Abstract
First described in 1958, the human monkeypox virus (hMPXV) is a neglected zoonotic pathogen closely associated with the smallpox virus. The virus usually spreads via close contact with the infected animal or human and has been endemic mostly in parts of the African continent. However, with the recent increase in trade, tourism, and travel, the virus has caused outbreaks in countries outside Africa. The recent outbreak in 2022 has been puzzling given the lack of epidemiological connection and the possible sexual transmission of the virus. Furthermore, there is limited understanding of the structural and pathogenetic mechanisms that are employed by the virus to invade the host cells. Henceforth, it is critical to understand the working apparatus governing the viral-immune interactions to develop effective therapeutical and prophylactic modalities. Hence, in the present short communication, we summarize the previously reported research findings regarding the virology of the human monkeypox virus.
Keywords: Monkeypox virus, Global, Sexual transmission, Virology, Pandemic, Infections, hMPXV
Abbreviations: MPXV, monkeypox virus; WHO, World Health Organization; CA Clade, Central African Clade; WA Clade, West African Clade
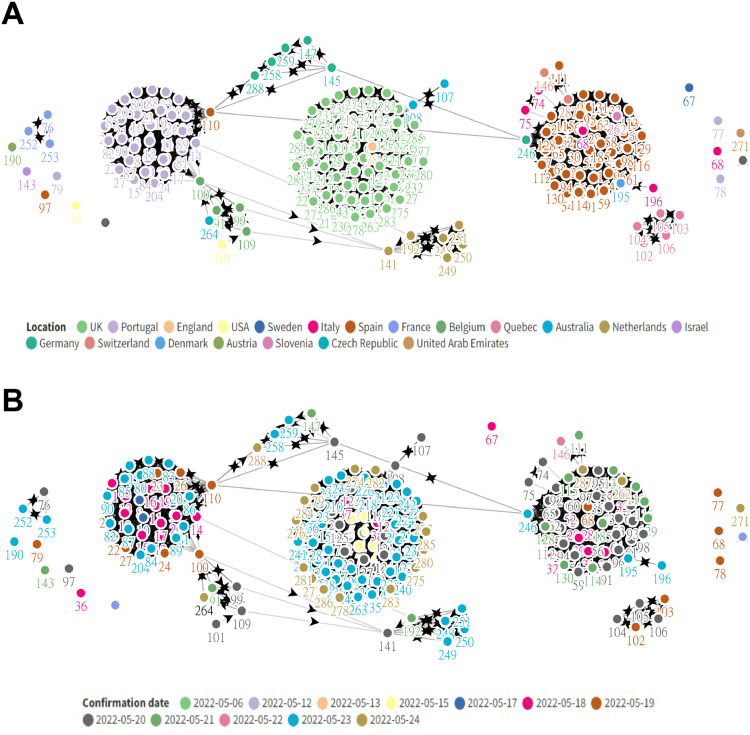
On the 7th of May 2022, the UKHSA (United Kingdom Health Security Agency) confirmed the first case of the human Monkeypox Virus (hMPXV) in a case travelling back from Nigeria. The patient had developed rashes a few days ago before travelling to the UK but presented to the hospital on the day of his arrival in the UK. A reverse transcriptase polymerase chain reaction (RT-PCR) on a vesicular swab was performed and hMPXV infection was confirmed (World Health Organization (WHO), 2022). Since then, more than 1500 cases in 45 countries have been suspected and/or confirmed (as of 11th June 2022) (Kraemer, 2022), prompting the World Health Organization (WHO) to convene an emergency meeting to curb the rampant spread of the virus (Fig. 1 ).
Fig. 1.
Cluster network charts showing the spread of the human Monkeypox Virus (hMPXV) as of 25th May 2022. (A) Clustering based on country and contact tracing; (B) Clustering based on date of case confirmation and contact tracing.
Monkeypox virus is a zoonotic virus belonging to the Orthopoxvirus genus. It is a linear double-stranded DNA virus belonging to the Poxviridae family that has been listed by the WHO in its list of diseases with epidemic or pandemic potential. The subset includes Smallpox (variola), Vaccinia, and Cowpox viruses. hMPXV is a 200 to 250 nm large, brick-shaped, enveloped, cytoplasmic virus that binds to glycosaminoglycans to enter the host cells (Swiss Institute of Bioinformatics (SIB), 2022). As an enveloped virus, it has been postulated to alternatively employ the classical apoptotic mimicry mechanism for entry into the host cells (Swiss Institute of Bioinformatics (SIB), 2022; Amara and Mercer, 2015).
Recent bibliometric analyses have shed light on the limited literature that is available on the pathogenesis, origins, and treatment of hMPXV (Cheng et al., 2022; Rodríguez-Morales et al., 2022). This is extremely concerning since the virus is characterized as Biosafety Level 3 (high threat) pathogen in the EU and is on the list of select agents in the United States (Central African clade) (Tian and Zheng, 2014; Sklenovská and Van Ranst, 2018). Hence in the present paper, we summarize the latest advances that have been reported regarding the virology of hMPXV.
1. Origins of the virus
The virus was first isolated in 1958 from smallpox-like vesiculopustular lesions amongst the captive imported monkeys (Java macaques) at the State Serum Institute in Copenhagen, Denmark (von Magnus, Andersen and Petersen, 1959). The monkeys were reported to suffer from a spontaneous outbreak of fever and rash. Over the next few years, similar outbreaks were reported in monkeys elsewhere. In 1966, the virus was identified as the causative agent behind a widespread outbreak at a zoo in Rotterdam (Dumbell and Smith, 1999). The virus was believed to have first affected the South American giant anteaters before spreading to various species of apes and monkeys. It was in 1970 that two separate incidents of smallpox-like disease in individuals from the Democratic Republic of Congo and Liberia were reported that led to the recognition of hMPXV as a distinct virus (Dumbell and Smith, 1999).
From 1970 to 2003, the virus was endemic to the rainforests of central and western African countries. In 2003, the first report of an outbreak outside Africa was reported in the US, when a shipment of exotic animals including nine different species of mammals was imported from Ghana (Ligon, 2004). The imported mammals were transported to different states with other animals, causing widespread infection of animals. Only after a 3-year-old girl was brought to the emergency after being bitten by a prairie dog, source tracing indicated a spill-over event (Ligon, 2004). Since then, sporadic outbreaks have been noted across the globe, all having traceable origins in the endemic African regions.
Given its capability to infect multiple mammalian taxa, its natural reservoir is still under investigation. Although monkeys have been eliminated as candidates for being the natural reservoir for the virus (Moss, 2020), rope squirrels and sooty mangabey are considered potential natural viral reservoirs (Khodakevich et al., 1986; Radonić et al., 2014).
2. MPXV clades – similarities & differences
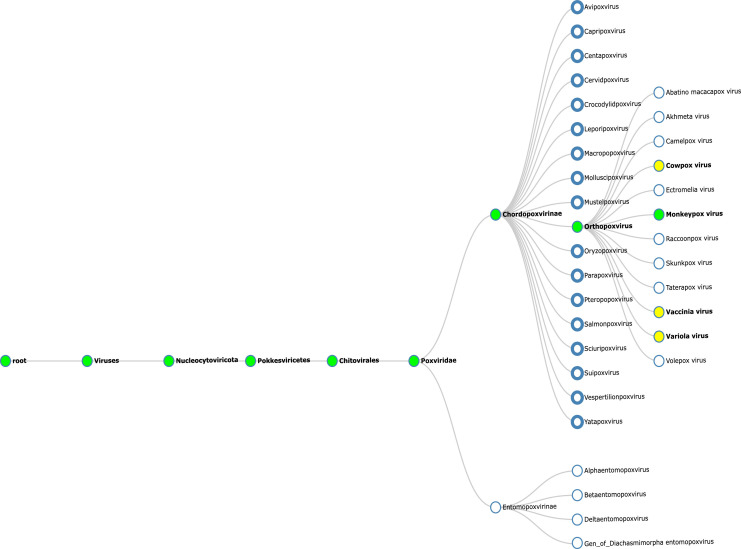
The Orthopoxvirus are antigenically and genetically similar (Fig. 2 ), with open reading frames (ORFs) having >90% sequence identity amongst its members (Shchelkunov et al., 2002). These viruses are undergoing evolutionary changes that are driven by progressive gene loss primarily at the terminal ends of the genome (Hendrickson et al., 2010; Kugelman et al., 2014). These changes could be accelerated by selective pressure from a host species (Hendrickson et al., 2010; Kugelman et al., 2014). Variations in gene copy number is another theory explaining the increasing viral fitness towards human infection and transmission (Elde et al., 2012). The 197 kb linear DNA genome of hMPXV contains close to 190 non-overlapping ORFs each longer than 60 amino acid residues (Hendrickson et al., 2010; Shchelkunov et al., 2001). The highly conserved central coding region sequence (CRS) is located at nucleotide positions 56,000–120,000 and is flanked by variable ends containing the inverted terminal repeats (ITRs) (Kugelman et al., 2014; Moss, 2001). There are at least four known ORFs in the ITR region of the hMPXV genome (Shchelkunov et al., 2001).
Fig. 2.
Phylogenetic relationship tree based on NCBI taxonomic classes. Green nodes track the taxonomic classification of the human Monkeypox Virus (hMPXV) whilst yellow nodes represent the closely associated viruses with hMPXV. The tree was created using Taxallnomy (available at http://bioinfo.icb.ufmg.br/taxallnomy/; accessed 25th May 2022).
The hMPXV has two described clades – the Central African/Congo Basin (CA) and West African (WA) clades. Similar to SARS-COV-2 (Covid-19), calls have been raised to rename the two clades to Clade 1 (CA) and Clades 2 and 3 (WA), in line with best international practices to move away from stigmatizing and discriminating nomenclature typically associated with geographic regions, nations, economies, and people (Happi et al., 2022). Accordingly, it has been recommended to assign hMPXV.B.1 to the lineage behind the current outbreak in Europe and the Global North (Happi et al., 2022).
The CA clade has estimated case mortality of 10% in non-vaccinated individuals with known human-human transmission whilst the WA clade has been associated with a milder form of the disease with lower mortality and less evident human-human transmission (Bunge et al., 2022). Genomic comparative studies have revealed a 0.55–0.56% nucleotide difference between the two clades (Chen et al., 2005), with the CA clade possessing 173 functional unique genes compared to 171 of the WA clade. The two clades were found to be 99.4% identical, sharing 170 orthologs at the protein level with no significant differences in the transcriptional regulatory sequences (Chen et al., 2005; Weaver and Isaacs, 2008). Amongst the virulence genes, 53 out of 56 were found in both clades and showcased 61 conservative, 93 non-conservative, and 121 silent amino acid changes (Chen et al., 2005).
The differences in virulence between the two clades have been postulated to stem from the differences in the gene orthologs - BR-203 (virulence protein), BR-209 (IL-1β binding protein), and COP-C3L (inhibitor of complement enzymes) (Chen et al., 2005; Likos et al., 2005). Other candidate gene orthologs include the WA clade-specific COP-A49R (unknown function) and COP-A52R (Bifunctional Toll-IL-1-receptor protein). In terms of CA clade-specific orthologs, candidates include BR-19 and BR-20 (unknown function) (Weaver and Isaacs, 2008). Another crucial gene responsible for the difference in virulence in clades is the D14R gene-coded inhibitor of complement-binding protein (MOPICE), an important anti-inflammatory factor that is absent from the hMPXV WA clade (Chen et al., 2005; Lopera et al., 2015; Liszewski et al., 2006). However, these genes are not the only factors responsible for virulence, with many more candidates yet to be identified.
Recently, four genomes of the hMPXV isolated during the 2022 outbreak (Germany, USA, Portugal, and Belgium) were published online by various groups of researchers (Isidro et al., 2022; Gigante et al., 2022; Antwerpen et al., 2022; Selhorst et al., 2022). A compiled version of the genomes for visualization is available at https://nextclade.org/monkeypox (Accessed 29th May 2022). Analysis of the genomes preliminarily hints at a very strong bias in mutations of bases Guanine (G) to Adenine (A) and Cytosine (C) to Thymine (T). The enzyme APOBEC3 (Apolipoprotein B Editing Complex), a cytidine deaminase, has been postulated to be responsible for these mutations (Rambaut, 2022). A genomic comparison from viral isolates from 2015 to 2022 showed a 30-T base long sequence in the middle of the viral genome, the role of which is yet to be determined (Perez and Lounnas, 2022).
3. Viral pathogenesis
The pathogenesis of the hMPXV involves viral entry, fusion, replication, and release, during which the virus produces two infectious forms - extracellular enveloped virions (EV) and intracellular mature virions (MV). Whilst MVs are single membrane-bound that are released only during host cell lysis, EVs are specialized MVs that are bound by an antigenically distinct triple membrane (the double membrane is gained by translocation to Golgi bodies) (Realegeno et al., 2020; Mucker et al., 2022a). It has been demonstrated that vaccines and antibodies that fail to produce or target the EV antigens provide lower protection than those including both (Golden et al., 2011; Lustig et al., 2005).
In the context of Orthopoxviruses, two multi-subunit complexes are crucial for completing the viral infectious cycle – GARP (Golgi-Associated Retrograde Protein) and COG (Conserved Oligomeric Golgi) complex (Realegeno et al., 2020). The GARP complex, responsible for retrograde endosomal transport, comprising four vacuolar protein sorting (VPS) genes – VPS51, VPS52, VPS53, and VPS54, all of which were found to be enriched in both CA and WA clades (except VPS53 which is specifically enriched in CA clade) (Realegeno et al., 2017). VPS52 and VPS54 knockout cells infected with MPXV have shown a significant decrease in EV yield, thereby confirming their prominent function in virus egress and cell-to-cell spread (Realegeno et al., 2017).
The COG complex, required for maintaining Golgi structure and intra-Golgi traffic regulation, comprises two lobes, each with four associated subunits (Lobe A – COG1, COG2, COG3, COG4, and Lobe B – COG5, COG6, COG7, COG8) (Realegeno et al., 2020). COG7 and COG8 are enriched in both clades whilst COG3 and COG4 are enriched only in the CA clade (Realegeno et al., 2017). COG4 and COG7 are thought to be the most important subunits for viral fusion (Realegeno et al., 2020).
Bioinformatic analysis of the hMPXV CA genome revealed two distinct regions (R1 – Open Reading Frame 17 to 32 and R2 – Open Reading Frame 179 to 193), deletion of which could attenuate the virulence of hMPXV (Lopera et al., 2015). Deletion of either or both regions led to reduced mice mortality and morbidity whilst attenuating viral replication (Lopera et al., 2015). The authors further were able to attribute genes in region R1 to be responsible for viral replication whilst those in region R2 to be responsible for viral pathogenicity (Lopera et al., 2015). A dual deletion of R1 and R2 was the most effective in suppressing viral virulence (Lopera et al., 2015).
Microarray analysis has revealed that hMPXV induces histone posttranslational modification in the host cell by the downregulation of histone expression regulation factors and upregulation of core histones (Alkhalil et al., 2010). These changes could indicate the dependence of viral DNA compaction and nucleosome formation on the expression of host cell histones (Alkhalil et al., 2010). Additionally, the authors reported a downregulation in the expression of cell membrane ion channels coupled with gene regulation favouring G2 phase cell arrest and S phase progression (Alkhalil et al., 2010).
4. Cytokine storm
Similar to the cytokine storm that is induced by Covid-19 infection (Ragab et al., 2020), a study performed cytokine profiling on 19 patients with PCR-confirmed hMPXV infection and reported on the correlation between the degree of cytokine modulation and the severity of hMPXV infection in patients (Johnston et al., 2015). The cytokine storm seemed to be Th2 mediated with interleukins IL-4,−5,−6, and −10 (IL-10 is a strong marker for cytokine storm) being significantly above the normal levels with an associated depression of Th1 associated cytokines IL-2, −12, TNF-α (tumor necrosis factor-alpha), and IFN-α and -γ (interferon alpha and gamma) (Johnston et al., 2015).
It has been demonstrated that Orthopoxvirus infection including hMPXV induces a cascade of complementary B cell responses that encodes for multiple cross-species reactive antigen clones (Gilchuk et al., 2016). The six crucial identified monoclonal antibodies (mABs) were targeted against H3, A27, D8, and L1 (MV antigens) and B5 and A33 (EV antigens). A cocktail of A33, B5, L1, and A27 was demonstrated to be the primary provider of protection against orthopoxviral respiratory tract infection whilst a cocktail of all six mABs was responsible for protection against systemic infection (Gilchuk et al., 2016). Such cocktails were also found to be more therapeutically effective than VIGIV (Vaccinia Immune Globulin Intravenous) (Gilchuk et al., 2016).
5. Advances in diagnostic modalities for hMPXV
The major diagnostic modalities for hMPXV include PCR, viral culture and isolation, negative staining electron microscopy of the rash, immunohistochemistry for orthopoxviral specific antigens, and IgM/IgG serology testing (Brown and Leggat, 2016). A rapid lateral flow-based screening test called Tetracore Orthopox BioThreat Alert® is available to prioritize sample testing for hMPXV (Brown and Leggat, 2016). An even more sensitive point-of-care diagnostic tool based on gravity-driven flow-through antigen capture ELISA has been also developed (Stern et al., 2016). Known as the ABICAP (Antibody Immuno Column for Analytical Processes) immunofiltration tool, it can detect all zoonotic orthopoxviruses. Recently, a quick under seven minutes recombinase polymerase amplification (RPA) assay was developed which targets the G2R gene of the virus (Davi et al., 2019). The assay demonstrated 100% specificity with 95% sensitivity in detecting both clades of the virus.
Serological and protein-based diagnostic tests suffer from the limitation of cross-reactivity between different Orthopoxvirus species. To overcome this limitation, research groups have tried to identify highly specific antibodies that could be used for screening and diagnostic testing. One such identified antibody is 69–126–3–7 which binds to the A27 protein of the hMPXV (Hughes et al., 2014). Other potential antibodies have been described by Keasey et al. (2010)) Animal studies have shown that evaluation of lymph node size and metabolic activity is a reliable predictor of hMPXV disease severity and course. Positron emission tomography/computed tomography (PET/CT) imaging with [18F]-fluorodeoxyglucose (FDG) has been demonstrated to predict the course of hMPXV disease in non-human primate models (Dyall et al., 2017). Increased FDG uptake with minimal lymph node enlargement in the early stages of the disease was found to be negatively associated with survival (Dyall et al., 2017).
6. Advances and challenges in treatment and prophylaxis
It has been demonstrated that 24 hours after exposure to lethal hMPXV in monkeys, initiation of antiviral treatment with cidofovir or with a related acyclic nucleoside phosphonate analogue is more effective than smallpox vaccination (Stittelaar et al., 2006). The antiviral regimen significantly reduces mortality and the number of MPX lesions (Stittelaar et al., 2006). In this regard, Tecovirimat (ST-246, TPOXX®) remains the first and only approved antiviral drug indicated for the treatment of Orthopoxvirus (Chan-Tack et al., 2019). The drug targets the F13L gene leading to impediments in the envelopment of virions and their subsequent release from the infected cells (Yang et al., 2005). The F13L protein interacts and co-localizes with B5R transmembrane glycoprotein forming the F13L-B5R complex (Husain and Moss, 2001) that is needed for viral wrapping.
However, mutations in the F13L gene have been described in a mutant clade of Cowpox virus (CPXV) that led to an 800-fold increase in Tecovirimat resistance (Yang et al., 2005). A case of Progressive Vaccinia (PV) treated with ST-246 and VIGIV has also been described who developed ST-246 resistance during the late stages of the disease (Lederman et al., 2012). A 50-fold increase in EC50 (half maximal effective concentration) was noted within one month of isolation of the virus from the patient sample (Lederman et al., 2012). The patient was subsequently administered CMX001, an orally bioavailable prodrug of cidofovir.
Additionally, the protective efficacy of the vaccine measured by the survival of lethal hMPXV challenge is largely unaffected by concurrent TPOXX treatment, but the humoral response to the vaccine may be adversely affected. The diminished humoral responses may have contributed to increased morbidity in ACAM2000+tecovirimat-treated animals lethally challenged with hMPXV (Russo et al., 2020; Berhanu et al., 2010).
To overcome the potential development of resistance against available antivirals, Priyamvada et al., demonstrated that PAV-164, a derivate of methylene blue, is a potent inhibitor of MPX viral replication (Priyamvada et al., 2021). Resveratrol, a natural polyphenol found in grapes, berries, etc., has also been shown in vitro to significantly reduce hMPXV replication (both clades) by suppressing DNA synthesis and associated downstream gene expression (Cao et al., 2017). Another candidate antiviral is based on NIOCH-14, a derivative of tricyclodicarboxylic acid, which causes a significant reduction in hMPXV viral production in the lungs of mice and marmots challenged with the virus (Mazurkov et al., 2016).
The recently published recommendations from the Advisory Committee on Immunization Practices (ACIP) in 2022, recommended JYNNEOS, a replication-deficient modified Vaccinia virus Ankara (MVA), as an alternative to ACAM2000 for primary vaccination (Rao et al., 2022). The vaccine does not pose risk for autoinoculation or inadvertent inoculation due to the absence of a major cutaneous reaction (“take”). The vaccine is administered in a 2-dose regimen set 28 days apart. A booster every 2 years for people with contact with more virulent orthopoxviruses and every 10 years for people with contact with low virulent orthopoxviruses has been recommended (Rao et al., 2022). Additionally, the ACIP recommended the use of JYNNEOS as a booster for those with ACAM2000 as primary vaccination. However, in a sero-study, the authors demonstrated that childhood vaccination does not provide complete protection against hMPXV, though the disease severity is reduced (Karem et al., 2007).
With the recent advances in gene-based vaccines, a DNA vaccine called 4pox has been developed and tested in different animal challenge models. The vaccine has been shown to inhibit viral shedding in vaccinated animals and prevented lethal effects of the viral infection (Golden et al., 2012; Mucker et al., 2022b). A recombinant Vaccinia virus immunoglobulin (rVIG) has been shown to possess higher neutralizing activity in vitro and in vivo when given both prophylactically as well as therapeutically (Parker et al., 2021). Another model in marmosets used a prophylactic dose of two human-chimeric monoclonal antibodies – c7D11 (against MV) and c8A (against EV) to protect against a lethal dose of MPXV (Mucker et al., 2018).
CRISPR/Cas9-based targeted antivirals delivered using adeno-associated virus (AAV) have been shown to reduce Orthopoxvirus viral titre by more than 90% in human embryonic kidney cells (Siegrist et al., 2020). Two specific genes were targeted by the authors which were found to be universally conserved across all Orthopoxvirus species – 12 L (production of mature virus, telomere binding, host-cell entry), and A17L (early viral envelope protein). A third gene E3L (affects host innate immune response) was also identified by the authors but was not tested (Siegrist et al., 2020).
Interestingly, the E3L gene has been long a front-runner candidate for any deletion mutant or subunit vaccine. However, in animal models, E3L-gene deleted Vaccinia vaccine (NYCBHΔE3L) failed to prevent lethal infection of MPXV, though it promoted pro-inflammatory signal transduction (Denzler et al., 2011). The lack of efficacy of such a mutant vaccine could be attributed to the need for E3L in the induction of protective T-cell epitopes (Ando et al., 2020). Whilst E3L-specific CD4+ T cells could not lyse infected cells, E3L-specific CD8+ T cells were able to lyse the infected cells if added within the first two hours of infection (due to the early expression of E3L – within 30 min of infection) (Ando et al., 2020). hMPXV contains a truncated homologue of E3 called F3L, both of which binds to the double-stranded RNA and move it away from the cellular pattern recognition receptors (PRR) (Watson et al., 1991). However, the presence of a truncated homologue confers hMPXV phenotype which is similar to wild-type Vaccina virus, thereby pointing toward the presence of other extragenic genes that help the virus to overcome the effects of truncation (Arndt et al., 2015). Interestingly, it has been shown that in MPXV-immune adults, the virus can escape both CD4+ and CD8+ T cell-mounted antiviral response by inducing a state of unresponsiveness in the T cells (Hammarlund et al., 2008). Yet, the recovered adults can mount a potent antiviral response possibly due to either alternative antigen presentation, cross-priming, and/or the ability of some cells to evade the immunomodulatory effects of MPXV (Hammarlund et al., 2008). hMPXV has been shown to possess IFN resistance and plaque formation remained unaffected by increasing concentrations of IFN (Arndt et al., 2016), which is interesting since the IFN-α and IFN-γ levels in hMPXV+ sero-profiled patients were found to be comparable to the normal levels (Realegeno et al., 2017). Furthermore, hMPXV is known to produce less double-stranded RNA than Vaccinia virus (Arndt et al., 2016) which confers it resistance to antipox antivirals like isatin-beta-thiosemicarbazone (Arndt et al., 2016; Pennington, 1977).
With countries like the US, UK, and Canada already starting ring vaccination (vaccination of close contacts of confirmed cases), it is essential to develop safer and easier-to-distribute vaccines. To aid in the future development of vaccines against Orthopoxviruses, an online resource EPIPOX (available at: http://imed.med.ucm.es/epipox/; Accessed 12th June 2022) has been developed that allows immunoinformatic characterization of shared T-cell epitopes between Orthopoxviruses which could be used in computational reverse vaccinology (Molero-Abraham et al., 2015).
7. The puzzling mystery of 2022 outbreak
What makes the recent 2022 global outbreak interesting though is the lack of known epidemiological links to central or western Africa, indicating a possible semi-endemic equilibrium. The transmission of the virus sexually, especially amongst men who have sex with men (MSM) community poses another interesting question. Interestingly, the clade associated with the current outbreak has been identified as the WA clade which is less virulent and less transmissible amongst humans. The lack of a high degree of changes in the genetic structure of the virus (as compared with Covid-19) makes the current events interesting to observe and follow as they unfold. With climate change and increased human-animal interactions due to migration of species northwards, deforestation, petting, zoos, etc., accelerating zoonotic spillover events, it becomes even more important for public health and epidemiological institutions to detect, manage, and guide society.
8. Conclusions
Human monkeypox virus (hMPXV) has long been a neglected zoonotic pathogen with a potential for misuse as a bioweapon and/or spill-over caused pandemics. The recent outbreaks across the globe have once again highlighted the need for constant and vigilant monitoring along with the development of novel prophylactic and therapeutic modalities. The selected administration of the smallpox vaccine and waning immunity in the population can accelerate the spread of the virus. Lessons learned from the Covid-19 pandemic shall be employed in the management and containment of the human monkeypox virus.
Declarations
Ethical approval
Not applicable. All data presented in the study has been collected from open-source platforms with proper citation and/or from media sources.
Data statement
All data presented in the present review is available online and can be accessed from the appropriate reference in the reference list.
Funding
The present review didn't receive any external funding and was self-supported by the authors.
Author contributions
NJ and EL conceptualized the present paper, whilst all authors were involved in data curation, formal analysis, and preparation of the initial draft of the manuscript. Funding acquisition was done by NJ and AR. Investigations and Methodology was led by NJ and EL. Project administration was done by NJ. Resources and software were led by NJ and AR. Supervision was done by SL and AR. Visualizations was done by NJ and EL. Validation was done by SL and AR. N.J. was responsible for review and editing of the final draft. All authors have read and agreed to the final version of the paper for publication.
Declaration of Competing Interest
The authors declare no conflicts of interest in regards with the present paper.
Acknowledgments
The support of ECOMSIR (European Collaboration of Medical Students in Research) and Riga Stradiņš University (RSU) is greatly acknowledged.
Data Availability
No data was used for the research described in the article.
References
- World Health Organization (WHO); 2022. Monkeypox - United Kingdom of Great Britain and Northern Ireland.https://www.who.int/emergencies/disease-outbreak-news/item/2022-DON381 [online] 16th MayAvailable from. Accessed 20th May 2022. [Google Scholar]
- Kraemer M., and Global health Monkeypox. Twitter post. 19th May 2022 1:36 AM. https://twitter.com/MOUGK/status/1527055553876348928?s=20&t=_bXBbMFBDKmR6drFjGfZZw (Accessed 21st May 2022).
- Swiss Institute of Bioinformatics (SIB). Orthopoxvirus [Internet]. Viral Zone. Available from: https://viralzone.expasy.org/149?outline=all_by_species (Accessed 25th May 2022).
- Amara A., Mercer J. Viral apoptotic mimicry. Nat. Rev. Microbiol. 2015;13(8):461–469. doi: 10.1038/nrmicro3469. AugEpub 2015 Jun 8. PMID: 26052667; PMCID: PMC7097103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng K., Zhou Y., Wu H. Bibliometric analysis of global research trends on monkeypox: are we ready to face this challenge? J. Med. Virol. 2022 doi: 10.1002/jmv.27892. May 29Epub ahead of print. PMID: 35644836. [DOI] [PubMed] [Google Scholar]
- Tian D., Zheng T. Comparison and analysis of biological agent category lists based on biosafety and biodefense. PLoS ONE. 2014;9(6) doi: 10.1371/journal.pone.0101163. Jun 30PMID: 24979754; PMCID: PMC4076228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Magnus P., Andersen E.K., Petersen K.B., et al. A pox-like disease in Cynomolgus monkeys. Acta Pathol. Microbiol. Scand. 1959;46:156–176. [Google Scholar]
- Dumbell K., Smith G.L. In: Encyclopedia of Virology. 2nd Edition. Granoff A., Webster R.G., editors. Elsevier; 1999. smallpox and Monkeypox viruses (Poxviridae) p. 1672. pageeditors. editorsISBN 9780122270307. [DOI] [Google Scholar]
- Ligon B.L. Monkeypox: a review of the history and emergence in the Western hemisphere. Semin. Pediatr. Infect. Dis. 2004;15(4):280–287. doi: 10.1053/j.spid.2004.09.001. OctPMID: 15494953; PMCID: PMC7129998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss W.J. In: Hunter's Tropical Medicine and Emerging Infectious Diseases. 10th Edition. Ryan E.T., Hill D.R., Solomon T., Aronson N.E., Endy T.P., editors. Elsevier; 2020. Viral Infections With Cutaneous Lesions (Chapter 32) p. 273. pageeditors. editorsISBN 9780323555128. [DOI] [Google Scholar]
- Khodakevich L., Jezek Z., Kinzanzka K. Isolation of monkeypox virus from wild squirrel infected in nature. Lancet. 1986;1:98–99. doi: 10.1016/S0140-6736(86)90748-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radonić A., Metzger S., Dabrowski P.W., Couacy-Hymann E., Schuenadel L., Kurth A., et al. Fatal monkeypox in wild-living sooty mangabey, Côte d'Ivoire, 2012. Emerg. Infect. Dis. 2014;20:1009–1011. doi: 10.3201/eid2006.131329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shchelkunov S.N., Totmenin A.V., Safronov P.F., Mikheev M.V., Gutorov V.V., Ryazankina O.I., Petrov N.A., et al. Analysis of the monkeypox virus genome. Virology. 2002;297(2):172–194. doi: 10.1006/viro.2002.1446. Jun 5PMID: 12083817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson R.C., Wang C., Hatcher E.L., Lefkowitz E.J. Orthopoxvirus genome evolution: the role of gene loss. Viruses. 2010;2(9):1933–1967. doi: 10.3390/v2091933. SepEpub 2010 Sep 15. PMID: 21994715; PMCID: PMC3185746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugelman J.R., Johnston S.C., Mulembakani P.M., Kisalu N., Lee M.S., Koroleva G., McCarthy S.E., et al. Genomic variability of monkeypox virus among humans, Democratic Republic of the Congo. Emerg. Infect. Dis. 2014;20(2):232–239. doi: 10.3201/eid2002.130118. FebPMID: 24457084; PMCID: PMC3901482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elde N.C., Child S.J., Eickbush M.T., Kitzman J.O., Rogers K.S., Shendure J., Geballe A.P., Malik H.S. Poxviruses deploy genomic accordions to adapt rapidly against host antiviral defenses. Cell. 2012;150(4):831–841. doi: 10.1016/j.cell.2012.05.049. Aug 17PMID: 22901812; PMCID: PMC3499626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shchelkunov S.N., Totmenin A.V., Babkin I.V., Safronov P.F., Ryazankina O.I., Petrov N.A., Gutorov V.V., Uvarova E.A., Mikheev M.V., Sisler J.R., Esposito J.J., Jahrling P.B., Moss B., Sandakhchiev L.S. Human monkeypox and smallpox viruses: genomic comparison. FEBS Lett. 2001;509(1):66–70. doi: 10.1016/s0014-5793(01)03144-1. Nov 30PMID: 11734207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss B. In: Field's Virology. 4th ed. Knipe DM, Howley PM., editors. Lippincott-Raven; Philadelphia: 2001. Poxviridae: the viruses and their replication; pp. 2849–2883. [Google Scholar]
- Happi C., Adetifa I., Mbala P., Njouom R., et al. 2022. Urgent Need For a Non-Discriminatory and Non-Stigmatizing Nomenclature For Monkeypox Virus.https://virological.org/t/urgent-need-for-a-non-discriminatory-and-non-stigmatizing-nomenclature-for-monkeypox-virus/853 June 10th[online]. Available from. Accessed 11th June 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunge E.M., Hoet B., Chen L., Lienert F., Weidenthaler H., Baer L.R., Steffen R. The changing epidemiology ofhuman monkeypox-A potential threat? A systematic review. PLoS Negl. Trop. Dis. 2022;16(2) doi: 10.1371/journal.pntd.0010141. Feb11PMID: 35148313; PMCID: PMC8870502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Li G., Liszewski M.K., Atkinson J.P., Jahrling P.B., Feng Z., Schriewer J., Buck C., Wang C., Lefkowitz E.J., Esposito J.J., Harms T., Damon I.K., Roper R.L., Upton C., Buller R.M. Virulence differences between monkeypox virus isolates from West Africa and the Congo basin. Virology. 2005;340(1):46–63. doi: 10.1016/j.virol.2005.05.030. Sep 15PMID: 16023693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver J.R., Isaacs S.N. Monkeypox virus and insights into its immunomodulatory proteins. Immunol. Rev. 2008;225:96–113. doi: 10.1111/j.1600-065X.2008.00691.x. OctPMID: 18837778; PMCID: PMC2567051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Likos A.M., Sammons S.A., Olson V.A., Frace A.M., Li Y., Olsen-Rasmussen M., Davidson W., Galloway R., Khristova M.L., Reynolds M.G., Zhao H., Carroll D.S., Curns A., Formenty P., Esposito J.J., Regnery R.L., Damon I.K. A tale of two clades: monkeypox viruses. J. Gen. Virol. 2005;86:2661–2672. doi: 10.1099/vir.0.81215-0. OctPt 10PMID: 16186219. [DOI] [PubMed] [Google Scholar]
- Lopera J.G., Falendysz E.A., Rocke T.E., Osorio J.E. Attenuation of monkeypox virus by deletion of genomic regions. Virology. 2015;475:129–138. doi: 10.1016/j.virol.2014.11.009. Jan 15Epub 2014 Dec 1. PMID: 25462353; PMCID: PMC4720520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liszewski M.K., Leung M.K., Hauhart R., Buller R.M., Bertram P., Wang X., Rosengard A.M., Kotwal G.J., Atkinson J.P. Structure and regulatory profile of the monkeypox inhibitor of complement: comparison to homologs in vaccinia and variola and evidence for dimer formation. J. Immunol. 2006;176(6):3725–3734. doi: 10.4049/jimmunol.176.6.3725. Mar 15PMID: 16517741. [DOI] [PubMed] [Google Scholar]
- Isidro J., Borges V., Pinto M., Ferreira R., Sobral D., Nunes A., et al. 2022. Multi-Country Outbreak of Monkeypox Virus: Genetic Divergence and First Signs of Microevolution.https://virological.org/t/multi-country-outbreak-of-monkeypox-virus-genetic-divergence-and-first-signs-of-microevolution/806 [online] 23rd MayAvailable from. Accessed 28th May 2022. [Google Scholar]
- Gigante C.M., Smole S., Wilkins K., McCollum A., Hutson C., et al. 2022. Draft Monkeypox Virus Genome from Confirmed Monkeypox Case in Massachusetts, United States, May 2022.https://www.ncbi.nlm.nih.gov/nuccore/ON563414 Available from. Accessed 29th May. [Google Scholar]
- Antwerpen M.H., Lang D., Zange S., Walter M.C., Woelfel R. 2022. First German Genome Sequence of Monkeypox Virus Associated to Multi-country Outbreak in May 2022.https://www.ncbi.nlm.nih.gov/nuccore/ON568298 Available from. Accessed 29th May. [Google Scholar]
- Selhorst P., Rezende A.M., de Block T., Coppens S., Smet H., Mariën J., Hauner A., et al. 2022. Belgian Case of Monkeypox Virus Linked to Outbreak in Portugal.https://virological.org/t/belgian-case-of-monkeypox-virus-linked-to-outbreak-in-portugal/801 [online] 20th MayAvailable from. Accessed 29th May 2022. [Google Scholar]
- Rambaut A. ARTIC Network; 2022. Discussion of On-Going MPXV Genome Sequencing.https://virological.org/t/discussion-of-on-going-MPXV-genome-sequencing/802 [online] 21st MayAvailable from. Accessed 29th May 2022. [Google Scholar]
- Perez jean-claude, Lounnas Valère. 2022. May 2022: Peculiar Evolution of the Monkeypox Virus Genomes.https://www.researchgate.net/publication/360973596_May_2022_Peculiar_Evolution_of_the_Monkeypox_Virus_Genomes?fbclid=IwAR25RQpaXEA0zyc7IPfaYgSaw7jOPP1Ssi5Ac7KbAE348FMltaGQ0bPsIc8 Available from. Accessed 11th June 2022. [Google Scholar]
- Realegeno S., Priyamvada L., Kumar A., Blackburn J.B., Hartloge C., Puschnik A.S., Sambhara S., Olson V.A., Carette J.E., Lupashin V., Satheshkumar P.S. Conserved Oligomeric Golgi (COG) complex proteins facilitate orthopoxvirus entry, fusion and spread. Viruses. 2020;12(7):707. doi: 10.3390/v12070707. Jun 30PMID: 32629851; PMCID: PMC7411930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucker E.M., Thiele-Suess C., Baumhof P., Hooper J.W. Lipid nanoparticle delivery of unmodified mRNAs encoding multiple monoclonal antibodies targeting poxviruses in rabbits. Mol. Ther. Nucleic Acids. 2022;28:847–858. doi: 10.1016/j.omtn.2022.05.025. May 10PMID: 35664703; PMCID: PMC9149018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden J.W., Zaitseva M., Kapnick S., Fisher R.W., Mikolajczyk M.G., Ballantyne J., Golding H., Hooper J.W. Polyclonal antibody cocktails generated using DNA vaccine technology protect in murine models of orthopoxvirus disease. Virol. J. 2011;8:441. doi: 10.1186/1743-422X-8-441. Sep 20PMID: 21933385; PMCID: PMC3192780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig S., Fogg C., Whitbeck J.C., Eisenberg R.J., Cohen G.H., Moss B. Combinations of polyclonal or monoclonal antibodies to proteins of the outer membranes of the two infectious forms of vaccinia virus protect mice against a lethal respiratory challenge. J. Virol. 2005;79(21):13454–13462. doi: 10.1128/JVI.79.21.13454-13462.2005. NovPMID: 16227266; PMCID: PMC1262616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Realegeno S., Puschnik A.S., Kumar A., Goldsmith C., Burgado J., Sambhara S., Olson V.A., Carroll D., Damon I., Hirata T., Kinoshita T., Carette J.E., Satheshkumar P.S. Monkeypox virus host factor screen using haploid cells identifies essential role of GARP complex in extracellular virus formation. J. Virol. 2017;91(11):e00011–e00017. doi: 10.1128/JVI.00011-17. May 12PMID: 28331092; PMCID: PMC5432867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkhalil A., Hammamieh R., Hardick J., Ichou M.A., Jett M., Ibrahim S. Gene expression profiling of monkeypox virus-infected cells reveals novel interfaces for host-virus interactions. Virol. J. 2010;7:173. doi: 10.1186/1743-422X-7-173. Jul 28PMID: 20667104; PMCID: PMC2920256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragab D., Salah Eldin H., Taeimah M., Khattab R., Salem R. The COVID-19 cytokine storm; what we know so far. Front. Immunol. 2020;11:1446. doi: 10.3389/fimmu.2020.01446. Jun 16PMID: 32612617; PMCID: PMC7308649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston S.C., Johnson J.C., Stonier S.W., Lin K.L., Kisalu N.K., Hensley L.E., Rimoin A.W. Cytokine modulation correlates with severity of monkeypox disease in humans. J. Clin. Virol. 2015;63:42–45. doi: 10.1016/j.jcv.2014.12.001. FebEpub 2014 Dec 4. PMID: 25600603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchuk I., Gilchuk P., Sapparapu G., Lampley R., Singh V., Kose N., Blum D.L., Hughes L.J., Satheshkumar P.S., Townsend M.B., Kondas A.V., Reed Z., Weiner Z., Olson V.A., Hammarlund E., Raue H.P., Slifka M.K., Slaughter J.C., Graham B.S., Edwards K.M., Eisenberg R.J., Cohen G.H., Joyce S., Crowe J.E., Jr. Cross-neutralizing and protective human antibody specificities to poxvirus infections. Cell. 2016;167(3):684–694.e9. doi: 10.1016/j.cell.2016.09.049. Oct 20PMID: 27768891; PMCID: PMC5093772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K., Leggat P.A. Human monkeypox: current state of knowledge and implications for the future. Trop. Med. Infect. Dis. 2016;1(1):8. doi: 10.3390/tropicalmed1010008. Dec 20PMID: 30270859; PMCID: PMC6082047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklenovská N., Van Ranst M. Emergence of Monkeypox as the most important Orthopoxvirus infection in humans. Front. Public Health. 2018;6:241. doi: 10.3389/fpubh.2018.00241. Sep 4PMID: 30234087; PMCID: PMC6131633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D., Olson V.A., Smith S.K., Pietraszczyk M., Miller L., Miethe P., Dorner B.G., Nitsche A. Rapid and sensitive point-of-care detection of Orthopoxviruses by ABICAP immunofiltration. Virol. J. 2016;13(1):207. doi: 10.1186/s12985-016-0665-5. Dec 9PMID: 27938377; PMCID: PMC5148848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davi S.D., Kissenkötter J., Faye M., Böhlken-Fascher S., Stahl-Hennig C., Faye O., Faye O., Sall A.A., Weidmann M., Ademowo O.G., Hufert F.T., Czerny C.P. Abd El Wahed A. Recombinase polymerase amplification assay for rapid detection of Monkeypox virus. Diagn. Microbiol. Infect. Dis. 2019;95(1):41–45. doi: 10.1016/j.diagmicrobio.2019.03.015. SepEpub 2019 Apr 11. PMID: 31126795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes L.J., Goldstein J., Pohl J., Hooper J.W., Lee Pitts R., Townsend M.B., Bagarozzi D., Damon I.K., Karem K.L. A highly specific monoclonal antibody against monkeypox virus detects the heparin binding domain of A27. Virology. 2014;464-465:264–273. doi: 10.1016/j.virol.2014.06.039. SepEpub 2014 Aug 9. PMID: 25108113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keasey S., Pugh C., Tikhonov A., Chen G., Schweitzer B., Nalca A., Ulrich R.G. Proteomic basis of the antibody response to monkeypox virus infection examined in cynomolgus macaques and a comparison to human smallpox vaccination. PLoS ONE. 2010;5(12):e15547. doi: 10.1371/journal.pone.0015547. Dec 30PMID: 21209900; PMCID: PMC3012712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyall J., Johnson R.F., Chefer S., Leyson C., Thomasson D., Seidel J., Ragland D.R., Byrum R., Jett C., Cann J.A., St Claire M., Jagoda E., Reba R.C., Hammoud D., Blaney J.E., Jahrling P.B. [18F]-Fluorodeoxyglucose uptake in lymphoid tissue serves as a predictor of disease outcome in the nonhuman primate model of monkeypox virus infection. J. Virol. 2017;91(21):e00817–e00897. doi: 10.1128/JVI.00897-17. Oct 13PMID: 28814515; PMCID: PMC5640857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stittelaar K.J., Neyts J., Naesens L., van Amerongen G., van Lavieren R.F., Holý A., De Clercq E., et al. Antiviral treatment is more effective than smallpox vaccination upon lethal monkeypox virus infection. Nature. 2006;439(7077):745–748. doi: 10.1038/nature04295. Feb 9Epub 2005 Dec 11. PMID: 16341204. [DOI] [PubMed] [Google Scholar]
- Chan-Tack K.M., Harrington P.R., Choi S.Y., Myers L., O'Rear J., Seo S., McMillan D., Ghantous H., Birnkrant D., Sherwat A.I. Assessing a drug for an eradicated human disease: US Food and Drug Administration review of tecovirimat for the treatment of smallpox. Lancet Infect. Dis. 2019;19(6):e221–e224. doi: 10.1016/S1473-3099(18)30788-6. JunEpub 2019 Mar 7. PMID: 30853252. [DOI] [PubMed] [Google Scholar]
- Yang G., Pevear D.C., Davies M.H., Collett M.S., Bailey T., Rippen S., Barone L., Burns C., Rhodes G., et al. An orally bioavailable antipoxvirus compound (ST-246) inhibits extracellular virus formation and protects mice from lethal orthopoxvirus Challenge. J. Virol. 2005;79(20):13139–13149. doi: 10.1128/JVI.79.20.13139-13149.2005. OctPMID: 16189015; PMCID: PMC1235851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husain M., Moss B. Vaccinia virus F13L protein with a conserved phospholipase catalytic motif induces colocalization of the B5R envelope glycoprotein in post-Golgi vesicles. J. Virol. 2001;75(16):7528–7542. doi: 10.1128/JVI.75.16.7528-7542.2001. AugPMID: 11462025; PMCID: PMC114988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederman E.R., Davidson W., Groff H.L., Smith S.K., Warkentien T., Li Y., Wilkins K.A., Karem K.L., et al. Progressive vaccinia: case description and laboratory-guided therapy with vaccinia immune globulin, ST-246, and CMX001. J. Infect. Dis. 2012;206(9):1372–1385. doi: 10.1093/infdis/jis510. NovEpub 2012 Aug 16. PMID: 22904336; PMCID: PMC3529603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Morales A.J., Ortiz-Martínez Y., Bonilla-Aldana D.K. What has been researched about Monkeypox? A bibliometric analysis of an old zoonotic virus causing global concern. N. Microbes New Infect. 2022 doi: 10.1016/j.nmni.2022.100993. in press100993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo A.T., Berhanu A., Bigger C.B., Prigge J., Silvera P.M., Grosenbach D.W., Hruby D. Co-administration of tecovirimat and ACAM2000™ in non-human primates: effect of tecovirimat treatment on ACAM2000 immunogenicity and efficacy versus lethal monkeypox virus challenge. Vaccine. 2020;38(3):644–654. doi: 10.1016/j.vaccine.2019.10.049. Jan 16Epub 2019 Oct 31. PMID: 31677948; PMCID: PMC6954297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berhanu A., King D.S., Mosier S., Jordan R., Jones K.F., Hruby D.E., Grosenbach D.W. Impact of ST-246® on ACAM2000™ smallpox vaccine reactogenicity, immunogenicity, and protective efficacy in immunodeficient mice. Vaccine. 2010;29(2):289–303. doi: 10.1016/j.vaccine.2010.10.039. Dec 16Epub 2010 Oct 29. PMID: 21036130; PMCID: PMC3023305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priyamvada L., Burgado J., Baker-Wagner M., Kitaygorodskyy A., Olson V., Lingappa V.R., Satheshkumar P.S. New methylene blue derivatives suggest novel anti-orthopoxviral strategies. Antiviral Res. 2021;191 doi: 10.1016/j.antiviral.2021.105086. JulEpub 2021 May 13. PMID: 33992710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S., Realegeno S., Pant A., Satheshkumar P.S., Yang Z. Suppression of poxvirus replication by resveratrol. Front. Microbiol. 2017;8:2196. doi: 10.3389/fmicb.2017.02196. Nov 17PMID: 29204136; PMCID: PMC5698801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazurkov O.Y., Kabanov A.S., Shishkina L.N., Sergeev A.A., Skarnovich M.O., Bormotov N.I., Skarnovich M.A., Ovchinnikova A.S., Titova K.A., Galahova D.O., Bulychev L.E., Sergeev A.A., Taranov O.S., Selivanov B.A., Tikhonov A.Y., Zavjalov E.L., Agafonov A.P., Sergeev A.N. New effective chemically synthesized anti-smallpox compound NIOCH-14. J. Gen. Virol. 2016;97(5):1229–1239. doi: 10.1099/jgv.0.000422. MayEpub 2016 Feb 8. PMID: 26861777. [DOI] [PubMed] [Google Scholar]
- Rao A.K., Petersen B.W., Whitehill F., Razeq J.H., Isaacs S.N., Merchlinsky M.J., Campos-Outcalt D., Morgan R.L., Damon I., Sánchez P.J., Bell B.P. Use of JYNNEOS (Smallpox and monkeypox vaccine, live, nonreplicating) for preexposure vaccination of persons at risk for occupational exposure to orthopoxviruses: recommendations of the advisory committee on immunization practices - United States, 2022. MMWR Morb. Mortal. Wkly. Rep. 2022;71(22):734–742. doi: 10.15585/mmwr.mm7122e1. Jun 3PMID: 35653347; PMCID: PMC9169520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karem K.L., Reynolds M., Hughes C., Braden Z., Nigam P., Crotty S., Glidewell J., Ahmed R., Amara R., Damon I.K. Monkeypox-induced immunity and failure of childhood smallpox vaccination to provide complete protection. Clin. Vaccine Immunol. 2007;14(10):1318–1327. doi: 10.1128/CVI.00148-07. OctEpub 2007 Aug 22. PMID: 17715329; PMCID: PMC2168110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden J.W., Josleyn M., Mucker E.M., Hung C.F., Loudon P.T., Wu T.C., Hooper J.W. Side-by-side comparison of gene-based smallpox vaccine with MVA in nonhuman primates. PLoS ONE. 2012;7(7):e42353. doi: 10.1371/journal.pone.0042353. Epub 2012 Jul 31. PMID: 22860117; PMCID: PMC3409187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucker E.M., Golden J.W., Hammerbeck C.D., Kishimori J.M., Royals M., Joselyn M.D., Ballantyne J., Nalca A., Hooper J.W. A nucleic acid-based orthopoxvirus vaccine targeting the vaccinia virus L1, A27, B5, and A33 proteins protects rabbits against lethal rabbitpox virus aerosol challenge. J. Virol. 2022;96(3) doi: 10.1128/JVI.01504-21. Feb 9Epub 2021 Dec 1. PMID: 34851148; PMCID: PMC8826804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker S., D'Angelo J., Buller R.M., Smee D.F., Lantto J., Nielsen H., Jensen A., Prichard M., George S.L. A human recombinant analogue to plasma-derived vaccinia immunoglobulin prophylactically and therapeutically protects against lethal orthopoxvirus challenge. Antiviral Res. 2021;195 doi: 10.1016/j.antiviral.2021.105179. NovEpub 2021 Sep 13. PMID: 34530009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucker E.M., Wollen-Roberts S.E., Kimmel A., Shamblin J., Sampey D., Hooper J.W. Intranasal monkeypox marmoset model: prophylactic antibody treatment provides benefit against severe monkeypox virus disease. PLoS Negl. Trop. Dis. 2018;12(6) doi: 10.1371/journal.pntd.0006581. Jun 21PMID: 29927927; PMCID: PMC6029809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegrist C.M., Kinahan S.M., Settecerri T., Greene A.C., Santarpia J.L. CRISPR/Cas9 as an antiviral against Orthopoxviruses using an AAV vector. Sci. Rep. 2020;10(1):19307. doi: 10.1038/s41598-020-76449-9. Nov 9PMID: 33168908; PMCID: PMC7653928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denzler K.L., Babas T., Rippeon A., Huynh T., Fukushima N., Rhodes L., Silvera P.M., Jacobs B.L. Attenuated NYCBH vaccinia virus deleted for the E3L gene confers partial protection against lethal monkeypox virus disease in cynomolgus macaques. Vaccine. 2011;29(52):9684–9690. doi: 10.1016/j.vaccine.2011.09.135. Dec 6Epub 2011 Oct 12. PMID: 22001879; PMCID: PMC5001690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando J., Ngo M.C., Ando M., Leen A., Rooney C.M. Identification of protective T-cell antigens for smallpox vaccines. Cytotherapy. 2020;22(11):642–652. doi: 10.1016/j.jcyt.2020.04.098. NovEpub 2020 May 8. PMID: 32747299; PMCID: PMC7205715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson J.C., Chang H.W., Jacobs B.L. Characterization of a vaccinia virus-encoded double-stranded RNA-binding protein that may be involved in inhibition of the double-stranded RNA-dependent protein kinase. Virology. 1991;185(1):206–216. doi: 10.1016/0042-6822(91)90768-7. NovPMID: 1681618. [DOI] [PubMed] [Google Scholar]
- Arndt W.D., Cotsmire S., Trainor K., Harrington H., Hauns K., Kibler K.V., Huynh T.P., Jacobs B.L. Evasion of the innate immune type i interferon system by monkeypox virus. J. Virol. 2015;89(20):10489–10499. doi: 10.1128/JVI.00304-15. OctEpub 2015 Aug 5. PMID: 26246580; PMCID: PMC4580173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarlund E., Dasgupta A., Pinilla C., Norori P., Früh K., Slifka M.K. Proc Natl Acad Sci U S A. Vol. 105. 2008. Monkeypox virus evades antiviral CD4+ and CD8+ T cell responses by suppressing cognate T cell activation; pp. 14567–14572. Sep 23Epub 2008 Sep 16. PMID: 18796610; PMCID: PMC2567221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt W.D., White S.D., Johnson B.P., Huynh T., Liao J., Harrington H., Cotsmire S., Kibler K.V., Langland J., Jacobs B.L. Monkeypox virus induces the synthesis of less dsRNA than vaccinia virus, and is more resistant to the anti-poxvirus drug, IBT, than vaccinia virus. Virology. 2016;497:125–135. doi: 10.1016/j.virol.2016.07.016. OctEpub 2016 Jul 26. PMID: 27467578; PMCID: PMC5026613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennington T.H. Isatin-beta-thiosemicarbazone causes premature cessation of vaccinia virus-induced late post-replicative polypeptide synthesis. J. Gen. Virol. 1977;35:567–571. doi: 10.1099/0022-1317-35-3-567. [DOI] [PubMed] [Google Scholar]
- Molero-Abraham M., Glutting J.P., Flower D.R., Lafuente E.M., Reche P.A. EPIPOX: immunoinformatic characterization of the shared T-cell epitome between variola virus and related pathogenic orthopoxviruses. J. Immunol. Res. 2015;2015 doi: 10.1155/2015/738020. Epub 2015 Oct 28. PMID: 26605344; PMCID: PMC4641182. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.