Abstract
Cystic fibrosis (CF) is caused by mutations in the CF transmembrane conductance regulator (CFTR) gene. We sought to correct the multiple organ dysfunction of the F508del CF-causing mutation using systemic delivery of peptide nucleic acid gene editing technology mediated by biocompatible polymeric nanoparticles. We confirmed phenotypic and genotypic modification in vitro in primary nasal epithelial cells from F508del mice grown at air-liquid interface and in vivo in F508del mice following intravenous delivery. In vivo treatment resulted in a partial gain of CFTR function in epithelia as measured by in situ potential differences and Ussing chamber assays and correction of CFTR in both airway and GI tissues with no off-target effects above background. Our studies demonstrate that systemic gene editing is possible, and more specifically that intravenous delivery of PNA NPs designed to correct CF-causing mutations is a viable option to ameliorate CF in multiple affected organs.
Polymeric nanoparticles encapsulating PNA gene editing agents correct cystic fibrosis mutations in multiple organs in vivo.
INTRODUCTION
Cystic fibrosis (CF) is an autosomal recessive disorder caused by mutations in the CF transmembrane conductance regulator (CFTR) gene (1, 2). CFTR encodes a chloride channel key to balancing ion and water secretion and absorption in epithelial tissues. CF patients experience multiorgan dysfunction; for example, CFTR defects cause mucus in several organs to thicken, leading to lung infections, a reduction in lung function, and poor digestive function in the gastrointestinal (GI) tract (1, 3). While the respiratory tract is the primary organ system affected by the disease, CF patients also experience severe GI issues, particularly as their life expectancy has increased. Notably, nutrition also affects pulmonary function and overall mortality (3). A majority (85%) of CF patients are born with pancreatic insufficiency (1, 3). Other GI manifestations of CF include meconium ileus, gastroesophageal reflux disease, constipation, distal intestinal obstruction syndrome, recurrent pancreatitis, and cancer (1). These manifestations are currently managed by various interventions, such as pancreatic enzyme replacement therapy, nutritional management, and supportive care.
While there are more than 1700 different CF-causing mutations, the most common mutation is F508del (2). This is a 3–base pair (bp) deletion in the genomic sequence that results in improper protein folding and impaired transport to the plasma membrane. The recent advancement of modulator therapies designed to mitigate effects of the F508del mutation by increasing transport to the membrane and improving channel function holds great promise (4–7), but this regimen requires expensive and continuous treatment (2). Moreover, the benefits of these therapies on alleviating GI issues remain unclear (1). Gene editing approaches, on the other hand, could offer a one-time cure applicable to all CF mutations, including the 10% of patients with rare mutations who are not candidates for modulator therapies (2).
Efforts to precisely correct genomic mutations that underlie hereditary diseases such as CF for therapeutic benefit have advanced alongside the emergence and improvement of genome editing technologies. These methods fall under two main classes: nuclease-based platforms such as zinc finger nucleases, TALENs, as well as the widely used CRISPR-Cas9 systems including base and prime editors, and oligo/polynucleotide strategies such as triplex-forming oligonucleotides (TFOs) (8). Programmable RNA-guided Cas9 endonucleases not only enable efficient genome editing in both cells and organisms but also cause collateral damage throughout the genome in the form of off-target effects due to nuclease activity, although this can be mitigated by impairing catalytic activity (9–13). Moreover, in vivo delivery of the large constructs that make up the complex CRISPR-Cas9 system remains challenging (14). We have developed a non–nuclease-based approach to gene editing by using endogenous DNA repair stimulated by the binding of peptide nucleic acids (PNAs) to genomic DNA (gDNA) to create a PNA/DNA/PNA triplex structure via both Watson-Crick and Hoogsteen H-bonding with displacement of the nonbound DNA strand. PNAs have a peptide backbone but undergo base pairing with DNA and RNA (15). They also lack intrinsic nuclease activity. Triplex PNA structures can initiate an endogenous DNA repair response mediated by high-fidelity nucleotide excision repair (NER) and homology-directed repair (HDR) pathways (16, 17). When PNAs are introduced with a single-stranded “donor” DNA containing the desired sequence modification, site-specific modification of the genome occurs (18). In terms of delivery, PNA and donor DNA are small relative to Cas9 systems and can be readily encapsulated into polymeric vehicles in the form of nanoparticles (NPs) (17, 18). We have previously demonstrated the efficacy of poly(lactic-co-glycolic acid) (PLGA) NPs encapsulating PNA and DNA to achieve gene editing both ex vivo and in vivo (16, 19–22). Here, we demonstrate the utility of PNA NPs for the systemic treatment of F508del CF.
Gene therapies for CF have thus far have had limited success in part due to challenges in delivery to key organs affected by the disease (23), namely, the lung and GI tract. Recent in vivo gene editing therapies, primarily using adenovirus-derived vector systems, focus on targeted correction of the CFTR gene in the airway epithelium (24–26). Furthermore, the use of CRISPR-Cas9–based gene editing has been demonstrated for the treatment of CF in vitro, with additional studies providing a path for its potential application as a targeted therapy to the lungs in vivo (2, 27, 28). Our own studies have shown that polymeric NPs deliver TFO-based gene editing agents to the lung, with phenotypic correction (16). However, CF is a systemic disease with multiple affected organs that could potentially benefit from gene correction therapies; while the in vivo approaches for remedying CF via gene therapy have shown promise, most recent studies remain limited to a local scope and do not tackle the systemic breadth of the disease (2). Here, we use PNA NPs for systemic delivery to correct the F508del mutation. We demonstrate their use both in vitro in primary cells grown in a physiologically relevant air-liquid interface (ALI) culture model and in vivo in mice homozygous for the F508del mutation. This is the first report of systemic therapeutic gene editing to correct CF-causing mutations in vivo.
RESULTS
PNA design and polymeric NP characterization
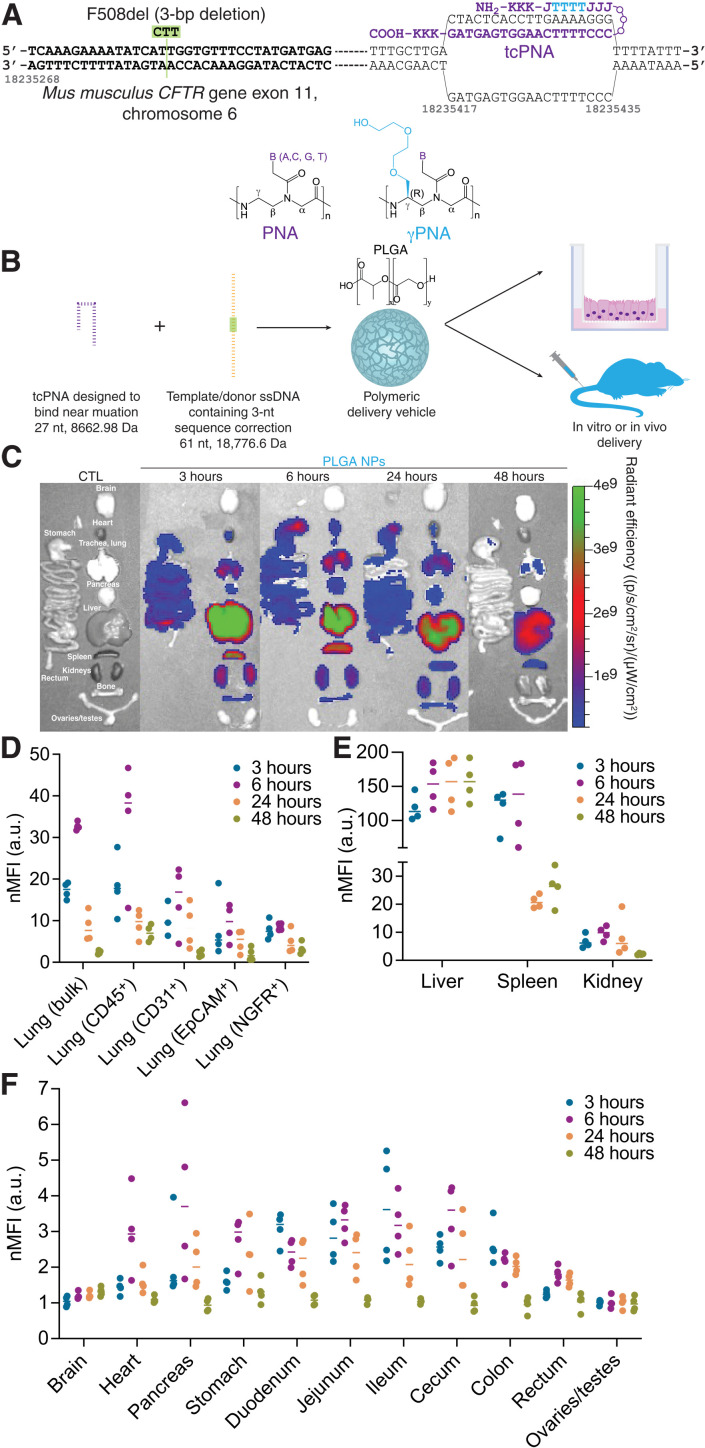
To determine the feasibility of systemic gene editing as a therapeutic approach for CF treatment, we used a murine CF model homozygous for the F508del mutation in exon 11. On the basis of our previous work with local intranasal delivery of PNA NPs in these mice (16), we designed tail-clamp PNA molecules that bind near the mutation site with homopurine/homopyrimidine stretches (Fig. 1A). Here, we incorporated modified PNA monomers with a mini–polyethylene glycol (PEG) group at the γ-position into the PNA sequence in the Hoogsteen domain. γPNAs exhibit enhanced DNA binding (29); we have previously demonstrated corresponding elevations in gene correction ex vivo and in vivo using these γ-modified PNAs in a β-thalassemia mouse (21) and also in human CF bronchial epithelial cells (CFBEs) homozygous for the F508del mutation (30). We also designed a single-stranded correcting donor DNA template to introduce 3 nucleotides (nt) consistent with the wild-type genomic sequence. Both PNA and donor DNA molecules were loaded into polymeric NPs in a 2:1 molar ratio (Fig. 1B) consisting of PLGA formulated using a double-emulsion solvent evaporation technique as described previously (17, 21). The resulting NPs were spherical and ~250 nm in diameter, as characterized by dynamic light scattering (DLS) (table S1) and scanning electron microscopy (SEM) (fig. S1). These NPs were administered both in vitro in ALI cultures of primary airway epithelial cells and in vivo via systemic intravenous injection to F508del-CFTR mice.
Fig. 1. PNA-based gene editing agents can be encapsulated into PLGA NPs, which exhibit accumulation in the lung and GI tract following systemic intravenous administration.
(A) Schematic of PNA design to correct F508del-CFTR, indicating the incorporation of γPNA monomers and the formation of the PNA/DNA/PNA triplex. (B) PNA and donor DNA in vitro and in vivo delivery strategy: Encapsulation into polymeric PLGA NPs. (C) Representative IVIS images indicating biodistribution of Cy5-conjugated PLGA NPs at 3, 6, 24, and 48 hours after intravenous administration in vivo compared to an untreated control animal (CTL). (D) Flow cytometry mean fluorescence intensity values normalized to untreated control animals (nMFI) for homogenized bulk lung and specific cell types (CD45+ macrophages, CD31+ endothelial cells, EpCAM+ epithelial cells, and NGFR+ basal cells) at 3, 6, 24, and 48 hours after intravenous administration of Cy5-conjugated PLGA NPs in vivo. Each dot represents data from one mouse. (E) Flow cytometry mean fluorescence intensity values normalized to untreated control animals (nMFI) for homogenized bulk liver, spleen, and kidney at 3, 6, 24, and 48 hours after intravenous administration of Cy5-conjugated PLGA NPs in vivo. Each dot represents data from one mouse. (F) Flow cytometry mean fluorescence intensity values normalized to untreated control animals (nMFI) for homogenized bulk brain, heart, pancreas, stomach, duodenum, jejunum, ileum, cecum, colon, rectum, and ovaries/testes at 3, 6, 24, and 48 hours after intravenous administration of Cy5-conjugated PLGA NPs in vivo. Each dot represents data from one mouse. a.u., arbitrary units.
As CF is a systemic disease with a heavy burden on both airway and GI epithelia, we first tested whether PLGA NPs could reach these organs when administered systemically. In previous work, we have found that NP size in particular is important for NP accumulation in multiple tissue types in vivo following intravenous administration (31). We assessed the biodistribution of Cy5-conjugated PLGA NPs at several time points after systemic intravenous administration. Whole-organ fluorescence was captured using the In Vivo Imaging System (IVIS) and indicated high levels of NP accumulation in the lung at 3, 6, and 24 hours after administration (Fig. 1C). Similar to previous reports of PLGA biodistribution, we also observed high accumulation in the liver and spleen (32), as well as lower levels of accumulation in other organ systems of interest for CF, including the GI tract. It is important to note, however, that IVIS analysis cannot determine which cell types have taken up the NPs, and so we also performed flow cytometry analyses to determine whether individual cells in multiple organs exhibit NP uptake. Consistent with the IVIS data, we observed an increase in lung NP accumulation by flow cytometry at 3 and 6 hours after administration (Fig. 1D). Notably, we observed NP uptake in multiple cell types in the lung, including CD45+ macrophages, CD31+ endothelial cells, EpCAM+ epithelial cells, and NGFR+ basal cells. In the context of gene editing, basal cells are the ideal target, as these are the presumptive stem cells of the airway epithelium (33). Flow cytometry analyses, as measured by the mean fluorescence intensity normalized to control samples, of other tissues including liver, spleen, kidney, brain, heart, pancreas, stomach, duodenum, jejunum, ileum, cecum, colon, rectum, and ovaries/testes were also consistent with the IVIS data. To account for any potential outliers skewing the mean intensity data, we also assessed median fluorescence intensity in flow cytometry samples, which yielded similar results (fig. S2). Last, we confirmed uptake in key tissues relevant to in vivo CFTR function assessment, namely, the nasal epithelium and rectum, by microscopy (fig. S3). In summary, these data suggest that PLGA-based NPs are able to reach target organs and cell types relevant to CF treatment after intravenous delivery. While the precise mechanisms of intracellular trafficking of PLGA NPs remain unclear, we and others have shown that uptake occurs primarily by adsorptive-type endocytosis (34, 35). Once inside cells, NPs are rapidly shuttled into the endolysosomal pathway and escape from the late endosome into the cytoplasm. In previous work with PNA/DNA NPs, we have shown that PLGA NPs readily associate with and are taken up by various cell types and that they accumulate in the perinuclear region (17).
In vitro correction of F508del-CFTR in primary cell ALI cultures
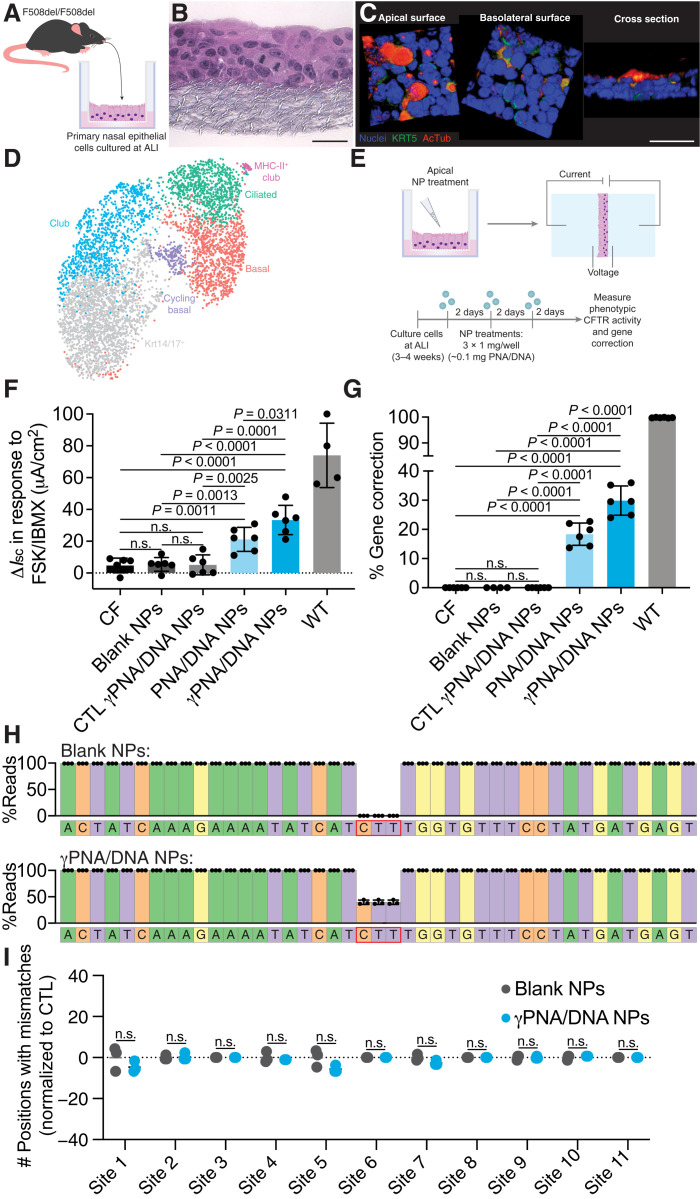
We next sought to develop a physiologically relevant in vitro model in which to test the therapeutic activity of our PNA NPs, taking advantage of ALI systems commonly used in CF research. In this case, we cultured primary nasal epithelial cells (NECs) to more closely mimic the in vivo environment. There are several benefits of this experimental system: (i) These cells can be expanded using feeder cells (fibroblasts) and provide an in vitro model in which we can test our gene editing reagents targeting the murine CFTR genomic locus; (ii) these are primary cells that mature into pseudostratified epithelia with multiple cell types; (iii) to culture these NECs, we adapted clinically relevant methods described to culture human nasal epithelial samples (36); and (iv) this system and protocol are comparable to those used for human NEC culture. To develop the culture system, primary NECs were isolated from F508del mice (Fig. 2A) and expanded using protocols recently described for the expansion of primary human airway epithelial cells (36). After expansion, cells were seeded on permeable supports in transwell inserts and transitioned to ALI over the course of several weeks. These cells formed pseudostratified epithelial layers similar to the epithelial structure in vivo (Fig. 2B) and were primarily composed of basal cells when seeded (Fig. 2C), with multiple cell types present in the polarized mature cultures (37). NEC ALI cultures exhibited an intact epithelium with resistance values similar to those reported in the literature (fig. S4) (36). To further characterize the model system, we performed single-cell RNA sequencing (scRNA-seq) analysis on ~105 primary NECs cultured at ALI using cell type markers (e.g., transcription factors and surface molecules) described in recent reports (38–41). The scRNA-seq data were visualized using a graph-based algorithm [Uniform Manifold Approximation and Projection (UMAP)] (42, 43) to facilitate the identification of distinct cell types. Cells were partitioned into distinct populations with unique gene expression signatures (Fig. 2D). Several cell types were identified, reminiscent of the in vivo environment: basal cells, cycling basal cells, ciliated cells, and club cells.
Fig. 2. PNA NP treatment results in phenotypic and genotypic CFTR correction in physiologically relevant primary NEC ALI cultures consisting of multiple cell types.
(A) Schematic of primary NEC isolation. (B) Hematoxylin and eosin–stained paraffin sections of NEC ALI cultures. Scale bar, 50 μm. (C) Three-dimensional renderings of confocal images of NEC cultures indicating a pseudostratified epithelium containing basal cells (Krt5; green) and ciliated cells [acetylated α-tubulin (AcTub); red]. Nuclei are shown in blue. Scale bar, 50 μm. (D) UMAP plot of scRNA-seq data for primary NEC ALI cultures indicating distinct cell types (basal, cycling basal, Krt14/17+, ciliated, club, and MHC-II+ club) (n = 3 cultures). (E) Schematic of NP treatment scheme in primary NEC ALI cultures. Cells were treated with three doses of 1 mg of blank or PNA/DNA PLGA NPs. PNA/DNA NPs contain ~2 μg/0.2 nmol of PNA and ~2 μg/0.1 nmol of DNA per milligram. (F) Phenotypic ΔIsc measurements following treatment of primary NEC ALI cultures with blank, CTL (nonspecific) γPNA/DNA, PNA/DNA, or γPNA/DNA NPs compared to CF or wild-type (WT) controls. (G) ddPCR gene correction assessments of NP-treated primary NEC ALI cultures. (H) Deep sequencing read analysis around the target site of NP-treated primary NEC ALI cultures. (I) Deep sequencing off-target analysis of blank NP– or γPNA/DNA NP–treated NEC ALI cultures at 11 genomic sites, with partial PNA binding site homology displayed as the number of positions with mismatches to the reference sequence normalized to untreated control samples.
We used this primary NEC ALI culture model to test the hypothesis that γ-modified PNAs could mediate enhanced gene correction compared to unmodified PNAs. PNA and γPNA NPs were administered to the primary NEC ALI cultures and assessed for both phenotypic and genotypic CFTR correction. To mimic the multidose in vivo treatment schemes that we have used in previous studies, each culture received a dose of 1 mg of NPs every 2 days for a total of three doses (Fig. 2E). Two days after the last treatment, CFTR activity was assessed via Ussing chamber measurements. We monitored CFTR-mediated ion transport as the change in short-circuit current (ΔIsc) in response to a CFTR-stimulating cocktail of forskolin (FSK) and 3-isobutyl-1-methylxanthine (IBMX). Representative traces are shown in fig. S5. Compared to untreated, blank NP–treated, and nonspecific control γPNA/DNA NP samples, we observed significant increases in ΔIsc for both PNA/DNA and γPNA/DNA NP–treated samples (Fig. 2F). The ΔIsc recorded for γPNA/DNA NP–treated samples was significantly higher than the ΔIsc for PNA/DNA NP–treated samples. After Ussing measurements, we next assessed the extent of genotypic correction giving rise to these phenotypic changes. To reduce potential polymerase chain reaction (PCR) bias, we developed a droplet digital PCR (ddPCR) assay to quantify the percentage of modified CFTR alleles in the NEC cultures (Fig. 2G and fig. S6). Consistent with the phenotypic changes we measured in the Ussing chamber, in apically treated NEC cultures, we observed ~18% correction after PNA/DNA NP treatment and ~30% correction after γPNA/DNA NP treatment. We also performed next-generation sequencing (NGS) and did not observe unintended indels at the target site (Fig. 2H). Furthermore, in 11 genomic sites with partial homology to the PNA or donor DNA binding sites identified using BLAST (table S2), the off-target mutation/error rates were similar to blank NP–treated controls with minimal variation from the reference sequence (Fig. 2I). Last, we tested the effects of PLGA NP treatment on cell viability in these cultures and did not observe any differences between treated cells and untreated controls (fig. S7). Together, these results suggest that PNA-based gene editors can produce both phenotypic and genotypic correction. Furthermore, γPNA/DNA NPs are significantly more effective for gene correction compared to PNA/DNA NPs in primary airway epithelial cells with a large basal cell population. These measurements are consistent with the results that we have observed in human CFBE cells and with the inhalational delivery of PNA versus γPNA/DNA NPs in the F508del mouse (30).
Phenotypic and genotypic CFTR correction in vivo by PNA/DNA NPs
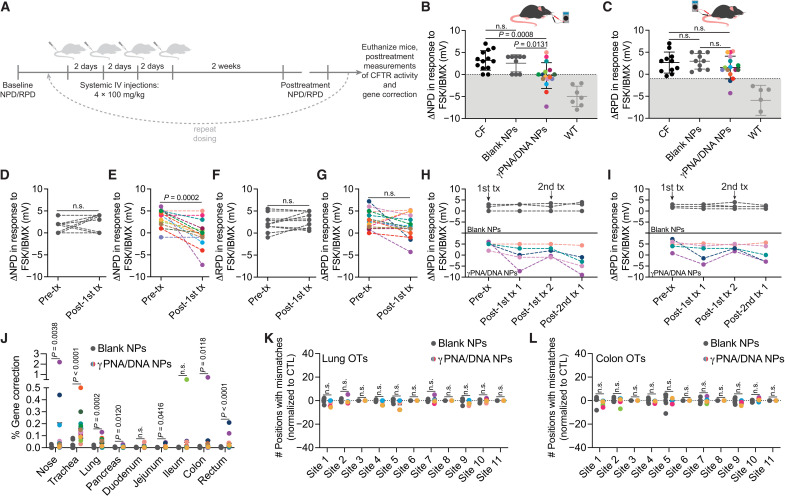
We next tested for phenotypic and genotypic correction of the F508del CF mutation in a murine disease model homozygous for the 3-bp deletion (F508del-CFTR). PLGA NPs encapsulating γPNA and donor DNA designed to correct the F508del mutation were formulated, and F508del-CFTR mice were treated with a 2-mg NP resuspension administered intravenously for four treatments over the course of 2 weeks (Fig. 3A). Both blank and γPNA/DNA NPs are well tolerated without indications of toxicity or inflammation (figs. S8 and S9 and table S3). Two weeks after the last treatment, in vivo phenotypic correction of the F508del mutation was assessed using noninvasive assays to detect CFTR activity in vivo: nasal potential difference (NPD) and rectal potential difference (RPD). The PD assay is a useful method to study ion transport if serial measurements are required (44, 45). Nasal and rectal epithelia in wild-type mice exhibit a robust cyclic adenosine monophosphate (cAMP)–stimulated chloride efflux, whereas CFTR dysfunction results in a lack of activation of cAMP-stimulated chloride flux. After one dosing round of intravenous-delivered γPNA NPs, we observed a partial amelioration of the impaired response to cAMP stimulation in the nasal epithelia, with some mice exhibiting hyperpolarized (more negative) ΔNPD in response to CFTR stimulation into the wild-type range (Fig. 3B), although the degree of hyperpolarization was variable with a subset of mice not responding to treatment. Untreated control and blank NP–treated mice exhibited typical CF NPD responses: a lack of hyperpolarization or a depolarization in response to a cAMP stimulation cocktail that is consistent with the absence of CFTR activity. Representative raw NPD traces are shown in fig. S10. Topical intranasal delivery of the same γPNA and donor DNA reagents using modified PLGA/PBAE [poly(β-amino ester)]/MPG NP formulations (as we have described previously) resulted in NPD responses closer to and not significantly different from wild-type values, consistent with our previous observations (fig. S11) (16), suggesting that our NEC ALI results (Fig. 2) are predictive of local in vivo delivery results. The RPD responses to cAMP-stimulating agents of systemically γPNA NP–treated mice were not significantly different from untreated and blank NP–treated controls, although a few mice exhibited hyperpolarization into the wild-type range (Fig. 3C). An examination of pretreatment and posttreatment NPD responses to cAMP stimulation for each mouse reveals an average percent change of −118% in the ΔNPD for γPNA/DNA NP–treated mice, whereas the average percent change in ΔNPD for blank NP–treated mice was +28% compared to baseline measurements (Fig. 3, D and E). A similar analysis of RPD responses shows an average percent change of −50% in ΔRPD for γPNA NP–treated mice and an average percent change of −6% in ΔRPD for blank NP–treated mice (Fig. 3, F and G). Animals treated with nonspecific γPNA/DNA control NPs exhibited similar responses to blank NP–treated mice (fig. S12). To assess the longevity of treatment responses, we performed additional NPD and RPD measurements for a subset of blank NP– and γPNA NP–treated mice 2 weeks after the first posttreatment measurements. We observed attenuation in the NPD response in the second measurement for the two mice that responded to γPNA NP treatment but no change in response for one mouse that did not exhibit a change in NPD following the first round of treatment (Fig. 3H). This attenuation in response is consistent with reports by others studying the longevity of gene editing, and specifically HDR repair events, with CRISPR-Cas9 (46). Following the second posttreatment measurements, we retreated each mouse with another round of four NP doses and measured the NPD response 2 weeks after the last treatment. After the second treatment round, the mice that had shown a response after the first treatment round exhibited NPD hyperpolarization in response to cAMP stimulation cocktail again, whereas the mice that did not respond to the first round of treatment did not respond to the second treatment. We observed similar trends for RPD measurements performed on the same mice (Fig. 3I). Blank NP–treated animals exhibited similar NPD measurements throughout the study, consistent with measurements of untreated animals from a variety of CF mouse models (44).
Fig. 3. Functional and genotypic correction of CFTR in vivo following PNA NP administration.
(A) Schematic of in vivo NP dosing scheme. IV, intravenous. (B) Nasal potential difference (NPD) and (C) rectal potential difference (RPD) measurements following either 4 × 2 mg of blank NP (dark gray circles) or 4 × 2 mg of γPNA/DNA NP (multicolored circles) treatment with CF (black circles) and wild-type (light gray circles) controls. Color coding of γPNA/DNA NP–treated animals is consistent throughout this figure, and each color represents a different mouse. γPNA/DNA NPs contain ~2 μg/0.2 nmol of PNA and ~2 μg/0.1 nmol of DNA per milligram; each animal received ~0.2 mg/kg of PNA and donor DNA per dose. Gray region indicates wild-type range. Pre- and posttreatment NPD measurements for F508del-CFTR mice treated with (D) blank NPs and (E) γPNA/DNA NPs. Pre- and posttreatment RPD measurements for F508del-CFTR mice treated with (F) blank NPs and (G) γPNA/DNA NPs. Serial (H) NPD and (I) RPD measurements performed over the course of two treatment rounds for a subset of animals in the γPNA/DNA NP–treated and blank NP–treated cohorts. Arrows indicate treatment round initiation. (J) Gene correction levels measured by ddPCR at the F508del locus for airway and GI organs from mice treated with either blank NPs or γPNA/DNA NPs. Deep sequencing off-target (OT) analysis of blank NP– or γPNA/DNA NP–treated F508del mice (K) lungs and (L) colons at 11 genomic sites, with partial PNA binding site homology displayed as the number of positions with mismatches to the reference sequence normalized to untreated control samples.
In addition to phenotypic measurements of CFTR correction in vivo, we also assessed gene correction in bulk tissues by ddPCR in several organs. We observed editing in tissues that make up the airways and GI tract, including the nasal epithelium (up to ~2%), trachea (up to ~0.3%), lung (up to ~0.1%), ileum (up to ~0.5%), colon (up to ~0.7%), and rectum (up to ~0.2%) (Fig. 3J). Each γPNA/DNA NP–treated mouse in this figure is color-coded such that phenotypic and genotypic results for each mouse can be correlated. There was a high degree of heterogeneity and variability in gene correction measurements among the treated mice, consistent with CRISPR-based editing approaches (47). However, the degree of editing for each mouse was consistent with the degree of electrophysiological response in NPD and RPD assays. NGS was also used to detect off-target editing events in the lung and colon (Fig. 3, K and L) with a focus on genomic regions with partial homology to the PNA or donor DNA binding sites. Detected mismatches were equivalent to those observed for control blank NP–treated samples with low variation from the reference sequence. As we observed high levels of NP accumulation in the liver and spleen in our biodistribution studies (Fig. 1), we also assessed the level of gene correction and off-target effects in these organs (fig. S13) and did not observe any significant editing or off-target modifications.
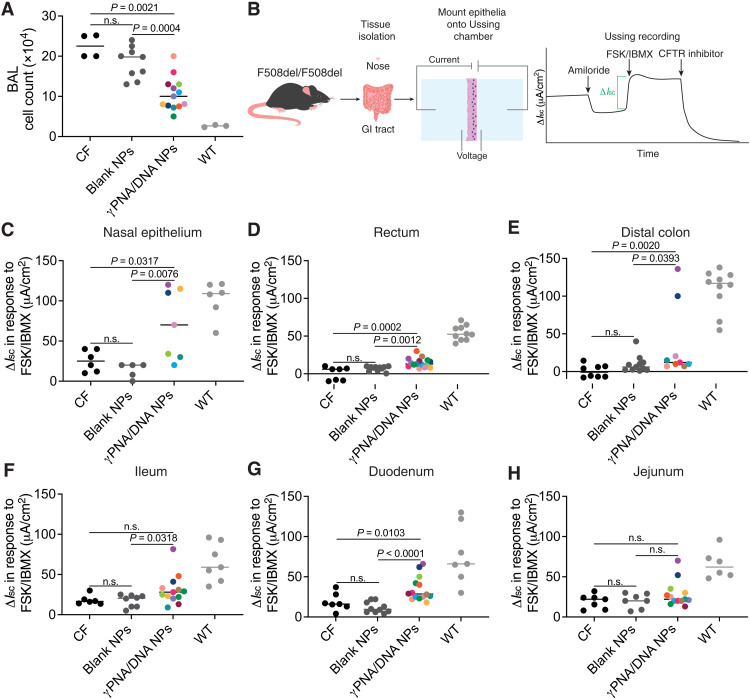
We further assessed long-term phenotypic disease amelioration by measuring cell counts in the bronchoalveolar lavage (BAL) fluid of treated mice, which typically includes alveolar macrophages and neutrophils (Fig. 4A) (48). As in Fig. 3, γPNA/DNA NP–treated mice throughout this figure panel are color-coded such that phenotypic and genotypic results for each mouse can be correlated. We observed consistently lower inflammatory cell counts in γPNA/DNA NP–treated mice, an additional indicator of lung health, suggesting partial restoration of CFTR function. In addition, we assessed CFTR function in treated mice by performing Ussing chamber measurements of ΔIsc in response to CFTR stimulation as described above on epithelial tissues ex vivo. Tissues (nasal epithelium, rectum, distal colon, ileum, duodenum, and jejunum) were immediately dissected after euthanasia and mounted into Ussing chambers (Fig. 4B). We recorded ΔIsc values for tissues for intact epithelia where possible. For several tissues including nasal epithelium, rectum, and distal colon, γPNA/DNA NP–treated mice exhibited ΔIsc measurements significantly different from both blank NP–treated animals and CF controls (Fig. 4, C to H). As expected, nonspecific control γPNA/DNA NP–treated animals exhibited similar phenotypic data to blank NP–treated animals (fig. S12). Furthermore, we observed again that the mice with more robust in vivo phenotypic responses had larger ΔIsc values. Variability in this assay and ddPCR results is likely due to heterogeneous gene editing throughout the tissues. As an additional measure of longer-term benefits of treatment, we also studied the weight gain per month of γPNA/DNA NP–treated mice and found that they were significantly higher that blank NP–treated animals (fig. S14).
Fig. 4. Functional correction of CFTR ex vivo following PNA NP administration.
(A) BAL cell counts following either blank NP (dark gray circles) or γPNA/DNA NP (multicolored circles) treatment with CF (black circles) and wild-type (light gray circles) controls. Color coding of γPNA/DNA NP–treated animals is consistent throughout this figure, and each color represents a different mouse. γPNA/DNA NPs contain ~2 μg/0.2 nmol of PNA and ~2 μg/0.1 nmol of DNA per milligram; each animal received ~0.2 mg/kg of PNA and donor DNA per dose. (B) Schematic of ex vivo tissue isolation and Ussing chamber assay. Phenotypic ΔIsc measurements in (C) nasal epithelium, (D) rectum, (E) distal colon, (F) ileum, (G) duodenum, and (H) jejunum following either 4 × 2 mg of blank NP (dark gray circles) or 4 × 2 mg of γPNA/DNA NP (multicolored circles) treatment with CF (black circles) and wild-type (light gray circles) controls.
DISCUSSION
The therapeutic approach for CF described here combines non–nuclease-based PNA gene editing technology with systemic delivery of biocompatible polymeric NPs to achieve gene correction in multiple tissue types in vitro and in vivo. Improved PNA molecules containing structure-modifying γ-substitutions demonstrated phenotypic correction and gene editing without unintended indel formation when co-delivered with donor DNA in vitro, to primary NECs isolated from mice harboring the F508del-associated CF mutation, and in vivo in the same mouse model. Enhanced gene editing with modified PNAs is consistent with our previous work in another disease context (21). We believe that this is the first evidence of in vivo systemic delivery of gene editing agents to correct a CF-causing mutation in multiple organs.
From a safety perspective, both the therapeutic PNA/donor DNA combination and the polymeric delivery vehicle did not exhibit detectable adverse effects. Low levels of off-target genomic mutations and well-tolerated systemic delivery are paramount to clinical translation of gene editing therapeutics. The intended correcting 3-bp CTT (cytosine-thymine-thymine) insertion was detected at the target site by both ddPCR and deep sequencing, with no unintended outcomes in the form of indels created in the flanking sequences. While much progress has been made with regard to methods for determining genome-wide off-target mutations for nuclease-based editing technologies (49–52), our ability to assess the potential off-target effects of PNA-based editing technology remains limited to sequence homology. As expected for PNA molecules with sequence specificity, deep sequencing of multiple sites with partial homology to the PNA binding site revealed no off-target effects above background mutation/read error rates in vitro in treated NECs and in vivo in the lungs and colons of treated mice. Additional concerns with systemically delivered gene editing therapeutics are the potential adverse consequences of the editors or the delivery vehicles (53). PNA NPs did not exhibit systemic toxicity as evidenced by serum chemistry analyses compared to untreated or blank NP controls. Last, particularly for intravenously injected therapeutics, the risk of on- or off-target or activity in inappropriate tissues highlights the need to ensure proper tissue tropism. While we observed widespread NP biodistribution to multiple tissues apart from airway and GI epithelia, we did not observe any pathology in tissues with particularly high NP accumulation, including the liver and spleen.
The work presented here provides a foundation for systemic in vivo gene editing to correct CF with PNA NPs. There are few studies to date that have described the systemic intravenous extrahepatic delivery of therapeutic gene editing agents, especially in disease animal models. One recent study reported 5 to 7.3% indels produced by Cas9 ribonucleoproteins encapsulated in lipid NPs (LNPs) in the liver following intravenous delivery in wild-type mice to edit the PCSK9 locus (54). Another study by Intellia Therapeutics similarly showed impressive liver editing by CRISPR-Cas9 LNPs up to ~60% in a preclinical study of transthyretin amyloidosis (55); clinical trial results were also promising (56). Rosenblum et al. (57) report ~80% editing in an ovarian tumor model, but by intraperitoneal administration of LNPs encapsulating CRISPR-Cas9 reagents targeting PLK1. It is important to note, however, the difference between precise correction achieved by our PNA/DNA NPs and gene disruption by CRISPR-Cas9. In the above examples of gene disruption by CRISPR-Cas9, editing is quantified by the sum of the frequencies of insertions, substitutions, and deletions at the target site, as opposed to the frequency of only a 3-bp insertion, as we have studied. We did not observe any unintended indels and are reporting only legitimate corrections to a functional genotype. While the response to PNA/DNA NP treatment across the cohort of mice studied was variable and the frequency of edited alleles was modest, we observed both phenotypic and genotypic correction of extrahepatic tissues in vivo. The variability in treatment response is likely due in part to variability in baseline phenotypic manifestations of disease. Moreover, as demonstrated in Fig. 1, NP delivery to target organs is also variable between animals. Last, to draw a parallel to human data, in patients with CF that are eligible for highly effective modulator therapy (Trikafta), even those with the same CF genotype exhibit variable responses to treatment (58). We found that elevated gene correction levels correlated well with performance in phenotypic assays. It has been estimated in several studies that correcting CFTR in ~5 to 15% of cells in epithelia should restore transepithelial chloride secretion to near wild-type levels, suggesting that in vivo correction of even a fraction of cells could provide therapeutic benefit (27, 59–63). While we did not observe editing in that range in our systemic in vivo studies, our results do suggest that even modest levels of editing can result in partial restoration of chloride transport. Compared to systemic delivery, local intranasal delivery of γPNA/DNA NPs resulted in phenotypes more consistently in the wild-type range. In light of these findings, local delivery to the airway epithelia might be combined with systemic delivery to maximize phenotypic correction across multiple organs affected by CF. Systemic delivery in particular can be further improved as a therapeutic strategy. For example, enhancements in vehicle design could be used to increase efficacy, including surface modifiers to target specific cell types as well as the use of different polymeric materials and formulation techniques to vary NP size, surface charge, and composition with the ultimate goal of modulating encapsulation efficiency and tissue tropism, particularly to the GI tract (64). Beyond NP engineering, a more in-depth parameter exploration of dosing and frequency of administration would also elucidate the limits of this approach.
The longevity of treatment response is also an important consideration for translation of gene editing therapeutics. We observed an attenuation in treatment response over time that was reversed upon additional treatment rounds. Optimization of the delivery to target stem cells will be key to obtaining a one-time cure.
MATERIALS AND METHODS
Experimental design
This section presents the materials and methods that allow us to establish causal relationships between PNA NP treatment of primary cells or animals and their resulting phenotypic and genotypic responses. First, we describe the formulation and characterization of PNA NPs. Second, we describe the generation, characterization, treatment, and assessment of ALI cultures of primary murine NECs. Third, we describe in vivo and ex vivo electrophysiological measurements of CFTR function. Fourth, we describe the polymeric NP biodistribution assessment. Fifth, we describe the assessment of genotypic correction by ddPCR and NGS. Last, we describe the statistical analyses used for all studies.
Materials
Boc-protected PNA monomers were purchased from ASM Research Chemicals. Mini-PEG γPNA monomers were prepared from Boc-2(2-(2-methoxyethoxy)ethyl)-l-serine as described previously (21, 29). PNA oligomers were synthesized on solid support [MBHA (4-methylbenzhydrylamine) resin] using Boc chemistry (29) and purified using high-performance liquid chromatography (HPLC). The CFTR tcPNA (tail-clamp PNA) sequence used in this study was H-KKK-JTTTTJJJ-OOO-CCCTTTTCAAGGTGAGTAG-KKK–NH2, with the underlined portion indicating the positions of γPNA monomers for the γPNA version of the sequence; K, lysine; J, pseudoisocytosine (a cytosine analog for improved PNA/DNA/PNA triplex formation at physiologic pH); O, 8-amino-2,6,10-trioxaoctanoic acid linkers connecting the Hoogsteen and Watson-Crick domains of the tcPNAs. In mismatched PNA control experiments, a γPNA molecule targeting the human β-globin gene was used, which contains 12 mismatches in the Watson-Crick domain relative to the CFTR γPNA molecule: H-KKK-JTTTJTTTJTJT-OOO-TCTCTTTCTTTCAGGGCA-KKK-NH2. The donor DNA oligonucleotide (61 nt) was synthesized by Midland Certified Reagent (Midland, TX) and end-protected with three phosphorothioate internucleotide linkages at both the 5′ and 3′ ends and purified by reversed-phase HPLC. This antisense donor DNA sequence matches the corresponding region in mouse CFTR exon 11, with the correcting 3-nt insertion underlined: 5′T(s)C(s)T(s)TATATCTGTACTCATCATAGGAAACACCAAAGATAATGTTCTCCTTGATAGTACC(s)C(s)G(s)G3′. PLGA polymer [50:50 DL-PLG [poly(DL-lactide-co-glycolide)], inherent viscosity: 0.55 to 0.75 dl/g] was purchased from Lactel and used as received. PLGA-Cy5 polymer (50:50 lactic acid: glycolic acid, Mn: 30,000 to 55,000 Da) was purchased from PolySciTech and used as received. Dichloromethane (DCM; HPLC grade, 99+%) was purchased from Sigma-Aldrich. DiD [DiIC18(5); 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanine, 4-chlorobenzenesulfonate salt] dye was purchased from Biotium and dissolved in dimethyl sulfoxide (DMSO) at 10 mg/ml before use. Heparin (1000 USP unit/ml) was purchased from Cardinal Health. Isoflurane was purchased from Sigma-Aldrich. Fisherbrand Superfrost Microscope slides were purchased from Thermo Fisher Scientific. Antibodies for flow cytometry were used as received: anti-mouse CD45 (clone 30-F11, Thermo Fisher Scientific), anti-mouse CD31 (clone 390, Thermo Fisher Scientific), anti-mouse EpCAM (clone G8.8, BioLegend), and anti-mouse NGFR (clone REA648, Miltenyi Biotec).
NP formulation and characterization
PLGA NPs encapsulating DiD were formulated using a single oil-in-water emulsion solvent evaporation technique as previously described (65). Briefly, 50 mg of polymer was dissolved in 900 μl of DCM overnight. One hundred microliters of DiD dye at 2.5 mg/ml in DMSO was added to the dissolved polymer immediately before formulation [0.5% (w/w)]. The polymer and dye solution was added dropwise under vortex into 2 ml of 5% (w/v) low–molecular weight polyvinyl alcohol (PVA) solution and sonicated with a probe tip sonicator to form an oil-in-water single emulsion and then diluted into 10 ml of 0.3% (w/v) PVA solution while mixing. The remaining organic solvent was evaporated using a rotary evaporator. The NPs were then washed twice in nuclease-free diH2O by centrifugation at 16,000g to remove excess PVA. NP size and zeta potential were measured via DLS. NP morphology was visualized by SEM. Blank PLGA NPs and NPs loaded with PNA and donor DNA were formulated using a water-in-oil-in-water double-emulsion, solvent evaporation technique as described previously (21). PNA and DNA were encapsulated at a 2:1 ratio with 2 nmol of PNA and 1 nmol of DNA loaded per milligram of polymer. Blank NPs were loaded with 150 ml of nuclease-free water, and PNA/DNA NPs were loaded with 100 μl of PNA and 50 μl of donor DNA each at 1 mM in nuclease-free diH2O. NP formulations were resuspended in nuclease-free diH2O with 30 mg of trehalose per 50-mg initial polymer weight, flash-frozen at −80°C, lyophilized for at least 48 hours, and stored at −20°C.
Isolation of primary NECs
Primary NECs from mice homozygous for the F508del mutation (fully backcrossed C57/BL6 background) were isolated on the basis of similar protocols described for human cells (36, 66–68). Briefly, noses were dissected and the nasal epithelium was digested with a protease mixture [1% protease and 0.01% deoxyribonuclease (DNase)] for 2 hours with agitation at 4°C. NECs were then collected and filtered through a 70-μm cell strainer and washed twice by centrifugation (1500 rpm, 10 min). Cells were collected by centrifugation and resuspended in F media as previously described (36) containing 5 mM Y-27632 and cultured on type I collagen–coated (Purecol; Advanced BioMatrix) tissue culture dishes with irradiated feeder cells. Differential trypsinization was used to separate feeder and epithelial cells during passaging as previously described (67).
ALI cell culture, NP treatments, and Ussing measurements
To initiate ALI cultures, ~1.5 × 106 cells were seeded on permeable polycarbonate supports with 0.4-mm pore size (Corning Costar 3801) in 250 ml of cell culture medium. These six-well plate inserts had a diameter of 12 mm and were coated with type I collagen (RatCol, Advanced BioMatrix) and exposed to ultraviolet radiation overnight. Three milliliters of culture medium was added to each well below the inserts. Cells were fed both apically and basolaterally three times per week over the course of 4 weeks, after which they were transitioned to ALI and fed only from the bottom. Cells were maintained at ALI for 3 to 4 weeks until cilia and mucus developed, and transepithelial resistance values were >300 ohm·cm2 (fig. S4) (36, 69). For in vitro PNA/DNA NP experiments, cells at ALI were treated either apically or basolaterally every 2 days with 1 mg of NPs (containing ~2 mg/0.2 nmol of PNA and ~2 mg/0.1 nmol of DNA) resuspended in 250 ml of cell culture medium by water bath sonication and vortexing for a total of three treatments. Ussing measurements were performed 2 days after the last treatment. Ussing experiments with ALI cultures were performed as previously described using the Easy Mount Ussing Chamber System (Physiologic Instruments) (70). Briefly, chambers were heated to 37°C, and current and voltage electrode tips were prepared by partially filling with 3% agar in 3 M KCl and then backfilling with 3 M KCl solution. The transwell inserts containing ALI cultures were loaded into P2300 snapwell chambers, filled with 6 ml of Kreb’s bicarbonate Ringer’s solution at 37°C (140 mM Na+, 120 mM Cl2, 5.2 mM K+, 1.2 mM Ca2+, 1.2 mM Mg2+, 25 mM HCO32−, 2.4 mM HPO42−, 0.4 mM H2PO42−, and 10 mM glucose at pH 7.4), and allowed to equilibrate for 20 min. A mixture of 95% O2 and 5% CO2 gas was bubbled through the solutions. Current-clamped Ussing experiments were performed with a bidirectional pulse of 1 mA of current for 3 s every 60 s, with transepithelial voltages and membrane resistances measured during each pulse. Amiloride (100 mM) was added apically with a 10-min equilibration and subsequently maintained. Short-circuit current (Isc) was then calculated using Ohm’s law when the apical chamber solution was replaced with a 0 Cl− solution containing FSK (10 mM) and IBMX (1 mM) with a 15-min equilibration. CFTR-specific inhibitor 172 (20 mM) was added apically after 20 min, followed by bilateral replacement of solution with Kreb’s bicarbonate Ringer’s solution.
scRNA-seq analysis
scRNA-seq was performed using scFTD-seq (single-cell freeze-thaw lysis directly toward 3’ mRNA sequencing) (71). Briefly, a polydimethylsiloxane microwell array chip was used for co-isolating single cells and uniquely barcoded mRNA capture beads. The design of the microwell arrays has been described previously (71). Each microchip has up to 25,000 wells and allows for barcoding of ~2500 cells in a single run to prepare a sequencing library. Library preparation and sequencing steps follow the same protocols as outlined previously (71). Transcriptome alignment to a reference transcriptome of the corresponding species including barcode/unique molecular identifier (UMI) identification and collapsing was performed as described in the scFTD-seq methods (71). The data analysis was performed using Seurat v3.1 on the normalized and log-transformed gene expression data (72, 73). A total of 5896 individual cells were used for single-cell data analysis. Cells with low expression of genes (<100 genes), high expression of genes (>2000), and a high percentage of mitochondrial genes (>15%) were digitally filtered out, resulting in 5302 single cells used for the subsequent analysis. After identifying the top 2000 variable genes, we performed principal components analysis and unsupervised clustering using UMAP (42), which was implemented in Seurat (72, 73). For cell type identification, support vector machine (SVM) with linear kernel was used (74). Briefly, the SVM model was first trained with labeled single-cell data from mouse tracheal epithelial cells (38) with 98.6% accuracy. Then, the trained model was applied to predict the cell types in this work.
Immunofluorescence and microscopy
To prepare paraffin-embedded NEC sections, cells were immediately fixed in 10% normal buffered formalin for 24 hours at room temperature before being transferred to phosphate-buffered saline (PBS) and kept at 4°C until paraffin embedding. For immunofluorescence, 5-mm sections were baked at 60°C for 1 hour and deparaffinized using standard procedures. Antigen retrieval was performed by steaming with citrate buffer (pH 6; Abcam), after which sections were rinsed with Dulbecco’s PBS (DPBS) and blocked with 10% normal goat serum for 30 min at room temperature. Primary antibodies in blocking buffer were added overnight at 4°C. Sections were then washed three times with DPBS for 5 min, and secondary antibody diluted in blocking buffer was added for 1 hour at room temperature. Sections were washed again with PBS three times for 5 min. Hoechst 33342 diluted 1:1000 in PBS was then added for 30 s.
In vivo NP administration and NPD/RPD measurements
All animal procedures were performed in accordance with the guidelines and policies of the Yale Animal Resource Center and approved by the Institutional Animal Care and Use Committee of Yale University. Male and female mice homozygous for the F508del mutation (fully backcrossed C57/BL6 background) primarily aged 3 to 6 months old were used. All mice were genotyped before use. Mice were anesthetized using isoflurane. Once respirations reduced to 1 breath per second, NPs were administered intravenously via retro-orbital injection. Two milligrams of NPs was resuspended in 150 μl of DPBS and sonicated before injection. Four total doses of 2 mg of NPs were given over the course of 2 weeks. γPNA/DNA NPs contain ~2 mg/0.2 nmol of PNA and ~2 mg/0.1 nmol of DNA per milligram; each animal received ~0.2 mg/kg of PNA and donor DNA per dose. NPD and RPD measurements as indicators of ion transport across respiratory and GI epithelia in vivo were performed as previously described (75). One baseline measurement and up to three post-NP treatment measurements were done for each mouse at least 2 weeks after the last treatment or measurement. Briefly, mice were anesthetized with ketamine/xylazine, after which an electrode probe is placed into one nostril (NPD) or the rectum (RPD) at 3 mm with a reference electrode, with 3% agar in Ringer’s solution placed subcutaneously in the tail. Saline flow through the probing electrode is controlled by a microperfusion pump at 23 ml min−1 for NPD and 0.5 ml hour−1 for RPD. Both electrodes are connected to a voltmeter to measure potential differences, which were recorded following a course of solutions: control Ringer’s solution, Ringer’s solution containing 100 μM amiloride, chloride-free solution with amiloride, and then chloride-free solution with amiloride and FSK/IBMX. The Δamiloride NPD responses for a representative subset of blank NP– and γPNA/DNA NP–treated animals are shown in table S4. The 0 Cl− solution used for all GI tract tissues contained 5 mM barium hydroxide to block potassium currents.
Ex vivo Ussing measurements, BAL fluid analysis, and histology
Following euthanasia of F508del/F508del mice with a lethal dose of ketamine/xylazine, blood was collected via retro-orbital eye bleed or cardiac puncture, and animals were heart-perfused with heparinized DPBS to remove remaining blood from circulation. Lungs were filled with 2 ml of DPBS containing protease inhibitors and 0.5 M EDTA at pH 8 for BAL, and this fluid was collected for further analysis (cell count and cytokine levels). The right lobes of the lung were tied off, and the left lobe was collected for histopathology by first inflating with 0.5% low-melt agarose at 37°C at constant pressure and then fixing in 10% neutral buffered formalin solution. Following fixation, lungs were embedded in paraffin, sectioned, and stained with hematoxylin and eosin before imaging. Samples of epithelia from nose, ileum, duodenum, jejunum, distal colon, and rectum were mounted in Ussing chambers, and Ussing analyses were performed as described by Grubb (76). Nasal and rectal epithelia are particularly delicate to manipulate and mount appropriately in the Ussing chambers, and so we only report values for intact epithelia of sufficient size without tears or holes that we are able to dissect (76). The 0 Cl− solution used for all GI tract tissues contained 5 mM barium hydroxide to block potassium currents.
Biodistribution assessment
After intravenous injection of one 2-mg dose of PLGA-Cy5 NPs (100 mg/kg) resuspended in 150 μl of DPBS, animals were euthanized at 3, 6, 24, or 48 hours. All animals were perfused transcardially with heparinized DPBS (100 USP/ml). Organs (brain, heart, trachea, lungs, GI tract, liver, pancreas, spleen, kidneys, bone, and gonads) were harvested. Fluorescent agent accumulation in the organs was visualized using an IVIS (Perkin Elmer). Tissues apart from the lung were then homogenized into a single-cell suspensions through a 70-μm cell strainer and washed twice with DPBS by centrifugation and resuspended in PBS containing 2% bovine serum albumin (BSA). Uptake of PLGA-Cy5 NPs in single cells was then analyzed by flow cytometry (BD LSRII) and compared to cells harvested from untreated control animals. Lung tissue was minced and then digested with a solution of collagenase I (5 mg/ml) and DNase (1 mg/ml) in Hanks’ balanced salt solution for 30 min while shaking at 37°C. The resulting slurry was further homogenized by shearing through an 18-gauge needle and filtered through a 70-μm cell strainer. The cells were collected by centrifugation at 330g and washed twice with DPBS by centrifugation. The cell pellet was then suspended in 2 ml of red blood cell lysis buffer for 2 min, rinsed with 5 ml of DPBS containing 2% BSA, and collected by centrifugation. Lung cells were then stained for cell-specific markers using fluorescently labeled antibodies. Cells (1 × 106) were stained with 5 μl of antibody for 1 hour on ice. Any unbound antibodies were rinsed by diluting in 1 ml of DPBS with 2% BSA and collected by centrifugation. Stained cells were resuspended in DPBS with 2% BSA.
Toxicity and immunogenicity
Twenty-four hours after one 2-mg dose (100 mg/kg) of NP treatment (γPNA/DNA or blank in 150 μl of DPBS), animals were euthanized and blood was collected by cardiac puncture. Aliquots of collected blood were separated into serum and plasma by centrifugation, and serum was analyzed for cytokine levels using a bead-based multiplex assay (MILLIPLEX, Millipore) for Luminex. Serum was also collected and sent to Antech Diagnostics for blood chemistry analyses per standard protocols.
gDNA extraction
gDNA was harvested from cells and tissues using the ReliaPrep gDNA Tissue Miniprep System (Promega) according to the manufacturer’s instructions. Tissues were first homogenized into a single-cell suspension before gDNA extraction.
Droplet digital PCR and deep sequencing
ddPCR was used to quantify gene editing at the target site. The concentration of gDNA samples was measured using NanoDrop, and 80 to 100 ng of gDNA were used for each ddPCR. ddPCRs were set up as follows: 11 ml of 2×ddPCR supermix for probes (no deoxyuridine triphosphate) (Bio-Rad), 0.2 ml of forward primer (100 mM), 0.2 ml of reverse primer (100 mM), 0.053 ml of CF (HEX) probe (100 mM), 0.053 ml of wild-type (edit) probe (100 mM) (Integrated DNA Technologies), 0.5 ml of Eco RI, and 10 ml of gDNA and nuclease-free dH2O combined. The primers used were 5′-TGCTCTCAATTTTCTTGGAT-3′ (forward) and 5′-GGCAAGCTTTGACAACA-3′ (reverse). The ddPCR probes were 5′-ATCATAGGAAACACCAATGATAT-3′ [CF (5′HEX)] and 5′-CATCATAGGAAACACCAAAGAT-3′ [wild type (5′FAM)].
ddPCR droplets were generated using an automated Droplet Generator (AutoDG, Bio-Rad). Thermocycler conditions were as follows: 95°C 10 min, (94°C 30 s, 53.7°C 2 min—ramp 2°C/s) × 40 cycles, 98°C 10 min, hold at 4°C. Following the PCR, droplets were left at 4°C for at least 30 min and read using the QX200 Droplet Reader (Bio-Rad). ddPCR data were analyzed using QuantaSoft software. Data are represented as the fractional abundance of the edited CFTR allele. gDNA from in vitro or in vivo samples was amplified by PCR to detect both on-target and off-target editing. PCRs were performed using KAPA HiFi HotStart ReadyMix (Roche) according to the manufacturer’s instructions. PCR primers for the on-target region were 5′-TCTGCTCTCAATTTTCTTGGA-3′ (forward) and 5′-GGCAAGCTTTGACAACACTC-3′ (reverse).
Thermocycler conditions were as follows: 95°C 3 min, (98°C 20s, 55°C 15 s, 72°C 15 s) × 35 cycles, 72°C 10 min, hold at 4°C. To study off-target effects, we looked for alterations in regions of partial homology to both the PNA and the donor DNA. The 11 off-target sites were chosen by searching for sites in the C57BL/6 mouse genome with high (>94%) sequence homology to the PNA binding site (matching 18 bp of the 19-bp PNA binding site). Primers were designed to amplify each of these sites, and the PCR amplicons were submitted for deep sequencing (NGS). Primers and barcodes for off-targets are shown in table S2. PCR products were purified using the QIAquick PCR Purification Kit (Qiagen). PCR products were prepared for NGS by end repair and adapter ligation according to Illumina protocols, and samples were sequenced by Illumina NovaSeq 6000 with 100 or 150 paired-end reads at the Yale Center for Genome Analysis. This instrument has a Q30 score of >85%, indicating that at least 85% of the bases have an error rate of 0.1% or less. This value is an average across the whole read length, and the error rate increases toward the end of the reads. Sequencing data were mapped to corresponding reference sequences and analyzed using CRISPResso2. Potential PCR artifacts of the donor DNA were eliminated by using AMPure XP beads (Beckman Coulter) to purify gDNA according to the manufacturer’s instructions before PCR amplification in a subset of samples.
Statistical analysis
Results were analyzed using GraphPad Prism (version 9.3.1). Data are presented as individual data points or as means ± SEM. To compare in vitro Ussing readouts and ddPCR editing frequencies in ALI cultures, we used analysis of variance (ANOVA) with Tukey’s posttest for multiple comparisons. To compare in vitro and in vivo off-target effects analyzed by NGS and in vivo ddPCR editing frequencies among treatment conditions, we used the Mann-Whitney test on off-target sites and multiple organs. We also used the Mann-Whitney test to compare NPD, RPD, Ussing, and BAL readouts following in vivo treatment among treatment conditions. To compare pre- and posttreatment in vivo NPD and RPD values, we used the Wilcoxon matched-pairs signed-rank test. For all statistical tests, P < 0.05 was considered to be statistically significant.
Acknowledgments
We thank biostatisticians D. Zelterman and W. Wei for their assistance with statistical analyses and Trucode Gene Repair Inc. for providing a portion of the PNAs used in this study.
Funding: This work was supported by NIH grant UG3HL147352 (to P.M.G. and W.M.S.), NIH grant R01 HL125892 (to P.M.G., W.M.S., and M.E.E.), NIH grant T32GM86287 (to A.S.P.-D.), NIH grant F32HL142144 (to A.S.P.-D.), NIH grant K99HL151806 (to A.S.P.-D.), Cystic Fibrosis Foundation grant EGAN641558 (to P.M.G., W.M.S., and M.E.E.), Cystic Fibrosis Foundation grant PIOTRO20F0 (to A.S.P.-D.), and Cystic Fibrosis Foundation grant PIOTRO21F5 (to A.S.P.-D.).
Author contributions: Conceptualization: A.S.P.-D., C.B., P.M.G., W.M.S., and M.E.E. Methodology: A.S.P.-D., C.B., A.S.R., R.P., R.F., P.M.G., W.M.S., and M.E.E. Investigation: A.S.P.-D., C.B., C.-Y.L., Y.D., D.W., T.C.B., E.X., A.Ga., R.N., and A.Gu. Visualization: A.S.P.-D., Y.D., and M.E.E. Supervision: P.M.G., W.M.S., and M.E.E. Writing—original draft: A.S.P.-D. Writing—review and editing: A.S.P.-D., P.M.G., W.M.S., and M.E.E.
Competing interests: During the time of writing this manuscript, A.S.P.-D., A.S.R., P.M.G., W.M.S., and M.E.E. were consultants to Trucode Gene Repair Inc. P.M.G. and W.M.S. are inventors on a patent related to this work filed by Yale University (no. 11,136,597, filed on 16 February 2017, issued on 5 October 2021). A.S.P.-D., A.S.R., P.M.G., and W.M.S. are inventors on a pending patent related to this work filed by Yale University (no. 16/603,152, filed on 4 October 2019). R.F. is a scientific founder and an adviser for IsoPlexis, Singleron Biotechnologies, and AtlasXomics, none of which are directly related to this work but may be perceived as a potential conflict of interest. The interests of A.S.P.-D., A.S.R., R.F., P.M.G., W.M.S., and M.E.E. were reviewed and managed by Yale University Provost’s Office in accordance with the University’s conflict of interest policies. The authors declare no other competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S14
Tables S1 to S4
REFERENCES AND NOTES
- 1.Ley D., Turck D., Digestive outcomes in cystic fibrosis. Best Pract. Res. Clin. Gastroenterol. 56–57, 101788 (2022). [DOI] [PubMed] [Google Scholar]
- 2.Hodges C. A., Conlon R. A., Delivering on the promise of gene editing for cystic fibrosis. Genes Dis. 6, 97–108 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sabharwal S., Gastrointestinal manifestations of cystic fibrosis. Gastroenterol. Hepatol. 12, 43–47 (2016). [PMC free article] [PubMed] [Google Scholar]
- 4.Taylor-Cousar J. L., Munck A., McKone E. F., van der Ent C. K., Moeller A., Simard C., Wang L. T., Ingenito E. P., McKee C., Lu Y., Lekstrom-Himes J., Elborn J. S., Tezacaftor-ivacaftor in patients with cystic fibrosis homozygous for Phe508del. N. Engl. J. Med. 377, 2013–2023 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Wainwright C. E., Elborn J. S., Ramsey B. W., Marigowda G., Huang X., Cipolli M., Colombo C., Davies J. C., De Boeck K., Flume P. A., Konstan M. W., McColley S. A., McCoy K., McKone E. F., Munck A., Ratjen F., Rowe S. M., Waltz D., Boyle M. P., Lumacaftor-ivacaftor in patients with cystic fibrosis homozygous for Phe508del CFTR. N. Engl. J. Med. 373, 220–231 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davies J. C., Moskowitz S. M., Brown C., Horsley A., Mall M. A., McKone E. F., Plant B. J., Prais D., Ramsey B. W., Taylor-Cousar J. L., Tullis E., Uluer A., McKee C. M., Robertson S., Shilling R. A., Simard C., Van Goor F., Waltz D., Xuan F., Young T., Rowe S. M.; VX16-659-101 Study Group , VX-659-tezacaftor-ivacaftor in patients with cystic fibrosis and one or two Phe508del alleles. N. Engl. J. Med. 379, 1599–1611 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keating D., Marigowda G., Burr L., Daines C., Mall M. A., McKone E. F., Ramsey B. W., Rowe S. M., Sass L. A., Tullis E., McKee C. M., Moskowitz S. M., Robertson S., Savage J., Simard C., Van Goor F., Waltz D., Xuan F., Young T., Taylor-Cousar J. L., VX-445-tezacaftor-ivacaftor in patients with cystic fibrosis and one or two Phe508del alleles. N. Engl. J. Med. 379, 1612–1620 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Piotrowski-Daspit A. S., Glazer P. M., Saltzman W. M., Debugging the genetic code: Non-viral in vivo delivery of therapeutic genome editing technologies. Curr. Opin. Biomed. Eng. 7, 24–32 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu Y., Foden J. A., Khayter C., Maeder M. L., Reyon D., Joung J. K., Sander J. D., High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 31, 822–826 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pattanayak V., Lin S., Guilinger J. P., Ma E., Doudna J. A., Liu D. R., High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat. Biotechnol. 31, 839–843 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cameron P., Fuller C. K., Donohoue P. D., Jones B. N., Thompson M. S., Carter M. M., Gradia S., Vidal B., Garner E., Slorach E. M., Lau E., Banh L. M., Lied A. M., Edwards L. S., Settle A. H., Capurso D., Llaca V., Deschamps S., Cigan M., Young J. K., May A. P., Mapping the genomic landscape of CRISPR-Cas9 cleavage. Nat. Methods 14, 600–606 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Anzalone A. V., Randolph P. B., Davis J. R., Sousa A. A., Koblan L. W., Levy J. M., Chen P. J., Wilson C., Newby G. A., Raguram A., Liu D. R., Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rees H. A., Liu D. R., Base editing: Precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 19, 770–788 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang L., Wu J., Fang W., Liu G.-H., Belmonte J. C. I., Regenerative medicine: Targeted genome editing in vivo. Cell Res. 25, 271–272 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.P. E. Nielsen, Peptide Nucleic Acids: Protocols and Applications (Horizon Bioscience, 2004). [Google Scholar]
- 16.McNeer N. A., Anandalingam K., Fields R. J., Caputo C., Kopic S., Gupta A., Quijano E., Polikoff L., Kong Y., Bahal R., Geibel J. P., Glazer P. M., Saltzman W. M., Egan M. E., Nanoparticles that deliver triplex-forming peptide nucleic acid molecules correct F508del CFTR in airway epithelium. Nat. Commun. 6, 6952 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McNeer N. A., Chin J. Y., Schleifman E. B., Fields R. J., Glazer P. M., Saltzman W. M., Nanoparticles deliver triplex-forming PNAs for site-specific genomic recombination in CD34+ human hematopoietic progenitors. Mol. Ther. 19, 172–180 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McNeer N. A., Schleifman E. B., Glazer P. M., Saltzman W. M., Polymer delivery systems for site-specific genome editing. J. Control. Release 155, 312–316 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schleifman E. B., McNeer N. A., Jackson A., Tamtich J., Leif J., Schulz L. D., Greiner D. L., Kumar P., Saltzman W. M., Glazer P. M., Site-specific genome editing of PBMCs with PLGA nanoparticle-delivered PNAs confers HIV-1 resistance in humanized mice. Mol. Ther. Nucleic Acids 19, e135 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McNeer N. A., Schleifman E. B., Cuthbert A., Brehm M., Jackson A., Greinert D. L., Shultz L. D., Kumar P., Cheng C., Saltzman W. M., Glazer P. M., Systemic delivery of triplix-forming PNA and donor DNA by nanoparticles mediates site-specific genome editing of human hematopoietic cells in vivo. Gene Ther. 20, 658–669 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bahal R., Ali McNeer N., Quijano E., Liu Y., Sulkowski P., Turchick A., Lu Y.-C., Bhunia D. C., Manna A., Greiner D. L., Brehm M. A., Cheng C. J., López-Giráldez F., Ricciardi A., Beloor J., Krause D. S., Kumar P., Gallagher P. G., Braddock D. T., Mark Saltzman W., Ly D. H., Glazer P. M., In vivo correction of anaemia in β-thalassemic mice by γPNA-mediated gene editing with nanoparticle delivery. Nat. Commun. 7, 13304 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ricciardi A. S., Bahal R., Farrelly J. S., Quijano E., Bianchi A. H., Luks V. L., Putman R., López-Giráldez F., Coşkun S., Song E., Liu Y., Hsieh W.-C., Ly D. H., Stitelman D. H., Glazer P. M., Saltzman W. M., In utero nanoparticle delivery for site-specific genome editing. Nat. Commun. 9, 2481 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koehler D. R., Hitt M. M., Hu J., Challenges and strategies for cystic fibrosis lung gene therapy. Mol. Ther. 4, 84–91 (2001). [DOI] [PubMed] [Google Scholar]
- 24.Steines B., Dickey D. D., Bergen J., Excoffon K. J. D. A., Weinstein J. R., Li X., Yan Z., Alaiwa M. H. A., Shah V. S., Bouzek D. C., Powers L. S., Gansemer N. D., Ostedgaard L. S., Engelhardt J. F., Stoltz D. A., Welsh M. J., Sinn P. L., Schaffer D. V., Zabner J., CFTR gene transfer with AAV improves early cystic fibrosis pig phenotypes. JCI Insight 1, e88728 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cao H., Ouyang H., Grasemann H., Bartlett C., Du K., Duan R., Shi F., Estrada M., Seigel K. E., Coates A. L., Yeger H., Bear C. E., Gonska T., Moraes T. J., Hu J., Transducing airway basal cells with a helper-dependent adenoviral vector for lung gene therapy. Hum. Gene Ther. 29, 643–652 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Cooney A. L., Singh B. K., Loza L. M., Thornell I. M., Hippee C. E., Powers L. S., Ostedgaard L. S., Meyerholz D. K., Wohlford-Lenane C., Stoltz D. A., McCray P. B. Jr., Sinn P. L., Widespread airway distribution and short-term phenotypic correction of cystic fibrosis pigs following aerosol delivery of piggyBac/adenovirus. Nucleic Acids Res. 46, 9591–9600 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krishnamurthy S., Wohlford-Lenane C., Kandimalla S., Sartre G., Meyerholz D. K., Théberge V., Hallée S., Duperré A.-M., Del’Guidice T., Lepetit-Stoffaes J.-P., Barbeau X., Guay D., McCray P. B., Engineered amphiphilic peptides enable delivery of proteins and CRISPR-associated nucleases to airway epithelia. Nat. Commun. 10, 4906 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou Z. P., Yang L. L., Cao H., Chen Z. R., Zhang Y., Wen X.-Y., Hu J., In vitro validation of a CRISPR-Mediated CFTR correction strategy for preclinical translation in pigs. Hum. Gene Ther. 30, 1101–1116 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Bahal R., Sahu B., Rapireddy S., Lee C. M., Ly D. H., Sequence-unrestricted, Watson-Crick recognition of double helical B-DNA by (R)-MiniPEG-γPNAs. Chembiochem 13, 56–60 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.H. Jamali, A. Gupta, C. Barone, A. Riccardi, E. Quijano, A. Piotrowski-Daspit, M. Saltzman, P. Glazer, M. Egan, in Pediatric Pulmonology (Wiley, 2018), vol. 53, p. 151. [Google Scholar]
- 31.Mandl H. K., Quijano E., Suh H. W., Sparago E., Oeck S., Grun M., Glazer P. M., Saltzman W. M., Optimizing biodegradable nanoparticle size for tissue-specific delivery. J. Control. Release 314, 92–101 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bracaglia L. G., Piotrowski-Daspit A. S., Lin C. Y., Moscato Z. M., Wang Y., Tietjen G. T., Saltzman W. M., High-throughput quantitative microscopy-based half-life measurements of intravenously injected agents. Proc. Natl. Acad. Sci. U.S.A. 117, 3502–3508 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rock J. R., Randell S. H., Hogan B. L., Airway basal stem cells: A perspective on their roles in epithelial homeostasis and remodeling. Dis. Model. Mech. 3, 545–556 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qaddoumi M. G., Ueda H., Yang J., Davda J., Labhasetwar V., Lee V. H., The characteristics and mechanisms of uptake of PLGA nanoparticles in rabbit conjunctival epithelial cell layers. Pharm. Res. 21, 641–648 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Cartiera M. S., Johnson K. M., Rajendran V., Caplan M. J., Saltzman W. M., The uptake and intracellular fate of PLGA nanoparticles in epithelial cells. Biomaterials 30, 2790–2798 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gentzsch M., Boyles S. E., Cheluvaraju C., Chaudhry I. G., Quinney N. L., Cho C., Dang H., Liu X., Schlegel R., Randell S. H., Pharmacological rescue of conditionally reprogrammed cystic fibrosis bronchial epithelial cells. Am. J. Respir. Cell Mol. Biol. 56, 568–574 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H., Sheppard D. N., Hug M. J., Transepithelial electrical measurements with the Ussing chamber. J. Cyst. Fibros. 3, 123–126 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Plasschaert L. W., Žilionis R., Choo-Wing R., Savova V., Knehr J., Roma G., Klein A. M., Jaffe A. B., A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature 560, 377–381 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Montoro D. T., Haber A. L., Biton M., Vinarsky V., Lin B., Birket S. E., Yuan F., Chen S., Leung H. M., Villoria J., Rogel N., Burgin G., Tsankov A. M., Waghray A., Slyper M., Waldman J., Nguyen L., Dionne D., Rozenblatt-Rosen O., Tata P. R., Mou H., Shivaraju M., Bihler H., Mense M., Tearney G. J., Rowe S. M., Engelhardt J. F., Regev A., Rajagopal J., A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature 560, 319–324 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strunz M., Simon L. M., Ansari M., Kathiriya J. J., Angelidis I., Mayr C. H., Tsidiridis G., Lange M., Mattner L. F., Yee M., Ogar P., Sengupta A., Kukhtevich I., Schneider R., Zhao Z., Voss C., Stoeger T., Neumann J. H. L., Hilgendorff A., Behr J., O’Reilly M., Lehmann M., Burgstaller G., Königshoff M., Chapman H. A., Theis F. J., Schiller H. B., Alveolar regeneration through a Krt8+ transitional stem cell state that persists in human lung fibrosis. Nat. Commun. 11, 3559 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kathiriya J. J., Wang C., Zhou M., Brumwell A., Cassandras M., Le Saux C. J., Cohen M., Alysandratos K.-D., Wang B., Wolters P., Matthay M., Kotton D. N., Chapman H. A., Peng T., Human alveolar type 2 epithelium transdifferentiates into metaplastic KRT5+ basal cells. Nat. Cell Biol. 24, 10–23 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.L. McInnes, J. Healy, J. Melville, UMAP: Uniform manifold approximation and projection for dimension reduction. arXiv:1802.03426 [stat.ML] (9 February 2018).
- 43.Weinreb C., Wolock S., Klein A. M., SPRING: A kinetic interface for visualizing high dimensional single-cell expression data. Bioinformatics 34, 1246–1248 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weiner S. A., Caputo C., Bruscia E., Ferreira E. C., Price J. E., Krause D. S., Egan M. E., Rectal potential difference and the functional expression of CFTR in the gastrointestinal epithelia in cystic fibrosis mouse models. Pediatr. Res. 63, 73–78 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Cartiera M. S., Ferreira E. C., Caputo C., Egan M. E., Caplan M. J., Saltzman W. M., Partial correction of cystic fibrosis defects with PLGA nanoparticles encapsulating curcumin. Mol. Pharm. 7, 86–93 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nelson C. E., Wu Y., Gemberling M. P., Oliver M. L., Waller M. A., Bohning J. D., Robinson-Hamm J. N., Bulaklak K., Rivera R. M. C., Collier J. H., Asokan A., Gersbach C. A., Long-term evaluation of AAV-CRISPR genome editing for Duchenne muscular dystrophy. Nat. Med. 25, 427–432 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mehravar M., Shirazi A., Nazari M., Banan M., Mosaicism in CRISPR/Cas9-mediated genome editing. Dev. Biol. 445, 156–162 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Bruscia E. M., Zhang P.-X., Ferreira E., Caputo C., Emerson J. W., Tuck D., Krause D. S., Egan M. E., Macrophages directly contribute to the exaggerated inflammatory response in cystic fibrosis transmembrane conductance regulator−/− mice. Am. J. Respir. Cell Mol. Biol. 40, 295–304 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsai S. Q., Zheng Z., Nguyen N. T., Liebers M., Topkar V. V., Thapar V., Wyvekens N., Khayter C., Iafrate A. J., Le L. P., Aryee M. J., Joung J. K., GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 33, 187–197 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim D., Bae S., Park J., Kim E., Kim S., Yu H. R., Hwang J., Kim J.-I., Kim J.-S., Digenome-seq: Genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat. Methods 12, 237–243 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Lazzarotto C. R., Nguyen N. T., Tang X., Malagon-Lopez J., Guo J. A., Aryee M. J., Joung J. K., Tsai S. Q., Defining CRISPR-Cas9 genome-wide nuclease activities with CIRCLE-seq. Nat. Protoc. 13, 2615–2642 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wienert B., Wyman S. K., Richardson C. D., Yeh C. D., Akcakaya P., Porritt M. J., Morlock M., Vu J. T., Kazane K. R., Watry H. L., Judge L. M., Conklin B. R., Maresca M., Corn J. E., Unbiased detection of CRISPR off-targets in vivo using DISCOVER-Seq. Science 364, 286–289 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saha K., Sontheimer E. J., Brooks P. J., Dwinell M. R., Gersbach C. A., Liu D. R., Murray S. A., Tsai S. Q., Wilson R. C., Anderson D. G., Asokan A., Banfield J. F., Bankiewicz K. S., Bao G., Bulte J. W. M., Bursac N., Campbell J. M., Carlson D. F., Chaikof E. L., Chen Z.-Y., Cheng R. H., Clark K. J., Curiel D. T., Dahlman J. E., Deverman B. E., Dickinson M. E., Doudna J. A., Ekker S. C., Emborg M. E., Feng G., Freedman B. S., Gamm D. M., Gao G., Ghiran I. C., Glazer P. M., Gong S., Heaney J. D., Hennebold J. D., Hinson J. T., Khvorova A., Kiani S., Lagor W. R., Lam K. S., Leong K. W., Levine J. E., Lewis J. A., Lutz C. M., Ly D. H., Maragh S., McCray P. B., McDevitt T. C., Mirochnitchenko O., Morizane R., Murthy N., Prather R. S., Ronald J. A., Roy S., Roy S., Sabbisetti V., Saltzman W. M., Santangelo P. J., Segal D. J., Shimoyama M., Skala M. C., Tarantal A. F., Tilton J. C., Truskey G. A., Vandsburger M., Watts J. K., Wells K. D., Wolfe S. A., Xu Q., Xue W., Yi G., Zhou J.; SCGE Consortium , The NIH somatic cell genome editing program. Nature 592, 195–204 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wei T., Cheng Q., Min Y.-L., Olson E. N., Siegwart D. J., Systemic nanoparticle delivery of CRISPR-Cas9 ribonucleoproteins for effective tissue specific genome editing. Nat. Commun. 11, 3232 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Finn J. D., Smith A. R., Patel M. C., Shaw L., Youniss M. R., van Heteren J., Dirstine T., Ciullo C., Lescarbeau R., Seitzer J., Shah R. R., Shah A., Ling D., Growe J., Pink M., Rohde E., Wood K. M., Salomon W. E., Harrington W. F., Dombrowski C., Strapps W. R., Chang Y., Morrissey D. V., A single administration of CRISPR/Cas9 lipid nanoparticles achieves robust and persistent in vivo genome editing. Cell Rep. 22, 2227–2235 (2018). [DOI] [PubMed] [Google Scholar]
- 56.Gillmore J. D., Gane E., Taubel J., Kao J., Fontana M., Maitland M. L., Seitzer J., O’Connell D., Walsh K. R., Wood K., Phillips J., Xu Y., Amaral A., Boyd A. P., Cehelsky J. E., McKee M. D., Schiermeier A., Harari O., Murphy A., Kyratsous C. A., Zambrowicz B., Soltys R., Gutstein D. E., Leonard J., Sepp-Lorenzino L., Lebwohl D., CRISPR-Cas9 in vivo gene editing for transthyretin amyloidosis. N. Engl. J. Med. 385, 493–502 (2021). [DOI] [PubMed] [Google Scholar]
- 57.Rosenblum D., Gutkin A., Kedmi R., Ramishetti S., Veiga N., Jacobi A. M., Schubert M. S., Friedmann-Morvinski D., Cohen Z. R., Behlke M. A., Lieberman J., Peer D., CRISPR-Cas9 genome editing using targeted lipid nanoparticles for cancer therapy. Sci. Adv. 6, eabc9450 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lopes-Pacheco M., CFTR modulators: The changing face of cystic fibrosis in the era of precision medicine. Front. Pharmacol. 10, 1662 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Johnson L. G., Olsen J. C., Sarkadi B., Moore K. L., Swanstrom R., Boucher R. C., Efficiency of gene transfer for restoration of normal airway epithelial function in cystic fibrosis. Nat. Genet. 2, 21–25 (1992). [DOI] [PubMed] [Google Scholar]
- 60.Goldman M. J., Yang Y., Wilson J. M., Gene therapy in a xenograft model of cystic fibrosis lung corrects chloride transport more effectively than the sodium defect. Nat. Genet. 9, 126–131 (1995). [DOI] [PubMed] [Google Scholar]
- 61.Farmen S. L., Karp P. H., Ng P., Palmer D. J., Koehler D. R., Hu J., Beaudet A. L., Zabner J., Welsh M. J., Gene transfer of CFTR to airway epithelia: Low levels of expression are sufficient to correct Cl- transport and overexpression can generate basolateral CFTR. Am. J. Physiol. Lung Cell. Mol. Physiol. 289, L1123–L1130 (2005). [DOI] [PubMed] [Google Scholar]
- 62.Zhang L., Button B., Gabriel S. E., Burkett S., Yan Y., Skiadopoulos M. H., Dang Y. L., Vogel L. N., McKay T., Mengos A., Boucher R. C., Collins P. L., Pickles R. J., CFTR delivery to 25% of surface epithelial cells restores normal rates of mucus transport to human cystic fibrosis airway epithelium. PLOS Biol. 7, e1000155 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dannhoffer L., Blouquit-Laye S., Regnier A., Chinet T., Functional properties of mixed cystic fibrosis and normal bronchial epithelial cell cultures. Am. J. Respir. Cell Mol. Biol. 40, 717–723 (2009). [DOI] [PubMed] [Google Scholar]
- 64.Piotrowski-Daspit A. S., Kauffman A. C., Bracaglia L. G., Saltzman W. M., Polymeric vehicles for nucleic acid delivery. Adv. Drug Deliv. Rev. 156, 119–132 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McCall R. L., Sirianni R. W., PLGA nanoparticles formed by single- or double-emulsion with vitamin E-TPGS. J. Vis. Exp. 51015 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.M. L. Fulcher, S. H. Randell, Human nasal and tracheo-bronchial respiratory epithelial cell culture, in Epithelial Cell Culture Protocols: Second Edition, S. H. Randell, M. L. Fulcher, Eds. (Humana Press, 2013), pp. 109–121. [DOI] [PubMed] [Google Scholar]
- 67.Liu X., Ory V., Chapman S., Yuan H., Albanese C., Kallakury B., Timofeeva O. A., Nealon C., Dakic A., Simic V., Haddad B. R., Rhim J. S., Dritschilo A., Riegel A., McBride A., Schlegel R., ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. Am. J. Pathol. 180, 599–607 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Suprynowicz F. A., Upadhyay G., Krawczyk E., Kramer S. C., Hebert J. D., Liu X., Yuan H., Cheluvaraju C., Clapp P. W., Boucher R. C. Jr., Kamonjoh C. M., Randell S. H., Schlegel R., Conditionally reprogrammed cells represent a stem-like state of adult epithelial cells. Proc. Natl. Acad. Sci. U.S.A. 109, 20035–20040 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jiang D., Schaefer N., Chu H. W., Air-liquid interface culture of human and mouse airway epithelial cells. Methods Mol. Biol. 1809, 91–109 (2018). [DOI] [PubMed] [Google Scholar]
- 70.Bruscia E. M., Price J. E., Cheng E. C., Weiner S., Caputo C., Ferreira E. C., Egan M. E., Krause D. S., Assessment of cystic fibrosis transmembrane conductance regulator (CFTR) activity in CFTR-null mice after bone marrow transplantation. Proc. Natl. Acad. Sci. U.S.A. 103, 2965–2970 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dura B., Choi J.-Y., Zhang K., Damsky W., Thakral D., Bosenberg M., Craft J., Fan R., scFTD-seq: Freeze-thaw lysis based, portable approach toward highly distributed single-cell 3′ mRNA profiling. Nucleic Acids Res. 47, e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R., Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W. M. III, Hao Y., Stoeckius M., Smibert P., Satija R., Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Abdelaal T., Michielsen L., Cats D., Hoogduin D., Mei H., Reinders M. J., Mahfouz A., A comparison of automatic cell identification methods for single-cell RNA sequencing data. Genome Biol. 20, 194 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Egan M. E., Pearson M., Weiner S. A., Rajendran V., Rubin D., Glockner-Pagel J., Canny S., Du K., Lukacs G. L., Caplan M. J., Curcumin, a major constituent of turmeric, corrects cystic fibrosis defects. Science 304, 600–602 (2004). [DOI] [PubMed] [Google Scholar]
- 76.B. R. Grubb, Bioelectric measurement of CFTR function in mice, in Cystic Fibrosis Methods and Protocols, W. R. Skach, Ed. (Humana Press, 2002), pp. 525–535. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S14
Tables S1 to S4