Abstract
Rapid technological advances in molecular biology, including next-generation sequencing, have identified key genetic alterations in central nervous system (CNS) tumors. Accordingly, the fifth edition of the World Health Organization (WHO) CNS tumor classification was published in 2021. We analyzed 303 patients with diffuse glioma using an amplicon-based glioma-tailored gene panel for detecting 1p/19q codeletion and driver gene mutations such as IDH1/2, TERTp, EGFR, and CDKN2A/B on a single platform. Within glioblastomas (GBMs), the most commonly mutated genes were TERTp, TP53, PTEN, NF1, and PDGFRA, which was the most frequently mutated tyrosine kinase receptor in GBM, followed by EGFR. The genes that most commonly showed evidence of loss were PTEN, CDKN2A/B, and RB1, whereas the genes that most commonly showed evidence of gain/amplification were EGFR, PDGFRA, and CDK4. In 22 grade III oligodendroglial tumors, 3 (14%) patients had CDKN2A/B homozygous deletion, and 4 (18%) patients had ARID1A mutation. In grade III oligodendroglial tumors, an ARID1A mutation was associated with worse progression-free survival. Reclassification based on the WHO 2021 classification resulted in 62.5% of grade II/III isocitrate dehydrogenase (IDH)-wildtype astrocytomas being classified as IDH-wildtype GBM and 37.5% as not elsewhere classified. In summary, our glioma-tailored gene panel was applicable for molecular diagnosis in the WHO 2021 classification. In addition, we successfully reclassified the 303 diffuse glioma cases based on the WHO 2021 classification and clarified the genetic profile of diffuse gliomas in the Japanese population.
Keywords: gene panel, next-generation sequencing, molecular genetic profile, WHO 2021 classification
Introduction
Due to the implementation of the revised 2016 World Health Organization (WHO) classification, the diagnosis of central nervous system (CNS) tumors has changed from a histology-based approach to an integrated diagnosis that combines histology and molecular characteristics.1) In this integrated diagnosis, isocitrate dehydrogenase (IDH) 1 or 2 gene mutation (IDH mutation), codeletion of chromosomal arms 1p and 19q (1p/19q codeletion), and H3K27M mutation are genetic alterations that need to be evaluated.1) To date, it has been reported that there are biologically relevant alterations in some core pathways, namely, the p53 pathway (MDM2, MDM4, and TP53), the Rb pathway (CDK4, CDK6, CCND2, CDKN2A/B, and RB1), and various components influencing the PI3K pathway (PIK3CA, PIK3R1, PTEN, EGFR, PDGFRA, and NF1).2,3) Other genetic alterations are known to play an important role in gliomas such as TERTp, which may serve as diagnostic, prognostic, and therapeutic biomarkers.4,5)
Recently, WHO upgraded the classification scheme of CNS tumors. This 2021 WHO classification (5th edition) modified some important diagnostic criteria, integrating the important implications for tumor classification and patient care advocated by the Consortium to Inform Molecular and Practical Approaches to CNS Tumor Taxonomy.6-8) Importantly, the WHO 2021 classification allows for a diagnosis of glioblastoma (GBM), which is CNS WHO grade 4, in IDH-wildtype astrocytomas, even in the absence of high-grade histopathologic features, when at least one of the following molecular features is present: TERTp mutation, EGFR amplification, or concurrent +7/−10.6) Moreover, grade II or III IDH-mutant diffuse astrocytomas in the revised 2016 WHO classification would be diagnosed as grade 4 IDH-mutant astrocytomas if CDKN2A/B homozygous deletion is detected.6) Given that molecular diagnosis has gained more importance in the diagnosis of glioma, molecular platform to detect key molecular alteration is urgently needed.
We recently developed an amplicon-based glioma-tailored gene panel comprising 50 genes to detect driver gene mutations, such as those in IDH1/2, TERTp, EGFR, and CDKN2A/B, and the 1p/19q codeletion based on a single platform.9) This platform enables us to not only perform an integrated diagnosis but also detect driver gene mutation profiles in glioma.
Here, we successfully analyzed 303 cases of diffuse glioma using our glioma-tailored gene panel and revealed the molecular genetic profile in Japanese patients with diffuse gliomas and the distinct subgroups of IDH-wildtype GBM. In addition, we reclassified all the tumors integrated in this study according to the WHO 2021 classification and discussed the current problems of this new diagnostic scheme.
Materials and Methods
Diffuse glioma samples
Three hundred three formalin-fixed paraffin-embedded (FFPE) tumor tissue samples were collected from the Kagoshima University, Kyushu University, and University of Occupational and Environmental Health. The study was approved by the Institutional Review Board of Kagoshima University (approval no. 180104) and complied with the tenets of the Declaration of Helsinki. Informed consent was obtained from all patients. Resected tumors were fixed with phosphate-buffered 10% formalin within 24 h of sampling and routinely processed for paraffin embedding, followed by sectioning for hematoxylin and eosin staining. All tumors were originally classified according to the WHO 2016 classification. All tissues were histologically evaluated by board-certified pathologists (M.K. and A.T.) to ensure an estimated tumor cell content of 30% or more. In all patients, when analyzing copy number variations, we sequenced the leukocyte DNA for comparison against matched tumor DNA.
DNA extraction and quantification
For the DNA preparation of the FFPE samples, we used the Maxwell 16 FFPE Tissue LEV DNA Purification kit (Promega, Madison, USA) according to the manufacturer's instructions. Thereafter, the concentration of DNA was measured using a Qubit 3.0 Fluorometer dsDNA BR Assay kit (Life Technologies, Carlsbad, USA), and DNA quality was monitored using the QIAseq DNA QuantiMIZE kit (QIAGEN, Hilden, Germany). The extracted DNA was diluted to a concentration of 5-10 ng/μL as a template, and PCR was performed using the QIAseq DNA QuantiMIZE kit. DNA with a quality check score <0.04 was considered high-quality DNA.
Next-generation sequencing
Next-generation sequencing (NGS) was performed using an amplicon-based glioma-tailored gene panel, with 2244 primers for the regions of interest (161179 bp) and an average exon coverage of 99.95%, as described previously.9) Amplicon sequences were aligned to the human reference genome GRCh37 (hg19) in the target region of the sequence. Data were analyzed using the QIAGEN Web Portal service (https://www.qiagen.com/us/shop/genes-and-pathways/data-analysis-center-overview-page/) and Mitsubishi Space Software (Amagasaki, Hyogo, Japan, https://www.mss.co.jp/business/life-science/).
Data analysis
We used OncoPrinter (cbioportal.org/oncoprinter) and MutationMapper (cbioportal.org/mutation_mapper), which are tools in the cBioPortal for Cancer Genomics, to visualize and analyze our data.10,11) Cluster analysis was performed based on the Euclidean distance with the vegan package and Ward. D2 linkage with the ComplexHeatmap package. This analysis was performed using R open-source statistical computing language (v3.5.3) and the integrated development environment RStudio (v0.99.484) as well as the R packages nmf (v0.20.6), mass (v7.3-51.5), and stats (v3.2.2). Statistical analysis was performed with GraphPad Prism software (version 9.2.0). A difference was considered statistically significant at p < 0.05.
Results
Genetic features of diffuse gliomas
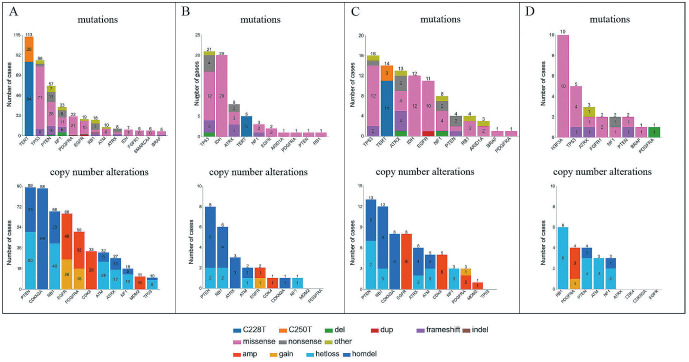
Among the 185 GBM cases, the average age of the patients was 63.5 years. IDH1 mutation was detected in 7 out of 185 cases (4%) (Fig. 1). No mutations and copy number alterations were detected in 4 (2.2%) cases of GBMs. The most commonly mutated genes were TERTp (61%), TP53 (46%), PTEN (31%), and NF1 (18%) (Fig. 2A), and PDGFRA (12%) was the most frequently mutated tyrosine kinase receptor in GBM, followed by EGFR (10%) (Fig. 1). The genes that most commonly showed evidence of a hemizygous or homozygous deletion were PTEN (48%), CDKN2A/B (48%), and RB1 (37%), whereas the most common genes showing evidence of gain/amplification were EGFR (gain, 14%; amplification, 22%), PDGFRA (gain, 10%; amplification, 17%), and CDK4 (amplification, 18%) (Fig. 2A). The representative mutual exclusivity was observed in the pairs of PDGFRA and TERTp (p < 0.001), PDGFRA and EGFR (p < 0.001), and CDKN2A and CDK4 (p < 0.001). The representative co-occurrence was observed in the pairs of ATM and RB1 (p < 0.001), ATM and ATRX (p < 0.001), CDK4 and MDM2 (p < 0.001), TERTp and PTEN (p < 0.001), and TERT and EGFR (p < 0.001). Patient clinical information and molecular status are listed in Supplementary Table S1.
Fig. 1.
Somatic alterations of driver genes in glioblastoma. Driver gene mutations and copy number alterations were generated and visualized using OncoPrinter, which is included in the cBioPortal for Cancer Genomics software suite.
Fig. 2.
Frequency of gene mutations (upper) and copy number alterations (lower) of glioblastoma (A), diffuse astrocytoma (B), anaplastic astrocytoma (C), and diffuse midline glioma (D).
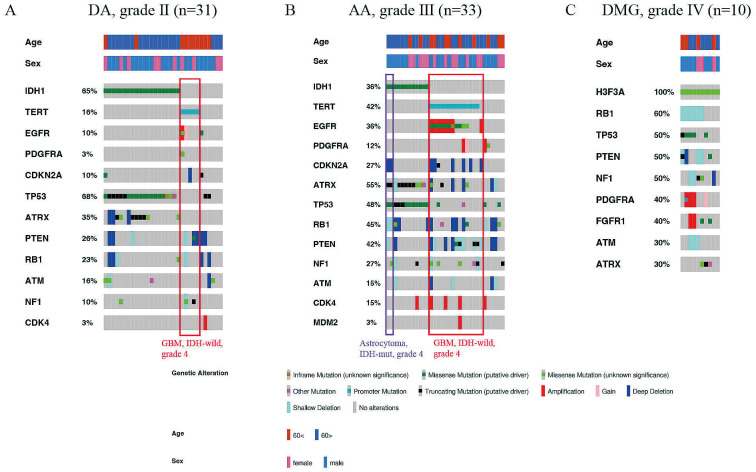
Among 31 diffuse astrocytoma (DA) cases, the average age of the patients was 47.5 years. The most commonly mutated genes were TP53 (68%), IDH1 (65%), and ATRX (26%) (Fig. 2B). The average age of patients with DA carrying IDH1 mutations was 40.3 years, whereas that of patients with DA without mutations averaged 60.5 years (p = 0.008). The genes that most commonly showed evidence of a hemizygous or homozygous deletion were PTEN (26%), RB1 (19%), and ATRX (10%) (Fig. 2B). There were no cases of IDH-mutant DA with CDKN2A/B homozygous deletion (Fig. 3A). TERTp mutation was detected in 5 out of 31 cases (16%) (Fig. 3A). One patient had EGFR amplification, and one had EGFR gain/PTEN loss, which was the surrogated marker of the combined whole chromosome 7 gain and whole chromosome 10 loss, both associated with TERTp mutations (Fig. 3A). DA lacking IDH1 and TERTp mutations showed TP53 mutations, PTEN hemizygous or homozygous deletions, and RB1 hemizygous or homozygous deletions (Fig. 3A). Patient clinical information and molecular status are listed in Supplementary Table S2.
Fig. 3.
Somatic alterations of driver genes in diffuse astrocytoma (A), anaplastic astrocytoma (B), and diffuse midline glioma (C). Driver gene mutations and copy number alterations were generated and visualized using OncoPrinter, which is included in the cBioPortal for Cancer Genomics software suite.
Among 33 anaplastic astrocytoma (AA) cases, the average age of the patients was 50.7 years. The most commonly mutated genes were TP53 (48%), TERTp (42%), and ATRX (39%) (Fig. 2C). The incidence of TERTp and EGFR mutations in AA was higher than that in DA (Fig. 3A, B). The genes that most commonly showed evidence of a hemizygous or homozygous deletion were PTEN (39%), RB1 (36%), and CDKN2A/B (24%), whereas the genes that most commonly showed evidence of amplification were EGFR (24%) and CDK4 (15%) (Fig. 2C). IDH1 mutation was detected in 12 out of 33 cases (36%) (Fig. 3B). The average age of patients with AA carrying IDH1 mutations was 40.6 years, whereas that of patients with AA without mutations averaged 56.5 years (p = 0.003). In IDH-mutant AA, two cases showed CDKN2A/B homozygous deletion (Fig. 3B). TERTp mutation was detected in 14 out of 33 cases (42%), EGFR amplification was detected in 8 out of 33 cases (24%), and EGFR gain/PTEN loss was not detected (Fig. 3B). The mutual exclusivity was observed in the pairs of IDH1 mutation and TERTp mutation. AA lacking IDH1 and TERTp mutations and EGFR amplification showed PDGFRA alterations and a lack of CDKN2A/B homozygous deletion (Fig. 3B). Patient clinical information and molecular status are listed in Supplementary Table S3.
Among 10 diffuse midline glioma (DMG) cases, the average age of the patients was 47.5 years. The most commonly mutated genes were H3F3A (100%), TP53 (50%), and ATRX (30%), whereas no TERTp mutations and EGFR alterations were detected (Fig. 2D). The genes that most commonly showed evidence of a hemizygous or homozygous deletion were RB1 (60%), PTEN (40%), ATM (30%), and NF1 (30%), whereas the gene that most commonly showed evidence of gain/amplification was PDGFRA (gain, 10%; amplification, 30%) (Fig. 2D). The incidence of PDGFRA and FGFR1 alterations in DMG was higher than that in other astrocytic tumors (Fig. 3C).
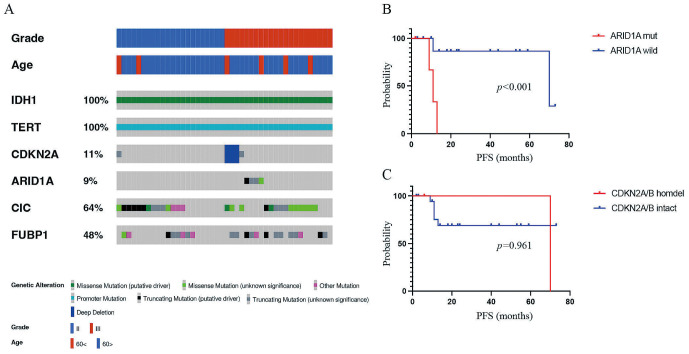
Among 44 oligodendroglial tumor cases (grade II, 22cases; grade III, 22 cases), the average age of the patients was 47.4 years. All patients had IDH1 mutations, 1p/19q codeletion, and TERTp mutations, and CIC was frequently mutated in oligodendroglial tumor, followed by FUBP1 (Fig. 4A). In 22 grade III oligodendroglial tumors, 3 (14%) patients had CDKN2A/B homozygous deletion, and 4 (18%) patients had ARID1A mutation (Fig. 4A). In 22 grade III oligodendroglial tumors, an ARID1A mutation was associated with worse progression-free survival (PFS) (Fig. 4B), whereas a CDKN2A/B homozygous deletion was not associated with PFS (Fig. 4C). Patient clinical information and molecular status are listed in Supplementary Table S4.
Fig. 4.
Somatic alterations of driver genes in oligodendroglial tumors. (A) Driver gene mutations and copy number alterations were generated and visualized using OncoPrinter, which is included in the cBioPortal for Cancer Genomics software suite. (B) Kaplan–Meier analysis of progression-free survival (PFS) of patients with grade III oligodendroglial tumors with and without ARID1A mutations. (C) Kaplan–Meier analysis of PFS of patients with grade III oligodendroglial tumors with and without CDKN2A/B homozygous deletions.
Mutation distributions of EGFR and PDGFRA in diffuse gliomas
Nineteen GBM cases, 11 AA cases, and 2 DA cases had EGFR mutations, and the frequency of EGFR mutations in AA (33%) was higher those that in GBM (10%) and DA (6%) (Fig. 3A-C). Five cases had two EGFR point mutations, and one case had three EGFR point mutations. We also identified mutations in the EGFR kinase domain in 19% of the sites (7/36 mutation sites) (Supplementary Table S5). EGFRA289D/T/V was the most common missense mutation, followed by EGFRR108K (Supplementary Table S5). On the other hand, 22 GBM cases, 1 AA case, 1 DA case, and 1 DMG case had PDGFRA mutations, and the frequency of PDGFRA mutations in GBM (12%) was higher than those in other astrocytic tumors (Fig. 2). Five cases had two PDGFRA point mutations, and one case had three PDGFRA point mutations. We also identified mutations in the PDGFRA kinase domain in 23% of the sites (7/31 mutation sites) (Supplementary Table S6). PDGFRAN468S was the most common missense mutation (Supplementary Table S6).
Reclassification of the retrospective cohort based on the WHO 2021 classification
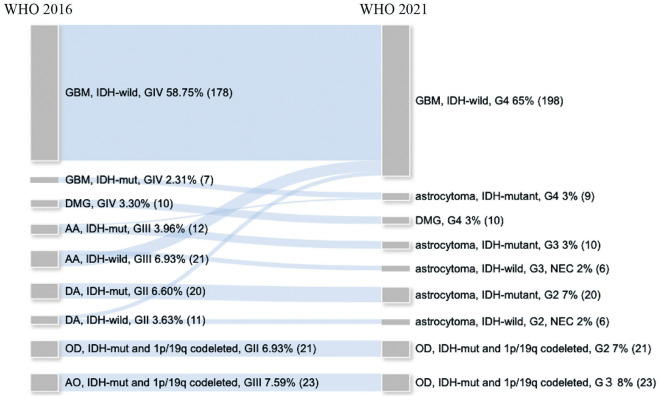
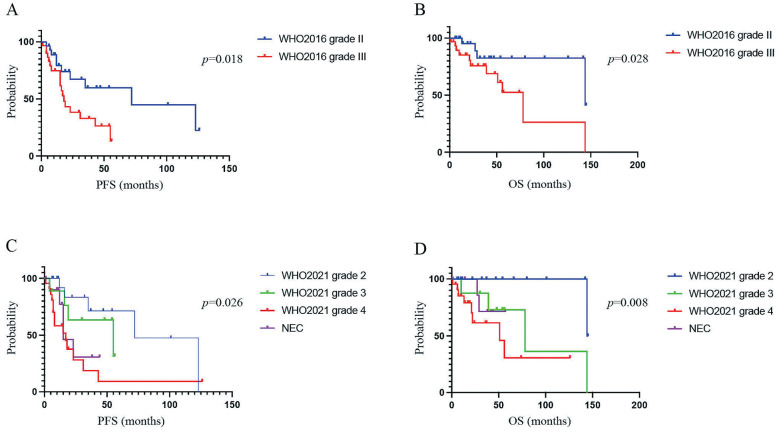
The analysis of the NGS data resulted in successful reclassification of all 303 diffuse gliomas according to the WHO 2021 classification. The reclassification resulted in a marked increase in the number of patients with IDH-wildtype GBMs (from 178 to 198 patients) (Fig. 5). No changes in patient numbers after reclassification were seen for WHO grade II and III oligodendroglial tumors and WHO grade IV DMGs (Fig. 5). Among the seven IDH-mutant GBMs, all cases were classified as IDH-mutant astrocytoma of WHO grade 4 (Fig. 5). Moreover, among the IDH-wildtype DA, 5 (45%) were classified into IDH-wildtype GBM, WHO grade 4, and 6 (55%) were classified into the not elsewhere classified (NEC) category (Fig. 5). Among the IDH-wildtype AAs, 15 (71%) were classified into IDH-wildtype GBM, WHO grade 4, and 6 (29%) were classified into the NEC category (Fig. 5). Among DAs and AAs, both PFS and overall survival (OS) were stratified by grade based on the WHO 2016 classification (Fig. 6A, B), whereas for the WHO 2021 classification, both PFS and OS were stratified more precisely by grade (Fig. 6C, D).
Fig. 5.
Results of the molecular reclassification of 303 diffuse gliomas from the retrospective cohort according to the World Health Organization (WHO) 2021 classification. The diagram shows the diagnostic change between the diagnosis based on the WHO 2016 classification (left) and the diagnosis based on the WHO 2021 classification (right).
Fig. 6.
Kaplan–Meier analysis of progression-free survival (PFS) and overall survival (OS) according to the World Health Organization (WHO) 2016 and 2021 classification based on patients with grade II and III astrocytic tumors. (A) Kaplan–Meier analysis of PFS according to the WHO 2016 classification for patients with grade II and III astrocytic tumors. (B) Kaplan–Meier analysis of OS according to the WHO 2016 classification for patients with grade II and III astrocytic tumors. (C) Kaplan–Meier analysis of PFS according to the WHO 2021 classification for patients with grade II and III astrocytic tumors. (D) Kaplan–Meier analysis of OS according to the WHO 2021 classification for patients with grade II and III astrocytic tumors.
Unsupervised hierarchical cluster analysis of IDH-wildtype GBM, grade 4
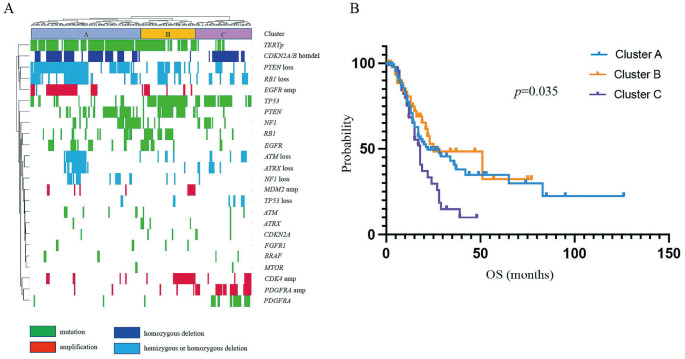
We next performed an unsupervised hierarchical cluster analysis of the 198 IDH-wildtype GBMs classified according to the WHO 2021 classification. This analysis revealed three major distinct groups of IDH-wildtype GBMs. One major cluster (Cluster A) was characterized by mutations in TERTp and EGFR, the amplification of EGFR, the homozygous deletion of CDKN2A/B, the hemizygous or homozygous deletion of PTEN, and a lack of TP53 mutations (Fig. 7A). A second major cluster (Cluster B) was classified by mutations in TERTp, PTEN, and TP53, as well as a lack of CDKN2A/B homozygous deletions (Fig. 7A). The third major cluster (Cluster C) was characterized by mutations in TP53 and PDGFRA, the amplification of PDGFRA, the homozygous deletion of CDKN2A/B, and a lack of TERTp and PTEN mutations (Fig. 7A). The OS was significantly shorter for Cluster C than for Clusters A and B (p = 0.035; Fig. 7B).
Fig. 7.
Unsupervised hierarchical clustering analysis. (A) Results of the unsupervised hierarchical clustering analysis of the 198 IDH-wildtype glioblastomas based on the World Health Organization (WHO) 2021 classification. (B) Survival analysis comparing Clusters A, B, and C by unsupervised hierarchical clustering analysis of the 198 IDH-wildtype glioblastomas.
Discussion
Here, we revealed the molecular genetic profile in Japanese patients with diffuse gliomas and two major distinct groups of IDH-wildtype GBMs using a glioma-tailored gene panel. Moreover, we successfully reclassified the diffuse gliomas according to the WHO 2021 classification.
One of the incorporated markers for the classification of diffuse gliomas is the TERT promoter mutation. Recent reports have indicated that 70%-80% of GBM genomes harbor TERTp mutations.12-14) On the contrary, lower frequencies were reported in the Japanese groups that were studied.5,15-17) In our cohort, 61% of GBM showed mutations in TERTp, suggesting that TERTp mutations may be less frequent in Japan than in other countries. The reason for this discrepancy in the frequency of TERTp mutation remains unknown. Another notable finding was that 22% of GBMs showed EGFR amplification. Lower EGFR amplification rates in patients with GBM from Asia were recently reported during a screening for the INTELLANCE1 and INTELLANCE2 randomized GBM trials as compared to that in other regions.18) Moreover, lower frequencies of EGFR amplifications were reported in the Japanese groups as compared to that in other populations, which was compatible with our results.16,17,19) Previous studies have reported a co-expression association between TERTp mutations and EGFR amplification,16,17) and the reason why the frequency of both TERTp and EGFR amplification is lower in the Japanese group than in other countries may be due to racial differences. Moreover, in our study, EGFRA289D/T/V was the most common missense mutation, which was comparable with the results of a previous study.2) A previous report indicated increased tumor invasion associated with EGFRA289D/T/V and revealed a significant reduction in the OS of patients with tumors harboring this variant, making it a potential therapeutic target.20)
In addition, we collected more samples than that in our previous study,9) and using hierarchical molecular classification of IDH-wildtype GBM, we revealed three distinct major groups. Notably, Cluster C was characterized by mutations in PDGFRA, the amplification of PDGFRA, and a lack of TERTp mutations and EGFR alterations. The OS was significantly shorter for Cluster C than for Clusters A and B, which was similar to the findings of our previous study.9) TERTp and EGFR are important markers for molecular diagnosis in the WHO 2021 classification and have been reported to be associated with prognosis.4,6,21) In addition, our results suggested that PDGFRA is an important driver gene in the molecular classification of GBM. PDGFRA mutations were associated with high proliferative activity via the platelet-derived growth factor receptor α and the cyclin-dependent kinase (CDK) 4/CDK6-cyclin D1 signaling pathways in a ligand-independent manner.22) In particular, PDGFRAN468S confers high sensitivity to multi-kinase inhibitors, receptor tyrosine kinase inhibitors, and CDK4/CDK6 inhibitors, making it a potential therapeutic target.22) Moreover, compared with that in grade II astrocytomas, grade III astrocytomas had a higher burden of molecular alterations, including TERTp mutation, EGFR alterations, and CDKN2A/B homozygous deletion.
In our study, 14% had CDKN2A/B homozygous deletion in grade III oligodendroglial tumors but not in grade II oligodendroglial tumors. Some studies revealed that the presence of CDKN2A/B homozygous deletion was a strong adverse prognostic factor for PFS and OS in grade III oligodendroglial tumors.23,24) Some studies did not observe any CDKN2A/B homozygous deletion in grade II oligodendroglial tumor, which was compatible with our results,23) whereas other studies reported CDKN2A/B homozygous deletion in some cases of grade II oligodendroglial tumors.25,26) In addition, 18% had ARID1A mutation in grade III oligodendroglial tumors but not in grade II oligodendroglial tumors. In grade III oligodendroglial tumors, an ARID1A mutation was associated with worse PFS. ARID1A functions as a tumor suppressor, wherein the majority of mutations are nonsense or frame-shift, resulting in a loss of protein expression.27) ARID1A mutations were more frequent in oligodendroglial tumors than in astrocytic tumors. Although there is one report of ARID1A mutations in oligodendrogliomas,28) to the best of our knowledge, our study is the first to demonstrate that ARID1A mutations are associated with worse PFS. The ARID1A mutation may be involved in the malignant transformation of oligodendroglial tumors, as it was found only in grade III oligodendrogliomas and not in grade II oligodendroglial tumors.
With the implementation of the WHO 2021 classification, molecular genetic information is an increasingly crucial component of the standard diagnostic work-up of diffuse gliomas.6) Our study found IDH mutations in 65% of patients with DA and 36% of patients with AA, both having a lower frequency of IDH mutations than that in previous reports.29) Based on the WHO 2021 classification, in grade II/III IDH-wildtype astrocytomas, 62.5% were classified into IDH-wildtype GBM, and 37.5% were classified into the NEC category. Fujimoto et al. reported that PDGFRA gain/amplification has a poor prognosis in IDH-wildtype TERTp-wildtype lower grade gliomas, which corresponds to the NEC category in the WHO 2021 classification.30) Therefore, PDGFRA gain/amplification is likely to serve as an additional marker to molecularly define GBM in the NEC category. However, a significant number of grade II/III IDH-wildtype astrocytomas were found to be classified into the NEC category, and further molecular classification of the NEC category is needed.
In summary, our glioma-tailored gene panel of 50 genes was applicable for molecular diagnosis, in accordance with the WHO 2021 classification. In addition, in the reclassification of 303 diffuse glioma cases based on the WHO 2021 classification, we revealed the molecular genetic profile of Japanese patients with diffuse gliomas.
Conflicts of Interest Disclosure
The authors declare that they have no competing interests.
Supplementary Material
References
- 1). Louis DN, Perry A, Reifenberger G, et al. : The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol 131: 803-820, 2016 [DOI] [PubMed] [Google Scholar]
- 2). Brennan CW, Verhaak RGW, McKenna A, et al. : The somatic genomic landscape of glioblastoma. Cell 155: 462-477, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3). Cancer Genome Atlas Research Network : Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455: 1061-1068, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4). Stichel D, Ebrahimi A, Reuss D, et al. : Distribution of EGFR amplification, combined chromosome 7 gain and chromosome 10 loss, and tert promoter mutation in brain tumors and their potential for the reclassification of IDHwt astrocytoma to glioblastoma. Acta Neuropathol 136: 793-803, 2018 [DOI] [PubMed] [Google Scholar]
- 5). Arita H, Yamasaki K, Matsushita Y, et al. : A combination of tert promoter mutation and MGMT methylation status predicts clinically relevant subgroups of newly diagnosed glioblastomas. Acta Neuropathol Commun 4: 79, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6). Louis DN, Perry A, Wesseling P, et al. : The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro-Oncology 23: 1231-1251, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7). Brat DJ, Aldape K, Colman H, et al. : cIMPACT-NOW update 3: recommended diagnostic criteria for “Diffuse astrocytic glioma, IDH-wildtype, with molecular features of glioblastoma, WHO grade IV”. Acta Neuropathol 136: 805-810, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8). Brat DJ, Aldape K, Colman H, et al. : cIMPACT-NOW update 5: recommended grading criteria and terminologies for IDH-mutant astrocytomas. Acta Neuropathol 139: 603-608, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9). Higa N, Akahane T, Yokoyama S, et al. : A tailored next-generation sequencing panel identified distinct subtypes of wildtype IDH and tert promoter glioblastomas. Cancer Sci 111: 3902-3911, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10). Cerami E, Gao J, Dogrusoz U, et al. : The cBio cancer genomics Portal: an open platform for exploring multidimensional cancer genomics data: Figure 1. Cancer Discov 2: 401-404, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11). Gao J, Aksoy BA, Dogrusoz U, et al. : Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6: pl1, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12). Spiegl-Kreinecker S, Lötsch D, Ghanim B, et al. : Prognostic quality of activating tert promoter mutations in glioblastoma: interaction with the rs2853669 polymorphism and patient age at diagnosis. Neuro-Oncology 17: 1231-1240, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13). Gramatzki D, Felsberg J, Hentschel B, et al. : Telomerase reverse transcriptase promoter mutation- and O6-methylguanine DNA methyltransferase promoter methylation-mediated sensitivity to temozolomide in isocitrate dehydrogenase-wild-type glioblastoma: is there a link? Eur J Cancer 147: 84-94, 2021 [DOI] [PubMed] [Google Scholar]
- 14). Nguyen HN, Lie A, Li T, et al. : Human TERT promoter mutation enables survival advantage from MGMT promoter methylation in IDH1 wild-type primary glioblastoma treated by standard chemoradiotherapy. Neuro-Oncol 19: 394-404, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15). Arita H, Ichimura K: Prognostic significance of tert promoter mutations in adult-type diffuse gliomas. Brain Tumor Pathol 39: 121-129, 2022 [DOI] [PubMed] [Google Scholar]
- 16). Kikuchi Z, Shibahara I, Yamaki T, et al. : Tert promoter mutation associated with multifocal phenotype and poor prognosis in patients with IDH wild-type glioblastoma. Neurooncol Adv 2: vdaa114, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17). Umehara T, Arita H, Yoshioka E, et al. : Distribution differences in prognostic copy number alteration profiles in IDH-wild-type glioblastoma cause survival discrepancies across cohorts. Acta Neuropathol Commun 7: 99, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18). Lassman AB, Aldape KD, Ansell PJ, et al. : Epidermal growth factor receptor (EGFR) amplification rates observed in screening patients for randomized trials in glioblastoma. J Neurooncol 144: 205-210, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19). Funakoshi Y, Hata N, Takigawa K, et al. : Clinical significance of CDKN2A homozygous deletion in combination with methylated MGMT status for IDH-wildtype glioblastoma. Cancer Med 10: 3177-3187, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20). Binder ZA, Thorne AH, Bakas S, et al. : Epidermal growth factor receptor extracellular domain mutations in glioblastoma present opportunities for clinical imaging and therapeutic development. Cancer Cell 34: 163-177.e7, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21). Tesileanu CMS, Dirven L, Wijnenga MMJ, et al. : Survival of diffuse astrocytic glioma, IDH1/2 wildtype, with molecular features of glioblastoma, WHO grade IV: a confirmation of the cIMPACT-NOW criteria. Neuro Oncol 22: 515-523, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22). Hamada T, Akahane T, Yokoyama S, et al. : An oncogenic splice variant of PDGFRα in adult glioblastoma as a therapeutic target for selective CDK4/6 inhibitors. Sci Rep 12: 1275, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23). Appay R, Dehais C, Maurage CA, et al. : CDKN2A homozygous deletion is a strong adverse prognosis factor in diffuse malignant IDH-mutant gliomas. Neuro Oncol 21: 1519-1528, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24). Alentorn A, Dehais C, Ducray F, et al. : Allelic loss of 9p21.3 is a prognostic factor in 1p/19q codeleted anaplastic gliomas. Neurology 85: 1325-1331, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25). Reis GF, Pekmezci M, Hansen HM, et al. : CDKN2A loss is associated with shortened overall survival in lower-grade (World Health Organization grades II-III) astrocytomas. J Neuropathol Exp Neurol 74: 442-452, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26). Cancer Genome Atlas Research Network, Brat DJ, Verhaak RG, et al. : Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med 372: 2481-2498, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27). Mullen J, Kato S, Sicklick JK, Kurzrock R: Targeting ARID1A mutations in cancer. Cancer Treat Rev 100: 102287, 2021 [DOI] [PubMed] [Google Scholar]
- 28). Padul V, Epari S, Moiyadi A, Shetty P, Shirsat NV: ETV/Pea3 family transcription factor-encoding genes are overexpressed in CIC-mutant oligodendrogliomas. Genes Chromosomes Cancer 54: 725-733, 2015 [DOI] [PubMed] [Google Scholar]
- 29). Hartmann C, Meyer J, Balss J, et al. : Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol 118: 469-474, 2009 [DOI] [PubMed] [Google Scholar]
- 30). Fujimoto K, Arita H, Satomi K, et al. : Tert promoter mutation status is necessary and sufficient to diagnose IDH-wildtype diffuse astrocytic glioma with molecular features of glioblastoma. Acta Neuropathol 142: 323-338, 2021 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.