Abstract
Recent molecular characterization of various microbial genomes has revealed differences in genome size and coding capacity between obligate symbionts and intracellular pathogens versus free-living organisms. Multiple symbiotic microorganisms have evolved with tsetse fly, the vector of African trypanosomes, over long evolutionary times. Although these symbionts are indispensable for tsetse fecundity, the biochemical and molecular basis of their functional significance is unknown. Here, we report on the genomic aspects of the secondary symbiont Sodalis glossinidius. The genome size of Sodalis is approximately 2 Mb. Its DNA is subject to extensive methylation and based on some of its conserved gene sequences has an A+T content of only 45%, compared to the typically AT-rich genomes of endosymbionts. Sodalis also harbors an extrachromosomal plasmid about 134 kb in size. We used a novel approach to gain insight into Sodalis genomic contents, i.e., hybridizing its DNA to macroarrays developed for Escherichia coli, a closely related enteric bacterium. In this analysis we detected 1,800 orthologous genes, corresponding to about 85% of the Sodalis genome. The Sodalis genome has apparently retained its genes for DNA replication, transcription, translation, transport, and the biosynthesis of amino acids, nucleic acids, vitamins, and cofactors. However, many genes involved in energy metabolism and carbon compound assimilation are apparently missing, which may indicate an adaptation to the energy sources available in the only nutrient of the tsetse host, blood. We present gene arrays as a rapid tool for comparative genomics in the absence of whole genome sequence to advance our understanding of closely related bacteria.
Tsetse flies are important insect vectors that transmit African trypanosomes, the causative agents of sleeping sickness disease in humans and nagana in animals. In addition to the parasites they transmit, tsetses harbor three different symbiotic microorganisms (2). Two of these organisms are members of the Enterobacteriaceae family and live in the gut tissue: the obligate primary symbiont (genus Wigglesworthia) (3, 5) and the secondary symbiont (genus Sodalis) (5, 12, 14). A third symbiont, a member of the Rickettsiaceae family, resides mainly in reproductive tissues and belongs to genus Wolbachia (28). The primary symbiont Wigglesworthia lives within the specialized epithelial cells (bacteriocytes) in the bacteriome tissue in the anterior midgut. Phylogenetic analysis has shown that Wigglesworthia displays concordant evolution with its host species, and its association with the tsetse ancestor is predicted to be about 50 to 80 million years old (11). Conversely, Sodalis is harbored both inter- and intracellularly in the tsetse midgut as well as in muscle, fat body, hemolymph, milk gland, and salivary gland tissues of certain species (12). While Sodalis is present in all tsetse species analyzed, its density in somatic tissues increases with the age of the fly and its prevalence varies in different species (12). Phylogenetic analysis has shown that Sodalis isolates from different tsetse species are almost identical, indicating either horizontal transfer events between tsetse species or recent independent acquisition of the bacterium by each species (11). During its intrauterine life, the tsetse larva receives nutrients along with both gut symbionts from its mother via milk gland secretions (4, 20), while Wolbachia is transmitted transovarially (28).
It has been difficult to study the functional role of the obligate endosymbionts in tsetse, as attempts to eliminate them have resulted in retarded growth of the insect and a decrease in egg production, preventing the aposymbiotic host from reproducing (19, 26, 32). The ability to reproduce, however, could be partially restored when the aposymbiotic flies were given a blood meal supplemented with B-complex vitamins (thiamine, pantothenic acid, pyridoxine, folic acid, and biotin), suggesting that the endosymbionts may play a role in metabolism that involves these compounds (25). While the functional significance of Sodalis is unknown, it has been implicated in the susceptibility of tsetse for trypanosome transmission (34). Unlike obligate symbionts, it has been possible to culture Sodalis in vitro and achieve genetic transformation using the broad-host-range replicon oriV derived from a Pseudomonas aeruginosa plasmid (6, 14, 35). The recombinant Sodalis transformed with the green fluorescent protein marker gene was acquired successfully by the intrauterine progeny when microinjected into the mother's hemolymph. The symbionts were also transmitted to F1 and F2 flies, where they expressed the green fluorescent protein (12). Since Sodalis lives in close proximity to the pathogenic trypanosomes in the tsetse gut, the constitutive expression of foreign antitrypanosomal gene products in Sodalis could provide a unique approach to interfere with trypanosome viability.
Recent characterization of intracellular genomes has shown that they have undergone significant size reductions and presumably loss of gene function. To date, the only mutualistic genome that has been completely sequenced is that of Buchnera, the symbiont of aphids (31). Its genome is about 640 kb, significantly smaller than those of the free-living enteric bacteria such as Escherichia coli (7). In addition, analysis of the genome sequences of intracellular organisms has shown a high A+T bias, with Buchnera being about 65 to 70% A+T rich. Recently, we have shown that the mutualist Wigglesworthia in tsetse also has a reduced genome size of less than 740 kb and a high A+T content (1). Here we report on the genomic characteristics of Sodalis, in particular on its genome size, A+T bias, and overall coding capacity. We determined the size of the Sodalis genome and the large plasmid it harbors by contour-clamped homogeneous electric field (CHEF) gel electrophoresis analysis and evaluated its DNA methylation status. Since the free-living bacterium E. coli is a close relative of Sodalis, we used the gene arrays which contain the 4,290 PCR-amplified open reading frames (ORFs) identified in the sequenced E. coli genome to examine the overall coding capability of Sodalis. We discuss both the size and the nature of the contents of Sodalis genome in the light of the symbiotic life it has established in tsetse and in comparison to those of intracellular obligate bacteria as well as free-living organisms closely related to Sodalis.
MATERIALS AND METHODS
Maintenance of Sodalis culture in vitro.
Sodalis was cultured from tsetse as described previously (6, 35) and maintained in vitro in Mitsuhashi-Maramorosch medium (Sigma, St. Louis, Mo.) supplemented with 5% heat-inactivated fetal bovine serum (American Bioanalytical, Natick, Mass.) at 25°C.
Determination of Sodalis genomic and plasmid DNA size.
Genomic DNA was prepared as described by Charles and Ishikawa (10). Approximately 109 Sodalis cells/ml were embedded in agarose plugs. The plugs were treated overnight in EC solution (6 mM Tris-HCl [pH 7.6], 100 mM EDTA, 1 M NaCl, 0.5% Brij 58, 0.2% deoxycholate, and 0.5% N-lauroylsarcosine in the presence of lysozyme [1 mg/ml] and RNase [20 μg/ml]) at 37°C as described for Buchnera (10). The EC solution was replaced with ESP (0.5 M EDTA [pH 8], 1% N-lauroylsarcosine, 1 mg of proteinase K per ml) and incubated at 50°C for 2 days. These plugs contained both the genomic and plasmid DNAs. To obtain pure chromosomal DNA devoid of plasmids, the plugs were subjected to CHEF gel electrophoresis (Bio-Rad, Hercules, Calif.) using a 150- to 200-s pulse time for 20 h at 200 V. Under these conditions, the plasmid(s) migrates into the gel while intact genomic DNA remains in the plug. Subsequently, the plugs were removed from the wells and incubated overnight with PmeI and PacI at 37°C and with SwaI at 25°C. CHEF gel electrophoresis was performed at 200 V at various ramping pulse and run times, depending on the resolution requirements. Plasmid DNA was prepared by the alkaline extraction protocol (29), further purified on CsCl gradients, digested overnight with EcoRI, HindIII, and PstI at 37°C, and analyzed by CHEF gel electrophoresis at 170 V, using a 2-s pulse time for 12 h.
Sequencing of Sodalis DNA.
Two protein-coding genes in Sodalis, groEL and ftsZ, were PCR amplified using E. coli-specific primers (Genosys Biotechnologies Inc., The Woodlands, Tex.). The amplification products were cloned into pGEM-T vector (Promega) and sequenced at the Keck Sequencing Center at Yale University.
Hybridization to E. coli macroarrays.
Sodalis genomic DNA was separated from plasmids as described above, using CHEF electrophoresis. The agarose plugs were then digested with FseI, and the digested DNA was purified using a QIAquick gel extraction kit (Qiagen Inc. Chatsworth, Calif.). DNA was radioactively labeled with [α-33P]ATP by using a polymerase I/DNase I nick translation kit (GIBCO catalog no. 18160-010). Panorama macroarrays (Genosys Biotechnologies) were prehybridized and hybridized in a 45% formamide–5× Denhardt's solution–5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.5% sodium dodecyl sulfate (SDS) buffer at 45°C. The arrays were washed at 42°C in 2× SSC–0.1% SDS and 0.1× SSC–0.1% SDS followed by 0.1× SSC–0.5% SDS. Arrays were exposed to maximum-resolution films (BMR; Eastman Kodak Company, Rochester, N.Y.), and signals were scored as strong (53%), medium (44%), or weak (3%). There were no cases where duplicate spots gave contradictory results.
Analysis of DNA modifications associated with chromosomal and plasmid DNAs.
Total Sodalis DNA was purified according to standard protocols, using proteinase K (100 μg/ml) and SDS (1%). The plasmid DNA was purified via ultracentrifugation on CsCl gradients. All purified DNAs were digested overnight with EcoRII, Sau3AI, and MboI at 37°C and with BstNI at 60°C, respectively. The digestion products were analyzed by conventional agarose gel electrophoresis.
Nucleotide sequence accession numbers.
The GenBank accession numbers are AF326971 for groEL and AY024353 for ftsZ.
RESULTS
Size of Sodalis genome and plasmid(s).
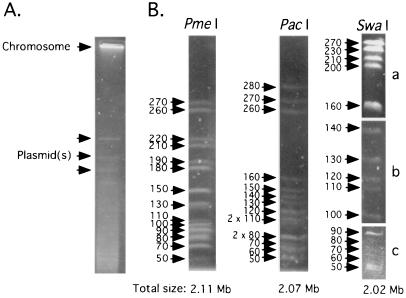
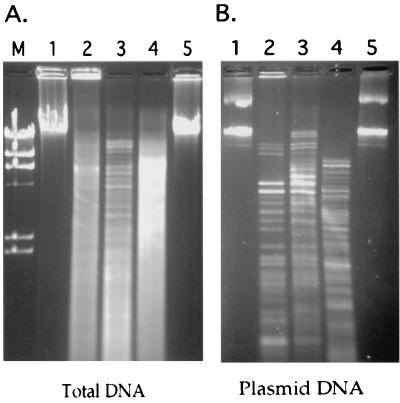
Since Sodalis contains multicopy extrachromosomal DNAs, agarose plugs containing total bacterial DNA were subjected to an initial CHEF electrophoresis that allowed the plasmid DNA to enter the gel while the intact chromosomal DNA remained in the wells (Fig. 1A). Subsequently, the plugs were removed from the wells, and chromosomal DNA was digested with one of the restriction enzymes PmeI, PacI, and SwaI. The restriction fragments were analyzed by CHEF electrophoresis at different pulse times to achieve resolution of desired size ranges (Fig. 1B). The sizes of all restriction fragments were determined and compiled to obtain the total size of Sodalis chromosome, which was found to be approximately 2.11, 2.07, and 2.02 Mb by PmeI, PacI, and SwaI digestions, respectively.
FIG. 1.
Sodalis genome size determination. (A) Bacterial DNA samples embedded in agarose plugs were subjected to CHEF electrophoresis using pulse times of 150 to 200 s for 20 h at 200 V and 14°C to obtain chromosomal DNA devoid of plasmids. Sizes on the left are indicated in kilobases. (B) Sodalis genomic DNA devoid of its plasmid was analyzed by CHEF gel electrophoresis. PmeI and PacI fragments were resolved at pulse times of 18.3 to 26.3 s over 35 h at 200 V. Three different pulse times were used to resolve the SwaI fragments in different size ranges: 18.3 to 26.3 s for 35 h (a), 6.8 to 12.9 s for 33 h (b), and 1 to 6 s and 6 to 15 s for 15 h each (c).
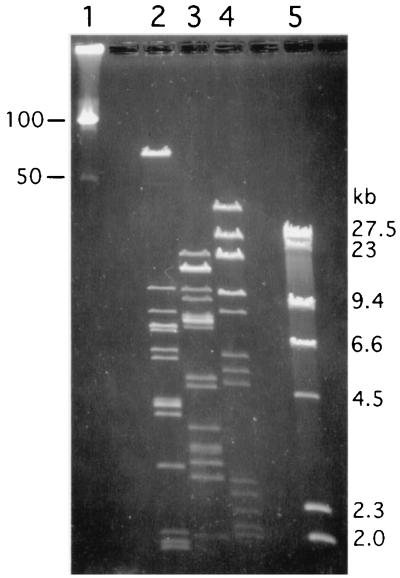
The total sizes of the generated plasmid DNA restriction fragments analyzed by CHEF elecrophoresis indicated the plasmid size to be about 134 kb (Fig. 2). Based on the intensity of the DNA fragments after staining with ethidium bromide, two fragments were consistently observed to be less abundant. Hence, Sodalis may contain at least one additional plasmid around 10 kb in size that is present in fewer copies (data not shown).
FIG. 2.
Sodalis plasmid size determination. Plasmid DNA fragments were resolved by CHEF gel electrophoresis at 2-s pulse time for 12 h at 170 V. Lanes 1 and 5, molecular weight markers (lambda ladder and lambda/HindIII, respectively); lanes 2 to 4, purified Sodalis plasmid DNA digested with restriction enzymes EcoRI, HindIII, and PstI, respectively.
A+T content of Sodalis genome.
We analyzed the coding sequences for two conserved genes, groEL and ftsZ, to examine the A+T content of the Sodalis genome. Both gene sequences have been extensively studied in other bacteria and hence can be used in comparative analysis with related organisms. Analysis of the groEL gene from Sodalis has shown that it is 44% A+T, while the ftsZ gene was found to be 41% A+T rich. The same loci characterized from E. coli are 47 and 46% A+T, respectively. In comparison, the groEL sequences characterized from the strict intracellular symbionts Wigglesworthia and Buchnera are 63% A+T in both organisms (GenBank accession no. AF321516 and AP001118, respectively). The ftsZ sequences from Wigglesworthia and Buchnera were similarly high in A+T content, i.e., 66% (GenBank accession no. AY024354 and AF012886, respectively).
Genome contents of Sodalis inferred from E. coli macroarray hybridizations.
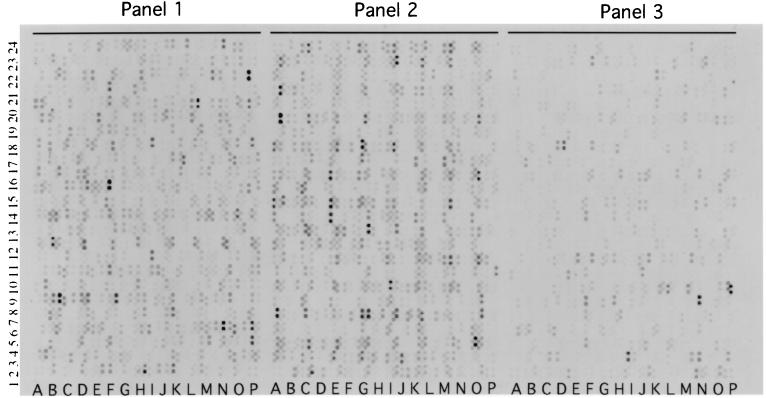
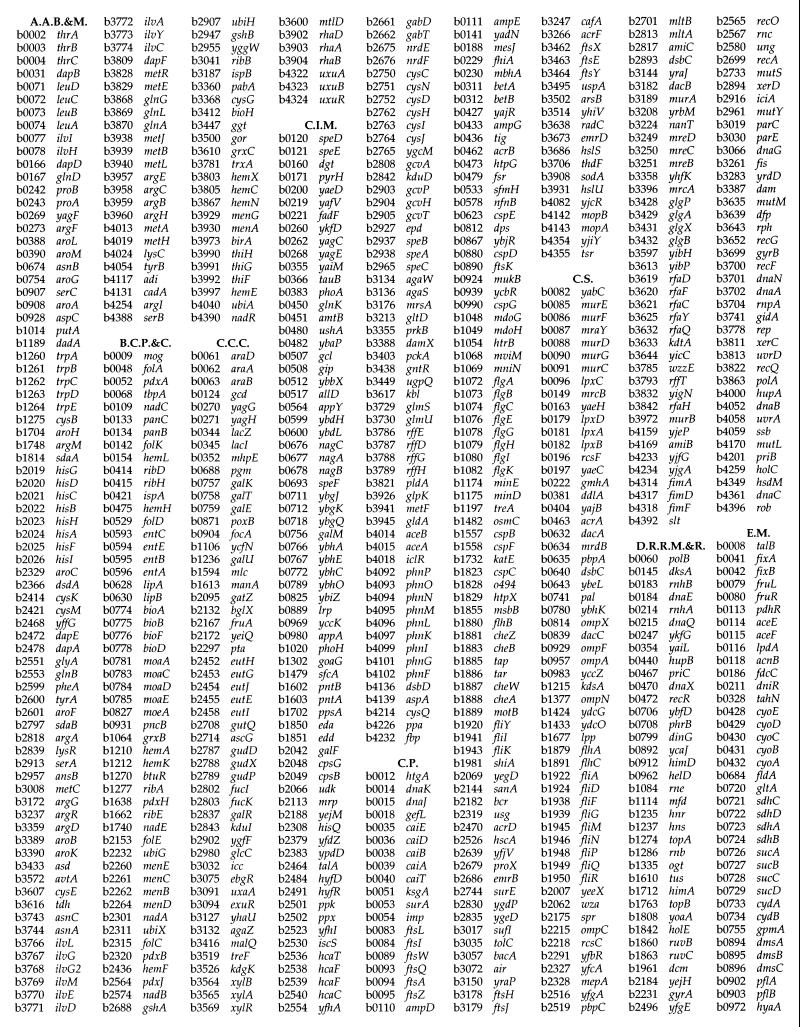
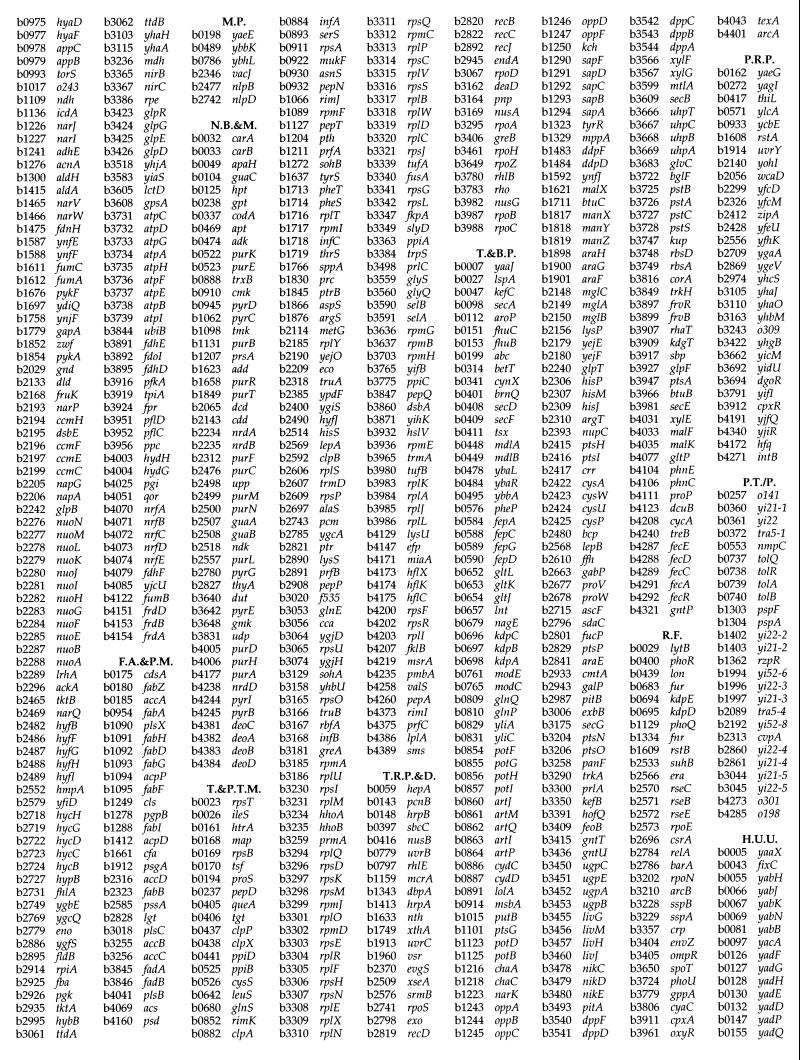
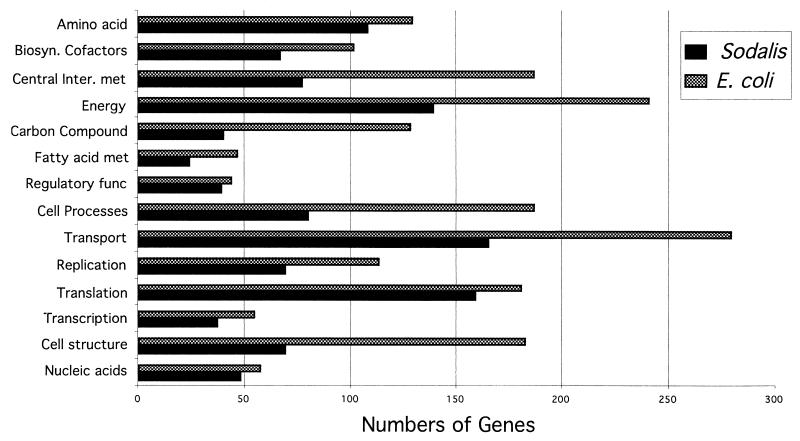
Hybridization of Sodalis genomic DNA devoid of plasmids to E. coli macroarrays revealed the presence of 1,800 orthologs (Fig. 3) which represent about 85% of the Sodalis genome, assuming an average size of 1 kb per gene (31). There are 4,290 ORFs represented on the E. coli array, and functional roles have been assigned to 1,938 of these. Of the 1,800 genes detected from Sodalis, 1,158 had functional roles assigned in E. coli, while the remaining 642 genes detected corresponded to genes with hypothetical functions (Fig. 4). Orthologs were grouped according to their known functions, and the number of genes in each group was compared to those present in the E. coli genome (Fig. 5). Although the Sodalis genome is about half the size of that of E. coli, this comparative analysis has revealed that it contains a high proportion of the genes for amino acid biosynthesis, regulatory functions, translation, transcription, and nucleic acid biosynthesis. Almost all of the genes necessary to synthesize each amino acid and for the de novo synthesis of nucleic acids could be detected in Sodalis via array hybridization. We were able to detect a complete set of genes involved in many metabolic pathways such as those associated with amino acid biosynthesis (e.g., trpABCDE for tryptophan, hisABCDFGHI for histidine, and thrABC, metL, lysC, and asd for threonine biosynthesis) and the tricarboxylic acid cycle (sdhABCD, sucABCD, fumABC, acnAB, gltA, icdA, and mdh) in addition to all of the genes coding for ribosomal subunit proteins, further validating the results of the orthologous array analysis (Fig. 4). Many genes involved in the biosynthesis of cofactors, replication, and transport functions were also found to be present. Most of the DNA repair and recombinase orthologs of E. coli involved in direct damage reversal, base excision repair, mismatch repair, recombinase pathways, and nucleotide excision repair were found to be retained. However, genes involved in carbon compound catabolism, central intermediary metabolism, fatty acid phospholipid metabolism, cell processes, and cell structure were fewer in numbers in comparison to the E. coli genome. Based on hybridization analysis, Sodalis appears to have respiratory oxidases, NADH dehydrogenase complex enzymes and a complete tricarboxylic acid cycle. It has the capability to grow on several sugars including galactose, fructose, and raffinose as well as the amino sugars N-acetyl-d-glucosamine, the methylpentoses l-fucose, l-rhamnose, l-arabinose, and xylose. Sodalis appears to have the ability to convert fatty acids to acetyl coenzyme A using the glyoxylate cycle enzymes. Twenty-six genes detected in Sodalis were grouped as phage/transposon or plasmid-like sequences in E. coli.
FIG. 3.
E. coli gene array hybridization analysis of Sodalis DNA. The autoradiogram shows the 1,800 signals detected by hybridization of Sodalis chromosomal DNA to Panorama macroarrays containing 4,290 E. coli ORFs. Each gene is spotted in duplicate over three panels.
FIG. 4.
List of genes detected in Sodalis by E. coli array hybridization analysis. A.A.B.&M., amino acid biosynthesis and metabolism; B.C.P.&C., biosynthesis of cofactors, prosthetic groups, and carriers; C.C.C., carbon compound catabolism; C.I.M., central intermediary metabolism; C.P., cell processes; C.S., cell structure; D.R.R.M.&R., DNA replication, recombination, modification, and repair; E.M., energy metabolism; F.A.&P.M., fatty acid and phospholipid metabolism; M.P., membrane proteins; N.B.&M., nucleotide biosynthesis and metabolism; T.&P.T.M., translation and posttranslational modification; T.R.P.&D., transcription, RNA processing, and degradation; T.&B.P., transport and binding proteins, R.F., regulatory function; P.R.P., putative regulatory proteins; P.T./P., phage, transposon, or plasmid; H.U.U., hypothetical, unclassified, unknown.
FIG. 5.
Numbers of genes in different functional categories in the known genome of E. coli compared to the numbers of putative genes detected in Sodalis on the basis of gene array analysis.
The array analysis was also repeated with purified Sodalis plasmid DNA (data not shown). Thirty-six genes were detected, with none corresponding to the genes detected with Sodalis chromosomal DNA, indicating that the genes reported in Fig. 4 are indeed of chromosomal origin. Among the genes detected were those coding for a membrane usher protein (yraJ) and an RNA helicase (dbpA). The remaining genes either were hypothetical with no known functions in E. coli or corresponded to phage/transposon-like sequences.
DNA methylation in Sodalis.
Of interest were two genes detected by array hybridization analysis, coding for DNA adenine (Dam) and cytosine (Dcm) methylase. DNA methylation in bacteria is thought to be involved in protection against foreign DNA in addition to regulatory functions for gene expression and replication. The functional presence of these genes was investigated by DNA restriction analysis using isoschizomers with different methylation requirements. Two pairs of isoschizomers that are diagnostic for Dcm (BstNI and EcoRII) and Dam (Sau3AI and MboI) methylation status of DNA were used to digest total chromosomal and plasmid DNA preparations (Fig. 6). Neither the plasmid nor the chromosomal DNA could be digested with Dam-sensitive restriction enzyme MboI (Fig. 6, lanes 5), while the same DNAs were cleaved with its isoschizomer Sau3AI (Fig. 6, lanes 4), indicating that Sodalis genomic as well as plasmid DNAs are extensively methylated at the adenine residues. Under the same digestion conditions, Wigglesworthia DNA could be completely digested with MboI (data not shown). Although both total and plasmid DNAs could be digested with BstNI (Fig. 6, lanes 2) and EcoRII (Fig. 6, lanes 3), we observed a difference in the plasmid digestion fragments, suggesting that this DNA may be hemimethylated at cytosine residues (Fig. 6B, lane 2 versus lane 3).
FIG. 6.
Methylation status of Sodalis DNA. Two pairs of isoschizomers that are diagnostic for Dcm (BstNI and EcoRII) and Dam (Sau3AI and MboI) methylation status of DNA were used to digest total (A) and plasmid (B) DNA preparations. (A) M, lambda/HindIII molecular weight marker; lane 1, Sodalis total DNA uncut; lanes 2 to 5, Sodalis total DNA digested with BstNI, EcoRII, Sau3AI, and MboI, respectively. (B) Lane 1, Sodalis plasmid DNA uncut; lanes 2 to 5, Sodalis plasmid DNA digested with BstNI, EcoRII, Sau3AI, and MboI, respectively.
DISCUSSION
Symbiotic associations with microorganisms are common in insects and form a continuum from obligate relationships required for host nutrition and fecundity to parasitic infections with selfish organisms which manipulate host physiology for their own benefit. The genome analysis of mutualists and intracellular pathogens has shown several hallmarks such as reduced genome size, increased A+T bias in coding sequences, and faster polypeptide evolution (21). We studied the genomic aspects of the secondary symbiont of tsetse, Sodalis, to better understand the functional nature of its symbiotic association with tsetse.
Genome size reductions have been observed for intracellular pathogens such as Chlamydia trachomatis (1.04 Mb), Treponema pallidum (1.14 Mb), Mycoplasma genitalium (0.58 kb), and Rickettsia prowazekii (1.1 Mb) (22). Recently, the genome of the obligate endosymbiont Buchnera from aphids has been characterized as 640 kb (10, 31), and the genome of the obligate Wigglesworthia from tsetse is found to be smaller than 750 kb (1), both apparently approaching the size of that of M. genitalium, the smallest bacterial genome reported thus far. Both Buchnera and Wigglesworthia are intracellular and live within specialized insect cells (bacteriocytes) which make up a defined organ (bacteriome). It has not been possible to culture either organism in vitro. The genome reductions imply genetic and presumably functional loss and may reflect the increased exploitation and dependence of these organisms on their host cells, unlike free-living organisms. In contrast, free-living bacteria such as E. coli and Salmonella have been found to have significantly larger genomes, around 4.5 Mb. The genome size of Sodalis is shown here to be about 2 Mb, significantly larger than those of the intracellular pathogens and obligate symbionts but smaller than those of the closely related free-living enterics. Genome-wide sequence analysis is necessary to understand the full spectrum of genes that have been lost from the enteric ancestor during symbiosis or to identify genes that may have been since acquired to mediate its symbiotic association. In the absence of this information, however, hybridization of its DNA to macroarrays of a closely related microorganism, E. coli, has provided rapid insight into its genome composition. While E. coli arrays have been useful for documenting gene inventories in different strains (27), data presented here show a different application which can provide a cost-effective and fast alternative to genome sequencing for broad comparative analysis of closely related organisms. The future availability of gene arrays from distant organisms and similar applications stand to improve the efficacy of this approach.
Based on its genomic composition revealed by array analysis, Sodalis has many of the capabilities of free-living bacteria. In fact, establishment of an in vitro culture for this organism supports the notion that it can synthesize all of the metabolites it needs for survival outside of host insect cells (6, 35). It appears to have retained many genes involved in transcription, translation, regulation, and nucleic acid and amino acid biosynthetic pathways. Meanwhile, Sodalis might have lost genes in carbon compound catabolism, central intermediary metabolism, and fatty acid phospholipid metabolism. While the absence of certain genes and pathways will need to be confirmed by complete genome sequencing, our findings represent an adaptation by Sodalis to its energy-rich environment, the single diet of tsetse, blood. Under in vitro conditions, Sodalis has been found to assimilate N-acetyl-d-glucosamine and raffinose (14). The symbionts of blood-feeding insects are thought to provide cofactors and vitamin metabolites to supplement the restricted diets of their host insects (8). Many genes involved in the biosynthesis of cofactors and vitamins were detected in Sodalis. Thus, Sodalis might indeed benefit its tsetse host via the synthesis of compounds such as biotin and lipoic acid, molybdenum cofactor, thiamine, riboflavin, and folic acid. In a similar study with Wigglesworthia, we have applied the E. coli arrays to understand the general aspects of its much reduced genome contents and found that it too has maintained many of the biosynthetic pathways for vitamin and cofactor synthesis, possibly indicating their significance for host tsetse biology (1). While this study provides a general understanding of the genomic coding capacity of Sodalis, it lacks information on loci not represented in the E. coli genome. There are at least two such examples; the first is a chitinase gene characterized from Sodalis that is absent in the E. coli genome (34), and the second is the recently described pathogenicity island genes, which may help Sodalis invade insect cells (15).
The overall A+T contents of the genomes of intracellular pathogens R. prowazekii and M. genitalium are 71 and 68%, respectively. Similarly, the genomes of mutualists are also A+T rich; for example, that of Buchnera was found to be 75% A+T (31). Genome analysis of intracellular pathogens and obligates indicate that loci encoding for DNA repair and recombination functions have been lost or limited in many of these organisms (22), and this loss of the repair functions may have led to their high A+T bias. In contrast, the genome of the free-living bacterium E. coli does not exhibit such a bias, and its overall A+T content is about 50%. The A+T content of Sodalis groEL and ftsZ gene sequences is less than 45%, another hallmark of free-living organisms. Unlike genomes of obligate intracellular bacteria, the Sodalis genome appears to have retained almost all of the genes involved in DNA repair and recombination functions.
Phylogenetic characterization of the obligate symbionts from various insects has shown that they display concordance with their host phylogenies including the symbionts from tsetse (5), aphids (23), whiteflies (13), mealybugs (24), and carpenter ants (30). Unlike these obligates, the phylogenetic analysis of the secondary symbionts such as Sodalis from tsetse and the symbionts of psyllids and aphids has shown them to be identical among distant species of each insect taxa (11, 16, 33). Based on 16S rRNA gene analysis, Sodalis forms a distinct lineage with the primary symbiont of the rice weevil Sitophilus oryzae, SOPE (4). Comparative analysis of their groEL sequences indicates 98% identity, indicating that they are close members of one bacterial taxon. The genome size of SOPE is 3 Mb, significantly larger than those of the intracellular obligates (9), and the A+T content of its groEL gene is about 45%, similar to that of Sodalis (17). Like Sodalis, it harbors large extracellular plasmids (17). In contrast to their shared evolutionary and molecular characteristics, the biology of SOPE in its weevil host is different from that of Sodalis. SOPE has been shown to reside within bacteriocytes in the weevil (18), similar to Wigglesworthia in tsetse. Its symbiosis in the weevil host is thought to be obligate in nature, and its elimination has been found to impair many physiological traits of its host, including fecundity (18). In tsetse, it has been difficult to disassociate the functional significance of Wigglesworthia from that of Sodalis since antibiotic treatment of flies eliminates both organisms. However, since the prevalence of Sodalis varies extensively in different tsetse species, its association may be considered commensal in nature (12). The transmission modes of Sodalis and SOPE are also different. SOPE is transovarially transmitted to insect progeny (18), while Sodalis is absent in reproductive tissues but is transmitted vertically to the intrauterine larva through the mother's milk (12, 20). It appears that upon association with the hosts, the common ancestor of SOPE and Sodalis adapted to the distinct functional biologies of the host insects. While SOPE is restricted to an intracellular association in the weevil, Sodalis can replicate in various tissues of tsetse and can replicate outside the host insect cells. It remains to be seen whether the different functional roles they display in their hosts result from host-derived factors or from variations in their genotypes. One precedent for such an association is Wolbachia, a parasitic Rickettsiaceae which has been shown to invade a wide range of insects where it displays many different phenotypes, ranging from reproductive incompatibilities to age-shortening effects. Further genome-wide comparative analysis between the closely related Sodalis and SOPE will undoubtedly shed light on the mechanistic as well as the functional basis of symbiosis in their hosts.
ACKNOWLEDGMENTS
L.A. and R.R. contributed equally to this report.
This work was supported by NIH/NIAID grant AI-34033 to S.A. L.A. is the recipient of a James Hudson Brown-Alexander Brown Coxe fellowship.
REFERENCES
- 1.Akman L, Aksoy S. Escherichia coli gene array analysis provides insight into the biology of the obligate endosymbiont of tsetse flies, Wigglesworthia glossinidia. Proc Natl Acad Sci USA. 2001;98:7546–7551. doi: 10.1073/pnas.131057498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aksoy S. Tsetse: a haven for microorganisms. Parasitol Today. 2000;16:114–119. doi: 10.1016/s0169-4758(99)01606-3. [DOI] [PubMed] [Google Scholar]
- 3.Aksoy S. Wigglesworthia gen. nov. and Wigglesworthia glossinidia sp. nov., taxa consisting of the mycetocyte-associated, primary endosymbionts of tsetse flies. Int J Syst Bacteriol. 1995;45:848–851. doi: 10.1099/00207713-45-4-848. [DOI] [PubMed] [Google Scholar]
- 4.Aksoy S, Chen X, Hypsa V. Phylogeny and potential transmission routes of midgut-associated endosymbionts of tsetse (Diptera:Glossinidae) Insect Mol Biol. 1997;6:183–190. doi: 10.1111/j.1365-2583.1997.tb00086.x. [DOI] [PubMed] [Google Scholar]
- 5.Aksoy S, Pourhosseini A A, Chow A. Mycetome endosymbionts of tsetse flies constitute a distinct lineage related to Enterobacteriaceae. Insect Mol Biol. 1995;4:15–22. doi: 10.1111/j.1365-2583.1995.tb00003.x. [DOI] [PubMed] [Google Scholar]
- 6.Beard C B, O'Neill S L, Mason P, Mandelco L, Woese C R, Tesh R B, Richards F F, Aksoy S. Genetic transformation and phylogeny of bacterial symbionts from tsetse. Insect Mol Biol. 1993;1:123–131. doi: 10.1111/j.1365-2583.1993.tb00113.x. [DOI] [PubMed] [Google Scholar]
- 7.Blattner F, Plunkett G R, Bloch C, Perna N, Burland V, Riley M, Collado-Vides J, Glasner J, Rode C, Mayhew G, Davis G J N, Kirkpatrick H, Goeden M, Rose D, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 8.Buchner P. Endosymbiosis of animals with plant micro-organisms. New York, N.Y: Interscience Publishers Inc.; 1965. pp. 210–338. [Google Scholar]
- 9.Charles H, Condemine G, Nardon C, Nardon P. Genome size characterization of the principal endocellular symbiotic bacteria of the weevil Sitophilus oryzae, using pulse field gel electrophoresis. Insect Biochem Mol Biol. 1997;27:345–350. [Google Scholar]
- 10.Charles H, Ishikawa H. Physical and genetic map of the genome of Buchnera, the primary endosymbiont of the pea aphid Acrythosiphon pisum. J Mol Evol. 1999;48:142–150. doi: 10.1007/pl00006452. [DOI] [PubMed] [Google Scholar]
- 11.Chen X, Song L, Aksoy S. Concordant evolution of a symbiont with its host insect species: molecular phylogeny of genus Glossina and its bacteriome-associated endosymbiont, Wigglesworthia glossinidia. J Mol Evol. 1999;48:49–58. doi: 10.1007/pl00006444. [DOI] [PubMed] [Google Scholar]
- 12.Cheng Q, Aksoy S. Tissue tropism, transmission and expression of foreign genes in vivo in midgut symbionts of tsetse flies. Insect Mol Biol. 1999;8:125–132. doi: 10.1046/j.1365-2583.1999.810125.x. [DOI] [PubMed] [Google Scholar]
- 13.Clark M A, Baumann L, Munson M A, Baumann P, Campbell B C, Duffus J E, Osborne L S, Moran N A. The eubacterial endosymbionts of whiteflies (Homoptera: Aleyrodoidea) constitute a lineage distinct from the endosymbionts of aphids and mealybugs. Curr Microbiol. 1992;25:119–123. [Google Scholar]
- 14.Dale C, Maudlin I. Sodalis gen. nov. and Sodalis glossinidius sp. nov., a microaerophilic secondary endosymbiont of the tsetse fly Glossina morsitans morsitans. Int J Syst Bacteriol. 1999;49:267–275. doi: 10.1099/00207713-49-1-267. [DOI] [PubMed] [Google Scholar]
- 15.Dale C, Young S A, Haydon D T, Welburn S C. The insect endosymbiont Sodalis glossinidius utilizes a type III secretion system for cell invasion. Proc Natl Acad Sci USA. 2001;98:1883–1888. doi: 10.1073/pnas.021450998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fukatsu T, Nikoh N, Kawai R, Koga R. The secondary endosymbiotic bacterium of the pea aphid Acyrthosiphon pisum (Insecta: Homoptera) Appl Environ Microbiol. 2000;66:2748–2758. doi: 10.1128/aem.66.7.2748-2758.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heddi A, Charles H, Khatchadourian C, Bonnot G, Nardon P. Molecular characterization of the principal symbiotic bacteria of the weevil Sitophilus oryzae: a peculiar G+C contents of an endocytobiotic DNA. J Mol Evol. 1998;47:52–61. doi: 10.1007/pl00006362. [DOI] [PubMed] [Google Scholar]
- 18.Heddi A, Grenier A M, Khatchadourian C, Charles H, Nardon P. Four intracellular genomes direct weevil biology: nuclear, mitochondrial, principal endosymbiont, and Wolbachia. Proc Natl Acad Sci USA. 1999;96:6814–6819. doi: 10.1073/pnas.96.12.6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hill P D S, Campbell J A. The production of symbiont-free Glossina morsitans and an associated loss of female fertility. Trans R Soc Trop Med Hyg. 1973;67:727–728. doi: 10.1016/0035-9203(73)90051-5. [DOI] [PubMed] [Google Scholar]
- 20.Ma W-C, Denlinger D L. Secretory discharge and microflora of milk gland in tsetse flies. Nature. 1974;247:301–303. [Google Scholar]
- 21.Moran N, Baumann P. Bacterial endosymbionts in animals. Curr Opin Microbiol. 2000;3:270–275. doi: 10.1016/s1369-5274(00)00088-6. [DOI] [PubMed] [Google Scholar]
- 22.Moran N, Wernegreen J. Lifestyle evolution in symbiotic baceria: insights from genomics. Trends Ecol Evol. 2000;15:321–326. doi: 10.1016/s0169-5347(00)01902-9. [DOI] [PubMed] [Google Scholar]
- 23.Munson M, Baumann P, Kinsey M. Buchnera gen. nov. and Buchnera aphidicola sp. nov., a taxon consisting of the mycetocyte-associated, primary endosymbionts of aphids. Int J Syst Bacteriol. 1991;41:566–568. [Google Scholar]
- 24.Munson M A, Baumann P, Moran N A. Phylogenetic relationships of the endosymbionts of mealybugs (Homoptera: Pseudococcidae) based on 16S rDNA sequences. Mol Phylogenet Evol. 1992;1:26–30. doi: 10.1016/1055-7903(92)90032-c. [DOI] [PubMed] [Google Scholar]
- 25.Nogge G. Significance of symbionts for the maintenance of an optional nutritional state for successful reproduction in hematophagous arthropods. Parasitology. 1981;82:101–104. [Google Scholar]
- 26.Nogge G. Sterility in tsetse flies (Glossina morsitans Westwood) caused by loss of symbionts. Experientia. 1976;32:995–996. doi: 10.1007/BF01933932. [DOI] [PubMed] [Google Scholar]
- 27.Ochman H, Jones I B. Evolutionary dynamics of full genome content in Escherichia coli. EMBO J. 2000;19:6637–6643. doi: 10.1093/emboj/19.24.6637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Neill S L, Gooding R H, Aksoy S. Phylogenetically distant symbiotic microorganisms reside in Glossina midgut and ovary tissues. Med Vet Entomol. 1993;7:377–383. doi: 10.1111/j.1365-2915.1993.tb00709.x. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Schroder D, Deppisch H, Obermayer M, Krohne G, Stackebrandt E, Holldobler B, Goebel W, Gross R. Intracellular endosymbiotic bacteria of Camponotus species (carpenter ants): systematics, evolution and ultrastructural characterization. Mol Microbiol. 1996;21:479–489. doi: 10.1111/j.1365-2958.1996.tb02557.x. [DOI] [PubMed] [Google Scholar]
- 31.Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature. 2000;407:81–86. doi: 10.1038/35024074. [DOI] [PubMed] [Google Scholar]
- 32.Southwood T R, Khalaf S, Sinden R E. The micro-organisms of tsetse flies. Acta Trop. 1975;32:259–266. [PubMed] [Google Scholar]
- 33.Thao M L, Clark M A, Baumann L, Brennan E B, Moran N A, Baumann P. Secondary endosymbionts of psyllids have been acquired multiple times. Curr Microbiol. 2000;41:300–304. doi: 10.1007/s002840010138. [DOI] [PubMed] [Google Scholar]
- 34.Welburn S C, Arnold K, Maudlin I, Gooday G W. Rickettsia-like organisms and chitinase production in relation to transmission of trypanosomes by tsetse flies. Parasitology. 1993;107:141–145. doi: 10.1017/s003118200006724x. [DOI] [PubMed] [Google Scholar]
- 35.Welburn S C, Maudlin I, Ellis D S. In vitro cultivation of rickettsia-like-organisms from Glossina spp. Ann Trop Med Parasitol. 1987;81:331–335. doi: 10.1080/00034983.1987.11812127. [DOI] [PubMed] [Google Scholar]