Objective:
To evaluate whether circulating micro ribonucleic acids (miRNAs) predict response to neoadjuvant chemotherapy (NAC) and inform decision-making in breast cancer patients.
Introduction:
Deciphering response to NAC remains a challenge. Those unlikely to respond may benefit from NAC de-escalation before completion, while “responders” should complete treatment. Establishing biomarkers which identify response to NAC is imperative to personalize treatment strategies. miRNAs are small noncoding RNA molecules which modulate genetic expression. miRNAs are believed to inform response to NAC.
Methods:
This prospective, multicenter trial (NCT01722851) recruited 120 patients treated with NAC across 8 Irish treatment sites. Predetermined miRNAs were quantified from patient whole bloods using relative quantification polymerase chain reactiond. Venous sampling was performed at diagnosis and midway during NAC. Trends in miRNA expression between timepoints were correlated with treatment response. Data analysis was performed using R 3.2.3.
Results:
A total of 120 patients were included (median age: 55 years). Overall, 49.2% had luminal breast cancers (59/120), 17.5% luminal B (L/HER2) (21/120), 12.5% human epidermal growth factor receptor-2 positive (HER2+) (15/120), and 20.8% triple negative disease (25/120). In total, 46.7% of patients responded to NAC (56/125) and 26.7% achieved a pathological complete response (pCR) (32/120). For patients with L/HER2, increased Let-7a predicted response to NAC (P=0.049), while decreased miR-145 predicted response to NAC in HER2+ (P=0.033). For patients with luminal breast cancers, reduced Let-7a predicted achieving a pCR (P=0.037) and reduced miR-145 predicted achieving a pCR to NAC in HER2+ (P=0.027).
Conclusions:
This study illustrates the potential value of circulatory miRNA measurement in predicting response to NAC. Further interrogation of these findings may see miRNAs personalize therapeutic decision-making for patients undergoing NAC for early breast cancer.
Keywords: breast cancer, miRNA, neoadjuvant therapies, personalized medicine, precision oncology
Although breast cancer incidence is increasing, prognosis has improved significantly with 5‐year survival outcomes improving from 40% to 90% over the past 5 decades.1 Breast cancer is a molecular diverse disease with at least 4 biologically distinct biological subtypes, each with individual clinical characteristics, therapeutic strategies, and prognoses.2 The St. Gallen Expert Consensus Panel advocate use of multigene expression assays [eg, OncotypeDX Recurrence Score (RS), Genomic Health Inc., Redwood City, CA] as the gold standard for molecular substratification2,3 and immunohistochemical evaluation of estrogen (ER), progesterone (PgR), and human epidermal growth factor receptor‐2 (HER2) receptors act as surrogate phenotypical biomarkers to decipher biological subtypes.2,4 Moreover, appraising receptor status is crucial for guiding therapeutic decision making.5,6
Breast cancer survival is equivalent for those treated with adjuvant and neoadjuvant chemotherapy (NAC).7,8 Nevertheless, advantages of NAC include potential tumor downstaging, increased patient eligibility for breast conservation surgery (BCS), improved resectability,9–11 and the generation of in vivo data regarding tumour sensitivity to systemic therapies, which carry prognostic significance.12,13 In particular, predicting response to NAC translates into important disease outcomes in the setting of HER2-positive (HER2+) and triple negative molecular subtypes.12,13 Unfortunately, predicting response is challenging due to variability in host and tumour factors.
Micro ribonucleic acids (or miRNAs) are small (19–25 nucleotides in length), noncoding ribonucleic acid (RNA) molecules which modulate gene expression by post-transcriptional degradation or translational inhibition of messenger RNA.14–17 Aberrant expression of miRNAs regulate breast cancer development and miRNAs have the ability to maintain stability in several biological tissues.18–21 Moreover, miRNA profiling may be performed relatively simply and inexpensively using real-time quantitative reverse transcriptase polymerase chain reaction,22–24 supporting their suitability as clinical biomarkers.
Translational research efforts are focused on identifying novel biomarkers capable of predicting response to NAC to tailor treatment strategies to each patient’s needs. The Cancer Trials Ireland – Irish Clinical Oncology Research Group 10/11 (CTRIAL ICORG10/11) is a prospective, multicenter trial which recruited 120 patients treated with NAC for breast cancer. Expression levels of a predetermined miRNA panel were relatively quantified from bloods samples using relative quantification polymerase chain reactiond at several predetermined timepoints during NAC.
The aims of this study were:
To determine whether circulating miRNA expression levels successfully predict response to NAC.
To establish whether subtype specific miRNA biomarkers predict response to NAC.
METHODS
Study Design
This prospective, multicenter trial was overseen by Clinical Trials Ireland (CTRIAL ICORG10/11—NCT1722851), with recruitment at 8 treatment sites in the Republic of Ireland. Local ethical approval was granted in February 2008 (C.A.151) and in January 2014 (C.A.1012).
MiRNA expression profiles were measured from liquid biopsies taken at diagnosis (Timepoint 1, or T1) and halfway during NAC (Timepoint 2, or T2) (Fig. 1). Thereafter, the changes in miRNA expression levels between these timepoints were determined (T2 minus T1) and correlated with response to NAC.
FIGURE 1.
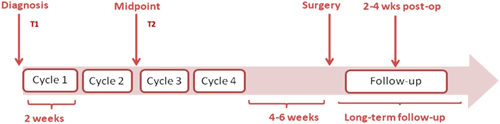
Scheme of timepoints at which venous sampling occurred during this study: Timepoint 1 (T1) which involved venous sampling at breast cancer diagnosis (and before standard-of-care with neoadjuvant chemotherapy), and Timepoint 2 (T2) at the halfway point during neoadjuvant chemotherapy).
Inclusion and Exclusion Criteria
Consecutive female patients aged 18 years or older indicated to undergo standard-of-care NAC for breast cancer were considered for inclusion. Patients had to be capable of providing informed written consent. Patients failing to meet these inclusion criteria were not considered for inclusion.
Histopathology and Molecular Subtyping
Breast cancer molecular subtypes were classified using the 12th St. Gallen Expert Consensus panel.25 In brief, tumors were evaluated for ER and PgR using immunohistochemistry as per the 2010 American Society of Clinical Oncology/College of American Pathologists (ASCO/CAP) consensus.26 HER2 status was identified by Herceptest (DAKO Agilent pathology solutions, Santa Clara, CA), with a score of 3+ considered positive. Any 2+ inconclusive results were confirmed using fluorescent in situ hybridization.27,28 Ki-67 evaluation was performed in just a few cases using MIB1 antibody testing.29,30 Luminal disease [luminal breast cancers (LBC)] was classified as ER+/HER2−, Luminal/HER2+ disease (L/HER2) was classified as ER+/HER2+, HER2 disease (HER2+) was classified as ER−/PgR−/HER2+, and triple negative breast cancer (TNBC) was classified as ER−/PgR−/HER2−.25 American Joint Committee on Cancer (AJCC) version 8 was used for staging.31
Response to Chemotherapy
Response to NAC was based on Miller-Payne classification.32 In addition, patients were classified as “responders” and “nonresponders”; “responders” were patients who achieved pathological complete response (pCR) or a >90% reduction in tumor cellularity on their specimen (Miller-Payne grades 4/5), while those with >90% were classified as “nonresponders” (Miller-Payne grades 1–3). Further subclassification was performed with respect to those who achieved a pCR (Miller-Payne grade 5) versus those who did not (Miller-Payne grades 1–4).32
Venous Blood Sampling
Venous samples from the 120 patients were collected during a 3-year period (May 2011–April 2014). Whole blood liquid biopsies were collected at 2 independent timepoints:
Timepoint 1 (T1) – at breast cancer diagnosis, before treatment with NAC.
Timepoint 2 (T2 – halfway during NAC) (Fig. 1).
Venous blood samples were collected in 3 mL ethylenediaminetetraacetic acid (EDTA) tubes and stored at the local Surgery Cancer Biobank. A study enrolment diagram illustrating recruitment is outlined in Supplementary Appendix 1. A (Supplemental Digital Content 1, http://links.lww.com/SLA/E85).
MiRNA Expression Panel
Following literature review, a 5 miRNA panel was selected for evaluation (Let-7a, miR-21, miR-145, miR-155, and miR-195) based on their relevance in breast cancer at the time of trial design.20,21,33 Two additional miRNAs were used as validated endogenous controls (miR-16 and miR-345), with the intention of standardizing miRNA expression.34 Table 1 outlines the relevance of each miRNA selected for evaluation.
TABLE 1.
The Relevance of the 5 Target-miRNA and 2 Endogenous Controls Included in our Predetermined miRNA Panel
| Target | References | MiRNA Function |
|---|---|---|
| Let-7a | Heneghan et al20,21 | Increased expression in treatment naive breast cancer patients versus controls and postresection |
| miR-21 | Heneghan et al20,21 | Known as a well described oncogenic miRNA |
| miR-145 | Heneghan et al20,21 | Increased expression levels in breast cancers relative to other malignancies and controls |
| miR-155 | Heneghan et al20,21 | Differentiated expression levels in breast cancers relative to other malignancies and controls |
| miR-195 | Heneghan et al20,21 | Increased expression in treatment naive breast cancer patients versus controls and other cancers |
| miR-16 | McDermott et al33 | Endogenous control in human circulation |
| miR-425 | McDermott et al33 | Endogenous control in human circulation |
RNA Isolation and Storage
Total RNA was extracted from whole blood (1 mL) using Trizol (as per the manufacturer’s instructions). RNA concentrations were determined using spectrophotometry (NanoDrop ND-1000 Technologies Inc., Wilmington, DE), as previously described.20 RNA was then transferred to storage tubes, labelled, and stored at −70°C in our Cancer Biobank.
Analysis of miRNA Expression Levels
For each sample, miRNAs were relatively quantified using quantitative reverse transcriptase polymerase chain reaction. TaqMan assays were used for the amplification of target miRNAs, as per manufacturer’s instructions (TaqMan Fast Universal Master Mix (2X), No AmpErase UNG: Applied biosystems, Foster City, CA).14,34 Assays were performed using AB7900HT (Applied Biosystems), as per the manufacturer’s instructions. Reactions were initiated with a 10-minute incubation at 95°C, followed by 40 cycles at 95°C for 15 seconds, and 60°C for 60 seconds. We utilised miR-26b as an interassay control from a breast cancer cell line, which was included on each plate. All reactions were performed in triplicate. The threshold SD for intra-assay and inter-assay replicates was 0.3. The percentage polymerase chain reaction amplification efficiencies (E) for each assay were calculated using the slope of the semi-log regression plot of cycle threshold versus log input of cDNA (10-fold dilution series of 5 points), with the following equation, and a threshold of 10% above or below 100% efficiency was applied: E=(10−1/slope−1)×100. MiRNA expression levels were calibrated and normalized using endogenous controls, before expression levels were calculated using QbasePlus© software (Biogazelle, Gent, Belgium) using the geNorm method. MiRNA analysis was performed blinded to clinical information.
Statistical Analysis
Data were analysed using R version 3.2.3. Differences in miRNA expression profiles between T2 and T1 were calculated. The Shapiro-Wilks test was used to assess the distribution of data. Nonparametric analyses [ie, Kruskal-Wallis test (used to compare medians among multiple groups) and the 2-sample Wilcoxon rank sum test (used for all 2-sample comparisons)] were used as appropriate to correlate miRNA expression with response to NAC. All analyses were 2-tailed and statistical significance was defined as P<0.050.
RESULTS
Clinicopathological and Surgical Data
Overall, 120 patients were included. The median age at diagnosis was 55.0 [interquartile range (IQR): 48.0–63.0] and tumor size was 38.0 mm (IQR: 28.0–54.0 mm). Almost 50.0% of included patients had LBC (49.2%, 59/120), 20.8% had TNBC (25/120), 17.5% had L/HER2 (21/120), and 12.5% had HER2+ (15/120). Most patients underwent BCS (55.8%, 67/120) and axillary lymph node dissection (70.8%, 85/120). Table 2 illustrates clinicopathological and surgical data for included patients.
TABLE 2.
Clinicopathological and Surgical Data for all 120 Included Patients With Correlations With Response to Neoadjuvant Chemotherapy
| Parameter | Variable | Total | “Responders”, n (%) | “Nonresponders”, n (%) | P | pCR, n (%) | Non-pCR, n (%) | P |
|---|---|---|---|---|---|---|---|---|
| Total number | — | 120 (100.0) | 56 (46.7) | 64 (53.3) | — | 32 (26.7) | 88 (73.3) | — |
| Age (y) | Median (IQR) | 55 (48–63) | 55 (48–64) | 55 (48.5–61.5) | 0.895 | 53 (47–63) | 55 (48–64) | 0.531 |
| Tumor size (mm) | Median (IQR) | 38 (28–54) | 36 (27–52) | 39 (30–54) | 0.674 | 35 (25–48) | 40 (30–55) | 0.176 |
| Nodal involvement | Negative | 43 (35.8) | 21 (48.8) | 22 (51.2) | 0.919 | 16 (37.2) | 26 (60.5) | 0.069 |
| Positive | 76 (64.2) | 35 (46.1) | 41 (53.9) | 16 (21.5) | 61 (80.3) | |||
| Missing | 1 (0.8) | 0 (0.0) | 1 (100.0) | 0 (0.0) | 1 (100.0) | |||
| Tumor grade | Grade 1 | 1 (0.8) | 0 (0.0) | 1 (100.0) | 0.003* | 0 (0.0) | 1 (100.0) | 0.002* |
| Grade 2 | 64 (53.3) | 21 (32.8) | 43 (67.2) | 9 (14.1) | 55 (85.9) | |||
| Grade 3 | 54 (45.0) | 34 (63.0) | 20 (37.0) | 23 (42.6) | 31 (57.4) | |||
| Missing | 1 (0.8) | 1 (100.0) | 0 (0.0) | 0 (0.0) | 1 (100.0) | |||
| Estrogen receptor | Positive | 78 (65.0) | 28 (35.9) | 50 (64.1) | 0.002* | 12 (15.4) | 67 (84.6) | <0.001* |
| Negative | 42 (35.0) | 28 (66.7) | 14 (33.3) | 20 (47.6) | 21 (52.4) | |||
| Progesterone receptor | Positive | 63 (52.5) | 21 (33.3) | 42 (66.7) | 0.004* | 9 (14.3) | 54 (85.7) | 0.002* |
| Negative | 57 (47.5) | 35 (61.4) | 22 (38.6) | 23 (40.4) | 34 (59.6) | |||
| HER2 receptor | Positive | 36 (30.0) | 24 (66.7) | 12 (33.3) | 0.007* | 15 (41.7) | 21 (58.3) | 0.027* |
| Negative | 84 (70.0) | 32 (38.1) | 52 (61.9) | 17 (20.2) | 67 (79.8) | |||
| Molecular Subtype | LBC | 59 (49.2) | 16 (27.1) | 43 (72.9) | <0.001* | 6 (10.2) | 53 (89.8) | <0.001* |
| L/HER2 | 21 (17.5) | 13 (61.9) | 8 (38.1) | 6 (28.6) | 15 (71.4) | |||
| HER2+ | 15 (12.5) | 11 (73.2) | 4 (26.8) | 9 (60.0) | 6 (40.0) | |||
| TNBC | 25 (20.8) | 16 (64.0) | 9 (36.0) | 11 (44.0) | 14 (56.0) | |||
| Surgery | BCS | 67 (55.8) | 37 (55.2) | 30 (44.8) | 0.054 | 24 (35.8) | 43 (64.2) | 0.014* |
| Mastectomy | 53 (44.2) | 19 (35.8) | 34 (64.2) | 8 (15.1) | 45 (84.9) |
Denotes statistical significance.
Response to NAC
Overall, 46.7% were “responders” to NAC (56/120). “Responders” were likely to have grade 3 (P<0.001), ER− (P=0.002), PgR− (P=0.004), HER2+ (P=0.002), and be L/HER2, HER2+ or TNBC molecular subtypes (P<0.001) (Table 2).
In the overall cohort, no miRNAs correlated with response to NAC (Supplementary Appendix 1.B, Supplemental Digital Content 1, http://links.lww.com/SLA/E85). In L/HER2, increased Let-7a expression identified “responders” to NAC (P=0.049). Similarly, decreased miR-21 expression trended toward significance for identifying “responders” in L/HER2 (P=0.064). For HER2+, decreased miR-145 expression identified “responders” to NAC (P=0.033) and decreased Let-7a expression trended toward significance for identifying “responders” (P=0.052). Furthermore, increased miR-21 expression identified “responders” in TNBC (P=0.089) (Fig. 2).
FIGURE 2.
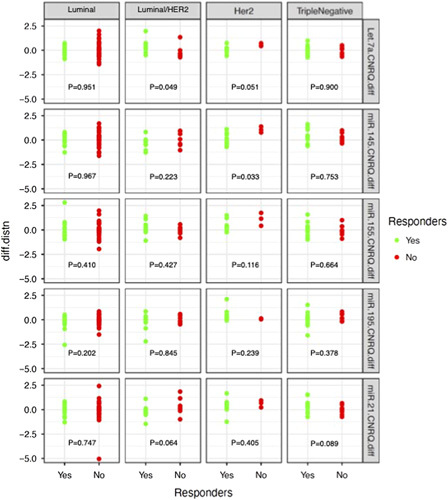
The difference in miRNA expression profiles between Timepoint 2 minus Timepoint 1 and the correlation with response to neoadjuvant chemotherapy for each of the breast cancer molecular subtypes.
Pathological Complete Response to NAC
Overall, 26.7% of patients achieved a pCR (32/120). Those achieving a pCR were likely to be node negative (P<0.001), grade 3 (P<0.001), ER− (P<0.001), PgR− (P=0.002), HER2+ (P=0.027), and be L/HER2, HER2+ or TNBC molecular subtypes (P<0.001). Patients achieving a pCR were likely to undergo BCS (P=0.014) (Table 2).
In the overall cohort, no miRNAs correlated with achieving a pCR (Supplementary Appendix 1.C, Supplemental Digital Content 1, http://links.lww.com/SLA/E85). In LBC, reduced Let-7a expression predicted achieving a pCR (P=0.037), while increased miR-145 expression trended toward significance in predicting a pCR (P=0.072). In HER2+, reduced miR-145 expression predicted achieving a pCR (P=0.027), while reduced Let-7a expression trended toward significance in predicting a pCR (P=0.062), In L/HER2, reduced miR-21 expression trended toward significance in predicting a pCR (P=0.058) (Fig. 3).
FIGURE 3.
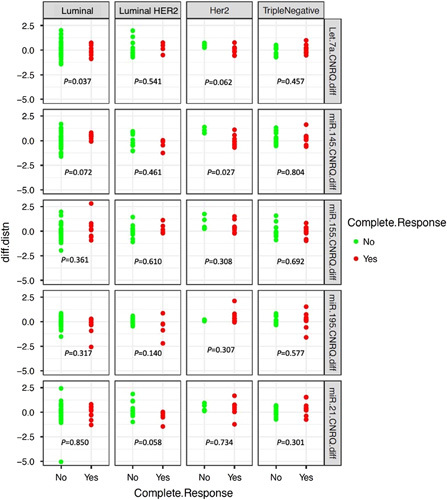
The difference in miRNA expression profiles between Timepoint 2 minus Timepoint 1 and the correlation with pathological complete response to neoadjuvant chemotherapy for each of the breast cancer molecular subtypes.
DISCUSSION
Breast cancer’s subclassification into distinct molecular subtypes is essential for personalizing treatment strategies.2,25 Once oncological control is established, efforts are focused on promoting personalized treatment. This represents the ideology of precision oncology and the aim of ICORG10/11 was to identify biomarkers of host response. Such a biomarker could allow individualization of therapy by identifying responders who should continue the course of treatment as planned, unlike their nonresponding counterparts who may benefit from halting NAC and proceeding to surgery. While Luminal A is consistently endocrine responsive,35 there was a traditional tendency to deliver chemotherapy to all patients with locally advanced and node-positive LBC.36 ICORG 10/11 pragmatically included LBC patients receiving NAC to elucidate responsible biomarkers. Unsurprisingly, pCR rates, the proportion of “responders,” and miRNA profiles which correlated with treatment response varied among molecular subgroups, therefore substratifying these subgroups based on their sensitivity to NAC, while laying the foundation for the discovery of novel miRNA-related molecular subtypes.37
Reduced miR-145 expression predicted patients who achieved a pCR to NAC in HER2+ (ER−/HER2+) disease (P=0.027), coinciding with the results of the translational research NeoALTTO trial.38 In NeoALTTO, Di Cosimo and colleagues described increased plasma miR-145 expression of those achieving a pCR after 2 weeks treatment with Trastuzumab. Moreover, reduced miR-145 expression differentiated “responders” from “nonresponders” following NAC in this study (P=0.033). Recent ASCO/CAP guidelines recommend NAC for patients with HER2+ cancers, with exceptions limited to those with T1a-b/N0 disease.39 Therefore, further validation of these preliminary results assessing the utility of miR-145 in delineating response to NAC is crucial before miR-145 may be used to guide therapeutic decision-making regarding NAC in clinical practice. Furthermore, such a biomarker may be useful in dividing HER2+ patients into two narrower molecular subtypes: Those likely to respond (and derive benefit from continuing NAC) and those unlikely to respond (and may be beneficiaries from early NAC cessation and progression to surgery). However, further interrogation of this biomarker in the next generation of translational research trials is mandated before fully establishing its value in predicting tumour sensitivity to NAC in HER2+ disease.
Increased expression of Let-7a segregated “responders” from “nonresponders” in L/HER2 (ER+/HER2+) (P=0.049). Interestingly, reduced Let-7a expression differentiated those achieving a pCR (P=0.062) and “responders” in HER2+ (ER-/HER2+) disease (P=0.052). While the current paradigm encourages the division of HER2+ breast cancers based on ER status,2 the use of Let-7a may provide further subdivision based on therapeutic response to NAC, as observed in ICORG 10/11 in correlating reduced miR-145 with the reduction in tumor volume in response to NAC. Furthermore, response to NAC predicts long-term oncological outcomes,12,13 therefore, it is unsurprising that Fuso et al40 demonstrated a survival advantage with increased Let-7a-5p expression in HER2+ disease (event-free survival: 61 vs. 36 mo, P=0.050). Thus, this study supports the potential use of Let-7a profiling to inform response to NAC which may translate indirectly into long-term survival outcomes.
This study provides preliminary data regarding reduced miR-21 in L/HER2 in predicting a pCR (P=0.058) and aberrant expression of miR-21 in deciphering “responders” from “nonresponders” in L/HER2 (P=0.064) and TNBC (P=0.089). This is a potentially anticipated finding: in previous ICORG10/11 data published from McGuire et al,41 miR-21 expression measured at T1 in isolation acted as an independent predictor of response to NAC (P=0.036). Similarly in the Geparquinto trial, Müller et al42 reported increased baseline miR-21 expression differentiated “responders” from “nonresponders” in HER2+ disease, while Rodriquez-Martinez et al43 outlined the discriminative ability of miR-21 to predict pCR to NAC. Interestingly, upregulation of miR-21 via signaling of the MAPK pathway correlates with increased HER2 signaling,44 providing the rationale for changes in miR-21 expression to predict NAC response in HER2+ disease in ICORG10/11.41–43 Moreover, miR-21 is a promotor of proliferation, invasion and initiating epithelial-mesenchymal transition in TNBC,45,46 supporting the reduction in miR-21 correlating with “response” to NAC in TNBC.
In LBC, decreased Let-7a expression correlated with achieving a pCR (P=0.037), while miR-195 expression failed to indicate sensitivity to NAC. These results refute the results from NeoALTTO, where reduced miR-195 identified those who achieved a pCR to combined Trastuzamab and Lapatinib in HER2+ disease.38 Furthermore, mir-195 levels have correlated with lower grade, reduced proliferation, and “True” LBC.20 While McGuire et al described reduced miR-145 to correlate with “responders” in LBC,41 increased miR-145 expression trended toward significance for predicting pCR in LBC (P=0.064). This emphasizes the importance of serial miRNA measurement to aid prediction of response to NAC, as demonstrated in the in vivo data from ICORG10/11 and NeoALTTO.
This study has several limitations: firstly, patients recruited were all habitants of a unique cultural European region, facilitating unavoidable selection from a small genetic pool, inherently limiting the translation of these results to a global level. Secondly, the predetermined miRNA panel evaluated included targets of most interest at the time of trial design. During the time elapsed between study initiation and completion, newer miRNA targets have been discovered. Thirdly, while this study provides insights into predicting response to NAC for each biomolecular subtype, this substratification of tumors into 4 subtypes was based on the 12th St. Gallen Expert Consensus.25 More recently, the 13th Expert Consensus Panel revised this subclassification to encompass 5 molecular subtypes.2 In addition, the seminal TAILORx and RxPONDER trials now facilitate RS to identify candidates with ER+/HER2− disease who may be spared systemic chemotherapies, whether in the neoadjuvant or adjuvant settings.47–50 Finally, although the goal of this study is to individualize NAC prescription based on circulating miRNA are promising, there are of course preliminary results which will inevitably require validation before having actual influence upon therapeutic decision making in clinical practice.
The ICORG10/11 study is the first prospective, multicenter, neoadjuvant translational research trial conducted evaluating and illustrating the potential value of circulatory miRNA measurement in predicting response to NAC in an Irish population. This study supports the ideology that circulatory miRNAs may personalize therapeutic decision-making for patients indicated to receive standard-of-care NAC. The next generation of translational research trials should focus on interrogating the use of circulatory miRNAs to inform tumor sensitivity to NAC, before adaptations to current therapeutic strategies may be made in clinical practice.
Supplementary Material
DISCUSSANTS
Catalin Vasilescu (Bucharest, Romania)
The manuscript is based on a prospective, multicenter clinical trial, addressing an original and exciting topic, namely the fact that the measurement of circulating miRNAs may predict the response to NAC, aiding in the informed decision-making process regarding treatment de-escalation in breast cancer. To my knowledge, the literature published on this subject is scarce so far. The manuscript has several strengths.
First, the fact that the trial is prospective and truly multi-institutional, involving 8 independent sites, is a strength. Most of the microRNAs biomarkers studies are published based on retrospective sets of data usually using a couple of sites on the training/validation design. Therefore, the data from this manuscript are more reliable.
Second, the authors used liquid biopsy at the site of miRNAs profiling by a simple and less expensive method: qRT-PCR. Therefore, the methodology is easy to perform not only within a research setting but also within a medical setting, so it has a high clinical applicability.
Third, the selected microRNAs to be tested are already well-known players in the pathogeny of breast cancer. Therefore, their significance for developing new therapeutics is high.
Finally, the statistical analyses are appropriate (presented and conducted) but there are some aspects that should be taken into account for completions: (I) The authors should mention, briefly (in the “statistical analysis” section) why the continuous variables are also investigated with nonparametric tests. (II) In statistics, it is very well known that the Kruskal-Wallis test is a nonparametric method for testing whether samples have their origins in the same distribution; it extends the Mann-Whitney U test to more than 2 groups. The null hypothesis of the Kruskal-Wallis test means that the mean ranks of the groups are the same. The alternative hypothesis of the test is not that one of the distributions has a different median. It is that one of the distributions has exactly the same shape as the others, but is shifted up-wards or down-wards – so you cannot conclude that there is a difference in median just because you reject the null hypothesis (with a P-value less than your threshold). You might also reject the null hypothesis because of a lack of independence, or because distributions do not have the same shapes. As a preliminary analysis, the shapes of the distributions should be investigated, and if there are no significant differences in shape, then the Kruskal-Wallis test might reveal the differences in medians. In short, these aspects should be presented in the “statistical analysis” section.
Response from Matthew G. Davey (Galway, Ireland)
In response to the first part of your discussion, this trial was designed over 10 years ago, when microRNAs were considered to be novel biomarkers. At the time of trial design, perhaps 100 to 300 biomarkers had been previously described. Currently, we know of 3000 to 4000 microRNAs. At the time of the trial design, we pragmatically included the ones that were most relevant to breast cancer pathogeny, and these were the ones we which perceived to be the most likely to inform therapeutic decision-making at that time. We are currently designing the next phase of neoadjuvant translational breast cancer research trials and have learned that predetermining the biomarkers of interest at the time of trial design may, in fact, not be the best way to identify biomarkers for evaluation in such a trial.
With respect to the performance of the Kruskal-Wallis analysis, the Department of Surgery has collaborated with our local Department of Mathematics and Statistics, which was responsible for performing the data analytics for the ICORG 10/11 trial. Our colleagues in the Department of Mathematics and Statistics have reassured us that they used the Shapiro-Wilk test to determine whether microRNA expression data was in a parametric or nonparametric distribution before non-parametric data analytics were performed.
Bas Wijnhoven (Rotterdam, The Netherlands)
You also perhaps had the opportunity to get some tissue samples. If you think that these microRNAs are shedding from the tumor, it might be nice to have a look at the tissue before neoadjuvant chemotherapy. Most of them had surgery; did you get tissue biopsies from after the operation as well, and try to see whether the differences make sense in the tissue and whether they correlated with the serum? What are the target genes of these microRNAs, and can you explain, to some extent, the observations by looking at the target genes or whether the microRNAs influence some tumor characteristics?
Response from Matthew G. Davey (Galway, Ireland)
That’s a very interesting point and it is something that we have since considered. As I previously mentioned, we are now designing the next generation of microRNA clinical trials to evaluate the role in neoadjuvant chemotherapy decision-making. We do intend to take a core biopsy at diagnosis, and then, at the time of resections, as well as blood samples along the way. It’s well established that microRNA expressions in the blood don’t necessarily correlate with that in the tumor, which not only speaks to the ubiquitous nature of microRNA expression but also how non-specific their behavior may be. It will be very exciting to compare the biopsies, from the initial diagnostic biopsy to the one at resection, to see what the microRNA profiles are like and correlate them with the blood. What that will inform, however, is yet to be seen. With respect to the targets of these microRNAs, this analysis was unfortunately not captured within the objectives or results of the ICORG 10/11 trial.
John Reynolds (Dublin, Ireland)
Regarding the association between microRNA 145 and HER-2 positive tumors, is that a random hit or is there a biological paradigm that might underpin it?
Response from Matthew G. Davey (Galway, Ireland)
I don’t think that this result was a random hit for two reasons. First, the NeoALTTO study, which is an even larger, though similar, prospective neoadjuvant translational research trial of 450 HER-2 positive patients, identified that differential expression of miR-145-5p, which is of a similar structure to miR-145, correlated with achieving a pathological complete response. These findings correlate directly with the results we found in this study. We understand miR-145 to be an oncomir, meaning it is a pro-oncogenic biomarker. Taking that into account, as a tumor is hopefully responding and downsizing to treatment, we would then expect to see reduced miR-145 expression that correlates with a reduction in the measurement of miR-145 in the circulation. That would be a reasonable hypothesis as to why we have observed these results in this study, and why this has been previously seen in another prospective clinical trial.
Footnotes
H.M.H., N.M., and M.J. Kerin: circulating miR-195 as a biomarker patent. All miRNA were measured on blinded samples and the unblinded analysis was performed by independent study statisticians.
The study was supported by Clinical Trials Ireland (formerly the All-Ireland Cooperative Oncology Research Group; ICORG) and the National Breast Cancer Research Institute (Ireland).
The authors report no conflicts of interest.
Supplemental Digital Content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's website, www.annalsofsurgery.com.
Contributor Information
Matthew G. Davey, Email: mattgdavey1@gmail.com.
Maire Caitlin Casey, Email: maireccasey@rcsi.ie.
Andrew McGuire, Email: andymcg963@gmail.com;Susanne.Gaal@usz.ch.
Ronan M. Waldron, Email: ronanwaldy@gmail.com;Susanne.Gaal@usz.ch.
Maxwell Paganga, Email: M.Paganga1@nuigalway.ie.
Emma Holian, Email: eemma.holian@nuigalway.ie;Susanne.Gaal@usz.ch.
John Newell, Email: john.newell@nuigalway.ie;Susanne.Gaal@usz.ch.
Helen M. Heneghan, Email: helen.heneghan@nuigalway.ie;Susanne.Gaal@usz.ch.
Ailbhe M. McDermott, Email: ailbhemcdermott@gmail.com.
Maccon M. Keane, Email: maccon.keane@hse.ie;Susanne.Gaal@usz.ch.
Aoife J. Lowery, Email: aoife.lowery@nuigalway.ie;Susanne.Gaal@usz.ch.
Nicola Miller, Email: nicola.miller@nuigalway.ie.
Michael J. Kerin, Email: michael.kerin@nuigalway.ie;Susanne.Gaal@usz.ch.
REFERENCES
- 1.Nardin S, et al. Breast cancer survivorship, quality of life, and late toxicities. Front Oncol. 2020;10:864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goldhirsch A, et al. Personalizing the treatment of women with early breast cancer: highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann Oncol. 2013;24:2206–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paik S, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. [DOI] [PubMed] [Google Scholar]
- 4.Perou CM, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. [DOI] [PubMed] [Google Scholar]
- 5.Fallahpour S, et al. Breast cancer survival by molecular subtype: a population-based analysis of cancer registry data. CMAJ open. 2017;5:E734–E739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomssen C, et al. St. Gallen/Vienna 2021: a brief summary of the consensus discussion on customizing therapies for women with early breast cancer. Breast Care (Basel, Switzerland). 2021;16:135–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mauri D, Pavlidis N, Ioannidis JP. Neoadjuvant versus adjuvant systemic treatment in breast cancer: a meta-analysis. J Natl Cancer Inst. 2005;97:188–194. [DOI] [PubMed] [Google Scholar]
- 8.Asselain B, et al. Long-term outcomes for neoadjuvant versus adjuvant chemotherapy in early breast cancer: meta-analysis of individual patient data from ten randomised trials. Lancet Oncol. 2018;19:27–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schott AF, Hayes DF. Defining the benefits of neoadjuvant chemotherapy for breast cancer. J Clin Oncol. 2012;30:1747–1749. [DOI] [PubMed] [Google Scholar]
- 10.Mougalian SS, et al. Use of neoadjuvant chemotherapy for patients with stage I to III breast cancer in the United States. Cancer. 2015;121:2544–2552. [DOI] [PubMed] [Google Scholar]
- 11.Vugts G, et al. Patterns of care in the administration of neo-adjuvant chemotherapy for breast cancer. A population-based study. Breast J. 2016;22:316–321. [DOI] [PubMed] [Google Scholar]
- 12.Davey MG, et al. Clinicopathological response to neoadjuvant therapies and pathological complete response as a biomarker of survival in human epidermal growth factor receptor-2 enriched breast cancer—a retrospective cohort study. Breast. 2021;59:67–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spring LM, et al. Pathologic complete response after neoadjuvant chemotherapy and impact on breast cancer recurrence and survival: a comprehensive meta-analysis. Clin Cancer Res. 2020;26:2838–2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davey MG, et al. MicroRNA expression profiles and breast cancer chemotherapy. Int J Mol Sci. 2021;22:10812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davey MG, et al. The role of MicroRNA as clinical biomarkers for breast cancer surgery and treatment. Int J Mol Sci. 2021;22:8290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richard V, et al. MicroRNAs in molecular classification and pathogenesis of breast tumors. Cancers (Basel). 2021;13:5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richard V, et al. The double agents in liquid biopsy: promoter and informant biomarkers of early metastases in breast cancer. Mol Cancer. 2022;21:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oliveto S, et al. Role of microRNAs in translation regulation and cancer. World J Biol Chem. 2017;8:45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Erson AE, Petty EM. MicroRNAs in development and disease. Clin Genet. 2008;74:296–306. [DOI] [PubMed] [Google Scholar]
- 20.Heneghan HM, et al. Systemic miRNA-195 differentiates breast cancer from other malignancies and is a potential biomarker for detecting noninvasive and early stage disease. Oncologist. 2010;15:673–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heneghan HM, et al. Circulating microRNAs as novel minimally invasive biomarkers for breast cancer. Ann Surg. 2010;251:499–505. [DOI] [PubMed] [Google Scholar]
- 22.Waldron RM, et al. MicroRNAs as biomarkers of multimodal treatment for rectal cancer. Br J Surg. 2021;108:e260–e261. [DOI] [PubMed] [Google Scholar]
- 23.Wan G, Lim QE, Too HP. High-performance quantification of mature microRNAs by real-time RT-PCR using deoxyuridine-incorporated oligonucleotides and hemi-nested primers. Rna. 2010;16:1436–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davey MG, et al. Overview of MicroRNA expression in predicting response to neoadjuvant therapies in human epidermal growth receptor-2 enriched breast cancer—a systematic review. Breast Cancer (Auckl). 2022;16:11782234221086684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goldhirsch A, et al. Strategies for subtypes—dealing with the diversity of breast cancer: highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann Oncol. 2011;22:1736–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mahlow J, et al. What to expect from the new ASCO/CAP guideline recommendations for hormone receptor testing in breast cancer: a national reference laboratory experience. Appl Immunohistochem Mol Morphol. 2021;29:245–250. [DOI] [PubMed] [Google Scholar]
- 27.Moelans CB, et al. Current technologies for HER2 testing in breast cancer. Crit Rev Oncol Hematol. 2011;80:380–392. [DOI] [PubMed] [Google Scholar]
- 28.Kostopoulou E, et al. Comparative evaluation of non-informative HER-2 immunoreactions (2+) in breast carcinomas with FISH, CISH and QRT-PCR. Breast. 2007;16:615–624. [DOI] [PubMed] [Google Scholar]
- 29.Dowsett M, et al. Assessment of Ki67 in breast cancer: recommendations from the International Ki67 in Breast Cancer working group. J Natl Cancer Inst. 2011;103:1656–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davey MG, et al. Ki-67 as a prognostic biomarker in invasive breast cancer. Cancers (Basel). 2021;13:4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amin MB, et al. The Eighth Edition AJCC Cancer Staging Manual: continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA Cancer J Clin. 2017;67:93–99. [DOI] [PubMed] [Google Scholar]
- 32.Ogston KN, et al. A new histological grading system to assess response of breast cancers to primary chemotherapy: prognostic significance and survival. Breast. 2003;12:320–327. [DOI] [PubMed] [Google Scholar]
- 33.Lowery AJ, et al. MicroRNA signatures predict oestrogen receptor, progesterone receptor and HER2/neu receptor status in breast cancer. Breast Cancer Res. 2009;11:R27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McDermott AM, Kerin MJ, Miller N. Identification and validation of miRNAs as endogenous controls for RQ-PCR in blood specimens for breast cancer studies. PLoS One. 2013;8:e83718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davey MG, et al. Disease recurrence and oncological outcome of patients treated surgically with curative intent for estrogen receptor positive, lymph node negative breast cancer. Surg Oncol. 2021;37:101531. [DOI] [PubMed] [Google Scholar]
- 36.Fisher B, et al. Tamoxifen and chemotherapy for lymph node-negative, estrogen receptor-positive breast cancer. J Natl Cancer Inst. 1997;89:1673–1682. [DOI] [PubMed] [Google Scholar]
- 37.Søkilde R, et al. Refinement of breast cancer molecular classification by miRNA expression profiles. BMC Genomics. 2019;20:503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Di Cosimo S, et al. Plasma miRNA levels for predicting therapeutic response to neoadjuvant treatment in HER2-positive breast cancer: results from the NeoALTTO Trial. Clin Cancer Res. 2019;25:3887. [DOI] [PubMed] [Google Scholar]
- 39.Korde LA, et al. Neoadjuvant chemotherapy, endocrine therapy, and targeted therapy for breast cancer: ASCO guideline. J Clin Oncol. 2021;39:1485–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fuso P, et al. Let-7a-5p, miR-100-5p, miR-101-3p, and miR-199a-3p hyperexpression as potential predictive biomarkers in early breast cancer patients. J Pers Med. 2021;11:816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McGuire A, et al. Prospective assessment of systemic MicroRNAs as markers of response to neoadjuvant chemotherapy in breast cancer. Cancers. 2020;12:1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Müller V, et al. Changes in serum levels of miR-21, miR-210, and miR-373 in HER2-positive breast cancer patients undergoing neoadjuvant therapy: a translational research project within the Geparquinto trial. Breast Cancer Res Treat. 2014;147:61–68. [DOI] [PubMed] [Google Scholar]
- 43.Rodríguez-Martínez A, et al. Exosomal miRNA profile as complementary tool in the diagnostic and prediction of treatment response in localized breast cancer under neoadjuvant chemotherapy. Breast Cancer Res. 2019;21:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang TH, et al. Up-regulation of miR-21 by HER2/neu signaling promotes cell invasion. J Biol Chem. 2009;284:18515–18524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arisan ED, et al. MiR-21 is required for the epithelial-mesenchymal transition in MDA-MB-231 breast cancer cells. Int J Mol Sci. 2021;22:1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fang H, et al. miRNA-21 promotes proliferation and invasion of triple-negative breast cancer cells through targeting PTEN. Am J Transl Res. 2017;9:953–961. [PMC free article] [PubMed] [Google Scholar]
- 47.Kalinsky K, et al. 21-Gene assay to inform chemotherapy benefit in node-positive breast cancer. N Engl J Med. 2021;385:2336–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sparano JA, et al. Adjuvant chemotherapy guided by a 21-gene expression assay in breast cancer. N Engl J Med. 2018;379:111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boland MR, et al. Value of a 21-gene expression assay on core biopsy to predict neoadjuvant chemotherapy response in breast cancer: systematic review and meta-analysis. Br J Surg. 2021;108:24–31. [DOI] [PubMed] [Google Scholar]
- 50.Davey MG, et al. Clinical utility of the 21-gene assay in predicting response to neoadjuvant endocrine therapy in breast cancer: a systematic review and meta-analysis. Breast. 2021;58:113–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


