Abstract
Iron is mostly devoted to the hemoglobinization of erythrocytes for oxygen transport. However, emerging evidence points to a broader role for the metal in hematopoiesis, including the formation of the immune system. Iron availability in mammalian cells is controlled by iron-regulatory protein 1 (IRP1) and IRP2. We report that global disruption of both IRP1 and IRP2 in adult mice impairs neutrophil development and differentiation in the bone marrow, yielding immature neutrophils with abnormally high glycolytic and autophagic activity, resulting in neutropenia. IRPs promote neutrophil differentiation in a cell intrinsic manner by securing cellular iron supply together with transcriptional control of neutropoiesis to facilitate differentiation to fully mature neutrophils. Unlike neutrophils, monocyte count was not affected by IRP and iron deficiency, suggesting a lineage-specific effect of iron on myeloid output. This study unveils the previously unrecognized importance of IRPs and iron metabolism in the formation of a major branch of the innate immune system.
Maintenance of iron supply in hematopoietic cells is necessary for the production of properly differentiated neutrophils.
INTRODUCTION
As an integral component of heme, iron must be supplied in adequate amounts to secure hemoglobin synthesis and prevent anemia. Anemia due to iron deficiency is highly prevalent (1). It is often due to insufficient dietary iron intake or to inhibition of iron delivery into the plasma under conditions of inflammation (2, 3). Iron overload resulting from, e.g., ineffective erythropoiesis or mutations in iron homeostasis genes, is also deleterious, affecting organs such as the liver and heart and promoting infections.
Body iron levels and distribution are controlled by the peptide hepcidin [or HAMP (hepcidin antimicrobial peptide)]. Hepcidin reduces the release of iron into the plasma from iron exporting cells by binding to and inhibiting the iron exporter ferroportin (FPN; also known as SLC40A1). Its levels augment in response to immune activation or when body iron levels are high. Conversely, stimulation of red blood cell (RBC) production suppresses hepcidin via the erythroblast-derived hormone erythroferrone (ERFE; also FAM132B), allowing for iron export via FPN to match erythroid iron needs (4).
At the cellular level, iron homeostasis is orchestrated by the interaction of iron regulatory protein 1 (IRP1) and IRP2 (also known as ACO1 and IREB2, respectively) with cis-regulatory iron responsive elements (IREs) present in the untranslated region (UTR) of mRNAs encoding iron metabolism molecules. When iron is scarce, IRPs bind to the 5′-UTR IRE of mRNAs encoding the ferritin-H and -L (FTH1 and FTL1, respectively) iron storage proteins, the iron exporter FPN, the erythroid aminolevulinate synthase (ALAS2), or the transcription hypoxia inducible factor 2 alpha (HIF2A; also known as EPAS1) and inhibit their translation. IRP binding to 3′-UTR IREs in the transferrin receptor 1 (TFRC) mRNA protects the transcript against the Regnase-1 (ZC3H12A) and Roquin-1 (RC3H1) nucleases (5, 6), thereby promoting iron uptake. A single 3′ IRE is also present in the divalent metal transporter 1 (DMT1; also known as SLC11A2) mRNA. In iron- and oxygen-replete cells, IRP2 is displaced from IRE RNA by F-box and leucine-rich repeat protein 5 (FBXL5) and is targeted for degradation (7), whereas IRP1 assembles an iron-sulfur cluster (ISC) and functions as an aconitase (8).
Studies in patients and animal models revealed that inadequate IRP activity is detrimental. Abnormal stimulation of IRP activity due to defects in ISC biogenesis impairs heme synthesis in erythroid cells (9), and aberrant gain of IRP2 function in FBXL5-null mice was found to be lethal (10). Conversely, constitutive, systemic deletion of IRP2 in mice results in microcytic anemia, diabetes, and neurological symptoms of varying severity depending on the mouse strain examined (11, 12). IRP1 deficiency results in transient polycythemia during early life, due to derepression of Hif2a translation in kidney cells and subsequent stimulation of erythropoietin (Epo). Despite these phenotypic traits, mice lacking either of the two IRPs remain viable and fertile. This is in distinct contrast with the early embryonic lethality resulting from double IRP deficiency (11) and shows that while the IRP/IRE network is essential, the two IRPs can largely compensate for each other.
We set out to determine the functions of the IRP/IRE regulatory network during adult life. To circumvent the early lethality due to compound IRP deficiency, we took advantage of conditional Irp alleles to acutely disrupt both IRP1 and IRP2 in the entire body of adult mice. Our study reveals that IRPs are critically important for granulocyte development and differentiation in the bone marrow (BM), unveiling a previously unrecognized role for iron metabolism and the IRP/IRE regulatory network in hematopoiesis and formation of the immune system.
RESULTS
Systemic acute loss of IRP function causes erythropenia and myelopenia
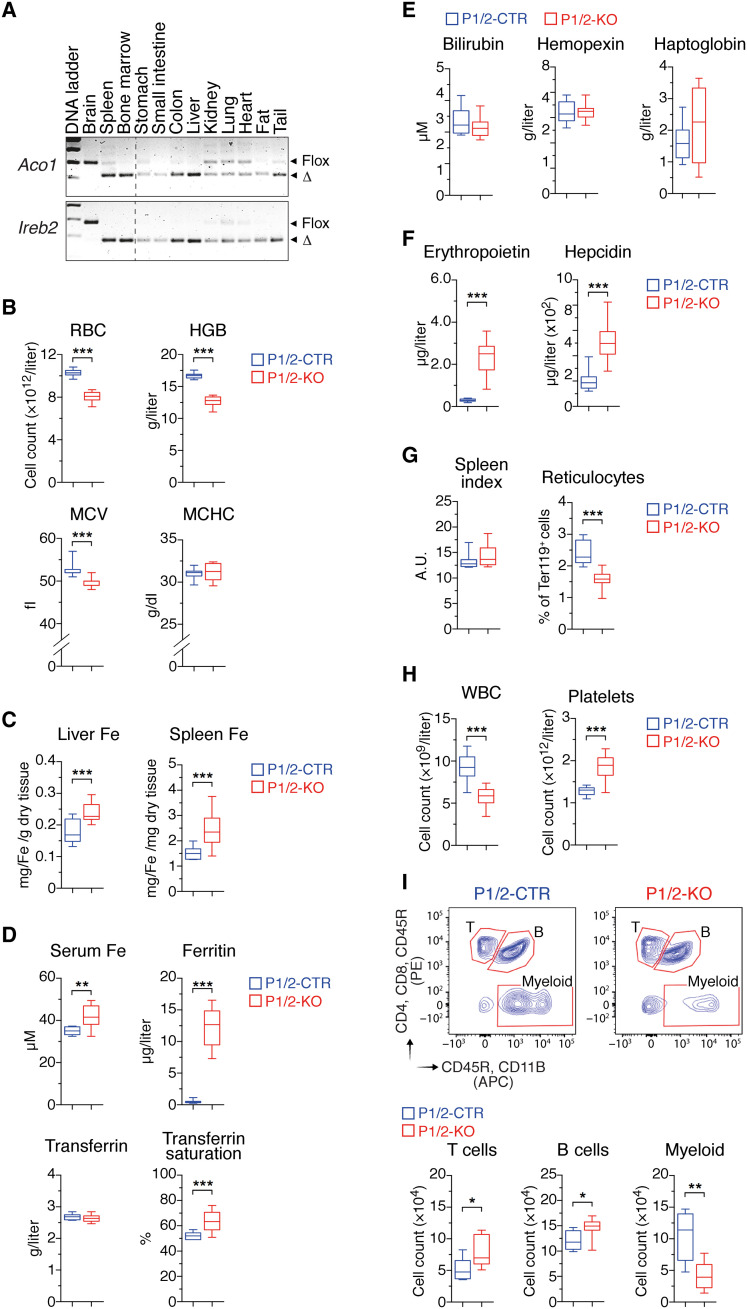
To study the impact of IRP deficiency during adult life, we generated mice that carry floxed Aco1 and Ireb2 alleles (13) and express a tamoxifen-inducible CRE recombinase (CreER) under the control of the Rosa26 promoter (14). The resulting mice are designated P1/2–knockout (KO); control littermates lacking CreER are designated P1/2–control (CTR). To control for potential effects of CRE (15), we also analyzed animals carrying the CreER sequence alone (CreER) and their wild-type (WT) littermates. Adult P1/2-KO mice treated with two nonconsecutive low doses of tamoxifen displayed efficient recombination of the floxed Aco1 and Ireb2 alleles in most tissues analyzed except for the brain (Fig. 1A).
Fig. 1. Acute IRP ablation during adulthood causes erythropenia and myelopenia.
P1/2-CTR and P1/2-KO littermates received tamoxifen on days 1 and 3 to induce IRP ablation. Mice were analyzed on day 10. (A) Genomic polymerase chain reaction (PCR) analysis of the Irp1 (Aco1) and Irp2 (Ireb2) alleles in tissues from P1/2-KO mice. The floxed (flox) and truncated (Δ) alleles are indicated. (B) RBC parameters (P1/2-CTR, n = 16; P1/2-KO, n = 18). HGB, hemoglobin; MCV, mean corpuscular volume; MCHC, mean corpuscular hemoglobin concentration. (C) Hepatic and splenic iron levels (n = 11). (D) Serum iron parameters (n = 10). (E) RBC decay markers (n = 10). (F) Serum levels of EPO (P1/2-CTR, n = 18; P1/2-KO, n = 17) and hepcidin (P1/2-CTR, n = 18; P1/2-KO, n = 16). (G) Spleen index = √[(100 × spleen weight in milligrams per body weight in grams)] (n = 7), and reticulocyte frequency in peripheral blood (PB) (P1/2-CTR, n = 7; P1/2-KO, n = 9). A.U., arbitrary units. (H) White blood cell (WBC) and platelet counts in PB (P1/2-CTR, n = 16; P1/2-KO, n = 18). (I) Flow cytometry (FCM) analysis of major WBC populations in PB. The gating strategy is shown on contour plots on the left. The histograms display cell counts for 3 × 105 events recorded (n = 8). (B to I) The results are presented as box plots (minimum to maximum values). Unpaired, two-tailed t test between P1/2-CTR and P1/2-KO. *P < 0.05; **P < 0.01; ***P < 0.001. APC, allophycocyanin; PE, phycoerythrin.
On day 10 after the commencement of tamoxifen treatment, P1/2-KO mice exhibited normal overall posture and activity pattern, and their body weight was unchanged. However, these mice displayed a substantial decrease in the number and size of RBCs, as well as low hemoglobin levels (Fig. 1B). The effect of combined IRP1/2 deficiency on RBC parameters was approximately twice that of single IRP2 ablation, suggesting that IRP1 can partially compensate for the absence of IRP2 (fig. S1). The anemia was accompanied by iron accumulation in the liver and spleen, elevated plasma iron and ferritin levels, and high transferrin saturation (Fig. 1, C and D). Despite high iron levels, IRP-deficient mice were anemic, suggesting that the anemia may be due to hemolysis and/or deficient RBC production. However, hemolysis is unlikely because serum bilirubin, haptoglobin, and hemopexin levels were unchanged (Fig. 1E). Notably, serum EPO concentration was markedly increased in P1/2-KO animals (Fig. 1F). High EPO levels are predicted to stimulate erythropoiesis and suppress hepcidin via ERFE. However, we observed a decrease in blood reticulocytes and no sign of extramedullary erythropoiesis (Fig. 1G). Moreover, ERFE levels remained below the level of detection, and hepcidin concentration was increased (Fig. 1F). Together, these results indicated that loss of IRP function suppresses erythropoiesis, thereby decreasing iron utilization and leading to iron accumulation in the body.
Unexpectedly, IRP ablation not only altered RBC numbers but also resulted in a substantial reduction in white blood cells (WBCs; Fig. 1H), a feature not observed in mice with single IRP1 or IRP2 ablation (fig. S1); platelets counts were increased (Fig. 1H). To better define what type of WBC is affected by IRP deficiency, we analyzed the major leukocyte populations in peripheral blood (PB) by flow cytometry (FCM). P1/2-KO animals displayed a 60% decrease in the number of myeloid cells, whereas B and T cells were slightly increased (Fig. 1I). The alterations in erythrocyte and leukocyte parameters were not consequences of CRE activation, as CreER animals subjected to the same regimen were largely asymptomatic (fig. S2, A to F). Collectively, these results revealed that, in addition to their critical role in erythropoiesis, IRPs are essential for maintaining physiological levels of myeloid cells during adult life.
Loss of IRP function impairs the production of erythrocytes and granulocytes
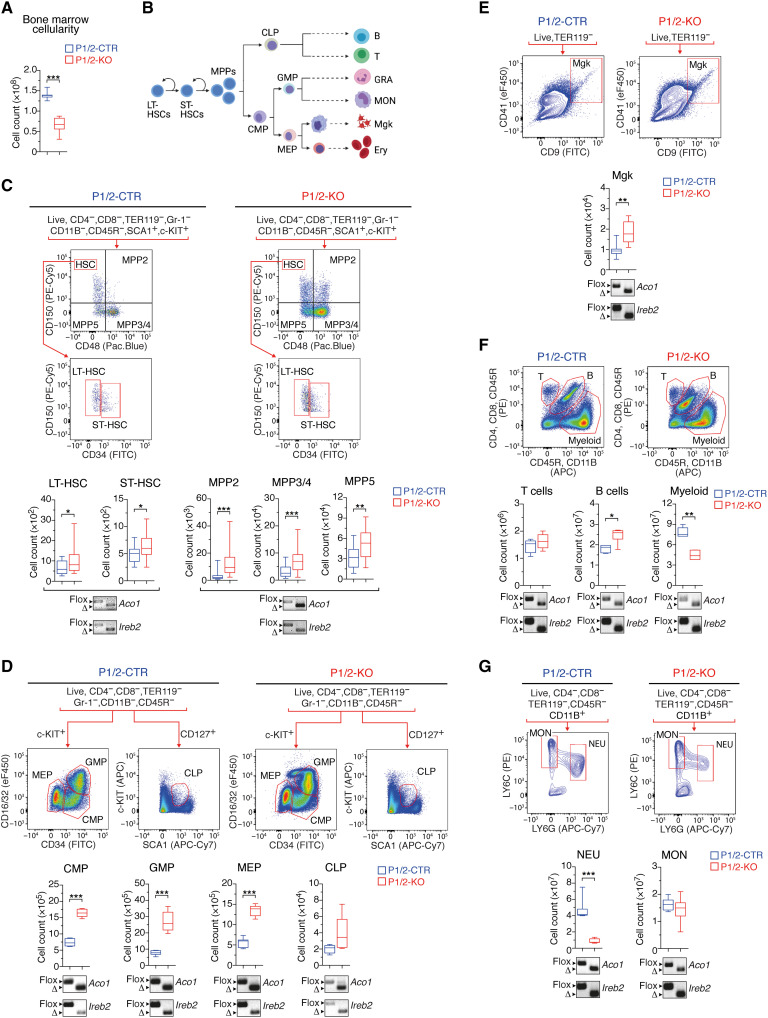
We speculated that the reduction in erythroid and myeloid cells in P1/2-KO animals might be due to impaired hematopoiesis in the BM. Consistent with this hypothesis, we observed a 50% decrease in BM cellularity in P1/2-KO versus P1/2-CTR animals (Fig. 2A). Hematopoiesis is typically depicted as a hierarchical process, in which self-renewing multipotent long-term (LT) hematopoietic stem cells (HSCs) give rise to short-term (ST)–HSCs that progress to lineage-committed progenitors with increasingly limited differentiation and self-renewal capacity, and lastly to terminally differentiated hematopoietic cells (Fig. 2B). P1/2-KO mice exhibited a clear expansion of both LT- and ST-HSCs, accompanied by an increase in all subtypes of multipotent progenitors (MPPs; Fig. 2C). The amount of common myeloid progenitor (CMP), megakaryocyte (Mgk)/erythroid progenitor (MEP), granulocyte/monocyte progenitor (GMP), and common lymphoid progenitor (CLP) was also elevated (Fig. 2D).
Fig. 2. Immuno-phenotyping of BM cell populations in adult mice with acute loss of IRP function.
(A) BM cellularity (n = 8). (B) Schematic representation of the hematopoietic system (created with BioRender.com) with self-renewing multipotent LT-HSCs giving rise to ST-HSCs that progress to lineage-committed progenitors with increasingly limited differentiation and self-renewal capacity and lastly to terminally differentiated hematopoietic cells. (C to G) FCM analysis of BM cell populations: (C) LT- and ST-HSCs (P1/2-CTR, n = 25; P1/2-KO, n = 26) and MPPs (P1/2-CTR, n = 18; P1/2-KO, n = 20); (D) CMP (n = 10), CLP (P1/2-CTR, n = 5; P1/2-KO, n = 6), MEP (n = 10), and GMP (n = 10); (E) Mgk (P1/2-CTR, n = 7; P1/2-KO, n = 9); (F) T and B cells, and myeloid cells (granulocytes/monocytes) (n = 8); (G) monocytes (MON) versus neutrophils (NEU; n = 7). The gating strategy is indicated for each cell population analyzed. Box plots (minimum to maximum values) show the number of cells in the BM of both hindlimbs. The data are presented as box plots (minimum to maximum values). Unpaired, two-tailed t test between P1/2-CTR and P1/2-KO. *P < 0.05; **P < 0.01; ***P < 0.001. Genomic PCR panels below the box plots show recombination of the Irp1 (Aco1) and Irp2 (Ireb2) alleles in cell populations isolated from the BM of P1/2-KO (right) versus P1/2-CTR (left) mice by FCM-activated cell sorting (floxed allele, flox; truncated allele, Δ). FITC, fluorescein isothiocyanate.
Despite the expansion of lineage-negative (LIN−) cells, the number of differentiated erythroid (Fig. 3A) and myeloid (Fig. 2F) cells was substantially reduced. Consistent with the PB phenotype, there was an increase in Mgk (Fig. 2E) and a slight elevation in B cells (Fig. 2F) in the BM. Hence, IRP ablation stimulates hematopoietic progenitor cells in the BM, while simultaneously suppressing erythroid and myeloid cells. CRE activation alone did not alter BM cell populations (fig. S2, G to L). To verify that the expanding LIN− cells did not escape CRE recombination, we isolated stem and progenitor cells by FACS (fluorescence-activated cell sorting) and analyzed the recombination of the Aco1 and Ireb2 alleles by genomic polymerase chain reaction (PCR). Both alleles were efficiently recombined in all cell types tested (Fig. 2, C and D). Irp genes were also efficiently inactivated in Mgk and in B and T cells (Fig. 2, E and F).
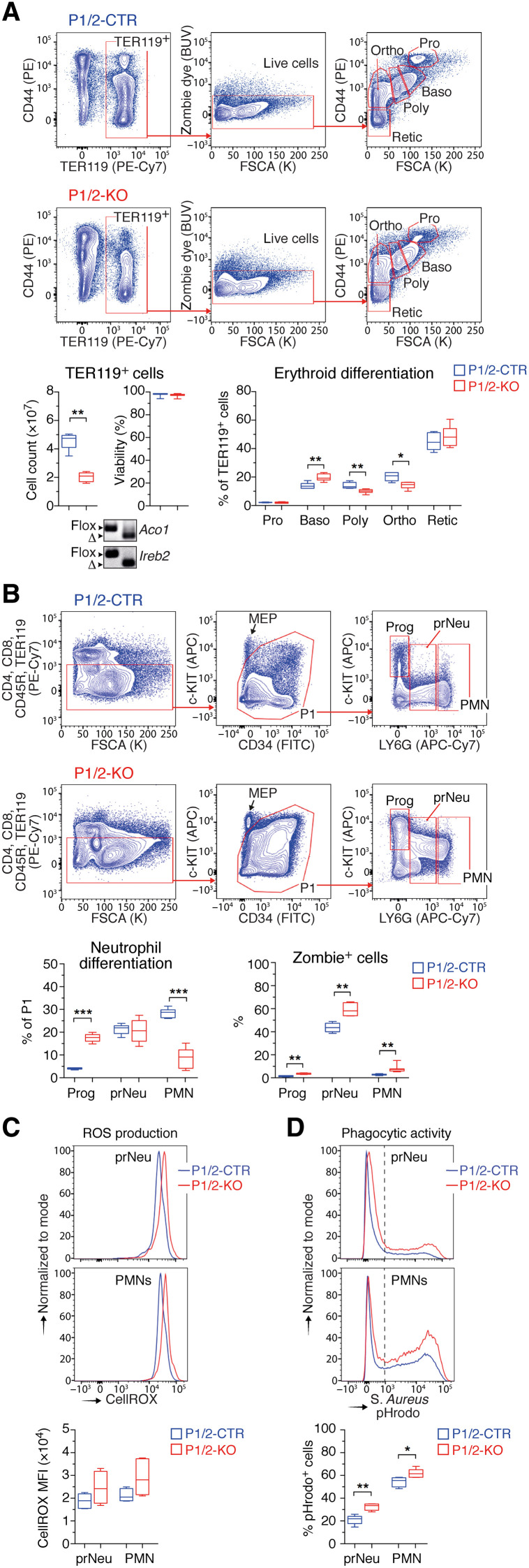
Fig. 3. IRP deficiency impairs neutropoiesis.
(A) FCM analysis of terminal erythroid differentiation (FSCA, forward side scatter area) in the BM (Pro: pro-erythroblasts; Baso, Poly, and Ortho: baso, poly-, and ortho-chromatic cells, respectively; Retic: reticulocytes). Box plots (minimum to maximum values, n = 5 to 6) show the number of TER119+ in the BM of both hindlimbs (left), the viability of TER119+ cells (middle), and the frequency of cells at different stages of erythroid differentiation (right). The panels below the box plots show the recombination of Irp1 (Aco1) and Irp2 (Ireb2) alleles in P1/2-KO (right) versus P1/2-CTR (left) mice. BUV, brilliant ultraviolet. (B) FCM analysis of neutrophil differentiation in the BM. c-KIThighLY6G− represent progenitor (Prog) cells; c-KIT−LY6Ghigh, polymorphonuclear (PMN) neutrophils; c-KIT−LY6Glow, preneutrophils (prNeu). Box plots (minimum and maximum values) show the overall frequency of Prog, prNeu, and PMN cells in the BM (left), and the corresponding fraction of Zombie dye-positive cells (right) (P1/2-CTR, n = 6; P1/2-KO, n = 7). (C and D) FCM analysis of (C) ROS (reactive oxygen species) production and (D) engulfment of pHrodo-labeled S. aureus particles. Top: Representative FCM plots. Bottom: Data obtained from five mice (box plots with minimum to maximum values). In (D), the dashed line delimits pHrodo-positive and -negative cells. (A to D) P values correspond to separate pairwise comparisons (unpaired two-tailed t test) between P1/2-CTR and P1/2-KO for each parameter and cell population analyzed (*P < 0.05; **P < 0.01; ***P < 0.001).
We next investigated whether the diminution of myeloid cells in the BM of P1/2-KO mice was confined to a specific subtype of cells. Despite efficient recombination of the floxed Irp alleles in both granulocytes and monocytes, IRP deficiency selectively suppressed neutrophils (defined as c-KIT−CD11B+LY6ClowLY6Ghigh cells), which represent the major granulocyte subfraction (Fig. 2G). Similar to neutrophils, eosinophils were also diminished to very low numbers (fig. S3), and basophils remained below the detection level. Monocytes (c-KIT−CD11B+LY6ChighLY6G−) were unchanged (Fig. 2G). Overall, the IRP/IRE system does not appear to be strictly essential for the expansion of LIN− hematopoietic progenitors in the BM; however, it is required for the efficient production of erythroid cells and granulocytes.
The IRP/IRE system is critical for neutrophil differentiation and survival in the BM
We investigated the possible causes of the low number of erythrocytes and granulocytes in IRP-mutant mice. The survival of erythroid (TER119+) cells did not appear to be compromised (Fig. 3A). Instead, we observed a defect in terminal erythroid differentiation, with impaired progression of erythroblasts from the basophilic to polychromatic stage. As erythroid differentiation is iron dependent (4), this defect could reflect a state of functional iron deficiency in P1/2-KO erythroblasts. Notably, CreER control animals exhibited normal erythropoiesis (fig. S4A).
Similarly, we examined the differentiation of neutrophils. c-KIThighLY6G− progenitors (designated Prog) progressively lose the c-KIT marker and acquire LY6G expression as they differentiate into c-KIT−LY6Ghigh polymorphonuclear neutrophils (PMNs), with an intermediate stage (designated prNeu) characterized by intermediate LY6G expression and low to undetectable c-KIT levels (Fig. 3B, top) (16). Consistent with the decrease in BM neutrophil counts (Fig. 2G), P1/2-KO animals exhibited a marked decrease in PMN numbers, associated with an accumulation of Prog cells and the presence of immature prNeu cells positive for both c-KIT and LY6G (c-KITlowLY6Ghigh; Fig. 3B). CRE activation alone had negligible effects on neutrophil differentiation compared to IRP ablation (fig. S4B). The neutropoiesis defect in P1/2-KO mice was accompanied by a slight increase in the proportion of Prog cells in the S phase of the cell cycle, associated with a reduction of cells in G0-G1, and the presence of cells able to incorporate 5-bromo-2′-deoxyuridine (BrdU) but stopped in the S phase (fig. S5). It was also associated with an augmentation of cell death (Fig. 3B), which may contribute to the low abundance of LY6G+ cells.
We examined two of the key immune functions of LY6G+ neutrophils: their capacity to produce reactive oxygen species (ROS) and to engulf foreign particles upon stimulation with lipopolysaccharide (LPS) (17). Unexpectedly, IRP deficiency did not impair ROS production (Fig. 3C) and, in fact, slightly increased the phagocytic activity of both prNeu and LY6Ghigh cells (Fig. 3D). Cumulatively, these results show that IRP deficiency impairs neutrophil differentiation and survival in the BM, yielding relatively immature neutrophils whose functionality appears to be preserved.
IRPs promote neutropoiesis in a cell intrinsic manner
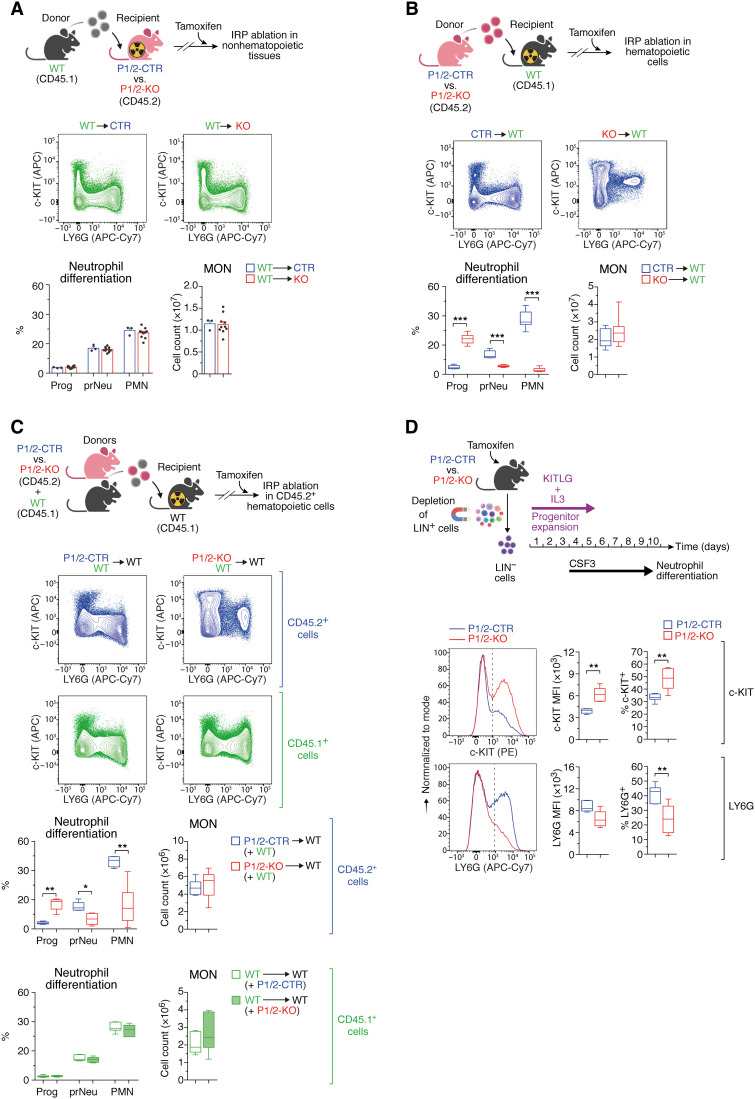
Physiological neutrophil differentiation could depend on IRP expression in cells of the hematopoietic niche or in the hematopoietic cells themselves. To distinguish between these two possibilities, we transplanted whole BM cells from WT mice into sublethally irradiated, untreated P1/2-KO recipients. Following stable engraftment, the resulting chimeras (designated WT➔KO) were treated with tamoxifen to ablate the IRPs in nonhematopoietic tissues. As a control, WT BM cells were transplanted into P1/2-CTR recipients (WT➔CTR). We found that IRP inactivation in nonhematopoietic cells does not alter neutrophil differentiation (Fig. 4A). We next generated reverse chimeras where WT recipients were transplanted with BM cells from either P1/2-KO (KO➔WT) or P1/2-CTR (CTR➔WT) mice. KO➔WT chimeras lacking IRP expression in hematopoietic cells reproduced the BM phenotype of P1/2-KO animals with systemic loss of IRP function. This included (i) the reduction in RBC indices and concomitant increase in body iron, EPO, and hepcidin levels (fig. S6, A to C); (ii) the expansion of stem/progenitor cells (fig. S6D); and (iii) the reduction in the number of PMNs in the BM, associated with the accumulation of c-KIThigh Prog cells and the presence of immature prNeus positive for both c-KIT and LY6G (Fig. 4B). Together, the hematopoietic abnormalities and particularly the defects in neutropoiesis observed in P1/2-KO animals are BM-intrinsic.
Fig. 4. IRPs promote neutropoiesis in a cell-autonomous manner.
(A) Top: CD45.1+ BM cells from WT mice transplanted into irradiated P1/2-CTR (n = 3) versus P1/2-KO (n = 10) recipients. (B) Top: CD45.2+ BM cells from P1/2-CTR (n = 9) versus P1/2-KO (n = 10) mice transplanted into WT recipients. (C) Top: WT recipients transplanted with a 1:1 mixture of BM cells from WT and either P1/2-KO (n = 6) or P1/2-CTR (n = 7) mice. (A to C) Following stable engraftment, chimeras were treated with tamoxifen on days 1 and 3 and were analyzed on day 10. Representative FCM plots based on c-KIT and LY6G markers are shown (same gating strategy as in Fig. 3B). Bar graphs (means + SEM) and box plots (minimum to maximum values) display monocyte counts in the BM of both hindlimbs and the frequency of Prog, prNeu, and PMN cells. (D) LIN− BM cells from tamoxifen-treated P1/2-CTR versus P1/2-KO mice were expanded ex vivo with KIT ligand (KITLG) and interleukin-3 (IL-3) and then differentiated into LY6G+ neutrophils with G-CSF/CSF3. Bottom: Representative FCM plot analysis of c-KIT (top) and LY6G (bottom); the dashed line delimits marker-positive and -negative cells. Box plots (minimum to maximum values, n = 7) display the median fluorescence intensity (MFI) for c-KIT and LY6G, respectively, and the percentage of cells positive for those markers. Comparisons between WT➔CTR and WT➔KO (A), P1/2-CTR(+WT)➔WT and P1/2-KO(+WT)➔WT (C), or WT➔WT(+P1/2-CTR) and WT➔WT(+P1/2-KO) (C) were made using the Mann-Whitney test. Comparisons between CTR➔WT and KO➔WT (B) were made using unpaired two-tailed t test. *P < 0.05; **P < 0.01; ***P < 0.001. (A to D) Top schemes created with BioRender.com.
To further investigate whether the neutropoietic phenotype of P1/2-KO mice is cell autonomous, we transplanted a 1:1 mixture of BM cells from WT mice (CD45.1+) and untreated P1/2-KO (CD45.2+) donors (or P1/2-CTR as control) into irradiated WT recipients (CD45.1+) and treated the mixed chimeras with tamoxifen. Hematopoietic cells derived from P1/2-KO donors show similar defects in neutropoiesis as those observed in P1/2-KO mice or KO➔WT chimeras (Fig. 4C). Furthermore, the presence of IRP-deficient cells did not affect the differentiation of precursor cells originating from WT donors (Fig. 4C).
In addition, we exposed primary cultures of hematopoietic precursor cells from tamoxifen-treated P1/2-KO versus P1/2-CTR mice to G-CSF (granulocyte colony-stimulating factor; also known as CSF3) to mimic neutrophil differentiation ex vivo (18). Similar to the neutropoiesis defect seen in vivo in P1/2-KO mice, we observed an increase in c-KIT levels and an accumulation of c-KIT+ cells to the detriment of LY6G+ cells (Fig. 4D). Notably, the effect of IRP deficiency on cell development seems neutrophil lineage–specific, as monocyte counts in the BM of transplanted mice are unaffected (Fig. 4, B and C) and the differentiation of P1/2-KO precursors into LY6C+ monocytes ex vivo is normal (fig. S7A). Together, these results demonstrate that IRPs are required cell-intrinsically for neutropoiesis.
IRPs are required to complete terminal neutrophil differentiation in the BM
To better understand how IRP deficiency alters neutrophil differentiation, we analyzed the transcriptome of Prog, prNeu, and PMN cell populations isolated from P1/2-KO versus P1/2-CTR mice. Most changes in gene expression can be accounted for by the differentiation stage of the cell, regardless of the IRP status (Fig. 5A). Nonetheless, IRP-deficient cells cluster separately from control at all stages of differentiation (Fig. 5A). We compared Prog, prNeu, and PMN cell populations in either P1/2-CTR or P1/2-KO mice and identified the differentially expressed genes (DEGs; table S1). DEGs were then grouped in four main clusters (table S2) reflecting their overall expression dynamics in P1/2-CTR cells (Fig. 5B). Consistent with the acquisition of specialized cellular functions (19), a majority of DEGs were either down-regulated (cluster 1) or up-regulated (cluster 3) during neutropoiesis (Fig. 5, B and C). Other genes exhibited a transient decrease (cluster 2) or peak (cluster 4) in expression at the intermediate differentiation stage (Fig. 5B). The expression pattern of genes coding for transcription factors required for myeloid development (20) was very similar in P1/2-KO and P1/2-CTR cells (fig. S8A). This included Cebpa (CCAAT enhancer binding protein α), which is needed for the generation of GMPs from HSCs, as well as Cebpe and Gfi1 (growth factor independent 1 transcriptional repressor), both of which are important for neutrophil development (21, 22). Likewise, the expression trajectory of genes typically associated with neutrophil differentiation, such as those encoding the antimicrobial factors contained in azurophilic granules (AGs), specific granules (SGs), and gelatinase granules (GGs), was largely preserved in P1/2-KO cells (fig. S8A). This suggests that the transcriptional programs driving neutropoiesis are not completely disrupted in IRP deficiency.
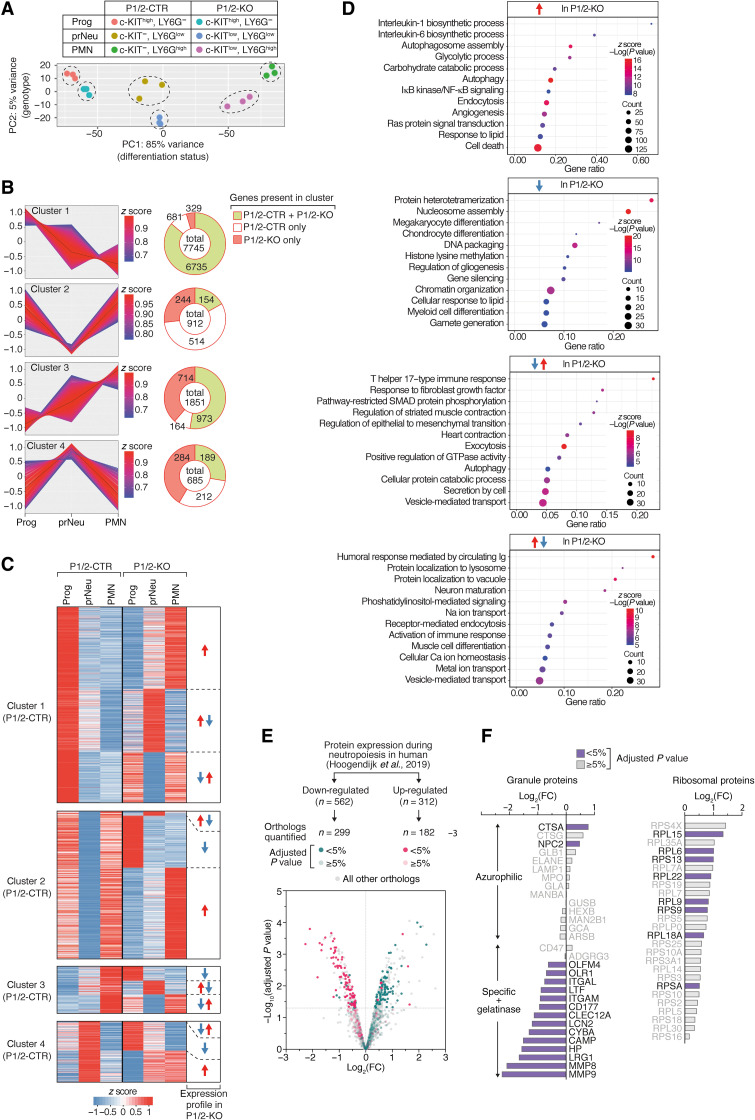
Fig. 5. Impact of IRP deficiency on gene expression dynamics during neutropoiesis.
(A to D) RNA sequencing (RNA-seq) analysis of the transcriptome of Prog, prNeu, and PMN cell populations isolated from the BM of P1/2-CTR versus P1/2-KO mice (n = 3). (A) Principal components (PC) analysis. (B) Genes differentially expressed during neutropoiesis were grouped in four main categories using k-means clustering (left). The pie charts (right) indicate (i) the total number of genes in a cluster, (ii) the number of genes found in that cluster only in P1/2-CTR (white) or in P1/2-KO (orange) cells, respectively, and (iii) the number of genes with similar expression patterns in both P1/2-CTR and P1/2-KO cells (green). (C) Heatmaps display expression profiles that differ between P1/2-CTR and P1/2-KO cells. For each cluster in P1/2-CTR cells, the expression trajectory of the corresponding genes in P1/2-KO cells is shown. (D) GO enrichment analysis of genes dysregulated during neutropoiesis in P1/2-KO cells. The dot plots show enrichment for biological processes. NF-κB, nuclear factor κB; IκB, inhibitor of nuclear factor κB; GTPase, guanosine triphosphatase. (E) Proteome analysis of whole BM LY6G+ cells from P1/2-KO versus P1/2-CTR mice (n = 3). The colors highlight proteins whose orthologs have been reported to be either down- or up-regulated during neutrophil differentiation in human BM (24). FC, fold change, P1/2-KO–P1/2-CTR. (F) Bar graphs: FC expression for selected granule (left) and ribosomal (right) proteins. In (E) and (F), the color code indicates the P value adjusted with the Benjamini-Hochberg method for multiple testing.
Sixteen percent of all DEGs displayed a distinct expression pattern in P1/2-KO cells when compared to P1/2-CTR (Fig. 5, B and C). Among those 1571 genes, nearly half (714) were up-regulated in P1/2-KO cells only, whereas 329 were down-regulated (Fig. 5B). Quantitative reverse transcription PCR analysis of selected transcripts confirmed these expression profiles and showed that those molecular features were due to IRP deficiency and not mere consequences of CRE activation (fig. S8B). Notably, the gene expression profiles in P1/2-CTR cells did not predict expression profiles in IRP-mutant cells. That is, genes belonging to a given cluster of P1/2-CTR cells were found to cluster in any of the other three groups of mutant cells (for example, the expression of genes normally up-regulated during neutrophil development may either increase or transiently peak/drop in IRP deficiency; Fig. 5C and table S2). A GO (gene ontology) enrichment analysis (table S3) revealed that genes displaying a transient increase (n = 284) or decrease (n = 244) in expression in P1/2-KO cells were related to cellular vesicles biology, secretion, immune responses, signal transduction, and cytoskeleton organization (Fig. 5D and table S3). mRNAs whose expression is abnormally down-regulated in IRP deficiency (n = 329) were mainly involved in cell differentiation programs and chromatin organization (Fig. 5D and table S3). Changes in chromatin architecture play a key role in the regulation of neutrophil differentiation genes (23), suggesting that these alterations in gene expression may reflect the inability of P1/2-KO cells to undergo normal neutrophil differentiation. Transcripts displaying increased expression in IRP deficiency (n = 714) encoded, among others, proteins involved in glycolysis and autophagy, as discussed later.
To complement our transcriptome study and assess how mRNA dysregulation in IRP-deficient neutrophils affects protein expression, we analyzed the proteome of total LY6G+ BM cells from P1/2-KO and P1/2-CTR mice. We focused on the 1942 proteins that yielded a signal in 70% of the samples in at least one of the genotypes analyzed and could thus be quantified with high confidence (table S4). We cross-referenced our data with a published dataset describing the proteomic changes that accompany the differentiation of premyelocytes toward mature neutrophils in humans (24). Of the 312 proteins reported to be up-regulated during neutropoiesis in humans, 182 mouse orthologs are present in our dataset. Most of these 182 proteins were less abundant in Ly6G+ cells from P1/2-KO mice (Fig. 5E). These proteins were mainly constituents of the SGs/GGs (Fig. 5F) that mark the final stages of neutrophil development in the BM (24). In comparison, the abundance of AG protein markers expressed during early neutropoiesis remained unchanged, suggesting that IRP deficiency impairs intermediate stages of neutrophil development. Conversely, most of the 299 proteins whose human orthologs were suppressed during neutropoiesis showed a trend toward higher expression in IRP deficiency (Fig. 5E and table S4). Several of these proteins are components of the ribosome (Fig. 5F). Biosynthetic processes usually decline as cells acquire neutrophil functions (24). Hence, the elevation of ribosomal proteins in LY6G+ cells from P1/2-KO mice is an additional sign that neutrophil differentiation is incomplete.
Overall, IRP deficiency does not seem to disrupt the core transcriptional program that drives neutropoiesis. Our data rather indicate that IRPs act in parallel with the gene regulators governing neutrophil development to facilitate the production of fully differentiated neutrophils in the BM.
IRP deficiency impairs metabolic rewiring during neutrophil development
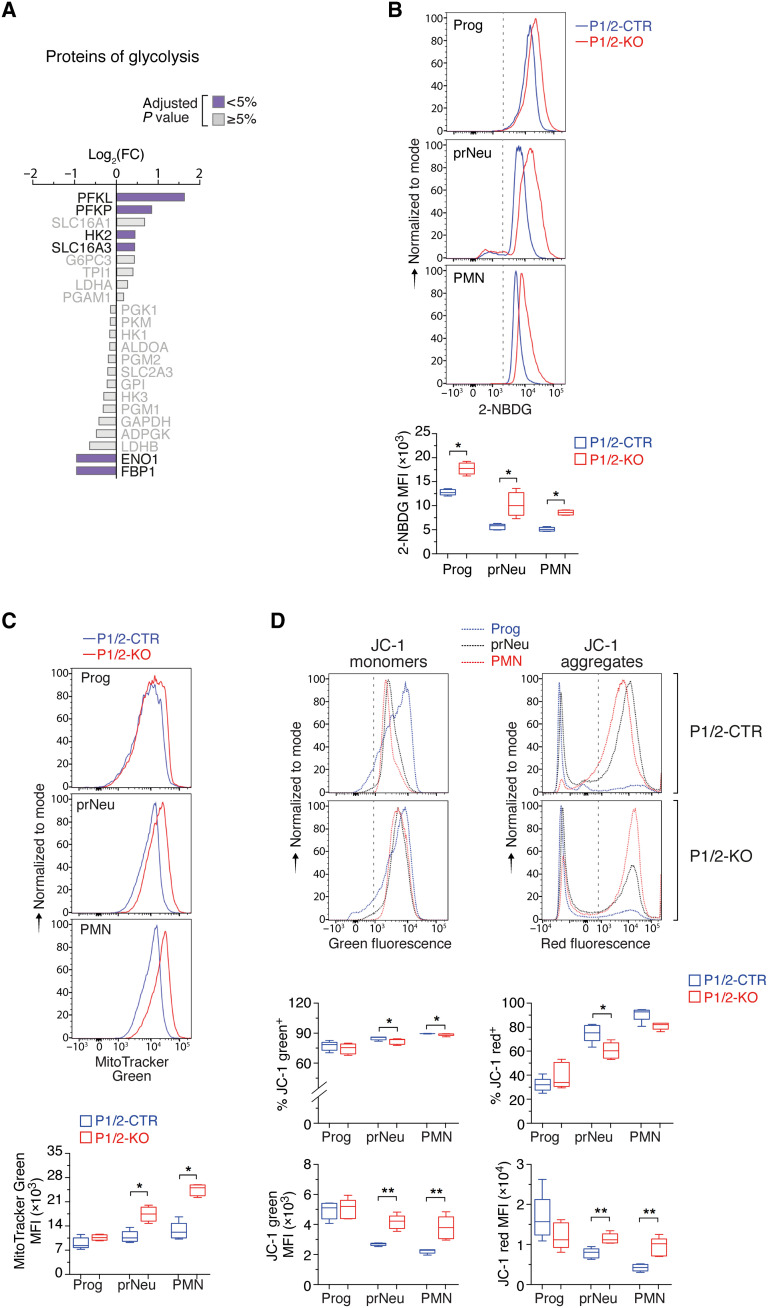
A substantial fraction of mRNAs dysregulated in IRP-deficient cells are abnormally stimulated (n = 714) during neutrophil development (Fig. 5, B and C, and table S2). These transcripts encode proteins mainly associated with GO terms related to autophagy, glucose catabolism, and cell death (Fig. 5D and table S3). The up-regulation of cell death–related genes is consistent with the increased frequency of Zombie dye+ cells in P1/2-KO mice (Fig. 3B). However, the up-regulation of genes involved in glycolysis and autophagy was unexpected, as both processes have been shown to decline during neutrophil development in the BM (16, 25). We further interrogated our proteomics data based on the idea that the protein level of rate-limiting enzymes determines the activity of metabolic pathways in the cell (26). LY6G+ cells from P1/2-KO mice expressed high levels of the phosphofructokinase isoenzymes (PFKL and PFKP), which promote glycolysis (27), and down-regulated antiglycolytic enzyme fructose bisphosphatase (FBP1; Fig. 6A). Furthermore, we observed that the uptake of the 2-deoxy-2-[(7-nitro-2,1,3-benzoxadiazol-4-yl)amino]-d-glucose (2-NBDG) glucose analog during neutrophil development was higher in P1/2-KO mice than in P1/2-CTR mice, which is further evidence for increased glycolytic flux (Fig. 6B). Similarly, FCM analysis of autophagic vacuole formation confirmed the augmentation of autophagic activity in LY6G+ cells of IRP-mutant animals (fig. S8C). RNA sequencing (RNA-seq) data indicated that the pathway “hypoxia” (fig. S8D) and the transcription factor HIF1A (table S5) were particularly active in PMN cells from P1/2-KO mice. It is possible that IRP deficiency enhances glycolysis and autophagy in LY6G+ cells, in part, through stimulation of HIF1.
Fig. 6. IRP deficiency causes abnormal metabolic remodeling during neutrophil development and differentiation.
(A) Expression of selected glycolysis proteins in whole BM LY6G+ cells (same representation as Fig. 5F). The color code indicates the P value adjusted with the Benjamini-Hochberg method for multiple testing. (B to D) FCM analysis of (B) 2-NBDG uptake, (C) mitochondrial mass (with MitoTracker green dye), and (D) mitochondrial membrane potential (MMP) (with JC-1 dye) during neutropoiesis. Top: Representative FCM plots (the dashed line separates stained from unstained cells). Bottom: Box plot with maximum to minimum MFI values [(A) and (B): P1/2-CTR, n = 5; P1/2-KO, n = 4; (D): n = 5]. Middle box plot in (D): Percentage of JC-1–positive cells (maximum to minimum values, n = 5). Prog, prNeu, and PMN cell populations were gated using the same strategy as in Fig. 2. P values (Mann-Whitney) correspond to separate pairwise comparisons between P1/2-CTR and P1/2-KO for each parameter and cell population analyzed. *P < 0.05; **P < 0.01.
We also noticed “response to lipids” as a GO term associated with genes that are either up- or down-regulated in IRP deficiency (Fig. 5D and table S3). FCM analysis with the BODIPY 493/503 dye revealed the accumulation of neutral lipids at all stages of neutrophil differentiation (fig. S8E). This suggests that IRP deficiency may impair lipid metabolism or transport or trigger lipid droplet formation as a protective measure (28).
Mitochondria in fully differentiated blood neutrophils are present in small numbers and serve to control cell survival rather than to produce energy (24). However, in the BM, adequate neutrophil differentiation is dependent on a metabolic shift that limits glycolysis while increasing mitochondrial content and respiration (16). Mitochondria use iron to produce energy via heme and ISC proteins (9). Mitochondrial integrity has been shown to be compromised in IRP-deficient hepatocytes (29). Furthermore, iron deprivation in deferoxamine (DFO)-treated cells was shown to selectively down-regulate components of the electron transport chain (ETC), more particularly ISC proteins present in complexes I and II (30). We, therefore, hypothesized that IRP ablation might alter neutropoiesis by reducing the content and/or activity of mitochondria in hematopoietic cells, resulting in a decrease in mitochondrial proteins.
Our proteomics data revealed a strong enrichment of GO terms related to the cellular component mitochondria, suggesting changes in mitochondrial protein abundance (table S6). Of the proteins listed in the mouse MitocCarta database (31), 225 could be quantified in LY6G+ cells. Most of these proteins were expressed at a higher level in P1/2-KO cells compared to P1/2-CTR cells and accounted for as much as one-fourth of all proteins significantly up-regulated in IRP deficiency (fig. S8F and table S4). In contrast, only two mitochondrial proteins were significantly reduced in P1/2-KO cells (4% of all down-regulated proteins). Our transcriptome data also did not reveal the ETC signature previously reported for DFO-treated cells (30). Hence, contrary to expectations, the mitochondrial content of LY6G+ cells appeared to be increased in IRP deficiency. Accordingly, FCM analysis with MitoTracker green dye revealed that the mitochondrial mass is markedly augmented in prNeu and PMN cells from P1/2-KO mice (Fig. 6C).
To test whether IRP deficiency could lead to the accumulation of unhealthy mitochondria, we analyzed the mitochondrial membrane potential (MMP) using the mitochondrial dye JC-1. JC-1 monomers emit green fluorescence in cells with low MMP, but JC-1 forms aggregates producing orange/red fluorescence in cells with high MMP; changes in the intensity of red versus green signal reflect changes in the cell’s MMP (32). In P1/2-CTR mice, the proportion of JC-1 red+ cells increased from the Prog to PMN stage, while the ratio of red to green signal intensity was globally preserved (Fig. 6D). Similarly, the percentage of JC-1 red+ cells in P1/2-KO mice increased during neutropoiesis, albeit to a slightly lesser extent at the prNeu stage (Fig. 6D). Both red and green signal intensities were elevated in prNeu and PMN cells (Fig. 6D), possibly reflecting the expansion of mitochondrial mass and corresponding increase in JC-1 dye uptake. However, we did not detect a notable gain in green versus red fluorescence intensity, which could have indicated membrane depolarization. Acute IRP ablation thus appeared to increase the amount of apparently healthy mitochondria during neutropoiesis. Together, these data suggest that IRP deficiency interferes with the metabolic remodeling that accompanies neutrophil development and differentiation in the BM, possibly reflecting a situation of stress characterized by high glycolytic and autophagic activities, lipid accumulation, and expansion of the mitochondrial content of the cell.
The IRP/IRE network supports neutropoiesis by securing iron bioavailability
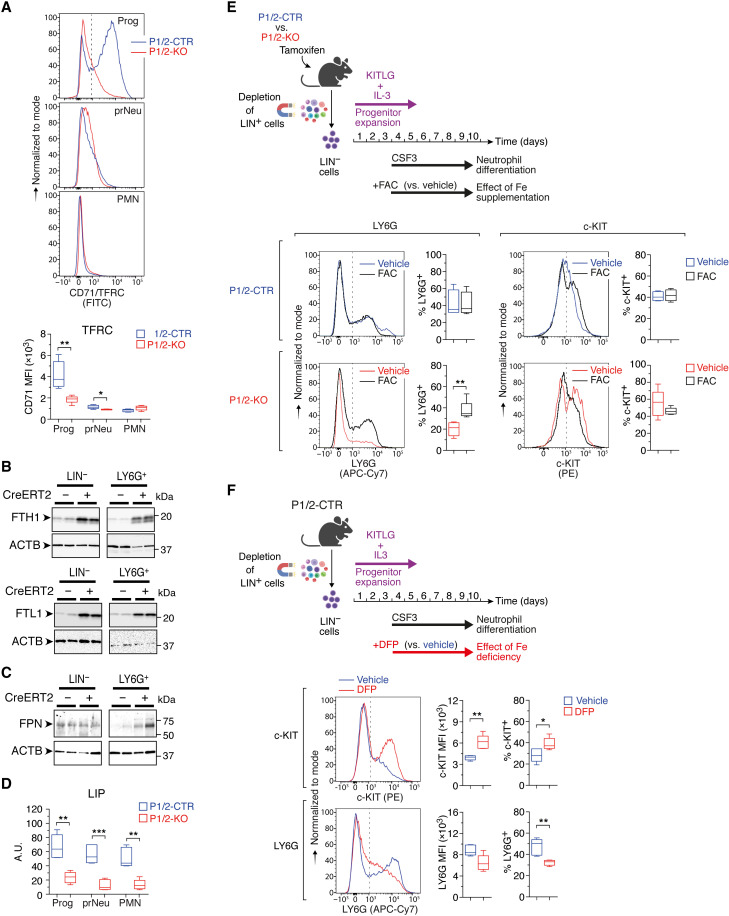
A transcriptome-wide search for IRP-interacting mRNAs (33) and recent in cellulo RNA foot-printing data (34) suggest that the IRP regulome extends beyond iron metabolism genes. This raised the question of whether the neutropoiesis defect of IRP-deficient mice is caused by alterations in iron metabolism or iron-independent IRP functions. We therefore examined how IRP deficiency affects the cellular iron status during neutropoiesis. IRPs normally inhibit the decay of Tfrc and Dmt1 mRNAs and hamper the translation of Ftl1, Fth1, and Fpn transcripts. Accordingly, Prog cells from P1/2-KO mice exhibited a strong decrease in TFRC expression compared with P1/2-CTR; TFRC levels in LY6Ghigh cells remained below detection regardless of genotype (Fig. 7A). IRP ablation did not affect Dmt1, most likely because the cells predominantly express the variant of Dmt1 mRNA that lacks the 3′IRE (fig. S9).
Fig. 7. IRPs support neutrophil differentiation by securing iron bioavailability.
(A) FCM analysis of CD71/TFRC expression during neutropoiesis in vivo. Top: Representative FCM plots. Bottom: Box plot with maximum to minimum MFI values (P1/2-CTR, n = 5; P1/2-KO, n = 6). Prog, prNeu, and PMN cell populations were gated using the same strategy as in Fig. 2. (B and C) Western blot analysis of ferritin (B) and FPN (C) expression in lineage negative (LIN−) versus LY6G+ cells from the BM of P1/2-CTR (CreERT2−) versus P1/2-KO (CreERT2+) mice. Loading control, ACTB (actin-β). (D) Box plot (maximum to minimum values) showing reduced LIP in BM cells of P1/2-KO versus P1/2-CTR mice during neutropoiesis in vivo (P1/2-CTR, n = 5; P1/2-KO, n = 6). (E and F) Top schemes (created with BioRender.com): Ex vivo differentiation of LIN− BM cells as in Fig. 4D, in the presence of either (E) FAC or (F) DFP; control cells were treated with vehicle. Bottom: Representative FCM plots showing LY6G and c-KIT levels, together with box plots (maximum to minimum values) displaying the MFI (F) or percentage of cells positive (E and F) for either marker [(E): P1/2-CTR, n = 4; P1/2-KO, n = 5; (F): n = 5]. The dashed lines delineate marker-positive and -negative cells. Comparisons were made between P1/2-CTR and P1/2-KO (A and D), vehicle and FAC (E), or vehicle and DFP (F). P values (Mann-Whitney test), *P < 0.01; **P < 0.01; ***P < 0.001.
Conversely, ferritin expression was higher in IRP deficiency in both LIN− and LY6G+ cells (Fig. 7B). FPN expression was elevated in LY6G+ cells, but not in progenitor cells (Fig. 7C). Down-regulation of TFRC together with stimulation of the ferritin iron sequestering molecules and FPN iron exporter (in LY6G+ cells) is predicted to lower cellular iron availability.
P1/2-KO BM cells showed a marked reduction in the metabolically active labile iron pool (LIP) during neutropoiesis compared to P1/2-CTR cells (Fig. 7D). A similar reduction in the LIP was observed in P1/2-KO cells undergoing ex vivo differentiation (fig. S10). These observations prompted us to test whether an extra source of iron could restore normal neutropoiesis. For this, we used our ex vivo neutrophil differentiation assay and exposed cells from tamoxifen-treated P1/2-KO mice to ferric ammonium citrate (FAC), a form of iron that cells can take up independently of TFRC (Fig. 7E, top). Compared to vehicle, P1/2-KO cells treated with FAC (10 μg/ml) differentiated more efficiently, whereas the same amount of FAC had no effect on cells from P1/2-CTR mice (Fig. 7E). These results strongly suggest that IRP ablation impairs neutropoiesis due to iron shortage.
As a complementary approach to the iron rescue experiment, we exposed cells from untreated P1/2-CTR mice to the iron chelator deferiprone (DFP; Fig. 7F, top). DFP treatment mimicked the effect of IRP ablation (Fig. 4D), with stimulation of c-KIT expression and accumulation of c-KIT+ cells at the expense of LY6G+ cells (Fig. 7F). In contrast to neutrophils, DFP stimulated the differentiation of hematopoietic progenitors toward LYC6+ monocytes (fig. S7B). This indicates that neutrophil differentiation is particularly sensitive to iron deprivation. In vivo, chronic treatment with Ferriprox, a DFP oral formulation, had only a minor effect on steady-state neutropoiesis (fig. S11, A to E). This could potentially be explained by relatively modest iron deprivation in BM cells compared to other cells, as the compound appears to have uneven effects on iron. DFP could deplete the liver iron store very effectively but had no measurable effect on spleen iron (fig. S11B). Nevertheless, Ferriprox altered neutrophil differentiation when mice were additionally treated with CSF2/G-CSF (in the form of Filgrastim/Neupogen) to mimic conditions of emergency granulopoiesis, with a significant increase in c-KIT levels (fig. S11, C and D) and accumulation of immature prNeu cells at the expense of LY6Ghigh PMN cells (fig. S11E). This reinforces the notion that iron bioavailability influences neutrophil differentiation in the BM.
DISCUSSION
Most of iron in the human body is contained in hemoglobin and dedicated to oxygen transport by erythrocytes (1). While the importance of iron for RBC formation is undisputed, its role in other facets of blood cell biology is only beginning to be recognized, with notably recent reports showing a role for the metal in adaptive immunity (35–37). Through genetic disruption of a key iron homeostatic machinery in mice, our work revealed that maintenance of cellular iron bioavailability is critical for granulocyte development and differentiation in the BM, establishing a previously underappreciated link between iron metabolism and a major arm of the innate immune system.
Studies in mice demonstrated that the IRP/IRE regulatory network is essential during early embryonic development (11). The hepatocytic and intestinal functions of the IRP/IRE system are also essential during perinatal life (11). Moreover, constitutive loss of both IRP1 and IRP2 in the heart or macrophages, respectively, remains asymptomatic under standard laboratory conditions (38, 39). These observations suggest that the IRP/IRE system exerts particularly important functions during pre- and perinatal life. Here, we used a temporally controlled CRE system enabling acute disruption of IRP1 and/or IRP2 in nearly the entire body of adult mice. Two evident short-term manifestations of acute IRP ablation in adult mice were microcytic anemia and neutropenia. The neutropenia was not observed in mice harboring a single loss of either IRP1 or IRP2, therefore reflecting overlapping functions between the two proteins. The anemia, on the other hand, was attributable to IRP2 deficiency and corroborates previous findings in standard mouse models with constitutive loss of IRP2 function (11). In this study, we focused on the role of the IRPs in hematopoiesis, but acute IRP deficiency may have additional effects yet to be investigated. Future work may uncover a role for the IRP/IRE network in other organs, at different stages of adult life (e.g., during aging), or in response to challenge.
Because of the importance of iron in oxygen transport, research on iron metabolism has focused primarily on the biology of RBCs. The role of the metal in overall hematopoiesis and, in particular, HSCs remains poorly understood. Iron homeostasis can affect HSCs in different manners. Iron overload due to ineffective erythropoiesis or RBC transfusion in patients with hematological diseases is known to impair HSC functions (40). Iron excess can be detrimental to cells of the hematopoietic niche (41, 42); it can also damage the HSCs themselves, as illustrated by the stem cell exhaustion phenotype of FBXL5-null mice with selective iron accumulation in HSCs (43). However, the impact of iron deficiency on HSCs is less clear. Iron deprivation with DFO diminished the proliferation and survival of hematopoietic progenitors in an in vitro model of mouse embryonic hematopoiesis (44). Whereas iron chelation in human and mouse HSCs with eltrombopag was shown to stimulate stem cell self-renewal and multilineage hematopoiesis (45). Our study revealed that hematopoietic stem/progenitor cells can expand in the absence of a fully functional IRP/IRE system and withstand the ensuing cellular iron deficiency, at least over a short period of time. Although we cannot exclude that IRP ablation may impair the stem/progenitor cell compartment in the long term, these data suggested that the iron requirements of hematopoietic stem/progenitor cells are relatively modest. Similarly, IRP-mediated iron homeostasis was dispensable for lymphoid development in the upper part of the hematopoietic hierarchy, at least during the time frame of the study. This may reflect the fact that the flow of MPPs to the lymphoid lineage is low compared to the erythro-myeloid lineage, as shown by in vivo fate mapping (46). This does not imply, however, that iron is not important for lymphoid compartment expansion at later stages of differentiation and iron availability has been shown to be critical during lymphocyte activation (37, 47).
Both RBCs and platelets are derived from MEPs. While IRP ablation suppressed RBCs, it caused an increase in platelet counts. The expansion of Mgk in the BM of IRP-mutant mice may reflect increased commitment of MEPs toward the Mgk lineage under conditions of iron deficiency (48). Mgk might also bypass the MEP progenitors and arise directly from the enlarged MPP2 population (49).
The most profound and unexpected effect of acute IRP ablation was the substantial decrease in neutrophils. This effect was, in part, due to functional iron deficiency in hematopoietic cells. While iron overload and high heme levels are known to impair the function of mature neutrophils (50, 51), how iron scarcity specifically affects neutrophils is not well understood (52). Cases of neutropenia associated with iron deficiency and reversed upon iron therapy have been reported (53, 54). In addition, DFP, an iron chelator mainly used to treat iron overload in patients with thalassemia, has been shown to cause neutropenia via a yet unclear mechanism (55). Although the cases are rare, such a side effect can increase the risk and severity of infection (56). Accordingly, we found that DFP limits neutrophil differentiation ex vivo. In vivo, DFP could alter neutrophil development upon pharmacological stimulation of neutropoiesis with G-CSF. Under the experimental conditions used in our study, the effect of DFP in mice was rather mild compared to the profound granulopoiesis defect seen in IRP-mutant animals, possibly due to insufficient access of the compound to the iron pool of BM cells.
IRP ablation suppressed granulocytes in vivo but did not reduce the abundance of monocytes, the other major constituent of the myeloid compartment. This was remarkable because both cell types are derived from GMPs, are relatively short-lived, and reside only briefly in the BM (57). We found that DFP promotes the differentiation of hematopoietic stem/progenitor cells into monocytes ex vivo. This is consistent with previous observations showing a positive effect of iron deprivation on the differentiation of cord blood cells and leukemic cell lines toward the monocytic lineage (58). Iron deficiency thus impairs granulopoiesis but, at the same time, appears to favor monopoiesis, suggesting that iron bioavailability may be critical for shaping the innate defense system. Assessing the differential effect of iron on granulopoiesis versus monopoiesis and determining which specific differentiation node is modulated by iron availability will be exciting questions to explore.
Iron metabolism is not needed to initiate neutropoiesis per se, as the expression trajectory of granule genes and transcription factors important for neutrophil differentiation is globally preserved in IRP deficiency. Iron rather operates in parallel to the gene regulators that drive neutropoiesis to enable the production of fully differentiated neutrophils. A similar role of metabolism has been reported for autophagy. Differentiating neutrophils mobilize cellular lipids through lipophagy to fuel fatty acid oxidation, thereby promoting a shift from glycolysis to oxidative phosphorylation that is critical for neutrophil differentiation (16). IRP ablation led to high glycolytic and autophagic activity, lipid droplet accumulation, and increased mitochondrial content, indicating that the metabolic rewiring that accompanies neutrophil differentiation cannot operate normally in IRP deficiency. Among other possibilities, elevated glycolysis and high mitochondrial content could be a compensatory response to an energy stress, whereas lipid droplets could form because of defective fatty acid oxidation or to avoid lipotoxicity during autophagy (28). To what extent these metabolic alterations may contribute to the neutropoietic defect of P1/2-KO mice and how they are linked to iron metabolism are not clear. Hundreds of proteins in the cell bind iron either as heme, ISC, or iron ions (59), and hence, multiple iron-dependent pathways may act together to support neutropoiesis. Future research should aim at deciphering the exact mechanism through which IRPs and iron promote neutrophil differentiation and survival in the BM.
Overall, our study reinforces the notion that iron metabolism influences multiple aspects of hematopoiesis and plays important roles in shaping the immune system. We showed that IRP-mediated iron homeostasis is critical for neutrophil development in the BM. This finding could be relevant, for instance, in the context of infection/inflammation-induced hypoferremia. This process of iron sequestration in iron stores, which is considered a mean for the host to restrict access of iron to circulating siderophilic pathogens, can limit the supply of iron to erythroblasts and cause anemia (2, 3). Our study suggests that acute inflammatory hypoferremia may also alter neutrophil development in the BM, which could potentially compromise host defenses. The influence of iron on neutropoiesis may also have implications for G-CSF–based therapies in patients receiving myelosuppressive antineoplastic treatments or BM transplants, as our study suggests that the iron status of the patient and/or transplanted cells may affect the efficacy of the treatment.
MATERIALS AND METHODS
Mouse models
Husbandry
The mouse lines carrying floxed Aco1 (Aco1tm1.1Mwh, MGI:3613495) and Ireb2 (Ireb2tm1.1Mwh, MGI:3613523) alleles and the Gt(ROSA)26Sortm1(cre/ERT)Nat deletor strain (JAX #004847) have been described (13, 14). Aco1flox/floxIreb2flox/floxRosa26+/CreERT2 (P1/2-KO) and Aco1flox/floxIreb2flox/floxRosa26+/+ (P1/2-CTR) male littermates derived from several breeding pairs were used in the study (to avoid cage effects, mice were not grouped according to genotype). P1/2-CTR and P1/2-KO mice were housed randomly in the same cage.
To control for potential effects of CRE, Aco1+/+Ireb2+/+Rosa26+/CreERT2 (CreER) and WT Aco1+/+Ireb2+/+Rosa26+/+ littermates were also analyzed. To generate mixed chimeras (see below), CD45.1+ BM cells were obtained from Ly5.1, B6 Cd45.1, B6.SJL-Ptprca Pepcb/BoyJ (JAX #002014) donors. Mice treated with Ferriprox (see below) were obtained from JANVIER Laboratories. Male mice were used throughout the study.
All mice were congenic on a homogenous C57BL6/J genetic background and were housed under specific pathogen–free and light-, temperature (21°C)–, and humidity (50 to 60% relative humidity)–controlled conditions. Food (diet #3437 from KLIBA NAFAG, containing around 200 parts per million of iron) and water were available ad libitum. Blood was collected by cardiac puncture after deep anesthesia of the mice with a ketamine/xylazine mixture. If not used immediately, then tissues were flash-frozen in liquid nitrogen and stored at −80°C.
Treatments
To induce CRE activity during adulthood, 8- to 12-week-old male mice were injected intraperitoneally with a low dose of tamoxifen [40 mg/kg of body weight in a 1:9 ethanol (EtOH)–sunflower seed oil mix, all from Sigma-Aldrich] on days 1 and 3. Mice in which recombination of Irp alleles clearly failed (assessed by genomic PCR analysis of whole BM samples, see below) were excluded from the study. To decrease body iron availability in adult animals, 8-week-old C57BL6/J WT mice received DFP (Ferriprox from Apotex Europe B.V.) in the drinking water at a dose of 0.4 μg/kg of body weight per day for a period of 2 months. They were subsequently treated daily with G-CSF (Neupogen from Amgen) at a dose of 250 μg/kg of body weight on three consecutive days to stimulate granulopoiesis; the animals were euthanized 1 day after the last G-CSF injection for analysis. To label proliferating cells, the mice were injected (intraperitoneally) with BrdU (BD Biosciences) at a dose of 50 μg/kg of body weight 2 hours before euthanasia.
BM transplantation and generation of chimeric mice
The mice were exposed to a total of 10 grays (in two doses separated by a 3- to 4-hour interval) from a 137Cs source. Following a 2-hour recovery period, the mice were injected (intravenously) with 6 × 106 total BM cells from at least two donor animals. As a prophylactic measure, the transplanted animals were treated with carprofen (subcutaneously, 5 mg/kg of body weight) once a day for the first 3 days and received sulfamethoxazole/trimethoprim (90 mg/kg per day) in the drinking water for 3 weeks. They were additionally given soft food. To assess engraftment efficiency, small blood samples were collected from the submandibular vein into EDTA tubes (Sarstedt) once a month, and the samples were analyzed by FCM as described below. Successful engraftment was typically obtained 12 weeks after transplantation.
Ethical statement
Animal care, husbandry, and killing were performed according to national guidelines and were approved by an institutional review board headed by the local animal welfare officers. Animal experiments were carried out according to project licenses G-270/18 and G-21/21, as approved by the Regiergrungspräsidium of Karlsruhe (Baden-Württemberg, Germany).
Hematology and serum parameters
Blood profiles and hemoglobin content were determined using an ABC Vet apparatus (HORIBA ABX SAS). Serum samples were prepared using Z-gel–containing microvette tubes (Sarstedt). EPO concentration was determined with the Mouse Erythropoietin Quantikine ELISA (enzyme-linked immunosorbent assay) Kit (MEP00B, Bio-Techne). Hepcidin was measured using a Hepcidin Murine-Compete ELISA kit (HMC-001, Intrinsic LifeSciences). Haptoglobin and hemopxin levels were analyzed using the Mouse Haptoglobin (ab157714) and Hemopexin (ab157716) ELISA kits from Abcam. Serum concentration of iron, ferritin, and transferrin was determined at the “Centre de Recherche sur l’Inflammation” (Paris, France) using an Olympus 400 analyzer. Samples were assigned noninformative codes during measurement. Samples partially coagulated (blood profiles) or hemolyzed (serum parameters) were excluded from the analysis.
Tissue non-heme iron levels
Iron was extracted from dried tissues in 10% tricarboxylic acid, 10% HCl, and iron concentration was determined against an iron atomic absorption standard solution (305952, Merck) using the bathophenanthroline chromogene procedure (60). Samples were assigned noninformative codes during preparation and analysis.
DNA analysis
DNA was isolated using the Extracta DNA Prep Kit (Quantabio). The genomic status of the Aco1 and Ireb2 alleles was analyzed by PCR using PCR Master Mix reagents (Merck) together with primers specific for Aco1 (forward 1, 5′-TACTGTAGCAAAAATGCTTTGTCTCTG-3′; forward 2, 5′-GTCATTTTTCTCATTTCTTGAGCATTAG-3′; reverse, 5′-TCTATCCCTGAGGTCGGTAGGC-3′) or Ireb2 (forward 1, 5′-CTGAAAGACTGACCCTTCTTGTTC-3′; forward 2, 5′-TGAGTGGTGCCTGCATTTTAAG-3′; reverse, 5′-GGCTTCAATAGTCTTCATACCACG-3′). Amplicon sizes are as follows: Aco1 allele, WT 243 base pairs (bp), floxed 296 bp, truncated 214 bp; Ireb2 allele, WT 245 bp, floxed 288 bp, truncated 199 bp.
RNA extraction
Total RNA from sorted Prog, prNeu, and PMN cell populations was extracted using the Arcturus PicoPure RNA Isolation Kit (KIT0204, Thermo Fisher Scientific) and treated with ribonuclease-free deoxyribonuclease I (DNAse I) (79254, QIAGEN) to eliminate genomic DNA. RNA concentration was measured with a Qubit Assay (Thermo Fisher Scientific), and RNA integrity was assessed using an RNA 6000 pico kit (5067-1513) and Bioanalyzer apparatus from Agilent (Agilent Technologies).
Quantitative reverse transcription polymerase chain reaction
Total RNA was reverse-transcribed using the SuperScript VILO Master Kit (11754050, Thermo Fisher Scientific), and RNA levels were determined on a CFX Connect Real-Time PCR System (denaturation at 95°C for 10 min, followed by 45 cycles of 15 s at 95°C, 15 s at 60°C, and 15 s at 72°C) using the iTaq Universal SYBR Green Supermix (1725125, Bio-Rad Laboratories) together with assays from Bio-Rad Laboratories (Ubxn4, qMmuCID0023115; Psmc2, qMmuCIP0034703; Abcg1, qMmuCID0005670; Bhlhe40, qMmuCID0013865; Clec7a, qMmuCID0013926; Egln3, qMmuCID0019903; Mylip, qMmuCID0010376; Pfkl, qMmuCID0018472; Pfkp, qMmuCID0023075; Slc2a1, qMmuCED0026836) or self-made assays for Gusb (forward, 5′-AAAATCACCCTGCGGTTGT-3′; reverse, 5′-TGTGGGTGATCAGCGTCTT-3′) and Ppib (forward, 5′-GGAGATGGCACAGGAGGA-3′; reverse, 5′-GGTGTCTTTGCCTGCATTG-3′). For each assay, primer efficiency (E) was determined using a serial dilution of a mix of cDNA from a pool of all samples analyzed. Semiquantification of a gene of interest (GOI) was done using the ΔΔCt method after calibration to the average expression of four standard genes (Std) as indicated in figure legends. The expression fold change was calculated as [EGOI*(ΔCt GOI)]/[EStd*(ΔCt Std)].
RNA sequencing
Ultralow RNA-seq libraries were prepared from 2.85 ng of input RNA using the SMART-Seq v4 Ultra Low Input RNA Kit for Sequencing (R400752, Takara Bio Europe SAS), combined with shearing on a Covaris apparatus (Covaris Ltd., Brighton, UK) and the NEBNext ChIP-Seq Library Prep Master Mix Set for Illumina (New England Biolabs) according to the manufacturer’s protocol. The libraries were quality-controlled using an Agilent 4200 Tape Station System (Agilent Technologies) and a Qubit ds DNA HS Assay kit (Thermo Fisher Scientific). On the basis of Qubit quantification and sizing, libraries were normalized, pooled, and clustered on a cBot system (Illumina) with a final concentration of 250 pM (spiked with 1% PhiX control v3 from Illumina). Fifty–base pair single-read sequencing was performed on a HiSeq 4000 instrument (Illumina) using standard protocols. Samples were assigned noninformative codes during library preparation and sequencing.
Demultiplexed Fastq files were checked for quality with the FastQC program (www.bioinformatics.babraham.ac.uk/projects/fastqc/) and mapped against the mm10 mouse genome using the STAR (Spliced Transcripts Alignment to a Reference) package (https://github.com/alexdobin/STAR/releases) to generate bam files (61). Sequencing reads were annotated against the GRCm38.p5 reference gene set from Ensembl (www.ensembl.org) and counted using the htseq-count package (62). DEGs were identified with the DESeq2 package (www.bioconductor.org/packages/release/bioc/html/DESeq2.html) (63), using the Benjamini-Hochberg adjustment for false discovery rate (FDR) calculation (64). Principal components analysis (PCA) plot was generated using the plotPCA function of the DESeq2 package. The splicing pattern of Dmt1 was visualized using the Sashimi plot function of the Integrative Genomics Viewer software (https://software.broadinstitute.org/software/igv/).
Western blotting
Magnetically sorted LIN− versus LY6G+ cells were lysed in a radioimmunoprecipitation assay buffer [50 mM tris-HCl (pH 7.5), 150 mM NaCl, 1% Triton X-100, 0.5% Na deoxycholate, 0.1% SDS, and 1 mM dithiothreitol] supplemented with the Halt Protease (87786) and Halt Phosphatase (78427) Inhibitor Cocktails (Thermo Fisher Scientific). The samples were incubated on ice for 30 min with occasional vortexing, and the debris was pelleted at 10,000g for 10 min at 4°C. Protein concentration in the supernatant was determined using the Pierce BCA protein Assay Kit (Thermo Fisher Scientific). Equal amounts of protein were mixed with Laemmli sample buffer, resolved onto 4 to 15% Criterion TGX midi protein gels, and transferred onto polyvinylidene difluoride (PVDF) membranes using a Trans-Blot Turbo 0.2-μm PVDF transfer pack together with a Trans-Blot Turbo transfer system (all from Bio-Rad Laboratories). The membranes were incubated in tris-buffered saline with 0.1% Tween 20 containing 5% (w/v) powder milk and primary antibodies against FPN (MTP11-A from Alpha Diagnostics, RRID: AB_1619475), FTL1 (clone EPR18878, ab69090 from Abcam, RRID: AB_1523609), FTH1 (ab183781, Abcam), or ACTB (clone AC-15, A1978 from Sigma-Alrich, RRID: AB_476692). Immune complexes were visualized using the Clarity Western ECL substrate (Bio-Rad Laboratories) together with a Chemocam Imager system equipped with the ChemoStar Imager software (INTAS Science Imaging Instruments GmbH).
Proteomics analysis
An amount of 10 μg of protein per condition was used for a short SDS–polyacrylamide gel electrophoresis separation (0.5 cm). After Commassie staining, the total protein sample was excised and subjected to trypsin digestion on a DigestPro MSi robotic system (CEM GmbH). The liquid chromatography–tandem mass spectrometry (MS/MS) analysis was performed on an Ultimate 3000 UPLC system directly connected to an Orbitrap Exploris 480 mass spectrometer (both from Thermo Fisher Scientific). Peptides were desalted on an Acclaim PepMap300 C18 trap cartridge (5 μm, 300-Å-wide pore from Thermo Fisher Scientific) for 3 min using a flow (30 μl/min) of 0.05% trifluoroacetic acid in water. The analytical multistep gradient was carried out on a nanoEase MZ Peptide analytical column (300 Å, 1.7 μm, 75 μm by 200 mm) from Waters GmbH using 0.1% formic acid in water as solvent A and 0.1% formic acid, 80% acetonitrile in water as solvent B. Solvent B concentration was linearly increased from 2 to 38% during 134 min, followed by a quick raise to 95% for 2 min. Solvent B concentration was then lowered to 2% and a 10-min equilibration step was added. Eluting peptides were analyzed in the mass spectrometer using a data depend acquisition mode. A full scan at 60,000 resolution [380 to 1400 mass/charge ratio (m/z), 300% AGC target, and 45-ms maxIT] was followed by up to 2 s of MS/MS scans. Peptide features were isolated with a window of 1.4 m/z, fragmented using 26% normalized collision energy. Fragment spectra were recorded at 15,000 resolution (100% automatic gain control target, 54-ms maxIT). Unassigned and singly charged eluting features were excluded from fragmentation, and dynamic exclusion was set to 35 s. Samples were assigned noninformative codes during preparation and analysis.
Data analysis was carried out by MaxQuant version 1.6.14.0 (www.maxquant.org/maxquant/) (65) using an organism-specific database extracted from Uniprot.org under default settings. Identification FDR cutoffs were 0.01 on peptide level and 0.01 on protein level. Match between runs option was enabled but restricted to transfer peptide identifications across RAW files based on accurate retention time and mass/charge ratio only within replicates.
Protein quantification was done using a label-free quantification (LFQ) approach based on the MaxLFQ algorithm (66). A minimum of two quantified peptides per protein was required for protein quantification.
Flow cytometry
Cell preparation and surface marker staining
To analyze BM cells, the femur, tibiae, and ilia were disinfected with 70% EtOH and crushed in Iscove’s modified Dulbecco medium (IMDM) (Thermo Fisher Scientific) using a mortar and pestle. The BM cell suspension obtained was filtered through a 40-μm cell strainer (Greiner Bio-One), and nucleated cells were counted using a Hemavet apparatus (Drew Scientific). Following a phosphate-buffered saline (PBS) wash, cells were stained for 20 min at room temperature with a Zombie dye (423108, BioLegend) to discriminate live versus dead cells. For surface marker staining, cells were washed in PBS and subsequently incubated for 20 min at 4°C in PBS containing 2% fetal bovine serum (FBS; Thermo Fisher Scientific) and antibody mixes as listed in table S7 (panels P1 to P21). To get rid of red cells in prior analysis, the samples were incubated in ACK red cell lysis buffer (882090, Biozym Scientific GmbH) for 10 min at room temperature and then washed in PBS. For ex vivo differentiation experiments (see below), the cells were washed and stained as described above, using the antibody panels P22 to P24 (table S7); DAPI (4′,6-diamidino-2-phenylindole; D1306, Thermo Fisher Scientific) was added to the cells shortly before FCM. The samples were analyzed on an LSRFortessa device (BD Biosciences) and the data processed using the Flow-Jo software v.10 from Tree Star.
Reticulocyte counts in blood
PB cells were washed and resuspended in PBS + 2% FBS containing anti-CD45 and anti-TER119 antibodies (panel P8, table S7) and were incubated for 30 min on ice. Following a PBS wash, the cells were resuspended in PBS + 2% FBS complemented with thiazol orange (1.7 μg/ml; 390062, Sigma-Aldrich) to stain the residual RNA present in reticulocytes. After a 30-min incubation on ice, the cells were directly analyzed by FCM.
Assessment of BM cell engraftment
PB cells (collected from the submandibular vein) were incubated for 20 min on ice in PBS + 2% FBS containing fluorochrome-conjugated antibodies against leukocyte antigens and the CD45.1/CD45.2 polymorphism markers (panels P5 and P6, table S7). In prior analysis, RBCs were eliminated using the ACK buffer as described above.
Cell cycle analysis
BM cells were incubated with antibodies against cell surface markers (panel P16, table S7) and were additionally stained for BrdU using the Fluorescein Isothiocyanate BrdU Flow Kit from BD Biosciences (559619; RRID: AB_2617060) according to the manufacturer’s instructions. Briefly, cells were washed in PBS + 2% FBS, fixed/permeabilized in cytofix/cytoperm solution, washed with Perm/Wash, and incubated with the anti-BrdU antibody overnight at 4°C. DNase treatment was used to expose the BrdU epitope. To determine the DNA content, BrdU-stained cells were incubated with DAPI.
Labile iron pool
BM cells washed in PBS were incubated for 1 hour at 37°C in serum-free IMDM (21980032, Thermo Fisher Scientific) supplemented with 5 μM BioTracker Far-Red Labile Ferrous Iron Dye (or FerroFarRed, SCT037, Merck KGaA). To analyze the LIP during neutrophil differentiation ex vivo, cells were collected at day 10 and spun at 300g for 5 min and incubated in IMDM + 5 μM BioTracker Far-Red Labile Ferrous Iron Dye for 1 hour at 37°C. Cells were then washed in PBS and stained for cell surface markers (panel P14 for BM cells and panel P24 for cells differentiated ex vivo, table S7) as described above.
Neutral lipids
BM cells washed in PBS were incubated 30 min at 37°C in IMDM containing 1 μM fluorescent neutral lipid dye BODIPY 493/503 (4,4-difluoro-1,3,5,7,8-pentamethyl-4-bora-3a,4a-diaza-s-indacene; D3922, Thermo Fisher Scientific). Cells were then washed in PBS and stained for cell surface markers (panel P15, table S7) as described above.
Autophagy
LIN− and LY6G+ cells were magnetically isolated as described below. The sorted cells were washed and then incubated for 30 min at 37°C in assay buffer containing 1:1000 of the CYTO-ID dye (ENZ-51031-0050, Enzo Life Sciences GmbH), a cationic amphiphilic tracer that labels autophagic vacuoles. The cells were analyzed by FCM after a PBS wash.
2-NBDG glucose uptake
BM cells washed in PBS were incubated for 1 hour at 37°C in glucose-free RPMI containing 100 μM 2-NBDG (1860768, PeproTech). Cells were then washed in PBS and stained for cell surface markers (panel P17, table S7) as described above.
ROS production
BM cells in serum-free IMDM were stimulated with LPS (L4641, Sigma-Aldrich) at a concentration of 2.5 μg/ml. After a 1-hour incubation at 37°C, the cells were washed in PBS, pelleted at 350g for 5 min, and then resuspended in serum-free IMDM containing 5 μM CellROX green reagent (C10444, Thermo Fisher Scientific). The cells were then incubated for 20 min at 37°C. Cells were washed in PBS and stained for cell surface markers (panel 20, table S7) as described above. ROS detection was validated using cells treated with tert-butyl hydroperoxide as positive control.
Phagocytic activity
BM cells were incubated for 1 hour at 37°C in serum-free IMDM in the presence of LPS (2.5 μg/ml) and pHrodo Green Staphylococcus aureus Bioparticles Conjugate (P35367, Thermo Fisher Scientific) at a 1:10 ratio. The cells were then washed in PBS, centrifuged at 350g for 5 min, washed again in PBS, and stained for cell surface markers (panel 21, table S7) as described above. As controls, the assay was performed either in the presence of 10 μM cytochalasin D (sc-201442, Santa Cruz Biotechnology) to block F-actin polymerization and phagocytosis or with cells incubated on ice.
Mitochondrial status
BM cells were washed in PBS and pelleted. Cells were then incubated at 37°C for 45 min in IMDM containing 200 nM MitoTracker Green dye (M7514, Thermo Fisher Scientific) to assess mitochondrial mass. To analyze the MMP, the cells were incubated at 37°C for 15 min in IMDM supplemented with JC-1 reagent (ENZ-52304, Enzo Life Sciences) at a concentration of 10 μg/ml. After a PBS wash, the cells were stained for surface markers (panels P18 and P19, table S7) as described above. To control for JC-1 red fluorescence specificity, cells were pretreated for 45 min with 250 nM depolarizing agent FCCP (carbonyl cyanide p-trifluoromethoxyphenylhydrazone; C2920, Sigma-Aldrich).
Fluorescence-activated cell sorting
BM cells were prepared and stained with antibodies against cell surface markers as described above for standard FCM. The cell populations of interest were isolated using a FACSAria Cell Sorter (BD Biosciences). A fraction of the sorted cells was reanalyzed by FCM to ascertain the purity (typically >90%) of the cell populations obtained. For DNA and RNA extraction, respectively, the sorted cells were collected directly into the corresponding lysis buffers.
Magnetic cell sorting
BM cell suspensions were prepared as described for FCM analysis. Hematopoietic progenitor cells were isolated using the Direct Lineage Depletion Kit for mouse (130-110-470, Miltenyi Biotec). A total of 2 × 107 BM cells in 80 μl of MACS buffer (PBS + 0.5% FBS + 2 mM EDTA) were incubated for 10 min at 4°C with 20 μl of microbeads conjugated to antibodies against various differentiation markers. The labeled cells were then loaded onto LS columns (Miltenyi Biotec) pre-equilibrated with MACS buffer and placed on a QuadroMACS Separator (Miltenyi Biotec). The flow through containing progenitor cells was collected for downstream applications.
To isolate LY6G+ cells, 107 BM cells in 90 μl of MACS buffer were incubated for 10 min at 4°C with 10 μl of Anti-LY6G MicroBeads UltraPure for mouse (130-120-337, Miltenyi Biotech). The cells were then loaded onto LS columns to eliminate the LY6G− cells present in the flow through. The columns were subsequently washed with MACS buffer, and LY6G+ cells were eluted in the same buffer.
Cell culture
BM progenitor cells were isolated using the Direct Lineage Depletion Kit as described earlier. The cells were seeded at a density of 2 × 105 cells/ml in progenitor growth medium (PGM) containing IMDM, 10% heat-inactivated and low endotoxin FBS (VWR International), 1% penicillin/streptomycin (Thermo Fisher Scientific), and 5% l-glutamine (VWR International). To enable the maintenance and expansion of progenitor cells, the culture medium was supplemented with interleukin-3 (IL-3) (213-13, PeproTech) and stem cell factor (SCF; also known as KITL) (250-03, PeproTech). Cells were incubated at 37°C in 5% CO2. Following 3 days of expansion, the cells were cryopreserved in 90% FBS + 10% dimethyl sulfoxide for later use.
To study cell differentiation, progenitor cells were thawed in PGM containing IL-3 + SCF and cultivated for 2 to 3 days in six-well plates. After this recovery phase, the cells were seeded in 12-well plates and grown in the same medium for another 3 days. To initiate neutrophil differentiation, the cells were washed in PBS and cultivated for 2 days in PGM containing IL-3 + SCF and complemented with G-CSF (250-05, PeproTech). For the final differentiation phase, the cells were incubated for another 5 days in PGM containing G-CSF only. To trigger differentiation toward monocytes, the cells were first grown for 3 days in PGM containing IL-3 + SCF. They were then incubated for 2 days in the presence of macrophage CSF (M-CSF) (315-02, PeproTech) and granulocyte/macrophage CSF [GM-CSF (315-03)] and lastly cultivated for 5 days in the presence of M-CSF only. Cell density was maintained at 2 × 105 cells/ml throughout the first 5 days of culture, both for neutrophil and monocyte differentiation. All cytokines were used at a final concentration of 50 ng/ml.
To study the effect of iron complementation on the differentiation of P1/2-KO progenitor cells, FAC (F5879, Sigma-Aldrich) was added to the culture medium during the G-CSF + IL-3 and G-CSF alone treatment periods. To study the consequences of iron deprivation on P1/2-CTR cell differentiation, DFP (379409, Sigma-Aldrich) was added to the culture medium also during the G-CSF + IL-3 and G-CSF exposure phases. Control cells not subjected to iron treatments were exposed to vehicle.
Statistical analysis of RNA-seq data
Three biological replicates were used for each genotype. Protein coding genes with a log2(fold change) of >1 or <−1 (FDR, 10%) in any of the pairwise comparisons tested (PMN versus Pro, PrNeu versus Pro, PMN versus PrNeu in P1/2-CTR samples and separately in P1/2-KO) were selected for subsequent analysis. For each gene, TPM (transcripts per million) values across the three cell populations tested were scaled to generate z scores. The z scores were then used to define specific expression patterns across the Pro, prNeu, and PMN differentiation stages (i.e., during differentiation), and the genes were grouped in four distinct clusters using the k-means method with the Hartigan-Wong algorithm (67). Genes whose expression in P1/2-KO cells did not follow the same pattern as in P1/2-CTR were subsequently analyzed for GO-term enrichment using the topGO package (DOI: 10.18129/B9.bioc.topGO) and were plotted using the ggplot2 package (Bioconductor, https://CRAN.R-project.org/package=ggplot2). Heatmaps were generated on scaled expression values using the heatmap.2 function of the ggplot2 package. Transcription factor and pathway activities were analyzed on the basis of the Wald statistic values (log2FC/SE) of the DEGs identified with DESeq2, using the weighted mean function of the decoupleR statistic tool (68) coupled with, respectively, the PROGENy (69) and DoRothEA (70) R packages.
Statistical analysis of proteomics data
The statistical analysis (three biological replicates per genotype) was performed with the R package “limma” (71). The LFQ (65) values were used for the statistical analysis. Adapted from the Perseus (www.maxquant.org/perseus/) recommendations (72), protein groups with nonzero intensity values in 70% of the samples of at least one of the conditions were used, and for missing values being completely absent under one condition, imputation with random values drawn from a downshifted (2.2 SD) and narrowed (0.3 SD) intensity distribution of the individual sample was applied. For missing values with no complete absence under one condition, the R package missForest was used for imputation (73). The P values were adjusted with the Benjamini-Hochberg method for multiple testing (64).
Perseus (version 1.6.15.0) was used for mouse protein annotation with GO and Kyoto Encyclopedia of Genes and Genomes and one-dimensional (1D) enrichment analysis (72). The 1D enrichment was performed with a Benjamini-Hochberg FDR of 0.02. The 1D enrichment on log fold changes accepting changes to both sides and the 1D enrichment on adjusted P values accepting only changes to lower.
Statistical analysis of other data
The values are presented as box plots with minimum to maximum values, or as means ± SEM, as indicated in figure legends. The normality of two independent groups of data was first assessed using the D’Agostino-Pearson test and then compared using either a two-tailed unpaired Student’s t test or a Mann-Whitney test. In all statistical analyses, the null hypothesis was rejected for P values of less than 0.05. All analyses were performed using the Prism application (v9) from GraphPad (GraphPad Software). Throughout the manuscript, the sample size (n) refers to biological replicates.
Acknowledgments
We thank the Center for Preclinical Research and the Flow Cytometry and the Genomics and Proteomics core facilities of the DKFZ for support. We wish to thank M. W. Hentze (EMBL) for sharing the floxed Irp mouse lines and A. Ruggieri (Heidelberg University) for critical reading of the manuscript.
Funding: This work was supported by a grant from the Deutsche Forschungsgemeinschaft (GA 2075/6-1) to B.G.
Author contributions: M.B. and B.G designed the research. M.B., E.T., C.K., M.Q., G.P., M.P.-S., W.R., A.E., N.A., and B.G performed the experiments. M.B., S.A., M.S., D.H., R.B., M.D.M., and B.G analyzed the data. M.B. and B.G. wrote and reviewed the manuscript with the help of all co-authors.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. RNA-seq data have been deposited in the ArrayExpress database at EMBL-EBI (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-10663. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (www.ebi.ac.uk/pride) with the dataset identifier PXD032077.
Supplementary Materials
This PDF file includes:
Figs. S1 to S11
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S7
REFERENCES AND NOTES
- 1.Pasricha S. R., Tye-Din J., Muckenthaler M. U., Swinkels D. W., Iron deficiency. Lancet 397, 233–248 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Ganz T., Anemia of inflammation. N. Engl. J. Med. 381, 1148–1157 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Haschka D., Hoffmann A., Weiss G., Iron in immune cell function and host defense. Semin. Cell Dev. Biol. 115, 27–36 (2021). [DOI] [PubMed] [Google Scholar]
- 4.Muckenthaler M. U., Rivella S., Hentze M. W., Galy B., A red carpet for iron metabolism. Cell 168, 344–361 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoshinaga M., Nakatsuka Y., Vandenbon A., Ori D., Uehata T., Tsujimura T., Suzuki Y., Mino T., Takeuchi O., Regnase-1 maintains iron homeostasis via the degradation of transferrin receptor 1 and prolyl-hydroxylase-domain-containing protein 3 mRNAs. Cell Rep. 19, 1614–1630 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Corral V. M., Schultz E. R., Eisenstein R. S., Connell G. J., Roquin is a major mediator of iron-regulated changes to transferrin receptor-1 mRNA stability. iScience 24, 102360 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang H., Shi H., Rajan M., Canarie E. R., Hong S., Simoneschi D., Pagano M., Bush M. F., Stoll S., Leibold E. A., Zheng N., FBXL5 regulates IRP2 stability in iron homeostasis via an oxygen-responsive [2Fe2S] cluster. Mol. Cell 78, 31–41.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rouault T. A., Maio N., Biogenesis and functions of mammalian iron-sulfur proteins in the regulation of iron homeostasis and pivotal metabolic pathways. J. Biol. Chem. 292, 12744–12753 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ward D. M., Cloonan S. M., Mitochondrial iron in human health and disease. Annu. Rev. Physiol. 81, 453–482 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moroishi T., Nishiyama M., Takeda Y., Iwai K., Nakayama K. I., The FBXL5-IRP2 axis is integral to control of iron metabolism in vivo. Cell Metab. 14, 339–351 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Wilkinson N., Pantopoulos K., The IRP/IRE system in vivo: Insights from mouse models. Front. Pharmacol. 5, 176 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Santos M. C. F., Anderson C. P., Neschen S., Zumbrennen-Bullough K. B., Romney S. J., Kahle-Stephan M., Rathkolb B., Gailus-Durner V., Fuchs H., Wolf E., Rozman J., de Angelis M. H., Cai W. M., Rajan M., Hu J., Dedon P. C., Leibold E. A., Irp2 regulates insulin production through iron-mediated Cdkal1-catalyzed tRNA modification. Nat. Commun. 11, 296 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galy B., Ferring D., Hentze M. W., Generation of conditional alleles of the murine iron regulatory protein (IRP)-1 and −2 genes. Genesis 43, 181–188 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Badea T. C., Wang Y., Nathans J. A., A noninvasive genetic/pharmacologic strategy for visualizing cell morphology and clonal relationships in the mouse. J. Neurosci. 23, 2314–2322 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hameyer D., Loonstra A., Eshkind L., Schmitt S., Antunes C., Groen A., Bindels E., Jonkers J., Krimpenfort P., Meuwissen R., Rijswijk L., Bex A., Berns A., Bockamp E., Toxicity of ligand-dependent Cre recombinases and generation of a conditional Cre deleter mouse allowing mosaic recombination in peripheral tissues. Physiol. Genomics 31, 32–41 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Riffelmacher T., Clarke A., Richter F. C., Stranks A., Pandey S., Danielli S., Hublitz P., Yu Z., Johnson E., Schwerd T., McCullagh J., Uhlig H., Jacobsen S. E. W., Simon A. K., Autophagy-dependent generation of free fatty acids is critical for normal neutrophil differentiation. Immunity 47, 466–480.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Németh T., Sperandio M., Mócsai A., Neutrophils as emerging therapeutic targets. Nat. Rev. Drug Discov. 19, 253–275 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Pelletier M. G. H., Szymczak K., Barbeau A. M., Prata G. N., O’Fallon K. S., Gaines P., Characterization of neutrophils and macrophages from ex vivo-cultured murine bone marrow for morphologic maturation and functional responses by imaging flow cytometry. Methods 112, 124–146 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grassi L., Pourfarzad F., Ullrich S., Merkel A., Were F., Carrillo-de-Santa-Pau E., Yi G., Hiemstra I. H. L., Tool A. T. J., Mul E., Perner J., Janssen-Megens E., Berentsen K., Kerstens H., Habibi E., Gut M., Yaspo M. L., Linser M., Lowy E., Datta A., Clarke L., Flicek P., Vingron M., Roos D., van den Berg T. K., Heath S., Rico D., Frontini M., Kostadima M., Gut I., Valencia A., Ouwehand W. H., Stunnenberg H. G., Martens J. H. A., Kuijpers T. W., Dynamics of transcription regulation in human bone marrow myeloid differentiation to mature blood neutrophils. Cell Rep. 24, 2784–2794 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Evrard M., Kwok I. W. H., Chong S. Z., Teng K. W. W., Becht E., Chen J., Sieow J. L., Penny H. L., Ching G. C., Devi S., Adrover J. M., Li J. L. Y., Liong K. H., Tan L., Poon Z., Foo S., Chua J. W., Su I. H., Balabanian K., Bachelerie F., Biswas S. K., Larbi A., Hwang W. Y. K., Madan V., Koeffler H. P., Wong S. C., Newell E. W., Hidalgo A., Ginhoux F., Ng L. G., Developmental analysis of bone marrow neutrophils reveals populations specialized in expansion, trafficking, and effector functions. Immunity 48, 364–379.e8 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Lawrence S. M., Corriden R., Nizet V., The ontogeny of a neutrophil: Mechanisms of granulopoiesis and homeostasis. Microbiol. Mol. Biol. Rev. 82, e00057-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ai Z., Udalova I. A., Transcriptional regulation of neutrophil differentiation and function during inflammation. J. Leukoc. Biol. 107, 419–430 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Manley H. R., Keightley M. C., Lieschke G. J., The neutrophil nucleus: An important influence on neutrophil migration and function. Front. Immunol. 9, 2867 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoogendijk A. J., Pourfarzad F., Aarts C. E. M., Tool A. T. J., Hiemstra I. H., Grassi L., Frontini M., Meijer A. B., van den Biggelaar M., Kuijpers T. W., Dynamic transcriptome-proteome correlation networks reveal human myeloid differentiation and neutrophil-specific programming. Cell Rep. 29, 2505–2519.e4 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Kumar S., Dikshit M., Metabolic insight of neutrophils in health and disease. Front. Immunol. 10, 2099 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahl P. J., Hopkins R. A., Xiang W. W., Au B., Kaliaperumal N., Fairhurst A. M., Connolly J. E., Met-flow, a strategy for single-cell metabolic analysis highlights dynamic changes in immune subpopulations. Commun. Biol. 3, 305 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanner L. B., Goglia A. G., Wei M. H., Sehgal T., Parsons L. R., Park J. O., White E., Toettcher J. E., Rabinowitz J. D., Four key steps control glycolytic flux in mammalian cells. Cell Syst. 7, 49–62.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olzmann J. A., Carvalho P., Dynamics and functions of lipid droplets. Nat. Rev. Mol. Cell Biol. 20, 137–155 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galy B., Ferring-Appel D., Sauer S. W., Kaden S., Lyoumi S., Puy H., Kölker S., Gröne H. J., Hentze M. W., Iron regulatory proteins secure mitochondrial iron sufficiency and function. Cell Metab. 12, 194–201 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Rensvold J. W., Ong S. E., Jeevananthan A., Carr S. A., Mootha V. K., Pagliarini D. J., Complementary RNA and protein profiling identifies iron as a key regulator of mitochondrial biogenesis. Cell Rep. 3, 237–245 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rath S., Sharma R., Gupta R., Ast T., Chan C., Durham T. J., Goodman R. P., Grabarek Z., Haas M. E., Hung W. H. W., Joshi P. R., Jourdain A. A., Kim S. H., Kotrys A. V., Lam S. S., McCoy J. G., Meisel J. D., Miranda M., Panda A., Patgiri A., Rogers R., Sadre S., Shah H., Skinner O. S., To T. L., Walker M. A., Wang H., Ward P. S., Wengrod J., Yuan C. C., Calvo S. E., Mootha V. K., MitoCarta3.0: An updated mitochondrial proteome now with sub-organelle localization and pathway annotations. Nucleic Acids Res. 49, D1541–D1547 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Troiano L., Ferraresi R., Lugli E., Nemes E., Roat E., Nasi M., Pinti M., Cossarizza A., Multiparametric analysis of cells with different mitochondrial membrane potential during apoptosis by polychromatic flow cytometry. Nat. Protoc. 2, 2719–2727 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Sanchez M., Galy B., Schwanhaeusser B., Blake J., Bähr-Ivacevic T., Benes V., Selbach M., Muckenthaler M. U., Hentze M. W., Iron regulatory protein-1 and -2: Transcriptome-wide definition of binding mRNAs and shaping of the cellular proteome by iron regulatory proteins. Blood 118, e168–e179 (2011). [DOI] [PubMed] [Google Scholar]
- 34.Corley M., Flynn R. A., Lee B., Blue S. M., Chang H. Y., Yeo G. W., Footprinting SHAPE-eCLIP reveals transcriptome-wide hydrogen bonds at RNA-protein interfaces. Mol. Cell 80, 903–914.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jabara H. H., Boyden S. E., Chou J., Ramesh N., Massaad M. J., Benson H., Bainter W., Fraulino D., Rahimov F., Sieff C., Liu Z. J., Alshemmari S. H., Al-Ramadi B. K., Al-Dhekri H., Arnaout R., Abu-Shukair M., Vatsayan A., Silver E., Ahuja S., Davies E. G., Sola-Visner M., Ohsumi T. K., Andrews N. C., Notarangelo L. D., Fleming M. D., Al-Herz W., Kunkel L. M., Geha R. S., A missense mutation in TFRC, encoding transferrin receptor 1, causes combined immunodeficiency. Nat. Genet. 48, 74–78 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang Y., Li C., Wu Q., An P., Huang L., Wang J., Chen C., Chen X., Zhang F., Ma L., Liu S., He H., Xie S., Sun Y., Liu H., Zhan Y., Tao Y., Liu Z., Sun X., Hu Y., Wang Q., Ye D., Zhang J., Zou S., Wang Y., Wei G., Liu Y., Shi Y., Chin Y. E., Hao Y., Wang F., Zhang X., Iron-dependent histone 3 lysine 9 demethylation controls B cell proliferation and humoral immune responses. Nat. Commun. 10, 2935 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frost J. N., Tan T. K., Abbas M., Wideman S. K., Bonadonna M., Stoffel N. U., Wray K., Kronsteiner B., Smits G., Campagna D. R., Duarte T. L., Lopes J. M., Shah A., Armitage A. E., Arezes J., Lim P. J., Preston A. E., Ahern D., Teh M., Naylor C., Salio M., Gileadi U., Andrews S. C., Dunachie S. J., Zimmermann M. B., van der Klis F. R. M., Cerundolo V., Bannard O., Draper S. J., Townsend A. R. M., Galy B., Fleming M. D., Lewis M. C., Drakesmith H., Hepcidin-mediated hypoferremia disrupts immune responses to vaccination and infection. Med 2, 164–179.e12 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haddad S., Wang Y., Galy B., Korf-Klingebiel M., Hirsch V., Baru A. M., Rostami F., Reboll M. R., Heineke J., Flögel U., Groos S., Renner A., Toischer K., Zimmermann F., Engeli S., Jordan J., Bauersachs J., Hentze M. W., Wollert K. C., Kempf T., Iron-regulatory proteins secure iron availability in cardiomyocytes to prevent heart failure. Eur. Heart J. 38, 362–372 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Nairz M., Ferring-Appel D., Casarrubea D., Sonnweber T., Viatte L., Schroll A., Haschka D., Fang F. C., Hentze M. W., Weiss G., Galy B., Iron regulatory proteins mediate host resistance to salmonella infection. Cell Host Microbe 18, 254–261 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weber S., Parmon A., Kurrle N., Schnütgen F., Serve H., The clinical significance of iron overload and iron metabolism in myelodysplastic syndrome and acute myeloid leukemia. Front. Immunol. 11, 627662 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu W., Zhao M., Rajbhandary S., Xie F., Chai X., Mu J., Meng J., Liu Y., Jiang Y., Xu X., Meng A., Free iron catalyzes oxidative damage to hematopoietic cells/mesenchymal stem cells in vitro and suppresses hematopoiesis in iron overload patients. Eur. J. Haematol. 91, 249–261 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Tanaka H., Espinoza J. L., Fujiwara R., Rai S., Morita Y., Ashida T., Kanakura Y., Matsumura I., Excessive reactive iron impairs hematopoiesis by affecting both immature hematopoietic cells and stromal cells. Cells 8, 226 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muto Y., Nishiyama M., Nita A., Moroishi T., Nakayama K. I., Essential role of FBXL5-mediated cellular iron homeostasis in maintenance of hematopoietic stem cells. Nat. Commun. 8, 16114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shvartsman M., Bilican S., Lancrin C., Iron deficiency disrupts embryonic haematopoiesis but not the endothelial to haematopoietic transition. Sci. Rep. 9, 6414 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kao Y. R., Chen J., Narayanagari S. R., Todorova T. I., Aivalioti M. M., Ferreira M., Ramos P. M., Pallaud C., Mantzaris I., Shastri A., Bussel J. B., Verma A., Steidl U., Will B., Thrombopoietin receptor-independent stimulation of hematopoietic stem cells by eltrombopag. Sci. Transl. Med. 10, eaas9563 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Höfer T., Busch K., Klapproth K., Rodewald H. R., Fate mapping and quantitation of hematopoiesis in vivo. Annu. Rev. Immunol. 34, 449–478 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Teh M. R., Frost J. N., Armitage A. E., Drakesmith H., Analysis of iron and iron-interacting protein dynamics during T-cell activation. Front. Immunol. 12, 714613 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xavier-Ferrucio J., Scanlon V., Li X., Zhang P. X., Lozovatsky L., Ayala-Lopez N., Tebaldi T., Halene S., Cao C., Fleming M. D., Finberg K. E., Krause D. S., Low iron promotes megakaryocytic commitment of megakaryocytic-erythroid progenitors in humans and mice. Blood 134, 1547–1557 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Psaila B., Mead A. J., Single-cell approaches reveal novel cellular pathways for megakaryocyte and erythroid differentiation. Blood 133, 1427–1435 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martins R., Maier J., Gorki A. D., Huber K. V. M., Sharif O., Starkl P., Saluzzo S., Quattrone F., Gawish R., Lakovits K., Aichinger M. C., Radic-Sarikas B., Lardeau C. H., Hladik A., Korosec A., Brown M., Vaahtomeri K., Duggan M., Kerjaschki D., Esterbauer H., Colinge J., Eisenbarth S. C., Decker T., Bennett K. L., Kubicek S., Sixt M., Superti-Furga G., Knapp S., Heme drives hemolysis-induced susceptibility to infection via disruption of phagocyte functions. Nat. Immunol. 17, 1361–1372 (2016). [DOI] [PubMed] [Google Scholar]
- 51.Renassia C., Louis S., Cuvellier S., Boussetta N., Deschemin J. C., Borderie D., Bailly K., Poupon J., Dang P. M. C., El-Benna J., Manceau S., Lefrère F., Vaulont S., Peyssonnaux C., Neutrophils from hereditary hemochromatosis patients are protected from iron excess and are primed. Blood Adv. 4, 3853–3863 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abuga K. M., Muriuki J. M., Williams T. N., Atkinson S. H., How severe anaemia might influence the risk of invasive bacterial infections in African children. Int. J. Mol. Sci. 21, 6976 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abuirmeileh A., Bahnassi A., Abuirmeileh A., Unexplained chronic leukopenia treated with oral iron supplements. Int. J. Clin. Pharm. 36, 264–267 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abdelmahmuod E., Yassin M. A., Ahmed M., Iron deficiency anemia-induced neutropenia in adult female. Cureus 12, e8899 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kontoghiorghes G. J., Kleanthous M., Kontoghiorghe C. N., The history of deferiprone (L1) and the paradigm of the complete treatment of iron overload in thalassaemia. Mediterr. J. Hematol. Infect. Dis. 12, e2020011 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tricta F., Uetrecht J., Galanello R., Connelly J., Rozova A., Spino M., Palmblad J., Deferiprone-induced agranulocytosis: 20 years of clinical observations. Am. J. Hematol. 91, 1026–1031 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yona S., Kim K. W., Wolf Y., Mildner A., Varol D., Breker M., Strauss-Ayali D., Viukov S., Guilliams M., Misharin A., Hume D. A., Perlman H., Malissen B., Zelzer E., Jung S., Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity 38, 79–91 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Callens C., Coulon S., Naudin J., Radford-Weiss I., Boissel N., Raffoux E., Wang P. H. M., Agarwal S., Tamouza H., Paubelle E., Asnafi V., Ribeil J. A., Dessen P., Canioni D., Chandesris O., Rubio M. T., Beaumont C., Benhamou M., Dombret H., Macintyre E., Monteiro R. C., Moura I. C., Hermine O., Targeting iron homeostasis induces cellular differentiation and synergizes with differentiating agents in acute myeloid leukemia. J. Exp. Med. 207, 731–750 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Andreini C., Putignano V., Rosato A., Banci L., The human iron-proteome. Metallomics 10, 1223–1231 (2018). [DOI] [PubMed] [Google Scholar]
- 60.Qatato M., Bonadonna M., Palais G., Ertl A., Schmidt G., Polycarpou-Schwarz M., Karim Z., Galy B., IRE-dependent regulation of intestinal Dmt1 prevails during chronic dietary iron deficiency but is dispensable in conditions of acute erythropoietic stress. HemaSphere 6, e693 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Anders S., Pyl P. T., Huber W., HTSeq--A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Benjamini Y., Hochberg Y., Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 57, 289–300 (1995). [Google Scholar]
- 65.Tyanova S., Temu T., Cox J., The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 11, 2301–2319 (2016). [DOI] [PubMed] [Google Scholar]
- 66.Cox J., Hein M. Y., Luber C. A., Paron I., Nagaraj N., Mann M., Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 13, 2513–2526 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hartigan J. A., Wong M. A., Algorithm AS 136: A K-means clustering algorithm. J. R. Stat. Soc. Ser. C. Appl. Stat. 28, 100–108 (1979). [Google Scholar]
- 68.Badia-i-Mompel P., Vélez Santiago J., Braunger J., Geiss C., Dimitrov D., Müller-Dott S., Taus P., Dugourd A., Holland C. H., Flores R. O. R., Saez-Rodriguez J., decoupleR: Ensemble of computational methods to infer biological activities from omics data. Bioinform. Adv. 2, vbac016 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schubert M., Klinger B., Klünemann M., Sieber A., Uhlitz F., Sauer S., Garnett M. J., Blüthgen N., Saez-Rodriguez J., Perturbation-response genes reveal signaling footprints in cancer gene expression. Nat. Commun. 9, 20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Garcia-Alonso L., Holland C. H., Ibrahim M. M., Turei D., Saez-Rodriguez J., Benchmark and integration of resources for the estimation of human transcription factor activities. Genome Res. 29, 1363–1375 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ritchie M. E., Phipson B., Wu D., Hu Y., Law C. W., Shi W., Smyth G. K., limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M. Y., Geiger T., Mann M., Cox J., The perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740 (2016). [DOI] [PubMed] [Google Scholar]
- 73.Stekhoven D. J., Bühlmann P., MissForest—Non-parametric missing value imputation for mixed-type data. Bioinformatics 28, 112–118 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S11
Tables S1 to S7