Abstract
Missense mutations at the three hotspots in the guanosine triphosphatase (GTPase) RAS—Gly12, Gly13, and Gln61 (commonly known as G12, G13 and Q61, respectively)—occur differentially among the three RAS isoforms. Q61 mutations in KRAS are infrequent and differ markedly in occurrence. Q61H is the predominant mutant (at 57%), followed by Q61R/L/K (collectively 40%), and Q61P and Q61E are the rarest (2% and 1%, respectively). Probability analysis suggested that mutational susceptibility to different DNA base changes cannot account for this distribution. Therefore, we investigated whether these frequencies might be explained by differences in the biochemical, structural, and biological properties of KRASQ61 mutants. Expression of KRASQ61 mutants in NIH/3T3 fibroblasts and RIE-1 epithelial cells caused various alterations in morphology, growth transformation, effector signaling, and metabolism. The relatively rare KRASQ61E mutant stimulated actin stress fiber formation, a phenotype distinct from that of KRASQ61H/R/L/P, which disrupted actin cytoskeletal organization. The crystal structure of KRASQ61E was unexpectedly similar to that of wild-type KRAS, a potential basis for its weak oncogenicity. KRASQ61H/L/R-mutant pancreatic ductal adenocarcinoma (PDAC) cell lines exhibited KRAS-dependent growth and, as observed with KRASG12-mutant PDAC, were susceptible to concurrent inhibition of ERK–MAPK signaling and of autophagy. Our results uncover phenotypic heterogeneity among KRASQ61 mutants and support the potential utility of therapeutic strategies that target KRASQ61 mutant-specific signaling and cellular output.
INTRODUCTION
The three RAS family genes (HRAS, KRAS and NRAS) comprise the most frequently mutated oncogene family in human cancer (~19%) and have been a major focus of cancer research for more than four decades (1). The encoded proteins—guanosine triphosphatases (GTPases) HRAS, KRAS4A/4B, and NRAS—function as molecular switches that cycle between active guanosine triphosphate (GTP)-bound and inactive guanosine diphosphate (GDP)-bound states (2). In the activated state, RAS proteins interact with a host of downstream effectors and act as key regulators of signal transduction pathways mediating cellular growth and proliferation. As GTPases, RAS proteins bind guanine nucleotides with high affinity and are capable of hydrolyzing GTP to GDP. Activation of RAS by cycling of bound GDP for GTP is mediated by guanine nucleotide exchange factors (RASGEFs). GTP hydrolysis results in RAS inactivation and is aided by GTPase-activating proteins (RASGAPs) that engage with RAS and greatly stimulate the slow intrinsic rate of nucleotide hydrolysis. Within the protein, RAS contains two regions called switch I (residues 30-40) and switch II (residues 60-76) that are conformationally dynamic, taking on different conformations depending on whether RAS is GDP- or GTP-bound. Dysregulation of RAS by oncogenic mutations results in aberrant activation of the protein and its downstream signaling pathways, leading to unchecked cellular growth and tumorigenesis (3).
Cancer-associated missense mutations in RAS genes are clustered predominantly at codons Gly12 (commonly referred to as G12), Gly13 (G13), and Gln61 (Q61), collectively at 99% (4). Based largely on the study of HRASG12V, the widely held perception is that these mutant RAS proteins are impaired in GTPase-activating protein (GAP)-mediated GTP hydrolysis, thereby disrupting the GDP-GTP cycle and populating RAS in the active GTP-bound state (5). However, studies have demonstrated that different point mutations, even at the same hotspot residue, do not have the same structural and biochemical consequences (6). Understanding how these mutant-specific biochemical consequences translate to differences in RAS signaling and oncogenic potential may yield new therapeutic opportunities.
There are striking differences in the frequencies of occurrence of specific mutations across the RAS isoforms, both with respect to mutated residue and to amino acid substitution (7). Across all cancer types, the predominant mutation hotspot in KRAS is at codon G12, which accounts for 83% of all KRAS mutations, but only 23% of all NRAS mutations (4). Further, though mutations at Q61 are extremely rare in KRAS (2%), Q61 is the predominant hotspot for cancer-associated mutations in NRAS (62%). In KRAS, the predominant substitution at Q61 is Q61H (57%), which appears rarely in both HRAS and NRAS (5% and 6% respectively). Instead, the dominant Q61 substitution in both NRAS and HRAS is Q61R (47% and 43% respectively).
Unlike for KRASG12 mutations, studies evaluating the role of KRASQ61 mutations in oncogenesis have been far more limited (8). Analyses in NIH/3T3 mouse fibroblasts found that KRASQ61H was reduced in transforming activity compared with KRASG12D/V/C and caused a substantially different gene transcription profile (9). Ectopic expression of different KRAS mutants in the zebrafish pancreas showed that KRASQ61L/R potently caused tumorigenesis, but also determined that KRAS mutants that are not found in pancreatic cancer (for example, A146T) were equally potent (10). An evaluation of KRAS-mutant PDAC patients demonstrated that KRASQ61H/R/K mutants correlated strikingly with more favorable survival and lower levels of activated ERK, yet paradoxically showed histologic features of PDAC with poor survival (11). A mouse model of carcinogen-induced lung cancer initiation, normally associated with Kras Q61R/K mutations, shifted to G12D/V mutations when Kras was overexpressed (12). It was concluded that Q61 mutants more potently activated ERK and ineffectively initiated lung cancer formation when overexpressed due to induction of cellular senescence.
Given the limited assessment and inconsistent observations regarding KRASQ61 mutations to date, in this study we evaluated the role of KRASQ61 mutants in supporting cancer growth. We compared the effects of frequent (Q61H, Q61L, and Q61R) and rare (Q61E and Q61P) mutations on KRAS-dependent growth and signaling. Our results suggest that the cancer-associated frequencies of specific Q61 mutations are driven more by their functional consequences for KRAS biological activities than by the probability of specific DNA mutations.
RESULTS
The observed incidence of KRAS Q61 mutations in cancer differs from the expected incidence
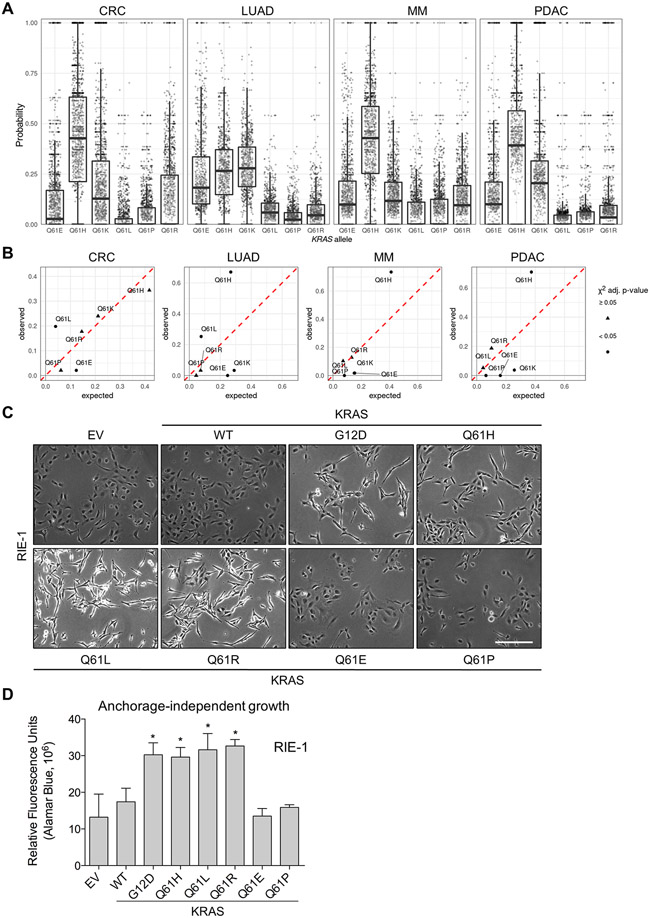
The six possible single base missense substitutions at KRAS codon 61 in all cancers occur at highly variable frequencies (COSMIC v92). Q61H mutations alone comprise 57% of all Q61 mutations, followed by Q61R, Q61L and Q61K; Q61E and Q61P are rare in comparison (fig. S1, A and B).
One possible explanation for these differences is that they represent the probabilistic results of the activity of latent mutational processes in the affected tissues. To address this, we used mutational signatures to estimate the activity of various mutagenic processes in human tumor samples of the four cancer types with the highest frequency of KRAS mutations: colon adenocarcinoma (COAD), lung adenocarcinoma (LUAD), multiple myeloma (MM) and pancreatic ductal adenocarcinoma (PDAC) (13). The compositions of the mutational signatures identified in each tumor sample were used to calculate a probability for each possible Q61 mutation in the tumor (Fig. 1A). In COAD, MM and PDAC, the most probable mutation on average was Q61H, which can occur via two different single-nucleotide base substitutions. In LUAD, Q61E, H, and K all had, on average, similar probabilities and were more probable than Q61L, P, and R in most tumor samples.
Fig. 1. The probability of observing each possible KRASQ61 mutation in tumor samples.
(A) The probability of observing each possible mutation from a single-nucleotide base substitution (SBS) at KRAS Q61 in individual tumor samples. Each point represents a tumor sample and each tumor sample of a given cancer type appears in the box-plot of each possible KRAS mutation (meaning a single tumor sample is represented by a point in each possible KRAS mutation). (B) The expected vs. observed frequencies of KRASQ61 mutations. The frequencies of all possible mutations to codon 61 of KRAS by SBS as predicted by the mutational signatures against the observed frequencies. A χ-squared test was used to detect if there was a difference between the predicted and observed frequency for each allele; triangles indicate where adjusted P ≥ 0.05; circles, where P < 0.05. (C) Representative brightfield images of RIE-1 cells ectopically expressing different KRAS mutations. Images were collected at 10x magnification 3 days after antibiotic selection. χ-squared test and P-values were adjusted for multiple hypothesis testing using the Benjamini–Hochberg method (14)). Scale bar, 500 μm. (D) Anchorage-independent colony formation of RIE-1 cells expressing KRASQ61 mutants. Cells were cultured for seven days in soft agar and developed using AlamarBlue reagent. Representative data is shown and quantified as mean ± S.E.M. from three independent experiments. * P ≤ 0.05 by one-way ANOVA.
From these probabilities, we calculated the expected frequency of each KRAS Q61 mutation relative to all Q61 mutations in a given cancer type by averaging over the population and then determined whether these matched the frequencies observed in that cancer (Fig. 1B and fig. S1C). This analysis demonstrated that some mutations occurred at the same frequency as expected from the mutational signatures, whereas others deviated substantially. For example, the expected frequencies of Q61P were at only single-digit levels, and this mutation was rarely (COAD) or never observed (LUAD, MM and PDAC). In contrast, the observed frequencies for Q61L were significantly higher than expected in COAD and LUAD (20 vs. 4%, 25 vs. 8%, respectively). Likewise, the observed frequencies for Q61H were twice the expected rates in LUAD, MM and PDAC (67 vs. 27%, 74 vs. 41% and 71 vs. 37%, respectively). Moreover, whereas Q61E was predicted to occur at rates between 12% (COAD) and 25% (LUAD), it was observed only rarely (2%) in COAD and MM and was not observed in LUAD or PDAC. The frequency of Q61K was well estimated in COAD and MM (24 vs. 21% and 17 vs. 15%, respectively), but poorly in LUAD and PDAC (3 vs. 30% and 4 vs. 26%, respectively). We conclude that the observed frequencies likely result from a combination of the probability of occurrence and oncogenic fitness.
KRASQ61E and KRASQ61P exhibit impaired morphologic and growth transforming activities
We then evaluated the possibility that the frequencies of different Q61 mutations are driven strongly by distinct biological consequences of each amino acid substitution. To address this hypothesis, we evaluated the biological properties of the three most prevalent (Q61H, Q61R and Q61L) and the two most infrequent (Q61P and Q61E) KRASQ61 mutants. We previously reported that the activities of the atypical mutant KRASG12R were masked by co-occurring events when evaluated in PDAC cell lines and revealed only when evaluated in a clean genetic background (6). Furthermore, not all Q61 mutants are found in established cancer cell lines. Therefore, we chose to evaluate the biological properties of KRASQ61 mutants in two well-characterized cell models that are highly sensitive to mutant RAS-driven growth and morphologic transformation (15, 16).
We stably expressed each KRASQ61 mutant in spontaneously immortalized NIH/3T3 mouse fibroblasts, a model that enabled the first identification of activated KRAS oncogenes in human cancer cells (15). For comparison, we also established NIH/3T3 cells stably expressing wild-type KRAS (KRASWT) and the most frequent KRAS mutation, G12D. After selecting for puromycin-resistant cells, expression of each KRAS protein was confirmed by immunoblot analyses (fig. S1D). The steady-state protein level of KRASWT was substantially lower than the Q61 mutant proteins, and KRASQ61E was expressed lower than the other mutants. All the resulting NIH/3T3 cell lines exhibited similar rates of anchorage-dependent proliferation (fig. S1E). As expected, expression of activated KRASG12D resulted in robust morphological transformation to an elongated and highly refractile phenotype (fig. S1F). Ectopic expression of the more prevalent KRASQ61H, KRASQ61L, and KRASQ61R mutants induced morphological transformation similar to KRASG12D. In contrast, cells expressing the infrequent KRASQ61E and KRASQ61P proteins resembled empty vector control or cells expressing exogeneous KRASWT. The relative potencies of the KRASQ61 mutants to cause morphological transformation are similar to those we reported previously in our focus formation analyses of 17 different Q61 mutants of HRAS when stably expressed in NIH/3T3 cells (17). In that study, of the six possible single base substitutions at Q61, only HRASQ61E and HRASQ61P were similar to HRASWT.
We next determined if the ability of each KRAS mutant to cause morphologic transformation correlated with activation of the key KRAS effector pathway, the RAF-MEK-ERK MAPK cascade (fig. S1D). We observed no direct relationship between morphological transformation and steady-state levels of phosphorylated and activated ERK (pERK). Although the common KRASG12D mutant, which is highly transforming, did not cause substantial ERK activation, cells expressing the KRASQ61 mutants exhibited increased pERK levels regardless of morphological transforming ability. That Q61 mutants more potently activate ERK is consistent with analyses of carcinogen-induced lung tumor formation in mouse models, where KrasQ61L/R mutants exhibited stronger induction of ERK activation compared with KrasG12D/V (12).
We showed previously that mutant RAS can potently cause transformed and tumorigenic growth transformation of RIE-1 rat intestinal epithelial cells (16). To determine if the variable potencies of the different KRASQ61 mutants to cause morphologic transformation in NIH/3T3 cells were seen in another cell type, we ectopically expressed the panel of KRAS mutants in RIE-1 cells and confirmed expression by immunoblot (fig. S1G). As we observed in NIH/3T3 cells, only RIE-1 cells expressing KRASQ61H, KRASQ61L, or KRASQ61R exhibited morphological transformation, whereas the morphologies of cells expressing KRASQ61E or KRASQ61P were indistinguishable from KRASWT and empty vector cells (Fig. 1C). However, as we also observed in NIH/3T3 cells, KRASQ61E was expressed at a substantially lower level than the other Q61 mutants. This reduced expression, which may be due to decreased protein stability (18), could contribute to the inability of this mutant to cause morphologic transformation.
RIE-1 cells stably expressing KRASG12D and all five of the KRASQ61 mutants proliferated at similar rates under anchorage-dependent conditions (fig. S1H). However, when suspended in soft agar, KRASQ61H, KRASQ61L, and KRASQ61R, but not KRASQ61E or KRASQ61P, enhanced multicellular colony formation compared with empty vector or ectopic KRASWT (Fig. 1D), correlating with their relative ability to cause morphological transformation.
KRAS Q61 mutants also displayed heterogeneity in effector signaling similar to the growth phenotypes observed, as assessed by immunoblot analyses (fig. S1G). Like NIH/3T3 cells, RIE-1 cells expressing KRASG12D did not exhibit a substantial increase in pERK. In contrast, cells stably expressing KRASQ61L and KRASQ61R displayed increased levels of pERK and of phosphorylated and activated AKT (pAKT) compared with cells expressing KRASQ61E. Notably, as we also found in NIH/3T3 cells, KRASQ61E was expressed at a level more comparable to KRASWT and substantially lower compared to the other Q61 mutants, again supporting that its reduced potency may be attributable in part to reduced steady-state levels.
KRASQ61E and KRASQ61P exhibit reduced formation of GTP-bound protein in live cells
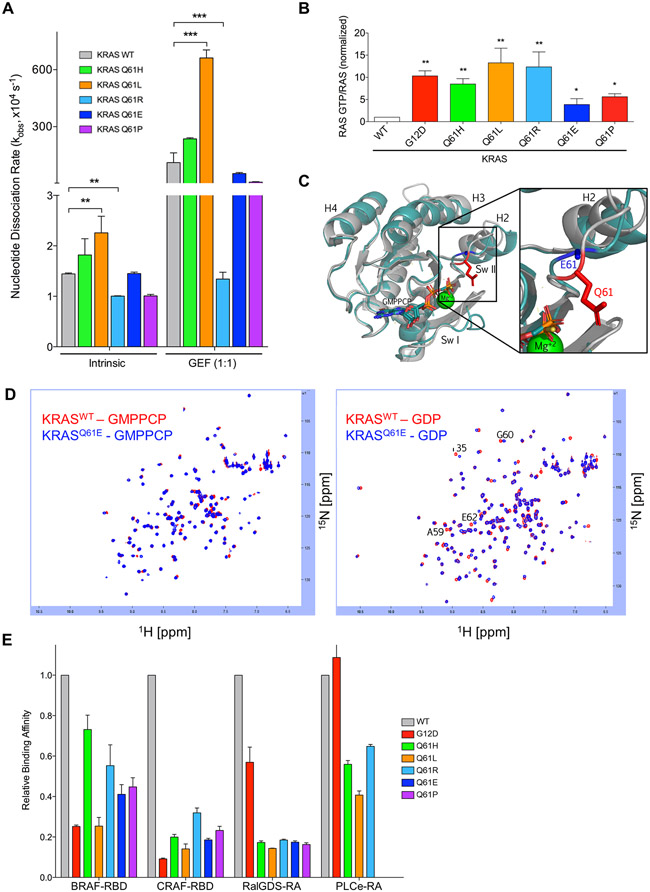
The Q61 residue in KRAS is the catalytic residue essential for proper coordination of GTP hydrolysis (19). Accordingly, it has been reported that RASQ61 mutants demonstrate decreased intrinsic GTPase activity (17, 20), and all substitutions at this position are expected to result in impaired intrinsic and GAP-stimulated GTP hydrolysis (21). Therefore, we addressed the possibility that the distinct functional consequences of KRASQ61E and KRASQ61P may instead reflect differential perturbations in nucleotide exchange. To this end, we measured both the intrinsic and GEF-stimulated rates of nucleotide dissociation in vitro of E. coli-expressed recombinant KRAS4B proteins with or without recombinant protein corresponding to the isolated catalytic domain of the RASGEF RASGRP1. Similar to what has been described previously for Q61L and Q61H mutations in KRAS (22), all Q61 mutant proteins exhibited small changes in their rates of intrinsic nucleotide exchange as compared to KRASWT (Fig. 2A). KRASQ61R nucleotide exchange was not stimulated upon GEF addition. However, the remaining Q61 mutants were responsive to GEF stimulation, comparably to KRASWT. Thus, differences in intrinsic and GEF-stimulated nucleotide exchange do not account for the impaired transforming activities of KRASQ61E and KRASQ61P.
Fig. 2. KRASQ61 proteins show distinct biochemical phenotypes.
(A) Quantification of nucleotide exchange rates of recombinant KRASQ61 mutant proteins (amino acids 2-169) in the absence (left) and in the presence of equimolar concentration of the catalytic domain of RASGRP1. Data are mean ± S.E.M. from three or more independent experiments. ** P ≤ 0.01 and *** P ≤ 0.001 by one-way ANOVA. (B) Quantification of pulldown assay for KRAS-GTP levels in RIE-1 cells using CRAF-RBD shown as mean ± S.E.M. of three independent experiments. * P ≤ 0.05 **, P ≤ 0.01, and *** P ≤ 0.001 by one-way ANOVA. Error bars, (C) Ribbon diagram showing X-ray structural overlays of KRASQ61E (teal, 7LZ5) with KRASWT (silver, 4DSO). Proteins were crystallized bound to non-hydrolyzable GMPPCP. The Q61 sidechain is indicated in red and the E61 sidechain is indicated in blue. (D) KRASQ61E NMR chemical shifts resemble KRASWT. 1H-15N HSQC NMR overlay of KRASWT (red) and KRASQ61E (blue) in the GMPPCP-bound (left) and GDP-bound (right) states. Data are representative of two biological replicates. (E) Relative binding affinities of KRASQ61 proteins to select RAS binding (RBD) and association (RA) domains. Values were normalized to KRASWT for each indicated effector. Data are averages from three or more independent experiments.
Given that KRAS GDP-GTP regulation in vivo is dictated by the activities of multiple GEFs and GAPs, the consequences of Q61 mutations on the activity of any one GEF or GAP in vitro will not reliably predict steady-state levels of KRAS-GTP in the cellular environment. Therefore, to determine the GTP-bound state of KRASQ61 mutants in cells, we performed RAS-GTP pulldown analyses in RIE-1 cells ectopically expressing each KRASQ61 mutant (Fig. 2B). We found increased levels of GTP-bound protein in all KRASQ61 mutants compared with KRAS WT. However, those levels were increased ~10-fold relative to WT in the more potent Q61 mutants, comparable to G12D, whereas they were increased ~5-fold in both Q61E and Q61P. Thus, the ineffective transforming potencies of these two mutants may be due, in part, to reduced formation and/or maintenance of the active GTP-bound state.
The reduced level of steady state GTP-bound KRASQ61E in cells compared to the more potent Q61 mutants was not expected, given that any substitution at Q61 will disrupt both intrinsic and GAP-stimulated GTP hydrolysis activities and favor formation of GTP-bound protein. To elucidate a molecular basis for this difference, we determined the crystal structure of KRASQ61E (PDB accession code 7LZ5) bound to the nonhydrolyzable GTP analog guanosine-5'-[(β,γ)-methyleno]triphosphate (GMPPCP) (Fig. 2C and table S1). In agreement with their similar biochemical characteristics, we found that the overall structure of KRASQ61E overlaid well on that of KRASWT (PDB accession code 4DSO). Despite the location of the Q61E mutation in the flexible Switch II region of RAS, the structures of GTP-bound Q61E and WT display similar conformations at these switch regions. The full Glu61 (E61) sidechain could not be resolved in the crystal structure, with electron density only supporting the coordinates for the Cβ atom, suggesting that the E61 sidechain is conformationally dynamic. In agreement with these data, 15N-1H HSQC nuclear magnetic resonance (NMR) analysis also revealed a similar overall signature of cross-peaks between KRASWT and KRASQ61E in both active GMPPCP-bound and inactive GDP-bound states (Fig. 2D). In contrast, we found that there were large spectral perturbations between KRASWT and KRASQ61H 15N-1H HSQC analysis in both the GMPPCP- and GDP-bound states (fig. S2A). Though this crystal structure captures only one of the conformations that the KRASQ61E protein samples, similarities to the KRASWT structure and biochemical characteristics suggest that the limited structural perturbations caused by the Q61E substitution may contribute to the weaker biological potency of KRASQ61E.
KRASQ61 mutations cause heterogeneous signaling activities
Another potential basis for the differential consequences of Q61 mutations to signaling and transforming activities may be distinct consequences for interactions with and activation of downstream effectors. To address this possibility, we first measured the binding affinities of KRAS mutants to the isolated RAS binding (RBD) and RAS-associating (RA) domains of KRAS effectors RAF, RALGDS and PLCε. Although the different G12/Q61 mutants displayed variable degrees of binding affinities compared to KRASWT, no substantial order-of-magnitude alterations (>10-fold decrease) were observed among the different KRASQ61 mutants (Fig. 2E and fig. S2B). We conclude that the reduced transforming potencies of KRASQ61E and KRASQ61P are not associated with impaired interactions with the RBD or RA domains of these RAS effectors. Furthermore, the variable affinities for RAF RBDs do not correlate with the differential activation of ERK1/2 by the mutants (figs. S1, D and G).
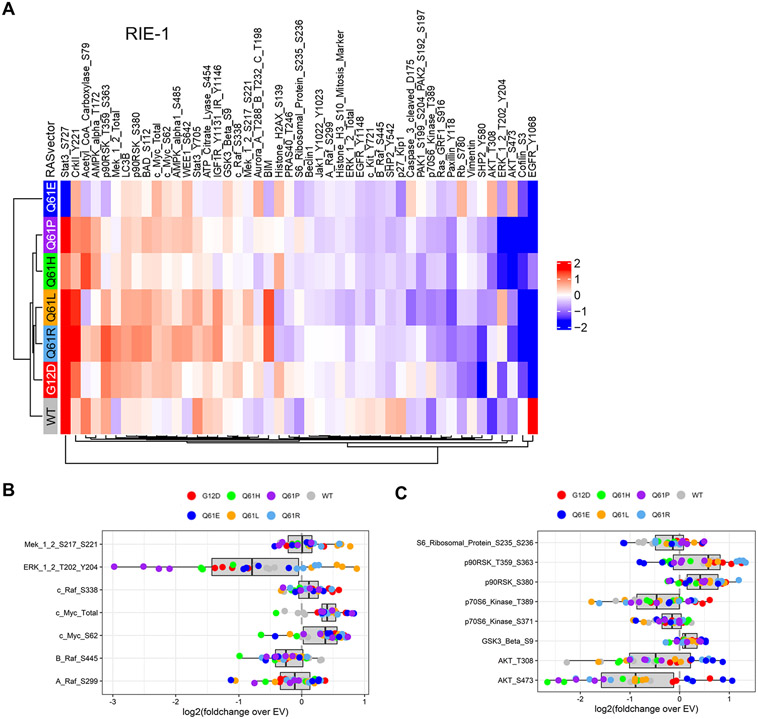
Next, we utilized reverse phase protein array (RPPA) analyses to profile signaling activities in RIE-1 cells expressing the different KRASQ61 mutants (Fig. 3A and fig. S3A). Unexpectedly, we found substantial heterogeneity in signaling activities among the KRASQ61 mutants tested despite the similar in vitro binding affinities to known RAS effectors (Fig. 2E). Generally, clustering analyses associated with our analyses of morphologic and growth transformation, but with significant divergence as well. The strongly transforming mutants, G12D, Q61R, and Q61L, clustered together. In contrast, despite their shared nontransforming phenotypes, Q61P and Q61E showed distinct signaling profiles, with Q61P more similar with the strongly transforming mutant Q61H. Thus, though signaling differences likely contribute to the differential transforming activities of Q61 mutants, additional functional differences likely contribute to the overall biological potency.
Fig. 3. RIE-1 cells expressing KRASQ61 mutants demonstrate heterogeneous signaling patterns.
(A) Reverse phase protein array (RPPA) pathway activation analysis of RIE-1 cells. Cell lysates were stained with either phospho-specific (site indicated) or total protein antibodies. Heat map represents four biological replicates for each mutant. RPPA data were log2 transformed and medians are presented normalized to EV RIE-1 cells. Phospho- or total protein levels were arranged by hierarchical clustering. Red, increased signal; blue, decreased signal. (B and C) Box plots of ERK MAPK and PI3K-associated signaling changes from RPPA analysis. Shown are individual replicates for indicated KRAS samples normalized to the EV condition.
Consistent with the immunoblot analyses, RPPA data showed greater levels of phosphorylation of MEK1/2 and ERK1/2 in cells expressing KRASQ61L/R than in those expressing KRASG12D and KRASQ61E/P (Fig. 3B and fig. S3B). However, because we have previously found that pERK levels may not accurately reflect ERK-dependent signaling activity (23, 24), we also evaluated ERK substrate phosphorylation. ERK directly phosphorylates the cytoplasmic p90RSK serine/threonine kinases, promoting their nuclear translocation. KRAS promotes increased levels of MYC protein, in part through ERK-stimulated increases in gene transcription and in part through increased protein stability upon ERK phosphorylation of MYC at residue Ser62 (25). Despite variable MEK-ERK activation, we observed increased phosphorylation of p90RSK, increased MYC protein levels, and correspondingly increased phosphorylation of MYC at Ser62 in RIE-1 cells stably expressing all KRASQ61 mutants, including KRASQ61E and KRASQ61P (Fig. 3C). Thus, differences in ERK signaling alone cannot account for the inability of KRASQ61E and KRASQ61P to transform RIE-1 cells.
KRASQ61 mutants cause distinct alterations in metabolic functions
Oncogenic KRAS drives cancer growth, in part, through upregulating metabolic activities that support the enhanced metabolic needs of cancer cells (26). We therefore determined whether the various KRASQ61 mutants drive variable metabolic perturbations. First, we evaluated mitochondrial morphology. Oncogenic RAS causes mitochondrial fragmentation through ERK phosphorylation of DRP1 (27, 28), and loss of DRP1 function causes accumulation of fused mitochondria and impairs cancer cell growth. Despite their different abilities to stimulate ERK activation, cells expressing KRASQ61H, KRASQ61L, or KRASQ61R exhibited fragmented mitochondria, similar to KRASG12D. In contrast, cells expressing KRASQ61E or KRASQ61P exhibited fused mitochondria, comparable to that of control empty vector cells (Fig. 4A). Consistent with the different degrees of mitochondrial fragmentation, we observed increased oxygen consumption as well as ATP production in cells expressing KRASQ61L or KRASQ61R compared to KRASQ61E or KRASQ61P (fig. S4). Thus, reduced upregulation of metabolic activities may also contribute to the impaired oncogenic potential of Q61E and Q61P.
Fig. 4. KRASQ61 mutants drive distinct metabolic phenotypes.
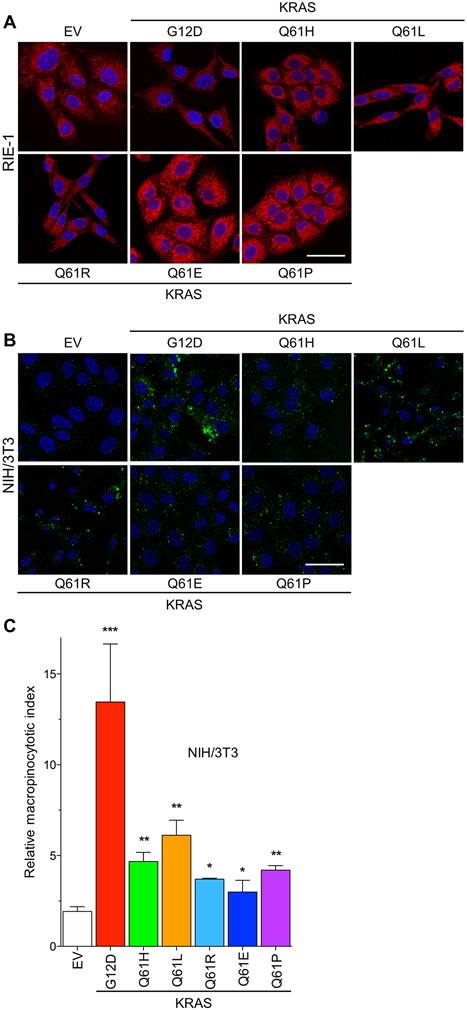
(A) Representative images of mitochondrial staining of RIE-1 cells expressing KRASQ61 mutants. Red, Mitotracker Red; blue, DAPI. Scale bar, 50 μm. (B) Representative images of FITC-dextran-labeled macropinosomes of NIH/3T3 cells expressing KRASQ61 mutants (green, FITC-Dextran; blue, DAPI). (C) Quantification of macropinocytosis in NIH/3T3 cells stably expressing KRASWT and mutants. Data are representative and, where quantified, mean ± S.E.M. from three independent experiments. * P ≤ 0.05, ** P ≤ 0.01, and *** P ≤ 0.001 by one-way ANOVA.
We showed recently that KRASG12D and KRASG12V but not KRASG12R can drive PI3K-dependent macropinocytosis (6). We therefore evaluated the macropinocytotic activity in NIH/3T3 cells expressing different KRAS mutants by monitoring the uptake of fluorescein isothiocyanate (FITC)-tagged dextran (Fig. 4, B and C). As we determined previously, KRASG12D stimulated a robust increase in macropinocytosis compared with control cells (~13-fold). Although all the Q61 mutants also stimulated macropinocytosis, they did so to a significantly lower extent than G12D, particularly Q61E. Because macropinocytosis is a metabolic activity essential for PDAC tumorigenic growth (6, 29), the reduced macropinocytotic capabilities of the Q61 mutants may provide an additional mechanistic basis for their infrequent occurrence in PDAC.
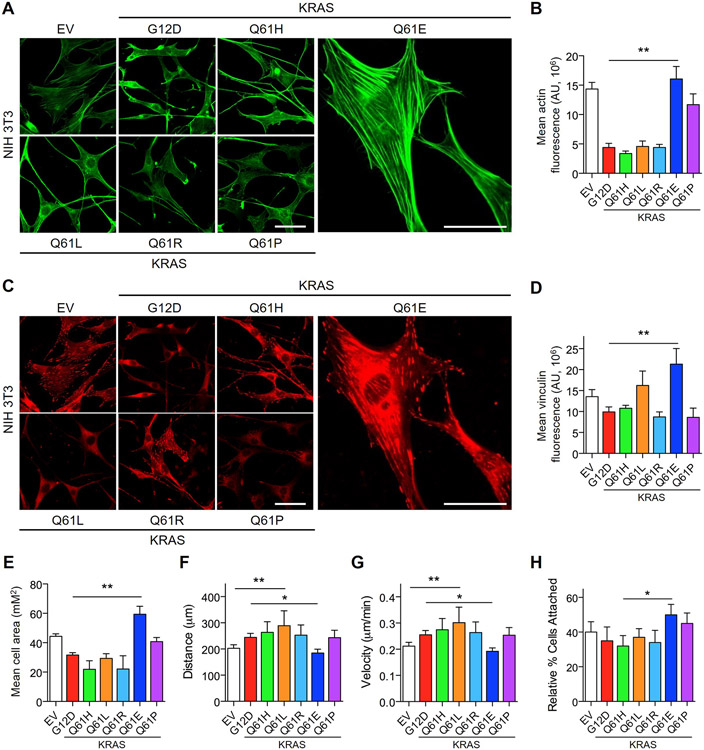
KRASQ61E promotes F-actin formation and cell adhesion and reduces migration
Previously, it has been shown that some of the morphological changes induced by RAS transformation of NIH/3T3 fibroblasts result from suppression of F-actin stress fiber formation (30). This cytoskeletal rearrangement is classically linked to activation of RHOA, and overexpression of activated RHOA mutants results in a potent increase in stress fiber formation (30, 31). Based on the morphological differences observed upon KRASQ61E and KRASQ61P expression in NIH/3T3 cells and the cytoskeletal signaling changes revealed by RPPA, we performed immunofluorescence imaging to interrogate the levels and organization of F-actin. Consistent with their similar transformed phenotype, KRASQ61H, KRASQ61L, and KRASQ61R all caused similar suppression of F-actin stress fibers (Fig. 5, A and B). In contrast, KRASQ61E unexpectedly and uniquely caused an increase in stress fiber formation; despite the morphological similarities between cells expressing KRASQ61E and KRASQ61P, KRASQ61P did not. Cells expressing KRASQ61E also displayed increased levels of vinculin (Fig. 5, C and D), a marker for focal adhesions that connect the actin cytoskeleton with the extracellular matrix (32). Moreover, the elevated vinculin signal was associated with increased actin organization and a corresponding increase in cell area in KRASQ61E-transformed cells (Fig. 5E). This gain-of-function phenotype of KRASQ61E, while opposite of that seen in highly oncogenic KRAS mutants, is similar to the gain-of-function phenotype that we described recently for the gastric cancer-associated oncogenic mutant RHOAY42C (33).
Fig. 5. KRASQ61E induces unique effects on the actin cytoskeleton and cell motility.
(A and B) Representative immunofluorescence images of F-actin stained with phalloidin (A, green) and corresponding quantification of normalized F-actin signal (B) in NIH/3T3 cells ectopically expressing KRASG12D, KRASQ61H or KRASQ61E. Cells were plated on glass coverslips coated with 10 μg/mL fibronectin. Scale bar, 50 μm. Data are representative of >20 cells per condition and quantified as mean ± S.E.M. from three independent experiments. ** P ≤ 0.01 by t-test. (C and D) Representative immunofluorescence images of the focal adhesion marker vinculin (C, in red) and corresponding quantification of normalized vinculin signal (D) of NIH/3T3 cells expressing KRASWT or mutant proteins. Scale bar, 50 μm. Data are representative of > 20 cells per condition and shown as mean ± S.E.M. from three independent experiments. ** P ≤ 0.01 by t-test. (E) Quantification of average cell area of NIH/3T3 cells as in panels (A and C). Data are representative of >20 cells per condition and shown as mean ± S.E.M. from three independent experiments. ** P ≤ 0.01 by t-test. (F and G) Random cell migration patterns showing total distance migrated (F) and average velocity (G) of NIH/3T3 cells expressing KRASWT or mutant proteins. Cells were plated on glass coverslips coated with 10 μg/mL fibronectin and monitored for 16 hours. Data are representative of >10 cells per condition and shown as mean ± S.E.M. from three independent experiments. * P ≤ 0.05 and ** P ≤ 0.01 by t-test. (H) Quantification of adhesion of NIH/3T3 cells to fibronectin-coated dishes. Cells were trypsinized, labeled with CellTracker Green viability dye for 10 min and allowed to rest for 30 min before plating. Percentage attached was normalized to total cells plated. Data shown are mean ± S.E.M. of three independent experiments. * P ≤ 0.05 by t-test.
Stress fiber formation promotes cell adhesion, which in turn can cause reduced cell motility (32). To determine if KRASQ61E-induced actin reorganization impacted these cellular properties, we performed analyses of random cell migration. NIH/3T3 cells expressing KRASQ61E displayed reduced random migration compared with empty vector cells, whereas all other Q61 mutants drove increased cell migration (Fig. 5, F and G). Further, this decreased cell motility was correlated with increased cell adhesion (Fig. 5H), providing additional defective phenotypes for Q61E and providing additional rationale for why this mutant is so rare in cancer.
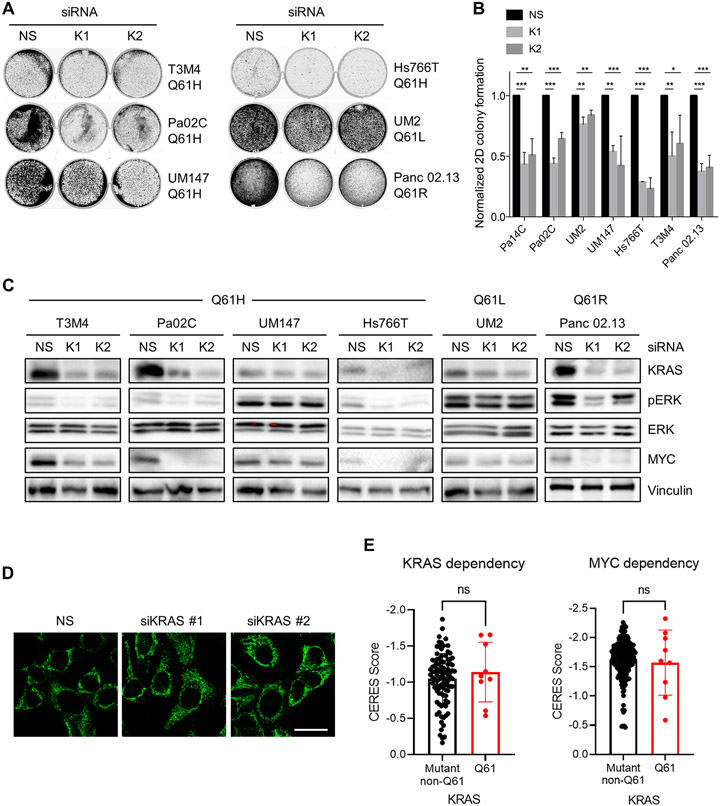
KRASQ61-mutant PDAC cell lines are dependent on KRAS for growth
We next evaluated the activities of endogenous KRASQ61 mutations in supporting the growth of PDAC cell lines. Q61 mutations comprise ~5% of KRAS mutations in PDAC, where Q61H is the predominant mutation (~80%) at this residue (34). Therefore, we obtained four cell lines harboring KRASQ61H, one harboring KRASQ61R and one harboring KRASQ61L for these analyses (fig. S5A). We determined previously that all KRASG12-mutant PDAC cell lines evaluated exhibited KRAS-dependent growth (23, 35). Likewise, siRNA suppression of KRAS also reduced the proliferation and clonogenic growth of KRASQ61-mutant PDAC lines (Fig. 6, A and B). We have previously showed that knockdown of KRAS in KRASG12-mutant PDAC cell lines caused loss of MYC protein through the impaired ERK-dependent suppression of MYC protein degradation (23). In the four cell lines where KRAS siRNA strongly suppressed KRAS expression, we observed substantial reductions in pERK as well as in MYC protein levels (Fig. 6C), indicating a shared role of MYC in the oncogenic function of both G12 and Q61 mutants. Furthermore, consistent with the relative ability of KRASQ61 mutants to drive mitochondrial fragmentation in model cell lines, we observed differential induction of mitochondrial fusion upon silencing of endogenous KRASQ61 mutants in PDAC cell lines (Fig. 6D). Finally, we evaluated data from Project Achilles reported at the Cancer Dependency Map (DepMap), where CRISPR-based KRAS and MYC knockouts showed comparable dependencies in KRAS Q61 and non-Q61 mutant cancers (Fig. 6E and data file S1). Thus, the degree of KRAS dependency cannot account for the low frequency of Q61 mutations in these cancers.
Fig. 6. Dependence of PDAC cell lines on KRASQ61 mutants with respect to growth, signaling and mitochondrial morphology.
(A) Anchorage-dependent colony formation of KRASQ61-mutant PDAC cell lines after silencing using a non-specific (NS) and two KRAS-targeting siRNAs. Cells were cultured for seven days and stained with crystal violet. (B) Quantification of colony formation described in (A). Data are mean ± S.E.M. from four independent experiments. * P ≤ 0.05, ** P ≤ 0.01, and *** P ≤ 0.001 by one-way ANOVA with Dunnett’s post-test. (C) Immunoblot analysis of the knockdown of KRAS protein levels and of effector signaling to ERK and MYC after transfection with KRAS siRNA. Blots are representative of three independent biological replicates. (D) Changes in mitochondrial morphology detected with Mitotracker Green in Pa02C PDAC cells after siRNA-mediated KRAS knockdown compared to controls (scrambled siRNA). Images are representative of three independent experiments. Scale bar, 50 μm. (E) Cellular dependency on KRAS or MYC expression as determined by CRISPR gene knockout (CERES scores, DepMap) in KRAS-mutant pancreas, colon, and lung cancer cell lines. Each dot represents an individual cell line with a KRASQ61 or non-Q61 activating oncogenic mutation (data file S1). The more negative a value, the greater dependency on KRAS or MYC expression. Data are mean ± SD; P values from unpaired t-test.
Combined inhibition of ERK and autophagy potently suppresses proliferation of KRASQ61-mutant PDAC cell lines
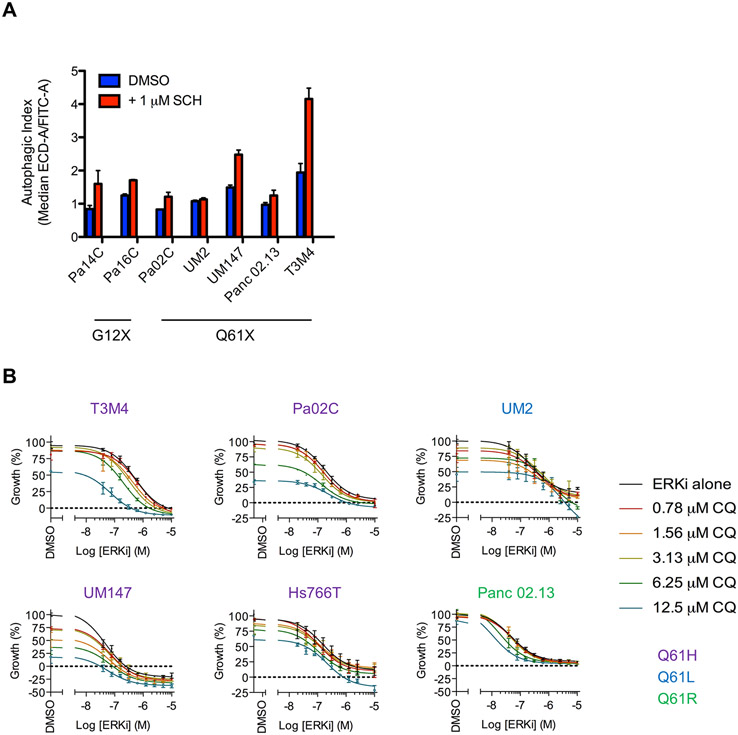
Our previous analyses of KRASG12D/V-mutant PDAC lines showed that pharmacologic inhibition of ERK suppressed glycolysis and mitochondrial function, leading to compensatory upregulation of autophagy (36). Exploiting this greater dependency on autophagy, we then found that concurrent inhibition of ERK1/2 and autophagy significantly suppressed the proliferation of KRAS-mutant PDAC cell lines, leading to our initiation of Phase I/II clinical trial evaluations of this combination in PDAC patients (NCT04132505 and NCT04386057). We therefore extended these analyses to KRASQ61-mutant PDAC lines. First, to determine if ERKi treatment also enhanced autophagy in these lines, we stably expressed the tandem fluorescence reporter mCherry-EGFP-LC3B in PDAC cell lines to assess autophagic flux, as previously described (36). As we observed in KRASG12D/V-mutant PDAC (36), short-term ERKi treatment also increased autophagic flux in KRASQ61-mutant PDAC lines (Fig. 7A).
Fig. 7. Response of KRASQ61-mutant PDAC cell lines to combined inhibition of ERK MAPK cascade and autophagy.
(A) PDAC cell lines were stably infected with a lentiviral construct encoding mCherry-EGFP-LC3B and then treated with SCH772984 (ERKi, 1 μM) or DMSO for 24 hours. Fluorescence intensities of mCherry and EGFP were monitored using FACS analysis, and autophagic index is plotted, indicating the ratio of the median fluorescence of mCherry to EGFP. Data are the average of three independent biological replicates. (B) Cell viability of KRASQ61-mutant PDAC cell lines co-treated with SCH772984 (ERKi) and chloroquine (CQ) as assessed by CellTiter Glo. Data are representative of three independent experiments.
Next, in agreement with the critical role of ERK1/2 in the proliferation of KRASG12-mutant PDAC (23), we observed that treatment with the ERK1/2-selective inhibitor SCH772984 (ERKi) caused potent dose-dependent reduction in proliferation (Fig. 7B). We then determined that the autophagy inhibitor chloroquine (CQ) reduced viability of most of the cell lines tested (Fig. 7B and fig. S5A). Concurrent ERKi and CQ treatment showed enhanced growth suppression in all lines, as indicated by the decreased GI50 for ERKi. Bliss analyses indicated modest synergy between the two agents (fig. S5B), largely due to the efficacy of single agent treatment with CQ alone. In UM2 and UM147 cells this effect was particularly strong, resulting in comparatively poor synergy with concurrent ERKi treatment. In summary, the therapeutic approach of using concurrent ERK MAPK blockade and autophagy inhibition can be effective in both KRASG12- and KRASQ61-mutant PDAC.
DISCUSSION
The three mutational hotspots at codons 12, 13, and 61 are shared among the three RAS genes, but there are strikingly distinct cancer- and RAS isoform-specific patterns in the observed frequencies of the six possible single base missense mutations at each hotspot (4). One simple explanation may be that the distinct occurrences of these mutations simply reflect tissue-specific DNA accessibility and/or carcinogen-specific mutagenicity. However, emerging evidence supports a substantial contribution of the biological potency of specific RAS mutations in driving the initiation, progression, and maintenance of cancer (37-41). In the present study, we focused on mutations at Q61, the least frequent hotspot for KRAS, representing only 2% of KRAS mutations in all cancers (COSMIC v92) and only 5% in PDAC (34). We found that different Q61 mutants exhibit surprisingly divergent properties, and we observed distinct functional differences between G12 and Q61 mutants (summarized in table S2) that may provide a rationale for the lower frequency occurrence of Q61 mutations in cancer. We conclude that oncogenic potency provides a strong basis for the limited occurrence of different KRASQ61 mutations in cancer. Finally, we determined that the rare KRASQ61-mutant PDAC are, like the common KRASG12-mutant PDAC, KRAS-addicted and responsive to ERK and autophagy inhibition.
To address a role for DNA mutational frequencies, we applied a computational approach that we described recently to evaluate the most prevalent cancer-associated mutations at G12, G13, and Q61 (42). We utilized the composition of the mutational signatures identified in the four cancer types with the highest frequency of KRAS mutations (lung, colon, pancreatic adenocarcinoma, and multiple myeloma) to calculate a probability for the six possible single-base missense mutations at codon 61 in each cancer type and compared these predicted frequencies with the observed frequencies. Although some predicted frequencies coincided strongly with the observed frequencies, there were significant outliers. Particularly striking, the KRASQ61E mutation was predicted to occur at rates between 12% to 25%, yet it appears rarely in colon or myeloid cancers and is not seen in lung and pancreatic cancers. Our previous studies comparing mutational profiles suggested that biological selection plays a substantial role in the underlying tissue-specific mutation frequencies observed (42). KRASQ61E represents a mutation that is expected but not observed. In agreement with biology driving mutational selection, ectopic expression of KRASQ61E failed to drive morphological transformation or anchorage independent proliferation, suggesting that the impaired biological function limits the prevalence of this mutation in human cancers. We determined that KRASQ61E exhibits structural, biochemical, and biological properties that are distinct from the highly transforming KRASQ61H/L/R mutants and instead are more similar to those of KRAS WT. Conversely, KRASQ61E displayed an unanticipated gain-of-function phenotype, promotion of actin stress fibers and focal adhesion assembly, an activity that is the polar opposite of that seen in highly oncogenic KRASG12/KRASQ61 mutants. Notably, this phenotype is similar to that we have described for a gain-of-function oncogenic phenotype of the gastric cancer-associated RHOAY42C mutant (33). Together, these findings support the poor oncogenic potency of KRASQ61E as a basis for its rare occurrence in cancer.
The biochemical properties of RASQ61E are unique among RAS mutants. HRASQ61E was originally described to have a slow hydrolysis rate (17). Notably, HRASQ61E has been described as having increased exchange and hydrolysis rates when treated with increasing concentrations of free nucleotide, making these observations difficult to compare to the single-turnover experiments that are typically performed. To our knowledge, no reported study has evaluated KRASQ61E hydrolysis rates or (as done here) intrinsic and GEF-mediated exchange. Interestingly, whereas E.coli-expressed HRASQ61E was described previously as unstable when purified for biochemical studies (18), we did not observe this with KRASQ61E purified from E. coli for our biochemical analysis. However, we did observe that the KRASQ61E mutant was poorly expressed in cells, when compared with all other KRAS mutants, which may also contribute to its rarity in cancer.
In contrast to the predicted occurrence of the Q61E missense mutation, the Q61P missense mutation has a very low predicted frequency of occurrence. However, although our analyses found that the Q61P mutant is biochemically similar to strongly transforming Q61 mutants, we found that KRASQ61P was, like KRASQ61E, nontransforming in our assays. This suggests that these mutants require appropriate co-occurring mutations to drive human cancers. Thus, the rarity of KRASQ61P may be driven by both low mutational frequency and poor oncogenic strength.
Our functional comparison of the potently transforming KRASQ61H/L/R mutants with the most frequent KRASG12D mutant provides another possible mechanistic basis for the lower occurrence of KRASQ61 mutations in cancer overall. The highly transforming KRASQ61 mutants caused greater ERK activation in NIH/3T3 and RIE-1 cells than did KRASG12D. In agreement with cellular ERK activation and our biochemical studies, a recent structural analysis of KRASQ61R interaction with the RAF RBD-CRD showed that the Q61R side chain sits within an empty pocket and does not interfere with the binding interface (43). These findings are also consistent with the observations of Counter and colleagues (12): that KrasQ61L/R mutants exhibited more potent ERK activation compared with the KrasG12D mutant in a mouse model of urethane-induced lung cancer and that higher levels of KRAS expression favored induction of tumors harboring the more weakly ERK-activating KRASG12D mutant. The more potent ERK activation by KrasQ61L/R was proposed to increase senescence, which may select against these mutations during cancer initiation and progression. That more robust ERK activation is deleterious to cancer development is also supported by the nonoverlapping occurrence of RAS mutations with mutations in BRAF, NF1, or EGFR (cBioPortal). The Goldilocks principle of ERK signaling, which is well-supported by genetic studies in mouse models of cancer (44), proposes that there is a sweet spot for driving cancer growth, and overactivation can lead to senescence or apoptosis. Thus, strong ERK activation by KRASQ61H/L/R may contribute to the rarity of KRASQ61 mutations in cancer. Conversely, it was observed that pERK levels in KRASQ61 mutant PDAC patient tumors was lower (11), suggesting an alternative basis where Q61 mutant proteins lack potent ERK activation to effectively drive PDAC growth.
Of note, all KRASQ61 mutants tested herein showed markedly weaker stimulation of macropinocytosis compared to KRASG12D. The ability to induce this metabolic process is critical for sustained tumor growth in pancreatic cancer (29). This may be another aspect of KRASQ61 distinctive biology that contributes to the rarity of these mutations in PDAC, where instead KRASG12 mutations are nearly universal.
As we found previously for KRASG12 mutant PDAC cell lines (6, 36), we found here that KRASQ61-mutant PDAC lines also demonstrated KRAS dependency. We speculated that our recently identified therapeutic strategy combining inhibition of ERK MAPK and autophagy (36, 45) may also prove effective for KRASQ61-mutant PDAC. That we observed potent growth suppression with concurrent ERKi and autophagy inhibition, as well as ERKi-dependent changes in autophagic flux, suggest that Q61 mutation status may not be a criterion for exclusion from this treatment. However, we have lower statistical power for Q61 versus G12 mutants due to the rarity of KRASQ61-mutant PDAC cell lines, and further studies will be required to reliably address this notion.
In summary, our study further emphasizes that there are consequences of different Q61 mutations on KRAS function that drive their overall and specific frequencies of occurrence in cancer. We note that our study was limited to in vitro analyses and therefore does not factor in the influence of the tumor microenvironment on the oncogenic potency of KRASQ61 mutant KRAS. To date, the vast majority of studies evaluating the ability of KRAS to initiate cancer development have focused on G12 mutations. Extension of such studies to evaluate the oncogenic potency of Q61 mutants in vivo will be needed to address the possibility that Q61 mutants may be too potent, or alternatively too weak, and are consequently “poor” oncogenes.
MATERIALS AND METHODS
Calculating the probabilities of KRAS alleles
The data collection and methods for calculating the probabilities of the different KRAS mutations were conducted as described previously (42). The mutational signatures are linear combinations of the 96-dimension spectrum of possible single-nucleotide base substitution (SBS) mutations (13). Thus, assuming that the prevalence of active mutational processes alone determines the frequency of KRAS alleles and the processes act uniformly throughout the genome, the probability of a tumor sample to acquire a specific KRASQ61 allele was calculated as the frequency of the same mutation across the entire genome.
Predicting KRAS allele frequency
The expected frequencies of the KRAS alleles were calculated as the mean probability of obtaining the KRAS allele across all tumor samples of a cancer type (see “Calculating the probabilities of KRAS alleles” above). The 95% confidence intervals around the mean were bootstrapped using the “boot” R package and the “percentile” method (46, 47). A χ-squared test was used to test the null hypothesis that there is no difference between the predicted and observed frequency for each KRAS allele. The p-values were adjusted for multiple hypothesis testing using the Benjamini–Hochberg method (14).
Cell culture
UM2 (RRID:CVCL_VH01) and UM147 are human pancreatic cancer patient-derived (PDX) cell lines (University of Michigan) (48). Pa02C, Pa14C and Pa16C cell lines were provided by A. Maitra (MD Anderson Cancer Center). NIH/3T3 mouse cells (RRID:CVCL_0594) were provided by G. Cooper (Dana-Farber Cancer Institute) and RIE-1 rat cells (RRID:CVCL_6723) were provided by R. Coffey (Vanderbilt University). The remaining PDAC cell lines and human HEK293T cells (RRID:CVCL_0063) were obtained from the American Type Culture Collection (ATCC) and were maintained in either Dulbecco’s Modified Eagle Medium (DMEM) or RPMI 1640 supplemented with fetal bovine serum (FBS) at either 15% [for Hs766T cells (RRID:CVCL_0334) and Panc 02.13 cells (RRID:CVCL_1634)] or 10% (for all other lines). RIE-1 were all maintained in DMEM supplemented with 10% FBS. NIH/3T3 was maintained in DMEM supplemented with 10% calf serum (Colorado Serum Company). All cell lines were maintained in a humidified chamber with 5% CO2 at 37°C. All cell line identities were verified by short tandem-repeat (STR) profiling and all lines were regularly monitored for mycoplasma contamination.
Antibodies and reagents
For immunoblot analysis, the following phospho-specific and total protein antibodies were obtained from Cell Signaling Technology: pAKT (9271, RRID:AB_329825), AKT (9272, RRID:AB_329827), pERK1/2 (4370, RRID:AB_2315112), ERK1/2 (9102, RRID:AB_330744), MYC (5605, RRID:AB_1903938), GST (2625, RRID:AB_490796). Antibodies to HA (H3663, RRID:AB_262051) and vinculin (V9131, RRID:AB_477629) were obtained from Sigma Aldrich. For immunofluorescence staining, the monoclonal antibody against vinculin (V9131) was obtained from Sigma Aldrich and the Alexa-Fluor-568 secondary antibody (A-11004, RRID:AB_2534072) was from Invitrogen. DAPI stain was obtained from ThermoFisher. F-actin was visualized by Phalloidin-conjugated with an Alexa-488 fluorophore (Invitrogen, A12379). For immunochemical labeling of mitochondria, MitoTracker CMXRos (ThermoFisher) was applied following the manufacturer’s recommended protocol. siRNA against scrambled (Negative Control No. 1) and KRAS (s7940, s7939) sequences were obtained from Invitrogen and transfected into cells by using Lipofectamine RNAiMAX following the manufacturer’s recommended protocol. Additional chemical reagents used included bafilomycin A1, oligomycin A, rotenone, antimycin, FCCP, CCP, doxycycline, MTT, chloroquine diphosphate (Sigma Aldrich) and SCH772984 (provided by Merck).
Retroviral and lentiviral vector infections
Human KRAS4B mutant proteins were ectopically expressed from the pBabe retroviral expression vector in RIE-1 and NIH/3T3 cells. The pBabe vector encoding mCherry-EGFP-LC3B was provided by J. Debnath (University of California at San Francisco, Addgene plasmid #22418) (49). Viral particles were generated by transient transfection of each expression vector into HEK293T cells using Fugene6 (Promega) with the PCL-10A1 packaging system for retrovirus or the psPAX2 and pMD2.G packaging system for lentivirus according to the manufacturer’s recommended protocol. Infection of cell lines was performed in growth medium supplemented with 8 mg/ml polybrene, with antibiotic selection beginning 48 hours after transduction.
Growth assays
To assess the effect of KRAS knockdown on growth, cells were treated for 48 hours with a KRAS-specific siRNA. To measure growth on plastic, cells were plated in duplicate in 6-well dishes. Plates were developed after seven days by removing the medium, washing and fixing cells with a 4% paraformaldehyde and crystal violet solution. To monitor 3D growth, 50 μL 0.6% bacto agar per well was placed into clear-bottom 96-well plates. Cells were mixed into a 1% SeaPrep agarose solution and plated. To quantify cell number, cell viability was determined by staining with AlamarBlue after seven days according to the manufacturer’s protocol. A matched 6-well plate was seeded for immunoblot analysis to confirm KRAS knockdown.
Immunoblot analyses
Cells were washed twice with ice-cold PBS and lysed in 1% NP-40 buffer (25 mM Tris-HCl, pH 7.4, 100 mM NaCl, 10 mM MgCl2, 1% NP-40) supplemented with phosphatase (Sigma) and protease (Roche) inhibitors. Lysates were scraped, collected in chilled Eppendorf tubes and clarified by centrifugation at 15,000 g for 15 min at 4°C. Protein concentrations were determined using Bradford reagent (Bio-Rad). Standard immunoblotting procedures were performed. Membranes were blocked in 5% milk diluted in TBS with 0.05% Tween 20 (TBST). To determine the levels of activated proteins, blot analyses utilized phospho-specific antibodies as described above with corresponding antibodies recognizing total proteins.
Protein purification
The cDNA sequence encoding truncated human KRAS4B (residues 2–169) was cloned into a pET21 bacterial expression vector containing an amino-terminal 6x-His purification tag followed by a Tobacco Etch Virus (TEV) protease cleavage site. The recombinant catalytic fragment of RASGRP1 (residues 50–468, pET28a) was described previously (50). Bacterial expression vectors encoding KRAS-GTP effector interacting domains included BRAF-RBD (residues 149-232, pET28a), CRAF-RBD (residues 54–131, pQlinkH), RGL2-RA (residues 647–736, pGEX3T-2) (51) and PLCε-RA (residues 2113–2221, pTriEx4) (52). All 6x-His-tagged proteins were expressed in BL21 (DE3) Rosetta2 cells and purified following the Qiagen Nickel NTA purification protocol with the 6x-His tags removed using TEV protease. For pGEX vectors, proteins were purified following the Glutathione Sepharose™ 4B purification protocol (Amersham Pharmacia Biotech) with the GST-tag removed using thrombin protease. If necessary, the proteins were further purified by size exclusion chromatography (Superdex-75 10/300 GL column; GE Life Sciences) and judged greater than 95% pure by SDS-PAGE analysis.
Guanine nucleotide exchange and protein binding assays
For nucleotide exchange and effector binding assays, KRAS was loaded with various nucleotide analogs. For the loading of GTPγS, the nonhydrolyzable GTP analog guanosine-5'-[(β,γ)-methyleno]triphosphate (GMPPCP), or mGMPPCP (Jena Biosciences), KRAS was incubated with alkaline phosphatase beads and 5-fold excess nucleotide for three hours with gentle rotation. Alkaline phosphatase and excess nucleotide removed with buffer exchange through a desalting column. The purified protein was checked for >95% nucleotide loading by HPLC (53). Exchange for mGDP (Jena Biosciences) was performed following previously published methods (54). Nucleotide exchange assays were performed using a Cary Eclipse Fluorescence Spectrophotometer (Agilent), as previously described (54). The minimal catalytic fragment of the RASGEF RASGRP1cat was used to stimulate nucleotide dissociation with the addition of 1000-fold excess of unlabeled nucleotide. All experiments were performed in triplicate.
For quantitative binding to isolated RAS effector RBD and RA domains, the fluorescent RAS-GTP-binding assay was adapted from a previous protocol (55). Briefly, KRAS loaded with mGMPPCP (1.5 μM) was incubated with increasing concentrations of effector proteins in reaction buffer (20 mM HEPES, 50 mM NaCl and 5 mM MgCl2 at pH 7.4). Nucleotide dissociation was initiated by the addition of 1000-fold excess of unlabeled GDP at 25°C. The rate of dissociation was monitored by the change in fluorescence at an excitation wavelength of 365 nm and emission at 435 nm using a SpectraMax M5 plate reader. Fluorescent nucleotide dissociation curves were fit to a one-phase exponential decay equation using GraphPad Prism. The dissociation rates were fit against the ligand concentration using previously published methods (56). All experiments were performed in triplicate.
NMR analyses
To generate 15N-enriched KRASQ61E, the protein was recombinantly expressed as above, using minimal media containing 1 g/L 15NH4Cl (Cambridge Isotope Laboratories) as the sole source of nitrogen. Purification of 15N-enriched KRASQ61E required no modifications to the purification protocol described above. To produce the nonhydrolyzable GTP analog GMPPCP-bound KRASQ61E protein, nucleotide loading was performed utilizing the alkaline phosphatase bead incubation method, as described above. Nucleotide loading was verified via HPLC analysis to exceed 95% for the desired state. For NMR analysis, 15N-enriched KRASQ61E (100 μM) was equilibrated in a buffer containing 20 mM NaH2PO4 (pH 6.8), 50 mM NaCl and 5 mM MgCl2, supplemented with 5% (v/v) D2O. Two-dimensional NMR 1H-15N HSQC spectra of 15N-labelled KRASQ61E were acquired on a Bruker Avance 850 MHz (19.97 T field strength) spectrometer at 25°C, using a cryogenic (TCI) 5 mm triple-resonance probe equipped with z-axis gradient. NMR data were collected using a spectral width of 16 ppm and 38 ppm and complex points of 2048 and 128 along the 1H and 15N dimension, respectively. The NMR data were processed using TopSpin (v3.6.1, Bruker) and the spectra were visualized using SPARKY (57). These NMR data were collected on both the inactive GDP- and active GMPPCP-bound states.
Protein crystallization and X-ray diffraction studies
The KRASQ61E GMPPCP-bound protein was brought to a concentration of 1.0 mM in crystallization buffer (20 mM HEPES, 50 mM NaCl, 5 mM MgCl2, pH 7.4). Crystals were obtained through the sitting-drop vapor diffusion method. Using a 96-well plate set-up, three 0.3 μL drops were equilibrated against a reservoir volume of 30 μL. KRASQ61E-GMPPCP crystals were obtained from a mother liquor solution of 0.2 M ammonium sulfate, 25% (w/v) PEG 3350 and 0.1 M HEPES-NaOH at pH 7.5, equilibrated against an identical reservoir solution at 20 °C for 6 weeks. The crystals were cryo-protected by briefly dipping them in reservoir solution enhanced with 15% ethylene glycol and then flash frozen in liquid N2. Data were collected at 100 K on the SER-CAT ID22 beamline at the APS synchrotron facility, utilizing a wavelength of 1.00 Å. All data were scaled and integrated using HKL2000 (59), molecular replacement was performed with Phaser-MR (60) from the PHENIX (61) suite of programs, utilizing protomer A of the KRASWT GTP-bound crystal structure (PDB accession code 5VQ2 (62)) as the search model for MR. Model building, manual placement of waters and alignment calculations utilized COOT (63) and refinement was carried out using phenix.refine (64). X-ray data collection and refinement statistics are provided in table S1.
Sample preparation and reverse phase protein microarray (RPPA)
Samples for RPPA analyses were prepared and arrays were constructed as previously described (65). In brief, cells were grown in 6-well plates, lysed, and immobilized alongside internal controls onto nitrocellulose-coated glass slides using an Aushon 2470 automated system (Quanterix). Total protein concentration was quantified in selected arrays using Sypro Ruby Protein Blot Stain (Molecular Probes) following manufacturer’s instructions. The remaining arrays were pre-treated with Reblot Antibody Stripping solution (MilliporeSigma) for 15 min at RT, followed by two washes with PBS and incubated for 5 hours in I-block (Applied Biosystems) before antibody staining (66). Using an automated system (DakoCytomation), arrays were incubated with 3% hydrogen peroxide, blocked in biotin and subsequently treated with an additional serum-free protein block to reduce nonspecific protein binding. Each array was probed for 30 min with one antibody targeting the protein of interest. Arrays were probed with >160 antibodies targeting phospho- and total proteins. All antibodies were evaluated previously for their specificity (67). Biotinylated anti-rabbit (Vector Laboratories, Inc.), anti-mouse secondary antibody (DakoCytomation) and a commercially available tyramide-based avidin/biotin amplification system (Catalyzed Signal Amplification System (CSA), DakoCytomation) were used for signal amplification. Fluorescent detection was read out using the IRDye 680RD Streptavidin (LI-COR Biosciences) system. Sypro Ruby- and antibody-stained slides were scanned on a Tecan laser scanner (TECAN) using the 580 nm and 620 nm channels. Images were analyzed using commercially available software (MicroVigene Version 5.1.0.0, Vigenetech) as previously described (68). Supervised hierarchical clustering was performed using R (version 3.4.1). Heatmaps were generated using the ComplexHeatmap package from Bioconductor. The RPPA standardized intensity data were log2 transformed, and the median of four independent biological replicates was determined for each feature.
Macropinocytosis
Macropinocytosis was performed as previously described (29). Cells were incubated for 30 min with 100 μg/ml FITC-dextran followed by a 90-min chase in serum-free DMEM/RPMI before fixation. Approximately 50–100 cells in >10 fields-of-view per condition were imaged on a Zeiss 700 confocal microscope (63x, 1.4 numerical aperture objective). For overall macropinocytosis levels, power and gain levels were set using the KRAS G12D condition in each experiment. The cell outline was mapped using a differential interference contrast image. Macropinocytotic index was quantified using ImageJ by taking the total macropinosome particle area divided by the total cell area and multiplying by 1000 (69).
Immunofluorescence studies
All cells were plated in glass-bottom dishes (MatTEK Corporation) or glass coverslips and imaged on a Zeiss 700 confocal microscope (63x, 1.4 numerical aperture objective). For antibody staining, cells were washed with room temperature (RT) PBS containing 1 mM CaCl2 and 0.5 mM MgCl2, and fixed with 3.7% (w/v) formaldehyde for 15 min. After fixation, cells were permeabilized in PBS with 0.5% Triton-X 100 (v/v) for 2 min. Non-specific signals were blocked using 2% BSA (Sigma) in PBS for 30 min at RT. Cells were incubated with indicated primary antibodies diluted in 2% BSA-PBS for 60 min at RT. After washing three times with PBS, cells were incubated with indicated secondary antibodies diluted in 2% BSA-PBS for 45 min at RT and washed three times with PBS. Cells on glass coverslips were mounted with Mowiol (Sigma). For quantification of F-actin and vinculin fluorescence, total cell fluorescence corrected for background was quantified and normalized to cell area using ImageJ. For each biological replicate, >5 representative images were collected with 10 cells per image for each indicated condition.
Cell migration assays
Cells were seeded at low density and cultured on fibronectin-coated (10 μg/ml) glass bottom dishes (MatTEK Corporation) and allowed to adhere overnight. For long-term time lapse imaging, cells were imaged on an Olympus VivaView Incubator fluorescence microscope (10x magnification) over a 16-hour period with 1 frame collected every 10 min. Single cells were tracked. Cells were excluded if they collided with other cells or debris, underwent mitosis, or migrated out of the field of view. For each biological replicate, >5 fields of view were imaged per cell line. The average distance and velocity of each KRAS cell lines was quantified based on the average of >60 individual cells per condition in ImageJ.
Cell adhesion assays
Prior to incubation, 96-well plates were coated with 10 μg/ml fibronectin and blocked with 1% BSA/PBS for 30 min. Cells were incubated with 2 μM CellTracker CMFDA (5-chloromethylfluorescein diacetate) for 10 min, added to wells (2.5 x 104 cells per well) in triplicate, allowed to adhere for one hour, and washed with PBS. The fluorescence signal from adhered cells was quantified using a SpectraMax i3x plate reader (Molecular Devices) at an excitation wavelength of 485 nm and emission of 535 nm.
Oxygen consumption assay
Cells were harvested and seeded in XF96 cell culture microplates (Seahorse Bioscience). To measure oxygen consumption rate (OCR), culture medium was exchanged for Seahorse assay medium containing 25 mM glucose, 1 mM glutamine and 1 mM sodium pyruvate. Oligomycin, FCCP, antimycin and rotenone (XF Cell Mito Stress Test Kit, Seahorse Bioscience) were injected sequentially. Experiments were performed using an XF96 analyzer, with raw data normalized to cell number as measured by live cell counting and imaging with a SpectraMax MiniMax 300 imaging cytometer (Molecular Devices).
Flow cytometry
To quantify autophagic flux, cells expressing mCherry-EGFP-LC3B were cultured and treated with 1 μM SCH772984 for 24 hours. Cells were trypsinized and washed with growth medium after centrifugation at 500 x g for 5 min. Samples were analyzed on a Beckman Coulter tabletop flow cytometer, with data collected and exported using Kaluza. Forward scatter and side scatter were used to gate for live and single cells in addition to setting an “autophagy” gate for cells expressing the fluorescent LC3B reporter. The ratio of the gated fluorescence of mCherry to the total fluorescence of EGFP is reported as the autophagic index.
Drug response testing
Cells (1,000–2,500 cells per well depending on cell line) were seeded in 96-well plates and incubated for 24 hours before addition of inhibitor. Increasing concentrations of drug, with DMSO as a negative control, were added to the wells in three replicates using a digital dispenser liquid handling device (TECAN D300e). Plates were incubated at 37 °C for 5 days after the addition of drug, stained and analyzed with Calcein AM (Invitrogen) according to the manufacturers recommended protocol, and counted using a SpectraMax MiniMax 300 imaging cytometer. For normalization to day 0 conditions, the average of six DMSO-treated wells at day 5 was used to determine maximum viability. All data were analyzed using SoftMax version 5 and GraphPad Prism using a 4-parameter drug response curve.
DepMap analyses
Genetic dependency scores for KRAS and MYC after CRISPR gene silencing were obtained from DepMap (21Q2 Public+Score, CERES). Mutation data for KRAS was also obtained from DepMap (21Q2) (70). All pancreas, lung, and colorectal cancer-derived cell lines with activating KRAS mutations were included for analysis. The dependency of KRAS or MYC was shown and grouped by mutant KRAS status (Q61 vs. non-Q61). More negative CERES scores indicate greater dependency on the indicated protein.
Statistical analysis
Data were analyzed by GraphPad Prism built-in tests (ANOVA for data of more than two groups, student’s two-tailed t-test otherwise). Data are presented relative to their respective control and quantified as mean ± S.E.M. for n ≥ 3 independent experiments (except where noted). P-values on graphs are denoted by * P ≤ 0.5; ** P ≤ 0.01; *** P ≤ 0.001 as determined in GraphPad Prism. The number of samples analyzed per experiment and whether the data presented are representative or an average is indicated in the respective figure legends.
Supplementary Material
Acknowledgements:
We thank Anirban Maitra and Jason Fleming for PDAC cell lines and Elisa Baldelli for assistance with RPPA analyses.
Funding:
M.V.H. was supported by the Slomo and Cindy Silvian Foundation. G.A.H. was supported by National Institute of General Medical Sciences grant P20GM130457 and National Cancer Institute (NCI) grants F32CA200313 and T32CA009156. L.M.C. was supported by NCI T32CA009156 and American Cancer Society PF-20-140-01 – CDD. J.N.D. was supported by the Slomo and Cindy Silvian Foundation, NCI T32CA071341, and F30CA243253. A.M.W. was supported by a fellowship from the American Cancer Society (F-18-061). R.D.T. was supported by NCI T32CA009156. S.L.C. and E.F.P. were supported by NCI CA203657. Support was provided by grants from the NCI to A.D.C. and/or C.J.D. (CA42978, R01CA175747, R01CA223775, P50CA196510, U01CA199235 and P01CA203657 and R35CA232113), and from the Pancreatic Cancer Action Network/AACR (15-90-25-DER).
Footnotes
Competing interests: C.J.D. is an advisory board member for Anchiano Therapeutics, Boragen, Deciphera Pharmaceuticals, Mirati Therapeutics, Revolution Medicines and SHY Therapeutics; has received research funding support from Boragen, Deciphera Pharmaceuticals, Mirati Therapeutics and SpringWorks Therapeutics; and has consulted for Day One Biotherapeutics, Eli Lilly, Jazz Therapeutics, Ribometrix, Sanofi, and Turning Point Therapeutics. A.D.C. has consulted for Eli Lilly and Mirati Therapeutics. E.F.P. and M.P. are consultants for and receive royalties from Avant Diagnostics and TheraLink Technologies, Inc. E.F.P. is a consultant to and shareholder of Avant Diagnostics, Inc and Perthera, Inc., and received funding support from Mirati Therapeutics, Genentech, Inc., and Abbvie, Inc.
Data and materials availability: Plasmids generated for this study will be deposited at Addgene. The crystal structure of KRASQ61E has been deposited into the Protein Data Bank (PDB accession code 7LZ5). All other data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
References and Notes
- 1.Prior IA, Hood FE, Hartley JL, The Frequency of Ras Mutations in Cancer. Cancer Res. 80, 2969–2974 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hobbs GA, Der CJ, Rossman KL, RAS isoforms and mutations in cancer at a glance. J Cell Sci. 129, 1287–1292 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Papke B, Der CJ, Drugging RAS: Know the enemy. Science. 355, 1158–1163 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ, Drugging the undruggable RAS: Mission possible? Nat Rev Drug Discov. 13, 828–851 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prior IA, Lewis PD, Mattos C, A comprehensive survey of Ras mutations in cancer. Cancer Res. 72, 2457–2467 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hobbs GA, Baker NM, Miermont AM, Thurman RD, Pierobon M, Tran TH, Anderson AO, Waters AM, Diehl JN, Papke B, Hodge RG, Klomp JE, Goodwin CM, DeLiberty JM, Wang J, Ng RWS, Gautam P, Bryant KL, Esposito D, Campbell SL, Petricoin EF, Simanshu DK, Aguirre AJ, Wolpin BM, Wennerberg K, Rudloff U, Cox AD, Der CJ, Atypical KRAS G12R Mutant Is Impaired in PI3K Signaling and Macropinocytosis in Pancreatic Cancer. Cancer Discov. 10, 104–123 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moore AR, Rosenberg SC, McCormick F, Malek S, RAS-targeted therapies: is the undruggable drugged? Nat Rev Drug Discov. 19, 533–552 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haigis KM, KRAS Alleles: The Devil Is in the Detail. Trends Cancer. 3, 686–697 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith G, Bounds R, Wolf H, Steele RJC, Carey FA, Wolf CR, Activating K-Ras mutations outwith “hotspot” codons in sporadic colorectal tumours - implications for personalised cancer medicine. Br J Cancer. 102, 693–703 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park JT, Johnson N, Liu S, Levesque M, Wang YJ, Ho H, Huso D, Maitra A, Parsons MJ, Prescott JD, Leach SD, Differential in vivo tumorigenicity of diverse KRAS mutations in vertebrate pancreas: A comprehensive survey. Oncogene. 34, 2801–2806 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Witkiewicz AK, McMillan EA, Balaji U, Baek G, Lin W-C, Mansour J, Mollaee M, Wagner K-U, Koduru P, Yopp A, Choti MA, Yeo CJ, McCue P, White MA, Knudsen ES, Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat Commun. 6, 6744 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pershing NLK, Lampson BL, Belsky JA, Kaltenbrun E, MacAlpine DM, Counter CM, Rare codons capacitate Kras-driven de novo tumorigenesis. J. Clin. Invest 125, 222–233 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alexandrov LB, Nik-Zainal S, Wedge DC, Campbell PJ, Stratton MR, Deciphering signatures of mutational processes operative in human cancer. Cell Rep. 3, 246–259 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benjamini Yoav, Hochberg Yosef, Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. 57, 289–300 (1995). [Google Scholar]
- 15.Der CJ, Krontiris TG, Cooper GM, Transforming genes of human bladder and lung carcinoma cell lines are homologous to the ras genes of Harvey and Kirsten sarcoma viruses. Proc Natl Acad Sci U S A. 79, 3637–3640 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oldham SM, Clark GJ, Gangarosa LM, Coffey RJ, Der CJ, Activation of the Raf-1/MAP kinase cascade is not sufficient for Ras transformation of RIE-1 epithelial cells. Proc Natl Acad Sci U S A. 93, 6924–6928 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Der CJ, Finkel T, Cooper GM, Biological and biochemical properties of human rasH genes mutated at codon 61. Cell. 44, 167–176 (1986). [DOI] [PubMed] [Google Scholar]
- 18.Frech M, Darden TA, Pedersen LG, Foley CK, Charifson PS, Anderson MW, Wittinghofer A, Role of glutamine-61 in the hydrolysis of GTP by p21H-ras: an experimental and theoretical study. Biochemistry. 33, 3237–3244 (1994). [DOI] [PubMed] [Google Scholar]
- 19.Scheffzek K, The Ras-RasGAP Complex: Structural Basis for GTPase Activation and Its Loss in Oncogenic Ras Mutants. Science. 277, 333–338 (1997). [DOI] [PubMed] [Google Scholar]
- 20.Buhrman G, Holzapfel G, Fetics S, Mattos C, Allosteric modulation of Ras positions Q61 for a direct role in catalysis. Proc. Natl. Acad. Sci. U.S.A 107, 4931–4936 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scheidig AJ, Burmester C, Goody RS, The pre-hydrolysis state of p21(ras) in complex with GTP: new insights into the role of water molecules in the GTP hydrolysis reaction of ras-like proteins. Structure. 7, 1311–1324 (1999). [DOI] [PubMed] [Google Scholar]
- 22.Hunter JC, Manandhar A, Carrasco MA, Gurbani D, Gondi S, Westover KD, Biochemical and Structural Analysis of Common Cancer-Associated KRAS Mutations. Mol. Cancer Res 13, 1325–1335 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Hayes TK, Neel NF, Hu C, Gautam P, Chenard M, Long B, Aziz M, Kassner M, Bryant KL, Pierobon M, Marayati R, Kher S, George SD, Xu M, Wang-Gillam A, Samatar AA, Maitra A, Wennerberg K, Petricoin EF, Yin HH, Nelkin B, Cox AD, Yeh JJ, Der CJ, Long-Term ERK Inhibition in KRAS-Mutant Pancreatic Cancer Is Associated with MYC Degradation and Senescence-like Growth Suppression. Cancer Cell. 29, 75–89 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vaseva AV, Blake DR, Gilbert TSK, Ng S, Hostetter G, Azam SH, Ozkan-Dagliyan I, Gautam P, Bryant KL, Pearce KH, Herring LE, Han H, Graves LM, Witkiewicz AK, Knudsen ES, Pecot CV, Rashid N, Houghton PJ, Wennerberg K, Cox AD, Der CJ, KRAS Suppression-Induced Degradation of MYC Is Antagonized by a MEK5-ERK5 Compensatory Mechanism. Cancer Cell. 34, 807–822.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Farrell AS, Sears RC, MYC degradation. Cold Spring Harb Perspect Med. 4, a014365 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bryant KL, Mancias JD, Kimmelman AC, Der CJ, KRAS: feeding pancreatic cancer proliferation. Trends Biochem Sci. 39, 91–100 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kashatus JA, Nascimento A, Myers LJ, Sher A, Byrne FL, Hoehn KL, Counter CM, Kashatus DF, Erk2 phosphorylation of Drp1 promotes mitochondrial fission and MAPK-driven tumor growth. Mol Cell. 57, 537–551 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagdas S, Kashatus JA, Nascimento A, Hussain SS, Trainor RE, Pollock SR, Adair SJ, Michaels AD, Sesaki H, Stelow EB, Bauer TW, Kashatus DF, Drp1 Promotes KRas-Driven Metabolic Changes to Drive Pancreatic Tumor Growth. Cell Rep. 28, 1845–1859.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Commisso C, Davidson SM, Soydaner-Azeloglu RG, Parker SJ, Kamphorst JJ, Hackett S, Grabocka E, Nofal M, Drebin JA, Thompson CB, Rabinowitz JD, Metallo CM, Vander Heiden MG, Bar-Sagi D, Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature. 497, 633–637 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khosravi-Far R, Chrzanowska-Wodnicka M, Solski PA, Eva A, Burridge K, Der CJ, Dbl and Vav mediate transformation via mitogen-activated protein kinase pathways that are distinct from those activated by oncogenic Ras. Mol Cell Biol. 14, 6848–6857 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paterson HF, Self AJ, Garrett MD, Just I, Aktories K, Hall A, Microinjection of recombinant p21rho induces rapid changes in cell morphology. J Cell Biol. 111, 1001–1007 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seetharaman S, Etienne-Manneville S, Cytoskeletal Crosstalk in Cell Migration. Trends Cell Biol. 30, 720–735 (2020). [DOI] [PubMed] [Google Scholar]
- 33.Zhang H, Schaefer A, Wang Y, Hodge RG, Blake DR, Diehl JN, Papageorge AG, Stachler MD, Liao J, Zhou J, Wu Z, Akarca FG, de Klerk LK, Derks S, Pierobon M, Hoadley KA, Wang TC, Church G, Wong K-K, Petricoin EF, Cox AD, Lowy DR, Der CJ, Bass AJ, Gain-of-Function RHOA Mutations Promote Focal Adhesion Kinase Activation and Dependency in Diffuse Gastric Cancer. Cancer Discov. 10, 288–305 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Waters AM, Der CJ, KRAS: The Critical Driver and Therapeutic Target for Pancreatic Cancer. Cold Spring Harb Perspect Med. 8 (2018), doi: 10.1101/cshperspect.a031435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ozkan-Dagliyan I, Diehl JN, George SD, Schaefer A, Papke B, Klotz-Noack K, Waters AM, Goodwin CM, Gautam P, Pierobon M, Peng S, Gilbert TSK, Lin KH, Dagliyan O, Wennerberg K, Petricoin EF, Tran NL, Bhagwat SV, Tiu RV, Peng S-B, Herring LE, Graves LM, Sers C, Wood KC, Cox AD, Der CJ, Low-Dose Vertical Inhibition of the RAF-MEK-ERK Cascade Causes Apoptotic Death of KRAS Mutant Cancers. Cell Rep. 31, 107764 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bryant KL, Stalnecker CA, Zeitouni D, Klomp JE, Peng S, Tikunov AP, Gunda V, Pierobon M, Waters AM, George SD, Tomar G, Papke B, Hobbs GA, Yan L, Hayes TK, Diehl JN, Goode GD, Chaika NV, Wang Y, Zhang G-F, Witkiewicz AK, Knudsen ES, Petricoin EF, Singh PK, Macdonald JM, Tran NL, Lyssiotis CA, Ying H, Kimmelman AC, Cox AD, Der CJ, Combination of ERK and autophagy inhibition as a treatment approach for pancreatic cancer. Nat. Med 25, 628–640 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burd CE, Liu W, Huynh MV, Waqas MA, Gillahan JE, Clark KS, Fu K, Martin BL, Jeck WR, Souroullas GP, Darr DB, Zedek DC, Miley MJ, Baguley BC, Campbell SL, Sharpless NE, Mutation-specific RAS oncogenicity explains NRAS codon 61 selection in melanoma. Cancer Discov. 4, 1418–1429 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Winters IP, Chiou S-H, Paulk NK, McFarland CD, Lalgudi PV, Ma RK, Lisowski L, Connolly AJ, Petrov DA, Kay MA, Winslow MM, Multiplexed in vivo homology-directed repair and tumor barcoding enables parallel quantification of Kras variant oncogenicity. Nat Commun. 8, 2053 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poulin EJ, Bera AK, Lu J, Lin Y-J, Strasser SD, Paulo JA, Huang TQ, Morales C, Yan W, Cook J, Nowak JA, Brubaker DK, Joughin BA, Johnson CW, DeStefanis RA, Ghazi PC, Gondi S, Wales TE, Iacob RE, Bogdanova L, Gierut JJ, Li Y, Engen JR, Perez-Mancera PA, Braun BS, Gygi SP, Lauffenburger DA, Westover KD, Haigis KM, Tissue-Specific Oncogenic Activity of KRASA146T. Cancer Discov. 9, 738–755 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson CW, Lin Y-J, Reid D, Parker J, Pavlopoulos S, Dischinger P, Graveel C, Aguirre AJ, Steensma M, Haigis KM, Mattos C, Isoform-Specific Destabilization of the Active Site Reveals a Molecular Mechanism of Intrinsic Activation of KRas G13D. Cell Rep. 28, 1538–1550.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zafra MP, Parsons MJ, Kim J, Alonso-Curbelo D, Goswami S, Schatoff EM, Han T, Katti A, Fernandez MTC, Wilkinson JE, Piskounova E, Dow LE, An In Vivo Kras Allelic Series Reveals Distinct Phenotypes of Common Oncogenic Variants. Cancer Discov. 10, 1654–1671 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cook JH, Melloni GEM, Gulhan DC, Park PJ, Haigis KM, The origins and genetic interactions of KRAS mutations are allele- and tissue-specific. Nat Commun. 12, 1808 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tran TH, Chan AH, Young LC, Bindu L, Neale C, Messing S, Dharmaiah S, Taylor T, Denson J-P, Esposito D, Nissley DV, Stephen AG, McCormick F, Simanshu DK, KRAS interaction with RAF1 RAS-binding domain and cysteine-rich domain provides insights into RAS-mediated RAF activation. Nat Commun. 12, 1176 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klomp JE, Klomp JA, Der CJ, The ERK mitogen-activated protein kinase signaling network: the final frontier in RAS signal transduction. Biochem Soc Trans. 49, 253–267 (2021). [DOI] [PubMed] [Google Scholar]
- 45.Kinsey CG, Camolotto SA, Boespflug AM, Guillen KP, Foth M, Truong A, Schuman SS, Shea JE, Seipp MT, Yap JT, Burrell LD, Lum DH, Whisenant JR, Gilcrease GW, Cavalieri CC, Rehbein KM, Cutler SL, Affolter KE, Welm AL, Welm BE, Scaife CL, Snyder EL, McMahon M, Protective autophagy elicited by RAF→MEK→ERK inhibition suggests a treatment strategy for RAS-driven cancers. Nat Med. 25, 620–627 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davison AC, Hinkley DV, Bootstrap Methods and their Application (Cambridge University Press, ed. 1, 1997; https://www.cambridge.org/core/product/identifier/9780511802843/type/book). [Google Scholar]
- 47.Canty Angelo, Ripley Brian, Boot: Bootstrap Functions (Originally by Angelo Canty for S; ) (2019), (available at https://CRAN.R-project.org/package=boot). [Google Scholar]
- 48.Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, Wicha M, Clarke MF, Simeone DM, Identification of pancreatic cancer stem cells. Cancer Res. 67, 1030–1037 (2007). [DOI] [PubMed] [Google Scholar]
- 49.N’Diaye E-N, Kajihara KK, Hsieh I, Morisaki H, Debnath J, Brown EJ, PLIC proteins or ubiquilins regulate autophagy-dependent cell survival during nutrient starvation. EMBO Rep. 10, 173–179 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iwig JS, Vercoulen Y, Das R, Barros T, Limnander A, Che Y, Pelton JG, Wemmer DE, Roose JP, Kuriyan J, Structural analysis of autoinhibition in the Ras-specific exchange factor RasGRP1. Elife. 2, e00813 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith MJ, Ikura M, Integrated RAS signaling defined by parallel NMR detection of effectors and regulators. Nat Chem Biol. 10, 223–230 (2014). [DOI] [PubMed] [Google Scholar]
- 52.Bunney TD, Harris R, Gandarillas NL, Josephs MB, Roe SM, Sorli SC, Paterson HF, Rodrigues-Lima F, Esposito D, Ponting CP, Gierschik P, Pearl LH, Driscoll PC, Katan M, Structural and mechanistic insights into ras association domains of phospholipase C epsilon. Mol Cell. 21, 495–507 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Wakizaka A, Kurosaka K, Okuhara E, Rapid separation of DNA constituents, bases, nucleosides and nucleotides, under the same chromatographic conditions using high-performance liquid chromatography with a reversed-phase column. J Chromatogr. 162, 319–326 (1979). [DOI] [PubMed] [Google Scholar]
- 54.Lenzen C, Cool RH, Wittinghofer A, Analysis of intrinsic and CDC25-stimulated guanine nucleotide exchange of p21ras-nucleotide complexes by fluorescence measurements. Methods Enzymol. 255, 95–109 (1995). [DOI] [PubMed] [Google Scholar]
- 55.Block C, Janknecht R, Herrmann C, Nassar N, Wittinghofer A, Quantitative structure-activity analysis correlating Ras/Raf interaction in vitro to Raf activation in vivo. Nat Struct Biol. 3, 244–251 (1996). [DOI] [PubMed] [Google Scholar]
- 56.Herrmann C, Horn G, Spaargaren M, Wittinghofer A, Differential interaction of the ras family GTP-binding proteins H-Ras, Rap1A, and R-Ras with the putative effector molecules Raf kinase and Ral-guanine nucleotide exchange factor. J Biol Chem. 271, 6794–6800 (1996). [DOI] [PubMed] [Google Scholar]
- 57.Lee W, Tonelli M, Markley JL, NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics. 31, 1325–1327 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee W, Tonelli M, Markley JL, NMRFAM-SPARKY: Enhanced software for biomolecular NMR spectroscopy. Bioinformatics. 31, 1325–1327 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Otwinowski Z, Minor W, [20] Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997). [DOI] [PubMed] [Google Scholar]
- 60.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ, Phaser crystallographic software. J Appl Crystallogr. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH, PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu S, Long BN, Boris GH, Chen A, Ni S, Kennedy MA, Structural insight into the rearrangement of the switch I region in GTP-bound G12A K-Ras. Acta Crystallogr D Struct Biol. 73, 970–984 (2017). [DOI] [PubMed] [Google Scholar]
- 63.Emsley P, Lohkamp B, Scott WG, Cowtan K, Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD, Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 68, 352–367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Baldelli E, Calvert V, Hodge A, VanMeter A, Petricoin EF, Pierobon M, Reverse Phase Protein Microarrays. Methods Mol Biol. 1606, 149–169 (2017). [DOI] [PubMed] [Google Scholar]
- 66.Pin E, Federici G, Petricoin EF, Curr Protoc Protein Sci, in press, doi: 10.1002/0471140864.ps2707s75. [DOI] [PubMed] [Google Scholar]
- 67.Signore M, Reeder KA, Antibody validation by Western blotting. Methods Mol Biol. 823, 139–155 (2012). [DOI] [PubMed] [Google Scholar]
- 68.Baldelli E, Bellezza G, Haura EB, Crinó L, Cress WD, Deng J, Ludovini V, Sidoni A, Schabath MB, Puma F, Vannucci J, Siggillino A, Liotta LA, Petricoin EF, Pierobon M, Functional signaling pathway analysis of lung adenocarcinomas identifies novel therapeutic targets for KRAS mutant tumors. Oncotarget. 6, 32368–32379 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Commisso C, Flinn RJ, Bar-Sagi D, Determining the macropinocytic index of cells through a quantitative image-based assay. Nat Protoc. 9, 182–192 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Meyers RM, Bryan JG, McFarland JM, Weir BA, Sizemore AE, Xu H, Dharia NV, Montgomery PG, Cowley GS, Pantel S, Goodale A, Lee Y, Ali LD, Jiang G, Lubonja R, Harrington WF, Strickland M, Wu T, Hawes DC, Zhivich VA, Wyatt MR, Kalani Z, Chang JJ, Okamoto M, Stegmaier K, Golub TR, Boehm JS, Vazquez F, Root DE, Hahn WC, Tsherniak A, Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat Genet. 49, 1779–1784 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.