Abstract
An extracellular enzyme activity in the culture supernatant of the acarbose producer Actinoplanes sp. strain SE50 catalyzes the transfer of the acarviosyl moiety of acarbose to malto-oligosaccharides. This acarviosyl transferase (ATase) is encoded by a gene, acbD, in the putative biosynthetic gene cluster for the α-glucosidase inhibitor acarbose. The acbD gene was cloned and heterologously produced in Streptomyces lividans TK23. The recombinant protein was analyzed by enzyme assays. The AcbD protein (724 amino acids) displays all of the features of extracellular α-glucosidases and/or transglycosylases of the α-amylase family and exhibits the highest similarities to several cyclodextrin glucanotransferases (CGTases). However, AcbD had neither α-amylase nor CGTase activity. The AcbD protein was purified to homogeneity, and it was identified by partial protein sequencing of tryptic peptides. AcbD had an apparent molecular mass of 76 kDa and an isoelectric point of 5.0 and required Ca2+ ions for activity. The enzyme displayed maximal activity at 30°C and between pH 6.2 and 6.9. The Km values of the ATase for acarbose (donor substrate) and maltose (acceptor substrate) are 0.65 and 0.96 mM, respectively. A wide range of additional donor and acceptor substrates were determined for the enzyme. Acceptors revealed a structural requirement for glucose-analogous structures conserving only the overall stereochemistry, except for the anomeric C atom, and the hydroxyl groups at positions 2, 3, and 4 of d-glucose. We discuss here the function of the enzyme in the extracellular formation of the series of acarbose-homologous compounds produced by Actinoplanes sp. strain SE50.
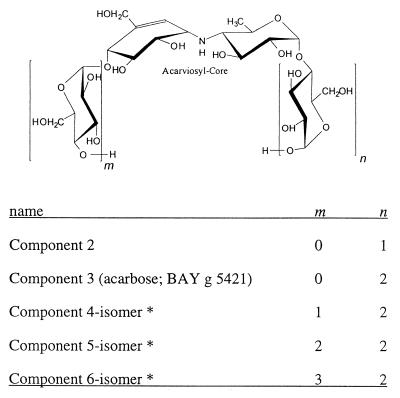
Several aminoglycosidic α-glycosidase inhibitors with C7-cyclitol moieties have been found in the culture broth of various actinomycetes (20, 30). These include the α-glucosidase and trehalase inhibitors of the acarbose-amylostatin group of compounds produced by Actinoplanes sp. and Streptomyces sp. (9, 21, 38) (Fig. 1), the oligostatins (26), adiposins (24, 25), and trestatins (42). Another member group of this class of compounds are the chitinase inhibitors validamycins and validoxylamines produced by Streptomyces hygroscopicus var. limoneus (13). They all contain 1 or 2 U of a valiolol-derived cyclitol. Since 1990 the α-glucosidase inhibitor acarbose is used in the therapy of non-insulin-dependent diabetes mellitus. The oral antidiabetic agent is produced by fermentation of the actinomycete Actinoplanes sp. strain SE50. Besides acarbose, the organism produces an extensive series of acarviosyl {4-N-4,6-didesoxy-4-([4,5,6-trihydroxy-3-hydroxymethyl-2-cyclohexen-1-yl]amino)-α-d-glucopyranose} containing pseudo-oligosaccharides (Fig. 1). These compounds differ in the number of glucose units connected among each other by α-1,4 glycosidic bonds which are attached to the acarviosyl core at the reducing and nonreducing ends. The number of glucose units determines the inhibitory specificity against different α-glycosidases. In addition, some compounds show variations in the type of the terminal glycosidic bond or in the nature of the terminal sugar moiety (Table 1).
FIG. 1.
Chemical structures of the acarbose and amylostatin family of α-glucosidase inhibitors from Actinoplanes sp. strain 50/110. For components marked with an asterisk, the main ingredient of the isomer mixture with m + n is 3 (4 or 5).
TABLE 1.
Names and compositions of acarviosyl-containing compoundsa
| Name | Composition |
|---|---|
| Acarbose (component 3) | Acarviosyl-1-4-Glc-1-4-Glc |
| Component A | Acarviosyl-1-4-Glc-1-4-Fru |
| Component B | Acarviosyl-1-4-Glc-1-4-Val |
| Component C | Acarviosyl-1-4-Glc-1-1-Glc |
| Component D | Acarviosyl-1-4-Glc-1-4-Man |
| Component 4a | Acarviosyl-1-4-Glc-1-4-Glc-1-4-Fru |
| Component 4b | Acarviosyl-1-4-Glc-1-4-Glc-1-4-Glc |
| Component 4c | Acarviosyl-1-4-Glc-1-4-Glc-1-1-Glc |
| Pseudo-acarbose | Acarviosyl-1-4-(6-desoxy)Glc-1-4-Glc |
Structural differences compared to acarbose or component 4b, respectively, are indicated in boldface. Glc, glucose; Fru, fructose; Man, mannose; Val, 1-epi, 2-epi-valienol.
The different homologues are formed dependent on the sugar source in the culture broth (34). If glucose or maltose are supplied as the sole carbon source inhibitors with a small number of glucose units are produced, preferentially acarbose (strong inhibition of disaccharidases), while addition of starch leads to compounds with a higher number of glucose units (strong inhibition of amylases). Now it is known that acarbose in contact with α-amylases and cyclodextrin glucanotransferases (CGTases) in the presence or absence of maltooligodextrins becomes converted to longer-chain derivatives containing at least two acarviosyl residues, as shown in crystallized enzyme-inhibitor complexes (10, 15, 31, 36). Therefore, acarbose can be regarded as a prodrug which forms more active inhibitors by the catalytic activity of its target site.
Fermentation experiments with Actinoplanes sp. in the presence of [U-14C]maltose showed that the maltosyl unit of acarbose is derived directly from maltose (K. Goeke and H. Pape, unpublished results). These findings point to an enzymatic activity responsible for many variations found in acarbose homologues. We describe here the purification and characterization of the enzyme AcbD (acarviosyl transferase [ATase]) from the supernatant of Actinoplanes sp. strain SN223/29 cultures. In addition, the location of the corresponding gene, encoding the AcbD protein, within the recently identified biosynthetic gene cluster for acarbose (35) is shown, and the protein was heterologously produced in Streptomyces lividans TK23.
MATERIALS AND METHODS
Bacterial strains, plasmids, medium, and culture conditions.
The Actinoplanes sp. strain SN223/29 used in this study is an improved acarbose producer developed from the wild-type strain SE 50/110. For purification of the AcbD protein, the organism was cultivated in a two-stage complex medium. For the preculture stage (72 h), defatted soy flour (Henselwerk, Magstadt, Germany) at 2%, glycerol at 2%, and CaCO3 at 0.2% in tap water were used; the pH was adjusted to 7.2 before sterilization. For the inoculation stage, a 4% (vol/vol) inoculum was obtained from a frozen stock. The main culture (125 ml) was cultivated for 120 h in soluble starch at 3%, defatted soy flour at 1%, and CaCO3 at 0.2% in tap water, with a 4% (vol/vol) preculture inoculum. The cultivation was carried out in Erlenmeyer flasks at 28°C and 150 rpm in a rotary shaking incubator. S. lividans 66 strain TK23 (12) was used as the host strain for the heterologous expression of the AcbD protein. The strain was routinely cultured at 28°C on SMA agar plates (7), and liquid cultures were carried out in TSB medium (12). The strain Actinoplanes SE50/110 was cultivated in acarbose-production medium [MD-50(maltodextrins), 70 g; (NH4)SO4, 5 g; yeast extract, 2 g; K2HPO4, 1 g; KH2PO4, 1 g; Tri-sodiumcitrat, 5 g; MgCl2 · 6H2O, 1 g; FeCl3 · 6H2O, 0.25 g; and CaCl2 · 2H2O, 2 g, all dissolved in 1,000 ml of H2O, with pH adjusted to 6.8, and sterilized by filtration]. The isolation of the plasmids pAS5 and pAS6, with 10.7-kb SstI and the 12.4-kb BglII inserts of Actinoplanes sp. DNA, respectively, was described earlier (35) (Fig. 2). Cloning experiments with Escherichia coli were performed using the plasmids pUC18 (41) and pUWL201 (8) and the host strain DH5α [F− φ80d lacZΔM15 endA1 recA1 hsdR17 (rK− mK+) supE44 thi-1 λ gyrA96 relA1 Δ(lacZYA-argF)U169 (11)], which was grown at 37°C in Luria-Bertani (LB) broth or on LB agar plates supplemented with ampicillin (100 μg/ml) (33). To maintain the pUWL201 derivatives in the corresponding S. lividans strains, the media were supplemented with thiostrepton (25 μg/ml).
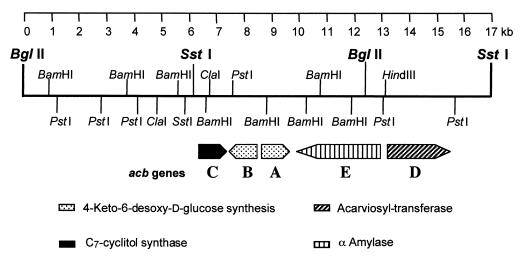
FIG. 2.
Map location of the acbD gene within the putative biosynthetic gene cluster for acarbose. The restriction sites flanking the inserts of 10.7-kb SstI and 12.4-kb BglII of Actinoplanes sp. DNA in plasmids pAS5 and pAS6 (35), respectively, are given in bold face. The transcriptional direction and the relative sizes of the predicted open reading frames are indicated by bars with arrowheads. The DNA sequence for this segment is available from the databases under accession no. AJ293724.
Purification of ATase.
The culture broth (500 ml) of Actinoplanes sp. strain SN223/29, grown in main culture medium, was centrifuged at 2,500 × g for 10 min to remove the cells. The supernatant was brought to 20% ammonium sulfate saturation in the cold by adding solid ammonium sulfate. After 2 h the solution was centrifuged at 25,000 × g for 30 min. The supernatant was brought to 40% ammonium sulfate saturation and again centrifuged. The resulting precipitate was dissolved overnight in 100 ml of 25 mM Tris-HCl buffer (pH 8.5), containing 10% glycerin–1 mM CaCl2 and then cleared by centrifugation. The enzyme solution was applied to a Fractogel DEAE anion exchanger (Merck, Darmstadt, Germany) column (2 by 16 cm) previously equilibrated with 25 mM Tris-HCl buffer (pH 8.5). Under these conditions the enzyme did not bind to the anion exchanger. The column was washed with 50 ml of the same buffer. The combined effluent was dialyzed overnight against the same buffer. The desalted enzyme solution was again applied to the same anion exchanger, and the column was washed with the equilibrating buffer. Proteins were eluted by applying a linear NaCl gradient in the same buffer (300 ml, 0 to 1 M NaCl, flow rate of 0.5 ml/min). The active fractions (0.15 to 0.3 M NaCl) collected from the column were pooled, 100 mg of prepared starch (see below) per ml was added, and the mixture was stirred overnight and then centrifuged at 40,000 × g for 1 h. The resulting pellet was homogenized in 30 ml of buffer (25 mM Tris-HCl [pH 7.5] plus 1 mM CaCl2) containing 25 mM acarbose, stirred at 20°C for 2 h, and then centrifuged (as described above). The supernatant was dialyzed against 10 mM Tris-HCl (pH 7.5) and 1 mM CaCl2 for 2 days with several changes of the buffer and then treated again with the prepared starch. From the resulting pellet, the enzyme was desorbed by incubation with a 250 mM maltose solution in 10 mM Tris-HCl (pH 7.5) and 1 mM CaCl2 at 4°C overnight. The resulting supernatant after centrifugation (as described above) was dialyzed against 0.1 mM Tris-HCl buffer (pH 7.2) and 0.01 mM CaCl2 for 2 days with several changes of buffer.
For the preparation of starch, soluble starch (100 mg/ml of water) was incubated for 20 min at 121°C and then precipitated for 4 days at 4°C. After centrifugation at 40,000 × g for 1 h, the pellet was homogenized in Tris-HCl buffer (25 mM at pH 8.5 containing 10% glycerin and 1 mM CaCl2) and centrifuged. The pellet was used for the starch adsorption of the ATase.
Determination of ATase activity.
The enzyme assay used for ATase activity determination is based upon the transfer of the acarviosyl moiety of acarbose (donor) to radioactive maltose (acceptor) (reaction A, see below). The standard assay used for determination of the ATase activity is based on this exchange reaction. With other sugars the catalyzed reaction has to be formulated as in reaction B.
(A) acarbose + maltose∗ ⇌ acarbose∗ + maltose
(B) acarbose + ROH ⇌ acarviosyl-OR + maltose
where an asterisk indicates a radioactively labeled sugar. After the incubation of unlabeled acarbose with [14C]maltose, the amount of radioactivity found in acarbose depends on the incubation time and the enzyme activity in the added protein solution.
A 10-μl volume of appropriately diluted enzyme solution was mixed with 4 μl of the reaction solution consisting of 0.25 M Tris-maleinate buffer (pH 6.3), 19.6 mM acarbose, and 1.75 mM [U-14C]maltose (44,000 dpm). This mixture was incubated at 30°C for normally 15 min. The reaction was stopped by adding 100 μl of ethanol. After centrifugation to remove precipitated protein, the radioactive acarbose was separated from radioactive maltose by adding the supernatant to a suspension (500 μl) of Dowex 50 WX 4 (Serva, Heidelberg, Germany), H+-form, in demineralized water (1:1; vol/vol). To remove the unbound radioactive maltose, the supernatant was taken off, the resin was washed three times with 500 μl of water, and the washings were collected. Acarbose was eluted from the cation exchanger with 0.5 M ammonia solution (three times with 500 μl). In both fractions the radioactivity was measured by scintillation counting. The relative amount of radioactivity in the acarbose fraction (the exchange rate, typically between 5 and 50% of the total radioactivity applied per assay, was corrected by the exchange rate of a control assay stopped directly with ethanol after addition of the reaction solution) is used as measure for enzyme activity.
Protein concentration was determined according to the method of Bradford (5).
Substrate specificity and enzyme kinetics.
The Km and relative Vmax values of ATase for acarbose and maltose were calculated from Lineweaver-Burk plots. The data were obtained through standard incubation tests (see above), with a constant concentration (10 mM) of one substrate and varied concentrations (0.1 to 6 mM) of the other substrate. Assays for the determination of acceptor specifity consisted of ATase preparation (15 nkat/ml) in buffer (0.1 mM Tris-HCl, 0.01 CaCl2; pH 7.2), acarbose (23 mM), and different acceptor substrates (23 mM each) in a total volume of 30 μl. The incubation was stopped after 18 h by adding 70 μl of ethanol. After centrifugation the supernatant was analyzed by thin-layer chromatography (TLC) and high-pressure liquid chromatography (HPLC) (see below). The donor specificity was tested by use of the purified ATase in similar reaction mixtures with the same buffer system and volume, but with maltose (200 mM) and different mixtures of acarbose homologues (16.6 mg/ml) as acceptor and donor substrates, respectively. The incubation was stopped after 0 and 24 h by adding ethanol, and the supernatant obtained after centrifugation was analyzed by HPLC (see below).
Further enzyme characterization.
For the determination of temperature and pH stability of the ATase activity, the assay mixture was incubated at various temperatures (18 to 67°C) and pH values (70 mM Tris-maleinate buffer [pH 5 to 8.4] at 30°C). The thermostability of the enzyme was estimated through preincubation of the ATase preparation at different temperatures (see above) for 15 min, followed by standard enzyme assay at 30°C. The effect of metal ions was analyzed for 1 mM additions of CaCl2, MgCl2, MnCl2, FeCl2, FeCl3, CoCl2, CuSO4, ZnSO4, and EDTA after incubation at 4°C for 72 h. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was done by the method of Laemmli (16). Marker proteins with molecular weights ranging from 205,000 to 29,000 (Fluka, Buchs, Switzerland) were used for estimation of the molecular weight of ATase. The isoelectric point was determined by isoelectric focusing (IEF) using ampholyte (Serva) for a pH gradient from 3 to 10 and marker proteins with isoelectric points from 3.6 to 9.3 (Sigma, Deisenhofen, Germany).
TLC.
The acceptor specifity assays were analyzed by TLC with silica gel plates (Merck, Darmstadt, Germany), which were developed with butanol-ethanol-H2O (5:3:2 [vol/vol/vol]). The spots were visualized by spraying the plate with ceric sulfate reagent [phosphomolybdic acid · H2O (25 g), cer(IV)-sulfate · 4H2O (10 g), concentrated H2SO4 (60 ml), demineralized water (940 ml)] followed by heating (15 min at 110°C).
HPLC.
The acceptor specificity assays were analyzed by HPLC on a Carbopac PA 1 anion-exchange column (4 by 250 mm; Dionex, Idstein, Germany), with a gradient formed by solvent A (0.1 M NaOH) and solvent B (0.1 M NaOH, 0.5 M sodium acetate) as eluent at a flow rate of 1 ml/min (time:%A values = 0:100, 10:92, 30:90, 45:35, 46:0, 48:0, 49:100, and 58:100). Peaks were recorded with a pulsed electrochemical detector (Model HP 1049 A; Hewlett-Packard, Ratingen, Germany) with measuring voltage of 0.1 V 0.2 s and a pulse voltage of 0.6 V 0.08 s and −0.6 V 0.08 s.
The donor specificity assays were analyzed with an NH2 column (4 by 250 mm; Shandon Hypersil APS 1 [5 μm]; refill by Muder und Wochele, Berlin, Germany), with an eluent formed by 75% acetonitrile and 25% 5 mM potassium hydrogen phosphate buffer (pH 6.8) at a flow rate of 2.5 ml/min and at 35°C. Acarbose homologues peaks were recorded at 210 nm with a UV 1000 detector (Thermo Separation Products, Darmstadt, Germany).
General DNA manipulation techniques.
Restriction enzymes and T4 DNA ligase were purchased from Life Technologies (Eggenstein, Germany) and used in accordance with the manufacturer's instructions. DNA fragments were recovered from agarose gels using the JetSorb Kit (Genomed, Bad Oeynhausen, Germany). DNA manipulations of E. coli were done as described by Sambrook et al. (33); transformations of E. coli were carried out by the method of Hanahan (11). Protoplast preparation and plasmid transformations techniques for S. lividans were performed according to published procedures (2, 12).
Identification, cloning, and expression of the acbD gene.
The acbD gene was identified as a part of the biosynthetic gene cluster for acarbose in Actinoplanes strain SE50/110 by using the recombinant plasmid pAS5. A 2.6-kb HindIII/PstI DNA fragment was isolated from pAS5 and ligated into pUC18 in E. coli DH5α. The resulting recombinant plasmid pAS5/15.1 was analyzed by DNA sequencing. One open reading frame of 2,172 bp was identified and named acbD. For the expression of acbD in S. lividans TK23 under the control of the ermEp promoter, the 2.6-kb HindIII/PstI DNA fragment was cloned into pUWL201 HindIII/PstI, resulting in pAS9. To remove the 126-bp sequence upstream of the GTG start codon of the acbD gene (Fig. 3), pAS9 was hydrolyzed with the enzyme EcoNI. After treatment with the Klenow enzyme (Roche, Mannheim, Germany) and hydrolysis with BamHI, a 2.5-kb DNA fragment was isolated and cloned in pUC18 HincII/BamHI resulting in pAS5/15.1.1. The deletion of the 126 bp was controlled by DNA sequencing. The shortened acbD DNA fragment was reisolated as a HindIII/BamH1 fragment and ligated into pUWL201 HindIII/BamHI, resulting in pAS9-2. The strains S. lividans TK23/pAS9 and S. lividans TK23/pAS9-2 were cultivated in 10 ml of TSB medium for 3 to 4 days, and the supernatants were dialyzed for 12 h at 4°C against buffer (5 mM Tris-HCl, 1 mM CaCl2; pH 7.5). To test the extracellular production of the AcbD protein, 500 μl of the desalted supernatants was concentrated under vacuum and analyzed by SDS-PAGE (8% polyacrylamide [PAA] gel).
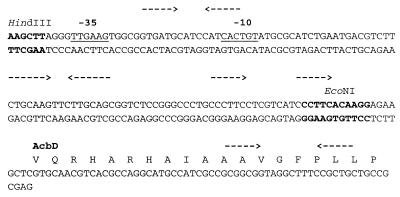
FIG. 3.
Putative regulatory region upstream of the acbD reading frame. Inverted- and direct-repeat structures are marked by arrows. The possible ribosome-binding site and a possible promoter region (−10, −35) with similarity to E. coli ς70-like Streptomyces sp. promoters are marked by underlining. The recognition sites for restriction enzymes are indicated.
DNA sequencing, computer analysis of DNA sequences, and accession number of the nucleotide sequence.
Various overlapping restriction fragments of the 2.6-kb HindIII/PstI DNA fragment insert in pAS5/15.1 were subcloned in pUC18 and sequenced by the dideoxynucleotide chain termination method (32) using an AutoRead sequencing kit and an A.L.F. DNA sequencer (Amersham Pharmacia Biotech, Freiburg, Germany). The entire sequences of both strands were determined from double-stranded plasmid DNAs prepared by the alkaline lysis method (33). The DNA sequences were analyzed using DNA-Strider 1.2 (19) and BrujeneII sequence analysis software. Homology searches were performed against EBI, GenBank, and SWISSPROT data libraries using BLAST (1) and FASTA 1.4×2 (27) software. The accession number of the nucleotide sequence is AJ293724.
Protein sequencing.
About 500 μg of the purified AcbD protein was dissolved in 1 ml of 6 M guanidinium hydrochloride–0.5 M Tris-(hydroxymethyl)-aminomethane (pH 8.6). Then, 5 μl of 1 M dithiothreitol was added, and the sample was reduced at 54°C for about 18 h. Next, 10 μl of 2 M sodium iodoacetate solution was added, and the sample was incubated in the dark for 35 min. After dialysis against 0.5 M urea–0.1 M ammonium hydrogen carbonate solution (buffer change after 2 and 4 h; total time of dialysis about 6 h; dialysis tube cutoff, 3.5 kDa). The sample was digested by adding 5 μg of bovine trypsin (sequence grade) at 37°C for 18 h. The sample was concentrated before HPLC analysis to about 500 μl by centrifugal evaporation in a Speedvac. The tryptic peptides were separated by HPLC using an HP1090 system equipped with a diode array detector 1040A, a chemstation, and a fraction collector. The HPLC components were from Hewlett-Packard (Waldbronn, Germany), and the fraction collector, Superrac 2211, was from Pharmacia (Freiburg, Germany). A Nucleosil RP-18 column (250 by 4.6 mm; 5-μm spheres; 300-Å pore diameter) from CS-Chromatographie Service (Langerwehe, Germany) was used for peptide separation. The collected peptides were used for sequence analysis. N-terminal sequence analyses of ATase samples were performed using the gas-liquid-solid-phase protein sequencer 473A from Applied Biosystems (Foster City, Calif.). The standard sequencer program FAST Normal was used. The sequencer, the different running programs, and the cycles, as well as the phenylthiohydantoin (PTH) separation system, are described in detail in the respective user manuals (i.e., for protein sequencing system model 473A [1989]; Applied Biosystems). The detection of PTH amino acids were performed online using an RP18-PTH column (220 by 2 mm, 5-μm spheres) from Applied Biosystems.
RESULTS
Production, purification, and properties of the ATase of Actinoplanes sp. strain SN223/29.
The extracellular ATase is optimally produced in starch-containing media. The enzyme was purified from the supernatant of soyflour starch cultures by a five-step procedure (as described in Materials and Methods): precipitation with ammonium sulfate, two ion-exchange chromatography steps, and two adsorption-elution steps using starch as the adsorbent and acarbose or maltose as the desorbing agents. Although the latter two steps are effective in obtaining a purification of ATase, the nature of the interaction between starch and enzyme is currently not clear. Perhaps, starch, maltose, and acarbose interact with the active center or a starch-binding site of the enzyme, similar to the raw-starch-binding sites described for CGTases (17).
The ATase was purified 18-fold to give a homogeneous preparation (as shown by SDS-PAGE and IEF) with a specific activity of 77 nkat/mg (Table 2). The molecular mass of the enzyme was 76 kDa, and it had an isoelectric point of 5.0. The addition of Ca2+ ions led to an enhanced enzyme activity corresponding to the total inhibition by addition of EDTA. The enzyme is thermostable up to 40°C and displays a slight maximum of activity at 30°C. The optimal pH for enzyme activity was determined to be between pH 6.2 and 6.9. The Km values of ATase for acarbose and maltose are 0.65 and 0.96 mM, respectively.
TABLE 2.
Purification of the ATase of Actinoplanes sp. strain SN223/29
| Stage | Total protein (mg) | Total activity (nkat) | Sp act (nkat/mg of protein) | Recovery (%) | Purification factor |
|---|---|---|---|---|---|
| Culture filtrate | 400 | 1,740 | 4.3 | 100 | 1.0 |
| Ammonium sulfate precipitation | 139 | 1,147 | 7.7 | 66 | 1.8 |
| DEAE-chromatography | |||||
| Step 1 | 89 | 1,078 | 12.1 | 62 | 2.8 |
| Step 2 | 20 | 789 | 38.8 | 45 | 8.9 |
| Starch adsorption-elution techniques | |||||
| Acarbose elution | 10 | 538 | 55.5 | 31 | 12.8 |
| Maltose elution | 6 | 425 | 77.3 | 24 | 17.8 |
The partial amino acid sequence of seven peptides out of the purified ATase were determined by digestion with trypsin and subsequent sequencing by Edman degradation (Fig. 4). These tryptic peptides were identified in the deduced amino acid sequence of the acbD gene encoding the ATase (see below).
FIG. 4.
Alignment of the protein sequences of the ATase AcbD with those of several CGTases. The proteins aligned were from the following organisms (with GenBank and EBI database accession codes in parentheses): Ac., Actinoplanes sp. (AJ293724); T. ther., Thermoanaerobacterium thermosulfurigenes (P26827); B. cir., Bacillus circulans 251 (P43379); P. mar., Paenibacillus macerans (P31835); and K. pneu., Klebsiella pneumoniae (P08704). The three acidic amino acid residues of the active site (catalytic triade), which are also conserved in most of the proteins of the amylase family, are shown in boldface (15). The amino acid residues identified to participate in the binding of acarbose to the CGTase are boxed (36); the aromatic amino acid residue identified to be crucial for the CGTases (28), which is changed to A in AcbD, is marked by a delta symbol (Δ). Residues identical in all four sequences are marked by an asterisk; those where side chains are replaced by similar ones are labeled by a dot. The partial protein sequences as determined from tryptic peptides of the purified ATase from Actinoplanes sp. strain 223/29 are underlined.
Donor and acceptor substrates of the ATase.
The ATase accepted a wide range of substrates, including glucose, maltooligosaccharides, dextrin, and amylopectin (Table 3). In standard ATase assays, using unlabeled maltose or the other compounds as competing substrates for radioactively labeled maltose, none of these compounds was as efficient an acarviosyl acceptor as was maltose itself. Interestingly, the glucosides of aliphatic alcohols were also good acceptor substrates for the ATase, with the highest rate measured for nonyl-α-glucopyranoside. As donor substrates, component 2 (acarviosyl glucose) and component 4b (acarviosyl maltotriose) were identified. The incubation of these substrates with maltose in the presence of purified ATase resulted in the formation of acarbose (Table 4). Only components A and C (Table 1) did not act as acarviosyl donors in an ATase-catalyzed reaction. In addition, component C (acarviosyl trehalose) was not formed by the ATase in the presence of acarbose and trehalose. So this component, which reaches high concentrations in fermentations of Actinoplanes sp., probably does not originate from the activity of the ATase. Component A arises by chemical modification reactions during the purification procedure of acarbose from the culture broth (Bayer AG, unpublished results), and so it is not a pseudo-oligosaccharide biosynthesized by the bacterium.
TABLE 3.
Acceptor specificity of ATasea
| Acceptor substrates of the ATaseb | Specificity valvesc
|
|
|---|---|---|
| TLC | HPLC (%) | |
| Nonyl-α-d-glucopyranoside | + | 28.1 |
| Xylobiose | + | 20.8 |
| Maltotetraose | + | 19.9 |
| Maltotriose | + | 19.8 |
| Maltoheptaose | + | 18.9 |
| Maltopentaose | + | 17.9 |
| Cellobiose | − | 17.2 |
| Maltohexaose | + | 17.2 |
| Octyl-α-d-maltopyranoside | + | 16.5 |
| Laminaribiose | + | 16.3 |
| Sophorose | + | 16.3 |
| Decyl-α-d-maltopyranoside | + | 16.2 |
| d-(−)-Salicin | + | 14.8 |
| Phenyl-α-d-glucopyranoside | + | 14.8 |
| Panose | + | 14.0 |
| Octyl-d-glucopyranoside | + | 14.0 |
| Methyl-α-d-Glucopyranoside | + | 11.5 |
| l-(−)-Xylose | + | 11.0 |
| Dextrin | + | 10.0 |
| Isomaltose | + | 9.7 |
| Palatinose | − | 9.3 |
| Nigerose | + | 8.8 |
| 4-Nitrophenyl-α-d-glucopyranoside | + | 8.5 |
| d-(+)-Glucose | + | 7.6 |
| d-(+)-Xylose | + | 5.7 |
| Isomaltotriose | + | 5.4 |
| α-d-(+)-Maltose-1-phosphate | + | 5.2 |
| 6-Deoxy-glucose | + | 4.9 |
| l-(−)-Glucose | + | 4.7 |
| 5-Thio-d-glucose | + | 4.4 |
| d-(−)-Gluconolactone | + | 3.7 |
| myo-Inosit | + | 3.2 |
| Maltitol | + | 3.0 |
| Amygdalin | + | |
| Amylopektin | + | |
| 4-Nitrophenyl-α-d-xylopyranoside | + | |
The various substrates (each at 23 mM) were incubated with acarbose (23 mM) and purified ATase (15 nkat/ml) in 0.1 mM Tris-HCl buffer (pH 7.2) plus 0.01 mM CaCl2 for 18 h.
No formation of maltose and of new compounds was measured (via TLC and/or HPLC) for the following substrates: l-arabinose (TLC, HPLC), d-arabinose (TLC, HPLC), d-lyxose (TLC, HPLC), d-ribose (TLC, HPLC), 1.6-anhydro-d-glucose (TLC, HPLC), N-acetyglucosamine (TLC, HPLC), 2-deoxy-d-glucose (TLC, HPLC), d-(−)-fructose (TLC, HPLC), l-(−)-fucose (TLC, HPLC), d-(+)-galactose (TLC, HPLC), glucosamine (TLC, HPLC), d-glucuronic acid (TLC, HPLC), α-d-glucose-6-phosphate (TLC, HPLC), 3-O-methyl-α-glucopyranoside (TLC), d-(+)-mannose (TLC, HPLC), lactose (HPLC), melibiose (HPLC), saccharose (HPLC), trehalose (HPLC), turanose (HPLC), melezitose (HPLC), raffinose (HPLC), cyclo-α-dextrin (TLC, HPLC), and valienamin (TLC).
Product formation was determined by HPLC (i.e., the relative percent signal intensity of the maltose peak [100% = the sum of all recorded peaks]) or by TLC (“+” [formation of maltose and a new compound detected by TLC] or “−” [no maltose formation]).
TABLE 4.
Donor specificity of ATasea
| Homologue or homologue mixture | % Decrease of component | Increase in acarbose concn |
|---|---|---|
| Mixture | ||
| Component 4a | 91 | |
| Component 4b | 91 | Yes |
| Component 4c | 46 | |
| Component 2 | 33 | Yes |
| Mixture | ||
| Component B | 80 | No |
| Pseudo-acarbose | 100 | |
| Component A | 8 | No |
| Component C | 8 | No |
Various acarbose homologous or homologue mixtures (16.6 mg/ml) were incubated with maltose (200 mM) and purified ATase (15 nkat/ml) in 0.1 mM Tris-HCl (pH 7.2) plus 0.01 mM CaCl2 for 0 and 24 h. The resulting concentration differences are presented. (See Table 1 for a definition of the homologues other than acarbose).
Identification and characterization of the gene for ATase in Actinoplanes sp.
The acbD gene encoding the ATase was identified in the DNA region adjacent to the already-characterized 3.4-kb genomic region (accession no. Y18523) of the biosynthetic gene cluster for the aminoglycoside acarbose in Actinoplanes sp. strain 50/110 (Fig. 2). An internal 2.6-kb HindIII/PstI DNA fragment of the plasmid pAS5 contained a single reading frame of 2,172 bp showing the typical codon bias found for genes from actinomycetes (4). In the deduced amino acid sequence, the seven tryptic peptides out of the purified ATase were identified (Fig. 4). This clearly indicates the described reading frame as the acbD gene.
Heterologous expression of the ATase in S. lividans.
The acbD gene was expressed in S. lividans under the control of the constitutive ermEup promoter by using the vector pAS9. In the dialyzed supernatants of a cultivation of S. lividans/pAS9 a strong additional protein band was visible in comparison to the vector control experiment (Fig. 5, lanes 1 and 2, respectively). As a standard, the dialyzed supernatant of Actinoplanes strain SE50/110 after cultivation in acarbose production medium is shown (Fig. 5, lane 3). To prove the heterologous expression of acbD, the ATase activity in the corresponding supernatants was measured. The specific activity of the recombinant ATase was 2.9 nkat/mg, and for the parental enzyme it was 0.65 nkat/mg (Fig. 5, lanes 2 and 3, respectively). In the supernatants of the vector control (Fig. 5, lane 1), no ATase activity was detectable.
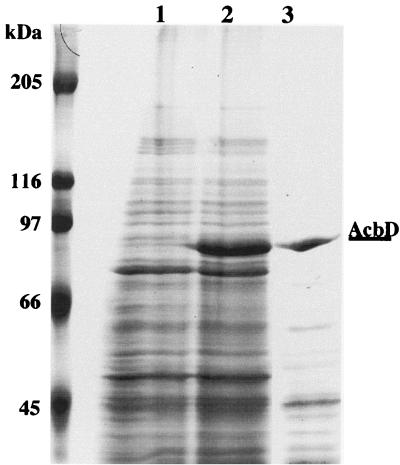
FIG. 5.
Extracellular expression of AcbD in S. lividans TK23. The lanes of the gel (0.1% SDS–8% polyacrylamide) contained samples of lyophilized protein from 500 μl of dialyzed culture supernatant from each of the following strains (for details of conditions, see Materials and Methods): lane 1, S. lividans TK23/pUWL201 (cultured in TSB medium); lane 2, S. lividans TK23/pAS9 (cultured in TSB medium); lane 3, Actinoplanes sp. strain SE50/110 (cultured in acarbose production medium); left lane, molecular weight marker. The location of the AcbD band is indicated.
The expression of acbD using the ermEup promoter shows an unstable phenotype in S. lividans. In order to reproduce the heterologous expression of acbD, the plasmid pAS9 has to be transformed anew into protoplasts of S. lividans and even so only a few of the resulting S. lividans/pAS9 strains again produced the AcbD protein. To overcome this problem, the putative regulatory region upstream acbD was eliminated by leaving the putative ribosome-binding site (Fig. 3). The resulting S. lividans/pAS9-2 strain did not produce the AcbD protein. Both plasmids pAS9 and pAS9-2 were still present in the corresponding strains, and no changes or DNA rearrangements were detectable by analysis using different restriction enzymes (data not shown).
Structural properties of the ATase and comparison to CGTases.
The AcbD protein shows significant similarity to members of the α-amylase superfamily of proteins. Among these groups the similarity is highest to the CGTases. AcbD retains the domain structure of CGTases, including the domains D and E for raw-starch binding (14, 29, 40). There are no actinomycete CGTases known so far. Therefore, the closest relatives are from other bacterial orders. The highest similarity scores (39 to 42%) were found with CGTases from the low-G+C gram-positive bacteria Bacillus circulans, Paenibacillus macerans, and Thermoanaerobacterium thermosulfurigenes and from the gram-negative bacterium Klebsiella pneumoniae (Fig. 4). The three acidic amino acid residues of the active site, e.g., the catalytic triade of D229, E257, and D331 in the CGTase of B. circulans 251 (15), which are also conserved in most of the proteins of the amylase family, are also present in the AcbD protein (Fig. 4). Most of the amino acid residues identified to participate in the binding of substrates and of the inhibitor acarbose in amylase and CGTase proteins of various organisms are also conserved (10, 18, 36, 37). However, the crucial aromatic amino acid residue in CGTases (Y or F at position 195; numbering system taken from the B. circulans 251 CGTase; Fig. 4) for the cyclization reaction of malto-oligosaccharides to α-, β-, and γ-cyclodextrins (23, 28) has been replaced by an A in AcbD. In this feature the ATase is more similar to α-amylases, in which this position is also occupied by smaller amino acid residues (G, S, T, V, or L) (40).
DISCUSSION
Structure-function relationships of the ATase in comparison to CGTases.
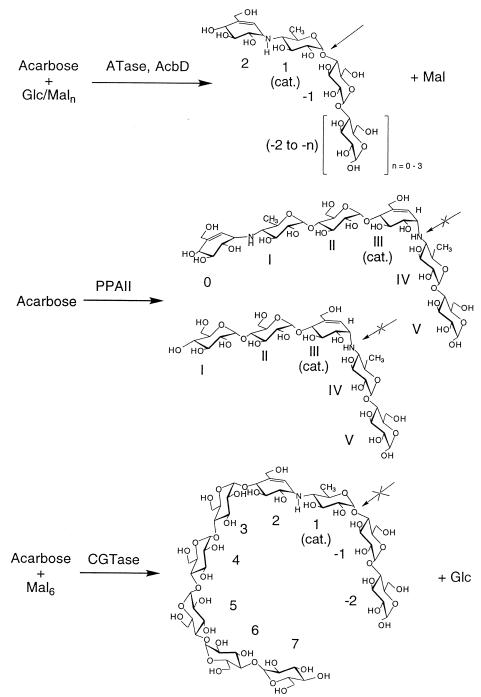
The ATase, as a member of the α-amylase superfamily, adds an interesting new type of catalytic specificity and substrate selectivity to this group of proteins: all members of the α-amylase family tested thus far bind acarbose and are inhibited by it but do not convert acarbose quantitatively. However, the ATase AcbD has neither α-amylase nor CGTase activity. The AcbD protein clearly belongs to the CGTase subfamily, exhibits solely transglycosylation activity on donor substrates containing the acarviosyl moiety, and accepts a wide range of acceptor substrates as shown in Fig. 6. Thus, the stronger transglycosylating activity of the CGTases, catalyzing a mixture of hydrolytic and group transfer reactions on various substrate lengths of α-1,4-glucosidic chains (22, 40), has been shifted during the evolution of AcbD toward a restriction of transferase activity and to substrates with smaller chain lengths. However, the strong binding of AcbD protein to starch and the structure of the protein chain suggest that the raw-starch-binding domain E is retained fully active but probably does not participate in catalysis or substrate binding as in the CGTases (29). Of the three types of transglycosylating activities which can be distinguished on the CGTases (22), only the so-called disproportionation reaction on α-1,4-glucosidic donors and acceptors resembles part of the ATase-catalyzed reactions and, therefore, could also follow a ping-pong mechanism (3, 39). The overall activities of both α-amylases and CGTases in the presence of acarbose seem, however, to be similar, since both enzyme groups use acarbose as donors and catalyze transglycosylation of parts of acarbose to either itself or other acceptors (10, 31, 36, 37) (Fig. 7). By this means, elongated and firmly bound inhibitors with a much higher inhibitor activity are formed, but with significant differences in their binding specificity in the catalytic site: (i) in the porcine pancreatic amylase II, these are positioned with the bridging pseudoglycosidic imino group of an acarviosyl moiety; and (ii) in the CGTase of B. circulans 251, the formed maltononaose inhibitor is positioned with the glycosidic bond between the reducing end of the acarviosyl moiety and the maltose. Based on these observations and the results presented here, a model for the binding specificity of the acarviosyl residue in the catalytic site of the ATase at positions +1 and +2 can be derived (Fig. 7).
FIG. 6.
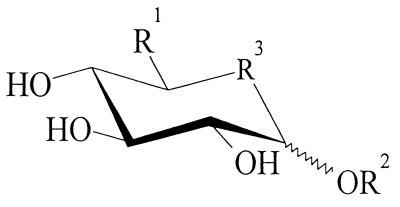
Acceptor specificity of the ATase (for details, see the text). R1 = H; OH; CH2PH; CH3. R2 = H; (CH2)mCH3, m = 0 to 9; pyranoses [α(1→2); α(1→3); α(1→4); α(1→6); β(1→2); β(1→3); β(1→4)]; furanoses [α(1→6)]; glucit; phenyl-; nitrophenyl-; etc. R3 = O; S; CHOH.
FIG. 7.
Comparison of catalytic mechanisms of α-amylase, CGTase, and ATase in the presence of acarbose alone or together with d-glucose (Glc) or maltooligodextrins (Maln) as transglycosylation donors or acceptors. The location of the substrate or inhibitor product, respectively, relative to the catalytic site (cat.) is indicated. The plain or crossed arrows indicate the ability or inability, respectively, to split the glycosidic bonds. The enzymes and data are as follows: PPA11, porcine pancreatic amylase II (10, 18); CGTase, CGTase from B. circulans 251 (28); ATase, AcbD, ATase (This study).
Donor and acceptor specificity of the ATase.
The reaction measured by the standard enzyme assay between acarbose and maltose probably does not represent a physiologically significant role of the ATase. Glucose, maltooligosaccharides, dextrin, and amylopectin were accepted with distinct transfer rates. In addition, the glucosides of aliphatic alcohols were also good acceptor substrates for the ATase, with the highest rate measured for nonyl-α-glucopyranoside. It is tempting to speculate that the enzyme may be responsible for the final step in the acarviosyl export: the transfer of an acarviosyl moiety from a membrane carrier (perhaps anchored in the membrane by an aliphatic moiety) to extracellular saccharides. However, the fact that other sugars are accepted as substrates indicates a central role of the ATase for the formation of the observed diversity of acarbose-related pseudo-oligosaccharides by Actinoplanes sp.
Taken together, the data in Table 3 allow us to draw a generalized structure for acceptor substrates of ATase (Fig. 6). This general model ignores the different acceptor efficiencies of the substrates. The basic structure is a pyranose ring with equatorial hydroxyl groups at C2, C3, and C4. At three positions substitution of the pyranose ring appears to be tolerated, as indicated in Fig. 6. The heteroatom in the pyran ring could be replaced by C or S. A preference for α- over β-glycosidic binding of the sugar, polyol, or aromatic residues at the anomeric hydroxyl was observed. The structures of donor substrates used by ATase (Table 4) correspond to this general model. Incubation of component 2 (acarviosyl glucose) and component 4b (acarviosyl maltotriose) with maltose resulted in the formation of acarbose (Table 4). Therefore, we generalize that the 4-hydroxyl group at the nonreducing end of the maltosaccharides can accept the acarviosyl moiety by the action of ATase.
Possible physiological role for ATase and acarbose in the ecology of Actinoplanes sp. strain SE50.
The ATase is probably responsible for the formation of the various pseudooligosaccharides observed in cultures of Actinoplanes sp., containing oligosaccharide chains of different lengths and compositions at the “reducing end” of the acarviosyl moiety, with the exception of components A and C. The dominant acceptor substrates in the extracellular matrix are the malto-oligosaccharides, originating from starch utilization by Actinoplanes. Taking into consideration the inhibitory action of acarbose and related pseudo-oligosaccharides toward α-glucosidases, one could envisage a central role of ATase in sugar utilization by the acarbose producer. The extracellular starch degradation by amylases from Actinoplanes sp. (or competing organisms in more natural environments) leads to the production of dextrins and malto-oligosaccharides. ATase transfers acarviosyl moieties to these saccharides, generating effective inhibitors for starch-degrading “foreign” enzymes, such as the acarbose-sensitive bacterial and fungal α-amylases of microbial competitors (20, 38). This system would be even more efficient for the producer if the acarbose family of inhibitors would also prevent the uptake of malto-oligodextrins by other microbes. In this context, the efficient inhibition of the maltose-binding protein MalE from E. coli by acarbose is interesting to note (6). Since the acarbose producer still grows on starch in the presence of even high acarbose concentrations, it must contain an acarbose-insensitive starch-degrading enzyme system. Together, this could result in a successful competition for a needed carbon source. This seems to be the case, since at least one extracellular α-amylase, AcbE, of Actinoplanes sp. is acarbose resistant (A. Stratmann and W. Piepersberg, unpublished results).
ACKNOWLEDGMENT
We thank Udo F. Wehmeier for comments on the manuscript.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipmann D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Babcock M J, Kendrick K E. Cloning of DNA involved in sporulation of Streptomyces griseus. J Bacteriol. 1988;170:2802–2808. doi: 10.1128/jb.170.6.2802-2808.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bender H. Studies of the mechanism of the cyclisation reaction catalysed by the wildtype and a truncated α-cyclodextrin glycosyltransferase from Klebsiella pneumoniae strain M5, and the β-cyclodextrin glycosyltransferase from Bacillus circulans strain 8. Carbohydr Res. 1990;206:257–267. doi: 10.1016/0008-6215(90)80065-b. [DOI] [PubMed] [Google Scholar]
- 4.Bibb M J, Findlay P R, Johnson M W. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene. 1984;30:157–166. doi: 10.1016/0378-1119(84)90116-1. [DOI] [PubMed] [Google Scholar]
- 5.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 6.Brunkhorst C, Andersen C, Schneider E. Acarbose, a pseudooligosaccharide, is transported but not metabolized by the maltose/maltodextrin system of Escherichia coli. J Bacteriol. 1999;181:2612–2619. doi: 10.1128/jb.181.8.2612-2619.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Distler J, Mansouri K, Piepersberg W. Streptomycin biosynthesis in Streptomyces griseus. II. adjacent genomic location of biosynthetic genes and one of two streptomycin resistance genes. FEMS Microbiol Lett. 1985;30:151–154. [Google Scholar]
- 8.Doumith M, Weingarten P, Wehmeier U F, Salah-Bey K, Benhamou B, Capdevilla C, Michel J-M, Piepersberg W, Raynal M-C. Analysis of genes involved in 6-deoxyhexose biosynthesis and transfer in Saccharopolyspora erythraea. Mol Gen Genet. 2000;264:477–485. doi: 10.1007/s004380000329. [DOI] [PubMed] [Google Scholar]
- 9.Fukuhara K, Murai H, Murao S. Isolation and structure-activity relationship of some amylostatins (F-1b fraction) produced by Streptomyces diastaticus subsp. amylostaticus. Agric Biol Chem. 1982;46:1941–1945. [Google Scholar]
- 10.Gilles C, Astier J P, Marchis-Mouren G, Cambillau C, Payan F. Crystal structure of pig pancreatic alpha-amylase isoenzyme II, in complex with the carbohydrate inhibitor acarbose. Eur J Biochem. 1996;238:561–569. doi: 10.1111/j.1432-1033.1996.0561z.x. [DOI] [PubMed] [Google Scholar]
- 11.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 12.Hopwood D A, Bibb M J, Chater K F, Kieser T, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Ward J M, Schrempf H. Genetic manipulation of Streptomyces: a laboratory manual. Norwich, United Kingdom: The John Innes Foundation; 1985. [Google Scholar]
- 13.Horii S, Kameda Y. Structure of the antibiotic validamycin A. J Chem Commun. 1972;1972:747–748. [Google Scholar]
- 14.Jespersen H M, MacGregor E A, Sierks M R, Svensson B. Comparison of the domain-level organization of starch hydrolases and related enzymes. Biochem J. 1991;280:51–55. doi: 10.1042/bj2800051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knegtel R M A, Strokopytov B, Penninga D, Faber O G, Rozoboom H J, Kalk K H, Dijkhuizen L, Dijkstra B W. Crystallographic studies on the interaction of cyclodextrin glycosyltransferase from Bacillus circulans strain 251 with natural substrates and products. J Biol Chem. 1995;270:29256–29264. doi: 10.1074/jbc.270.49.29256. [DOI] [PubMed] [Google Scholar]
- 16.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 17.Lawson C L, Van Montfort R, Strokopytov B, Rozeboom H J, Kalk K H, De Vries G E, Penninga D, Dijkhuizen L, Dijkstra B W. Nucleotide sequence and X-ray structure of cyclodextrin glycosyltransferase from Bacillus circulans strain 251 in a maltose-dependent crystal form. J Mol Biol. 1994;236:590–600. doi: 10.1006/jmbi.1994.1168. [DOI] [PubMed] [Google Scholar]
- 18.Machius M, Vertesy L, Huber R, Wiegand G. Carbohydrate and protein-based inhibitors of porcine pancreatic alpha-amylase: structure analysis and comparison of their binding characteristics. J Mol Biol. 1996;260:409–421. doi: 10.1006/jmbi.1996.0410. [DOI] [PubMed] [Google Scholar]
- 19.Mark C. “DNA-Strider”: a C program for the fast analysis of DNA and protein sequences on the Macintosh family of computers. Nucleic Acids Res. 1988;16:1829–1836. doi: 10.1093/nar/16.5.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Müller L. Chemistry, biochemistry and therapeutic potential of microbial α-glucosidase inhibitors. In: Demain A L, Somkuti G A, Hunter-Creva J C, Rossmoore H W, editors. Novel microbial products for medicine and agriculture. Amsterdam, The Netherlands: Elsevier Science Publishers; 1989. pp. 109–116. [Google Scholar]
- 21.Murao S, Ohyama K. New amylase inhibitor (S-I) from Streptomyces diastaticus var. amylostaticus no. 2476. Agr Biol Chem. 1975;39:2271–2273. [Google Scholar]
- 22.Nakamura A, Haga K, Yamane K. Three histidine residues in the active centre of cyclodextrin glucanotransferase from alkalophilic Bacillus sp. 1011: effects of the replacement on pH dependence and transition-state stabilization. Biochemistry. 1993;32:6624–6631. doi: 10.1021/bi00077a015. [DOI] [PubMed] [Google Scholar]
- 23.Nakamura A, Haga K, Yamane K. Four aromatic residues in the active center of cyclodextrin glucanotransferase from alkalophilic Bacillus sp. 1011: effects of replacements on substrate binding and cyclization characteristics. Biochemistry. 1994;33:9929–9936. doi: 10.1021/bi00199a015. [DOI] [PubMed] [Google Scholar]
- 24.Namiki S, Kangouri K, Nagate T, Hara H, Sugita K, Omura S. Studies on the alpha-glucoside hydrolase inhibitor, adiposin. II. Taxonomic studies on the producing micoorganism. J Antibiot. 1982;35:1156–1159. doi: 10.7164/antibiotics.35.1156. [DOI] [PubMed] [Google Scholar]
- 25.Namiki S, Kangouri K, Nagate T, Hara H, Sugita K, Omura S. Studies on the alpha-glucoside hydrolase inhibitor, adiposin. I. Isolation and physicochemical properties. J Antibiot. 1982;35:1234–1236. doi: 10.7164/antibiotics.35.1234. [DOI] [PubMed] [Google Scholar]
- 26.Omoto S, Itoh J, Ogino H, Iwamatsu K. Oligostatins, new antibiotics with amylase inhibitory activity. II. Structures of oligostatins C, D and E. J Antibiot. 1981;34:1429–1433. doi: 10.7164/antibiotics.34.1429. [DOI] [PubMed] [Google Scholar]
- 27.Pearson W R, Lipman D J. Improved tools for biological sequence analysis. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Penninga D, Strokopytov B, Rozeboom H J, Lawson L, Dijkstra B W, Bergsma J, Dijkhuizen L. Site-directed mutations in tyrosine 195 of cyclodextrin glycosyltransferase from Bacillus circulans strain 251 affect activity and product specificity. Biochemistry. 1995;34:3368–3376. doi: 10.1021/bi00010a028. [DOI] [PubMed] [Google Scholar]
- 29.Penninga D, van der Veen B A, Knegtel R M, van Hijum S A, Rozoboom H J, Kalk K H, Dijkstra B W, Dijkhuizen L. The raw starch binding domain of cyclodextrin glycosyltransferase from Bacillus circulans strain 251. J Biol Chem. 1995;271:32777–32784. doi: 10.1074/jbc.271.51.32777. [DOI] [PubMed] [Google Scholar]
- 30.Piepersberg W, Distler J. Aminoglycosides and sugar components in other secondary metabolites. In: Rehm H-J, Reed G, editors. Biotechnology. 2nd ed. 7. Products of secondary metabolism. WeinheimWeinheim, Germany: VCH; 1997. pp. 397–488. [Google Scholar]
- 31.Qian M, Haser R, Buisson G, Duée E, Payan F. The active center of a mammalian α-amylase. Structure of the complex of a pancreatic α-amylase with a carbohydrate inhibitor refined to 2.2-Å resolution. Biochemistry. 1994;33:6284–6294. doi: 10.1021/bi00186a031. [DOI] [PubMed] [Google Scholar]
- 32.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 34.Schmidt D D, Frommer W, Junge B, Müller L, Wingender W, Truscheit E. α-Glucosidase inhibitors: new complex oligosaccharides of microbial origin. Naturwissenschaften. 1977;64:535–536. doi: 10.1007/BF00483561. [DOI] [PubMed] [Google Scholar]
- 35.Stratmann A, Mahmud T, Lee S, Distler J, Floss H G, Piepersberg W. The AcbC protein from Actinoplanes species is a C7-cyclitol synthase related to 3-dehydroquinate synthases and is involved in the biosynthesis of the α-glucosidase inhibitor acarbose. J Biol Chem. 1999;274:10889–10896. doi: 10.1074/jbc.274.16.10889. [DOI] [PubMed] [Google Scholar]
- 36.Strokopytov B, Penninga D, Rozoboom H J, Kalk K H, Dijkhuizen L, Dijkstra B W. Structure of cyclodextrin glycosyltransferase complexed with acarbose. Implications for the catalytic mechanism of glycosidases. Biochemistry. 1995;34:2234–2240. doi: 10.1021/bi00007a018. [DOI] [PubMed] [Google Scholar]
- 37.Strokopytov B, Knegtel R M, Penninga D, Rozoboom H J, Kalk K H, Dijkhuizen L, Dijkstra B W. X-ray structure of cyclodextrin glycosyltransferase complexed with a maltononaose inhibitor at 2.6 angstrom resolution. Biochemistry. 1996;35:4241–4249. doi: 10.1021/bi952339h. [DOI] [PubMed] [Google Scholar]
- 38.Truscheit E, Frommer W, Junge B, Müller L, Schmidt D D, Wingender W. Chemistry and biochemistry of α-glucosidase inhibitors. Angew Chem Int Ed. 1981;20:744–761. [Google Scholar]
- 39.van der Veen B A, van Alebeek G J, Uitdehaag J C, Dijkstra B W, Dijkhuizen L. The three transglycosylation reactions catalyzed by cyclodextrin glycosyltransferase from Bacillus circulans (strain 251) proceed via different kinetic mechanisms. Eur J Biochem. 2000;267:658–665. doi: 10.1046/j.1432-1327.2000.01031.x. [DOI] [PubMed] [Google Scholar]
- 40.Wind R D, Buitelaar R M, Dijkhuizen L. Engineering of factors determining alpha-amylase and cyclodextrin glycosyltransferase specificity in the cyclodextrin glycosyltransferase from Thermoanaerobacterium thermosulfurigenes EM1. Eur J Biochem. 1998;253:598–605. doi: 10.1046/j.1432-1327.1998.2530598.x. [DOI] [PubMed] [Google Scholar]
- 41.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequence of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 42.Yokose K, Ogawa K, Sano T, Watanabe K, Maruyama H B, Suhara Y. New α-amylase inhibitor, trestatins. II. Structure determination of trestatins A, B and C. J Antibiot. 1983;36:1166–1175. doi: 10.7164/antibiotics.36.1166. [DOI] [PubMed] [Google Scholar]