SUMMARY
Quorum sensing fundamentally alters the interaction of Vibrio cholerae with aquatic environments, environmental hosts, and the human intestine. At high cell density, the quorum-sensing regulator HapR represses not only expression of cholera toxin and the toxin co-regulated pilus, virulence factors essential in human infection, but also synthesis of the Vibrio polysaccharide (VPS) exopolysaccharide-based matrix required for abiotic and biotic surface attachment. Here, we describe a feature of V. cholerae quorum sensing that shifts the host-pathogen interaction toward commensalism. By repressing pathogen consumptive anabolic metabolism and, in particular, tryptophan uptake, V. cholerae HapR stimulates host intestinal serotonin production. This, in turn, activates host intestinal innate immune signaling to promote host survival.
Graphical Abstract
In brief
Here, Jugder et al. show that the V. cholerae master high cell density regulator HapR represses pathogen tryptophan consumption. This provides host enterocytes precursors for serotonin synthesis, which activates intestinal innate immune signaling and prolongs host survival. Thus, V. cholerae HapR promotes a commensal relationship with the host.
INTRODUCTION
Regulatory quorum sensing circuits are encoded in the chromosomes of most if not all bacteria (Kareb and Aider, 2020; Papenfort and Bassler, 2016). These circuits allow bacteria to measure the density of their neighbors through receptors that bind specific diffusible signals that are synthesized and excreted into the environment. These quorum sensing receptors then activate a signaling cascade leading to global transcriptional changes.
Bacterial quorum sensing circuits play disparate roles in the host-microbe interaction. In the case of the symbiont Vibrio fischeri, quorum sensing plays an integral role in the development and survival of its squid host Euprymna scolopes in the environment (Essock-Burns et al., 2021; Lupp et al., 2003). In the human pathogen Pseudomonas aeruginosa, high cell density (HCD) quorum sensing increases expression of virulence factors to the detriment of the host (Chadha et al., 2021). The plant pathogen Agrobacterium tumefaciens uses HCD quorum sensing regulation to increase the copy number of its virulence plasmid and promote conjugal virulence plasmid transfer (Platt et al., 2014). The multiple quorum sensing circuits of the Gram-negative halophilic bacterium Vibrio cholerae have been intensively studied (Jung et al., 2016; Mashruwala and Bassler, 2020). Although only a subset of V. cholerae strains are human pathogens that express the principal intestinal colonization factor toxin co-regulated pilus (TCP) and the diarrheal agent cholera toxin (CTX), the genomes of pathogenic and environmental V. cholerae strains encode quorum sensing circuits. Given the heterogeneity of V. cholerae strains and diversity of environments they inhabit, the overarching role of quorum sensing in the life cycle of V. cholerae remains opaque.
A particularly complex V. cholerae quorum sensing circuit includes four quorum sensing receptors that respond to four unique ligands (Jung et al., 2015, 2016). At low cell density, these quorum sensing receptors are kinases that phosphorylate the response regulator LuxO. Phosphorylated LuxO activates transcription of four small regulatory RNAs, Qrr1–Qrr4, that promote translation of the low cell density transcription factor AphA and inhibit translation of the HCD transcription factor HapR. When their ligand is abundant, any one of these quorum sensing receptors can dephosphorylate LuxO to increase HapR protein levels (Jung et al., 2015). As the master regulator of adaptation to HCD, HapR increases transcription of hapA, encoding the secreted hemagglutinin protease responsible for most of the protease activity in V. cholerae supernatants (Wang et al., 2011). It also represses expression of the VPS biofilm exopolysaccharide synthesis enzymes and the virulence factors CTX and TCP (Beyhan et al., 2007; Waters et al., 2008; Zhu and Mekalanos, 2003; Zhu et al., 2002). Because TCP is the principal colonization factor in the small intestine, investigators have proposed that, at HCD, HapR mediates V. cholerae detachment from the human intestine and environmental dispersal (Fong et al., 2010; Tacket et al., 1998; Zhu et al., 2002). However, the ubiquity of these circuits in non-pathogenic Vibrio suggest that they may also play an important role outside of the human intestine.
Because V. cholerae is found in close association with arthropods in the environment (Broza et al., 2005; Halpern et al., 2004; Sack et al., 2003), we previously proposed the common fruit fly Drosophila melanogaster as a model arthropod host for V. cholerae (Blow et al., 2005). The fly intestine is comprised of a foregut, midgut, hindgut, and rectum (Capo et al., 2019). The midgut, which is the seat of digestion and nutrient absorption, is comprised of an anterior region (anterior midgut [AMG]) that houses the microbiota, an acidic middle region, and a relatively microbe-free posterior midgut (PMG) (Wong et al., 2016). Its epithelium includes enterocytes (ECs), enteroendocrine cells (EECs), and stem cells. Although all of these cell types encode components of the tumor necrosis factor (TNF)-like immune deficiency (IMD) innate immune signaling pathway, this pathway likely has a unique regulon in each cell type (Ferguson and Foley, 2021; Kamareddine et al., 2018a; Ryu et al., 2006; Wang et al., 2013).
Systemic IMD signaling is initiated by peptidoglycan monomers and polymers that are sensed by two peptidoglycan receptors, peptidoglycan recognition protein (PGRP)-LE and PGRP-LC. IMD signaling terminates in phosphorylation and cleavage of the transcription factor Relish, which is homologous to nuclear factor κB (NF-κB) of mammals. The N-terminal fragment of Relish, Rel68, then translocates to the nucleus to activate expression of several antimicrobial peptides (AMPs) as well as other genes, many of which have unknown significance in innate immunity (Kleino and Silverman, 2014). A subset of EEC in the Drosophila AMG and PMG express and secrete the enteroendocrine peptides Tachykinin (Tk), diuretic hormone 31, and neuropeptide F (Dutta et al., 2015; Guo et al., 2019). These cells coordinate EC AMP expression in response to commensal and pathogenic bacteria through transcriptional activation and release of Tk (Kamareddine et al., 2018a). Because Tk also represses intestinal lipid synthesis, decreased expression of Tk in EECs results in lipid accumulation in ECs (Song et al., 2014). Therefore, Tk may act as a metabolic regulator and as a cytokine (Jugder et al., 2021).
In oral infection of Drosophila, V. cholerae passes through the AMG, experiences a bottleneck in the acidic middle midgut, and accumulates in the PMG and rectum. Host death is the ultimate outcome. Although TCP and CTX are not major virulence factors in this infection, ingested V. cholerae activates transcription of the genes required for VPS exopolysaccharide-dependent biofilm formation, and this biofilm is essential for colonization of the fly intestine (Purdy and Watnick, 2011). As the infection progresses, the intestinal biofilm reaches densities that activate HCD quorum sensing and repress VPS biosynthesis genes in a HapR-dependent manner (Kamareddine et al., 2018b). However, in continuous feeding experiments, which do not depend on colonization, WT V. cholerae and VPS mutants are equally lethal to the Drosophila host (Kamareddine et al., 2018b).
We have reported previously that HapR attenuates V. cholerae virulence in the Drosophila host. Here we demonstrate that repression of tryptophan uptake by V. cholerae HapR fuels the conversion of tryptophan to serotonin in host ECs and that activation of host intestinal IMD signaling by serotonin prolongs host survival of infection.
In the mammalian host, modulation of dietary tryptophan by commensal microbes alters serotonin levels, and this affects intestinal innate immunity and inflammation (Gao et al., 2018; Haq et al., 2021; Liu et al., 2021). We propose that, in human and Drosophila infection, a decrease in V. cholerae tryptophan consumption as pathogen burden increases may attenuate intestinal inflammation and improve metabolic homeostasis.
RESULTS
A V. cholerae ΔhapR mutant demonstrates increased virulence
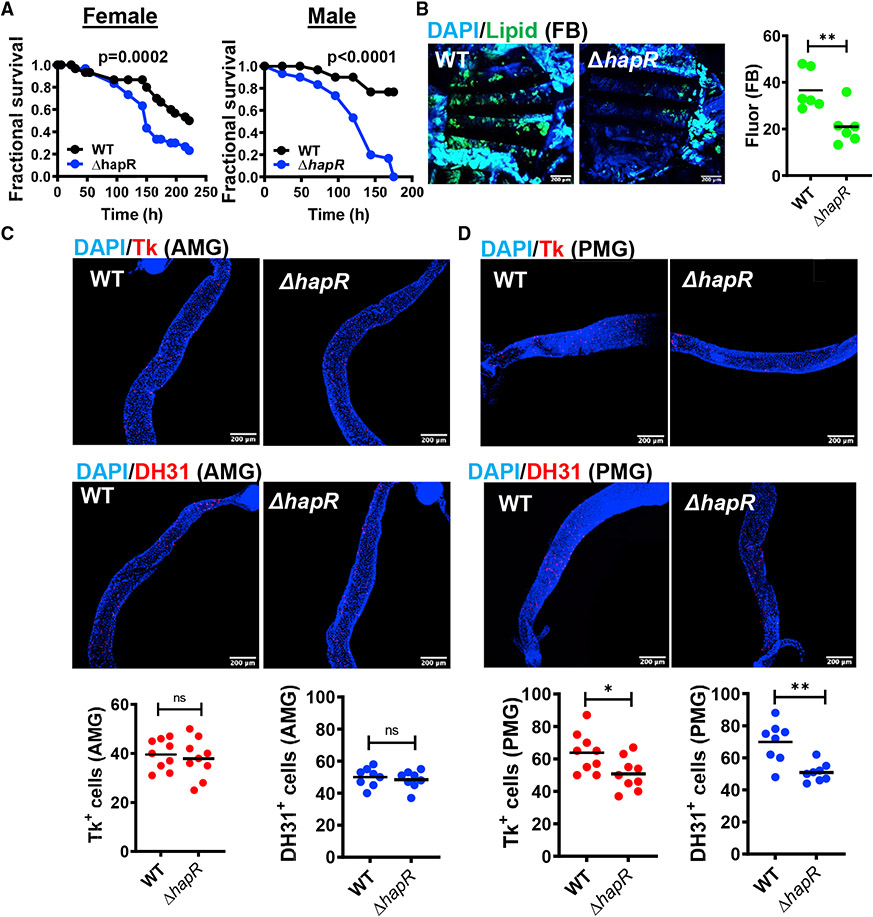
We have reported previously that a V. cholerae ΔhapR mutant is more virulent in a fly model of oral infection (Kamareddine et al., 2018b). Increased virulence has also been observed for a ΔhapR ΔvpsA mutant that is defective for biofilm formation. This more virulent phenotype was documented in infection of Oregon R (OreR) males and females (Figure 1A). Depletion of lipid stores from the main lipid repository, the fat body, was accelerated during a ΔhapR mutant infection (Figure 1B).
Figure 1. HCD V. cholerae quorum sensing promotes host survival, preserves host lipid stores, and modulates EEP expression in the PMG.
(A) Fractional survival over time of OreR flies fed WT V. cholerae or a ΔhapR mutant. Significance was calculated by log rank analysis.
(B) Representative micrographs and fluorescence quantification (lipid) in fat bodies stained with Bodipy and DAPI.
(C and D) Representative micrographs of Tachykinin (Tk) and diuretic hormone 31 (DH31) immunofluorescence in (C) the anterior midgut (AMG) or (D) posterior midgut (PMG) of flies analyzed in (B). Flies were dissected after 48 h of infection with the indicated V. cholerae strain. Scale bars, 200 μm.
For (B)–(D), at least 6 fat bodies or guts were examined. The mean is shown. Error bars represent the standard deviation. A Student’s t test was used to assess significance. **p < 0.01, *p < 0.05. See also Figure S1.
Because the PMG is stably colonized by V. cholerae (Purdy and Watnick, 2011), we questioned whether infection with a ΔhapR mutant might differentially alter Tk and DH31 expression in this intestinal compartment. In fact, compared with the intestines of flies infected with wild-type (WT) V. cholerae, we noted a significant decrease in Tk+ and DH31+ cells in the PMG but not AMG of OreR flies infected with a ΔhapR mutant (Figures 1C and 1D). Total intestinal Tk expression was also slightly but significantly decreased (Figure S1A). However, in contrast to the observation in TK knockdown flies, no increase in lipid accumulation was observed in the AMG or PMG (Figure S1B; Song et al., 2014). This suggests that Tk+ cells in the AMG and PMG respond to distinct microbial signals and that ΔhapR mutant infection does not decrease Tk expression sufficiently to derepress intestinal lipid synthesis.
V. cholerae HCD quorum sensing signaling activates ecdysone-dependent gene regulation, IMD signaling, and cuticular synthesis in the host intestine
We have reported previously that activation of V. cholerae HCD quorum sensing diminishes virulence in the model arthropod host D. melanogaster in part because of differential utilization of environmental succinate by the V. cholerae ΔhapR mutant (Kamareddine et al., 2018b). To better understand this process, we interrogated the host intestine transcriptional response to V. cholerae HCD quorum sensing by RNA sequencing (RNA-seq) analysis using the following infection model. Flies were fed V. cholerae in Luria-Bertani (LB) broth for 48 h, followed by a 24-h washout in PBS. We have found previously that, after this washout period, V. cholerae is principally present in the PMG and rectum (Purdy and Watnick, 2011). Colonization was preserved during the washout period, and we did not observe a significant difference in colonization between WT V. cholerae and a ΔhapR mutant (Figure S2A). As shown in Figures S2B and S3A, under these conditions, transcription of V. cholerae vpsL, a gene essential for VPS synthesis, was increased in ΔhapR mutant infection, whereas transcription of V. cholerae hapA was decreased. Therefore, in this experimental design, V. cholerae is stably associated with the Drosophila intestine, colonization by WT V. cholerae and a ΔhapR mutant is comparable, and HCD quorum sensing is activated.
Using this model, a transcriptomic comparison of WT V. cholerae and ΔhapR-infected fly intestines identified 782 differentially regulated fly genes (Table S1). Of these, the transcription of 672 genes was increased in WT infection, whereas transcription of 110 was increased in ΔhapR infection. This demonstrates that the Drosophila intestine actively responds to the changes imposed on the V. cholerae transcriptome by HapR regulation.
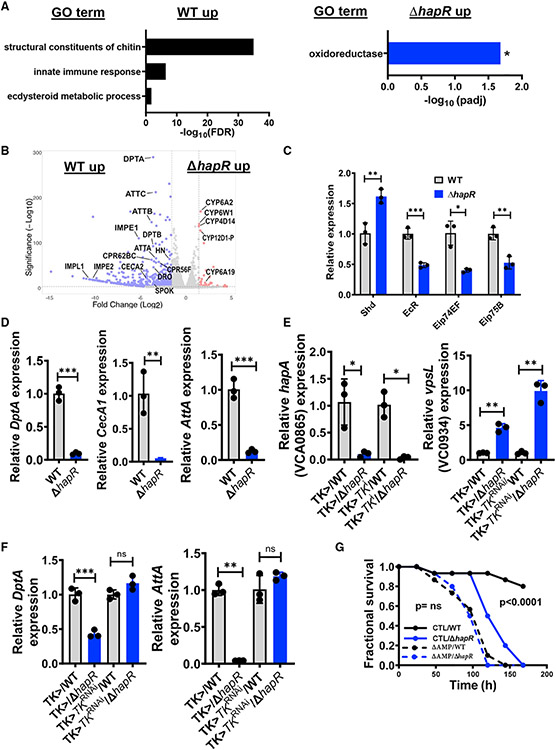
To uncover broad categories of differentially represented genes, we performed Gene Ontology (GO) analysis. The Gene Ontology Terms (GOTERMS) representing structural constituents of chitin or the cuticle, the IMD innate immune response, and the ecdysteroid metabolic process were significantly overrepresented in the list of Drosophila genes activated by WT V. cholerae infection, and IMD-regulated AMPs, ecdysone-responsive genes, and cuticular genes were among the most highly and significantly differentially upregulated in WT infection (Figures 2A and 2B). In contrast, only GOTERMs related to oxidation-reduction were significantly overrepresented in the list of host genes with increased transcription in ΔhapR mutant infection (Figures 2A and 2B).
Figure 2. Oral infection with a HCD quorum-sensing-defective V. cholerae mutant leads to decreased activation of ecdysone-responsive, innate immune, and epithelial repair genes.
(A and B) Gene Ontology (GO) analysis and (B) volcano plot of significantly regulated genes in the fly intestine during oral infection with WT V. cholerae compared with a ΔhapR mutant. Analysis is based on an RNA-seq experiment with biological triplicates that included an expression fold change greater than 2 with adjusted p < 0.05 to be significant. The calculated false discovery rate (FDR) is shown.
(C and D) qRT-PCR analysis of (C) the indicated 20E-regulated genes and (D) transcription of the IMD pathway-regulated AMPs DptA, CecA1, and AttA in the intestines of OreR flies orally infected with WT V. cholerae or a ΔhapR mutant.
(E and F) qRT-PCR transcriptional analysis of (E) the V. cholerae HapR-regulated genes hapA and vpsL and (F) the indicated IMD pathway-regulated AMPs in the intestines of Tk > driver only and Tk > TkRNAi flies orally infected with WT V. cholerae or a ΔhapR mutant. Measurements for ΔhapR-infected flies were normalized to that of WT V. cholerae-infected flies of similar genotype. For qRT-PCR results, the mean of biological triplicates is shown. Error bars represent the standard deviation. A Student’s t test was used to assess significance.
(G) Fractional survival over time of isogenic control and mutant flies with deletions in genes encoding the AMPs Def, AttC, Dro, AttA, AttB, DptA, Drs, and AttD fed WT V. cholerae or a ΔhapR mutant.
Significance was calculated by log-rank analysis. **p < 0.01, *p < 0.05, **p < 0.01, ***p < 0.001. See also Figures S2 and S3 and Table S1.
The steroid hormone 20-hydroxyecdysone (20E) regulates cuticle synthesis during development and potentiates Drosophila IMD signaling in many organ systems of the adult fly, including the intestine (Ali and Takaki, 2020; Flatt et al., 2008; Jugder et al., 2021; Nunes et al., 2021; Rus et al., 2013; Zheng et al., 2018). We observed differential activation of several 20E-regulated genes in the intestines of flies infected with WT V. cholerae (Figures 2B and S3B; Table S1). In addition, 46 genes associated with cuticle formation and at least 20 known IMD pathway-regulated genes, including 10 AMPs, were differentially activated in response to WT V. cholerae infection (Figure 2B; Table S1).
Some cuticular proteins we identified by GO analysis are expressed in all intestinal cell types, others are most highly expressed in EECs, and most are induced in response to infection (Dutta et al., 2015). A study in the silk moth Bombyx mori has demonstrated regulation of cuticular genes by the IMD pathway (Liang et al., 2015). We hypothesize that, when synthesized by intestinal epithelial cells, these proteins participate in peritrophic matrix synthesis, whose renewal would be a logical component of the innate immune response to intestinal infection. We propose that 20E-responsive, IMD-regulated, and cuticular genes fall within one regulatory pathway.
Activation of the intestinal IMD pathway by HCD quorum sensing follows a pathway similar to that utilized by the commensal microbiota
We previously showed that the commensal microbiota activates intestinal IMD signaling by amplifying 20E-dependent regulation via chromatin remodeling in Tk-expressing EECs (Jugder et al., 2021). IMD signaling in these cells controls expression of Tk, which then serves as a cytokine to activate IMD signaling in other cell types. To establish that WT V. cholerae activates IMD signaling via a similar route, we allowed OreR flies to ingest WT V. cholerae or a ΔhapR mutant in LB broth for 48 h, washed out non-adherent bacteria, and then harvested intestinal mRNA. We observed a pattern of 20E-regulated gene transcription similar to that observed in response to microbiota-derived acetate and consistent with activation by WT infection downstream of 20E synthesis (Figure 2C). We also corroborated increased transcription of DptA, CecA1, and AttA in WT infection (Figure 2D).
To determine whether Tk participates in IMD signaling in response to V. cholerae HCD quorum sensing, we administered WT V. cholerae or a ΔhapR mutant to Tk > driver-only and Tk > TkRNAi flies. V. cholerae HCD regulation was observed in the intestines of control and RNAi flies (Figure 2E). Despite this, infection with WT V. cholerae compared with a ΔhapR mutant yielded increased intestinal DptA and AttA expression only in driver control but not in Tk RNAi flies (Figure 2F). This suggests that increased AMP expression in response to V. cholerae HCD quorum sensing is dependent on Tk expression.
The commensal microbiota activates the intestinal innate immune response but does not kill the host. We questioned whether differential activation of IMD pathway signaling in a WT V. cholerae infection might account for decreased host susceptibility to infection. To test this, we infected parental flies and flies with deletions of eight IMD-regulated AMPs (ΔAMP) with WT or ΔhapR mutant V. cholerae and then monitored survival. As shown in Figure 2G, parental flies died significantly faster after infection with a ΔhapR mutant compared with infection with WT V. cholerae, whereas there was no difference in survival of ΔAMP flies after infection with WT V. cholerae or a ΔhapR mutant. These results suggest that WT V. cholerae prolongs host survival by activating intestinal IMD signaling through a pathway related to that employed by the commensal microbiota. Based on these findings, we propose that V. cholerae HCD quorum sensing promotes a commensal interaction with this host.
V. cholerae HapR regulates pathogen metabolism at HCD
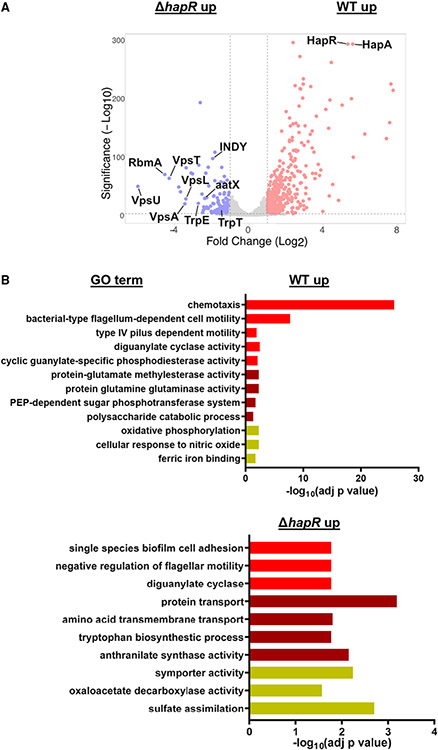
To identify differences in the bacterial transcriptome between WT V. cholerae and a ΔhapR mutant, we performed an RNA-seq experiment on planktonic bacteria grown in culture to early stationary phase. We included a ΔhapRΔvpsA mutant to assess the effect of increased VPS biosynthesis on the ΔhapR transcriptome. The conditions of this experiment differ from those of previously published microarray studies that focused on mid-exponential phase, AKI medium, or biofilm cultures (Yildiz et al., 2004; Zhu and Mekalanos, 2003; Zhu et al., 2002). A total of 501 genes were differentially regulated by HapR, including 345 genes with increased transcription in WT V. cholerae and 156 with increased transcription in a ΔhapR mutant (Table S2). Only expression of the vpsA gene was significantly different between the ΔhapR and ΔhapR ΔvpsA mutants. This suggests that, in early stationary phase, secondary effects of biofilm matrix synthesis on the ΔhapR transcriptome are minimal.
HapR has been reported previously to repress the canonical virulence factors TCP and CTX as well as biofilm formation (Vance et al., 2003; Zhu and Mekalanos, 2003; Zhu et al., 2002). As proof of principle, we noted that biofilm genes as well as the genes encoding the hemagglutinin protease were some of the most differentially and significantly regulated (Figure 3A). GO analyses of differentially regulated genes are shown in Figure 3B. As expected, we found that genes involved in flagellar and type IV pilus motility were differentially represented in the set of genes upregulated in WT V. cholerae compared with a ΔhapR mutant. In contrast, biofilm adhesion genes and those involved in repression of motility were differentially represented in the set of genes upregulated in a ΔhapR mutant.
Figure 3. HCD quorum sensing regulates amino acid and carbohydrate metabolism in addition to biofilm formation.
(A and B) Volcano plot (A) and GO analysis (B) of genes differentially expressed in 8-h LB cultures of WT V. cholerae and a ΔhapR mutant. Analysis is based on an RNA-seq experiment with bioiogical triplicates in which the fold change in expression was required to be greater than or equal to 2 and adjusted p < 0.05 was taken to be significant. The calculated adjusted p value is shown. See also Figures S4 and S5.
GO analysis also revealed a role of HapR in regulation of metabolism. In particular, although phosphoenolpyruvate phosphotransferase system (PTS)-dependent transport of trehalose, cellobiose, galactose, mannose, and glucose and oxidative phosphorylation were differentially represented in genes upregulated in WT V. cholerae, genes involved in the transport of amino acids such as alanine, glutamate, serine, threonine, and tryptophan as well as dicarboxylic acids were differentially represented in the group of genes upregulated in a ΔhapR mutant (Figures 3B, S4A, and S4B). We confirmed differential regulation of the three dicarboxylic acid transporters by qRT-PCR (Figure S4C). To document the functional relevance of these changes in transcription, we also demonstrated improved growth of WT V. cholerae on glucose and of the ΔhapR mutant on threonine, glutamate, and succinate (Figure S4D).
V. cholerae HapR represses the glyoxylate shunt
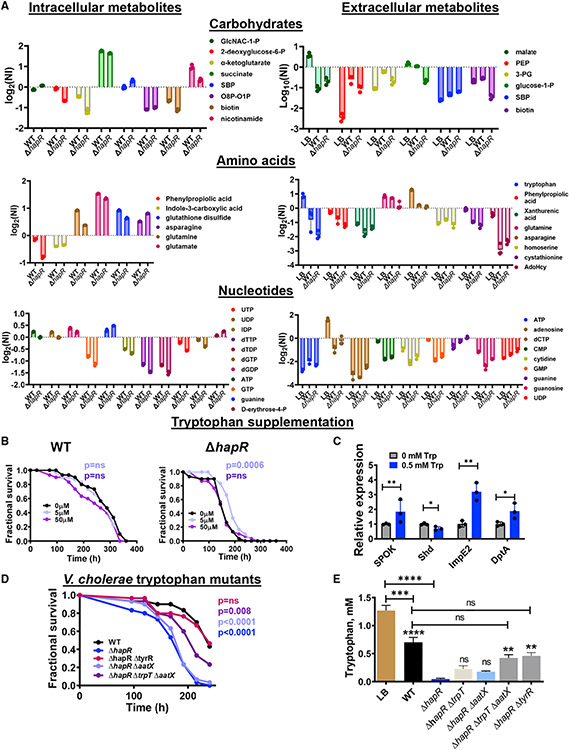
To further understand the role of HapR in regulation of V. cholerae metabolism, we analyzed the intracellular and extracellular polar metabolites of WT V. cholerae and a ΔhapR mutant in early stationary phase cultures (Tables S3 and S4). WT V. cholerae and a ΔhapR mutant consumed carbohydrates, amino acids, and nucleotides present in LB broth (Figure 4A). However, the ΔhapR mutant consumed and utilized these nutrients more avidly.
Figure 4. The decreased virulence of WT V. cholerae is recapitulated in a ΔhaprR mutant through tryptophan supplementation or inhibition of tryptophan uptake.
(A) Polar metabolomics of LB broth as well as the intracellular and extracellular milieus of WT V. cholerae and the ΔhapR mutant cultured for 8 h in LB broth. The mean of log2 of the normalized intensity (NI) of biological triplicates is shown. A one-way ANOVA with Fisher’s least significant difference (LSD) post hoc analysis was used to assess significance. Only significantly different metabolites with between WT and ΔhapR FDR < 0.05 are shown.
(B) Fractional survival of OreR flies infected with WT V. cholerae or a ΔhapR mutant and supplemented with the indicated concentrations of tryptophan. Log-rank analysis was used to calculate statistical significance.
(C) qRT-PCR analysis of 20E and IMD pathway-regulated genes in the intestines of uninfected OreR flies fed LB broth supplemented with the indicated concentrations of tryptophan. The mean of biological triplicates is shown. Error bars represent the standard deviation. A Student’s t test was used to calculate significance.
(D) Fractional survival of OreR flies infected with WT V. cholerae or a ΔhapR mutant alone or with deletions of the indicated genes hypothesized to be involved in tryptophan transport.
(E) High-performance liquid chromatography (HPLC) analysis of tryptophan concentrations in LB broth or the supernatants of the indicated V. cholerae strains harvested after 8 h of culture in LB broth. The mean of biological triplicates is shown. Error bars represent the standard deviation. A one-way ANOVA with Tukey’s multiple comparisons test was used to calculate statistical significance.
Floating symbols indicate significance with respect to the ΔhapR strain. ****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.05; ns, not significant. See also Figure S6.
Because the ΔhapR mutant does not activate HCD regulation, we hypothesized that it might be locked in a consumptive, anabolic state that would ordinarily be used to fuel log-phase growth. One characteristic of anabolic growth is transcriptional activation of the glyoxylate shunt (Figure S5A). Isocitrate lyase and malate synthase, the two enzymes involved in the glyoxylate shunt, enable nutrients catabolized through the tricarboxylic acid cycle to bypass steps in which carbon is lost as CO2, preserving these moieties for synthesis of essential cellular components (Dolan and Welch, 2018). As shown in Figure S5B, in early stationary-phase LB broth cultures of WT V. cholerae or a ΔhapR mutant, transcription of aceB, the gene encoding malate synthase, was significantly increased, whereas the increase in transcription of aceA, the gene encoding isocitrate lyase, did not reach statistical significance. In the absence of the glyoxylate shunt, growth on acetate is not possible because both carbons are lost as CO2 and, therefore, unavailable for synthesis of essential cellular components. We hypothesized that, if the glyoxylate shunt were repressed by HapR, then WT V. cholerae should grow more poorly than the ΔhapR mutant in minimal medium containing only acetate as a carbon source. We first established that transcription of aceA and aceB was differentially activated in a ΔhapR mutant during growth on acetate (Figure S5C). As predicted, the ΔhapR mutant grew to higher cell densities than WT V. cholerae in minimal medium containing acetate (Figure S5D). We conclude that, as V. cholerae growth slows at HCD, HapR actively represses the glyoxylate shunt as well as uptake of many nutrients, including dicarboxylic acids and amino acids.
Tryptophan uptake is correlated with virulence in the Drosophila model
V. cholerae uptake and excretion of metabolites dramatically alters the outcome of infection in a Drosophila model (Blow et al., 2005; Hang et al., 2014; Kamareddine et al., 2018b; Vanhove et al., 2017). In our metabolomic analysis, we noted significant differences in the intracellular and extracellular concentrations of aromatic amino acid metabolites, including phenylpropiolic acid, indole-3-carboxylic acid, tryptophan, and xanthurenic acid (Figure 4A). Tryptophan and its metabolites, such as indole-3-carboxylic acid and xanthurenic acid, are known bioactive compounds that are affected by the commensal intestinal microbiota of the fly (Lesperance and Broderick, 2020). In mammals, these metabolites have been shown to modulate intestinal innate immunity and inflammation (Ala, 2021; Gao et al., 2018). Therefore, we questioned whether differential utilization of tryptophan by a ΔhapR mutant might contribute to its increased virulence. To test this, we first supplemented the diets of WT V. cholerae or a ΔhapR-infected OreR flies with tryptophan. As shown in Figure 4B, low concentrations of tryptophan significantly improved survival of flies infected with a ΔhapR mutant but not WT V. cholerae. High tryptophan concentrations have been reported previously to be toxic to flies (Lesperance and Broderick, 2020), and this was also true for flies infected with WT V. cholerae or a ΔhapR mutant. The intestines of flies infected with a ΔhapR mutant display differential transcription of genes involved in 20E synthesis and signaling. If this were dependent on the intestinal tryptophan supply, we reasoned, then provision of supplemental tryptophan in the diet would have an inverse effect. In fact, we noted that supplementation of the diet with 0.5 mM tryptophan decreased transcription of the 20E synthesis gene shd but increased transcription of ImpE2 and DptA, a pattern opposite to that observed in a ΔhapR mutant infection (Figure 4C).
We noted that trpE, which encodes anthranilate synthase, the first enzyme in tryptophan synthesis, was upregulated approximately 16-fold in LB cultures of a ΔhapR mutant compared with that of WT V. cholerae, and this could be rescued by provision of hapR in trans (Table S2; Figure S6A). trpT, encoding a predicted tryptophan permease, was upregulated approximately 2-fold (Table S2; Figure S6A). This is consistent with the consumptive, anabolic program of the ΔhapR mutant. To determine whether V. cholerae tryptophan metabolism might play a role in pathogenesis, we constructed in-frame deletions in trpE, trpT, and the tryptophan repressor trpR in the parental and ΔhapR backgrounds and tested the virulence of these strains. As shown in Figure S6B, mutation of these genes singly in either background had only minor effects on host survival, and the ΔtrpT and ΔtrpR mutants grew well in M9 medium supplemented only with glucose (Figure S6C). The ΔtrpE and ΔtrpEΔtrpT mutants were also able to grow in M9/glucose medium, albeit with significant growth defects (Figure S6C). This suggests that there is redundancy in the V. cholerae tryptophan synthesis pathway. Redundancy in amino acid transport is also common (Vanhove et al., 2020). We found that supplementation of M9/glucose medium with 2 mM tryptophan enabled the ΔtrpEΔtrpT double mutant to grow at a rate similar to that of WT V. cholerae (Figure S6C). This suggests that there is redundancy in tryptophan transport as well. We noted that VC2283, which was significantly repressed by HapR, was predicted to be a TrpR-regulated tryptophan transporter in the RegPrecise database (Table S2; http://regprecise.sbpdiscovery.org:8080/WebRegPrecise/index.jsp; Novichkov et al., 2013). We named this gene aatX (aromatic amino acid transporter X). aatX demonstrated increased transcription in a ΔhapR mutant, and transcription was repressed by provision of hapR in trans (Figure S6A). aatX was also repressed by TrpR, whereas deletion of trpR had little effect on expression of TrpT (Figure S6D). Deletion of aatX alone had a small effect on virulence, whereas deletion of trpT and aatX significantly decreased the virulence of a ΔhapR mutant (Figure 4D). The transcription factor TyrR, which activates tryptophan transport in Escherichia coli in response to tyrosine and phenylalanine, was identified in a previous transposon screen for V. cholerae virulence genes in the Drosophila model (Hang et al., 2014). Deletion of tyrR also decreased the virulence of a ΔhapR mutant (Figure 4D). However, qRT-PCR suggested that TyrR does not regulate trpE, trpT, or aatX at HCD (Figure S6E).
HapR represses expression of tryptophan transporters, but we have no evidence that this is the case for TyrR. Therefore, as a more direct measurement of tryptophan uptake, we used high-performance liquid chromatography (HPLC) to measure tryptophan concentrations in the supernatants of 8-h LB broth cultures of WT V. cholerae, a ΔhapR mutant, and tryptophan uptake mutants. As shown in Figure 4E, the concentration of tryptophan in ΔhapR supernatants was more than 15-fold lower than that in WT supernatants. Although deletion of trpT or aatX singly did not significantly increase extracellular tryptophan concentrations in ΔhapR supernatants, the ΔhapRΔtrpTΔaatX and ΔhapRΔtyrR mutants consumed significantly less tryptophan than the ΔhapR mutant.
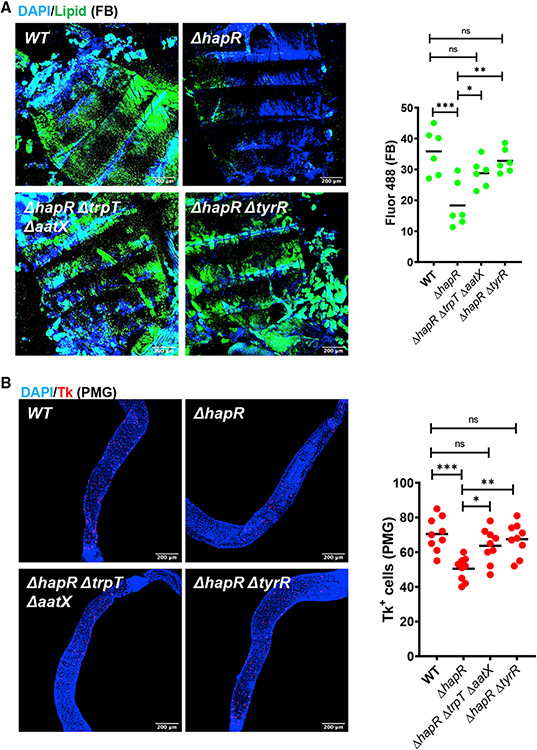
ΔhapR mutant infection results in decreased fat body lipid stores and decreased Tk+ cells in the host PMG. Deletion of the V. cholerae tryptophan uptake genes or tyrR in the ΔhapR mutant background also rescued these phenotypes (Figure 5). We conclude that impairment of tryptophan uptake in a V. cholerae ΔhapR mutant is correlated with reduced virulence and reduced disruption of lipid storage in the Drosophila host.
Figure 5. Decreased tryptophan uptake by the V. cholerae ΔhapR mutant preserves host fat stores and increases Tk expression in the PMG during infection.
(A) Representative micrographs and fluorescence quantification (lipid) in fat bodies of OreR flies infected with the indicated V. cholerae strains and stained with Bodipy and DAPI.
(B) Representative micrographs of Tk immunofluorescence in the PMG of flies analyzed in (A). Flies were dissected after 48 h of infection with the indicated V. cholerae strain. Scale bars, 200 μm. At least six intestines were evaluated. The mean is shown. A one-way ANOVA with Tukey’s multiple comparisons test was used to calculate statistical significance. ***p < 0.001, **p < 0.01, *p < 0.05.
Evidence that dietary serotonin activates intestinal innate immune signaling and improves survival of V. cholerae infection
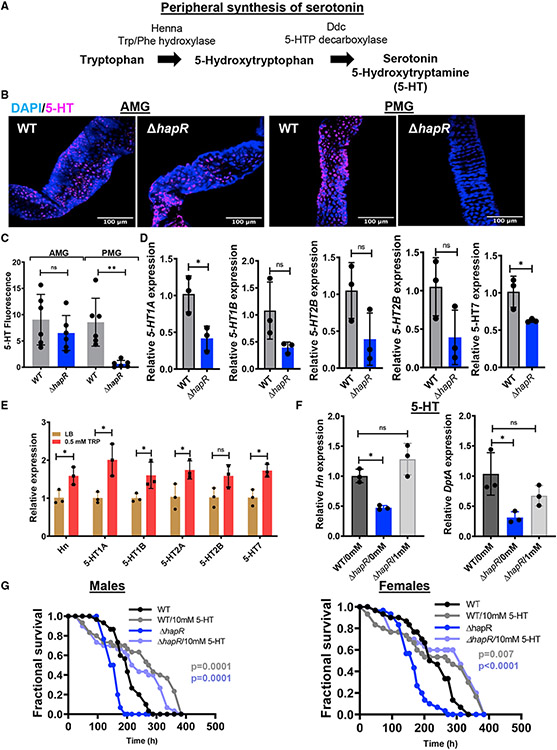
In mammals, intestinal tryptophan fuels synthesis of the neurotransmitter serotonin, which, in turn, modulates intestinal inflammation and epithelial barrier renewal (Liu et al., 2021; Morris et al., 2017). Tryptophan conversion to 5-hydroxytryptophan by tryptophan hydroxylase is the rate-limiting step in serotonin synthesis (Figure 6A). 5-hydroxytryptophan decarboxylase then converts 5-hydroxytryptophan to 5-hydroxytryptamine (5-HT) or serotonin. Mammals and Drosophila have two distinct tryptophan hydroxylases; one is expressed in the central nervous system and the other in peripheral tissues such as the intestine (Neckameyer et al., 2007; Walther and Bader, 2003; Zhang et al., 2004). The Drosophila peripheral tryptophan hydroxylase is named Henna (Hn) for its role in pigmentation. It is expressed in every cell type in the gut (Dutta et al., 2015). Our RNA-seq experiment showed increased transcription of Hn in the intestines of flies infected with WT V. cholerae, supporting a connection between intestinal tryptophan supply and serotonin synthesis (Figure 2B; Table S1).
Figure 6. Evidence that V. cholerae-derived tryptophan is converted to serotonin in the host intestine.
(A) Enzymes and intermediates involved in peripheral conversion of tryptophan to serotonin in Drosophila.
(B and C) Representative micrographs (B) and quantification (C) of serotonin immunofluorescence in the AMG and PMG of OreR flies infected with WT V. cholerae or a ΔhapR mutant for 48 h, followed by a 24-h PBS washout. Scale bars, 100 μm. At least six intestines were evaluated. The mean is shown. A Student’s t test was used to calculate significance.
(D and E) qRT-PCR analysis of serotonin receptor expression in the intestines of OreR flies (D) infected with WT V. cholerae or a ΔhapR mutant or (E) supplemented with tryptophan.
(F) qRT-PCR analysis of Hn and DptA expression in the intestines of OreR flies infected with WT or ΔhapR mutant V. cholerae alone or with serotonin (5-HT) supplementation.
(C–F) For qRT-PCR, the mean of biological triplicates is shown. Error bars represent the standard deviation. Significance was calculated using a Student’s t test (C–E) or a one-way ANOVA with Dunnett’s multiple comparisons test (F).
(G) Fractional survival of male and female OreR flies orally infected with WT or ΔhapR mutant V. cholerae alone or with serotonin (5-HT) supplementation. Significance was calculated by log rank analysis.
**p < 0.01, *p < 0.05. See also Table S5.
We hypothesized that increased intestinal tryptophan availability during infection with WT V. cholerae might result in increased serotonin synthesis. To assess intestinal tryptophan concentrations, we first performed metabolomics on the intestines of Drosophila continuously infected with WT V. cholerae or a ΔhapR mutant. However, no difference in intestinal tryptophan concentrations was detected (Table S5). We then considered the possibility that available tryptophan might be rapidly transported into host ECs and converted to serotonin. To examine this, we infected Drosophila with WT V. cholerae or a ΔhapR mutant for 48 h, followed by a 24-h washout period. We then detected serotonin in Drosophila intestines by immunofluorescence. As shown in Figure 6B and quantified in Figure 6C, serotonin was greatly decreased in the PMG of flies infected with the ΔhapR mutant compared with WT V. cholerae, whereas there was very little difference in the AMG. As further evidence that differential production of serotonin might underlie the differences in the intestinal transcriptome during WT and ΔhapR mutant infection, we measured intestinal transcription of several genes encoding serotonin receptors in the setting of infection. All showed a trend toward decreased expression in ΔhapR mutant infection (Figure 6D). However, statistical significance was only reached for 5-HT1A and 5-HT7. Tryptophan supplementation of the diets of uninfected Drosophila resulted in significantly increased transcription of Hn as well as the serotonin receptors (Figure 6E). Serotonin supplementation in the setting of a ΔhapR infection increased Hn and DptA expression to that measured in the intestines of flies infected with WT V. cholerae (Figure 6F). Serotonin supplementation also increased survival of male and female flies infected with WT V. cholerae or the ΔhapR mutant (Figure 6G). These data provide additional evidence of the role of increased serotonin synthesis in the transcriptional differences observed in the intestines of flies infected with WT V. cholerae or a ΔhapR mutant.
Serotonin synthesis in the intestine activates innate immune signaling and improves host survival of infection
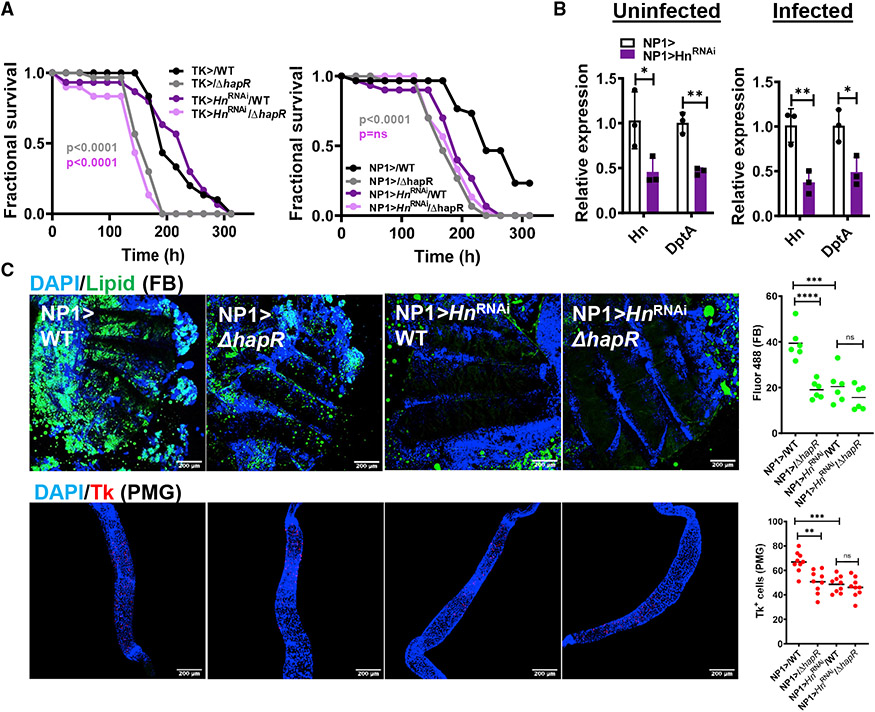
To directly test whether conversion of tryptophan to serotonin is involved in resistance to WT V. cholerae infection, we drove HnRNAi to Tk-expressing EECs and ECs. As shown in Figure 7A, mortality of Tk > HnRNAi flies was not significantly different from that of driver-only controls, whereas HnRNAi in ECs resulted in reduced survival of WT V. cholerae infection. We then documented reduced Hn transcription in the intestines of infected or uninfected NP1> HnRNAi compared with driver-only control flies (Figure 7B). DptA expression was also reduced in both, suggesting that serotonin synthesis controls IMD signaling. Expression of HnRNAi in ECs also decreased Tk+ cells in the PMG and lipid storage in the fat body of infected flies (Figure 7C). The most highly expressed intestinal serotonin receptor is 5-HT1A, and it is principally expressed in EECs (Dutta et al., 2015). However, knockdown of this receptor in EECs yielded only a partial phenotype (Figure S7). These observations suggest that diminished tryptophan uptake by WT V. cholerae provides this amino acid to host ECs for conversion to serotonin. This, in turn, amplifies expression of innate immune genes in the setting of infection and promotes host survival of V. cholerae infection.
Figure 7. Serotonin synthesis by Hn in ECs is responsible for the differential virulence of WT V. cholerae and a ΔhapR mutant in the Drosophila host.
(A) Fractional survival of EEC driver control (Tk>), EC driver control (NP1>), or cell-type-specific Hn knockdown flies orally infected with WT V. cholerae or a ΔhapR mutant. Log rank analysis was used to calculate significance.
(B) qRT-PCR analysis of expression of the indicated genes in EC driver control (NP1>) or NP1> HnRNAi uninfected or WT V. cholerae-infected flies. A washout step was performed prior to dissection. The mean of biological triplicates is shown. Error bars represent the standard deviation.
(C) Representative micrographs and fluorescence quantification (lipid) in fat bodies stained with Bodipy and DAPI and PMG immunofluorescence imaging and quantification of Tk in the intestines of Drosophila with the indicated genotypes and infected with the indicated V. cholerae strains. Flies were dissected after 48 h of infection with the indicated V. cholerae strain. Scale bars, 200 μm. At least six intestines were evaluated. The mean is shown. A Student’s t test was used to calculate statistical significance.
****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.05. See also Figure S7.
DISCUSSION
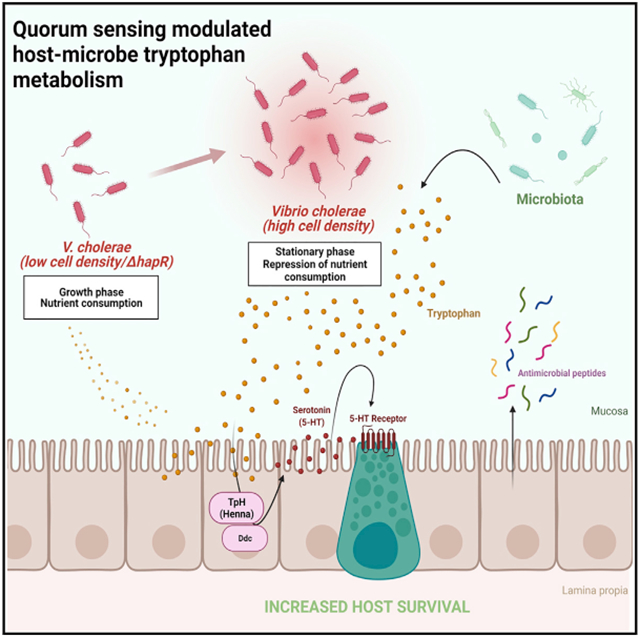
Here we show that the HCD quorum sensing master regulator HapR represses consumptive, anabolic metabolism in the diarrheal pathogen V. cholerae. Repression of V. cholerae tryptophan uptake, in particular, fuels host intestinal serotonin synthesis. This activates intestinal innate immune signaling and slows expenditure of host lipid stores. Thus, V. cholerae HCD signaling alters the outcome of infection by reducing the competition for tryptophan between host and pathogen.
Although V. cholerae HCD quorum sensing is thought to mitigate infection by repressing the canonical virulence factors, here we show that it also provides the host with an unexpected means of limiting intestinal colonization. Tryptophan is an essential amino acid for flies and mammals, the majority of which is obtained from the diet (Agus et al., 2018). In contrast, most bacteria can synthesize tryptophan via endogenous pathways but prioritize transport from the intestinal environment (Lingens, 1968). Tryptophan may be converted into serotonin by the host intestine, into indole by intestinal bacteria, or into kynurenine and its metabolites by both (Agus et al., 2018). Intestinal conversion of tryptophan to serotonin plays a key role in many disease states, including irritable bowel syndrome, intestinal inflammation, inflammatory bowel disease, colorectal cancer, and autism spectrum disorder (Haq et al., 2021; Mishima and Ishihara, 2021; Settanni et al., 2021). Here we provide evidence that serotonin synthesized by the Drosophila intestine in response to V. cholerae HCD regulation protects the host by activating the intestinal innate immune response.
Repression of V. cholerae virulence by the HCD master regulator HapR has been conceived as an adaptation that accelerates dissemination of V. cholerae into the environment (Merrell et al., 2002; Tsou et al., 2008; Zhu et al., 2002). However, V. cholerae HCD regulation is also believed to repress pathogen accumulation in the host intestine via multiple mechanisms. First, decreased synthesis of TCP and VPS limits V. cholerae colonization of the intestinal epithelium. Second, CTX increases the concentration of sodium in the small intestine from 20–30 mM to 90–120 mM, which is ideal for growth of a halophile such as V. cholerae (Molla et al., 1981; Spiller, 1994). Therefore, repression of CTX expression by HapR creates an unfavorable intestinal environment. Third, decreased expression of myriad transporters and anabolic pathways limits the ability of V. cholerae to synthesize the building blocks necessary for pathogen proliferation. Last, decreased V. cholerae tryptophan consumption shuttles this precursor of serotonin to the host. Serotonin synthesis, in turn, activates the host innate immune response and increases expression of AMPs to reduce pathogen burden. Because all of these functions prolong host survival, we hypothesize that HCD quorum sensing regulation in V. cholerae promotes cooperation between host and pathogen.
Limitations of the study
Animal models for the study of V. cholerae pathogenesis have strengths and weaknesses. With the exception of humans, intestinal expression of V. cholerae virulence factors, colonization of the small intestine, and osmotic diarrhea have only been observed in neonatal mammals (Attridge et al., 1996; Thelin and Taylor, 1996). These models permit elucidation of V. cholerae virulence factor regulation in vivo. However, V. cholerae HCD quorum sensing is not activated in the neonatal mouse intestine (Zhu et al., 2002). To enable a study of the effect of HCD quorum sensing and serotonin synthesis on the host intestine, here we use a Drosophila model. In this model, the canonical virulence factors do not play a major role in pathogenesis, and the intestinal environment and response to microbes may differ from that of the mature mammalian intestine (Blow et al., 2005). Therefore, a direct confirmation of our findings awaits a human study or development of an improved mammalian model.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for reagents may be directed to and will be fulfilled by the lead contact, Dr. Paula I. Watnick (paula.watnick@childrens.harvard.edu).
Materials availability
All mutants and plasmids generated in this study are available from the lead contact without restriction.
Data and code availability
RNA-seq data for WT V. cholerae and a ΔhapR mutant grown to early stationary phase (NCBI: PRJNA810852) and Drosophila intestines infected with these two strains (NCBI: PRJNA811240) have been deposited in the NCBI repository. No code was generated as a part of these studies. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Drosophila Husbandry and strains
Flies were reared on standard fly food (16.5 g L−1 yeast, 9.5 g L−1 soy flour, 71 g L−1 cornmeal, 5.5 g L−1 agar, 5.5 g L−1 malt, 7.5% corn syrup and 0.4% propionic acid) and kept in incubators at 25°C, 70% humidity on a 12 h light/dark cycle. The following fly lines were obtained from the Bloomington Drosophila Stock Center (http://flystocks.bio.indiana.edu/): Ore R-C (BL 5), yw (BL 1495), HnRNAi (BL 60025), and 5-HT1ARNAi (BL 33885). The TK-Gal4 and NP1-Gal4 (Myo1A-Gal4) driver flies were kind gifts from Norbert Perrimon. For all experiments, 5–7 day old flies were used. Female flies were used unless otherwise noted.
V. cholerae strains
Wild-type and mutant V. cholerae all derive from wild-type V. cholerae strain HC-494 isolated from a cholera patient in year 2013 of the Haitian V. cholerae epidemic (Azarian et al., 2014; Kamareddine et al., 2018b). This strain is quorum sensing-competent and resistant to streptomycin. For all experiments, V. cholerae strains were prepared by culturing in LB broth supplemented with 100 μg/mL streptomycin (Sm100).
METHOD DETAILS
Vibrio cholerae mutant construction and rescue
V. cholerae mutants were constructed by double homologous recombination as previously described (Vanhove et al., 2020) in the background of the HC494 strain collected from Haiti in 2013 (Azarian et al., 2014). Suicide plasmids for gene deletions were constructed using the primers listed in Table S6. The hapR gene was amplified using the primers listed in Table S6 and then cloned into the pFLAG expression vector under IPTG control. For rescue experiments, expression was induced with 1 mM IPTG.
Vibrio cholerae growth measurements
Starter cultures were inoculated into LB broth from single colonies harvested from LB-agar plates supplemented with 100 μg/mL of streptomycin and grown overnight. The cells were then washed three times with phosphate-buffered saline (PBS) to remove all traces of LB broth and then resuspended in 1 mL of M9 medium. This cell suspension was diluted 1:100 into M9 medium supplemented with the indicated carbon source. 100 ul aliquots were then dispensed into a 96 well plate in triplicate, and the optical density at 600 was measured using an Infinite M Nano Microplate Reader (Tecan).
Vibrio cholerae infections
Oral Vibrio cholerae infections were carried out in an arthropod containment level 2 facility in a 25°C incubator with 12 h light/dark cycles. Survival was measured as previously described (Jugder et al., 2021). For RT-qPCR and bacterial burden experiments, midguts were harvested either directly after a 48-h infection or after an additional 24-h wash-out period on sterile PBS as indicated.
Immunofluorescence
The midguts and fat bodies of 5- to 7-day-old female flies were dissected and fixed at room temperature in PBS+ 1% Tween 20 (PBS-T) containing 4% formaldehyde (Thermo Fisher Scientific NC9658705) for at least 30 min. After fixation, tissues were washed three times with fresh PBS -T for 10 min at room temperature, followed by incubation for 1 h at room temperature in a blocking buffer comprised of PBS-T with 2% BSA and 0.1% Triton X-100 (Sigma-Aldrich 9002-93-1). Guts were then incubated overnight at 4°C with the primary Rabbit anti-Tk, Rabbit anti-DH31, or Rat anti-serotonin antibody (Abcam, AB6336) at a dilution of 1:500 in blocking buffer. Samples were then washed three times for 10 min in PBS-T, incubated in blocking buffer supplemented with an Alexa 594-conjugated anti-rabbit secondary antibody (1:500, Thermo Fisher Scientific A-11012) or Alexa 488-conjugated anti-Rat secondary antibody (1:1000, Invitrogen, A48262), DAPI (1:1000, Invitrogen D1306), and BODIPY 493/503 (1 mg mL−1, Invitrogen D3922) for 2 h at room temperature, and washed again in PBS-T three times for 10 min. The tissues were then mounted in Vectashield mounting medium (Vector Laboratories H-1000). Confocal images were acquired using a Zeiss LSM 780 inverted confocal microscope with a 90.1 μM pin hole diameter, 1.11 μM pixel size, a 1.58 μsec/pixel scan rate, and 566.8 μM × 566.8 μM scan area. An Argon laser was used for illumination with 594 nm, 488 nm, and 405 nm laser lines. A 10X oil objective for Tk and lipid imaging and a 40X oil objective for serotonin imaging. Zen Blue v. 1.0 acquisition software was used. Tachykinin-expressing cells in the anterior and posterior midgut were counted manually. For assessment of lipid accumulation in fat bodies or intestines, ImageJ (FIJI) was used to quantify total fluorescence. This was normalized by dividing by the total area included in the measurement. Background fluorescence was subtracted from this measurement. A minimum of 6 midguts or fat bodies were quantified for each condition and genotype.
Drosophila RNA extraction, RNAseq, and RT-qPCR
Total RNA was extracted from a minimum of 10 midguts or whole flies using TRIzol reagent (Thermo Fisher Scientific 15596026) followed by further purification using the Direct-zol RNA MiniPrep Plus kit (Zymo Research R2070) according to the supplier’s instructions. RNA was quantified using a NanoDrop 1000 Spectrophotometer. cDNA was synthesized from 500 ng of total RNA using the iScript cDNA Synthesis Kit (Bio-Rad 1708891) as described by the manufacturer.
RT-qPCR was performed using iTaq SYBR Green (Bio-Rad 1725121) on a StepOnePlus real-time PCR system (Applied Biosystems). Three biological replicates with technical duplicates of each were performed for each experiment. Relative expression of target genes was calculated using the comparative CT method normalized to rp49. Primers used for RT-qPCR analyses are listed in Table S6.
Library construction, paired-end sequencing on the Illumina NovaSeq platform, and statistical analyses were performed by the Molecular Biology Core Facilities (MBCF) at the Dana-Farber Cancer institute (http://mbcf.dfci.harvard.edu/genomics/next-generation-sequencing).
Vibrio cholerae RNA extraction, RNAseq and RT-qPCR
Overnight cultures of V. cholerae in LB broth were diluted in a ratio of 1:100 into fresh LB broth. After growth for 8 h at 27°C with shaking, the cell pellet from 1 mL of culture was resuspended in TRIzol reagent and heated for 10 min at 60°C or lysed using FastPrep tubes containing 0.1 mm Zirconia/Silica beads (MP Biomedicals MP115076600). Samples were either stored at −80°C or immediately purified with the Direct-zol RNA Miniprep kit according to the manufacturer’s instructions. For some experiments, following elution into 100 μL of water, samples were subjected to Turbo DNase treatment for 45 min at 37°C with shaking, a second purification step with the Direct-zol RNA Miniprep kit and elution in 70 μL of water.
For RNAseq and statistical analysis, these sample were submitted to the The Microbial ‘Omics Core (MOC) at the Broad Institute (https://www.broadinstitute.org/infectious-disease-and-microbiome/microbial-’omics-core). cDNA was prepared for qRT-PCR using iScript Reverse Transcription Supermix (Bio-Rad 1708841) according to the manufacturer’s protocol. PowerUP SYBR Green Master Mix (Applied Biosystems 10002984) was used in a StepOnePlus Real-Time PCR System (Applied Biosystems). Three biological replicates with duplicate technical measurements were performed for each experiment. Relative expression of target genes was calculated using the comparative CT method normalized to clpX. Primers for RT-qPCR are listed in Table S6.
Sample preparation for V. cholerae metabolomics
Single colonies were harvested from V. cholerae cultured overnight on LB agar plates supplemented with Sm100 and resuspended in 1 mL of LB broth supplemented with Sm100. This suspension was diluted to yield a starting OD600 of 0.01 and cultured for 8 h at 27°C. The culture was then centrifuged, and cell and supernatants were separated. The supernatant was filtered through a 0.22 uM filter (Fisher) and combined with methanol in a ratio of 1:4 supernatant:methanol. Cell pellets were resuspended in 1 mL 80% methanol. Both pellets and supernatant mixtures were incubated overnight at −80°C and then centrifuged to remove cell debris, transferred to clean tubes, and dried.
Sample preparation for metabolomics of the Drosophila intestine.
Intestines were prepared for polar metabolomic analysis as previously described (Vanhove et al., 2017). Briefly, the intestines of 15 y1w1 V. cholerae-infected flies for each of three biological replicates derived from independent vials were dissected and homogenized in 500 μL of a cold methanol:water solution (80:20). After incubation for 2 h at −80°C, samples were centrifuged for ten minutes at 14,000 × g, transferred to a new vial, re-extracted with methanol:water solution (400 μL), and incubated for 6 h at −80°C. Supernatants from the two extractions were combined and desiccated at room temperature in a Speedvac concentrator.
Targeted mass spectrometry
Samples were re-suspended in 20 mL HPLC grade water for mass spectrometry. 5–7 μL were injected and analyzed using a hybrid 6500 QTRAP triple quadrupole mass spectrometer (AB/SCIEX) coupled to a Prominence UFLC HPLC system (Shimadzu) via selected reaction monitoring (SRM) of a total of 298 endogenous water-soluble metabolites for steady-state analyses of samples. Some metabolites were targeted in both positive and negative ion mode for a total of 309 SRM transitions using positive/negative ion polarity switching. ESI voltage was +4950V in positive ion mode and −4500V in negative ion mode. The dwell time was 3 ms per SRM transition and the total cycle time was 1.55 s. Approximately 9–12 data points were acquired per detected metabolite. Samples were delivered to the mass spectrometer via hydrophilic interaction chromatography (HILIC) using a 4.6 mm i.d x 10 cm Amide XBridge column (Waters) at 400 μL/min. Gradients were run starting from 85% buffer B (HPLC grade acetonitrile) to 42% B from 0-5 min; 42% B to 0% B from 5-16 min; 0% B was held from 16-24 min; 0% B to 85% B from 24-25 min; 85% B was held for 7 min to re-equilibrate the column. Buffer A was comprised of 20 mM ammonium hydroxide/20 mM ammonium acetate (pH = 9.0) in 95:5 water:acetonitrile. Peak areas from the total ion current for each metabolite SRM transition were integrated using MultiQuant v3.0 software (AB/SCIEX).
Quantification of tryptophan in supernatants by HPLC-MS
Supernatants were prepared as described for metabolomics. L-tryptophan (L-Trp) quantification was carried out on an Agilent 1200 HPLC instrument equipped with a Luna 5 μm C18(2) 100 Å column (Phenomenex, 250 mm × 4.6 mm) and coupled with an Agilent LC/MS 6130 Quadrupole analyzer. The mobile phase was composed of 0.1% formic acid in water (v/v) (A) and 0.1% formic acid in acetonitrile (v/v) (B). Linear gradient elution was performed at 0.5 mL/min starting at 1% B increasing to 80% B at 15 min, and finally returning to 1% B at 18 min for column re-equilibration, which was completed at 20 min. Chromatographic data were processed using Agilent OpenLab ChemStation software. The retention time of L-Trp was 11 min, and the wavelength was 225 nm. For the MS detector, data acquisition was performed in single ion monitoring mode [m/z]+ 205 for L-Trp.
QUANTIFICATION AND STATISTICAL ANALYSIS
GraphPad Prism 9.0 software was used for generation of all graphs and for statistical analyses with the exception of transcriptomic and metabolomic analyses. Measurements represent the mean of at least three biological replicates in all graphs. Error bars represent mean ± the standard deviation. As appropriate, a two-tailed Students t-test, ordinary one-way ANOVA or Brown-Forsythe ANOVA with post-hoc Dunnett’s test was used to calculate significance. The statistical test applied is noted in the figure legend. For all tests, statistical significance was calculated as a p value. Asterisks indicate the following p value ranges: * for p < 0.05, ** for p < 0.01, *** for p < 0.001, **** for p < 0.0001, and ns (non-significant) for p > 0.05.
For Drosophila and V. cholerae transcriptomic experiments, DEseq was used to calculate fold change and statistical significance. For both, data were filtered to select genes whose transcription was altered by 2-fold or more and yielded an adjusted p-value of ≤ 0.05. For Drosophila transcriptomic data, lists of genes with increased expression in the intestines of flies infected with WT V. cholerae or a ΔhapR mutant were subjected to GO analysis separately using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) v6.8 (Huang da et al., 2009a; b). Categories that were differentially represented with a p value of ≤0.05 were considered significant. For V. cholerae transcriptomic data, lists of genes with increased expression in WT V. cholerae cultures and ΔhapR mutant cultures were subjected to GO analysis separately using the GSEA-PRO v3 online tool (http://gseapro.molgenrug.nl/). Volcano plots were generated using VolcaNoseR (Goedhart and Luijsterburg, 2020). Metabolomics data were processed using the open access program Metaboanalyst 5.0 (https://www.metaboanalyst.ca/) (Pang et al., 2021; Xia et al., 2009) as follows. Samples were first normalized by sum and then Log2 transformed. To calculate significance, a Student’s t test was used for intracellular metabolites and a one-way analysis of variance (ANOVA) with a Fisher’s LSD post hoc test was used for supernatants. A false discovery rate cut-off of 0.05 was used in both cases.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat polyclonal anti-Rabbit IgG (H + L) Secondary Antibody, Alexa Fluor® 594 conjugate | Thermo Fisher Scientific | Cat# A-11012; RRID:AB_2534079 |
| Goat anti-rat IgG (H+L) Secondary Antibody, Alexa Fluor® 488 conjugate | Fisher Scientific, Invitrogen | A48262; RRID:AB_2896330 |
| Rabbit anti-Tachykinin (DTK) | Kind Gift from Jan Adrianus Veenstra | https://www.ncbi.nlm.nih.gov/pubmed/18972134; RRID:AB_2923320 |
| Rabbit anti-DH31 | Kind Gift from Jan Adrianus Veenstra | https://www.ncbi.nlm.nih.gov/pubmed/18972134; RRID:AB_2923321 |
| Rat anti-Serotonin | Abcam | AB6336; RRID:AB_449517 |
| Chemicals, peptides, and recombinant proteins | ||
| Bacto-Agar | VWR | Cat# 214010 |
| LB Broth | Fisher Scientific (BD Difco) | DF0446-17-3 |
| Sodium Phosphate, Dibasic | Sigma-Aldrich | 567550 |
| Potassium phosphate monobasic | Sigma-Aldrich | P5655 |
| Sodium chloride | Sigma-Aldrich | S5886 |
| Ammonium chloride | Sigma-Aldrich | A9434 |
| Glucose | Sigma-Aldrich | 1083421000 |
| MgSO4 | Sigma-Aldrich | 208094 |
| CaCl2 | Sigma-Aldrich | C5670 |
| Alanine | Sigma-Aldrich | A7469 |
| Arginine monohydrochloride | Sigma-Aldrich | A6969 |
| Cysteine | Sigma-Aldrich | C7352 |
| Glycine | Sigma-Aldrich | G8790 |
| Histidine hydrochloride monohydrate | Sigma-Aldrich | H5659 |
| Isoleucine | Sigma-Aldrich | I7403 |
| Leucine | Sigma-Aldrich | L8912 |
| Lysine | Sigma-Aldrich | L8662 |
| Methionine | Sigma-Aldrich | M2768 |
| Phenylalanine | Sigma-Aldrich | P5482 |
| Serine | Sigma-Aldrich | S4311 |
| Threonine | Sigma-Aldrich | T8441 |
| Tyrosine | Sigma-Aldrich | T8566 |
| Valine | Sigma-Aldrich | V0513 |
| L-tryptophan | T0254 | |
| Phosphate buffered saline (PBS) | TEKnova | Cat# P0191 |
| Bovine Serum Albumin | Sigma-Aldrich | Cat# A8022 |
| Triton™ X-100 | Sigma-Aldrich | Cat# 9002-93-1 |
| DAPI (4′,6-diamidino-2-phenylindole,Dihydrochloride) | Invitrogen | Cat# D1306 |
| BODIPY® 493/503 (4,4-Difluoro-1,3,5,7,8-Pentamethyl- 4-Bora-3a,4a-Diaza-s-Indacene) | Invitrogen | Cat# D3922 |
| Vectashield Antifade Mounting Medium | Vector Laboratories | Cat# H-1000 |
| Molecular Biology Grade Ethanol | Thermo Fisher Scientific | Cat# BP28184 |
| Methanol | Sigma-Aldrich | 34860 |
| Tween20 | American Bioanalytical | Cat# 9005-64-5 |
| TRIzol Reagent | Thermo Fisher Scientific | Cat# 15596026 |
| Paraformaldehyde | EMS | Cat# 19208 |
| Sodium Acetate | Thermo Fisher Scientific | Cat# 127-09-3 |
| Ampicillin, Sodium Salt | Thermo Fisher Scientific | Cat# 69-52-3 |
| Streptomycin, Sulfate | Sigma-Aldrich | Cat# 3810-74-0 |
| Critical commercial assays | ||
| Direct-zol RNA MiniPrep Plus | Zymo Research | Cat# R2070 |
| iScript cDNA Synthesis Kit | Bio-Rad | Cat# 1708891 |
| iTaq Universal SYBR Green Supermix | Bio-Rad | Cat# 1725121 |
| PowerUP SYBR Green Master Mix | Applied Biosystems | 10002984 |
| Turbo DNase | Thermo Fisher Scientific | Cat# AM2238 |
| Deposited data | ||
| WT V. cholerae vs a ΔhapR mutant | Boston Children’s Hospital | NCBI: PRJNA810852 |
| Ore R Drosophila colonized with WT V. cholerae vs a ΔhapR mutant | Boston Children’s Hospital | NCBI: PRJNA811240 |
| Experimental models: Organisms/strains | ||
| OreR-C | Bloomington Drosophila Stock Center (BDSC) | BL 5 |
| y1w1 | BDSC | BL 1495 |
| HnRNAi line | BDSC | BL 60025 |
| 5-HT1ARNAi line | BDSC | BL 33885 |
| delta-AMPs line (iso DrosDel w1118; DefSK3, AttCMi, DroAttABSK2, MtkR1, DptSK1; DrsR1, AttDSK1) | Kind gift from Bruno Lemaitre | https://www.ncbi.nlm.nih.gov/pubmed/30803481 |
| Tk-Gal4 | Kind gift from Norbert Perrimon | N/A |
| Myo1A-Gal4 | Kind gift from Norbert Perrimon | N/A |
| AstC-Gal4 | BDSC | BL52017 |
| Software and algorithms | ||
| Graphpad Prism 7.00 | Boston Children’s Hospital | N/A |
| Fiji (ImageJ) | https://imagej.net/software/fiji | |
| Database for Annotation, Visualization and Integrated Discovery (DAVID) | https://david.ncifcrf.gov/ | N/A |
| Metaboanalyst v5.0 | https://www.metaboanalyst.ca/ | N/A |
Highlights.
V. cholerae HapR represses pathogen tryptophan consumption
Host enterocytes convert dietary tryptophan to serotonin
Intestinal serotonin synthesis activates IMD signaling and prolongs host survival
HapR orchestrates a commensal interaction between host and pathogen
ACKNOWLEDGMENTS
This work was supported by NIH R01 162701 (to P.I.W.) and Harvard Bacteriology PhD Training Program T32 AI132120 (to J.A.G.). Anti-TK and anti-DH31 antibodies were generously provided by Jan Veenstra, and AMP-deficient flies and the corresponding parental strain were generously provided by Bruno Lemaitre. The TK-Gal4 and NP1-Gal4 (Myo1A-Gal4) driver flies were kind gifts from Norbert Perrimon. Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study. The graphical abstract was created with BioRender. Microscopy images were acquired at the Microscopy Resources on the North Quad (MicRoN) core at Harvard Medical School. We thank Paola Montero-Lopis at the MicRoN core for providing expertise with image acquisition and quantification and Wayne Lencer for helpful discussions.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111368.
REFERENCES
- Agus A, Planchais J, and Sokol H (2018). Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host Microbe 23, 716–724. 10.1016/j.chom.2018.05.003. [DOI] [PubMed] [Google Scholar]
- Ala M (2021). Tryptophan metabolites modulate inflammatory bowel disease and colorectal cancer by affecting immune system. Int. Rev. Immunol 41, 326–345. 10.1080/08830185.2021.1954638. [DOI] [PubMed] [Google Scholar]
- Ali MS, and Takaki K (2020). Transcriptional regulation of cuticular genes during insect metamorphosis. Front. Biosci 25, 106–117. 10.2741/4796. [DOI] [PubMed] [Google Scholar]
- Attridge SR, Manning PA, Holmgren J, and Jonson G (1996). Relative significance of mannose-sensitive hemagglutinin and toxin- coregulated pili in colonization of infant mice by Vibrio cholerae El Tor. Infect. Immun 64, 3369–3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azarian T, Ali A, Johnson JA, Mohr D, Prosperi M, Veras NM, Jubair M, Strickland SL, Rashid MH, Alam MT, et al. (2014). Phylodynamic analysis of clinical and environmental Vibrio cholerae isolates from Haiti reveals diversification driven by positive selection. mBio 5, e01824–14. 10.1128/mBio.01824-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyhan S, Bilecen K, Salama SR, Casper-Lindley C, and Yildiz FH (2007). Regulation of rugosity and biofilm formation in Vibrio cholerae: comparison of VpsT and VpsR regulons and epistasis analysis of vpsT, vpsR, and hapR. J. Bacteriol 189, 388–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow NS, Salomon RN, Garrity K, Reveillaud I, Kopin A, Jackson FR, and Watnick PI (2005). Vibrio cholerae infection of Drosophila melanogaster mimics the human disease cholera. PLoS Pathog. 1, e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broza M, Gancz H, Halpern M, and Kashi Y (2005). Adult non-biting midges: possible windborne carriers of Vibrio cholerae non-O1 non-O139. Environ. Microbiol 7, 576–585. [DOI] [PubMed] [Google Scholar]
- Capo F, Wilson A, and Di Cara F (2019). The intestine of Drosophila melanogaster: an emerging versatile model system to study intestinal epithelial homeostasis and host-microbial interactions in humans. Microorganisms 7, E336. 10.3390/microorganisms7090336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadha J, Harjai K, and Chhibber S (2021). Revisiting the virulence hallmarks of Pseudomonas aeruginosa: a chronicle through the perspective of quorum sensing. Environ. Microbiol 10.1111/1462-2920.15784. [DOI] [PubMed] [Google Scholar]
- Dolan SK, and Welch M (2018). The glyoxylate shunt, 60 Years on. Annu. Rev. Microbiol 72, 309–330. 10.1146/annurev-micro-090817-062257. [DOI] [PubMed] [Google Scholar]
- Dutta D, Dobson AJ, Houtz PL, Gläßer C, Revah J, Korzelius J, Patel PH, Edgar BA, and Buchon N (2015). Regional cell-specific transcriptome mapping reveals regulatory complexity in the adult Drosophila midgut. Cell Rep. 12, 346–358. 10.1016/j.celrep.2015.06.009. [DOI] [PubMed] [Google Scholar]
- Essock-Burns T, Bennett BD, Arencibia D, Moriano-Gutierrez S, Medeiros M, McFall-Ngai MJ, and Ruby EG (2021). Bacterial quorum-sensing regulation induces morphological change in a key host tissue during the Euprymna scolopes-Vibrio fischeri symbiosis. mBio 12, e0240221. 10.1128/mBio.02402-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson M, and Foley E (2021). Microbial recognition regulates intestinal epithelial growth in homeostasis and disease. FEBS J. 289, 3666–3691. 10.1111/febs.15910. [DOI] [PubMed] [Google Scholar]
- Flatt T, Heyland A, Rus F, Porpiglia E, Sherlock C, Yamamoto R, Garbuzov A, Palli SR, Tatar M, and Silverman N (2008). Hormonal regulation of the humoral innate immune response in Drosophila melanogaster. J. Exp. Biol 211, 2712–2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong JCN, Syed KA, Klose KE, and Yildiz FH (2010). Role of Vibrio polysaccharide (vps) genes in VPS production, biofilm formation and Vibrio cholerae pathogenesis. Microbiology 156, 2757–2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Xu K, Liu H, Liu G, Bai M, Peng C, Li T, and Yin Y (2018). Impact of the gut microbiota on intestinal immunity mediated by tryptophan metabolism. Front. Cell. Infect. Microbiol 8, 13. 10.3389/fcimb.2018.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedhart J, and Luijsterburg MS (2020). VolcaNoseR is a web app for creating, exploring, labeling and sharing volcano plots. Sci. Rep 10, 20560. 10.1038/s41598-020-76603-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Yin C, Yang F, Zhang Y, Huang H, Wang J, Deng B, Cai T, Rao Y, and Xi R (2019). The cellular diversity and transcription factor code of Drosophila enteroendocrine cells. Cell Rep. 29, 4172–4185.e5. 10.1016/j.celrep.2019.11.048. [DOI] [PubMed] [Google Scholar]
- Halpern M, Broza YB, Mittler S, Arakawa E, and Broza M (2004). Chironomid egg masses as a natural reservoir of Vibrio cholerae non-O1 and non-O139 in freshwater habitats. Microb. Ecol 47, 341–349. [DOI] [PubMed] [Google Scholar]
- Hang S, Purdy AE, Robins WP, Wang Z, Mandal M, Chang S, Mekalanos JJ, and Watnick PI (2014). The acetate switch of an intestinal pathogen disrupts host insulin signaling and lipid metabolism. Cell Host Microbe 16, 592–604. 10.1016/j.chom.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haq S, Grondin JA, and Khan WI (2021). Tryptophan-derived serotonin-kynurenine balance in immune activation and intestinal inflammation. FASEB J 35, e21888. 10.1096/fj.202100702R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, and Lempicki RA (2009a). Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13. 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, and Lempicki RA (2009b). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc 4, 44–57. 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Jugder BE, Kamareddine L, and Watnick PI (2021). Microbiota-derived acetate activates intestinal innate immunity via the Tip60 histone acetyltransferase complex. Immunity 54, 1683–1697.e3. 10.1016/j.immuni.2021.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung SA, Chapman CA, and Ng WL (2015). Quadruple quorum-sensing inputs control Vibrio cholerae virulence and maintain system robustness. PLoS Pathog. 11, e1004837. 10.1371/journal.ppat.1004837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung SA, Hawver LA, and Ng WL (2016). Parallel quorum sensing signaling pathways in Vibrio cholerae. Curr. Genet 62, 255–260. 10.1007/s00294-015-0532-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamareddine L, Robins WP, Berkey CD, Mekalanos JJ, and Watnick PI (2018a). The Drosophila immune deficiency pathway modulates enteroendocrine function and host metabolism. Cell Metab. 28, 449–462.e5. 10.1016/j.cmet.2018.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamareddine L, Wong ACN, Vanhove AS, Hang S, Purdy AE, Kierek-Pearson K, Asara JM, Ali A, Morris JG Jr., and Watnick PI (2018b). Activation of Vibrio cholerae quorum sensing promotes survival of an arthropod host. Nat. Microbiol 3, 243–252. 10.1038/s41564-017-0065-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kareb O, and Aïder M (2020). Quorum sensing circuits in the communicating mechanisms of bacteria and its implication in the biosynthesis of bacteriocins by lactic acid bacteria: a review. Probiotics Antimicrob. Proteins 12, 5–17. 10.1007/s12602-019-09555-4. [DOI] [PubMed] [Google Scholar]
- Kleino A, and Silverman N (2014). The Drosophila IMD pathway in the activation of the humoral immune response. Dev. Comp. Immunol 42, 25–35. 10.1016/j.dci.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesperance DNA, and Broderick NA (2020). Gut bacteria mediate nutrient availability in Drosophila diets. Appl. Environ. Microbiol 87, e01401–20. 10.1128/AEM.01401-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Wang T, Xiang Z, and He N (2015). Tweedle cuticular protein BmCPT1 is involved in innate immunity by participating in recognition of Escherichia coli. Insect Biochem. Mol. Biol 58, 76–88. 10.1016/j.ibmb.2014.11.004. [DOI] [PubMed] [Google Scholar]
- Lingens F (1968). The biosynthesis of aromatic amino acids and its regulation. Angew. Chem. Int. Ed. Engl 7, 350–360. 10.1002/anie.196803501. [DOI] [PubMed] [Google Scholar]
- Liu N, Sun S, Wang P, Sun Y, Hu Q, and Wang X (2021). The mechanism of secretion and metabolism of gut-derived 5-hydroxytryptamine. Int. J. Mol. Sci 22, 7931. 10.3390/ijms22157931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupp C, Urbanowski M, Greenberg EP, and Ruby EG (2003). The Vibrio fischeri quorum-sensing systems ain and lux sequentially induce luminescence gene expression and are important for persistence in the squid host. Mol. Microbiol 50, 319–331. 10.1046/j.1365-2958.2003.t01-1-03585.x. [DOI] [PubMed] [Google Scholar]
- Mashruwala AA, and Bassler BL (2020). The Vibrio cholerae quorum-sensing protein VqmA integrates cell density, environmental, and host-derived cues into the control of virulence. mBio 11, e01572–20. 10.1128/mBio.01572-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrell DS, Butler SM, Qadri F, Dolganov NA, Alam A, Cohen MB, Calderwood SB, Schoolnik GK, and Camilli A (2002). Host-induced epidemic spread of the cholera bacterium. Nature 417, 642–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima Y, and Ishihara S (2021). Enteric microbiota-mediated serotonergic signaling in pathogenesis of irritable bowel syndrome. Int. J. Mol. Sci 22, 10235. 10.3390/ijms221910235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molla AM, Rahman M, Sarker SA, Sack DA, and Molla A (1981). Stool electrolyte content and purging rates in diarrhea caused by rotavirus, enterotoxigenic E. coli, and V. cholerae in children. J. Pediatr 98, 835–838. 10.1016/s0022-3476(81)80863-3. [DOI] [PubMed] [Google Scholar]
- Morris G, Berk M, Carvalho A, Caso JR, Sanz Y, Walder K, and Maes M (2017). The role of the microbial metabolites including tryptophan catabolites and short chain fatty acids in the pathophysiology of immune-inflammatory and neuroimmune disease. Mol. Neurobiol 54, 4432–4451. 10.1007/s12035-016-0004-2. [DOI] [PubMed] [Google Scholar]
- Neckameyer WS, Coleman CM, Eadie S, and Goodwin SF (2007). Compartmentalization of neuronal and peripheral serotonin synthesis in Drosophila melanogaster. Genes Brain Behav. 6, 756–769. 10.1111/j.1601-183X.2007.00307.x. [DOI] [PubMed] [Google Scholar]
- Novichkov PS, Kazakov AE, Ravcheev DA, Leyn SA, Kovaleva GY, Sutormin RA, Kazanov MD, Riehl W, Arkin AP, Dubchak I, and Rodionov DA (2013). RegPrecise 3.0–a resource for genome-scale exploration of transcriptional regulation in bacteria. BMC Genom. 14, 745. 10.1186/1471-2164-14-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes C, Sucena É, and Koyama T (2021). Endocrine regulation of immunity in insects. FEBS J. 288, 3928–3947. 10.1111/febs.15581. [DOI] [PubMed] [Google Scholar]
- Pang Z, Chong J, Zhou G, de Lima Morais DA, Chang L, Barrette M, Gauthier C, Jacques PÉ, Li S, and Xia J (2021). MetaboAnalyst 5.0: narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 49, W388–W396. 10.1093/nar/gkab382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort K, and Bassler BL (2016). Quorum sensing signal-response systems in Gram-negative bacteria. Nat. Rev. Microbiol 14, 576–588. 10.1038/nrmicro.2016.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt TG, Morton ER, Barton IS, Bever JD, and Fuqua C (2014). Ecological dynamics and complex interactions of Agrobacterium megaplasmids. Front. Plant Sci 5, 635. 10.3389/fpls.2014.00635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purdy AE, and Watnick PI (2011). Spatially selective colonization of the arthropod intestine through activation of Vibrio cholerae biofilm formation. Proc. Natl. Acad. Sci. USA 108, 19737–19742. 1111530108 [pii] 10.1073/pnas.1111530108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rus F, Flatt T, Tong M, Aggarwal K, Okuda K, Kleino A, Yates E, Tatar M, and Silverman N (2013). Ecdysone triggered PGRP-LC expression controls Drosophila innate immunity. EMBO J. 32, 1626–1638. 10.1038/emboj.2013.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu JH, Ha EM, Oh CT, Seol JH, Brey PT, Jin I, Lee DG, Kim J, Lee D, and Lee WJ (2006). An essential complementary role of NF-kappaB pathway to microbicidal oxidants in Drosophila gut immunity. Embo J 25, 3693–3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sack RB, Siddique AK, Longini IM Jr., Nizam A, Yunus M, Islam MS, Morris JG Jr., Ali A, Huq A, Nair GB, et al. (2003). A 4-year study of the epidemiology of Vibrio cholerae in four rural areas of Bangladesh. J. Infect. Dis 187, 96–101. [DOI] [PubMed] [Google Scholar]
- Settanni CR, Bibbò S, Ianiro G, Rinninella E, Cintoni M, Mele MC, Cammarota G, and Gasbarrini A (2021). Gastrointestinal involvement of autism spectrum disorder: focus on gut microbiota. Expert Rev. Gastroenterol. Hepatol 15, 599–622. 10.1080/17474124.2021.1869938. [DOI] [PubMed] [Google Scholar]
- Song W, Veenstra JA, and Perrimon N (2014). Control of lipid metabolism by tachykinin in Drosophila. Cell Rep. 9, 40–47. 10.1016/j.cel-rep.2014.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiller RC (1994). Intestinal absorptive function. Gut 35, S5–S9. 10.1136/gut.35.1_suppl.s5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacket CO, Taylor RK, Losonsky G, Lim Y, Nataro JP, Kaper JB, and Levine MM (1998). Investigation of the roles of toxin-coregulated pili and mannose- sensitive hemagglutinin pili in the pathogenesis of Vibrio cholerae O139 infection. Infect. Immun 66, 692–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelin KH, and Taylor RK (1996). Toxin-coregulated pilus, but not mannose-sensitive hemagglutinin, is required for colonization by Vibrio cholerae O1 El Tor biotype and O139 strains. Infect. Immun 64, 2853–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou AM, Frey EM, Hsiao A, Liu Z, and Zhu J (2008). Coordinated regulation of virulence by quorum sensing and motility pathways during the initial stages of Vibrio cholerae infection. Commun. Integr. Biol 1, 42–44. 10.4161/cib.1.1.6662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance RE, Zhu J, and Mekalanos JJ (2003).A constitutively active variant of the quorum-sensing regulator LuxO affects protease production and biofilm formation in Vibrio cholerae. Infect. Immun 71, 2571–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhove AS, Hang S, Vijayakumar V, Wong AC, Asara JM, and Watnick PI (2017). Vibrio cholerae ensures function of host proteins required for virulence through consumption of luminal methionine sulfoxide. PLoS Pathog. 13, e1006428. 10.1371/journal.ppat.1006428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhove AS, Jugder BE, Barraza D, and Watnick PI (2020). Methionine availability in the arthropod intestine is elucidated through identification of Vibrio cholerae methionine acquisition systems. Appl. Environ. Microbiol 86, e00371–20. 10.1128/AEM.00371-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther DJ, and Bader M (2003). A unique central tryptophan hydroxylase isoform. Biochem. Pharmacol 66, 1673–1680. 10.1016/s0006-2952(03)00556-2. [DOI] [PubMed] [Google Scholar]
- Wang H, Wu JH, Ayala JC, Benitez JA, and Silva AJ (2011). Interplay among cyclic diguanylate, HapR, and the general stress response regulator (RpoS) in the regulation of Vibrio cholerae hemagglutinin/protease. J. Bacteriol 193, 6529–6538. 10.1128/JB.05166-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Hang S, Purdy AE, and Watnick PI (2013). Mutations in the IMD pathway and mustard counter Vibrio cholerae suppression of intestinal stem cell division in Drosophila. mBio 4, e00337–00313. 10.1128/mBio.00337-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters CM, Lu W, Rabinowitz JD, and Bassler BL (2008). Quorum sensing controls biofilm formation in Vibrio cholerae through modulation of cyclic di-GMP levels and repression of vpsT. J. Bacteriol 190, 2527–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong ACN, Vanhove AS, and Watnick PI (2016). The interplay between intestinal bacteria and host metabolism in health and disease: lessons from Drosophila melanogaster. Dis. Model. Mech 9, 271–281. 10.1242/dmm.023408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Psychogios N, Young N, and Wishart DS (2009). MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 37, W652–W660. 10.1093/nar/gkp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz FH, Liu XS, Heydorn A, and Schoolnik GK (2004). Molecular analysis of rugosity in a Vibrio cholerae O1 El Tor phase variant. Mol. Microbiol 53, 497–515. [DOI] [PubMed] [Google Scholar]
- Zhang X, Beaulieu JM, Sotnikova TD, Gainetdinov RR, and Caron MG (2004). Tryptophan hydroxylase-2 controls brain serotonin synthesis. Science 305, 217. 10.1126/science.1097540. [DOI] [PubMed] [Google Scholar]
- Zheng W, Rus F, Hernandez A, Kang P, Goldman W, Silverman N, and Tatar M (2018). Dehydration triggers ecdysone-mediated recognition-protein priming and elevated anti-bacterial immune responses in Drosophila Malpighian tubule renal cells. BMC Biol. 16, 60. 10.1186/s12915-018-0532-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, and Mekalanos JJ (2003). Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Dev. Cell 5, 647–656. [DOI] [PubMed] [Google Scholar]
- Zhu J, Miller MB, Vance RE, Dziejman M, Bassler BL, and Mekalanos JJ (2002). Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 99, 3129–3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data for WT V. cholerae and a ΔhapR mutant grown to early stationary phase (NCBI: PRJNA810852) and Drosophila intestines infected with these two strains (NCBI: PRJNA811240) have been deposited in the NCBI repository. No code was generated as a part of these studies. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.