To the Editor
In this journal, Jolly and Scaria, reported distinct phylogenetic cluster of monkeypox virus (MPXV) genomes suggesting an early spread of virus.1 Since May 2022, monkeypox cases have been reported in more than 102 countries indicating expansion of its geographic range. Recent studies have also reported microevolution of MPXV genome of 2022 outbreak compared to earlier outbreaks.2, 3, 4, 5 Here, we report the complete genome analysis of monkeypox cases detected in India.
The clinical specimens i.e., orophryngeal swab, nasopharyngeal swab, lesion crust and lesion fluids of 96 suspected Monkeypox cases were referred to ICMR-National Institute of Virology, Pune, India for diagnosis of Monkeypox. Of all the cases, MPXV infection was confirmed in ten cases (Kerala=5, Delhi=5) using Monkeypox specific real time PCR.6 Cases from Delhi had no international travel history; while cases from Kerala had travel history from United Arab Emirates to India.5 All the cases were immunocompetent with no comorbidities and their clinical presentations are described in supplementary Table 1.
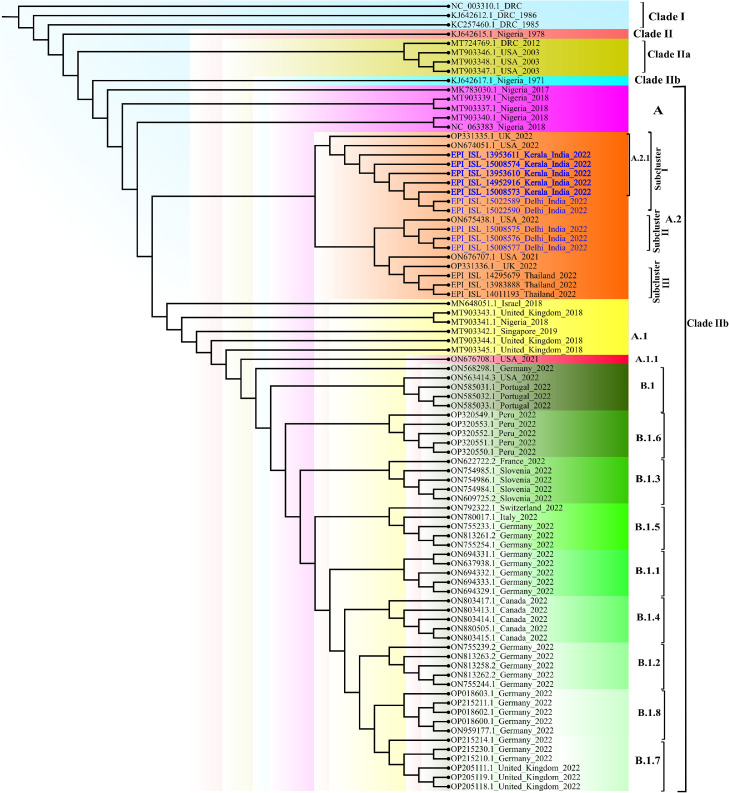
The genomic characterization of these MPXV positive samples were carried out using next generation sequencing.7 The Maximum-Likelihood phylogenetic tree analysis placed ten genome sequences (Retrieval 90 to 99%) from India (highlighted in blue) and eight genomes from USA (n = 3), UK (n = 2) and Thailand (n = 3) under lineage A.2 of clade IIb (Fig. 1 ). Further, they diverge into three sub clusters of A.2 lineage consist of total eighteen sequences. The seven sequences (Kerala n = 5, Delhi n = 2) grouped into sub cluster I showing highest similarity with MPXV_USA_2022_FL001. In this sub cluster, five sequences from Kerala were designated as A.2.1 based on the lineage defining mutations in the position C 25072 T, A 140492 C, C 179537 T. Two sequences from Delhi are lacking these three mutations hence still defined into A.2 lineage. These mutations were also lacking in the 3 sequences of Delhi from sub cluster II which aligned with sequences of lineage A.2 reported from USA 2022 (USA_2022_VA001). Apparently, Delhi MPXV sequences in sub cluster I and II are showing divergence which needs to be further explored.
Fig. 1.
The maximum-likelihood phylogenetic tree of MPxV/hMPxV genome constructed using software IQ TREE with 1000 ultra bootstrap replication cycle. The retrieved sequences from ten monkeypox cases from India belong to A.2 lineage of Clade IIb (Highlighted in red color).
The sub cluster III has monkeypox sequences reported from UK, Thailand during current outbreak of 2022 and USA during 2021. These sequences have many shared mutations that separated them from other two sub cluster including India (A.2) and travel-associated cases from 2017 to 2021 of other lineages (A, A.1, A.1.1). The A.2 lineage MPXV sequences from India showed a divergence from the MPXV sequences reported from Germany, Italy, Portugal, Switzerland and France (lineage B.1) and earlier outbreak sequences from Nigeria, Israel and Singapore 2017/18 (lineage A.1). The findings of our study are in concordance with the recent study of Gigante et al, which reported the circulation of both A.2 and B.1 lineages in current outbreak in USA with similarity to MPXV sequences of traveler from Nigeria to Texas in 2021.3
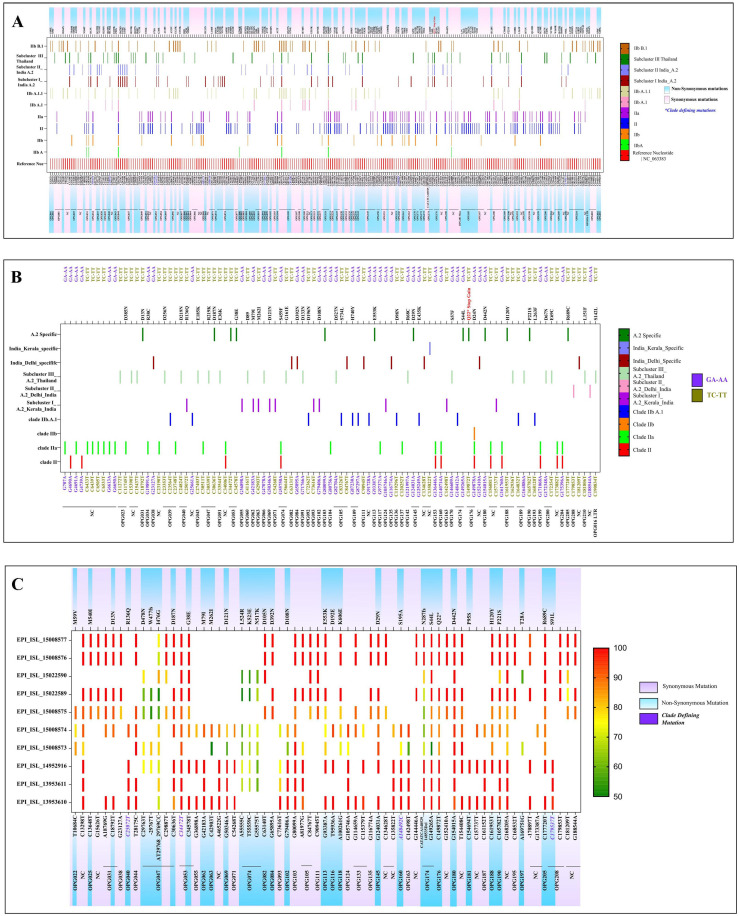
Analysis of Clusters of Orthologous Genes for orthopox viruses (OPG) revealed that the sub cluster I of seven sequences from India showed 07 synonymous mutations (OPG055, OPG071, OPG074, OPG093, OPG124, OPG163, OPG187) and 11 non-synonymous mutations (OPG040, OPG062, OPG063, OPG069, OPG074, OPG116, OPG160 and OPG208). Sub cluster II included three sequences from India which indicated 04 non synonymous mutations (OPG025, OPG082, OPG084, OPG099) and 08 synonymous mutations (OPG037, OPG038, OPG105, OPG111, and OPG135). Mapping the A.2 sequences with the reference genome (NC_063383 strain) indicated total fifteen mutations (OPG031, OPG047, OPG053, OPG103, OPG113, OPG145, OPG174, OPG176, OPG180, OPG188, OPG190 and OPG205) which were common throughout all the three sub clusters of the A.2 lineage (Fig. 2 A). We have also noted a seven-nucleotide deletion in all three A.2 sub clusters in OPG 174 gene. It is known to play role in influencing virulence by suppressing immune system; further studies are needed to determine the impact of this deletion on virulence of A.2 lineage.
Fig. 2.
(A) The variant analysis of MPXV genome was done using software MEGA 10 w.r.t reference sequence NC_063383. The depicted figure showed the synonymous and non-synonymous mutations in Clade II and lineages. The A.2 lineage is further characterized into sub-cluster I, II and III. All the clade defining mutations are marked with blue color positions. (B) The APOBEC 3 mutation analysis was done using software MEGA 10.0. The mutations (GA-AA, TC-TT) were marked for all the clades. (C) The variant analysis of MPXV genome of all the retrieved sequences was done using CLC genomics workbench with 50% frequency. The reference positions marked with synonymous and non-synonymous mutations for all the sequences with the respective frequencies.
We have also observed single nucleotide polymorphisms (SNPs) specific to sub cluster III compared to reference (NC 063383.1) which were not found in the sequences of sub cluster I and II. There were total 27 non synonymous changes observed in Thailand sequences.
APOBEC 3 mutation analysis indicated presence of 13 mutations in A.2 sequences from the current MPXV outbreak 2022. These could be A.2 lineage defining mutations apart from OPG053; C 34472 T reported during earlier studies (https://master.clades.nextstrain.org) (Fig. 2B). Further, we identified 25 additional APOBEC 3 mutations from the MPXV strain circulating in India. Besides this, 21 synonymous and non-synonymous APOBEC-3 mutations were also noted in sub cluster III. Recently O'Toole and Rambaut reported that the APOBEC3 with cytidine deaminase activity could be significant factor in the short-term evolution of MPXV since 2017.8 Our APOBEC3 mutation analysis supported a strong inclination for GA to AA and TC to TT mutations, indicating cytosine deaminase functioning reported only in Clade II since 2017 and not in Clade I.4 A recent study Isidro et al. also demonstrated microevolution and divergence of MPXV sequences of 2022 outbreak pertaining to APOBEC3 and other proteins.2
Variant analysis of all sequences of A.2 lineage indicated a total of 34/67 synonymous mutations and 33/67 non synonymous mutations. Fourteen mutations are found in the non coding region and 53 mutations were observed in coding region of different ORFs. Most of the mutations are observed in gene OPG 047 which is closer to the middle of the genome followed by OPF 053, OPG 074 and OPG 105. Earlier reported clade defining mutation was observed (C 34472 T) in all the retrieved sequences from India. During complete genome analysis, we have observed insertions; one in OPG 047 (insertion of T at 29767), and other in non coding region (insertion of T at 170897). A deletion of CATATCA was also noted at 148529- 148535 in gene OPG 174 9 (Fig. 2C).
Interestingly, one di-nucleotide substitution of AT-CC in OPG 047 at position 29768 leading to amino acid change (I 476 G) was observed in the retrieved sequences of ten monkeypox cases. Of 67, a total 63 substitution mutations were observed in cluster from India in which 58 are transitions and 05 are transversion (Non synonymous 30 and synonymous 33). One substitution mutation which led to stop gain at position 149872 was also recorded in gene OPG 176. No mutations were noted on H3L (OPG 108), glycosil transferase which plays important role in pox virus entry into host cell. We have also observed four non synonymous and two synonymous SNPs in OPG 31 and OPG 174 genes that are predicted to modulate the host immune response.
The 2022 Monkeypox outbreak demonstrates accelerated microevolution of MPXV leading to divergence in viral phylogeny.2, 3, 4, 5 Gigante et al., demonstrated 80 nucleotide changes in lineage A.2 compared to the B.1 lineage which has been predominant lineage of 2022 suggesting an independent virus strain emergence.3 The genomic research on the 2022 MPXV outbreak has also grabbed attention depicting divergence of lineage B.1 from lineage A.1 of 2018–2019 outbreaks.2 Hence, genome evolution mechanisms and importance of gene functions needs to be studied further to understand evolution of the MPXV genome. As B.1 is found to be the predominant lineage of 2022 Monkeypox outbreak globally, the introduction event of A.2 lineage specifically in the USA, UK, India and Thailand is the question of further exploration.
Ethical approval
The study was approved by the Institutional Human Ethics Committee of ICMR-NIV, Pune, India under the project ‘Providing diagnostic support for referred samples of viral hemorrhagic fever and other unknown etiology and outbreak investigation’. The clinical data collected were anonymized. The informed consents were obtained for the use of the clinical details in the study.
Financial support & sponsorship
The intramural grant was provided from ICMR-National Institute of Virology, Pune for conducting this study.
Supplementary information
Supplementary Table-1. Clinico-demographic, viral load and outcome of ten monkey pox cases from India, July-August 2022
CRediT authorship contribution statement
Anita M. Shete: Visualization, Formal analysis, Methodology, Writing – original draft, Writing – review & editing, Writing – review & editing. Pragya D. Yadav: Visualization, Formal analysis, Methodology, Writing – original draft, Writing – review & editing, Writing – review & editing. Abhinendra Kumar: Data curation, Methodology, Writing – review & editing. Savita Patil: Data curation, Methodology, Writing – review & editing. Deepak Y. Patil: Data curation, Methodology, Writing – review & editing, Writing – review & editing. Yash Joshi: Data curation, Methodology, Writing – review & editing. Triparna Majumdar: Data curation, Methodology, Writing – review & editing. Vineet Relhan: Data curation, Methodology, Writing – review & editing. Rima R. Sahay: Data curation, Methodology, Writing – review & editing, Writing – review & editing. Meenakshy Vasu: Data curation, Methodology, Writing – review & editing. Pranita Gawande: Data curation, Methodology, Writing – review & editing. Ajay Verma: Data curation, Methodology, Writing – review & editing. Arbind Kumar: Data curation, Methodology, Writing – review & editing, Writing – review & editing. Shivram Dhakad: Data curation, Methodology, Writing – review & editing. Anukumar Bala Krishnan: Data curation, Methodology, Writing – review & editing. Shubin Chenayil: Data curation, Methodology, Writing – review & editing. Suresh Kumar: Data curation, Methodology, Writing – review & editing. Priya Abraham: Writing – review & editing, Writing – review & editing.
Declaration of Competing Interest
Authors do not have a conflict of interest among themselves.
Acknowledgement
Authors extend gratitude to Smt. Veena George [Hon'ble Minister for Health and Family Welfare, Kerala], for efficient coordination of the monkeypox virus disease control activities, and “The Team Kerala Health,” the district administration. We would also extend our gratitude towards Dr. Asha Thomas, Additional Chief secretary of Medical Education, Mrs. Tinku Biswal, Principal Secretary for Health Kerala, Dr. Thomas Mathew, Director of Medical Education and Dr. Preetha PP, Director of Health Services, Kerala. The authors would like to thank Dr. Lakshmi Geetha Gopalkrishnan, State Epidemiologist, and District Surveillance Officers of Thiruvananthapuram [Dr. Preethi James], Kannur [Dr. Shaj MK], Kollam [Dr. Sandhya], and Thrissur [Dr. Anoop TK and Dr. Kavya Karunakaran]. Dr. Kala Kesavan P, Principal; Dr. Nizarudeen A, Medical superintendent; Dr. Aravind Reghukumar, HOD Infectious Diseases and Dr. Manjusree Suresh, from Government Medical College Thiruvananthapuram. The authors are thankful to Dr. Prathap Somanath, Principal, Dr. Sudeep, HOD Infectious Diseases; Dr. Manasi Ravindranath from Government Medical College Kannur; Dr. Shinas, Government Medical College, Manjeri. The authors also acknowledge the support from Dr. Fazil Abubaker from Daya General Hospital and Specialty Surgical Centre, Thrissur.
Authors extend sincere thanks to Dr. Bijayalaxmi Sahoo, Professor and Head, and resident doctors Dr. Abhinav Kumar, Dr. Aneet Kaur, Dr. Bhawna Solanki, Dr. Anjali Bagrodia of Dermatology; Dr. Sonal Saxena, Professor, and Head, Department of Microbiology from Maulana Azad Medical College and Lok Nayak Hospital, New Delhi for providing support for sample collection and transportation. We are also grateful to Dr. Lalit Dar, Professor, Department of Microbiology; Dr. Aashish Choudhary, Dr. Megha Brijwal, from All India Institute of Medical Sciences, New Delhi. The authors are thankful to Dr. Avdesh Kumar, State Surveillance Officer, New Delhi and his team for coordination. The authors are extremely grateful to Dr. Nivedita Gupta, Scientist ‘F’ and Head, Epidemiology and Communicable Diseases, ICMR, New Delhi for her constant support.
We also acknowledge the excellent technical support from Dr. Kannan Sabarinath PS, Dr. Rajlaxmi Jain, Ms. Jyoti Yemul, Mr. Sunil Shelkande, Ms. Pratiksha Vedhpathak, Mrs. Shubhangi Sathe, Ms. Vaishnavi Kumari, Ms. Nandini Shende, Mr. Raj Hawale for the diagnosis and data management for the diagnosis and data management. The authors also would like to thank and express immense gratitude to the monkeypox cases and family members, who willingly agreed and provided consent to be part of the study.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jinf.2022.09.024.
Appendix. Supplementary materials
References
- 1.Jolly B., Scaria V. A distinct phylogenetic cluster of Monkeypox genomes suggests an early and cryptic spread of the virus. J Infect. 2022 doi: 10.1016/j.jinf.2022.08.013. Aug 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Isidro J., Borges V., Pinto M., Sobra D., Santos J.D, Nunes A., et al. Phylogenomic characterization and signs of microevolution in the 2022 multi-country outbreak of monkeypox virus. Nat Med. 2022:1–4. doi: 10.1038/s41591-022-01907-y. Jun 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gigante C.M., Korber B., Seabolt M.H., Wilkins K., Davidson W., Rao A.K., et al. Multiple lineages of Monkeypox virus detected in the United States, 2021-2022. BioRxiv. 2022 doi: 10.1126/science.add4153. Jan 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O'Toole A., Rambaut A. ARTIC Network; 2022. Initial observations about putative APOBEC3 deaminase editing driving short-term evolution of MPXV since 2017. [Google Scholar]
- 5.Yadav P.D., Reghukumar A., Sahay R.R., Sudeep K., Shete A.M, Raman A., et al. First two cases of Monkeypox virus infection in travellers returned from UAE to India, July 2022. J Infect. 2022;S0163-4453(22):00471–00476. doi: 10.1016/j.jinf.2022.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y., Olson V.A., Laue T., Laker M.T., Damon I.K. Detection of monkeypox virus with real-time PCR assays. J Clin Virol. 2006;36(3):194–203. doi: 10.1016/j.jcv.2006.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yadav P.D., Nyayanit D.A., Shete A.M., Jain S., Majumdar T., Chaubal G.Y, et al. Complete genome sequencing of Kaisodi virus isolated from ticks in India belonging to Phlebovirus genus, family Phenuiviridae. Ticks Tick-Borne Dis. 2019;10(1):23–33. doi: 10.1016/j.ttbdis.2018.08.012. [DOI] [PubMed] [Google Scholar]
- 8.Senkevich T.G., Yutin N., Wolf Y.I., Koonin E.V, Moss B. Ancient gene capture and recent gene loss shape the evolution of orthopoxvirus-host interaction genes. Mbio. 2021;12(4) doi: 10.1128/mBio.01495-21. e01495-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nguyen L.T., Schmidt H.A., Von Haeseler A., Minh B.Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evolut. 2015;32(1):268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.