Abstract
Low plasma iron (hypoferremia) induced by hepcidin is a conserved inflammatory response that protects against infections but inhibits erythropoiesis. How hypoferremia influences leukocytogenesis is unclear. Using proteomic data, we predicted that neutrophil production would be profoundly more iron-demanding than generation of other white blood cell types. Accordingly in mice, hepcidin-mediated hypoferremia substantially reduced numbers of granulocytes but not monocytes, lymphocytes, or dendritic cells. Neutrophil rebound after anti-Gr-1–induced neutropenia was blunted during hypoferremia but was rescued by supplemental iron. Similarly, hypoferremia markedly inhibited pharmacologically stimulated granulopoiesis mediated by granulocyte colony-stimulating factor and inflammation-induced accumulation of neutrophils in the spleen and peritoneal cavity. Furthermore, hypoferremia specifically altered neutrophil effector functions, suppressing antibacterial mechanisms but enhancing mitochondrial reactive oxygen species–dependent NETosis associated with chronic inflammation. Notably, antagonizing endogenous hepcidin during acute inflammation enhanced production of neutrophils. We propose plasma iron modulates the profile of innate immunity by controlling monocyte-to-neutrophil ratio and neutrophil activity in a therapeutically targetable system.
Low plasma iron controls cellular innate immunity by limiting neutrophil production and altering function including NETosis.
INTRODUCTION
Iron is required for cellular biochemistry, supporting processes such as oxidative metabolism, DNA synthesis, and epigenetic remodeling (1, 2). Sequestration of iron from plasma, termed hypoferremia, commonly occurs during the acute phase of infection, driven by inflammatory induction of the iron regulatory hormone, hepcidin (3). Hepcidin blocks iron recycling by hemophagocytic macrophages and iron absorption by duodenal enterocytes, suppressing serum iron concentrations (4). This response can protect against certain siderophilic bacterial pathogens, constituting an acute “nutritional immune” mechanism but, if prolonged, leads to inflammatory anemia and can inhibit proliferative lymphocyte responses to immunization and infection (5–7).
Of particular note, inflammatory hypoferremia during acute infection frequently coincides with a need to remodel hematopoiesis (8) and to support metabolically demanding innate cell effector functions—both potentially iron-dependent processes. While studies have highlighted that the neutrophil oxidative burst is impaired by iron deficiency, how cellular innate immunity is affected by plasma iron status and extracellular iron availability more broadly remains unclear (9–16). Here, we show, via pharmacological manipulation of plasma iron availability in mice, that neutrophil production and functionality are highly iron-sensitive processes, thereby providing insights into how physiologically relevant systemic shifts in nutrient availability modulate innate immunity.
RESULTS
Neutrophil production is iron-requiring and sensitive to low iron availability
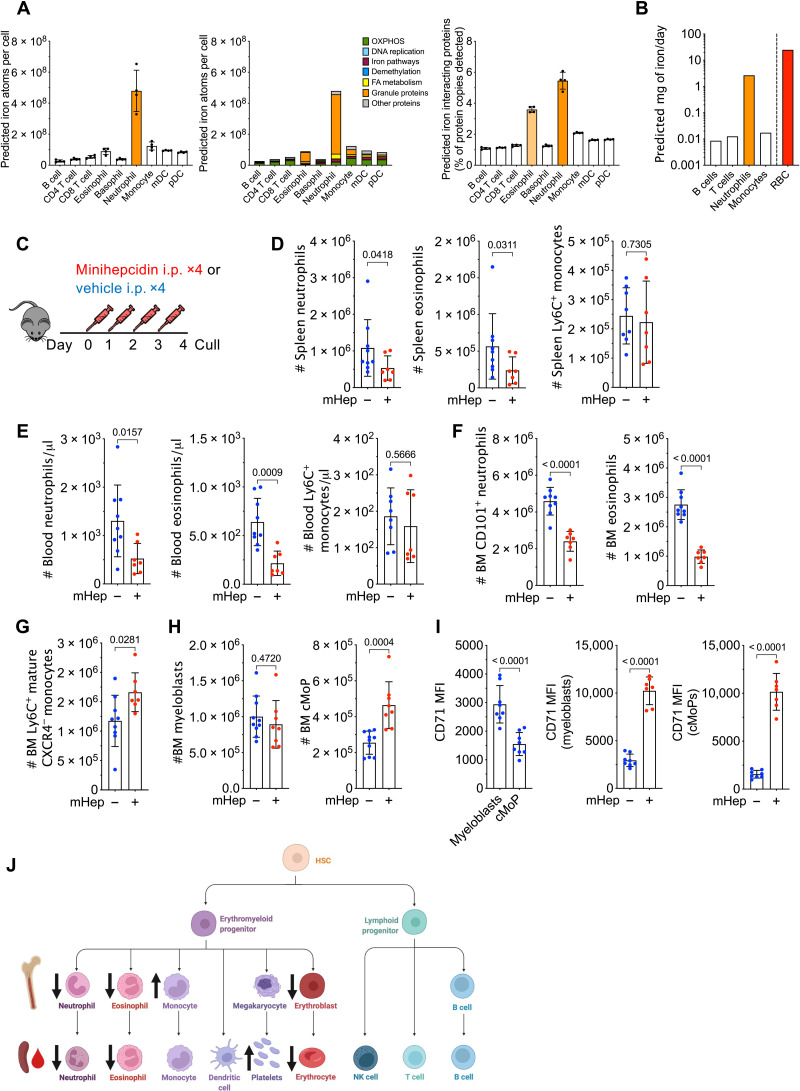
Erythropoiesis consumes ~25 mg of iron per day in humans (17). However, the iron needs of leukocytes are unknown. Having previously modeled iron demands during T cell activation (2), we applied the same method to a published proteomic resource (18) to estimate the iron content of resting human peripheral blood leukocytes. We predict that neutrophils have substantially higher iron content per cell than other leukocytes (Fig. 1A). Moreover, a larger proportion of the neutrophil and eosinophil proteomes is “iron-interacting” compared to other leukocytes (Fig. 1A). Using estimates for the number of leukocytes generated per day (19), we predict neutrophil production uses ~100-fold more iron than B cells, T cells, or monocytes and only 10-fold less iron than estimated for erythropoiesis (Fig. 1B) (17). Thus, neutrophil production is likely to be particularly sensitive to changes in serum iron.
Fig. 1. Hepcidin reduces systemic neutrophil numbers.
(A) Predicted absolute number of iron atoms in total and grouped by pathway, as well as percentage of detected proteins that are iron-interacting proteins in resting human peripheral blood leukocytes. Means ± SD. mDC, myeloid dendritic cell; pDC, plasmacytoid dendritic cell; FA, fatty acid. (B) Predicted iron requirements (in milligrams per day) for hematopoietic production of the specified lineage, compared to literature-derived value for red blood cell (RBC) production. (C) Experiment scheme to test effect of mHep on hematopoiesis. i.p., intraperitoneally. (D) Number of splenic myeloid subsets in experiment described in (C). Representative of three independent replicates. Neutrophils and eosinophils, Mann-Whitney test; monocytes, unpaired t test. Means ± SD. (E) Blood myeloid subsets (numbers per microliter) in experiment described in (C). Representative of three independent replicates. Neutrophils, Welch’s unpaired t test; eosinophils and monocytes, unpaired t test. Means ± SD. (F) Numbers of mature neutrophils and eosinophils in the bone marrow (BM) in the experiment described in (C). Representative of three independent replicates. Unpaired t test. Means ± SD. (G) Numbers of mature monocytes in the BM in experiment described in (C). Representative to three independent replicates. Unpaired t test. Means ± SD. (H) Numbers of committed granulocyte progenitors and committed monocyte progenitors (cMoPs) in experiment described in (C). Representative of three independent replicates. Unpaired t test. Means ± SD. (I) Comparison of CD71 expression between myeloblasts (granulocyte progenitors) and cMoPs in experiment described in (C). Paired t test. Effect of mHep treatment on CD71 expression by myeloblasts and cMoPs. Unpaired t test. Means ± SD. MFI, median fluorescence intensity. (J) Graphical summary of changes in cell numbers observed after mHep treatment. NK, natural killer.
To evaluate the sensitivity of leukocyte subsets to physiological variation in plasma iron availability experimentally, we administered a mimetic of the iron regulatory hormone hepcidin—minihepcidin (mHep)—daily for 4 days to C57BL/6 mice (Fig. 1C) (6). This caused hypoferremia and suppressed endogenous hepatic hepcidin expression (fig. S1A) (20). Mice given mHep had fewer neutrophils and eosinophils, but unchanged Ly6C+ and Ly6C− monocyte frequency, in the spleen and blood (Fig. 1, D and E, and fig. S1D). Splenic basophils, dendritic cells and lymphocyte subsets (fig. S1, B and C), and bone marrow (BM) B cells (fig. S1C) were also unaffected. In addition, fewer mature neutrophils [CD101+ Ly6G+ (21)] and Siglec F+ eosinophils were present in the BM after mHep treatment (Fig. 1F); in contrast, mature BM monocytes [CXCR4− CD115+ (22)] were increased in treated mice (Fig. 1G). BM neutrophil and monocyte production are driven by highly proliferative committed progenitors: Ly6C+ c-kit+ CD115− CXCR4+ granulocytic myeloblasts and Ly6C+ c-kit+ CD115+ CXCR4+ committed monocyte progenitors (cMoPs), respectively (21, 23). Following mHep treatment, we observed no change in myeloblasts but increased cMoP numbers (Fig. 1H) that could be driving the accumulation of mature BM monocytes.
To probe the differential sensitivity of neutrophil and monocyte lineages to low iron, we quantified CD71 expression (transferrin receptor, which mediates cellular iron acquisition) on committed progenitors. Myeloblasts had significantly higher CD71 expression than cMoPs in control mice, consistent with our in silico prediction of greater neutrophil iron demand (Fig. 1I). Moreover, mHep treatment markedly increased CD71 expression by both progenitor subsets, likely indicating cellular iron deficiency (Fig. 1I). A slight, but significant, increase in the proportion of myeloblasts in S phase was observed in mHep-treated animals, potentially indicating an iron-associated cell cycle perturbation that could link to reduced BM neutrophil production (fig. S1E) (24). No difference in cMoP cell cycle profile was observed (fig. S1E). mHep also suppressed erythropoiesis and increased circulating platelet counts (fig. S1, F and G) as previously described under low iron conditions (25, 26).
Alongside a lineage-intrinsic dependence of granulopoiesis on iron, low iron could also indirectly influence granulocyte production. Granulocyte colony-stimulating factor (G-CSF) and interleukin-5 (IL-5) are key cell-extrinsic hematopoietic cytokines controlling neutrophil and eosinophil production, respectively (27, 28). We found no evidence of reduced serum G-CSF or IL-5 mHep-treated mice (fig. S1H). G-CSF was elevated in hypoferremic animals, consistent with the reduced peripheral neutrophil frequency (29).
Together, these data indicate that mHep-induced hypoferremia associates with altered hematopoietic output, particularly reduced circulating frequencies of neutrophils and eosinophils (Fig. 1J), in line with the predicted lineage-specific iron demands. In contrast, we observed an expansion of monocytes and their precursors in the BM.
Hepcidin-driven hypoferremia impairs enhanced granulopoiesis
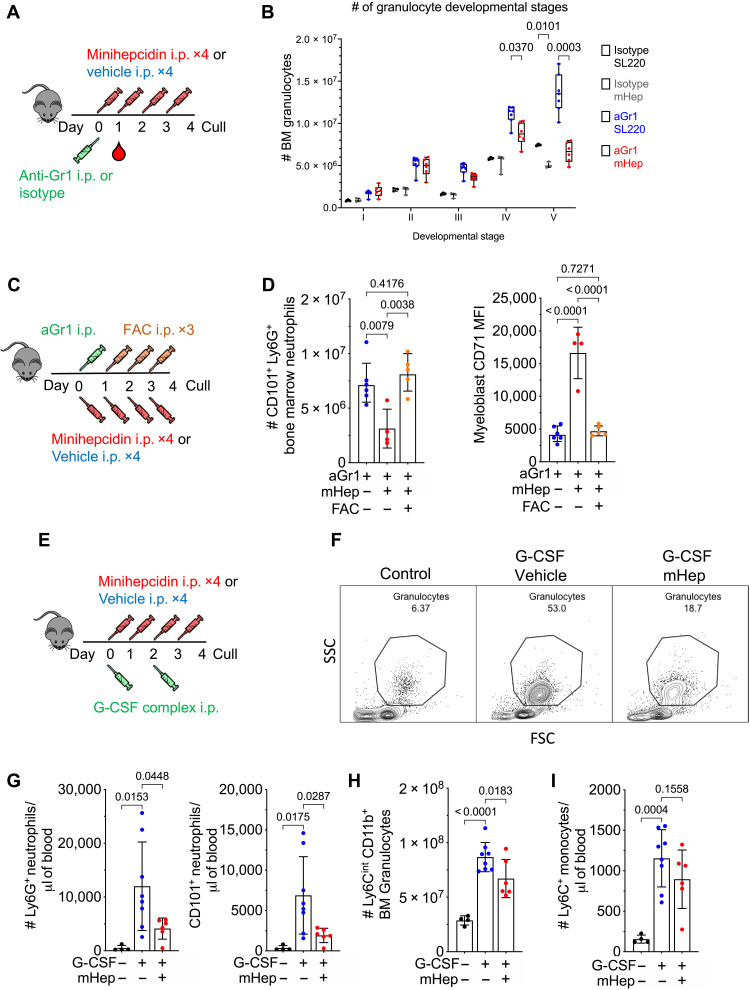
We next considered whether acute hypoferremia would affect the response to enhanced granulopoiesis. We administered low dose anti–Gr-1 to mice to transiently and selectively deplete mature neutrophil numbers indicated by peripheral blood cell frequencies 24 hours after antibody treatment (fig. S2A), followed by four daily mHep doses to induce hypoferremia during granulopoietic recovery (Fig. 2A). Differentiation from committed granulocyte progenitor to mature neutrophil can be defined by decreased c-kit and increased Ly6G expression (fig. S2B) (30). After 4 days of mHep administration, mature Ly6G-high BM neutrophil production was suppressed, while more immature cells accumulated (Fig. 2B and fig. S2C); blood neutrophil counts were also reduced (fig. S2D).
Fig. 2. Control of enhanced myelopoiesis by hepcidin-driven iron deficiency.
(A) Experimental scheme for effect of mHep treatment on recovery after neutrophil depletion. (B) BM granulocyte differentiation trajectory in mice treated as in (A), representative to two independent experiments. Two-way analysis of variance (ANOVA). Mean, quartiles, and range. (C) Experimental scheme for studying the effect of mHep-induced hypoferremia and its amelioration with FAC on recovery from aGr1-mediated neutrophil depletion. All mice received anti–Gr-1. (D) Number of mature BM CD101+ neutrophils and myeloblast CD71 expression from mice treated as in (C). One-way ANOVA. Means ± SD. (E) Experimental scheme for studying the effect of mHep on G-CSF complex driven granulocyte expansion. (F) Example flow cytometry scatter plot indicating frequency of neutrophils in the blood from experiment outlined in (E). SSC, side scatter; FSC; forwrd scatter. (G) Number of blood neutrophils in the blood from experiment outlined in (E). One-way ANOVA. Means ± SD. (H) Total BM granulocytes and CXCR4+ c-kit+ granulocyte precursors in the BM from experiment outlined in (E). One-way ANOVA. Means ± SD. (I) Number of blood monocytes in the blood from experiment outlined in (E). One-way ANOVA. Means ± SD.
To test whether mHep-mediated suppression of neutrophil production was directly related to iron limitation, mice were administered ferric ammonium citrate (FAC; an exogenous iron source) concurrently with mHep following anti–Gr-1 treatment (Fig. 2C). FAC rescued CD101+ Ly6G+ BM neutrophil numbers in mHep-treated mice and reduced myeloblast CD71 expression, suggesting that cellular iron deficiency was ameliorated (Fig. 2D); FAC also rescued splenic eosinophil numbers (fig. S2E). Therefore, suppression of granulopoiesis by mHep is iron dependent.
During inflammation, cytokines such as G-CSF drive enhanced neutrophil production, termed emergency granulopoiesis (31). G-CSF is also used clinically to mobilize neutrophils after chemotherapy. We injected mice with G-CSF/anti–G-CSF complex [more potent than G-CSF alone (32)] to stimulate granulopoiesis, with or without mHep as above (Fig. 2E). mHep-treated mice were hypoferremic, while mice treated with G-CSF complex alone had elevated serum iron, likely because of granulopoiesis displacing more iron-requiring erythropoiesis (fig. S2, F and G). The expansion of blood and splenic Ly6G+ and mature CD101+ Ly6G+ neutrophils in G-CSF complex–treated mice was suppressed by mHep (Fig. 2, F and G, and fig. S2H). Although total circulating and BM granulocytes were reduced by mHep treatment (Fig. 2H and fig. S2I), a greater proportion of blood granulocytes had an immature CXCR4+ phenotype, and the number of CXCR4+ c-kit+ BM progenitors was elevated (fig. S2, J and K). mHep treatment also led to a modest, but significant, accumulation of CXCR4+ c-kit+ BM granulocyte progenitors in S phase, which may contribute to reduced production (fig. S2L). In contrast, we found that mHep treatment did not prevent the G-CSF complex–driven increase in monocytes in the blood, spleen, and BM (Fig. 2I and fig. S2M). Thus, mHep-mediated granulopoietic suppression is still observed even in the presence of excess exogenous G-CSF complex, further supporting the hypothesis that the suppressive effect is not due to inhibition of inductive signals.
Functional impairment of neutrophils by plasma iron deficiency
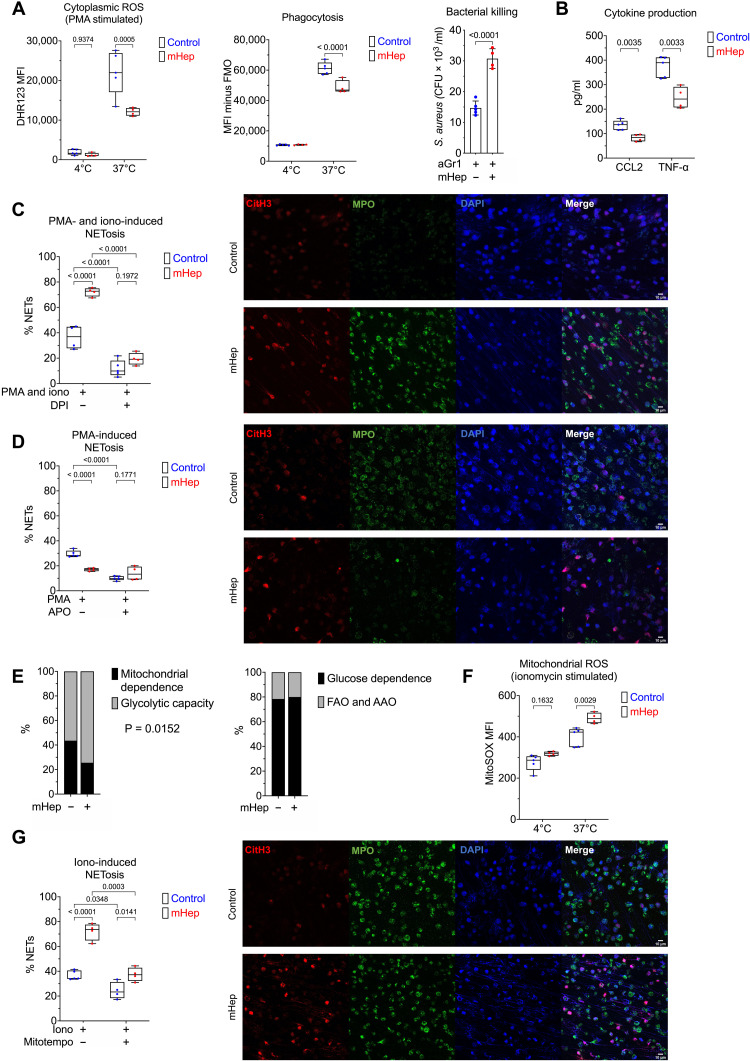
To examine whether in vivo iron restriction influenced neutrophil effector functions, we characterized isolated BM neutrophils (fig. S3A) following anti–Gr-1 depletion/recovery with and without mHep, as above (fig. S2A). Isolated neutrophils from hypoferremic mice displayed blunted phorbol 12-myristate 13-acetate (PMA)–induced reactive oxygen species (ROS) production, impaired phagocytosis of fluorescently labeled Escherichia coli and reduced Staphylococcus aureus killing (Fig. 3A). Production of C-C motif chemokine ligand 2 (CCL2) and tumor necrosis factor–α (TNF-α) upon ex vivo stimulation with zymosan was also suppressed (Fig. 3B).
Fig. 3. Functional alteration of neutrophils in serum iron deficiency.
(A) Phenotyping ex vivo of isolated BM neutrophils (obtained from mice treated as in Fig. 2A). Dihydrorhodamine 123 (DHR123) fluorescence as a reporter for cytoplasmic ROS in PMA-stimulated cells. Phagocytosis of GFP-labeled E. coli. FMO, fluorescence minus one. Two-way ANOVA. Mean, quartiles, and range. Bacterial killing of S. aureus, resolved as colony-forming units of S. aureus recovered after coculture with neutrophils. Unpaired t test. Means ± SD. (B) Supernatant CCL2 and TNF levels produced by zymosan-stimulated neutrophils measured by enzyme-linked immunosorbent assay (ELISA). Unpaired t test. Mean, quartiles, and range. (C) NETosis was evaluated in isolated BM neutrophils after stimulation ex vivo with PMA and ionomycin ± DPI. A minimum of 250 cells from two replicates per sample counted. Two-way ANOVA. Mean, quartiles, and range. Representative microscopy images taken at ×20 magnification. (D) NETosis was evaluated in isolated BM neutrophils after stimulation ex vivo with PMA ± apocynin (APO). A minimum of 250 cells from two replicates per sample counted. Two-way ANOVA. Mean, quartiles, and range. Representative microscopy images taken at ×20 magnification. (E) SCENITH (single-cell metabolism by profiling translation inhibition) analysis of Ly6G+ BM neutrophils from experiment described in Fig. 1A, analyzing proportional shifts in O-propargyl-puromycin (OPP) incorporation after ex vivo treatment with metabolic inhibitors as detailed in Materials and Methods. Two-way ANOVA with matching for sample. Reported P value is the effect of mHep treatment. Mean. FAO, fatty acid oxidation; AAO,amino acid oxidation. (F) MitoSOX fluorescence as a reporter for mitoROS in ionomycin stimulated isolated BM neutrophils. Two-way ANOVA. Mean, quartiles, and range. (G) NETosis was evaluated in isolated BM neutrophils after stimulation ex vivo with ionomycin (iono) ± mitoTEMPO. A minimum of 250 cells from two replicates per sample counted. Two-way ANOVA. Mean, quartiles, and range. Representative microscopy images taken at ×20 magnification.
We then evaluated NETosis, an important neutrophil effector function playing a central role in both host defense and immunopathology (33). NETosis was absent in unstimulated neutrophils (fig. S3B), but, unexpectedly, given the reduced ROS production of PMA-stimulated iron restricted neutrophils (Fig. 3A), neutrophils from hypoferremic mice showed a heightened propensity to undergo NETosis in response to PMA-ionomycin stimulation (Fig. 3C). We confirmed the ROS dependence of NETosis in our hands using combined cytoplasmic and mitochondrial ROS (mitoROS) inhibitor diphenyleneiodonium chloride (DPI; Fig. 3C and fig. S3C) (34). Attempting to reconcile these NETosis measurements with our observations of the reduced ROS production of PMA-stimulated iron restricted neutrophils (Fig. 3A), we measured NETosis after PMA stimulation alone. Consistent with our PMA-induced ROS measurements and the known catalytic role of iron in reduced form of nicotinamide adenine dinucleotide phosphate (NADPH) oxidase (NOX) (35) and myeloperoxidase (36) function, iron-depleted neutrophils underwent NOX-dependent NETosis less efficiently in response to PMA (Fig. 3D and fig. S3D).
While PMA drives NOX-dependent ROS production and NETosis, ionomycin induces NETosis dependent on mitoROS (37). We evaluated whether iron deficiency disrupted mitochondrial function by assessing neutrophil energy metabolism in vivo using SCENITH (single-cell metabolism by profiling translation inhibition) (38). Mitochondrial-dependent adenosine 5′-triphosphate (ATP) production was reduced in Ly6G+ granulocytes from mHep-treated mice, while proportional glucose dependence was unaffected (Fig. 3E). In parallel to reduced mitochondrial ATP production, iron-restricted neutrophils produced greater amounts of mitoROS (Fig. 3F) and had an increased propensity to undergo mitoROS-dependent NETosis in response to ionomycin (Fig. 3G and fig. S3E).
Monopoiesis is relatively resistant to perturbation by hypoferremia (Fig. 1, G and H). Therefore, we asked whether monocyte function was also preserved. In splenic monocytes from mHep-treated mice, secretion of IL-12, TNF, and IL-6 in response to ex vivo lipopolysaccharide (LPS) stimulation was unchanged compared to monocytes from control mice (fig. S3D).
Counteracting inflammatory hepcidin induction improves neutrophil production
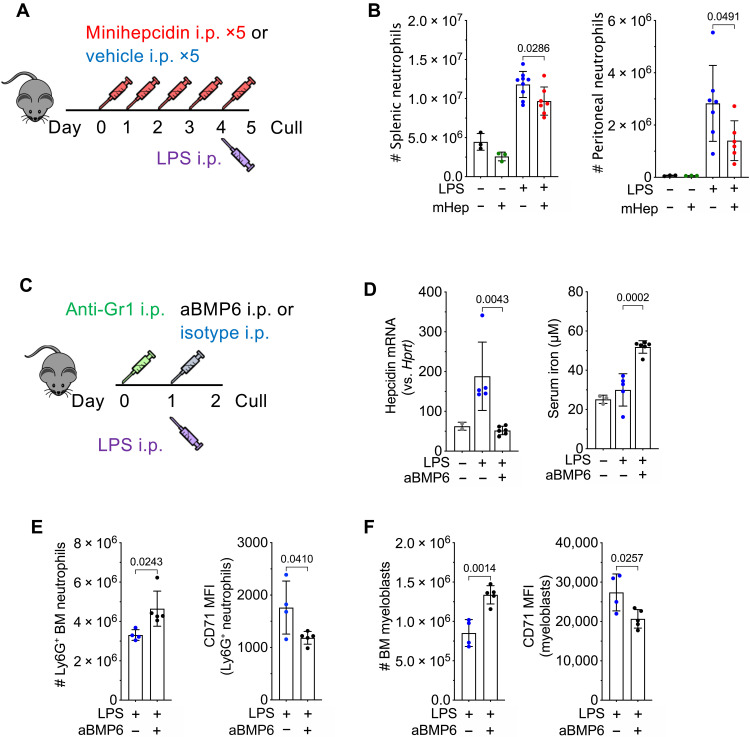
Low plasma iron and raised hepcidin are commonly observed in the context of inflammation (39). Having demonstrated iron-dependent granulopoietic suppression under noninflammatory conditions, we challenged mice after 4 days of mHep treatment with LPS to interrogate how intensified hypoferremia alters the neutrophil response in the context of inflammation (Fig. 4A). Hypoferremia reduced inflammatory accumulation of neutrophils in the spleen and peritoneal cavity (Fig. 4B) consistent with published reduced splenic neutrophil counts in iron-deficient animals infected with Salmonella (40); however, no difference in the number of Ly6G+ neutrophils remaining in the BM was observed (fig. S4A). This is possibly because inflammatory mobilization out of the BM dominated over production under these conditions.
Fig. 4. Modulation of hepcidin during inflammation alters neutrophil production.
(A) Experimental scheme for investigation of effect of sustained hypoferremia on neutrophil response to sterile inflammatory challenge. (B) Number of mature Ly6G+ neutrophils in the spleen and peritoneal cavity in mice treated as in (A). t test. Means ± SD. (C) Experimental scheme for investigation of effect of endogenous hepcidin response on granulopoiesis. (D) Liver hepcidin expression (Mann-Whitney test) and serum iron from mice treated as in (C). Means ± SD. Non-LPS–treated control group was treated with aGr1 24 hours before culling to reflect the systemic iron environment before LPS injection. Representative of two independent experiments. (E) Number of BM Ly6G+ neutrophils and CD71 MFI on Ly6G+ neutrophils in mice treated as in (C). t test. Means ± SD. Representative of three independent experiments. (F) Number of BM c-kit+ CD115− Ly6G− myeloblasts and CD71 MFI on c-kit+ myeloblasts in mice treated as in (C). t test. Means ± SD. Representative of three independent experiments.
We next considered whether endogenous hepcidin would suppress granulopoiesis during inflammation. Hepatic hepcidin transcription is induced during inflammation by IL-6 via a signaling axis that also requires sustained tonic bone morphogenetic protein 6 (BMP6)/SMAD signaling (20). After depleting neutrophils using anti–Gr-1 to enhance granulopoietic demand, we administered mice with LPS as a hepcidin-stimulatory inflammatory signal, with or without anti-BMP6 antibody (Fig. 4C). As previously shown (41), BMP6 neutralization suppressed the endogenous hepcidin response to inflammation and increased plasma iron at 24 hours after LPS injection (Fig. 4D). The LPS dose used here previously caused transient hepcidin-dependent hypoferremia at 6 hours after injection that was resolved by 24 hours (42). Accordingly, in the present experiment, although serum iron in animals treated with LPS alone was similar to that of controls at 24 hours, these mice had likely experienced transient inflammatory hypoferremia (Fig. 4D). Although anti-BMP6 increased serum iron, it did not alter other aspects of systemic LPS-mediated inflammation, as determined by weight loss (fig. S4B), liver Saa1 and Fga mRNA expression (fig. S4C), and serum cytokine levels (fig. S4D).
Focusing on granulopoiesis, anti-BMP6 increased numbers of mature Ly6G+ neutrophils and CD115−ve c-kit+ neutrophil progenitors in the BM of inflamed mice (Fig. 4, E and F). Furthermore, CD71 expression by both Ly6G+ cells and progenitors was decreased in anti-BMP6 treated mice, indicating that improved iron acquisition by granulopoiesis during inflammation was associated with increased production of neutrophils (Fig. 4, E and F).
DISCUSSION
Systemic hypoferremia is a feature of the acute phase response of human infections (43–46) and likely protects against extracellular siderophilic infections. However, beyond the well-characterized anemia of inflammation (39), further consequences of hypoferremia for host physiology are poorly defined. We show that granulopoiesis is particularly sensitive to iron availability, commensurate with the relatively high production of neutrophils containing a relatively iron-rich proteome.
In contrast to granulopoiesis, steady-state and G-CSF–stimulated monocyte production appeared resistant to hypoferremia, with evidence of increased numbers of BM monocyte and their progenitors in resting mice. Monocyte cytokine production ex vivo was also preserved. This is a notable result as, similar to neutrophils, monocytes are an innate cell type exhibiting rapid turnover in vivo (21, 47), supporting our hypothesis that iron may be particularly important to neutrophils beyond the requirement to support the production of a cell type with a short half-life. Notably, the “iron resistance” of the monocyte lineage is supported by observations of normal NO2 production by LPS-stimulated monocytes from iron-deficient individuals (15).
Our results suggest that neutrophil progenitors have higher baseline iron requirements than monocyte progenitors and that iron restriction does not perturb monocyte progenitor cell cycle profiles. Functionally, hypoferremia could modulate the overall profile of innate immunity at baseline and during stress myelopoiesis by controlling the ratio of monocytes to neutrophils. In addition to these systemic effects, local skin–derived hepcidin also influences innate immunity by facilitating mature neutrophil chemotaxis (48).
We found that neutrophils produced during hypoferremia exhibited reduced ability to phagocytose fluorescent E. coli produce cytokines and kill S. aureus. We also observed suppression of NOX-dependent NETosis, linking systemic nutrient availability to NETosis and building on previous reports of inhibited NOX-dependent ROS production during iron deficiency (10, 13, 15). Although nonphysiological iron chelation has previously been linked to NETosis with conflicting results (49, 50), we observe that physiological iron restriction greatly enhanced mitoROS production and mitoROS-dependent NETosis. The importance of different NETosis pathways in response to different stimuli is complex and an ongoing area of study (51). However, notably, mitoROS-dependent NETosis may play a dominant role in immune complex–driven NETosis during lupus-like autoimmunity (52), where NETs play a key role in driving pathology (53). These patients frequently present with raised hepcidin and low serum iron (54). Our results prompt the question of whether prolonged iron restriction due to raised hepcidin under chronic inflammatory conditions contributes to neutrophil dysfunction, NETosis, and propagation of immunopathology in autoimmunity.
Consistent with our observations in T cells (6), we show that iron restriction changes hematopoietic cell metabolism in vivo, suppressing oxidative mitochondrial metabolism in BM granulocytes. Disrupted mitochondrial metabolism may be linked to increased mitoROS production. While mature neutrophils are extremely reliant on glycolysis for ATP production (55), oxidative mitochondrial metabolism, which is a highly iron-dependent process, has been suggested to be important for neutrophil differentiation and development in the BM (30). The impaired metabolism, differentiation, and function of iron-deficient neutrophils in our study are consistent with this. Further studies will be required to elucidate how iron mechanistically supports neutrophil differentiation, biosynthesis of effector proteins, metabolism, and progenitor proliferation.
Blocking hepcidin and increasing serum iron in a mouse model of acute inflammatory hypoferremia increased the number of BM granulocytes without altering markers of inflammation. We hypothesize that therapeutic hepcidin suppression could be leveraged to control neutrophil numbers and functionality during infection with pathogens insensitive to hepcidin-mediated hypoferremia (56) or during autoimmunity where raised hepcidin might contribute to neutrophil dysfunction and proinflammatory NETosis. However, given results from iron-loaded hemochromatosis and dialysis patients, it will be important to avoid extreme iron loading of neutrophils as this may also lead to dysfunction (16, 57, 58).
Relatively little is known regarding how changes in systemic nutrient availability, including hypoferremia, modulate innate immune cell function in vivo. Our results highlight that neutrophil production is a particularly iron-dependent aspect of hematopoiesis, sensitive to rapid physiological changes in serum iron concentration. We propose that hepcidin-mediated hypoferremia is a previously unidentified immune axis regulating myelopoiesis, specifically negatively regulating granulopoiesis, resulting in fewer neutrophils with altered effector functions. In contrast, monopoiesis is unscathed by hypoferremia. These findings have implications for understanding inflammatory pathology and for the clinical use of granulopoiesis-mobilizing agents in patients at risk of infection and iron deficiency.
MATERIALS AND METHODS
Experimental design
We used computational methods to extract the iron-associated protein proteome from a published proteomics dataset and then predicted the number of iron atoms within by human hematopoietic cells. Mice were also injected with mHep to induce hypoferremia and the sensitivity of various immune cell subsets, detected by flow cytometry, was determined at baseline, during inflammation and in response to G-CSF. The capacity of iron supplementation to rescue the effect of mHep was also determined. BM neutrophils were isolated from mice treated with mHep, and their function was evaluated using ex vivo assays. Last, hepcidin blockade was used to determine whether neutrophil production was improved when endogenous hepcidin-mediated hypoferremia was ameliorated. Age- and sex-matched wild-type (WT) mice were used, and treatments were randomized within cages. Evaluation of NETosis histologically was blinded, but other aspects of analysis were not.
Mice
Unless otherwise stated, animal procedures were performed under the authority of U.K. Home Office project and personal licenses in accordance with the Animals (Scientific Procedures) Act 1986 and were approved by the University of Oxford ethical review committee. Mice were housed in individually ventilated cages and fed ad libitum with a standard diet containing 188 parts per million of iron (SDS Dietex Services, diet 801161). Mice were euthanized in increasing CO2 concentrations. All mice were sex-matched and age-matched (to within 2 weeks) within individual experiment. WT C57BL/6JOlaHsd mice were ordered from Envigo (057). Mice were used between 6 and 10 weeks of age when initiating each experiment. Female mice were used for all experiments, aside from experiment described in Fig. 4C where male mice were used because of their lower liver iron stores and more pronounced acute phase response. Sample size was determined according to prior experience of effect sizes and variability in response to mHep-driven hypoferremia. Within each experiment, mice were randomly allocated to treatment groups such that an equal number of mice in each cage received each treatment. Investigators were not blinded aside from for microscopy analysis of NETosis.
Injected substances
An appropriate mass of mHep PR73 (da-TH-Dpa-bhPro-RCR-bhPhe-Ahx-Ida(Hexadecylamine)-NH2; Chinese Peptide Company) was dissolved in 80% ethanol and then mixed with 60 mg of Purebright SL-220/Sunbright DSPE-020CN (NOF). The control solution was Purebright SL-220 dissolved in ethanol. The ethanol was evaporated off using a vacuum chamber warmed to 50°C. The resultant gel was stored up to 24 hours at 4°C and redissolved in an appropriate volume of water to give an mHep concentration of 1 mM: 100 nM mHep in 100 μl of water was injected per mouse per dose.
FAC (Sigma-Aldrich, F5879) was dissolved to 3 mg/ml in phosphate-buffered saline (PBS). A total of 150 μl was intraperitoneally injected into mice. In anti–Gr-1–mediated neutrophil depletion, 10 μg (female mice) or 20 μg (male mice) of anti–Gr-1/mouse (Ultra-LEAF, clone RB6-8C5, BioLegend, 108436, RRID AB_11147952) diluted in sterile low endotoxin PBS (Gibco, 10010015) was intraperitoneally injected into mice. E. coli LPS-O55:B5 (Sigma-Aldrich, L2880) was injected at a dose of 1 mg/kg of body weight diluted in sterile low endotoxin PBS (42) and intraperitoneally injected into mice. Anti-BMP6 and isotype control were injected at a dose of 10 mg/kg of body weight diluted in sterile low endotoxin PBS and intraperitoneally injected into mice. G-CSF complexes were prepared as described in (32) using recombinant human G-CSF (Filgrastim, Sandoz, 05050650066012) and anti-human G-CSF (clone BVD11-37G10, SouthernBiotech, 10128-0, RRID AB_2794186). A total of 7.5 μg of anti-G-CSF and 1.5 μg of G-CSF were complexed, diluted in sterile low endotoxin PBS, and injected intraperitoneally. A total of 7.5 μg of anti–G-CSF alone was used as the control injection to control for trace endotoxin contamination of the antibody and direct effects of the antibody.
Flow cytometry staining
Single-cell suspensions of the spleen were made by mechanical dissociation through a 40-μm cell strainer. Red cells were lysed in spleen suspensions using tris ammonium chloride (ACT) red cell lysis buffer [tris base (2.06 g/liter), NH4Cl (7.47 g/liter), and 1 liter of H20, adjusted to pH 7.2]. BM was flushed though a 70-μm filter to make a single-cell suspension. Peritoneal lavage was obtained by flushing the peritoneal cavity with a BD 23-gauge needle and syringe filled with 8 ml of cold PBS.
For analysis of murine peripheral blood leukocytes, 50 to 100 μl of whole blood collected by tail bleed into a BD microtainer EDTA tube (BD, 365974) was mixed with 1 ml of ACT red cell lysis buffer and incubated at room temperature (RT) for 20 min. The blood solution was spun down at 400g for 5 min, supernatant was removed, and the leukocyte pellet was transferred to a round bottom 96-well plate for flow cytometric staining.
Tissue cell suspensions or in vitro–activated cells at appropriate time points after activation cells were transferred to a 96-well round bottom plate, spun down, and washed with 200 μl of PBS. Cells were stained with appropriate concentrations of FC receptor block (TruStain FcX, BioLegend, 101319), fluorophore-conjugated antibodies, and a fixable live dead dye (LIVE/DEAD Fixable Near-IR stain, Invitrogen, 15519340, or Zombie Aqua Fixable Viability Kit, BioLegend, 423101) in 40 μl of PBS for 20 min at 4°C in the dark. Cells were washed and ran directly on an Invitrogen Attune or BD LSRFortessa, and analysis was carried out using FlowJo 10.8.1. For some experiments, cells were fixed for 10 min in 100 μl of 4% paraformaldehyde (Fixation Buffer, BioLegend, 420801), at 4°C in the dark before washing and resuspension for analysis or resuspended in saponin-based perm buffer (BioLegend, 421002) to stain intracellular antigens. Intracellular Ly6G staining was used 24 hours after anti–Gr-1 treatment to accurately resolve neutrophil depletion despite masking of surface epitopes (59). Cell numbers were calculated using both total counts from tissues and Precision Count Beads (BioLegend, 424902) where appropriate. DAPI (4′,6-diamidino-2-phenylindole; 1 μg/ml; Sigma-Aldrich, D9542) was added before running, to fixed and permeabilized cells, for DNA content staining.
Cell culture
All cells were cultured at 37°C with 20% O2 and 5% CO2. For monocyte intracellular cytokine production, 2 million whole splenocytes (in a round bottom 96-well plate) were activated for 3 hours in the presence of brefeldin A (5 μg/ml; BioLegend, 420601) with/without E. coli LPS-O55:B5 (5 ng/ml; Sigma-Aldrich, L2880) in iron-free medium. Iron-free medium [RPMI 1640 (Gibco, 21875034), glutamine (2 mM; Sigma-Aldrich, G7513-100ML), penicillin (100 U/ml), streptomycin (0.1 mg/ml; Sigma-Aldrich, P0781-100ML), and β-mercaptoethanol (55 μM; Gibco, 31350010)] to preserve in vivo iron status fetal bovine serum was replaced with 10% (v/v) Panexin NTS serum substitute (Pan Biotech, P04-95080, custom order). Splenocytes were then washed and stained for surface epitopes and intracellular cytokines as described in flow cytometry.
Neutrophil isolation
Neutrophils were negatively isolated from whole BM of mHep-treated and control-treated animals as per the manufacturer’s instructions (Neutrophil Isolation Kit, Miltenyi Biotec, 130-097-658).
Flow cytometric ROS measurement
To identify dihydrorhodamine 123 (DHR123) staining of predominantly cytoplasmic ROS, isolated neutrophils were incubated with 7 μM DHR123 (2.5 μg/ml; Thermo Fisher Scientific, D23806) in complete RPMI 1640 medium (Gibco, 21875034, and Sigma-Aldrich, F9665-500ML) and stimulated by 50 nM PMA (Sigma-Aldrich, P1585) for 20 min at 37°C. Cells were subsequently washed with PBS, and the fluorescence intensities of each subset/cell were measured by flow cytometry. A 4°C incubation was used as a control.
MitoROS was measured using mitoSOX red (Thermo Fisher Scientific, M36008). Isolated neutrophils in complete RPMI 1640 medium were incubated with 5 mM mitoSOX red for 20 min at 37°C in the presence of 10 mM ionomycin (Sigma-Aldrich, I9657). Cells were subsequently washed with PBS, and the fluorescence intensity as the indicator of mitoROS generation was measured by fluorescence-activated cell sorting. A 4°C incubation was used as a control.
Phagocytosis
The phagocytosis capacity of BM neutrophils was measured by flow cytometry–based method using fluorescent E. coli (American Type Culture Collection, 25922GFP). Isolated neutrophils were incubated with fluorescent E. coli at a multiplicity of infection (MOI) of 10 in complete RPMI 1640 medium for 15 min at 37°C. Neutrophils were subsequently washed with PBS, and the fluorescence intensity was measured by flow cytometry.
Bacterial killing assay
Bacterial killing assay was performed with S. aureus (Heatley Oxford, NCTC 6571), which was used at an MOI of 10. For the bacterial killing assay, isolated neutrophils were interacted with S. aureus for 2 hours at 37°C in 5% CO2 tissue culture incubator and then lysed in 1% triton buffer. The lysates were then plated on agar plates. Bacterial culture plates were incubated at 37°C overnight, and the colony number on each plate was counted the following morning as an absolute colony-forming unit count.
Measurement of cytokine production
For the cytokine array, 2 × 106 isolated neutrophils seeded in 2 ml of RPMI 1640 were stimulated with Zymosan (50 μg/ml; Sigma-Aldrich, Z4250) or dimethyl sulfoxide vehicle (Sigma-Aldrich, 41639-100ML) for 2 hours. After incubation, Mouse CCL2 DuoSet enzyme-linked immunosorbent assay (ELISA; R&D Systems, DY479-05) and Mouse TNF-α DuoSet ELISA (R&D Systems, MTA00B) were used to detect the levels of CCL2 and TNF-α, respectively, in the supernatants of stimulated isolated neutrophils, following the manufacturer’s instruction. For readout, signals were detected by chemical luminescence and subsequently measured with the CLARIOstar Microplate Reader (BMG Labtech).
NETosis
To induce NETosis, isolated neutrophils were seeded into eight-well Lab-Tek II chamber slide (VWR International, 734-2050 l) coated with 2% poly-l-lysine (Sigma-Aldrich, P4707) at a volume of 1 ml at the density of 1 × 106/ml. Neutrophils were stimulated with 10 μM ionomycin and 10 μM PMA together or individually, overnight at 37°C in 5% CO2 tissue culture incubator, and were subsequently fixed with 4% paraformaldehyde (Sigma-Aldrich, 47608) in Dulbecco’s PBS (D-PBS) (Gibco, 14190144) for 30 min at RT. To investigate the effect of ROS on NETosis, different ROS inhibitors were used. A total of 25 μM DPI (Sigma-Aldrich, D2926) was used to block total ROS generation from ionomycin and PMA, 100 μM apocynin (Abcam, ab120615) was used to block ROS generation via NOX2 complex by PMA, and 5 μM mitoTEMPO (Cayman Chemical, 16621) was used to inhibit mitoROS production. Afterward, cells were washed three times with D-PBS and incubated with blocking buffer (2% bovine serum albumin in tris-buffered saline with 0.1% Tween 20; Sigma-Aldrich, A7030, and Sigma-Aldrich, P9416) for 20 min. Following blocking, primary antibodies [rabbit anticitrullinated histone H3 (polyclonal, Abcam, ab5103, RRID AB_304752) and mouse anti-mouse myeloperoxidase (MPO) (clone 8F4, Hycult, HM1051BT, RRID AB_1953633) at 1:100 dilution in blocking buffer] were added and incubated for 2 hours at RT or overnight at 4°C. Cells were washed with D-PBS before adding secondary antibodies: goat anti-rabbit AF647 (Thermo Fisher Scientific, #A-21244) and goat anti-mouse AF488 (Thermo Fisher Scientific, #A-11001) at 1:300 dilution in blocking buffer for a 1-hour incubation at RT in the dark. Subsequently, cells were washed with D-PBS. Before imaging, the chambers were removed from the slides, and cells were covered with ProLong gold antifade mounting media with DAPI (Thermo Fisher Scientific, P36931). Images were obtained by Zeiss LSM 980 confocal microscope. Neutrophils with a clear formation of fibers, together with a diffuse nucleus stained by DAPI and colocalization with MPO and citrullinated histone 3, were counted as neutrophils under NETosis. For each condition, 100 to 200 cells of each sample have been counted from three different fields of two independent experiments. ImageJ was used for image analysis and presentation.
SCENITH analysis
SCENITH is a flow cytometry approach in which translation rate is quantified by measuring puromycin incorporation into protein (38, 60). Translation is highly ATP dependent. Therefore, changes in the rate of translation after the addition of different metabolic inhibitors can be used to estimate cellular glucose dependence, mitochondrial dependence, glycolytic capacity, and fatty acid/amino acid oxidation capacity. The SCENITH protocol was adapted from the original method (38) for use with the Invitrogen Click-iT Plus O-propargyl-puromycin (OPP) Protein Synthesis Assay Kit (Invitrogen, C10456). To prevent cells from iron loading ex vivo during short-term culture, all steps were conducted in iron-free media. BM cells were plated at 3 × 106 cells per well in a 96-well plate. Matched samples were treated in parallel with 100 μl of iron-free media at 37°C with 5% CO2 with five different conditions: media alone, 2-deoxy-l-glucose (100 mM; Alfa Aesar, L07338.03), oligomycin (1 μM; MP Biomedicals, 151786), 2-deoxyglucose (100 mM) + oligomycin (1 μM), and harringtonine (2 μg/ml; Cayman Chemicals, 15361). Thirty minutes into inhibitor treatment, 100 μl of OPP was added on top for a final OPP dilution of 1:1000 and incubated for a further 30 min at 37°C with 5% CO2. Cells were then washed, surface-stained, fixed, and permeabilized as described in flow cytometry. Following permeabilization, intracellular OPP was labeled according to the manufacturer’s instructions. Analysis was conducted as described by Argüello et al. (38).
Blood measurements
Full murine red blood cell indices and platelet counts were performed on 100 μl of cardiac whole-blood taken into a BD microtainer EDTA tube (Becton Dickinson, 365974) and measured on a Sysmex KN-21 blood analyzer.
For serum, analysis of murine samples up to 400 μl of blood obtained by cardiac puncture was placed in a BD microtainer SST tube (Becton Dickinson, 365967). Serum was obtained by spinning the clotted blood sample was spun at 8000g for 5 min and stored at −80°C. Iron measurements were determined using an ABX Pentra instrument. Serum cytokine levels were measured by Luminex (IL-5 and G-CSF, Life Technologies) or LEGENDplex (M1 macrophage panel, with added G-CSF capture beads, BioLegend, 740848) in accordance with the manufacturer’s instructions.
Liver gene expression analysis by quantitative polymerase chain reaction
Liver explants were stored in RNAlater (Invitrogen, AM7020). RNA was extracted using the RNeasy Mini Plus Kit (QIAGEN, 74136), and cDNA was synthesized using the High-Capacity RNA-to-cDNA Kit (Applied Biosystems, 4387406), following the manufacturer’s protocols. Quantitative polymerase chain reaction was performed on the QuantStudio 7 Flex System (Applied Biosystems, 4485701), using TaqMan Gene Expression Master Mix (Applied Biosystems, 4369016) and TaqMan Gene Expression Assays (Applied Biosystems, 4331182), specifically Mm01545399_m1 (Hprt), Mm04231240_s1 (Hamp), Mm00656927_g1 (Saa1), and Mm00802584_m1 (Fga). Gene expression was evaluated by 2([CT of Hprt] – [CT of gene of interest]).
Mathematic modeling
Iron counts per cell were calculated using the method described (2). Human iron-interacting proteins were identified in human immune cell proteomes as described (18) by cross-referencing against the list of human iron interacting proteins identified (1). Hemoglobin proteins (HBA1, HBB, HBD, HBG1, HBG2, HBM, HBQ1, and HBZ) were removed as these proteins should not be expressed in immune cells and their presence is likely indicative of red cell contamination. Protein copy number values for each iron-interacting protein species were multiplied against the protein:iron atom stoichiometry values previously curated as described (2) to give the number of iron atoms per protein species. Total cellular iron counts were calculated as the sum of iron atoms attributed across all detected iron-interacting protein species.
We assigned iron-interacting protein species to pathways using pre-established gene ontology and Reactome gene sets: Oxidative phosphorylation (OXPHOS), GOBP_AEROBIC_RESPIRATION; DNA replication, GOBP_DNA_REPLICATION; iron pathways, pooled GOBP_CELLULAR_IRON_ION_HOMEOSTASIS and GOBP_IRON_SULFUR_CLUSTER_ASSEMBLY; demethylation, GOBP_DEMETHYLATION; fatty acid metabolism, GOBP_FATTY_ACID_METABOLIC_PROCESS; granule proteins, REACTOME_NEUTROPHIL_DEGRANULATION. Where overlaps in gene sets were present, we assigned pathways according to best fit given the current literature (see scripts for specific designations). Atoms attributed to protein species were in turn assigned to each protein’s corresponding pathway.
Statistical analysis
GraphPad Prism (version 9.3.0) was used for to perform statistical tests and generate P values. Raw P values are presented throughout, and the absence of a P value or “ns” designates a P > 0.05. Data are summarized as bar graphs, scatter plots, or box and whisker as appropriate with description of what the bars depict for individual graphs in the figure legends. When two groups were compared, an unpaired t test, unpaired t test with Welch’s correction when SD were different, or Mann-Whitney U test for nonparametric data was carried out as appropriate and noted in the figure legend. For comparing multiple groups, a one-way or two-way analysis of variance was used with either a Tukey or Sidak’s multiple comparison test (respectively).
Acknowledgments
We thank the staff of the Department of Biomedical Services, University of Oxford, for animal husbandry; Y.-L. Chen for assistance running the Luminex assay; and G. Mohammad and S. Lakhal-Littleton for assistance with serum iron measurements.
Funding: U.K. Medical Research Council (MRC Human Immunology Unit core funding to HD, award no. MCU_12010/10) (to J.N.F., A.E.P., A.E.A., and H.D.); Wellcome Trust Infection, Immunology and Translational Medicine doctoral program (grant no. 108869/Z/15/Z). (to S.K.W.); Clarendon Fund and the Corpus Christi College A. E. Haigh graduate scholarship (to M.R.T.); Oxford-BMS Fellowship (to A.C.); Deutsche Forschungsgemeinschaft (GA2075/5-1 and GA2075/6.1) (to B.G. and M.B.); Wellcome Trust (211072/Z/18/Z) (to A.J.C.); Novo Nordisk Foundation (Tripartite Immunometabolism Consortium to L.W.); MeRIAD Consortium (to I.A.U.); Chinese Science Council (to Z.A.); Wellcome Trust (Investigator Award 209422/Z/17/Z) (to I.A.U.).
Author contributions: Conceptualization: J.N.F. and H.D. Methodology: J.N.F., M.R.T., Z.A., L.W., M.B., A.C., Y.Y., A.J.C., I.A.U., and H.D. Investigation: J.N.F., S.K.W., A.E.P., M.R.T., M.B., Z.A., L.W., A.C., and N.W. Visualization: J.N.F. and L.W. Supervision: B.G., I.A.U., and H.D. Writing—original draft: J.N.F. and H.D. Writing—review and editing: J.N.F., L.W., M.B., B.G., A.E.A., I.A.U., and H.D.
Competing interests: These authors declare that they do not have any competing interests.
Data and materials availability: All data are available in the main text or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S4
Table S1
REFERENCES AND NOTES
- 1.Andreini C., Putignano V., Rosato A., Banci L., The human iron-proteome. Metallomics 10, 1223–1231 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Teh M. R., Frost J. N., Armitage A. E., Drakesmith H., Analysis of iron and iron-interacting protein dynamics during T-cell activation. Front. Immunol. 12, 714613 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drakesmith H., Prentice A. M., Hepcidin and the iron-infection axis. Science 338, 768–772 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Drakesmith H., Nemeth E., Ganz T., Ironing out ferroportin. Cell Metab. 22, 777–787 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arezes J., Jung G., Gabayan V., Valore E., Ruchala P., Gulig P. A., Ganz T., Nemeth E., Bulut Y., Hepcidin-induced hypoferremia is a critical host defense mechanism against the siderophilic bacterium Vibrio vulnificus. Cell Host Microbe 17, 47–57 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frost J. N., Tan T. K., Abbas M., Wideman S. K., Bonadonna M., Stoffel N. U., Wray K., Kronsteiner B., Smits G., Campagna D. R., Duarte T. L., Lopes J. M., Shah A., Armitage A. E., Arezes J., Lim P. J., Preston A. E., Ahern D., Teh M., Naylor C., Salio M., Gileadi U., Andrews S. C., Dunachie S. J., Zimmermann M. B., van der Klis F. R. M., Cerundolo V., Bannard O., Draper S. J., Townsend A. R. M., Galy B., Fleming M. D., Lewis M. C., Drakesmith H., Hepcidin-mediated hypoferremia disrupts immune responses to vaccination and infection. Med (N. Y.) 2, 164–179.e12 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Littwitz-Salomon E., Moreira D., Frost J. N., Choi C., Liou K. T., Ahern D. K., O’Shaughnessy S., Wagner B., Biron C. A., Drakesmith H., Dittmer U., Finlay D. K., Metabolic requirements of NK cells during the acute response against retroviral infection. Nat. Commun. 12, 5376 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boettcher S., Manz M. G., Regulation of inflammation- and infection-driven hematopoiesis. Trends Immunol. 38, 345–357 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Cronin S. J. F., Woolf C. J., Weiss G., Penninger J. M., The role of iron regulation in immunometabolism and immune-related disease. Front. Mol. Biosci. 6, 116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahluwalia N., Sun J., Krause D., Mastro A., Handte G., Immune function is impaired in iron-deficient, homebound, older women. Am. J. Clin. Nutr. 79, 516–521 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Egeli A. K., Framstad T., Morberg H., Clinical biochemistry, haematology and body weight in piglets. Acta Vet. Scand. 39, 381–393 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hassan T. H., Badr M. A., Karam N. A., Zkaria M., el Saadany H. F., Abdel Rahman D. M., Shahbah D. A., al Morshedy S. M., Fathy M., Esh A. M. H., Selim A. M., Impact of iron deficiency anemia on the function of the immune system in children. Medicine 95, e5395 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurtoglu E., Ugur A., Baltaci A. K., Mogolkoc R., Undar L., Activity of neutrophil NADPH oxidase in iron-deficient anemia. Biol. Trace Elem. Res. 96, 109–116 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Monteith A. J., Skaar E. P., The impact of metal availability on immune function during infection. Trends Endocrinol. Metab. 32, 916–928 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paino I. M. M., Miranda J. C., Marzocchi-Machado C. M., Cesarino E. J., de Castro F. A., de Souza A. M., Phagocytosis, oxidative burst, and produced reactive species are affected by iron deficiency anemia and anemia of chronic diseases in elderly. Biol. Trace Elem. Res. 129, 116–125 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Renassia C., Louis S., Cuvellier S., Boussetta N., Deschemin J. C., Borderie D., Bailly K., Poupon J., Dang P. M. C., el-Benna J., Manceau S., Lefrère F., Vaulont S., Peyssonnaux C., Neutrophils from hereditary hemochromatosis patients are protected from iron excess and are primed. Blood Adv. 4, 3853–3863 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muckenthaler M. U., Rivella S., Hentze M. W., Galy B., A red carpet for iron metabolism. Cell 168, 344–361 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rieckmann J. C., Geiger R., Hornburg D., Wolf T., Kveler K., Jarrossay D., Sallusto F., Shen-Orr S. S., Lanzavecchia A., Mann M., Meissner F., Social network architecture of human immune cells unveiled by quantitative proteomics. Nat. Immunol. 18, 583–593 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Cosgrove J., Hustin L. S. P., de Boer R. J., Perié L., Hematopoiesis in numbers. Trends Immunol. 42, 1100–1112 (2021). [DOI] [PubMed] [Google Scholar]
- 20.Wang C.-Y., Babitt J. L., Liver iron sensing and body iron homeostasis. Blood 133, 18–29 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Evrard M., Kwok I. W. H., Chong S. Z., Teng K. W. W., Becht E., Chen J., Sieow J. L., Penny H. L., Ching G. C., Devi S., Adrover J. M., Li J. L. Y., Liong K. H., Tan L., Poon Z., Foo S., Chua J. W., Su I.-H., Balabanian K., Bachelerie F., Biswas S. K., Larbi A., Hwang W. Y. K., Madan V., Koeffler H. P., Wong S. C., Newell E. W., Hidalgo A., Ginhoux F., Ng L. G., Developmental analysis of bone marrow neutrophils reveals populations specialized in expansion, trafficking, and effector functions. Immunity 48, 364–379.e8 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Chong S. Z., Evrard M., Devi S., Chen J., Lim J. Y., See P., Zhang Y., Adrover J. M., Lee B., Tan L., Li J. L. Y., Liong K. H., Phua C., Balachander A., Boey A., Liebl D., Tan S. M., Chan J. K. Y., Balabanian K., Harris J. E., Bianchini M., Weber C., Duchene J., Lum J., Poidinger M., Chen Q., Rénia L., Wang C. I., Larbi A., Randolph G. J., Weninger W., Looney M. R., Krummel M. F., Biswas S. K., Ginhoux F., Hidalgo A., Bachelerie F., Ng L. G., CXCR4 identifies transitional bone marrow premonocytes that replenish the mature monocyte pool for peripheral responses. J. Exp. Med. 213, 2293–2314 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hettinger J., Richards D. M., Hansson J., Barra M. M., Joschko A. C., Krijgsveld J., Feuerer M., Origin of monocytes and macrophages in a committed progenitor. Nat. Immunol. 14, 821–830 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Yu Y., Kovacevic Z., Richardson D. R., Tuning cell cycle regulation with an iron key. Cell Cycle 6, 1982–1994 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Xavier-Ferrucio J., Scanlon V., Li X., Zhang P. X., Lozovatsky L., Ayala-Lopez N., Tebaldi T., Halene S., Cao C., Fleming M. D., Finberg K. E., Krause D. S., Low iron promotes megakaryocytic commitment of megakaryocytic-erythroid progenitors in humans and mice. Blood 134, 1547–1557 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Evstatiev R., Bukaty A., Jimenez K., Kulnigg-Dabsch S., Surman L., Schmid W., Eferl R., Lippert K., Scheiber-Mojdehkar B., Michael Kvasnicka H., Khare V., Gasche C., Iron deficiency alters megakaryopoiesis and platelet phenotype independent of thrombopoietin. Am. J. Hematol. 89, 524–529 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kopf M., Brombacher F., Hodgkin P. D., Ramsay A. J., Milbourne E. A., Dai W. J., Ovington K. S., Behm C. A., Köhler G., Young I. G., Matthaei K. I., IL-5-deficient mice have a developmental defect in CD5+ B-1 cells and lack eosinophilia but have normal antibody and cytotoxic T cell responses. Immunity 4, 15–24 (1996). [DOI] [PubMed] [Google Scholar]
- 28.Lieschke G. J., Grail D., Hodgson G., Metcalf D., Stanley E., Cheers C., Fowler K. J., Basu S., Zhan Y. F., Dunn A. R., Mice lacking granulocyte colony-stimulating factor have chronic neutropenia, granulocyte and macrophage progenitor cell deficiency, and impaired neutrophil mobilization. Blood 84, 1737–1746 (1994). [PubMed] [Google Scholar]
- 29.Stark M. A., Huo Y., Burcin T. L., Morris M. A., Olson T. S., Ley K., Phagocytosis of apoptotic neutrophils regulates granulopoiesis via IL-23 and IL-17. Immunity 22, 285–294 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Riffelmacher T., Clarke A., Richter F. C., Stranks A., Pandey S., Danielli S., Hublitz P., Yu Z., Johnson E., Schwerd T., Cullagh J. M., Uhlig H., Jacobsen S. E. W., Simon A. K., Autophagy-dependent generation of free fatty acids is critical for normal neutrophil differentiation. Immunity 47, 466–480.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manz M. G., Boettcher S., Emergency granulopoiesis. Nat. Rev. Immunol. 14, 302–314 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Rubinstein M. P., Salem M. L., Doedens A. L., Moore C. J., Chiuzan C., Rivell G. L., Cole D. J., Goldrath A. W., G-CSF/anti-G-CSF antibody complexes drive the potent recovery and expansion of CD11b+Gr-1+ myeloid cells without compromising CD8+ T cell immune responses. J. Hematol. Oncol. 6, 75 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Papayannopoulos V., Neutrophil extracellular traps in immunity and disease. Nat. Rev. Immunol. 18, 134–147 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Li Y., Trush M. A., Diphenyleneiodonium, an NAD(P)H oxidase inhibitor, also potently inhibits mitochondrial reactive oxygen species production. Biochem. Biophys. Res. Commun. 253, 295–299 (1998). [DOI] [PubMed] [Google Scholar]
- 35.Sumimoto H., Structure, regulation and evolution of Nox-family NADPH oxidases that produce reactive oxygen species. FEBS J. 275, 3249–3277 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Davies M. J., Myeloperoxidase-derived oxidation: Mechanisms of biological damage and its prevention. J. Clin. Biochem. Nutr. 48, 8–19 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Douda D. N., Khan M. A., Grasemann H., Palaniyar N., SK3 channel and mitochondrial ROS mediate NADPH oxidase-independent NETosis induced by calcium influx. Proc. Natl. Acad. Sci. U.S.A. 112, 2817–2822 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Argüello R. J., Combes A. J., Char R., Gigan J.-P., Baaziz A. I., Bousiquot E., Camosseto V., Samad B., Tsui J., Yan P., Boissonneau S., Figarella-Branger D., Gatti E., Tabouret E., Krummel M. F., Pierre P., SCENITH: A flow cytometry-based method to functionally profile energy metabolism with single-cell resolution. Cell Metab. 32, 1063–1075.e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weiss G., Ganz T., Goodnough L. T., Anemia of inflammation. Blood 133, 40–50 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoffmann A., Haschka D., Valente de Souza L., Tymoszuk P., Seifert M., von Raffay L., Hilbe R., Petzer V., Moser P. L., Nairz M., Weiss G., Baseline iron status and presence of anaemia determine the course of systemic Salmonella infection following oral iron supplementation in mice. EBioMedicine 71, 103568 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petzer V., Tymoszuk P., Asshoff M., Carvalho J., Papworth J., Deantonio C., Bayliss L., Wake M. S., Seifert M., Brigo N., Valente de Souza L., Hilbe R., Grubwieser P., Demetz E., Dichtl S., Volani C., Berger S., Böhm F., Hoffmann A., Pfeifhofer-Obermair C., von Raffay L., Sopper S., Arndt S., Bosserhoff A., Kautz L., Perrier P., Nairz M., Wolf D., Weiss G., Germaschewski V., Theurl I., A fully human anti-BMP6 antibody reduces the need for erythropoietin in rodent models of the anemia of chronic disease. Blood 136, 1080–1090 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Armitage A. E., Lim P. J., Frost J. N., Pasricha S. R., Soilleux E. J., Evans E., Morovat A., Santos A., Diaz R., Biggs D., Davies B., Gileadi U., Robbins P. A., Lakhal-Littleton S., Drakesmith H., Induced disruption of the iron-regulatory hormone hepcidin inhibits acute inflammatory hypoferraemia. J. Innate Immun. 8, 517–528 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Armitage A. E., Stacey A. R., Giannoulatou E., Marshall E., Sturges P., Chatha K., Smith N. M. G., Huang X. J., Xu X. N., Pasricha S. R., Li N., Wu H., Webster C., Prentice A. M., Pellegrino P., Williams I., Norris P. J., Drakesmith H., Borrow P., Distinct patterns of hepcidin and iron regulation during HIV-1, HBV, and HCV infections. Proc. Natl. Acad. Sci. U.S.A. 111, 12187–12192 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Darton T. C., Blohmke C. J., Giannoulatou E., Waddington C. S., Jones C., Sturges P., Webster C., Drakesmith H., Pollard A. J., Armitage A. E., Rapidly escalating hepcidin and associated serum iron starvation are features of the acute response to typhoid infection in humans. PLOS Negl. Trop. Dis. 9, e0004029 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shah A., Frost J. N., Aaron L., Donovan K., Drakesmith H.; Collaborators, McKechnie S. R., Stanworth S. J., Systemic hypoferremia and severity of hypoxemic respiratory failure in COVID-19. Crit. Care 24, 320 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Williams A. M., Ladva C. N., Leon J. S., Lopman B. A., Tangpricha V., Whitehead R. D. Jr., Armitage A. E., Wray K., Morovat A., Pasricha S. R., Thurnham D., Tanumihardjo S. A., Shahab-Ferdows S., Allen L., Flores-Ayala R. C., Suchdev P. S., Changes in micronutrient and inflammation serum biomarker concentrations after a norovirus human challenge. Am. J. Clin. Nutr. 110, 1456–1464 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yona S., Kim K. W., Wolf Y., Mildner A., Varol D., Breker M., Strauss-Ayali D., Viukov S., Guilliams M., Misharin A., Hume D. A., Perlman H., Malissen B., Zelzer E., Jung S., Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity 38, 79–91 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Malerba M., Louis S., Cuvellier S., Shambat S. M., Hua C., Gomart C., Fouet A., Ortonne N., Decousser J. W., Zinkernagel A. S., Mathieu J. R., Peyssonnaux C., Epidermal hepcidin is required for neutrophil response to bacterial infection. J. Clin. Invest. 130, 329–334 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kono M., Saigo K., Yamamoto S., Shirai K., Iwamoto S., Uematsu T., Takahashi T., Imoto S., Hashimoto M., Minami Y., Wada A., Takenokuchi M., Kawano S., Iron-chelating agent, deferasirox, inhibits neutrophil activation and extracellular trap formation. Clin. Exp. Pharmacol. Physiol. 43, 915–920 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Völlger L., Akong-Moore K., Cox L., Goldmann O., Wang Y., Schäfer S. T., Naim H. Y., Nizet V., von Köckritz-Blickwede M., Iron-chelating agent desferrioxamine stimulates formation of neutrophil extracellular traps (NETs) in human blood-derived neutrophils. Biosci. Rep. 36, e00333 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kenny E. F., Herzig A., Krüger R., Muth A., Mondal S., Thompson P. R., Brinkmann V., von Bernuth H., Zychlinsky A., Diverse stimuli engage different neutrophil extracellular trap pathways. eLife 6, e24437 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lood C., Blanco L. P., Purmalek M. M., Carmona-Rivera C., de Ravin S. S., Smith C. K., Malech H. L., Ledbetter J. A., Elkon K. B., Kaplan M. J., Neutrophil extracellular traps enriched in oxidized mitochondrial DNA are interferogenic and contribute to lupus-like disease. Nat. Med. 22, 146–153 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gupta S., Kaplan M. J., Bite of the wolf: Innate immune responses propagate autoimmunity in lupus. J. Clin. Invest. 131, e144918 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kunireddy N., Jacob R., Khan S. A., Yadagiri B., Sai Baba K. S. S., Rajendra Vara Prasad I., Mohan I. K., Hepcidin and ferritin: Important mediators in inflammation associated anemia in systemic lupus erythematosus patients. Indian J. Clin. Biochem. 33, 406–413 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Borregaard N., Herlin T., Energy metabolism of human neutrophils during phagocytosis. J. Clin. Invest. 70, 550–557 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stefanova D., Raychev A., Arezes J., Ruchala P., Gabayan V., Skurnik M., Dillon B. J., Horwitz M. A., Ganz T., Bulut Y., Nemeth E., Endogenous hepcidin and its agonist mediate resistance to selected infections by clearing non–transferrin-bound iron. Blood 130, 245–257 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Patruta S. I., Edlinger R., Sunder-Plassmann G., Hörl W. H., Neutrophil impairment associated with iron therapy in hemodialysis patients with functional iron deficiency. J. Am. Soc. Nephrol. 9, 655–663 (1998). [DOI] [PubMed] [Google Scholar]
- 58.Cantinieaux B., Boelaert J. R., De Meuleneire J., Kerrels V., Fondu P., Neutrophils from patients with secondary haemosiderosis contain excessive amounts of autotoxic iron. Eur. J. Haematol. 51, 161–165 (1993). [DOI] [PubMed] [Google Scholar]
- 59.Boivin G., Faget J., Ancey P.-B., Gkasti A., Mussard J., Engblom C., Pfirschke C., Contat C., Pascual J., Vazquez J., Bendriss-Vermare N., Caux C., Vozenin M.-C., Pittet M. J., Gunzer M., Meylan E., Durable and controlled depletion of neutrophils in mice. Nat. Commun. 11, 2762 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lopes N., McIntyre C., Martin S., Raverdeau M., Sumaria N., Kohlgruber A. C., Fiala G. J., Agudelo L. Z., Dyck L., Kane H., Douglas A., Cunningham S., Prendeville H., Loftus R., Carmody C., Pierre P., Kellis M., Brenner M., Argüello R. J., Silva-Santos B., Pennington D. J., Lynch L., Distinct metabolic programs established in the thymus control effector functions of γδ T cell subsets in tumor microenvironments. Nat. Immunol. 22, 179–192 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S4
Table S1