Abstract
Escherichia coli microcin J25 (MccJ25) is a plasmid-encoded, cyclic peptide antibiotic consisting of 21 unmodified amino acid residues. It is primarily active on gram-negative bacteria related to the producer strain, inducing cell filamentation in an SOS-independent way. A mutation causing resistance to MccJ25 was isolated. Genetic analysis indicated that it resided in the rpoC gene, encoding the β′ subunit of RNA polymerase, at 90 min on the E. coli genetic map. The mutation was genetically crossed on to a plasmid containing the wild-type rpoC gene. The presence of the recombinant plasmid conferred complete resistance to otherwise sensitive strains. Nucleotide sequencing of the plasmid-borne, mutant rpoC gene revealed a ACC (Thr)-to-ATC (Ile) change at codon 931, within homology block G, an evolutionarily conserved region in the large subunits of all RNA polymerases. MccJ25 decreased RNA synthesis both in vivo and in vitro. These results point to the RNA polymerase as the target of microcin action. We favor the possibility that the filamentous phenotype induced by MccJ25 results from impaired transcription of genes coding for cell division proteins. As far as we know, MccJ25 is the first peptide antibiotic shown to affect RNA polymerase.
Escherichia coli microcin J25 (MccJ25) is a plasmid-encoded, cyclic peptide antibiotic consisting of 21 unmodified amino acid residues (3, 21). Production of MccJ25 is induced at the onset of stationary growth phase and is optimal in iron-depleted medium (21, 23). The growth-phase-dependent expression may be explained by the concerted action of the positively acting transition state regulators guanosine 3′,5′-bispyrophosphate (ppGpp), leucine-responsive regulatory protein (Lrp), and integration host factor (5). MccJ25 is primarily active on gram-negative bacteria related to the producer strain, inducing cell filamentation in an SOS-independent way (21); some pathogenic bacteria, including Salmonella and Shigella species, are hypersensitive to MccJ25. We have previously reported the molecular characterization of the four plasmid genes, mcjA, -B, -C, and -D, involved in MccJ25 synthesis and immunity (29, 30). McjB and McjC most likely take part in MccJ25 maturation, which would imply the removal of an N-terminal leader of 37 amino acids from the 58-residue precursor McjA, followed by the head-tail cyclization of the 21-residue C-terminal propeptide (3, 30). The microcin immunity protein, McjD, which is highly similar to many ABC exporters, was found to be required for MccJ25 secretion (30). Thus, the immunity conferred by McjD could well be mediated by active efflux of the peptide, which would keep its intracellular concentration below a critical level. Also, we have found that the E. coli outer membrane protein TolC may be implicated in the secretion of MccJ25, possibly by forming an export complex with McjD (7).
Previously, we have isolated and characterized four classes of E. coli MccJ25-resistant mutants affected in genes encoding the cell envelope proteins FhuA, TonB, ExbB, and SbmA (22, 24). Most likely, all of these mutants are impaired in the uptake of the antibiotic. To enhance our understanding of the mechanism of action of MccJ25, in the present work we looked for mutants affected in the target of the antibiotic. Selection of this class of mutant was complicated by the high frequency with which mutants resistant to exogenous MccJ25 (fhuA, tonB, exbB, and sbmA) arise. However, we succeeded in isolating a novel class of MccJ25-resistant mutant. Genetic tests demonstrated that the mutation was located in rpoC, the gene encoding the β′ subunit of E. coli RNA polymerase (RNAP). The mutation in this strain was cloned by in vivo recombination into an rpoC+ plasmid. The presence of the recombinant plasmid conferred complete resistance to otherwise sensitive strains. Nucleotide sequence analysis of the plasmid-borne, mutant rpoC gene showed that the mutational change involved an evolutionarily conserved residue of the protein. These results, along with the observation that MccJ25 inhibits in vivo and in vitro RNA synthesis, provide convincing evidence that RNA polymerase is the target for MccJ25 action.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains, all E. coli K-12 derivatives, and plasmids used in this study are listed in Table 1.
TABLE 1.
Bacteria and plasmids used
| Strain or plasmid | Genotype or descriptiona | Reference or sourceb |
|---|---|---|
| Strains | ||
| AB259 | supQ80λ− relA1 spoT1 thi-1 | CGSC |
| SBG231 | AB259 sjmA1; spontaneous MccJ25r mutant | This study |
| SBG231recA | SBG231 Δ(recA-srl)306 srl-301::Tn10-84 | This study |
| χ342 | proC29 relA1 spoT1 metB1 bglF18::IS150 | CGSC |
| AB1133 | F−thr-1 ara-14 leuB6 lacY1 Δ(gpt-proA)62 supE44 galK2 λ− rac hisG4 rfbD1 rpsL31 kdgK51 xyl-5 mtl-1 argE3 thi-1 | CGSC |
| SBG232 | SBG231 rpoB | This study |
| CSH126 | Δ(recA-srl)306 srl-301::Tn10-84 | CSHL |
| Plasmids | ||
| pDJJ11 | pBR322 carrying rplJL rpoB | C. Gross |
| pDJJ12 | pBR322 carrying rplJL rpoB rpoC htrC | C. Gross |
| pMADC2 | pDJJ12 carrying the rpoC931 mutation (MccJ25r) | This study |
| pGC01 | pBR322 carrying fhuA | J. Coulton |
| pTUC347 | pUC18 carrying mcjABC; MccJ25+ ImmJ25− | 30 |
| pJS200 | pACYC184 carrying mcjD; MccJ25− ImmJ25+ | 29 |
| pGEM-4Z | Apr; cloning vector | Promega |
MccJ25r, resistant to microcin J25; MccJ25+, MccJ25 producer; ImmJ25+, immune to MccJ25.
CGSC, E. coli Genetic Stock Center; CSHL, Cold Spring Harbor Laboratory.
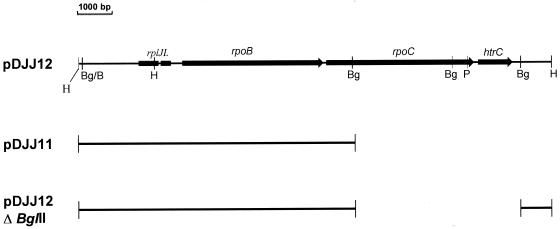
Plasmids pDJJ11 and pDJJ12 have been described (14). pDJJ11 is a pBR322 derivative carrying the rplJL and rpoB genes driven by the strong rplJ promoter. pDJJ12 carries, in addition, the rpoC gene. These plasmids and the pDJJ12 deletion derivative constructed in this work are depicted in Fig. 1.
FIG. 1.
Schematic representation of the plasmids used in this work. The portion of the rpoBC region contained in the various plasmids is indicated. Only relevant restriction sites are included: BglII (Bg); BamHI (B), HindIII (H); and PstI (P).
Media and growth conditions.
The standard Luria-Bertani (LB) broth and M9 minimal medium have been previously described (19). M9 medium was supplemented with glucose (0.2%) and thiamine (1 μg/ml). Solid media contained 1.5% agar. Antibiotics were added at the following concentrations: ampicillin, 50 μg/ml; tetracycline, 15 μg/ml; kanamycin, 50 μg/ml; and rifampin, 100 μg/ml. Liquid cultures were grown with aeration by gyrotory shaking. Growth was monitored by measuring the optical density at 600 nm (OD600). All cultures were incubated at 37°C.
MccJ25 sensitivity test.
For a reliable comparison of the sensitivities of different strains, we used a spot-on-lawn test for determining the MIC of MccJ25 for each strain, as follows. MccJ25 was purified as previously described (3). Doubling dilutions of the pure microcin preparation (1 mg/ml) were spotted (10 μl) onto LB plates and dried. Aliquots (50 μl) of cultures to be tested for sensitivity, in stationary phase, were mixed with 3 ml of top agar (0.7% agar) and overlaid onto the plates. After overnight incubation, the plates were examined for different degrees of inhibition; the higher the last dilution which produced a spot, the more sensitive the strain tested.
Genetic methods.
Phage P1 vir was used for routine transduction of genetic markers (19). Spontaneous (rifampin-resistant) Rifr mutants were isolated by plating 2 × 108 to 5 × 108 stationary-phase cells on LB plates containing rifampin, which were then incubated for 24 h.
Transposon Tn5 insertion mutagenesis of the rpoC gene was done as follows. Plasmid pDJJ12 was transformed into a Tn5 donor strain harboring a chromosomal copy of the transposon, selecting for ampicillin resistance (Apr). One transformant was grown overnight in LB medium supplemented with ampicillin and kanamycin, and a 0.2-ml sample of the culture was plated on LB medium containing neomycin at 300 μg/ml to favor the growth of cells carrying plasmids with transposon insertions. After incubation for 48 h, plasmid DNA was isolated from the pool of colonies and used to transform SBG231recA cells. The mixture was plated on LB medium containing ampicillin and MccJ25, selecting for Apr MccJ25r clones. The rationale of this selection was that cells that had received plasmids in which the gene complementing the mutation had been inactivated by Tn5 insertion should retain the microcin-resistant phenotype and, consequently, would be able to grow on the MccJ25-containing medium. Plasmid DNA was extracted from four resistant derivatives and physically analyzed to locate the insertions.
DNA manipulations and nucleotide sequencing.
Plasmid DNA was isolated with the Wizard miniprep DNA purification system (Promega). Digestions with restriction endonucleases, ligation with T4 DNA ligase, transformation of competent cells by the CaCl2 procedure, and agarose gel electrophoresis were done as described previously (25).
Automated DNA sequencing was carried out by dideoxy termination using primer walking. Nucleotide sequences of rpoC alleles were compared to that of GenBank using the online BLAST Network Service at the National Center for Biotechnology Information, National Institutes of Health, Bethesda, Md.
Incorporation of labeled leucine and uridine into macromolecules.
The MccJ25-sensitive strain AB259 and its resistant derivative SBG231 were grown in M9-glucose (10 ml) to early exponential phase (OD600 = 0.2 to 0.3), and each culture was split into two 5-ml portions. One of them received MccJ25 at a final concentration of 0.8 μM, and the other served as a control. The concentration of MccJ25 used is sufficient to induce extensive cell filamentation. After a 15-min preincubation, 15 μg of leucine per ml and 0.1 μCi of [3H]leucine were added to the four cultures. At the times indicated, portions of 0.5 ml were removed from the flasks, mixed with 1.5 ml of 5% cold trichloroacetic acid (TCA), chilled on ice for 1 h, and then boiled for 15 min. Each sample was filtered through a Millipore HAWP02500 filter and washed with 10 ml of cold TCA. The radioactivity retained on the dried filters was estimated in a Beckman LS-1801 liquid scintillation counter. Triplicate samples were taken at each time point.
Determination of RNA synthesis in microcin-treated AB259 and SBG231 cells was done in a similar way, except that 15 μg of uridine per ml and 0.1 μCi of [3H]uridine were added to the cells and the heating step before filtration was omitted.
In vitro transcription assay.
RNAP activity was determined essentially as described by Gross et al. (9), with some modifications. Purified plasmid pBR322 (2 μg, quantified by OD determinations and by gel electrophoresis) served as template, and [33P]UTP was used instead of [3H]UTP. Assay mixtures (50 μl) contained no MccJ25 (control), or increasing concentrations of MccJ25. The reaction was initiated by the addition of 1.5 U of purified E. coli RNA polymerase holoenzyme (Pharmacia) and allowed to proceed at 37°C for 15 min before precipitation with 1 ml of cold 10% TCA. After 1 h of incubation on ice, the precipitates were collected on glass fiber filters (Millipore type APFC), washed with 10 ml of cold 10% TCA, and dried, and the radioactivity retained on the filters was estimated in a Beckman LS-1801 liquid scintillation counter. Results are the averages of triplicate assays.
RESULTS
Isolation of a novel MccJ25-resistant mutant.
For the isolation of MccJ25r mutants, 100 μl of an overnight culture of E. coli AB259 was plated onto LB agar containing MccJ25 as selective agent. Fifty microcin-insensitive colonies were picked up, purified, and retested for the microcin-resistant phenotype. As expected, most of the clones isolated belonged to the already known classes of resistant mutants (i.e., sbmA, tonB, exbB, and fhuA) (22, 24). However, one of them showed a distinct phenotype, since the mutation was not complemented by plasmids bearing the wild-type sbmA, tonB, exbB, or fhuA genes. Therefore, the strain represented a new type of mutant and was designated SBG231. The mutation carried by SBG231 was provisionally named sjmA1 (sensitivity to J25 microcin, locus A).
Physiological characterization of the mutant.
The growth rates of wild-type AB259 and the isogenic MccJ25r mutant strain SBG231 were measured in LB medium. SBG231 grew at a rate that was indistinguishable from that of the parent strain and showed no morphological defect. The degree of resistance to exogenous MccJ25 conferred by the mutation was estimated as described in Materials and Methods. The mutant SBG231 was fully resistant against MccJ25 at the highest concentration available (103 μg/ml), whereas for the parent strain AB259 the MIC was 16 μg/ml. We then tried to increase the amount of FhuA, the outer membrane receptor of MccJ25, to see whether an enhanced MccJ25 entry led to such a high intracellular microcin concentration that the resistance conferred by the mutation could be overcome. Overproduction of FhuA can be induced by transformation of E. coli cells with plasmids carrying the fhuA gene (6, 8). Strains AB259 and SBG231 were transformed with plasmid pGC01 (fhuA+). The sensitivity of the wild-type AB259 carrying the plasmid was increased 16-fold (MIC, 1 μg/ml), whereas SBG231 (pGC01) still remained insensitive to the antibiotic at 103 μg/ml. Thus, mutant cells expressed a very high level of microcin resistance.
The sjmA1 mutation affects an intracellular MccJ25 target.
We presumed that SBG231 carried a microcin target mutation and reasoned that such a mutation should overcome the inhibitory effect of endogenous MccJ25 in the absence of the immunity gene mcjD. To confirm this hypothesis, we used plasmid pTUC347 (30), a pUC18 derivative containing the entire MccJ25 genetic system but which has a deletion that removes the 117 amino acid residues of the C end of the immunity protein McjD, including the highly conserved Walker's motif B, which is essential for function of the exporter. The deletion does not affect the mcjABC microcin synthesis genes. Because of the lack of immunity, this construct is lethal to nonimmune host cells and must be maintained in cells harboring pJS200 (29), a pACYC184-based compatible plasmid which provides an intact immunity gene in trans. The mutant SBG231 was transformed with a plasmid preparation containing both pTUC347 (Apr) and pJS200 (tetracycline resistant [Tcr]), selecting only for ampicillin resistance. Several clones harboring only pTUC347 were obtained. These transformants grew normally, indicating that the mutation overcame the inhibitory effect of endogenously produced microcin and suggesting that it could be indeed a target mutation.
Genetic mapping of the sjmA1 mutation.
To determine the approximate map location of the mutation, we used a set of Hfrs containing Tn10 insertions at known positions 20 to 30 min from the origin of transfer. The strategy was to mate the Hfrs to our mutant and to score the Tcr exconjugants for loss of the microcin-resistant phenotype. These experiments indicated that the altered gene was located somewhere between min 88 and 92 on the chromosome. More precise mapping of the mutation was accomplished by a series of two-factor crosses mediated by P1. Bacteriophage P1 propagated on SBG231 was used to transduce strains χ342 (metB1) and AB1133 (argE) to MetB+ and Arg+, respectively, and the transductants were examined for microcin resistance. We found that sjmA1 was linked to both metB (13% cotransduction) and argE (38% cotransduction), which lie at 89 and 89.5 min, respectively, on the E. coli genetic map. These results located the mutation at 90 min and showed that it was tightly linked to rpoB, the gene encoding the β subunit of RNAP. To determine the relative order of these markers, we first isolated spontaneous Rifr mutants of SBG231 as indicated in Materials and Methods. One of these mutants, designated SBG232 (Arg+ MccJ25r Rifr), was purified and used as donor in a P1-mediated three-point cross with strain AB1133 (Arg− MccJ25s Rifs) as the recipient. The results, which are presented in Table 2, indicated that the sjmA1 allele mapped very close to and downstream of rpoB, being 90.4% cotransducible with Rifr, and that the relative sequence in clockwise orientation was argE-rpoB-sjmA1.
TABLE 2.
Transductional mapping of the sjmA1 mutation
| Donor (relevant markers) | Recipient (relevant markers) | Selected marker | Distribution of unselected markers
|
|
|---|---|---|---|---|
| Class | No. | |||
| SBG232 (sjmA RifrargE+) | AB1133 (sjmA+ RifsargE) | Arg+ | RifssjmA+ | 66/146 |
| RifrsjmA+ | 11/146 | |||
| RifrsjmA | 66/146 | |||
| RifssjmA | 3/146 | |||
It is of note that in the course of the mapping procedures the sjmA1 mutation was repeatedly transferred by transduction to clean backgrounds, where it created the full resistance level observed with donor strain SBG231. Conversely, when the mutant allele in SBG231 was replaced by the wild-type one by transduction, no residual resistance to MccJ25 was seen in all transductants examined, which were as sensitive to the antibiotic as the wild-type parent AB259. These results make it unlikely that, in addition to the sjmA1 mutation in SBG231, another change occurred which acts synergistically or additively to give high-level resistance to MccJ25 and also demonstrate that the phenotype is not strain dependent. Moreover, the SBG231 mutant appeared spontaneously on a plate containing a high concentration of MccJ25, and its colony size was comparable to that of the other common resistant mutants after 16 h of incubation. This points to a single-step, high-level resistant mutant. Altogether, these results indicate that the sjmA1 mutation alone is necessary and sufficient for conferring the microcin resistance phenotype.
Complementation tests show that the sjmA1 mutation is an rpoC allele.
The linkage of sjmA1 and Rifr suggested that sjmA1 may be in the rpoC gene, encoding the β′ subunit of RNAP, although the possibility that rpoB was affected could not be dismissed. These possibilities were examined by complementation tests. First, we constructed a recA derivative of SBG231 by P1 transduction, using a recA srl::Tn10 strain (CSH126) as the donor. SBG231recA cells were then transformed with plasmid pDJJ12, which carries a chromosomal fragment including the rpoB, rpoC, rplJ, and rplL genes (the two latter coding for ribosomal proteins L10 and L12, respectively) (Fig. 1). The recA mutation prevented marker rescue by recombination between plasmid and chromosomal DNA. Transformation of the plasmid into sjmA cells fully reversed the microcin resistance phenotype (i.e., cells became completely sensitive to the antibiotic). In contrast, plasmid pDJJ11, which carries the rpoB, rplJ, and rplL genes, but not rpoC, did not correct the mutant phenotype when transformed into SBG231, indicating that complementation of the mutation by pDJJ12 was not due to the β subunit of RNAP or to the rplJ and rplL gene products. These results suggested that the mutation could be located in rpoC. To further test this possibility, we disrupted the rpoC gene of pDJJ12 by deleting two BglII fragments which comprise the C-terminal two-thirds of the gene (Fig. 1). The deletion derivative, pDJJ12ΔBglII, no longer complemented the chromosomal sjmA1 mutation in SBG231 (i.e., the cells remained resistant to MccJ25). Plasmid pDJJ12 was further manipulated by insertion of Tn5, as indicated in Materials and Methods. In all cases, Tn5 insertions eliminating the complementing capacity of pDJJ12 mapped within the rpoC gene. This finding lends further support to the assumption that the sjmA1 mutation carried by strain SBG231 is an rpoC allele. However, it is worth remarking that plasmid pDJJ12 also carries a small open reading frame of 120 amino acid equivalents located immediately downstream of rpoC (Fig. 1), and the deletion in pDJJ12ΔBglII that suppresses the complementing ability of pDJJ12 also eliminates this open reading frame (Fig. 1). Thus, the question arose as to whether it was this gene, and not rpoC, which complemented the mutation. Based on codon usage analysis, this sequence was first proposed to be not highly expressed, or not expressed at all, in vivo (31). However, Raina and Georgopoulos (20) demonstrated that this gene, which they called htrC, encodes a heat shock protein which is dispensable for growth at 30°C but is essential at temperatures above 42°C. Null mutations in the htrC gene lead to extensive filamentation at temperatures between 37 and 42°C. Thus, the htrC gene product seems to play an important role in cell division at a stage prior to septum initiation. Since the most conspicuous effect of MccJ25 is a perturbation of cell division (21), we suspected that the htrC product could be a candidate for being the target of the antibiotic and that it could be altered in the resistant mutant. To test this possibility, a 1.1-kb PstI-BglII fragment from pDJJ12 containing the wild-type htrC gene (20) (Fig. 1) was subcloned into pGEM-4Z (Promega). Introduction of the resulting recombinant plasmid into SBG231 did not complement the chromosomal mutation, suggesting that the htrC gene was not involved. This result, taken together with those from the complementation analysis and deletion-insertion mutagenesis, strongly indicated that the sjmA1 mutation was in the rpoC gene.
Introduction of the sjmA1 mutation onto rpoB+ rpoC+ plasmid pDJJ12.
The sjmA1 mutation was genetically crossed on to rpoB+ rpoC+ plasmid pDJJ12, as follows. The microcin-resistant mutant SBG231 was transformed with pDJJ12, and transformants were selected on LB-ampicillin plates. One of these transformants (which, as expected, had become microcin sensitive) was grown in LB medium for about 20 generations (three serial overnight cultures started by 1:100 dilution of the preceding culture) in the presence of ampicillin to maintain the plasmid. This allowed the plasmid-borne rpoC genes to recombine with the chromosomal mutant allele, thereby acquiring the MccJ25r locus from the chromosome, and the subsequent multiplication of the recombinant plasmids in the cells. A 0.1-ml portion of the last culture was spread onto LB agar containing ampicillin and MccJ25. The rationale for such a selection was that colonies in which a substantial fraction of the plasmid population had acquired the MccJ25r allele could become MccJ25r. Plasmid DNA was prepared from the pool of colonies and used to retransform strain SBG231, selecting for resistance to ampicillin (Apr). Two of ten transformants screened expressed resistance to MccJ25, indicating that they harbored mutant plasmids which were unable to complement the chromosomal MccJ25r mutation. Plasmid DNAs were isolated from these clones and introduced into the microcin-sensitive strain DH5α. Both plasmids conferred a MccJ25r phenotype. This result confirmed the presence of the sjmA1 mutation in the plasmids and the transdominance of the mutant allele when present in a high number of copies. One of these recombinant plasmids, designated pMADC2, was selected for further study. Restriction endonuclease analysis revealed the same fragment pattern as that of its wild-type counterpart pDJJ12. When introduced into several sensitive E. coli strains, pMADC2 rendered them fully resistant to the microcin, resulting in tolerances to concentrations of the antibiotic of as high as 1 mg/ml. To verify that the mutation isolated in pMADC2 was in rpoC, we made a BglII deletion which, as noted above, removes most of rpoC, while leaving rpoB intact. The deletion destroyed the ability of the plasmid to confer the mutant phenotype, showing that the mutation was indeed located in rpoC.
Nucleotide sequencing of the mutant rpoC allele.
To identify the putative alteration in rpoC, this gene was entirely sequenced in the mutated plasmid pMADC2 and the parental pDJJ12. We also sequenced the htrC gene in pMADC2 and found no change with respect to the published sequence (31), providing definitive evidence that this gene was not involved in the MccJ25r phenotype. Comparison of the rpoC sequences revealed a single nucleotide difference between pDJJ12 and the mutant plasmid. The latter had an ATC at codon position 931, while pDJJ12 had an ACC at that position, as does the published sequence in the current databases. This C-to-T transition would give rise to a substitution of isoleucine for threonine at amino acid residue 931 of the β′ subunit. We renamed the MccJ25r mutation rpoC931 and henceforth will refer to it by that name.
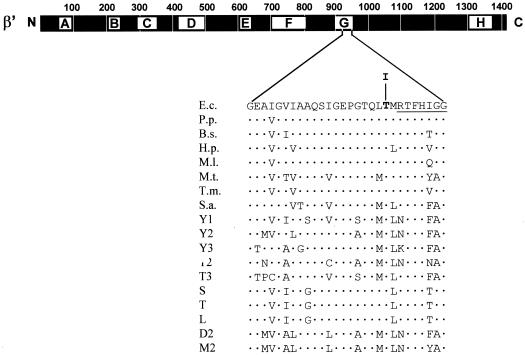
Alignment of prokaryotic and eukaryotic RNAP sequences has defined segments of substantial sequence conservation (1, 33), termed homology regions (15), which may represent domains with conserved functions. For the β′ subunit of prokaryotic RNAP, eight segments with a particularly high degree of conservation have been designated A through H (Fig. 2). The mutant described here exhibits a change in segment G (Fig. 2).
FIG. 2.
Genetic context and sequence alteration of the rpoC931 mutation. The heavy bar represents the 1,407-amino-acid β′ subunit of E. coli RNAP. Lettered boxes (A to H) symbolize evolutionarily conserved segments of β′ (33). A relevant sequence stretch (Gly912 to Gly939) from segment G of β′ (E.c.) is expanded underneath. The position of the amino acid alteration (a Thr-to-Ile change at codon 931) caused by the rpoC931 mutation is highlighted by boldface. The top sequence from E. coli is aligned with the corresponding portions of eubacterial, archaeal, and eukaryotic homologues: Pseudomonas putida (P.p.); Bacillus subtilis (B.s.); Helicobacter pylori (H.p.); Mycobacterium leprae (M.l.); Methanobacterium thermoautotrophicum (M.t.); Thermotoga maritima (T.m.); Sulfolobus acidocaldarium (S.a.); Saccharomyces cerevisiae RNAP I, II, and III (Y1, Y2, and Y3, respectively); RNAP II and III of Trypanosoma (T2 and T3, respectively); chloroplasts from spinach (S), tobacco (T), and liverwort (L); RNAP II of Drosophila melanogaster (D2); and RNAP II of mouse (M2). The dots symbolize amino acid sequence identity. The highly conserved 7-amino-acid sequence stretch (RTFHIGG) shown by Borukhov et al. (4) to contact the nascent RNA product is underlined.
Effect of MccJ25 on the in vivo incorporation of labeled uridine and leucine on RNA and proteins.
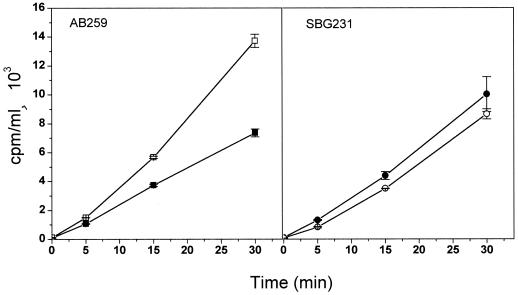
We treated exponentially growing cells of SBG231 (MccJ25r) and its parent AB259 with 0.8 μM microcin and measured RNA and protein syntheses as described in Materials and Methods. RNA accumulation (as measured by determining the incorporation of [3H]uridine into acid-insoluble material) in strain AB259 was reduced by 50% after a 30-min treatment (Fig. 3). Under these conditions, the mutant SBG231 continued to accumulate RNA to a level comparable with the parent strain (Fig. 3). Thus, the mutation prevented the inhibition of RNA synthesis by MccJ25. As shown in Fig. 4, overall protein synthesis (as measured by the incorporation of [3H]leucine) was not significantly affected by the antibiotic within the same time frame.
FIG. 3.
Effect of MccJ25 on RNA synthesis of parent AB259 and mutant SBG231 strains. RNA synthesis was measured by monitoring incorporation of [3H]uridine into TCA-precipitable material as described in Materials and Methods. Accumulated RNA in the absence (□) and in the presence (■) of microcin is shown.
FIG. 4.
Effect of MccJ25 on protein synthesis of parent AB259 and mutant SBG231 strains. Incorporation of [3H]leucine in the absence (□) and in the presence (■) of microcin is shown and was determined as described in Materials and Methods.
Effect of MccJ25 on in vitro transcription.
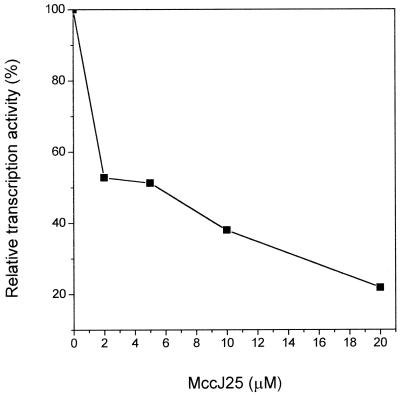
We studied the influence of MccJ25 on the activity of purified RNAP in vitro. As shown in Fig. 5, in vitro RNA synthesis was inhibited by MccJ25, and this inhibition was dose dependent. Total RNA accumulation was diminished by 2 μM MccJ25 to 50% of that of the control without the antibiotic. This result pointed to a direct action of MccJ25 on RNAP.
FIG. 5.
Inhibition of RNA polymerase by MccJ25 in vitro. The transcription assay was done as indicated in Materials and Methods. The amount of transcription activity relative to the uninhibited enzyme is indicated as a function of increasing MccJ25 concentration.
DISCUSSION
In this report, a mutation causing resistance to both exogenous and endogenous MccJ25 was isolated. Transduction experiments and complementation analyses indicated that the mutation maps within rpoC, the gene encoding the β′ subunit of E. coli RNAP. The chromosomal mutation was cloned by in vivo recombination into an rpoC+ plasmid. Nucleotide sequencing of the mutant, plasmid-borne rpoC gene revealed only a ACC-to-ATC change at codon 931. The C→T transition would result in the replacement of threonine at position 931 by isoleucine. The mutation, designated rpoC931, lies in homology block G, the seventh of eight stretches of amino acids which share extensive sequence similarity with β′ homologues from eubacteria, archaea, and eukaryotes (1, 15, 33). Notably, the residue affected is conserved in all prokaryotic and eukaryotic homologues examined (Fig. 2), suggesting that it plays an important role in RNAP function. There is good evidence indicating that region G is part of the catalytic center of the enzyme. For example, cross-linking experiments with E. coli RNAP have indicated that an approximately 90-amino-acid β′ fragment including the C-terminal end of segment G contacts the nascent transcript. Within this fragment there is a highly conserved 7-amino-acid region (RTFHIGG) (Fig. 2) where the cross-link between RNAP and 8-azido AMP present at the 3′ end of the nascent RNA was mapped (4). The target amino acid identified in the current study lies in close proximity to this conserved sequence stretch (Fig. 2). Other findings implicate segment G directly in transcript elongation, transcriptional arrest, and transcript cleavage (17, 18, 34, 35). Interestingly, the location and nature of rpoC931 correspond exactly to the rpb1-502 mutation isolated by Hekmatpanah and Young (12) in the Saccharomyces cerevisiae RNAP II large-subunit gene RPB1. The rpb1-502 mutation affects the accuracy of mRNA start site selection by producing a small but detectable increase in the 5′-end heterogeneity of transcripts. It is unlikely that our mutation exerts the same effect, since in E. coli the site of transcription initiation is fixed by the interaction of the ς subunit of RNAP with conserved promoter sequences and the purine nucleoside triphosphate preference of the active site of the enzyme.
The rpoC931 alteration occurred immediately adjacent to a cluster of substitutions affecting residues 933, 934, and 936 in region G that alter the termination and elongation properties of RNAP (34). All of these changes are recessive lethal. Despite its close proximity to these sites, rpoC931 does not have major effects on RNAP function in vivo, since the mutant derivative SBG231 is viable in the haploid state and grows as well as the parent strain. Therefore, it seems that the mutational change is well tolerated by the polymerase.
Based on the available data, we propose that the amino acid residue changed by rpoC931 is part of the microcin target site, which would be located within the catalytic center of the polymerase, and that the capacity of this site to bind the antibiotic is prevented or reduced by the change. Alternatively, there is the possibility that the mutational lesion results in a conformational change in the enzyme which affects a distant microcin-binding site indirectly. To us, however, this seems unlikely. It is worth noting that the putative target site for MccJ25 lies within a region spanning amino acids 842 to 1140 (thus containing segment G), which has been shown to be exposed on the surface of RNAP (16).
If the target site for MccJ25 binding lies in the catalytic center of RNAP, one could anticipate some effect of the antibiotic on transcription. This presumption was confirmed by our observation that MccJ25 has an inhibitory effect on transcription both in vivo and in vitro. RNA accumulation in sensitive cells was 50% reduced within 30 min after antibiotic addition. In contrast, in the SBG231 mutant cells the RNA synthesis remains unaffected. Over the same time period, protein synthesis is virtually unchanged. Only 2% of bacterial RNA is mRNA, and it does not accumulate, since it is metabolically unstable (2). It is therefore most likely that the decrease in RNA accumulation reflects mainly a reduced synthesis from stable RNA promoters. On the other hand, in vitro transcription experiments show that microcin can inhibit mRNA synthesis from pBR322 promoters. Collectively, these results point to a global effect of MccJ25 on transcription. The question arises as to how the reduced transcriptional activity may account for the division block, leading to the filamentous phenotype, caused by MccJ25. It is relevant to note that E. coli K-12 cells treated with the MIC of MccJ25 form filaments extensively but lose viability very slowly. Even in the presence of MccJ25 concentrations as high as eight times the MIC, the viable-cell count drops to 25% of the initial value only 2 h after addition of the antibiotic (21). We interpret this to mean that MccJ25 has relatively little effect on cell metabolism, affecting mainly the division process. Cell death probably results from long-term expression of division inhibition, as has been reported for the overproduction of the SfiA (or SulA) protein, a well-known division inhibitor (13). Taken together, these observations suggest that, by binding to RNAP, MccJ25 might preferentially inhibit the transcription of genes encoding either cell division proteins or regulatory factors required for expression of cell division genes.
It can be argued that RNAP is not the direct target of MccJ25 and that the mutant RNAP confers resistance to microcin by selectively changing the expression of a gene or genes whose product(s) directly confers microcin resistance. In this case, however, a direct action of MccJ25 on RNAP in vitro would not have been expected. Alternatively, MccJ25 could associate with some factor which, in turn, might interact with RNAP to modulate its function. The rpoC931 mutation could then interfere with the binding of this factor to the enzyme. Again, since MccJ25 inhibits RNA synthesis in vitro without special factors, the existence of such a factor seems highly questionable.
The observation that overproduction of wild-type β′ subunits from a multicopy plasmid relieves the mutant phenotype can be explained by assuming that, under these conditions, only a low proportion of RNAPs would contain mutated (resistant), chromosome-encoded, β′ subunits. Enough wild-type, microcin-susceptible, polymerase enzymes will be formed from the plasmid to restore the sensitivity to the antibiotic. In other words, we propose that the ratio of microcin-sensitive to microcin-insensitive polymerases in the polymerase cell pool must be the determining factor in cell susceptibility to microcin J25. It should be noted that a similar phenomenon is observed with rpoB mutants transformed with a high-copy-number plasmid containing a cloned, wild-type rpoB gene (14). In this case, the presence of the plasmid confers a Rifs phenotype to the otherwise Rifr strains due to the overexpression of the wild-type rpoB gene from the multicopy plasmid. The transdominance of the mutant allele when present in a high copy number would also be consistent with this argument. In this case, in wild-type cells overexpressing the mutant rpoC allele most enzymes would contain mutated subunits and, consequently, should not bind microcin.
Certain antibiotics have been shown to act directly on E. coli RNAP. These are rifampin (10), which inhibits initiation of RNA synthesis, and streptolydigin (28), which blocks transcription upon addition to elongating, as well as initiating, RNAP. All known mutations leading to rifampin resistance map to the rpoB gene (reference 14 and references cited therein), while mutations causing streptolydigin resistance have been found in both β and β′ subunits (11, 26, 27). Recently, rpoB and rpoC mutants of E. coli resistant to a novel aminopolyol antibiotic, zwittermicin A, have been described (32). However, all of these antibiotics are structurally very different from MccJ25, which appears as the first peptide antibiotic affecting RNAP.
Finally, it is remarkable that only 1 of 100 microcin-insensitive clones selected in our screens (this study and reference 24) carried a mutation in RNAP. In all likelihood, there are additional residues on β′ involved in binding microcin, so it might be expected that other substitutions leading to resistance would have been isolated. One explanation for the infrequent isolation of RNAP mutants could be that some substitutions have lethal consequences and thus may have escaped our screens. We are in the process of generating microcin-resistant RNAP by chemical mutagenesis or by systematically changing residues surrounding the one identified in the present study. Such mutants will allow us to define the anatomy of the MccJ25 binding site.
ACKNOWLEDGMENTS
We are indebted to Barbara Bachmann, Carol Gross, and James Coulton for gifts of bacterial strains and plasmids.
Financial support was provided by FONCYT (grant 01-00132-02291), CABBIO (grant 11Ar/12Br), and CIUNT (grant 26/D114). M.A.D. was the recipient of a CONICET fellowship, and R.N.F. was a career investigator of CONICET.
REFERENCES
- 1.Allison L A, Moyle M, Shales M, Ingles C J. Extensive homology among the largest subunits of eukaryotic and prokaryotic RNA polymerases. Cell. 1985;42:599–610. doi: 10.1016/0092-8674(85)90117-5. [DOI] [PubMed] [Google Scholar]
- 2.Baracchini E, Bremer H. Determination of synthesis rate and lifetime of bacterial mRNAs. Anal Biochem. 1987;167:245–260. doi: 10.1016/0003-2697(87)90160-6. [DOI] [PubMed] [Google Scholar]
- 3.Blond A, Péduzzi J, Goulard C, Chiuchiolo M J, Barthélémy M, Prigent Y, Salomón R A, Farías R N, Moreno F, Rebuffat S. The cyclic structure of microcin J25, a 21-residue peptide antibiotic from Escherichia coli. Eur J Biochem. 1999;259:747–755. doi: 10.1046/j.1432-1327.1999.00085.x. [DOI] [PubMed] [Google Scholar]
- 4.Borukhov S, Lee J, Goldfarb A. Mapping of a contact for RNA 3′-terminus in the largest subunit of RNA polymerase. J Biol Chem. 1991;266:23932–23935. [PubMed] [Google Scholar]
- 5.Chiuchiolo M J, Delgado M A, Farías R N, Salomón R A. Growth-phase-dependent expression of the cyclopeptide antibiotic microcin J25. J Bacteriol. 2001;183:1755–1764. doi: 10.1128/JB.183.5.1755-1764.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coulton J W, Mason P, DuBow M S. Molecular cloning of the ferrichrome-iron receptor of Escherichia coli K-12. J Bacteriol. 1983;156:1315–1321. doi: 10.1128/jb.156.3.1315-1321.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delgado M A, Solbiati J O, Chiuchiolo M J, Farías R N, Salomón R A. Escherichia coli outer membrane protein TolC is involved in production of the peptide antibiotic microcin J25. J Bacteriol. 1999;181:1968–1970. doi: 10.1128/jb.181.6.1968-1970.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fecker L, Braun V. Cloning and expression of the fhu genes involved in iron(III)-hydroxamate uptake by Escherichia coli. J Bacteriol. 1983;156:1301–1314. doi: 10.1128/jb.156.3.1301-1314.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gross C, Engbaek F, Flammang T, Burgess R. Rapid micromethod for the purification of Escherichia coli ribonucleic acid polymerase and the preparation of bacterial extracts active in ribonucleic acid synthesis. J Bacteriol. 1976;128:382–389. doi: 10.1128/jb.128.1.382-389.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartmann G, Honikel K O, Knüsel F, Nüesch J. The specific inhibition of the DNA-directed RNA synthesis by rifamycin. Biochim Biophys Acta. 1967;145:843–844. doi: 10.1016/0005-2787(67)90147-5. [DOI] [PubMed] [Google Scholar]
- 11.Heisler L M, Suzuki H, Landick R, Gross C A. Four contiguous amino acids define the target for streptolydigin resistance in the β subunit of Escherichia coli RNA polymerase. J Biol Chem. 1993;268:25369–25375. [PubMed] [Google Scholar]
- 12.Hekmatpanah D S, Young R A. Mutations in a conserved region of RNA polymerase II influence the accuracy of mRNA start site selection. Mol Cell Biol. 1991;11:5781–5791. doi: 10.1128/mcb.11.11.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huisman O, D'Ari R, Gottesman S. Cell division control in Escherichia coli: specific induction of the SOS function SfiA protein is sufficient to block septation. Proc Natl Acad Sci USA. 1984;81:4490–4494. doi: 10.1073/pnas.81.14.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin D J, Gross C A. Mapping and sequencing of mutations in the Escherichia coli rpoB gene that lead to rifampicin resistance. J Mol Biol. 1988;202:45–58. doi: 10.1016/0022-2836(88)90517-7. [DOI] [PubMed] [Google Scholar]
- 15.Jokerst R S, Weeks J R, Zehring W A, Greenleaf A L. Analysis of the gene encoding the largest subunit in RNA polymerase II in Drosophila. Mol Gen Genet. 1989;215:266–275. doi: 10.1007/BF00339727. [DOI] [PubMed] [Google Scholar]
- 16.Katayama A, Fujita N, Ishihama A. Mapping of subunit-subunit contact surfaces on the β′ subunit of Escherichia coli RNA polymerase. J Biol Chem. 2000;275:3583–3592. doi: 10.1074/jbc.275.5.3583. [DOI] [PubMed] [Google Scholar]
- 17.Koulich D, Orlova M, Malhotra A, Sali A, Darst S A, Borukhov S. Domain organization of Escherichia coli transcript cleavage factors GreA and GreB. J Biol Chem. 1997;272:7201–7210. doi: 10.1074/jbc.272.11.7201. [DOI] [PubMed] [Google Scholar]
- 18.Markovtsov V, Mustaev A, Goldfarb A. Protein-RNA interactions in the active center of transcription elongation complex. Proc Natl Acad Sci USA. 1996;93:3221–3226. doi: 10.1073/pnas.93.8.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller J H. A short course in bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1992. [Google Scholar]
- 20.Raina S, Georgopoulos C. A new Escherichia coli heat shock gene, htrC, whose product is essential for viability only at high temperatures. J Bacteriol. 1990;172:3417–3426. doi: 10.1128/jb.172.6.3417-3426.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salomón R A, Farías R N. Microcin 25, a novel antimicrobial peptide produced by Escherichia coli. J Bacteriol. 1992;174:7428–7435. doi: 10.1128/jb.174.22.7428-7435.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salomón R A, Farías R N. The FhuA protein is involved in microcin 25 uptake. J Bacteriol. 1993;175:7741–7742. doi: 10.1128/jb.175.23.7741-7742.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salomón R A, Farías R N. Influence of iron on microcin 25 production. FEMS Microbiol Lett. 1994;121:275–280. doi: 10.1016/0378-1097(94)90303-4. [DOI] [PubMed] [Google Scholar]
- 24.Salomón R A, Farías R N. The peptide antibiotic microcin 25 is imported through the TonB pathway and the SbmA protein. J Bacteriol. 1995;177:3323–3325. doi: 10.1128/jb.177.11.3323-3325.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 26.Severinov K, Soushko M, Goldfarb A, Nikiforov V. Rifampicin region revisited. New rifampicin-resistant and streptolydigin-resistant mutants in the β subunit of Escherichia coli RNA polymerase. J Biol Chem. 1993;268:14820–14825. [PubMed] [Google Scholar]
- 27.Severinov K, Markov D, Severinova E, Nikiforov V, Landick R, Darst S A, Goldfarb A. Streptolydigin-resistant mutants in an evolutionarily conserved region of the β′ subunit of Escherichia coli RNA polymerase. J Biol Chem. 1995;270:23926–23929. doi: 10.1074/jbc.270.41.23926. [DOI] [PubMed] [Google Scholar]
- 28.Siddhikol C, Erbstoeszer J W, Weisblum B. Mode of action of streptolydigin. J Bacteriol. 1969;99:151–155. doi: 10.1128/jb.99.1.151-155.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Solbiati J O, Ciaccio M, Farías R N, Salomón R A. Genetic analysis of plasmid determinants for microcin J25 production and immunity. J Bacteriol. 1996;178:3661–3663. doi: 10.1128/jb.178.12.3661-3663.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Solbiati J O, Ciaccio M, Farías R N, González-Pastor E, Moreno F, Salomón R A. Sequence analysis of the four plasmid genes required to produce the circular peptide antibiotic microcin J25. J Bacteriol. 1999;181:2659–2662. doi: 10.1128/jb.181.8.2659-2662.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Squires C, Krainer A, Barry G, Chen W-F, Squires C L. Nucleotide sequence at the end of the gene for the RNA polymerase β′ subunit (rpoC) Nucleic Acids Res. 1981;9:6827–6840. doi: 10.1093/nar/9.24.6827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stabb E V, Handelsman J. Genetic analysis of zwittermicin A resistance in Escherichia coli: effects on membrane potential and RNA polymerase. Mol Microbiol. 1998;27:311–322. doi: 10.1046/j.1365-2958.1998.00678.x. [DOI] [PubMed] [Google Scholar]
- 33.Sweetser D, Nonet M, Young R A. Prokaryotic and eukaryotic RNA polymerases have homologous core subunits. Proc Natl Acad Sci USA. 1987;84:1192–1196. doi: 10.1073/pnas.84.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weilbaecher R, Hebron C, Feng G, Landick R. Termination-altering amino acid substitutions in the β′ subunit of Escherichia coli RNA polymerase identify regions involved in RNA chain elongation. Genes Dev. 1994;8:2913–2927. doi: 10.1101/gad.8.23.2913. [DOI] [PubMed] [Google Scholar]
- 35.Zakharova N, Bass I, Arsenieva E, Nikiforov V, Severinov K. Mutations in and monoclonal antibody binding to evolutionary hypervariable region of Escherichia coli RNA polymerase β′ subunit inhibit transcript cleavage and transcript elongation. J Biol Chem. 1998;273:24912–24920. doi: 10.1074/jbc.273.38.24912. [DOI] [PubMed] [Google Scholar]