Abstract
Small cell lung cancer (SCLC), accounting for around 13% of all lung cancers, often results in rapid tumor growth, early metastasis, and acquired therapeutic resistance. The POU class 2 homeobox 3 (POU2F3) is a master regulator of tuft cell identity and defines the SCLC-P subtype that lacks the neuroendocrine markers. Here, we have identified a previously uncharacterized protein, C11orf53, which is coexpressed with POU2F3 in both SCLC cell lines and patient samples. Mechanistically, C11orf53 directly interacts with POU2F3 and is recruited to chromatin by POU2F3. Depletion of C11orf53 reduced enhancer H3K27ac levels and chromatin accessibility, resulting in a reduction of POU2F3-dependent gene expression. On the basis of the molecular function of C11orf53, we renamed it as “POU Class 2 Homeobox Associating Factor 2” (POU2AF2). In summary, our study has identified a new coactivator of POU2F3 and sheds light on the therapeutic potential of targeting POU2AF2/POU2F3 heterodimer in human SCLC.
C11orf53 is a new coactivator of POU2F3 that is critical for enhancer activity, chromatin accessibility, and SCLC cell viability.
INTRODUCTION
Lung cancer is one of the leading causes of cancer death worldwide (1). Specifically, small cell lung cancer (SCLC) is a highly aggressive and lethal form of malignancy that develops within tissues of the lungs and is typically diagnosed at later stages when it has already metastasized (2–4). Although SCLC consists of approximately 13% of all lung cancer cases, it has one of the lowest 5-year survival rates and a very poor prognosis (5).
Recent studies have made advancements in the classification of SCLC defined by relative expression of four major molecular subtypes of key lineage-specific transcription and cotranscription regulators: achaete-scute homolog 1 (ASCL1; SCLC-A), neurogenic differentiation factor 1 (NEUROD1; SCLC-N), yes-associated protein 1 (YAP1; SCLC-Y), and POU class 2 homeobox 3 (POU2F3; SCLC-P) (6). Analysis of the morphological features have further indicated that SCLC can be classified into neuroendocrine (NE)–high (SCLC-A and SCLC-N) or NE-low (SCLC-Y and SCLC-P) subtypes on the basis of the expression pattern of different NE markers and the diversity in genetic alterations, growth properties, and immune infiltration (7–10).
Previously, we have identified an essential epigenetic coregulator and biomarker, additional sex combs-like protein 3 (ASXL3), which is associated with the SCLC-A molecular subtype. ASXL3 functions as a scaffold protein that links histone H2A deubiquitinase BAP1 and the bromodomain-containing protein BRD4 at active enhancer and drives lineage-specific transcriptional programming (11). Therefore, our study suggested that it is necessary to globally identify and study these subtype-specific essential factors to gain a more in-depth understanding of underlying mechanisms regulating SCLC tumorigenesis that may potentially assist in treatment decisions for SCLC and create a much-needed individualized strategic approach.
For instance, the SCLC-P subtype, which is defined by the expression of a master transcription factor, POU2F3, is known to be a variant form of SCLC that lacks NE features (12). However, identifying other contributing factors in the progression of SCLC-P, specifically a cofactor of POU2F3 function, has yet to be determined. Through our current dependency data analysis and subsequent biochemical and genetic studies, we have identified a previously uncharacterized gene, C11orf53, as a codependent gene of POU2F3. In these studies, we have comprehensively uncovered an essential role of C11orf53 as a coactivator of POU2F3 in regulatory gene expression to establish the cell identity of the SCLC-P subtype.
RESULTS
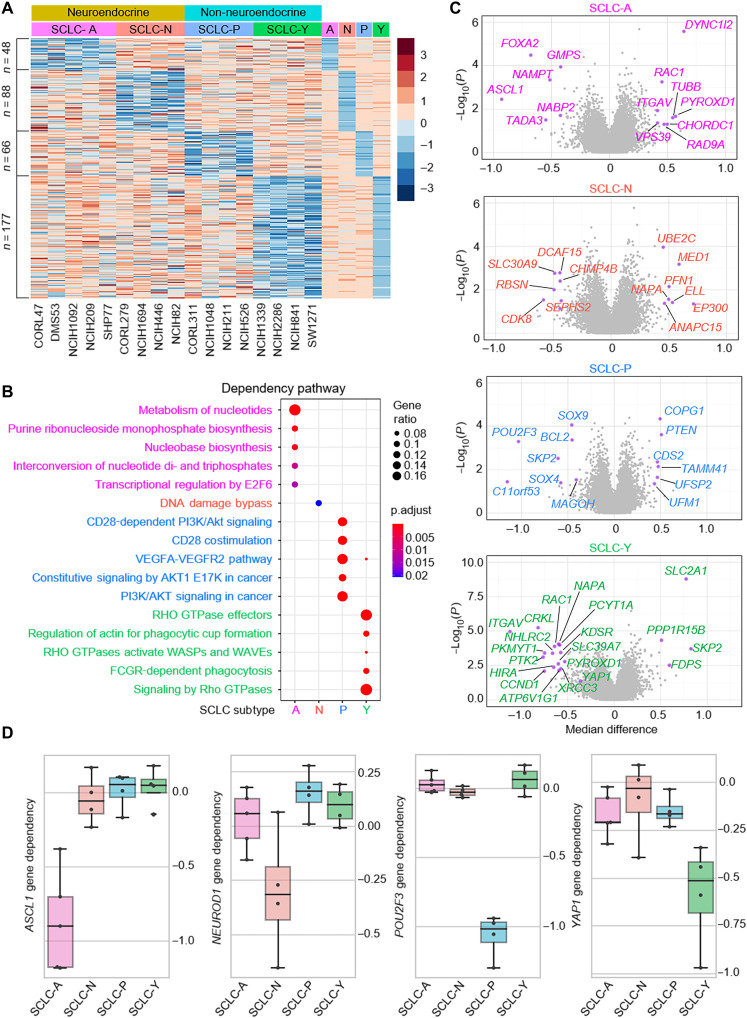
Landscape of SCLC subtype–specific dependency
Global gene expression profiling has been widely used to define molecular subtypes in human cancers (13, 14), including lung cancer (6). However, not all highly expressed genes are essential for tumor cell growth or cell viability. Therefore, to identify the functional subtype-specific dependent factor within the four SCLC molecular subtypes, we used Cancer Cell Line Encyclopedia (CCLE) SCLC cell lines (15) encompassing all four subtypes (6) defined previously and explored the genes that are most selectively essential to each subtype compared to the other classifications. Following the criteria for target gene effect score (≤−0.5; median difference <0.2 versus median value of the average of all other groups), we identified 48 genes for SCLC-A, 88 genes for SCLC-N, 66 genes for SCLC-P, and 177 genes for SCLC-Y subtypes defined as being selectively essential in each SCLC subtype (Fig. 1A and table S1). The expression levels of these genes were compared with no global up- or down-regulation in each subgroup (fig. S1A). The essential genes in each subtype are enriched in distinct pathways (Fig. 1B), suggesting that each subtype may depend on a unique set of signaling pathways for cell survivability and proliferation. Next, we highlighted the top genes that are essential or redundant in each subtype (Fig. 1C), revealing the nonoverlapping essentiality profiles in these subtypes. The gene effect scores of the four markers—ASCL1, NEUROD1, POU2F3, and YAP1—showed the selective dependency (Fig. 1D), which is consistent with previous studies for the function of each gene.
Fig. 1. Landscape of SCLC subtype–specific dependency.
(A) Gene effect scores of all genes were retrieved from DepMap Public 21Q3 datasets for the following 17 SCLC cell lines: A-subtype, CORL47, DMS53, NCI-H1092, NCI-H209, and SHP77; N-subtype, CORL279, NCI-H1694, NCI-H446, and NCI-H82; P-subtype, CORL311, NCI-H1048, NCI-H211, and NCI-H526; Y-subtype, NCI-H1339, NCI-H2286, NCI-H841, and SW1271. A- and N-subtypes were classified as NE and P- and Y-subtypes were classified as non-NE groups. For each subtype, the median gene effect scores of each subtype were calculated, and the z-score heatmap showed genes selectively essential in each group with the criteria that target gene effect score is ≤ −0.5 and the median difference is 0.2 less than the median value of the average of all other groups. (B) Metascape pathway analysis of the essential genes in each subtype as identified above. (C) Volcano plots of the gene essentiality in the four subtypes. X axis is the median difference between the indicated group and others, and y axis is negative log10 P value calculated with t test for the means of two independent samples of scores. Highlighted genes have P < 0.05 and median difference > 0.4 for SCLC-A, SCLC-N, and SCLC-P; P < 0.01, median difference > 0.5 for SCLC-Y subtypes. (D) Box plots showed the ASCL1, NEUROD1, POU2F3, and YAP1 gene dependency in each subtype.
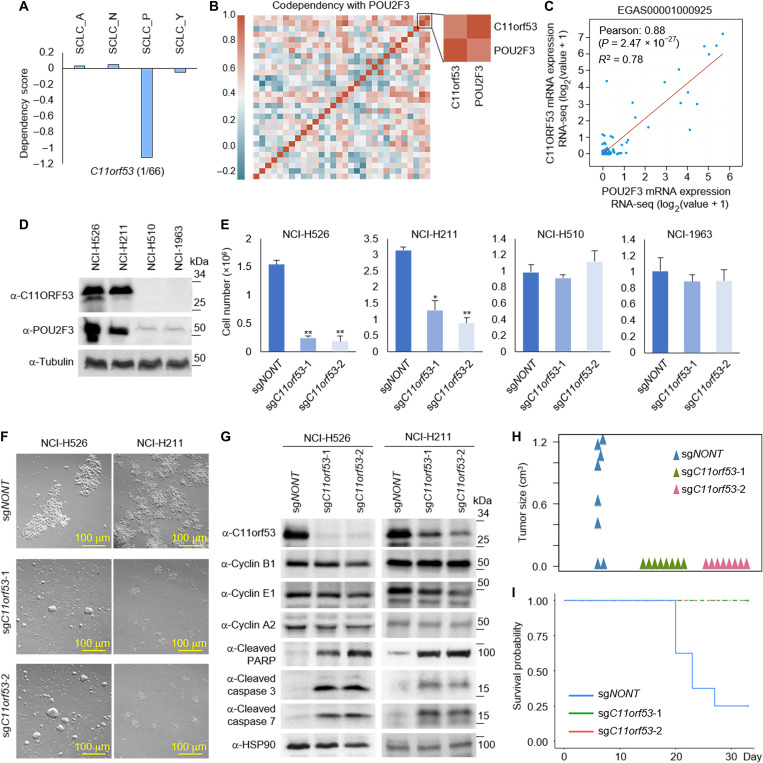
Identification of C11orf53 as a new marker for SCLC-P subtype
Previous studies have identified that the transcription factor POU2F3 is a master regulator of a tuft cell–like variant of small cell lung cancer (12). Therefore, POU2F3 has been widely used to define the SCLC-P subtype. On the basis of our dependency data analysis in Fig. 1A, we noted that the gene dependency score related to a gene called chromosome 11 open reading frame 53 (C11orf53; fig. S2A) is selectively higher in rank (1 of 66) versus POU2F3 (2 of 66) in its significance to maintain the integrity of SCLC-P subtype (Fig. 2A). C11orf53 is an uncharacterized gene that is an evolutionarily conserved sequence from zebrafish to human (fig. S2, B and C) and is expressed at high levels in all four SCLC-P cell lines examined (fig. S2D).
Fig. 2. Identification of C11orf53 as a new marker for SCLC-P subtype.
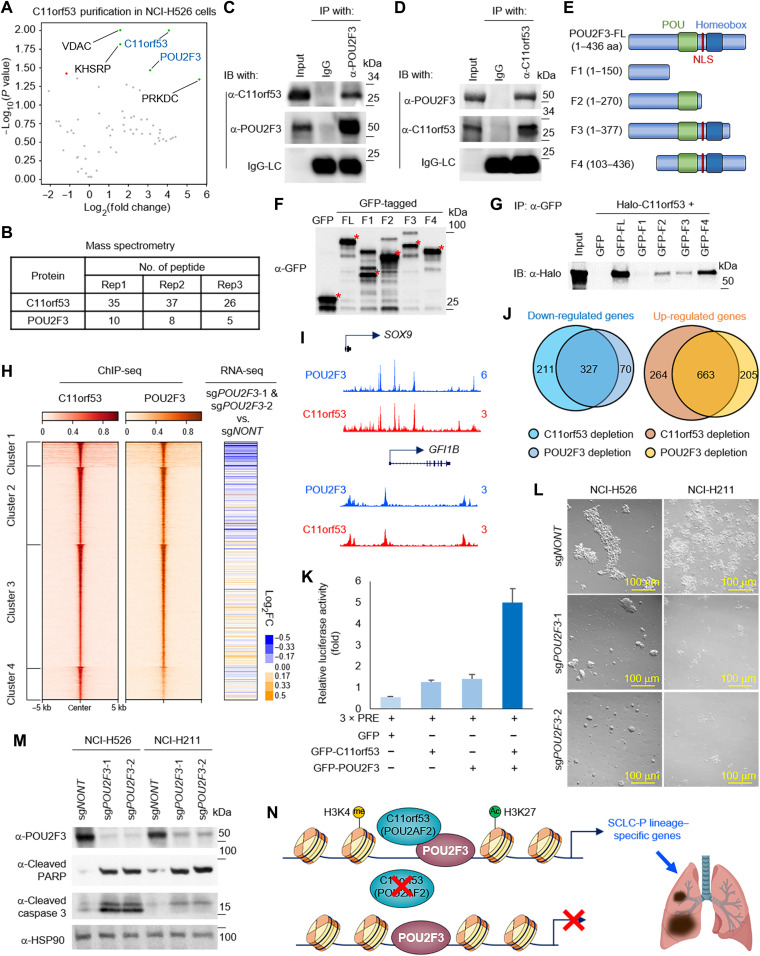
(A) Box plots showed the C11orf53 gene dependency in each subtype. (B) Codependency matrix showing the Pearson correlation coefficient values of the top 30 genes selectively essential in SCLC-P subtype. (C) The scatter plot shows the correlation between POU2F3 and C11orf53 mRNA levels in 110 patient samples (EGAS00001000925). RNA-seq, RNA-sequencing. (D) The protein levels of C11orf53 were determined by Western blot in four different SCLC cell lines. (E) Four different SCLC cell lines—NCI-H526 (SCLC-P), NCI-H211 (SCLC-P), NCI-H510 (non–SCLC-P), and NCI-H1963 (non–SCLC-P)—were transduced with either nontargeting CRISPR sgRNA or C11orf53-specific sgRNAs for 4 days. The cell viability was determined by cell counting assay, n = 3, two-tailed unpaired Student’s t test. **P < 0.01; *P < 0.05. (F) NCI-H526 and NCI-H211 cell lines were transduced with either nontargeting CRISPR sgRNA or C11orf53-specific sgRNAs for 4 days. The cell morphology was shown under bright field microscopy. (G) The protein levels of C11orf53, cyclin B1, cyclin E1, cyclin A2, cleaved PARP, cleaved caspase 3, and cleaved caspase 7 were determined by Western blot in NCI-H526 and NCI-H211 cell lines with C11orf53 CRISPR depletion. HSP90 was used as an internal control. A total of 1 × 106 of NCI-H526 SCLC cells were transduced with either nontargeting sgRNAs or two distinct C11orf53 sgRNAs that were then inoculated into the right flank of athymic nude mice, n = 8 per group. The tumor growth was measured using a calibrated caliper every 2 to 3 days. Weltch’s t test was used for statistical analysis (H). When each tumor reached 1 cm3, a mouse was euthanized and the survival probability was shown in (I). Log-rank test was performed.
The C11orf53 gene encodes for a 288–amino acid protein product without any known functional domain/motif or cellular localization signal. When analyzing the codependency scores of top genes that are selectively essential in SCLC-P subtype, POU2F3 and C11orf53 have an overall correlation coefficient of 0.812 in the SCLC-P subtype compared to 0.357 in all 17 SCLC cell lines (Fig. 2B). This prompted us to investigate the functional relationship between C11orf53 and POU2F3 in P-subtype SCLC. In published SCLC patient samples (EGAS00001000925), we have detected a strongly positive correlation between the expression levels of POU2F3 and C11orf53 (Fig. 2C). Therefore, this gene may function as a new biomarker for SCLC-P subtype cells.
To study the function of C11orf53 in SCLC cells, we generated our homemade polyclonal antibody for C11orf53 (fig. S2E) and detected the protein levels of C11orf53 in four different SCLC cell lines. As shown in Fig. 2D, we have detected very high levels of C11orf53 protein in the NCI-H526 and NCI-H211 SCLC-P subtype cell lines. In contrast, there were no detectable signals from NCI-H510 nor NCI-H1963 non–SCLC-P subtype cell lines. Genetic depletion of C11orf53 by two distinct CRISPR single guide RNAs (sgRNAs) in SCLC cells markedly reduced cell viability (Fig. 2, E and F). To further determine how this gene contributes to cell viability, we examined the protein levels of several factors involved in both cell cycle regulatory checkpoints and apoptosis from two different SCLC-P cell lines. As shown in Fig. 2G, in both SCLC-P cell lines, we found that genetic depletion of C11orf53 via CRISPR knockout could induce cleavage of poly(adenosine diphosphate–ribose) polymerase (PARP), caspase 3, and caspase 7. In addition, loss of C11orf53 protein also induces a notable cell cycle arrest, as shown by fluorescence-activated cell sorting analysis in NCI-H526 cells (fig. S2F). In summary, our results suggest that the C11orf53 gene is critical for the maintenance of SCLC-P cell viability. Consistent with the in vitro cell growth results, genetic depletion of C11orf53 in the in vivo xenograft model significantly repressed tumor growth (P = 0.0033; Fig. 2H) and delayed further progression of disease in mice (P = 0.0023; Fig. 2I).
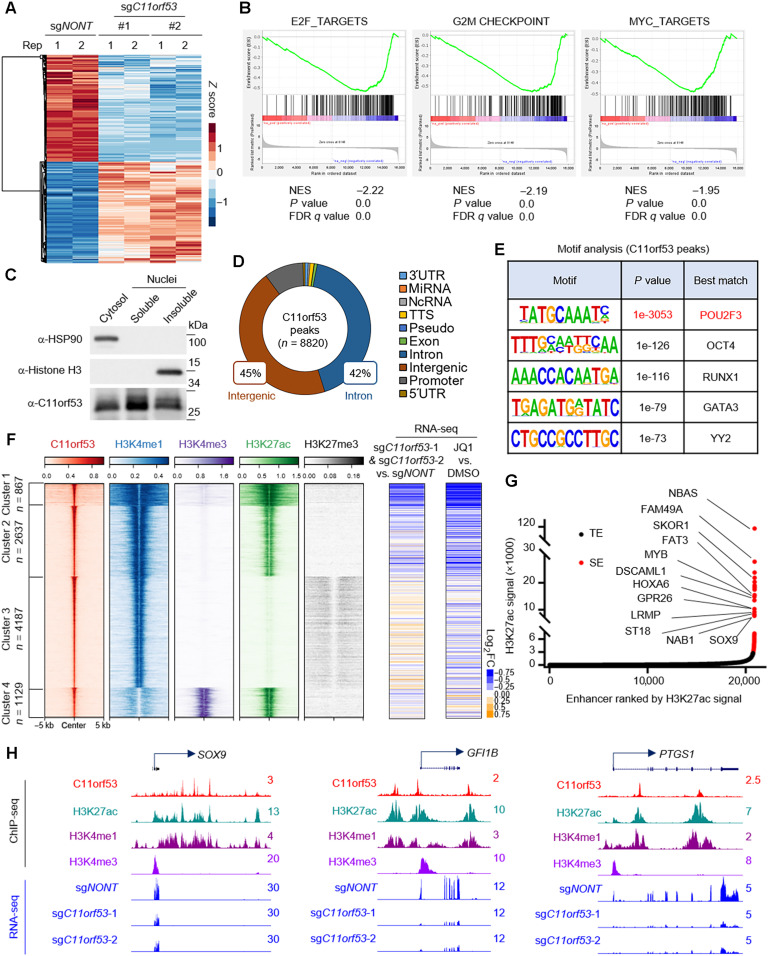
C11orf53 regulates lineage-specific genes expression at super enhancers
To understand how C11orf53 globally affects gene expression in SCLC cells, we conducted RNA sequencing (RNA-seq) in the NCI-H526 cell line transduced with either nontargeting CRISPR sgRNA or two distinct C11orf53-specific sgRNAs. As shown in Fig. 3A, we found a total of 3719 and 3259 genes that were significantly down- and up-regulated, respectively, upon genetic depletion of C11orf53. Moreover, gene set enrichment analysis (GSEA) pathway analysis had identified several key cellular growth biological processes, such as Myc and E2F signaling pathway–dependent gene signatures, which were markedly down-regulated upon genetic depletion of C11orf53 (Fig. 3B). To understand how this small protein regulates gene expression, we first sought to determine the cellular localization of C11orf53 by cell fractionation assay. Unexpectedly, we found that the C11orf53 protein could be detected in the cytosol, soluble nuclear, and chromatin (insoluble nuclear) fractions (Fig. 3C), although it obviously has no nuclear localization signal. To further investigate how this protein regulates gene expression at the chromatin, we conducted chromatin immunoprecipitation sequencing (ChIP-seq) with two of our homemade polyclonal antibodies. On the basis of our ChIP-seq analysis, both of our antibodies showed a consistent performance and the peaks are highly correlated (fig. S3, A and B). In general, we have detected 8820 C11orf53-specific peaks in NCI-H526 cells (Fig. 3D). We found that most of these peaks were localized at intergenic or intron regions, indicating that C11orf53 may regulate gene expression at distal enhancer elements. In addition, the motif analysis has identified POU2F3 motif being matched the top hit (P value, 1 × 10−3053) at C11orf53 peaks (Fig. 3E). This result implies that there might be a similar functionality or genetic interaction between POU2F3 and C11orf53 within SCLC-P subtype cells.
Fig. 3. C11orf53 regulates lineage-specific gene expression at super enhancers.
(A) RNA-seq was conducted with NCI-H526 cell lines transduced with either nontargeting CRISPR sgRNA or C11orf53-specific sgRNAs for 4 days. The heatmap shows the differentially regulated genes, n = 2. (B) The GSEA plot shows the enrichment of E2F, G2M checkpoint, and MYC pathways genes enriched in the down-regulated genes with C11orf53 depletion. NES, normalized enrichment score; FDR, false discovery rate. (C) The protein levels of C11orf53 in cytosol, soluble nuclear fraction, and insoluble nuclear fraction was determined by Western blot. HSP90 was used as cytoplasmic protein control, and the histone H3 was used as nuclear insoluble protein control. (D) The pie plot shows the annotation and distribution of C11orf53 peaks at the genome. (E) Motif analysis shows the best-matched motifs occupied by C11orf53. (F) The total C11orf53 peaks were divided into four clusters based on k-means clustering. The histone marks were further centered on C11orf53 peaks in each cluster (left). RNA-seq was conducted with cells transduced with either nontargeting CRISPR sgRNA or C11orf53-specific sgRNAs. The log2 fold-change (FC) heatmaps shows the expression change of nearest genes to C11orf53 peaks (middle), n = 2. RNA-seq was conducted with cells treated with either dimethyl sulfoxide (DMSO) or JQ1 (1 μM). The log2 fold-change heatmaps shows the expression change of nearest genes to C11ORF53 peaks (right), n = 2. (G) The Histone H3 lysine 27 acetylation (H3K27ac) signals from chromatin immunoprecipitation (ChIP) sequencing identifies putative super enhancers (SEs) in NCI-H526 cells. Hockey-stick plot representing the normalized rank and signals of H3K27ac. Representative SE-associated genes that are occupied by C11orf53 are labeled. TE, typical enhancers. (H) Representative tracks showing the enhancer binding of C11orf53, which contributes to activation of gene expression.
To further characterize the function of C11orf53 at the genome-wide scale and understand how C11orf53 regulates gene expression at distal enhancers, we divided all C11orf53 peaks into four clusters based on k-means clustering. As shown in Fig. 3F, we have detected a very significant overlap between enhancer marker H3K4me1 and C11orf53 peaks. Both of cluster 1 and 2 peaks are also enriched with active enhancer marker H3K27ac, suggesting that C11orf53 may function as a transcriptional activator at active enhancers. We further integrated our RNA-seq data with the ChIP-seq data and identified the expression change of the genes that are nearest to C11orf53 peaks (table S2). As shown in the middle panel of Fig. 3F and fig. S3C, we found that genes nearest to cluster 1 peaks are markedly reduced upon C11orf53 depletion, where very broad H3K4me1 peaks were detected. As shown in Fig. 3G, most of the super enhancer (SE)–associated genes (table S3) are reduced after C11orf53 depletion. In general, around 66% (71 of 108) SE-associated genes are occupied by C11orf53 (table S4), in which 73.2% (52 of 71) genes were down-regulated upon C11orf53 depletion (fig. S3D). Pathway analysis with genes nearest to cluster 1 peaks show a significant enrichment in several neuronal function and differentiation pathways (fig. S3E). Next, to further confirm the role of C11orf53 involvement in regulating SE-associated genes, we determined the gene expression profiles within cells treated with either dimethyl sulfoxide or JQ1 (table S5). As shown in the right panel of Fig. 3 (F and G) and fig. S3 (F to I), the JQ1 treatment has induced a very similar change in the gene expression profile as C11orf53 depletion. The SCLC-P subtype cells are tuft cell–like variant of small cell lung cancer, which express tuft cell–like specific markers, such as SOX9, GFI1B, and PTGS1. Notably, these tuft cell–like specific genes were down-regulated after C11orf53 depletion or JQ1 treatment (Fig. 3H and fig. S3J).
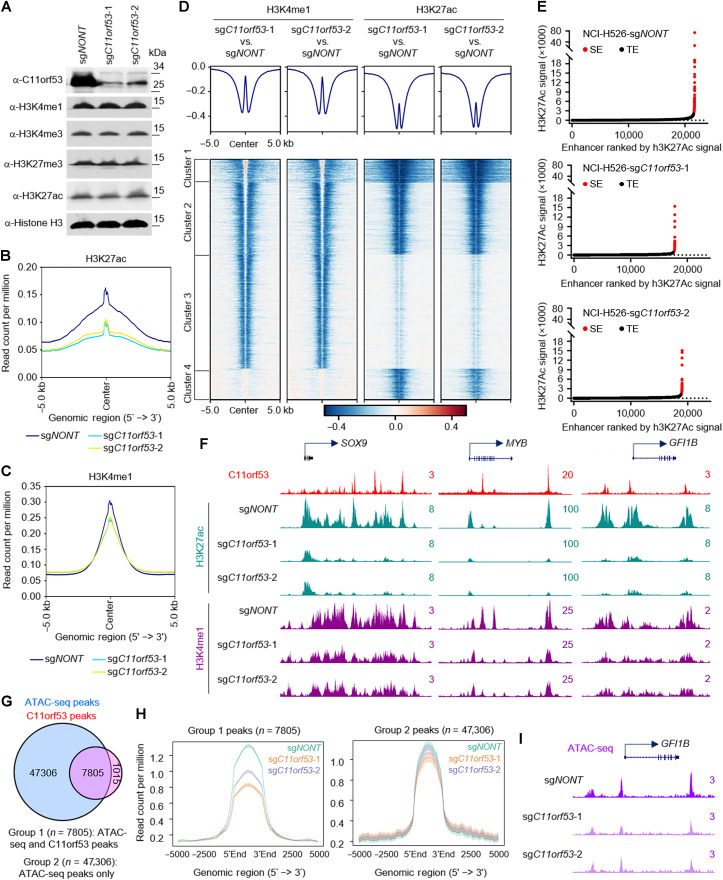
Loss of C11orf53 reduces enhancer activity and chromatin accessibility
Because C11orf53 occupies active enhancers and is critical for maintenance in the expression levels of the enhancer-nearby genes, we asked whether loss of C11orf53 affects the protein levels of enhancer histone marks, such as H3K4me1 and H3K27ac. We conducted Western blot in cells transduced with either nontargeting CRISPR sgRNA or two distinct C11orf53 sgRNAs. As a result, we found that loss of C11orf53 expression does not affect the bulk of histone modifications (Fig. 4A). Next, we conducted ChIP-seq to determine whether there were locus-specific changes in active enhancer marks (H3K4me1/H3K27ac levels). As shown in Fig. 4 (B and C), C11orf53 genetic depletion strongly reduced H3K27ac levels and a moderate reduction in H3K4me1 levels. Then, we centered the log2 fold change of H3K4me1/H3K27ac levels at the previously defined four C11orf53-specific clusters in Fig. 3F. As shown in Fig. 4D, we have detected a very strong reduction of H3K27ac and H3K4me1 histone marks near C11orf53 peaks. Consistently, there is also a marked reduction of SE signals in C11orf53-depleted cells (Fig. 4, E and F). Last, we conducted transposase-accessible chromatin with sequencing (ATAC-seq) experiments to determine whether the chromatin accessibility has been altered in C11orf53-depleted cells. As a result, we have detected a total of 55,111 ATAC-seq peaks with 7805 of those peaks being occupied by C11orf53 (Fig. 4G). We further separated the total ATAC-seq peaks into two groups, depending on whether they were cobound with C11orf53 or not. As shown in Fig. 4H, we found that group 1 peaks (ATAC-seq+/C11orf53+) were strongly reduced upon both sgRNA treatments [Fig. 4, H (left) and I, and fig. S4, A and B], while group 2 peaks (ATAC-seq+/C11orf53−) were not significantly reduced upon C11orf53 genetic depletion (Fig. 4H, right, and fig. S4C). In summary, our results have shown a novel function of C11orf53 involved in chromatin accessibility and maintenance of enhancer-driven transcriptional program in SCLC-P subtype cells.
Fig. 4. Loss of C11orf53 reduces enhancer activity and chromatin accessibility.
(A) NCI-H526 cell lines transduced with either nontargeting CRISPR sgRNA or C11orf53-specific sgRNAs for 4 days. The protein levels of C11orf53, H3K4me1, H3K4me3, H3K27me3, and H3K27ac levels were determined by Western blot. Total histone H3 was used as internal control. The average plot shows the total H3K27ac (B), and H3K4me1 (C) peaks in NCI-H526 cell lines transduced with either nontargeting CRISPR sgRNA or C11orf53-specific sgRNAs. Chromatin from Drosophila S2 cells (10%) was used as spike-in. IgG, immunoglubulin G. (D) The log2 fold-change heatmap shows the loss of H3K4me1 and H3K27ac signal at C11ORF53 peaks after C11ORF53 depletion. (E) Histone H3K27ac signals from ChIP-seq identifies putative SEs in NCI-H526 cells transduced with either nontargeting CRISPR sgRNA (top) or two distinct C11orf53-specific sgRNAs (middle and bottom). Hockey-stick plot representing the normalized rank and signals of H3K27ac. Representative of top-ranked SE-associated genes from each group are labeled. aa, amino acids. (F) Representative track examples have shown the H3K27ac levels at SOX9, GFI1B, and PTGS1 gene loci in cells transduced with either CRISPR sgRNA or C11orf53-specific sgRNAs. (G) The Venn diagram shows the overlap between ATAC-seq peaks and C11orf53 peaks in NCI-H526 cells. (H) The average plot shows the ATAC-seq signal between cells transduced with either CRISPR sgRNA or C11orf53-specific sgRNAs. The group 1 peaks are ATAC-seq/C11orf53 common peaks. Group 2 peaks are ATAC-seq alone peaks. (I) Representative tracks showing ATAC-seq peaks at GFI1B gene loci in cells transduced with either CRISPR sgRNA or C11orf53-specific sgRNAs.
C11orf53 is a coactivator of POU2F3 in SCLC cells
As an uncharacterized 288–amino acid protein, there is no obvious functional domain/motif within C11orf53. To study the underlying molecular basis of how C11orf53 is recruited to chromatin and regulates gene expression, we purified green fluorescent protein (GFP)–tagged proteins from SCLC cell line NCI-H526 that stably express either GFP or GFP-tagged C11orf53. By mass spectrometry analysis, we found POU2F3 as one of the top candidates that interacted with C11orf53 (Fig. 5, A and B). We further performed IP experiments with whole-cell lysate from NCI-H526 cells (Fig. 5, C and D) and NCI-H211 cells (fig. S5, A and B) to confirm the protein-protein interaction between the endogenous C11orf53 and POU2F3. To understand how C11orf53 interacts with POU2F3, we truncated POU2F3 into several different truncated fragments (Fig. 5E) and purified each of these GFP-tagged fragments in human embryonic kidney (HEK) 293T cells cotransfected with Halo-tagged C11orf53 (Fig. 5F). As shown in Fig. 5G, we found that the POU domain within the POU2F3 protein is critical for the interaction with C11orf53. To determine a potential cofunction between C11orf53 and POU2F3, we conducted ChIP-seq to determine the chromatin occupancy of POU2F3 in NCI-H526 cells. As we expected, there is a significant overlap between POU2F3 and C11orf53 occupancy across the genome in both NCI-H526 [Fig. 5, H (left) and I, and fig. S5F] and NCI-H211 (fig. S5, C to E and G) cells.
Fig. 5. C11orf53 is a coactivator of POU2F3 in SCLC cells.
(A) The volcano plot shows the significant enriched protein during GFP-tagged C11orf53 purification from NCI-H526 cells, n = 3. Peptide numbers of POU2F3 and C11orf53 were shown (B). IP of endogenous POU2F3 from NCI-H526 cells followed by immunoblotting (IB) for POU2F3 and C11orf53 (C) and vice versa (D). (E) Schematic diagram depicting the domain organization of the human POU2F3 protein and the GFP-tagged fusion truncations. (F) Whole-cell lysates were used for Western blot with GFP antibodies in cells transfected with empty vector (GFP) or different POU2F3 fragments in (E). (G) IP with GFP antibodies followed by IB for Halo-tag in cells cotransfected with Halo-tagged C11orf53, and either empty vector (GFP) or POU2F3 fragments in (E). (H) The heatmaps show the co-occupancy of C11orf53 and POU2F3 in NCI-H526 SCLC cells. All rows are centered on C11orf53 four clusters as divided by k-means clustering (left). The log2 fold-change heatmaps shows the expression change of nearest genes to C11orf53 peaks (middle) after POU2F3 depletion, n = 2 (right). (I) The representative tracks show the colocalization of POU2F3 and C11orf53 at SOX9 and GFI1B gene loci. (J) The Venn diagram shows the overlap of targeted genes between POU2F3 and C11orf53. (K) HEK293T cells were transfected with GFP-tagged C11orf53 and/or POU2F3 in the presence of 3 × PRE (POU2F3 Response Element) reporter plasmid. Luciferase assay was performed 48 hours after transfection, n = 3. (L) NCI-H526 and NCI-H211 cell lines were transduced with either nontargeting CRISPR sgRNA or POU2F3-specific sgRNAs for 4 days. The cell morphology was shown under bright field. (M) The protein levels of POU2F3, cleaved PARP, and cleaved caspase 3 were determined by Western blot. (N) Model of C11orf53 functions as coactivator of POU2F3 in SCLC cells.
Because C11orf53 protein has no known DNA or histone binding domain, we hypothesized that it may be transported into the nuclei and recruited to chromatin by POU2F3. To test this hypothesis, we transfected HEK293T cells (which express neither POU2F3 nor C11orf53) with Halo-tag or Halo-tagged POU2F3 in the presence of GFP-tagged C11orf53. As shown in fig. S6 (A and B), the chromatin bound C11orf53 was markedly increased in the presence of POU2F3, as determined by Western blot and ChIP-seq results. To confirm this observation in SCLC cells, we depleted POU2F3 by CRISPR knockout in NCI-H526 cells for 2 days and determined the chromatin-bound C11orf53 levels. As shown in fig. S6 (C and D), genetic depletion of POU2F3 could also reduce the chromatin-bound C11orf53, as determined by Western blot and ChIP–quantitative polymerase chain reaction (qPCR) results. These results imply that C11orf53 might function as a coactivator that is recruited to chromatin by POU2F3 (16).
In NCI-H526 cells, POU2F3 genetic depletion reduced the expression of group 1–clustered genes, which is similar compared to C11orf53 genetic depletion (Fig. 5H, right, and table S6). In total, there are 327 genes that are co–down-regulated by C11orf53 or POU2F3 genetic depletion (fold change >2), in which we have identified numerous tuft cell lineage genes—which has been demonstrated to be targeted genes of POU2F3 (Fig. 5J and fig. S6E). Because the chromatin binding motif between C11orf53 and POU2F3 is extremely similar, we have introduced a luciferase assay to further confirm the cofunction between these two factors. As shown in Fig. 5K, we found that cotransfection of C11orf53 and POU2F3 has the strongest effect on inducing luciferase expression. Similar to C11orf53 genetic depletion, a loss in the expression of POU2F3 also markedly induces apoptosis and reduces cell viability in two different SCLC-P cell lines (Fig. 5, L and M). In summary, our model has shown that this previous uncharacterized protein, C11orf53, functions as a coactivator of POU2F3 and maintains chromatin accessibility at POU2F3-targeted genes in SCLC cells (Fig. 5N).
DISCUSSION
The POU family proteins are a large class of DNA binding transcription factors that are divided into six classes, with POU2F3 belonging to the POU II class, which includes POU2F1 (Oct-1) and POU2F2 (Oct-2) (17). As the only known binding partner of POU II class, POU2AF1 (POU Class 2 Homeobox Associating Factor 1) functions as coactivator of POU2F1, and to a lesser extent than POU2F1, as a coactivator of POU2F2 (18–21). Our current study has provided evidence for a previously uncharacterized protein, C11orf53, functioning as a coactivator of POU2F3 (also known as Oct-11). Therefore, we are defining this gene on the basis of its novel function and classifying it as the POU Class 2 Homeobox Associating Factor 2 (or POU2AF2). On the basis of our mass spectrometry analysis of purified C11orf53, we did not detect POU2F1 or POU2F2, which are both expressed in NCI-H526 cells based on our RNA-seq data. This result suggested that C11orf53 is quite a unique coactivator of POU2F3. It is known that POU2AF1 binds to POU2F1 and POU2F2 using its α-helical segment, while C11orf53 does not display a similar structure (22). Therefore, it would be interesting to determine the crystal structure of C11orf53 bounded to POU2F3 but also challenging because of the disordered nature of C11orf53 structure.
The function of POU2F3 in small cell lung cancer has been identified by genome-wide CRISPR screening in previous studies (12). Therefore, it has been widely used as the biomarker that defines the SCLC-P subtype, which expresses markers of the chemosensory lineage instead of NE markers. Mechanistically, POU2F3 binds to distal enhancer elements and drives the expression of the lineage-specific genes. As a nonhistone/DNA binding coactivator, C11orf53 is recruited to chromatin by the DNA binding transcription factor, POU2F3, and maintains openness of the chromatin to regulate the expression of POU2F3-targeted genes. On the basis of our model (Fig. 5N), C11orf53 may not function as a canonical pioneer factor that recruits transcription factor(s) to chromatin.
On the basis of our genome-wide RNA-seq analysis in Fig. 5J, there are approximately 32% of C11orf53-targeted genes that are not affected upon POU2F3 genetic depletion, which suggests that C11orf53 may have a POU2F3-independent function. On the basis of our cellular fractionation assay (Fig. 3C and fig. S6C), a significant proportion of C11of53 proteins have been detected in the cytoplasm, while a vast majority of POU2F3 could only be detected in the nuclei. Therefore, future studies may be focused on the cytoplasmic function of C11orf53 in SCLC cells.
Dysregulation or mutations within enhancer binding factors have been identified as direct drivers for many types of cancers (23, 24). Our current studies have shown a critical role of C11orf53 at SEs. Consequently, loss of C11orf53 leads to a loci-specific reduction of H3K27ac levels and gene expression, which is similar to the effects of JQ1 treatment. Our result is consistent with previous reports that SCLC cells are more sensitive to BET inhibitor treatment (11, 25, 26).
In summary, our study sheds light on the potential impact of targeting C11orf53 or C11orf53/POU2F3 heterodimer for new therapeutic approaches against SCLC-P subtype because of its specificity and high dependency in this particular cancer type, which is in agreement with a recent work published by Vakoc’s group (27).Therefore, establishing small-molecule inhibitors or peptide drugs that can disrupt C11orf53/POU2F3 interaction may specifically inhibit POU2F3-dependent transcriptional programming in SCLC cells and thus aid in the development of a more personalized approach to SCLC therapy.
MATERIALS AND METHODS
Antibodies and reagents
POU2F3 (no. 36135), H3K27ac (no. 8173S), H3K4me1 (no. 5326S), H3K4me3 (no. 9751), H3K27me3 (no. 9733), histone H3 (no. 4499S), cyclin B1 (no. 12231), cyclin E1 (no. 20808), cyclin A2 (no. 91500), cleaved PARP (no. 5625), cleaved caspase 3 (no. 9664), and cleaved caspase 7 (no. 8438) antibodies were purchased from Cell Signaling Technology. Tubulin antibody (E7) was purchased from Developmental Studies Hybridoma Bank. Heat shock protein 90 (HSP90) (sc-7947) and GFP (sc-9996) antibodies were purchased from Santa Cruz Biotechnology. Halo-tag (G9211) antibody was purchased from Promega. The C11orf53 antibody was produced in rabbit in house by using full-length C11orf53 recombinant protein as antigen.
Cell lines
HEK293T cells were obtained from American Type Culture Collection (ATCC) and then maintained with Dulbecco’s modified Eagle’s medium (Gibco, Gaithersburg, MD) containing 10% fetal bovine serum (FBS; Sigma-Aldrich). The SCLC cell lines NCI-H526, NCI-H211, NCI0H510, and NCI-H1963 were obtained from ATCC and were maintained with ATCC-formulated RPMI 1640 medium containing 10% FBS (Sigma-Aldrich). The Drosophila S2 cells were maintained in HyClone SFX-Insect Cell Culture Media containing 10% FBS (Sigma-Aldrich).
Mouse experiments
All mouse work was performed in accordance with protocols approved by The Center for Comparative Medicine of Northwestern University. Five- to 6-week-old athymic nude mice were used for xenograft experiments. The human NCI-H526 SCLC cell line was transduced with either nontargeting CRISPR sgRNA or two distinct C11orf53-specific CRISPR sgRNAs that we used for all the other biochemical studies, and further selected with puromycin (1 μg/ml) for 48 hours. Then, we determined the cell viability by trypan blue staining, and injected 1 × 106 of living cells from each group were inoculated into the right flank of nude mice, respectively. Tumor growth was monitored every 3 days for 2 weeks after inoculation.
Immunoprecipitation
The IP experiment was performed as previously described (11). Briefly, the cells were lysed in the lysis buffer [50 mM tris (pH 8.0), 150 mM NaCl, 0.5% Triton X-100, 10% glycerol, protease inhibitors, and benzonase]. After centrifugation at maximum speed at 4°C for 15 min, the supernatants (1-mg protein lysate for each sample) were collected and incubated with the primary antibody and immobilized Protein A/G (Santa Cruz Biotechnology) at 4°C overnight with rotation. Then, the samples were washed with lysis buffer four times and boiled in 5× SDS sample loading buffer.
RNA interference, CRISPR-mediated knockouts, and real-time PCR
Designed sgRNAs were cloned into lentiCRISPR v2 (Addgene, 52961) vector. The lentiviral-mediated CRISPR-Cas9 knockout was described previously (28). Oligo sequences used in this manuscript were as follows: sgNONT (GCTGAAAAAGGAAGGAGTTGA), sgC11orf53-1 (GTGACGTCTACACCTCCAGCG), sgC11orf53-2 (GAGAGGCAACTCGTGCTGGG), sgPOU2F3-1 (GCCCACGCTTAGGGAGATGTG), and sgPOU2F3-2 (GTCCTACCAAATACTTCACTG). Primer sequences for ChIP-qPCR used in this manuscript were as follows: SOX9-E1-F: 5′-CTTCCAACCCTACTCCAGGC-3′, SOX9-E1-R: 5′-TTGGGACAAGGGTAGGCTCT-3′; SOX9-E2-F: 5′-CTGGAAAGGGCTAGAGGCTG-3′, SOX9-E1-R: 5′-TCCCCTGCTAGTTTTATGGGC-3′; GFI1B-E1-F: 5′-ACCACTTTCACAAGCCCACT-3′, SOX9-E1-R: 5′-GGCCGCCTTCGGAGATTTTA-3′; GFI1B-E2-F: 5′-CGGGATTTCAGCCACTTCTG-3′, GFI1B-E2-R: 5′-CGCCAAACCTCAGTCGACAA-3′; and IRAG2-E1-F: 5′-GTATCCAAGACACCTGGTCCC-3′, IRAG2-E1-R: 5′-ACATGGCAAAACAAGTTGGCT-3′.
RNA-seq and analysis
RNA-seq was conducted as previously described. Paramagnetic beads coupled with oligo (dT) are combined with total RNA to isolate polyadenylated [poly (A)+] transcripts based on NEBNext Poly(A) mRNA Magnetic Isolation Module manual. All remaining steps for library construction were used according to the manufacturer’s recommendations. Samples were pooled and sequenced on a HiSeq with a read length configuration of 150 paired-end (PE). Gene counts were computed by HTSeq and used as an input for edgeR 3.0.85257. Genes with Benjamini-Hochburg–adjusted P values less than 0.01 were considered to be differentially expressed (unless otherwise specified).
ChIP-seq assay and analysis
ChIP-seq was performed as described previously (11). For histone modifications, 10% of Drosophila chromatin was used as spike-in control. For ChIP-seq analysis, all the peaks were called with the MACS v2.1.0 software using default parameters and corresponding input samples. Metaplots and heatmaps were generated using the ngsplot database to display ChIP-seq signals. Peak annotation, motif analysis, and SE analysis were performed with HOMER and ChIPseeker. Pathway analysis was performed with Metascape and ChIPseeker.
ATAC-seq and analysis
ATAC-seq was performed as described previously (29). In brief, frozen cells were thawed and washed once with phosphate-buffered saline (PBS) and then resuspended in 500 μl of cold ATAC lysis buffer. The cell number was assessed by Cellometer Auto 2000 (Nexcelom Bioscience). Nuclei (50 to 100K) were then centrifuged (prechilled) at 500g for 10 min. Supernatant was then removed, and the nuclei were resuspended in 50 μl of tagmentated DNA by pipetting up and down six times. The reactions were incubated at 37°C for 30 min in a thermomixer shaking at 1000 rpm and then cleaned up by the MinElute Reaction Cleanup Kit (Qiagen). Tagmentated DNA was amplified with barcode primers. Library quality and quantity were assessed with Qubit 2.0 DNA HS Assay (Thermo Fisher Scientific, Massachusetts, USA), Tapestation High Sensitivity D1000 Assay (Agilent Technologies, California, USA), and QuantStudio 5 System (Applied Biosystems, California, USA). Equimolar pooling of libraries was performed on the basis of quality control (QC) values and sequenced on an Illumina HiSeq (Illumina, California, USA) with a read-length configuration of 150 PE for 50M PE reads (25M in each direction) per sample. ATAC-seq reads are shifted +4 bp and −5 bp for positive and negative strands, respectively, using the alignmentSieve function from the deepTools package. ATAC-seq peaks are called with Macs v2.1.0. sgNONT ATAC-seq peaks were intersected with C11orf53 peaks using bedtools.
Mass spectrometry sample preparation and analysis
Mass spectrometry was performed as described previously. Protein pellet was denatured in 50 μl of 8 M urea/0.4 M ammonium bicarbonate followed by reduction in 2 μl of 100 mM dithiothreitol. The digests were acidified to 0.5% trifluoroacetic acid and the peptides were then desalted on C18 Sep-Paks (Waters). The pooled extracts were dried in a vacuum concentrator and resuspended in 30 μl of 5% ACN/0.1% FA for liquid chromatography (LC) mass spectrometry analysis. Peptides were analyzed by LC–tandem mass spectrometry using a Dionex UltiMate 3000 Rapid Separation LC (RSLC) system and a linear ion trap-Orbitrap hybrid Elite mass spectrometer (Thermo Fisher Scientific, San Jose, CA).
Statistical analyses
For statistical analyses, GraphPad Prism 7, Microsoft Excel, and R were used. All data being reported that met the criteria to use the appropriate statistical tests involved a statistical analysis; for the normal distribution of data, the empirical rule was used to infer the distribution. For growth curves and time course, RNA-seq t tests were calculated between the area-under-the-curve values. Statistical tests used are reported in the figure legends.
Acknowledgments
We would like to thank B. Braschi and HUGO Gene Nomenclature Committee (HGNC) for working with us to rename C11orf53 gene as POU2AF2 officially. We would like to thank Y. Shi and C. Vakoc for critical discussion. We would like to thank F. Zhang for the gifts of the Px330 and lentiCRISPR v2 vectors.
Funding: L.W. was supported by NIH grant R35GM146979 and the Research Scholar Grant (RSG-22-039-01-DMC) from the American Cancer Society. L.W. was also supported by Lynn Sage Scholar Award. R.H. was supported by the U.S. National Institutes of Health (R01NS126513).
Author contributions: L.W. and Z.Z. designed the study. A.P.S., N.T., and L.W. performed all the biochemistry and sequencing experiments. N.T. performed the animal experiments. Z.Z. performed the bioinformatics analysis. A.P.S., N.T., and L.W. wrote the manuscript. N.T., A.P.S., J.W., R.H., and L.W. revised the manuscript. All authors read and approved the final manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: NGS data generated for this study are available at the Gene Expression Omnibus (GEO) under accession number GSE197426. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S6
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S6
REFERENCE AND NOTES
- 1.Sung H., Ferlay J., Siegel R. L., Laversanne M., Soerjomataram I., Jemal A., Bray F., Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J. Clin. 71, 209–249 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Gardner E. E., Lok B. H., Schneeberger V. E., Desmeules P., Miles L. A., Arnold P. K., Ni A., Khodos I., de Stanchina E., Nguyen T., Sage J., Campbell J. E., Ribich S., Rekhtman N., Dowlati A., Massion P. P., Rudin C. M., Poirier J. T., Chemosensitive relapse in small cell lung cancer proceeds through an EZH2-SLFN11 Axis. Cancer Cell 31, 286–299 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gazdar A. F., Bunn P. A., Minna J. D., Small-cell lung cancer: What we know, what we need to know and the path forward. Nat. Rev. Cancer 17, 725–737 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Rudin C. M., Brambilla E., Faivre-Finn C., Sage J., Small-cell lung cancer. Nat. Rev. Dis. Primers. 7, 3 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller K. D., Nogueira L., Mariotto A. B., Rowland J. H., Yabroff K. R., Alfano C. M., Jemal A., Kramer J. L., Siegel R. L., Cancer treatment and survivorship statistics, 2019. Cancer J. Clin. 69, 363–385 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Rudin C. M., Poirier J. T., Byers L. A., Dive C., Dowlati A., George J., Heymach J. V., Johnson J. E., Lehman J. M., MacPherson D., Massion P. P., Minna J. D., Oliver T. G., Quaranta V., Sage J., Thomas R. K., Vakoc C. R., Gazdar A. F., Author Correction: Molecular subtypes of small cell lung cancer: A synthesis of human and mouse model data. Nat. Rev. Cancer 19, 415–415 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Zhang W., Girard L., Zhang Y.-A., Haruki T., Papari-Zareei M., Stastny V., Ghayee H. K., Pacak K., Oliver T. G., Minna J. D., Gazdar A. F., Small cell lung cancer tumors and preclinical models display heterogeneity of neuroendocrine phenotypes. Transl. Lung Cancer Res. 7, 32–49 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang D., Denny S. K., Greenside P. G., Chaikovsky A. C., Brady J. J., Ouadah Y., Granja J. M., Jahchan N. S., Lim J. S., Kwok S., Kong C. S., Berghoff A. S., Schmitt A., Reinhardt H. C., Park K. S., Preusser M., Kundaje A., Greenleaf W. J., Sage J., Winslow M. M., Intertumoral heterogeneity in SCLC is influenced by the cell type of origin. Cancer Discov. 8, 1316–1331 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwendenwein A., Megyesfalvi Z., Barany N., Valko Z., Bugyik E., Lang C., Ferencz B., Paku S., Lantos A., Fillinger J., Rezeli M., Marko-Varga G., Bogos K., Galffy G., Renyi-Vamos F., Hoda M. A., Klepetko W., Hoetzenecker K., Laszlo V., Dome B., Molecular profiles of small cell lung cancer subtypes: Therapeutic implications. Mol. Ther. Oncolytics 20, 470–483 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bebber C. M., Thomas E. S., Stroh J., Chen Z., Androulidaki A., Schmitt A., Höhne M. N., Stüker L., de Pádua Alves C., Khonsari A., Dammert M. A., Parmaksiz F., Tumbrink H. L., Beleggia F., Sos M. L., Riemer J., George J., Brodesser S., Thomas R. K., Reinhardt H. C., von Karstedt S., Ferroptosis response segregates small cell lung cancer (SCLC) neuroendocrine subtypes. Nat. Commun. 12, 2048 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szczepanski A. P., Zhao Z., Sosnowski T., Goo Y. A., Bartom E. T., Wang L., ASXL3 bridges BRD4 to BAP1 complex and governs enhancer activity in small cell lung cancer. Genome Med. 12, 63 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang Y. H., Klingbeil O., He X. Y., Wu X. S., Arun G., Lu B., Somerville T. D. D., Milazzo J. P., Wilkinson J. E., Demerdash O. E., Spector D. L., Egeblad M., Shi J., Vakoc C. R., POU2F3 is a master regulator of a tuft cell-like variant of small cell lung cancer. Gene Dev. 32, 915–928 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shai R., Shi T., Kremen T. J., Horvath S., Liau L. M., Cloughesy T. F., Mischel P. S., Nelson S. F., Gene expression profiling identifies molecular subtypes of gliomas (vol 22, pg 4918, 2003). Oncogene 25, 4256–4256 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Bertucci F., Finetti P., Rougemont J., Charafe-Jauffret E., Cervera N., Tarpin C., Nguyen C., Xerri L., Houlgatte R., Jacquemier J., Viens P., Birnbaum D., Gene expression profiling identifies molecular subtypes of inflammatory breast cancer. Cancer Res. 65, 2170–2178 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Tlemsani C., Pongor L., Elloumi F., Girard L., Huffman K. E., Roper N., Varma S., Luna A., Rajapakse V. N., Sebastian R., Kohn K. W., Krushkal J., Aladjem M. I., Teicher B. A., Meltzer P. S., Reinhold W. C., Minna J. D., Thomas A., Pommier Y., SCLC-CellMiner: A resource for small cell lung cancer cell line genomics and pharmacology based on genomic signatures. Cell Rep. 33, 108296 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moritsugu R., Tamai K., Nakano H., Aizu T., Nakajima K., Yamazaki T., Sawamura D., Functional analysis of the nuclear localization signal of the POU transcription factor Skn-1a in epidermal keratinocytes. Int. J. Mol. Med. 34, 539–544 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Ryan A. K., Rosenfeld M. G., POU domain family values: Flexibility, partnerships, and developmental codes. Gene Dev. 11, 1207–1225 (1997). [DOI] [PubMed] [Google Scholar]
- 18.Gstaiger M., Knoepfel L., Georgiev O., Schaffner W., Hovens C. M., A B-cell coactivator of octamer-binding transcription factors. Nature 373, 360–362 (1995). [DOI] [PubMed] [Google Scholar]
- 19.Strubin M., Newell J. W., Matthias P., Obf-1, a novel B-cell-specific coactivator that stimulates immunoglobulin promoter activity through association with octamer-binding proteins. Cell 80, 497–506 (1995). [DOI] [PubMed] [Google Scholar]
- 20.Shore P., Dietrich W., Corcoran L. M., Oct-2 regulates CD36 gene expression via a consensus octamer, which excludes the co-activator OBF-1. Nucleic Acids Res. 30, 1767–1773 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shakya A., Goren A., Shalek A., German C. N., Snook J., Kuchroo V. K., Yosef N., Chan R. C., Regev A., Williams M. A., Tantin D., Oct1 and OCA-B are selectively required for CD4 memory T cell function. J. Exp. Med. 212, 2115–2131 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chasman D., Cepek K., Sharp P. A., Pabo C. O., Crystal structure of an OCA-B peptide bound to an Oct-1 POU domain/octamer DNA complex: Specific recognition of a protein-DNA interface. Gene Dev. 13, 2650–2657 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Plass C., Pfister S. M., Lindroth A. M., Bogatyrova O., Claus R., Lichter P., Mutations in regulators of the epigenome and their connections to global chromatin patterns in cancer. Nat. Rev. Genet. 14, 765–780 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Khan P., Siddiqui J. A., Maurya S. K., Lakshmanan I., Jain M., Ganti A. K., Salgia R., Batra S. K., Nasser M. W., Epigenetic landscape of small cell lung cancer: Small image of a giant recalcitrant disease. Semin. Cancer Biol., 57–76 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lenhart R., Kirov S., Desilva H., Cao J., Lei M., Johnston K., Peterson R., Schweizer L., Purandare A., Ross-Macdonald P., Fairchild C., Wong T., Wee S., Sensitivity of small cell lung cancer to BET inhibition is mediated by regulation of ASCL1 gene expression. Mol. Cancer Ther. 14, 2167–2174 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Lam L. T., Lin X., Faivre E. J., Yang Z., Huang X., Wilcox D. M., Bellin R. J., Jin S., Tahir S. K., Mitten M., Magoc T., Bhathena A., Kati W. M., Albert D. H., Shen Y., Uziel T., Vulnerability of small-cell lung cancer to apoptosis induced by the combination of BET bromodomain proteins and BCL2 inhibitors. Mol. Cancer Ther. 16, 1511–1520 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Wu X. S., He X.-Y., Ipsaro J. J., Huang Y.-H., Preall J. B., Ng D., Shue Y. T., Sage J., Egebald M., Joshua-Tor L., Vakoc C. R., OCA-T1 and OCA-T2 are coactivators of POU2F3 in the tuft cell lineage. Nature 607, 169–175 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang L., Zhao Z., Ozark P. A., Fantini D., Marshall S. A., Rendleman E. J., Cozzolino K. A., Louis N., He X., Morgan M. A., Takahashi Y.-H., Collings C. K., Smith E. R., Ntziachristos P., Savas J. N., Zou L., Hashizume R., Meeks J. J., Shilatifard A., Resetting the epigenetic balance of Polycomb and COMPASS function at enhancers for cancer therapy. Nat. Med. 24, 758–769 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao Z. B., Szczepanski A. P., Tsuboyama N., Abdala-Valencia H., Goo Y. A., Singer B. D., Bartom E. T., Yue F., Wang L., PAX9 determines epigenetic state transition and cell fate in cancer. Cancer Res. 81, 4696–4708 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S6
Tables S1 to S6