Objective:
To propose a new decision algorithm combining biomarkers measured in a tumor biopsy with clinical variables, to predict recurrence after liver transplantation (LT).
Background:
Liver cancer is one of the most frequent causes of cancer-related mortality. LT is the best treatment for hepatocellular carcinoma (HCC) patients but the scarcity of organs makes patient selection a critical step. In addition, clinical criteria widely applied in patient eligibility decisions miss potentially curable patients while selecting patients that relapse after transplantation.
Methods:
A literature systematic review singled out candidate biomarkers whose RNA levels were assessed by quantitative PCR in tumor tissue from 138 HCC patients submitted to LT (>5 years follow up, 32% beyond Milan criteria). The resulting 4 gene signature was combined with clinical variables to develop a decision algorithm using machine learning approaches. The method was named HepatoPredict.
Results:
HepatoPredict identifies 99% disease-free patients (>5 year) from a retrospective cohort, including many outside clinical criteria (16%–24%), thus reducing the false negative rate. This increased sensitivity is accompanied by an increased positive predictive value (88.5%–94.4%) without any loss of long-term overall survival or recurrence rates for patients deemed eligible by HepatoPredict; those deemed ineligible display marked reduction of survival and increased recurrence in the short and long term.
Conclusions:
HepatoPredict outperforms conventional clinical-pathologic selection criteria (Milan, UCSF), providing superior prognostic information. Accurately identifying which patients most likely benefit from LT enables an objective stratification of waiting lists and information-based allocation of optimal versus suboptimal organs.
Key Words: hepatocellular carcinoma, liver transplant, gene expression signature, prognostic, algorithm
Hepatocellular carcinoma (HCC) is the most common primary malignancy of the liver with over 900,000 new cases diagnosed annually worldwide.1 It is the third leading cause of cancer-related mortality, killing more than 800,000 patients/year.1 Clinical decision making in the setting of a multidisciplinary tumor board, made by an experienced team able to select the proper treatment for each individual patient, is key in the management of HCC. Due to organ scarcity, partial hepatectomy is accepted in many centers as the first-line treatment for HCC.2 However, poor underlying liver function, as well as tumor number and location, preclude hepatic resection in many patients.3 Liver transplantation (LT) in adequately selected patients was popularized by the works of Bismuth et al4 and of Mazzaferro et al,5 that subsequently introduced the Milan criteria (MC). Presently, the 5-year overall survival (OS) after LT for HCC within the MC varies from 61% up to 94%,6,7 equivalent to indications for benign disease, making LT the treatment of choice for HCC in selected patients.
The limitations of the MC are, however, well recognized by the community.8–15 On one hand it still selects a significant number of patients for transplant that relapse afterwards, resulting in allocation of organs to patients that proved not to benefit from this procedure. On the other, many patients that fall outside the MC can have a good prognosis if transplanted (false negative according to MC). This situation is illustrated by our own experience at the Portuguese reference transplant center of Curry Cabral Hospital (CCH): an OS rate of >50% in patients transplanted beyond MC, even with suboptimal organs.16 Hence, many patients who could be potentially treated have a transplant denied as they fall outside the current criteria. Improved criteria for transplantation are thus necessary to ensure that the limiting number of organs available for transplant are used more efficiently, with higher survival rates and reduced overall costs per successful transplant. Several authors proposed expansion of these criteria, essentially based on morphological features17–20 or in the presence of serologic markers such as alpha-fetoprotein (AFP),21–23 neutrophil to lymphocyte ratio24 or protein induced by vitamin K absence-II.25 All these criteria differentiate patients with low risk of recurrence and patients with higher risk, but all failed to reach a global consensus. AFP in particular has been extensively validated and even adopted in several national guidelines,26 but accumulating clinical data suggests that AFP measurements lack sensitivity and reproducibility mostly due to absence of universally established cut off.23,26,27
Several groups have suggested downstaging approaches, where a patient with intrahepatic disease noneligible for transplantation can become eligible by reduction of the tumor through locoregional or systemic approaches.28 These have shown great promise with downstaged patients having survival rates after transplantation comparable to patients originally meeting transplantation criteria.29 However, there is currently no standard definition of successful downstaging. In addition, downstaging requires waiting periods and/or adequate response to therapy, leading many times to disease complications and dropouts.30
Advances in genomic technologies have fueled a major change in biomarker research and the discovery and clinical translation of molecular biomarkers for a variety of cancer types and medical decisions.31,32 Gene expression signatures have gained increasing impact in oncology as prognostic markers. For example multiple gene expression signatures are now routinely used and integrate international guidelines for patient stratification and therapy selection in breast cancer33 and prostate cancer.34 However, despite the enthusiasm driving the search for molecular biomarkers for HCC,35–45 the lack of multicohort validation, relevant predictive power, together with the lack of focus on specific clinical decisions have precluded their widespread adoption.
Here we propose a new gene expression signature measured in the tumor which, integrated in a decision algorithm with clinical variables, predicts low recurrence rates and survival in cirrhotic patients with HCC following orthotopic LT.
METHODS
Systematic Review of Biomarkers
We performed a systematic literature review aiming to identify molecular biomarkers prognostic of LT in HCC (details in the Supplemental Digital Content Tables 1, 2 Supplemental Digital Content 1, http://links.lww.com/SLA/E111).
Study Cohort and Design
We analyzed a retrospective collection of 138 HCC patients submitted to LT in our Center CCH, out of 301 LT HCC patients from September 1992 to February 2014 (Supplemental Digital Content Table 3, Supplemental Digital Content 1, http://links.lww.com/SLA/E111, and details in the Supplemental Digital Content Methods, Supplemental Digital Content 1, http://links.lww.com/SLA/E111). Patient exclusion criteria included: age above 18 years; absence of cirrhosis; Fibrolamellar or Hepatocholangiocarcinoma; extra-hepatic-invasion or residual disease. All patients received whole liver grafts for their first elective transplantation. We considered recurrence our main outcome measure. A subset of cases (26) within/outside MC or with/without recurrence, selected by random sampling, was initially used as a pilot set to test candidate biomarkers.
The first part of the study consisted of analyzing the differential expression of candidate biomarkers using the pilot set of samples and correlating the results with recurrence and within/outside criteria status. The top 4 candidates with significant differential expression were further selected and subsequently tested with the full cohort.
Both the CCH medical ethical review committee and the ethical review board of NOVA Medical School approved this study.
Gene Expression Measurements
Histopathological characterization and area selection (mimicking needle biopsy of viable tumor area) of archived formalin-fixed, paraffin embedded (FFPE) surgical explants was carried out under the supervision of experienced pathologists. Total RNA was extracted from selected areas of 2 consecutive 20 mm2, 5 μm thick FFPE sections per sample, equivalent to the material obtained in a needle biopsy, using the RNeasy FFPE kit (#73504, Qiagen) after deparaffinization with #19093 (Qiagen) according to manufacturer’s instructions (exception: proteinase K incubation performed overnight). Expression levels of the genes of interest, reference genes, and residual genomic DNA contamination (Chr3) were assessed by independent 1-step RT-qPCR reactions. Primers and probes were designed for an amplicon length of 70 to 108 bp (Supplemental Digital Content Table 4A and B, Supplemental Digital Content 1, http://links.lww.com/SLA/E111). All samples were analyzed in duplicate using TaqPath 1-step RT-qPCR Master Mix (#A15300, Thermo Fisher Scientific) in a final reaction volume of 10 μL containing template (1 μL), 0.5 μM each primer (0.25 μM for Chr3) and 0.25 μM probe (0.125 μM for Chr3). All reactions were processed using QuantStudio 5 Real-Time PCR System, 96-well, 0.1 mL (#A34322, ThermoFisher Scientific) with the cycling program: 25°C, 2 minutes and 50°C, 15 minutes (RT) followed by 95°C, 2 minutes and 40 cycles of 95°C, 3 s and 58°C, 30 s (qPCR). The geometric mean of the cycle threshold (Cq) of the reference genes was subtracted to the Cq values of the genes of interests for normalization.46
Statistical Analysis and Machine Learning Modeling
Statistical analysis was performed with R language for Statistical Computing. The outcome variables were recurrence and death (OS). Time to outcome was calculated using the LT date until date of the event, or until date of last follow-up time (for patients without event). Kaplan-Meier survival curves were constructed for outcome analysis after transplantation.
The development of a machine learning supervised model was built on the scikit-learn framework in Python (V1.0.1). We systematically tested all 10 algorithms present in scikit-learn (Supplemental Digital Content Methods, Supplemental Digital Content 1, http://links.lww.com/SLA/E111), feeding each model 4 molecular and 32 clinical variables. Each model was trained on a subset of data and then tested using jackknifing to minimize overfitting, for predictive power for recurrence of machine learning algorithms. We systematically investigated both continuous and discrete variables, and individual and ensembles of predictors. We selected for further investigation models with highest average precision and/or recall, and reiterated the process systematically searching for parameters and variable combinations that optimized predictive power. Feature selection was informed both by each feature mutual information with relapse, and its contribution to the predictive power of the algorithm.
RESULTS
Identification of relevant published molecular biomarkers was performed through a systematic review of the literature (Supplemental Digital Content Table 1, Supplemental Digital Content 1, http://links.lww.com/SLA/E111). Most relevant candidate genes (N=16, Supplemental Digital Content Table 2, Supplemental Digital Content 1, http://links.lww.com/SLA/E111) were analyzed for significant differential expression using a pilot set of cases and correlated to clinical data (eg, recurrence status, inside/outside criteria) to further trim down the number of candidates. From 16 genes whose expression was tested in the pilot set (Supplemental Digital Content Table 5, Supplemental Digital Content 1, http://links.lww.com/SLA/E111), we selected four that gave strongest and consistent expression differences (CLU, DPT, SPRY2, CAPSN1—see Supplemental Digital Content Table 5, Supplemental Digital Content 1, http://links.lww.com/SLA/E111 and Supplemental Digital Content Figure 1 for details, Supplemental Digital Content 1, http://links.lww.com/SLA/E111). A systematic testing of combinations of clinical variables measurable in the pretransplant setting, resulted in the choice of 3 variables which maximized the combined predictive power of the algorithms: largest nodule size, total tumor volume and number of nodules. Other variables had individually predictive power but did not bring additional when in combination.
Our systematic, brute force search for features and algorithms, revealed that by combining 2 distinct algorithms, 1 optimized for precision and another for recall, we could outperform other combinations as well as current clinical criteria and address distinct clinical problems, as discussed below. HepatoPredict is then a 2-level Linear Support-Vector Machine predictor: the first level (Class l), works with highest precision while the second level (Class II) with higher recall, is another Linear Support-Vector Machine that forecasts on the negative predictions of the first level, with different variables and cut off values.
When used at highest precision (Class I), HepatoPredict proposes for transplantation ~57% and ~51% of the patients, compared with Milan and UCSF, respectively, but with a positive predictive value (PPV) of 94.3% (Fig. 1A). At highest sensitivity mode (Class I+II) it proposes for transplantation ~31% more patients than MC, and more ~17% than UCSF criteria (Fig. 1A) with no loss in positive predictive value (PPVHepatopredict=88,5% vs. PPVMilan=88,2%, PPVUCSF=86,5%) and can identify all good prognosis patients identified under Milan or UCSF criteria (Fig. 1B). Addition of Milan or UCSF positive predictions to HepatoPredict either in Class I alone or Class I+II brought no additional sensitivity but increased the rate of erroneous predictions (Supplemental Digital Content Figure 2, Supplemental Digital Content 1, http://links.lww.com/SLA/E111). Finally, improvement is observed in comparison to other patient selection models for both recall and precision (Table 1).
FIGURE 1.
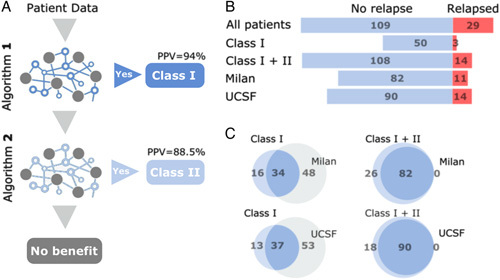
HepatoPredict 2-step algorithm. A, First algorithm identifies good prognosis patients with a PPV=94% (Class I); the remaining patients go through a second algorithm that identifies further good prognosis patients at a PPV=88,5% (Class II). For the remaining patients HepatoPredict does not predict any benefit in liver transplantation. B, Patients proposed for transplantation and their outcome after 5 years according to HepatoPredict Class I and Class I+II versus Milan Criteria (Milan) or UCSF Criteria (UCSF); a positive outcome (No relapse) represented in light blue versus a negative outcome (Relapsed) in red. B, Overlap of transplanted patients that did not relapse (darker blue) with a positive prognosis according to HepatoPredict Class I or Class I+II (light blue), Milan or UCSF criteria (light gray).
TABLE 1.
Comparison of HepatoPredict Predictive Power With Other Models
| Precision (PPV) (%) | Recall (%) | Accuracy (%) | FPR (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Criteria | HP | Criteria | HP | Criteria | HP | Criteria | HP | n | |
| Milan | 88.1 | 88.5 | 75.2 | 99.1 | 72.5 | 89.1 | 37.9 | 48.3 | 138 |
| UCSF | 86.5 | 88.5 | 82.6 | 99.1 | 76.1 | 89.1 | 37.9 | 48.3 | 138 |
| Up to 7 | 80.3 | 88.5 | 93.6 | 99.1 | 76.8 | 89.1 | 86.2 | 48.3 | 138 |
| AFP criteria | 90.0 | 92.3 | 90.0 | 100 | 82.0 | 92.5 | 85.7 | 71.4 | 67 |
| Metroticket 2.0 | 90.1 | 92.3 | 91.7 | 100 | 83.5 | 92.5 | 85.7 | 71.4 | 67 |
| TTV | 82.9 | 88.7 | 95.1 | 100 | 80.0 | 89.6 | 87.0 | 56.5 | 125 |
| TTV AFP | 91.7 | 92.2 | 93.2 | 100 | 86.4 | 92.4 | 71.4 | 71.4 | 66 |
Criteria represents the criteria under comparison, and HP stands for HepatoPredict. Computations are made over the sample size for which all data were available for the calculation (column n). Precision is the same as positive predictive value [TP/(TP+FP), recall is calculated as TP/(TP+FN), accuracy as (TP+TN)/(P+N), and FPR is the false positive rate, calculated as FP/N, where T/F stand for true/false and P/N stand for positive/negative].
HepatoPredict (Class I+II-based selection) presents increased sensitivity, that is, identifies more eligible patients for transplantation without increased recurrence rates over 5 years (Fig. 2 and Figure 5A, Supplemental Digital Content 1, http://links.lww.com/SLA/E111). Recurrence rates under Class I+II are significantly different and more expressive than under Milan or UCSF criteria (Figs. 2A, B), independently of the population of patients being originally noneligible (outside) or eligible (within) for a transplant according to Milan (Figs. 2C, E) and UCSF criteria (Figs. 2D, F). In addition, HepatoPredict appears to have increased negative predictive power as the patients it rejects (outside Class I+II) have higher recurrence rates. In terms of OS, the same picture emerges: HepatoPredict (Class I+II) identifies more patients that have equivalent OS at 5 years (Supplemental Digital Content Figure 3, Supplemental Digital Content 1, http://links.lww.com/SLA/E111) and 15 years (Fig. 3 and Figure 5A, Supplemental Digital Content 1, http://links.lww.com/SLA/E111) and rejects more patients with higher death rates over the same periods when compared to Milan (Supplemental Digital Content Figure 3A, Supplemental Digital Content 1, http://links.lww.com/SLA/E111) and UCSF (Supplemental Digital Content Figure 3B, Supplemental Digital Content 1, http://links.lww.com/SLA/E111) criteria. The observations on HepatoPredict positive and negative predictive power hold for patients both outside and within Milan (Figs. 2C, E, Supplemental Digital Content Figures 3C and 3E, Supplemental Digital Content 1, http://links.lww.com/SLA/E111) and UCSF (Figs. 2D, F, Supplemental Digital Content Figures 3D and 3F, Supplemental Digital Content 1, http://links.lww.com/SLA/E111) criteria. The benefit lasts over 16 years (Fig. 3).
FIGURE 2.
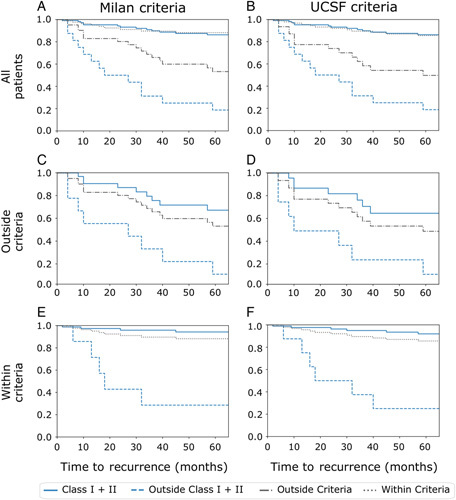
Recurrence curves of transplanted liver cancer patients at 5 years (60 months), according to HepatoPredict Class I+II (blue lines) versus Milan or UCSF Criteria (gray lines, left and right columns, respectively). Cumulative recurrence curves for the entire population of transplanted patients compared with Milan (A) and UCSF (B) criteria, for the subpopulations of patients originally classified outside Milan (C) and UCSF (D) criteria, and for the subpopulations of patients originally classified within (E) Milan and (F) UCSF criteria.
FIGURE 3.
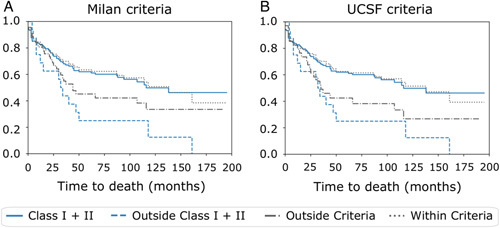
Overall survival curves of transplanted liver cancer patients at ~15 years according to the original Milan or UCSF selection criteria (left and right columns, respectively). Cumulative overall survival curves for the entire population of transplanted patients divided according to (A) Milan or (B) UCSF criteria. The curves represent the overall survival prediction according to HepatoPredict Class I+II (blue lines) versus Milan or UCSF Criteria (gray lines).
DISCUSSION
Surgical approaches and in particular LT are the best treatment for HCC patients. However, the number of transplantable organs has not significantly changed over the past few years while the indications for LT are expanding,47 making patient selection for transplantation critical. We proposed a new prognostic tool to predict success of LT for HCC patients. This tool is based on the assessment of a gene expression signature plus clinical variables using a machine learning-based decision algorithm. The gene expression signature is measured using RT-qPCR in a FFPE viable tumor needle biopsy material, which simplifies the adoption of this approach by molecular pathology laboratories. Gene expression signatures measured directly in tumor tissue are recommended in international guidelines and routinely used to determine the prognosis and/ treatment decisions of different cancers, for example, breast33 and prostate.34
The proposed tool has an overall better performance for a higher number of patients eligible for transplantation with a higher precision than clinical criteria (Fig. 1, Table 1). It performs exceptionally well in patients beyond Milan or SF Criteria, achieving comparable recurrence rates and OS as patients eligible for transplantation under Milan and SF criteria (Figs. 2, 3). This suggests that an immediate gain for patient survival would be to test patients not currently eligible for LT. We are currently evaluating this use under a prospective clinical trial (NCT04499833), as well as extending to other retrospective cohorts. HepatoPredict 2-step nature offers an objective way to support differential organ availability and quality, distinguishing the best prognosis patients within those eligible (Fig. 2, Supplemental Digital Content Figure 3, Supplemental Digital Content 1, http://links.lww.com/SLA/E111).
Intratumoral spatial and temporal heterogeneity has been observed in HCC.48,49 Here we used posttransplant surgical specimens, but propose an implementation based on a needle biopsy of the tumor pretransplantation. The laboratory implementation proposed is compatible with the minute amounts of tissue sampled using this approach, and our preliminary data (Supplemental Digital Content Table 6, Supplemental Digital Content 1, http://links.lww.com/SLA/E111) showed uniformity of the intranodule and internodule expression, independently of the analyzed tumor areas, consistent with the results of Villanueva et al50 and Losic et al,49 but further data will be needed. Moreover, the evolution of the gene expression signature over time from diagnosis to transplant, with or without downstaging will need to be addressed. Utility of the proposed approach is dependent on the demonstration that the gene expression signature has prognostic power at transplant listing. Our prospective study (NCT04499833) currently collecting single-needle biopsies at transplantation listing will allow us to assess both spatial and temporal representation of the tumor biopsy. Finally, diagnosis of rare cases of hepatocholangiocarcinoma is difficult and may negatively impact the performance of the proposed model.
Several HCC prognostic biomarkers, including molecular signatures based on mRNA, microRNAs and other genes have been reported.35–45 However, few specifically address HCC patients submitted to LT50,51 and are outperformed by the method proposed here, with the advantages of implementation simplicity and clinical objectivity. The most accepted prognostic biomarker with predictive value for recurrence and reduced post LT survival is the high level of preoperative serum AFP (>1000 ng/mL) which, when used in conjunction with MC improves its discrimination capability.21 Current consensus guidelines recommend monitoring of listed patients, in the context of bridging therapies, every 3 months using imaging and AFP measurements, to identify those who develop disease progression.52 However, about one third of HCCs are negative for AFP,53,54 and the best cut off for AFP level (alone or in combination with other clinical factor) is under debate; AFP levels can be modulated by non-HCC factors (eg, active hepatitis, liver regeneration, inflammation, etc.) which can mask a patient with good prognosis and finally a long waiting time in LT lists in some centers may render an AFP negative HCC patient to become AFP positive.55 Incomplete clinical records for patients referred from other clinical centers, precluded consideration of AFP for the proposed prognostic tool, but our preliminary analysis suggests that AFP levels are not different in HepatoPredict good and bad prognosis patients, suggesting that the biology that it captures is already considered in the variables included in our model (Supplemental Digital Content Figure 4, Supplemental Digital Content 1, http://links.lww.com/SLA/E111), and this is clear in the comparison between HepatoPredict and AFP-based models (Table 1).
The proposed approach requires a needle biopsy of a viable tumor, an invasive procedure with potential adverse events. A liver biopsy is often required when clinically important information about the diagnosis, prognosis or management of a patient cannot be obtained from noninvasive methods.56,57 Liver biopsies are currently performed under established minimally invasive techniques and, overall, liver biopsy procedures are safe,58 with minor morbidity,59 a low risk of needle tract seeding of tumor cells56 and extremely rare severe complications,60 being recommended by the EASL guidelines (2018) in certain situations. Adoption of the proposed approach will demand the demonstration that the small and manageable risk associated with obtaining the needle biopsy is outweighed by the benefit in more efficient organ allocation. Furthermore, monitoring of patients from disease presentation to surgery date will require tools that can provide a dynamic picture of the evolution of the disease, such as liquid biopsies. This is particularly relevant in face of downstaging procedures that can result in extensive necrosis.
With our approach we aimed to identify a robust set of prognostic genes for LT decisions in HCC. As already learnt from gene expression signatures for other cancers, these are not necessarily the only predictive genes.61 Extensive multicohort validation will be needed to test the clinical utility of the proposed gene expression signature and associated algorithm, including the intended use in a pretransplant setting; several retrospective and one prospective study are already ongoing. We show that incorporating biomarkers through a gene expression signature can support HCC patient selection for LT.
Supplementary Material
ACKNOWLEDGMENTS
The authors wish to thank the Centro de Medicina Laboratorial Germano de Sousa for continued support of this project, and to the Ophiomics team. They further wish to thank all those that over the years have built the Hospital Curry Cabral Biobank on which this research was based.
DISCUSSANT
Mickaël Lesurtel (Clichy France)
Thank you to the ESA for the privilege of discussing this interesting paper. In this single-center retrospective study, the authors should be complimented for trying refining selection criteria before LT for HCC, using the gene expression signature of the tumor.
The subject of the study is of importance, as refining selection criteria before LT for HCC would allow us to reduce tumor recurrence and spare liver grafts in this setting. At this stage, this is only a proof of concept that needs to be validated prospectively, or in external cohorts. I have the following questions:
First, gene expression was assessed on a surgical specimen, but not on a pre-transplant biopsy. How could you convince us that a pre-LT biopsy would be sufficient to determine genetic profiling in the context of the heterogeneity of HCC with necrotic parts, multinodular tumors, and neoadjuvant treatment?
Second, a limitation of your study lies in the absence of internal or external validation. The chosen genes were selected on a pilot set and the proposed algorithm was built based on retrospective survival data belonging to the same monocentric series. Don’t you think it would be prudent to wait for the results of your prospective study and validations?
Finally, if your proof of concept is confirmed in the future, how will you select HCC patients in practice? Which thresholds would you set, in terms of recurrence and survival, and which cut-off values, in terms of tumor variables, would you use?
Response From Sílvia Gomes Da Silva (Lisboa, Portugal)
Thank you for your comments and questions. First, regarding the sample size, we have already performed a technical validation, and we can find gene expression in a very small amount of tissue. A needle biopsy for diagnosis is enough. Next, regarding the heterogeneity, I pointed out 14 (7 of each) analyses in this issue, but we are now working with another center to enlarge this cohort, and this information will be more robust.
Regarding the issue of necrosis is of uttermost importance, as we cannot find gene expression in necrotic tissue. We can only find gene expression in viable tissue. Therefore, we recommend performing the biopsy at the moment of diagnosis, and if the patient is beyond Millan Criteria, we perform HepatoPredict. For patients, who have already undergone downstaging of their tumor, our interventional radiologists only perform biopsies in viable tissue. Following this, our pathologists analyze the biopsies, and we only perform the signature if HCC is identified. Evidently, the entire team has to be involved in this process, and if every element knows what to look for, we can obtain good results.
With regard to your remark on the change in biology with downstaging, we are already addressing it. We have started an active collaboration with La Fe Hospital, in Valencia, and are hoping to receive the first samples to enlarge our retrospective cohorts soon. Also, we are currently analyzing this variable in the prospective study.
Moving on to your second question regarding the timing of results, as I mentioned previously, this is a proof of principle study, which is very precise and easy to assess. We are aware that we need external and internal validation; however, in order to obtain validations, we need collaborations. That is why it is so pivotal that we share this idea and our results with the wider transplantation community.
Finally, regarding the applicability of the model, we have already started putting it into practice. We can perform needle biopsies with a small amount of tissue, which is followed by gene analysis and the application of the algorithm. These patients wouldn’t be transplanted, if it weren’t for these signatures, and the benefit for the patient is clearly higher than the risk of a needle biopsy.
Footnotes
H.P.-M. and J.C. contributed equally and share first authorship.
This work was partly funded by a grant from the European Innovation Council under the EIC Accelerator scheme (contract no. 946364).
The work described here is subject to patent WO 2021/064230 A1; J.P.L., J.C.V., E.B., and H.P.M. declare an ownership interest in the company Ophiomics.
The authors report no conflicts of interest.
Supplemental Digital Content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's website, www.annalsofsurgery.com.
Contributor Information
Hugo Pinto-Marques, Email: hugoscpm@gmail.com.
Joana Cardoso, Email: jvaz@ophiomics.com.
João L. Neto, Email: jneto@ophiomics.com.
Maria Gonçalves-Reis, Email: mreis@ophiomics.com.
Daniela Proença, Email: dproenca@ophiomics.com.
Marta Mesquita, Email: martamesquita78@gmail.com;Susanne.Gaal@usz.ch.
André Manso, Email: a-manso@hotmail.com.
Sara Carapeta, Email: sara.carapeta@germanodesousa.com.
Mafalda Sobral, Email: mafaldasnsobral@gmail.com.
Antonio Figueiredo, Email: antonio.figueiredo@chlc.min-saude.pt.
Clara Rodrigues, Email: maria.rodrigues@chlc.min-saude.pt.
Adelaide Milheiro, Email: maria.milheiro@chlc.min-saude.pt.
Ana Carvalho, Email: ana.carvalho5@chlc.min-saude.pt.
Rui Perdigoto, Email: rui.perdigoto@chlc.min-saude.pt;Susanne.Gaal@usz.ch.
Eduardo Barroso, Email: drebarroso@gmail.com.
José B. Pereira-Leal, Email: jleal@ophiomics.com.
REFERENCES
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- 2.Saito R, Amemiya H, Hosomura N, et al. Prognostic factors for post-recurrent survival in hepatocellular carcinoma after curative resection. Anticancer Res. 2019;39:3033–3038. [DOI] [PubMed] [Google Scholar]
- 3.Golabi P, Fazel S, Otgonsuren M, et al. Mortality assessment of patients with hepatocellular carcinoma according to underlying disease and treatment modalities. Medicine. 2017;96:e5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bismuth H, Chiche L, Adam R, et al. Liver resection versus transplantation for hepatocellular carcinoma in cirrhotic patients. Ann Surg. 1993;218:145–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mazzaferro V, Regalia E, Doci R, et al. Liver transplantation for the treatment of small hepatocellular carcinomas in patients with cirrhosis. N Engl J Med. 1996;334:693–700. [DOI] [PubMed] [Google Scholar]
- 6.Silva MF, Sherman M. Criteria for liver transplantation for HCC: what should the limits be? J Hepatol. 2011;55:1137–1147. [DOI] [PubMed] [Google Scholar]
- 7.Dhir M, Melin AA, Douaiher J, et al. A review and update of treatment options and controversies in the management of hepatocellular carcinoma. Ann Surg. 2016;263:1112–1125. [DOI] [PubMed] [Google Scholar]
- 8.Llovet J, Schwartz M, Fuster J, et al. Expanded criteria for hepatocellular carcinoma through down-staging prior to liver transplantation: not yet there. Semin Liver Dis. 2006;26:248–253. [DOI] [PubMed] [Google Scholar]
- 9.Yao F. Expanded criteria for hepatocellular carcinoma: down-staging with a view to liver transplantation-yes. Semin Liver Dis. 2006;26:239–247. [DOI] [PubMed] [Google Scholar]
- 10.Zaydfudim VM, Vachharajani N, Klintmalm GB, et al. Liver resection and transplantation for patients with hepatocellular carcinoma beyond milan criteria. Ann Surg. 2016;264:650–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim Y, Stahl CC, Makramalla A, et al. Downstaging therapy followed by liver transplantation for hepatocellular carcinoma beyond Milan criteria. Surgery. 2017;162:1250–1258. [DOI] [PubMed] [Google Scholar]
- 12.Rudnick SR, Russo MW. Liver transplantation beyond or downstaging within the Milan criteria for hepatocellular carcinoma. Expert Rev Gastroenterol Hepatol. 2018;12:265–275. [DOI] [PubMed] [Google Scholar]
- 13.Pavel M-C, Fuster J. Expansion of the hepatocellular carcinoma Milan criteria in liver transplantation: future directions. WJG. 2018;24:3626–3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Halazun KJ, Sapisochin G, von Ahrens D, et al. Predictors of outcome after liver transplantation for hepatocellular carcinoma (HCC) beyond Milan criteria. Int J Surg. 2020;82:61–69. [DOI] [PubMed] [Google Scholar]
- 15.Kardashian A, Florman SS, Haydel B, et al. Liver transplantation outcomes in a US. multicenter cohort of 789 patients with hepatocellular carcinoma presenting beyond Milan Criteria. Hepatology. 2020;72:2014–2028. [DOI] [PubMed] [Google Scholar]
- 16.Marques HP, Ribeiro V, Almeida T, et al. Long-term results of domino liver transplantation for hepatocellular carcinoma using the “double piggy-back” technique: a 13-year experience. Ann Surg. 2015;262:749–756. [DOI] [PubMed] [Google Scholar]
- 17.Yao F. Liver transplantation for hepatocellular carcinoma: expansion of the tumor size limits does not adversely impact survival. Hepatology. 2001;33:1394–1403. [DOI] [PubMed] [Google Scholar]
- 18.Mazzaferro V, Llovet JM, Miceli R, et al. Predicting survival after liver transplantation in patients with hepatocellular carcinoma beyond the Milan criteria: a retrospective, exploratory analysis. Lancet Oncol. 2009;10:9. [DOI] [PubMed] [Google Scholar]
- 19.Prasad KR, Young RS, Burra P, et al. Summary of candidate selection and expanded criteria for liver transplantation for hepatocellular carcinoma: a review and consensus statement: Candidate Selection and Expanded Criteria. Liver Transpl. 2011;17:S81–S89. [DOI] [PubMed] [Google Scholar]
- 20.Kaido T, Ogawa K, Mori A, et al. Usefulness of the Kyoto criteria as expanded selection criteria for liver transplantation for hepatocellular carcinoma. Surgery. 2013;154:1053–1060. [DOI] [PubMed] [Google Scholar]
- 21.Duvoux C, Roudot–Thoraval F, Decaens T, et al. Liver transplantation for hepatocellular carcinoma: a model including α-fetoprotein improves the performance of Milan criteria. Gastroenterology. 2012;143:986–994.e3. [DOI] [PubMed] [Google Scholar]
- 22.Mazzaferro V, Sposito C, Zhou J, et al. Metroticket 2.0 model for analysis of competing risks of death after liver transplantation for hepatocellular carcinoma. Gastroenterology. 2018;154:128–139. [DOI] [PubMed] [Google Scholar]
- 23.Brusset B, Dumortier J, Cherqui D, et al. Liver transplantation for hepatocellular carcinoma: a real-life comparison of Milan criteria and AFP model. Cancers. 2021;13:2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Halazun KJ, Najjar M, Abdelmessih RM, et al. Recurrence after liver transplantation for hepatocellular carcinoma: a new MORAL to the story. Ann Surg. 2017;265:557–564. [DOI] [PubMed] [Google Scholar]
- 25.Asman Y, Evenson AR, Even-Sapir E, et al. [18 F]fludeoxyglucose positron emission tomography and computed tomography as a prognostic tool before liver transplantation, resection, and loco-ablative therapies for hepatocellular carcinoma: PET for Prognosis of HCC Invasive Therapies. Liver Transpl. 2015;21:572–580. [DOI] [PubMed] [Google Scholar]
- 26.Toniutto P, Fumolo E, Fornasiere E, et al. Liver transplantation in patients with hepatocellular carcinoma beyond the Milan criteria: a comprehensive review. J Clin Med. 2021;10:3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mahmud N, John B, Taddei TH, et al. Pre‐transplant alpha‐fetoprotein is associated with post‐transplant hepatocellular carcinoma recurrence mortality. Clin Transplant. 2019;33:e13634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Llovet JM, De Baere T, Kulik L, et al. Locoregional therapies in the era of molecular and immune treatments for hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2021;18:293–313. [DOI] [PubMed] [Google Scholar]
- 29.Mazzaferro V, Citterio D, Bhoori S, et al. Liver transplantation in hepatocellular carcinoma after tumour downstaging (XXL): a randomised, controlled, phase 2b/3 trial. The Lancet Oncology. 2020;21:947–956. [DOI] [PubMed] [Google Scholar]
- 30.Parikh ND, Waljee AK, Singal AG. Downstaging hepatocellular carcinoma: a systematic review and pooled analysis: down-staging hepatocellular carcinoma. Liver Transpl. 2015;21:1142–1152. [DOI] [PubMed] [Google Scholar]
- 31.Patel LR, Nykter M, Chen K, et al. Cancer genome sequencing: understanding malignancy as a disease of the genome, its conformation, and its evolution. Cancer Lett. 2013;340:152–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chakravarty D, Solit DB. Clinical cancer genomic profiling. Nat Rev Genet. 2021;22:483–501. [DOI] [PubMed] [Google Scholar]
- 33.Varnier R, Sajous C, de Talhouet S, et al. Using breast cancer gene expression signatures in clinical practice: unsolved issues, ongoing trials and future perspectives. Cancers. 2021;13:4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choudhury A, West CML. Translating prognostic prostate cancer gene signatures into the clinic. Transl Cancer Res. 2017;6:S405–S408. [Google Scholar]
- 35.Hoshida Y, Villanueva A, Kobayashi M, et al. Gene expression in fixed tissues and outcome in hepatocellular carcinoma. N Engl J Med. 2008;359:1995–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee J-S, Chu I-S, Heo J, et al. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology. 2004;40:667–676. [DOI] [PubMed] [Google Scholar]
- 37.Singhal A, Jayaraman M, Dhanasekaran DN, et al. Molecular and serum markers in hepatocellular carcinoma: predictive tools for prognosis and recurrence. Critical Reviews in Oncology/Hematology. 2012;82:116–140. [DOI] [PubMed] [Google Scholar]
- 38.Boyault S, Rickman DS, de Reyniès A, et al. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology. 2007;45:42–52. [DOI] [PubMed] [Google Scholar]
- 39.Ke K, Chen G, Cai Z, et al. Evaluation and prediction of hepatocellular carcinoma prognosis based on molecular classification. CMAR. 2018;10:5291–5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhai X, Xue Q, Liu Q, et al. Classifier of cross talk genes predicts the prognosis of hepatocellular carcinoma. Mol Med Rep. 2017;16:3253–3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu G, Xie W, Zhang C, et al. Identification of a four‐gene metabolic signature predicting overall survival for hepatocellular carcinoma. J Cell Physiol. 2020;235:1624–1636. [DOI] [PubMed] [Google Scholar]
- 42.Zhao Q-J, Zhang J, Xu L, et al. Identification of a five-long non-coding RNA signature to improve the prognosis prediction for patients with hepatocellular carcinoma. WJG. 2018;24:3426–3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qiao G, Chen L, Wu J, et al. Identification of an eight-gene signature for survival prediction for patients with hepatocellular carcinoma based on integrated bioinformatics analysis. Peer J. 2019;7:e6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gu J-X, Zhang X, Miao R-C, et al. Six-long non-coding RNA signature predicts recurrence-free survival in hepatocellular carcinoma. World J Gastroenterol. 2019;25:220–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng Y, Liu Y, Zhao S, et al. Large-scale analysis reveals a novel risk score to predict overall survival in hepatocellular carcinoma. Cancer Manag Res. 2018;10:6079–6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vandesompele J, Preter KD, Roy NV, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:research0034.1-0034.11. [DOI] [PMC free article] [PubMed]
- 47.Hughes CB, Humar A. Liver transplantation: current and future. Abdom Radiol. 2021;46:2–8. [DOI] [PubMed] [Google Scholar]
- 48.Zhang Q, Lou Y, Bai X-L, et al. Intratumoral heterogeneity of hepatocellular carcinoma: From single-cell to population-based studies. World J Gastroenterol. 2020;26:3720–3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Losic B, Craig AJ, Villacorta-Martin C, et al. Intratumoral heterogeneity and clonal evolution in liver cancer. Nat Commun. 2020;11:291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Villanueva A, Hoshida Y, Battiston C, et al. Combining clinical, pathology, and gene expression data to predict recurrence of hepatocellular carcinoma. Gastroenterology. 2011;140:1501–1512.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dvorchik I, Schwartz M, Fiel MI, et al. Fractional allelic imbalance could allow for the development of an equitable transplant selection policy for patients with hepatocellular carcinoma. Liver Transpl. 2008;14:443–450. [DOI] [PubMed] [Google Scholar]
- 52.Clavien P-A, Lesurtel M, Bossuyt PM, et al. Recommendations for liver transplantation for hepatocellular carcinoma: an international consensus conference report. Lancet Oncol. 2012;13:e11–e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Agopian VG, Harlander-Locke MP, Markovic D, et al. Evaluation of patients with hepatocellular carcinomas that do not produce α-fetoprotein. JAMA Surg. 2017;152:55. [DOI] [PubMed] [Google Scholar]
- 54.Carr BI, Akkiz H, Üsküdar O, et al. HCC with low- and normal-serum alpha-fetoprotein levels. Clin Pract (Lond). 2018;15:453–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lerut J, Iesari S, Foguenne M, et al. Hepatocellular cancer and liver transplantation: necessity to go from chaos to order. Al'm Klin Med. 2018;46:552–559. [Google Scholar]
- 56.Tommaso LD, Spadaccini M, Donadon M, et al. Role of liver biopsy in hepatocellular carcinoma. World J Gastroenterol. 2019;25:6041–6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rastogi A. Changing role of histopathology in the diagnosis and management of hepatocellular carcinoma. World J Gastroenterol. 2018;24:4000–4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Neuberger J, Patel J, Caldwell H, et al. Guidelines on the use of liver biopsy in clinical practice from the British Society of Gastroenterology, the Royal College of Radiologists and the Royal College of Pathology. Gut. 2020;69:1382–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eisenberg E, Konopniki M, Veitsman E, et al. Prevalence and characteristics of pain induced by percutaneous liver biopsy. Anesth Analg. 2003;96:1392–1396. [DOI] [PubMed] [Google Scholar]
- 60.McCarty TR, Bazarbashi AN, Njei B, et al. Endoscopic ultrasound-guided, percutaneous, and transjugular liver biopsy: a comparative systematic review and meta-analysis. Clin Endosc. 2020;53:583–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Itzel T, Spang R, Maass T, et al. Random gene sets in predicting survival of patients with hepatocellular carcinoma. J Mol Med. 2019;97:879–888. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


