Sandakhchiev L.S. FEBS Lett. 1993;319:80–83. doi: 10.1016/0014-5793(93)80041-r. [DOI] [PubMed] [Google Scholar]
- 38.Shchelkunov S.N. Virus Genes. 1995;10:53–71. doi: 10.1007/BF01724297. [DOI] [PubMed] [Google Scholar]
- 39.Alcami A., Smith G.L. Proc. Natl. Acad. Sci. USA. 1996;93:11029–11034. doi: 10.1073/pnas.93.20.11029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shchelkunov S.N., Blinov V.M., Sandakhchiev L.S. FEBS Lett. 1993;319:163–165. doi: 10.1016/0014-5793(93)80059-4. [DOI] [PubMed] [Google Scholar]
- 41.Douglass N.J., Dumbell K.R. J. Gen. Virol. 1996;77:947–951. doi: 10.1099/0022-1317-77-5-947. [DOI] [PubMed] [Google Scholar]


