Highlights
► The Tetracore Orthopox BioThreat® Lateral Flow Assay was evaluated. ► The assay detected reproducibly laboratory grown vaccinia and monkeypox viruses containing 107 pfu/ml. ► In a blind study, 9 of 11 orthopoxvirus clinical samples were identified accurately. ► In a blind study, only 1 of 11 non-orthopoxvirus clinical samples (including varicella, pseudocowpox, Orf and HSV-1) produced a false positive result.
Keywords: Monkeypox, Orthopoxvirus, BioThreat, Diagnostic, Rapid
Abstract
The commercially available Orthopox BioThreat® Alert assay for orthopoxvirus (OPV) detection is piloted. This antibody-based lateral-flow assay labels and captures OPV viral agents to detect their presence. Serial dilutions of cultured Vaccinia virus (VACV) and Monkeypox virus (MPXV) were used to evaluate the sensitivity of the Tetracore assay by visual and quantitative determinations; specificity was assessed using a small but diverse set of diagnostically relevant blinded samples from viral lesions submitted for routine OPV diagnostic testing. The BioThreat® Alert assay reproducibly detected samples at concentrations of 107 pfu/ml for VACV and MPXV and positively identified samples containing 106 pfu/ml in 4 of 7 independent experiments. The assay correctly identified 9 of 11 OPV clinical samples and had only one false positive when testing 11 non-OPV samples. Results suggest applicability for use of the BioThreat® Alert assay as a rapid screening assay and point of care diagnosis for suspect human monkeypox cases.
1. Introduction
More than 30 years after the eradication of smallpox, Orthopoxviruses remain an emerging threat. In the past decade, zoonotic outbreaks of MPXV in central Africa and the US, cowpox virus in Europe, and VACV in Brazil highlight the emerging potential of these viruses (Centers for Disease Control and Prevention, 2003, Learned et al., 2005, Trindade et al., 2006, Vorou et al., 2008). Monkeypox is a zoonotic disease found in Central and West Africa that produces a pustular rash illness in humans. Although human contact with infected animals is the most common route of infection, extended human-to-human transmission chains can occur (Learned et al., 2005). Two genetic clades of MPXV have been characterized (Likos et al., 2005) and include the West African (mortality rate less than 1%) and Central African (up to 10% mortality) clades (Jezek et al., 1987). MPXV was also the cause of a 2003 outbreak in the United States. Captive black tailed prairie dogs transmitted the virus to humans after being co-housed with MPXV infected African rodents (Reed et al., 2004). Thirty-seven confirmed and ten probable human cases resulted (Reynolds et al., 2006). More recently an outbreak of monkeypox occurred in southern Sudan where MPXV disease had previously never been reported (Damon et al., 2006, Formenty et al., 2010). Historical outbreaks of monkeypox in the Congo basin and West Africa, along with the emergence of this disease in the US and southern Sudan emphasize the importance of MPXV as an emerging infectious agent of global scale.
In addition to a clinical appearance similar to that of smallpox, human monkeypox is often confused with other rash illnesses; up to 50% of suspected monkeypox cases in the Democratic Republic of Congo are actually Varicella-Zoster virus (VZV) infections (Jezek et al., 1988, Meyer et al., 2002). Early detection of OPV infection is important for making informed decisions regarding patient treatment, defining epidemiology of disease and implementing disease control measures. Current state of the art diagnoses of acute OPV infections largely utilize non-commercial PCR-based methods. Real-time PCR (RT-PCR) assays allow rapid detection and species level identification of multiple different OPVs in high-throughput, high-sensitivity formats (Kulesh et al., 2004, Li et al., 2006, Olson et al., 2004, Shchelkunov et al., 2011). In these assays, viral genomic material is isolated from swabs of lesions or from homogenized tissues prior to diagnostic testing. RT-PCR assays are highly sensitive and can detect as few as 10–100 genomes (Olson et al., 2004, Shchelkunov et al., 2011) and thus are state of the art for virus identification. However, requirements for skilled technicians, expensive instrumentation and strong laboratory controls to prevent cross-contamination limit the use of this technology in resource-poor areas such as rural Africa where rapid diagnosis of monkeypox would be beneficial.
Several alternative in-house assays have also been developed for diagnosis of OPV infection. A shell vial culture assay and direct fluorescent antibody (DFA) assay were tested with patient samples from recent VACV vaccines and detected virus as early as 4 days after vaccination (Fedorko et al., 2005). The DFA assay was the least sensitive (40% positive results OPX samples), but the most rapid and could be completed within an hour. The shell vial culture was sensitive to 89% of the samples tested but required up to 2 days to obtain results. By comparison, the RT-PCR assay used in that study detected virus in 100% of the samples tested. Both assays require fluorescent probes, expensive instrumentation and extensive laboratory equipment and expertise to perform. An in-house IgM ELISA (Karem et al., 2005) affords detection of antibody markers of recent OPV infection by examining humoral immune response to infection. It uses equipment found in most clinical laboratories, but requires laboratory space, specialized reagents, and technical expertise to perform the assay and interpret the results. It is also necessary to wait at least 5 days after rash onset to obtain predictive results.
Despite advances in nucleic acid diagnostic assays and serologic assays for post-exposure surveillance, rapid point-of-care OPV diagnostics remain limited. This limitation is particularly acute in rural Africa where the orthopox MPXV is endemic and resources for diagnostics are scarce. A rapid, simple, point-of-care diagnostic assay would eliminate many of these limitations, allowing local health workers to quickly confirm suspected monkeypox cases and enact measures to prevent further spread of the disease. The Tetracore Orthopox BioThreat Alert ® assay is the first commercially available lateral-flow based detection assay for OPV. Viral agent detection is accomplished through a combination of antibodies raised against VACV (O’Brien, 2012). One set of antibodies is conjugated with colloidal gold for labeling, while the second set of antibodies is bound at the detection line to capture labeled agents for visualization. Prior evaluation was performed at Plum Island with positive visual responses for Orthopoxviruses Horsepox and Camelpox, and negative visual responses for Suipoxvirus Swinepox, and Capripoxviruses Sheeppox and Goatpox (O’Brien, 2012). Serologic cross-reactivity between various OPV is well known (Mercer et al., 2007), and as such the manufacturer insert suggests the assay should be sensitive to MPXV. However, this has not been verified. The current study addresses sensitivity and specificity of the Tetracore assay for VACV and MPXV, and examines its potential as a method for diagnosing human monkeypox.
2. Materials and methods
2.1. Tetracore assay protocol
The Tetracore assay can be utilized with a variety of sample types including both solid and liquid samples. Manufacturer instructions for dry or solid material include directly resuspending tissue or swab samples in Tetracore sample buffer. Liquid samples are to be diluted in Tetracore assay sample buffer by at least 1:2 ratio. Instructions then detail the application of approximately 150 μl of sample to the strips, which re-hydrates test reagents, and allows binding of OPV agents by labeled antibody. Test results are to be read after 15 min. A positive result is indicated by the formation of a colored line at the sample line window. The control line contains antibodies directed against labeled antibody and verifies the test was run appropriately; this line should appear regardless of the presence or absence of OPV. The test is invalid if no control line appears. A sample test strip image is shown in Fig. 1 .
Fig. 1.
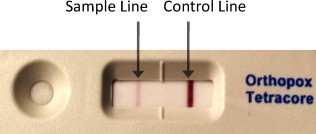
Tetracore Orthopox cassette image and strip layout showing application port, the sample test line for the orthopox detection, and a control line to indicate the test was successfully completed.
2.2. Samples utilized
2.2.1. Laboratory viral isolates
Crude Wyeth Dryvax® VACV and MPXV-ROC Congo Basin strain 2003-385 (Hutson et al., 2009) virus stocks were grown in BSC-40 cells for 2–3 days, purified from cellular debris by centrifugation, and resuspended in 10 mM Tris pH 9.0 (Likos et al., 2005). Purified virus was evaluated in a cell-culture plaque forming assay to determine pfu/ml. Briefly, extracted samples were applied to BSC-40 cell monolayers in 10-fold dilutions, incubated at 36 °C/6% CO2 for 48–72 h and subsequently fixed and stained with formalin and crystal violet to reveal plaques. Uninfected BSC-40 cells were processed through the virus purification procedure and collected for use as a control (BSC-40 lysate).
Crude VZV (Webster strain) was grown in human lung fibroblast (HLF) cells, lysed with radio immunoprecipitation assay (RIPA) buffer (50 mM Tris–HCl pH 8.0, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS), centrifuged to remove cellular debris, and the supernatant containing the virus was collected (Behrman et al., 2003). The virus was used without further characterization. Uninfected HLF cells were processed with this same protocol and used as a control (HLF lysate). Purified VZV (vaccine strain vOka) was also grown in HLF cells. Infected cells were rinsed with PBS, removed by agitation with glass beads in DMEM media supplemented with 10% sorbitol, and vortexed after transfer to a 50 ml conical tube. A low speed spin pelleted the glass beads and cellular debris. Supernatant was then further centrifuged to enhance purity (Rahaus et al., 2003, Schmidt and Lennette, 1975), then aliquoted and frozen at −70 °C. Virus titer was determined with an in-house colorimetric assay. Briefly, 48-well plates of HLF cells were infected with serial dilutions of the virus stock and fixed in 2% paraformaldehyde after 48 h. Wells were blocked with PBS containing 10% BSA, 0.5% FBS and 0.1% tween-20. Primary antibody (MAb to VZV gpII, Biodesign, Saco, ME) was diluted 1:1000 in blocking buffer and applied to each well for 1 h, followed by a 1:1000 diluted goat anti-mouse HRP (Invitrogen) for 30 min. BM Blue POD (Roche, Indianapolis, IN) was applied until color developed. Wells were washed with PBS to stop the reaction and color stabilization buffer (50 mM Tris pH 6.8, 100 mM NaCl, 1 mM EDTA) was added (Chen et al., 2007).
2.2.2. Clinical specimens
Samples submitted to the CDC poxvirus program for OPV testing are held in a clinical samples database after diagnostics are complete and include swabs of lesions or vesicle fluid, or scab material. Swabs are resuspended in 400 μl 1× PBS (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4 and 1.47 mM KH2PO4), extracted using SETS (Swab Extraction Tube System, Roche Applied Science, Indianapolis, IN) then sonicated. Scab material is resuspended with 300 μl of 1× PBS, then frozen in a dry-ice/ethanol bath, ground using a disposable pestle, vortexed, and sonicated. After diagnostic testing is complete, remaining sample is stored frozen at −80 °C. In this study, rash lesion clinical samples were used to test the efficacy of the BioThreat Alert strips for detection of viral antigen in clinical samples. Samples were blinded and included human specimens from monkeypox, vaccinia, herpesvirus (Varicella and HSV-1) and parapoxvirus infections. Specimen remainders from 22 human cases were chosen; 11 were from OPV infections (monkeypox and vaccinia) and 11 were non-OPV (VZV, pseudocowpox, Orf and HSV1). By design, OPV clinical samples with early RT-PCR cycle threshold (Ct) values were selected to ensure antigen presence; earlier Ct values indicate higher concentration of genomic DNA. Genome equivalents were extrapolated by comparison with standard curves of Ct values generated from 10-fold dilutions of purified viral DNA (17.5 fg to 1.75 ng) using an E9L RT-PCR assay (Li et al., 2006). A generic genome size of 200,000 base pairs was used to calculate individual genome mass, and thus genome equivalents. All orthopox samples utilized had Ct values less than 24, or approximately 8 × 104 genome equivalents. Infectious virus was then evaluated by plaque assay (described in Section 2.2.1). The CDC's Human Research Protection Office determined that use of de-identified diagnostic remainder for this activity was exempt from consideration under 45 CFR 46.101(4)(b) (3/11/2009).
2.3. Tetracore BioThreat testing procedures
2.3.1. Sensitivity to viral isolates
Tetracore Orthopox BioThreat Alert ® assay (catalog number TC-8001-025) was provided by Tetracore (Rockville, MD). The BioThreat Alert assay was tested for sensitivity to lab grown OPV strains on duplicate strips by two users. Viruses were diluted in Tetracore sample buffer in 10-fold dilution series to final concentrations ranging from 102 to 108 pfu/ml of VACV, or 104 to 108 pfu/ml of MPXV, and 150 μl aliquots applied to the test strips (1.5 × 101–1.5 × 107 pfu/strip). Initial dilutions of VACV were 1:70 (from a stock concentration of 6.8 × 109 to 108 pfu/ml), so BSC-40 lysate was also diluted at 1:70, and Crude VZV and HLF cell lysates were diluted at 1:100. The 150 μl samples were applied to each test strip and results were visually scored as positive or negative after 15 min. For strips used to test VACV, detection was also performed using the ESEQuant Lateral Flow Reader (Qiagen, Valencia, CA) and analyzed with Lateral Flow Studio (Qiagen) to quantify line intensity. The reader measures reflectance, where increased line intensity reduces reflectance, and thus reduced signal (mV) received at the detector; the resulting line measurement is displayed as an inverted peak (i.e., a trough). Lateral Flow Studio software (Qiagen) calculates peak height, and along with the number of data point measurements quantifies the peak area. For MPXV, restrictions in moving select agent materials from our laboratory prevented ESEQuant analysis of these samples (reader is in another laboratory), thus MPXV isolates were only analyzed visually. A second study assessed improved sensitivity of later measurement time points and utilized the field portable BioThreat Alert Reader. This reader provides quantitative signal value (SV) measurements of sample line intensity. Purified VZV and DMEM media used for VZV resuspension were diluted at 1:2 in Tetracore buffer prior to application. Strips were analyzed between 15 and 37 min after sample application and those with an SV above the instrument default threshold value (TV) of 0.01 were scored positive.
2.3.2. Specificity with clinical samples
The specificity experiment was performed in duplicate on 22 unique clinical samples; 44 BioThreat Alert strips were utilized. Samples were supplied in 75 μl of PBS and diluted at 1:2 with Tetracore sample buffer (final 150 μl volume) and run on the Orthopox BioThreat Alert strips according to manufacturer instructions. The blind study samples were examined 15 min after sample application and visually scored, including qualitative notations of intensities for positive samples of strong, medium, weak or faint. “Strong” line intensity was defined as intensity equivalent to the control line. Signal for “medium” and “weak” notations had reduced intensity and a score of “faint” was given to indicate the weakest detectible line by naked eye. Although the control line can vary in intensity due to improper storage of the testing reagents or test age, no qualitative differences in intensity were noted throughout the testing period.
3. Results
3.1. Detection of orthopox viral isolates using the Tetracore assay
Visual analysis of laboratory preparations of VACV resulted in positive identifications at concentrations of 108 and 107 pfu/ml, but had varying success at 106 pfu/ml concentration with two samples identified as positive and three as negative (Table 1 ). Dilutions below 105 were negative. Crude VZV and uninfected HLF and BSC-40 cell lysates controls were negative. Quantitative peak area measurements using the ESEQuant® Lateral Flow Reader show the 108–107 pfu/ml peak area 100-fold above control sample peak areas and were 2248 ± 446 or 2346 ± 85, respectively. The 106 pfu/ml peak area average was almost 10-fold higher than lysate controls, and averaged 165 ± 69 units. Control values (e.g., VZV and cell lysates) were between 20 and 26 units based on peak area measurement. MPXV preparations were only analyzed visually (Table 1), but yielded similar results with 108–106 pfu/ml positive, and the 106 pfu/ml scoring weakly positive by comparison to the higher concentrations. Overall, a strong correlation was noted between visual analysis and quantitative measurements.
Table 1.
Visual and quantitative results using laboratory preparations of orthopoxviruses.
| Vaccinia virus |
Monkeypox virus |
|||||||
|---|---|---|---|---|---|---|---|---|
| Visual IDa |
Quantitative results (test 1 and 2 only) |
Visual IDa |
||||||
| Test 1 | Test 2 | Test 3 | Test 4 | Test 5 | Peak areab, c | Test 1 | Test 2 | |
| 108 pfu/ml | Y | Y | Y | Y | Y | 2248 ± 446 | Y | Y |
| 107 pfu/ml | Y | Y | Y | Y | Y | 2346 ± 85 | Y | Y |
| 106 pfu/ml | Y | N | N | N | Y | 165 ± 69 | Y | Y |
| 105 pfu/ml | N | N | N | N | N | 18 ± 6 | N | N |
| 104 pfu/ml | N | N | N | N | N | 32 ± 9 | N | N |
| Crude VZV | N | N | – | – | – | 26 | – | – |
| HLF lysate | N | N | – | – | – | 22 | – | – |
| BSC-40 lysate | N | N | – | – | – | 26 | – | – |
Identified positive (Y) or negative (N) by two independent users 15 min after sample application.
Peak area measurements are defined in Section 2.3.1.
Average and standard deviation measurements, no cutoff was defined by the ESE reader.
To assess the utility of the Tetracore BioThreat field portable reader and potential higher signal at later time points, VACV dilutions were run in duplicate and scanned at approximately 7.5 min intervals, beginning at 15 min after sample application (Fig. 2a). Variability in processing time resulted in some adjustments in measurement time resulting in three measurement windows from 15 to 19 min, 21 to 27 min and 29 to 37 min. Measurements for all concentrations show increases in signal with additional incubation time. Response between 15 and 19 min revealed 108, 107, and 106 pfu/ml virus samples positive, although one of the two 106 pfu/ml sample was nominally above the 0.01 cutoff value. At later time points the 108, 107 and 106 pfu/ml virus samples produced greater signal. One of the 105 pfu/ml samples had increasing signal throughout the time course but remained negative, however, the second 105 sample became positive after 33 min. Lysates from BSC-40 cells were negative (Fig. 2b). Unlike the crude VZV preparations tested with the ESE Quant Reader, purified VZV was above the positive cutoff; however, the DMEM media used to resuspend VZV after purification was also positive. When diluted an additional 10-fold, both samples were negative throughout the measurement timeframe (Fig. 2b). Additionally, intensities for the purified VZV and DMEM media samples did not change over time indicating that the reaction observed is non-specific (i.e. does not titrate in this specific assay).
Fig. 2.
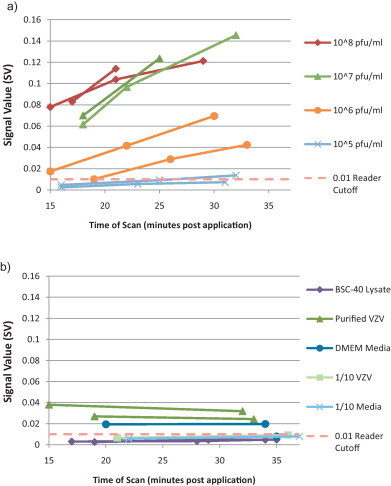
Sensitivity of BioThreat Alert reader scanned strips to (a) lab grown vaccinia virus and (b) negative controls. The manufacturer cutoff for the reader was 0.01. Samples were applied in duplicate to test strips and measured at approximately 7.5 min intervals. Not all samples had measurements taken at each interval.
3.2. Blind study clinical sample screening with the Tetracore assay
Using visual analysis only, 9 of 11 clinical OPV samples were identified positively (Table 2 ). All samples were tested in duplicate resulting in four false negative tests (4 of 22 strip tests, or 18%). Among OPV positive samples some correlation between real time PCR Ct value and correct visual identification of OPV was seen as samples 3 and 8 had among the later Ct values (21 and 22, respectively) and were associated with the false negatives. There was however no correlation between the Ct value and line intensity. For example, sample 1 had a late Ct value (22) but produced the strongest intensity line while samples 10 and 11 had very early Ct values (17 and 15, respectively) but produced the weakest detectible lines with the assay. Modest correlation was seen between line intensity and titer; samples 1, 2, and 4 had titers above 4.6 × 103 pfu/ml and were positive with medium or weak line intensities, and samples 8, 9 and 11 had titers below 2.8 × 103 pfu/ml and were only weakly positive or negative. Sample 3, which contained 4.9 × 104 pfu/ml, was the exception as one strip became positive after the defined 15 min measurement time and the replicate was scored negative. VACV samples were identified as positive as few as 3 days post rash onset; days post rash were unavailable for MPXV samples. Non-OPV samples including VZV, pseudocowpox, Orf and HSV were also tested, with 10 of 11 samples correctly identified as negative (Supplementary Table 1). One positive was noted for one of the duplicate strip test for a single VZV sample; that sample was unusual visually, but marked positive as it had a weak intensity line at the sample line location.
Table 2.
Blind study results for Orthopox diagnostic remainder clinical specimens (tested in duplicate).
| Sample | Visual ID | Line intensity | Sample type | Days post rash onset | Virus | CT | PFU/ml | Correct? |
|---|---|---|---|---|---|---|---|---|
| 1 | + | Med | Vesicle/skin | 3 | Vaccinia | 22 | 4.6E + 03 | ✓ |
| + | Med | ✓ | ||||||
| 2 | + | Med | Swab | Unknown | Vaccinia | Nd | 6.1E + 05 | ✓ |
| + | Med | ✓ | ||||||
| 3 | −/+a | Faint | Swab | 11 | Vaccinia | 21 | 4.9E + 04 | × |
| − | – | × | ||||||
| 4 | + | Weak | Swab | 12 | Vaccinia | 19 | 1.2E + 05 | ✓ |
| + | Weak | ✓ | ||||||
| 5 | + | Faint | Vesicle/skin | 3 | Vaccinia | 22 | Ndb | ✓ |
| + | Weak | ✓ | ||||||
| 6 | + | Weak | Scab | Unknown | Monkeypox | 15 | Nd | ✓ |
| + | Med | ✓ | ||||||
| 7 | + | Faint | Swab | Unknown | Monkeypox | 18 | Nd | ✓ |
| + | Faint | ✓ | ||||||
| 8c | − | – | Swab | Unknown | Monkeypox | 22 | 1.0E + 01 | × |
| − | – | × | ||||||
| 9 | + | Med | Scab | Unknown | Monkeypox | 16 | Nd | ✓ |
| + | Med | ✓ | ||||||
| 10 | + | Faint | Swab | Unknown | Monkeypox | 17 | 2.8E + 03 | ✓ |
| + | Faint | ✓ | ||||||
| 11c | + | Weak | Swab | Unknown | Monkeypox | 15 | 1.0E + 01 | ✓ |
| + | Faint | ✓ | ||||||
Sample became positive after scheduled 15 min read time.
Not determined.
Re-extracted samples.
4. Discussion
The Tetracore Orthopox BioThreat Alert assay is marketed as a rapid, point-of-care diagnostic for first responders in the event of a bioterrorism attack. Its ease of use and stability at ambient temperature suggest that the test would be applicable for use under field conditions which might be encountered by first-responders, or by rural health workers in geographic areas such as Central Africa where MPXV is endemic. In these situations, specimen transport and on-site laboratory capabilities would likely be limited. This is the first study to assess the BioThreat Alert assay utility for identifying laboratory OPV isolates and clinical samples.
Visual analysis was preferred for its simplicity and used to evaluate initially assay performance. In all cases, the assay reproducibly allowed detection of 107 pfu/ml of VACV or MPXV, with no disagreement between replicates and independent users. Although only two OPVs were tested herein, they are known to possess broad cross-reactivity (Mercer et al., 2007) and results should be similar for all OPV. Assay sensitivity, as defined for non-quantitative measurements as the threshold level, below which there is no detectable response and above which a response occurs (Delfert et al., 1987), would be 107 pfu/ml; all laboratory isolates tested at this concentration produced a positive response. Quantitative analysis with the ESEQuant® reader confirmed these results. Peak areas, as reported by the ESEQuant® were nearly equivalent for the 108 and 107 pfu/ml samples, and were 50–100-fold higher than negative control samples; the 106 pfu/ml VACV sample was only 5–10-fold above negatives. Thus, reproducible positive results can be anticipated with 107 pfu/ml samples, with some positives observed at the 106 pfu/ml concentration.
All visual experiments were read at 15 min after sample application according to the manufacturer instructions, which included a notation that samples that become positive after 20 min may not be valid. Extended run times are known to increase assay sensitivity but also can increase false positives (Wong and Tse, 2009). Observations suggested that a later measurement time point could increase sensitivity without sacrificing specificity. To test this assumption a dilution series of VACV was utilized and read with the field portable BioThreat reader. This reader provides quantitative measurements in fewer than 45 s and outputs positive or negative identification based on a default cutoff value of 0.01. Extending the assay incubation time from 15 to over 30 min improved VACV reader output as much as 4-fold, but did not increase background signals from control samples (Fig. 2a and b). Thus, sensitivity could be augmented by allowing longer incubation times as increases in signal were specific for VACV. Specifically, samples in the 105–106 range would likely benefit from additional incubation time. The 106 pfu/ml sample with measurements at 19 min (Fig. 2a) was visually negative at 15 min (Table 1, VACV Test 3), but was positive quantitatively by 19 min (SV = 0.0101), and was then easily identified as positive by 27 min (SV = 0.0289) post sample application. This correlates with visual analysis results where after 15 min many strips were determined to be negative, but would have been positive if read at a later time point. Furthermore, one of the two 105 samples became positive at 33 min. Additionally, while false positives were noted with purified lab grown VZV and the DMEM media used to resuspend the VZV virus, use of a longer incubation prior to test measurement would not have effected test outcome in these samples. However, a thorough testing of extended run times with clinical specimens is necessary to determine the maximum time allowed without significant increases in false positive results.
VZV has been difficult historically to grow to high titers, particularly when used in cell free preparations (Harper et al., 1998). In this case, the stock used was only 103 pfu/ml. Accounting for infective/inactive particles (Carpenter et al., 2009), the likely total virions in this cell-free purification were less than 107 pfu/ml. To address lower number of virions present relative to OPV samples, purified VZV and resuspension buffer were added to the strips after only a 1:2 dilution. Both resulted in false positives. Dilution of VZV virus or DMEM an additional 10-fold (to a 1:20 final dilution) reduced signals below the positive cutoff and eliminated these false positives. Thus, a component of the virus resuspension media and application of samples at such a minimal dilution was likely the reason for the false positives. Examination of signal from these false positives indicated no increase in signal over time. As a precaution for field screening assays, a minimum sample dilution should be established or limits placed on acceptable sample types. Any concerns of false positives can be minimized by adding multiple measurement times after sample application to observe specificity of the reaction indicated by increasing signal over time; field portable instruments such as the BioThreat Reader would allow quantization of multiple time point measurements.
Prior to testing clinical samples, the relationship between virus found in laboratory isolates and a real-time PCR E9L Assay (Li et al., 2006) was assessed. The E9L assay is the current standard for identification of OPV in clinical specimens. A correlation between the real-time PCR extrapolated genome equivalents with the assay sensitivity to laboratory virus was established. Estimates for total particles to infectious ones range from 2.4 to 50:1 (Contreras and Ohlbaum, 1968, Overman and Sharp, 1959). Based on 107 pfu/ml sensitivity to laboratory grown virus, and using an intermediate value of 10:1 virus particles to infectious units suggests a detection limit of approximately 108 particles/ml, or assuming one particle contains one genome, 108 genomes/ml. In the 150 μl sample applied to each strip, this equates to 1.5 × 107 genomes. Vesiculo-pustular samples obtained between 5 and 13 days post rash onset during the 2003 monkeypox outbreak had genome copies ranging from 6 × 103 to 3 × 109. Therefore, for higher concentration clinical samples sensitivity would be adequate.
Within the context of the specificity determination for the Orthopox BioThreat Alert ® assay, OPV samples with Ct values ranging from 15 to 22, or 1.5 × 107 to 2.3 × 105 genome equivalents were selected for testing. Orthopox BioThreat Alert false negative results with samples 3 and 8 had later Ct values of 21 and 22. Additionally, sample 8 had been re-extracted during initial diagnostic testing. Although this re-extracted sample Ct value was still strongly positive, it is possible that the integrity of some antigens was altered during re-processing which reduced binding of antigen to the Orthopox BioThreat Alert assay, but viral genomes were left intact. Variability of sample quality may have also affected the results and subsequent correlation to PCR Ct values. It is also of use to note that samples 1 and 5 had Ct values of 22, or approximately 2.3 × 105 genome equivalents. Based on estimations of non-infectious to infectious virions (10:1), this correlates with 2.3 × 104 pfu/ml. This concentration was negative with all tested laboratory viruses. This further suggests that sample integrity or quality may directly impact test results. As such, assay sensitivity to clinical specimens may prove to be better with freshly prepared field samples, although this remains to be proven. Additionally, while the number of clinical samples tested was limited, the diversity of samples as well as their well characterized nature provides a good starting point for determining assay specificity.
5. Conclusion
In conclusion, VACV and MPXV samples with 107 pfu/ml were identified reproducibly by visual and quantitative methods when applied to the BioThreat strips, and after extending the manufacturers recommended incubation time, concentrations as low as 105 pfu/ml produced positive results. While visual analysis was sufficient to identify positive readings, the availability of a field portable reader such as the BioThreat Reader would allow documentation of intensity and standardize readings. When testing clinical samples there was variability in correlation with other methods to identify viruses such as RT-PCR or virus culture, but indications suggest the assay works well for higher concentration samples and discriminates well against other pustular rash causing viruses. These studies suggest that the Orthopox BioThreat Alert assay may be a valuable method for screening and identification of human monkeypox. Rapid diagnosis would allow prioritization of samples for further testing and would be crucial for implementation of infection control, outbreak detection, and clinical care.
Acknowledgments
The authors thank Dr. Scott Schmid, Nancy Jensen, Denise Brown of the CDC Herpervirus program for providing VZV stocks and technical information regarding those stocks. We also thank Dr. Tim Granade and Shon Workman of the CDC Division of HIV/AIDS Prevention Laboratory Branch for use of the ESEQuant Reader and software, and Dr. Tom O’Brien and Jennifer Walker of Tetracore for providing the BioThreat Alert Reader.
The findings and conclusions in this report are those of the author(s) and do not necessarily represent the views of the funding agency.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.jviromet.2012.08.023.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- Behrman A., Schmid D.S., Crivaro A., Watson B. A cluster of primary varicella cases among healthcare workers with false-positive varicella zoster virus titers. Infection Control and Hospital Epidemiology: The Official Journal of the Society of Hospital Epidemiologists of America. 2003;24:202–206. doi: 10.1086/502187. [DOI] [PubMed] [Google Scholar]
- Carpenter J.E., Henderson E.P., Grose C. Enumeration of an extremely high particle-to-pfu ratio for varicella-zoster virus. Journal of Virology. 2009;83:6917–6921. doi: 10.1128/JVI.00081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention Multistate outbreak of monkeypox – Illinois, Indiana, and Wisconsin, 2003, MMWR. Morbidity and Mortality Weekly Report. 2003;52:537–540. [PubMed] [Google Scholar]
- Chen M.H., Zhu Z., Zhang Y., Favors S., Xu W.B., Featherstone D.A., Icenogle J.P. An indirect immunocolorimetric assay to detect rubella virus infected cells. Journal of Virological Methods. 2007;146:414–418. doi: 10.1016/j.jviromet.2007.08.021. [DOI] [PubMed] [Google Scholar]
- Contreras G., Ohlbaum A. Target volume analysis of vaccinia virus: influence of virus dispersion and noninfectious particles. Journal of Virology. 1968;2:1102–1106. doi: 10.1128/jvi.2.10.1102-1106.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damon I.K., Roth C.E., Chowdhary V. Discovery of monkeypox in Sudan. New England Journal of Medicine. 2006;355:962–963. doi: 10.1056/NEJMc060792. [DOI] [PubMed] [Google Scholar]
- Delfert D.M., Rea M.R., Kessler G., Siegfried B.A., Valdes R. Criteria for evaluating nonquantitative assays – application to serum choriogonadotropin. Clinical Chemistry. 1987;33:150–153. [PubMed] [Google Scholar]
- Fedorko D.P., Preuss J.C., Fahle G.A., Li L., Fischer S.H., Hohman P., Cohen J.I. Comparison of methods for detection of vaccinia virus in patient specimens. Journal of Clinical Microbiology. 2005;43:4602–4606. doi: 10.1128/JCM.43.9.4602-4606.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formenty P., Muntasir M.O., Damon I., Chowdhary V., Opoka M.L., Monimart C., Mutasim E.M., Manuguerra J.C., Davidson W.B., Karem K.L., Cabeza J., Wang S., Malik M.R., Durand T., Khalid A., Rioton T., Kuong-Ruay A., Babiker A.A., Karsani M.E., Abdalla M.S. Human monkeypox outbreak caused by novel virus belonging to Congo Basin clade, Sudan, 2005. Emerging Infectious Diseases. 2010;16:1539–1545. doi: 10.3201/eid1610.100713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper D.R., Mathieu N., Mullarkey J. High-titre, cryostable cell-free varicella zoster virus. Archives of Virology. 1998;143:1163–1170. doi: 10.1007/s007050050364. [DOI] [PubMed] [Google Scholar]
- Hutson C.L., Olson V.A., Carroll D.S., Abel J.A., Hughes C.M., Braden Z.H., Weiss S., Self J., Osorio J.E., Hudson P.N., Dillon M., Karem K.L., Damon I.K., Regnery R.L. A prairie dog animal model of systemic orthopoxvirus disease using West African and Congo Basin strains of monkeypox virus. Journal of General Virology. 2009;90:323–333. doi: 10.1099/vir.0.005108-0. [DOI] [PubMed] [Google Scholar]
- Jezek Z., Szczeniowski M., Paluku K.M., Mutombo M. Human monkeypox: clinical features of 282 patients. Journal of Infectious Diseases. 1987;156:293–298. doi: 10.1093/infdis/156.2.293. [DOI] [PubMed] [Google Scholar]
- Jezek Z., Szczeniowski M., Paluku K.M., Mutombo M., Grab B. Human monkeypox: confusion with chickenpox. Acta Tropica. 1988;45:297–307. [PubMed] [Google Scholar]
- Karem K.L., Reynolds M., Braden Z., Lou G., Bernard N., Patton J., Damon I.K. Characterization of acute-phase humoral immunity to monkeypox: use of immunoglobulin M enzyme-linked immunosorbent assay for detection of monkeypox infection during the 2003 North American outbreak. Clinical and Diagnostic Laboratory Immunology. 2005;12:867–872. doi: 10.1128/CDLI.12.7.867-872.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulesh D.A., Loveless B.M., Norwood D., Garrison J., Whitehouse C.A., Hartmann C., Mucker E., Miller D., Wasieloski L.P., Jr., Huggins J., Huhn G., Miser L.L., Imig C., Martinez M., Larsen T., Rossi C.A., Ludwig G.V. Monkeypox virus detection in rodents using real-time 3′-minor groove binder TaqMan assays on the Roche LightCycler. Laboratory Investigation. 2004;84:1200–1208. doi: 10.1038/labinvest.3700143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Learned L.A., Reynolds M.G., Wassa D.W., Li Y., Olson V.A., Karem K., Stempora L.L., Braden Z.H., Kline R., Likos A., Libama F., Moudzeo H., Bolanda J.D., Tarangonia P., Boumandoki P., Formenty P., Harvey J.M., Damon I.K. Extended interhuman transmission of monkeypox in a hospital community in the Republic of the Congo, 2003. American Journal of Tropical Medicine and Hygiene. 2005;73:428–434. [PubMed] [Google Scholar]
- Li Y., Olson V.A., Laue T., Laker M.T., Damon I.K. Detection of monkeypox virus with real-time PCR assays. Journal of Clinical Virology. 2006;36:194–203. doi: 10.1016/j.jcv.2006.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Likos A.M., Sammons S.A., Olson V.A., Frace A.M., Li Y., Olsen-Rasmussen M., Davidson W., Galloway R., Khristova M.L., Reynolds M.G., Zhao H., Carroll D.S., Curns A., Formenty P., Esposito J.J., Regnery R.L., Damon I.K. A tale of two clades: monkeypox viruses. Journal of General Virology. 2005;86:2661–2672. doi: 10.1099/vir.0.81215-0. [DOI] [PubMed] [Google Scholar]
- Mercer A.A., Schmidt A., Weber O.F. Basel; Boston: 2007. Poxviruses. [Google Scholar]
- Meyer H., Perrichot M., Stemmler M., Emmerich P., Schmitz H., Varaine F., Shungu R., Tshioko F., Formenty P. Outbreaks of disease suspected of being due to human monkeypox virus infection in the Democratic Republic of Congo in 2001. Journal of Clinical Microbiology. 2002;40:2919–2921. doi: 10.1128/JCM.40.8.2919-2921.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien, T., 2012. Personal Communication, Tetracore, Inc.
- Olson V.A., Laue T., Laker M.T., Babkin I.V., Drosten C., Shchelkunov S.N., Niedrig M., Damon I.K., Meyer H. Real-time PCR system for detection of orthopoxviruses and simultaneous identification of smallpox virus. Journal of Clinical Microbiology. 2004;42:1940–1946. doi: 10.1128/JCM.42.5.1940-1946.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overman J.R., Sharp D.G. Ratios of vaccinia virus particles to virus infectious units; studies of ratio changes during growth and adaptation in eggs, guinea pigs, and rabbits. Journal of Experimental Medicine. 1959;110:461–480. doi: 10.1084/jem.110.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahaus M., Desloges N., Wolff M.H. Development of a multiplex RT-PCR to detect transcription of varicella-zoster virus encoded genes. Journal of Virological Methods. 2003;107:257–260. doi: 10.1016/s0166-0934(02)00250-1. [DOI] [PubMed] [Google Scholar]
- Reed K.D., Melski J.W., Graham M.B., Regnery R.L., Sotir M.J., Wegner M.V., Kazmierczak J.J., Stratman E.J., Li Y., Fairley J.A., Swain G.R., Olson V.A., Sargent E.K., Kehl S.C., Frace M.A., Kline R., Foldy S.L., Davis J.P., Damon I.K. The detection of monkeypox in humans in the Western Hemisphere. New England Journal of Medicine. 2004;350:342–350. doi: 10.1056/NEJMoa032299. [DOI] [PubMed] [Google Scholar]
- Reynolds M.G., Yorita K.L., Kuehnert M.J., Davidson W.B., Huhn G.D., Holman R.C., Damon I.K. Clinical manifestations of human monkeypox influenced by route of infection. Journal of Infectious Diseases. 2006;194:773–780. doi: 10.1086/505880. [DOI] [PubMed] [Google Scholar]
- Schmidt N.J., Lennette E.H. Neutralizing antibody responses to varicella-zoster virus. Infection and Immunity. 1975;12:606–613. doi: 10.1128/iai.12.3.606-613.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shchelkunov S.N., Shcherbakov D.N., Maksyutov R.A., Gavrilova E.V. Species-specific identification of variola, monkeypox, cowpox, and vaccinia viruses by multiplex real-time PCR assay. Journal of Virological Methods. 2011;175:163–169. doi: 10.1016/j.jviromet.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trindade G.S., Lobato Z.I., Drumond B.P., Leite J.A., Trigueiro R.C., Guedes M.I., da Fonseca F.G., dos Santos J.R., Bonjardim C.A., Ferreira P.C., Kroon E.G. Short report: isolation of two vaccinia virus strains from a single bovine vaccinia outbreak in rural area from Brazil: implications on the emergence of zoonotic orthopoxviruses. American Journal of Tropical Medicine and Hygiene. 2006;75:486–490. [PubMed] [Google Scholar]
- Vorou R.M., Papavassiliou V.G., Pierroutsakos I.N. Cowpox virus infection: an emerging health threat. Current Opinion in Infectious Diseases. 2008;21:153–156. doi: 10.1097/QCO.0b013e3282f44c74. [DOI] [PubMed] [Google Scholar]
- Wong R.C., Tse H.Y. Springer; New York, NY: 2009. Lateral Flow Immunoassay. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


