Abstract
NADPH is an intermediate in the oxidation of organic compounds coupled to Fe(III) reduction in Geobacter species, but Fe(III) reduction with NADPH as the electron donor has not been studied in these organisms. Crude extracts of Geobacter sulfurreducens catalyzed the NADPH-dependent reduction of Fe(III)-nitrilotriacetic acid (NTA). The responsible enzyme, which was recovered in the soluble protein fraction, was purified to apparent homogeneity in a four-step procedure. Its specific activity for Fe(III) reduction was 65 μmol · min−1 · mg−1. The soluble Fe(III) reductase was specific for NADPH and did not utilize NADH as an electron donor. Although the enzyme reduced several forms of Fe(III), Fe(III)-NTA was the preferred electron acceptor. The protein possessed methyl viologen:NADP+ oxidoreductase activity and catalyzed the reduction of NADP+ with reduced methyl viologen as electron donor at a rate of 385 U/mg. The enzyme consisted of two subunits with molecular masses of 87 and 78 kDa and had a native molecular mass of 320 kDa, as determined by gel filtration. The purified enzyme contained 28.9 mol of Fe, 17.4 mol of acid-labile sulfur, and 0.7 mol of flavin adenine dinucleotide per mol of protein. The genes encoding the two subunits were identified in the complete sequence of the G. sulfurreducens genome from the N-terminal amino acid sequences derived from the subunits of the purified protein. The sequences of the two subunits had about 30% amino acid identity to the respective subunits of the formate dehydrogenase from Moorella thermoacetica, but the soluble Fe(III) reductase did not possess formate dehydrogenase activity. This soluble Fe(III) reductase differs significantly from previously characterized dissimilatory and assimilatory Fe(III) reductases in its molecular composition and cofactor content.
Dissimilatory Fe(III) reduction plays an important role in the degradation of natural and contaminant organic matter under anaerobic conditions (29). The ability to couple growth with the reduction of Fe(III) has been found in phylogenetically diverse members of the Bacteria and Archaea (25, 27), and geological and microbiological evidence suggests that Fe(III) reduction was one of the earliest forms of microbial respiration (26, 50). Studies on the physiology and biochemistry of dissimilatory Fe(III) reduction have focused largely on members of the genera Shewanella and Geobacter. Shewanella species are gram-negative facultative anaerobes that can use a variety of electron acceptors including Fe(III) (36, 37). Geobacter species are strictly anaerobic, gram-negative organisms that can couple the complete oxidation of organic matter to the reduction of Fe(III) (27). Members of the genus Geobacter are found in a variety of freshwater and sedimentary environments in which Fe(III) reduction is a dominant electron-accepting process (9, 47).
The mechanisms for electron transfer to Fe(III) in dissimilatory Fe(III)-reducing microorganisms have yet to be determined. In most environments, much of the Fe(III) is present as insoluble Fe(III) oxide, and it has generally been considered that Fe(III)-reducing organisms must be in close contact with this insoluble Fe(III) in order to reduce it (28). A number of studies have identified membrane-associated c-type cytochromes in Shewanella (12, 18, 36) and Geobacter (16, 24, 33) species which can reduce Fe(III) in vitro, and it has been suggested that these cytochromes are involved in electron transport to extracellular Fe(III). However, it has yet to be proven that the Fe(III) derived from Fe(III) oxide is reduced extracellularly. Furthermore, soluble Fe(III) is present in a variety of sedimentary environments in which microbial Fe(III) reduction is important (32, 38, 43), and the presence of chelators that solubilize Fe(III) can greatly stimulate Fe(III) reduction (31). This suggests that soluble Fe(III) could also serve as an electron acceptor.
Recent studies on acetate oxidation in Geobacter species demonstrated that NADPH is an intermediate of the tricarboxcylic acid cycle (7, 15). In this study we describe the purification and characterization of an enzyme from G. sulfurreducens that catalyzes the reduction of soluble Fe(III) with NADPH as the electron donor.
MATERIALS AND METHODS
Cell growth.
G. sulfurreducens strain PCA (ATCC 51573) was obtained from our laboratory culture collection and cultivated in an anaerobic freshwater medium, containing 20 mM sodium acetate as the electron donor and either 50 mM ferric citrate or 40 mM fumarate as the electron acceptor, under an atmosphere of N2-CO2 (80:20) (6). Cells were grown in 1-liter Pyrex bottles filled with 0.9 liter of medium or, for mass cultivation, in 10-liter carboys. The cells were harvested at the end of the exponential growth phase by centrifugation and washed in anaerobic 50 mM HEPES (pH 7.0) containing 5 mM dithiothreitol and 1 mM MgCl2. The washed cells were suspended in 1 ml of anaerobic 50 mM HEPES (pH 7.0) per g of wet cells and stored at −80°C.
Enzyme assays.
All enzyme assays were performed at 30°C in a UV2401-PC dual-beam spectrophotometer (Shimadzu, Baltimore, Md.) equipped with a temperature-controlled cuvette holder. Fe(III) reductase activity was routinely measured aerobically by monitoring the reduction of Fe(III) to Fe(II) using the colorimetric Fe(II) capture agent ferrozine [3-(2-pyridyl)-5,6-(4-phenylsulfonic acid)-1,2,4-triazine]. Reaction mixtures (1.0 ml) contained 50 mM MES (pH 5.5), 1.0 mM ferrozine, 2 mM Fe(III)-nitrilotriacetic acid [Fe(III)-NTA], and 200 μM NADPH. The reactions were started by the addition of either NADPH or protein (1 to 200 μg), and the formation of the Fe(II)-ferrozine complex was monitored at 562 nm (ɛ = 28 mM−1 · cm−1). One unit is the amount of enzyme catalyzing the reduction of 1 μmol of Fe(III) per min. In the absence of NADPH, the addition of crude extract to the assay mixture resulted in the reduction of 5 to 10 μM Fe(III) over 2 to 3 min. Therefore, enzymatic assays on crude extracts were initiated by the addition of NADPH after this nonspecific reduction of Fe(III) ceased. Reduction of other forms of Fe(III) was determined similarly by replacing Fe(III)-NTA with 1 mM Fe(III)-EDTA, 1 mM Fe(III)-citrate, or 0.5 mmol of Fe(III)-oxyhydroxide per liter. The pH optimum of the enzyme was determined using the following buffers: 100 mM acetate at pH 4.0, 4.5 and 5.0; 50 mM 2-[N-morpholino]ethanesulfonic acid-NaOH (MES) at pH 5.0, 5.5, 6.0, and 6.5; 50 mM 3-(N-morpholino]propanesulfonic acid)-NaOH (MOPS) at pH 6.5 and 7.0; and 50 mM Tris-HCl at pH 7.0, 7.5, and 8.0.
The reduction of menadione (2-methyl-1,4-naphthoquinone) was assayed by monitoring the oxidation of NADPH at 366 nm (ɛ = 3.3 mM−1 · cm−1). The reaction mixtures (1 ml) contained 50 mM MES (pH 5.5), 0.3 mM NADPH, and 0.5 mM menadione. The reactions were started by the addition of enzyme (5 to 20 μg) or menadione.
Methyl viologen:NADP+ oxidoreductase activity was measured under anaerobic conditions in an assay mixture (1 ml) containing 50 mM Tris-HCl (pH 7.5), 10 mM methyl viologen, and 0.3 mM NADP+. Before the reaction was initiated by adding either enzyme (1 to 5 μg) or NADP+, the reaction mixture was reduced chemically to an absorption at 578 nm of 1.5 by adding 0.1 M Ti(III) citrate. Oxidation of methyl viologen was monitored at 578 nm (ɛ = 9.7 mM−1 · cm−1). Malate dehydrogenase activity was measured by monitoring the oxidation of NADH at 340 nm (ɛ = 6.22 mM−1 · cm−1) in the presence of oxaloacetate (44). Reaction mixtures (1 ml) contained 50 mM HEPES-NaOH (pH 7.0), 0.2 mM NADH, and 0.4 mM oxaloacetate. Reactions were started by the addition of crude extract (10 to 30 μg) or oxaloacetate. Formate dehydrogenase was assayed under anaerobic conditions in a reaction mixture (1 ml) containing 50 mM Tris-HCl (pH 7.5), 3 mM dithiothreitol, 10 mM formate, and 10 mM methyl viologen. The reaction mixture was reduced to give a slight blue color by adding 0.1 M Ti(III)-citrate. Reactions were started by adding enzyme (5 to 20 μg) or formate, and the reduction of methyl viologen was monitored at 578 nm (ɛ = 9.7 mM−1 · cm−1).
Preparation of subcellular fractions.
All manipulations were carried out under anaerobic conditions in a glove bag under an atmosphere of 7% CO2–5% H2 balanced with N2. Solutions were made anaerobic by repeatedly evacuating and flushing them with oxygen-free N2 passed over a heated copper column. Subcellular fractions were prepared using a modification of the method of Gaspard et al. (16). Freshly harvested cells (about 1 g [wet weight]) were washed in 100 ml of anaerobic 50 mM HEPES buffer (pH 7.0) and resuspended in 20 ml of 100 mM Tris-HCl (pH 8.0) containing 25% (wt/vol) sucrose. Lysozyme (20 mg) was added, and the suspension was stirred for 20 min. Following the addition of Na2-EDTA (pH 8.0) to a final concentration of 5 mM and stirring for 15 min, MgCl2 was added to a final concentration of 13 mM and the suspension was stirred for another 15 min. Microscopic examination of the suspension revealed that almost all the cells had been converted to spheroplasts by this treatment. After centrifugation for 30 min at 20,000 × g at 4°C to pellet the spheroplasts, the supernatant was reserved as the periplasmic fraction. The pellet was resuspended in 20 ml of 10 mM HEPES (pH 7.0). To lyse the spheroplasts, a few crystals of DNase I were added and the suspension was frozen and thawed twice. The resulting crude extract was centrifuged for 30 min at 20,000 × g to remove unlysed cells and cell debris. The supernatant was subsequently centrifuged for 1 h at 100,000 × g to pellet the membranes. The final supernatant, the cytoplasmic fraction, was reserved for further analysis, and the membrane pellet was suspended in 5 ml of 50 mM HEPES (pH 7.0).
Purification of soluble Fe(III) reductase.
Wet cells (10 to 20 g) were suspended in 10 to 20 ml of 50 mM HEPES (pH 7.0), amended with a few crystals of DNase I, and disrupted by passage through a French pressure cell at 40,000 kPa. The crude extract was centrifuged for 1 h at 100,000 × g at 4°C to pellet cell debris and membranes. Soluble Fe(III) reductase was purified in an anaerobic glove bag at 20°C. All buffers were made anaerobic as described above. The ultracentrifuged crude extract (ca. 30 ml) was filtered (0.2-μm-pore-size filters), diluted to 90 ml with 25 mM MOPS (pH 6.5), and applied to a Q-Sepharose HP column (1.6 by 10 cm; Amersham Pharmacia Biotech, Piscataway, N.J.) equilibrated with 25 mM MOPS (pH 6.5). Protein was eluted with a linear gradient of 0 to 0.4 M NaCl (240 ml). Fractions containing Fe(III) reductase activity eluted at about 0.22 M NaCl. The pooled fractions were amended with 50 mM Tris-HCl (pH 7.5) containing 3.2 M ammonium sulfate to give a final concentration of 0.7 M ammonium sulfate and applied to a butyl-Sepharose HiPrep column (1.6 by 10 cm; Amersham Pharmacia Biotech) equilibrated with 50 mM Tris-HCl (pH 7.5) containing 0.7 M ammonium sulfate. Bound protein was eluted in a linear gradient of 0.7 to 0 M ammonium sulfate (200 ml). Fractions containing Fe(III) reductase activity, which eluted at 0.5 M ammonium sulfate, were pooled and concentrated to ca. 1.5 ml using Ultrafree-15 (30-kDa cutoff) centrifugal filter devices (Millipore, Bedford, Mass.). The concentrated pool was filtered (0.2-μm-pore-size filters) and applied to a Superdex 200 prep grade column (1.6 by 60 cm; Amersham Pharmacia Biotech) equilibrated with 50 mM Tris-HCl (pH 7.5), and the protein was eluted with the same buffer. Fe(III) reductase-containing fractions were pooled and applied to a MonoQ column (0.5 by 5 cm; Amersham Pharmacia Biotech) equilibrated with 50 mM Tris-HCl (pH 7.5) containing 20% (vol/vol) ethylene glycol. Protein was eluted with a gradient of 0.1 to 0.3 M NaCl, and the soluble Fe(III) reductase eluted at 0.2 M NaCl.
Molecular weight determination.
The native Mr of the soluble Fe(III) reductase was determined by gel filtration on a Superdex 200 column (1.6 by 60 cm; Amersham Pharmacia Biotech) with 50 mM Tris-HCl (pH 7.5) containing 0.15 M NaCl as the elution buffer. Thyroglobulin (Mr 669,000), apoferritin (Mr 443,000), β-amylase (Mr 200,000), alcohol dehydrogenase (Mr 150,000), and bovine serum albumin (Mr 66,000) were used as molecular weight standards.
Gel electrophoresis.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out by the method of Laemmli (23). Phosphorylase b (Mr 97,000), bovine serum albumin (Mr 66,000), ovalbumin (Mr 45,000), carbonic anhydrase (Mr 30,000), trypsin inhibitor (Mr 20, 000), and α-lactalbumin (Mr 14,400) were used as molecular weight standards. Proteins were separated on 10% polyacrylamide gels and stained with Coomasssie brilliant blue.
Determination of flavin, iron, and sulfur contents.
To determine the flavin cofactor content, purified Fe(III) reductase (0.5 to 1 μM) was air oxidized and extracted with 5% (wt/vol) trichloroacetic acid. Denatured protein was removed by centrifugation, and the pH of the supernatant was adjusted to 6.0 with 2 M K2HPO4. Then 20 μl of supernatant was analyzed by high-pressure liquid chromatography using a Supelcosil-LCPAH column (Supelco, Bellefonte, Pa.). The mobile phase was 20 mM ammonium acetate buffer (pH 6.0) containing 21% acetonitrile and was used at a flow rate of 0.5 ml/min (retention times: flavin adenine dinucleotide [FAD], 3.2 min; flavin mononucleotide [FMN], 4.2 min). Flavins were detected with a HP1312A fluorescence detector (Hewlett-Packard, Wilmington, Del.) by their emission at 525 nm following excitation at 260 nm (2).
Iron content was determined as described by Fish (13), using an iron volumetric standard (Aldrich, Milwaukee, Wis.). Acid-labile sulfur was estimated by the method of Rabinowitz (42) with ferredoxin from Clostridium pasteurianum as a reference. The presence of iron, molybdenum, tungsten, and selenium was tested by inductively coupled plasma mass spectrometry (ICP-MS) analysis by A. Siripiny from the Department of Chemistry at the University of Massachusetts, Amherst, Mass., using a Sciex Elan 6000 instrument (Perkin-Elmer, Norwalk, Conn.). Total protein was determined by the bicinchoninic acid method (46) with bovine serum albumin as standard. The cofactor content was calculated based on protein determination and a native molecular mass of 320 kDa.
Determination of N-terminal amino acid sequences.
For N-terminal amino acid sequencing, the purified protein was separated by SDS-PAGE and transferred to a polyvinylidene difluoride membrane. The amino acid sequences were determined by automated Edman degradation (J. Leszyk, University of Massachusetts Medical School, Worcester, Mass.). The N-terminal amino acid sequences were used to search the preliminary sequence data of the G. sulfurreducens genome available at The Institute for Genomic Research (http://www.tigr.org).
RESULTS
Localization of NADPH-dependent Fe(III) reductase.
Crude extracts of G. sulfurreducens catalyzed the reduction of Fe(III)-NTA with NADPH as the electron donor at a rate of 0.24 μmol · min−1 · mg−1. More than 90% of the NADPH-dependent Fe(III) reductase activity was localized in the cytoplasmic fraction (Table 1). A similar distribution was found for malate dehydrogenase, an enzyme known to reside in the cytoplasm (53). These results suggest that the NADPH-dependent Fe(III) reductase is a soluble enzyme present either in the cytoplasm or only loosely associated with the inner membrane.
TABLE 1.
Localization of Fe(III) reductase
| Fraction | Ferric reductase
|
Malate dehydrogenase
|
||
|---|---|---|---|---|
| Sp act (U/mg) | Total activity (U) (% of total) | Sp act (U/mg) | Total activity (U) (% of total) | |
| Periplasm | 0.06 | 0.83 (6) | 6 | 84 (13) |
| Membrane | 0.05 | 0.32 (2) | 0.41 | 2.5 (0.4) |
| Cytoplasm | 0.24 | 12.87 (92) | 10.2 | 542.6 (86) |
Purification and properties of soluble Fe(III) reductase.
Crude extracts prepared from cells grown with fumarate as the electron acceptor exhibited the same level of soluble Fe(III) reductase activity as did those prepared from cells grown on Fe(III) citrate (data not shown). This suggests that G. sulfurreducens constitutively produces this enzyme. Therefore, the enzyme was purified from cells grown on fumarate because it was technically simpler to mass culture the organism on fumarate.
Incubation of the crude extract under air for 2 h resulted in the loss of about 75% of the Fe(III) reductase activity. Due to this apparent oxygen sensitivity, the enzyme was purified under strictly anaerobic conditions. Using a four-step procedure, the enzyme was purified to apparent homogeneity with a yield of about 20% (Table 2). The specific activities of the final preparation varied from 40 to 70 U/mg.
TABLE 2.
Purification of soluble Fe(III) reductase
| Purification step | Amt of protein (mg) | Activity (U) | Sp act (U/mg) | Yield (%) | Purification (fold) |
|---|---|---|---|---|---|
| Crude extract | 493.8 | 54.8 | 0.11 | 100 | 1 |
| Q-Sepharose | 38.4 | 37.8 | 0.98 | 69 | 9 |
| Butyl-Sepharose | 5.4 | 21.3 | 3.94 | 39 | 36 |
| Superdex 200 | 0.5 | 15.7 | 31.40 | 29 | 282 |
| MonoQ | 0.17 | 11.1 | 65.30 | 20 | 594 |
Physical characterization.
The native molecular mass of the soluble Fe(III) reductase was determined by gel filtration to be 320 kDa. In SDS-PAGE, two subunits with relative molecular masses of 87 (α) and 78 (β) kDa were detected in equal amounts (Fig. 1). This suggests an α2β2 configuration for the native protein.
FIG. 1.
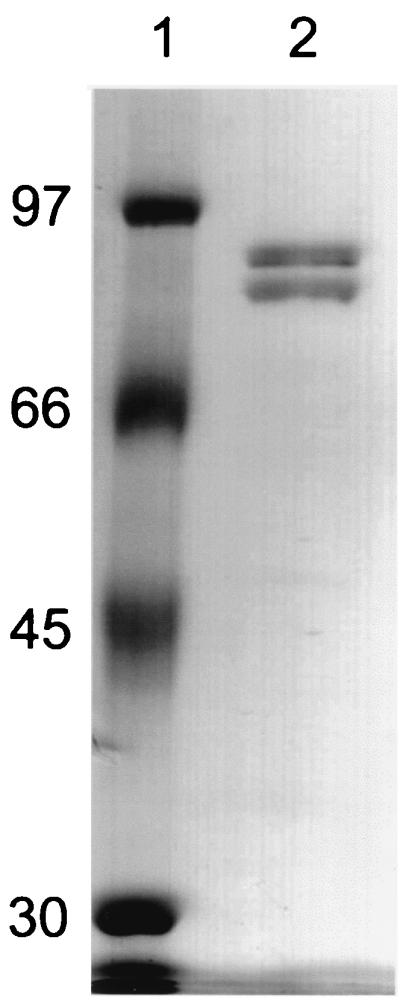
SDS-PAGE of purified soluble Fe(III) reductase. Lanes: 1, molecular mass markers (in kilodaltons); 2, 0.7 μg of soluble Fe(III) reductase. Proteins were resolved on a 10% polyacrylamide gel and stained with Coomassie brilliant blue.
The purified protein was yellowish brown, and the UV-visible spectrum had a broad absorption ranging from 360 to 500 nm with a peak at 390 nm and a slight shoulder at 450 nm. The absorption peak at 390 nm indicated the possible presence of iron-sulfur clusters (40).
Using ICP-MS, 28.9 ± 0.4 (mean ± standard deviation; n = 2) mol of iron per mol of protein was detected in two different preparations of the enzyme. The same value was obtained with a third enzyme preparation using ferrozine to determine the iron content colorimetrically (13). Since the sequence of soluble Fe(III) reductase showed the highest similarity to formate dehydrogenases (see below), which can contain W, Mo, and Se, the presence of these metals in the protein was also investigated. However, no W, Mo, or Se could be detected by ICP-MS. Determination of acid-labile sulfur gave a value of 17.4 ± 2.5 (n = 2) mol per mol of protein.
A trichloroacetic acid extract of the protein had a UV-visible spectrum typical for flavins (data not shown). High-pressure liquid chromatography analysis combined with fluorescence detection revealed the presence of 0.7 mol of FAD per mol of protein. Small amounts of FMN (<0.2 mol/mol) were also detected.
Enzymatic properties.
The Fe(III) reductase had a pH optimum of 5.5, and the highest activity was found at 45°C. The enzyme had a strict specificity for NADPH and did not utilize NADH as an electron donor. In contrast to previously described soluble Fe(III) reductases (14, 48), the addition of FAD or FMN to the enzyme assay mixture did not stimulate its activity. The apparent Km values for NADPH and Fe(III)-NTA were estimated to be 25 μM and 1 mM, respectively.
The enzyme preferentially reduced Fe(III)-NTA over the other forms of Fe(III) that were evaluated. Fe(III) complexed with EDTA or citrate was reduced at rates less than 5% of that observed with Fe(III)-NTA. Synthetic Fe(III)-oxyhydroxide was reduced by the enzyme at a rate of 2.5% compared to Fe(III)-NTA.
The enzyme did not reduce menadione with NADPH as the electron donor, suggesting that it cannot transfer electrons to quinones. The protein catalyzed the reduction of NADP+ with reduced methyl viologen as the electron donor at a rate of 385 U/mg.
Because of the sequence similarity of soluble Fe(III) reductase to a formate dehydrogenase (see below), the formate dehydrogenase activity of the protein was examined. However, there was no formate dehydrogenase activity with formate as the electron donor and methyl viologen as the electron acceptor. Furthermore, formate was not an electron donor for the reduction of Fe(III)-NTA.
Amino acid sequence analysis of soluble Fe(III) reductase.
The N-terminal sequence of the α subunit as determined by automated Edman degradation was Met-Val-Ser-Leu-Thr-Ile-Asp-Gly-Lys-Asp-Ile-Thr-Val-Ala-Lys-Glu-Thr-Thr-Ile-Leu. The N-terminal sequence of the β subunit was Ala-Gln-Val-Val-Phe-Ser-Ser-Trp-Gly-Arg-Thr-Ile-Val-Asp-Asn-Arg-Lys-Gly-Gly-Glu. These sequences were used to search the preliminary G. sulfurreducens genome sequence database of The Institute of Genomic Research, and 100% matches were found. The open reading frame containing the N-terminal sequence of the α subunit encoded a polypeptide of 844 amino acids with a predicted molecular weight of 89,386. This molecular weight was consistent with the molecular weight of the α subunit determined by SDS-PAGE (Fig. 1). Thus, the gene encoding this protein was designated sfrA (for “soluble Fe(III) reductase alpha subunit”). An open reading frame containing the N-terminal sequence of the beta subunit of the soluble Fe(III) reductase was located upstream of sfrA. The encoded protein consisted of 671 amino acids and had a predicted molecular weight of 74,142, which was consistent with the molecular weight determined for the β subunit of the purified enzyme. This gene was therefore designated sfrB. The two genes were separated by 126 bp and translated in the same reading frame. Hypothetical proteins encoded by open reading frames identified in the flanking regions of SfrA and SfrB did not show significant sequence similarities to proteins of known function (data not shown).
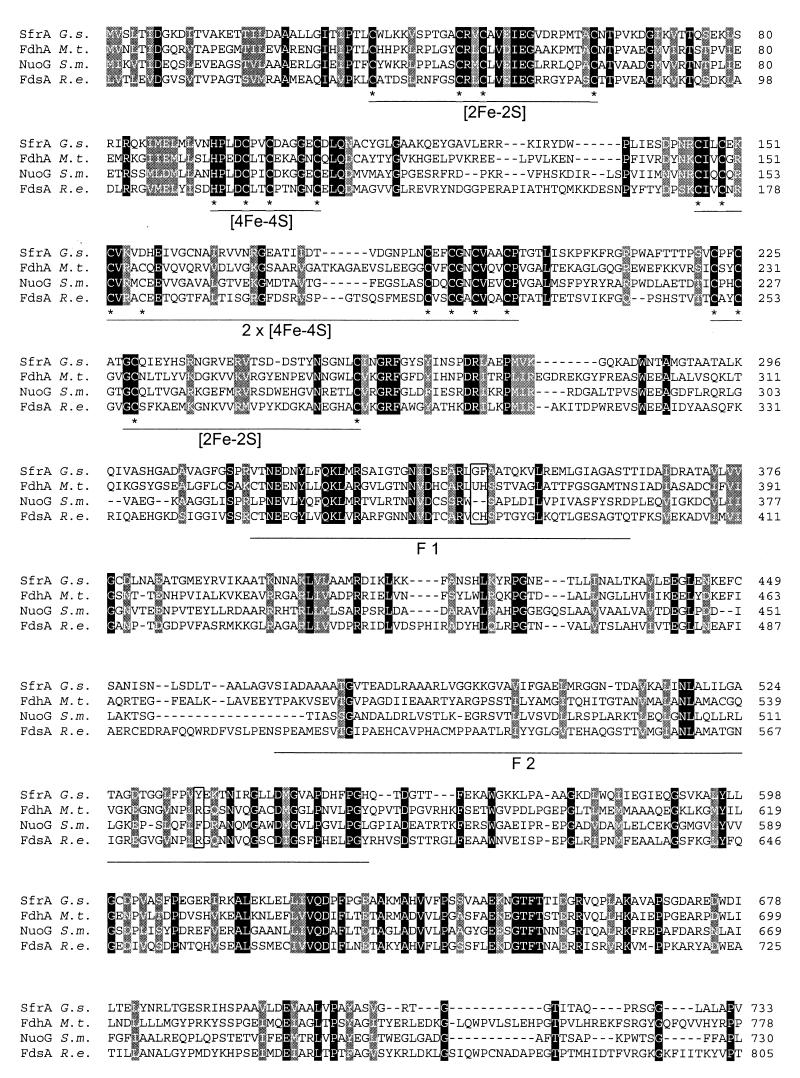
Databases were searched for similar proteins by using the BLAST algorithm (1). SfrA had the highest overall sequence similarity (32% identical and 49% similar amino acids) to the large subunit of the tungsten- and selenium-containing NADP+-dependent formate dehydrogenase (FdhA) from Moorella thermoacetica (52). Other similar proteins included chain G2 of NADH dehydrogenase I (NuoG) from Sinorhizobium meliloti (GenBank accession number P56914) (31% identical and 50% similar amino acids) and the large subunit of molybdenum-containing formate dehydrogenase (FdsA) from Ralstonia eutropha (39) (29% identical and 45% similar amino acids). The N-terminal part (residues 1 to 240) of SfrA, FhaA, NuoG, and FdsA contains five clusters of Cys residues (Fig. 2) that are thought to be involved in the binding of two [2Fe-2S] and three [4Fe-4S] clusters (39). The arrangement of the iron-sulfur cluster binding motifs in SfrA is similar to that in chain G of NADH dehydrogenase from various organisms and is not conserved in formate dehydrogenase sequences except for FdhA from M. thermoacetica and FdsA from R. eutropha (39). Structural information from formate dehydrogenase H from Escherichia coli (4) and sequence comparison of formate dehydrogenases (39) demonstrated that two conserved regions within the large subunit (F1 and F2 in Fig. 2) form the active site for formate oxidation. While some conservation is seen in these regions between SfrA and the formate dehydrogenases of M. thermoacetica and R. eutropha, SfrA lacks the residues that are catalytically important for formate oxidation. These residues (boxed in Fig. 2) include a Cys (or SeCys) residue in region F1 which is a ligand to the heavy-metal atom (Mo or W) of the molybdopterin cofactor, a neighboring His residue, and an Arg residue in region F2 (4).
FIG. 2.
Alignment of SfrA with related proteins. Identical and conservatively substituted residues are highlighted by black and gray backgrounds, respectively. Putative binding regions for iron-sulfur clusters are indicated, and the potential cluster-ligating residues are highlighted by asterisks. F1 and F2 indicate regions that are conserved in formate dehydrogenase sequences, with the active-site residues in boxes (see the text). Abbreviations: SfrA G.s., soluble Fe(III) reductase α subunit from G. sulfurreducens; FdhA M.t., formate dehydrogenase α subunit from M. thermoacetica (GenBank accession number U73807); NuoG S.m., NADH dehydrogenase I chain G 2 from S. meliloti (GenBank accession number P56914); FdsA R.e., soluble formate dehydrogenase α subunit from R. eutropha (39).
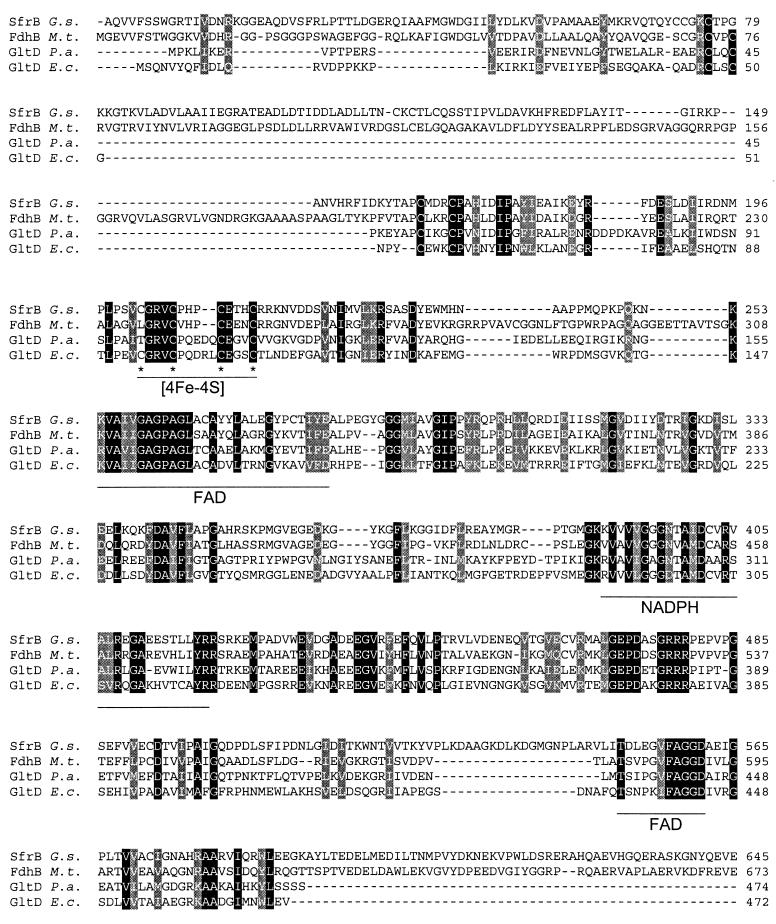
SfrB is most similar to the beta subunit of formate dehydrogenase (FdhB) from M. thermoacetica (38% identity and 50% similarity) and to the small subunit of glutamate synthase (GltD) (about 35% identity and 48% similarity) from different organisms (Pyrococcus abyssi, Thermotoga maritima, Aquifex aeolicus, and E. coli). An alignment of SfrB with FdhB from M. thermoacetica and GltD from P. abyssi and E. coli is provided in Fig. 3. Residues 386 to 420 resemble a consensus sequence proposed to be responsible for NAD(P)H binding in glutamate synthase (41). Residues 254 to 282 resemble a consensus sequence for the formation of an ADP binding fold in NAD(P)H-dependent and FAD-containing oxidoreductases (34, 51) that is thought to be involved in FAD binding. In the C-terminal part of SfrB, another conserved region is present (residues 550 to 560) that is indicative of FAD binding (11). The N-terminal part of SfrB (residues 1 to 220, 12 Cys) is similar to the corresponding region of GltD which is thought to provide Cys ligands for two [4Fe-4S] clusters (49). Residues 202 to 214 of SfrB resemble one of the proposed binding motives for a [4Fe-4S] cluster in GltD (21). In addition, the N-terminal region of SfrA showed similarity to iron-sulfur cluster binding domains found in hydrogenases (e.g., the HoxF subunit from the cyanobacteria Anabaena variabilis and Synechococcus strain PCC6301 [45]) and chain F of NADH dehydrogenase I. Although these similarities and the large number of Cys residues suggest that SfrB might bind more than one [Fe-S] cluster, sequence comparison did not indicate potential binding sites for these clusters.
FIG. 3.
Alignment of SfrB with related proteins. Identical and conservatively substituted residues are highlighted by black and gray backgrounds, respectively. Putative binding regions for an iron-sulfur cluster are indicated, and the potential cluster-ligating residues are highlighted by asterisks. Potential binding sites for FAD and NADPH are indicated (see the text). Abbreviations: SfrB G.s., soluble Fe(III) reductase β subunit from G. sulfurreducens; FdhB M.t., formate dehydrogenase β subunit from M. thermoacetica (GenBank accession number U73807); GltD P.a., glutamate synthase β subunit from P. abyssi (GenBank accession number E75069); GltD E.c., glutamate synthase β subunit from E. coli (19).
There was some discrepancy between the amounts of iron (29 mol) and acid-labile sulfur (17 mol) that were determined analytically and the amounts of iron and sulfur (40 mol each) that would be predicted for the α2β2 holoenzyme based on the potential binding sites for two [2Fe-2S] and three [4Fe-4S] clusters in the sequence of the α subunit and a potential binding site for at least one [4Fe-4S] cluster in the sequence of the β subunit (Fig. 2 and 3). It is possible that not all of the potential binding sites are actually ligating [Fe-S] centers or that iron and especially sulfur were not released quantitatively during chemical analysis.
DISCUSSION
The data presented above expand the known diversity of enzymes that may be involved in Fe(III) reduction in dissimilatory Fe(III)-reducing microorganisms. The Fe(III) reductase described here represents the first description of a purified soluble Fe(III) reductase from an organism that conserves energy to support growth from Fe(III) reduction. As discussed in detail below, the discovery of significant soluble NADPH-dependent Fe(III) reductase activity in G. sulfurreducens, coupled with the recent report of a similar activity in the dissimilatory Fe(III)-reducing archaeon Pyrobaculum islandicum (8), suggests that significant soluble Fe(III) reductase activity may be found in a wide phylogenetic diversity of dissimilatory Fe(III)-reducing microorganisms.
Comparison with other Fe(III) reductases in dissimilatory Fe(III)-reducing microorganisms.
The soluble Fe(III) reductase described here is significantly different from the membrane-bound Fe(III) reductases previously described in dissimilatory Fe(III)-reducing bacteria. Previous studies on Fe(III) reduction in G. sulfurreducens (16, 33) and the closely related G. metallireducens (17) focused on the NADH-dependent Fe(III) reductase activity, which was localized primarily in the membrane fraction (16, 17, 33). A similar membrane localization of Fe(III) reductase activity has also been noted in Shewanella species (10, 35). In general, cytochromes have been considered to be important electron carriers associated with these membrane-bound Fe(III) reductase activities (3, 12, 16–18, 30, 33, 36). The Fe(III) reductase described here not only was recovered in the soluble protein fraction but also differed from the membrane-bound Fe(III) reductases in the use of NADPH as the electron donor and the lack of a c-type cytochrome in the Fe(III) reductase complex.
The soluble NADPH-dependent Fe(III) reductase in G. sulfurreducens has similarities to the Fe(III) reductase activity found in P. islandicum (8). Although P. islandicum does not contain c-type cytochromes (8), it can conserve energy via dissimilatory Fe(III) oxide reduction (22). NADPH was the preferred electron donor for the reduction of Fe(III) in crude extracts of P. islandicum, and the Fe(III) reductase activity was found in the soluble fraction (8). No membrane-bound Fe(III) reductase activity could be detected in P. islandicum. Detailed comparisons between the soluble Fe(III) reductase in G. sulfurreducens and the one in P. islandicum are not yet possible because the enzyme from P. islandicum has yet to be purified and characterized.
Comparison with known soluble assimilatory Fe(III) reductases.
Most previously described soluble Fe(III) reductases have been recovered from aerobic or facultative microorganisms in which the Fe(III) reductase is considered to play a role in iron assimilation (5, 20). These enzymes require the addition of exogenous flavins to reduce Fe(III). Since reduced flavins can nonenzymatically reduce Fe(III), it has been suggested that these enzymes should be referred to as flavin reductases rather than Fe(III) reductases (14). A recent example of such an enzyme is the soluble Fe(III) reductase from Archaeoglobus fulgidus, which requires exogenous FAD or FMN for activity (48).
The soluble Fe(III) reductase described here did not require the addition of flavin for Fe(III) reductase activity, and the addition of flavins did not stimulate Fe(III) reduction. The Fe(III) reductase does contain about 1 mol of FAD per mol of enzyme, and analysis of the gene sequence for the Fe(III) reductase revealed the presence of two conserved regions that could form a flavin binding site in the β subunit of the soluble Fe(III) reductase. This bound flavin is likely to play a role in the transfer of electrons to Fe(III).
Potential physiological role.
The available evidence suggests that the enzyme described here is a redox-active protein with the potential to reduce soluble forms of Fe(III). However, on the basis of the current evidence, it cannot be determined whether this represents the physiological function of the protein. The specific activity of NADPH-dependent Fe(III) reduction in crude extracts is orders of magnitude higher than that of the NADH-dependent activity described previously for G. sulfurreducens (33). The same also holds true when the specific activity of the purified soluble Fe(III) reductase is compared to the enriched NADH-dependent membrane complex (33). This suggests that NADPH-dependent reduction of soluble Fe(III) might be important during dissimilatory reduction of Fe(III). On the other hand, the cytoplasmic location of soluble Fe(III) reductase and the fact that the protein in expressed at the same level during growth on fumarate compared to growth on Fe(III) as the electron acceptor may argue against a role of the protein in dissimilatory Fe(III) reduction. However, a similar situation is found in the dissimilatory Fe(III)-reducing archaeon P. islandicum. This organism readily grows via Fe(III) reduction (22), while Fe(III) reductase activity in cell-free preparations is found predominantly in the cytoplasmic fraction (8).
A role of soluble Fe(III) reductase in assimilatory Fe(III) reduction seems doubtful, since G. sulfurreducens grows only under environmental conditions in which dissolved Fe(II) is likely to be abundantly available. Furthermore, it might be expected that the synthesis of an assimilatory Fe(III) reductase would be regulated by the availability of soluble iron.
Sequence analysis did not clarify the physiological function of soluble Fe(III) reductase. Although both subunits of the soluble Fe(III) reductase have the highest sequence similarity to the corresponding subunits of the tungsten- and selenium-containing NADPH-dependent formate dehydrogenase of M. thermoacetica, it is not a formate dehydrogenase. The enzyme did not have formate dehydrogenase activity, formate was not utilized as an electron donor for the reduction of Fe(III), and the enzyme did not contain tungsten or selenium. Furthermore, sequence alignments indicated that it lacked the active-site residues conserved in formate dehydrogenases. The amino acid sequence of the β subunit of soluble Fe(III) reductase was similar to the β subunit of glutamate synthase from different sources. Bacterial glutamate synthase consists of two subunits and contains FAD, FMN, and three iron-sulfur clusters (49). The α subunit is the site of glutamate synthesis, whereas the β subunit catalyzes the oxidation of NADPH (49). The α subunit of soluble Fe(III) reductase had no similarity to the α subunit of glutamate synthase, which makes it unlikely that soluble Fe(III) reductase has a function in glutamate synthesis. The similarity of the β subunit of soluble Fe(III) reductase to the β subunit of glutamate synthase indicates that this subunit might contain the site for NADPH oxidation.
Another possibility is that the protein is involved in redox reactions not related to the reduction of Fe(III). Studies on the citric acid cycle in G. metallireducens (7) and G. sulfurreducens (15) demonstrated that the oxidation of 2-oxoglutarate can be measured with viologen as the electron acceptor rather than NAD(P)+. This suggests that in vivo, a ferredoxin is the electron acceptor for 2-oxoglutarate dehydrogenase. Ferredoxin is probably reoxidized by a ferredoxin:NADP+ oxidoreductase, and the presence of such an enzyme was postulated (15). The purified soluble Fe(III) reductase possessed high methyl viologen:NADP+ oxidoreductase activity, and it could be hypothesized that the enzyme is involved in transferring electrons from reduced ferredoxin to NADP+.
In summary, the NADPH-dependent soluble Fe(III) reductase from G. sulfurreducens is unlike any previously described enzyme with the capacity for Fe(III) reduction. Genetic studies are under way to further evaluate the physiological role of this enzyme.
ACKNOWLEDGMENTS
We thank G. R. Voight for technical assistance. We thank A. Siripiny and R. M. Barnes from the Department of Chemistry at the University of Massachusetts, Amherst, Mass., for performing ICP-MS analyses, and we thank M. Coppi for critical reading of the manuscript.
Sequencing of the complete genome of G. sulfurreducens was accomplished with support from the Department of Energy. This research was funded by the National Science Foundation grant MCB-9727840 and the Department of Energy NABIR program grant DE-FG02-97ER62475.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Andres-Lacueva C, Mattivi F, Tonon D. Determination of riboflavin, flavin mononucleotide and flavin-adenine dinucleotide in wine and other beverages by high-performance liquid chromatography with fluorescence detection. J Chromatogr. 1998;823:355–363. doi: 10.1016/s0021-9673(98)00585-8. [DOI] [PubMed] [Google Scholar]
- 3.Beliaev A S, Saffarini D A. Shewanella putrefaciens mtrB encodes an outer membrane protein required for Fe(III) and Mn(IV) reduction. J Bacteriol. 1998;180:6292–6297. doi: 10.1128/jb.180.23.6292-6297.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyington J C, Gladyshev V N, Khangulov S V, Stadtman T C, Sun P D. Crystal structure of formate dehydrogenase H: catalysis involving Mo, molybdopterin, selenocysteine, and an Fe4S4 cluster. Science. 1997;275:1305–1308. doi: 10.1126/science.275.5304.1305. [DOI] [PubMed] [Google Scholar]
- 5.Braun V, Killmann H. Bacterial solutions to the iron-supply problem. Trends Biochem Sci. 1999;24:104–109. doi: 10.1016/s0968-0004(99)01359-6. [DOI] [PubMed] [Google Scholar]
- 6.Caccavo F, Lonergan D J, Lovley D R, Davis M, Stolz J F, Mclnerney M J. Geobacter sulfurreducens sp. nov., a hydrogen- and acetate-oxidizing dissimilatory metal-reducing microorganism. Appl Environ Microbiol. 1993;60:3752–3759. doi: 10.1128/aem.60.10.3752-3759.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Champine J E, Underhill B, Johnston J M, Lilly W W, Goodwin S. Electron transfer in the dissimilatory iron-reducing bacterium Geobacter metallireducens. Anaerobe. 2000;6:187–196. [Google Scholar]
- 8.Childers S E, Lovley D R. Differences in Fe(III) reduction in the hyperthermophilic archaeon, Pyrobaculum islandicum, versus mesophilic Fe(III)-reducing bacteria. FEMS Microbiol Lett. 2001;195:253–258. doi: 10.1111/j.1574-6968.2001.tb10529.x. [DOI] [PubMed] [Google Scholar]
- 9.Coates J D, Phillips E J, Lonergan D J, Jenter H, Lovley D R. Isolation of Geobacter species from diverse sedimentary environments. Appl Environ Microbiol. 1996;62:1531–1536. doi: 10.1128/aem.62.5.1531-1536.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dobbin P S, Powell A K, McEwan A G, Richardson D J. The influence of chelating-agents upon the dissimilatory reduction of Fe(III) by Shewanella putrefaciens. Biometals. 1995;8:163–173. [Google Scholar]
- 11.Eggink G, Engel H, Vriend G, Terpstra P, Witholt B. Rubredoxin reductase of Pseudomonas oleovorans. Structural relationship to other flavoprotein oxidoreductases based on one NAD and two FAD fingerprints. J Mol Biol. 1990;212:135–142. doi: 10.1016/0022-2836(90)90310-I. [DOI] [PubMed] [Google Scholar]
- 12.Field S J, Dobbin P S, Cheesman M R, Watmough N J, Thomson A J, Richardson D J. Purification and magneto-optical spectroscopic characterization of cytoplasmic membrane and outer membrane c-type cytochromes from Shewanella frigidimarina NCIMB400. J Biol Chem. 2000;275:8518–8522. doi: 10.1074/jbc.275.12.8515. [DOI] [PubMed] [Google Scholar]
- 13.Fish W W. Rapid colorimetric micromethod for the quantitation of complexed iron in biological samples. Methods Enzymol. 1988;158:357–364. doi: 10.1016/0076-6879(88)58067-9. [DOI] [PubMed] [Google Scholar]
- 14.Fontecave M, Covès J, Pierre J-L. Ferric reductases or flavin reductases? BioMetals. 1994;7:3–8. doi: 10.1007/BF00205187. [DOI] [PubMed] [Google Scholar]
- 15.Galushko A S, Schink B. Oxidation of acetate through reactions of the citric acid cycle by Geobacter sulfurreducens in pure culture and in syntrophic coculture. Arch Microbiol. 2000;174:314–321. doi: 10.1007/s002030000208. [DOI] [PubMed] [Google Scholar]
- 16.Gaspard S, Vazquez F, Holliger C. Localization and solubilization of the iron(III) reductase of Geobacter sulfurreducens. Appl Environ Microbiol. 1998;64:3188–3194. doi: 10.1128/aem.64.9.3188-3194.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gorby Y A, Lovley D R. Electron transport in the dissimilatory iron reducer, GS-15. Appl Environ Microbiol. 1991;57:867–870. doi: 10.1128/aem.57.3.867-870.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gordon E H, Pike A D, Hill A E, Cuthbertson P M, Chapman S K, Reid G A. Identification and characterization of a novel cytochrome c3 from Shewanella frigidimarina that is involved in Fe(III) respiration. Biochem J. 2000;349:153–158. doi: 10.1042/0264-6021:3490153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gosset G, Merino E, Recillas F, Oliver G, Becerril B, Bolivar F. Amino acid sequence analysis of the glutamate synthase enzyme from Escherichia coli K-12. Protein Sequences Data Anal. 1989;2:9–16. [PubMed] [Google Scholar]
- 20.Guerinot M L. Microbial iron transport. Annu Rev Microbiol. 1994;48:743–772. doi: 10.1146/annurev.mi.48.100194.003523. [DOI] [PubMed] [Google Scholar]
- 21.Hagen W R, Vanoni M A, Rosenbaum K, Schnackerz K D. On the iron-sulfur clusters in the complex redox enzyme dihydropyrimidine dehydrogenase. Eur J Biochem. 2000;267:3640–3646. doi: 10.1046/j.1432-1327.2000.01393.x. [DOI] [PubMed] [Google Scholar]
- 22.Kashefi K, Lovley D R. Reduction of Fe(III), Mn(IV), and toxic metals at 100°C by Pyrobaculum islandicum. Appl Environ Microbiol. 2000;66:1050–1056. doi: 10.1128/aem.66.3.1050-1056.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.Lloyd J R, Blunt-Harris E L, Lovley D R. The periplasmic 9.6-kilodalton c-type cytochrome of Geobacter sulfurreducens is not an electron shuttle to Fe(III) J Bacteriol. 1999;181:7647–7649. doi: 10.1128/jb.181.24.7647-7649.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lonergan D J, Jenter H L, Coates J D, Phillips E J, Schmidt T M, Lovley D R. Phylogenetic analysis of dissimilatory Fe(III)-reducing bacteria. J Bacteriol. 1996;178:2402–2408. doi: 10.1128/jb.178.8.2402-2408.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lovley D R. Fe(III) and Mn(IV) reduction. In: Lovley D R, editor. Environmental microbe-metal interactions. Washington, D.C.: ASM Press; 2000. pp. 3–30. [Google Scholar]
- 27.Lovley D R. Fe(III)- and Mn(IV)-reducing prokaryotes. In: Dworkin M, et al., editors. The prokaryotes. New York, N.Y: Springer-Verlag, Inc.; 2000. [Google Scholar]
- 28.Lovley D R. Microbial reduction of iron, manganese, and other metals. Adv Agron. 1995;54:175–231. [Google Scholar]
- 29.Lovley D R, Coates J D. Novel forms of anaerobic respiration of environmental relevance. Curr Opin Microbiol. 2000;3:252–256. doi: 10.1016/s1369-5274(00)00085-0. [DOI] [PubMed] [Google Scholar]
- 30.Lovley D R, Giovannoni S J, White D C, Champine J E, Phillips E J, Gorby Y A, Goodwin S. Geobacter metallireducens gen. nov. sp. nov., a microorganism capable of coupling the complete oxidation of organic compounds to the reduction of iron and other metals. Arch Microbiol. 1993;159:336–344. doi: 10.1007/BF00290916. [DOI] [PubMed] [Google Scholar]
- 31.Lovley D R, Woodward J C, Chapelle F H. Stimulated anoxic biodegradation of aromatic hydrocarbons using Fe(III) ligands. Nature. 1994;370:128–131. doi: 10.1038/370128a0. [DOI] [PubMed] [Google Scholar]
- 32.Luther G W, Shellenbarger P A, Brendel P J. Dissolved organic Fe(III) and Fe(II) complexes in salt marsh porewaters. Geochim Cosmochim Acta. 1996;60:951–960. [Google Scholar]
- 33.Magnuson T S, Hodges-Myerson A L, Lovley D R. Characterization of the membrane-bound NADH-dependent Fe3+ reductase from the dissimilatory Fe3+ -reducing bacterium Geobacter sulfurreducens. FEMS Microbiol Lett. 2000;185:205–211. doi: 10.1111/j.1574-6968.2000.tb09063.x. [DOI] [PubMed] [Google Scholar]
- 34.McKie J H, Douglas K T. Evidence for gene duplication forming similar fold for NAD(P)H and FAD in pyridine nucleotide-dependent flavoproteins. FEBS Lett. 1991;279:5–6. doi: 10.1016/0014-5793(91)80236-v. [DOI] [PubMed] [Google Scholar]
- 35.Myers C R, Myers J M. Ferric reductase is associated with the membranes of anaerobically grown Shewanella putrefaciens MR-1. FEMS Microbiol Lett. 1993;108:15–22. [Google Scholar]
- 36.Myers C R, Myers J M. Outer membrane cytochromes of Shewanella putrefaciens MR-1: spectral analysis, and purification of the 83-kDa c-type cytochrome. Biochim Biophys Acta. 1997;1326:307–318. doi: 10.1016/s0005-2736(97)00034-5. [DOI] [PubMed] [Google Scholar]
- 37.Nealson K H, Saffarini D. Iron and manganese in anaerobic respiration: environmental significance, physiology, and regulation. Annu Rev Microbiol. 1994;48:311–343. doi: 10.1146/annurev.mi.48.100194.001523. [DOI] [PubMed] [Google Scholar]
- 38.Nevin K P, Lovley D R. Lack of production of electron-shuttling compounds or solubilization of Fe(III) during reduction of insoluble Fe(III) oxide by Geobacter metallireducens. Appl Environ Microbiol. 2000;66:2248–2251. doi: 10.1128/aem.66.5.2248-2251.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oh J-I, Bowien B. Structural analysis of the fds operon encoding the NAD+-linked formate dehydrogenase from Ralstonia eutropha. J Biol Chem. 1998;273:26349–26360. doi: 10.1074/jbc.273.41.26349. [DOI] [PubMed] [Google Scholar]
- 40.Orme-Johnson W H, Orme-Johnson N R. Iron-sulfur proteins: the problem of determining the cluster type. In: Spiro T G, editor. Iron-sulfur proteins. New York, N.Y: Wiley-Interscience; 1982. [Google Scholar]
- 41.Pelanda R, Vanoni M A, Perego M, Piubelli L, Galizzi A, Curti B, Zanetti G. Glutamate synthase genes of the diazotroph Azospirillum brasilense. Cloning, sequencing, and analysis of functional domains. J Biol Chem. 1993;268:3099–3106. [PubMed] [Google Scholar]
- 42.Rabinowitz J C. Analysis of acid-labile sulfide and sulfhydryl groups. Methods Enzymol. 1978;53:275–277. doi: 10.1016/s0076-6879(78)53033-4. [DOI] [PubMed] [Google Scholar]
- 43.Ratering S, Schnell S. Localization of iron-reducing activity in paddy soil by profile studies. Biogeochemistry. 2000;48:341–365. [Google Scholar]
- 44.Reeves H C, Rabin R, Wegener W S, Ajl J. Assays of enzymes of the tricaboxylic acid and glyoxylate cycles. Methods Microbiol. 1971;6A:425–462. [Google Scholar]
- 45.Schmitz O, Boison G, Hilscher R, Hundeshagen B, Zimmer W, Lottspeich F, Bothe H. Molecular biological analysis of a bidirectional hydrogenase from cyanobacteria. Eur J Biochem. 1995;233:266–276. doi: 10.1111/j.1432-1033.1995.266_1.x. [DOI] [PubMed] [Google Scholar]
- 46.Smith P K, Krohn R I, Hermanson G T, Mallia A K, Gartner F H, Provenzano M D, Fujimoto E K, Goeke N M, Olson B J, Klenk D C. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. . (Erratum, 163:279, 1987.) [DOI] [PubMed] [Google Scholar]
- 47.Snoeyenbos-West O L, Nevin K P, Anderson R T, Lovley D R. Enrichment of Geobacter species in response to stimulation of Fe(III) reduction in sandy aquifer sediments. Microb Ecol. 2000;39:153–167. doi: 10.1007/s002480000018. [DOI] [PubMed] [Google Scholar]
- 48.Vadas A, Monbouquette H G, Johnson E, Schroeder I. Identification and characterization of a novel ferric reductase from the hyperthermophilic Archaeon Archaeoglobus fulgidus. J Biol Chem. 1999;274:36715–36721. doi: 10.1074/jbc.274.51.36715. [DOI] [PubMed] [Google Scholar]
- 49.Vanoni M A, Curti B. Glutamate synthase: a complex iron-sulfur flavoprotein. Cell Mol Life Sci. 1999;55:617–38. doi: 10.1007/s000180050319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vargas M, Kashefi K, Blunt-Harris E L, Lovley D R. Microbiological evidence for Fe(III) reduction on early Earth. Nature. 1998;395:65–67. doi: 10.1038/25720. [DOI] [PubMed] [Google Scholar]
- 51.Wierenga R K, Terpstra P, Hol W G. Prediction of the occurrence of the ADP-binding beta alpha beta-fold in proteins, using an amino acid sequence fingerprint. J Mol Biol. 1986;187:101–107. doi: 10.1016/0022-2836(86)90409-2. [DOI] [PubMed] [Google Scholar]
- 52.Yamamoto I, Saiki T, Liu S-M, Ljungdahl L G. Purification and properties of NADP-dependent formate dehydrogenase from Clostridium thermoaceticum, a tungsten-selenium-iron protein. J Biol Chem. 1983;258:1826–1832. [PubMed] [Google Scholar]
- 53.Zhu Z, Sun D, Davidson V L. Localization of periplasmic redox proteins of Alcaligenes faecalis by a modified general method for fractionating gram-negative bacteria. J Bacteriol. 1999;181:6540–6542. doi: 10.1128/jb.181.20.6540-6542.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]