Abstract
An imbalance in the activities of the Polycomb and Trithorax complexes underlies numerous human pathologies, including cancer. The BRCA1 associated protein-1 (BAP1) deubiquitinase negatively regulates Polycomb activity and recruits the Trithorax histone H3K4 methyltransferase, mixed-lineage leukemia protein 3 (MLL3) within Complex Proteins Associated with Set1 (COMPASS), to the enhancers of tumor suppressor genes. We previously demonstrated that the BAP1-MLL3 pathway is mutated in several cancers, yet how BAP1 recruits MLL3 to its target loci remains an important unanswered question. We demonstrate that the ASXL2 subunit of the BAP1 complex mediates a direct interaction with MLL3/COMPASS. ASXL2 loss results in decreased MLL3 occupancy at enhancers and reduced BAP1-MLL3 target gene expression. Interaction between ASXL2 and MLL3 is negatively regulated by protein arginine methyltransferase 4 (PRMT4/CARM1), which methylates ASXL2 at R639/R641. ASXL2 methylation blocks binding to MLL3 and impairs the expression of MLL3/COMPASS-dependent genes. This previously unidentified transcriptional repressive function of CARM1 provides insight into the BAP1/MLL3-COMPASS axis and reveals a potential cancer therapeutic target.
Arginine methylation of ASXL2 inhibits the transcriptional activation function of the BAP1-MLL3/COMPASS complex.
INTRODUCTION
Genetic alterations of epigenetic factors occur frequently in human cancer (1, 2), making these proteins potential therapeutic targets (3). MLL3 (KMT2C)/COMPASS catalyzes histone H3K4 monomethylation at intergenic enhancers (4, 5) and is commonly mutated in cancer (6, 7). MLL3 mutations occur in a variety of human tumors, including hepatocellular carcinoma, colon cancer, bladder cancer, myelodysplastic syndromes, and acute myeloid leukemia (AML) (8–10). In human breast cancer, the frequency of MLL3 mutations (8.41%) is remarkably higher than those found in other cancer types (11). In addition, in HR+/HER2− breast cancer, MLL3 mutation rates are higher in metastatic breast cancer than in early breast cancer (12). Precisely how these MLL3 mutations drive tumorigenesis is currently unknown.
We previously identified a mutational hotspot in MLL3, located within the plant homeodomain (PHD) finger repeats (13). Cancer-associated mutations in the PHD domain correlate with reduced breast cancer survival rates, suggesting that this mutational hotspot alters MLL3 functionality. We demonstrated that the MLL3 PHD fingers directly interact with the BAP1 complex, a major histone H2A deubiquitinase and tumor suppressor. Moreover, cancer-associated mutations in the PHD fingers disrupt the interaction between MLL3 and BAP1 (13). The enhancer occupancies of MLL3 and an additional COMPASS subunit, lysine-specific demethylase 6A (KDM6A; also known as UTX), are reduced in the absence of BAP1 (13). Collectively, these results support a model that the BAP1 complex is an essential cofactor for MLL3 function that interacts through the MLL3 PHD domain and an as of yet uncharacterized region of the BAP1 complex. On the basis of these findings, and because of the dynamics of histone modifications, gene expression, and epigenetic reprogramming, we established a new model for resetting the epigenetic balance of Polycomb/COMPASS function for cancer therapy, and this concept is under a phase 1 study at Northwestern Medicine for the treatment of human cancer [Cancer Therapy Evaluation Program (CTEP)/NCI Experimental Therapeutics Clinical Trials Network (ETCTN) trial #10183].
The BAP1 complex contains as many as 10 distinct subunits, including ASXL1/2/3, FOXK1/2, MBD5/6, and LSD2 (14–18). The BAP1 complex functions as a general transcriptional activator by catalyzing the removal of monoubiquitin from histone H2AK119 (19, 20). However, the specific subunit(s) within the BAP1 complex that mediates the interaction between BAP1 and the MLL3-PHD domain has not been identified or molecularly characterized. This is an important question in defining how this complex controls gene expression and remains an unknown mechanism in how this master regulator is involved in maintaining the epigenetic balance. Thus, dysregulation or mutations of this factor would affect MLL3’s recruitment to enhancer chromatin binding sites, which will consequently affect the expression of downstream tumor suppressor genes.
Here, we identify additional sex combs-like 2 (ASXL2) as the factor that links MLL3 to the BAP1 complex. We further characterize the MLL3 binding domain within ASXL2, which is negatively regulated by PRMT4/CARM1-mediated arginine methylation. Depletion of CARM1 function by CRISPR or catalytic inhibitors increases MLL3 chromatin recruitment and activates MLL3/BAP1-dependent tumor suppressors. Last, we established a novel combinatorial treatment targeting both EZH2 and CARM1 activity with small molecular inhibitors. In summary, our study provides a full genetic and molecular characterization that addresses a previously unexplored question related to dysregulated epigenetic balances within human cancers.
RESULTS
ASXL2 links BAP1 complex to MLL3/COMPASS
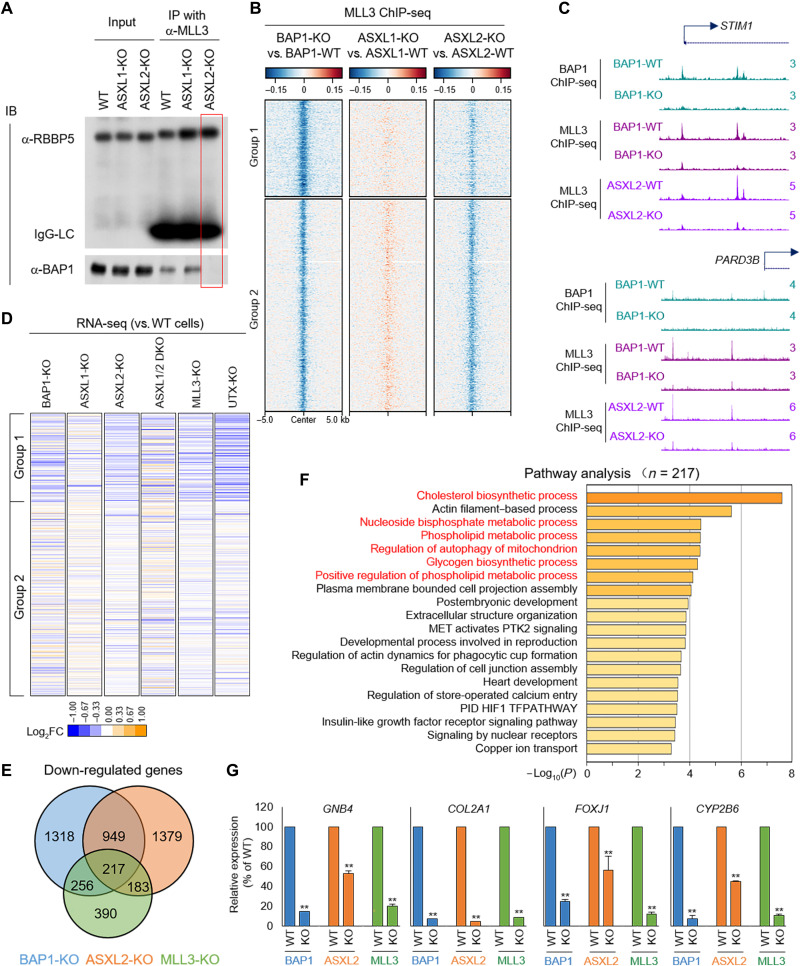
Previously, we demonstrated that the BAP1 complex recruits MLL3/COMPASS to enhancers (13). Depletion of BAP1 by CRISPR reduces MLL3 recruitment and function at enhancers, leading to decreased expression of tumor suppressors regulated by these enhancers. There are two major BAP1 complexes in mammalian cells: PR-DUB.1 and PR.DUB.2, which contain ASXL1 (21) or ASXL2 (22), respectively (fig. S1A). To determine which PR-DUB interacts with MLL3/COMPASS, we eliminated ASXL1 and ASXL2 individually, or in combination [double knockout (DKO)], by CRISPR in CAL51 cells (fig. S1, B and C). Using RNA sequencing (RNA-seq) analysis, we confirmed the absence of ASXL1 and/or ASXL2 mRNA in these cells (fig. S1, B and C). Notably, BAP1 protein levels are almost undetectable by Western blotting in ASXL1/2-DKO (23–25), whereas levels in ASXL1 and ASXL2 single KO cells are similar to wild type (WT) (fig. S1D). We performed immunoprecipitation (IP) experiments in WT, ASXL1-KO, and ASXL2-KO CAL51 cells and found that depletion of ASXL2, but not ASXL1, completely blocks the interaction between BAP1 and MLL3, suggesting that ASXL2 is the scaffold protein directly linking BAP1 to the MLL3 COMPASS (Fig. 1A). To determine whether loss of ASXL2 phenocopies loss of BAP1 in the chromatin recruitment of MLL3/COMPASS, we performed MLL3 chromatin IP sequencing (ChIP-seq) in WT, ASXL1-KO, and ASXL2-KO CAL51 cells. For our bioinformatic analysis, we used a set of BAP1 ChIP-seq peaks, which were divided into two groups based on our previous studies (fig. S1E) (13). Depletion of BAP1 and ASXL2, but not ASXL1, leads to a substantial reduction of MLL3 occupancy at the chromatin (Fig. 1, B and C, and fig. S1F) without affecting the protein levels of MLL3 (fig. S1G). Analysis of RNA-seq data revealed that ASXL2-KO, MLL3-KO, and BAP1-KO exhibit similar alterations in gene expression (Fig. 1, D and E). These results suggest that ASXL2 depletion leads to reduced expression of genes that have MLL3/BAP1-dependent enhancers. Pathway analysis by Metascape suggests that most of MLL3, BAP1, and ASXL2 target genes are involved in metabolic pathways (Fig. 1F), which is consistent with a recent work reporting ASXL2 as a key factor for controlling energy expenditure by macrophages in response to dietary factors (26). Furthermore, the expression levels of tumor suppressors such as GNB4, COL2A1, FOXJ1, and CYP2B6 were significantly reduced in BAP1-KO, ASXL2-KO, and MLL3-KO cells (Fig. 1G).
Fig. 1. ASXL2 links BAP1 complex to MLL3/COMPASS.
(A) Endogenous IPs with MLL3 antibody from WT, ASXL1-KO, and ASXL2-KO CAL51 cells were subjected to Western blotting with BAP1 and RBBP5 (a conserved core subunit of MLL3 that regulates its histone methyltransferase activity); n = 2. IB, immunoblot; IgG-LC, IgG-light chain. (B) The log2 fold change (log2FC) heatmap shows the occupancy of MLL3 in BAP1/ASXL1/ASXL2 KO cells versus WT cells. (C) Representative track examples from ChIP-seq showing enhancer occupancies of MLL3 in WT cells, BAP1-KO cells, and ASXL2-KO CAL51 cells. (D) The log2 fold change heatmap shows the gene expression change in BAP1-KO, MLL3-KO, ASXL1-KO, ASXL2-KO, and ASXL1/2 DKO CAL51 cells. (E) The Venn diagram shows the overlapped down-regulated genes in MLL3-KO, BAP1-KO, and ASXL2-KO CAL51 cells. (F) Pathway analysis with Metascape shows the most significantly enriched pathways involved with MLL3/BAP1/ASXL2 coregulated genes. (G) Relative mRNA levels of GNB4, COL2A1, FOXJ1, and CYP2B6 were determined by real-time polymerase chain reaction in WT, MLL3-KO, BAP1-KO, and ASXL2-KO cells; n = 3. **P < 0.01.
Identification of LOCAP: A linker domain of COMPASS and PR-DUB of ASXL2
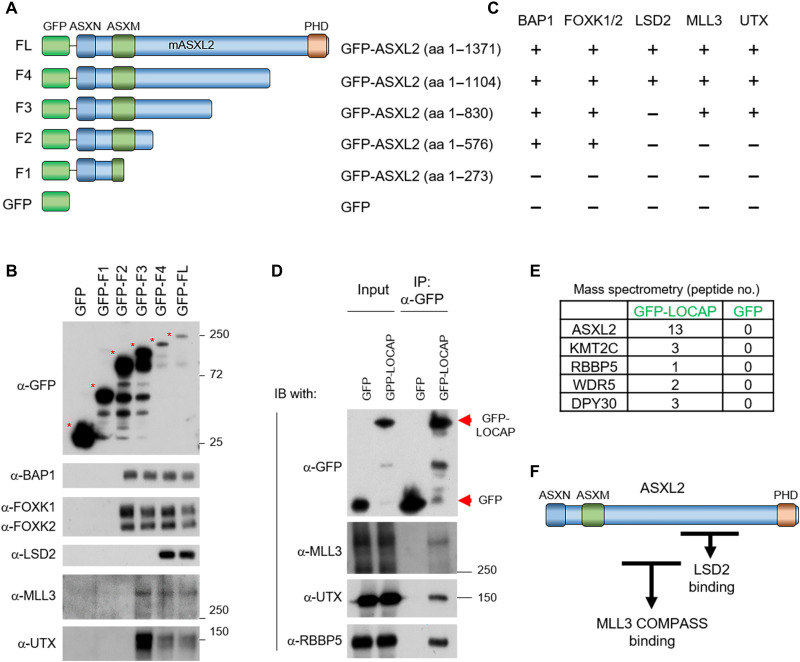
To further understand the molecular mechanism of how ASXL2 interacts with MLL3, we truncated ASXL2 into five distinct fragments [including green fluorescent protein (GFP) as a control] and purified each of the GFP-tagged fragments from human embryonic kidney (HEK) 293T cells (Fig. 2A). Both MLL3 and UTX interact with ASXL2-F3 but not ASXL2-F2 (Fig. 2, B and C). Both of these fragments include the ASXN and ASXM N-terminal domains, suggesting that only amino acids 576 to 830 of ASXL2 are necessary for MLL3 COMPASS binding. LSD2/KDM1B is a histone-modifying enzyme that demethylates histone H3 lysine residue 4 dimethylation (H3K4me2) that was recently been reported as a unique component of the PR-DUB.2 complex (24). We found that LSD2 interacts with ASXL2-F4 but not ASXL2-F3, suggesting that the LSD2 binding domain is contained within ASXL2 amino acids 830 to 1104 (Fig. 2, B and C). Thus, the MLL3 and LSD2 binding regions in ASXL2 are separate, but adjacent, sequences (Fig. 2F). On the basis of these molecular studies, we named the region within ASXL2 (amino acids 576 to 830) that interacts with MLL3 LOCAP (linker of COMPASS and PR-DUB) domain. Consistent with the results from our biochemistry studies, protein alignment of ASXL1-3 revealed that the LOCAP sequence is unique and evolutionarily conserved in ASXL2 homologs (fig. S2A). To determine whether this domain is sufficient to interact with MLL3 COMPASS in cells, we transfected HEK293T cells with either GFP or GFP-tagged LOCAP domain. We found that the LOCAP domain alone is able to pull down endogenous MLL3 (Fig. 2D). To further confirm the interaction between the ASXL2-LOCAP domain and MLL3 COMPASS, we immunoprecipitated the LOCAP domain from HEK293T cells and subjected the enriched proteins to mass spectrometry analysis. We found that MLL3 COMPASS is one of the top interactors with the LOCAP domain of ASXL2 (Fig. 2E and table S1). Collectively, our results reveal that ASXL2 acts as a scaffold protein that binds to both MLL3 and LSD2. Last, we purified LSD2 from HEK293T cells to determine whether MLL3, ASXL2, and LSD2 could potentially form an integrated complex with one another (fig. S2B and table S2). However, we did not detect MLL3 COMPASS in our LSD2 purification, indicating that LSD2 and MLL3 might be mutually exclusive binding partners of the BAP1/ASXL2 complex (Fig. 2F). Notably, we have detected a number of truncating mutations within ASXL2 N terminus in human cancers based on cBioPortal (fig. S2, C and D), suggesting that loss of MLL3-ASXL2/BAP1 contact might be a causal event in these cancers.
Fig. 2. Identification of LOCAP domain in ASXL2 that mediates MLL3/BAP1 interaction.
(A) A schematic diagram of full-length ASXL2 and its truncated derivatives. Both N-terminal domains (ASXN and ASXM) and a C-terminal domain (PHD) of ASXL2 are shown. (B) Plasmids expressing ASXL2 cDNA derivatives that correspond to each of the fragments shown in (A) were transfected into HEK293T cells for 24 hours. The GFP-tagged ASXL2 truncations were then purified and displayed interactions with BAP1, FOXK1/2, LSD2, MLL3, and UTX detected by Western blot. (C) The interaction between each ASXL2 fragments and BAP1, FOXK1/2, UTX, MLL3, and LSD2 is shown. The GFP-LOCAP domain fragment was expressed in HEK293T cells and then subjected to GFP purification from nuclear extracts. The protein-protein interaction between the ASXL2-LOCAP domain and MLL3 COMPASS was determined by Western blot (D) and mass spectrometry analysis (E); n = 3. (F) Protein interactome map of ASXL2 with MLL3 COMPASS [amino acids (aa) 576 to 830] and LSD2 (amino acids 830 to 1104) binding domains. MLL3 COMPASS binding domain is identified as the LOCAP domain.
CARM1 methylates ASXL2-LOCAP domain at R639/R641
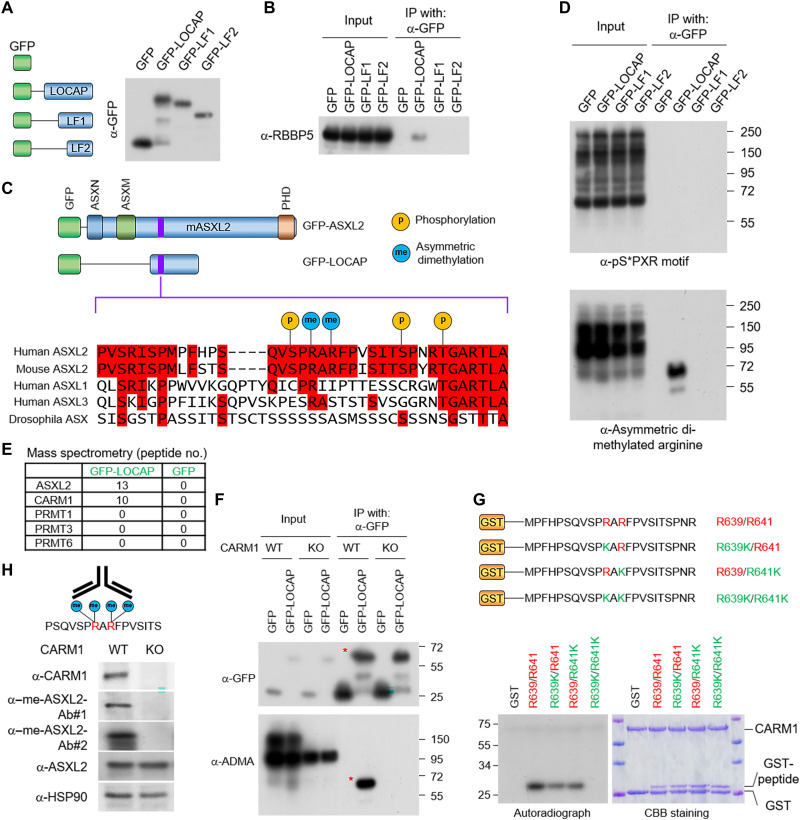
To further narrow down the interaction interface between MLL3 and ASXL2-LOCAP domain, we compared the full LOCAP region with two truncated fragments and identified a 35–amino acid region in the first segment of the LOCAP domain critical for MLL3 COMPASS binding (Fig. 3, A and B). Examination of the PhosphoSitePlus database (27) revealed that this small region contains potential multiple posttranslational modifications (PTMs), such as phosphorylation at serine residues and methylation at arginine residues (Fig. 3C). To determine whether the LOCAP domain is posttranslationally modified in cells, we transfected HEK293T cells with a GFP-tagged LOCAP domain and performed IP–Western blotting using phosphoserine (pS*PXR) and asymmetric dimethyl arginine (ADMA) antibodies. We found that the full-length LOCAP domain is methylated but is not detectably phosphorylated (Fig. 3D). Notably, both truncated constructs completely lack arginine methylation, demonstrating that the methylation site resides within the first 35 amino acids of the LOCAP sequence. To identify candidate ASXL2 arginine methyltransferases, we purified LOCAP from HEK293T cells and subjected the enriched proteins to mass spectrometry analysis. We found that CARM1 (also known as PRMT4) is the only arginine methyltransferase that binds to the LOCAP domain (Fig. 3E). To determine whether CARM1 directly methylates the LOCAP domain, we immunoprecipitated a GFP-tagged ASXL2-LOCAP from WT and CARM1-KO cells and performed Western blotting analysis using an anti-ADMA antibody. This experiment revealed that methylation of the LOCAP region is completely abolished in the absence of CARM1 (Fig. 3F). In further support of this observation, we performed in vitro methyltransferase assays and found that CARM1 directly methylates arginine residues 639 and 641 within the LOCAP domain (Fig. 3G). To investigate the prevalence of endogenous ASXL2 dimethylation in cells, we generated a rabbit polyclonal antibody specific to asymmetric dimethylated ASXL2 peptide (Fig. 3H, left). These methyl-ASXL2 antibodies specifically detect a methyl-ASXL2 peptide in vitro (fig. S3A) and also detect methylation of endogenous ASXL2 in CARM1-WT cells but not in CARM1-KO cells (Fig. 3H, right). Although methyl-specific antibodies may cross-react with some other CARM1 substrates, we found that methyl-ASXL2 is the major target of this antibody (fig. S3B). Overall, our study has demonstrated that the endogenous ASXL2 could be methylated by CARM1.
Fig. 3. Arginine methyltransferase CARM1 methylates ASXL2-LOCAP domain at R639/R641.
(A) A schematic diagram of the full-length ASXL2-LOCAP domain and its two truncated derivatives. The GFP-tagged LOCAP-FL, LOCAP-F1, and LOCAP-F2 were purified from HEK293T cells. (B) Western blot showing the interactions between each of the LOCAP domain truncations and RBBP5, one of the core subunits of MLL3 COMPASS. (C) PTMs at the LOCAP domain based on the PhosphoSitePlus database (www.phosphosite.org/homeAction.action). (D) IP of the GFP-tagged LOCAP derivatives coupled with Western blot detecting serine phosphorylation (via anti-serine phosphorylated antibody) (top) and arginine methylation (via anti-ADMA antibody) (bottom). The Western blot shows two arginine residues within LOCAP domain that are methylated in cells. (E) GFP-LOCAP was expressed in HEK293T cells and subjected to GFP purification from nuclear extracts and mass spectrometry analysis to identify which arginine methyltransferase is associated with ASXL2-LOCAP. The peptide number from all type II arginine methyltransferases is shown. (F) GFP-LOCAP was expressed in CARM1-WT/KO cells, and the methylation levels of LOCAP domain were determined by Western blot using anti-ADMA antibody. (G) Autoradiograph (left) and Coomassie brilliant blue staining (right) of in vitro methylated glutathione S-transferase (GST)–peptide fusion peptides by CARM1 in the presence of adenosyl-l-methionine, S-[methyl-3H] (SAM[3H]). Modifications to both arginine residues of ASXL2 (R639/R641) lead to loss of CARM1 protein-protein interaction. (H) Antigen peptide used for generation of methyl-specific antibody (left). Western blot analysis of the protein levels of CARM1, ASXL2, and me-ASXL2 (recognized by two antibodies) in CARM1-WT and KO cells (right). me-ASXL2, methylated ASXL2.
CARM1 negatively regulates MLL3 interaction with ASXL2/BAP1 and impairs its chromatin binding and gene activation function
Arginine methylation is a prevalent PTM, which functions as an essential epigenetic regulator of transcription, mRNA splicing, signaling transduction, and cell fate decisions (28–30). Arginine methylation of proteins can either inhibit (31) or promote protein-protein interactions (32, 33), depending on the type of methylation (34–36). Thus, the dual transcriptional function of CARM1 has clear implications for a multitude of physiological and pathological conditions, including breast cancer (37). To determine whether methylation within the LOCAP domain affects MLL3/ASXL2 interaction, we first mutated both of the arginine residues of ASXL2 (R639/R641) to lysine (R to K; which mimics unmethylated arginine residue) or to phenylalanine (R to F; which mimics methylated arginine residue) (38). We transfected cells with plasmids expressing LOCAP-WT, LOCAP-R639K/R641K, and LOCAP-R639F/R641F and found that arginine-to-phenylalanine mutation at both residues strongly inhibits the interaction between MLL3 and ASXL2 (Fig. 4A). Consistent with this result, we detected an increased interaction between MLL3 and BAP1/ASXL2 in CARM1-KO cells, compared with CARM1-WT cells (Fig. 4B). These results suggest that ASXL2 methylation by CARM1 may also inhibit MLL3 recruitment to enhancer regions in chromatin. To test this hypothesis, we conducted MLL3 ChIP-seq analysis in both CARM1-WT and CARM1-KO cells. Depletion of CARM1 markedly increases MLL3 recruitment to chromatin, especially at BAP1-dependent enhancer regions (Fig. 4, C to E). In further support of these ChIP-seq data, loss of BAP1 or MLL3 reduces the expression levels of genes near BAP1-dependent enhancers, whereas CARM1 depletion markedly increases expression of these genes, most likely due to increased recruitment of MLL3 (Fig. 4, F and G). In addition, KO of CARM1 does not increase the total protein levels of MLL3 or other subunits within MLL3 COMPASS (fig. S4A). These results have revealed an unexpected function of CARM1 as a transcription repressor through negative regulation of MLL3 function. We found 305 genes that are down-regulated in MLL3-KO cells, which are up-regulated in CARM1-KO cells (fig. S4A), many of which are involved in cell growth, development, and cancer (fig. S4, B to D).
Fig. 4. CARM1 negatively regulates MLL3 interaction with ASXL2/BAP1 and impairs its chromatin binding and gene expression.
(A) Plasmids expressing the WT and mutant LOCAP domain were transfected in HEK293T cells for 24 hours. The protein-protein interaction between MLL3 and the LOCAP domain was determined by an IP experiment. (B) Endogenous IPs with MLL3 antibody from CARM1-WT and CARM1-KO cells were subjected to Western blotting with BAP1 and ASXL2 antibody; n = 2. (C) The log2FC heatmap showing MLL3 occupancy in BAP1-KO and CARM1-KO cells. (D) The average plot compares MLL3 occupancy between WT cells, BAP1-KO cells, and CARM1-KO cells. (E) The ChIP-seq track example shows an increased in MLL3 occupancy at enhancers due to CARM1 depletion (CARM1-KO). The RNA-seq heatmap (F) and track example (G) shows the gene expression change between BAP1-, MLL3-, and CARM1-depleted cells.
Pharmaceutical inhibition of CARM1 enhances MLL3 function
CARM1 has been demonstrated to function as an oncogenic arginine methyltransferase in multiple human cancers, thus making it a promising therapeutic target (39). For example, inhibition of CARM1 catalytic activity via small molecules inhibits tumor growth in vitro and in vivo (40). To determine whether CARM1 catalytic activity is involved in MLL3 chromatin recruitment, we treated CAL51 cells with the CARM1 inhibitor, EZM2302, and conducted MLL3 ChIP-seq. We found that inhibition of CARM1 activity leads to an increase of MLL3 occupancy at BAP1-dependent enhancers (group 1), without affecting MLL3 recruitment at promoters (group 2) (Fig. 5, A and B). In support of this model, we generated ASXL2 knock-in cells carrying R639K/R641K and R639F/R641F mutations using CRISPR-Cas9 (fig. S5A). We then conducted MLL3 ChIP-seq in cells expressing either nonmethylated ASXL2 (R639K/R641K) or R639F/R641F mutant ASXL2. MLL3 binding to chromatin is markedly increased in CAL51 cells expressing nonmethyl-ASXL2 compared to the methyl mimic ASXL2 (fig. S5, B and C). We further conducted an RNA-seq experiment to determine whether the recruitment of MLL3 by the nonmethylated ASXL2 could activate gene expression. Increasing MLL3 occupancy contributes to a marked increase in gene expression by RNA-seq analysis (fig. S5B). On the basis of our previous model, the epigenetic balance between Polycomb repressive complex 2 (PRC2) and COMPASS is critical for the transcriptional activation of numerous tumor suppressors, and EZH2 catalytic inhibition can restore the expression of genes that are down-regulated by MLL3/UTX/BAP1 depletion (13). Here, we sought to test whether increasing MLL3 recruitment and function by CARM1 inhibition and reducing PRC2 function with EZH2 inhibitor, GSK126, would cause a synergistic effect on tumor cell growth. After confirming that EZH2i does not affect global arginine asymmetric demethylation and EZM2302 does not affect global H3K27me3 levels (Fig. 5C), we treated two breast cancer cell lines (MCF7 and CAL51) with both epigenetic inhibitors. Treatment with both epigenetic inhibitors shows strong combinational effects on both cell lines by cell counting assay (Fig. 5, D and E). Moreover, the results from RNA-seq analysis of cells treated with either dimethyl sulfoxide (DMSO), EZM2302 (CARM1 inhibitor), GSK126 (EZH2 inhibitor), or both inhibitors show a similar synergistic effect on gene expression (Fig. 5F). Pathway analysis revealed that apoptosis- and differentiation-related genes are commonly up-regulated by either CARM1 inhibitor or EZH2 inhibitor treatment (Fig. 5G and fig. S5D). These results suggested that inhibition of CARM1 may at least partially rescue the expression of the tumor-suppressive genes controlled by ASXL2/MLL3/UTX axis.
Fig. 5. Pharmaceutical inhibition of CARM1 enhances MLL3 function.
(A) MLL3 ChIP-seq was conducted in CAL51 cells that were treated with either DMSO or CARM1 inhibitor EZM2302 for 72 hours. The average plot shows the occupancy of MLL3 at group 1 and group 2 peaks. (B) The ChIP-seq track examples show increases in MLL3 occupancy at enhancers induced by CARM1 inhibitor treatment. (C) The breast cancer cell line was treated with DMSO, EZM2302 (CARM1 inhibitor), GSK126 (EZH2 inhibitor), or both inhibitors for 96 hours. Western blot showing the total protein levels of ADMA and histone H3K27me3; n = 3. CAL51 and MCF7 cells were treated with combinations of GSK126 and EZH2302 for 4 days. The cell counting assay shows the percentage of cell viability in each group; n = 3 (D). The synergistic effect in (D) was quantified by the SynergyFinder software (E). ZIP, zero interaction potency. (F) The RNA-seq heatmap showing the synergistic effect of CARM1/EZH2 inhibitors on gene expression in CAL51 cells. (G) The Venn-diagram shows the overlap between EZH2302- and GSK126-targeted genes.
DISCUSSION
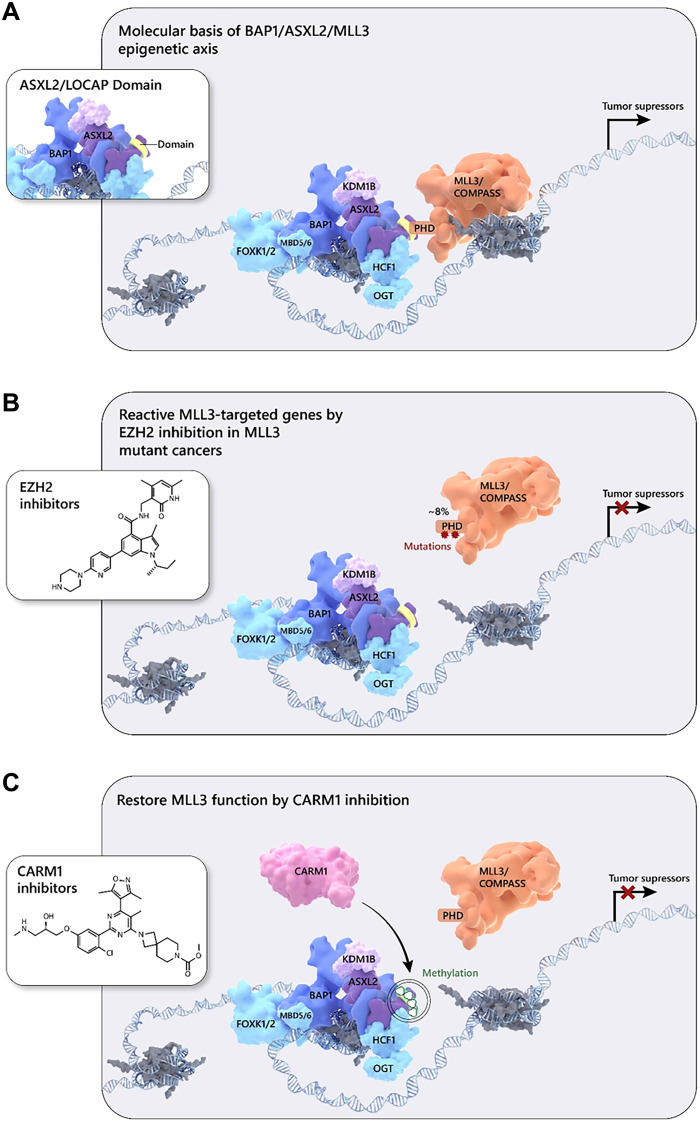
Loss of BAP1 has been implicated as a direct driver of human solid tumors (41), as well as leukemia (42). However, emerging studies suggest that BAP1 can function as an oncogene, especially when the core subunit ASXL1 is mutated in leukemia (43). These contradictory results imply nuances to the function of the BAP1 complex, which may be due to the diverse protein composition of its various subunits. As a close homolog of ASXL1, ASXL2 is also crucial for BAP1 function and activity in mammalian cells. ASXL2 is essential for hematopoiesis and acts as a haploinsufficient tumor suppressor in leukemia where ASXL2 deficiency promotes AML1-ETO–driven leukemogenesis (44). However, the mechanism of how ASXL2 functions as a tumor suppressor remains to be elucidated. In our current study, we have provided an epigenetic balance model to explain how ASXL2 can activate tumor suppressor gene expression by functioning as a scaffold protein that links MLL3 and BAP1 together at BAP1-dependent chromatin enhancers (Fig. 6A).
Fig. 6. Model: BAP1/ASXL2/MLL3 as a central axis for cancer therapy using EZH2 and CARM1 inhibitors.
(A) In normal cells, ASXL2 bridges the MLL3 COMPASS and BAP1 complexes to maintain proper expression levels of tumor suppressor genes. (B) At around 6 to 8% of tumor cells that harbor mutations in the MLL3 PHD domain, MLL3-COMPASS is not recruited to BAP1-associated enhancer regions, leading to a silencing of the downstream genes. (C) In malignant tumors with WT MLL3, increased levels of the arginine methyltransferase CARM1 directly methylate ASXL2 at two arginine residues, which prevents MLL3 binding and blocks MLL3 recruitment to enhancers.
MLL3 has been previously identified as a binding protein in ASXL2 IPs (18); however, MLL3 has not been observed to be significantly enriched in mass spectrometry studies using BAP1 as a bait (15, 20, 45). This is expected because most of the BAP1 core subunits, such as ASXL1-3 and FOXK1/2, are present at stoichiometric levels within the BAP1 complex and likely dominate mass spectrometry analysis. Moreover, the LOCAP domain is unique in ASXL2; therefore, BAP1/ASXL1 (PR-DUB1) and BAP1/ASXL3 (PR-DUB3) do not interact with MLL3.
In comparison to ASXL1, ASXL2 is rarely mutated in human cancers, suggesting that there might be other mechanisms that regulate the oncogenic function of ASXL2. In our current study, we mapped a small region within the LOCAP domain of ASXL2 that is critical for MLL3 binding (Fig. 3C). On the basis of our biochemical studies, we have identified that dimethylation at two arginine residues (R639/R641) by CARM1 impairs MLL3 binding (Fig. 3G). There are no recurrent or germline mutations associated with this region in any type of human cancers; instead, multiple phosphorylation modification (S637, S648, and T652) was observed at the flanking R639/R641, suggesting that there might be other mechanisms that control the activity of the LOCAP domain within ASXL2.
Arginine methylation has been reported to block certain protein-protein interactions while promoting others (30). For instance, interactions mediated by SH3 domains are demonstrated to be sensitive to arginine methylation, while WW and Tudor domain interactions are unaffected or increased by arginine methylation (46–48). Here, we demonstrated that arginine methylation at R639/R641 of ASXL2 changes the hydrophobic status of this region and blocks MLL3 binding. Therefore, this finding supports evidence toward arginine methylation function in blocking protein-protein interaction, instead of promoting protein-protein interaction. CARM1 was demonstrated to be an oncogenic methyltransferase in multiple human cancers, including prostate cancer (49), breast cancer (50, 51), and leukemia (52). Originally, CARM1 was proposed to function as a transcriptional activator by methylating histone H3R17 (53). However, on the basis of the RNA-seq analysis from our study and studies from other groups (33), the loss of CARM1 may also lead to an increase in gene expression. Here, we demonstrated that CARM1 directly methylates ASXL2 at R639/R641, which further blocks MLL3 recruitment at enhancer chromatin, leading to the silencing of nearby genes. Our results highlight a model for how CARM1 represses gene expression in mammalian cells. Consistent with our biochemistry results, we found that CARM1 inhibition can restore MLL3 recruitment at BAP1 loci (e.g., HOX cluster genes), suggesting that it is possible to recover loss of MLL3 COMPASS function by inhibiting CARM1 as a potential therapeutic strategy for cancer treatment.
LSD2 (or AOF1) was initially identified as a histone H3K4 demethylase in human cells (54). On the basis of published mass spectrometry results, LSD2 has numerous interactors in mammalian cells (55). We found that both MLL3 (Fig. 2E) and LSD2 (fig. S2B) interact with ASXL2 in mammalian cells. However, LSD2 does not directly interact with MLL3 (fig. S2B), suggesting that there might be two different subcomplexes, BAP1/ASXL2/MLL3 and BAP1/ASXL2/LSD2. It will be interesting to further investigate the epigenetic function between these two different complexes in regulating enhancer activity, histone modifications, and gene expression.
MLL3 is one of the most frequently mutated genes in human cancers (13). On average, MLL3 is mutated in around 6% of all human cancers. In our previous studies, we proposed an epigenetic balancing model (13). Inhibition of EZH2 by small-molecule inhibitors could activate the genes that were initially lost due to MLL3 loss-of-function mutations (Fig. 6, A and B). On the other hand, more than 90% of tumor cells are WT or heterozygous for MLL3. An increase in the abundance of arginine methyltransferase CARM1 could directly methylate ASXL2 at the LOCAP domain and subsequently block the MLL3-BAP1 interaction, leading to a similar phenotype seen in MLL3 mutations (Fig. 6C). Our current study has identified a dominant mechanism for arginine methylation in controlling the epigenetic balance between PRC2 and COMPASS. Therefore, to maximize the rescue in the expression of MLL3 downstream genes, we combined both EZH2 and CARM1 inhibitors, which resulted in a strong synergistic effect that could be a potential novel therapeutic for clinical cancer treatments, especially for breast cancer associated with MLL3 mutations.
MATERIALS AND METHODS
Antibodies and reagents
BAP1 (#13271S), UTX (#33510), and ADMA (#13522) antibodies were purchased from Cell Signaling Technology. HSP90 (sc-7947) and GFP (sc-9996) antibodies were purchased from Santa Cruz Biotechnology. ASXL2 (A302-037A), FOXK1 (A301-728A), and FOXK2 (A301-729A) antibodies were purchased from Bethyl Laboratories. Tubulin antibody (E7) was purchased from the Developmental Studies Hybridoma Bank. Flag (F3165) antibody was purchased from Sigma-Aldrich. LSD2 (ab234863) antibody was purchased from Abcam. BAP1, UTX, and MLL3 ChIP-seq antibodies were homemade, as previously described (13).
Cell lines
HEK293T, MCF7, and CAL51 cells were obtained from American Type Culture Collection and then maintained with Dulbecco’s modified Eagle’s medium (Gibco, Gaithersburg, MD) containing 10% fetal bovine serum (Sigma-Aldrich).
CRISPR-mediated KOs
Designed single guide RNAs were cloned into lentiCRISPR v2 (Addgene, 52961) vector. The lentiviral-mediated CRISPR-Cas9 KO was described previously (13). For the lentiviral mediated CRISPR knock-in, the targeting vector and single-stranded DNA donor were cotransfected in cells for 24 hours and followed by 2 days of puromycin selection. Targeted single-cell clones were lysed, and genomic DNA was purified and screened by polymerase chain reaction.
IP and Western blot
The cells were lysed in the lysis buffer [50 mM tris (pH 8.0), 150 mM NaCl, 0.5% Triton X-100, 10% glycerol, 1 mM dithiothreitol (DTT), protease inhibitors, and benzonase]. After centrifugation at maximum speed for 20 min, the supernatants were collected and incubated with each designated primary antibodies at 4°C overnight with rotation. After incubation with immobilized protein A/G (Santa Cruz Biotechnology), the protein A/G beads were washed with lysis buffer and were resuspended in 5× SDS sample loading buffer and then subjected to SDS–polyacrylamide gel electrophoresis. The resolved proteins were either transferred to nitrocellulose membranes for immunoblotting or stained with Coomassie brilliant blue.
Normal goat serum sample preparation
ChIP-seq libraries were prepared using the KAPA HTP Library Preparation Kit, complemented with NEXTflex DNA Barcodes from Bioo Scientific. Ten nanograms of DNA was used as starting material for input and IP samples. Libraries were then amplified using 13 cycles on the thermocycler. Postamplification libraries were size-selected at 250 to 450 base pairs in length using Agencourt AMPure XP beads from Beckman Coulter. Libraries were validated using the Agilent High-Sensitivity DNA Kit. RNA-seq libraries were prepared using the Illumina TruSeq Stranded Total RNA Preparation Kit with Ribo-Depletion. Input RNA quality was validated using the Agilent RNA 6000 Nano Kit. One microgram of total RNA was used as starting material. Libraries then were validated using the Agilent DNA 1000 Kit.
RNA-seq analysis
Gene counts were computed by HTSeq (56) and used as input for edgeR 3.0.852 (57). Genes with Benjamini-Hochburg adjusted P values of less than 0.01 were considered to be differentially expressed, unless otherwise specified. RNA-seq heatmaps adjacent to ChIP-seq heatmaps display log2 fold change values of genes corresponding to transcription start sites nearest to ChIP-seq peaks and were displayed using Java TreeView. Gene Ontology functional analysis was carried out using Metascape with default parameters.
ChIP-seq analysis
The ChIP-seq was performed as previously described before (16). For ChIP-seq analysis, all the peaks were called with the MACS v1.4.2 (58) software using default parameters and corresponding input samples. Metaplots and heatmaps were generated using ngs.plot (59) display ChIP-seq signals aligned to peaks.
Mass spectrometry sample preparation and data analysis
The mass spectrometry experiment was performed as described previously (16). Protein pellets were denatured in 50 μl of 8 M urea/0.4 M ammonium bicarbonate followed by reduction in 2 μl of 100 mM DTT. Protein was alkylated with 18 mM iodoacetamide for 30 min at room temperature in the dark. Samples were then diluted with water four times the volume to bring urea concentration to 1.8 M. Sequencing-grade trypsin (Promega) was added at 1:100 (enzyme:substrate) and incubated at 37°C overnight. The digests were acidified to 0.5% trifluoroacetic acid (TFA), and the peptides were desalted on C18 Sep-Paks (Waters). Peptides were eluted with 2× 50 μl of 80% acetone/0.1% TFA to ensure complete recovery. The pooled extracts were dried in a vacuum concentrator and resuspended in 30 μl of 5% acetonitrile/0.1% TFA for liquid chromatography–mass spectrometry analysis. Proteins were identified from the mass spectrometry raw files using Mascot search engine (Matrix Science, London, UK; version 2.5.1). Tandem mass spectrometry spectra were searched against the SwissProt human database. All searches included carbamidomethyl cysteine as a fixed modification and oxidized Met, deamidated Asn and Gln, and acetylation of protein N-terminus as variable modifications. Three missed tryptic cleavages were allowed. The MS1 precursor mass tolerance was set to 10 parts per million, and the MS2 tolerance was set to 0.6 Da. A 1% false discovery rate cutoff was applied at the peptide level. Only proteins with a minimum of two peptides above the cutoff were considered for further studies.
Statistical analyses
For statistical analyses, GraphPad Prism 7, Microsoft Excel, and R were used. All data involving a statistical analysis being reported met the criteria to use the appropriate statistical tests; for the normal distribution of data, the empirical rule was used to infer the distribution. For growth curves and time course, RNA-seq t tests were calculated between the area under the curve values. Statistical tests used are reported in the figure legends.
Acknowledgments
We thank E. Smith for critical scientific discussion and editorial assistance, D. Zha for help with NGS sample preparation, and N. Tsuboyama for help with manuscript revision. We thank F. Zheng for the gifts of the lentiCRISPR v2 vector (plasmid #52961).
Funding: Studies in the Shilatifard laboratory related to this project are supported by National Cancer Institute’s Outstanding Investigator Award R35CA197569.
Author contributions: A.S. and L.W. designed the study. L.W., Z.Z., and A.P.S. performed all the biochemistry and sequencing experiments. Z.Z. performed the bioinformatics analysis. E.J.R. sequenced all the NGS samples. L.W., Z.Z., M.A.M., and A.S. wrote the manuscript and the revised manuscript. L.W., A.S., Z.Z., A.P.S., and M.A.M. revised the manuscript. All authors read and approved the final manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. NGS data generated for this study are available at the Gene Expression Omnibus (GEO) under accession number GSE182625.
Supplementary Materials
This PDF file includes:
Figs. S1 to S5
REFERENCES AND NOTES
- 1.Morgan M. A., Shilatifard A., Chromatin signatures of cancer. Gene Dev 29, 238–249 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Plass C., Pfister S. M., Lindroth A. M., Bogatyrova O., Claus R., Lichter P., Mutations in regulators of the epigenome and their connections to global chromatin patterns in cancer. Nat. Rev. Genet. 14, 765–780 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Black J. C., Van Rechem C., Whetstine J. R., Histone lysine methylation dynamics: Establishment, regulation, and biological impact. Mol. Cell 48, 491–507 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herz H. M., Mohan M., Garruss A. S., Liang K., Takahashi Y. H., Mickey K., Voets O., Verrijzer C. P., Shilatifard A., Enhancer-associated H3K4 monomethylation by Trithorax-related, the Drosophila homolog of mammalian Mll3/Mll4. Genes Dev. 26, 2604–2620 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu D. Q., Gao X., Morgan M. A., Herz H.-M., Smith E. R., Shilatifard A., The MLL3/MLL4 branches of the COMPASS family function as major histone H3K4 monomethylases at enhancers. Mol. Cell. Biol. 33, 4745–4754 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujimoto A., Totoki Y., Abe T., Boroevich K. A., Hosoda F., Nguyen H. H., Aoki M., Hosono N., Kubo M., Miya F., Arai Y., Takahashi H., Shirakihara T., Nagasaki M., Shibuya T., Nakano K., Watanabe-Makino K., Tanaka H., Nakamura H., Kusuda J., Ojima H., Shimada K., Okusaka T., Ueno M., Shigekawa Y., Kawakami Y., Arihiro K., Ohdan H., Gotoh K., Ishikawa O., Ariizumi S. I., Yamamoto M., Yamada T., Chayama K., Kosuge T., Yamaue H., Kamatani N., Miyano S., Nakagama H., Nakamura Y., Tsunoda T., Shibata T., Nakagawa H., Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat. Genet. 44, 760–764 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Kandoth C., McLellan M. D., Vandin F., Ye K., Niu B., Lu C., Xie M., Zhang Q., McMichael J. F., Wyczalkowski M. A., Leiserson M. D. M., Miller C. A., Welch J. S., Walter M. J., Wendl M. C., Ley T. J., Wilson R. K., Raphael B. J., Ding L., Mutational landscape and significance across 12 major cancer types. Nature 502, 333–339 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gui Y., Guo G., Huang Y., Hu X., Tang A., Gao S., Wu R., Chen C., Li X., Zhou L., He M., Li Z., Sun X., Jia W., Chen J., Yang S., Zhou F., Zhao X., Wan S., Ye R., Liang C., Liu Z., Huang P., Liu C., Jiang H., Wang Y., Zheng H., Sun L., Liu X., Jiang Z., Feng D., Chen J., Wu S., Zou J., Zhang Z., Yang R., Zhao J., Xu C., Yin W., Guan Z., Ye J., Zhang H., Li J., Kristiansen K., Nickerson M. L., Theodorescu D., Li Y., Zhang X., Li S., Wang J., Yang H., Wang J., Cai Z., Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat. Genet. 43, 875–878 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y., Chen L., Nie C. J., Zeng T. T., Liu H., Mao X., Qin Y., Zhu Y. H., Fu L., Guan X. Y., Downregulation of RBMS3 is associated with poor prognosis in esophageal squamous cell carcinoma. Cancer Res. 71, 6106–6115 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Ellis M. J., Ding L., Shen D., Luo J., Suman V. J., Wallis J. W., van Tine B. A., Hoog J., Goiffon R. J., Goldstein T. C., Ng S., Lin L., Crowder R., Snider J., Ballman K., Weber J., Chen K., Koboldt D. C., Kandoth C., Schierding W. S., McMichael J. F., Miller C. A., Lu C., Harris C. C., McLellan M. D., Wendl M. C., DeSchryver K., Allred D. C., Esserman L., Unzeitig G., Margenthaler J., Babiera G. V., Marcom P. K., Guenther J. M., Leitch M., Hunt K., Olson J., Tao Y., Maher C. A., Fulton L. L., Fulton R. S., Harrison M., Oberkfell B., du F., Demeter R., Vickery T. L., Elhammali A., Piwnica-Worms H., McDonald S., Watson M., Dooling D. J., Ota D., Chang L. W., Bose R., Ley T. J., Piwnica-Worms D., Stuart J. M., Wilson R. K., Mardis E. R., Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature 486, 353–360 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bertucci F., Ng C. K. Y., Patsouris A., Droin N., Piscuoglio S., Carbuccia N., Soria J. C., Dien A. T., Adnani Y., Kamal M., Garnier S., Meurice G., Jimenez M., Dogan S., Verret B., Chaffanet M., Bachelot T., Campone M., Lefeuvre C., Bonnefoi H., Dalenc F., Jacquet A., de Filippo M. R., Babbar N., Birnbaum D., Filleron T., le Tourneau C., André F., Author correction: Genomic characterization of metastatic breast cancers. Nature 572, E7–E7 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Bertucci F., Ng C. K. Y., Patsouris A., Droin N., Piscuoglio S., Carbuccia N., Soria J. C., Dien A. T., Adnani Y., Kamal M., Garnier S., Meurice G., Jimenez M., Dogan S., Verret B., Chaffanet M., Bachelot T., Campone M., Lefeuvre C., Bonnefoi H., Dalenc F., Jacquet A., De Filippo M. R., Babbar N., Birnbaum D., Filleron T., Le Tourneau C., André F., Genomic characterization of metastatic breast cancers. Nature 569, 560–564 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Wang L., Zhao Z., Ozark P. A., Fantini D., Marshall S. A., Rendleman E. J., Cozzolino K. A., Louis N., He X., Morgan M. A., Takahashi Y. H., Collings C. K., Smith E. R., Ntziachristos P., Savas J. N., Zou L., Hashizume R., Meeks J. J., Shilatifard A., Resetting the epigenetic balance of Polycomb and COMPASS function at enhancers for cancer therapy. Nat. Med. 24, 758–769 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carbone M., Yang H., Pass H. I., Krausz T., Testa J. R., Gaudino G., BAP1 and cancer. Nat. Rev. Cancer 13, 153–159 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dey A., Seshasayee D., Noubade R., French D. M., Liu J., Chaurushiya M. S., Kirkpatrick D. S., Pham V. C., Lill J. R., Bakalarski C. E., Wu J., Phu L., Katavolos P., LaFave L. M., Abdel-Wahab O., Modrusan Z., Seshagiri S., Dong K., Lin Z., Balazs M., Suriben R., Newton K., Hymowitz S., Garcia-Manero G., Martin F., Levine R. L., Dixit V. M., Loss of the tumor suppressor BAP1 causes myeloid transformation. Science 337, 1541–1546 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szczepanski A. P., Zhao Z., Sosnowski T., Goo Y. A., Bartom E. T., Wang L., ASXL3 bridges BRD4 to BAP1 complex and governs enhancer activity in small cell lung cancer. Genome Med. 12, 63 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hauri S., Comoglio F., Seimiya M., Gerstung M., Glatter T., Hansen K., Aebersold R., Paro R., Gstaiger M., Beisel C., A high-density map for navigating the human polycomb complexome. Cell Rep. 17, 583–595 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Baymaz H. I., Fournier A., Laget S., Ji Z., Jansen P. W. T. C., Smits A. H., Ferry L., Mensinga A., Poser I., Sharrocks A., Defossez P. A., Vermeulen M., MBD5 and MBD6 interact with the human PR-DUB complex through their methyl-CpG-binding domain. Proteomics 14, 2179–2189 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Fursova N. A., Turberfield A. H., Blackledge N. P., Findlater E. L., Lastuvkova A., Huseyin M. K., Dobrinić P., Klose R. J., BAP1 constrains pervasive H2AK119ub1 to control the transcriptional potential of the genome. Gene Dev. 35, 749–770 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Conway E., Rossi F., Fernandez-Perez D., Ponzo E., Ferrari K. J., Zanotti M., Manganaro D., Rodighiero S., Tamburri S., Pasini D., BAP1 enhances Polycomb repression by counteracting widespread H2AK119ub1 deposition and chromatin condensation. Mol. Cell 81, 3526–3541.e8 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sahtoe D. D., van Dijk W. J., Ekkebus R., Ovaa H., Sixma T. K., BAP1/ASXL1 recruitment and activation for H2A deubiquitination. Nat. Commun. 7, 10292 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lai H. L., Wang Q. T., Additional sex combs-like 2 is Required for polycomb repressive complex 2 binding at select targets. PLOS ONE 8, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Daou S., Hammond-Martel I., Mashtalir N., Barbour H., Gagnon J., Iannantuono N. V. G., Nkwe N. S., Motorina A., Pak H., Yu H., Wurtele H., Milot E., Mallette F. A., Carbone M., Affar E. B., The BAP1/ASXL2 histone H2A deubiquitinase complex regulates cell proliferation and is disrupted in cancer. J. Biol. Chem. 290, 28643–28663 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Campagne A., Lee M. K., Zielinski D., Michaud A., le Corre S., Dingli F., Chen H., Shahidian L. Z., Vassilev I., Servant N., Loew D., Pasmant E., Postel-Vinay S., Wassef M., Margueron R., BAP1 complex promotes transcription by opposing PRC1-mediated H2A ubiquitylation. Nat. Commun. 10, 348 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kolovos P., Nishimura K., Sankar A., Sidoli S., Cloos P. A., Helin K., Christensen J., PR-DUB maintains the expression of critical genes through FOXK1/2- and ASXL1/2/3-dependent recruitment to chromatin and H2AK119ub1 deubiquitination. Genome Res. 30, 1119–1130 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zou W., Rohatgi N., Brestoff J. R., Moley J. R., Li Y., Williams J. W., Alippe Y., Pan H., Pietka T. A., Mbalaviele G., Newberry E. P., Davidson N. O., Dey A., Shoghi K. I., Head R. D., Wickline S. A., Randolph G. J., Abumrad N. A., Teitelbaum S. L., Myeloid-specific Asxl2 deletion limits diet-induced obesity by regulating energy expenditure. J. Clin. Invest. 130, 2644–2656 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hornbeck P. V., Kornhauser J. M., Tkachev S., Zhang B., Skrzypek E., Murray B., Latham V., Sullivan M., PhosphoSitePlus: A comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 40, D261–D270 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bedford M. T., Arginine methylation at a glance. J. Cell Sci. 120, 4243–4246 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Blanc R. S., Richard S., Arginine methylation: The coming of age. Mol. Cell 65, 8–24 (2017). [DOI] [PubMed] [Google Scholar]
- 30.McBride A. E., Silver P. A., State of the Arg. Cell 106, 5–8 (2001). [DOI] [PubMed] [Google Scholar]
- 31.Xu W., Chen H., du K., Asahara H., Tini M., Emerson B. M., Montminy M., Evans R. M., A transcriptional switch mediated by cofactor methylation. Science 294, 2507–2511 (2001). [DOI] [PubMed] [Google Scholar]
- 32.Kawabe Y. I., Wang Y. X., McKinnell I. W., Bedford M. T., Rudnicki M. A., Carm1 regulates Pax7 transcriptional activity through MLL1/2 recruitment during asymmetric satellite stem cell divisions. Cell Stem Cell 11, 333–345 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nie M., Wang Y., Guo C., Li X., Wang Y., Deng Y., Yao B., Gui T., Ma C., Liu M., Wang P., Wang R., Tan R., Fang M., Chen B., He Y., Huang D. C. S., Ju J., Zhao Q., CARM1-mediated methylation of protein arginine methyltransferase 5 represses human γ-globin gene expression in erythroleukemia cells. J. Biol. Chem. 293, 17454–17463 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bedford M. T., Clarke S. G., Protein arginine methylation in mammals: Who, what, and why. Mol. Cell 33, 1–13 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee Y. H., Stallcup M. R., Minireview: Protein arginine methylation of nonhistone proteins in transcriptional regulation. Mol. Endocrinol. 23, 425–433 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raposo A. E., Piller S. C., Protein arginine methylation: An emerging regulator of the cell cycle. Cell Division 13, 3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guccione E., Richard S., The regulation, functions and clinical relevance of arginine methylation. Nat. Rev. Mol. Cell Biol. 20, 642–657 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Mostaqul Huq M. D., Gupta P., Tsai N. P., White R., Parker M. G., Wei L. N., Suppression of receptor interacting protein 140 repressive activity by protein arginine methylation. EMBO J. 25, 5094–5104 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith E., Zhou W., Shindiapina P., Sif S., Li C., Baiocchi R. A., Recent advances in targeting protein arginine methyltransferase enzymes in cancer therapy. Expert Opin. Ther. Targets 22, 527–545 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu H., Qian K., Ho M. C., Zheng Y. G., Small molecule inhibitors of protein arginine methyltransferases. Expert Opin. Investig. Drugs 25, 335–358 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carbone M., Ferris L. K., Baumann F., Napolitano A., Lum C. A., Flores E. G., Gaudino G., Powers A., Bryant-Greenwood P., Krausz T., Hyjek E., Tate R., Friedberg J., Weigel T., Pass H. I., Yang H., BAP1 cancer syndrome: Malignant mesothelioma, uveal and cutaneous melanoma, and MBAITs. J. Transl. Med. 10, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dkhissi F., Aggoune D., Pontis J., Sorel N., Piccirilli N., LeCorf A., Guilhot F., Chomel J. C., Ait-Si-Ali S., Turhan A. G., The downregulation of BAP1 expression by BCR-ABL reduces the stability of BRCA1 in chronic myeloid leukemia. Exp. Hematol. 43, 775–780 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Gelsi-Boyer V., Brecqueville M., Devillier R., Murati A., Mozziconacci M. J., Birnbaum D., Mutations in ASXL1 are associated with poor prognosis across the spectrum of malignant myeloid diseases. J. Hematol. Oncol. 5, 12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Micol J. B., Pastore A., Inoue D., Duployez N., Kim E., Lee S. C. W., Durham B. H., Chung Y. R., Cho H., Zhang X. J., Yoshimi A., Krivtsov A., Koche R., Solary E., Sinha A., Preudhomme C., Abdel-Wahab O., ASXL2 is essential for haematopoiesis and acts as a haploinsufficient tumour suppressor in leukemia. Nat. Commun. 8, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu H., Mashtalir N., Daou S., Hammond-Martel I., Ross J., Sui G., Hart G. W., Rauscher F. J. III, Drobetsky E., Milot E., Shi Y., Affar E. B., The ubiquitin carboxyl hydrolase BAP1 forms a ternary complex with YY1 and HCF-1 and is a critical regulator of gene expression. Mol. Cell. Biol. 30, 5071–5085 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bedford M. T., Frankel A., Yaffe M. B., Clarke S., Leder P., Richard S., Arginine methylation inhibits the binding of proline-rich ligands to Src homology 3, but not WW, domains. J. Biol. Chem. 275, 16030–16036 (2000). [DOI] [PubMed] [Google Scholar]
- 47.Friesen W. J., Massenet S., Paushkin S., Wyce A., Dreyfuss G., SMN, the product of the spinal muscular atrophy gene, binds preferentially to dimethylarginine-containing protein targets. Mol. Cell 7, 1111–1117 (2001). [DOI] [PubMed] [Google Scholar]
- 48.Selenko P., Sprangers R., Stier G., Bühler D., Fischer U., Sattler M., SMN tudor domain structure and its interaction with the Sm proteins. Nat. Struct. Biol. 8, 27–31 (2001). [DOI] [PubMed] [Google Scholar]
- 49.Lee E., Madar A., David G., Garabedian M. J., DasGupta R., Logan S. K., Inhibition of androgen receptor and β-catenin activity in prostate cancer. Proc. Natl. Acad. Sci. U.S.A. 110, 15710–15715 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang L., Zhao Z., Meyer M. B., Saha S., Yu M., Guo A., Wisinski K. B., Huang W., Cai W., Pike J. W., Yuan M., Ahlquist P., Xu W., CARM1 methylates chromatin remodeling factor BAF155 to enhance tumor progression and metastasis. Cancer Cell 25, 21–36 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang L., Zeng H., Wang Q., Zhao Z., Boyer T. G., Bian X., Xu W., MED12 methylation by CARM1 sensitizes human breast cancer cells to chemotherapy drugs. Sci. Adv. 1, e1500463 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Greenblatt S. M., Man N., Hamard P.-J., Asai T., Karl D., Martinez C., Bilbao D., Stathias V., Jermakowicz A. M., Duffort S., Tadi M., Blumenthal E., Newman S., Vu L., Xu Y., Liu F., Schurer S. C., McCabe M. T., Kruger R. G., Xu M., Yang F.-C., Tenen D. G., Watts J., Vega F., Nimer S. D., CARM1 is essential for myeloid leukemogenesis but dispensable for normal hematopoiesis (vol 33, pg 1111, 2018). Cancer Cell 34, 888–888 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu J., Xu W., Histone H3R17me2a mark recruits human RNA polymerase-associated factor 1 complex to activate transcription. Proc. Natl. Acad. Sci. U.S.A. 109, 5675–5680 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Karytinos A., Forneris F., Profumo A., Ciossani G., Battaglioli E., Binda C., Mattevi A., A novel mammalian flavin-dependent histone demethylase. J. Biol. Chem. 284, 17775–17782 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Machida Y. J., Machida Y., Vashisht A. A., Wohlschlegel J. A., Dutta A., The deubiquitinating enzyme BAP1 regulates cell growth via interaction with HCF-1. J. Biol. Chem. 284, 34179–34188 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Anders S., Pyl P. T., Huber W., HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nusbaum C., Myers R. M., Brown M., Li W., Liu X. S., Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shen L., Shao N., Liu X., Nestler E., ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics 15, 284 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S5