Abstract
Proteus mirabilis urease catalyzes the hydrolysis of urea to CO2 and NH3, resulting in urinary stone formation in individuals with complicated urinary tract infections. UreR, a member of the AraC family, activates transcription of the genes encoding urease enzyme subunits and accessory proteins, ureDABCEFG, as well as its own transcription in the presence of urea. Based on sequence homology with AraC, we hypothesized that UreR contains both a dimerization domain and a DNA-binding domain. A translational fusion of the leucine zipper dimerization domain (amino acids 302 to 350) of C/EBP and the C-terminal half of UreR (amino acids 164 to 293) activated transcription from the ureD promoter (pureD) and bound to a 60-bp fragment containing pureD, as analyzed by gel shift. These results were consistent with the DNA-binding specificity residing in the C-terminal half of UreR and dimerization being required for activity. To localize the dimerization domain of UreR, a translational fusion of the DNA-binding domain of the LexA repressor (amino acids 1 to 87) and the N-terminal half of UreR (amino acids 1 to 182) was constructed and found to repress transcription from psulA-lacZ (sulA is repressed by LexA) and bind to the sulA operator site, as analyzed by gel shift. Since LexA binds this site only as a dimer, the UreR1–182-LexA1–87 fusion also must dimerize to bind psulA. Indeed, purified UreR-Myc-His eluted from a gel filtration column as a dimer. Therefore, we conclude that the dimerization domain of UreR is located within the N-terminal half of UreR. UreR contains three leucines that mimic the leucines that contribute to dimerization of AraC. Mutagenesis of Leu147, Leu148, or L158 alone did not significantly affect UreR function. In contrast, mutagenesis of both Leu147 and Leu148 or all three Leu residues resulted in a 85 or 94% decrease, respectively, in UreR function in the presence of urea (P < 0.001). On the contrary, His102 and His175 mutations of UreR resulted in constitutive induction in the absence of urea. We conclude that a dimerization domain resides in the N-terminal half of the polypeptide, that Leu residues may contribute to this function, and that sequences within the C-terminal half of UreR are responsible for DNA binding to the urease promoter regions. Selected His residues also contribute significantly to UreR function.
Proteus mirabilis infects the urinary tract of humans and is most commonly responsible for causing disease in individuals with structural abnormalities of the urinary tract or in patients who undergo long-term catheterization (16). Cystitis, acute pyelonephritis, and urinary stone formation are all possible consequences of P. mirabilis infection (17).
P. mirabilis produces a urea-inducible urease, a high-molecular-weight, multimeric, cytoplasmic nickel metalloenzyme. Urease catalyzes the hydrolysis of urea to ammonia and carbon dioxide (18). During the course of infection, the production of ammonia by urea hydrolysis raises the pH in the local environment, subsequently precipitating polyvalent ions that are normally soluble in urine. The result is the formation of urinary stones. The elevated pH also creates an environment that is more favorable for growth of this species (4). Increased ammonia production can also lead to acute inflammation with possible tissue necrosis (18).
The P. mirabilis urease gene cluster is found in single copy on the chromosome and consists of eight contiguous genes, ureRDABCEFG (12, 19, 24). The ureA (UreA, 11 kDa), ureB (UreB, 12 kDa), and ureC (UreC, 61 kDa) genes encode the structural polypeptides required for the assembly of a catalytically inactive urease apoenzyme (18). The accessory genes, ureD (UreD, 31 kDa), ureE (UreE, 18 kDa), ureF (UreF, 23 kDa), and ureG (UreG, 22 kDa), encode proteins required for insertion of nickel ions into the metalloenzyme resulting in catalytically active urease (18). The urease gene cluster is regulated by the gene product of ureR (UreR, 33 kDa).
P. mirabilis UreR and the plasmid-encoded UreR found in Escherichia coli are positive transcriptional activators of the urease genes. The two proteins share 70% amino acid identity (6) and are functionally interchangeable in the activation of transcription from the ureR (pureR) and ureD (pureD) promoters in both the P. mirabilis and plasmid-encoded urease gene clusters (6). The UreR binding sites of both promoters have the consensus sequence T(A/G)(T/C)(A/T)(T/G)(C/T)T(A/T)(T/A)ATTG (25). Both UreR proteins have been shown to activate transcription from pureD in the presence of urea (11, 6). In addition, UreR regulates its own transcription in the presence of urea from pureR in the direction opposite the rest of the gene cluster (6). In the absence of urea induction, H-NS represses ureR expression (3). Because UreR activates transcription in a urea-inducible manner, it is hypothesized that UreR binds urea; however, this has not been directly demonstrated.
UreR is a member of the AraC family of transcriptional regulators and contains a putative helix-turn-helix in addition to an AraC signature sequence (5, 19). The AraC signature sequence, found within all AraC family members, is a second helix-turn-helix that is hypothesized to also bind DNA (7). Moreover, UreR also contains three conserved leucine residues (Leu147, Leu148, and Leu158) in the same relative location with the same spatial distance relative to each other as in AraC (Leu150, Leu151, and Leu161). These leucine residues are critical for AraC dimerization (23), and we therefore also hypothesize that UreR dimerizes via this mechanism. In the presence of arabinose, AraC uses these three critical leucines for dimerization via an antiparallel coiled-coil in a “knobs-into-holes” manner, as elucidated by X-ray crystallographic studies (23). This coiled-coil is also the primary dimerization face in the absence of arabinose, shown by both size exclusion chromatography and sedimentation velocity analytical ultracentrifugation of an AraC mutant with mutations in Leu150, Leu151, Asn154, and Leu161 (15). A secondary dimerization face in the β barrel of AraC is evident; however, it does not appear to represent the primary means of dimer interaction (15).
AraC contains two separate and independent domains, each with a distinct function, namely, dimerization and DNA binding; UreR is predicted to have similar domains with similar functions. Previously, chimeric proteins containing the two domains of AraC to characterize each of the domain's functions were synthesized (2). The predicted AraC DNA-binding domain was fused to C/EBP, a known eukaryotic transcriptional activator that dimerizes via a leucine zipper. The C/EBP-AraC fusion was found to bind to pBAD and activate transcription (2). The hypothesized AraC dimerization domain was fused to the LexA DNA-binding domain. This fusion was predicted to mimic full-length LexA and demonstrated the need for dimerization in order to repress transcription of genes normally turned off by LexA. A chromosomal transcriptional fusion of psulA to lacZ was repressed in the presence of the AraC-LexA fusion protein. This strategy was used to identify both domains of AraC (2).
In this study, we constructed fusion proteins to identify putative domains of UreR and assign dimerization and DNA-binding functions to each of the domains as well as identifying key amino acid residues involved in dimerization and urea induction.
MATERIALS AND METHODS
Chemicals and enzymes.
All enzymes were purchased from Life Technologies (Rockville, Md.) or New England Biolabs (Beverly, Mass.). All chemicals were purchased from Sigma-Aldrich (St. Louis, Mo.) unless otherwise noted. The DIG (digoxigenin) gel shift kit was obtained from Amersham Pharmacia Biotech (Piscataway, N.J.).
Bacterial strains and growth conditions.
Bacterial strains used in this study are listed in Table 1. Bacteria were grown either in Luria-Bertani (LB) broth at 37°C with aeration in a shaking incubator (200 rpm) or on LB plates containing 1.5% agar at 37°C. Plates and media were supplemented with the antibiotic chloramphenicol (10 μg/ml), tetracycline (7.5 μg/ml), ampicillin (100 μg/ml), or kanamycin (50 μg/ml).
TABLE 1.
E. coli strains and plasmids used in this study
| Strain/plasmid | Description | Reference or source |
|---|---|---|
| E. coli strains | ||
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF) U169 deoR recA 1 endA 1 hsdR17 (rk− mk+) phoA supE44 λ −thi-1 gyrA96 relA1 | |
| Top10 | F−mcrAΔ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL(Strr) endA1 nupG | |
| JL1436 | F′ laclqlacZΔM15::Tn9/lexA71::Tn5 recA+sulA211 (λsulA::lacZ cl ind−) | 2 |
| Plasmids | ||
| pSE380 | Invitrogen | |
| pBAD/Myc-His A | Invitrogen | |
| pCC042 | pureD-lacZ reporter construct | This work |
| pSE380-lexA | 2 | |
| pSE380-lexA-C/EBP | 2 | |
| pCP015 | C/EBP302–350-UreR164–293-pBAD | This work |
| pCP016 | Full UreR-pBAD | This work |
| pCP018 | Full UreR-pSE380 | This work |
| pCP019 | UreR1–182-LexA1–87-pSE380 | This work |
| pCP025 | LexA1–87-pSE380 | This work |
| pCP026 | C/EBP302–350-pBAD | This work |
| pCP029 | L147A-L148A UreR | This work |
| pCP030 | L147A UreR | This work |
| pCP031 | L148A UreR | This work |
| pCP051 | UreR164–293-pBAD | This work |
| pCP056 | UreRHis175Ala | This work |
| pCP059 | UreR-Myc-His-pBAD | This work |
| pCP063 | UreRHis102Ala | This work |
| pCP071 | UreR164–293-Myc-His-pBAD | This work |
| pCP072 | C/EBP302–350-Myc-His-pBAD | This work |
| pCP073 | LexA1–87-Myc-His-pSE380 | This work |
| pCP079 | L147A-L148A UreR-Myc-His | This work |
| pCP086 | L158A UreR | This work |
| pCP088 | L147A-L148A-L158A UreR | This work |
| pCP089 | L147A-L148A-L158A UreR-Myc-His | This work |
PCR amplification of DNA used to make fusion proteins.
PCR primers are listed in Table 2. The amplification protocol for PCRs was as follows: denaturation, 94°C, 3 min; annealing, 50 to 55°C, 45 s; elongation, 72°C, 1 min; for 30 cycles.
TABLE 2.
PCR primers used in this study
| Primer no. | Primer sequence | Gene | Enzyme cleaved | Construct |
|---|---|---|---|---|
| 906 | 5′ CGAAATACGGGCAGACATGG3′ | pureD-lacZ reporter (ureR-ureD intergenic) | ||
| 915 | 5′ AGGCAAGTTCAAAATGAACATGG3′ | pureD-lacZ reporter (ureR-ureD intergenic) | ||
| 1126 | 5′ CCATGGAATACAAACACATACTTTCTTCTAAC 3′ | ureR | 5′ NcoI | UreR |
| UreR-Myc-His | ||||
| UreR1–182-LexA1–87 | ||||
| L147A-L148A | ||||
| L147A | ||||
| L148A | ||||
| L147A-L148A-Myc-His | ||||
| UreRHis175Ala | ||||
| UreRHis102Ala | ||||
| L158A | ||||
| L147A-L148A-L158A | ||||
| L147A-L148A-L158A-Myc-His | ||||
| 1128 | 5′ GGATCCATGAAAGCGTTAACGGCCAGGCAACAA3′ | lexA | 5′ BamHI | UreR1–182-LexA1–87 |
| 1129 | 5′ CTCGAGCTATGGTTCACCGGCAGCCACACGACCTAC 3′ | lexA | 3′ Xhol | UreR1–182-LexA1–87 |
| LexA1–87 | ||||
| 1132 | 5′ CCATGGAGAAAGCCAAACAGCGCAACGTGGAGACG 3′ | C/EBP | 5′ Ncol | C/EBP302–350-UreR164–293 |
| C/EBP302–350 | ||||
| C/EBP302–350-Myc-His | ||||
| 1133 | 5′ GGATCCCAAGGAGCTCTCAGGCAGCTGGCGGAA 3′ | C/EBP | 3′ BamHI | C/EBP302–350-UreR164–293 |
| C/EBP302–350 | ||||
| 1594 | 5′ GGATCCTTGCGGATCTTGTGTTATTAGATGAGT 3′ | ureR | 3′ BamHI | UreR1–182-LexA1–87 |
| 1595 | 5′ GGATCCAATTATGATGAGCCAAAAAATCAGGCG 3′ | ureR | 5′ BamHI | C/EBP302–350-UreR164–293 |
| 1596 | 5′ CTCGAGTTAAAATACTTTTTTTATTGATTCGTC 3′ | ureR | 3′ Xhol | C/EBP302–350-UreR164–293 |
| UreR | ||||
| L147A and L148A | ||||
| L147A | ||||
| L148A | ||||
| L158A | ||||
| L147A-L148A-L158A | ||||
| UreRHis175Ala | ||||
| UreRHis102Ala | ||||
| UreR164–293 | ||||
| 1634 | 5′ AGCCTCTTTTTTATTGCGGCGGCGGTTTATCACGAA 3′ | ureR | L147A-L148A | |
| L147A-L148A-Myc-His | ||||
| L147A-L148A-L158A | ||||
| L147A-L148A-L158A-Myc-His | ||||
| 1635 | 5′ TTCGTGATAAACCGCCGCCGCAATAAAAAAGAGGCT 3′ | ureR | L147A-L148A | |
| L147A-L148A-Myc-His | ||||
| L147A-L148A-L158A | ||||
| L147A-L148A-L158A-Myc-His | ||||
| 1636 | 5′ AGCCTCTTTTTTATTGCGCTGGCGGTTTATCAC 3′ | ureR | L147A | |
| 1637 | 5′ GTGATAAACCGCCAGCGCAATAAAAAAGAGGCT 3′ | ureR | L147A | |
| 1638 | 5′ CTCTTTTTTATTTTGGCGGCGGTTTATCACGAA 3′ | ureR | L148A | |
| 1639 | 5′ TTCGTGATAAACCGCCGCCAAAATAAAAAAGAG 3′ | ureR | L148A | |
| 1646 | 5′ CCATGGCAAAAGCGTTAACGGCCAGGCAACAA 3′ | lexA | 5′ NcoI | LexA1–87 |
| 1647 | 5′ CTCGAGCAAGGAGCTCTCAGGCAGCTGGCGGAA 3′ | C/EBP | 3′ Xhol | C/EBP302–350 |
| 1663 | 5′ AATCAGGCGATCACTGCTCTAATAACACAAGAT 3′ | ureR | UreRHis175Ala | |
| 1664 | 5′ ATCTTGTGTTATTAGAGCAGTGATCGCCTGATT 3′ | ureR | UreRHis175Ala | |
| 1712 | 5′ CCATGGCAAATTATGATGAGCCAAAAAATCAGGCG 3′ | ureR | 5′ NcoI | UreR164–293 |
| UreR164–293-Myc-His | ||||
| 1729 | 5′AAGCTTAAATACTTTTTTTATTGATTCGTC 3′ | ureR | 3′ HindIII | UreR-Myc-His |
| L147A-L148A-Myc-His | ||||
| L147A-L148A-L158A-Myc-His | ||||
| 1739 | 5′ ATTTTACACCGAGTTTCAAAAAATGCGTTTTAAATAAACAGCAAT 3′ | pureD oligo | ||
| 1740 | 5′ ATTTTTTCTAAACAAATTGCTGTTTATTTAAAACGCATTTTTTGA3′ | pureD oligo | ||
| 1749 | 5′ AATCAATCCAGCCCCTGTGAGTTACTGTATGGATGTACAGTACATCCAGTG 3′ | psulA oligo | ||
| 1750 | 5′ TGATCTTTGTTGTCACTGGATGTACTGTACATCCATACAGTAACTCACAGG 3′ | psulA oligo | ||
| 1797 | 5′ GCGCCGATTACTCGTGCTCTTCCAGATTATCAT 3′ | ureR | UreRHis102Ala | |
| 1798 | 5′ ATGATAATCTGGAAGAGCACGAGTAATCGGCGC 3′ | ureR | UreRHis102Ala | |
| 1833 | 5′ TTTTTTAAGCTTAAATACTTTTTTTATTGATTCGTC 3′ | ureR | 3′ HindIII | UreR164–293-Myc-His |
| 1834 | 5′ TTTTTTAAGCTTTGGTTCACCGGCAGCCACACGACCTAC 3′ | lexA | 3′ HindIII | LexA1–87-Myc-His |
| 1848 | 5′ AAAAAACCATGGAGAAAGCCAAACA 3′ | C/EBP | 5′ NcoI | C/EBP302–350-Myc-His |
| 1851 | 5′ AAAAAACCATGGCAAAAGCGTTAACGGCCAGGCAACAA 3′ | lexA | 5′ NcoI | LexA1–87-Myc-His |
| 1852 | 5′ TTTTTTCTCGAGATGATGATGATGATGATGGTC 3′ | lexA | 3′ XhoI | LexA1–87-Myc-His |
| 1855 | 5′ AAAAAAAAGCTTCAAGGAGCTCTCAGGCAGCTGGCGGAA 3′ | C/EBP | 3′ HindIII | C/EBP302–350-Myc-His |
| 1883 | 5′ GGGGTCGATATTGCTAATATTTTTCGT 3′ | ureR | L158A | |
| L147A-L148A-L158A | ||||
| L147A-L148A-L158A-Myc-His | ||||
| 1884 | 5′ ACGAAAAATATTAGCAATATCGACCCC 3′ | ureR | L158A | |
| L147A-L148A-L158A | ||||
| L147A-L148A-L158A-Myc-His |
Cloning of PCR products.
PCR products were ligated into pCR-BluntII-TOPO (Invitrogen, Carlsbad, Calif.). Inserts were excised using the appropriate restriction enzymes (either NcoI-BamHI, BamHI-XhoI, NcoI-XhoI, or NcoI-HindIII) and separated by agarose gel electrophoresis. PCR amplification products for the inserts encoding C/EBP302–350-Myc-His and LexA1–87-Myc-His were digested with restriction enzymes; Table 2 lists inserts and restriction enzymes used. All constructs use an NcoI site to ligate into the vector at the gene sequence encoding the start codon. A BamHI site is at the junction of the gene sequences encoding the domains in both C/EBP302–350-UreR164–293 and UreR1–182-LexA1–87. All constructs except the gene sequences encoding UreR-Myc-His, UreR164–293-Myc-His, C/EBP302–350-Myc-His, LexA1–87-Myc-His, L147A-L148-Myc-His, and L147A-L148A-L158A-Myc-His, which use a 3′ HindIII site, contain a XhoI site at the 3′ end for cloning into the expression vector. Inserts were purified using a Qiaquick gel extraction kit (Qiagen) and ligated into either pBAD/MHA or pSE380 (Invitrogen). Plasmids were introduced into the corresponding laboratory strain by CaCl2 transformation (21). The LexA1–87-Myc-His gene sequence was ligated into pBAD/MHA to take advantage of the Myc-His epitopes. After transformation, the pBAD vector containing the gene sequence encoding LexA1–87-Myc-His was purified using a Qiagen miniprep kit. The plasmid was then used in a PCR with primers to amplify the gene sequence for LexA1–87-Myc-His. The PCR product was then cut with NcoI and XhoI and ligated into the NcoI and XhoI sites in pSE380.
Construction of pureD-lacZ reporter plasmid.
A low-copy-number ureD-lacZ fusion reporter plasmid compatible with pBAD-Myc-His was constructed. Primers MOB906 and MOB915 were used to PCR amplify an approximately 4.3 kb DNA fragment from plasmid pΔR10 ureD-lacZ (11) under the following conditions: 95°C denaturation, 54°C annealing, and 72°C elongation for 30 cycles, using Vent DNA polymerase in the presence of 1 mM MgSO4. The PCR product was gel purified and ligated to PCR-Blunt (Invitrogen), forming pCC026. pCC026 was digested with EcoRI, and the 4.3-kb DNA fragment encoding ureD-lacZ was ligated to an EcoRI-digested derivative of pACYC184 that had previously been cut with PvuII and religated (thus, it does not encode a functional chloramphenicol acetyltransferase). The resulting recombinant plasmid, pCC042, carries a tetracycline resistance marker and encodes a ureD-lacZ transcriptional fusion that is activated in the presence of UreR and urea.
Sequencing.
Both strands of plasmid constructs were sequenced across each junction and throughout the insert. Sequencing was done by the Biopolymer Laboratory at the University of Maryland, Baltimore.
β-Galactosidase expression assays.
Fresh medium was inoculated with a single colony from LB-agar plates containing the appropriate antibiotics and cultured at 37°C overnight. Overnight cultures were used to inoculate fresh medium. Cultures were monitored until they reached an optical density at 600 nm (OD600) of ∼0.4 to 0.6, at which time inducer (isopropyl-β-d-thiogalactopyranoside [IPTG], arabinose, or urea) was added, and cultures were incubated for an additional hour. Cultures were placed on ice, and the OD600 was measured. Chloroform (100 μl) and 0.1% sodium dodecyl sulfate (SDS; 50 μl) were added, and cultures were vortexed. The suspension of permeabilized cells (10 μl) was added to 990 μl of Z buffer and 200 μl of o-nitrophenyl-β-d-galactopyranoside (4 mg/ml in H2O). Timed reactions were stopped with 500 μl of 1 M Na2CO3. OD420 and OD550 measurements were recorded, and Miller units were calculated (20). All constructs were assayed in three or more independent experiments.
Western blot analysis.
Overnight cultures were used to inoculate 4 ml of fresh LB medium containing the appropriate antibiotics and allowed to grow to mid-exponential phase. Cultures were induced with the appropriate inducer and incubated for the indicated time. Cultures were placed on ice. OD600 was determined, and all cultures were adjusted to the same reading. Bacteria were harvested from 1 ml of culture by centrifugation and resuspended in equivalent amounts of Laemmli sample buffer. Samples were boiled for 5 min and placed on ice. The sample volume listed for each experiment was loaded onto a 3.75% stacking and either a 12.5 or 15% SDS-polyacrylamide gel by the method of Laemmli (13). SDS running buffer was used for electrophoresis. Gels were transferred onto Immobilon P membranes in a transfer chamber containing transfer buffer. Transfer occurred overnight at ∼12 V at 4°C. Membranes were blocked in 5% dry milk in 0.1% TTBS (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.1% Tween 20) for 1 h. Primary antibodies were added to 0.1% TTBS (anti-LexA, anti-Myc and anti-His, diluted 1:5,000), and membranes were exposed to antibodies for 2 h at room temperature. Membranes were washed in 0.1% TTBS three times for 15 min each. Secondary antibodies (anti-mouse immunoglobulin G coupled to alkaline phosphatase detected anti-Myc and anti-His antibodies; anti-rabbit immunoglobulin G coupled to alkaline phosphatase detected anti-LexA antibodies) were placed in 0.1% TTBS (dilution 1:2,000), and membranes were incubated in the secondary antibodies for 1 h. Membranes were washed three times for 15 min each in 0.1% TTBS and developed with nitroblue tetrazolium–5-bromo-4-chloro-3-indolylphosphate dissolved in H2O (according to the Western blot protocol as described in reference 1).
Gel shift analysis.
Gel shift experiments used either a 60-bp (DNA-binding domain study) or a 64-bp (dimerization study) double-stranded oligonucleotide that was synthesized from smaller overlapping oligonucleotide fragments that were allowed to anneal and extend using Vent polymerase. One microliter of each reaction was run on a gel for quantification purposes. Both double-stranded oligonucleotides (3.85 pmol of each) were labeled by the DIG gel shift (Amersham Pharmacia) protocol. Each double-stranded oligonucleotide was diluted to 30 fmol for the binding study.
Overnight cultures were used to inoculate fresh LB medium containing appropriate antibiotics. These cultures were grown to mid-exponential phase. The appropriate inducer molecule was added and incubated further. The cultures were placed on ice, and OD600 measurements were taken. OD600 for all cultures was adjusted to the same reading by using LB medium. Equivalent volumes of each culture were centrifuged, and the pellet was resuspended in 1 ml of TEN buffer (10 mM Tris, 1 mM EDTA, 0.1 M NaCl [pH 8.0]). Bacterial suspensions were disrupted by passage through a French press (18,000 lb/in2). Lysates were centrifuged (12,000 × g, 10 min, 4°C). Supernatants were collected and used as the extract for the binding assay. The binding assay was done as instructed by the DIG gel shift kit manufacturer (Amersham Pharmacia), using binding buffer, 1 μg of poly(dI-dC), 1 μg of poly-l-lysine, specific extract volume, and 2 μl of the labeled DNA. Binding reactions for pBAD, UreR, C/EBP302–350-UreR164–293, and UreR-Myc-His also included 100 mM urea (final concentration). The binding reactions were run on a preelectrophoresed 6% native polyacrylamide gel in TAE buffer (0.04 M Tris acetate, 0.001 M EDTA) at 90 V for 1.5 h. The gel was placed on a Hybond N+ membrane and transferred for 30 min at 400 mA in 1× TAE buffer. The membrane was developed according to the instructions of the DIG gel shift kit manufacturer (Amersham Pharmacia).
Purification of UreR derivatives.
E. coli Top10 transformed with pCP016 or pCP088 was grown in Luria broth at 37°C. Expression of UreR-Myc-His6 was induced with 0.2% arabinose when an OD600 of ∼0.6 was reached. Following 3 h of induction, cells were collected by centrifugation and lysed by two passages through a French pressure cell (18,000 lb/in2). Single-step purification of UreR-Myc-His6 was performed by nickel-chelating nitrilotriacetic acid affinity chromatography. A single polypeptide of approximately 33 kDa was eluted from the column with 250 mM imidazole as seen on a Coomassie blue-stained 12% SDS-polyacrylamide gel (data not shown). In a similar experiment, purified protein was electrophoresed, transferred to a nitrocellulose membrane, and reacted with rabbit antiserum specific for the Myc epitope. Western blot anlysis showed that arabinose-induced E. coli Top10 transformed with either pCP016 or pCP088 produced a single species of 33 kDa, consistent with the predicted size for UreR-Myc-His6 (see Fig. 3). The band was absent from the vector control under identical conditions.
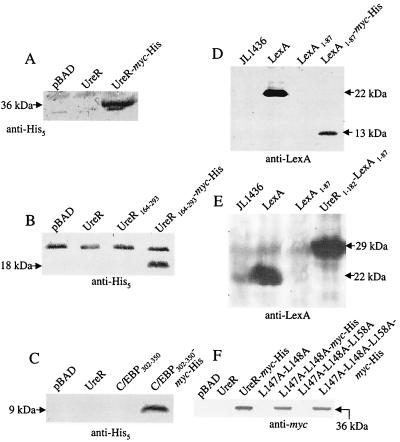
FIG. 3.
Western blot analysis of fusion constructs. E. coli Top10 (A, B, C, and F) or E. coli JL1436 (D and E), transformed with vector or clones expressing UreR or chimeric proteins, was cultured in LB medium to mid-exponential phase and induced with 2% arabinose for 4 h (unless otherwise noted). Samples (10 μl) of each suspension were boiled for 5 min in gel sample buffer and electrophoresed on an SDS-polyacrylamide gel. Membranes were incubated with antiserum or monoclonal antibodies and developed with secondary antibodies conjugated to alkaline phosphotase (Sigma). (A) pBAD, UreR, UreR-Myc-His; 12.5% SDS-polyacrylamide gel. Membranes were reacted with mouse anti-His antibody (diluted 1:1,000) (Invitrogen). (B) pBAD, UreR, UreR164–293, or UreR164–293-Myc-His; 15% SDS-polyacrylamide gel. Membranes were reacted with mouse anti-His antibody (diluted 1:1,000) (Invitrogen). (C) pBAD, UreR, C/EBP302–350, C/EBP302–350-Myc-His; 15% SDS-polyacrylamide gel. Membranes were reacted with mouse anti-His antibody (diluted 1:1,000) (Invitrogen). (D) LexA, LexA1–87, LexA1–87-Myc-His; induced with 2 mM IPTG. Samples (2 μl) of each were electrophoresed on a 15% SDS-polyacrylamide gel. Membranes were hybridized with rabbit polyclonal anti-LexA antibody (diluted 1:5,000) (Invitrogen). (E) LexA, LexA1–87, UreR1–182-LexA1–87; induced with 2 mM IPTG. Samples (1.25 μl) of each, including strain only, were electrophoresed on a 12.5% SDS-polyacrylamide gel. Membranes were reacted with rabbit polyclonal anti-LexA antibody (diluted 1:5,000) (Invitrogen). (F) pBAD, UreR, UreR-Myc-His, UreR L147A-L148A, UreR L147A-L148A-Myc-His, UreR L147A-L148A-L158A, and UreR L147A-L148A-L158A-Myc-His; induced with 2% arabinose for 4 h. Samples (10 μl) of each were electrophoresed on a 12.5% SDS-polyacrylamide gel. Membranes were hybridized with mouse anti-Myc antibody (diluted 1:5,000) (Invitrogen).
Gel filtration chromatography.
Gel filtration chromatography was performed at room temperature on a Sephadex G-75 column (1 by 35 cm) equilibrated with running buffer (50 mM phosphate [pH 7.5], 150 mM NaCl) at a flow rate of 0.5 ml/min. Molecular weight standards bovine serum albumin (68 kDa) and carbonic anhydrase (34.5 kDa) were used to calibrate the column. Purified wild-type and mutant UreR-Myc-His6 proteins (100 μl of 0.1 mg/ml) were injected onto the column, and 0.5-ml fractions were collected. The protein elution profile was monitored by absorbance at 280 nm. An aliquot (100 μl) of each fraction was transferred to an Immobilon P membrane, and immunoblot analysis performed with anti-Myc antibodies.
RESULTS
Localization of the UreR DNA-binding domain.
To localize the UreR DNA-binding domain, a protein chimera was constructed by fusing the gene sequence encoding the C-terminal half of UreR to the gene sequence encoding the leucine zipper dimerization domain of C/EBP (Fig. 1; primers are listed in Table 2). All translational fusion constructs were cloned into the E. coli arabinose-inducible expression vector pBAD/Myc-His A (Invitrogen) (Table 1). A pureD-lacZ plasmid reporter was constructed in pACYC184 to assay specific induction of the urease gene cluster, using β-galactosidase activity as the readout. Both reporter plasmid and pBAD constructs with and without insert were transformed into E. coli Top10.
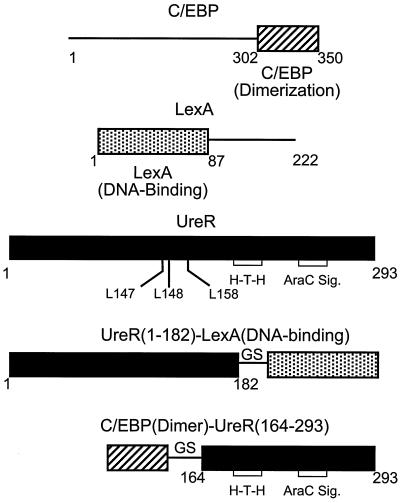
FIG. 1.
Schematic of fusion constructs. The domain structure of each of the chimeric proteins is shown; the amino acid residue number is labeled at each boundary. A BamHI restriction site (coding for a Gly-Ser amino acid linker (GS)) was inserted between gene sequences encoding each domain. Key leucine residues and helix-turn-helix motifs (H-T-H) are indicated. Sig., signature.
The C/EBP302–350-UreR164–293 fusion chimera was tested for its ability to bind to pureD and activate transcription. The C/EBP302–350-UreR164–293 fusion activated transcription from this promoter in both the absence (264% of uninduced wild-type UreR level) and presence (42% of urea-induced wild-type UreR level) of urea (Fig. 2). The level of activation of pureD, mediated by C/EBP302–350-UreR164–293 and measured by the β-galactosidase assay, was not significantly different in the presence and absence of urea. This result is consistent with the urea-binding domain not residing in the C-terminal half of UreR or urea binding not affecting DNA binding in the chimera.
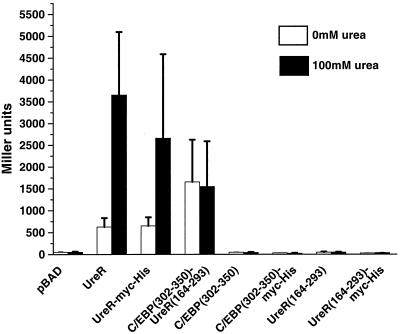
FIG. 2.
β-Galactosidase assays using pureD-lacZ reporter construct. β-Galactosidase activity was measured in E. coli Top10 transformed with the pureD-lacZ reporter construct and the chimeric fusion constructs. Cultures (4 ml) were grown in LB medium to mid-exponential phase and induced with 0.02% arabinose with or without 100 mM urea for 1 h. β-Galactosidase activity is expressed in Miller units (18). All constructs were assayed in duplicate in at least three different experiments.
A UreR-Myc-His fusion was constructed so that UreR could be readily detected on Western blots and in gel mobility shift assays. The UreR-Myc-His fusion induced β-galactosidase expression using the pureD-lacZ reporter construct and elicited levels of expression that were not significantly different from wild-type UreR level (104 and 73% of wild-type UreR activation in the absence and presence of urea, respectively) (Fig. 2). Since we observed similar β-galactosidase expression from pureD-lacZ for both UreR and UreR-Myc-His, we considered these two proteins to be interchangeable in their ability to regulate transcription. This translational fusion contains a c-Myc epitope and His6 tail that are recognized by anti-Myc (Invitrogen) and anti-His5 (Qiagen) monoclonal antibodies, respectively. Expression of the UreR-Myc-His protein product was verified by Western blotting. The translational fusion protein was detected as a band corresponding to the expected molecular size of ∼36 kDa with both anti-Myc (Fig. 3F) and anti-His (Fig. 3A) antibodies.
Neither the putative UreR DNA-binding domain alone (amino acids 164 to 293) nor C/EBP302–350 alone activated transcription from pureD in the presence or absence of urea; values were below levels detected for the vector plasmid DNA alone (Fig. 2). The stability of UreR164–293 and C/EBP302–350 was examined by translationally fusing Myc-His to the C-terminal end of both truncated proteins. Both the UreR164–293 and the C/EBP302–350 proteins showed negligible β-galactosidase activities that were not significantly different from their Myc-His fusion counterparts. Western blot analysis using monoclonal anti-His5 (Qiagen) revealed that both the UreR164–293-Myc-His (18 kDa) and the C/EBP302–350-Myc-His (9 kDa) are produced (Fig. 3B and C).
DNA binding by UreR constructs assessed by gel shift.
Gel shift experiments were performed to verify that C/EBP302–350-UreR164–293 used in the β-galactosidase assays could bind to pureD. A 60-bp double-stranded oligonucleotide that contained the DNA sequence for pureD (−66 to −6 upstream of the transcriptional start of ureD) was synthesized and labeled with DIG-11-ddUTP by terminal transferase.
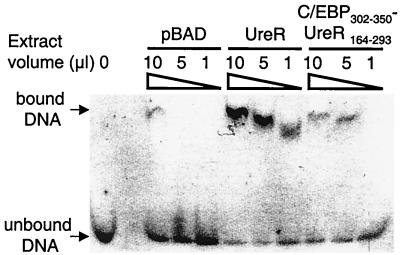
Binding assays were performed with the labeled double-stranded oligonucleotide, whole-cell extracts containing each of the overexpressed protein products, and 100 mM urea. A cell extract containing full-length UreR retarded the migration of the double-stranded DNA (dsDNA) fragment, indicating that UreR protein bound to DNA (Fig. 4). The fusion protein, C/EBP302–350-UreR164–293, also bound the labeled DNA, reflected by a somewhat less intense shifted band. The UreR-Myc-His fusion protein bound and retarded the labeled DNA as well as full-length UreR (data not shown). The pBAD vector control lane showed a faint band at the highest concentration of lysate; however, there are no other bands evident in the pBAD lanes containing lower concentrations of protein. The limited nonspecific binding seen in the vector control lane can likely be explained by the use of whole-cell extracts incubated with the target DNA (14). These results demonstrate that full-length UreR, C/EBP302–350-UreR164–293, and UreR-Myc-His bind to the pureD DNA double-stranded oligonucleotide and are likely responsible for the activation from pureD.
FIG. 4.
Gel mobility shift assay for interaction of UreR with the P. mirabilis pureD. A 60-bp double-stranded oligonucleotide was synthesized based on the sequence of pureD. Whole-cell extracts of E. coli Top10 transformed with either pBAD or vector expressing C/EBP302–350-UreR164–293 were obtained by inducing a 4-ml culture in mid-exponential phase with 2% arabinose for 4 h. Whole-cell extracts of E. coli Top10 expressing UreR and UreR-Myc-His were obtained by inducing a 4-ml culture in mid-exponential phase with 2% arabinose and 100 mM urea for 4 h. Bacterial suspensions were adjusted to the same OD, and 3 ml of each culture was harvested by centrifugation (10,000 × g, 5 min, 4°C). Bacteria were resuspended in 1 ml of TEN buffer and lysed in a French press (18,000 1b/in2). Binding reactions with the 60-bp pureD dsDNA fragment were carried out in the presence of 100 mM urea with various amounts of the extracts.
Localization of the UreR dimerization domain.
Protein chimeras were constructed to localize the UreR dimerization domain. The gene sequence encoding the N-terminal half (amino acids 1 to 182) of UreR was fused to the gene sequence encoding the LexA DNA-binding domain (amino acids 1 to 87), forming the UreR1–182-LexA1–87 fusion protein (Fig. 1; primers are listed in Table 2). All fusion constructs used in these experiments were cloned into pSE380 (Invitrogen) (Table 1) under the control of an IPTG-inducible promoter. The reporter strain, JL1436 (Table 1), originally described by Bustos and Schleif (2), consisted of a chromosomal transcriptional fusion of psulA with lacZ placed downstream; the sulA gene is repressed by dimerized LexA (2).
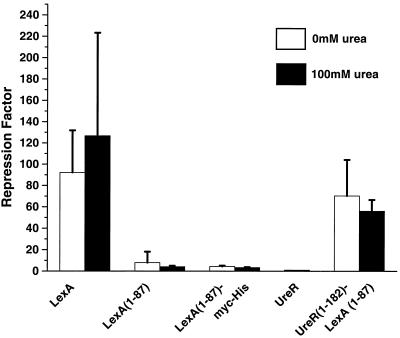
The fusion proteins were assayed for the ability to repress transcription of lacZ by binding to psulA. As expected, both untransformed E. coli JL1436 and JL1436 transformed with full-length ureR showed no repression of β-galactosidase expression (Fig. 5). The UreR1–182-LexA1–87 fusion repressed transcription from psulA by factors of 70 and 56 in the absence and presence of urea, respectively, comparable to the results for full-length LexA (the repression factor is defined as the Miller units of the strain only divided by the Miller units of the strain carrying the expression vector). LexA has repression factors of 92 and 127 in the absence and presence of urea, respectively. The LexA DNA-binding domain alone (LexA1–87) did not repress, indicated by repression factors of only 8 in the absence of urea and 4 in the presence of urea. As expected, the repression factors seen for both UreR1–182-LexA1–87 and full-length LexA are not significantly different in the absence of urea, showing that they are similar in the ability to repress transcription from psulA. However, both LexA and UreR1–182-LexA1–87 had repression factors that were significantly different (P ≤ 0.001) from those of the LexA1–87 control in the presence and absence of urea, suggesting that the LexA1–87 lacks a dimerization domain that was provided by UreR1–182 to repress transcription. UreR1–182-LexA1–87 did not repress transcription in a urea-inducible manner.
FIG. 5.
β-Galactosidase reporter activity of the psulA-lacZ chromosomal fusion reporter construct in strain JL1436. E. coli JL1436 (carries a single copy of a psulA-lacZ chromosomal fusion) transformed with the chimeric fusion constructs was cultured in LB medium (4 ml) to mid-exponential phase and induced with 2 mM IPTG with or without 100 mM urea for 1 h. β-Galactosidase levels measured in Miller units (18) as an index of expression from psulA-lacZ. The Repression factor was calculated by dividing the Miller units for strain only (JL1436) by Miller units for each construct in JL1436. All constructs were assayed in duplicate for at least three independent experiments.
We noted that LexA1–87 was not stable on Western blots. Thus, to examine whether lack of repression at the sulA reporter was due to the lack of stability, we constructed another control. LexA1–87 protein was translationally fused to Myc-His at its C-terminal end. LexA1–87-Myc-His protein also did not repress transcription from psulA, as evidenced by β-galactosidase assays. LexA1–87, either tagged or not with the Myc-His epitope, showed negligible repression and values were not significantly different.
Expression of both full-length LexA and the UreR1–182-LexA1–87 used in the β-galactosidase assay was also verified by Western blotting using an anti-LexA polyclonal antibody (Invitrogen) (Fig. 3E). Strong signals were observed in lanes containing LexA and UreR1–182-LexA1–87. Western blot analysis using anti-LexA showed a 13-kDa band corresponding to LexA1-87-Myc-His (Fig. 3D). Therefore, the repression of β-galactosidase expression observed in the UreR1–182-LexA1–87 β-galactosidase assay was likely due to the expression of the fusion protein.
Binding of the UreR-LexA fusions to target DNA assayed by gel shift.
Gel shift experiments were performed to show that the fusion proteins as well as the LexA control bound to psulA, providing an explanation for the inhibition of transcription initiation from psulA-lacZ. A 64-bp double-stranded oligonucleotide comprising the LexA binding site upstream of sulA (+26 to −38 relative to the transcriptional start of sulA) was synthesized and labeled with DIG-11-ddUTP by terminal transferase.
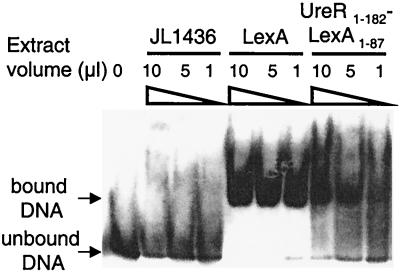
Lanes containing LexA revealed a strong signal representing a shifted band (Fig. 6). The UreR1–182-LexA1–87 lanes contained a retarded band that was not as strong in intensity. Cell extracts from JL1436 carrying no plasmid (Fig. 6) or JL1436 transformed with the LexA1–87-Myc-His construct alone (data not shown) were unable to retard the mobility of the dsDNA fragment.
FIG. 6.
Gel mobility shift assay for chimeric protein and the LexA DNA-binding site upstream of psulA. A 64-bp double-stranded oligonucleotide was synthesized based on the nucleotide sequence of psulA. Whole-cell extracts were obtained by inducing a 100-ml culture, in mid-exponential phase, with 2 mM IPTG for 3 h. Culture (50 ml) was centrifuged, and bacteria were resuspended in 1 ml of TEN buffer and lysed in a French press (18,000 lb/in2). All lanes contained the labeled 64-bp dsDNA fragment. The lane containing no extract contained labeled DNA only. JL1436, LexA, and UreR1–182-LexA1–87 whole-cell extracts were added at volumes of 10, 5, and 1 μl.
PCR site-directed mutagenesis of leucines in the putative UreR dimerization domain.
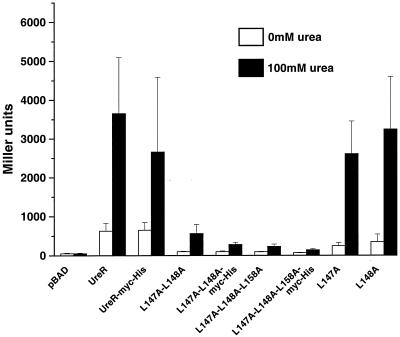
AraC uses three critical leucines (Leu150, Leu151, and Leu161) for dimerization via an antiparallel coiled-coil in the absence and presence of arabinose (15). Due to the conservation of these three leucines in both overall location and spatial orientation within UreR, we hypothesized that these leucine residues are important for dimerization of UreR. To directly test biologically whether these Leu residues are required for dimerization of UreR monomers, PCR site-directed mutagenesis (9) was used to create leucine to alanine mutants of UreR in Leu147 (L147A) alone, Leu148 (L148A) alone, Leu158 (L158A) alone, Leu147 and Leu148 (L147A-L148A), and Leu147, Leu148, and Leu158 (L147A-L148A-L158A). β-Galactosidase expression from the pureD-lacZ reporter plasmid was measured for each of the UreR leucine mutants in the presence and absence of urea (Fig. 7). In the presence of urea, the L147A, L148A, and L158A single mutants had levels of expression that were 71, 89, and 137%, respectively, of the wild-type UreR level. Thus, dimerization, required for activity, was not dramatically altered. The L147A-L148A double mutant, however, had an expression level that was only 15% of the wild-type UreR level in the presence of urea, a significant drop in activity (P < 0.001). The expression level observed for the L147A-L148A-L158A triple mutant, 6% of the wild-type UreR level (P < 0.001) in the presence of urea, tended to be even lower than that of the double mutant (P = 0.052).
FIG. 7.
β-Galactosidase assays using the pureD-lacZ reporter construct to measure activation by the UreR leucine mutants. E. coli transformed with the Leu mutant constructs was cultured to mid-log phase in LB medium (4 ml) and induced with 0.02% arabinose with or without 100 mM urea for 1 h. β-Galactosidase activity represented expression from pureD-lacZ and is expressed in Miller units. All constructs were assayed in duplicate for at least three independent experiments.
To verify that both the double and triple mutant proteins were expressed and stable, L147A-L148A and L147A-L148A-L158A were translationally fused to Myc-His. The L147A-L148-Myc-His protein activated from pureD to the same degree (15 and 8% of the wild-type UreR level in the absence and presence of urea, respectively) as seen for L147A-L148A protein in the β-galactosidase assay. The L147A-L148A-L158A-Myc-His protein activated from pureD to the same degree (11 and 4% of the wild-type UreR level in the absence and presence of urea, respectively) as seen for L147A-L148A-L158A protein in the β-galactosidase assays. Western blot analysis using monoclonal anti-Myc antibodies (Invitrogen) confirmed the stable expression of two 36-kDa bands corresponding to L147A-L148A-Myc-His and L147A-L148A-L158A-Myc-His (Fig. 3F).
Dimer formation in UreR derivatives.
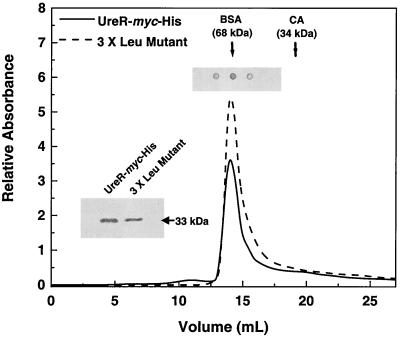
To determine whether the low activity of the triple Leu mutant of UreR-Myc-His was due a loss of dimerization, we compared the elution profile of this protein to that of UreR-Myc-His on a Sephadex G-75 gel filtration column. The two proteins, purified from induced cell lysates on a Ni-nitrilotriacetic acid column, were applied to the column. Both UreR-Myc-His and the L147A-L148A-L158A-Myc-His derivative of UreR eluted at fractions corresponding to the dimerized protein (Fig. 8). Thus, loss of activity in the site-directed mutant was not due to the inability to dimerize.
FIG. 8.
Gel filtration chromatography of UreR-Myc-His and its triple Leu mutant. Purified UreR-Myc-His and UreR-L147A-L148A-L158A-Myc-His (3 X Leu mutant) (approximately 10 μg of protein) were applied to a Sephadex G-75 column. Protein elution was monitored at 280 nm. Fractions (0.5 ml) were collected, and aliquots of the 3 X Leu mutant (0.15 ml) were assayed by immunoblotting using anti-Myc antibodies (positioned above peak fractions). The elution volumes of bovine serum albumin (BSA) and carbonic anhydrase (CA) are indicated by arrows. The inset shows Western blot of lysates used for purification, run on a denaturing gel, and developed using anti-Myc antibodies. The apparent molecular size is noted.
Constitutive induction of urease genes by His mutants of UreR.
We did not observe urea inducibility using the chimeric fusion proteins. We had reasoned earlier, however, that histidine residues were likely involved in either urea binding or transmission of a structural alteration induced by urea activation to the DNA-binding domain. Indeed, the active site of urease coordinates urea by interaction with four His, one Cys, one Asp, and one Lys residue (18). Thus, urea could be coordinated by similarly configured His residues within UreR. Using site-directed mutagenesis, eight His residues were changed to Ala. Six mutations (H5A, H73A, H107A, H186A, H129A, and H152A) did not significantly alter urea inducibility of the mutated UreR (data not shown). Two of eight His-to-Ala mutants tested, however, H102A and H175A, constitutively induced urease genes (i.e., in the absence of urea), as assayed using the ureD-lacZ translational fusion to levels that were not significantly different from the wild-type UreR level in the presence of urea (Table 3). The His102 residue resides in the N-terminal domain, the region predicted to bind urea. Interestingly, the His175 residue resides in the linker region that joins the dimerization domain and DNA-binding domain.
TABLE 3.
Induction of Urease Genes by His mutants of UreR
| E. coli Top10 (pureD-lacZ) cotransformed with: | β-Galactosidase activity (Miller units ± SD)
|
|
|---|---|---|
| 0 mM urea | 100 mM urea | |
| pBAD (vector control) | 53 ± 15 | 51 ± 17 |
| pCP016 (UreR) | 627 ± 204 | 3,654 ± 1442 |
| pCP063 (UreRHis102Ala) | 2,414 ± 522ab | 4,229 ± 296c |
| pCP056 (UreRHis175Ala) | 2,871 ± 43ac | 4,770 ± 262c |
P < 0.001 compared to uninduced (0 mM urea) UreR.
P = 0.16, not significantly different from induced (100 mM urea) UreR.
P > 0.2, not significantly different from induced (100 mM urea) UreR.
DISCUSSION
UreR chimeric proteins were constructed to identify and localize functional domains of the AraC-like transcriptional activator of the P. mirabilis urease gene cluster. Our studies led us to conclude that the N-terminal half of UreR contains the dimerization domain and the C-terminal half of UreR serves as the DNA-binding domain. Leucine residues in the putative dimerization domain of UreR, conserved with respect to AraC and other UreR homologues, were required to fully activate transcription. Site-directed mutagenesis studies were consistent with their involvement in dimerization. While it is proposed that urea binding by UreR is required for activation, construction of the chimeric proteins did not allow us to elucidate this role. Two His mutants of UreR, however, displayed constitutive induction of urease genes.
Our experimental results support the hypothesis that the dimerization domain of UreR localizes to the N-terminal half of UreR. Repression of psulA-lacZ by UreR1–182-LexA1–87 is consistent with the presence of a dimerization domain supplied by UreR. LexA1–87 alone was unable to repress transcription from psulA-lacZ (Fig. 5). Only when the N terminus of UreR was fused to LexA1–87 was repression evident, indicating the requirement for dimerization. Other studies have demonstrated that the LexA1–87 as well as the λ repressor requires a dimerization domain for full function (8, 10, 22). That dimerization occurs in the fusion protein is further supported by the observation that UreR1–182-LexA1–87 is capable of retarding the mobility of a double-stranded oligonucleotide containing the sulA promoter in a gel shift assay (Fig. 6). LexA1–87-Myc-His, a stably expressed protein, does not retard the mobility of the target DNA. Thus, the addition of a dimerization domain, provided by the UreR N-terminal amino acid sequences, to LexA1–87 is necessary and sufficient to restore the DNA-binding capability of LexA1–87 for its target, psulA.
A number of observations also led us to conclude that the DNA-binding domain resides in the C-terminal half of UreR. We demonstrated that a functional chimeric protein, C/EBP302-350-UreR164–293, binds to a double-stranded oligonucleotide containing the mapped UreR-binding site (25) within pureD (Fig. 4). The UreR-binding site, mapped by Thomas and Collins, is −57 to −34 upstream of the transcriptional start of ureD in the P. mirabilis ureR-ureD intergenic region (25). Interestingly, C/EBP302–350-UreR164–293 activated transcription from pureD to the same degree in both the absence and presence of urea (Fig. 2). This finding is consistent with the hypothesis that the putative urea-binding site resides in the nonhomologous N-terminal domain. The C/EBP302–350-UreR164–293 fusion has slightly less than optimal activation in comparison to UreR. This may have resulted from constraints imparted on the UreR164–293 by the heterologous C/EBP dimerization domain. The ability of the C/EBP302–350-UreR164–293 to bind and activate transcription from pureD is consistent with DNA-binding specificity residing in the putative UreR DNA-binding domain. This assertion is further supported by the observation that in the absence of a dimerizing mechanism, the UreR DNA-binding domain alone was unable to activate transcription from pureD-lacZ even though UreR164–293 could recognize its target sequence (Fig. 2) when dimerized. Furthermore, these results suggest that a dimerization domain, provided by C/EBP in this case, is necessary and sufficient to allow the binding of the C-terminal portion of UreR to the ureD promoter. Taken together, these results suggest that, as in other AraC family members, dimerization of UreR is required for activation of urease promoters and that the DNA-binding domain, rather than the dimerization domain, resides in the C-terminal portion of UreR.
UreR activates transcription from both pureD and pureR in a urea-inducible manner (6), leading to the hypothesis that UreR binds urea. Likely, this mechanism involves a structural change induced by urea binding. We also hypothesized that because the AraC family of transcriptional regulators contains little or no homology in the N-terminal portion of the proteins, binding specificity for an inducer molecule would likely reside in the N-terminal portion of the protein (7). Although UreR1–182-LexA1–87 represses transcription from psulA-lacZ, the hypothesized urea requirement for UreR function was not observed in the β-galactosidase assays (Fig. 5). The tertiary structure of the UreR1–182-LexA1–87 fusion appears to allow for UreR dimerization to occur but clearly eliminates the urea-inducible mechanism. If UreR dimerizes via two different conformations depending on whether urea is present or not, then possibly only one of the dimerization conformations is attainable when fused to LexA1–87, allowing for binding of the UreR1–182-LexA1–87 fusion to psulA-lacZ. This may account for the unresponsiveness of UreR1–182-LexA1–87 to urea.
Nevertheless, some clues as to the mechanism of urea inducibility by UreR were uncovered by site-directed mutagenesis. Interestingly, among eight His-to-Ala mutants of UreR tested for urea inducibility, two resulted in constitutive transcriptional activation from the ureD promoter in the absence of urea. His102 resides in the putative urea-binding (N-terminal) domain. His175 resides in the amino acid sequence that comprises a linker region between the N-terminal and C-terminal domains. It could be speculated that this latter residue transmits the structural alteration that follows urea binding to the C-terminal DNA-binding domain, resulting in binding to specific DNA sequences within the ureR-ureD intergenic region.
AraC dimerizes via an antiparallel coiled-coil containing three leucines that fits into a knobs-into-holes conformation at both ends of the coil (23, 15). UreR retains these three conserved leucines in the same spatial orientation and relative location. PCR site-directed mutagenesis of Leu147, Leu148, and Leu158 in UreR demonstrates the requirement of these residues for transcriptional activation of pureD-lacZ (Fig. 7). The levels of transcriptional activation for each of the single leucine mutants of UreR are not significantly different from the wild-type UreR level in the presence of urea. The double and triple leucine mutants, however, show a dramatic decrease in activation in comparison to both wild-type UreR and the single leucine mutants in both the presence and absence of urea (as little as 6% of native UreR activity). These results indicate that alteration of each one of the leucines alone is not sufficient to disrupt the protein structure to any degree in the presence of urea. However, because both Leu147 and Leu148 from one monomer may flank Leu158 from the other monomer and act as a dimerization anchor at each end of the coiled-coil helix, the mutation of both Leu147 and Leu148 would be expected to perturb the native dimerization state. A mutation in one of the leucines still may allow for the interaction between the nonmutated leucine and Leu158 from the other monomer. However, mutation of both Leu147 and Leu148 may eliminate the pocket into which Leu158, from the other monomer, is anchored. Surprisingly, while the native dimerization state may have been altered, the triple Leu mutant remained a dimer. The purified Myc-His derivative of the triple mutant eluted at the identical fraction as purified UreR-Myc-His on a gel filtration column (Fig. 8), indicating that under physiologic conditions the two monomers remained associated. More drastic amino acid substitutions of the homologous residues (Leu to Lys in combination with Leu to Ser) in a construct expressing the N-terminal half of AraC did disrupt the dimeric state to yield the monomers (15). Likely, this would be the case for UreR as well. In this study, the Leu residues were conservatively changed to only Ala. While this study is the first to show the biological relevance of these critical leucine residues in UreR, clearly additional studies are required to substantiate their precise role.
The UreR-Myc-His fusion was constructed to detect and follow a protein that mimics wild-type UreR in vivo and in vitro. Although UreR-Myc-His contains the c-Myc and His6 epitopes at the C terminus of UreR, β-galactosidase reporter activity demonstrated that UreR-Myc-His activated transcription from pureD-lacZ as well as wild-type UreR (Fig. 2). Overexpression of UreR-Myc-His and detection by anti-Myc and anti-His antibodies on a Western blot show that the fusion protein was produced (Fig. 3). Furthermore, gel mobility shift assays demonstrated that UreR-Myc-His was able to bind to pureD, which resulted in a strong shift of labeled target DNA comparable to the shift seen with wild-type UreR (data not shown).
Using protein fusion technology, we have provided evidence that the AraC-like transcriptional activator UreR consists of dimerization and DNA-binding domains. We have also elucidated a possible dimerization mechanism based on the knobs-into-holes conformation facilitated by three leucine residues that are conserved within both UreR and AraC. Our chimeric proteins were unresponsive to urea activation, and thus a urea-binding domain could not be assigned although we identified two key His residues that were involved in urea inducibility.
ACKNOWLEDGMENTS
This work was supported in part by National Institutes of Health Public Health Service grant AI23328.
We thank Robert Schleif for the gifts of strains and plasmids. We thank Magdeline Spence for skillful technical assistance. We thank Xin Li for helpful discussions, technical advice, and critical review. We also thank David Rasko, Susan Heimer, Janette Harro, and Angela Jansen for critical review of the manuscript.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1995. pp. 10.8.1–10.8.17. [Google Scholar]
- 2.Bustos S A, Schleif R F. Functional domains of the AraC protein. Proc Natl Acad Sci USA. 1993;90:5638–5642. doi: 10.1073/pnas.90.12.5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coker C, Bakare O O, Mobley H L T. H-NS is a repressor of the Proteus mirabilis urease transcriptional activator gene ureR. J Bacteriol. 2000;182:2649–2653. doi: 10.1128/jb.182.9.2649-2653.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins C M, D'Orazio S E F. Bacterial ureases: structure, regulation of expression and role in pathogenesis. Mol Microbiol. 1993;9:907–913. doi: 10.1111/j.1365-2958.1993.tb01220.x. [DOI] [PubMed] [Google Scholar]
- 5.D'Orazio S E F, Collins C M. The plasmid-encoded urease gene cluster of the family Enterobacteriaceae is positively regulated by UreR, a member of the AraC family of transcriptional activators. J Bacteriol. 1993;175:3459–3467. doi: 10.1128/jb.175.11.3459-3467.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.D'Orazio S E F, Thomas V, Collins C M. Activation of transcription at divergent urea-dependent promoters by the urease gene regulator UreR. Mol Microbiol. 1996;21:643–655. doi: 10.1111/j.1365-2958.1996.tb02572.x. [DOI] [PubMed] [Google Scholar]
- 7.Gallegos M, Schleif R, Bairoch A, Hofmann K, Ramos J L. AraC/XylS family of transcriptional regulators. Microbiol Mol Biol Rev. 1997;61:393–410. doi: 10.1128/mmbr.61.4.393-410.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Granger-Schnarr M, Benusiglio E, Schnarr M, Sassone-Corsi P. Transformation and transactivation suppressor activity of the c-Jun leucine zipper fused to a bacterial repressor. Proc Natl Acad Sci USA. 1992;89:4236–4239. doi: 10.1073/pnas.89.10.4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 10.Hu J C, O'Shea E K, Kim P S, Sauer R T. Sequence requirements for coiled-coils: analysis with lambda repressor-GCN4 leucine zipper fusions. Science. 1990;250:1400–1403. doi: 10.1126/science.2147779. [DOI] [PubMed] [Google Scholar]
- 11.Island M D, Mobley H L T. Proteus mirabilis urease: operon fusion and linker insertion analysis of ure gene organization, regulation, and function. J Bacteriol. 1995;177:5653–5660. doi: 10.1128/jb.177.19.5653-5660.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones B D, Mobley H L T. Proteus mirabilis urease: nucleotide sequence determination and comparison with jack bean urease. J Bacteriol. 1989;171:6414–6422. doi: 10.1128/jb.171.12.6414-6422.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 14.Lane D, Prentki P, Chandler M. Use of gel retardation to analyze protein-nucleic acid interactions. Microbiol Rev. 1992;56:509–528. doi: 10.1128/mr.56.4.509-528.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.LaRonde-LeBlanc N, Wolberger C. Characterization of the oligomeric states of wild type and mutant AraC. Biochemistry. 2000;39:11593–11601. doi: 10.1021/bi001262g. [DOI] [PubMed] [Google Scholar]
- 16.Mobley H L T, Warren J W. Urease-positive bacteriuria and obstruction of long-term urinary catheters. J Clin Microbiol. 1987;25:2216–2217. doi: 10.1128/jcm.25.11.2216-2217.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mobley H L T, Belas R. Swarming and pathogenicity of Proteus mirabilis in the urinary tract. Trends Microbiol. 1995;3:280–284. doi: 10.1016/s0966-842x(00)88945-3. [DOI] [PubMed] [Google Scholar]
- 18.Mobley H L T, Island M D, Hausinger R P. Molecular biology of microbial ureases. Microbiol Rev. 1995;59:451–480. doi: 10.1128/mr.59.3.451-480.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nicholson E B, Concaugh E A, Foxall P A, Island M D, Mobley H L T. Proteus mirabilis urease: transcriptional regulation by UreR. J Bacteriol. 1993;175:465–473. doi: 10.1128/jb.175.2.465-473.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Platt T, Meuler-Hill B, Miller J. Assays of β-galactosidase activity. In: Miller J, editor. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1972. pp. 352–355. [Google Scholar]
- 21.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1989. [Google Scholar]
- 22.Schmidt-Dörr T, Oertel-Buchheit P, Pernelle C, Bracco L, Schnarr M, Granger-Schnarr M. Construction, purification, and characterization of a hybrid protein comprising DNA binding domain of the LexA repressor and the Jun leucine zipper: a circular dichroism and mutagenesis study. Biochemistry. 1991;30:9657–9664. doi: 10.1021/bi00104a013. [DOI] [PubMed] [Google Scholar]
- 23.Soisson S M, MacDougall-Shackleton B, Schleif R, Wolberger C. Structural basis for ligand-regulated oligomerization of AraC. Science. 1997;276:421–425. doi: 10.1126/science.276.5311.421. [DOI] [PubMed] [Google Scholar]
- 24.Sriwanthana B, Island M D, Mobley H L T. Sequence of the Proteus mirabilis urease accessory gene ureG. Gene. 1993;129:103–106. doi: 10.1016/0378-1119(93)90703-6. [DOI] [PubMed] [Google Scholar]
- 25.Thomas V, Collins C M. Identification of UreR binding sites in the Enterobacteriaceae plasmid-encoded and Proteus mirabilis urease gene operons. Mol Microbiol. 1999;31:1417–1428. doi: 10.1046/j.1365-2958.1999.01283.x. [DOI] [PubMed] [Google Scholar]