Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged at the end of 2019 and caused the devastating global pandemic of coronavirus disease 2019 (COVID-19), in part because of its ability to effectively suppress host cell responses1–3. In rare cases, viral proteins dampen antiviral responses by mimicking critical regions of human histone proteins4–8, particularly those containing post-translational modifications required for transcriptional regulation9–11. Recent work has demonstrated that SARS-CoV-2 markedly disrupts host cell epigenetic regulation12–14. However, how SARS-CoV-2 controls the host cell epigenome and whether it uses histone mimicry to do so remain unclear. Here we show that the SARS-CoV-2 protein encoded by ORF8 (ORF8) functions as a histone mimic of the ARKS motifs in histone H3 to disrupt host cell epigenetic regulation. ORF8 is associated with chromatin, disrupts regulation of critical histone post-translational modifications and promotes chromatin compaction. Deletion of either the ORF8 gene or the histone mimic site attenuates the ability of SARS-CoV-2 to disrupt host cell chromatin, affects the transcriptional response to infection and attenuates viral genome copy number. These findings demonstrate a new function of ORF8 and a mechanism through which SARS-CoV-2 disrupts host cell epigenetic regulation. Further, this work provides a molecular basis for the finding that SARS-CoV-2 lacking ORF8 is associated with decreased severity of COVID-19.
Subject terms: Epigenetics, Infectious diseases
The SARS-CoV-2 protein ORF8 functions as a mimic of histone H3 to disrupt host cell epigenetic regulation.
Main
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has proven to be a highly virulent virus resulting in a devastating and global pandemic. While recent findings have suggested that SARS-CoV-2 infection disrupts epigenetic regulation12–14 and suppresses the innate antiviral host cell response1–3, it is unclear how this occurs. In rare cases, other highly virulent viruses interfere with host cell epigenetic regulation through mimicry of host cell proteins15–17, particularly histones4–8. Histones function by wrapping DNA into complex structures and, in doing so, control access to the genome. Histone proteins are modified by a wide range of post-translational modifications (PTMs) that are dynamically regulated to control gene expression9–11. Histone mimicry allows viruses to disrupt the host cell’s ability to regulate gene expression and respond to infection effectively. However, no validated cases of histone mimicry have previously been reported within coronaviruses. Although SARS-CoV-2 probably uses many mechanisms to interfere with host cell functions, we examined whether it uses histone mimicry to disrupt chromatin regulation and the transcriptional response to infection.
ORF8 contains a histone H3 mimic
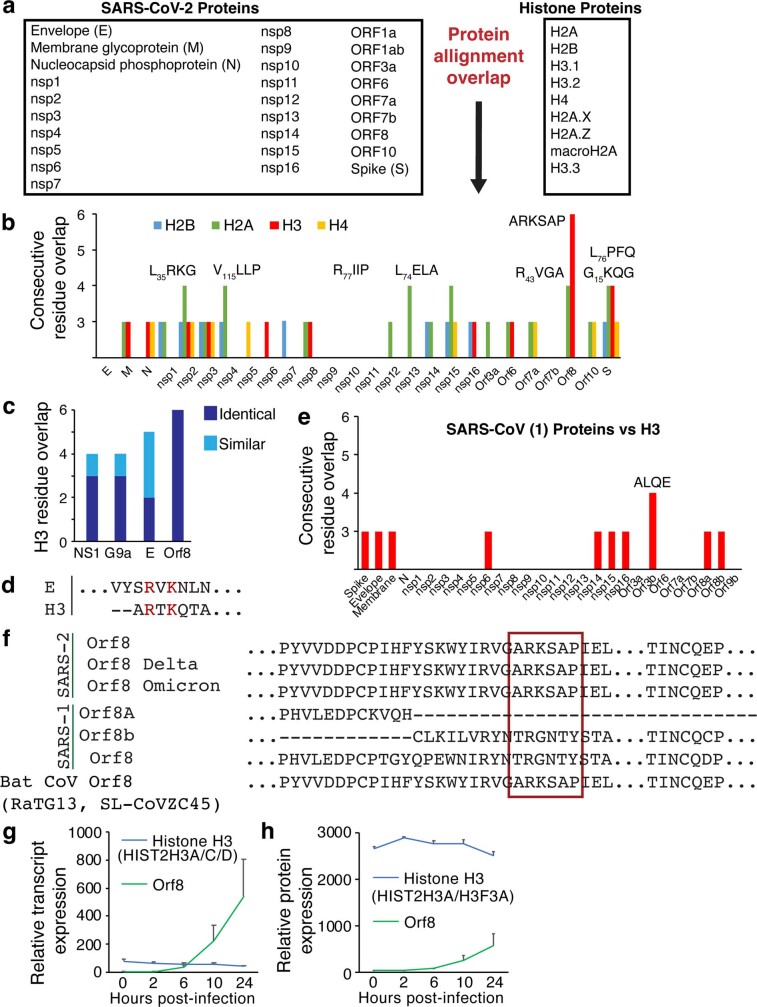
To determine whether histone mimicry is used by SARS-CoV-2, we first performed a bioinformatic comparison of all SARS-CoV-2 viral proteins18 with all human histone proteins (Extended Data Fig. 1a,b). Most SARS-CoV-2 proteins are highly similar to those in the coronavirus strain that caused the previous major SARS-CoV outbreak with the notable exception of the proteins encoded by ORF3b and ORF8, of which ORF8 is the most divergent in SARS-CoV-2 (refs 19,20). Notably, we detected an identical match between amino acids 50–55 of the protein encoded by ORF8 and critical regions within the histone H3 N-terminal tail (Fig. 1a). Furthermore, ORF8 aligns to a longer sequential set of amino acids (six residues) than in any previously described and validated case of histone mimicry4–7,21 or a putative histone mimic in the SARS-CoV-2 envelope protein22,23 (Extended Data Fig. 1c,d). On the basis of a crystal structure of ORF8, these residues are located in a disordered region on the surface of the protein in an ORF8 monomer24. Most compellingly, the motif contains the ‘ARKS’ sequence, which is found at two distinct sites in the histone H3 tail (Fig. 1a) and is well established as one of the most critical regulatory regions within H3. Both H3 ARKS sites are modified with multiple PTMs, including mono-, di- and trimethylation and acetylation at H3 lysine 9 (H3K9me and H3K9ac) and at H3 lysine 27 (H3K27me and H3K27ac). This amino acid stretch is absent from the previous SARS-CoV ORF8-encoded protein both before and after a deletion generated ORF8a and ORF8b25 but is present in bat SARS-CoV-2 and variants of concern (Extended Data Fig. 1e,f). ORF8 is highly expressed during infection26,27, with ORF8 transcript expressed at higher levels than histone H3 and ORF8 protein expressed at over 20% above the level of the most abundant histone H3 protein within 24 h of infection28 (Extended Data Fig. 1g,h). Finally, proteomic characterization of SARS-CoV-2 protein binding partners indicates that ORF8 binds DNA methyltransferase 1 (DNMT1)22,29.
Extended Data Fig. 1. Orf8 is a putative histone mimic.
(a) Alignments performed to identify putative histone mimic sites within the SARS-CoV-2 genome. (b) The number of exact sequential overlapping amino acids found between SARS-CoV-2 proteins and histone proteins. (c) The overlap of Orf8 and H3 compared to other proposed cases of histone mimicry. Exact amino acid overlap shown in dark blue and structurally similar amino acid overlap shown in light blue. NS1 is from Influenza A H3N2. Protein E is a previously proposed mimic in SARS-CoV-2. G9a is a human protein that mimics H3. (d) Previously proposed histone H3 mimic in SARS-CoV-2 protein E. (e) Overlap of SARS-CoV proteins with H3. (f) The ARKS motif is present in Bat SARS-CoV Orf8 and variants of concern but is not found with SARS-CoV Orb8a/b or the SARS-CoV precursor Orf8 before a mutation generated two distinct proteins. (g-h) Orf8 transcript (g) and protein (h) expression in SARS-CoV-2 infected Caco-2 cells from published datasets. MOI = 1. Plots indicate mean ± SEM.
Fig. 1. ORF8 associates with chromatin.
a, ORF8 contains an ARKS motif at amino acid 50 that matches the histone H3 tail. b, Lamin A/C staining of HEK293T cells transfected to express Strep–ORF8. c, ORF8 and lamin A/C staining of SARS-CoV-2-infected A549ACE2 cells at MOI = 1, 48 h after infection. d, Sequential salt extraction of HEK293T cells expressing ORF8 or ORF8ΔARKSAP. e, Gene tracks for ORF8 ChIP–seq normalized to input controls. f, Targeted mass spectrometry analysis of trypsin-digested ORF8 showing that ORF8 is acetylated at lysine 52. The intact peptide or precursor at 879.9508 m/z with a 2+ charge was isolated and fragmented. Tandem mass spectrometry spectra show unfragmented precursor (green) with matching product ions within a mass error of 10 ppm. Fragment intensity is relative to that for the ion with the highest intensity across the m/z range. The colour, letter and number for each fragment indicate the sequence that fragment contains within the larger peptide (top). y (red) and b (blue) fragments indicate C- and N-terminus-matched fragments, respectively. g, ORF8 expression results in decreased levels of KAT2A. Scale bars, 10 μm. For gel source data, see Supplementary Fig. 1b,l.
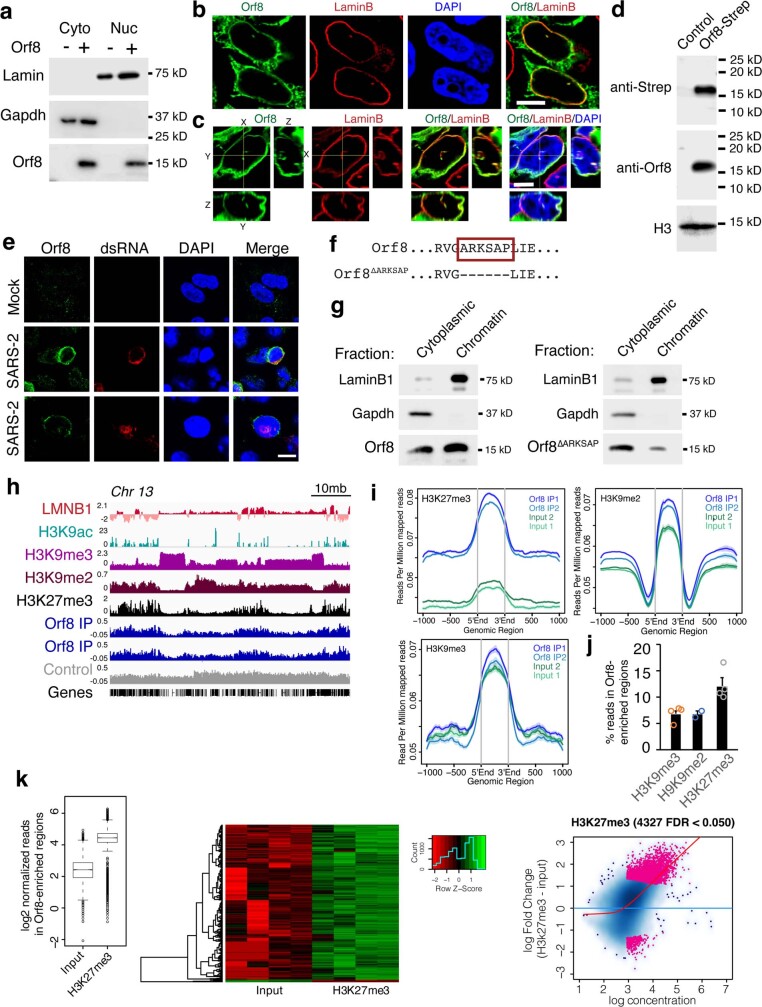
To determine whether ORF8 functions as a histone mimic, we began by examining its intracellular localization. Although ORF8 does not have a well-defined nuclear localization sequence, it is 15 kDa in size and thus small enough to diffuse into the nucleus. We transfected HEK293T cells with a construct encoding Strep-tagged ORF8 and visualized ORF8 with a Strep-Tactin-conjugated fluorescent probe. Although ORF8 localization was variable in appearance, ORF8 was typically located in the cytoplasm and at the periphery of the nucleus when using immunofluorescence (Fig. 1b), as previously reported30, and in both the cytoplasm and nucleus when using cell fractionation (Extended Data Fig. 2a). Given the observed expression pattern, we next asked whether ORF8 colocalizes with lamin proteins. We found that ORF8 colocalized with lamin B1 and lamin A/C in cells transfected to express ORF8 (Fig. 1b and Extended Data Fig. 2b,c). Next, we infected an A549 lung epithelial-derived cell line expressing the ACE2 receptor (A549ACE2) with SARS-CoV-2, stained cells with an antiserum specific to ORF8 (Extended Data Fig. 2d,e) and confirmed a similar expression pattern in infected cells (Fig. 1c). Notably, while other functions have been proposed for ORF8 (refs 30–37), a potential role for ORF8 in the nucleus of host cells and specifically in regulating chromatin has not been explored.
Extended Data Fig. 2. Orf8 localization.
(a). Subcellular fractionation of HEK293T cells transfected with Strep-Orf8 indicates Orf8 is present in the cytoplasm and nucleus. (b) HEK293T cells expressing Orf8 co-stained with Lamin B and streptactin to detect Orf8. (c) Rotation of z-stacks (right and bottom panel for each stain) indicate colocalization of Orf8 and Lamin B. (d) Orf8 antiserum specifically detects Strep-tagged Orf8 by western blot In HEK293T cells transfected with Strep-Orf8. (e) Orf8 antiserum specifically stains infected A549ACE cells 48 h after SARS-CoV-2 infection with no staining observed in mock infection. (f) Deletion construct used to test effects of ARKSAP motif in Orf8. (g) Cellular fractions generated for use in subsequent chromatin-immunoprecipitation for ChIP-sequencing shows lower ratio of Orf8ΔARKSAP present in chromatin fraction then Orf8. (h) Gene tracks for Orf8 ChIP-sequencing normalized to input controls. Control indicates negative control IgG ChIP-sequencing. (i) Orf8 is enriched in genomic regions with high H3K27me3 and H3K9me2 relative to input controls with lower enrichment at regions with high H3K9me3. (j) Percentage of reads within Orf8-enriched regions from ENCODE datasets. N = 4 samples from ENCSR000FCJ and ENCSR179BUC for H3K9me3, N = 2 samples from ENCSR55LYM for H3K9me2, and N = 4 samples from ENCSR000AKD for H3K27me3. (k) Diffbind analysis of H3K27me3 vs input controls in Orf8 enriched regions. Scale bar = 5μM in a-b, 10μM in d. For gel source data, see Supplementary Fig. 1a, f, k. Bar plots indicate mean ± SEM.
We next tested whether ORF8 is associated with chromatin by using increasing salt concentrations to examine chromatin binding. We found that ORF8 dissociated from the chromatin fraction at salt concentrations similar to those at which lamin and histone proteins dissociate (Fig. 1d). By contrast, ORF8 with a deletion of the ARKSAP motif (ORF8ΔARKSAP) dissociated at lower salt concentrations and was present at lower levels in the chromatin fraction in comparison to ORF8 with this motif (Fig. 1d and Extended Data Fig. 2f,g), indicating that the putative histone mimic site affects the strength of ORF8’s association with chromatin. We next performed chromatin immunoprecipitation with sequencing (ChIP–seq) for ORF8 to determine whether and where ORF8 associates with genomic DNA. Although ORF8 did not have clearly defined peaks, ORF8 immunoprecipitation showed enrichment over input (Fig. 1e) and ORF8 was enriched within specific genomic regions, particularly those associated with H3K27me3 (Extended Data Fig. 2h–k).
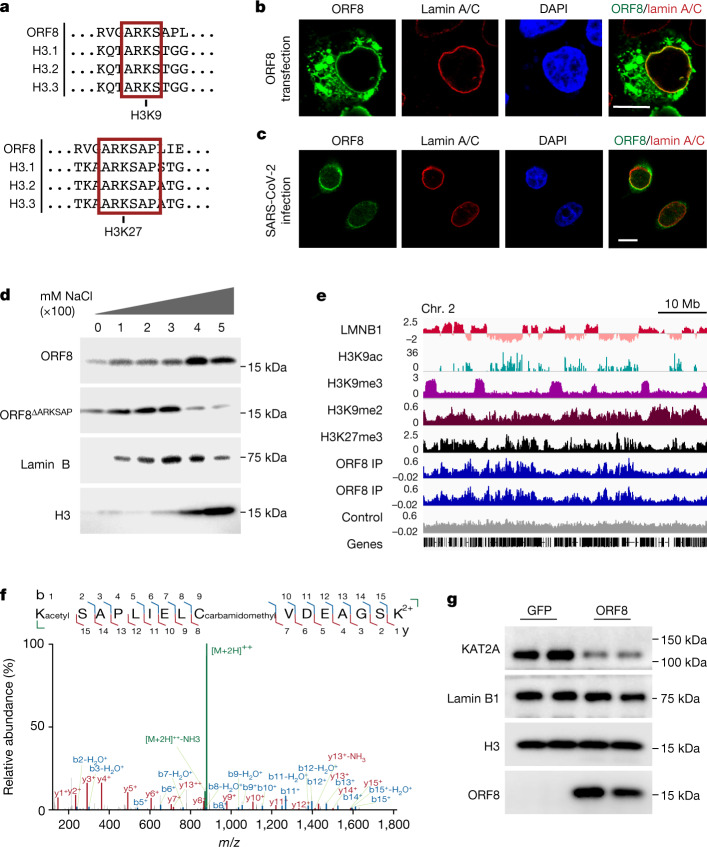
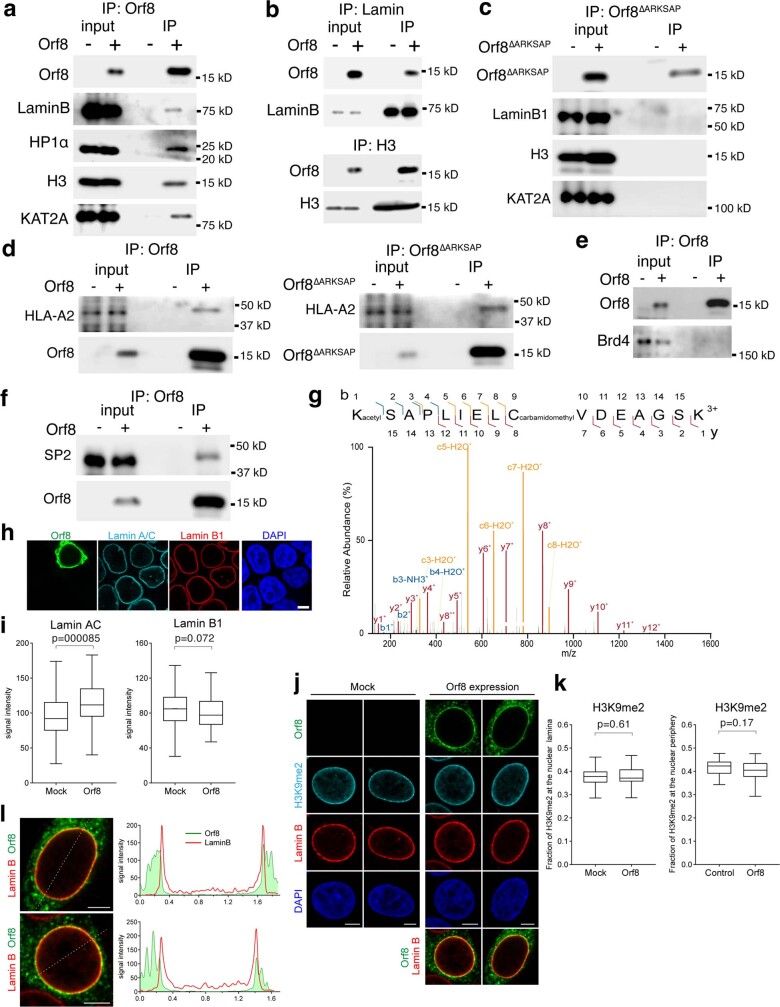
On the basis of the localization of ORF8 to the periphery of the nucleus and its association with chromatin (observed using both biochemical and sequencing approaches), we further tested whether ORF8 associates with lamin-complex proteins. We found that ORF8 co-immunoprecipitated with lamin B1, histone H3 and HP1α, a protein associated with both lamin proteins and histones (Extended Data Fig. 3a). Reciprocal co-immunoprecipitation for lamin B1 and histone H3 confirmed ORF8 binding (Extended Data Fig. 3b). Next, we tested whether ORF8 also co-immunoprecipitates with the histone-modifying enzymes that target the ARKS motif within histone H3. We found that ORF8 was associated with the histone acetyltransferase KAT2A (also known as GCN5), which targets H3K9 (Fig. 1f). Although both ORF8 and ORF8ΔARKSAP immunoprecipitated with a previously established cytoplasmic binding partner, HLA-A2 (ref. 30), we did not detect ORF8ΔARKSAP association with chromatin proteins, indicating that the ARKSAP motif strengthens ORF8’s association with chromatin proteins (Extended Data Fig. 3c,d). Further, ORF8 did not bind to BRD4, which preferentially binds acetylated histone H4 (Extended Data Fig. 3e). Finally, we used mass spectrometry to identify additional binding partners beyond those found through a candidate approach focused on chromatin modifiers (Supplementary Table 1). Whole-cell lysate that was largely depleted of chromatin proteins was used in a complementary approach in which mainly cytoplasmic proteins were therefore identified. However, the transcription factor SP2 was detected and confirmed to bind to ORF8 by co-immunoprecipitation (Extended Data Fig. 3f).
Extended Data Fig. 3. Orf8 protein interactions and modifications.
(a) Orf8 co-immunoprecipitates with Lamin complex-associated proteins including LaminB, HP1α, and H3, and KAT2A. ‘-‘ indicates cells that are not expressing Orf8 for negative control IPs performed in parallel. (b) Reciprocal co-immunoprecipitation for Lamin and H3 confirm Orf8 binding. (c) Orf8ΔARKSAP co-immunoprecipitation with LaminB, HP1α, and H3, and KAT2A is not detected. (d) Orf8 and Orf8ΔARKSAP co-immunoprecipitates with MHC1 complex protein HLA-A2. (e) Orf8 co-immunoprecipitation with Brd4 is not detected. (f) Confirmation of mass spectrometry analysis for transcription factor SP2. Orf8 was immunoprecipitated with Streptactin beads and resulting lysates were probed for SP2 and Streptactin. Proteins purified for mass spectrometry analysis were isolated using whole cell lysate conditions rather than through enrichment of the chromatin fraction. This approach (described in methods) captured predominantly cytoplasmic binding partners and a limited numbers of chromatin-associated proteins. (g) Targeted mass spectrometry analysis of trypsin-digested Orf8 of the peptide containing the proposed histone mimic in Orf8. Orf8 acetylation at K52 is detected in the 3+ charged peptide by targeted mass spectrometry. MS/MS spectra with matching product ions (b ions in blue, y ions in red, c ions in yellow) within 10ppm mass error. (h) Representative confocal images of HEK293T cells transfected with orf8 expression construct (green) and stained for Lamin A/C (cyan) and Lamin B1 (red). DAPI counterstain shown in blue. Scale bar 5μm. (i) Bar graphs show distributions of total Lamin A/C or Lamin B1 signal intensity per cell in Mock and Orf8-expressing cells. n = 129 and 86 cells per sample. Statistical analysis was performed using Mann-Whitney test. (j) Representative confocal images of HEK293T cells transfected with Orf8 expression construct (green) and stained for H3K9me2 (red) and Lamin B (cyan). DAPI counterstain shown in blue. Scale bar 5μm. (k) Bar graph show fraction of H3K9me2-marked chromatin at the nuclear lamina/periphery of control and Orf8-expressing cells. n ≥ 32 cells per sample. (l) Representative confocal images shown in j of HEK293T cells transfected with Orf8 expression construct (green) and stained for Lamin B1 (red) with Orf8 and Lamin B1 enrichment through the cell plotted along the dotted lines shown in image. Scale bar 5μm. Statistical analysis was performed using Welch 2-sided t test. Box plots show median, 25th and 75th percentiles. Whiskers show Tukey confidence intervals. For gel source data, see Supplementary Fig. 1c, d, g–j.
On the basis of the observation that ORF8 associates with KAT2A, we used targeted mass spectrometry to determine whether the proposed ORF8 histone mimic site is modified similarly to histones. Using a bottom–up approach, ORF8 was purified from cells, reduced, alkylated and digested. Separation with liquid chromatography was followed by parallel reaction monitoring mass spectrometry (LC–PRM-MS) targeting possible unmodified and modified forms of ORF8 commonly found for histones, including serine phosphorylation and lysine monomethylation, dimethylation, trimethylation and acetylation. Of these targets, unmodified and acetylated lysine were identified. The acetylated peptide contained a mass shift of +42 Da and demonstrated almost complete coverage of all possible product ions. High-resolution mass spectrometry differentiated the precursor from the trimethylated peptide and matched all product ions within a mass error of 10 ppm (Fig. 1f and Extended Data Fig. 3g). This demonstrates that ORF8 is acetylated on the lysine within the proposed ARKS histone mimic site, similarly to histone H3. Notably, presence of acetylated lysine within the ARKSAP motif is probably incompatible with dimerization of ORF8, which involves a hydrogen-bond interaction at this residue24, and thus suggests that ORF8 can exist as a monomer within cells. Finally, given that ORF8 promotes lysosomal degradation of another binding partner30,38, we examined whether ORF8 similarly affects chromatin-associated proteins. ORF8 expression resulted in a marked decrease in the abundance of KAT2A (Fig. 1g), whereas levels of nuclear lamina proteins and lamina-associated heterochromatin were unchanged or slightly increased (Extended Data Fig. 3h–l). These findings suggest that not only does ORF8 associate with proteins such as acetyltransferases, but it probably also is modified by them similarly to histone H3 and induces their degradation. Taken together, these findings demonstrate that ORF8 is well positioned to act as a histone mimic on the basis of its association with chromatin and chromatin-modifying enzymes and its ability to deplete the histone acetyltransferase KAT2A.
ORF8 disrupts chromatin regulation
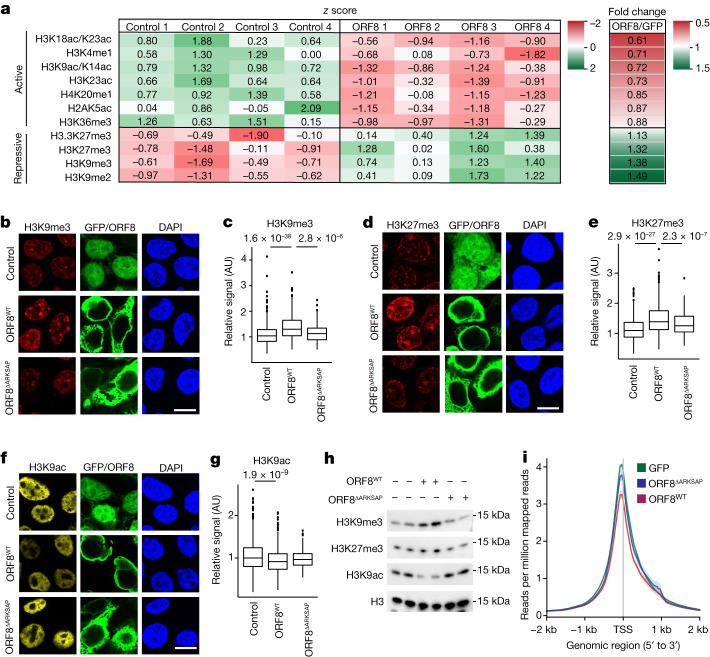
We next examined whether ORF8 expression disrupts histone PTMs using an unbiased mass spectrometry approach. HEK293T cells were transfected with a control plasmid encoding GFP or with a plasmid encoding ORF8 with a Strep tag. Transfected cells, identified by GFP fluorescence or by a Strep-Tactin-conjugated fluorescent probe, were isolated using fluorescence-activated cell sorting (FACS). Histones were purified through acid extraction, and bottom–up unbiased mass spectrometry was performed to quantify all detected histone PTMs. Notably, histone modifications associated with transcriptional repression were increased while numerous histone modifications associated with active gene expression were depleted in cells expressing ORF8 (Fig. 2a). In particular, modifications within the H3 ARKS motifs were highly disrupted. For example, the peptides containing methylated H3K9 and H3K27, which are associated with transcriptional repression, showed robustly increased abundance in response to ORF8 expression. Conversely, the peptide containing both H3K9ac and H3K14ac, both of which have a well-established link to active gene expression, showed decreased abundance in response to ORF8 expression. These data support a role for ORF8 as a putative histone mimic and demonstrate that it is capable of disrupting histone PTM regulation at numerous critical sites within histones.
Fig. 2. ORF8 function in histone PTM regulation.
a, Mass spectrometry analysis of histone PTMs in control (GFP-expressing) or ORF8-expressing HEK293T cells isolated by FACS. The z score and fold change are shown for modifications that were significantly changed in response to ORF8 expression, were detected in over 1% of the total peptide abundance and have well-established functions (full results shown in Supplementary Table 2). b–g, Immunofluorescence analysis of HEK293T cells transfected to express GFP or Strep–ORF8 showing that ORF8 expression increases H3K9me3 (b,c) and H3K27me3 (d,e) while decreasing H3K9ac (f,g). Conversely, ORF8 with deletion of the histone mimic site ARKSAP (ORF8ΔARKSAP) does not affect these histone PTMs. n = 614 (GFP), 497 (ORF8) and 170 (ORF8ΔARKSAP) cells for H3K9me3; 616, 550 and 154 cells for H3K27me3; and 666, 568 and 170 cells for H3K9ac compiled from three independent transfections. One-way ANOVA with post hoc two-sided t test and Bonferroni correction. h, Western blot analysis of histones isolated from FACS-sorted transfected cells. i, ATAC-seq of HEK293T cells expressing GFP, ORF8 or ORF8ΔARKSAP isolated by FACS. Reads per million mapped reads surrounding the transcription start site (TSS) of all expressed genes were averaged. n = 2 independent replicates. Original blots shown in Supplementary Fig. 1. Scale bars, 10 μm. The FACS gating strategy and cell numbers isolated are shown in Supplementary Fig. 2. For gel source data, see Supplementary Fig. 1e. Box plots are centred on the median with bounds at the 25th and 75th percentile, the minimum and maximum defined as the median ± 1.5× the interquartile range and whiskers extending to the lowest and highest values in the range.
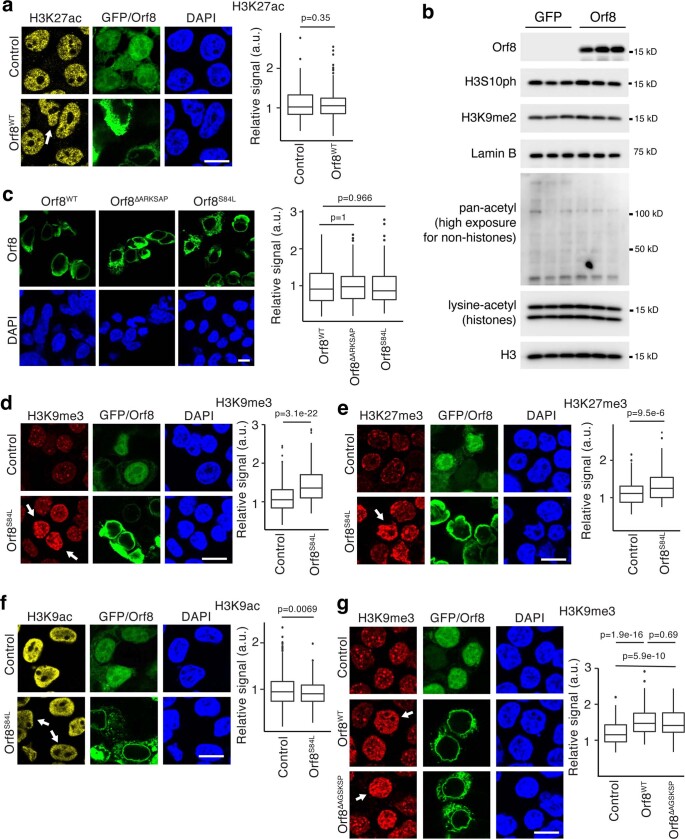
To confirm the mass spectrometry findings, we used immunofluorescence imaging to measure methylated and acetylated H3K9 and H3K27. We found that cells expressing ORF8 exhibited increased H3K9me3 and H3K27me3 and decreased H3K9ac staining compared with those transfected with control plasmid (Fig. 2b–g). ORF8 expression did not significantly disrupt H3K27ac, global acetylation, H3S10 phosphorylation, H3K9me2 or lamin B (Extended Data Fig. 4a,b). Although ORF8ΔARKSAP was expressed at similar levels to ORF8 (Extended Data Fig. 4c), it did not increase H3K9me3 or H3K27me3 and had a non-significant intermediate effect on H3K9ac (Fig. 2b–g). Next, we examined an acquired mutation in ORF8 commonly found in SARS-CoV-2 strains encoding an S84L substitution (ORF8S84L). This site is unlikely to affect protein stability31,39 and lies outside the histone mimic region, and the substitution is thus not expected to affect the ability of ORF8 to regulate histone PTMs. Expression of ORF8S84L also increased H3K9me3 and H3K27me3 levels while decreasing H3K9ac (Extended Data Fig. 4d–f), indicating that, as predicted, this common variant does not alter the histone mimic function of ORF8. Similarly, a six-residue deletion in another unstructured region of ORF8 with similar amino acid make-up but a different sequence (AGSKSP) as the histone mimic site did not affect the ability of ORF8 to disrupt histone regulation (Extended Data Fig. 4g).
Extended Data Fig. 4. Orf8 effects on histone PTMs.
(a) Orf8 does not significantly affect H3K27ac. N = 616 (GFP), 550 (Orf8) from 3 independent transfections. (b) H3 serine 10 phosphorylation (H3S10ph), H3K9 dimethylation (H3K9me2), Lamin B and global acetylation (of histones or of non-histone proteins) in GFP or Orf8 expressing cells isolated by FACS. (c) Orf8 constructs tested show similar levels of expression in HEK293T cells. N = 137 (Orf8), 87 (Orf8ΔARKSAP), 120 (S84L) from 4 independent transfections. (d–f) Orf8 with a mutation commonly found in the SARS-CoV-2 genome, Orf8-S84L, shows the same effects on histone PTMs H3K9me3 (d), H3K27me3 (e), and H3K9ac (f). N = 332 (GFP), 237 (S84L) cells for H3K9me3; 186, 166 cells for H3K27me3; 332, 237 cells for H3K9ac from 2 independent transfections. (g) Orf8 with a 6 amino acid deletion outside of the ARKSAP motif shows the same effects on H3K9me3. N = 216 (GFP), 120 (Orf8), 88 (AGSKSP) cells from 2 independent transfections. (a,g) 1-way ANOVA with post-hoc 2-sided t-test and Bonferroni correction. (b,d,e,f) 2-sided t-test. Scale bar = 10μM. For gel source data, see Supplementary Fig. 1n-m.
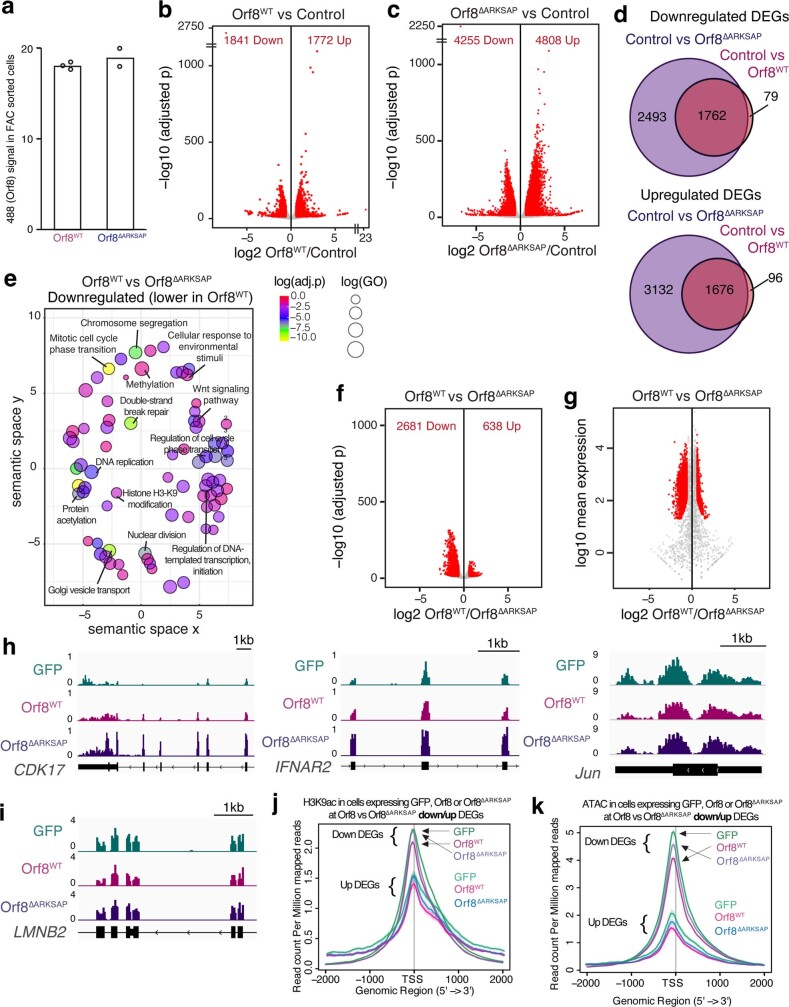
We next sought to confirm these findings using independent methods. To ensure equal levels of expression of ORF8 and ORF8ΔARKSAP, we isolated transfected cells by FACS (Extended Data Fig. 5a). We then isolated histones through acid extraction and confirmed that ORF8 increased H3K9me3 and H3K27me3 and deceased H3K9ac in an ARKSAP-dependent manner by western blot analysis (Fig. 2h). Similarly, CUT&Tag sequencing of H3K9ac demonstrated that ORF8, but not ORF8ΔARKSAP, deceased H3K9ac (Extended Data Fig. 5b,c). Finally, assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq) demonstrated that ORF8, but not ORF8ΔARKSAP, decreased chromatin accessibility (Extended Data Fig. 5d and Fig. 2i). The changes in both H3K9ac and chromatin accessibility were largely global but were particularly evident for genes with intermediate to high expression (Extended Data Fig. 5e–h).
Extended Data Fig. 5. Orf8 effects on histone PTMs and chromatin accessibility.
(a) Expression of WT Orf8 and Orf8ΔARKSAP from FAC sorted cells used for western blot analysis. (b) Expression of WT Orf8 and Orf8ΔARKSAP from FAC sorted cells used for H3K9ac CUT&TAG. (c) H3K9ac CUT&TAG sequencing average profile of all expressed genes. (d) Expression of WT Orf8 and Orf8ΔARKSAP from FAC sorted cells used for ATAC-seq. (e) H3K9ac CUT&TAG average profiles for high, mid, low, and non-expressing genes. (f) ATAC-seq average profiles for high, mid, low, and non-expressing genes. (g) H3K9ac CUT&TAG and ATAC-seq gene tracks of genes relevant to viral responses in HEK 293T cells expression a control plasmid, Orf8, or Orf8ΔARKSAP. (h) Gene track example with limited changes between Orf8 and Orf8ΔARKSAP in H3K9ac CUT&TAG and ATAC-seq. N = 2 biologically independent samples for a,b,d. Bar plots indicate mean values. FACS gating strategy and cell numbers isolated shown in Fig. S2.
To determine how these chromatin disruptions affect gene expression, we used RNA sequencing (RNA-seq) to define differentially expressed genes in transfected cells (Extended Data Fig. 6a–c). While ORF8 and ORF8ΔARKSAP shared a subset of differentially expressed genes, the presence of the histone mimic motif resulted in less dynamic gene expression changes. Distinct gene groups were also differentially expressed between ORF8 and ORF8ΔARKSAP, with ORF8 decreasing gene expression relative to ORF8ΔARKSAP, particularly highly expressed genes (Extended Data Fig. 6d–i and Supplementary Table 3). Genes that were downregulated in response to ORF8 expression relative to ORF8ΔARKSAP also had higher basal levels of H3K9ac and greater accessibility than genes that were upregulated (Extended Data Fig. 6j,k), suggesting that they may be more sensitive to depletion of H3K9ac. Together, these results support a model in which ORF8 has multiple functions as previously proposed30–33,40 and activates a number of gene expression pathways, particularly in the absence of the ARKSAP motif. However, presence of the ARKSAP motif dampens the host cell transcriptional response and decreases expression of genes with high accessibility and H3K9ac. Together, these data define a role for ORF8 in disruption of host cell histone PTMs through a new case of histone mimicry of the ARKS motifs in histone H3.
Extended Data Fig. 7. Orf8 mediates SARS-CoV-2 effects on repressive histone PTMs.
(a) qRT-PCR analysis of expression of SARS-CoV-2 gene RdRp analysis of viral titer in A549ACE pulmonary cells at 24 and 48 h after infection with SARS-CoV-2WT or SARS-CoV-2ΔOrf8 at MOI = 1. (b) Plaque assay analysis of viral titer in A549ACE pulmonary cells at 24 and 48 h after infection with SARS-CoV-2WT or SARS-CoV-2ΔOrf8 at MOI = 1. N = 3 replicates from infection done in parallel. (c-d) ChIP-RX for H3K9me3 (c) and H3K27me3 (d) of A549ACE cells with SARS-CoV-2WT, SARS-CoV-2ΔOrf8, or mock infection at MOI = 1, 48 h after infection. N = 3. Peaks shown in right panels of c-d indicate SICER peak regions from merged ChIP-RX data. Grey lines indicate peak borders and within-peak distances anchored by each end of the peak. (e) ChIP-seq gene tracks of genes in signaling pathways relevant to viral response. P values indicate 2-way ANOVA with Šidák’s multiple comparisons correction. Bar plots indicate mean ± SEM. RPM, reads per million mapped reads.
Extended Data Fig. 6. Orf8 disrupts gene expression.
(a) Expression of Orf8WT and Orf8ΔARKSAP from FAC sorted cells used for RNA-seq. (b) Volcano plot of differential gene expression analysis of Orf8WT expressing cells compared to GFP expressing cells. (c) Volcano plot of differential gene expression analysis of Orf8ΔARKSAP expressing cells compared to GFP expressing cells. Red indicates significantly differentially expressed genes. (d) Overlap of genes down or upregulated by Orf8 and Orf8ΔARKSAP compared to GFP expressing cells. (e) Gene ontology analysis of genes that are downregulated by Orf8 compared to Orf8ΔARKSAP. (f-g) Volcano plot of differential gene expression analysis of Orf8WT expressing cells compared to Orf8ΔARKSAP expressing cells graphed by p value (f) and mean expression (g). (h) Gene tracks of genes that are induced by Orf8ΔARKSAP but show a dampened response to Orf8. (i) Gene tracks of a gene that is not disrupted by Orf8 expression. (j) H3K9ac CUT&Tag reads at down and upregulated DEGs in Orf8 verses Orf8ΔARKSAP. (k) ATAC-seq reads at down and upregulated DEGs in Orf8 verses Orf8ΔARKSAP. DEG: differentially expressed gene. Volcano plots show differentially expressed genes in DESeq2 analysis with multiple comparison corrections. Gene ontology significance based on clusterProfiler analysis with Benjamini-Hochberg adjusted p-values. For RNA-seq, N = 2 for GFP and Orf8ΔARKSAP and N = 3 for Orf8WT. FACS gating strategy and cell numbers isolated shown in Supplemental Fig. 2.
SARS-CoV-2 disrupts chromatin regulation
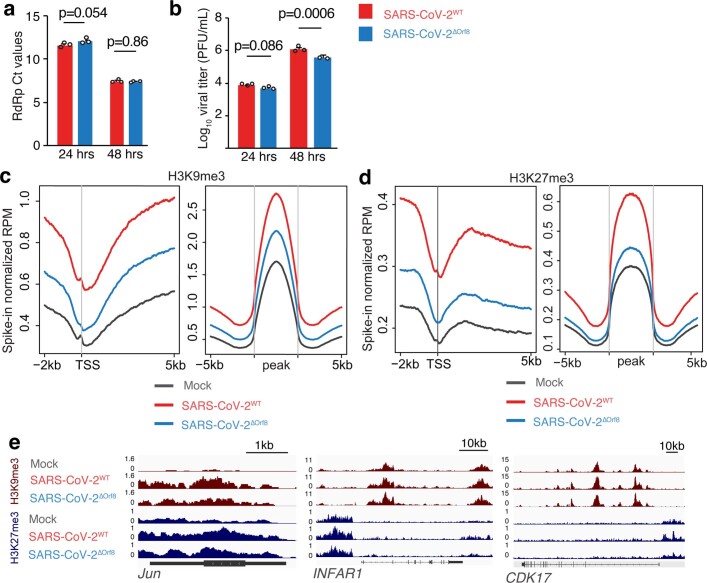
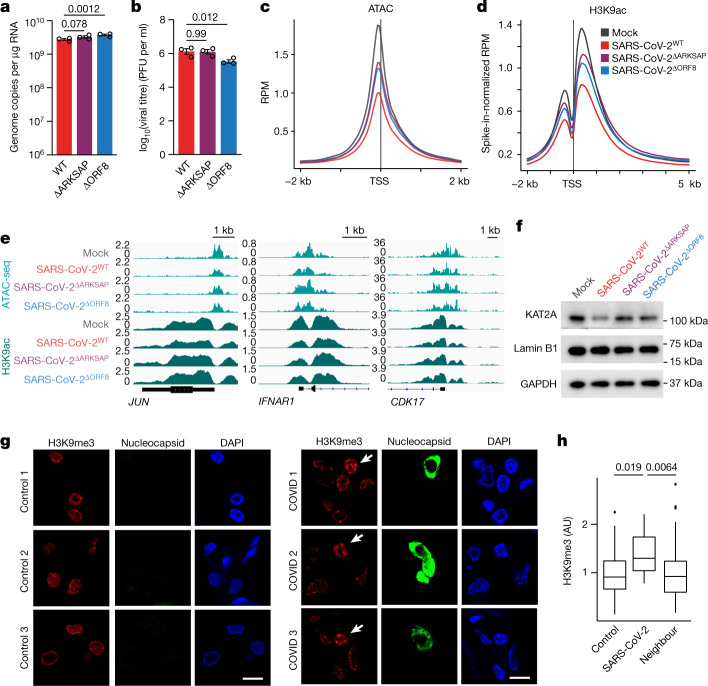
Having shown that ORF8 alone is sufficient to disrupt chromatin regulation, we next examined the effect of ORF8 on histone PTM regulation in the context of viral infection. We generated a recombinant mutant SARS-CoV-2 virus with a deletion of ORF8 (SARS-CoV-2ΔORF8) using a cDNA reverse genetics system41,42. We infected A549ACE2 cells with SARS-CoV-2 or SARS-CoV-2ΔORF8 and compared the levels of the viral genomes and infectious virus production in the presence and absence of ORF8. Because of their overexpression of the ACE2 receptor, these cells are readily and rapidly infected by SARS-CoV-2 and thus provide an ideal system in which to compare the cellular responses to mutant forms of the virus without the confounding factor of different rates of infection. No differences in genome copy number or viral titre were detected at 24 h, and only subtle differences were observed at 48 h (Extended Data Fig. 7a,b and Fig. 3a,b), allowing for direct comparison of these two viruses at these early time points after infection.We therefore infected A549ACE cells with SARS-CoV-2 or SARS-CoV-2ΔORF8 and used ChIP–seq with spike-in normalization (ChIP-RX) to allow for the detection of global changes in histone PTMs. We found that SARS-CoV-2 infection resulted in robust increases in H3K9me3 and H3K27me3 compared with mock-infected cells (Extended Data Fig. 7c–e), mirroring the effects of ORF8 expression. However, deletion of ORF8 substantially attenuated this effect, indicating that the effect of SARS-CoV-2 on repressive histone modifications is partly due to ORF8 expression. Similarly, ATAC-seq demonstrated that infection with wild-type SARS-CoV-2 resulted in substantial chromatin condensation whereas infection with SARS-CoV-2ΔORF8 resulted in an intermediate phenotype. Finally, ChIP-RX indicated that SARS-CoV-2 infection resulted in decreased H3K9ac, and this effect was again attenuated in cells infected with SARS-CoV-2ΔORF8 (Fig. 3d,e). These data demonstrate that ORF8 contributes to the effects of SARS-CoV-2 infection on chromatin accessibility and histone modifications in host cells.
Fig. 3. SARS-CoV-2 infection affects histone PTMs.
a,b, Reverse transcription with quantitative PCR (qRT–PCR) analysis of expression of the SARS-CoV-2 gene RDRP (a) and plaque assay analysis of viral titre (b) in A549ACE cells 48 h after infection with wild-type SARS-CoV-2 (SARS-CoV-2WT), SARS-CoV-2ΔARKSAP or SARS-CoV-2ΔORF8 at MOI = 1. Two-way ANOVA with Dunnett’s multiple-comparison test (additional time points shown in Supplementary Table 4). Representative of two independent infections. PFU, plaque-forming units. c,d, ATAC-seq (c) and H3K9ac ChIP-RX (d) of A549ACE cells with SARS-CoV-2WT, SARS-CoV-2ΔARKSAP, SARS-CoV-2ΔORF8 or mock infection 48 h after infection. MOI = 1. n = 3 for ATAC-seq except n = 2 for SARS-CoV-2ΔARKSAP. n = 3 for ChIP-RX except n = 2 for SARS-CoV-2WT. RPM, reads per million. e, ChIP–seq and ATAC-seq gene tracks of genes in signalling pathways relevant to viral response. f, Western blot analysis of KAT2A in A549ACE cells following infection with wild-type or mutant SARS-CoV-2 viruses. g, Post-mortem lung tissue from patients with COVID-19 stained for H3K9me3 and nucleocapsid protein to identify SARS-CoV-2-infected cells. Arrows indicate infected cells. h, Quantification of H3K9me3 in infected cells compared with neighbouring cells and with control tissue. n = 12 infected cells and 131 uninfected neighbouring cells from three patients with COVID-19 and 60 cells from three control individuals. One-way ANOVA with post hoc two-sided t test and Bonferroni correction. Scale bars, 10 μm. For gel source data, see Supplementary Fig. 1o. Box plots are centred on the median with bounds at the 25th and 75th percentiles, the minimum and maximum defined as the median ± 1.5× the interquartile range and whiskers extending to the lowest and highest values in the range.
Because it is likely that ORF8 has multiple effects on cellular function, on the basis of both recent puplications30–33,40 and our mechanistic data, we also sought to determine whether these effects were specifically due to the histone mimic motif. To do this, we generated a mutant form of SARS-CoV-2 with a deletion of only the ARKSAP motif (SARS-CoV-2ΔARKSAP). In A549ACE2 cells, SARS-CoV-2ΔARKSAP replicated similarly to wild-type virus (Fig. 3a,b) but substantially alleviated the effect of infection on chromatin accessibly and H3K9ac, matching the effects of ORF8 deletion (Fig. 3c–e). Given the robust effects of SARS-CoV-2 on H3K9ac and the ability of ORF8 to deplete KAT2A (Fig. 1g), we also examined the effect of infection on KAT2A levels. Wild-type SARS-CoV-2 infection reduced KAT2A expression, whereas infection with SARS-CoV-2ΔORF8 or SARS-CoV-2ΔARKSAP did not (Fig. 3f). These data indicate that ORF8, and specifically the ARKSAP motif within ORF8, contributes to the effects of SARS-CoV-2 on the host cell epigenome.
To ensure that the differences observed in host cell chromatin regulation following SARS-CoV-2 and SARS-CoV-2ΔORF8 infection are not due to any subtle difference in rates of infection between viruses, we sought to further confirm these finding using an approach that is independent of the number of cells infected. We used immunocytochemistry to stain for histone modifications of interest, using staining for double-stranded RNA (dsRNA) to identify and specifically examine infected cells. At 24 h after infection, cells infected with SARS-CoV-2 had increased H3K9me3 and H3K27me3 and decreased H3K9ac compared with either mock-infected cells or uninfected neighbouring cells (Extended Data Fig. 8a–f). As observed in ChIP–seq data, this effect was largely lost with deletion of ORF8.
Extended Data Fig. 8. Orf8 mediates SARS-CoV-2 effects on histone PTMs.
(a,c,e) H3K9me3 (a), H3K27me3 (c) or H3K9ac (e) staining of A549ACE cells 24 h after SARS-CoV-2WT, SARS-CoV-2ΔOrf8, or mock infection at MOI = 1. (b,d,f) Quantification of H3K9me3 (b), H3K27me3 (d) or H3K9ac (f). For H3K9me3 N = 39 (SARS-CoV-2), 35 (SARS-CoV-2ΔOrf8), for H3K27me3 N = 48 (SARS-CoV-2), 37 (SARS-CoV-2ΔOrf8), for H3K9ac N = 94 (SARS-CoV-2), 120 (SARS-CoV-2ΔOrf8) cells normalized to uninfected neighbor cells from 1-2 independent infections. Dotted line indicates relative signal in Mock infected condition. (g) Quantification of H3K9me3 in infected cells and neighboring cells from the same tissue slice for each individual patient sample shown separately, relative to control samples. For sample 1, N = 3 infected and 48 uninfected, sample 2, N = 7 infected and 125 uninfected, sample 3, N = 5 infected and 55 uninfected cells. (b,d,f) 1-way ANOVA with post-hoc 2-sided t-test and Bonferroni correction. Scale bar = 10μM. Box plots centered on median, bounds at 25th and 75th percentile, minimum and maximum defined as median ± 1.5x interquartile range, with whiskers extended to lowest/highest value in range.
To determine whether similar effects also occur in the context of a patient population, we obtained post-mortem lung tissue samples from three patients with coronavirus disease 2019 (COVID-19) and matched controls. We stained tissue for H3K9me3 as well as for SARS-CoV-2 nucleocapsid protein to identify infected cells. We found that, in all patient samples, infected cells showed increased H3K9me3 staining compared with neighbouring cells within the same tissue, as well as compared with control tissue (Fig. 3g,h and Extended Data Fig. 8g). While sample availability limits the conclusions that can be drawn from this assay, this finding indicates that histone PTMs are also disrupted in patients with severe COVID-19 disease. In summary, we found that the effects of SARS-CoV-2 infection on histone PTMs and chromatin compaction require ORF8 expression and mirror the ARKSAP-dependent effects of ORF8.
SARS-CoV-2 effects on transcription
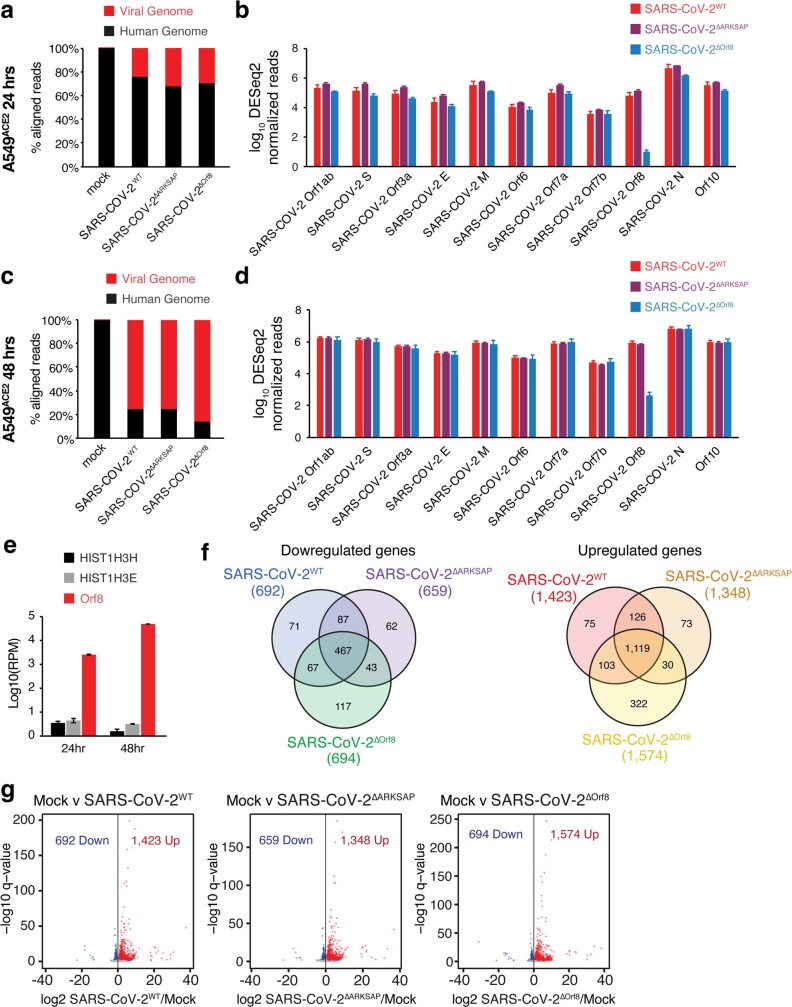
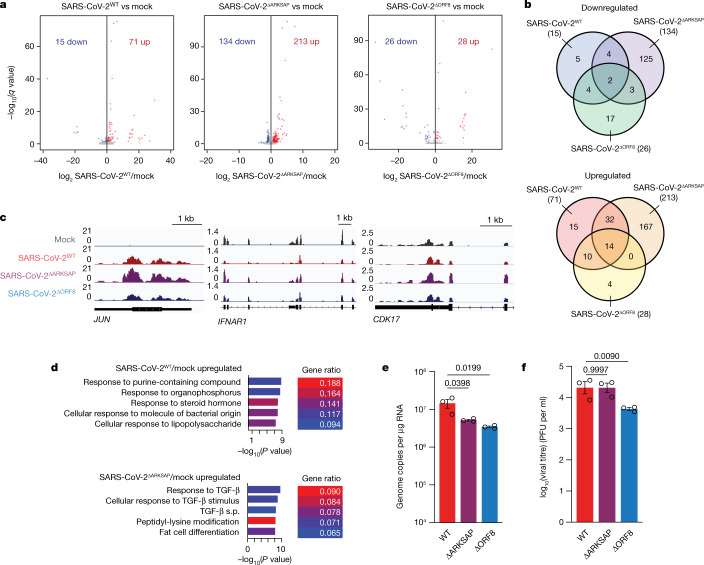
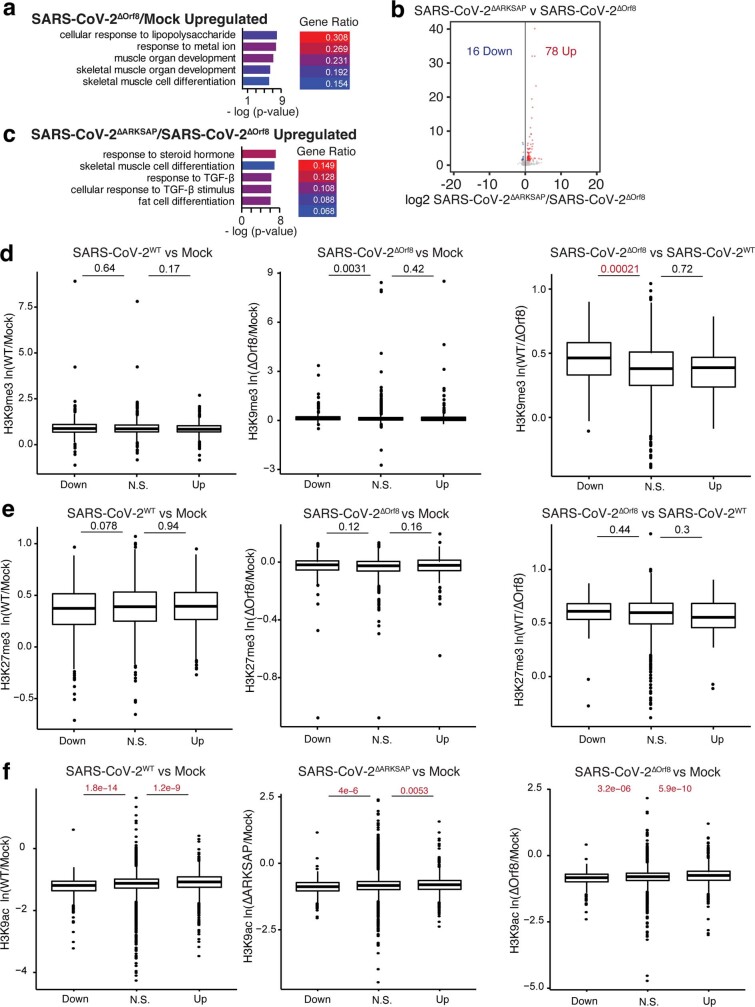
Next, we examined how the changes in histone PTMs detected through ChIP–seq relate to gene expression using RNA-seq. All viruses contained similar numbers of reads, and the only difference in SARS-CoV-2 transcript expression was for ORF8 in SARS-CoV-2ΔORF8 (Extended Data Fig. 9a–d). However, in wild-type virus, ORF8 transcript was highly expressed and more abundant than histone H3-encoding transcripts (Extended Data Fig. 9e). Interestingly, early in infection, the three viruses tested each disrupted a distinct set of genes, indicating that presence of the histone mimic motif changes the transcriptional response to infection (Fig. 4a–c). By 48 h after infection, all three viruses made up the vast majority of the mapped reads and resulted in robust changes in gene expression compared with mock-infected cells (Extended Data Fig. 9c,f,g). The functional groups of genes most induced by infection also differed among the three viruses, indicating distinct host cell responses at early time points (Fig. 4d and Extended Data Fig. 10a). This is notable given that wild-type SARS-CoV-2 and SARS-CoV-2ΔARKSAP had nearly identical copy numbers and replication rates in A549ACE2 cells (Fig. 3a,b), and thus the different transcriptional responses are unlikely to be due to differences in the number of cells infected or the viral load within infected cells. Interestingly, direct comparison of SARS-CoV-2ΔORF8 and SARS-CoV-2ΔARKSAP also showed distinct gene expression changes and functional group enrichment (Extended Data Fig. 10b,c), indicating again that ORF8 probably has multiple functions beyond those mediated by the ARKSAP domain. In addition, gene expression changes in response to infection were correlated with changes in H3K9ac (Extended Data Fig. 10d–f). Notably, these data further support recent findings indicating that SARS-CoV-2 results in a limited early transcriptional response1,2,43 and demonstrate that the ORF8 ARKSAP domain is linked to changes in gene expression.
Extended Data Fig. 9. RNA-sequencing read comparisons and 48 hour DEG.
(a) Reads aligned to the human and SARS-CoV-2 genomes 24 h after infection. (b) Levels of SARS-CoV-2 transcripts in cells infected with SARS-CoV-2WT, SARS-CoV-2ΔARKSAP, or SARS-CoV-2ΔOrf8 24 h after infection. (c) Reads aligned to the human and SARS-CoV-2 genomes 48 h after infection. (d) Levels of SARS-CoV-2 transcripts in cells infected with SARS-CoV-2WT, SARS-CoV-2ΔOrf8, or SARS-CoV-2ΔARKSAP 48 h after infection. (e) Normalized reads of SARS-CoV-2 Orf8 and human transcripts of histone H3 genes in cells infected with SARS-CoV-2WT or SARS-CoV-2ΔOrf8. (f) Overlap of differentially expressed genes in response to SARS-CoV-2WT, SARS-CoV-2ΔOrf8, or SARS-CoV-2ΔARKSAP compared to mock infection at MOI = 1, 48 h after infection. (g) Differential gene expression analysis by RNA-seq of A549ACE cells 48 h after SARS-CoV-2WT, SARS-CoV-2ΔOrf8, or SARS-CoV-2ΔARKSAP, compared to mock infection at MOI = 1. Significantly differentially expressed genes (relative to mock infection) are shown in blue (down) and red (up). N = 3. Volcano plots show differentially expressed genes in DESeq2 analysis with multiple comparison corrections. Bar plots indicate mean ± SEM.
Fig. 4. ORF8 affects gene expression and viral replication during SARS-CoV-2 infection.
a, Differential gene expression analysis by RNA-seq of A549ACE2 cells 24 h after infection with SARS-CoV-2WT, SARS-CoV-2ΔORF8 or SARS-CoV-2ΔARKSAP, compared with mock infection. MOI = 1. Significantly differentially expressed genes are shown in blue (downregulated) and red (upregulated). n = 3. Significance based on DESeq2 analysis with multiple-comparison correction. b, Overlap of differentially expressed genes in response to infection with SARS-CoV-2WT, SARS-CoV-2ΔORF8 or SARS-CoV-2ΔARKSAP. c, Gene tracks of genes in signalling pathways relevant to viral response. d, Top gene ontology (GO) terms for genes upregulated by SARS-CoV-2WT and SARS-CoV-2ΔARKSAP infection. Significance based on clusterProfiler analysis with Benjamini–Hochberg-adjusted P values. e,f, qRT–PCR analysis of expression of the SARS-CoV-2 gene RDRP (e) and plaque assay analysis of viral titre (f) in iAT2 pulmonary cells at 48 h after infection with SARS-CoV-2WT, SARS-CoV-2ΔORF8 or SARS-CoV-2ΔARKSAP at MOI = 1. n = 3 replicates. One-way (e) or two-way (f) ANOVA with Dunnett’s multiple-comparison test (additional time points for f and all replicates shown in Supplementary Table 4). Bar plots indicate mean ± s.e.m.
Extended Data Fig. 10. Comparison of ChIP-sequencing and RNA-sequencing data in A549ACE cells.
(a) Gene ontology analysis of genes upregulated by SARS-CoV-2ΔOrf8 infection. (b) Genes expression changes between SARS-CoV-2ΔOrf8 and SARS-CoV-2ΔARKSAP. (c) Gene ontology analysis of genes upregulated by SARS-CoV-2ΔARKSAP compared to SARS-CoV-2ΔOrf8. (d) Mean peak difference for H3K9me3 in genes that are down, unchanged, or upregulated in responses to infection. (e) Mean peak difference for H3K27me3 in genes that are down, unchanged, or upregulated in responses to infection. (f) Mean peak difference for H3K9ac in genes that differentially expressed in responses to infection. Peak values determined by diffbind for the most significant peak intersecting the TSS of a gene. N = 3 ChIP-sequencing for each modification except N = 2 for H3K9ac wildtype SARS-CoV-2WT. P values indicate results from a Kruskal-Wallis test followed by a post-hoc Dunn’s test and Bonferroni correction. N.S. indicates genes that are not significant differentially expressed between groups. Box plots centered on median, bounds at 25th and 75th percentile, minimum and maximum defined as median ± 1.5x interquartile range, with whiskers extended to lowest/highest value in range.
Given the robust effects of ORF8 deletion on host cell chromatin regulation and the transcriptional response to infection, we sought to test whether ORF8 mediates the replication of SARS-CoV-2 using a physiologically relevant cell type. Induced human pluripotent stem cell-derived lung alveolar type II (iAT2) pulmonary cells44 were infected with SARS-CoV-2, SARS-CoV-2ΔORF8 or SARS-CoV-2ΔARKSAP (multiplicity of infection (MOI) = 1). Notably, we observed that both mutant viruses had decreased genome copy numbers at 48 h after infection in most replicates (Fig. 4e and Supplementary Table 4), suggesting that ORF8, and specifically the ARKSAP domain, affects SARS-CoV-2 genome replication in a host cell. However, viral titres measured through plaque assays demonstrated that SARS-CoV-2ΔORF8 generated fewer infectious particles than wild-type SARS-CoV-2 while SARS-CoV-2ΔARKSAP appeared similar to wild-type virus and in some cases even showed more plaque formation (Fig. 4f and Supplementary Table 4). Fitting with previous work indicating that ORF8 affects endoplasmic reticulum (ER) stress pathways32, this suggests that ORF8 probably has an ARKSAP-independent function that may promote viral particle formation. Taken together, this work presents a link between a specific SARS-CoV-2 protein and the epigenetic disruptions that occur in response to infection and provides a mechanistic explanation for mounting evidence12,13,45 that epigenetic disruptions contribute to the severity of COVID-19.
Discussion
The work described here identifies a new case of histone mimicry during infection by SARS-CoV-2 and defines a mechanism through which SARS-CoV-2 acts to disrupt host cell chromatin regulation. We found that the protein encoded by the SARS-CoV-2 ORF8 gene contains an ARKS motif and that ORF8 expression disrupts histone PTM regulation. ORF8 is associated with chromatin-associated proteins, histones and the nuclear lamina and is itself acetylated within the histone mimic motif similarly to histones. ORF8 expression disrupts multiple critical histone PTMs and promotes chromatin compaction, whereas ORF8 lacking the histone mimic motif does not. Further, SARS-CoV-2 infection in human cell lines and post-mortem patient lung tissue causes similar global disruptions to chromatin acting in part through the histone mimic. In addition, deletion of the ORF8 gene or the sequence encoding the histone mimic affects the host cell transcriptional response to SARS-CoV-2 infection. Finally, loss of ORF8 decreases the replication of SARS-CoV-2 in human induced pluripotent stem cell-derived iAT2 pulmonary cells while loss of the histone mimic motif specifically affects viral genome copy number.
Notably, the role of ORF8 in chromatin disruption early in infection is not inconsistent with other proposed roles for ORF8 in other cellular compartments or at later stages of infection30–32,34,46 and does not preclude other proposed mechanisms of transcriptional disruption in response to SARS-CoV-2 (ref. 23). In fact, our data point towards a model in which ORF8 has multiple functions, including acting as a histone mimic motif. The effects of deletion of accessory proteins from SARS-CoV-2 in a transgenic mouse model appear complex, with ORF8 loss causing decreases in replication and viral load but having limited effects on survival47. However, data from patients with COVID-19 were used to examine a rare 382-nucleotide deletion variation in SARS-CoV-2 isolated in Singapore that results in the loss of a small portion of ORF7B and the majority of the ORF8 gene. This work found that this SARS-CoV-2 variant is associated with a milder infection in patients with COVID-19 and an improved interferon response48,49. Our findings in human iAT2 pulmonary cells point towards the loss of ORF8 as a possible cause for these differences and provide an epigenetic mechanism underlying the role of ORF8 in promoting SARS-CoV-2 virulence within the patient population. Finally, the work described here has critical implications for understanding emerging viral strains carrying deletions and mutations in the ORF8 gene50 and COVID-19 pathogenesis in patients.
Methods
A549ACE cells
ACE2-expressing A549 cells were generated as previously described3. A549ACE2 cells were grown in RPMI-1640 with 10% FBS and 1% penicillin-streptomycin and were maintained free of mycoplasma. Cells were infected at an MOI of 1 and fixed or lysed at 24 or 48 h after infection.
HEK293T cells
HEK293T cells were obtained from the American Type Culture Collection (ATCC), cultured in DMEM (with 4.5 g L–1 glucose, l-glutamine and sodium pyruvate) supplemented with 10% FBS (Sigma-Aldrich, F2442-500ML) and 1% penicillin-streptomycin (Gibco, 15140122) and maintained free of mycoplasma. Calcium phosphate transfection was used to introduce plasmid DNA encoding GFP, ORF8 and mutant ORF8 into HEK293T cells. For immunocytochemistry experiments, cells were plated on poly(d-lysine)-coated coverslips. Cells were washed 24 h after transfection with culture medium and fixed or pelleted and flash frozen 48 h after transfection. Cells were fixed using 4% paraformaldehyde (PFA) in PBS for 8 min. To pellet cells, cells were detached from the culture plate using TrypLE Express (Gibco, 12605010) dissociation reagent, spun down for 5 min at 180 g and flash frozen in liquid nitrogen.
iAT2 cells
Generation of human-derived induced alveolar epithelial type II-like (iAT2) cells was performed as described44. To maintain a stable and pure culture of the iAT2 cell line, SFTPCtdTomato+ cells were sorted and serially passaged every 14 d. Cells were grown in organoid format using 90% Matrigel with a cell density of 400 cells per µl. Cells were fed using CK+DCI medium + Rock inhibitor for the first 48 h after splitting and then changed to K+DCI medium for 5 d followed by CK+DCI medium for 7 d. Every 14 d, alveolosphere organoids were passaged, organoids were released from Matrigel using 2 mg ml–1 Dispase for 1 h at 37 °C and single cells were then generated using 0.05% trypsin for 15 min at 37 °C. Cell number and viability were assessed using Trypan blue, and cells were finally passaged to new Matrigel drops left to polymerize for 30 min at 37 °C in a 5% CO2 incubator, after which cells in solidified Matrigel were fed according to plate format.
For the generation of two-dimensional (2D) alveolar cells for virus infection, when alveolosphere organoids were passaged, cells were plated on precoated 1:30 Matrigel plates at a cell density of 125,000 cells per cm2 using CK+DCI medium + Rock inhibitor for the first 48 h, and the medium was then changed to CK+DCI medium. Seventy-two hours after cell plating, cells were infected with SARS-CoV-2 virus using an MOI of 1 for 48 h.
Cell line validation and testing
Cell lines were authenticated as previously described3. HEK293T and Vero E6 cells were obtained from ATCC at the onset of this project. All cell lines used were confirmed to be negative for mycoplasma and are retested twice annually.
ORF8 constructs
The ORF8 expression plasmid was obtained from Addgene, pLVX-EF1alpha-SARS-CoV-2-orf8-2xStrep-IRES-Puro (Addgene plasmid 141390). ORF8 deletion constructs were produced on the ORF8 backbone using Pfu Turbo HotStart DNA polymerase (Agilent, 600322-51), and primers were created using the DNA-based primer design feature of the online PrimerX tool. Constructs were verified by Sanger sequencing.
SARS-CoV-2 infection
Virus generation
SARS-CoV-2 (USA-WA1/2020 strain) was obtained from BEI and propagated in Vero E6 cells. The genome RNA was sequenced and found to be identical to GenBank MN985325.1. Mutant viruses were generated using the cDNA reverse genetics system as previously described42.
Infections
Cells were infected with wild-type or mutant SARS-CoV-2 at an MOI of 1 PFU per cell (A549ACE2) or 5 PFU per cell (iAT2) as previously described3. Virus was added to cells for 1 h at 37 °C and was then removed and replaced with medium. Cells were lysed at 48 h after infection and RNA was isolated. All infections and virus manipulations were conducted in a Biosafety Level 3 (BSL3) laboratory using appropriate protective equipment and protocols.
Viral growth kinetics and plaque assays
Growth kinetics analysis and plaque assays were performed as previously described3. In brief, at the indicated time points, 200 µl of supernatant was collected from cells and stored at −80 °C for titration of infectious virus. Samples were diluted in serum-free DMEM and adsorbed onto Vero E6 cells at 37 °C for 1 h before a liquid overlay was added (DMEM with 2% FBS, 1× sodium pyruvate and 0.1% agarose). After 3 d, the overlay was removed and cells were fixed with 4% PFA and stained with crystal violet for plaque visualization and counting. All plaque assays were performed in biological triplicate and technical duplicate.
Viral genome quantification by qRT–PCR
RNA collection, qRT–PCR and viral genome quantification were performed as previously described3. In brief, at the indicated time points, infected cells were lysed using RLT Plus Buffer, genomic DNA was removed and RNA was extracted using the Qiagen RNeasy Mini kit (Qiagen, 74134). cDNA was generated using a High-Capacity cDNA Reverse Transcriptase kit (Applied Biosystems, 4368814). cDNA was amplified using specific qRT–PCR primers targeting viral NSP12 (forward, 5′-GGTAACTGGTATGATTTCG-3′; reverse, 5′-CTGGTCAAGGTTAATATAGG-3′), iQ SYBR Green Supermix (Bio-Rad, 1708880) and the QuantStudio 3 PCR system (Thermo Fisher). Quantification of SARS-CoV-2 genome copies was performed using a standard curve generated by serially diluting a known concentration of the pcDNA6B-nCoV NSP12-FLAG construct encoding the RDRP gene (a gift from G. Stark, Cleveland Clinic) after digestion with XhoI. Genome copy numbers were determined using standard curve analysis in QuantStudio 3 software, and copy numbers per microgram of RNA were calculated using the cDNA reaction volumes and input RNA for the cDNA reactions.
Cell fractionation
Pelleted cells were briefly thawed on ice. Buffer 1 (15 mM Tris-HCl (pH 7.5), 60 mM KCl, 15 mM NaCl, 5 mM MgCl2, 1 mM CaCl2 and 0.25 M sucrose with 1 mM PMSF, 1 mM DTT and a Complete Protease Inhibitor cocktail tablet added immediately before use) was added to the pellet at roughly five times the volume of the pellet and gently pipetted up and down to dissociate the pellet. Samples were incubated on ice for 5 min, followed by addition of an equal volume of buffer 1 with 0.4% NP-40 to the sample. Samples were then mixed by inversion for 5 min at 4 °C. Samples were spun at 200 g for 10 min in a prechilled centrifuge to pellet nuclei. The supernatant (cytoplasmic fraction) was transferred to a new tube. Pellets were resuspended gently in 0.5 ml buffer 1 to wash the nuclei and then pelleted again with the supernatant discarded. Nuclear pellet solubilization buffer (150 mM NaCl, 50 mM Tris-HCl (pH 8.0), 1% NP-40 and 5 mM MgCl2 with 1 mM PMSF, 1 mM DTT and Benzonase enzyme at 250 U µl–1 added shortly before use) was added to the pellet at half the volume of buffer 1 used. Samples were then incubated at room temperature in a thermoshaker until the pellet was fully dissolved. The amount of Benzonase enzyme was doubled in samples with undissolved material left after 20 min. Samples were then centrifuged at 13,000 r.p.m. for 20 min at 4 °C. Supernatant (nuclei fraction) was collected. Sample concentrations were determined by BCA assay, and samples were boiled in a western loading buffer for 10 min before analysis by western blotting.
Chromatin sequential salt extraction
Salt extractions were performed as described51. In brief, a 2× RIPA solution was made (100 mM Tris (pH 8.0), 2% NP-40 and 0.5% sodium deoxycholate) and mixed with varying concentrations of a 5 M NaCl solution to generate RIPA containing 0, 100, 200, 300, 400 and 500 mM NaCl. Pelleted cells were resuspended in buffer A with protease inhibitors (0.3 M sucrose, 60 mM KCl, 60 mM Tris (pH 8.0), 2 mM EDTA and 0.5% NP-40) and rotated at 4 °C for 10 min. Nuclei were pelleted by centrifugation at 6,000 g for 5 min at 4 °C. Supernatant was removed and saved, and 200 µl of RIPA with 0 mM NaCl and protease inhibitors was added to the sample. Samples were mixed by pipetting 15 times and incubated on ice for 3 min before centrifuging at 6,500 g for 3 min at 4 °C. Supernatant was saved and the RIPA steps were repeated for all NaCl concentrations. Samples were then boiled and sonicated before analysis by western blotting.
ATAC-seq
HEK293T cells were stained and sorted to isolate transfected cells using the same method as described below. Sorted cells were resuspended in cold lysis buffer (10 µl per 10,000 cells; 10 mM Tris-Cl (pH 7.5), 10 mM NaCl, 3 mM MgCl2, 0.1% (vol/vol) NP-40, 0.1% (vol/vol) Tween-20 and 0.01% (vol/vol) digitonin) and washed in wash buffer (10 mM Tris-Cl (pH 7.5), 10 mM NaCl, 3 mM MgCl2 and 0.1% (vol/vol) Tween-20). Transposition was performed with Tagment DNA TDE1 (Illumina, 15027865). Transposition reactions were cleaned with AMPure XP beads (Beckman, A63880), and libraries were generated by PCR with NEBNext High-Fidelity 2× PCR Master Mix (NEB, M0541). Library size was confirmed on a Bioanalyzer before sequencing on the NextSeq 550 platform (40-bp read length, paired end).
Infected A549ACE cells were fixed before collection for ATAC-seq. The protocol was performed as above except with 0.05% Igepal CA-630 added to the lysis buffer. In addition, after the transposase reaction, a reverse cross-linking solution was added (with a final concentration of 50 mM Tris-Cl, 1 mM EDTA, 1% SDS, 0.2 M NaCl and 5 ng ml–1 proteinase K) up to 200 μl. The mixture was incubated at 65 °C with shaking at 1,000 r.p.m. in a heat block overnight and then purified as above.
For ATAC-seq analysis, alignments were performed with Bowtie2 (2.1.0)52 using the hg38 genome with the pipeline at https://github.com/shenlab-sinai/chip-seq_preprocess. Reads were mapped using NGS plot. For HEK293T cell ATAC-seq, genes with high, intermediate, low and no expression were defined by DESeq2 normalized basemean values from HEK293T cell RNA-seq data with under 2 basemean as non-expressing genes and the remaining genes binned into three groups for low, intermediate and high expression. For A549ACE cell ATAC-seq, three biological replicates each with 2–3 technical replicates were performed. Ten million reads from each individual technical replicate were subsetted (SAMtools v1.9, seed 1) and merged, and each condition was then merged across biological replicates. For average profile plots, each condition was downsampled to 40 million reads and plotted against all genes identified by DESeq2 as expressed over 1 from A549ACE RNA-seq data.
ChIP–seq
For ORF8 ChIP–seq, 2 d after transfection, cells were fixed for 5 min with 1% PFA in PBS and the reaction was then quenched with 2.5 M glycine. Cells were washed twice, collected in PBS with protease and phosphatase inhibitors and then pelleted at 1,200 r.p.m. for 5 min. Cells were then rotated in lysis buffer 1 (50 mM HEPES-KOH (pH 7.5), 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40 and 0.25% Triton X-100) for 10 min at 4 °C and spun at 1,350 g for 5 min at 4 °C to isolate nuclei. Supernatant was discarded and cells were resuspended in lysis buffer 2 (10 mM Tris-HCl (pH 8), 200 mM NaCl, 1 mM EDTA and 0.5 mM EGTA) to lyse nuclei. Samples were rotated for 10 min at room temperature and were spun again at 1,350 g for 5 min at 4 °C. The supernatant was discarded and the pellet was resuspended in lysis buffer 3 (10 mM Tris-HCl (pH 8), 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% EDTA and 0.5% N-lauroylsarcosine). Lysates were sonicated on a Covaris sonicator for 40 min (200 cycles per burst). Triton X-100 was added to reach a final concentration of 1%, and lysates were spun at 20,000 g for 10 min at 4 °C. Strep-Tactin magnetic beads (MagStrep type 3 XT beads; IBA, 2-4090-002) were added to the lysates overnight with rotation at 4 °C. Beads were then washed with a low-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris (pH 8) and 150 mM NaCl), a high-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris (pH 8) and 500 mM NaCl), a LiCl wash buffer (150 mM LiCl, 1% NP-40, 1% sodium deoxycholate, 1 mM EDTA and 10 mM Tris (pH 8)) and then TE with 50 mM NaCl. Chromatin was eluted from beads for 30 min with shaking at room temperature in 55 µl BXT elution buffer (IBA, 2-1042-025) followed by the addition of 150 µl elution buffer (50 mM Tris-HCl (pH 8.0), 10 mM EDTA and 1% SDS) for 30 min at 65 °C. Samples were removed from beads and cross-linking was reversed by further incubating chromatin overnight at 65 °C. RNA was digested with RNase for 1 h at 37 °C, and protein was digested with proteinase K for 30 min at 55 °C. DNA was then purified with the Zymo PCR purification kit. The Illumina TruSeq ChIP purification kit was used to prepare samples for sequencing on an Illumina NextSeq 500 instrument (42-bp read length, paired end).
For ORF8 ChIP–seq analysis, alignments were performed with Bowtie2 (2.1.0)52 using the hg38 genome with a ChIP–seq pipeline (https://github.com/shenlab-sinai/chip-seq_preprocess). ORF8 reads were mapped using NGS plot. For comparison with histone modification ChIP–seq datasets, ENCODE and 4D nucleome data were used for H3K9ac (experiment ENCSR000ASV), lamin (4DN experiment set 4DNES24XA7U8), H3K9me3 (experiments ENCSR000FCJ and ENCSR179BUC), H3K9me2 (experiment ENCSR55LYM) and H3K27me3 (experiment ENCSR000AKD). To define ORF8-enriched regions, HiddenDomains was used for each of two ORF8 ChIP–seq experiments normalized to input. Output files were merged with bedtools (v2.18.1) intersect to select the subset of enriched regions found in both replicates. DiffBind (3.4.11) was used to examine H3K27me3 enrichment within ORF8-enriched regions. The Deeptools (3.3.0) plotEnrichment tool was used to count percentages of reads of histone modification ENCODE ChIP–seq datasets that were within ORF8-enriched regions. ngs.plot.r (2.63) was used to generate plots of ORF8 enrichment within genomic regions of interest.
For histone PTM ChIP–seq, 4–10 million cells were resuspended in 1 ml of lysis buffer 1 (50 mM HEPES-KOH (pH 7.5), 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40 and 0.25% Triton X-100) and rotated at 4 °C for 10 min, followed by centrifugation and removal of supernatant. Cells were then resuspended in 1 ml of lysis buffer 2 (10 mM Tris-HCl (pH 8.0), 200 mM NaCl, 1 mM EDTA and 0.5 mM EGTA) and rotated for 10 min at 4 °C, followed by centrifugation and removal of supernatant. Cells were then resuspended in 1 ml of lysis buffer 3 (10 mM Tris-HCl (pH 8.0), 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% sodium deoxycholate and 0.5% N-lauroylsarcosine) and rotated again for 10 min at 4 °C. Cells were then sonicated with a Covaris S220 sonicator for 35 min (peak incident power, 140; duty factor, 5%; cycles per burst, 200). This was followed by addition of 110 µl Triton X-100 and centrifugation at maximum speed (20,000 g) for 15 min at 4 °C to clear the lysate. The lysate chromatin concentration was then equalized according to DNA content (as measured with a Qubit fluorometer). Following this, 5% of equivalently treated chromatin from Camponotus floridanus pupae was added to all samples according to chromatin concentration, and 50 µl of lysate was saved as input shearing control. Then, 250 µl of equalized lysate was added to washed, antibody-conjugated Protein A/G Dynabeads (2 µg of antibody conjugated to 15 µl of Protein A/G Dynabeads, resuspended in 50 µl per immunoprecipitation), and immunoprecipitations were rotated overnight at 4 °C in a final volume of 300 µl. The following day, immunoprecipitations were washed five times in RIPA wash buffer (50 mM HEPES-KOH (pH 7.5), 500 mM LiCl, 1 mM EDTA, 1% NP-40 and 0.7% sodium deoxycholate) and once in TE (pH 8.0). Washes were followed by two elutions into 75 µl of elution buffer (50 mM Tris-HCl (pH 8.0), 10 mM EDTA and 1% SDS) at 65 °C for 45 min with shaking (1,100 r.p.m.). DNA was purified by phenol:chloroform:isoamyl alcohol (25:24:1) extraction followed by ethanol precipitation. Pelleted DNA was resuspended in 25 µl TE. Libraries for sequencing were prepared using the NEBNext Ultra II DNA Library Prep Kit for Illumina (NEB, E7645), as described by the manufacturer but using half the volume for all reagents and starting material. For PCR amplification, the optimal number of PCR cycles was determined using a qPCR side reaction with 10% of the adaptor-ligated, size-selected DNA. Seven to ten cycles of PCR were used for histone PTM libraries and 5 cycles were used for input controls. Samples were sequenced on a NextSeq 500 instrument (42-bp read length, paired end).
For analysis of histone PTM ChIP–seq data, reads were demultiplexed using bcl2fastq2 (Illumina) with the options ‘--mask-short-adapter-reads 20 --minimum-trimmed-read-length 20 --no-lane-splitting --barcode-mismatches 0’. Reads were trimmed using TRIMMOMATIC53 with the options ‘ILLUMINACLIP:[adapter.fa]:2:30:10 LEADING:5 TRAILING:5 SLIDINGWINDOW:4:15 MINLEN:15’ and aligned to a hybrid hg38 + C. floridanus (v7.5, RefSeq) genome assembly using bowtie2 (v2.2.6)52 with the option ‘--sensitive-local’. Alignments with a mapping quality below 5 (using SAMtools) and duplicated reads were removed. Peaks were called using MACS2 (v2.1.1.20160309)54 with the options ‘--call-summits --nomodel --B’. Differential ChIP peaks were called using DiffBind55 with the options ‘bFullLibrarySize=FALSE, bSubControl=TRUE, bTagwise=FALSE’ for dba.analyze(). For DiffBind testing, the DESeq2 algorithm with blocking was used, and ChIP replicate was used as the blocking factor while testing for differences between mock and infected samples. For ChIP signal tracks, individual replicate tracks were produced for RPM and fold enrichment over input control, merged and averaged.
To account for potential global differences in histone PTM abundance that would otherwise be missed by more standard quantile normalization-type approaches, high-quality deduplicated read counts were produced for both human- and C. floridanus-mapping reads, resulting in proportions of reads mapping to the exogenous genome for each histone PTM. Input controls were also treated in this way to account for potential differences in initial spike-in addition between samples. For each histone PTM, the proportion of spike-in reads was normalized by the appropriate input control value. Because spike-ins should be inversely proportional to target chromatin concentration, a ratio of SARS-CoV-2/mock values was produced for each histone PTM × replicate, and for SARS-CoV-2 samples resulting signal values were divided by this ratio. This resulted in per-base-pair signal values adjusted by the degree of global difference in a given histone PTM’s level between sample types.
All antibodies are described in Supplementary Table 6.
RNA-seq
RNA was extracted using a Qiagen RNA purification kit. Samples were prepared for sequencing using the Illumina TruSeq purification kit and sequenced on an Illumina NextSeq 500 instrument (75-bp read length, single read). Library size was confirmed on a Bioanalyzer before sequencing on the NextSeq 550 platform (single end, 75 cycles).
For RNA-seq analysis for SARS-CoV-2 infection experiments, a reference genome for alignment was built by concatenating the human (GRCh38 assembly) and SARS-CoV-2 (WA-CDC-WA1/2020 assembly; MN985325.1) genomes. For RNA-seq analysis for HEK293T cell experiments, the GRCh38 assembly was used. For all RNA-seq, reads were aligned using STAR (v2.6.1a) with default parameters and only uniquely mapped reads were retained for downstream analysis. TDF files were generated using IGVtools. Reads were counted towards human genes (GENCODE v35) and SARS-CoV-2 genes (WA-CDC-WA1/2020 assembly; MN985325.1) using featureCounts (v1.6.2). Low-count genes were filtered out so that only genes with counts per million (CPM) values greater than 1 in at least three samples were used. Data normalization and differential gene expression analysis were performed using the DESeq2 R package (v1.26.0). We defined genes as significant using a false discovery rate (FDR) cut-off of 0.05 and 1.5× fold change. GO enrichment analysis for differentially expressed genes was implemented with the clusterProfiler R package (v3.14.3), using the human genome annotation record in the org.Hs.eg.db R package (v3.10.0) and a Benjamini–Hochberg-adjusted P value of 0.05 as the cut-off.
Immunoprecipitation
Anti-Strep tag affinity purification, whole-cell lysate and cytoplasmic HLA-A2 co-immunoprecipitation
Protein and binding partners were purified with affinity Strep tag purification. For ORF8 PTM analysis and mass spectrometry binding partner analysis, whole-cell lysates were prepared as described below. Frozen cell pellets were thawed briefly and suspended in lysis buffer (immunoprecipitation (IP) buffer (50 mM Tris-HCl (pH 7.5) at 4 °C, 150 mM NaCl, 1 mM EDTA and 10 mM sodium butyrate) supplemented with 0.5% Nonidet P 40 Substitute (NP-40; Fluka Analytical) and cOmplete mini EDTA-free protease and PhosSTOP phosphatase inhibitor cocktails (Roche)). Samples were incubated on a tube rotator for 30 min at 4 °C. Debris was pelleted by centrifugation at 13,000 g for 15 min at 4 °C. Lysates were then incubated with Strep-Tactin magnetic beads (40 µl; MagStrep type 3 XT beads; IBA, 2-4090-002) for 2 h with rotation at 4 °C. Beads were washed three times with 1 ml wash buffer (IP buffer supplemented with 0.05% NP-40) and then once with 1 ml IP buffer. Strep-tagged ORF8 complexes were eluted from beads in BXT buffer (IBA, 2-1042-025) with shaking at 1,100 r.p.m. for 30 min.
Anti-Strep tag affinity purification for chromatin binding partners
Cells were rotated in lysis buffer 1 (50 mM HEPES-KOH (pH 7.5), 140 mM NaCl, 10 mM sodium butyrate, 1 mM EDTA, 10% glycerol, 0.5% NP-40 and 0.25% Triton X-100) supplemented with 0.5% Nonidet P 40 Substitute (NP-40; Fluka Analytical) and cOmplete mini EDTA-free protease and PhosSTOP phosphatase inhibitor cocktails (Roche) for 10 min at 4 °C and spun at 1,350 g for 5 min at 4 °C to isolate nuclei. Supernatant was discarded and cells were resuspended in lysis buffer 2 (10 mM Tris-HCl (pH 8), 200 mM NaCl, 10 mM sodium butyrate, 1 mM EDTA and 0.5 mM EGTA) to lyse nuclei. Cells were rotated for 10 min at room temperature and were spun again at 1,350 g for 5 min at 4 °C. The supernatant was discarded and the chromatin pellet was resuspended in lysis buffer 3 (10 mM Tris-HCl (pH 8), 100 mM NaCl, 10 mM sodium butyrate, 1 mM EDTA, 0.5 mM EGTA, 0.1% EDTA and 0.5% N-lauroylsarcosine). Lysates were sonicated using a tip sonicator with three 5-s bursts at 50% power with chilling on ice between bursts. After sonication, lysates were brought to a concentration of 1% Triton X-100 to disrupt lamina protein interactions. Debris was pelleted by centrifugation at 16,000 g at 4 °C, and the supernatant was incubated with Strep-Tactin magnetic beads (40 µl; MagStrep type 3 XT beads; Iba, 2-4090-002) for 2 h with rotation at 4 °C. Beads were washed three times with 1 ml wash buffer (IP buffer supplemented with 0.05% NP-40) and then once with 1 ml IP buffer. Strep-tagged ORF8 complexes were eluted from beads in BXT buffer (IBA, 2-1042-025) with shaking at 1,100 r.p.m. for 30 min. To analyse relative ORF8 construct levels in cytoplasmic versus chromatin fractions by western blotting, samples were taken from lysis buffer 1 and lysis buffer 3, respectively.
Reverse immunoprecipitation
Chromatin pellet lysate was obtained as described above for chromatin protein immunoprecipitation. Lysates were combined with antibody-conjugated Protein A Dynabeads (15 µg of antibody conjugated to 100 µl of Dynabeads) and rotated overnight at 4 °C. The following day, beads were washed three times with 1 ml wash buffer (IP buffer supplemented with 0.05% NP-40) and then once with 1 ml IP buffer. Chromatin protein complexes were eluted from beads in elution buffer (50 mM Tris-HCl (pH 8.0), 10 mM EDTA and 1% SDS) for 30 min with shaking at 65 °C.
All antibodies are described in Supplementary Table 6.
Immunocytochemistry
Fluorescence immunocytochemistry of HEK293T cells and A549ACE2 cells
Cells were fixed in 4% PFA for 10 min and washed with PBS. Fixed cells were permeabilized using 0.5% Triton X-100 in PBS for 20 min. Cells were blocked in blocking solution (PBS with 3% BSA, 2% serum and 0.1% Triton X-100) for at least 1 h and stained with designated primary antibody overnight at 4 °C. The following day, cell coverslips were washed with PBS and incubated with secondary antibody for 1 h at room temperature. For detection of Strep-tagged ORF8, Strep-Tactin DY-488 (IBA, 2-1562-050; 1:500) was added to the secondary antibody solution. Nuclei were stained with DAPI (1:1,000 in PBS) for 10 min with washing in PBS. Coverslips were mounted onto microscope slides using ProLong Gold antifade reagent (Thermo Fisher).
Fluorescence immunocytochemistry analysis of lamin B1, lamin A/C and H3K9me2
HEK293T cells were fixed with 2% PFA (Electron Microscopy Sciences, 15710) for 8 min at room temperature and washed three times with DPBS (Gibco, 14190-136). Cells were permeabilized with 0.25% Triton X-100 (Thermo Fisher, 28314) for 10 min, washed three times with DPBS for 5 min each wash and blocked in 1% BSA (Sigma, A4503) in PBST (DPBS with 0.05% Tween-20, pH 7.4 (Thermo Fisher, 28320)) for 60 min. Cells were incubated with primary antibody diluted in blocking buffer for 1 h, washed three times with PBST for 5 min each wash and incubated with secondary antibody diluted in blocking buffer for 60 min. Cells were washed twice with PBST and once with PBS for 5 min each wash and were then mounted on a slide using Duolink In Situ Mounting Medium with DAPI (Sigma, DUO82040-5ML). All procedures were performed at room temperature.
Immunohistological staining of patient lung tissue
Formalin-fixed, paraffin-embedded slides were obtained from Penn’s Pathology Clinical Service Center. Slides were deparaffinized and rehydrated as follows: incubation for 10 min with xylene (twice), 10 min with 100% ethanol (twice), 5 min with 95% ethanol, 5 min with 70% ethanol, 5 min with 50% ethanol and then running distilled water. Slides were then processed using heat-induced epitope retrieval (HIER). Slides were incubated in hot sodium citrate buffer (10 mM sodium citrate and 0.05% Tween-20, pH 6.0), placed in a pressure cooker and heated in a water bath for 25 min with high pressure settings. Slides were cooled at room temperature and washed twice in TBS. Membranes were permeabilized in TBS with 0.4% Triton X-100 for 20 min. Slides were then incubated in blocking solution (TBS with 10% goat serum, 1% BSA and 0.025% Triton X-100) for 2 h. Slides were incubated in mouse primary antibody solution containing anti-SARS-CoV-2 nucleocapsid and rabbit anti-H3K9me3 antibody solution overnight at 4 °C. The following day, slides were washed with TBS and incubated in secondary antibody solution. Nuclei were stained with DAPI (5 µg ml–1) in TBS for 10 min followed by washing with TBS. Coverslips were mounted with ProLong Gold antifade reagent (Thermo Fisher). All antibodies are described in Supplementary Table 6.
Image acquisition
Fluorescence immunocytochemistry of ORF8 and histone PTMs
Cells were imaged on an upright Leica DM 6000, TCS SP8 laser scanning confocal microscope with 405-nm, 488-nm, 552-nm and 638-nm lasers. The microscope uses two HyD detectors and three PMT detectors. The objective used was a ×63 HC PL APO CS2 oil objective with an NA of 1.40. Type F immersion liquid (Leica) was used for oil objectives. Images were 175.91 × 171.91 µm2, 1,024 × 1,024 pixels and 16 bits per pixel. For PTM quantification, HEK293T cells and human lung tissue were imaged at a single z plane and A549 cells were imaged with a z stack through the nucleus.
Fluorescence immunocytochemistry analysis of lamin B1, lamin A/C and H3K9me2
All confocal immunofluorescence images were acquired using a Leica SP8 laser scanning confocal system with a ×63/1.40-NA HC PL APO CS2 objective and HyD detectors in standard mode with 100% gain. For comparison of lamin A/C and lamin B1 signal intensities between mock and ORF8-positive cells, single-plane confocal images were acquired. All images were acquired with the same microscope settings (zoom, laser power, gain, etc.). For analysis of the organization of H3K9me2-marked chromatin at the nuclear lamina, three-dimensional (3D) images of the middle z plane of the nucleus were taken as z stacks using 0.1-μm intervals with a range of 1 μm per nucleus. Confocal 3D images were deconvoluted with Huygens Professional software using the microscope parameters, standard PSF and automatic settings for background estimation.
Image analysis
Images were analysed using ImageJ software (version 2.0.0-rc-69/1.52p, build 269a0ad53f). Single-z-plane images of HEK293T cells and human lung tissue and summed z stacks through A549 nuclei were used for PTM quantification. Regions of interest (ROIs) of in-focus nuclei were semi-automatically defined using the DAPI channel and the ‘analyze particles’ functionality with manual corrections. HEK293T histone PTMs were quantified in transfected cells and non-transfected neighbouring cells using mean grey values. Signal for Strep-tagged ORF8 constructs (Strep-Tactin-488) and GFP was used to define transfected cells, and the HEK293T histone PTM levels in transfected cells were relativized to the histone PTM levels in non-transfected neighbouring cells. Histone PTMs were quantified in A549 cells and human lung tissue using integrated density values. dsRNA and SARS-CoV-2 nucleocapsid signal was used to define infected A549 cells and human lung cells, respectively. The total fluorescence intensity of the lamin A/C and lamin B1 signal was measured from the whole nuclei of mock and ORF8-positive cells. Analysis of the peripheral heterochromatin organization was performed as a comparison of a fraction of H3K9me2-marked chromatin at the nuclear lamina/periphery of mock and ORF8-positive cells. A fraction of H3K9me2 signal at the nuclear lamina/periphery was measured using lamin B signal as a mask or DAPI signal to create a mask of a 0.6-μm-thick nuclear peripheral zone.
Protein alignment
To identify potential histone mimicry, SARS-CoV-2 protein sequences were aligned to human histone protein sequences (H2A, H2B, H3.1, H3.2, H4, H2A.X, H2A.Z, macroH2A and H3.3) using Multiple Sequence Comparison by Log-Expectation (MUSCLE) with default settings. SARS-CoV-2 protein sequences were obtained from protein sequences published for the first Wuhan isolate56.
FACS
HEK293T cell pellets were gently resuspended in 1 ml FACS buffer (Ca2+/Mg2+-free PBS with 2% BSA) and pelleted at 500 g for 5 min at 4 °C; the supernatant was removed. Cells transfected with ORF8 construct and non-transfected control cells were then gently resuspended in 1 ml FACS buffer with a 1:500 dilution of Strep-Tactin DY-488 and rotated at 4 °C for 1 h, protected from light. Cells were then washed twice in 1 ml FACS buffer, resuspended in 1 ml FACS buffer and filtered through a 35-µm mesh into FACS tubes. A BD Influx cell sorter was used to analyse cells. Strep-Tactin DY-488 and GFP were excited with a 488-nm laser and signal was collected with a 530/40-nm detector. Excluding doublets and cell debris, cells were gated on the Strep-Tactin DY-488 signal, where thresholds were set using non-transfected control cells such that <1% of control cells were considered positive for Strep-Tactin DY-488. Strep-Tactin DY-488-positives cells were collected in FACS buffer and pelleted for subsequent experiments. The FACS gating strategy and cell numbers isolated are shown in Supplementary Fig. 2.
Histone extraction
Transfected cells were isolated by FACS as described above. Sorted cells were pelleted, resuspended in 1 ml cold H2SO4 and rotated overnight at 4 °C. Following the overnight incubation, cells were pelleted at maximum speed and the supernatant was transferred to a fresh tube. Trichloroacetic acid was added to 25% by volume, and the cells were left on ice at 4 °C overnight. Cells were again pelleted at maximum speed, and the supernatant was discarded. Prechilled acetone was then used to gently wash the pellet twice. Following the second wash, the tubes were left to air dry before the pellet was resuspended in water. Samples were then broken up by alternating 10 min of sonication and 30 min of shaking at 50 °C until pellets were fully dissolved.
Mass spectrometry
Histone PTM analysis by quantitative mass spectrometry
Purification of histones was validated by SDS–PAGE followed by Coomassie staining demonstrating sufficient enrichment. A BCA assay (Thermo Fisher) was performed for protein estimation using the manufacturer’s instructions, and 20 µg of histone was used for chemical derivatization and digestion as described previously57. In brief, unmodified lysines were derivatized twice with a 1:3 ratio of acetonitrile to proprionic anhydride. Histones were then digested with trypsin in a 1:20 enzyme to protein ratio at 37 °C overnight. Digested histones with newly formed N termini were derivatized twice as done previously. Finally, histones were dried with a vacuum concentrator. The dried samples were reconstituted in 0.1% trifluoroacetic acid (TFA) and desalted with the C18 micro spin column (Harvard Apparatus). The column was prepared with 200 μl of 100% acetonitrile and equilibrated with 200 μl of loading buffer with 0.1% TFA. Peptides were loaded onto the column, washed with loading buffer and eluted with 200 μl of 70% acetonitrile in 0.1% formic acid. All steps for loading, washing and elution were carried out with benchtop centrifugation (300 g for 2 min). The eluted peptides were then dried in a centrifugal vacuum concentrator.
Dried histone peptides were reconstituted in 0.1% formic acid. A synthetic library of 93 heavy labelled and derivatized peptides containing commonly measured histone PTMs58 was spiked into the endogenous samples to a final concentration of approximately 100 ng µl–1 for endogenous peptides and 100 fmol µl–1 for each heavy labelled synthetic analyte. For each analysis, 1 µl of sample was injected onto the column for data-independent analysis on a Q-Exactive HF instrument (Thermo Scientific) attached to an Ultimate 3000 nano-UPLC system and Nanospray Flex ion source (Thermo Scientific). Using aqueous solution of 0.1% formic acid as buffer A and organic solution of 80% acetonitrile and 0.1% formic acid as buffer B, peptides were separated on a 63-min gradient at 400 nl min–1 starting at 4% buffer B and increasing to 32% buffer B over 58 min and then increasing to 98% buffer B over 5 min. The column was then washed at 98% buffer B over 5 min and equilibrated to 3% buffer B. Data-independent acquisition was performed with the following settings. A full MS1 scan from 300 to 950 m/z was acquired with a resolution of 60,000, an automatic gain control (AGC) target of 3 × 106 and a maximum injection time of 55 ms. Then, a series of 25 MS2 scans was acquired across the same mass range with sequential isolation windows of 24 m/z with a collision energy of 28, a resolution of 30,000, an AGC target of 1 × 106 and a maximum injection time of 55 ms. Data analysis and manual inspection using the synthetic library as a reference were performed with Skyline (MacCoss Lab). Ratios were generated using R Studio and statistical analysis was carried out in Excel as in previous histone analysis.
Trypsin and chymotrypsin digestion of ORF8 for identification of ORF8 modifications
The gel band containing ORF8 was destained with 50 mM ammonium bicarbonate with 50% acetonitrile. The band was then reduced in 10 mM DTT in 50 mM ammonium bicarbonate for 30 min at 55 °C. Next, the band was alkylated with 100 mM iodoacetamide in 50 mM ammonium bicarbonate at room temperature for 30 min in the dark. Protein was then digested by incubation with chymotrypsin or trypsin at an approximately 1:20 enzyme to protein ratio at 37 °C overnight. Following digestion, the supernatant was collected. To extract additional peptides from the gel, 150 μl of 50% acetonitrile and 1% TFA was added and samples were incubated with constant shaking for 30 min. The supernatant was collected and 100 μl of acetonitrile was added followed by incubation with constant shaking for 10 min. The final supernatant was collected. All three supernatants were combined and dried. The dried samples were then desalted as described above.
ORF8 versus control immunoprecipitation for identification of binding partners
ORF8 immunoprecipitation elutants were reduced and alkylated as described above. Proteins were then digested and desalted with mini S-Trap (Protifi) following the manufacturer’s instructions. In brief, 25 μl of elutant was combined with 25 μl of 10% SDS to a final SDS concentration of 5% after alkylation. Samples were then acidified with phosphoric acid and precipitated by adding 90% methanol in 100 mM triethylammonium bicarbonate (TEAB) in a 6:1 (vol/vol) ratio. Protein was then added to the trap with benchtop centrifugation (4,000 g for 1 min), washed and digested with trypsin at a 1:10 enzyme to protein ratio at 37 °C overnight. Following digestion, peptides were eluted from the trap with 40 μl of 100 mM TEAB, 40 μl of 0.2% formic acid and 40 μl of 50% acetonitrile in 0.2% formic acid. Combined elutant volumes were then dried.
Chymotrypsin LC–MS/MS and LC–PRM-MS analysis
Dried peptides were reconstituted with 0.1% formic acid, and 2 µg of each sample was injected. Chymotrypsin-digested ORF8 samples were analysed on a Q-Exactive (Thermo Scientific) coupled to an Easy nLC 1000 UHPLC system and Nanospray Flex ion source (Thermo Scientific). The LC instrument was equipped with a 75 µm × 20 cm column packed in house using Reprosil-Pur C18 AQ (2.4 µm; Dr. Maisch). Using the same column and buffer conditions as described previously, peptides were separated on an 85-min gradient at 400 nl min–1 starting at 3% buffer B and increasing to 32% buffer B over 79 min and then increasing to 50% buffer B over 5 min and finally increasing to 90% buffer B over 1 min. The column was then washed at 90% buffer B over 5 min and equilibrated to 3% buffer B. Data-dependent acquisition was performed with dynamic exclusion of 40 s. A full MS1 scan from 350 to 1,200 m/z was acquired with a resolution of 70,000, an AGC target of 1 × 106 and a maximum injection time of 50 ms. Then, a series of MS2 scans was acquired for the top 15 precursors with a charge state of 2–7, a collision energy of 28 and an isolation window of 2.0 m/z. Each MS2 scan was acquired with a resolution of 17,500, an AGC target of 2 × 105 and a maximum injection time of 50 ms. A database search was performed using the human SwissProt sequence and ORF8 sequence with Proteome Discoverer 2.3 or 2.4 (Thermo Scientific) using the following search criteria: carboxyamidomethylation at cysteine residues as a fixed modification; oxidation at methionine and acetylation at lysine as variable modifications; two maximum allowed missed cleavages; precursor MS tolerance of 10 ppm; a 0.02-Da MS/MS. An unscheduled parallel reaction-monitoring method59 was developed to identify or validate 45 possible modified and unmodified peptide targets of ORF8. Peptides were separated with the same LC gradient conditions. A full MS1 scan from 300 to 900 m/z was acquired with a resolution of 70,000, an AGC target of 1 × 106 and a maximum injection time of 50 ms. Then, a series of MS2 scans was acquired with a loop count of 23 precursors, a collision energy of 28 and an isolation window of 1.2 m/z. Each MS2 scan was acquired with a resolution of 17,500, an AGC target of 1 × 106 and a maximum injection time of 100 ms. Data analysis and manual inspection were performed with Skyline60 (MacCoss Lab) and IPSA61.
Trypsin ORF8 LC–MS/MS and LC–PRM/MS analysis and IP LC–MS/MS analysis
Dried peptides were reconstituted with 0.1% formic acid, and 2 µg of each sample was injected. Data-dependent acquisition runs were analysed on a Q-Exactive HF or HF-X (Thermo Scientific) attached to an Ultimate 3000 nano UPLC system and Nanospray Flex Ion Source (Thermo Scientific). Using the same column and buffer conditions as described above, peptides were separated on a 112-min gradient at 400 nl min–1 starting at 5% buffer B, increasing to 35% buffer B over 104 min and then increasing to 60% buffer B over 8 min. The column was then washed at 95% buffer B for 5 min and equilibrated to 5% buffer B. Data-dependent acquisition was performed with dynamic exclusion of 45 s. A full MS1 scan from 380 to 1,200 m/z was acquired with a resolution of 120,000, an AGC target of 3 × 106 and a maximum injection time of 32 ms. Then, a series of MS2 scans were acquired for the top 20 precursors with a charge state of 2–5, a collision energy of 28 and an isolation window of 1.2 m/z. Each MS2 scan was acquired with a resolution of 30,000, an AGC target of 1 × 106 and a maximum injection time of 32 ms (HF) or 55 ms (HFX). A database search was performed using the human SwissProt sequence and ORF8 sequence with Proteome Discoverer 2.3 or 2.4 (Thermo Scientific) with the following search criteria: carboxyamidomethylation at cysteine residues as a fixed modification; oxidation at methionine and acetylation at lysine as variable modifications; two maximum allowed missed cleavages; precursor MS1 tolerance of 10 ppm; a 0.02-Da MS2 tolerance. An unscheduled parallel reaction-monitoring method59 was developed to identify 16 possible modified and unmodified peptide targets of ORF8. Peptides were separated with the same LC gradient conditions. A full MS1 scan from 350 to 950 m/z was acquired with a resolution of 120,000, an AGC target of 3 × 106 and a maximum injection time of 100 ms. Then, a series of MS2 scans were acquired with a loop count of 16 precursors, a collision energy of 28 and an isolation window of 1.2 m/z. Each MS2 scan was acquired with a resolution of 30,000, an AGC target of 1 × 106 and a maximum injection time of 100 ms. Data analysis and manual inspection were performed with Skyline60 (MacCoss Lab) and IPSA61.
Statistics and reproducibility
Box-and-whisker plots show the median as the centre line, box limits for upper and lower quartiles, whiskers for 1.5× the interquartile range and points for outliers. ANOVA testing was performed and plots were generated with R. Bonferroni corrections were applied for multiple comparisons. Fiji was used for image analysis. Imaging and analysis were performed with the experimenter blinded to the experimental condition whenever possible. In some instances, such as for patient tissue imaging, analysis required targeted selection, imaging and analysis of infected cells compared with uninfected cells. This required the experimenter to be aware of cell infection status while imaging. However, in these cases, the measurement of interest (such as staining for a histone modification) was not viewed before choosing fields to avoid biasing selection.
Images are representative of multiple replicates as follows:
Figure 1b: >5 independent experiments.
Figure 1c: two independent experiments.
Figure 1d: three independent experiments.
Figure 1g: five independent samples from two separate runs of FACS sorting.
Figure 2b,d,f: exact cell numbers and replicates described in Fig. 2c,d,g.
Figure 2h: two shown of four independent samples from one FACS sort.
Figure 3f: three independent samples per condition from one infection.
Figure 3g: exact cell numbers and replicates described in Fig. 3h.
Extended Data Fig. 2a: three independent experiments.
Extended Data Fig. 2b,c: >5 independent experiments.
Extended Data Fig. 2d: two independent experiments.
Extended Data Fig. 2e: two independent experiments.
Extended Data Fig. 3b: two independent experiments.
Extended Data Fig. 4a: lamin and histone H3, three independent experiments; HP1α and KAT2A, two independent experiments.
Extended Data Fig. 4b: two independent experiments.
Extended Data Fig. 4c: two independent experiments.
Extended Data Fig. 4d: one independent experiment, repeating previously published data.
Extended Data Fig. 4e: two independent experiments.
Extended Data Fig. 4f: two independent experiments.
Extended Data Fig. 5a: exact cell numbers and replicates described in Extended Data Fig. 5b.
Extended Data Fig. 5c: exact cell numbers and replicates described in Extended Data Fig. 5d.
Extended Data Fig. 5e: same images as in Extended Data Fig. 5c.
Extended Data Fig. 6b: three shown of five independent samples from two runs of FACS sorting.
Extended Data Fig. 10a: exact cell numbers and replicates described in Extended D ata Fig. 10b.
Extended Data Fig. 10c: exact cell numbers and replicates described in Extended Data Fig. 10d.
Extended Data Fig. 10e: exact cell numbers and replicates described in Extended Data Fig. 10f.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at 10.1038/s41586-022-05282-z.
Supplementary information
This file contains Supplementary Fig. 1 (original western blot images), Supplementary Fig. 2 (FACS gating strategy) and the legends for Supplementary Tables 1–6 (see separate Excel files for the tables).
ORF8 binding partners.
Gene ontology analysis of gene expression changes following ORF8 expression.
Histone PTM changes with ORF8 expression.
All infection data.
Gene ontology analysis of gene expression changes following SARS-CoV-2 infection of A549ACE cells.
Antibodies used in the manuscript.
Acknowledgements
We thank and acknowledge M. Weitzman for feedback and suggestions, A. Stout for microscopy support, C. Comar for viral infections, R. Jain and P. Shaw for protocols, M. Feldman and K. Montone for providing patient samples, and S. Wolf and P. Korb for supervision and support. J.K., S.T., E.K. and research were supported by the Penn Center for Coronavirus and Other Emerging Pathogens and the Dean’s Innovation Fund from the Perelman School of Medicine at the University of Pennsylvania. E.K. was supported by the Sloan Research Fellowship, the Klingenstein-Simons Fellowship, the NARSAD Young Investigator Award and NIH grants R00MH111836 and 1DP2MH129985. Viral work was supported by NIH grant RO1AI140442 supplement for SARS-CoV-2 and funds from the Penn Center for Coronavirus Research and Other Emerging Pathogens. D.M.R. was supported in part by T32AI055400. A.P., A.A.K. and J.A.E. were supported by NIH grant R35HL140018. R.T. and W.Y. were supported in part by institutional funds from the University of Pennsylvania Perelman School of Medicine to the iPSC Core. iPSC cell line generation and sharing were supported by NIH grants NO175N92020C00005 and U01TR001810. Mass spectrometry was supported by AI118891. P.-Y.S. was supported by NIH grants HHSN272201600013C, U01AI151801 and U19AI171413 and by awards from the Sealy Smith Foundation, the Kleberg Foundation, the John S. Dunn Foundation, the Amon G. Carter Foundation, the Gillson Longenbaugh Foundation and the Summerfield Robert Foundation.
Extended data figures and tables
Source data
Source Data Figs. 2 and 3 and Extended Data Figs. 4, 7 and 8.
Author contributions
J.K. designed, performed and analysed the results for the majority of the experiments. S.T. generated cells, samples and DNA constructs and performed analysis. D.M.R. and Y. Li performed the SARS-CoV-2 viral infections and analysis of viral infections. K.G. and Z.Z. performed and analysed the results of ChIP–seq experiments. K.P. performed ATAC-seq and generated samples for histone PTM analysis. J.C. performed and analysed the results of mass spectrometry experiments with guidance from B.A.G. X.Z., X.X. and P.-Y.S. generated the deletion viruses. Y. Lan provided bioinformatic analysis. F.L.C.-D. and R.T. generated iAT2 cells with guidance from W.Y. and E.M. D.N.K. and K.D.A. provided stem cell lines. A.P., A.A.K. and J.A.E. performed ORF8 and nuclear lamina imaging and analysis. S.L.B. provided input and expertise and led ChIP–seq studies. S.R.W. provided input and expertise and led viral work. E.K. led the project and wrote the manuscript.
Peer review
Peer review information
Nature thanks the anonymous reviewers for their contribution to the peer review of this work.
Data availability
All genome-wide sequencing data are available under accession number GSE186628. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository62 with the dataset identifier PXD034379. Publicly available data used for analysis include ENCODE data ENCSR000ASV, ENCSR000FCJ, ENCSR179BUC, ENCSR55LYM and ENCSR000AKD and 4D nucleome dataset 4DNES24XA7U8.
Competing interests
The laboratory of P.-Y.S. has received funding support in sponsored research agreements from Pfizer, Gilead, Novartis, Merck, GSK, IGM Biosciences and Atea Pharmaceuticals. P.-Y.S. is a member of the scientific advisory boards of AbImmune and is founder of FlaviTech. S.R.W. is on the scientific advisory board of Ocugen and Immunome.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Samuel Thudium, David M. Renner, Karl Glastad
Change history
2/6/2023
A Correction to this paper has been published: 10.1038/s41586-023-05764-8
Change history
1/9/2023
A Correction to this paper has been published: 10.1038/s41586-022-05631-y
Extended data
is available for this paper at 10.1038/s41586-022-05282-z.
Supplementary information
The online version contains supplementary material available at 10.1038/s41586-022-05282-z.
References
- 1.Hadjadj J, et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blanco-Melo D, et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Y, et al. SARS-CoV-2 induces double-stranded RNA-mediated innate immune responses in respiratory epithelial-derived cells and cardiomyocytes. Proc. Natl Acad. Sci. USA. 2021;118:e2022643118. doi: 10.1073/pnas.2022643118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marazzi I, et al. Suppression of the antiviral response by an influenza histone mimic. Nature. 2012;483:428–433. doi: 10.1038/nature10892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schaefer U, Ho JSY, Prinjha RK, Tarakhovsky A. The ‘histone mimicry’ by pathogens. Cold Spring Harb. Symp. Quant. Biol. 2013;78:81–90. doi: 10.1101/sqb.2013.78.020339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Avgousti DC, et al. A core viral protein binds host nucleosomes to sequester immune danger signals. Nature. 2016;535:173–177. doi: 10.1038/nature18317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Avgousti DC, et al. Adenovirus core protein VII downregulates the DNA damage response on the host genome. J. Virol. 2017;91:e01089-17. doi: 10.1128/JVI.01089-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tarakhovsky A, Prinjha RK. Drawing on disorder: how viruses use histone mimicry to their advantage. J. Exp. Med. 2018;215:1777–1787. doi: 10.1084/jem.20180099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 10.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 11.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 12.Ho JSY, et al. TOP1 inhibition therapy protects against SARS-CoV-2-induced lethal inflammation. Cell. 2021;184:2618–2632. doi: 10.1016/j.cell.2021.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee S, et al. Virus-induced senescence is a driver and therapeutic target in COVID-19. Nature. 2021;599:283–289. doi: 10.1038/s41586-021-03995-1. [DOI] [PubMed] [Google Scholar]
- 14.Zazhytska M, et al. Non-cell-autonomous disruption of nuclear architecture as a potential cause of COVID-19-induced anosmia. Cell. 2022;185:1052–1064. doi: 10.1016/j.cell.2022.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elde NC, Malik HS. The evolutionary conundrum of pathogen mimicry. Nat. Rev. Microbiol. 2009;7:787–797. doi: 10.1038/nrmicro2222. [DOI] [PubMed] [Google Scholar]
- 16.Davey NE, Travé G, Gibson TJ. How viruses hijack cell regulation. Trends Biochem. Sci. 2011;36:159–169. doi: 10.1016/j.tibs.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 17.Chaurushiya MS, et al. Viral E3 ubiquitin ligase-mediated degradation of a cellular E3: viral mimicry of a cellular phosphorylation mark targets the RNF8 FHA domain. Mol. Cell. 2012;46:79–90. doi: 10.1016/j.molcel.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chan JF-W, et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu R, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuen K-S, Ye Z-W, Fung S-Y, Chan C-P, Jin D-Y. SARS-CoV-2 and COVID-19: the most important research questions. Cell Biosci. 2020;10:40. doi: 10.1186/s13578-020-00404-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sampath SC, et al. Methylation of a histone mimic within the histone methyltransferase G9a regulates protein complex assembly. Mol. Cell. 2007;27:596–608. doi: 10.1016/j.molcel.2007.06.026. [DOI] [PubMed] [Google Scholar]
- 22.Gordon DE, et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vann KR, et al. Binding of the SARS-CoV-2 envelope E protein to human BRD4 is essential for infection. Structure. 2022;30:1224–1232. doi: 10.1016/j.str.2022.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flower, T. G. et al. Structure of SARS-CoV-2 ORF8, a rapidly evolving immune evasion protein. Proc. Natl Acad. Sci. USA118, e2021785118 (2021). [DOI] [PMC free article] [PubMed]
- 25.Muth D, et al. Attenuation of replication by a 29 nucleotide deletion in SARS-coronavirus acquired during the early stages of human-to-human transmission. Sci. Rep. 2018;8:15177. doi: 10.1038/s41598-018-33487-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hachim A, et al. ORF8 and ORF3b antibodies are accurate serological markers of early and late SARS-CoV-2 infection. Nat. Immunol. 2020;21:1293–1301. doi: 10.1038/s41590-020-0773-7. [DOI] [PubMed] [Google Scholar]
- 27.Wang X, et al. Accurate diagnosis of COVID-19 by a novel immunogenic secreted SARS-CoV-2 orf8 protein. mBio. 2020;11:e02431-20. doi: 10.1128/mBio.02431-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bojkova D, et al. Proteomics of SARS-CoV-2-infected host cells reveals therapy targets. Nature. 2020;583:469–472. doi: 10.1038/s41586-020-2332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bouhaddou M, et al. The global phosphorylation landscape of SARS-CoV-2 infection. Cell. 2020;182:685–712. doi: 10.1016/j.cell.2020.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang, Y. et al. The ORF8 protein of SARS-CoV-2 mediates immune evasion through down-regulating MHC-Ι. Proc. Natl Acad. Sci. USA118, e2024202118 (2021). [DOI] [PMC free article] [PubMed]
- 31.Rashid F, et al. Mutations in SARS-CoV-2 ORF8 altered the bonding network with interferon regulatory factor 3 to evade host immune system. Front. Microbiol. 2021;12:703145. doi: 10.3389/fmicb.2021.703145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rashid F, Dzakah EE, Wang H, Tang S. The ORF8 protein of SARS-CoV-2 induced endoplasmic reticulum stress and mediated immune evasion by antagonizing production of interferon β. Virus Res. 2021;296:198350. doi: 10.1016/j.virusres.2021.198350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Geng H, et al. SARS-CoV-2 ORF8 forms intracellular aggregates and inhibits IFNγ-induced antiviral gene expression in human lung epithelial cells. Front. Immunol. 2021;12:679482. doi: 10.3389/fimmu.2021.679482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin X, et al. ORF8 contributes to cytokine storm during SARS-CoV-2 infection by activating IL-17 pathway. iScience. 2021;24:102293. doi: 10.1016/j.isci.2021.102293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu X, et al. Viral mimicry of interleukin-17A by SARS-CoV-2 ORF8. mBio. 2022;13:e0040222. doi: 10.1128/mbio.00402-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen J, et al. Severe acute respiratory syndrome coronavirus 2 ORF8 protein inhibits type I interferon production by targeting HSP90B1 signaling. Front. Cell. Infect. Microbiol. 2022;12:899546. doi: 10.3389/fcimb.2022.899546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li J-Y, et al. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Res. 2020;286:198074. doi: 10.1016/j.virusres.2020.198074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsuoka K, et al. SARS-CoV-2 accessory protein ORF8 is secreted extracellularly as a glycoprotein homodimer. J. Biol. Chem. 2022;298:101724. doi: 10.1016/j.jbc.2022.101724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohki S, Imamura T, Higashimura Y, Matsumoto K, Mori M. Similarities and differences in the conformational stability and reversibility of ORF8, an accessory protein of SARS-CoV-2, and its L84S variant. Biochem. Biophys. Res. Commun. 2021;563:92–97. doi: 10.1016/j.bbrc.2021.05.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zinzula, L. Lost in deletion: the enigmatic ORF8 protein of SARS-CoV-2. Biochem. Biophys. Res. Commun.10.1016/j.bbrc.2020.10.045 (2020). [DOI] [PMC free article] [PubMed]
- 41.Xie X, et al. An infectious cDNA clone of SARS-CoV-2. Cell Host Microbe. 2020;27:841–848. doi: 10.1016/j.chom.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xie X, et al. Engineering SARS-CoV-2 using a reverse genetic system. Nat. Protoc. 2021;16:1761–1784. doi: 10.1038/s41596-021-00491-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li, Y. et al. SARS-CoV-2 induces double-stranded RNA-mediated innate immune responses in respiratory epithelial-derived cells and cardiomyocytes. Proc. Natl Acad. Sci. USA118, e2022643118 (2021). [DOI] [PMC free article] [PubMed]
- 44.Jacob A, et al. Derivation of self-renewing lung alveolar epithelial type II cells from human pluripotent stem cells. Nat. Protoc. 2019;14:3303–3332. doi: 10.1038/s41596-019-0220-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, R. et al. SARS-CoV-2 restructures the host chromatin architecture. Preprint at bioRxiv10.1101/2021.07.20.453146 (2021).
- 46.Valcarcel A, Bensussen A, Álvarez-Buylla ER, Díaz J. Structural analysis of SARS-CoV-2 ORF8 protein: pathogenic and therapeutic implications. Front. Genet. 2021;12:693227. doi: 10.3389/fgene.2021.693227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Silvas JA, et al. Contribution of SARS-CoV-2 accessory proteins to viral pathogenicity in K18 human ACE2 transgenic mice. J. Virol. 2021;95:e0040221. doi: 10.1128/JVI.00402-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Young BE, et al. Effects of a major deletion in the SARS-CoV-2 genome on the severity of infection and the inflammatory response: an observational cohort study. Lancet. 2020;396:603–611. doi: 10.1016/S0140-6736(20)31757-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fong, S.-W. et al. Robust virus-specific adaptive immunity in COVID-19 patients with SARS-CoV-2 Δ382 variant infection. J. Clin. Immunol.10.1007/s10875-021-01142-z (2021). [DOI] [PMC free article] [PubMed]
- 50.DeRonde S, Deuling H, Parker J, Chen J. Identification of a novel SARS-CoV-2 variant with a truncated protein in ORF8 gene by next generation sequencing. Sci. Rep. 2022;12:4631. doi: 10.1038/s41598-022-08780-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Porter, E. G., Connelly, K. E. & Dykhuizen, E. C. Sequential salt extractions for the analysis of bulk chromatin binding properties of chromatin modifying complexes. J. Vis. Exp.10.3791/55369 (2017). [DOI] [PMC free article] [PubMed]
- 52.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stark, R. & Brown, G. DiffBind 10.18129/B9.BIOC.DIFFBIND (2017).
- 56.Wu F, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sidoli, S., Bhanu, N. V., Karch, K. R., Wang, X. & Garcia, B. A. Complete workflow for analysis of histone post-translational modifications using bottom-up mass spectrometry: from histone extraction to data analysis. J. Vis. Exp.10.3791/54112 (2016). [DOI] [PMC free article] [PubMed]
- 58.Lin S, et al. Stable-isotope-labeled histone peptide library for histone post-translational modification and variant quantification by mass spectrometry. Mol. Cell. Proteomics. 2014;13:2450–2466. doi: 10.1074/mcp.O113.036459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peterson AC, Russell JD, Bailey DJ, Westphall MS, Coon JJ. Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics. Mol. Cell. Proteomics. 2012;11:1475–1488. doi: 10.1074/mcp.O112.020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.MacLean B, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brademan DR, Riley NM, Kwiecien NW, Coon JJ. Interactive Peptide Spectral Annotator: a versatile web-based tool for proteomic applications. Mol. Cell. Proteomics. 2019;18:S193–S201. doi: 10.1074/mcp.TIR118.001209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Perez-Riverol Y, et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022;50:D543–D552. doi: 10.1093/nar/gkab1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This file contains Supplementary Fig. 1 (original western blot images), Supplementary Fig. 2 (FACS gating strategy) and the legends for Supplementary Tables 1–6 (see separate Excel files for the tables).
ORF8 binding partners.
Gene ontology analysis of gene expression changes following ORF8 expression.
Histone PTM changes with ORF8 expression.
All infection data.
Gene ontology analysis of gene expression changes following SARS-CoV-2 infection of A549ACE cells.
Antibodies used in the manuscript.
Data Availability Statement
All genome-wide sequencing data are available under accession number GSE186628. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository62 with the dataset identifier PXD034379. Publicly available data used for analysis include ENCODE data ENCSR000ASV, ENCSR000FCJ, ENCSR179BUC, ENCSR55LYM and ENCSR000AKD and 4D nucleome dataset 4DNES24XA7U8.