Abstract
Conservation of biodiversity is critical for the coexistence of humans and the sustenance of other living organisms within the ecosystem. Identification and prioritization of specific regions to be conserved are impossible without proper information about the sites. Advanced monitoring agencies like the Intergovernmental Science-Policy Platform on Biodiversity and Ecosystem Services (IPBES) had accredited that the sum total of species that are now threatened with extinction is higher than ever before in the past and are progressing toward extinct at an alarming rate. Besides this, the conceptualized global responses to these crises are still inadequate and entail drastic changes. Therefore, more sophisticated monitoring and conservation techniques are required which can simultaneously cover a larger surface area within a stipulated time frame and gather a large pool of data. Hence, this study is an overview of remote monitoring methods in biodiversity conservation via a survey of evidence-based reviews and related studies, wherein the description of the application of some technology for biodiversity conservation and monitoring is highlighted. Finally, the paper also describes various transformative smart technologies like artificial intelligence (AI) and/or machine learning algorithms for enhanced working efficiency of currently available techniques that will aid remote monitoring methods in biodiversity conservation.
Keywords: Artificial intelligence, Geographic information system, Molecular techniques, Remote sensing, Wildlife conservation
Introduction
Biological diversity “biodiversity” entails the assortment of earthly life forms heterogeneously ranging from genetic to ecosystem level. It can embrace the evolutionary, ecological, and cultural aspects that uphold life in various forms (McQuatters-Gollop et al. 2019). It fosters ecological functioning that paves the path for fundamental ecosystem services comprising food, water, preservation of soil fertility, and management of pests and diseases (Avigliano et al. 2019; Whitehorn et al. 2019). The plasticity of co-existence between mankind and nature is irreversible because of the symbiotic relationship that sustains the co-survival of humans with other living organisms (Arias-Maldonado 2016).
Biological diversity of forest originating from gene to ecosystem, through species, supports forest habitat that gives rise to fodders and other goods and services in a wide array of diverse biophysical and socio-economic ambience. Despite the applicability and significance of biodiversity, its conservation is vaguely acknowledged. Presently, human invasions have distorted around 75% of the land-based territory and about 66% of the marine ecosystem. Further to this, over a third of the global terrestrial regions are now devoted to domestic pursuit (FAO 2019). Moreover, since 1970, the significance of agricultural crop yield has increased by about 300%, and harvesting of raw timber has hiked by 45%. Moreover, renewable and non-renewable resources roughly of 60 billion tons are presently extracted annually across the globe. Exploitation of land has abridged the prolificacy of 23% of the global land area, annually, up to US$577 billion in worldwide crops are in jeopardy from pollinator loss, and about 100–300 million people are at elevated threat of natural disaster due to loss of coastal habitats and protection (IPBES 2019). If such trends continue then by 2050, the transformative change in nature can lead to an unprecedented devastating irreversible impact on mankind, which will take centuries to recover.
These atrocities of biodiversity need to be averted through proper monitoring and conservation measures. Based on the present advancements in technology, a combination of system-based smart techniques, remote sensing, and molecular approaches will be necessary for implementation of such ambitious conservation drives. Computer-based simulation techniques such as geographic information system (GIS), active and passive radio detection and ranging (RADAR) system, and light detection and ranging (LiDAR) system are playing a crucial role for monitoring biodiversity in real time (Bouvier et al. 2017; Bae et al. 2019; Bakx et al. 2019). Further to this, the application of recent advancements like artificial intelligence (AI) (Kwok 2019) and/or machine learning algorithms (Fernandes et al. 2020) have also been exploited for the same (Hu et al. 2015). These systems are not only reliable in monitoring biodiversity globally but can also help prevent further biodiversity loss worldwide. Besides monitoring tools, conservation of individual species and genetic biodiversity as a whole will require the use of recent molecular techniques. Conservation genomics revolves around the concept that genome-scale data will meliorate the competence of resource proprietors to conserve species. Despite the decades-long utilization of genetic approaches for conservation research, it has only recently been implied for generating genome-wide data which is functional for conservation (Supple and Shapiro 2018). The revolutionary molecular tools like restriction fragment length polymorphism (RFLP), amplified fragment length polymorphism (AFLP), random amplified polymorphic DNA (RAPD), sequence characterized amplified region (SCAR), microsatellites and mini-satellites, expressed sequence tags (ESTs), inter-simple sequence repeat (ISSR), and single nucleotide polymorphisms (SNPs) have transformed the hierarchy of biodiversity conservation to a higher level (Mosa et al. 2019).
Evidently, more sophisticated monitoring methods such as system-based simulation techniques, remote sensing, artificial intelligence, and geographic information system as well as molecular-based techniques facilitate the monitoring methods in biodiversity conservation and restoration. Therefore, the present study is an overview of remote monitoring methods in biodiversity conservation via a survey of evidence-based reviews and related studies, wherein the description of the application of some technology for biodiversity conservation and monitoring. Finally, the paper also describes various transformative smart technologies like artificial intelligence (AI) and/or machine learning algorithms for enhanced working efficiency of currently available techniques that will aid remote monitoring methods in biodiversity conservation.
Methodology of literature search
The relevant literature search was done electronically by using Google Search Engine, PubMed, ScienceDirect, SpringerLink, Frontiers Media, and MDPI databases. The most importantly searched keywords were biodiversity and potential threats, techniques to monitoring biodiversity, geographic information system, remote sensing, active remote sensing system, radio detection and ranging (RADAR) system, light detection and ranging (LiDAR) systems, passive remote sensing systems, techniques for identification and genetic conservation of species, restriction fragment length polymorphism (RFLP), amplified fragment length polymorphism (AFLP), random amplified polymorphic DNA (RAPD), sequence characterized amplified region (SCAR), mini- and micro-satellites, expressed sequence tags (ESTs), inter-simple sequence repeat (ISSR), single nucleotide polymorphisms (SNPs), and artificial intelligence in biodiversity monitoring which were used and placed repeatedly within the text.
Biodiversity and potential threats
Biodiversity in simple terms is a heterogenic distribution of flora and fauna throughout the world or in a particular niche (Naeem et al. 2016). The number of species described around the world as per IUCN (2020) accounts for 2,137,939, of which 72,327 are vertebrates, 1,501,581 are invertebrates, 422,756 are plants, and 141,275 are identified as fungi and protists. It is now acknowledged that biodiversity is a major indicator of community ecosystem fluctuations and functioning (Tilman et al. 2014). These include provisioning of food, pollination, cultural recreation, and supporting nutrient cycling (Harrison et al. 2014; Bartkowski et al. 2015). Biodiversity as a whole is represented by two major components that are species richness and species evenness. A biogeographic region with a significant level of endemic species and with a higher loss of habitat is generally depicted as a biodiversity hotspot (Marchese 2015). These areas have proven themselves as a tool for establishing conservation priorities and orchestrate vital rationale in decision-making for cost-effective tactics to safeguard biodiversity in its natural conserved state. Usually, the hotspots are marked by single or multiple species-based metrics or concentrate on phylogenetic and functional diversity to shield species that sustain exclusive and inimitable functions inside the ecosystem (Marchese 2015).
Currently, as per the IUCN Red List of Species 2020–2021, of the 2,137,939 species around the world, about 31,030 species are categorized as “threatened” species. Among these, plants with 16,460 numbers contribute the most followed by vertebrates (9063), invertebrates (5333), and fungi and protists (174) [IUCN 2020]. Many of the species are still not assessed due to a lack of reliable identification tools or techniques. Biodiversity is mostly threatened by over-population, habitat and landscape modification, indiscriminate exploitation of resources, pollution, and lack of proper documentation (Marchese 2015; Liu et al. 2020; Reid et al. 2019). Demographic changes can be considered an imperative module for assisting the indirect drivers of biodiversity alternations specifically associated with land use patterns (Newbold et al. 2015). Population explosion, central demographic developments, and urbanization impact both ecosystems and the species it harbors (Mehring et al. 2020). As the changing demographic pattern is associated with population explosion, this may pose a pessimistic impact on food availability, restricted emission of greenhouse gases, control of invasive species and diseases, etc. (Lampert 2019; Manisalidis et al. 2020; Hoban et al. 2020; Reid et al. 2019). To generate such massive data over a stipulated time frame and process them simultaneously to extrude applicable information requires cutting-edge tools and multidisciplinary scientific input (Randin et al. 2020). With recent advancements in mapping software, large-scale data processors, and monitoring tools and genetics, artificial intelligence for generating accurate data over a larger area as a part of a global monitoring strategy has now become feasible (Wetzel et al. 2015; Randin et al. 2020). Hence, an assortment of the above mention techniques and tools will be essential for the conservation and restoration of biodiversity.
Techniques for monitoring biodiversity
Mapping and monitoring techniques have been frontiers in predicting and modeling anthropogenic activities, habitat use, and pattern of land use over time in a particular region. These advanced physical techniques include GIS, LiDAR, and RADAR systems (Bouvier et al. 2017; Bae et al. 2019; Bakx et al. 2019).
Geographic information system (GIS)
Understanding functional geography and making intelligent decisions is widely beneficial for naturalists. GIS is a popular tool for analyzing possible and current spatial-temporal distribution, location, distribution patterns, population assessment, and identification of priority areas for their conservation and management (Krigas et al. 2012; Salehi and Ahmadian 2017). Currently, development of ecological niche models based on topographic, bioclimatic, soil, and land use variables was mapped and predicted for species such as Clinopodium nepeta, Thymbra capitata, Melissa officinalis, Micromeria juliana, Origanum dictamnus, O. vulgare, O. onites, Salvia fruticosa, S. pomifera, and Satureja thymbra (Bariotakis et al. 2019). With the assistance of digitally integrated video and audio-GIS (DIVA-GIS), actual geographic distribution and the future potential assortments of several Zingiber sp. like Z. mioga, Z. officinale, Z. striolatum, and Z. cochleariforme were analyzed (Huang et al. 2019). Most recently, important climatic inconsistencies distressing the geographical dispersion of wild Akebia trifoliate based on the formation of spatial database were successfully determined with the help of GIS (Wang et al. 2020). However, GIS possesses certain limitations such as expensive software, hardware, capturing GIS data, and difficulty in their use (Bearman et al. 2016) (Fig. 1, Table 1).
Fig. 1.
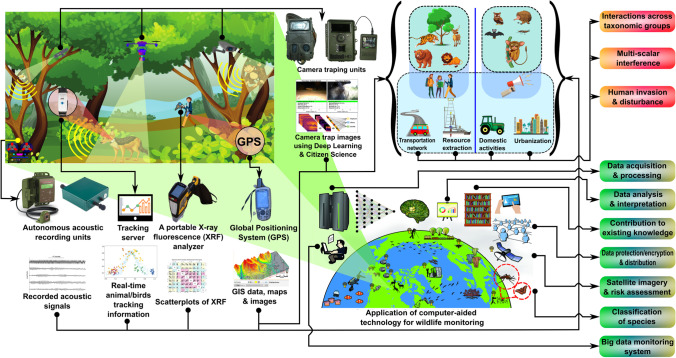
Application of Geographic Information Systems (GIS), remote sensing technologies like radio detection and ranging (RADAR) and satellite-based light detection and ranging (LiDAR) for wildlife monitoring in the forest ecosystem. The figure describes global forest distribution (Our World In Data 2020), wherein displayed GIS and different levels of GIS data, schismatic of passive remote sensing, active remote sensing segregated into primary RADAR system, and a block diagram of satellite-based LiDAR system for generation of DCHM (digital canopy height model) image and 3-D point cloud image of the whole organism is outlined. Global positioning system (GPS), Inertial measurement unit (IMU). The figure is inspired by the following sources: Omasa et al. (2007), Admin (2017), Bhatta and Priya (2017), Jahncke et al. (2018), Martone et al. (2018), Srivastava et al. (2020). The components of the figure are modifications of Portree (2006), Organikos (2012); Smithsonian’s National Zoo and Conservation Biology Institute Smithsonian’s National Zoo (2016), and Freepik (2021). Abbreviations: FP-mode, first-pulse mode; LP-mode, last-pulse mode; DEM, digital elevation model; DTM, digital terrain model
Table 1.
Application of geographic information system (GIS) in biodiversity monitoring
| Subject investigated | Tool | Objectives | Findings | References |
|---|---|---|---|---|
| Geographic information system (GIS) | ||||
| Clinopodium nepeta, Thymbra capitata, Melissa officinalis, Micromeria juliana, Origanum dictamnus, O. vulgare, O. onites, Salvia fruticosa, S. pomifera, Satureja thymbra | Geographic information system (GIS) |
• Predict and map the potential habitat • Investigate spatial patterns (niche similarities, suitability “hotspots”) • Development of database |
Ecological niche models were developed for each studied taxon, based on topographic, bioclimatic, soil, and land use variables | Bariotakis et al. (2019) |
| Zingiber mioga, Z. officinale, Z. striolatum, Z. cochleariforme, Z. kwangsiense, Z. linyunense, Z. confine, Z. kawagoii, Z. koshunensis, Z. laoticum, Z. roseum, Z. corallinum, Z. atrorubens, Z. pleiostachyum | Digitally integrated video and audio-GIS (DIVA-GIS) | • Analysis of ecological distribution and richness | Actual geographic distribution and the present and future potential distributions of Zingiber spp. | Huang et al. (2019) |
| Lippia graveolens | GIS | • Development of an analytical framework based on multi-criteria-multi objective analyses | New potential oregano harvesting areas were identified | Irina et al. (2019) |
| Panax quinquefolius | GIS | • Prediction of areas with potential ecological suitability for global medicinal plants | Ecologically suitable habitats and distribution for medicinal plant were predicted | Shen et al. (2019) |
| Brassica napus | GIS database | • Quantification of landscape composition and configuration | Landscape composition and configuration of B. napus were quantified | Reeth et al. (2019) |
| Tianzhu Mountain, National Forest Park, Fujian province, South China | GIS-analytic hierarchy process (AHP) | • Quantification and integration of the visual sensitivity and ecological sensitivity areas of the Tianzhu Mountain, National Forest Park | A comprehensive means to integrate the quantitative and objective analysis of the visual sensitivity and the ecological sensitivity was developed | Zheng et al. (2019) |
| Zea mays | GIS environment simulation | • Assessment of wildlife damage estimation methods in Z. mays with simulation in GIS environment | GIS simulations were tested as a suitable tool for testing the sampling methods in assessing wildlife damages to agriculture | Kovács et al. (2020) |
| Z. mays | GIS-AHP | • Assessment of land suitability for Z. mays farming | GIS modeling approach based on environmental and agro-ecological data was highly beneficiary for Z. mays farming and a good guide for future land-use management | Tashayo et al. (2020) |
| Ebenus armitagei, Periploca Angustifolia | GIS | Estimation of endangered plants and their distributive areas | GIS data predicted the most threatened locations, available soil dynamics, distribution, and vegetation coverage structure of the plants | Gamal et al. (2020) |
| Gentiana kurroo, Lilium polyphyllum, Saussurea costus, Aconitum chasmanthum | Global positioning system (GPS) | • To understand the distribution, ecology, and conservation implications of critically endangered endemic plants | The study revealed that the species are restricted to a small geographic range and their highly specific ecological niche makes them vulnerable to stochastic events and human interferences | Mir et al. (2020) |
| Akebia trifoliate | GIS | • Estimation of climatic variables affecting A. trifoliate growth from a macro perspective | Key climatic variables affecting the geographical distribution of wild A. trifoliate based on the establishment of spatial database were successfully determined | Wang et al. (2020) |
Remote sensing
The ability to extract information about the environment without physical contact from a large distance by a sensor that reflects and/ or emits electromagnetic spectrum (visible, infrared, and microwave spectra) is defined as remote sensing. Based on the source of radiation emitted, which comes in contact with the object, remote sensing can be categorized as active or passive remote sensing systems (Höppler et al. 2020). Remote sensing of biodiversity can be used for habitat mapping including species area curve and habitat heterogeneity, species mapping/distribution, plant functional diversity/ traits, spectral diversity including vegetation indices and spectral species (Cavender-Bares et al. 2020; Wang and Gamon 2019).
Active remote sensing system
An active remote sensor emits energy pulses and records the return time and amplitude of the backscattered energy pulses from the object to generate the required information about it (Vogeler and Cohen 2016). Currently used active remote sensing technologies like RADAR and LiDAR systems can be used to determine the location, speed, and direction of any wildlife form.
Radio detection and ranging (RADAR) system
As a sub-set of the active remote sensing system, RADAR, operates in the microwave of wavelengths of 1 mm to 1 m. Additionally, the modern RADAR systems are incorporated with software routines to mathematically enhance spatial resolution and manage multiple pictures of the same object, also as Synthetic Aperture RADARs (SARs) (Fig. 1). These systems can be used to determine the polarization of the emitted and receive electromagnetic rays which provides a better understanding of the analyzed surface properties (Hay 2000; Valbuena et al. 2020; Barlow and O’Neill 2020) (Table 2).
Table 2.
Biodiversity monitoring through remote sensing
| Subject investigated | Tool | Objectives | Findings | References |
|---|---|---|---|---|
| Radio detection and ranging (RADAR) | ||||
| Brazilian Amazon (Manaus, Santarém, Machadinho d’Oeste) | Single-date Landsat Thematic Mapper (TM), Advanced Land Observing Satellite (ALOS) Phased Arrayed L-band Synthetic Aperture RADAR (PALSAR) data |
• Investigation of the combined use of TM, ALOS, and PALSAR to • Discriminate mature forest, non-forest, and secondary forest • Retrieving the age of secondary forests |
The study resulted in highlighting that ASF maps can be generated without the need for extensive time-series analysis of Landsat and/or SAR data | Carreiras et al. (2017) |
| 52 least-disturbed sites along a climatic gradient between semi-arid and arid zones in the south-eastern Mediterranean | Polarimetric SAR (PolSAR) | • Determination of radar polarization and ecological pattern properties | Full polarimetric L-band PALSAR data was acquired and empirical assessments of the relationships between radar polarization properties and ecological pattern properties of climatic transition ecosystems were also acquired | Chang and Shoshany (2017) |
| Zürich | European Space Agency(ESA)-Sentinel-1A single C-band SAR, Sentinel-2A-MultiSpectral Instrument (MSI) | • Sentinel-1A SAR and Sentinel-2A MSI data fusion for urban ecosystem service mapping | Method and underlying data were effective for urban land cover and ecosystem service mapping | Haas and Ban (2017) |
| Broadleaf, needle leaf, and mixed forests in Northeastern China | ALOS/PALSAR | • Estimation of aboveground biomass | A nonlinear relationship between PALSAR backscatter coefficients and forest AGB of different forest types in NE China was constructed | Ma et al. (2017) |
| Howland Forest in Maine, Chequamegon-Nicolet National Forest in Wisconsin, Harvard Forest in Massachusetts | Light detection and ranging (LiDAR) systems, ALOS/ PALSAR-horizontal transmit, and vertical receive (HV), Shuttle Radar Topography Mission (SRTM) | • Modeling forest canopy height | As per the results, these techniques were potential for estimating canopy height in a different biome; furthermore, the biome-level model developed was efficient and can be used as a reliable estimator of biome-level forest structure | García et al. (2018) |
| Gran Chaco stretching across Argentina, Bolivia, Paraguay, and Brazil | Landsat-8 optical, Sentinel-1-SAR | • Mapping continuous fields of trees, shrubs, vegetation types, and savannas | Continuous fields of tree, shrub, vegetation types, and savannas covered across the entire Chaco were mapped; additionally, the relationship between tree and shrub cover and a suite of environmental and socio-economic variables were established | Baumann et al. (2018) |
| The tropical rainforests of the Western Ghats, India | Sentinel-2-MSI, SAR Sentinel-1 satellite | • Mapping and assessment of vegetation types | The magnitude of habitat fragmentation was effectively evaluated, high accuracy in classification was obtained, and vegetation indices and textures for vegetation type classification were mapped for prioritizing top conservation areas | Erinjery et al. (2018) |
| Nova Scotia |
Multi-beam RADARSAT-2 PALSAR, ALOS, and LiDAR |
• Mapping wetlands | The techniques were beneficial and generated improved wetland mapping | Jahncke et al. (2018) |
| Five villages of the Koningue commune: Dougan, Sukumba, Ngueguesso 1, Ngueguesso 2, Banesso | Sentinel-2-MSI, SRTM | • Estimation of smallholder crops production | The approach opened up new prospects for food security and agricultural performance monitoring and accuracy in smallholder farming systems | Lambert et al. (2018) |
| Tapajos National Forest | Japan Aerospace Exploration Agency’s Advanced Land Observing Satellite 2 (JAXA’s ALOS-2), Spaceborne SAR interferometry | • Detection and quantification of selective logging events | This method has been shown to detect and quantify forest disturbance at a large scale with fine spatial resolution. It also has the potential to provide a global coverage, efficient in observing prototype of forest disturbance | Lei et al. (2018) |
| Global forest/non-forest | TerraSAR-X add-on for Digital Elevation Measurement (TanDEM-X) | • Forest/non-forest mapping | Remote sensing data represented a highly valuable source of land classification purposes, through the study identification, and monitoring of vegetated areas for agriculture, forestry, global change research, and regional planning was possible | Martone et al. (2018) |
| Rubus cuneifolius | Sentinel-2 and Landsat 8 with SAR imagery | • Detection of invasive alien plant species and mapping | The technique can be used to improve discrimination and mapping | Rajah et al. (2018) |
| The state of Oklahoma, southern Great Plains, USA | ALOS/PALSAR | • Characterizing the encroachment of juniper forests into sub-humid and semi-arid prairies | Efficient analysis of the spatial dynamics of encroachment at state and county spatial scales | Wang et al. (2018) |
| Greater St. Lucia Wetland Park, South Africa |
Sentinel-1 and 2 combined with the System for Automated Geoscientific Analyses (SAGA) |
• Monitoring wetlands | An increase in overall accuracy was observed, promising ability to distinguish wetland environments, land use, and land cover classification in a location where both wetland and non-wetland classes exist was also achieved | Whyte et al. (2018) |
| Gembloux, Walloon region, Belgium | Drone-borne ground-penetrating radar (GPR) | • Soil moisture mapping | The developed tools are operational and provide consistent results in terms of spatial patterns as well as in terms of absolute soil moisture values | Wu et al. (2019a) |
| The Sahelian zone | SAR Sentinel-1 satellite, Sentinel-2-MSI | • Woody plants mapping in savannas |
High spatial resolution woody canopy map (10 m) of savanna Ecosystems were generated |
Zhang et al. 2019c) |
| Four reference sites (three located in the Amazon basin and one located in the Iberian Peninsula), 18 sites around the world (boreal forests to tropical and subtropical forests, savannas, and grasslands) |
Sentinel-1 dual-polarized backscatter images, Sentinel-2-MSI, Landsat-8 optical |
• Detection of burned area and mapping | Self-adapting to local scattering conditions, and the ability to detect burned areas during periods with no thermal anomalies were the major advantages of the technology | Belenguer-Plomer et al. (2019) |
| The eastern edge of the Tapajós National Forest region | TanDEM-X SAR interferometry | • Mapping forest successional stages | The approach showed a high performance with a low uncertainty with respect to other approaches | Bispo et al. (2019) |
| Eucalyptus forests of Western Australia | Radar Burn Ratio (RBR), SAR-based multi-temporal index, ALOS PALSAR-2 data | • Estimation of prescribed fire impacts and post-fire tree survival | The approach largely maintained the estimation accuracy of the original RBR framework while decreasing computational complexities | Fernandez-Carrillo et al. (2019) |
| Global tropical forests | ALOS-2/PALSAR-2 dual-polarization ScanSAR data | • Mapping the spatial-temporal variability of forest | Comprehensive forest variability maps for the tropics based on a homogeneous big data analysis was carried out, and the map produced were accurate and precise | Koyama et al. (2019) |
| West Kameng and Tawang districts of Arunachal Pradesh and its vicinity areas | ALOS, PALSAR, Sentinel-1A data | • Tree diversity assessment and aboveground forests biomass estimation | The study implies the potential use of the information in the reliable estimation of spatial aboveground biomass in sub-tropical to alpine forest regions | Kumar et al. (2019) |
| Gabon and Switzerland | Sentinel-2-MSI, Convolutional Neural Networks (CNN) | • Mapping of country-wide high-resolution vegetation height | The high spatial resolution, Sentinel-2 provides a new image every few days, which enables frequent updates, and helps to obtain good coverage even in regions with frequent clouds | Lang et al. (2019) |
| North Karelia, Białowieża, Hainich, Râsca, Colline, Metallifere, Alto Tajo, Boreal | Sentinel-2-MSI | • Assessment of variation in plant functional diversity | The technique was the potential in achieving better spatial and temporal remote sensing of the plant functional diversity | Ma et al. (2019) |
| Tanjung Karang, Sekincan, and Kuala Selangor on the west coast of Peninsular Malaysia | ALOS-2 PALSAR-2 L-band, Sentinel-1 C-band SAR backscatter | • Characterizing of oil palm production landscape on tropical peat lands | Findings suggested that the technology has great potential to discriminate oil palm production landscapes managed under different management systems | Oon et al. (2019) |
| Amazon basin | DEM Multi-Error-Removed-Improved-Terrain (MERIT) based SRTM | • Mapping of inland water in tropical areas under dense vegetation | The technique provided a better sensing ability of water underneath the vegetation compared with optical sensor systems | Parrens et al. (2019) |
| Hubbard Brook Experimental Forest, Teakettle Experimental Forest, La Selva Biological Station | TanDEM-X, InSAR data, simulated Global Ecosystem Dynamic Investigation (GEDI) LiDAR data | • Estimation of forest biomass | The technique yields an improved ability to map and monitor biomass globally | Qi et al. (2019) |
| Romanian forests | Sentinel-1 satellite constellation, ALOS PALSAR-2 | • Monitoring forest changes | This study exposed the strengths and shortcomings of currently available spaceborne SAR techniques and highlighted the need for developing novel processing and modeling techniques | Tanase et al. (2019) |
| Two pasture sites are located at EI Reno, central Oklahoma, USA | SAR, Sentinel-1, Landsat-8 and Sentinel-2 data | • Estimation of leaf area index and aboveground biomass of grazing pastures | Improvements in capturing the phenology stages, and combined ability to monitor the seasonal dynamics of grazing pastures | Wang et al. (2019a) |
| Global forest types (tropical forest, temperate broadleaf, mixed forest, boreal forest) | ALOS, PALSAR/PALSAR-2, Moderate Resolution Imaging Spectroradiometer (MODIS) NDVI (Normalized difference vegetation index) | • Mapping annual forest cover | The method produced an annual 25m forest maps during 2007–2016, filled the four-year gap in the ALOS and ALOS-2 time series, and enhanced the existing mapping activity | Zhang et al. (2019a, d ) |
| Thomas Fire (Dec. 2017), and Carr Fire (July 2018) in Southern California, USA |
Sentinel-1 C-band SAR sensors |
• Mapping of burnt area | The technique demonstrated better potential in highlighting burnt areas with higher accuracy | Zhang et al. (2019b) |
| Two 5 × 5-km sub-sites in northern France | SAR Sentinel-1 satellite, Sentinel-2-MSI | • Prediction of wheat and rapeseed phenological stages | Principal and secondary phenological stages of wheat and rapeseed were identified successfully | Mercier et al. (2020) |
| The St. Lucia wetlands, South Africa | SAR Sentinel-1 satellite, Sentinel-2-MSI | • Mapping and characterization of wetland | The technique yields highly accurate maps and detailed classification of the wetland | Slagter et al. (2020) |
| Combomune, Gaza Province, Southern Africa | Sentinel-2-MSI | • Monitoring intra- and inter-annual dynamics of forest degradation from charcoal production | The study showed an improvement in the spatial and temporal characterization of the main cause of forest degradation | Sedano et al. (2020) |
| Madre de Dios, a region located in southeastern Peru | MODIS NDVI, Advanced Spaceborne Thermal Emission and Reflection Radiometer (ASTER), DEM | • Monitoring tropical forest degradation | Major factors associated with forest degradation were identified categorized and explored the advanced techniques | Tarazona and Miyasiro-López (2020) |
| Long-Term Socio-Ecological Research site “Pyrénées-Garonne” located in southwest France | SAR Sentinel-1 satellite, Sentinel-2-MSI | • Prediction of plant diversity in grasslands | Improved significance of prediction accuracy was obtained from Sentinel-2 data in comparison to Sentinel-1 | Fauvel et al. (2020) |
| Matang Mangrove Forest Reserve locates at west coast of Peninsular Malaysia |
Interferometric SRTM X/C-band, TanDEM-X-band |
• Structural characterization of mangrove forests | The study provided a better understanding of mangrove states and dynamics; most mangroves in the productive zones have been harvested | Lucas et al. (2020) |
| University of New England SMART Farms located near Armidale | SAR Sentinel-1 satellite, Sentinel-2-MSI | • Discrimination of species composition types | The study shows that the combination of Sentinel-1 and Sentinel-2 data can support the measurement of pasture species composition in a grazing landscape | Crabbe et al. (2020) |
| Lantana camara | Landsat 8 Operational Land Imager (OLI), Sentinel-2-MSI satellite data | • Invasive assessment of a plant species | The feasibility of using the technology for operational mapping of the invasive species and monitoring its distributional response to changes in climate, management systems, land use, and land cover | Dube et al. (2020) |
| Central Argentinian semiarid Caldén savanna biome | SRTM | • Assessment of the relationships between landscape features, soil properties, and vegetation determine ecological sites | The methodology potentially allows creation of ES maps at a regional scale and facilitates identification of threats associated with land use change, and the prescription of management strategies | Buss et al. (2020) |
| Grassland of Federal Republic of Germany | Sentinel-2 MSI, Landsat-8 OLI | • Characterization of grassland | The approach allows mapping the number and timing of mowing events in grassland at a high spatial resolution | Griffiths et al. (2020) |
| Winter wheat fields, Göttingen, Germany | SAR Sentinel-1 | • Monitoring the phenology of winter wheat | The study confirmed the use of SAR sensors to detect phenological stages of crops based on time series metrics, and to monitor crops on a regular basis | Schlund and Erasmi (2020) |
| Southern and Central China | SAR Sentinel-1 | • Monitoring of flood | The study described a reliable and efficient methodology to monitoring floods, offering continuous information on daily flood dynamics | Zeng et al. (2020) |
| Light detection and ranging (LiDAR) | ||||
| Beibu Gulf, China | LiDAR | • Determination of the post-typhoon recovery of a meso macro-tidal beach | The temporal features of the beach recovery process were reviled, erosion/accretion degrees in various beach sections were examined, and control factors of the beach recovery process were explored | Ge et al. (2017) |
| Boreal Forest and Foothills Natural Regions | Airborne LiDAR systems | • Regional mapping of vegetation structure | Three-dimensional vegetation structure was accurately measured and mapped | Guo et al. (2017) |
| The middle Heihe River Basinin Zhangye City of Gansu Province northwest China | Airborne LiDAR data, hyperspectral imagery | • Estimation of aboveground and belowground forest biomass | Results demonstrated that biomass accuracy which was improved by the use of fused LiDAR and hyperspectral data | Luo et al. (2017) |
| Evergreen and deciduous species of Mediterranean forests of Campanian forests, Telesina valley in southern Apennines of Italy | Leaf-off dedicated Airborne LiDAR data | • Detailed land use mapping, high-resolution LiDAR data, and field surveys were developed to categorize the productive and non-productive mixed forests in terms of stand attributes and structural diversity | Eight out of the ten forest types in the Campanian region were identified by high-resolution LiDAR by INFC. | Teobaldelli et al. (2017) |
| Mediterranean-type dune ecosystem in Southwest Portugal, a part of the Natura 2000 network of protected areas in the European Union. | Airborne hyperspectral imagery, LiDAR | • Detection and quantification of Gross Primary Production (GPP)-related regime shifts after Acacia invasion by mapping invader patches, accessing spatio-temporal changes during invasion, and applicability of NIRv. |
Successful mapping of high-impact invader Acacia longifolia in a heterogenous dune ecosystem. NIRv index was identified as a standardized “model metric” for quantification of high impacts of invasion even in the early stages. |
Große-Stoltenberg et al. (2018) |
| The Batang Toru tropical rain forests in the Indonesian province of North Sumatra. | LiDAR-derived Canopy Height model by Airborne Laser scanning (ALS) using LiDAR |
• Identification and characterization of forest patches based on structural attributes • Topographic attributes of the cluster by LiDAR derived Canopy Height Model |
The tallest clusters were found at significantly higher elevation (>850m) and steeper slopes (> 26°) than others, which were found to be remnants of undisturbed forests important for conservation and habitat studies along with an understanding of carbon stock. | Alexander et al. (2018) |
| Bats in four ancient semi-natural broad-leaved woodlands in Dartmoor National park situated in southwest England | Ground-based field survey with Airborne LiDAR imagery |
• Daily tracking of bats on foot-to-roost trees using various radio receivers and antennas. • Location of tree cavities using directional antennas and binoculars from the ground |
Pregnant and lactating bats switched roosts less frequently in comparison to post lactating and nulliparous bats by selecting their cavities higher on trees may be to protect their offspring from predation, | Carr et al. (2018) |
| Rainforest stands in the Central Highlands region of Southeastern Australia | Airborne LiDAR detection and ranging | • Examining the utility of complete LiDAR PAVD (plant area volume density) profiles to classify stand types in south-eastern Australia | A study demonstrated that LiDAR PAVD is used in identifying and mapping the extent of rainforest and eucalyptus stand types across a heterogenous landscape. | Fedrigo et al. (2018) |
| Tropical forest on Barro Colorado Island, Panama | LiDAR with process-based forest model FORMIND | • Comparison of 29 different LiDAR metrics by using simulated stands for their utility as predictors of tropical forest biomass at different at various spatial scales | Some LiDAR metrics like height metrics showed good correlations for forest biomass even for an undisturbed forest. | Knapp et al. (2018) |
| Bavarian forest National park in the southern- eastern part of Germany | Airborne LiDAR | • Assessing the scan angle impact on gap fraction (Pgap) and vertical Pgap profile in several forest type using different scan angles of airborne LiDAR | Discontinuous plots, Pgap, and vertical Pgap were maximum when observed from the nadir direction and rapidly decrease with increased scan angle. This indicates large off-nadir direction should be avoided to ensure a more accurate Pgap and leaf area index (LAI) estimation | Liu et al. (2018) |
| Mixed coniferous forest of Northern Sierra Nevada, California, USA | Bitemporal airborne LiDAR with field survey | • Systematic assessment of uncertainties of satellite imagery-based vegetation indices forest structural changes induced due to fuel treatment using aboveground biomass (AGB) and canopy cover as a ground reference. | Differences in vegetation indices had relatively weaker correlations to biomass changes in forest with sparse or dense biomass than in forests with moderate density before disturbance | Ma et al. (2018) |
| Set of forest structural attributes for Lidar plots located across Canada’s forest-dominated ecosystem |
LiDAR plot-derived information, Landsat pixel-based composites (large area forest attribute imputed model) |
• Demonstration of the temporal and spatial extension of imputation model using time-series of annual surface reflectance image composites with samples of airborne LiDAR • Highlighting the potential of scientific insights related to growth and recovery over large areas. |
Extension of spatial and temporal scale of the modeling framework finally demonstrated its efficacy and robustness for the entire forest of Canada over a period of three decades Built confidence in portability of their models. |
Matasci et al. (2018) |
| Subtropical intertidal wetland located to the south of Zhangjiang estuary in the southeast China | Unmanned aerial vehicle (UAV) camera imagery and LiDAR data | • Exploring mangrove-inundation spatial patterns |
Results indicated that >90% of mangrove forests (mainly Kandelia abovate, Avicennia marina, and Aegiceras corniculatum) were situated within a 1-m elevation range between local mean sea level and higher high water Whereas, spatial distribution showed hump-shaped patterns along the inundation gradient |
Zhu et al. (2019) |
| Three seasonal semi-deciduous natural forest cover types (OG, SGpas, and SGeuc) in the Atlantic forest biome of Southern Brazil | ALS with portable ground LiDAR remote sensing as a proxy |
• Analysis of structural attributes of forest canopies undergoing restoration. • Assessing ability of the attributes to distinguish forest cover types to estimate aboveground woody biomass (AGB) |
A set of six canopy structure attributes were able to classify five cover types with an overall accuracy of 75% AGB was well predicted by canopy height and unprecedented “leaf area height volume” |
Almeida et al. (2019) |
| A widely distributed bamboo species Moso bamboo (Phyllostachys pubescens) in the subtropical forests of south China | Airborne LiDAR with Principal component analysis (PCA) | • Assessing estimation of canopy structure and biomass of Moso bamboo by PCA | The result showed LiDAR well predicted the AGB of Moso bamboo with the percentile heights and coefficient of variation of height (hcv) having the highest relative importance for estimating AGB and culm biomass | Cao et al. (2019) |
| An area characterized by uneven-aged mixed species forest called Lavarone (Trento) in the Italian Alps | Airborne LiDAR |
• Mapping fine-scale variation in aboveground carbon density (ACD) and its change over time across the landscape • Linking the changes in ACD to forest structural attributes, species composition, disturbance regime, and local topography |
Results indicated between 2007 and 2011 the majority of landscapes (61%) increased in ACD with a much smaller fraction of study areas exhibiting evidence of small-scale natural disturbances (3.7%) | Dalponte et al. (2019) |
| Two European forest types; a coniferous mountain forest in the Eastern Italian Alps and a mixed temperate forest in southern Germany | LiDAR with spectral variation hypothesis (SVH) | • Height variation hypothesis (HVH) is a concept of linking height heterogeneity (HH) of a forest and its tree diversity using the heterogeneity index Rao’s Q by Canopy Height Model (CHM) | HH was highly correlated to tree species diversity of the forest ecosystem in coniferous mountain forests than in mixed temperate forests when calculated with CHM resolution 2.5 m | Torresani et al. (2020) |
| Insects flying over Ostra Herrestad wind farm near the town Shimrishamn in southernmost Sweden | High resolution Scheimflug lidar | • Testing feasibility of a high resolution of a vertical Scheimflug lidar to resolve a small target over a wind farm independently of sunlight to test insect hypothesis | Insect swarms appeared at short intervals and varied in their time of formation, dispersal, density, and size of swarming; which was sometimes according to emergence time of bats | Jansson et al. (2020) |
| Municipalities of Nova Ubirita and Feliz Natal, Matto Grosso near the southern extent of closed-canopy forests in the Brazilian Amazon | Acoustic Space Occupancy Model with ecoacoustic and Airborne LiDAR data | • Evaluating the role of forest structure in explaining variability in acoustic community assembly by illustrating and investigating hypothesized synergies between 3D observations of acoustic space-filling and physical space filling | The findings underscore the important synergies between lidar and ecoacoustics for informing models of occupancy and detection and supporting future investigations towards the role of habitat structure in shaping habitat use | Rappaport et al. (2020) |
| Inner Mongolia autonomous region of China | ICESat-2 data, airborne LiDAR data |
• Develop a machine learning workflow for mapping the spatial pattern of the forest canopy height (Hcanopy) foot print products from ICES-2 and Sentinel satellite data. • Its validation by high-resolution canopy height from airborne LiDAR at different spatial scales • Performance comparison between two machine learning models (DL and RF), and between Sentinel, and LANDSAT satellites. |
Results revealed the reliability of ICES-2 vegetation height products as ICES-2 Hcanopy showed to have the highest correlation with airborne LiDAR canopy height at a spatial scale Performance comparison of DL and RF model revealed satisfactory accuracy on the up-scaling of ICES-2 Hcanopy |
Li et al. (2020b) |
| The Bialoweiza Forest UNESCO world heritage site of European forest | Hyperspectral and LiDAR data | • Derivation of information on tree species dominance and aboveground biomass in the forest by testing and evaluating hyperspectral and LiDAR data |
Outcomes showed vegetation indices from hyperspectral data can support species dominance detection. Accuracy of the level of species dominance in the forest plots varied when tested using the Multivariate Multiple Linear Regression model |
Laurin et al. (2020) |
| Six sites located across Canada cover a wide range of forest types, terrain conditions, species composition, stand structural characteristics, and disturbance regimes. | Landsat time series data, airborne laser scanning (ALS) LiDAR | • Investigation of accuracy of spectral indices and time series lengths for estimates of forest attributes as stand height, basal area, and stem volume across a range of conditions by ALS calibration |
An improvement in estimation accuracy in the six sites was seen with an increase in time series length Model accuracies plateaued at a time series length of 15 years for two sites |
Bolton et al. (2020) |
| Four spoil heaps originating from brown coal mining located in the north Bohemian Brown coal basin in the Czech Republic of Europe | Hyperspectral data, LiDAR data, and integrated approaches of both | • Comparing various approaches (hyperspectral data, LiDAR data, and integrative approach) in the extraction of open surface water bodies on spoil heaps from very high-resolution airborne data (<2 m) |
Individual approaches resulted in 2–22.4% underestimation of the water surface area and 0.4–1.8 % overestimation. The integrative approach improved discrimination over all other approaches |
Prošek et al. (2020) |
| Five forest types, where four forest study sites were part of the ForestGEO megaplot network (Moist lowland rainforest at Barro Colorado Island (BCI) in Panama of Central America, Rabi, Gabon a Central African lowland rainforest, SERC plot of USA, and Traunstein in Germany) and the fifth site was lowland tropical rainforest of Paracou at French Guiana in South America | Airborne discrete return LiDAR data | • Quantification of different aspects of forest structure, i.e., mean canopy height, maximum canopy height, stand density, vertical heterogeneity, and wood density different for each discretion. | Biomass predictions using the best general model were found to be almost as accurate as predictions using five site-specific models. | Knapp et al. (2020) |
There are two basic types of RADAR systems, namely, primary and secondary. In the primary system, the signal is transmitted in all directions however some of the signals are reflected back to the receiver after colliding with the target thereby defining or detecting the location of the target (Hirst 2008; Bhatta and Priya 2017). In this system, the transmitted signal needs to be of high power to ensure that the reflected signal is sufficient enough to provide accurate and precise information about the target (Bhatta and Priya 2017). Again, noise and signal attenuation due to some factors might disrupt the reflected signal which can also be regarded as a limitation (Bhatta and Priya 2017; Martone et al. 2018). In the secondary RADAR system, an active answering signal system has been installed for accuracy, where the transmitted signal is received by a compatible transponder that retrieves the signal and further sends a signal comprising the useful information in a coded form (Hirst 2008; Bhatta and Priya 2017). The receiver receives the coded signal, and after decryption of the code, the information about the target is transcribed, thereby providing information about the real-time spatial orientation (Bhatta and Priya 2017; Jahncke et al. 2018).
Further to this, ultra wideband (UWB) RADAR is one of the traditional methods used for life detection that analyzes the reflected/echo signal received after hitting the target. Micro-motions by humans, nearby environment, and clutter signals can modulate the reflected signal. As per evidence, it is a reasonable, effective, and complete non-invasive life detection method (Chunming and Guoliang 2012; Karthikeyan and Preethi 2018; Yin and Zhou 2019). With the growth of scientific innovation in the field of remote sensing, a hybrid (On-Chip Split-Ring-Based Sensor) RADAR system has emerged with high-resolution range and sensitivity. This system can easily detect multiple life forms simultaneously even across obstacles (Liu et al. 2016).
One of the major applications of RADAR is range detection and to date, the replacement of the sensing and detection efficiency and accuracy by RADAR has not been possible by any other electronic system (Bhatta and Priya 2017; Parrens et al. 2019). Extension of the sensing capability with respect to atmospheric conditions such as rain, snow, smoke, darkness, and fog and collecting the data makes it unexceptional and advantageous. At present, RADARs have broad areas of applications in defense and control systems, monitoring and forecast systems, astronomy, target-locating system and remote sensing, etc. (Bhatta and Priya 2017).
Light detection and ranging (LiDAR) systems
LiDAR is a widely recognized technology, especially the airborne laser scanner (ALS), which focuses on the emission and receipt of laser pulses. During field surveys, LiDAR technology offers the potential to establish variables, representing forest structures that are distinct from those detected or assessed. Bitemporal airborne LiDAR with field survey is widely used for systematic assessment of uncertainties in satellite imagery-based vegetation (Ma et al. 2018). Ground-based field survey with airborne LiDAR is applicable for daily tracking of bats on foot-to-roost trees using various radio receivers and antennas and the location of tree cavities using directional antennas and binoculars from the ground (Carr et al. 2018; Stephenson 2020) (Fig. 1, Table 2).
Unmanned aerial vehicle (UAV) camera with LiDAR data is used for surveying mangrove-inundation spatial patterns in a subtropical intertidal wetland in southeast China (Zhu et al. 2019). With the support of artificial intelligence (AI), LiDAR remote sensing is found to be successful in predicting models for efficient biodiversity study by covering more areas with a clear database in a very span of time. LiDAR plot extracted information and Landsat pixel-based composites along with time-series were effective in modeling sets of reflectance images of forest structure across Canada’s forest-dominated ecosystem (Matasci et al. 2018). More advanced LiDAR technologies have now overtaken the previous ones in terms of both efficacy and accuracy. Almeida et al. (2019) in their research on three seasonal semi-deciduous natural forest cover types in the Atlantic forest biome of Southern Brazil have considered ALS with portable ground LiDAR remote sensing as a proxy for analysis of structural hallmarks of forest canopies enduring restoration. Airborne LiDAR with principal component analysis (PCA) assessed the estimation of canopy structure and biomass of Moso bamboo (Phyllostachys pubescens) in widely distributed subtropical forests of south China (Cao et al. 2019). High-resolution vertical Scheimpflug LiDAR has proven to resolve hypothesis for insects flying over Ostra Herrestad wind farm near the town Shimrishamn in southern Sweden (Jansson et al. 2020).
To conserve plant diversity information on various forest attributes, aboveground biomass (AGB), canopy structure, canopy cover, and leaf area index (LAI) are considered important and can be assessed most efficiently with a technique like LiDAR (Bolton et al. 2020). Forest canopy height could be an important indicator of biodiversity, productivity, and carbon storage (Li et al. 2020b). LiDAR when combined with other RS techniques, i.e., high spatial resolution with hyperspectral sensors, thermal remote sensing, and satellite RS are yielding eye-catching results, particularly in biodiversity and ecosystem conservation.
Passive remote sensing systems
Passive remote sensing is often understood as a system that operates by passive sensors which can only be used for detection in presence of the natural source of energy, i.e., sunlight (visible to shortwave spectrum and infrared thermal radiation) (Srivastava et al. 2020). These sensors have a specialty to detect natural energy (radiation) that is either emitted or reflected from the source of energy or object (Earthdata 2021). Limitations of the applicability of passive remote sensing for biodiversity and ecosystem conservation are its dependency on sunlight as a source of radiation which is again dependent completely on the season, region, and climatic conditions (Fig. 1).
Techniques for identification and genetic conservation of species
Restriction fragment length polymorphism (RFLP)
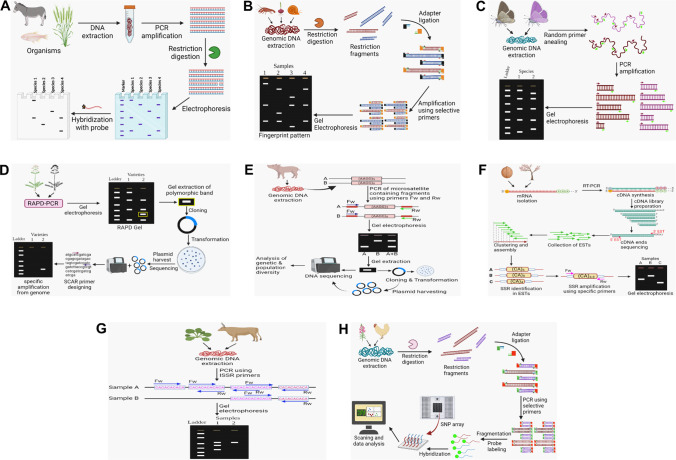
RFLP is a biallelic, polymorphic genetic marker characterized by hybrid labeled probes of DNA fragments and digested with restriction endonucleases for estimations of genetic diversity (Vignal et al. 2002; Amom and Nongdam 2017). Different restriction sites in DNA represent the genetic divergence between different populations or related species within a population. Özdil et al. (2018) demonstrated genetic diversity among 11 donkey species in Turkey by conducting PCR-RFLP of two genes. The restriction sites of DraII, MboI, and EagI on the lactoferrin gene (LTF) and PstI on the κ-casein gene (CSN3) have been validated to identify the polymorphism among the donkey population (Özdil et al. 2018). Meikasari et al. (2019) have also made elucidation of low genetic diversity among the seahorse (Hippocampus comes) found in Bintan waters. Another recent study indicates the utilization of PCR-RFLP in genetic-based sex determination of Sebastes rockfish (Vaux et al. 2020). The PCR-RFLP application in identification of durum (Triticum durum L.) and bread wheat (T. aestivum) species has also been studied by analyzing chloroplast DNA (Haider and Nabulsi 2020) (Table 3, Fig. 2A).
Table 3.
Molecular and Genomic techniques used for conservation of biodiversity
| Technique | Species | Purpose | Output | References | |
|---|---|---|---|---|---|
| Common name | Scientific name | ||||
| Restriction fragment length polymorphism (RFLP) | Japanese Rana | Rana japonica, R. ornativentris, R. tagoitagoi | Species identification | Successful in detecting Rana species in static water in both laboratory and field | Igawa et al. (2019) |
| Turkish donkey | Equus asinus | Genetic diversity of κ-casein (CSN3) and lactoferrin (LTF) genes | Novel single nucleotide polymorphism has been firstly detected in the LTF gene with the EagI restriction enzyme | Özdil et al. (2019) | |
| Seahorse | Hippocampus comes | Identification of allelic diversity | Genetic diversity of the population of seahorse species of H. comes was low | Meikasari et al. (2019) | |
| Cattle breeds | - | Assessment of the genetic structure based on functional gene polymorphism | Gene flow, little variation, and genetic isolation were observed due to close geographic proximity | Kasprzak-Filipek et al. (2019) | |
| White River beardtongue | Penstemon scariosus | Genetic diversity and differentiation patterns analysis | Most of the genetic variation is within populations and high proportion of the genetic variation is due to geographic distance | Rodríguez-Peña et al. (2018) | |
| Pau-branco | Picconia azorica | Genetic diversity and biotaxonomy analysis | Monophyly of Picconia and low genetic diversity and a weak genetic structure | Ferreira et al. (2011) | |
| Durum and bread wheat | Triticum durum L., and T. aestivum L. | Species identification | Provided sufficient variation for the identification | Haider and Nabulsi (2020) | |
| Brown trout | Salmo trutta | Genetic structure analysis | Showed a high percentage of allochthonous genes among the individuals | Molerović et al. (2019) | |
| Sebastes rockfish | Sebastes melanops, S. mystinus, S. pinniger, S. diaconus, S. entomelas, and S. flavidus | Sex identification | Indicated that the association of this restriction site with sex is species-dependent | Vaux et al. (2020) | |
| Egyptian Origanum and Thymus | Origanum vulgare, Origanumsyriacum (L.) subsp. sinaicum, Thymus vulgaris, T. capitatus L., and T. decassatus | Genetic diversity analysis | Elucidate the genetic variations and phylogenetic relationships within the studied species | El-Demerdash et al. (2019) | |
| Amplified fragment length polymorphism (AFLP) | Guazuma | Guazuma crinita | Genetic diversity analysis | Showed higher genetic diversity | Tuisima-Coral et al. (2020) |
| Ornamental onion | Allium stipitatum | Genetic characterization | High genetic diversity occurred among the population | Ebrahimi et al. (2019) | |
| Himalayan bitter gourd | Herpetospermum caudigerum | Genetic diversity and genetic structure analysis | Showed rich genetic diversities and greater genetic differentiation among populations | Xu et al. (2019) | |
| Wildrye | Elymus tangutorum | Genetic diversity and genetic structure analysis | A significant correlation between genetic and geographical distance was observed, also the significant ecological influence of average annual precipitation on genetic distance was revealed | Wu et al. (2019b) | |
| Noble crayfish | Astacus astacus L. | Genetic diversity analysis | Significant genetic diversity was revealed | Panicz et al. (2019) | |
| Anomura | Aegla georginae, A. ludwigi, and A. platensis | Genetic diversity analysis | Showed low levels of genetic diversity | Zimmermann et al. (2019) | |
| Freshwater snail | Melanopsis etrusca | Genetic diversity analysis | Showed two or perhaps three clusters of populations and there probably is some connectivity among populations in close geographic proximity | Neiber et al. (2020) | |
| Muga silkworm | Antherae aassamensis | Phylogeny and Genetic diversity analysis | Wide range of genetic distance was observed and sequences comprised of two major clade | Saikia et al. (2019) | |
| Random amplified polymorphic DNA (RAPD) | Wormwood | Artemisia judaica | Genetic diversity analysis | Presence of genetic variation | Al-Rawashdeh (2011) |
| Pa Co pine | Pinus kwangtungensis | Analysis of genetic diversity | High genetic variation was found | Thuy et al. (2020) | |
| Jongly Ada | Alpinia nigra | Genetic diversity analysis | Lower amount of gene flow was observed | Basak et al. (2019) | |
| white garland-lily | Hedychium coronarium | Population genetic structure and diversity analysis | High genetic diversity occurred among the population and showed similar grouping patterns | Ray et al. (2019) | |
| Old World tree frogs | Rhacophorus margaritifer | Genetic diversity analysis | Showed high genetic diversity and high locus variation | Sulistyahadi et al. (2020) | |
| Awassi sheep | - | Genetic polymorphism and diversity analysis | The study revealed substantial divergence among populations and displayed a remarkable degree of consistency between geographic origins, breeding histories, and the pattern of genetic differentiation | Al-Allak et al. (2020) | |
| Eri silkworm | Philosamia ricini | Genetic diversity analysis | revealed a low genetic diversity among the morphs | Saikia and Devi (2019) | |
| Banapsha | Viola serpens | Species identification | Revealed successful identification of the genuine samples | Jha et al. (2020) | |
| Sequence characterized amplified region (SCAR) | Oriental penthorum | Penthorumchinense | Genetic identification and authentication | Enabled reliable genetic identification | Mei et al. (2017) |
| - | Morusboninensis and Morusacidosa | Development of SCAR markers | All hybrid seedlings identified according to the molecular markers | Tani et al. (2003) | |
| Anglojap yew | Taxus media | Development of SCAR markers | Turned out to be a quick, efficient, and reliable tool for identification | Hao et al. (2018) | |
| Japanese honeysuckle | Lonicera japonica | Development of SCAR markers | Found as an efficient molecular marker to study the genetic variation of any organism | Yang et al. (2014) | |
| Okinawa Agu pigs | - | Evaluation of the genetic structure | Revealed a substantial loss of genetic diversity among Agu pigs due to inbreeding | Touma et al. (2019) | |
| Micro-satellites and mini-satellites | Robusta coffee | Coffeaca nephora | Genetic diversity and population structure analysis | Detected population genetic structure with distinct geographic distribution | Labouisse et al. (2020) |
| White-lipped peccaries | Tayassu pecari | Genetic diversity and population structure analysis | Detected a similar levels of genetic diversity in all localities and a pattern of isolation by distance | de Góes Maciel et al. (2019) | |
| Vilca | Anadenanthera colubrina | Demographic history and spatial genetic structure analysis | Revealed an ancient population expansion and found high levels of genetic diversity and high inbreeding | Goncalves et al. (2019) | |
| Fragrant rosewood | Dalbergia odorifera | Genetic diversity and population structure analysis | Medium genetic diversity level was inferred and genetic variation existed among populations | Liu et al. (2019a) | |
| Mediterranean swordfish | Xiphias gladius | Genetic diversity and population structure analysis | Revealed the presence of three genetic clusters and a high level of admixture within the Mediterranean Sea | Righi et al. (2020) | |
| Marsh orchid | Dactylorhiza majalis | Genetic diversity analysis | Showed a moderate level of genetic diversity and a significant correlation between the morphological and genetic distance matrices | Naczk and Ziętara (2019) | |
| Central American river turtle | Dermatemys mawii | Genetic diversity and population structure analysis | Showed significantly higher genetic diversity and highest genetic differentiation in wild population | Gallardo-Alvárez et al. (2019) | |
| Turkish sheep breeds | - | Genetic diversity and differentiation analysis | Showed some populations still hold enough genetic diversity despite decreasing in population sizes in the last few decades | Karsli et al. (2020) | |
| Iron walnut | Juglans regia | Population genetic structure and adaptive differentiation analysis | Revealed relatively low genetic diversity, high interpopulation genetic differentiation, and asymmetric interpopulation gene flow | Sun et al. (2019) | |
| Expressed sequence tags (ESTs) | Liquorice | Glycyrrhiza glabra L | Genetic structure and variation analysis | Variability was related to the intra-population level | Esmaeili et al. (2020) |
| Asunaro | Thujopsisdolabrata | Phylogeographic and genetic population relationships | Exhibited relatively uniform genetic diversity and found a significant and relatively high value of overall population differentiation among the populations | Inanaga et al. (2020) | |
| - | Magnolia patungensis | Genetic diversity analysis | Significant genetic differentiation and low gene flow among populations were discovered | Wagutu et al. (2020) | |
| Pearly everlasting | Anaphalis spp. | Genetic diversity analysis | Genetic diversity has a fairly low value that occurs within and between populations | Ade et al. (2019) | |
| Star magnolia | Magnolia sinostellata | Genetic diversity and population structure | Showed rich genetic diversity and serious genetic drift occurred within populations. Also, genetic differentiation is apparent among the populations | Wang et al. (2019b) | |
| Ironwood | Parrotia subaequalis | Genetic diversity analysis | A relatively high level of genetic diversity and genetic differentiation level was detected. Additionally, a high level of cross-transferability was displayed | Zhang et al. (2019e) | |
| Shrubby horsetail | Ephedra foliata | Analysis of genetic diversity and population genetic structure | Revealed higher levels of polymorphism, diversity index, and effective multiplex ratio | Meena et al. (2019) | |
| Inter-simple sequence repeat (ISSR) | Zarrin-Giah | Dracocephalumkotschyi | Genetic diversity analysis | Revealed a broad genetic variation among the populations | Sereshkeh et al. (2019) |
| Bergenia | Bergenia ciliata | Genetic diversity analysis | Revealed highest percentage of variation and low genetic diversity within populations | Tiwari et al. (2020) | |
| Peony | Paeonia decomposita | Analysis of genetic diversity and population genetic structure | Revealed moderate genetic diversity and a significant positive correlation between geographic and genetic distance among populations | Wang et al. (2020) | |
| Thunb | Huperzia serrata | Analysis of genetic diversity and variation | Genetic diversity was relatively high at population and species level | Minh et al. (2019) | |
| New Zealand white rabbit | Oryctolagus cuniculus | Genetic diversity analysis | No significant differences in the genetic diversity | Li et al. (2020a) | |
| Tunisian local cattle | - | Genetic diversity analysis | Genetic diversity was relatively high at population and species level | El Hentati et al. (2019) | |
| Korean-native Jeju horse | Equus caballus | Comprehensive genome and transcriptome analyses | Help in designing high-density SNP chip for studying other native horse breeds | Srikanth et al. (2019) | |
| Single nucleotide polymorphisms (SNPs) | Wildcats and domestic cats | - | To assess hybridization between wildcat and domestic cat populations | Showed a complete genetic continuum or hybrid swarm structure | Senn et al. (2019) |
| Lion | Panthera leo | Spatiotemporal genetic diversity | Gene flow and local subpopulations due to habitat fragmentation | Curry et al. (2021) | |
| Italian chicken breeds | - | Genetic diversity and population structure analysis | Showed most breeds formed non-overlapping clusters and were clearly separate populations, which indicated the presence of gene flow | Cendron et al. (2020) | |
| Ethiopian durum wheat | Triticum turgidum ssp. durum | Genetic diversity and population structure analysis | Showed higher genetic diversity and higher genetic admixture between landraces despite their geographic origin | Alemu et al. (2020) | |
| Dragonhead | Dracocephalumruy schiana | Genetic diversity and differentiation | Showed structuring and differentiation between populations | Kyrkjeeide et al. (2020) | |
| Star swertia | Swertia mussotii | Genetic differentiation | Showed genetic differentiations often occurred in genes known to be involved in various metabolic processes and revealed a significant positive correlation between the genetic similarity and geographical distance | Qiao et al. (2021) | |
| Tasmanian devil | Sarcophilus harrisii | Genetic diversity and population structure | Low genetic diversity within population was observed | Miller et al. (2011) | |
Fig. 2.
Molecular techniques for conservation of biodiversity. A Restriction fragment length polymorphism; the genomic DNA extracted from different organisms is PCR amplified and subjected to restriction digestion using specific restriction enzymes, then DNA fragments are separated by electrophoresis and hybridized with radiolabeled probes (Özdil et al. 2018; Chaudhary and Maurya 2019; Panigrahi et al. 2019; Haider and Nabulsi 2020). B Amplified fragment length polymorphism; the restriction fragments of genomic DNA was ligated with compatible adapters and PCR amplified using selective primers against adapters, and the amplified fragments were separated by electrophoresis for DNA fingerprint analysis (Blears et al. 1998; Malik et al. 2018; Wu et al. 2019b; Zimmermann et al. 2019; Neiber et al. 2020). C Random amplified polymorphic DNA; random PCR fragments were amplified from the genome of different species using primers with random sequences and separated by gel electrophoresis to determine the difference between species based on RAPD markers (Panigrahi et al. 2019; Saikia et al. 2019). D Sequence characterized amplified region; PCR products are generated using primers for RAPD markers from the genome of different varieties and separated in the gel. The polymorphic DNA band is gel extracted and processed for cloning and sequencing for getting specific amplification by designing SCAR primers (Yang et al. 2014; Bhagyawant 2015; Ganie et al. 2015; Cunha and Domingues 2017). E Micro-satellites; the microsatellite repeats were PCR amplified using primers that flank the repeated sequence and separated by gel electrophoresis. The individual bands were cloned and sequenced for analysis of genetic and population diversity (Kim 2019; Touma et al. 2019). F Expressed sequence tag-simple sequence repeat; cDNA library was prepared from cDNA synthesized from isolated mRNAs, and then end sequencing of cDNA library was performed by EST primers followed by its assembly. The SSR regions were amplified in assembled ESTs using specific primers and the products were analyzed in gel (Rudd 2003; Sun et al. 2019; Wagutu et al. 2020). G Inter-simple sequence repeat; the isolated genomic DNA from organisms was subjected to PCR amplification using primers specific to microsatellite, and the PCR products were separated by electrophoresis for analysis (Sarwat 2012; El Hentati et al. 2019; Tiwari et al. 2020). H Single nucleotide polymorphism; the genomic DNA from different samples was digested with suitable restriction enzyme followed by adapter ligation and PCR amplification using selective primers. Further to this, PCR products were fragmented with DNase I and labeled with fluorescent probes followed by hybridization in an SNP array. The wells were scanned and data were analyzed (Alsolami et al. 2013; Scionti et al. 2018; Cendron et al. 2020; Kyrkjeeide et al. 2020)
Amplified fragment length polymorphism (AFLP)
AFLP is considered an effective means of detecting polymorphism in DNA without having any prior information regarding the genome. Being a dominant marker, it can analyze multiple loci through amplification of DNA performing PCR reaction (Bryan et al. 2017). The method employs restriction digestion of DNA and amplification of fragments through ligation of adapters on both ends and using primers specific to adapters (Malik et al. 2018). Genetic differences can be identified from the disparity in the number and length of bands on electrophoretic separation. Its application ranges from the assessment of genetic diversity within species to generate of genetic maps for disease diagnosis and phylogenetic studies. AFLP data analysis study represents the genetic diversity in E. tangutorum population contributed by geographical and environmental factors (Wu et al. 2019b). The phylogenetic relationships and genetic distances among A. platensis populations and other distinct related species such as A. georginae and A. ludwigi in southern Brazil were also investigated through AFLP (Zimmermann et al. 2019). Population structure and differentiation among Melanopsis etrusca were clearly distinct between the eastern, western, and central regions populations in Italy (Neiber et al. 2020) (Table 3, Fig. 2B).
Random amplified polymorphic DNA (RAPD)
The RAPD is a PCR-based technique in which 8–10 short nucleotides comprise both forward and reverse primers that bind arbitrary nucleotide sequences of chromosomal DNA to generate random fragments. Due to this random nature of primers, no prior knowledge about genome sequence is needed. The annealing sites of these random primers vary for different species or individual to individual. Discrimination can be identified or determined from the amplified DNA fragments (RAPD markers) separated by agarose gel electrophoresis (Freigoun et al. 2020). RAPD markers are dominant and involved in various applications such as genome mapping, molecular evolutionary genetics, genetic diversity analysis, and population genetics as well as determining taxonomic identity (Qamer et al. 2021). Saikia et al. (2019) deduced genetic variation among the different morphs of muga silkworm of Northeast India through RAPD analysis. Moreover, Sulistyahadi et al. (2020) studied the locus diversity as well as genetic polymorphism of the endemic species Rhacophorus margaritifer population by this technique. It has also been used to elucidate the genetic variation in a medicinal plant species found in the south of Jordan named Artemisia judaica (Al-Rawashdeh 2011) (Table 3, Fig. 2C).
Sequence characterized amplified region (SCAR)
SCAR markers are DNA fragments generated by PCR amplification using specific 15–30-bp long primers derived from RAPD markers through cloning and sequencing (Bhagyawant 2015). Usually, RAPD markers are associated with low reproducibility and are dominant in nature, making it inappropriate for species identification (Sairkar et al. 2016). To overcome this disadvantage, RAPD markers are converted to SCAR markers which are locus-specific and co-dominant in nature (Bhagyawant 2015; Feng et al. 2018). Due to the specificity of primers, PCR amplification of SCARs is less sensitive to reaction condition and thus are easy to perform (Yuskianti and Shiraishi 2010). SCAR markers provide authenticate information both for species identification and population genetic diversity analysis. Researchers have successfully developed SCAR markers for the medicinal plant V. serpens using 1135-bp long amplicon through RAPD obtained by six accessions of the plant, thereby preventing it from extinction (Jha et al. 2020) (Table 3, Fig. 2D).
Mini- and micro-satellites
Mini-satellites (variable number of tandem repeats (VNTRs) 6–100 bp) and micro-satellites (1–6 bp) (simple sequence repeats (SSR) and short tandem repeats (STR)) are randomly repetitive DNA sequences widely dispersed in all eukaryotic species genomes. These multi-allelic markers are co-dominantly inherited with species-specific location and size within the genome (Vergnaud and Denoeud 2000; Vieira et al. 2016). Due to the high level of polymorphism associated with mini and microsatellites, it is extensively utilized in genetic analysis and population studies. Microsatellites are interspersed all over the genome and therefore represent high variability and their identification show great variation among species of the different population (Abdul-Muneer 2014). Its analysis includes PCR amplification of loci by using primers that flank the repeated sequence. By using microsatellite markers, genetic structure of Agu pigs has been elucidated along with its correlation with Ryukyu wild boar, two Chinese breeds and five European breeds (Touma et al. 2019). Similarly, De Góes Maciel et al. (2019) analyzed 13 microsatellite loci of 361 white-lipped peccaries for assessment of their population structure and level of genetic diversity (Table 3, Fig. 2E).
Expressed sequence tags (ESTs)
ESTs are small sequences of DNA usually 200 to 500 nucleotides long that act as tags for the expressed genes in certain cells, tissues, or organs. ESTs are generated by sequencing either the 3′ end or 5′ end of a segment derived from random clones from the cDNA library and long enough for the identity illustration of the expressed gene (Behera et al. 2013). ESTs are widely involved in gene discovery, determining the phylogenetic relationship between individuals, genetic diversity, and proteomic analysis as well as transcriptome profiling (Cai et al. 2015). EST-derived SSR markers are more informative than genomic SSRs for genetic diversity analysis due to several advantages such as high conserved nature, variation in coding sequence, and high heritability to closely related species (Parthiban et al. 2018). Sun et al. (2019) have conducted the structure analyses of expressed sequence tag-simple sequence repeat (EST-SSR) markers in Juglans sigillata and demonstrated the genetic structure based on its geographic feature. Moreover, EST-SSR analyses have provided information regarding the genetic distance between the J. regia and J. sigillata populations. By considering EST-SSRs and genotype sequencing data, they have interpreted iron walnut as the subspecies of J. regia (Sun et al. 2019). Investigation of evolutionary relations and genotypic relatedness are essential for the conservation of endangered species. Recently the genetic variability of an endangered species Magnolia patungensis was studied by analyzing the EST-SSR polymorphic markers (Wagutu et al. 2020) (Table 3, Fig. 2F).
Inter-simple sequence repeat (ISSR)
ISSR markers are used in diversified analyses such as species identification, evolutionary and taxonomic studies, genome mapping, genetic diversity, and gene tagging because of their high polymorphic nature (Arif et al. 2011; Abdelaziz et al. 2020). These multilocus markers are generated through PCR amplification by using microsatellites as primers. Prior sequence knowledge is not required for primer designing as repeat sequence is used to amplify these inter-microsatellite regions (Ng and Tan 2015). It overcomes all the limitations possessed by other markers such as RAPD and AFLP which are associated with low reproducibility (Najafzadeh et al. 2014). Genetic diversity and population structure analysis have been performed among 11 populations of Bergenia ciliata using 15 ISSR markers. The analysis shows a high level of polymorphism among this medicinal plant species, found in the Indian Himalayan Region (Tiwari et al. 2020). El Hentati et al. (2019) have studied genetic diversity and phylogenetic relationships among 20 samples of three geographical local cattle populations using ISSR primers. They found a significant variation and geographical separation among the cattle from the north, northeast, and northwest of Tunisia (Table 3, Fig. 2G).
Single nucleotide polymorphisms (SNPs)
Single nucleotide variation in genetic sequences defines the Single nucleotide polymorphism (SNP) among individuals, generated due to point mutation or replication errors, giving rise to different alleles within a locus (Van den Broeck et al. 2014). SNPs are the most common form of variation present extensively in the non-coding, coding, and inter-genic regions of DNA (Vallejos-Vidal et al. 2019). SNPs are mainly exploited for population structure, genetic diversity, genetic map construction, and identification of particular traits, etc. (Xia et al. 2019). Their abundance in coding regions makes them more attractive markers for the detection of mutations associated with diseases. SNP markers are however less polymorphic than SSR markers due to their biallelic or triallelic nature (Casci 2010; Mammadov et al. 2012). Cendron et al. (2020) demonstrated the population structure and genetic diversity of local Italian chicken breeds by using SNPs for conservation purposes which revealed lower genetic diversity among the local breeds. In another study, genetic diversity and differentiation among the D. ruyschiana populations of the Norwegian region were investigated by analyzing 96 SNPs derived from 43 sites that reported the existence of four distinct genetic groups within the population (Kyrkjeeide et al. 2020) (Table 3, Fig. 2H).
Artificial intelligence in biodiversity monitoring
With the growing performance of computing power and DL in recent years, machines had become significantly more intelligent and reliable than ever. Modern machines can handle more extensive data and more complex DL models than before (Dean 2019; Chen et al. 2020a). Through this progress, machines had achieved the ability to replicate human expertise (Liu et al. 2019b). Currently, several problems exist within our diverse planet. Researchers began to accelerate the development of several AI solutions with DL to preserve the earth for the later generations to come. In most studies, DL method’s employment provided an automated capability for machines to recognize, classify, and detect images, sounds, and behavior of animals, plants, and even humans (Abeßer 2020). According to Klein et al. (2015), one of the primary methods of preserving our biodiversity consists of monitoring and manual data collection. However, frequent conduct of such practices can become tedious and cause disturbances to sensitive wildlife habitats. With that said, monitoring became less reliable and brief (Table 4). AI-based methods have shown that even at its pre-mature level, biodiversity can have an improvement by reducing animal extinction, prolonged and in-depth monitoring of various life forms, unlocking and accessing unexplored areas, and faster and easier classification of species. With the continuing efforts in data duration, transparency, and research collaborations, these technology types may reach far beyond our expectations. These solutions, if appropriately handled, can yield a massive impact to preserve the planet and its resources without involving humans. Furthermore, the implementation of AI-based methods also extends humans’ capability to explore locations that our biological composition cannot handle, leading to discoveries of new species and life. Due to the accessibility of various capture devices, a wide range of collected data through images, videos, audio, and other forms of data fast-tracked DL and AI development. The problematic method and reliance on organic experts to perform a small to large-scale monitoring of animals, plants, and insects became less challenging as automation systems have improved significantly over a short period (Bergslien 2013; Buxton et al. 2018; Willi et al. 2018) (Fig. 3).
Table 4.
Involvement of AI-based methods at its pre-mature level for biodiversity monitoring
| Species | Subject investigated | Method | Objectives | Findings | References |
|---|---|---|---|---|---|
| Artificial intelligence | |||||
| Recent works based on vision (images and videos) | |||||
| Albatross | The paper employs the U-Net segmentation algorithm to detect albatrosses using satellite imagery captured from space | U-Net segmentation model with satellite imagery | Collect satellite imagery from WorldView-3 and annotate the albatrosses. Train the U-Net segmentation model and compare its performance against human efforts. | Lack of data made it difficult for the U-Net model to perform better classifications. Nonetheless, the model still showed promising results and potential improvements in the future with robust data. | Bowler et al. (2020) |
| Various birds | The authors investigated the automated detection and counting of bird populations based on a vision-based deep learning model, Faster R-CNN | Object detection using Faster R-CNN (bounding boxes) | Collection of geotagged images of birds to train a Faster R-CNN model to detect and classify birds in various locations | Faster R-CNN installed within a GIS tracking application resulted in a faster and more robust approach to monitoring and counting bird populations according to their locations and species with 94% precision. | Akçay et al. (2020) |
| Bears | The authors’ work involved the use of camera traps to observe the behavior and population of brown bears. With their deployed cameras, the authors incorporated a face recognition algorithm within their system to automate the process | Face recognition (BearID) | Acquisition of bear images captured using DSLR cameras and labeled for developing a face recognition model, the BearID, to monitor brown bears. | The BeadID had a 97.7% precision and 83.9% accuracy. The combination of camera traps and deep learning had shown a remarkable improvement in bear activity monitoring and classification of its species. | Clapham et al. (2020) |
| Deers | The study focused on monitoring the problem of invasive species. The authors decided to employ a real-time detector based on the YOLOv3 algorithm on an edge device connected to the internet to identify and count deer species existing species in a specific area. | YOLOv3 real-time detection (bounding box) | Collect and label deer images from the wild and train a YOLOv3 algorithm. Deploy camera traps with the trained model, solar panels, batteries, and regulators within a microcontroller connected to the internet. | Within the 14-day test and 170 videos, 87% of the images had no detections. Manual counting of deers consumed 18.5 hours. Their proposed model only had 0.3 hours, a 98% reduction of effort. | Arshad et al. (2020) |
| Elephants | The authors employed CNN on satellite imagery that focuses on the automated counting of elephants in the Savanna. | CNN for satellite imagery | Collect high-resolution image data from WorldView-3 and 4 satellite data from 2014 to 2019. Train a CNN model with the collected images and compare the performance with human cognition. | CNN attained F2 scores of 0.78 from heterogenous and 0.73 from homogeneous locations. In comparison, humans had 0.77, and 0.80, respectively. Justifying that CNN trained on satellite imagery can perform similarly to humans. | Duporge et al. (2020) |
| Fishes | The authors developed an automated detection of fish underwater based on a deep learning algorithm, Mask R-CNN, and image processing techniques. | Instance segmentation using Mask R-CNN | Capture video footage of life under the waters of Queensland and annotate fish species to train a Mask R-CNN model. Compare the performance against experts to evaluate the improvements. | Their model attained an F1-Score of 92.4% and a Mean Average Precision (mAP) of 92.5% that yielded a 7.1% improvement against marine experts and 13.4% against citizen scientists in determining fish abundance underwater. | Ditria et al. (2020) |
| Various insects | Researchers identified the importance of deep learning and sensors in counting various phenotypic traits and analyzing behavior, and insects’ interactions from an in-depth perspective. | Impact of computer vision, deep learning, and sensor technologies | The researchers identify future possibilities that may unlock new research pathways involving computer vision, sensor technology, and deep learning. | They identified that sensors and cameras equipped with deep learning models in various conditions could yield better monitoring approaches to insects and the potential to discover new species. | Høye et al. (2021) |
| Jellyfish | The researchers proposed a real-time counting and monitoring system for jellyfishes using an object detection algorithm based on the Inception ResNet V2 backbone with a Faster R-CNN detector. | Real-time detection using an Inception ResNet V2 model backbone with Faster R-CNN | Collect images of jellyfish and label each using the LabelImg tool to train compatibly with the desired DCNN backbone and detection algorithm. | Results achieved an F1 score of 95.2%. Further improvements can arise with the addition of reliable data. Nonetheless, their work acquired remarkable results that can aid in the monitoring of jellyfish. | Martin-Abadal et al. (2020) |
| Mosquitoes | The authors employed a YOLOv3 object detection algorithm and concatenated it with another to identify species, gender through the detection of vector mosquitoes in real-time. | Concatenated YOLOv3 model for real-time gender and species identification | Collect high-quality images of mosquitoes in a controlled environment. Annotate and provide meta-descriptions to each sample and train the YOLOv3 models with it. | Results achieved an mAP of 99% and a sensitivity of 92.4 with 100% classification accuracy for all 13 classes of mosquitoes, thus, showing effectiveness to control outbreaks and population of vector mosquitoes. | Kittichai et al. (2021) |
| Moths | The authors provided a solution to moth species monitoring’s difficult task by developing a light trap with a camera that can detect various moths through temporal dimension sequences and CNNs. | Light trap with CNN trained on images with temporal dimension sequences | Collect images of moths to train a CNN model and install it on a camera light trap to attract moths and perform automated classifications and counting. | During 48 nights, the device captured 250,000 moth images. They acquired a classification F1 score of 0.93 from 2000 live moths, while its tracking had a 0.79. The approach can emanate improvements in moth monitoring with less cost. | Bjerge et al. (2021) |
| Various animals | The researchers explored the capability of DL and traditional machine learning (ML) methods to classify various species of animals that may impact future studies in reducing animal extinction. | Image classification (InceptionV3 and K-NN) | The authors prepared and acquired various animals’ images to train a classification model built on an InceptionV3 with a K-NN classifier. | Achieved a 98.3% with the InceptionV3 model combined with a K-NN classifier and outperformed other models like the AlexNet, ResNet50, and VGG16 and 19 | Jamil et al. (2020) |
| Banana | The authors focused on employing ML algorithms like RF and PCA to detect and classify bananas captured from multi-level satellite images to monitor their health status | Satellite imagery with random forest (RF), principal component analysis (PCA), and vegetation indices (VI) | Acquire images from Sentinel 2, PlanetScope, and WorldView-2, and preprocess them through PCA to train an RF algorithm to conduct pixel-based classifications of bananas for large-scale automated monitoring. | The overall performance attained a 97% accuracy, fewer errors than 10%, and a Kappa coefficient of 0.96. The researchers proved its effectiveness in monitoring aerial images with the help of ML algorithms. | Selvaraj et al. (2020) |
| Coral reefs | The authors selected to use DCNNs to provide a feasible and less costly approach to monitoring coral reefs | DCNN classification from random point annotations | Collect coral reef data from a global dataset. Train a DCNN model that can detect and perform monitoring of diverse coral reefs. | The result had an agreed 97% performance between a human expert and the automated system. Such a solution can reduce delays in data handling and reporting from coral reef monitoring by about 200 times. | González-Rivero et al. (2020) |
| Mangroves | The researchers employed a Capsule U-Net, an improved segmentation algorithm to identify mangroves’ dynamic change over the past 25 years | Capsule U-Net segmentation algorithm for remote sensing data | Use LandSat data to train a DL model based on Capsule U-Net and determine the mangrove coverage’s dynamic change from 1990 to 2015. | The results from 1990, 2000, 2010, and 2015 had accuracies of 86.9%, 87.3%, 85.7%, and 88.7%, respectively. Showing a 0.96 annual rate of change from 1990 to 2015. They identified that loss became less over the past years. | Guo et al. (2021) |
| Orchids | The researchers developed an automated assessment for 13,910 orchid species using a novel deep neural network called IUC-NN to reduce specific species’ extinction. | IUC-NN for orchid species classification | Collect and label various orchid species and split the data into 90% train and 10% test data. Train the IUC-NN model to provide the classification of orchid species. | The results identified possibly threatened species with 84.3% accuracy and indicated its effectiveness in reducing orchid species extinction. | Zizka et al. (2020) |
| Various plants | The authors classified 26 variants of diseases from 14 species of plants using DCNN models to provide and improve the process of leaf disease diagnosis | State-of-the-art DCNN models | Acquire data from the PlantVillage dataset to train a wide range of DCNN models and test to identify its performance in classifying various leaf diseases from different plant species. | They acquired the highest performance of 99.81% F1 score from the Xception model trained with the Adam optimizer. The results proved that deep learning, specifically DCNNs, can yield valuable results to diagnose plant diseases better. | Saleem et al. (2020) |
| Various plants | The authors identified various plant traits through images captured from satellites using radiative transfer learning and various machine learning algorithms to improve biodiversity monitoring. | Spectral data with radiative transfer learning and ML | Collect data from Sentinel-2 and prepare a spectral dataset to train ML models with radiative transfer learning to predict three plant traits chlorophyll content, canopy chlorophyll, and area index. | The RTM trained model attained an LCC (R2 = 0.26, RMSE = 3.9 μg/cm2), CCC (R2 = 0.65, RMSE = 0.33 g/m2), and LAI (R2 = 0.47, RMSE = 0.73 m2/m2). While the RF algorithm had LCC (R2 = 0.34, RMSE = 4.06 μg/cm2), CCC (R2 = 0.65, RMSE = 0.34 g/m2), and LAI (R2 = 0.47, RMSE = 0.75 m2/m2). The results show a massive potential for the monitoring of the following traits in terms of plant species biodiversity. | Ali et al. (2021) |
| Recent works based on sound/audio | |||||
| Bats | The authors have collected a dataset of bat sounds and developed a novel software with a deep learning model to identify various bat species according to the sounds they produce. | WaveMan and BatNet for sound identification based on convolutional neural networks | Collect sound from bats calls and convert it from single-dimensional signals to a two-dimensional spectrogram image to train the CNN model. | The authors successfully attained results of a 0.94 accuracy with only 0.147 false positives, outperforming previous works. Justifying that DL can significantly assist in automating species identification of bats in the wild. | Chen et al. (2020b) |
| Frogs | The authors classified 41 forest frog species based on their produced sound. Their work involved converting audio recordings from frogs converted into 2D spectrograms to train a deep learning model. | Sound classification based on InceptionV3 | Convert a dataset from collected audio to 2D portable network graphics (PNG) image spectrograms labeled accordingly to a specific frog species to train the InceptionV3 classification model | They achieved accuracies of 94.3% for 20 species of frogs, 100% for 11, and 90% for 17. The lowest scores landed at 70% and 75% for 2, showing improvements in classification, and discovery. | Khalighifar et al. (2021) |
| Woodpecker | The authors developed an automated detection and classification of various woodpecker species based on their acoustic signals harnessed by an edge device imbued with ML and DL models. | Two-staged sound classification using artificial neural networks, and machine learning classifiers | Collect sounds from various woodpecker species and train a two-stage classification model composed of DL and ML algorithms employed on an edge device for automated monitorin | Results indicated the feasibility and effectiveness of an automated monitoring device for various woodpecker species from the sounds they produce with a performance of 94.02% accuracy. | Vidaña-Vila et al. (2020) |
Fig. 3.
Application of advanced computer-added physical instruments and associated smart technologies for biodiversity monitoring. The figure describes in a conserved forest ecosystem wildlife and forest ecosystem can be monitored by using autonomous acoustic recording units, animal tracking units (GIS-based), X-ray fluorescence (XRF) analyzer, global positioning system (GPS), and camera trapping units. Configuration of these data from multiple sources and data heterogeneity can be monitored, processed, interpreted, analyzed, and distributed (encrypted) via artificial intelligence and machine-learning-based technologies. Using these technologies wildlife morphology (camera), behavior, phenology, distribution, abundance, phylogeny (acoustic), and diversity with respect to human invasion can be observed. The figure is inspired by the following sources: Bergslien (2013), Buxton et al. (2018), Willi et al. (2018). The components of the figure are modifications of The RAFOS group at the Graduate School of Oceanography, University of Rhode Island, Kingston, RI 02881 (2001), Mudgineer (2011); Leapfrog (2021), Pixabay (2021), Pngtree (2021), and Vecteezy (2021a, b)
Recently, a wide range of low-cost yet powerful sensors, microphones, and cameras have become available, giving aid to alleviating the problem of collecting data. Such extensive data collections from the said technologies fueled DL models to learn more patterns that generated solutions to better monitor and manage biodiversity. The common uses include automated recognition, classification, and detection of people (Kim and Moon 2016), animals (Verma and Gupta 2018), plants (Saleem et al. 2019), fish (Jalal et al. 2020), and even insects (Xia et al. 2018) based on their sound or image (Christin et al. 2019). Even with DL’s promising capabilities, it still exhibits some caveats that limit its full potential in biodiversity monitoring, specifically in real time. Monitoring wildlife through video became an exponential and popular recent development that improved interpretability with less comprehension to researchers and the like (Chen et al. 2019). However, it became difficult and expensive due to the challenging deployment of capable computers or capture devices to perform the task (Willi et al. 2019). While operating with DL models in urban areas is relatively easy due to the availability of sufficient data on infrastructure, functioning in remote areas still relies on post-monitoring systems (He et al. 2016; Zhang et al. 2019a).
Researchers are also on for finding more efficient data collection techniques that will require less computational cost and fewer complexes. Currently, the computer on a hardware basis still rigorously improves and becomes more affordable and independently deployable. With that said, DL can become more efficient and reliable over time that can produce real-time wildlife monitoring in remote areas through a more visual aspect like videos without much constraint from the limited infrastructure.
Challenges and future prospects
Approximately, 1 million of the 10 million species that exist in the world are threatened with extinction (Bawa et al. 2020). Besides monitoring tools, a combination of efforts from varied disciplines will be essential for the safeguard of individual species and biodiversity as a whole. Computer model-based technologies like the GIS, RADAR, remote sensing, and LiDAR are actively used for the monitoring of habitats, state of threats, land uses, and conversion. Molecular approaches such as Mitochondrial DNA (Cyt b), SNPs, RFLP, microsatellites, etc. are also playing a pivotal role in identifying, tracking, and determining the impact of anthropogenic and environmental factors on wildlife (Krestoff et al. 2021; Gouda et al. 2020; Ridley et al. 2020). However, many of these techniques face challenges in form of cost-efficiency and expert handling and have single or limited focal species at the ecosystem level. Some of the possible changes and prospects in biodiversity monitoring systems that can be implemented in near future on broader aspects are discussed below.
The science of chorology with advances in GIS and remote sensing techniques in recent times has better presented the landscape as a functional unit for biodiversity management. Visualizations of spatial-temporal changes and development of biotic and abiotic threats to species also known as “threat maps” emerged as multipurpose techniques for the implementation of conservation activities at the ground level (Ridley et al. 2020). InVEST (Integrated Valuation of Ecosystem Services and Trade-offs) is a newly develop modeling software with set parameters for screening and quantification of ecosystem services such as carbon stock, changes in land use, landscape, forest cover, etc. SolVES is a modern-day ArcGIS-dependent tool that provides the user with easy access to several functions of the Ecosystem Services (ES), human perceptions associated with social and cultural beliefs, socio-economic values, usage of resources, etc. even without conducting questionnaires or other ground surveys of the local people and other stakeholders (Neugarten et al. 2020).
ARIES (ARtificial Intelligence for Ecosystem Services) is a series of algorithm processes which are generated through detecting or recognizing and keeping the track of living systems. It is a software-based platform that solves complex and arduous social or bio-geographical dimensions by integrating biodiversity data (Silvestro et al. 2022). It has been successfully tested for carbon emission, climate change, water levels, and ethnic/recreational values (Bagstad et al. 2018). Costing Nature is another easy-to-use rapid and reliable web-based technique used for screening protected areas, land use and land cover (LULC), trends of habitation, biodiversity assessment, and possible future threats using global database. It has been used for testing ES for timber, fuel wood, grazing/fodder, and non-wood forest products (Thessen 2016; Dominguez-Morales et al. 2021; Neugarten et al. 2018).
As rightly pointed out by Malavasi (2020), biodiversity maps are always selective and do not necessarily display all values that are known about any given region or ecosystem. They are often inevitably affected by personal views or scientific blindness and it is therefore important to strive and rate maps not only in terms of scientific accuracy but also on their “viability.” The use of Public Participation Geographic Information Systems (PPGIS) over conventional screening systems can act as a bottom-up approach to empower concern agencies about the threats and conservation priorities by providing visual tools. Similarly, the use of a counter map can prove as a possible substitute for mitigating the loss of biodiversity in a more “systemic” manner (Schägner et al. 2013; Malavasi 2020).
Genomics models and concepts are widely applied for biodiversity sustenance, from ideal seed selection for preservation to assessing the degree of impact at community-level effects. The concept of population genomics has provided valuable information on population size, demographic history, ability of the populations to evolve and adapt to the changing environment, etc. (Miraldo et al. 2016; Hu et al. 2020, 2021; Hohenlohe et al. 2021). They have been able to successfully develop large sets of markers that increase the ability to detect and quantify low levels of hybridization or admixture. Techniques such as intron sequences with assistance from Transcriptome Ortholog Alignment Sequence Tools (TOASTs), Next-Generation Sequencing (NGS), and Comparative Anchor Tagged Sequences (CATs) may represent a good proxy to assess functional adaptive potential or functional diversity in future genomic studies (Forcina et al. 2021).
Conclusion
With continuous advances in technology, more precise and reliable techniques have been designed for biodiversity conservation. However, association mapping and expanding knowledge on “omics” will help in identifying morphological traits and bring together intellectual minds to a platform for developing advanced gene traits. It also helps identify high biodiversity conservation priority areas or hotspots. Working closely with international agencies like the Convention on Biological Diversity (CBD) and UN Framework Convention on Climate Change (UNFCCC) and achieving its targets will be important for the conservation of biodiversity on the planet. Lastly, it is the human who understands the importance of coexistence and cohabitation with other forms of living beings that will help implement conservation measures and create a sense of protecting the ecosystem. Therefore, it is suggested that a combination of sophisticated monitoring methods including system-based smart techniques, transformative smart technologies, remote sensing, geographical information system, and artificial intelligence in combination with molecular approaches will smartly keep the track of living organisms and will help in biodiversity conservation and restoration.
Author contribution
RGK, FJPM, RD, and SP wrote the initial draft, GPM, GKM, VN, ABJ, KEU, RCJ, and SG edited and reviewed the whole manuscript, and SM and JRR comprehensively edited the manuscript which was then reviewed and approved by all authors.
Availability of data and materials
All data generated or analyzed during this study are included in this article.
Declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent to publish
Not applicable.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Rout George Kerry, Email: routgeorgekerry3@gmail.com.
Francis Jesmar Perez Montalbo, Email: francismontalbo@ieee.org.
Rajeswari Das, Email: das.rajeswari2008@gmail.com.
Sushmita Patra, Email: sushmitapatra282@gmail.com.
Gyana Prakash Mahapatra, Email: gyanamahapatra2@gmail.com.
Vinayak Nayak, Email: vn46520@gmail.com.
Atala Bihari Jena, Email: jena.atala@gmail.com.
Kingsley Eghonghon Ukhurebor, Email: ukeghonghon@gmail.com, Email: ukhurebor.kingsley@edouniversity.edu.ng.
Ram Chandra Jena, Email: rcjenauu@gmail.com.
Sushanto Gouda, Email: sushantogouda@gmail.com.
Sanatan Majhi, Email: sanatanm@gmail.com.
Jyoti Ranjan Rout, Email: routjr@aiph.ac.in, Email: routjr@gmail.com.
References
- Abdelaziz SM, Medraoui L, Alami M, Pakhrou O, Makkaoui M, Boukhary OMS, Filali-Maltouf A (2020) Inter simple sequence repeat markers to assess genetic diversity of the desert date (Balanites aegyptiaca Del.) for Sahelian ecosystem restoration. Sci Rep 10:14948 [DOI] [PMC free article] [PubMed]
- Abdul-Muneer PM. Application of microsatellite markers in conservation genetics and fisheries management: recent advances in population structure analysis and conservation strategies. Gen Res Int. 2014;2014:691759. doi: 10.1155/2014/691759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abeßer J. A review of deep learning based methods for acoustic scene classification. Appl Sci. 2020;10:2020. doi: 10.3390/app10062020. [DOI] [Google Scholar]
- Ade FY, Hakim L, Arumingtyas EL, Azrianingsih R. The detection of Anaphalis spp. genetic diversity based on molecular character (using ITS, ETS, and EST-SSR markers) Int J Adv Sci Eng Inform Technol. 2019;9:1695–1702. doi: 10.18517/ijaseit.9.5.9597. [DOI] [Google Scholar]
- Admin (2017) My blog, GIS Layers, Environmental Science and Resource Management. http://heleneloyan.cikeys.com/update/gis-layers/. Accessed 11 Aug 2021
- Akçay HG, Kabasakal B, Aksu D, Demir N, Öz M, Erdoğan A. Automated bird counting with deep learning for regional bird distribution mapping. Animals : an Open Access Journal from MDPI. 2020;10:1207. doi: 10.3390/ani10071207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Allak ZS, Dragh MA, Hussain AS. Genetic polymorphism and diversity of Iraqi Awassi sheep using PCR-RAPD technique. Basrah J Vet Res. 2020;19:147–154. doi: 10.23975/bjvetr.2020.170619. [DOI] [Google Scholar]
- Alemu A, Feyissa T, Letta T, Abeyo B. Genetic diversity and population structure analysis based on the high density SNP markers in Ethiopian durum wheat (Triticum turgidum ssp. durum) BMC Genet. 2020;21:18. doi: 10.1186/s12863-020-0825-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander C, Korstjens AH, Usher G, Nowak MG, Fredriksson G, Hill RA. LiDAR patch metrics for object-based clustering of forest types in a tropical rainforest. Int J Appl Earth Obs Geoinf. 2018;73:253–261. [Google Scholar]
- Ali AM, Darvishzadeh R, Skidmore A, Gara TW, Heurich M. Machine learning methods’ performance in radiative transfer model inversion to retrieve plant traits from Sentinel-2 data of a mixed mountain forest. Int J Digital Earth. 2021;14:106–120. doi: 10.1080/17538947.2020.1794064. [DOI] [Google Scholar]
- Almeida DRA, Broadbent EN, Zambrano AMA, Wilkinson BE, Ferreira ME, Chazdon R, Meli P, Gorgens EB, Silva CA, Stark SC, Valbuena R, Papa DA, Brancalion PHS. Monitoring the structure of forest restoration plantations with a drone-lidar system. Int J Appl Earth Obs Geoinf. 2019;79:192–198. [Google Scholar]
- Al-Rawashdeh IM. Genetic variability in a medicinal plant Artemisia judaica using random amplified polymorphic DNA (RAPD) markers. Int J Agr Biol. 2011;13:279–282. [Google Scholar]
- Alsolami R, Knight SJ, Schuh A. Clinical application of targeted and genome-wide technologies: can we predict treatment responses in chronic lymphocytic leukemia? Person Med. 2013;10:361–376. doi: 10.2217/pme.13.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amom T, Nongdam P. The use of molecular marker methods in plants: a review. Int J Curr Res Rev. 2017;9:01–07. [Google Scholar]
- Arias-Maldonado M. The anthropocenic turn: theorizing sustainability in a postnatural age. Sustainability. 2016;8:10. doi: 10.3390/su8010010. [DOI] [Google Scholar]
- Arif IA, Khan HA, Bahkali AH, Al Homaidan AA, Al Farhan AH, Al Sadoon M, Shobrak M. DNA marker technology for wildlife conservation. Saudi J Biol Sci. 2011;18:219–225. doi: 10.1016/j.sjbs.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arshad B, Barthelemy J, Pilton E, Perez P (2020) Where is my deer?-wildlife tracking and counting via edge computing and deep learning. In: 2020 IEEE SENSORS. IEEE, Rotterdam, Netherlands, pp 1–4
- Avigliano E, Rosso JJ, Lijtmaer D, Ondarza P, Piacentini L, Izquierdo M, Cirigliano A, Romano G, Nuñez Bustos E, Porta A, Mabragaña E, Grassi E, Palermo J, Bukowski B, Tubaro P, Schenone N. Biodiversity and threats in non-protected areas: a multidisciplinary and multi-taxa approach focused on the Atlantic Forest. Heliyon. 2019;5:e02292. doi: 10.1016/j.heliyon.2019.e02292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae S, Levick SR, Heidrich L, Magdon P, Leutner BF, Wöllauer S, Serebryanyk A, Nauss T, Krzystek P, Gossner MM, Schall P, Heibl C, Bässler C, Doerfler I, Schulze E-D, Krah F-S, Culmsee H, Jung K, Heurich M, Fischer M, Seibold S, Thorn S, Gerlach T, Hothorn T, Weisser WW, Müller J. Radar vision in the mapping of forest biodiversity from space. Nat Commun. 2019;10:4757. doi: 10.1038/s41467-019-12737-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagstad KJ, Cohen E, Ancona ZH, McNulty SG, Sun G. The sensitivity of ecosystem service models to choices of input data and spatial resolution. Appl Geogr. 2018;93:25–36. doi: 10.1016/j.apgeog.2018.02.005. [DOI] [Google Scholar]
- Bakx TRM, Koma Z, Seijmonsbergen AC, Kissling WD. Use and categorization of light detection and ranging vegetation metrics in avian diversity and species distribution research. Divers Distrib. 2019;25:1045–1059. doi: 10.1111/ddi.12915. [DOI] [Google Scholar]
- Bariotakis M, Georgescu L, Laina D, Oikonomou I, Ntagounakis G, Koufaki M-I, Souma M, Choreftakis M, Zormpa OG, Smykal P, Sourvinos G, Lionis C, Castanas E, Karousou R, Pirintsos SA. From wild harvest towards precision agriculture: use of ecological niche modelling to direct potential cultivation of wild medicinal plants in Crete. Sci Total Environ. 2019;694:133681. doi: 10.1016/j.scitotenv.2019.133681. [DOI] [PubMed] [Google Scholar]
- Barlow SE, O’Neill MA. Technological advances in field studies of pollinator ecology and the future of e-ecology. Curr Opin Insect Sci. 2020;38:15–25. doi: 10.1016/j.cois.2020.01.008. [DOI] [PubMed] [Google Scholar]
- Bartkowski B, Lienhoop N, Hansjürgens B. Capturing the complexity of biodiversity: a critical review of economic valuation studies of biological diversity. Ecol Econ. 2015;113:1–14. doi: 10.1016/j.ecolecon.2015.02.023. [DOI] [Google Scholar]
- Basak S, Chakrabartty I, Hedaoo V, Shelke RG, Rangan L. Assessment of genetic variation among wild Alpinia nigra (Zingiberaceae) population: an approach based on molecular phylogeny. Mol Biol Rep. 2019;46:177–189. doi: 10.1007/s11033-018-4458-3. [DOI] [PubMed] [Google Scholar]
- Baumann M, Levers C, Macchi L, Bluhm H, Waske B, Gasparri NI, Kuemmerle T. Mapping continuous fields of tree and shrub cover across the Gran Chaco using Landsat 8 and Sentinel-1 data. Remote Sens Environ. 2018;216:201–211. doi: 10.1016/j.rse.2018.06.044. [DOI] [Google Scholar]
- Bawa KS, Nawn N, Chellam R, Krishnaswamy J, Mathur V, Olsson SB, Pandit N, Rajagopal P, Sankaran M, Shaanker RU, Shankar D, Ramakrishnan U, Vanak AT, Quader S. Opinion: envisioning a biodiversity science for sustaining human well-being. Proc Natl Acad Sci. 2020;117:25951–25955. doi: 10.1073/pnas.2018436117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearman N, Jones N, André I, Cachinho HA, DeMers M. The future role of GIS education in creating critical spatial thinkers. J Geogr High Educ. 2016;40:394–408. doi: 10.1080/03098265.2016.1144729. [DOI] [Google Scholar]
- Behera PM, Behera DK, Panda A, Dixit A, Padhi P. In silico expressed sequence tag analysis in identification of probable diabetic genes as virtual therapeutic targets. Biomed Res Int. 2013;2013:704818. doi: 10.1155/2013/704818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belenguer-Plomer MA, Tanase MA, Fernandez-Carrillo A, Chuvieco E. Burned area detection and mapping using Sentinel-1 backscatter coefficient and thermal anomalies. Remote Sens Environ. 2019;233:111345. doi: 10.1016/j.rse.2019.111345. [DOI] [Google Scholar]
- Bergslien ET. X-ray diffraction and field portable X-ray fluorescence analysis and screening of soils: project design. Geol Soc Lond, Spec Publ. 2013;384:27–46. doi: 10.1144/SP384.14. [DOI] [Google Scholar]
- Bhagyawant SS. RAPD-SCAR Markers: an interface tool for authentication of traits. J Biosci Med. 2015;4:1–9. [Google Scholar]
- Bhatta NP, Priya MG. Radar and its applications. Int J Control Theory Appl. 2017;10:1–9. [Google Scholar]
- Bispo PDC, Pardini M, Papathanassiou KP, Kugler F, Balzter H, Rains D, dos Santos JR, Rizaev IG, Tansey K, dos Santos MN, Spinelli Araujo L. Mapping forest successional stages in the Brazilian Amazon using forest heights derived from TandEM-X SAR interferometry. Remote Sens Environ. 2019;232:111194. doi: 10.1016/j.rse.2019.05.013. [DOI] [Google Scholar]
- Bjerge K, Nielsen JB, Sepstrup MV, Helsing-Nielsen F, Høye TT. An automated light trap to monitor moths (lepidoptera) using computer vision-based tracking and deep learning. Sensors. 2021;21:343. doi: 10.3390/s21020343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blears MJ, De Grandis SA, Lee H, Trevors JT. Amplified fragment length polymorphism (AFLP): a review of the procedure and its applications. J Ind Microbiol Biotechnol. 1998;21:99–114. doi: 10.1038/sj.jim.2900537. [DOI] [Google Scholar]
- Bolton DK, Tompalski P, Coops NC, White JC, Wulder MA, Hermosilla T, Queinnec M, Luther JE, van Lier OR, Fournier RA, Woods M, Treitz PM, van Ewijk KY, Graham G, Quist L. Optimizing Landsat time series length for regional mapping of lidar-derived forest structure. Remote Sens Environ. 2020;239:111645. doi: 10.1016/j.rse.2020.111645. [DOI] [Google Scholar]
- Bouvier M, Durrieu S, Gosselin F, Herpigny B. Use of airborne lidar data to improve plant species richness and diversity monitoring in lowland and mountain forests. PLoS One. 2017;12:e0184524. doi: 10.1371/journal.pone.0184524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler E, Fretwell PT, French G, Mackiewicz M. Using deep learning to count albatrosses from space: assessing results in light of ground truth uncertainty. Remote Sens. 2020;12:2026. doi: 10.3390/rs12122026. [DOI] [Google Scholar]
- Bryan GJ, McLean K, Waugh R, Spooner DM. Levels of Intra-specific AFLP diversity in tuber-bearing potato species with different breeding systems and ploidy levels. Front Genet. 2017;8:119. doi: 10.3389/fgene.2017.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buss MEF, Leizica E, Peinetti R, Noellemeyer E. Relationships between landscape features, soil properties, and vegetation determine ecological sites in a semiarid savanna of central Argentina. J Arid Environ. 2020;173:104038. doi: 10.1016/j.jaridenv.2019.104038. [DOI] [Google Scholar]
- Buxton RT, Lendrum PE, Crooks KR, Wittemyer G. Pairing camera traps and acoustic recorders to monitor the ecological impact of human disturbance. Global Ecol Conserv. 2018;16:e00493. doi: 10.1016/j.gecco.2018.e00493. [DOI] [Google Scholar]
- Cai C, Yang Y, Cheng L, Tong C, Feng J. Development and assessment of EST-SSR marker for the genetic diversity among tobaccos (Nicotiana tabacum L.) Russ J Genet. 2015;51:591–600. doi: 10.1134/S1022795415020064. [DOI] [PubMed] [Google Scholar]
- Cao L, Coops NC, Sun Y, Ruan H, Wang G, Dai J, She G. Estimating canopy structure and biomass in bamboo forests using airborne LiDAR data. ISPRS J Photogramm Remote Sens. 2019;148:114–129. doi: 10.1016/j.isprsjprs.2018.12.006. [DOI] [Google Scholar]
- Carr A, Zeale MRK, Weatherall A, Froidevaux JSP, Jones G. Ground-based and LiDAR-derived measurements reveal scale-dependent selection of roost characteristics by the rare tree-dwelling bat Barbastella barbastellus. For Ecol Manag. 2018;417:237–246. doi: 10.1016/j.foreco.2018.02.041. [DOI] [Google Scholar]
- Carreiras JMB, Jones J, Lucas RM, Shimabukuro YE. Mapping major land cover types and retrieving the age of secondary forests in the Brazilian Amazon by combining single-date optical and radar remote sensing data. Remote Sens Environ. 2017;194:16–32. doi: 10.1016/j.rse.2017.03.016. [DOI] [Google Scholar]
- Casci T. SNPs that come in threes. Nat Rev Genet. 2010;11:8–8. doi: 10.1038/nrg2725. [DOI] [PubMed] [Google Scholar]
- Cavender-Bares J, Gamon JA, Townsend PA. The use of remote sensing to enhance biodiversity monitoring and detection: a critical challenge for the twenty-first century. In: Cavender-Bares J, Gamon JA, Townsend PA, editors. Remote sensing of plant biodiversity. Cham: Springer International Publishing; 2020. pp. 1–12. [Google Scholar]
- Cendron F, Perini F, Mastrangelo S, Tolone M, Criscione A, Bordonaro S, Iaffaldano N, Castellini C, Marzoni M, Buccioni A, Soglia D, Schiavone A, Cerolini S, Lasagna E, Cassandro M. Genome-wide SNP analysis reveals the population structure and the conservation status of 23 Italian chicken breeds. Animals : an Open Access Journal from MDPI. 2020;10:1441. doi: 10.3390/ani10081441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J, Shoshany M. Radar polarization and ecological pattern properties across Mediterranean-to-arid transition zone. Remote Sens Environ. 2017;200:368–377. doi: 10.1016/j.rse.2017.08.032. [DOI] [Google Scholar]
- Chaudhary R, Maurya GK. In: Restriction fragment length polymorphism. Encyclopedia of Animal Cognition and Behavior. Vonk J, Shackelford T, editors. Cham: Springer International Publishing; 2019. pp. 1–3. [Google Scholar]
- Chen R, Little R, Mihaylova L, Delahay R, Cox R. Wildlife surveillance using deep learning methods. Ecol Evol. 2019;9:9453–9466. doi: 10.1002/ece3.5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M-Y, Chiang H-S, Lughofer E, Egrioglu E. Deep learning: emerging trends, applications and research challenges. Soft Comput. 2020;24:7835–7838. doi: 10.1007/s00500-020-04939-z. [DOI] [Google Scholar]
- Chen X, Zhao J, Chen Y, Zhou W, Hughes AC. Automatic standardized processing and identification of tropical bat calls using deep learning approaches. Biol Conserv. 2020;241:108269. doi: 10.1016/j.biocon.2019.108269. [DOI] [Google Scholar]
- Christin S, Hervet É, Lecomte N. Applications for deep learning in ecology. Methods Ecol Evol. 2019;10:1632–1644. doi: 10.1111/2041-210X.13256. [DOI] [Google Scholar]
- Chunming W, Guoliang D. The study of UWB RADAR life-detection for searching human subjects. Energy Procedia. 2012;17:1028–1033. doi: 10.1016/j.egypro.2012.02.203. [DOI] [Google Scholar]
- Clapham M, Miller E, Nguyen M, Darimont CT. Automated facial recognition for wildlife that lack unique markings: a deep learning approach for brown bears. Ecol Evol. 2020;10:12883–12892. doi: 10.1002/ece3.6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe RA, Lamb D, Edwards C. Discrimination of species composition types of a grazed pasture landscape using Sentinel-1 and Sentinel-2 data. Int J Appl Earth Obs Geoinf. 2020;84:101978. [Google Scholar]
- Cunha JT, Domingues L. RAPD/SCAR Approaches for identification of adulterant breeds’ milk in dairy products. Methods Mol Biol (Clifton NJ) 2017;1620:183–193. doi: 10.1007/978-1-4939-7060-5_13. [DOI] [PubMed] [Google Scholar]
- Curry CJ, Davis BW, Bertola LD, White PA, Murphy WJ, Derr JN. Spatiotemporal genetic diversity of lions reveals the influence of habitat fragmentation across Africa. Mol Biol Evol. 2021;38:48–57. doi: 10.1093/molbev/msaa174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalponte M, Jucker T, Liu S, Frizzera L, Gianelle D. Characterizing forest carbon dynamics using multi-temporal lidar data. Remote Sens Environ. 2019;224:412–420. doi: 10.1016/j.rse.2019.02.018. [DOI] [Google Scholar]
- de Góes Maciel F, Rufo DA, Keuroghlian A, Russo AC, Brandt NM, Vieira NF, da Nóbrega BM, Nava A, Nardi MS, de Almeida Jácomo AT, Silveira L, Furtado MM, Tôrres NM, Miyaki CY, Tambosi LR, Biondo C. Genetic diversity and population structure of white-lipped peccaries (Tayassu pecari) in the Pantanal, Cerrado and Atlantic Forest from Brazil. Mamm Biol. 2019;95:85–92. doi: 10.1016/j.mambio.2019.03.001. [DOI] [Google Scholar]
- Dean J (2019) The deep learning revolution and its implications for computer architecture and chip design. http://arxiv.org/abs/1911.05289. Accessed 12 Aug 2021
- Ditria EM, Lopez-Marcano S, Sievers M, Jinks EL, Brown CJ, Connolly RM. Automating the analysis of fish abundance using object detection: optimizing animal ecology with deep learning. Front Mar Sci. 2020;7:429. doi: 10.3389/fmars.2020.00429. [DOI] [Google Scholar]
- Dominguez-Morales JP, Duran-Lopez L, Gutierrez-Galan D, Rios-Navarro A, Linares-Barranco A, Jimenez-Fernandez A. Wildlife monitoring on the edge: a performance evaluation of embedded neural networks on microcontrollers for animal behavior classification. Sensors. 2021;21:2975. doi: 10.3390/s21092975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dube T, Shoko C, Sibanda M, Madileng P, Maluleke XG, Mokwatedi VR, Tibane L, Tshebesebe T. Remote Sensing of Invasive Lantana camara (Verbenaceae) in Semiarid Savanna Rangeland Ecosystems of South Africa. Rangel Ecol Manag. 2020;73:411–419. doi: 10.1016/j.rama.2020.01.003. [DOI] [Google Scholar]
- Duporge I, Isupova O, Reece S, Macdonald DW, Wang T. Using very-high-resolution satellite imagery and deep learning to detect and count African elephants in heterogeneous landscapes. Remote Sens Ecol Conserv. 2020;7(3):369–381. doi: 10.1002/rse2.195. [DOI] [Google Scholar]
- Earthdata (2021) Remote Sensors. Earthdata.
- Ebrahimi R, Hassandokht MR, Zamani Z, Roldan-Ruiz I, Muylle H, Van Glabeke S, Van Bockstaele E, Kashi A. Genetic characterization of Allium stipitatum accessions: an economically wild edible Allium species with unique flavor. Braz J Bot. 2019;42:83–96. doi: 10.1007/s40415-018-0505-5. [DOI] [Google Scholar]
- El Hentati H, Thamri N, Derouich W, Hadhli M, Boukhorsa T. Study of genetic diversity in Tunisian local cattle populations using ISSR markers. J Anim Plant Sci. 2019;42(3):7296–7302. doi: 10.35759/JAnmPlSci.v42-3.2. [DOI] [Google Scholar]
- El-Demerdash E-SS, Elsherbeny EA, Salama YAM, Ahmed MZ. Genetic diversity analysis of some Egyptian origanum and Thymus species using AFLP markers. J Gen Eng Biotechnol. 2019;17:13. doi: 10.1186/s43141-019-0012-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erinjery JJ, Singh M, Kent R. Mapping and assessment of vegetation types in the tropical rainforests of the Western Ghats using multispectral Sentinel-2 and SAR Sentinel-1 satellite imagery. Remote Sens Environ. 2018;216:345–354. doi: 10.1016/j.rse.2018.07.006. [DOI] [Google Scholar]
- Esmaeili H, Karami A, Hadian J, Nejad Ebrahimi S, Otto L-G. Genetic structure and variation in Iranian licorice (Glycyrrhiza glabra L.) populations based on morphological, phytochemical and simple sequence repeats markers. Ind Crop Prod. 2020;145:112140. doi: 10.1016/j.indcrop.2020.112140. [DOI] [Google Scholar]
- FAO, A . 2019, Moving forward on food loss and waste reduction. Rome: Food and Agriculture Organization of the United Nations; 2019. The state of food and agriculture. 2019. [Google Scholar]
- Fauvel M, Lopes M, Dubo T, Rivers-Moore J, Frison P-L, Gross N, Ouin A. Prediction of plant diversity in grasslands using Sentinel-1 and -2 satellite image time series. Remote Sens Environ. 2020;237:111536. doi: 10.1016/j.rse.2019.111536. [DOI] [Google Scholar]
- Fedrigo M, Newnham GJ, Coops NC, Culvenor DS, Bolton DK, Nitschke CR. Predicting temperate forest stand types using only structural profiles from discrete return airborne lidar. ISPRS J Photogramm Remote Sens. 2018;136:106–119. doi: 10.1016/j.isprsjprs.2017.11.018. [DOI] [Google Scholar]
- Feng S, Zhu Y, Yu C, Jiao K, Jiang M, Lu J, Shen C, Ying Q, Wang H. Development of species-specific SCAR markers, based on a SCoT analysis, to authenticate Physalis (Solanaceae) species. Front Genet. 2018;9:192. doi: 10.3389/fgene.2018.00192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes ACM, Gonzalez RQ, Lenihan-Clarke MA, Trotter EFL, Arsanjani JJ. Machine learning for conservation planning in a changing climate. Sustainability. 2020;12:7657. doi: 10.3390/su12187657. [DOI] [Google Scholar]
- Fernandez-Carrillo A, McCaw L, Tanase MA. Estimating prescribed fire impacts and post-fire tree survival in eucalyptus forests of Western Australia with L-band SAR data. Remote Sens Environ. 2019;224:133–144. doi: 10.1016/j.rse.2019.02.005. [DOI] [Google Scholar]
- Ferreira RC, Piredda R, Bagnoli F, Bellarosa R, Attimonelli M, Fineschi S, Schirone B, Simeone MC. Phylogeography and conservation perspectives of an endangered macaronesian endemic: Picconia azorica (Tutin) Knobl. (Oleaceae) Eur J For Res. 2011;130:181–195. doi: 10.1007/s10342-010-0420-1. [DOI] [Google Scholar]
- Forcina G, Camacho-Sanchez M, Tuh FYY, Moreno S, Leonard JA. Markers for genetic change. Heliyon. 2021;7:e05583. doi: 10.1016/j.heliyon.2020.e05583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freepik (2021) Download 3d isometric terrain of a mountainous landscape for free. In: Freepik. href='https://www.freepik.com/photos/map'>Map photo created by kjpargeter - www.freepik.com. Accessed 9 Aug 2021
- Freigoun SAB, Elagib TY, Raddad EYA. Analysis of genetic diversity in four Sudanese provenances of Balanites aegyptiaca (L.) Del. based on random amplified polymorphic DNA (RAPD) marker. Afr J Biotechnol. 2020;19:408–414. doi: 10.5897/AJB2020.17128. [DOI] [Google Scholar]
- Gallardo-Alvárez MI, Lesher-Gordillo JM, Machkour-M’Rabet S, Zenteno-Ruiz CE, Olivera-Gómez LD, del Rosario Barragán-Vázquez M, Ríos-Rodas L, Valdés-Marín A, Vázquez-López HG, Arriaga-Weiss SL. Genetic diversity and population structure of founders from wildlife conservation management units and wild populations of critically endangered Dermatemys mawii. Global Ecol Conserv. 2019;19:e00616. doi: 10.1016/j.gecco.2019.e00616. [DOI] [Google Scholar]
- Gamal E, Khdery G, Morsy A, Ali M, Hashim A, Saleh H. Using GIS based modelling to aid conservation of two endangered plant species (Ebenus armitagei and Periploca angustifolia) at Wadi Al-Afreet, Egypt. Remote Sens Appl: Soc Environ. 2020;19:100336. [Google Scholar]
- Ganie SH, Upadhyay P, Das S, Prasad Sharma M. Authentication of medicinal plants by DNA markers. Plant Gene. 2015;4:83–99. doi: 10.1016/j.plgene.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García M, Saatchi S, Ustin S, Balzter H. Modelling forest canopy height by integrating airborne LiDAR samples with satellite Radar and multispectral imagery. Int J Appl Earth Obs Geoinf. 2018;66:159–173. [Google Scholar]
- Ge Z, Dai Z, Pang W, Li S, Wei W, Mei X, Huang H, Gu J. LIDAR-based detection of the post-typhoon recovery of a meso-macro-tidal beach in the Beibu Gulf, China. Mar Geol. 2017;391:127–143. doi: 10.1016/j.margeo.2017.08.008. [DOI] [Google Scholar]
- Goncalves AL, García MV, Heuertz M, González-Martínez SC. Demographic history and spatial genetic structure in a remnant population of the subtropical tree Anadenanthera colubrina var. cebil (Griseb.) Altschul (Fabaceae) Ann For Sci. 2019;76:18. doi: 10.1007/s13595-019-0797-z. [DOI] [Google Scholar]
- González-Rivero M, Beijbom O, Rodriguez-Ramirez A, Bryant DEP, Ganase A, Gonzalez-Marrero Y, Herrera-Reveles A, Kennedy EV, Kim CJS, Lopez-Marcano S, Markey K, Neal BP, Osborne K, Reyes-Nivia C, Sampayo EM, Stolberg K, Taylor A, Vercelloni J, Wyatt M, Hoegh-Guldberg O. Monitoring of coral reefs using artificial intelligence: a feasible and cost-effective approach. Remote Sens. 2020;12:489. doi: 10.3390/rs12030489. [DOI] [Google Scholar]
- Gouda S, Kerry RG, Das A, Chauhan NS. Wildlife forensics: a boon for species identification and conservation implications. Forensic Sci Int. 2020;317:110530. doi: 10.1016/j.forsciint.2020.110530. [DOI] [PubMed] [Google Scholar]
- Griffiths P, Nendel C, Pickert J, Hostert P. Towards national-scale characterization of grassland use intensity from integrated Sentinel-2 and Landsat time series. Remote Sens Environ. 2020;238:111124. doi: 10.1016/j.rse.2019.03.017. [DOI] [Google Scholar]
- Große-Stoltenberg A, Hellmann C, Thiele J, Werner C, Oldeland J. Early detection of GPP-related regime shifts after plant invasion by integrating imaging spectroscopy with airborne LiDAR. Remote Sens Environ. 2018;209:780–792. doi: 10.1016/j.rse.2018.02.038. [DOI] [Google Scholar]
- Guo X, Coops NC, Tompalski P, Nielsen SE, Bater CW, John Stadt J. Regional mapping of vegetation structure for biodiversity monitoring using airborne lidar data. Ecol Inform. 2017;38:50–61. doi: 10.1016/j.ecoinf.2017.01.005. [DOI] [Google Scholar]
- Guo Y, Liao J, Shen G. Mapping large-scale mangroves along the maritime silk road from 1990 to 2015 using a novel deep learning model and landsat data. Remote Sens. 2021;13:245. doi: 10.3390/rs13020245. [DOI] [Google Scholar]
- Haas J, Ban Y. Sentinel-1A SAR and sentinel-2A MSI data fusion for urban ecosystem service mapping. Remote Sens Appl: Soc Environ. 2017;8:41–53. [Google Scholar]
- Haider N, Nabulsi I. Identification of bread and durum wheats from their diploid ancestral species based on chloroplast DNA. Agriculture (Pol’nohospodárstvo) 2020;66:56–66. [Google Scholar]
- Hao J, Jiao K, Yu C, Guo H, Zhu Y, Yang X, Zhang S, Zhang L, Feng S, Song Y, Dong M, Wang H, Shen C. Development of SCoT-based SCAR marker for rapid authentication of Taxus media. Biochem Genet. 2018;56:255–266. doi: 10.1007/s10528-018-9842-0. [DOI] [PubMed] [Google Scholar]
- Harrison PA, Berry PM, Simpson G, Haslett JR, Blicharska M, Bucur M, Dunford R, Egoh B, Garcia-Llorente M, Geamănă N, Geertsema W, Lommelen E, Meiresonne L, Turkelboom F. Linkages between biodiversity attributes and ecosystem services: a systematic review. Ecosyst Serv. 2014;9:191–203. doi: 10.1016/j.ecoser.2014.05.006. [DOI] [Google Scholar]
- Hay SI. An overview of remote sensing and geodesy for epidemiology and public health application. Adv Parasitol. 2000;47:1–35. doi: 10.1016/S0065-308X(00)47005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, Kays R, Zhang Z, Ning G, Huang C, Han TX, Millspaugh J, Forrester T, McShea W. Visual informatics tools for supporting large-scale collaborative wildlife monitoring with citizen scientists. IEEE Circuits SystMag. 2016;16:73–86. doi: 10.1109/MCAS.2015.2510200. [DOI] [Google Scholar]
- Hirst M. Operational environment. The air transport system. Cambridge: Woodhead Publishing; 2008. pp. 72–101. [Google Scholar]
- Hoban S, Bruford M, D’Urban Jackson J, Lopes-Fernandes M, Heuertz M, Hohenlohe PA, Paz-Vinas I, Sjögren-Gulve P, Segelbacher G, Vernesi C, Aitken S, Bertola LD, Bloomer P, Breed M, Rodríguez-Correa H, Funk WC, Grueber CE, Hunter ME, Jaffe R, Liggins L, Mergeay J, Moharrek F, O’Brien D, Ogden R, Palma-Silva C, Pierson J, Ramakrishnan U, Simo-Droissart M, Tani N, Waits L, Laikre L. Genetic diversity targets and indicators in the CBD post-2020 Global biodiversity framework must be improved. Biol Conserv. 2020;248:108654. doi: 10.1016/j.biocon.2020.108654. [DOI] [Google Scholar]
- Hohenlohe PA, Funk WC, Rajora OP. Population genomics for wildlife conservation and management. Mol Ecol. 2021;30:62–82. doi: 10.1111/mec.15720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höppler L, Gödde F, Gutleben M et al (2020) Synergy of active- and passive remote sensing: An approach to reconstruct three-dimensional cloud macro- and microphysics. https://www.atmos-meas-tech-discuss.net/amt-2020-49/. Accessed 12 Aug 2021
- Høye TT, Ärje J, Bjerge K, Hansen OLP, Iosifidis A, Leese F, Mann HMR, Meissner K, Melvad C, Raitoharju J. Deep learning and computer vision will transform entomology. Proc Natl Acad Sci. 2021;118:e2002545117. doi: 10.1073/pnas.2002545117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Rampitsch C, Bykova NV. Advances in plant proteomics toward improvement of crop productivity and stress resistancex. Front Plant Sci. 2015;6:209. doi: 10.3389/fpls.2015.00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C, Pan T, Wu Y, Zhang C, Chen W, Chang Q. Spatial genetic structure and historical demography of East Asian wild boar. Anim Genet. 2020;51:557–567. doi: 10.1111/age.12955. [DOI] [PubMed] [Google Scholar]
- Hu C, Yuan S, Sun W, Chen W, Liu W, Li P, Chang Q. Spatial genetic structure and demographic history of the wild boar in the Qinling Mountains, China. Animals. 2021;11:346. doi: 10.3390/ani11020346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Xie L, Wang H, Zhong J, Li Y, Liu J, Ou Z, Liang X, Li Y, Huang H, Lin Z, Zhang K, Zhang L, Zheng X. Geographic distribution and impacts of climate change on the suitable habitats of Zingiber species in China. Ind Crop Prod. 2019;138:111429. doi: 10.1016/j.indcrop.2019.05.078. [DOI] [Google Scholar]
- Igawa T, Takahara T, Lau Q, Komaki S. An application of PCR-RFLP species identification assay for environmental DNA detection. PeerJ. 2019;7:e7597. doi: 10.7717/peerj.7597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inanaga M, Hasegawa Y, Mishima K, Takata K. Genetic diversity and structure of Japanese endemic genus Thujopsis (Cupressaceae) using EST-SSR markers. Forests. 2020;11:935. doi: 10.3390/f11090935. [DOI] [Google Scholar]
- IPBES (2019) The IPBES’ 2019 global assessment report on biodiversity and ecosystem services. UN Report: Nature’s Dangerous Decline “Unprecedented”; Species Extinction Rates “Accelerating.” In: United Nations Sustainable Development. https://www.un.org/sustainabledevelopment/blog/2019/05/nature-decline-unprecedentedreport. Accessed 7 Jul 2020
- Irina L-T, Javier B-P, Teresa C-BM, Eurídice L-A, María L, del Carmen C-I. Integrating ecological and socioeconomic criteria in a GIS-based multicriteria-multiobjective analysis to develop sustainable harvesting strategies for Mexican oregano Lippia graveolens Kunth, a non-timber forest product. Land Use Policy. 2019;81:668–679. doi: 10.1016/j.landusepol.2018.11.038. [DOI] [Google Scholar]
- IUCN (2020) IUCN 2020. The IUCN red list of threatened species. Version 2020-1. In: IUCN red list of threatened species. https://www.iucnredlist.org/en. Accessed 6 Jul 2020
- Jahncke R, Leblon B, Bush P, LaRocque A. Mapping wetlands in Nova Scotia with multi-beam RADARSAT-2 Polarimetric SAR, optical satellite imagery, and Lidar data. Int J Appl Earth Obs Geoinf. 2018;68:139–156. [Google Scholar]
- Jalal A, Salman A, Mian A, Shortis M, Shafait F. Fish detection and species classification in underwater environments using deep learning with temporal information. Ecol Inform. 2020;57:101088. doi: 10.1016/j.ecoinf.2020.101088. [DOI] [Google Scholar]
- Jamil S, Fawad, Abbas MS et al (2020) Deep learning and computer vision-based a novel framework for himalayan bear, marco polo sheep and snow leopard detection. In: 2020 International Conference on Information Science and Communication Technology (ICISCT). IEEE, Karachi, Pakistan, pp 1–6
- Jansson S, Malmqvist E, Brydegaard M, Åkesson S, Rydell J. A Scheimpflug lidar used to observe insect swarming at a wind turbine. Ecol Indic. 2020;117:106578. doi: 10.1016/j.ecolind.2020.106578. [DOI] [Google Scholar]
- Jha SR, Naz R, Asif A, Okla MK, Soufan W, Al-Ghamdi AA, Ahmad A. Development of an in vitro propagation protocol and a sequence characterized amplified region (SCAR) marker of Viola serpens Wall ex. Ging. Plants (Basel, Switzerland) 2020;9:246. doi: 10.3390/plants9020246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsli BA, Demir E, Fidan HG, Karsli T. Assessment of genetic diversity and differentiation among four indigenous Turkish sheep breeds using microsatellites. Arch Anim Breed. 2020;63:165–172. doi: 10.5194/aab-63-165-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthikeyan S, Preethi NSR. (2018) Life detection system using UWB RADAR during disaster. Second Int Conf Green Comput Interne Things (ICGCIoT) 2018;2:361–365. doi: 10.1109/ICGCIoT.2018.8752992. [DOI] [Google Scholar]
- Kasprzak-Filipek K, Sawicka-Zugaj W, Litwińczuk Z, Chabuz W, Šveistienė R, Bulla J. Assessment of the genetic structure of Central European cattle breeds based on functional gene polymorphism. Global Ecol Conserv. 2019;17:e00525. doi: 10.1016/j.gecco.2019.e00525. [DOI] [Google Scholar]
- Khalighifar A, Brown RM, Goyes Vallejos J, Peterson AT. Deep learning improves acoustic biodiversity monitoring and new candidate forest frog species identification (genus Platymantis) in the Philippines. Biodivers Conserv. 2021;30:643–657. doi: 10.1007/s10531-020-02107-1. [DOI] [Google Scholar]
- Kim S-K. Genetic diversity and DNA markers in fish. In: Kim S-K, editor. Essentials of Marine Biotechnology. Cham: Springer International Publishing; 2019. pp. 109–144. [Google Scholar]
- Kim Y, Moon T. Human detection and activity classification based on micro-doppler signatures using deep convolutional neural networks. IEEE Geosci Remote Sens Lett. 2016;13:8–12. doi: 10.1109/LGRS.2015.2491329. [DOI] [Google Scholar]
- Kittichai V, Pengsakul T, Chumchuen K, Samung Y, Sriwichai P, Phatthamolrat N, Tongloy T, Jaksukam K, Chuwongin S, Boonsang S. Deep learning approaches for challenging species and gender identification of mosquito vectors. Sci Rep. 2021;11:4838. doi: 10.1038/s41598-021-84219-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein DJ, McKown MW, Tershy BR. Deep learning for large scale biodiversity monitoring. New York: Bloomberg Data for Good; 2015. p. 7. [Google Scholar]
- Knapp N, Fischer R, Huth A. Linking lidar and forest modeling to assess biomass estimation across scales and disturbance states. Remote Sens Environ. 2018;205:199–209. doi: 10.1016/j.rse.2017.11.018. [DOI] [Google Scholar]
- Knapp N, Fischer R, Cazcarra-Bes V, Huth A. Structure metrics to generalize biomass estimation from lidar across forest types from different continents. Remote Sens Environ. 2020;237:111597. doi: 10.1016/j.rse.2019.111597. [DOI] [Google Scholar]
- Kovács I, Tóth B, Schally G, Csányi S, Bleier N. The assessment of wildlife damage estimation methods in maize with simulation in GIS environment. Crop Prot. 2020;127:104971. doi: 10.1016/j.cropro.2019.104971. [DOI] [Google Scholar]
- Koyama CN, Watanabe M, Hayashi M, Ogawa T, Shimada M. Mapping the spatial-temporal variability of tropical forests by ALOS-2 L-band SAR big data analysis. Remote Sens Environ. 2019;233:111372. doi: 10.1016/j.rse.2019.111372. [DOI] [Google Scholar]
- Krestoff ES, Creecy JP, Lord WD, Haynie ML, Coyer JA, Sampson K. Mitochondrial DNA evaluation and species identification of Kemp’s Ridley Sea Turtle (Lepidochelys kempii) bones after a 3-year exposure to submerged marine and terrestrial environments. Front Mar Sci. 2021;8:646455. doi: 10.3389/fmars.2021.646455. [DOI] [Google Scholar]
- Krigas N, Papadimitriou K, Mazaris AD (2012) GIS and ex situ plant conservation. In: Alam BM (ed) Application of Geographic Information Systems. IntechOpen, London, SW1P 1WG, UK
- Kumar A, Kishore BSPC, Saikia P, Deka J, Bharali S, Singha LB, Tripathi OP, Khan ML. Tree diversity assessment and above ground forests biomass estimation using SAR remote sensing: a case study of higher altitude vegetation of North-East Himalayas, India. Physics Chem Earth, Parts A/B/C. 2019;111:53–64. doi: 10.1016/j.pce.2019.03.007. [DOI] [Google Scholar]
- Kwok R. AI empowers conservation biology. Nature. 2019;567:133–134. doi: 10.1038/d41586-019-00746-1. [DOI] [PubMed] [Google Scholar]
- Kyrkjeeide MO, Westergaard KB, Kleven O, Evju M, Endrestøl A, Brandrud MK, Stabbetorp O. Conserving on the edge: genetic variation and structure in northern populations of the endangered plant Dracocephalum ruyschiana L. (Lamiaceae) Conserv Genet. 2020;21:707–718. doi: 10.1007/s10592-020-01281-7. [DOI] [Google Scholar]
- Labouisse J-P, Cubry P, Austerlitz F, Rivallan R, Nguyen HA. New insights on spatial genetic structure and diversity of Coffea canephora(Rubiaceae) in Upper Guinea based on old herbaria. Plant Ecol Evol. 2020;153:82–100. doi: 10.5091/plecevo.2020.1584. [DOI] [Google Scholar]
- Lambert M-J, Traoré PCS, Blaes X, Baret P, Defourny P. Estimating smallholder crops production at village level from Sentinel-2 time series in Mali’s cotton belt. Remote Sens Environ. 2018;216:647–657. doi: 10.1016/j.rse.2018.06.036. [DOI] [Google Scholar]
- Lampert A. Over-exploitation of natural resources is followed by inevitable declines in economic growth and discount rate. Nat Commun. 2019;10:1419. doi: 10.1038/s41467-019-09246-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang N, Schindler K, Wegner JD. Country-wide high-resolution vegetation height mapping with Sentinel-2. Remote Sens Environ. 2019;233:111347. doi: 10.1016/j.rse.2019.111347. [DOI] [Google Scholar]
- Laurin GV, Puletti N, Grotti M, Stereńczak K, Modzelewska A, Lisiewicz M, Sadkowski R, Kuberski Ł, Chirici G, Papale D. Species dominance and above ground biomass in the Białowieża Forest, Poland, described by airborne hyperspectral and lidar data. Int J Appl Earth Obs Geoinf. 2020;92:102178. [Google Scholar]
- Leapfrog (2021) GIS Data, Maps and Images. https://help.seequent.com/Geothermal/4.1/en-GB/Content/gisdata/gis-data.htm. Accessed 2 Aug 2021
- Lei Y, Treuhaft R, Keller M, dos-Santos M, Gonçalves F, Neumann M (2018) Quantification of selective logging in tropical forest with spaceborne SAR interferometry. Remote Sens Environ 211:167–183
- Li J, Zhao B, Chen Y, Zhao B, Yang N, Hu S, Shen J, Wu X. A genetic evaluation system for New Zealand white rabbit germplasm resources based on SSR markers. Animals : an Open Access Journal from MDPI. 2020;10:1258. doi: 10.3390/ani10081258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Niu Z, Shang R, Qin Y, Wang L, Chen H. High-resolution mapping of forest canopy height using machine learning by coupling ICESat-2 LiDAR with Sentinel-1, Sentinel-2 and Landsat-8 data. Int J Appl Earth Obs Geoinf. 2020;92:102163. [Google Scholar]
- Liu L, Guo C, Li J, Xu H, Zhang J, Wang B. Simultaneous life detection and localization using a wideband chaotic signal with an embedded tone. Sensors. 2016;16:1866. doi: 10.3390/s16111866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Skidmore AK, Jones S, Wang T, Heurich M, Zhu X, Shi Y. Large off-nadir scan angle of airborne LiDAR can severely affect the estimates of forest structure metrics. ISPRS J Photogramm Remote Sens. 2018;136:13–25. doi: 10.1016/j.isprsjprs.2017.12.004. [DOI] [Google Scholar]
- Liu F-M, Zhang N-N, Liu X-J, Yang Z-J, Jia H-Y, Xu D-P (2019a) Genetic diversity and population structure analysis of Dalbergia odorifera germplasm and development of a core collection using microsatellite markers. Genes 10 [DOI] [PMC free article] [PubMed]
- Liu X, Faes L, Kale AU, Wagner SK, Fu DJ, Bruynseels A, Mahendiran T, Moraes G, Shamdas M, Kern C, Ledsam JR, Schmid MK, Balaskas K, Topol EJ, Bachmann LM, Keane PA, Denniston AK. A comparison of deep learning performance against health-care professionals in detecting diseases from medical imaging: A systematic review and meta-analysis. Lancet Digit Health. 2019;1:e271–e297. doi: 10.1016/S2589-7500(19)30123-2. [DOI] [PubMed] [Google Scholar]
- Liu J, Yong DL, Choi C-Y, Gibson L (2020) Transboundary frontiers: An emerging priority for biodiversity conservation. Trends Ecol Evol 35:679–690. 10.1016/j.tree.2020.03.004 [DOI] [PubMed]
- Lucas R, Van De Kerchove R, Otero V, Lagomasino D, Fatoyinbo L, Omar H, Satyanarayana B, Dahdouh-Guebas F. Structural characterisation of mangrove forests achieved through combining multiple sources of remote sensing data. Remote Sens Environ. 2020;237:111543. doi: 10.1016/j.rse.2019.111543. [DOI] [Google Scholar]
- Luo S, Wang C, Xi X, Pan F, Peng D, Zou J, Nie S, Qin H. Fusion of airborne LiDAR data and hyperspectral imagery for aboveground and belowground forest biomass estimation. Ecol Indic. 2017;73:378–387. doi: 10.1016/j.ecolind.2016.10.001. [DOI] [Google Scholar]
- Ma J, Xiao X, Qin Y, Chen B, Hu Y, Li X, Zhao B. Estimating aboveground biomass of broadleaf, needleleaf, and mixed forests in Northeastern China through analysis of 25-m ALOS/PALSAR mosaic data. For Ecol Manag. 2017;389:199–210. doi: 10.1016/j.foreco.2016.12.020. [DOI] [Google Scholar]
- Ma Q, Su Y, Luo L, Li L, Kelly M, Guo Q. Evaluating the uncertainty of Landsat-derived vegetation indices in quantifying forest fuel treatments using bi-temporal LiDAR data. Ecol Indic. 2018;95:298–310. doi: 10.1016/j.ecolind.2018.07.050. [DOI] [Google Scholar]
- Ma X, Mahecha MD, Migliavacca M, van der Plas F, Benavides R, Ratcliffe S, Kattge J, Richter R, Musavi T, Baeten L, Barnoaiea I, Bohn FJ, Bouriaud O, Bussotti F, Coppi A, Domisch T, Huth A, Jaroszewicz B, Joswig J, Pabon-Moreno DE, Papale D, Selvi F, Laurin GV, Valladares F, Reichstein M, Wirth C. Inferring plant functional diversity from space: the potential of Sentinel-2. Remote Sens Environ. 2019;233:111368. doi: 10.1016/j.rse.2019.111368. [DOI] [Google Scholar]
- Malavasi M. The map of biodiversity mapping. Biol Conserv. 2020;252:108843. doi: 10.1016/j.biocon.2020.108843. [DOI] [Google Scholar]
- Malik MH, Moaeen-Ud-Din M, Bilal G, Ghaffar A, Muner RD, Raja GK, Khan WA. Development of amplified fragment length polymorphism (AFLP) markers for the identification of Cholistani cattle. Arch Anim Breed. 2018;61:387–394. doi: 10.5194/aab-61-387-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammadov J, Aggarwal R, Buyyarapu R, Kumpatla S. SNP Markers and their impact on plant breeding. Int J Plant Genomics. 2012;2012:728398. doi: 10.1155/2012/728398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manisalidis I, Stavropoulou E, Stavropoulos A, Bezirtzoglou E. Environmental and health impacts of air pollution: a review. Front Public Health. 2020;8:14. doi: 10.3389/fpubh.2020.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchese C. Biodiversity hotspots: a shortcut for a more complicated concept. Global Ecol Conserv. 2015;3:297–309. doi: 10.1016/j.gecco.2014.12.008. [DOI] [Google Scholar]
- Martin-Abadal M, Ruiz-Frau A, Hinz H, Gonzalez-Cid Y. Jellytoring: real-time jellyfish monitoring based on deep learning object detection. Sensors. 2020;20:1708. doi: 10.3390/s20061708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martone M, Rizzoli P, Wecklich C, González C, Bueso-Bello J-L, Valdo P, Schulze D, Zink M, Krieger G, Moreira A. The global forest/non-forest map from TandEM-X interferometric SAR data. Remote Sens Environ. 2018;205:352–373. doi: 10.1016/j.rse.2017.12.002. [DOI] [Google Scholar]
- Matasci G, Hermosilla T, Wulder MA, White JC, Coops NC, Hobart GW, Bolton DK, Tompalski P, Bater CW. Three decades of forest structural dynamics over Canada’s forested ecosystems using Landsat time-series and lidar plots. Remote Sens Environ. 2018;216:697–714. doi: 10.1016/j.rse.2018.07.024. [DOI] [Google Scholar]
- McQuatters-Gollop A, Mitchell I, Vina-Herbon C, Bedford J, Addison PFE, Lynam CP, Geetha PN, Vermeulan EA, Smit K, Bayley DTI, Morris-Webb E, Niner HJ, Otto SA. From science to evidence – how biodiversity indicators can be used for effective marine conservation policy and management. Front Mar Sci. 2019;6:109. doi: 10.3389/fmars.2019.00109. [DOI] [Google Scholar]
- Meena B, Singh N, Mahar KS, Sharma YK, Rana TS. Molecular analysis of genetic diversity and population genetic structure in Ephedra foliata: an endemic and threatened plant species of arid and semi-arid regions of India. Physiol Mol Biol Plants: An International Journal of Functional Plant Biology. 2019;25:753–764. doi: 10.1007/s12298-019-00648-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehring M, Mehlhaus N, Ott E, Hummel D. A systematic review of biodiversity and demographic change: a misinterpreted relationship? Ambio. 2020;49:1297–1312. doi: 10.1007/s13280-019-01276-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei Z, Khan MA, Zhang X, Fu J. Rapid and accurate genetic authentication of Penthorum chinense by improved RAPD-derived species-specific SCAR markers. Biodivers J Biol Diver. 2017;18:1243–1249. doi: 10.13057/biodiv/d180349. [DOI] [Google Scholar]
- Meikasari NS, Nurilmala M, Butet NA, Sudrajat AO. PCR-RFLP as a detection method of allelic diversity seahorse Hippocampus comes (Cantor, 1849) from Bintan waters, Riau Island. IOP Conf Series: Earth Environ Sci. 2019;404:012046. [Google Scholar]
- Mercier A, Betbeder J, Baudry J, Le Roux V, Spicher F, Lacoux J, Roger D, Hubert-Moy L. Evaluation of Sentinel-1 and 2 time series for predicting wheat and rapeseed phenological stages. ISPRS J Photogramm Remote Sens. 2020;163:231–256. doi: 10.1016/j.isprsjprs.2020.03.009. [DOI] [Google Scholar]
- Miller W, Hayes VM, Ratan A, Petersen DC, Wittekindt NE, Miller J, Walenz B, Knight J, Qi J, Zhao F, Wang Q, Bedoya-Reina OC, Katiyar N, Tomsho LP, Kasson LM, Hardie R-A, Woodbridge P, Tindall EA, Bertelsen MF, Dixon D, Pyecroft S, Helgen KM, Lesk AM, Pringle TH, Patterson N, Zhang Y, Kreiss A, Woods GM, Jones ME, Schuster SC. Genetic diversity and population structure of the endangered marsupial Sarcophilus harrisii (Tasmanian devil) Proc Natl Acad Sci. 2011;108:12348–12353. doi: 10.1073/pnas.1102838108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh NTA, Van TT, Hau HV, Trieu LN, Tien CV, Vinh TT, Van DN. Genetic diversity and variation of Huperzia serrata (Thunb. ex Murray) Trevis. population in Vietnam revealed by ISSR and SCoT markers. Biotechnol Biotechnol Equip. 2019;33:1525–1534. doi: 10.1080/13102818.2019.1671896. [DOI] [Google Scholar]
- Mir AH, Tyub S, Kamili AN. Ecology, distribution mapping and conservation implications of four critically endangered endemic plants of Kashmir Himalaya. Saudi J Biol Sci. 2020;27:2380–2389. doi: 10.1016/j.sjbs.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miraldo A, Li S, Borregaard MK, Flórez-Rodríguez A, Gopalakrishnan S, Rizvanovic M, Wang Z, Rahbek C, Marske KA, Nogués-Bravo D. An anthropocene map of genetic diversity. Science. 2016;353:1532–1535. doi: 10.1126/science.aaf4381. [DOI] [PubMed] [Google Scholar]
- Molerović N, Rašković B, Đedović R, Andrić OD, Marković Z, Marić S. Characterization of the genetic structure of the brown trout (Salmo trutta) from “Braduljica” fish farm, Serbia. Biotechnol Anim Husband. 2019;35:289–299. doi: 10.2298/BAH1903289M. [DOI] [Google Scholar]
- Mosa KA, Gairola S, Jamdade R, El-Keblawy A, Al Shaer KI, Al Harthi EK, Shabana HA, Mahmoud T. The promise of molecular and genomic techniques for biodiversity research and DNA barcoding of the arabian peninsula flora. Front Plant Sci. 2019;9:1929. doi: 10.3389/fpls.2018.01929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudgineer O jpg: *derivative work: (2011) Oil drilling rig, simple illustration. https://commons.wikimedia.org/wiki/File:Oil_Rig_NT8.svg. Accessed 9 Aug 2021
- Naczk AM and Ziętara MS (2019) Genetic diversity in Dactylorhiza majalis subsp. majalis populations (Orchidaceae) of northern Poland. Nordic J Bot 37. 10.1111/njb.01989 [DOI]
- Naeem S, Prager C, Weeks B, Varga A, Flynn DFB, Griffin K, Muscarella R, Palmer M, Wood S, Schuster W. Biodiversity as a multidimensional construct: a review, framework and case study of herbivory’s impact on plant biodiversity. Proc Biol Sci. 2016;283:20153005. doi: 10.1098/rspb.2015.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najafzadeh R, Arzani K, Bouzari N, Saei A. Genetic diversity assessment and identification of new sour cherry genotypes using intersimple sequence repeat markers. Int J Biodivers. 2014;2o14:308398. [Google Scholar]
- Neiber MT, Cianfanelli S, Bartolini F, Glaubrecht M. Not a marginal loss: genetic diversity of the endangered freshwater snail Melanopsis etrusca (Brot, 1862) from thermal springs in Tuscany, Italy. Conserv Genet. 2020;21:199–216. doi: 10.1007/s10592-019-01241-w. [DOI] [Google Scholar]
- Neugarten RA, Langhammer PF, Osipova E et al (2018) Tools for measuring, modelling, and valuing ecosystem services: guidance for key biodiversity areas, natural world heritage sites, and protected areas, 1st edn. (ed. by Groves C) IUCN, International Union for Conservation of Nature, USA
- Neugarten RA, Moull K, Martinez NA, Andriamaro L, Bernard C, Bonham C, Cano CA, Ceotto P, Cutter P, Farrell TA, Gibb M, Goedschalk J, Hole D, Honzák M, Kasecker T, Koenig K, Larsen TH, Ledezma JC, McKinnon M, Mulligan M, Nijbroek R, Olsson A, Rakotobe ZL, Rasolohery A, Saenz L, Steininger M, Wright TM, Turner W. Trends in protected area representation of biodiversity and ecosystem services in five tropical countries. Ecosyst Serv. 2020;42:101078. doi: 10.1016/j.ecoser.2020.101078. [DOI] [Google Scholar]
- Newbold T, Hudson LN, Hill SLL, Contu S, Lysenko I, Senior RA, Börger L, Bennett DJ, Choimes A, Collen B, Day J, De Palma A, Díaz S, Echeverria-Londoño S, Edgar MJ, Feldman A, Garon M, Harrison MLK, Alhusseini T, Ingram DJ, Itescu Y, Kattge J, Kemp V, Kirkpatrick L, Kleyer M, Correia DLP, Martin CD, Meiri S, Novosolov M, Pan Y, Phillips HRP, Purves DW, Robinson A, Simpson J, Tuck SL, Weiher E, White HJ, Ewers RM, Mace GM, Scharlemann JPW, Purvis A. Global effects of land use on local terrestrial biodiversity. Nature. 2015;520:45–50. doi: 10.1038/nature14324. [DOI] [PubMed] [Google Scholar]
- Ng WL, Tan SG. Inter-Simple Sequence Repeat (ISSR) markers: are we doing it right? ASM Sci J. 2015;9:30–39. [Google Scholar]
- Omasa K, Hosoi F, Konishi A. 3D lidar imaging for detecting and understanding plant responses and canopy structure. J Exp Bot. 2007;58:881–898. doi: 10.1093/jxb/erl142. [DOI] [PubMed] [Google Scholar]
- Oon A, Ngo KD, Azhar R, Ashton-Butt A, Lechner AM, Azhar B. Assessment of ALOS-2 PALSAR-2L-band and Sentinel-1 C-band SAR backscatter for discriminating between large-scale oil palm plantations and smallholdings on tropical peatlands. Remote Sens Appl: Soc Environ. 2019;13:183–190. [Google Scholar]
- Organikos (2012) Thermal imaging, elephant listening. In: Organikos. https://organikos.net/2012/09/23/thermalimaging-elephant-listening/. Accessed 9 Aug 2021
- Our World In Data (2020) Forest area as share of land area, Source: UN Food and Agriculture Organization (FAO). https://commons.wikimedia.org/wiki/File:Forest_area_as_share_of_land_area,_OWID.svg. Accessed 9 Aug 2021
- Özdil F, İlhan F, Işık R. Genetic characterization of some Turkish sheep breeds based on the sequencing of the Ovar-DRB1 gene in the major histocompatibility complex (MHC) gene region. Arch Anim Breed. 2018;61:475–480. doi: 10.5194/aab-61-475-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özdil F, Bulut H, Işık R. Genetic diversity of κ-casein (CSN3) and lactoferrin (LTF) genes in the endangered Turkish donkey (Equus asinus) populations. Arch Anim Breed. 2019;62:77–82. doi: 10.5194/aab-62-77-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panicz R, Napora-Rutkowski Ł, Keszka S, Skuza L, Szenejko M, Śmietana P. Genetic diversity in natural populations of noble crayfish (Astacus astacus L.) in north-western Poland on the basis of combined SSR and AFLP data. PeerJ. 2019;7:e7301. doi: 10.7717/peerj.7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi S, Velraj P, Subba Rao T. Chapter 21 - Functional microbial diversity in contaminated environment and application in bioremediation. In: Das S, Dash HR, editors. Microbial Diversity in the Genomic Era. London: Academic Press; 2019. pp. 359–385. [Google Scholar]
- Parrens M, Bitar AA, Frappart F, Paiva R, Wongchuig S, Papa F, Yamasaki D, Kerr Y. High resolution mapping of inundation area in the Amazon basin from a combination of L-band passive microwave, optical and radar datasets. Int J Appl Earth Obs Geoinf. 2019;81:58–71. [Google Scholar]
- Parthiban S, Govindaraj P, Senthilkumar S. Comparison of relative efficiency of genomic SSR and EST-SSR markers in estimating genetic diversity in sugarcane. 3. Biotech. 2018;8:144. doi: 10.1007/s13205-018-1172-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pixabay (2021) 110,000+ free vector stock art images, hand selected - pixabay. https://pixabay.com/vectors/. Accessed 11 Aug 2021
- Pngtree (2021) Millions of PNG images, backgrounds and vectors for free download. In: Pngtree. href='https://pngtree.com/so/shouting-horn'> shouting horn png from pngtree.com. Accessed 2 Aug 2021
- Portree D (2006) A diagram showing the orbital configuration of an Almaz radar satellite, a type of Soviet reconnaissance satellite based on the Almaz OPS space stations. https://commons.wikimedia.org/wiki/File:Almaz_radar_satellite.svg. Accessed 9 Aug 2021
- Prošek J, Gdulová K, Barták V, Vojar J, Solský M, Rocchini D, Moudrý V. Integration of hyperspectral and LiDAR data for mapping small water bodies. Int J Appl Earth Obs Geoinf. 2020;92:102181. [Google Scholar]
- Qamer S, Al-Abbadi AA, Sajid M, Asad F, Khan MF, Khan NA, Sthanadar AA, Akhtar MN, Mahmoud AH, Mohammed OB. Genetic analysis of honey bee, Apis dorsata populations using random amplified polymorphic DNA (RAPD) markers. J King Saud Univ - Sci. 2021;33:101218. doi: 10.1016/j.jksus.2020.10.015. [DOI] [Google Scholar]
- Qi W, Saarela S, Armston J, Ståhl G, Dubayah R. Forest biomass estimation over three distinct forest types using TandEM-X InSAR data and simulated GEDI lidar data. Remote Sens Environ. 2019;232:111283. doi: 10.1016/j.rse.2019.111283. [DOI] [Google Scholar]
- Qiao Y, Guo F, Huo N, Zhan L, Sun J, Zuo X, Guo Z, Gu YQ, Wang Y, Liu Y. Genotyping-by-sequencing to determine the genetic structure of a Tibetan medicinal plant Swertia mussotii Franch. Genet Resour Crop Evol. 2021;68:469–484. doi: 10.1007/s10722-020-00993-6. [DOI] [Google Scholar]
- Rajah P, Odindi J, Mutanga O. Feature level image fusion of optical imagery and Synthetic Aperture Radar (SAR) for invasive alien plant species detection and mapping. Remote Sens Appl: Soc Environ. 2018;10:198–208. [Google Scholar]
- Randin CF, Ashcroft MB, Bolliger J, Cavender-Bares J, Coops NC, Dullinger S, Dirnböck T, Eckert S, Ellis E, Fernández N, Giuliani G, Guisan A, Jetz W, Joost S, Karger D, Lembrechts J, Lenoir J, Luoto M, Morin X, Price B, Rocchini D, Schaepman M, Schmid B, Verburg P, Wilson A, Woodcock P, Yoccoz N, Payne D. Monitoring biodiversity in the Anthropocene using remote sensing in species distribution models. Remote Sens Environ. 2020;239:111626. doi: 10.1016/j.rse.2019.111626. [DOI] [Google Scholar]
- Rappaport DI, Royle JA, Morton DC. Acoustic space occupancy: combining ecoacoustics and lidar to model biodiversity variation and detection bias across heterogeneous landscapes. Ecol Indic. 2020;113:106172. doi: 10.1016/j.ecolind.2020.106172. [DOI] [Google Scholar]
- Ray A, Jena S, Haldar T, Sahoo A, Kar B, Patnaik J, Ghosh B, Chandra Panda P, Mahapatra N, Nayak S. Population genetic structure and diversity analysis in Hedychium coronarium populations using morphological, phytochemical and molecular markers. Ind Crop Prod. 2019;132:118–133. doi: 10.1016/j.indcrop.2019.02.015. [DOI] [Google Scholar]
- Reeth CV, Michel N, Bockstaller C, Caro G. Influences of oilseed rape area and aggregation on pollinator abundance and reproductive success of a co-flowering wild plant. Agric Ecosyst Environ. 2019;280:35–42. doi: 10.1016/j.agee.2019.04.025. [DOI] [Google Scholar]
- Reid AJ, Carlson AK, Creed IF, Eliason EJ, Gell PA, Johnson PTJ, Kidd KA, MacCormack TJ, Olden JD, Ormerod SJ, Smol JP, Taylor WW, Tockner K, Vermaire JC, Dudgeon D, Cooke SJ. Emerging threats and persistent conservation challenges for freshwater biodiversity. Cambridge Philos Soc. 2019;94:849–873. doi: 10.1111/brv.12480. [DOI] [PubMed] [Google Scholar]
- Ridley FA, McGowan PJ, Mair L. The scope and extent of literature that maps threats to species: a systematic map protocol. Environ Evid. 2020;9:23. doi: 10.1186/s13750-020-00206-8. [DOI] [Google Scholar]
- Righi T, Splendiani A, Fioravanti T, Petetta A, Candelma M, Gioacchini G, Gillespie K, Hanke A, Carnevali O, Caputo Barucchi V. Mediterranean swordfish (Xiphias gladius Linnaeus, 1758) population structure revealed by microsatellite DNA: genetic diversity masked by population mixing in shared areas. PeerJ. 2020;8:e9518. doi: 10.7717/peerj.9518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Peña RA, Johnson RL, Johnson LA, Anderson CD, Ricks NJ, Farley KM, Robbins MD, Wolfe AD, Stevens MR. Investigating the genetic diversity and differentiation patterns in the Penstemon scariosus species complex under different sample sizes using AFLPs and SSRs. Conserv Genet. 2018;19:1335–1348. doi: 10.1007/s10592-018-1103-6. [DOI] [Google Scholar]
- Rudd S. Expressed sequence tags: alternative or complement to whole genome sequences? Trends Plant Sci. 2003;8:321–329. doi: 10.1016/S1360-1385(03)00131-6. [DOI] [PubMed] [Google Scholar]
- Saikia M, Devi D. Analysis of genetic diversity and phylogeny of Philosamia ricini (Lepidoptera: Saturniidae) by using RAPD and internal transcribed spacer DNA1. Mol Biol Rep. 2019;46:3035–3048. doi: 10.1007/s11033-019-04740-8. [DOI] [PubMed] [Google Scholar]
- Saikia M, Haloi K, Nath R, Devi D. Genetic diversity among the morphs of Antheraea assamensis Helfer: study using RAPD and internal transcribed spacer DNA1. Indian J Exp Biol. 2019;57:418–426. [Google Scholar]
- Sairkar PK, Sharma A, Shukla NP (2016) SCAR marker for identification and discrimination of Commiphora wightii and C. myrrha. Mol Biol Int 2016:1482796. 10.1155/2016/1482796 [DOI] [PMC free article] [PubMed]
- Saleem MH, Potgieter J, Arif KM. Plant disease detection and classification by deep learning. Plants. 2019;8:468. doi: 10.3390/plants8110468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleem MH, Potgieter J, Arif KM. Plant disease classification: a comparative evaluation of convolutional neural networks and deep learning optimizers. Plants. 2020;9:1319. doi: 10.3390/plants9101319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehi F, Ahmadian L. The application of geographic information systems (GIS) in identifying the priority areas for maternal care and services. BMC Health Serv Res. 2017;17:482. doi: 10.1186/s12913-017-2423-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarwat M. ISSR: a reliable and cost-effective technique for detection of DNA polymorphism. In: Sucher NJ, Hennell JR, Carles MC, editors. Plant DNA Fingerprinting and Barcoding: Methods and Protocols Methods in Molecular Biology. Totowa: Humana Press; 2012. pp. 103–121. [DOI] [PubMed] [Google Scholar]
- Schägner JP, Brander L, Maes J, Hartje V. Mapping ecosystem services’ values: current practice and future prospects. Ecosyst Serv. 2013;4:33–46. doi: 10.1016/j.ecoser.2013.02.003. [DOI] [Google Scholar]
- Schlund M, Erasmi S. Sentinel-1 time series data for monitoring the phenology of winter wheat. Remote Sens Environ. 2020;246:111814. doi: 10.1016/j.rse.2020.111814. [DOI] [Google Scholar]
- Scionti F, Di Martino MT, Pensabene L, Bruni V, Concolino D. The cytoscan HD array in the diagnosis of neurodevelopmental disorders. High-Throughput. 2018;7:E28. doi: 10.3390/ht7030028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedano F, Lisboa S, Duncanson L, Ribeiro N, Sitoe A, Sahajpal R, Hurtt G, Tucker C. Monitoring intra and inter annual dynamics of forest degradation from charcoal production in Southern Africa with Sentinel – 2 imagery. Int J Appl Earth Obs Geoinf. 2020;92:102184. [Google Scholar]
- Selvaraj MG, Vergara A, Montenegro F, Alonso Ruiz H, Safari N, Raymaekers D, Ocimati W, Ntamwira J, Tits L, Omondi AB, Blomme G. Detection of banana plants and their major diseases through aerial images and machine learning methods: a case study in DR Congo and Republic of Benin. ISPRS J Photogramm Remote Sens. 2020;169:110–124. doi: 10.1016/j.isprsjprs.2020.08.025. [DOI] [Google Scholar]
- Senn HV, Ghazali M, Kaden J, Barclay D, Harrower B, Campbell RD, Macdonald DW, Kitchener AC. Distinguishing the victim from the threat: SNP-based methods reveal the extent of introgressive hybridization between wildcats and domestic cats in Scotland and inform future in situ and ex situ management options for species restoration. Evol Appl. 2019;12:399–414. doi: 10.1111/eva.12720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sereshkeh FM, Azizi A, Noroozisharaf A. Structure of genetic diversity among and within populations of the endemic Iranian plant Dracocephalum kotschyi. Hortic Environ Biotechnol. 2019;60:767–777. doi: 10.1007/s13580-019-00149-1. [DOI] [Google Scholar]
- Shen L, Li X-W, Meng X-X, Wu J, Tang H, Huang L-F, Xiao S-M, Xu J, Chen S-L. Prediction of the globally ecological suitability of Panax quinquefolius by the geographic information system for global medicinal plants (GMPGIS) Chin J Nat Med. 2019;17:481–489. doi: 10.1016/S1875-5364(19)30069-X. [DOI] [PubMed] [Google Scholar]
- Silvestro D, Goria S, Sterner T, Antonelli A. Improving biodiversity protection through artificial intelligence. Nat Sustain. 2022;5:415–424. doi: 10.1038/s41893-022-00851-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slagter B, Tsendbazar N-E, Vollrath A, Reiche J. Mapping wetland characteristics using temporally dense Sentinel-1 and Sentinel-2 data: a case study in the St. Lucia wetlands, South Africa. Int J Appl Earth Obs Geoinf. 2020;86:102009. [Google Scholar]
- Smithsonian’s National Zoo and Conservation Biology Institute Smithsonian’s National Zoo (2016) Asian elephant. In: Smithsonian’s National Zoo. https://nationalzoo.si.edu/animals/asian-elephant. Accessed 9 Aug 2021
- Srikanth K, Kim N-Y, Park W, Kim J-M, Kim K-D, Lee K-T, Son J-H, Chai H-H, Choi J-W, Jang G-W, Kim H, Ryu Y-C, Nam J-W, Park J-E, Kim J-M, Lim D. Comprehensive genome and transcriptome analyses reveal genetic relationship, selection signature, and transcriptome landscape of small-sized Korean native Jeju horse. Sci Rep. 2019;9:16672. doi: 10.1038/s41598-019-53102-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava PK, Malhi RKM, Pandey PC, Anand A, Singh P, Pandey MK, Gupta A. 1 - Revisiting hyperspectral remote sensing: origin, processing, applications and way forward. In: Pandey PC, Srivastava PK, Balzter H, Bhattacharya B, Petropoulos GP, editors. Hyperspectral Remote Sensing Earth Observation. Amsterdam: Elsevier; 2020. pp. 3–21. [Google Scholar]
- Stephenson PJ. Technological advances in biodiversity monitoring: applicability, opportunities and challenges. Curr Opin Environ Sustain. 2020;45:36–41. doi: 10.1016/j.cosust.2020.08.005. [DOI] [Google Scholar]
- Sulistyahadi FN, Puspitasari IGAAR, Nuryanto A. Diversity analysis of Rhacophorus margaritifer (Schlegel, 1837) in Baturraden based on RAPD markers. J Trop Biodiversi Biotechnol. 2020;5:44–52. doi: 10.22146/jtbb.49110. [DOI] [Google Scholar]
- Sun Y-W, Hou N, Woeste K, Zhang C, Yue M, Yuan X-Y, Zhao P. Population genetic structure and adaptive differentiation of iron walnut Juglans regia subsp. sigillata in southwestern China. Ecol Evol. 2019;9:14154–14166. doi: 10.1002/ece3.5850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supple MA, Shapiro B. Conservation of biodiversity in the genomics era. Genome Biol. 2018;19:131. doi: 10.1186/s13059-018-1520-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanase MA, Villard L, Pitar D, Apostol B, Petrila M, Chivulescu S, Leca S, Borlaf-Mena I, Pascu I-S, Dobre A-C, Pitar D, Guiman G, Lorent A, Anghelus C, Ciceu A, Nedea G, Stanculeanu R, Popescu F, Aponte C, Badea O. Synthetic aperture radar sensitivity to forest changes: a simulations-based study for the Romanian forests. Sci Total Environ. 2019;689:1104–1114. doi: 10.1016/j.scitotenv.2019.06.494. [DOI] [PubMed] [Google Scholar]
- Tani N, Kawahara T, Yoshimaru H, Hoshi Y. Development of SCAR markers distinguishing pure seedlings of the endangered species Morus boninensis from M. boninensis × M. acidosa hybrids for conservation in Bonin (Ogasawara) Islands. Conserv Genet. 2003;4:605–612. doi: 10.1023/A:1025655331429. [DOI] [Google Scholar]
- Tarazona Y, Miyasiro-López M. Monitoring tropical forest degradation using remote sensing. Challenges and opportunities in the Madre de Dios region, Peru. Remote Sens Appl: Soc Environ. 2020;19:100337. [Google Scholar]
- Tashayo B, Honarbakhsh A, Akbari M, Eftekhari M. Land suitability assessment for maize farming using a GIS-AHP method for a semi- arid region, Iran. J Saudi Soc Agric Sci. 2020;19(5):332–338. [Google Scholar]
- Teobaldelli M, Cona F, Saulino L, Migliozzi A, D’Urso G, Langella G, Manna P, Saracino A. Detection of diversity and stand parameters in Mediterranean forests using leaf-off discrete return LiDAR data. Remote Sens Environ. 2017;192:126–138. doi: 10.1016/j.rse.2017.02.008. [DOI] [Google Scholar]
- The RAFOS group at the Graduate School of Oceanography, University of Rhode Island, Kingston, RI 02881 (2001) The ideal signal received from moored SOFAR emitters and several recorded signals from the float. The arrival time can be measured very accurately. https://commons.wikimedia.org/wiki/File:Sound_wave_Correlation.jpg. Accessed 11 Aug 2021
- Thessen A. Adoption of machine learning techniques in ecology and earth science. One Ecosyst. 2016;1:e8621. doi: 10.3897/oneeco.1.e8621. [DOI] [Google Scholar]
- Thuy MTP, Ha TTT, Quang TH. Analysis of genetic diversity in Pa Co pine (Pinus kwangtungensis Chun ex Tsiang) using RAPD and ISSR markers. Vietnam J Sci Technol Eng. 2020;62:62–68. doi: 10.31276/VJSTE.62(1).62-68. [DOI] [Google Scholar]
- Tilman D, Isbell F, Cowles JM. Biodiversity and ecosystem functioning. Annu Rev Ecol Evol Syst. 2014;45:471–493. doi: 10.1146/annurev-ecolsys-120213-091917. [DOI] [Google Scholar]
- Tiwari V, Meena B, Nair NK, Rana TS. Molecular analyses of genetic variability in the populations of Bergenia ciliata in Indian Himalayan Region (IHR) Physiol Mol Biol Plants: An International Journal of Functional Plant Biology. 2020;26:975–984. doi: 10.1007/s12298-020-00797-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torresani M, Rocchini D, Sonnenschein R, Zebisch M, Hauffe HC, Heym M, Pretzsch H, Tonon G. Height variation hypothesis: a new approach for estimating forest species diversity with CHM LiDAR data. Ecol Indic. 2020;117:106520. doi: 10.1016/j.ecolind.2020.106520. [DOI] [Google Scholar]
- Touma S, Arakawa A, Oikawa T. Evaluation of the genetic structure of indigenous Okinawa Agu pigs using microsatellite markers. Asian Australas J Anim Sci. 2019;33:212–218. doi: 10.5713/ajas.19.0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuisima-Coral LL, Hlásná Čepková P, Weber JC, Lojka B. Preliminary evidence for domestication effects on the genetic diversity of Guazuma crinita in the Peruvian Amazon. Forests. 2020;11:795. doi: 10.3390/f11080795. [DOI] [Google Scholar]
- Valbuena R, O’Connor B, Zellweger F, Simonson W, Vihervaara P, Maltamo M, Silva CA, Almeida DRA, Danks F, Morsdorf F, Chirici G, Lucas R, Coomes DA, Coops NC. Standardizing ecosystem morphological traits from 3D information sources. Trends Ecol Evol. 2020;35(8):656–667. doi: 10.1016/j.tree.2020.03.006. [DOI] [PubMed] [Google Scholar]
- Vallejos-Vidal E, Reyes-Cerpa S, Rivas-Pardo JA, Maisey K, Yáñez JM, Valenzuela H, Cea PA, Castro-Fernandez V, Tort L, Sandino AM, Imarai M, Reyes-López FE. Single-nucleotide polymorphisms (SNP) mining and their effect on the tridimensional protein structure prediction in a set of immunity-related expressed sequence tags (EST) in Atlantic salmon (Salmo salar) Front Genet. 2019;10:1406. doi: 10.3389/fgene.2019.01406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Broeck T, Joniau S, Clinckemalie L, Helsen C, Prekovic S, Spans L, Tosco L, Van Poppel H, Claessens F. The role of single nucleotide polymorphisms in predicting prostate cancer risk and therapeutic decision making. Biomed Res Int. 2014;2014:627510. doi: 10.1155/2014/627510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaux F, Aycock HM, Bohn S, Rasmuson LK, O’Malley KG. Sex identification PCR–RFLP assay tested in eight species of Sebastes rockfish. Conserv Genet Resour. 2020;12:541–544. doi: 10.1007/s12686-020-01150-y. [DOI] [Google Scholar]
- Vecteezy (2021a) Set of wild animal. In: Vecteezy.com. href="https://www.vecteezy.com/freevector/vector"> Vector Vectors by Vecteezy. Accessed 25 May 2021
- Vecteezy (2021b) Forest scene with tall trees. In: Vecteezy.com. https://www.vecteezy.com/vector-art/298788-forestscene-with-tall-trees. Accessed 25 May 2021
- Vergnaud G, Denoeud F. Minisatellites: mutability and genome architecture. Genome Res. 2000;10:899–907. doi: 10.1101/gr.10.7.899. [DOI] [PubMed] [Google Scholar]
- Verma GK, Gupta P. Wild animal detection using deep convolutional neural network. In: Chaudhuri BB, Kankanhalli MS, Raman B, editors. Proceedings of 2nd International Conference on Computer Vision and Image Processing Advances in Intelligent Systems and Computing. Singapore: Springer; 2018. pp. 327–338. [Google Scholar]
- Vidaña-Vila E, Navarro J, Alsina-Pagès RM, Ramírez Á. A two-stage approach to automatically detect and classify woodpecker (Fam. Picidae) sounds. Appl Acoust. 2020;166:107312. doi: 10.1016/j.apacoust.2020.107312. [DOI] [Google Scholar]
- Vieira MLC, Santini L, Diniz AL, de Freitas Munhoz C. Microsatellite markers: what they mean and why they are so useful. Genet Mol Biol. 2016;39:312–328. doi: 10.1590/1678-4685-GMB-2016-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignal A, Milan D, SanCristobal M, Eggen A. A review on SNP and other types of molecular markers and their use in animal genetics. Gen Select Evol: GSE. 2002;34:275–305. doi: 10.1186/1297-9686-34-3-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogeler JC, Cohen WB. A review of the role of active remote sensing and data fusion for characterizing forest in wildlife habitat models. Span J Remote Sens. 2016;45:1–14. [Google Scholar]
- Wagutu GK, Fan X-R, Njeri HK, Wen X-Y, Liu Y-L, Chen Y-Y. Development and characterization of EST-SSR markers for the endangered tree Magnolia patungensis (Magnoliaceae) Ann Bot Fenn. 2020;57:97–107. doi: 10.5735/085.057.0114. [DOI] [Google Scholar]
- Wang R, Gamon JA. Remote sensing of terrestrial plant biodiversity. Remote Sens Environ. 2019;231:111218. doi: 10.1016/j.rse.2019.111218. [DOI] [Google Scholar]
- Wang J, Xiao X, Qin Y, Doughty RB, Dong J, Zou Z. Characterizing the encroachment of juniper forests into sub-humid and semi-arid prairies from 1984 to 2010 using PALSAR and Landsat data. Remote Sens Environ. 2018;205:166–179. doi: 10.1016/j.rse.2017.11.019. [DOI] [Google Scholar]
- Wang J, Xiao X, Bajgain R, Starks P, Steiner J, Doughty RB, Chang Q. Estimating leaf area index and aboveground biomass of grazing pastures using Sentinel-1, Sentinel-2 and Landsat images. ISPRS J Photogramm Remote Sens. 2019;154:189–201. doi: 10.1016/j.isprsjprs.2019.06.007. [DOI] [Google Scholar]
- Wang X, Chen W, Luo J, Yao Z, Yu Q, Wang Y, Zhang S, Liu Z, Zhang M, Shen Y. Development of EST-SSR markers and their application in an analysis of the genetic diversity of the endangered species Magnolia sinostellata. Mol Gen Genomics: MGG. 2019;294:135–147. doi: 10.1007/s00438-018-1493-7. [DOI] [PubMed] [Google Scholar]
- Wang L, Deng H, Qiu X, Wang P, Yang F. Determining the impact of key climatic factors on geographic distribution of wild Akebia trifoliata. Ecol Indic. 2020;112:106093. doi: 10.1016/j.ecolind.2020.106093. [DOI] [Google Scholar]
- Wetzel FT, Saarenmaa H, Regan E, Martin CS, Mergen P, Smirnova L, Tuama ÉÓ, Camacho FAG, Hoffmann A, Vohland K, Häuser CL. The roles and contributions of Biodiversity Observation Networks (BONs) in better tracking progress to 2020 biodiversity targets: a European case study. Biodiversity. 2015;16:137–149. doi: 10.1080/14888386.2015.1075902. [DOI] [Google Scholar]
- Whitehorn PR, Navarro LM, Schröter M, Fernandez M, Rotllan-Puig X, Marques A. Mainstreaming biodiversity: a review of national strategies. Biol Conserv. 2019;235:157–163. doi: 10.1016/j.biocon.2019.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte A, Ferentinos KP, Petropoulos GP. A new synergistic approach for monitoring wetlands using Sentinels-1 and 2 data with object-based machine learning algorithms. Environ Model Softw. 2018;104:40–54. doi: 10.1016/j.envsoft.2018.01.023. [DOI] [Google Scholar]
- Willi M, Pitman RT, Cardoso AW, Locke C, Swanson A, Boyer A, Veldthuis M, Fortson L (2018) Software, data and models used in “Identifying animal species in camera trap images using deep learning and citizen science
- Willi M, Pitman RT, Cardoso AW, Locke C, Swanson A, Boyer A, Veldthuis M, Fortson L. Identifying animal species in camera trap images using deep learning and citizen science. Methods Ecol Evol. 2019;10:80–91. doi: 10.1111/2041-210X.13099. [DOI] [Google Scholar]
- Wu K, Rodriguez GA, Zajc M, Jacquemin E, Clément M, De Coster A, Lambot S. A new drone-borne GPR for soil moisture mapping. Remote Sens Environ. 2019;235:111456. doi: 10.1016/j.rse.2019.111456. [DOI] [Google Scholar]
- Wu W-D, Liu W-H, Sun M, Zhou J-Q, Liu W, Zhang C-L, Zhang X-Q, Peng Y, Huang L-K, Ma X. Genetic diversity and structure of Elymus tangutorum accessions from western China as unraveled by AFLP markers. Hereditas. 2019;156:8. doi: 10.1186/s41065-019-0082-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia D, Chen P, Wang B, Zhang J, Xie C. Insect detection and classification based on an improved convolutional neural network. Basel: Sensors; 2018. p. 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia W, Luo T, Zhang W, Mason AS, Huang D, Huang X, Tang W, Dou Y, Zhang C, Xiao Y. Development of high density SNP markers and their application in evaluating genetic diversity and population structure in Elaeis guineensis. Front Plant Sci. 2019;10:130. doi: 10.3389/fpls.2019.00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Lei P, Jiang M, Sang L, Guan F, Meng F, Quan H. Genetic diversity of Herpetospermum caudigerum (Ser.) Baill using AFLP and chloroplast microsatellites. Biotechnol Biotechnol Equip. 2019;33:1260–1268. doi: 10.1080/13102818.2019.1642798. [DOI] [Google Scholar]
- Yang L, Khan MA, Mei Z, Yang M, Zhang T, Wei C, Yang W, Zhu L, Long Y, Fu J. Development of RAPD-SCAR markers for Lonicera japonica (Caprifolicaceae) variety authentication by improved RAPD and DNA cloning. Revista De Biol Trop. 2014;62:1649–1657. doi: 10.15517/rbt.v62i4.13493. [DOI] [PubMed] [Google Scholar]
- Yin L and Zhou YM (2019) Life detection strategy based on infrared vision and ultra-wideband radar data fusion. Elect Eng Syst Sci 1–7
- Yuskianti V, Shiraishi S. Sequence characterized amplified region (SCAR) markers in Sengon (Paraseriathes falcataria (L.)) Nielsen. Hayati J Biosci. 2010;17:167–172. doi: 10.4308/hjb.17.4.167. [DOI] [Google Scholar]
- Zeng Z, Gan Y, Kettner AJ, Yang Q, Zeng C, Brakenridge GR, Hong Y. Towards high resolution flood monitoring: an integrated methodology using passive microwave brightness temperatures and Sentinel synthetic aperture radar imagery. J Hydrol. 2020;582:124377. doi: 10.1016/j.jhydrol.2019.124377. [DOI] [Google Scholar]
- Zhang C, Patras P, Haddadi H. Deep learning in mobile and wireless networking: a survey. IEEE Commun Surv Tutor. 2019;21:2224–2287. doi: 10.1109/COMST.2019.2904897. [DOI] [Google Scholar]
- Zhang P, Nascetti A, Ban Y, Gong M. An implicit radar convolutional burn index for burnt area mapping with Sentinel-1 C-band SAR data. ISPRS J Photogramm Remote Sens. 2019;158:50–62. doi: 10.1016/j.isprsjprs.2019.09.013. [DOI] [Google Scholar]
- Zhang W, Brandt M, Wang Q, Prishchepov AV, Tucker CJ, Li Y, Lyu H, Fensholt R. From woody cover to woody canopies: how Sentinel-1 and Sentinel-2 data advance the mapping of woody plants in savannas. Remote Sens Environ. 2019;234:111465. doi: 10.1016/j.rse.2019.111465. [DOI] [Google Scholar]
- Zhang Y, Ling F, Foody GM, Ge Y, Boyd DS, Li X, Du Y, Atkinson PM. Mapping annual forest cover by fusing PALSAR/PALSAR-2 and MODIS NDVI during 2007–2016. Remote Sens Environ. 2019;224:74–91. doi: 10.1016/j.rse.2019.01.038. [DOI] [Google Scholar]
- Zhang Y, Zhang M, Hu Y, Zhuang X, Xu W, Li P, Wang Z. Mining and characterization of novel EST-SSR markers of Parrotia subaequalis (Hamamelidaceae) from the first Illumina-based transcriptome datasets. PLoS One. 2019;14:e0215874. doi: 10.1371/journal.pone.0215874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Lan S, Chen WY, Chen X, Xu X, Chen Y, Dong J. Visual sensitivity versus ecological sensitivity: an application of GIS in urban forest park planning. Urban For Urban Green. 2019;41:139–149. doi: 10.1016/j.ufug.2019.03.010. [DOI] [Google Scholar]
- Zhu X, Hou Y, Weng Q, Chen L. Integrating UAV optical imagery and LiDAR data for assessing the spatial relationship between mangrove and inundation across a subtropical estuarine wetland. ISPRS J Photogramm Remote Sens. 2019;149:146–156. doi: 10.1016/j.isprsjprs.2019.01.021. [DOI] [Google Scholar]
- Zimmermann BL, De Vargas Machado JV, Santos S, Bartholomei-Santos ML. Genetic diversity of three aegla species (Decapoda, Anomura) revealed by AFLP and mtDNA markers. Crustaceana. 2019;19:445–462. doi: 10.1163/15685403-00003873. [DOI] [Google Scholar]
- Zizka A, Silvestro D, Vitt P, Knight TM. Automated conservation assessment of the orchid family with deep learning. Conserv Biol. 2020;35:897–908. doi: 10.1111/cobi.13616. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this article.