Abstract
The genome-wide transcription profile of Escherichia coli cells treated with hydrogen peroxide was examined with a DNA microarray composed of 4,169 E. coli open reading frames. By measuring gene expression in isogenic wild-type and oxyR deletion strains, we confirmed that the peroxide response regulator OxyR activates most of the highly hydrogen peroxide-inducible genes. The DNA microarray measurements allowed the identification of several new OxyR-activated genes, including the hemH heme biosynthetic gene; the six-gene suf operon, which may participate in Fe-S cluster assembly or repair; and four genes of unknown function. We also identified several genes, including uxuA, encoding mannonate hydrolase, whose expression might be repressed by OxyR, since their expression was elevated in the ΔoxyR mutant strain. In addition, the induction of some genes was found to be OxyR independent, indicating the existence of other peroxide sensors and regulators in E. coli. For example, the isc operon, which specifies Fe-S cluster formation and repair activities, was induced by hydrogen peroxide in strains lacking either OxyR or the superoxide response regulators SoxRS. These results expand our understanding of the oxidative stress response and raise interesting questions regarding the nature of other regulators that modulate gene expression in response to hydrogen peroxide.
The Salmonella enterica serovar Typhimurium and Escherichia coli responses to hydrogen peroxide initially were analyzed 15 years ago using two-dimensional gel separation of proteins (10, 20, 33). These studies showed that the expression of approximately 30 proteins is induced by hydrogen peroxide treatment: 12 proteins are maximally induced within 10 min and 18 proteins are maximally induced between 10 and 30 min after the addition of hydrogen peroxide (10). Mutational studies led to discovery of the OxyR regulatory protein, which was shown to regulate the expression of 9 of the 12 rapidly induced proteins (10). A variety of approaches have led to the identification of some of the OxyR-activated genes, including katG (encoding hydroperoxidase I), ahpCF (encoding an alkyl hydroperoxide reductase), oxyS (encoding a small regulatory RNA), dps (encoding a nonspecific DNA binding protein), gorA (encoding glutathione reductase), grxA (encoding glutaredoxin 1), trxC (encoding thioredoxin 2), fur (encoding the Fur repressor of ferric ion uptake), and dsbG (encoding a disulfide chaperone-isomerase) (41; also reviewed in reference 30). OxyR also has been shown to be a repressor of its own expression as well as that of fhuF (encoding a ferric ion reductase) and flu (encoding the antigen 43 outer membrane protein). Nevertheless, the identity of many of the hydrogen peroxide-inducible proteins has remained unknown.
The recently developed microarray technology has allowed the parallel study of the expression of every gene in an organism. This approach has already been successfully used in studying E. coli gene expression under a number of different growth conditions (13, 26, 31, 35, 36). Here, we report a survey of gene expression in response to hydrogen peroxide. In addition to confirming the hydrogen peroxide induction of most known OxyR-regulated genes, we have identified several new members of the OxyR regulon. We also have found that many genes are induced by hydrogen peroxide in an OxyR-independent fashion, revealing complex regulation of the cellular response to oxidative stress.
MATERIALS AND METHODS
Plasmids and strains.
The DNA sequences and coordinates throughout the study are for E. coli from GenBank accession no. U00096 (5). The plasmids used in the study were constructed using fragments PCR amplified from chromosomal DNA. The sequences of all oligonucleotides are listed in Table 1. To generate the hemH promoter plasmids (pGSO131 and pGSO132), a 280-bp fragment produced using primers 819 and 821 was cloned into pCR2.1 (Invitrogen) in both orientations. To generate a sufA promoter plasmid (pGSO133), a 378-bp fragment produced using primers 820 and 825 was cloned into pCR2.1. To generate the isc promoter plasmid (pGSO135), a 500-bp fragment produced using primers 699 and 700 was cloned into the EcoRI and BamHI sites of pUC18. The ΔoxyR::kan (GSO9 [32]) mutant allele was moved into MG1655 (2) by P1 transduction (27) to generate GSO77. MC4100 (wild type), GSO47 (MC4100 ΔoxyR::kan), GSO71 (MC4100 ΔsoxRS), and GSO72 (MC4100 Δfur::kan) were described previously (40).
TABLE 1.
Sequences of oligonucleotides used in the study
| No. | Sequencea |
|---|---|
| 188 | 5′-GCAAAAGTTCACGTTGG |
| 686 | 5′-GCATTGCGGTCACGGCATAG |
| 699 | 5′-GCCAACGGATCCGGGCCCGCTTCA |
| 700 | 5′-GATGCGACGCGGAATTCGTCTTAT |
| 819 | 5′-TCGGGCGTACCCAGGTTTG |
| 820 | 5′-TCAGCGTTAAGCCTTGCCA |
| 821 | 5′-AAGCGCGCACGGACAGTC |
| 823 | 5′-TGAATAATTTTCTGATGGGACAT |
| 825 | 5′-AGCCTGTGTCGCACAGAC |
| 828 | 5′-CTGATGTGTGGGTTAAC |
Restriction sites are underlined.
RNA isolation.
Cultures were grown under aeration at 37°C in Luria-Bertani (LB) rich medium (27). Exponential-phase cultures (optical density at 600 nm = 0.2 to 0.5) were split into aliquots; one aliquot was left untreated, and the other aliquots were treated with the indicated amounts of hydrogen peroxide or paraquat. After 10 min, the cells from 5, 10, or 25 ml of culture were harvested and resuspended in 1 ml of Trizol equilibrated at 4°C (Gibco BRL). All subsequent purification steps were carried out according to the Trizol reagent manual (based on reference 9).
DNA microarray experiments.
Fabrication of the E. coli DNA microarray and procedures for cDNA labeling, hybridization, and array quantification were described previously (28, 35).
Primer extension assays.
Total RNA samples were subjected to primer extension assays as described previously (37), using primer 819 specific to hemH, primer 820 specific to sufA, primer 188 specific to oxyS, primer 823 specific to soxS, and primer 686 specific to the yfhP gene in the isc operon.
DNase I footprinting.
DNase I footprinting assays of purified OxyR binding to the hemH and sufA promoters were carried out as described previously (32).
RESULTS
DNA microarray measurements.
Wild-type (MG1655) cells and isogenic ΔoxyR (GSO77) mutant cells were grown to exponential phase in LB rich medium. The cultures were split, and half of each culture was treated with 1 mM hydrogen peroxide. After 10 min, total RNA was isolated from the untreated and treated cultures. To check whether the RNA samples showed a well-characterized peroxide stress response, we examined the expression of the oxyS, ahpC, katG, and fhuF genes using primer extension assays. As observed previously, oxyS, ahpC, and katG showed OxyR-dependent induction by hydrogen peroxide, and fhuF showed repression in the wild-type strain and slight induction in the oxyR deletion strain (data not shown).
Each of the RNA samples was used as a template for cDNA synthesis with attendant incorporation of either of two fluorescent dyes, Cy3 and Cy5. Pairs of differentially labeled untreated and treated cDNA samples from each strain then were hybridized to a glass slide on which two sets of the 4,169 E. coli open reading frames (ORFs) were printed. For each strain, two slides were used for hybridization: for one slide, the untreated sample was labeled with Cy3 and the treated sample was labeled with Cy5, and for the second slide, the dye-sample pairings were reversed. Thus, the expression for each gene was measured four times. The average of the four data points is reported here. Overall, the mRNA levels of 140 genes in the wild-type strain showed >4-fold induction after treatment with hydrogen peroxide, and the mRNA levels of 167 genes in the ΔoxyR strain showed >4-fold induction. The 30 genes whose expression was induced most strongly in the wild-type strain are listed in Table 2, and the 30 genes whose expression was induced most strongly in the ΔoxyR strain are listed in Table 3. All of these genes have induction ratios of >10-fold.
TABLE 2.
The 30 most strongly hydrogen peroxide-induced genes in the wild-type strain
| Gene | b no. | Induction ratioa | Functionb |
|---|---|---|---|
| dps | b0812 | 180 | Stress response DNA binding protein |
| yaiA | b0389 | 56 | Function unknown |
| katG | b3942 | 44 | Catalase hydrogen peroxidase I |
| grxA | b0849 | 37 | Glutaredoxin I |
| yfiA | b2597 | 36 | Function unknown |
| ibpA | b3687 | 29 | Chaperone, heat-inducible protein of HSP20 family |
| yjiD | b4326 | 29 | Function unknown |
| ycfR | b1112 | 26 | Function unknown |
| ahpF | b0606 | 22 | Alkyl hydroperoxide reductase large subunit |
| trxC | b2582 | 21 | Thioredoxin 2 |
| sufA | b1684 | 21 | Homology with IscA |
| ymgB | b1166 | 20 | Function unknown |
| ahpC | b0605 | 20 | Alkyl hydroperoxide reductase small subunit |
| ibpB | b3686 | 18 | Chaperone, heat-inducible protein of HSP20 family |
| yaaA | b0006 | 18 | Function unknown |
| tnaA | b3708 | 18 | Tryptophanase |
| fpr | b3924 | 17 | Ferredoxin NADP+ reductase |
| cysK | b2414 | 16 | Cysteine synthase |
| sufB | b1683 | 16 | Function unknown |
| dsdX | b2365 | 15 | Homology with gluconate permease |
| ybjM | b0848 | 15 | Function unknown |
| yeeD | b2012 | 14 | Function unknown |
| dsdA | b2366 | 13 | d-Serine deaminase |
| soxS | b4062 | 13 | Regulatory protein of soxRS regulon |
| sbp | b3917 | 12 | Periplasmic sulfate binding protein |
| sufC | b1682 | 12 | Putative ABC transporter |
| phoH | b1020 | 12 | Member of pho regulon |
| hemH | b0475 | 11 | Ferrochelatase |
| yljA | b0881 | 11 | Function unknown |
| ycgZ | b1164 | 11 | Function unknown |
Ratio of transcript levels for hydrogen peroxide-treated wild-type (MG1655) strain to transcript levels for untreated wild-type strain.
Function descriptions are taken from http://genolist.pasteur.fr/Colibri/.
TABLE 3.
The 30 most strongly hydrogen peroxide-induced genes in the ΔoxyR strain
| Gene | b no. | Induction ratioa | Functionb |
|---|---|---|---|
| yfiA | b2597 | 109 | Function unknown |
| ibpB | b3686 | 54 | Chaperone, heat-inducible protein of HSP20 family |
| ibpA | b3687 | 35 | Chaperone, heat-inducible protein of HSP20 family |
| tnaA | b3708 | 30 | Tryptophanase |
| yjiD | b4326 | 29 | Function unknown |
| cysK | b2414 | 25 | Cysteine synthase |
| uxuA | b4322 | 23 | Mannonate hydrolase |
| dsdX | b2365 | 22 | Homology with gluconate permease |
| ytfK | b4217 | 20 | Function unknown |
| recN | b2616 | 20 | Recombination and repair |
| soxS | b4062 | 19 | Regulatory protein of soxRS regulon |
| dsdA | b2366 | 18 | d-Serine deaminase |
| fpr | b3924 | 18 | Ferredoxin NADP+ reductase |
| ymgB | b1166 | 17 | Function unknown |
| yeeD | b2012 | 17 | Function unknown |
| ygaQ | b2654 | 17 | Function unknown |
| yaiA | b0389 | 16 | Function unknown |
| yceP | b1060 | 16 | Function unknown |
| glgS | b3049 | 15 | Glycogen synthesis protein |
| ydcH | b1426 | 15 | Function unknown |
| tnaL | b3707 | 15 | Regulatory leader peptide for tna operon |
| phoH | b1020 | 14 | Member of pho regulon |
| ymgA | b1165 | 14 | Function unknown |
| ydeN | b1498 | 14 | Function unknown |
| sbp | b3917 | 12 | Periplasmic sulfate binding protein |
| ynaF | b1376 | 12 | Function unknown |
| cysP | b2425 | 12 | Periplasmic sulfate binding protein |
| yaeH | b0163 | 12 | Function unknown |
| manX | b1817 | 11 | Mannose phosphotransferase system |
| ycgK | b1178 | 10 | Function unknown |
Ratio of transcript levels for hydrogen peroxide-treated ΔoxyR (GSO77) strain to transcript levels for untreated ΔoxyR strain.
Function descriptions are taken from http://genolist.pasteur.fr/Colibri/.
OxyR-dependent response.
A hallmark of the E. coli response to hydrogen peroxide is the rapid and strong induction of a set of OxyR-regulated genes, including dps, katG, grxA, ahpCF, and trxC. The observed >20-fold induction of all of these genes in the wild-type strain (Table 2) but not the ΔoxyR strain (Table 3) provided an internal validation of the microarray experiment. Of the other known OxyR-activated genes, fur and gorA were slightly induced and dsbG was unchanged in the wild-type strain treated with hydrogen peroxide. Of the known OxyR-repressed genes, flu was unchanged and fhuF was slightly repressed. A comparison of the induction ratios between the wild-type strain and the ΔoxyR strain indicated that, among the 30 most highly induced genes, 8 additional genes (hemH, sufABC, yaiA, yaaA, yljA, and ybjM [Table 4]) might be regulated by OxyR. The remaining 16 most highly induced genes showed approximately equal levels of hydrogen peroxide induction in the wild-type strain and the ΔoxyR strain.
TABLE 4.
OxyR regulon
| Gene | b no. | Basal level (ppm)a | Induction ratio in strain:
|
|
|---|---|---|---|---|
| Wild type | ΔoxyR | |||
| dps | b0812 | 330 | 180 | 2.0 |
| katG | b3942 | 270 | 44 | 2.1 |
| grxA | b0849 | 54 | 37 | 1.2 |
| ahpF | b0606 | 350 | 22 | 1.2 |
| trxC | b2582 | 73 | 21 | 2.1 |
| ahpC | b0605 | 2,000 | 20 | 2.2 |
| fur | b0683 | 390 | 2.9 | 1.1 |
| gor | b3500 | 160 | 2.1 | 0.9 |
| dsbG | b0604 | 93 | 0.7 | 0.7 |
| flu | b2000 | 110 | 1.0 | 1.9 |
| fhuF | b4367 | 200 | 0.4 | 5.7 |
| hemH | b0475 | 39 | 11 | 1.1 |
| sufA | b1684 | 270 | 21 | 3.6 |
| sufB | b1683 | 90 | 16 | 4.0 |
| sufC | b1682 | 120 | 12 | 3.4 |
| sufD | b1681 | 160 | 8.3 | 3.0 |
| sufS | b1680 | 220 | 3.5 | 1.5 |
| sufE | b1679 | 140 | 8.2 | 3.5 |
| yaaA | b0006 | 110 | 18 | 4.2 |
| yaiA | b0389 | 130 | 56 | 16 |
| ybjM | b0848 | 51 | 15.0 | 1.1 |
| yljA | b0881 | 250 | 11.0 | 4.6 |
Levels in cells during exponential growth in LB medium. The data are from unpublished data of Y. Wei and R. A. LaRossa.
(i) hemH.
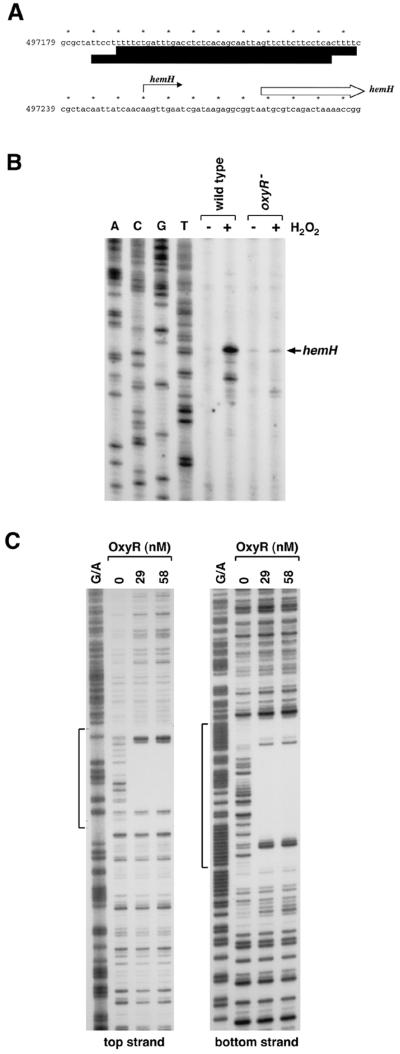
One gene, hemH (b0475), whose expression was induced 11-fold by hydrogen peroxide in the wild-type strain and <2-fold in the ΔoxyR strain, encodes a ferrochelatase that catalyzes the incorporation of ferrous ion into protoporphyrin IX in the final step in protoheme biosynthesis (3). The hemH locus also was named visA since some ferrochelatase mutants of E. coli were identified by virtue of having a photosensitive phenotype (19). It was determined previously that the photosensitivity was caused by the increased levels of protoporphyrin IX which accumulate in the mutants lacking ferrochelatase (21). To confirm OxyR regulation of hemH, we carried out primer extension assays. As shown in Fig. 1B, hemH induction by hydrogen peroxide in vivo was clearly dependent on OxyR. The start of the transcript corresponds to an A residue at position 497255, 24 bp upstream of the hemH start codon. An in vitro DNase I footprinting experiment showed that oxidized OxyR binds to the hemH promoter centered at 497211, overlapping and just upstream of the −35 region (Fig. 1C).
FIG. 1.
OxyR-dependent induction of hemH. (A) Sequence of the hemH promoter. The hemH transcription start is marked by the black arrow, and the start of the corresponding ORF is denoted by the white arrow. The DNase I footprints for OxyR binding are indicated by the dark boxes. (B) Primer extension assays of hemH expression in wild-type (MC4100) and ΔoxyR (GSO47) strains grown in LB medium. Exponential-phase cultures were split into two aliquots: one aliquot was left untreated, and the other was treated with 1 mM hydrogen peroxide. The cells were harvested after 10 min, total RNA was isolated, and primer extension assays were carried out with primer 819 specific to hemH. The neighboring sequencing reactions were carried out with the same primer. (C) DNase I footprinting assays of oxidized OxyR binding to the top and bottom strands relative to the hemH promoter. The regions protected by OxyR on both strands are indicated by the brackets. The plasmids carrying the hemH promoter fragment in both orientations were digested with NotI, labeled with 32P, and then digested with BamHI to give the labeled top and bottom strands. The samples were run in parallel with Maxam-Gilbert G/A sequencing ladders.
(ii) suf operon.
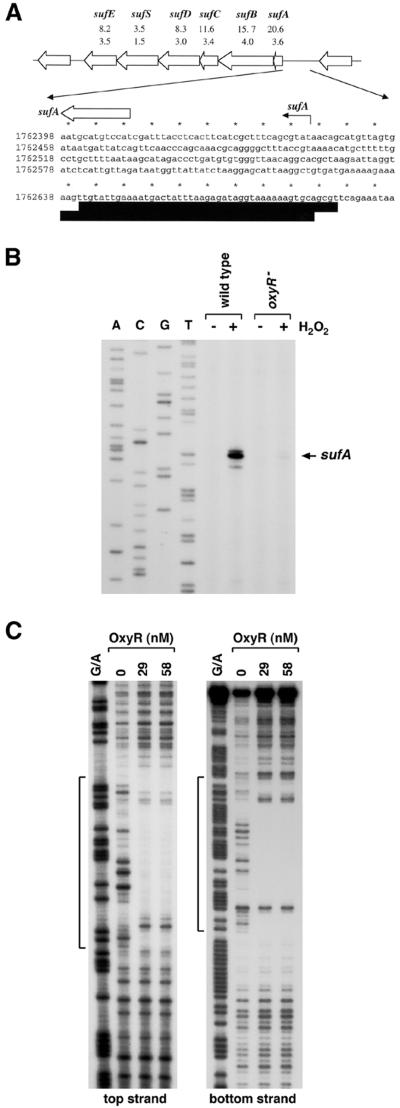
Three genes, sufA/ydiC (b1684), sufB/ynhE (b1683), and sufC/ynhD (b1682), whose expression was strongly induced in the wild-type strain but not the ΔoxyR strain, are members of a six-gene cluster (b1679 to b1684). These genes are transcribed in the same direction and show potential for translational coupling. Patzer and Hantke (24) suggested that the gene cluster forms an operon and, because of possible involvement in sulfur mobilization, named the genes sufA, sufB, sufC, sufD, sufS, and sufE. Of the six ORFs, the sufS gene and its product are the best characterized. This gene encodes one of three NifS homologs in E. coli and also has been named csdB (18). All three E. coli NifS homologs, IscS, CSD, and SufS/CsdB, catalyze the elimination of sulfur from l-cysteine and selenium from l-selenocysteine (18). However, the SufS/CsdB protein was shown to be 290 times more active on l-selenocysteine than on l-cysteine and was thus considered the E. coli counterpart of the mammalian selenocysteine lyase (18). The gene products of the remaining five suf genes are less well studied, but some show interesting homologies to other characterized proteins. sufA encodes a homolog of IscA which, together with the products of the iscS and iscU genes, is involved in Fe-S cluster formation and repair. The sufB, sufC, and sufD genes encode components of an ATP binding cassette (ABC) transporter. No biochemical data can be found with regard to these three genes. However, genetic studies have shown that E. coli sufC mutants have delayed soxR-dependent induction of a soxS-lacZ gene fusion (22), and the stability of the [2Fe-2S] ferric ion reductase protein encoded by fhuF is decreased in sufD mutants (24). The final gene in the operon, sufE, encodes a conserved oxidoreductase, a homolog of which is present downstream of the csd gene.
Although only sufA, sufB, and sufC are among the most highly induced genes, sufD (b1681), sufS (b1680), and sufE (b1679) also are induced with higher ratios in the wild-type strain than in the ΔoxyR strain (Table 4 and Fig. 2A). To confirm OxyR regulation of this operon and to map the start of the suf transcript, we carried out primer extension assays (Fig. 2B). Consistent with the microarray data, we detected OxyR-dependent induction of sufA. A strong primer extension product ending in a T residue at position 1762442, 32 bp upstream of the sufA start codon at position 1762410, was observed when RNA isolated from the wild-type strain was used as the template. We also carried out DNase I footprinting experiments to test for OxyR binding to the sufA promoter. In a computational scan of the E. coli genome, we predicted a putative OxyR site centered at 1762663, 253 bp upstream of the sufA start codon (41). The DNase I footprinting carried out using a 378-bp fragment from the sufA-ydiH intragenic region showed that oxidized OxyR exclusively bound to the predicted site (Fig. 2C). However, this single OxyR binding site was far upstream from the end of the primer extension product, and at all other known OxyR-activated promoters, the transcription factor binds at a position overlapping or directly upstream of the −35 sequence of the promoter. Possibly, the initial sufA transcript is longer and is either processed or folded into a complex secondary structure impervious to reverse transcriptase. Given the unusually high conservation of the sufA-ydiH intragenic region (34), it is likely that the regulation of sufA expression is complex. We did not observe strong Fur regulation of sufA expression under conditions used in our experiments (data not shown). However, two putative Fur binding sites, centered at positions 1762460 and 1762466, have been predicted in the suf promoter (K. Lewis, B. Doan, M. Zheng, G. Storz, and T. D. Schneider, unpublished data), and Fur-dependent expression of sufD-lacZ and sufS-lacZ fusions has been observed previously (24). More experiments are needed to fully understand the regulation of the suf operon.
FIG. 2.
OxyR-dependent induction of the suf operon. (A) Structure of the suf operon and sequence of the sufA promoter. The induction ratios observed for the wild-type and ΔoxyR mutant strains in the microarray experiment are given below each gene. The sufA transcription start is marked by the black arrow, and the start of the corresponding ORF is denoted by a white arrow. The DNase I footprints for OxyR binding are indicated by the dark boxes. Our computational search (41) predicted an OxyR binding site of 9.6 bits centered at position 922026. (B) Primer extension assays of sufA expression in wild-type (MC4100) and ΔoxyR (GSO47) strains grown in LB medium. Exponential-phase cultures were split into two aliquots: one aliquot was left untreated, and the other was treated with 1 mM hydrogen peroxide. The cells were harvested after 10 min, total RNA was isolated, and primer extension assays were carried out with primer 820 specific to sufA. The neighboring sequencing reactions were carried out with the same primer. (C) DNase I footprinting assays of oxidized OxyR binding to the top and bottom strands relative to the sufA promoter. The regions protected by OxyR on both strands are indicated by the brackets. For OxyR binding to the top strand, the 32P-labeled primer 828 and unlabeled primer 825 were used to PCR amplify a 187-bp fragment. For OxyR binding to the bottom strand, the 378-bp NotI-BamHI fragment of pGSO133 was labeled with 32P at the BamHI site. The samples were run in parallel with Maxam-Gilbert G/A sequencing ladders.
(iii) Genes of unknown function.
Four ORFs of unknown function, yaiA, yaaA, yljA, and ybjM, showed much stronger peroxide induction in the wild-type strain than in the oxyR deletion strain. yaaA and yljA are conserved ORFs, but experimental data about the function of the corresponding proteins cannot be found in the literature. Primer extension assays confirmed that transcripts initiating directly upstream of the predicted first codon of the yaiA, yaaA, and yljA ORFs are induced by hydrogen peroxide in an OxyR-dependent manner (data not shown). The computational search for OxyR binding sites (41) predicted a putative OxyR binding site centered at position 922026 in the yljA promoter. Interestingly, we were not able to detect a transcript that would encode the predicted 125-amino-acid (aa) YbjM ORF. Instead, Northern blots and primer extension analysis showed that the OxyR-regulated grxA transcript is approximately 600 nucleotides in length and extends into the strand opposite ybjM. Inspection of the ybjM antisense sequence revealed that this opposite strand also encodes an 81-aa ORF. The arrays used in our experiments do not distinguish between strands. Thus, we suggest that the signal detected for ybjM actually corresponds to OxyR activation of a gene which is encoded on the opposite strand and is likely to be in an operon with grxA.
(iv) Possible OxyR-repressed genes.
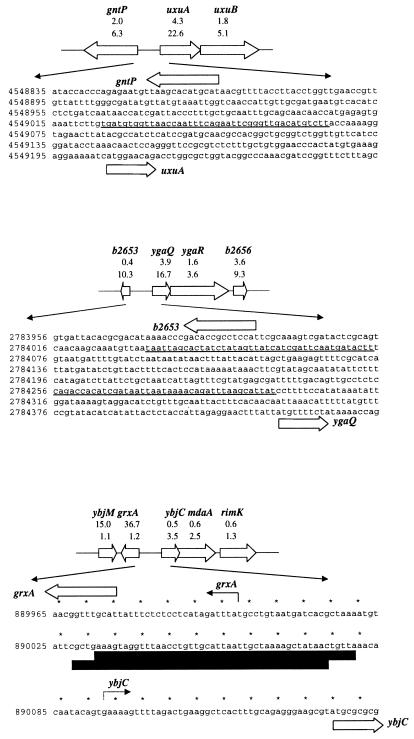
In comparing genes induced by hydrogen peroxide in the wild type (Table 2) and the ΔoxyR mutant (Table 3), we noted a number of genes, uxuA, ygaQ, ytfK, ydcH, ydeN, and yaeH, that were induced more strongly in the ΔoxyR background. OxyR is both an activator and a repressor. Thus, it is possible that OxyR represses these genes in response to oxidative stress. Our computational search predicted an OxyR binding site at position 4549044 between uxuAB (encoding mannonate hydrolase and mannonate oxidoreductase) and the divergent gntP gene (encoding a possible gluconate permease) (Fig. 3). Since uxuB and gntP also showed higher induction ratios in the absence of OxyR, it is intriguing to speculate that the two divergent promoters are repressed by oxidized OxyR. Similarly, there are two predicted OxyR binding sites at positions 2784053 and 2784276 between ygaQ and a putative divergent gene designated b2653, and both ygaQ and b2653 showed a higher induction ratio in the ΔoxyR mutant strain. In this context, it also is noteworthy that three genes, ybjC, nfsA/mdaA (encoding nitrofuran reductase I activity B), and rimK (encoding a ribosomal modification protein), divergent to grxA were induced in the ΔoxyR mutant but not the wild-type strain. No OxyR binding sites were predicted upstream of ytfK, ydcH, ydeN, and yaeH, so these genes may or may not be repressed directly by OxyR. OxyR binding to these promoters and the predicted sites will need to be tested experimentally.
FIG. 3.
Possible OxyR-dependent repression of gntP and uxuAB; b2653, ygaQ, ygaR, and b2656; and ybjC, nfsA/mdaA, and rimK. The gene organization of the corresponding operons is shown. Predicted OxyR binding sites (41) of 7.0 bits centered at position 4549044, 8.8 bits centered at position 2784053, and 5.4 bits centered at position 2784276 are underlined. The confirmed OxyR binding site upstream of the grxA promoter (39) is indicated by the black boxes. The transcription starts documented for grxA and predicted for ybjC are denoted by solid and dotted arrows, respectively. The induction ratios observed for the wild-type and ΔoxyR mutant strains in the microarray experiment are given below each gene.
OxyR-independent responses.
Below, we describe the expression of genes that are induced similarly by hydrogen peroxide in the wild-type strain and the ΔoxyR mutant strain. It is interesting that among genes induced moderately in both the wild-type strain and the ΔoxyR deletion strain were a number of heat shock genes (groEL, groES, grpE, dnaK, and htpG) including those encoding proteolytic activities (clpA, clpB, clpX, and clpP). SOS genes (recA, recN, lexA, and dinD); sulfate and cysteine metabolism genes (cysKAUPNDHJ and sbp); genes specifying tricarboxylic acid cycle enzymes (acnA and fumA); the nrdHIEF operon, which directs synthesis of a second ribonucleotide reductase system; and the universal stress gene uspA also were induced to some extent in both the wild-type strain and the mutant strain. In contrast, the expression of many ribosomal protein genes, cold shock genes, ATP synthase genes, and transporter genes was repressed.
(i) SoxRS regulon.
The SoxRS regulon was reported previously to be induced primarily by superoxide-generating compounds and not by hydrogen peroxide (23). Thus, we were surprised to find that several members of this regulon such as fpr (encoding ferredoxin-flavodoxin reductase) and sodA (with an induction ratio of 8 and encoding manganese superoxide dismutase), as well as soxS itself, were among the genes most strongly induced by 1 mM hydrogen peroxide in both the wild-type background and the ΔoxyR mutant strain background. To directly compare the induction of OxyR and SoxR target genes by both hydrogen peroxide and superoxide-generating compounds, we treated wild-type cells with either 0, 0.01, 0.03, 0.1, 0.3, and 1 mM hydrogen peroxide or the same concentrations of paraquat (methyl viologen), a standard inducer of the soxRS regulon. We then carried out primer extension assays to examine the expression of oxyS, a primary OxyR target, and soxS, the only known SoxR target (Fig. 4). As expected, oxyS was strongly induced by all concentrations of hydrogen peroxide and soxS was induced by all concentrations of paraquat, consistent with the notion that OxyR primarily senses hydrogen peroxide while SoxR primarily responds to superoxide-generating compounds. However, we also observed that soxS was partially induced by high concentrations of hydrogen peroxide and that oxyS was slightly induced by high concentrations of paraquat.
FIG. 4.
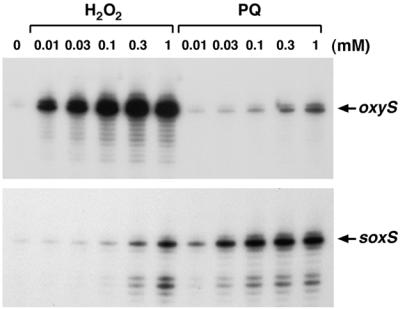
oxyS and soxS induction by hydrogen peroxide and paraquat. The figure shows the results of primer extension assays of oxyS and soxS transcript levels in wild-type (MC4100) cells grown in LB medium. An exponential-phase culture was split into aliquots: one aliquot was left untreated, and the other aliquots were exposed to the indicated concentrations of hydrogen peroxide and paraquat. The cells were harvested after 5 min, total RNA was isolated, and primer extension assays were carried out with primer 188 specific to oxyS and primer 823 specific to soxS.
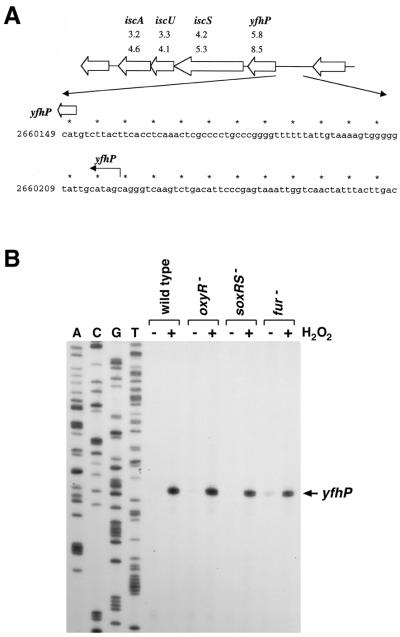
(ii) isc operon.
The isc gene cluster, yfhP (b2531), iscS/yfhO (b2530), iscU/yfhN (b2529), iscA/yfhF (b2528), hscB (b2527), hscA (b2526), fdx (b2525), and yfhJ (b2524), has received attention due to its role in Fe-S cluster formation and cysteine-related metabolism (12, 25, 38). The fact that Fe-S cluster damage is a major consequence of oxidative stress prompted us to examine the regulation of this cluster. The expression profiles indicated that the first four genes (yfhP, iscS, iscU, and iscA) in the cluster were modestly induced in both the wild-type strain and the oxyR deletion strain (Fig. 5A). The induction ratios for the last four genes (hscB, hscA, fdx, and yfhJ) were within the error of the experiment, suggesting that these four genes were not induced and that they are regulated differently. This result is consistent with a report that the hscBA genes are transcribed independently of the isc genes (15). Primer extension assays showed that isc operon induction by hydrogen peroxide (Fig. 5B) and paraquat (data not shown) was independent of both OxyR and SoxRS. These assays also allowed the transcription start to be mapped to a G residue (on the strand opposite the one shown in Fig. 5A) at position 2660219, 68 bp upstream of the yfhP start codon.
FIG. 5.
OxyR- and SoxRS-independent induction of the isc operon. (A) Structure of the isc operon and sequence of the yfhP promoter. The induction ratios observed for the wild-type and ΔoxyR mutant strains in the microarray experiment are given below each gene. The yfhP transcription start is marked by the black arrow, and the start of the corresponding ORF is denoted by a white arrow. (B) Primer extension assays of yfhP expression in wild-type (MC4100), ΔoxyR (GSO47), ΔsoxRS (GSO71), and Δfur (GSO72) strains grown in LB medium. Exponential-phase cultures were split into two aliquots: one aliquot was left untreated, and the other was treated with 1 mM hydrogen peroxide. The cells were harvested after 10 min, total RNA was isolated, and primer extension assays were carried out with primer 686 specific to yfhP. The neighboring sequencing reactions were carried out with the same primer.
(iii) Induction of other genes.
Among the other genes whose expression was strongly induced in an OxyR-independent manner were cysK, encoding cysteine synthase, which catalyzes the last step in cysteine synthesis; sbp and cysP, encoding periplasmic sulfate binding proteins; and dsdA, encoding a d-serine deaminase. We also observed modest induction of other genes in the cysteine biosynthesis pathway. The induction of these genes suggested a concerted effort to accumulate more cysteine in response to hydrogen peroxide treatment. Two other genes, tnaA and tnaL, that were highly induced in an OxyR-independent manner are involved in amino acid catabolism. The ibpA and ibpB genes encoding heat shock proteins also were strongly induced in both the wild-type and ΔoxyR strains (Tables 1 and 2). The observed induction of the genes encoding the HSP20 chaperones in response to oxidative stress was consistent with a recent report that ibpA-, ibpB-, and ibpAB-overexpressing strains are resistant not only to heat but also to paraquat treatment (14).
There are 10 genes of unknown function among the most highly induced genes whose expression is independent of OxyR. yfiA was strongly induced (36-fold) in the wild-type strain and became the most strongly induced (109-fold) gene in the ΔoxyR mutant strain. The function of YfiA is not clear. Recent reports have shown that the YfiA protein is associated with 70S ribosomes and stabilizes ribosomes against dissociation (1, 16). Nine other strongly induced genes encode relatively small ORFs: yjiD (133 aa), ymgB (88 aa), yeeD (75 aa), ycfR (85 aa, induced 9-fold in the ΔoxyR strain), ycgZ (78 aa, induced 9-fold in the ΔoxyR strain), ymgA (90 aa, induced 3-fold in the wild type), yceP (84 aa, induced 10-fold in the wild type), ynaF (144 aa, induced 8-fold in the wild type), and ycgK (133 aa, induced 9-fold in the wild type). Homology searches suggest that ynaF encodes a filament protein. YeeD and YcfR show homology to other unknown ORFs, but all of the other genes encoding the small ORFs at best have very low scores in BLAST searches. We note that the small size of many of these predicted proteins may have precluded them from being detected on the two-dimensional gels initially used to characterize the response to hydrogen peroxide.
DISCUSSION
Use of DNA microarrays to characterize the oxidative stress response.
DNA microarray technology already has been shown to be a useful tool in studying global expression patterns in response to a number of different growth conditions; here, we examine the E. coli response to oxidative stress. In the experiment that we have presented, we were able to confirm the induction of many oxidative stress genes that were laboriously identified over a period of almost 20 years. In addition, we were able to identify several new hydrogen peroxide-inducible genes: some new members of the OxyR regulon and others induced by an OxyR-independent mechanism. We do note a few limitations of our experiment. First, since the glass slides that we used carry only DNA corresponding to ORFs, we did not detect expression of the strongly induced OxyS RNA. Second, for reasons that are not understood, the induction of some of the OxyR-regulated ORFs such as dsbG was not detected. Thus, it has been an advantage to simultaneously carry out a computational search for additional OxyR binding sites (41). A third limitation is that the arrays are not strand specific. From the array data, we assumed that the annotated ybjM gene was induced by OxyR; however, primer extension and Northern experiments showed that hydrogen peroxide treatment actually leads to the induction of a transcript on the opposite strand. Despite the limitations listed above, future microarray experiments to examine the global gene response to hydrogen peroxide and other oxidants over a range of concentrations and times should give even further insight into the E. coli response to oxidative stress. Experiments to examine the gene expression profiles in specific mutants also should help to further delineate the roles of specific regulators. For example, by examining the hydrogen peroxide response in strains lacking the OxyS small RNA regulator, we may be able to differentiate those genes regulated directly by OxyR and those regulated indirectly through OxyS RNA.
Identification of OxyR-regulated genes.
We have identified several new OxyR-regulated genes. One example is hemH encoding the ferrochelatase that catalyzes the conversion of protoporphyrin IX to protoheme, the final step of protoheme biosynthesis. Heme is an essential cofactor for both the katG- and the katE-encoded hydroperoxidase enzymes, and it has long been known that katG transcription and hydroperoxidase I activity are strongly induced in response to oxidative stress. If ferrochelatase is the limiting step in protoheme biosynthesis, the induction of hemH may be needed to satisfy the need for increased heme levels associated with increased hydroperoxidase production. Protoporphyrin IX can generate reactive oxygen species in the presence of light (21). Thus, the induction of hemH may be important to reduce the concentration of the potentially toxic protoporphyrin IX intermediate. Since hydrogen peroxide oxidizes the pool of intracellular ferrous iron, increased ferrochelatase production also may be required to allow for the enzyme to compete for lowered levels of iron. Additional experiments are needed to distinguish between these possible explanations for the OxyR-dependent induction of hemH. It is interesting that, in Bacillus subtilis, an operon (hemAXCDBL) encoding enzymes for the early steps of heme biosynthesis is induced by hydrogen peroxide (6, 8), although our DNA microarray experiments did not provide evidence for the induction of the corresponding genes in E. coli.
We also discovered that the sufA, sufB, sufC, sufD, sufS, and sufE genes are part of the OxyR regulon. The observation that all of the genes were induced by hydrogen peroxide in an OxyR-dependent way is consistent with a previous proposal that these genes form an operon (24). Although the exact in vivo function of the suf operon is not clear, limited biochemical evidence suggests that suf-encoded proteins are involved in Fe-S cluster and/or S and/or Se metabolism. Since Fe-S clusters are one of the primary cellular targets of oxidative stress (reviewed in reference 29), there is a clear need to induce proteins that help to assemble or repair these clusters. Patzer and Hantke (24) isolated sufD and sufS mutants based on their requirement for the fhuF-encoded ferric ion reductase activity. Given this observation, it seems contradictory that the suf operon is induced by oxidized OxyR while fhuF is repressed by oxidized OxyR (41). We suggest that this opposing regulation indicates that the suf-encoded proteins are required for cellular functions in addition to FhuF. In general, since hemH and the suf operon, as well as some of the OxyR-regulated genes of unknown function, are highly conserved among prokaryotic species, future genetic and biochemical studies of these genes and their gene products will increase our understanding of cellular defenses against oxidative stress.
Overlap with other regulatory pathways.
We observed soxS induction by 1 mM hydrogen peroxide in our DNA microarray measurements and, by primer extension assays, confirmed that soxS could be induced by 0.3 and 1 mM hydrogen peroxide. Recent assays of a SoxRS-regulated micF::luxCDABE fusion (4) and measures of soxS transcript levels by multiplex reverse transcription-PCR (17) also suggest that SoxR is activated by high concentrations of hydrogen peroxide. These results expand the overlap between the superoxide and peroxide responses in E. coli. The observed soxS induction may be due to some SoxR oxidation by high peroxide concentrations. Alternatively, hydrogen peroxide might lead to the generation of another signal that activates SoxR or to inefficient SoxR reduction, by as yet uncharacterized mechanisms. Two known members of the SoxRS regulon, fpr and sodA, were among the most highly hydrogen peroxide-induced genes in both the wild-type and the ΔoxyR mutant cells. It is possible that the expression of other transcripts that are induced independently of OxyR is under the control of the SoxRS regulators.
Our DNA microarray results also show that there is overlap between the oxidative stress and heat shock and SOS responses. This overlap was presaged by the initial two-dimensional gel experiments (20, 33) as well as by assays of small panels of stress-responsive promoters fused to luxCDABE (4). A systematic identification of all the genes regulated by particular transcription factors should help to map out the complex genetic regulatory network among the different stress responses. Two recent studies have examined the whole-genome expression pattern in Saccharomyces cerevisiae cells exposed to a variety of stress conditions including oxidative stress and heat shock (7, 11). One difference between the bacterial and yeast responses to hydrogen peroxide is noteworthy; while there is a predominant, clearly defined OxyR-regulated response in E. coli, the S. cerevisiae response involves a large set of general stress proteins. In the ΔoxyR mutant strain, the induction of other general stress transcripts becomes more pronounced, making the peroxide response in the ΔoxyR strain more akin to the S. cerevisiae response. Possible parallels can be drawn between the response to oxidative stress and that to amino acid starvation. While wild-type E. coli cells induce specific operons in response to deprivation of specific amino acids (for example, Khodursky et al. [13] describe the specific response to tryptophan starvation), wild-type S. cerevisiae cells induce a generalized response upon encountering a limited supply of any one amino acid. Inactivation of an E. coli response to a particular amino acid-mediated regulatory circuit results in an emphasis upon the second, more generalized stringent response.
Our study points out that the activities of transcription factors, in addition to OxyR and SoxRS, may be modulated by oxidative stress. An example of such regulation is the isc operon. Both microarray and primer extension measurements showed that isc expression is induced by peroxide. Given the role of the isc gene products in Fe-S assembly, this induction is not surprising. However, it was unexpected to find that the induction is independent of both OxyR and SoxR, indicating the existence of an unidentified redox regulatory pathway. We also were intrigued by the strong OxyR-independent induction of the yfiA gene. Identification of these alternate pathways of hydrogen peroxide-dependent gene induction and the characterization of the redox sensing mechanisms involved are important directions for future studies.
ACKNOWLEDGMENTS
We thank L. Heineman and E. DeRose of DuPont for assistance in data processing and J. Imlay for useful comments on the manuscript.
The work in Bethesda was supported by the intramural programs of the National Institute of Child Health and Human Development and the National Cancer Institute and a fellowship from the American Cancer Society (M.Z.).
REFERENCES
- 1.Agafonov D E, Kolb V A, Nazimov I V, Spirin A S. A protein residing at the subunit interface of the bacterial ribosome. Proc Natl Acad Sci USA. 1999;96:12345–12349. doi: 10.1073/pnas.96.22.12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bachmann B J. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 2. Washington, D.C.: ASM Press; 1996. pp. 2460–2488. [Google Scholar]
- 3.Beale S I. Biosynthesis of hemes. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 2. Washington, D.C.: ASM Press; 1996. pp. 731–748. [Google Scholar]
- 4.Belkin S, Smulski D R, Dadon S, Vollmer A C, Van Dyk T K, LaRossa R A. A panel of stress-responsive luminous bacteria for the detection of selected classes of toxicants. Water Res. 1997;31:3009–3016. [Google Scholar]
- 5.Blattner F R, Plunkett G, Bloch C A, Perna N T, Burland V, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 6.Bsat N, Herbig A, Casillas-Martinez L, Setlow P, Helmann J D. Bacillus subtilis contains multiple Fur homologues: identification of the iron uptake (Fur) and peroxide regulon (PerR) repressors. Mol Microbiol. 1998;29:189–198. doi: 10.1046/j.1365-2958.1998.00921.x. [DOI] [PubMed] [Google Scholar]
- 7.Causton H C, Ren B, Koh S S, Harbison C T, Kanin E, Jennings E G, Lee T I, True H L, Lander E S, Young R A. Remodeling of yeast genome expression in response to environmental change. Mol Biol Cell. 2001;12:323–337. doi: 10.1091/mbc.12.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen L, Keramati L, Helmann J D. Coordinate regulation of Bacillus subtilis peroxide stress genes by hydrogen peroxide and metal ions. Proc Natl Acad Sci USA. 1995;92:8190–8194. doi: 10.1073/pnas.92.18.8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 10.Christman M F, Morgan R W, Jacobson F S, Ames B N. Positive control of a regulon for defenses against oxidative stress and some heat-shock proteins in Salmonella typhimurium. Cell. 1985;41:753–762. doi: 10.1016/s0092-8674(85)80056-8. [DOI] [PubMed] [Google Scholar]
- 11.Gasch A P, Spellman P T, Kao C M, Carmel-Harel O, Eisen M B, Storz G, Botstein D, Brown P O. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kambampati R, Lauhon C T. IscS is a sulfurtransferase for the in vitro biosynthesis of 4-thiouridine in Escherichia coli tRNA. Biochemistry. 1999;38:16561–16568. doi: 10.1021/bi991119r. [DOI] [PubMed] [Google Scholar]
- 13.Khodursky A B, Peter B J, Cozzarelli N R, Botstein D, Brown P O, Yanofsky C. DNA microarray analysis of gene expression in response to physiological and genetic changes that affect tryptophan metabolism in Escherichia coli. Proc Natl Acad Sci USA. 2000;97:12170–12175. doi: 10.1073/pnas.220414297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kitagawa M, Matsumura Y, Tsuchido T. Small heat shock proteins, IbpA and IbpB, are involved in resistances to heat and superoxide stresses in Escherichia coli. FEMS Microbiol Lett. 2000;184:165–171. doi: 10.1111/j.1574-6968.2000.tb09009.x. [DOI] [PubMed] [Google Scholar]
- 15.Lelivelt M J, Kawula T H. Hsc66, an Hsp70 homolog in Escherichia coli, is induced by cold shock but not heat shock. J Bacteriol. 1995;177:4900–4907. doi: 10.1128/jb.177.17.4900-4907.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maki Y, Yoshida H, Wada A. Two proteins, YfiA and YhbH, associated with resting ribosomes in stationary phase Escherichia coli. Genes Cells. 2000;5:965–974. doi: 10.1046/j.1365-2443.2000.00389.x. [DOI] [PubMed] [Google Scholar]
- 17.Manchado M, Michán C, Pueyo C. Hydrogen peroxide activates the SoxRS regulon in vivo. J Bacteriol. 2000;182:6842–6844. doi: 10.1128/jb.182.23.6842-6844.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mihara H, Maeda M, Fujii T, Kurihara T, Hata Y, Esaki N. A nifS-like gene, csdB, encodes an Escherichia coli counterpart of mammalian selenocysteine lyase. Gene cloning, purification, characterization and preliminary x-ray crystallographic studies. J Biol Chem. 1999;274:14768–14772. doi: 10.1074/jbc.274.21.14768. [DOI] [PubMed] [Google Scholar]
- 19.Miyamoto K, Nakahigashi K, Nishimura K, Inokuchi H. Isolation and characterization of visible light-sensitive mutants of Escherichia coli K12. J Mol Biol. 1991;219:393–398. doi: 10.1016/0022-2836(91)90180-e. [DOI] [PubMed] [Google Scholar]
- 20.Morgan R W, Christman M F, Jacobson F S, Storz G, Ames B N. Hydrogen peroxide-inducible proteins in Salmonella typhimurium overlap with heat shock and other stress proteins. Proc Natl Acad Sci USA. 1986;83:8059–8063. doi: 10.1073/pnas.83.21.8059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakahigashi K, Nishimura K, Miyamoto K, Inokuchi H. Photosensitivity of a protoporphyrin-accumulating, light-sensitive mutant (visA) of Escherichia coli K-12. Proc Natl Acad Sci USA. 1991;88:10520–10524. doi: 10.1073/pnas.88.23.10520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nachin L, El Hassouni M, Loiseau L, Expert D, Barras F. SoxR-dependent response to oxidative stress and virulence in Erwinia chrysanthemi: the key role of SufC, an orphan ABC ATPase. Mol Microbiol. 2001;39:960–972. doi: 10.1046/j.1365-2958.2001.02288.x. [DOI] [PubMed] [Google Scholar]
- 23.Nunoshiba T, Hidalgo E, Amábile Cuevas C F, Demple B. Two-stage control of an oxidative stress regulon: the Escherichia coli SoxR protein triggers redox-inducible expression of the soxS regulatory gene. J Bacteriol. 1992;174:6054–6060. doi: 10.1128/jb.174.19.6054-6060.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patzer S I, Hantke K. SufS is a NifS-like protein, and SufD is necessary for stability of the [2Fe-2S] FhuF protein in Escherichia coli. J Bacteriol. 1999;181:3307–3309. doi: 10.1128/jb.181.10.3307-3309.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwartz C J, Djaman O, Imlay J A, Kiley P J. The cysteine desulfurase, IscS, has a major role in in vivo Fe-S cluster formation in Escherichia coli. Proc Natl Acad Sci USA. 2000;97:9009–9014. doi: 10.1073/pnas.160261497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Selinger D W, Cheung K J, Mei R, Johansson E M, Richmond C S, Blattner F R, Lockhart D J, Church G M. RNA expression analysis using a 30 base pair resolution Escherichia coli genome array. Nat Biotechnol. 2000;18:1262–1268. doi: 10.1038/82367. [DOI] [PubMed] [Google Scholar]
- 27.Silhavy T J, Berman M L, Enquist L W. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1984. [Google Scholar]
- 28.Smulski D R, Huang L L, McCluskey M P, Gladnick Reeve M J, Vollmer A C, Van Dyk T K, LaRossa R A. Combined, functional genomic-biochemical approach to intermediary metabolism: interaction of acivicin, a glutamine amidotransferase inhibitor, with Escherichia coli K-12. J Bacteriol. 2001;183:3353–3364. doi: 10.1128/JB.183.11.3353-3364.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Storz G, Imlay J A. Oxidative stress. Curr Opin Microbiol. 1999;2:188–194. doi: 10.1016/s1369-5274(99)80033-2. [DOI] [PubMed] [Google Scholar]
- 30.Storz G, Zheng M. Oxidative stress. In: Storz G, Hengge-Aronis R, editors. Bacterial stress responses. Washington, D.C.: ASM Press; 2000. pp. 47–59. [Google Scholar]
- 31.Tao H, Bausch C, Richmond C, Blattner F R, Conway T. Functional genomics: expression analysis of Escherichia coli growing on minimal and rich media. J Bacteriol. 1999;181:6425–6440. doi: 10.1128/jb.181.20.6425-6440.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Toledano M B, Kullik I, Trinh F, Baird P T, Schneider T D, Storz G. Redox-dependent shift of OxyR-DNA contacts along an extended DNA-binding site: a mechanism for differential promoter selection. Cell. 1994;78:897–909. doi: 10.1016/s0092-8674(94)90702-1. [DOI] [PubMed] [Google Scholar]
- 33.VanBogelen R A, Kelley P M, Neidhardt F C. Differential induction of heat shock, SOS, and oxidative stress regulons and accumulation of nucleotides in Escherichia coli. J Bacteriol. 1987;169:26–32. doi: 10.1128/jb.169.1.26-32.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wassarman, K. M., F. Repoila, C. Rosenow, G. Storz, and S. Gottesman. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev., in press. [DOI] [PMC free article] [PubMed]
- 35.Wei Y, Lee J-M, Richmond C, Blattner F R, Rafalski J A, LaRossa R A. High-density microarray-mediated gene expression profiling of Escherichia coli. J Bacteriol. 2001;183:545–556. doi: 10.1128/JB.183.2.545-556.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wei Y, Lee J-M, Smulski D R, LaRossa R A. Global impact of sdiA amplification revealed by comprehensive gene expression profiling of Escherichia coli. J Bacteriol. 2001;183:2265–2272. doi: 10.1128/JB.183.7.2265-2272.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang A, Altuvia S, Tiwari A, Argaman L, Hengge-Aronis R, Storz G. The OxyS regulatory RNA represses rpoS translation and binds the Hfq (HF-I) protein. EMBO J. 1998;17:6061–6068. doi: 10.1093/emboj/17.20.6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng L, Cash V L, Flint D H, Dean D R. Assembly of iron-sulfur clusters. Identification of an iscSUA-hscBA-fdx gene cluster from Azotobacter vinelandii. J Biol Chem. 1998;273:13264–13272. doi: 10.1074/jbc.273.21.13264. [DOI] [PubMed] [Google Scholar]
- 39.Zheng M, Åslund F, Storz G. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science. 1998;279:1718–1721. doi: 10.1126/science.279.5357.1718. [DOI] [PubMed] [Google Scholar]
- 40.Zheng M, Doan B, Schneider T D, Storz G. OxyR and SoxRS regulation of fur. J Bacteriol. 1999;181:4639–4643. doi: 10.1128/jb.181.15.4639-4643.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng M, Wang X, Doan B, Lewis K A, Schneider T D, Storz G. Computation-directed identification of OxyR DNA binding sites in Escherichia coli. J Bacteriol. 2001;183:4571–4579. doi: 10.1128/JB.183.15.4571-4579.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]