Abstract
A monkeypox virus was detected from a human clinical case in 2018 in Cameroon; a country where no human cases were reported since 1989. The virus exhibited close genetic relatedness with another monkeypox virus isolated in Nigeria during the 2017–2018 outbreak. Although our molecular findings argue in favor of an extension of the monkeypox outbreak from Nigeria into Cameroon, the possibility that the monkeypox virus detected could be indigenous to Cameroon cannot be ruled out.
Keywords: Monkeypox virus, Phylogenetic relationships, West African clade, Cameroon
Human monkeypox is a rare zoonotic infection caused by Monkeypox virus (MPXV), which is a large, double-stranded DNA virus belonging to the Orthopox genus in the Poxviridae family. Since its original identification in 1958, MPXV-associated outbreaks have occurred primarily in rural rainforest areas of the Congo basin and West Africa. Phylogenetic studies have uncovered the segregation of MPXV strains into two main clades: “West African” and “Congo Basin” (Faye et al., 2018; Nakazawa et al., 2015). MPXV infections have a mortality rate ranging from 1 to 11%; with the higher rates (11%) in unvaccinated individuals (Durski et al., 2018). The West African clade is associated with milder disease, fewer deaths, and limited human-to-human transmission compared to the Congo Basin clade, which is more virulent (McCollum and Damon, 2014; Nakazawa et al., 2015). The Democratic Republic of Congo has consistently reported cases of human monkeypox each year for decades; however, increased reports have recently come from other endemic countries, which have not routinely reported cases (Durski et al., 2018).
In Cameroon, only two confirmed cases of human monkeypox have been documented, and the most recent one occurred in 1989 in Ekoumdouma in the Centre region (Harrigan et al., 2018; Nakazawa et al., 2015; Tchokoteu et al., 1991). Since then, no human cases have been recorded despite the fact that two MPXV-associated outbreaks occurred among captive chimpanzees (Pan troglodytes) in primate sanctuaries of the Centre region of Cameroon in 2014 and 2016 (Nakazawa et al., 2015).
From February to May 2018, seven suspected human cases of monkeypox were recorded in Cameroon, in the health district of Akwaya in the South West region (2 cases: 14V-4555 and 18V-4966) and the health district of Njikwa in the North West region (5 cases) (Table 1 ). All suspected cases had ongoing (or history of) fever, malaise, body aches and a generalized painful maculopapular rash. Cases 14V-4551, 14V-4552 and 14V-4554 specifically developed pharyngitis as well as sub-mandibular and cervical lymphadenopathy.
Table 1.
Characteristics of the seven suspected monkeypox cases and results for the orthopox (OPX), monkeypox (MPX) or varicella-zoster (VZ) specific PCR assays.
| Laboratory code | Region | Health District | Sexe | Age | Date of fever onset | Date of specimen collection | Type of specimen | PCR (orthopox virus) | PCR (monkeypox virus) | PCR (varicella-zoster virus) |
|---|---|---|---|---|---|---|---|---|---|---|
| 14V-4555 | South West | Akwaya | M | 58 yrs | 10/02/2018 | 10/05/2018 | Serum | Negative | NT | Negative |
| 18V-4966 | South West | Akwaya | F | 3 wks | 05/03/2018 | 17/05/2018 | Serum | Negative | NT | Negative |
| 14V-4554 | North West | Njikwa | M | 39 yrs | 09/03/2018 | 10/05/2018 | Serum | Negative | NT | Negative |
| 14V-4556 | North West | Njikwa | F | 20 yrs | 24/03/2018 | 04/05/2018 | Serum | Negative | NT | Negative |
| 04/05/2018 | Serum | Positive | Positive | Negative | ||||||
| 14V-4552 | North West | Njikwa | M | 20 yrs | 24/04/2018 | 04/05/2018 | Biopsy of skin lesions | Positive | Positive | Negative |
| 04/05/2018 | Swab of skin lesions | Positive | Positive | Negative | ||||||
| 14V-4551 | North West | Njikwa | M | 10 yrs | 01/05/2018 | 10/05/2018 | Serum | Negative | NT | Negative |
| 14V-4553 | North West | Njikwa | F | 13 yrs | 04/05/2018 | 09/05/2018 | Serum | Negative | NT | Negative |
NT, not tested; M, male; F, female; yrs., years; wks, weeks.
Since orthopox and varicella-zoster viruses are present in the blood for a short time period, “negative” PCR results do not necessarily mean that the patients did not have monkeypox or varicella-zoster; they possibly had no detectable viral DNA in serum at the time of blood collection.
The first case 14V-4555 was an animal guard at the Kagwene gorilla sanctuary where no infected or symptomatic primates has been reported. The second case 18V-4966 was a 3-weeks-old baby who had no relation or contact with the first case. The case 18V-4966 from Akwaya came into direct contact with the case 14V-4554 who was her healthcare provider in the neighboring district of Njikwa. This third case 14V-4554 also has direct contacts with his colleague 14V-4556 at the health centre. The case 14V-4551 came into direct contacts with his brother 14V-4552 during his illness but they reported no relation or interactions with previous cases. In the same manner, the last case 14V-4553 was a student who had no known contact with previous cases. All cases reported that they or their relatives have no travel history to the neighboring Nigeria in 2018. They also reported no known interactions with travelers from Nigeria.
Blood specimens were obtained from each of the seven suspected human monkeypox cases after informed consent. Moreover, a skin lesion biopsy and a swab of the exudate from maculopapular skin lesions were obtained from the case 14V-4552 (Table 1). Sera clarified by centrifugation of whole blood and other specimens were shipped to the Centre Pasteur of Cameroon where total DNA were purified from each specimen. Resulting DNA samples were subjected to a pan-orthopoxvirus real-time PCR Taqman assay described elsewhere (Kulesh et al., 2004). DNA specimens amplified with the pan-orthopox virus assay were confirmed by the generic monkeypox real-time PCR Taqman assay (Li et al., 2010).
Both serum and skin lesions specimens from the case 14V-4552 were positive for orthopox and monkeypox assays whereas sera from the six remaining patients were negative for orthopox (Table 1). All samples from the seven cases were negative for varicella zoster virus (VZV) specific PCR performed as previously described (Burrel et al., 2012). A limitation to efficient diagnosis in this study is that the time elapsed between the date of fever onset and the date of the specimen collection was greater than the 14–21 days during which symptoms are evident. Because of the short duration of orthopox and varicella-zoster viremia and timing of specimen collection, it is possible that the first four cases were false negatives who did not longer have detectable viral DNA in serum at the time of blood collection (Table 1). A complementary approach would have consisted to test convalescent-phase serum specimens from these patients for monkeypox virus specific IgM or IgG antibodies. Their detection in an unvaccinated individual with a history of severe illness and rash suggests an indirect diagnosis of monkeypox.
To determine whether the virus detected originated from an indigenous zoonotic spillover event or from an importation from a neighboring country, a portion of the sequence of the A-type Inclusion (ATI) gene from the monkeypox virus identified (14 V-4552) was determined. The resulting 639 nucleotides sequence was compared to MPXV sequences from other countries [including the only available sequence derived from the last human monkeypox case reported in Cameroon in 1989 (Nakazawa et al., 2015)], using Maximum Likelihood based phylogenetic analysis.
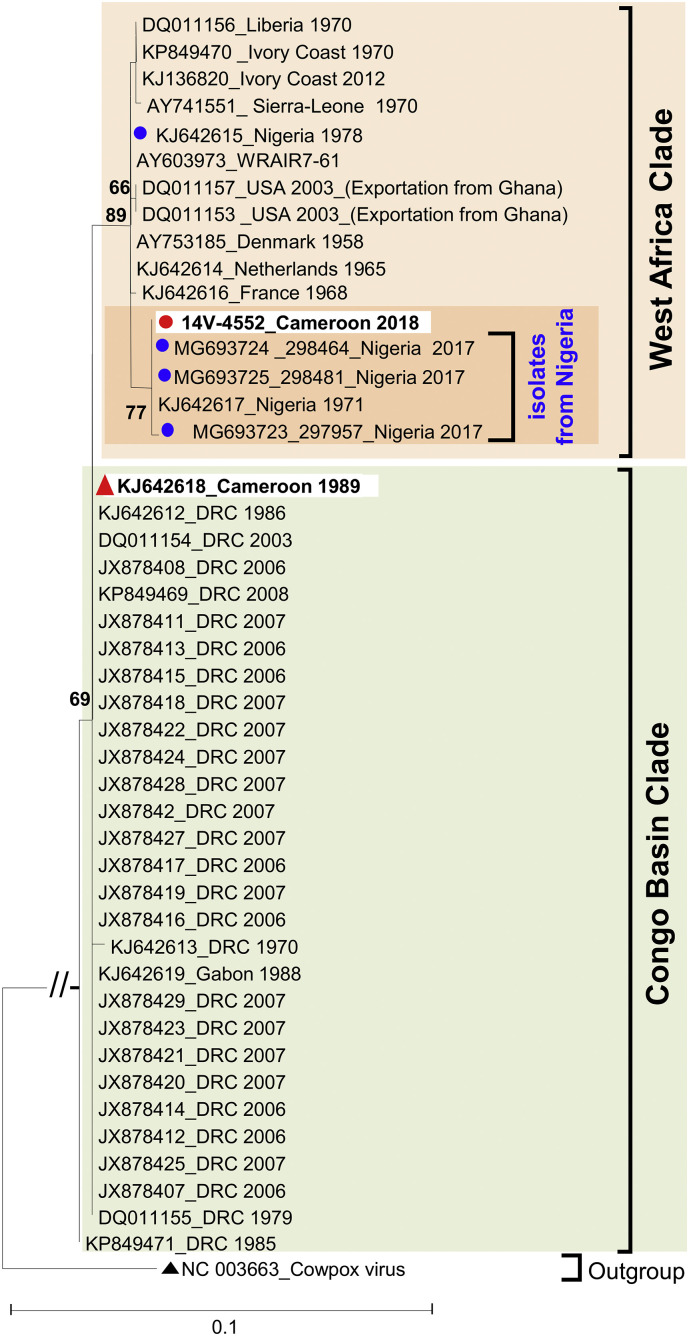
As result, the studied virus 14V-4552 belongs to the West Africa clade rather than the Congo Basin clade, similarly to the previously reported MPXV from Cameroon (Nakazawa et al., 2015; Tchokoteu et al., 1991) (Fig. 1 ). Interestingly, within the West Africa clade, isolate 14V-4552 featured the closest sequence similarities with Nigerian isolates (Fig. 1), including those recovered during the 2017–2018 outbreak in Nigeria (Eteng et al., 2018; Faye et al., 2018; Harrigan et al., 2018; Yinka-Ogunleye et al., 2018). Actually, the 14V-4552 sequence displayed 100% nucleotide identity with that of the virus 298464.
Fig. 1.
Phylogenetic relationships of the studied monkeypox virus isolate recovered from current infection in Cameroon. The phylogenetic tree was based on the multiple alignment of 639 nucleotides partial sequences of the A-type Inclusion (ATI) gene [nucleotides 126,536 to 127,174 according to sequence isolate 298,464 originating from Nigeria in 2017 (GenBank N° MG693724)]. Phylogenetic analysis was performed by the Maximum Likelihood method with the best-fit model identified by Smart Model Selection using the Bayesian Information Criterion (Lefort et al., 2017): Tamura-Nei with gamma-distributed rate heterogeneity and 4 rate categories (TN93 + G + Γ4). The studied and reference monkeypox virus isolates from Cameroon are highlighted by red circle (●) and red triangles (▲), respectively. The year and country of isolation of databases available isolates are indicated (DRC: Democratic Republic of the Congo; USA: United-States of America). The reliability of individual tree nodes was estimated using 1000 bootstrap pseudoreplicates and values less than 60% have been omitted. The scale bars indicate nucleotide evolutionary distance as substitutions per site. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Our molecular data unambiguously demonstrate that isolate 14 V-4552 does not derive from MPXV previously detected in Cameroon. Unfortunately, MPXV sequences from recent outbreaks among chimpanzees in Cameroon were not available for comparison with 14 V-4552. All cases were from rural areas where farming, hunting and bush meat consumption increase the risk of animal to human transmission. All cases reported that they or their relatives had neither travel history to the neighboring Nigeria nor known interactions with travelers from Nigeria in 2018. This suggests that the monkeypox virus detected in this study may have circulated in humans or animals in Cameroon for sometimes without being detected. It cannot be excluded that the same monkeypox virus strain was circulating in Cameroon and Nigeria, but was not previously detected in Cameroon as result of surveillance bias due unrecognition by health workers.
Although the hypothesis of a local spillover event involving an animal reservoir in Cameroon cannot be completely ruled out, it is possible that the human monkeypox case detected in Cameroon in 2018 resulted from an importation from the neighboring Nigeria. This is supported by the i) absence of human monkeypox in Cameroon since 1989, ii) striking genetic relatedness of the studied MPXV with those from Nigeria, iii) close geographic proximity between the regions affected in Cameroon and Nigeria, and iv) occurrence of monkeypox in Nigeria in 2017–2018 (Eteng et al., 2018; Harrigan et al., 2018; Yinka-Ogunleye et al., 2018). Cameroon and Nigeria share very permeable borders that allow extensive human populations movements between the two countries, especially since the beginning of a socio-political crisis affecting the Western regions of Cameroon. There is no geographical barrier, such as sea or river, which prevents the spread of MPXV reservoirs from Nigeria to Cameroon and vice versa. Further investigations, based on “One Health Approach”, would be of great interest to rule out potential endemicity of the “West African” genotype of MPXV in the western regions of Cameroon. Neither the studied cases nor their close relatives had a travel history to Nigeria. However, the striking genetic relationship between the studied isolate and Nigerian isolates and the absence of human cases in the western regions of Cameroon for decades suggest that the Nigerian outbreak may have extended into Cameroon. Such MPXV exportation from Nigeria has recently affected the United Kingdom, a more distant European Country (Vaughan et al., 2018)
Our findings highlight the need for risk-based awareness campaigns and local capacity strengthening for monkeypox surveillance and targeted human/animal surveys in the regions affected. These interventions are critical for prevention, early detection and control of monkeypox in Cameroon.
Acknowledgments
This work was supported by the Centre Pasteur of Cameroon. We are thankful to the patients or their legal guardians who agreed to be enrolled in this study. We are grateful to Mrs. Gwladys Monamele for English language review.
References
- Burrel S., Fovet C., Brunet C., Ovaguimian L., Hamm N., Conan F., Kalkias L., Agut H., Boutolleau D. Routine use of duplex real-time PCR assays including a commercial internal control for molecular diagnosis of opportunistic DNA virus infections. J. Virol. Methods. 2012;185:136–141. doi: 10.1016/j.jviromet.2012.05.031. [DOI] [PubMed] [Google Scholar]
- Durski K.N., McCollum A.M., Nakazawa Y., Petersen B.W., Reynolds M.G., Briand S., Djingarey M.H., Olson V., Damon I.K., Khalakdina A. Emergence of Monkeypox - West and Central Africa, 1970-2017. MMWR Morb. Mortal. Wkly Rep. 2018;67:306–310. doi: 10.15585/mmwr.mm6710a5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eteng W.E., Mandra A., Doty J., Yinka-Ogunleye A., Aruna S., Reynolds M.G., McCollum A.M., Davidson W., Wilkins K., Saleh M., Ipadeola O., Manneh L., Anebonam U., Abdulkareem Z., Okoli N., Agenyi J., Dan-Nwafor C., Mahmodu I., Ihekweazu C. Notes from the field: responding to an outbreak of monkeypox using the one health approach - Nigeria, 2017-2018. MMWR Morb. Mortal. Wkly Rep. 2018;67:1040–1041. doi: 10.15585/mmwr.mm6737a5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faye O., Pratt C.B., Faye M., Fall G., Chitty J.A., Diagne M.M., Wiley M.R., Yinka-Ogunleye A.F., Aruna S., Etebu E.N., Aworabhi N., Ogoina D., Numbere W., Mba N., Palacios G., Sall A.A., Ihekweazu C. Genomic characterisation of human monkeypox virus in Nigeria. Lancet Infect. Dis. 2018;18:246. doi: 10.1016/S1473-3099(18)30043-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrigan R.J., Thomassen H.A., Smith T.B. Emergence of monkeypox in West Africa and Central Africa, 1970-2017. R. Soc. Open Sci. 2018;93:125–132. [Google Scholar]
- Kulesh D.A., Baker R.O., Loveless B.M., Norwood D., Zwiers S.H., Mucker E., Hartmann C., Herrera R., Miller D., Christensen D., Wasieloski L.P., Jr., Huggins J., Jahrling P.B. Smallpox and pan-orthopox virus detection by real-time 3′-minor groove binder TaqMan assays on the roche LightCycler and the Cepheid smart Cycler platforms. J. Clin. Microbiol. 2004;42:601–609. doi: 10.1128/JCM.42.2.601-609.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefort V., Longueville J.E., Gascuel O. SMS: smart model selection in PhyML. Mol. Biol. Evol. 2017 doi: 10.1093/molbev/msx149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zhao H., Wilkins K., Hughes C., Damon I.K. Real-time PCR assays for the specific detection of monkeypox virus West African and Congo Basin strain DNA. J. Virol. Methods. 2010;169:223–227. doi: 10.1016/j.jviromet.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCollum A.M., Damon I.K. Human monkeypox. Clin. Infect. Dis. 2014;58:260–267. doi: 10.1093/cid/cit703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa Y., Mauldin M.R., Emerson G.L., Reynolds M.G., Lash R.R., Gao J., Zhao H., Li Y., Muyembe J.J., Kingebeni P.M., Wemakoy O., Malekani J., Karem K.L., Damon I.K., Carroll D.S. A phylogeographic investigation of African monkeypox. Viruses. 2015;7:2168–2184. doi: 10.3390/v7042168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchokoteu P.F., Kago I., Tetanye E., Ndoumbe P., Pignon D., Mbede J. Variola or a severe case of varicella? A case of human variola due to monkeypox virus in a child from the Cameroon. Ann. Soc. Med. Trop. 1991;71:123–128. [PubMed] [Google Scholar]
- Vaughan A., Aarons E., Astbury J., Balasegaram S., Beadsworth M., Beck C.R., Chand M., O'Connor C., Dunning J., Ghebrehewet S., Harper N., Howlett-Shipley R., Ihekweazu C., Jacobs M., Kaindama L., Katwa P., Khoo S., Lamb L., Mawdsley S., Morgan D., Palmer R., Phin N., Russell K., Said B., Simpson A., Vivancos R., Wade M., Walsh A., Wilburn J. Two cases of monkeypox imported to the United Kingdom, September 2018. Euro Surveill. 2018;23 doi: 10.2807/1560-7917.ES.2018.23.38.1800509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yinka-Ogunleye A., Aruna O., Ogoina D., Aworabhi N., Eteng W., Badaru S., Mohammed A., Agenyi J., Etebu E.N., Numbere T.W., Ndoreraho A., Nkunzimana E., Disu Y., Dalhat M., Nguku P., Mohammed A., Saleh M., McCollum A., Wilkins K., Faye O., Sall A., Happi C., Mba N., Ojo O., Ihekweazu C. Reemergence of Human Monkeypox in Nigeria, 2017. Emerg. Infect. Dis. 2018;24:1149–1151. doi: 10.3201/eid2406.180017. [DOI] [PMC free article] [PubMed] [Google Scholar]