Abstract
The yatapoxvirus genus contains three members: tanapox virus (TPV), yaba-like disease virus (YLDV) and yaba monkey tumor virus (YMTV), two of which (TPV and YLDV) may infect humans. However, only a very small number of patients have been diagnosed with TPV outside Africa. Given the increased international travel and the similarity of clinical signs during the early stages of a TPV/YLDV infection as compared to diseases caused by agents of potential biological warfare, such as smallpox, monkeypox, tularemia and anthrax, the rapid and reliable recognition of a TPV/YLDV infection is crucial. A real-time PCR assay using TaqMan® chemistry was developed in order to identify unambiguously TPV/YLDV. Primers and probe targeting a 101 bp region of the PstI L fragment of TPV, initial optimisations steps were carried out with YLDV DNA as template. Using probit regression analysis, the lower limit of detection was calculated to be ca. 8 copies per assay. A total of five TPV strains, one YDLV strain and scab-derived DNA from a patient with a TPV infection yielded specific amplification, whereas the DNA of YMTV was not amplified. Various viral and bacterial pathogens (n = 29) associated with rash-causing illnesses were not detected using this assay.
The genus yatapoxvirus belongs to the family poxviridae, subfamily chordopoxvirinae and consists currently of three members, tanapox virus (TPV), yaba-like disease virus (YLDV) and yaba monkey tumor virus (YMTV) (Büchen-Osmond, 2003). Infections of monkeys and humans with both, TPV and YLDV, have been reported, whereas YMTV has infected so far only non-human primates naturally (Grace and Mirand, 1965). The mode of transmission of yatapoxviruses is not well understood. Biting arthropods are suspected to act as mechanical vectors, however, yatapoxvirus has not been isolated so far from arthropods.
TPV infection is a rare zoonosis and was first observed in the human population in the flood plains of the Tana River in Kenya during two epidemics in 1957 and 1962 (Downie et al., 1971). However, it is believed that TPV is endemic throughout tropical Africa. TPV infection in humans results in an acute febrile illness with one or more localized skin lesions. A mild pre-eruptive fever that lasts 3–4 days is followed by severe headache, backache and lymphadenopathy. Itching occurs at the site where skin lesions ultimately develop. An initially small, pock-like lesion becomes papular and gradually enlarges to reach a maximum diameter of about 15 mm by the end of second week. During the third week, the skin lesion ulcerates and finally heals within 6 weeks, leaving a scar (Downie et al., 1971). Several authors pointed out that TPV-induced lesions in the early stages can be confused easily with smallpox or human monkeypox (Jezek et al., 1985, Downie et al., 1971, Stich et al., 2002, Dhar et al., 2004).
YLDV was first reported during epizootics in 1965 and 1966 in primate centers in the USA with animal handlers presenting identical clinical features as compared to TPV infection (Casey et al., 1967; Crandel et al., 1969; Downie and Espana, 1972). The third member of the genus, YMTV, was isolated in 1958 during an outbreak in a colony of captive rhesus monkeys in Yaba, Nigeria. Infected monkeys developed epidermal histocytomas with masses of polygonal mononuclear cell infiltrates and the development of a suppurative inflammatory reaction (Niven et al., 1961).
Comparison of YLDV with TPV sequences available in the databases demonstrated a high identity, for instance, the sequences of 12.5 and 10.7 kb from the left and right ends have an identity of >98.6% (Lee et al., 2001). It has been assumed that TPV and YLDV may be different strains of the same virus species. In contrast, comparison of the complete genome sequences of YLDV and YMTV revealed an identity of only about 75% (Brunetti et al., 2003).
Clinical diagnosis of TPV/YLDV infection in humans is based on the appearance of only a few nodular lesions, the absence of pustulation and a benign course. Investigation of scabs by electron microscopy cannot differentiate yatapoxviruses from orthopoxviruses (including smallpox and monkeypox virus). Although yatapoxviruses are distinct antigenically from all other poxviruses (Downie et al., 1971), the method of choice is PCR. Two PCR assays for the specific detection of TPV/YLDV have been published (Stich et al., 2002, Dhar et al., 2004).
Increased international travel has led to appearance of TPV infections outside Africa (Stich et al., 2002, Dhar et al., 2004). Given the fact that early stages of this infection can be mistaken for more serious conditions, such as monkeypox, smallpox, tularemia or anthrax, a rapid, unambiguous identification is extremely important to exclude the deliberate use of agents of biological warfare and bioterrorism.
This report describes the development of a rapid PCR assay to detect TPV and YLDV, both of which are human pathogens. The assay uses the closed-tube TaqMan® format that has several advantages over conventional gel-based PCR methods. Most important, the inclusion of a TaqMan® probe confirms the identity of the PCR product, thereby, eliminating time- and labour-intensive post-PCR processing steps prone to cause cross-contamination. Initially, two sets of primers and probes were selected based on the sequence of the PstI L fragment of TPV (Accession number AF 153912): the TP-TaqMan® assay, based on a conventional PCR (Stich et al., 2002) with an amplicon size of 333 bp and the IT-TaqMan® assay targeting a 101 bp region. Primers and probes (Table 1 ) had been selected with Primer Express software (Applied Biosystems, Germany) and the respective sequences did not show any significant homology to other published sequences in the GenBank and EMBL databases by use of the BLAST (www.ncbi.nlm.nih.gov/BLAST) and FASTA (www.ebi.ac.uk/fasta33) algorithms. The following protocol was used for real-time PCR assay: 12.5 μl of TaqMan® Universal PCR Master Mix (Applied Biosystems, Germany), 0.5 μl bovine serum albumin (20 mg/ml), 1 μl (12 μm) of primers IT1/IT1a or TP1/TP1a (Tib-Molbiol, Germany), 0.15 μl probe (30 μm) IT1s or TP1s (Tib-Molbiol, Germany), PCR-graded water to a final volume of 20 μl and 5 μl of template. PCR was performed on a MX3000 (Stratagene, Germany). Real-time PCR involved initial denaturation (95 °C for 10 min), followed by amplification and detection of the DNA target for 45 cycles (95 °C for 15 s and 55 °C for 1 min). R 2-values, slopes, efficiencies of amplification (Efficiency = 10(−1/slope) −1) and y-intercept were determined.
Table 1.
Nucleotide sequences of primers and probes
| Primer/probe | Sequence (5′–3′) | Positiona | Amplicon size (bp) |
|---|---|---|---|
| TP1b | ACGATGTTATATGCGATGTAAAATGGT | 91–113 | 333 |
| TP1ab | AAGTCTGCATCTTTAACTGTG | 403–423 | |
| TP1s | FAM-5′-AATTAAGAGCGTTGGAAGCTACGACAGAACAATAA-3′-TAMRA | 377–358 | |
| IT1 | CGTGTACGGATATTCACCTATTCATAA | 1586–1612 | 101 |
| IT1a | CGGTTTTGTCGTTAGGATTTGC | 1686–1665 | |
| IT1s | FAM-5′-AAACATTGACATTGTAAAAAGATGGCTGGATTACG-3′-TAMRA | 1628–1662 | |
| TP3c | AGTTAGTAAAGGAGCAAATG | 1337–1356 | 465 |
| TP3ac | GGAGTATTCCCATATTGATC | 1801–1782 |
Indicates the corresponding nucleotide position of the tanapox virus PstI L fragment (Accession number AF153912).
According to Stich et al. (2002).
Used to generate a cloned positive control spanning the IT1/IT1a amplicon.
Initial experiments were conducted using DNA of the YLDV strain Davis grown in African green monkey kidney cell line MA 104 at 34 °C. DNA was isolated from 100 μl aliquots by magnetic beads separation using MagNA Pure Compact Nucleic Acid Isolation Kit I on the Magna Pure Compact (Roche, Germany).
A comparison of the two real-time PCR assays using a serial 10-fold dilution of YLDV DNA demonstrated specific amplification for both assays. However, due to lower C t-values, a more efficient amplification (average 92.875%) and an almost optimal y-intercept (average 38.95) of the IT-TaqMan® assay led to this assay for further evaluation. It seems that selection of a shorter amplicon (101 bp versus 333 bp of the TP-TaqMan® assay) leads to a higher amplification efficiency.
DNA preparations from five TPV strains and one YMTV strain (kindly provided by J.J. Esposito, Centers for Disease Control, Atlanta, USA), one YLDV strain and scab-derived DNA from a German traveller with a TPV infection (Stich et al., 2002) were examined (Table 2 ). With the exception of YMTV, all preparations reacted positive in the IT-TaqMan assay (Fig. 1 ). Lack of amplification of YMTV is in agreement with available YMTV sequence data indicating several mismatches within the primer and probe binding site.
Table 2.
Origin and designation of viruses and DNA preparations used in this study
| Virus strain | Year | Host | Location | Amplification IT-TaqMan® assay |
|---|---|---|---|---|
| Yaba-like disease virus, Davies | Unknown | Monkey | USA | Positive |
| Tanapox virus, TANVWI-38a | 1957 | Human | Kenya | Positive |
| Tanapox virus, TANV#81-I-92a | 1981 | Female, age 32 years | Zaire | Positive |
| Tanapox virus, TANV#81-I-124a | 1981 | Female, age 11 years | Zaire | Positive |
| Tanapox virus, TANV#80-I-231a | 1980 | Male, age 7 years | Zaire | Positive |
| Tanapox virus, TANV#80-I-228a | 1980 | Female, age 39 years | Zaire | Positive |
| Yaba monkey tumor virusa | 1958 | Monkey | Nigeria | Negative |
| PM/99b | 1999 | Male, age 49 years | Germany | Positive |
DNA was kindly provided by J.J. Esposito, CDC, Atlanta, USA.
Scab-derived DNA preparation from a patient (Stich et al., 2002).
Fig. 1.
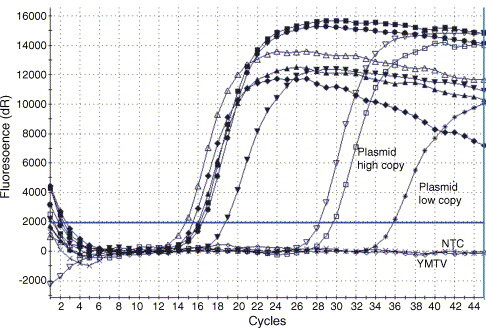
Detection of tanapox virus and yaba-like disease virus using the IT-TaqMan® PCR. Amplification plots show the results from five tanapox viruses (TNV#81-I-92, TNV#81-I-124, TNV#81-I-231, TNV#81-I-228 and TNVWI-38), one yaba-like disease virus and a scab-derived DNA preparation from a patient (PM/99). Plasmid preparations containing ca. 680 copies/μl (plasmid high copy) and 6.8 copies/μl (plasmid low copy) served as positive controls. Yaba monkey tumor virus (YMTV) and the non-template control (NTC) did not show any amplification.
To determine the analytical sensitivity, plasmid pCR 2.1 was generated. This plasmid contains a 465 bp PCR-amplified TPV sequence generated by using primer pair TP3/TP3a (Table 1) spanning the IT primers and probe. To this end, a standard PCR was carried out in a 50 μl volume containing 5 μl 10×PCR-Mix + MgCl2, 2 μl platinum Taq-polymerase (both Invitrogen, Germany), 3 μl primers (10 μM) TP3/TP3a (Tib-Molbiol, Germany), 1 μl PCR nucleotide Mix (10 mM dNTP each), 32.8 μl PCR-graded water and 5 μl template. Initial denaturation for 5 min at 94 °C was followed by 35 cycles consisting of a melting step (94 °C for 1 min), an annealing step (1 min at 52 °C), an elongation step (72 °C for 1 min) and a final step (20 min at 72 °C). A 5 μl aliquot of the PCR product was analysed by gel electrophoresis relative to a size marker (Roth, Germany). Cloning was carried out using standard procedures and DNA of the resulting plasmid pCR 2.1 was purified and photometrically quantified. Plasmid DNA was serially diluted in Tris–EDTA buffer (10 mM/1 mM) and tested in eight replicates on 3 independent days with DNA concentrations corresponding to 6.8e + 002, 6.8e + 001, 6.8, 0.68, 0.068 and 0.0068 copies/μl as inputs (Fig. 2 ). All amplification reactions were run eight-fold in three independent assays. A probit analysis as a model of non-linear regression was done with software SPSS 6.0 and Microcal Origin 4.0. The software determines a continuous 95% confidence interval of the probability of achieving a positive result at any given input DNA concentration within the concentration range of the experiment. The lower limit of detection (LOD) accounts for 8.341 copies/μl with a confidence interval of 95% (Fig. 3 ). Due to the rareness of TPV/YLDV infection, the test could only be performed with a scab-derived DNA preparation and with DNA from cell-cultured strains. But, because of the high sensitivity, it is assumed that TPV/YLDV can be diagnosed also from clinical samples taken in earlier stages of infection.
Fig. 2.
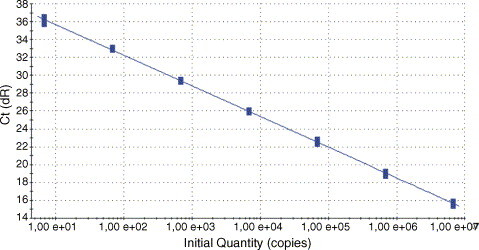
Sensitivity and linearity of the IT-TaqMan® PCR. Ten-fold serial dilutions of yaba-like disease virus DNA (6.8e + 006 − 6.8 copies/μl) were eight-fold amplified. The mean Ct-values were plotted against the copy number.
Fig. 3.
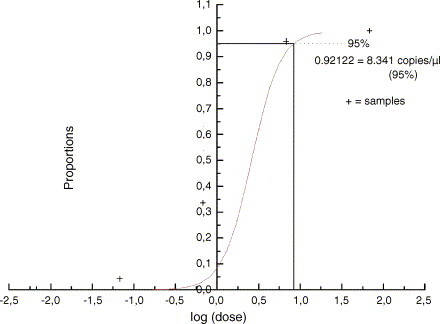
Predicted proportion of positive amplification results versus the input concentration of purified plasmid DNA as determined by probit regression analysis. The concentration at which 95% of results are expected to be positive was calculated to be ca. 8 copies/μl.
Specificity of the assay was investigated by testing 29 DNA preparations from various rash illness-causing viruses and bacteria including the following: vaccinia virus MVA and Elstree, cowpox virus Brighton, cat-pox 3, L97 and Norway, camelpox virus VD Mauretania, mousepox virus MP-1, parapoxvirus strain ORF D1701 and avipoxvirus strain HP-1. Varicella Zooster virus, a herpesvirus, causing a similar clinical entity was tested as well as bacterial agents: Bacillus anthracis, Brucella abortus and canis, Burkholderia mallei, pseudomallei and cepacia, Francisella tularensis, Pseudomonas aeruginosa and putida, Staphylococcus aureus and epidermidis, Stenotrophomonas maltophila, Vibrio parahaemolyticus, Yersinia pseudotuberculosis and pestis. However, no amplification was obtained with any of the preparations tested.
The use of this assay for examining clinical specimens was investigated using 40 DNA preparations from vesicles or crusts of human cases of suspected monkeypox (kindly provided by M. Tamfum, Kinshasa, Democratic Republic of Congo). Of these samples, eight had been confirmed previously as positive for monkeypox virus, while 22 were positive for Varicella Zoster virus (unpublished results). However, none of these samples was positive for TPV/YLDV.
In summary, a highly sensitive and specific TaqMan® assay was developed for the detection of human-pathogenic yatapoxviruses TPV and YDLV. Using this method, it is possible to obtain a reliable laboratory diagnosis within several hours after receipt of specimens. From the public health point of view, it is important to exclude infections which in the early stages might mimic infections caused by a possible bioterrorism agent and therefore pose serious public health consequences. This sensitive and specific test complements nicely the arsenal of existing methods used to discriminate more serious entities such as smallpox, monkeypox, tularemia and anthrax.
Acknowledgements
Technical support provided by D. Wenzel is gratefully acknowledged. DNA of various bacterial species were kindly provided by H. Tomaso, Bundeswehr Institute of Microbiology, Munich, Germany.
References
- Brunetti C.R., Amano H., Ueda Y., Qin J., Miyamura T., Suzuki T., Li X., Barrett J.W., McFadden G. Complete genomic sequence and comparative analysis of the tumorigenic poxvirus yaba monkey tumor virus. J. Virol. 2003;77(24):13335–13347. doi: 10.1128/JVI.77.24.13335-13347.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büchen-Osmond, C., 2003 (Ed.). 00.058.1.08. Yatapoxvirus. In: ICTVdB—The Universal Virus Database, Version 3. ICTVdB Management, Columbia University, NY, USA.
- Casey H.W., Woodruff J.M., Butcher W.I. Electron microscopy of a benign epidermal disease of rhesus monkeys. Am. J. Pathol. 1967;51:431–446. [PMC free article] [PubMed] [Google Scholar]
- Crandel R.A., Casey H.W., Brumlow W.B. Studies of a newly recognized poxvirus of monkeys. J. Infect. Dis. 1969;119:80–88. doi: 10.1093/infdis/119.1.80. [DOI] [PubMed] [Google Scholar]
- Dhar A.D., Werchniak A.E., Li Y., Brennick J.B., Goldsmith C.S., Kline R., Damon I., Klaus S.N. Tanapox infection in a college student. N. Engl. J. Med. 2004;350(4):361–366. doi: 10.1056/NEJMoa031467. [DOI] [PubMed] [Google Scholar]
- Downie A.W., Taylor-Robinson C.H., Caunt A.E., Nelson G.S., Manson-Bahr P.E., Matthews T.C. A new disease caused by a pox virus. Br. Med. J. 1971;1(745):363–368. doi: 10.1136/bmj.1.5745.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downie A.W., Espana C. Comparison of tanapox virus and yaba-like viruses causing epidemic disease in monkeys. J. Hyg. (Lond.) 1972;70(1):23–32. doi: 10.1017/s0022172400022051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grace J.T., Mirand E.A. Yaba virus infection in humans. Exp. Med. Surg. 1965;23(2):213–216. [PubMed] [Google Scholar]
- Jezek Z., Arita I., Szczeniowski M., Paluku K.M., Kalisa R., Nakano J.H. Human tanapox in Zaire: clinical and epidemiological observations on cases confirmed by laboratory studies. Bull. WHO. 1985;63:1027–1035. [PMC free article] [PubMed] [Google Scholar]
- Lee H.J., Essai K., Smith G.L. The genome sequence of yaba-like disease virus, a yatapoxviruses. Virology. 2001;281(2):170–192. doi: 10.1006/viro.2000.0761. [DOI] [PubMed] [Google Scholar]
- Niven J.S.F., Armstrong J.A., Andrewes C.H., Pereira H.G., Valentine R.C. Subcutaneous “growth” in monkeys produced by a poxvirus. J. Pathol. Bacteriol. 1961;81:1–14. [PubMed] [Google Scholar]
- Stich A., Meyer H., Kohler B., Fleischer K. Tanapox: first report in a European traveller and identification by PCR. Trans. R. Soc. Trop. Med. Hyg. 2002;96(2):178–179. doi: 10.1016/s0035-9203(02)90295-6. [DOI] [PubMed] [Google Scholar]


