Abstract
Oxygen is a major determinant of both survival and mortality of aerobic organisms. For the facultative anaerobe Lactococcus lactis, oxygen has negative effects on both growth and survival. We show here that oxygen can be beneficial to L. lactis if heme is present during aerated growth. The growth period is extended and long-term survival is markedly improved compared to results obtained under the usual fermentation conditions. We considered that improved growth and survival could be due to the capacity of L. lactis to undergo respiration. To test this idea, we confirmed that the metabolic behavior of lactococci in the presence of oxygen and hemin is consistent with respiration and is most pronounced late in growth. We then used a genetic approach to show the following. (i) The cydA gene, encoding cytochrome d oxidase, is required for respiration and plays a direct role in oxygen utilization. cydA expression is induced late in growth under respiration conditions. (ii) The hemZ gene, encoding ferrochelatase, which converts protoporphyrin IX to heme, is needed for respiration if the precursor, rather than the final heme product, is present in the medium. Surprisingly, survival improved by respiration is observed in a superoxide dismutase-deficient strain, a result which emphasizes the physiological differences between fermenting and respiring lactococci. These studies confirm respiratory metabolism in L. lactis and suggest that this organism may be better adapted to respiration than to traditional fermentative metabolism.
The toxic cellular effects of oxygen are a major factor in aging and mortality (3, 28). Oxygen toxicity is attributed to the activity of reactive oxygen species that attack proteins, lipids, and nucleic acids (15). Effects of oxygen have been extensively studied by use of bacterial models, principally with the facultatively respiring bacterium Escherichia coli (see references 7 and 13 for reviews). In this model, respiration itself is implicated as a source of oxidative damage in E. coli (8, 9, 18, 20, 27, and 36). It has been suggested that the shutdown of respiration in nutrient-limited conditions may reduce reactive oxygen species levels and thereby improve E. coli survival. Recent evidence further suggests that survival is favored by shifting cells to anaerobic conditions during entry into stationary phase (9).
Current information on the effects of oxygen is mainly based on respiring organisms. As such, the question of what anaerobes do in the presence of oxidative stress has been explored little. It is presumed that these organisms cope with stress in much the same way as aerobes, except that their defense systems, which may include superoxide dismutases (SODs) and catalases, may be more limited. However, there has been no demonstration to date that responses of anaerobes to an oxidative environment are predictable from the behavior of respiring bacteria.
The effects of oxygen have been examined with Lactococcus lactis, a gram-positive facultative anaerobe with a fermentative metabolism that can use different sugars to produce mainly l-(+)-lactic acid (19). Oxygenation of cultures results in an altered redox state and greater NADH oxidase activity (24, 25, 35); as a consequence, sugar fermentation is shifted toward mixed fermentation, and acetic acid, formic acid, CO2, ethanol, and acetoin, as well as lactic acid, are produced (25).
Despite its classification as an anaerobe and studies that have focused nearly entirely on its fermentative metabolism, results obtained about 30 years ago suggested that L. lactis is able to undergo respiratory growth, provided that heme is added to aerated cultures; this view was supported by a demonstration of altered metabolic end products, cytochrome formation, and hemin-dependent oxygen uptake (34). However, more recent studies of an L. lactis subsp. diacetylactis strain suggested that respiration does not occur under these conditions, as cytochromes could not be detected (21). To date this question has not been further explored, and the consequences of respiratory growth have not been analyzed.
The toxic effects of oxygen on L. lactis growth and survival have been revealed by several studies under fermentation conditions. Growth is reportedly inhibited by oxygen (5), and prolonged aeration of lactococcal cultures can lead to cell death and DNA degradation (10). Oxygen toxicity may be due to formation of hydrogen peroxide and hydroxyl radicals (1, 10). Unlike E. coli, L. lactis possesses a single SOD (31) and no catalase. It was found that the addition of exogenous catalase improved survival of L. lactis cells exposed to oxygen (10). These results suggest that L. lactis may not be fully equipped to withstand the toxic effects of an oxidative environment.
Our studies on oxygen toxicity led us to dissect the positive effects of the addition of exogenous catalase on growth and survival of L. lactis. As catalase contains a heme nucleus (in which iron is complexed with a porphyrin molecule), we first examined the effects of oxygen in the presence of heme. We confirmed that L. lactis is capable of respiratory growth, in agreement with earlier work (34). Respiration conditions result in improved growth and a spectacular increase in long-term survival compared to growth under conventional fermentation conditions. The observed phenotypes require an intact cydA gene, which encodes cytochrome d oxidase. Under respiration conditions, fermentation occurs during initial growth, while respiration is greatest during the late exponential phase.
(An initial oral communication of respiration and its effects on lactococci was first presented at the Lactic Acid Conference in Veldhoven, Holland, in September 1999.)
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
The strains used were L. lactis MG1363 (16); MG1363 sodA (31) and hemZ and cydA (this work) derivatives; and L. lactis subsp. lactis and L. lactis subsp. cremoris strains IL-1403, IL-582, IL801, IL896, Z105, Z106, and Z191 (CNRZ strain collection; kindly provided by P. Tailliez [37]). The two subspecies are closely related (over 80% identity between the subspecies strains, as determined using BLAST for gene analogues). Plasmid pKatE encodes the Bacillus subtilis katE gene (12) under the control of a constitutive promoter (P. Duwat and S. Sourice, unpublished data).
L. lactis strains were grown at 30°C in rich M17 medium (containing Bacto Tryptone, Bacto Soytone, meat digest, yeast digest, ascorbic acid, magnesium sulfate, and disodium β-glycerophosphate; Difco) supplemented with 1% glucose (M17-glu). Note that slight differences in L. lactis growth were observed between medium batches. Erythromycin at 2.5 μg/ml was added to sodA, hemZ, and cydA culture medium. Hemin (Sigma) stock solution (0.5 mg/ml) was prepared in alkaline water (0.05 N NaOH) and autoclaved; 20 μl of hemin was added per ml of medium. Protoporphyrin IX (PPIX; Sigma) stock solution (0.5 mg/ml) was prepared in alkaline water; 20 μl of PPIX was added per ml of medium. Precultures (routinely nonaerated cultures grown overnight in medium lacking hemin) were diluted 1/1,000 in fresh test medium in order to perform growth and survival experiments under the specified conditions. Nonaerated cultures were grown without agitation, and aerated cultures were maintained in Erlenmeyer flasks filled to less than 1/10-volume capacity in a shaking (250 rpm) water bath. Aliquots of prepared cells were removed at various times for plating on nonselective solid M17-glu, measurements of the optical density at 600 nm (OD600), and pH determinations.
Metabolic product determinations.
Biochemical measurements were performed with 24-h culture supernatants of cells grown as described above. Measurements of glucose, acetate, lactate, and ethanol were obtained according to kit supplier's instructions (Boehringer GmbH, Mannheim, Germany). Levels of diacetyl and acetoin were determined by gas chromatography (HRGC5160 Carlo Erba Instrument; ThermoQuest, Les Ulis, France) with an FFAP column (12 m; inner diameter, 0.52 mm; phase thickness, 1 μm; J and W Scientific, Folsom, Calif.). Conditions for static headspace analysis were as follows. Five milliliters of a culture supernatant saturated with NH2SO4 (5 g) and containing 0.25 ml of 22% H2SO4 plus 0.2 ml of a 0.15% aqueous solution of 2-pentanol as an internal standard was placed in a 22-ml vial fitted with a Minimert valve (Interchim). The mixture was warmed to 60°C for 15 min before samples (0.1 ml) were removed with a gas syringe. Chromatography was performed with an initial oven temperature of 55°C (2 min). The temperature was increased by 30°C min−1 up to 145°C and then by 5°C min−1 up to 150°C (kept there for 4 min). The flow rate of the carrier gas (H2) was 5 ml min−1. The temperature of the injector, in the splitless mode, was 145°C; that of the flame ionization detector was 200°C. Data were recorded with a Kontron PC integration pack. Concentrations were calculated from standard curves.
Oxygen detection.
Dissolved oxygen was determined using an oxygen meter (Ponselle, Versailles, France). Twenty-four-hour aerated (oxygenated) cultures grown with or without hemin (Ox/H or Ox/No H, respectively) were centrifuged, washed twice in saline buffer (50 mM Tris-HCl, 0.15 M NaCl [pH 7]), and resuspended at an OD600 of 0.5 in saline buffer containing 1% glucose. Aerated buffer devoid of bacteria was used as the reference for oxygen-saturated medium, and aerated water was used for calibrations. Thirty-milliliter samples were used for measurements. After cells were resuspended in buffer, all samples were vortexed and allowed to reequilibrate for 3 min before measurement at the first time point. Measurements were obtained at room temperature. Data represent the means of two independent experiments. Measurements varied by a maximum of 10%.
Mutant constructions.
Alignments of microbial cydA and hemZ genes (http://www.ncbi.nlm.nih.gov/BLAST/unfinishedgenome.html and http://www.ncbi.nlm.nih.gov/BLAST/) were used to design several pairs of degenerate primers predicted to isolate respective internal gene fragments. The primers used in the final experiments were as follows: 5′-CARTTYGGNATGAAYTGG-3′ and 5′-CATRATNCTRAANGTCCA-3′ for cydA (designed to amplify cydA open reading frame positions 75 through 334, as defined for ∼470-amino-acid full-length cytochrome d ubiquinol oxidase subunit I) and 5′-ATHTGGACNGAYGARGG-3′ and 5′-ARNGGDATNCCRTGRTA-3′ for hemZ (designed to amplify hemZ open reading frame positions 70 through 200, as defined for ∼310-amino-acid full-length ferrochelatase lyase). PCRs using Thermus aquaticus Taq polymerase were carried out as follows: 5 min at 96°C, followed by 30 cycles of 15 s at 96°C, 15 s at 50°C, and 15 s at 72°C. Fragment sizes were ∼780 bp for cydA and ∼390 bp for hemZ. After end filling in using T4 polymerase plus the Klenow fragment, purified fragments were cloned into the SmaI site of pRV300 (23). Sequences were confirmed to correspond to putative cydA and hemZ genes.
The cydA and hemZ genes were inactivated by single-crossover recombination. L. lactis MG1363 electrocompetent cells were transformed (10) with pRV300 derivatives containing the appropriate internal gene segments, with selection on M17-glu containing 2.5 μg of erythromycin/ml. Strain constructions were verified by Southern hybridization using appropriate gene fragments as probes.
Northern blotting experiments.
Cultures were inoculated with a 1/200 dilution of an overnight culture of MG1363 grown without aeration in M17-glu and then were grown under Ox/H and Ox/No H conditions. Samples were removed at various cell densities, corresponding to about 3, 4, 6, and 8 h after the start of growth. Cell concentrations were all adjusted to a cell density equal to an OD600 of 1 before cell samples were prepared. RNA preparation and Northern blot analyses were carried out as previously described (29). The cydA-specific probe corresponds to a 777-bp PCR-amplified internal fragment, as described above. A 16S ribosomal DNA (rDNA)-specific probe was used to ensure that the same amounts of total RNA were present in the wells (data not shown). Probes were labeled by nick translation using Ready-to Go (Pharmacia) and [32P]dCTP.
RESULTS
L. lactis growth period is extended in aerated medium containing heme.
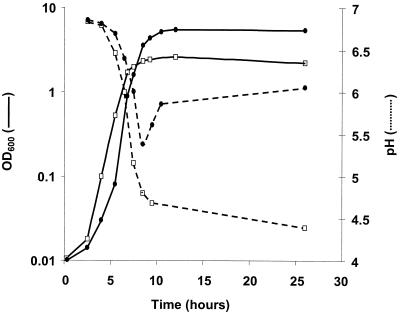
The growth of L. lactis strain MG1363 in M17-glu was compared under Ox/H or Ox/No H conditions and conditions of nonoxygenation with or without hemin (No Ox/H or No Ox/No H, respectively) (Fig. 1). Exponential-phase growth kinetics were similar for the first 8 h under all four conditions. After this time, the No Ox/H, Ox/No H, and No Ox/No H cultures entered stationary phase and attained a final pH of 4.4. In contrast, growth of the Ox/H culture continued, with a final biomass about twofold greater than those of the other three cultures and a final pH of ∼6. The pH decline during the first 8 h of growth was similar for all cultures; after this time, the Ox/H culture pH started to rise. These results suggest that L. lactis metabolism is altered by growth in Ox/H conditions; differences are first detected just before control cultures enter stationary phase.
FIG. 1.
Altered growth and pH of L. lactis grown under Ox/H conditions. An overnight 30°C MG1363 culture in M17-glu was used to inoculate media (at a 1/1,000 dilution) for growth and pH measurements under four conditions at 30°C: Ox/H, No Ox/H, Ox/No H, and No Ox/No H. As growth curves and final pHs for the No Ox/H, Ox/No H, and No Ox/No H conditions were similar, only data for the No Ox/No H (open squares; solid line for growth, broken line for pH) and Ox/H (closed circles; solid line for growth, broken line for pH) conditions are shown. Growth of all cultures was saturated at 24 h. Experiments were performed six times and yielded comparable results. Results of a representative experiment are shown.
Ten lactococcal strains representative of L. lactis species were examined for these effects of oxygen and hemin on growth. All exhibited increased biomass and a relative higher pH when grown in Ox/H conditions compared to the usual fermentation conditions (No Ox/No H) (data not shown). This result suggests that the capacity for improved growth and altered metabolism can be generalized to L. lactis.
Ox/H conditions result in markedly improved long-term survival.
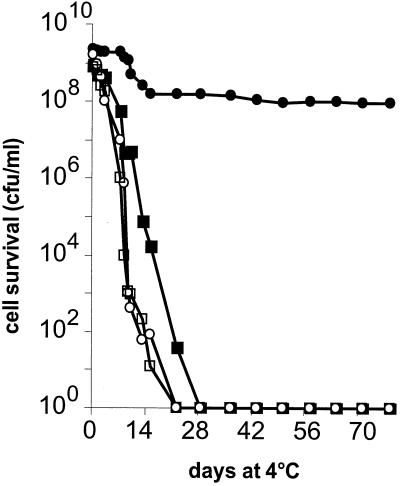
Strain MG1363 was cultured under Ox/H, No Ox/H, Ox/No H, and No Ox/No H conditions. The cell populations of the latter three cultures at 24 h reached ∼2 × 109 per ml, while that of the Ox/H culture was 7 × 109 per ml. After 80 h, survival of the three control cultures dropped 105-, 106-, and 108-fold for the No Ox/H, Ox/No H, and No Ox/No H cultures, respectively. In marked contrast, the viability of the Ox/H culture remained within twofold the initial cell number.
The effects of the different growth conditions on the long-term storage survival of L. lactis were examined. After 24 h of growth, cultures were stored at 4°C. After 10 days, the viability of cultures grown under No Ox/H, Ox/No H, and No Ox/No H conditions declined by more than 106-fold and continued to decline with time. In contrast, the Ox/H culture was nearly 100% viable; after 2 months of storage, viability was still about 10% (Fig. 2).
FIG. 2.
Improved long-term survival of L. lactis after growth under Ox/H conditions. MG1363 cells were grown under four conditions at 30°C as described in the legend to Fig. 1: Ox/H (closed circles), No Ox/H (closed squares), Ox/No H (open circles), and No Ox/No H (open squares). After 24 h, cultures were transferred to 4°C. Cell viability was determined by plating dilutions on solid M17-glu at the indicated times. Experiments were performed four times and yielded comparable results. Results of a representative experiment are shown.
As the final pH was higher in the Ox/H culture, it was possible that the greater survival was a consequence of the higher pH and differences in medium conditions. This possibility was addressed in two ways. First, the growth medium was strongly buffered (with 400 mM morpholinepropanesulfonic acid [MOPS]) and glucose was limited to 0.25%, so that the pH did not vary during the growth of Ox/H and No Ox/No H cultures. Under these conditions, the Ox/H culture showed a more modest improvement in biomass (the OD600 was 0.8, compared to 0.5 for the fermentation culture), as expected from the limiting glucose conditions. After 1 month at 4°C, the survival of the Ox/H culture was ∼50%, compared to only 0.0001% for the No Ox/No H culture. In a second approach, Ox/H and No Ox/No H cultures were transferred to Ringer's buffered solution after 24 h of growth to alleviate the possible influence of medium differences on survival. After 20 days at 4°C, the survival of the Ox/H culture was 50%, compared to 0.005% for the No Ox/No H culture. Both of these controls indicated that the change in cell physiology, rather than environmental factors, is the determining factor in the improved survival of the Ox/H culture.
These results demonstrate that the presence of heme in aerated L. lactis cultures results in a striking improvement in long-term survival.
Metabolism is altered under Ox/H compared to fermentative growth conditions, particularly late in growth.
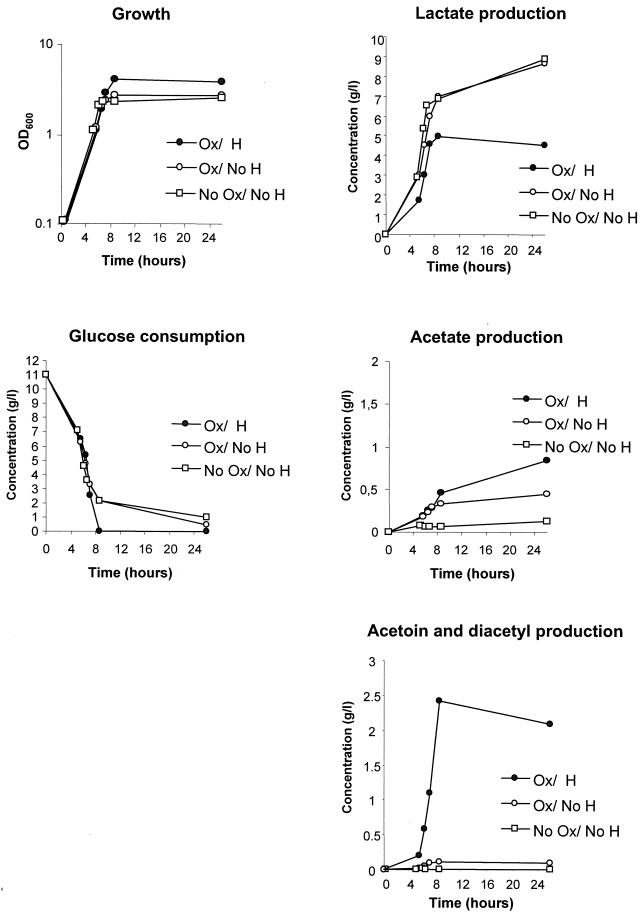
During L. lactis fermentation, glucose is metabolized to form mainly lactic acid, and the final pH is ∼4.5. However, while the Ox/H culture initially produced a pH decrease, the final pH was significantly higher (Fig. 1). As this result suggested altered metabolism, particularly late in growth, we compared glucose consumption and the amounts of expected fermentation products for Ox/H, Ox/No H, and No Ox/No H cultures (Fig. 3). The three cultures displayed similar glucose consumption and lactic acid accumulation during the first 6 to 7 h of growth, indicating that fermentation occurs in all cases. After this time, the Ox/H culture could be distinguished from the other cultures by continued rapid glucose consumption, arrest, and subsequent reduction in lactic acid accumulation. (Final amounts of lactic acid under Ox/H conditions were sometimes as low as 1 g/liter; data not shown.) Acetoin was observed to accumulate early during growth in the Ox/H culture, and the amounts were clearly different from those in the other cultures; the greatest increment in acetoin compared to that in the other cultures occurred after ∼8 h of growth. Similarly, higher acetate levels in the Ox/H culture showed the greatest difference late in growth. These results show that (i) the higher final pH of the Ox/H culture is correlated with reduced amounts of acid end products; (ii) the Ox/H culture undergoes fermentation in the beginning of growth, as reflected by lactic acid accumulation; and (iii) a metabolic shift in the Ox/H culture is most pronounced late in growth. The above results are consistent with the results of earlier studies (34) that correlated the altered metabolism with respiration. They further suggest that the respiration-like metabolism occurs late in growth.
FIG. 3.
Metabolic alterations in Ox/H cultures are greatest in late exponential phase. L. lactis MG1363 was grown aerobically in M17-glu in the presence or absence of hemin or anaerobically in the absence of hemin. Samples were taken at various intervals to monitor growth, glucose consumption, and lactate, acetate, acetoin, and diacetyl accumulation (see Materials and Methods). Determinations were performed at least twice, with a maximum deviation between measurements of 10%.
Genetic evidence for respiration.
Aerobic respiration requires a functional electron transfer chain formed by cytochromes with oxygen as a terminal electron acceptor, resulting in the production of ATP (17). In E. coli, two terminal oxidases may be produced when cells are grown under aerobic conditions. Cytochrome bo, encoded by the cyoABCDE operon, is induced under oxygen-rich conditions, and cytochrome bd, encoded by the cydAB operon, is induced in stationary phase or under oxygen-limited conditions (6). The above results suggested that respiratory-like growth in L. lactis occurs preferentially in late logarithmic phase. Furthermore, L. lactis may grow under oxygen-poor conditions in nature (14). We thus hypothesized that cytochrome bd, which is active under oxygen-limited conditions, may be present.
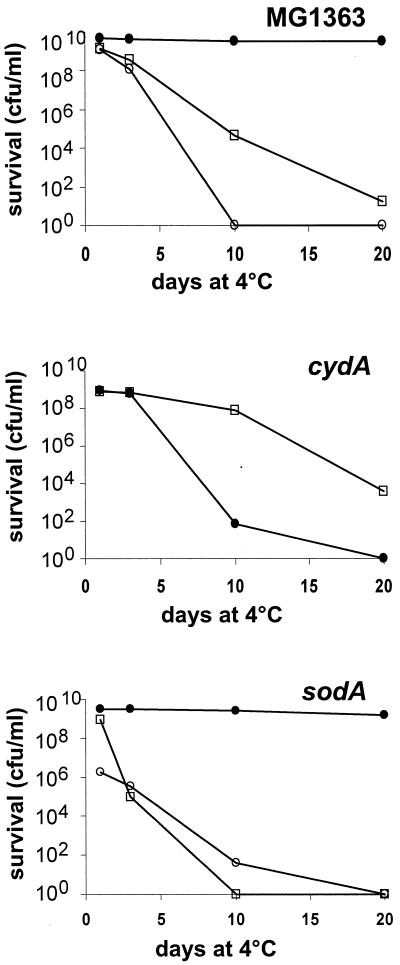
The existence of an active cydA homologue in L. lactis MG1363 was confirmed. Using degenerate primer amplification, an internal segment of the cydA gene was recovered and used to generate a cydA inactivation mutant. The mutant was tested for its capacity to undergo the respiration-like metabolism. Growth of the cydA strain under Ox/H conditions was fermentative; the final pH was 4.5, and the biomass was equivalent to that of fermentation cultures. Long-term survival of the cydA mutant under Ox/H conditions was poor; a 107-fold decrease in viability was observed after 10 days of storage, compared to full survival of the wild-type strain (Fig. 4). (Note that under fermentation conditions, survival at 10 days was better for the cydA mutant than for MG1363, although some variations in these values were seen between experiments.) The poor viability of the cydA mutant rules out the possibility that the heme molecule itself protects cells from oxidative damage (manganic porphyrins have a reported protective effect against superoxide radicals [2]). Therefore, the effects of hemin and oxygen on L. lactis growth depend on the cydA gene product. These results are strongly suggestive of a respiratory metabolism that requires the cydA gene product in L. lactis.
FIG. 4.
Effects of cydA and sodA mutations on L. lactis long-term survival under respiration conditions. MG1363 and cydA and sodA mutants were grown at 30°C as described in the legend to Fig. 2 under Ox/H (closed circles), Ox/No H (open circles; for MG1363 and sodA strains), and No Ox/No H (open squares) conditions. After 24 h, cultures were transferred to 4°C. Cell viability was determined by plating dilutions on solid M17-glu at the indicated times. Experiments were performed at least twice; the results of a representative experiment are shown.
cydA expression is induced in the late growth phase under respiration conditions.
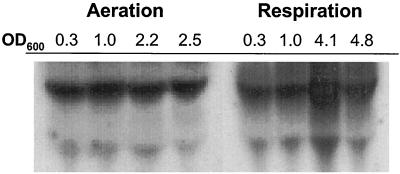
Our results indicate that respiratory growth is a late-exponential-phase event. We examined whether cydA expression is growth regulated in L. lactis. For this purpose, Northern blotting was performed with mRNA samples extracted at different times during growth from cells cultured under aeration or respiration conditions (Fig. 5). RNA concentrations were monitored using a 16S rDNA probe (data not shown). The cydA-specific probe revealed the presence of two cydA transcripts. As a potential cydA-cydB operon was revealed by diagnostic sequencing of L. lactis strain IL-1403 (4), we consider it likely that the two transcripts correspond to cydA mRNA and the bicistronic mRNA. The cydA transcripts were detected in all samples and under both growth conditions. However, the expression of cydA mRNA was induced under respiration conditions after ∼6 h of growth (corresponding to an OD600 of 4.1) but not under aeration conditions. This late induction of cydA is in keeping with our results suggesting that respiration is favored during the late growth phase. Nevertheless, the detection of cydA transcripts under other growth conditions may indicate that cytochrome activity is limited by the absence of a necessary cofactor; heme uptake also may be a growth-phase-regulated event.
FIG. 5.
Expression of cydA is induced under Ox/H conditions in late-exponential-phase cells. L. lactis strain MG1363 was grown under aeration and respiration conditions (see Materials and Methods), and samples were removed for total RNA extraction at different OD600s during growth (times, from left to right, correspond to approximately 3, 4, 6, and 8 h after the start of growth). Northern blot analysis was performed using an internal cydA fragment as a probe. Membranes were subsequently dehybridized and rehybridized with a 16S rDNA probe to verify that the amounts deposited in the wells were identical. Northern blotting was performed three times and yielded results similar to those shown here.
cydA is directly involved in L. lactis oxygen uptake.
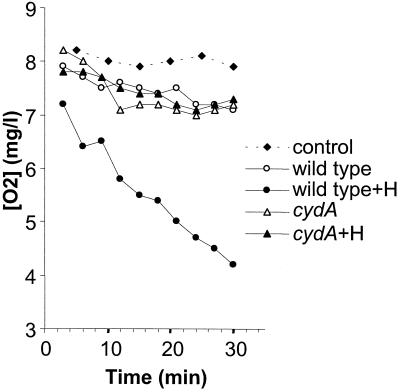
Respiration involves oxygen consumption from a medium. It was previously shown that oxygen is consumed by L. lactis and that oxygen uptake is abolished in the presence of KCN, a respiration inhibitor (34). If cytochrome d oxidase is required for respiration, then a cydA mutant would be expected to be deficient for oxygen uptake. To address this question, we compared oxygen depletion from a medium containing either wild-type or cydA resting cells that had been cultured under aeration conditions in the presence or absence of hemin (Fig. 6). Note that this experiment is designed to determine relative oxygen consumption between strains in a resting state and is not a measure of respiration rate. Rapid oxygen depletion from the medium was observed for wild-type cells that had been grown under Ox/H conditions but not under Ox/No H conditions. In contrast, no oxygen depletion was observed for cydA cells regardless of the growth conditions. These results show that cydA is required for oxygen utilization in L. lactis.
FIG. 6.
Heme-dependent oxygen consumption by L. lactis is abolished in the mutant cydA strain. Aerated cultures of MG1363 and the cydA mutant, grown with or without hemin (H), were harvested after overnight growth. Oxygen consumption was evaluated with cells resuspended in saline buffer containing 1% glucose (see Materials and Methods). Oxygen remaining in the medium was measured using an Oxymeter. Ox/H and Ox/No H conditions are represented by closed and open symbols, respectively. Duplicate samples showed less than a 10% difference in measurements. Experiments were performed twice and yielded comparable results.
It could be argued that cydA plays an indirect role in oxygen consumption, by affecting heme uptake. If the cydA mutation causes a defect in heme uptake, then a heme-requiring enzyme would be inactive in the mutant. To test this idea, we expressed a heterologous enzyme, the heme-requiring intracellular catalase (KatE) from B. subtilis (12), in wild-type and cydA mutant strains. (Note that L. lactis is catalase negative, even in the presence of heme.) Plasmid pKatE (encoding KatE) was established in both strains, and resuspended cell pellets of overnight cultures were tested for catalase activity. Both strains were catalase negative in the absence of heme and catalase positive when heme was present during growth. These results show that both cydA mutant and wild-type L. lactis strains are capable of assimilating heme.
Taken together, the above results demonstrate genetically that the cydA gene in L. lactis encodes an active product. Furthermore, this active cydA gene product participates directly in heme-mediated oxygen assimilation in L. lactis.
Ferrochelatase activity in L. lactis.
Respiratory growth requires the addition of a heme compound to the aerated medium. We observed that the addition of the heme precursor PPIX confers effects equivalent to those conferred by hemin on the growth and survival of L. lactis. As PPIX allows respiratory growth, we postulated that L. lactis must express ferrochelatase, which charges PPIX with reduced iron to form heme. Degenerate primers were used to amplify from L. lactis strain MG1363 a 390-bp fragment that, as determined by sequencing, corresponds to a hemZ (ferrochelatase-encoding) gene. The fragment was used to construct a hemZ-disrupted mutant of MG1363. The hemZ-disrupted mutant was capable of respiratory-like growth upon heme addition to the medium. However, respiration did not occur when PPIX was added. These results show that at least the last gene of the heme biosynthesis pathway is functional in L. lactis. They also suggest that L. lactis has transport systems that allow protoporphyrin and iron uptake.
SodA is not required for long-term survival under respiration conditions.
Superoxides are a major cause of stationary-phase mortality; for this reason, SODs are needed for bacterial survival in aerobic conditions (e.g., see reference 2). We examined the role of the unique SOD, SodA, in L. lactis. As expected, the sodA mutant survived very poorly under Ox/No H conditions (Fig. 4); only ∼104 to 106 cell survivors were present in a 24-h aerated culture. In striking contrast, the sodA mutant behaved like the wild-type strain under respiration conditions; 50% of the cells were viable after 20 days of storage. Thus, in L. lactis, SodA is required for survival under aeration conditions but not under respiration conditions. These results support our observations that heme has considerable effects on L. lactis physiology, such that cells are less susceptible to oxidative stress and hence do not require SodA for survival when grown in the presence of a heme compound.
DISCUSSION
Capacity of a fermenting anaerobe to undergo respiration.
The fermentative metabolism of L. lactis has been intensively studied, mainly due to its industrial relevance. Despite a past report suggesting that L. lactis can respire when oxygen and hemin are available (34), this observation has not been further documented since that time. We provide genetic and additional biochemical evidence for respiration and demonstrate for the first time that lactococci can be better adapted for growth and survival under these conditions than under fermentation conditions. The strong impact of respiration on lactococcal growth and long-term survival suggests that this mode of growth may be significant to lactococcal biology and should be further examined.
Our results revealed that cydA is present in wild-type L. lactis and that its activity is needed for the observed respiration. The cydA gene product is 46 and 32% identical to the cydA gene products of B. subtilis and E. coli, respectively. L. lactis sequence analysis also revealed the presence of menB, menC, menD, menE, and ubiE (menH) open reading frames, which are involved in biosynthesis of the electron carriers menaquinone and ubiquinone (4). The existence of other factors involved in respiration is suggested from recent genome sequence data (4, 38), as well as from the detection of specific enzyme activities of the Krebs cycle (22). At least the last enzyme of the heme biosynthesis pathway, encoded by hemZ, is present and active, as respiration in L. lactis can occur when a heme precursor (PPIX) is added to the medium. The hemZ open reading frame is 35% identical to its E. coli homologue. Putative hemK and hemN genes, involved in the transformation of coproporphyrinogen III to PPIX, are also present (4); homologues of B. subtilis hemA, hemB, hemC, and hemD genes were not detected by a BLAST search. The ability to utilize an external heme source indicates that a heme uptake mechanism must also be present in L. lactis.
Several metabolic changes are associated with respiration, including a large decrease in lactic acid accumulation and a large increase in acetoin accumulation. Oxidative conditions are expected to increase the NAD/NADH ratio, rerouting part of pyruvate metabolism toward acetoin production (24). As acetoin is also accumulated under nonrespiratory oxidative conditions, it is likely that its accumulation is not involved in the respiration process.
Previous studies suggested that another bacterium within the family Streptococcaceae, Streptococcus faecalis (now classified as Enterococcus faecalis), may also be capable of respiration (30, 40; see reference 39 for a review). In view of the diverse ecological niches and functions of the Streptococcaceae (e.g., as pathogens in animals and humans as well as in foods and on plants [11]), the existence of a respiration mechanism may be an important feature distinguishing these related organisms.
Respiration is a late-phase event.
Several lines of evidence indicate that lactococcal metabolism is biphasic under respiration conditions, as tested here, growing first mainly via fermentation and then via respiration. (i) Lactic acid accumulation and a pH decline are observed during the first ∼7 h of growth under respiration conditions; similar observations were previously described for an L. lactis subsp. diacetylactis strain, although respiration was ruled out (21). (ii) The cydA gene, which is required for respiration, is induced transcriptionally in late exponential phase in cells grown under respiration conditions. In E. coli, the corresponding cydA gene is also expressed in stationary phase (6). L. lactis genome analysis (4) has revealed the presence of cydA, cydB, cydC, and cydD open reading frames (the latter two are required for cytochrome d oxidase assembly [32]), while the cyoABCDE genes (required for cytochrome o oxidase synthesis) are absent. Note that although cydA transcription is induced late in Ox/H growth, transcripts are detected throughout growth. This finding suggests that other factors, e.g., hemin uptake, may limit cytochrome activity in early exponential phase.
The biphasic metabolism observed for L. lactis suggests that the balance between fermentation and respiration may be affected by the growth conditions used. It will be of interest to determine the role of medium components that may be involved in the balance between these very different modes of growth.
Respiration confers long-term survival on L. lactis.
The respiratory lifestyle of lactococci results in a greater growth yield and a remarkable improvement in survival (up to 108-fold) compared to that under fermentation conditions. These results indicate a strong link between respiratory metabolism and long-term survival. Studies with E. coli have revealed the importance of numerous functions associated with long-term survival, several of which are involved in defense against oxygen (26, 32, 33). For example, the cydCD genes, which are needed for cytochrome d oxidase formation, also affect long-term survival or exit from stationary phase (32, 33). In lactococci, improved long-term survival after growth under respiration conditions requires an active cydA gene. We suggest that cytochrome d oxidase activity may serve as an oxygen trap to reduce oxygen toxicity and may also be involved in the activation of other survival genes. This hypothesis may explain why lactococci are generally observed to have poor long-term survival under conventional fermentation conditions (10), where heme and oxygen are absent.
Paradoxically, the potentially toxic effects of respiration under conditions approaching starvation have been evoked in E. coli (8, 9, and 27); cell survival is greater if the culture is shifted to anaerobic conditions before starvation than if cells are left in aerobic conditions. In contrast, L. lactis survival is greatly improved when oxygen-mediated respiration is possible during the late growth phase. The distinct responses of these evolutionarily distant microorganisms suggest that the E. coli aerobic model may not be universally applicable for oxidative stress response and survival. This proposal is substantiated by our observations that under respiration conditions, a sodA L. lactis mutant exhibited long-term survival equivalent to that of the wild-type strain. In contrast, mutations in sodA genes of several fully respiratory organisms result in long-term survival defects (e.g., see reference 9). L. lactis may thus constitute a prototype for a class of organisms that undergo both fermentation and respiration during aerobic growth.
ACKNOWLEDGMENTS
We thank our colleagues Claus Maxel Henrikson, Stig Lykke Iversen, Dan Nilsson, and Egon Bech Hansen (Chr Hansen, Hørshlom, Denmark) for discussions and communication of unpublished data. We thank Pierre DeKinkelin for use of the oxygen meter and Alexandre Bolotin, Alexei Sorokin, and Dusko Ehrlich for access to the L. lactis database prior to publication. We are grateful to Philippe Bouloc, Anne Bravard, and Astrid Vrang for discussions; David Halpern and Matthieu Schaeffer for technical assistance; and Meriem El Karoui for valuable suggestions during this work. We thank Michel Desmazeaud and the Génétique Appliquée team for support throughout this work.
Part of this work was supported by a research grant from Chr Hansen.
Footnotes
This paper is dedicated to Patrick Duwat (died 5 January 2000), who was a driving force of this work.
REFERENCES
- 1.Anders R F, Hogg D M, Jago G R. Formation of hydrogen peroxide by group N streptococci and its effect on their growth and metabolism. Appl Environ Microbiol. 1970;19:608–612. doi: 10.1128/am.19.4.608-612.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benov L, Fridovich I. A superoxide dismutase mimic protects sodA sodB Escherichia coli against aerobic heating and stationary-phase death. Arch Biochem Biophys. 1995;322:291–294. doi: 10.1006/abbi.1995.1465. [DOI] [PubMed] [Google Scholar]
- 3.Berlett B S, Stadtman E R. Protein oxidation in aging, disease, and oxidative stress. J Biol Chem. 1997;272:20313–20316. doi: 10.1074/jbc.272.33.20313. [DOI] [PubMed] [Google Scholar]
- 4.Bolotin A, Mauger S, Malarme K, Ehrlich S D, Sorokin A. Low-redundancy sequencing of the entire Lactococcus lactis IL1403 genome. Antonie Leeuwenhoek. 1999;76:27–76. [PubMed] [Google Scholar]
- 5.Condon S. Responses of lactic acid bacteria to oxygen. FEMS Microbiol Rev. 1987;46:269–280. [Google Scholar]
- 6.Cotter P A, Chepuri V, Gennis R B, Gunsalus R P. Cytochrome o (cyoABCDE) and d (cydAB) oxidase gene expression in Escherichia coli is regulated by oxygen, pH, and the fnr gene product. J Bacteriol. 1990;172:6333–6338. doi: 10.1128/jb.172.11.6333-6338.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Demple B, Harrison L. Repair of oxidative damage to DNA: enzymology and biology. Annu Rev Biochem. 1996;63:915–948. doi: 10.1146/annurev.bi.63.070194.004411. [DOI] [PubMed] [Google Scholar]
- 8.Dukan S, Nystrom T. Bacterial senescence: stasis results in increased and differential oxidation of cytoplasmic proteins leading to developmental induction of the heat shock regulon. Genes Dev. 1998;12:3431–3441. doi: 10.1101/gad.12.21.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dukan S, Nystrom T. Oxidative stress defense and deterioration of growth-arrested Escherichia coli cells. J Biol Chem. 1999;274:26027–26032. doi: 10.1074/jbc.274.37.26027. [DOI] [PubMed] [Google Scholar]
- 10.Duwat P, Ehrlich S D, Gruss A. The recA gene of Lactococcus lactis: characterization and involvement in oxidative and thermal stress. Mol Microbiol. 1995;17:1121–1131. doi: 10.1111/j.1365-2958.1995.mmi_17061121.x. [DOI] [PubMed] [Google Scholar]
- 11.Duwat P, Hammer K, Bolotin A, Gruss A. Genetics of lactococci. In: Fischetti V, editor. Gram-positive pathogens. Washington, D.C.: ASM Press; 2000. pp. 295–306. [Google Scholar]
- 12.Engelmann S, Lindner C, Hecker M. Cloning, nucleotide sequence, and regulation of katE encoding a sigma B-dependent catalase in Bacillus subtilis. J Bacteriol. 1995;177:5598–5605. doi: 10.1128/jb.177.19.5598-5605.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farr S B, Kogoma T. Oxidative stress responses in Escherichia coli and Salmonella typhimurium. Microbiol Rev. 1991;55:561–585. doi: 10.1128/mr.55.4.561-585.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fenton M P. An investigation into the sources of lactic acid bacteria in grass silage. J Appl Bacteriol. 1987;62:181–188. [Google Scholar]
- 15.Fridovich I. Oxygen toxicity: a radical explanation. J Exp Biol. 1998;201:1203–1209. doi: 10.1242/jeb.201.8.1203. [DOI] [PubMed] [Google Scholar]
- 16.Gasson M J. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol. 1983;154:1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gennis R B, Stewart V. Respiration. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: ASM Press; 1996. pp. 217–261. [Google Scholar]
- 18.Gonzalez-Flecha B, Demple B. Metabolic sources of hydrogen peroxide in aerobically growing Escherichia coli. J Biol Chem. 1995;270:13681–13687. doi: 10.1074/jbc.270.23.13681. [DOI] [PubMed] [Google Scholar]
- 19.Holt J G, Krieg N R, Sneath P H A, Staley J T, Williams S T. Gram-positive cocci. In: Hensyl W R, editor. Bergey's manual of determinative microbiology. 9th ed. Baltimore, Md: Williams & Wilkins; 1994. pp. 527–558. [Google Scholar]
- 20.Imlay J A, Fridovich I. Assay of metabolic superoxide production in Escherichia coli. J Biol Chem. 1991;266:6957–6965. [PubMed] [Google Scholar]
- 21.Kaneko T, Tagahashi M, Suzuki H. Acetoin fermentation by citrate-positive Lactococcus lactis subsp. lactis 3022 grown aerobically in the presence of hemin or Cu2+ Appl Environ Microbiol. 1990;56:2644–2649. doi: 10.1128/aem.56.9.2644-2649.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lapujade P, Cocaign-Bousquet M, Loubière P. Glutamate biosynthesis in Lactococcus lactis subsp. lactis NCDO 2118. Appl Environ Microbiol. 1998;64:2485–2489. doi: 10.1128/aem.64.7.2485-2489.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leloup L, Ehrlich S D, Zagorec M, Morel-Deville F. Single-crossover integration in the Lactobacillus sake chromosome and insertional inactivation of the ptsI and lacL genes. Appl Environ Microbiol. 1997;63:2117–2123. doi: 10.1128/aem.63.6.2117-2123.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lopez de Felipe F, Kleerebezem M, de Vos W M, Hugenholtz J. Cofactor engineering: a novel approach to metabolic engineering in Lactococcus lactis by controlled expression of NADH oxidase. J Bacteriol. 1998;180:3804–3808. doi: 10.1128/jb.180.15.3804-3808.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lopez de Felipe F, Starrenburg M J C, Hugenholtz J. The role of NADH oxidation in acetoin and diacetyl production from glucose in Lactococcus lactis subsp. lactis MG1363. FEMS Microbiol Lett. 1997;156:15–19. [Google Scholar]
- 26.Martinez A, Kolter R. Protection of DNA during oxidative stress by the nonspecific DNA-binding protein Dps. J Bacteriol. 1997;179:5188–5194. doi: 10.1128/jb.179.16.5188-5194.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nystrom T, Larsson C, Gustafsson L. Bacterial defense against aging: role of the Escherichia coli ArcA regulator in gene expression, readjusted energy flux and survival during stasis. EMBO J. 1996;15:3219–3228. [PMC free article] [PubMed] [Google Scholar]
- 28.Orr W C, Sohal R S. Extension of life-span by overexpression of superoxide dismutase and catalase in Drosophila melanogaster. Science. 1994;263:1128–1130. doi: 10.1126/science.8108730. [DOI] [PubMed] [Google Scholar]
- 29.Raya R, Bardowski J, Andersen P S, Ehrlich S D, Chopin A. Multiple transcriptional control of the Lactococcus lactis trp operon. J Bacteriol. 1998;180:3174–3180. doi: 10.1128/jb.180.12.3174-3180.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ritchey T W, Seeley H W., Jr Distribution of cytochrome-like respiration in streptococci. J Gen Microbiol. 1976;93:195–203. doi: 10.1099/00221287-93-2-195. [DOI] [PubMed] [Google Scholar]
- 31.Sanders J W, Leenhouts K J, Haandrikman A J, Venema G, Kok J. Stress response in Lactococcus lactis: cloning, expression analysis, and mutation of the lactococcal superoxide dismutase gene. J Bacteriol. 1995;177:5254–5260. doi: 10.1128/jb.177.18.5254-5260.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siegele D A, Imlay K R C, Imlay J A. The stationary-phase-exit defect of cydC (surB) mutants is due to the lack of a functional terminal cytochrome oxidase. J Bacteriol. 1996;178:6091–6096. doi: 10.1128/jb.178.21.6091-6096.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siegele D A, Kolter R. Isolation and characterization of an Escherichia coli mutant defective in resuming growth after starvation. Genes Dev. 1993;7:2629–2640. doi: 10.1101/gad.7.12b.2629. [DOI] [PubMed] [Google Scholar]
- 34.Sijpesteijn A K. Induction of cytochrome formation and stimulation of oxidative dissimilation by hemin in Streptococcus lactis and Leuconostoc mesenteroides. Antonie Leeuwenhoek. 1970;36:335–348. doi: 10.1007/BF02069035. [DOI] [PubMed] [Google Scholar]
- 35.Smart J B, Thomas T D. Effect of oxygen on lactose metabolism in lactic streptococci. Appl Environ Microbiol. 1987;53:533–541. doi: 10.1128/aem.53.3.533-541.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Storz G, Imlay J A. Oxidative stress. Curr Opin Microbiol. 1999;2:188–194. doi: 10.1016/s1369-5274(99)80033-2. [DOI] [PubMed] [Google Scholar]
- 37.Tailliez P, Tremblay J, Ehrlich S D, Chopin A. Molecular diversity and relationship within Lactococcus lactis, as revealed by randomly amplified polymorphic DNA (RAPD) Syst Appl Microbiol. 1998;21:530–538. doi: 10.1016/S0723-2020(98)80065-9. [DOI] [PubMed] [Google Scholar]
- 38.Wang H, Baldwin K A, O'Sullivan D J, McKay L L. Identification of a gene cluster encoding Krebs cycle oxidative enzymes linked to the pyruvate carboxylase gene in Lactococcus lactis ssp. lactis C2. J Dairy Sci. 2000;83:1912–1918. doi: 10.3168/jds.S0022-0302(00)75066-1. [DOI] [PubMed] [Google Scholar]
- 39.Whittenbury R. Biochemical characteristics of Streptococcus species. Soc Appl Bacteriol Symp Ser. 1978;6:51–69. [PubMed] [Google Scholar]
- 40.Winstedt L, Frankenberg L, Hederstedt L, von Wachenfeldt C. Enterococcus faecalis V583 contains a cytochrome bd-type respiratory oxidase. J Bacteriol. 2000;182:3863–3866. doi: 10.1128/jb.182.13.3863-3866.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]