Abstract
A computational search was carried out to identify additional targets for the Escherichia coli OxyR transcription factor. This approach predicted OxyR binding sites upstream of dsbG, encoding a periplasmic disulfide bond chaperone-isomerase; upstream of fhuF, encoding a protein required for iron uptake; and within yfdI. DNase I footprinting assays confirmed that oxidized OxyR bound to the predicted site centered 54 bp upstream of the dsbG gene and 238 bp upstream of a known OxyR binding site in the promoter region of the divergently transcribed ahpC gene. Although the new binding site was near dsbG, Northern blotting and primer extension assays showed that OxyR binding to the dsbG-proximal site led to the induction of a second ahpCF transcript, while OxyR binding to the ahpCF-proximal site leads to the induction of both dsbG and ahpC transcripts. Oxidized OxyR binding to the predicted site centered 40 bp upstream of the fhuF gene was confirmed by DNase I footprinting, but these assays further revealed a second higher-affinity site in the fhuF promoter. Interestingly, the two OxyR sites in the fhuF promoter overlapped with two regions bound by the Fur repressor. Expression analysis revealed that fhuF was repressed by hydrogen peroxide in an OxyR-dependent manner. Finally, DNase I footprinting experiments showed OxyR binding to the site predicted to be within the coding sequence of yfdI. These results demonstrate the versatile modes of regulation by OxyR and illustrate the need to learn more about the ensembles of binding sites and transcripts in the E. coli genome.
The OxyR transcription factor is found in many prokaryotic organisms (reviewed in reference 17). This LysR-type regulator activates the expression of numerous genes in response to oxidative stress, in particular upon exposure to hydrogen peroxide, and has served as a paradigm for understanding cellular sensing of oxidative stress. Hydrogen peroxide directly activates OxyR through the formation of an intramolecular disulfide bond (22). Oxidized OxyR then activates transcription of antioxidant genes, including katG (encoding hydroperoxidase I), ahpCF (encoding an alkyl hydroperoxide reductase), dps (encoding a nonspecific DNA binding protein), gorA (encoding glutathione reductase), grxA (encoding glutaredoxin I), and oxyS (encoding a small regulatory RNA) (reviewed in reference 17).
Much has been learned about cellular defenses against oxidative stress through studies of the OxyR regulon. Thus, as part of a continuing effort to better define the physiological roles of OxyR, we initiated a computational approach to identify additional OxyR-regulated genes. We used an algorithm based on information theory (11) that uses previously identified OxyR binding site sequences as a model to search through the entire Escherichia coli genome for putative OxyR binding sites. Our computational approach predicted several sites that had not been identified previously by experimental means. One target gene, identified using this approach and experimentally confirmed to be regulated by OxyR, encodes the iron metabolism regulator Fur, demonstrating coordinate regulation between oxidative stress response and iron metabolism (23).
Here we report our computer-directed identification of three more OxyR binding sites: one upstream of dsbG, one upstream of fhuF, and one within yfdI. The dsbG gene encodes a periplasmic chaperone-isomerase that facilitates correct folding of disulfide bond proteins (1, 2, 14, 20). The fhuF gene encodes a 2Fe-2S protein whose likely function is ferric iron reduction, required for iron uptake by the cell (8). The yfdI-encoded gene product shares homology with ligases, but its biochemical function has not been experimentally demonstrated. We confirmed the presence of OxyR binding to the three sites by in vitro DNase I footprinting assays. We also examined expression of the dsbG and fhuF genes and found the regulation of these two genes to be unique.
MATERIALS AND METHODS
Computer search program.
The initial computational search was carried out as described elsewhere (23). Seven OxyR target sites identified by footprinting, E. coli oxyR (position 163 in GenBank entry JO4553), katG (position 68 in GenBank entry M21516), ahpC (position 116 in GenBank entry D13187), dps (position 202 in GenBank entry X69337), gor (position 60491 in GenBank entry U00039), and grxA (position 207 in GenBank entry M13449); Mu phage mom-1 (position 68 in GenBank entry VO1463); Salmonella orf; and two sites identified by homology, E. coli ahpC (position 116 in GenBank entry D13187) and a second Mu phage mom-2 site (position 59 in GenBank entry VO1463), were used to generate an individual information weight matrix (11). The matrix was scanned across the entire E. coli genomic sequence (3), and the identified sites were sorted by information content. Local regions of the genome surrounding the strongest sites were displayed using the Lister program (version 9.02) (see Fig. 1, 4, and 7) to show coding regions along with sequence walkers representing potential binding sites (12). Detailed Lister maps of the 20 sites with the highest information content are available online (http://www.lecb.ncifcrf.gov/∼toms/paper/zheng.storz2001/). A subsequent computational search was carried out using the OxyR target sites at E. coli oxyR, katG, ahpC, dps, gor, and grxA and at Mu phage mom-1 as well as at E. coli fur, dsbG, fhuF-1, fhuF-2, yfdI, trxC, flu, sufA, and hemH (positions 710103, 637851, 4603273, 4603357, 2467337, 2716640, 2069355, 1762663, and 497211 in GenBank entry U00096, respectively) to generate the individual information weight matrix.
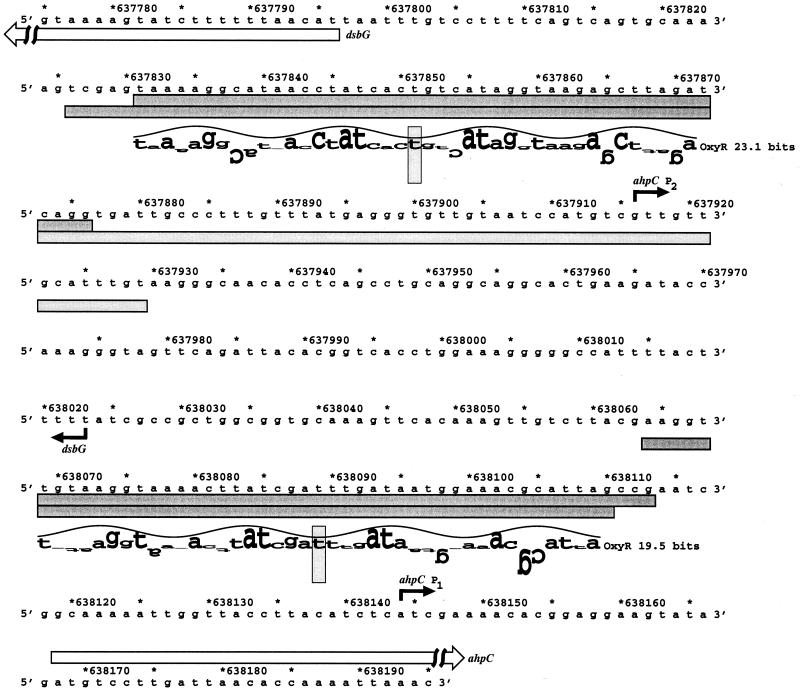
FIG. 1.
Sequence of the ahpC and dsbG promoters. The ahpC and dsbG transcription starts are marked by black arrows, and the starts of the corresponding ORFs are denoted by white arrows. The DNase I footprints for OxyR binding to the top and bottom strands are indicated by the dark gray boxes. The DNase I footprint for RNA polymerase binding adjacent to the dsbG-proximal OxyR binding site on the bottom strand is indicated by the light gray box. The locations of the two predicted oxyR sites are shown by sequence walkers (12), in which the rectangles surrounding the T's at positions 637851 and 638089 indicate the centers of the binding sites. A sequence walker consists of a string of letters in which the height of each letter shows the contribution that the corresponding base would make to the average sequence conservation shown by the sequence logo of all binding sites (10). The sequence walker is given on a scale of bits of information. The scale, from −3 bits up to the maximum conservation at +2 bits, is given by the rectangle surrounding the T. Positively contributing bases are above the zero line, and negatively contributing bases are below the line. By using bits, the heights of all letters can be added together to obtain the information content of a site.
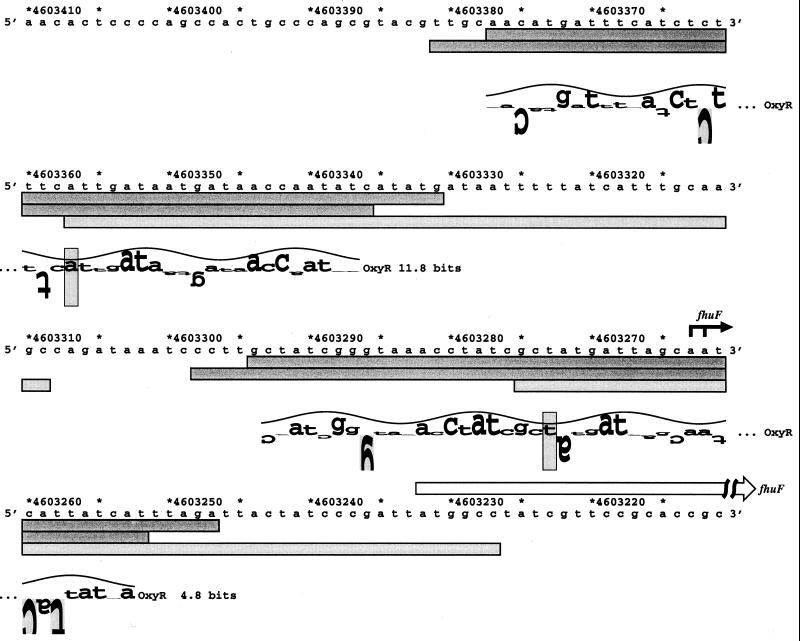
FIG. 4.
Sequence of the fhuF promoter. The fhuF transcription start is marked by the black arrow, and the start of the FhuF ORF is denoted by the white arrow. The DNase I footprints for OxyR binding to the top and bottom strands are indicated by the dark gray boxes. The DNase I footprints for Fur binding to the top strands (K. A. Lewis, B. Doan, M. Zheng, G. Storz, and T. D. Schneider, unpublished data) are indicated by the light gray boxes. The locations of the two predicted oxyR sites are shown by sequence walkers (12), in which the rectangles surrounding the A at position 4603357 and the T at position 4603273 indicate the centers of the binding sites. The site with an information content of 11.8 bits is designated fhuF-2, and the site with an information content of 4.8 bits is designated fhuF-1.
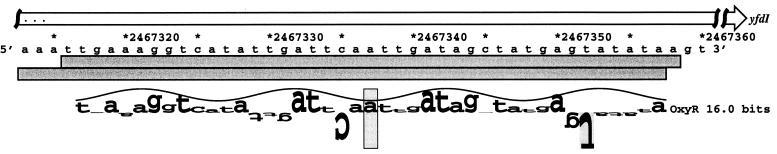
FIG. 7.
Sequence of the yfdI gene. The YfdI ORF, which extends from position 2467151 to 2468482, is indicated by the white arrow. The DNase I footprints for OxyR binding to the top and bottom strands are indicated by the dark gray boxes. The location of the predicted oxyR site is shown by a sequence walker (12), in which the rectangle surrounding the A at position 2467330 indicates the center of the binding site.
Plasmids and strains.
The DNA sequence and coordinates are for E. coli from GenBank accession no. U00096 (3). The plasmids used in the study were constructed using fragments PCR amplified from chromosomal DNA. The sequences of all oligonucleotides are listed in Table 1. To generate the dsbG promoter plasmid (pGSO123) used to test for OxyR binding, a 230-bp fragment generated using primers 433 and 434 was cloned into the EcoRI and BamHI sites of pUC8. To generate the ahpC-dsbG promoter plasmid (pGSO124) used to test for RNA polymerase binding in the presence of OxyR, a 430-bp fragment generated using primers 709 and 710 was cloned into the SmaI site of pRS415. To construct the fhuF promoter plasmid (pGSO129) used to test for OxyR binding, a 240-bp fragment generated using primers 473 and 474 was cloned into the EcoRI and BamHI sites of pUC18. To generate the ydfI plasmid (pGSO130) to test for OxyR binding, a 180-bp fragment generated using primers 475 and 476 was cloned into the EcoRI and BamHI sites of pUC18. The sequence of all inserts was verified. The strains used in the study, MC4100 (wild type), GSO47 (MC4100 ΔoxyR::kan), and GSO72 (MC4100 Δfur::kan), were described previously (23).
TABLE 1.
Sequence of oligonucleotides used in the study
| No. | Sequencea |
|---|---|
| 433 | 5′-TTTGAATTCTTCAGTGCCTGCCTGC |
| 434 | 5′-CTGGATCCGGAAGTTCCTCTGCG |
| 473 | 5′-GCGGCTGGAGATGAATTCGCCAGATG |
| 474 | 5′-GCCCTGCAATCAGGGATCCCGGCAGC |
| 475 | 5′-GTTAAATACATCTGAATTCGGAAGAGCC |
| 476 | 5′-CCCTAGGATCCAGATATTCAGTAAAAATGC |
| 610 | 5′-CAGTCAGTGCAAAAGTCGAG |
| 704 | 5′-GCGGCTGGAGATGCG |
| 706 | 5′-CGCCAGATGACATCTTC |
| 709 | 5′-GAATGCCCGGGTTTTTAAAAGGTTTA |
| 710 | 5′-GCA GACCCGGGAAAAGTATCTTTTT |
| 726 | 5′-CGGCGAATTCAAGTAAAATGGCCC |
Restriction sites are underlined.
Growth conditions.
Cultures were grown at 37°C in Luria-Bertani (LB) rich medium or M63 minimal medium supplemented with 2 mg of glucose/ml and 20 μg of vitamin B1/ml (15). 2,2′-Dipyridyl (0.5 mM) was added to some cultures to achieve iron depletion.
RNA isolation and primer extension assays.
Exponential-phase cultures (optical density at 600 nm = 0.2 to 0.5) were split into aliquots: one aliquot was left untreated, and the other aliquot was treated with the indicated amounts of hydrogen peroxide. After 10 min, the cells from 5, 10, or 25 ml of culture were harvested and resuspended in 1 ml of Trizol equilibrated at 4°C (Gibco BRL). All subsequent purification steps were carried out according to the Trizol reagent manual (based on reference 4). RNA samples were subjected to primer extension assays as described elsewhere (21), using primer 709 specific to ahpC, primer 610 specific to dsbG, and primers 704 and 706 specific to fhuF.
DNase I footprinting.
The DNase I footprinting assays of purified OxyR binding to the dsbG, fhuF, and yfdI fragments were carried out as described previously (19).
RESULTS
Initial search for new OxyR binding sites.
An initial OxyR binding site model was constructed using nine OxyR target sequences. Seven of these binding sites, upstream of the E. coli oxyR, katG, dps, gorA, and grxA genes; the Salmonella orf gene; and the Mu phage mom gene, were previously confirmed by DNase I footprinting. Two sites, a second Mu phage mom site and a site upstream of the E. coli ahpC gene, were included based on their homology to the confirmed binding sequences. The average information content of the nine sites used in this model is 16.7 ± 1.9 bits. Information content is a measure of the amount of pattern with respect to the model and is quantitated in bits, the choice between two equally likely possibilities. Our initial model was used to search the entire E. coli genome sequence for additional OxyR binding sites. Table 2 lists the 20 sites with the highest information content based on this initial model. The information content of the 20 sites ranges from 11.8 to 26.0 bits, and five of the six E. coli OxyR binding sites used in the model fell within the top 20 sequences. The sixth E. coli site, upstream of gorA, had an information content of 11.2 bits. In a previous study, we showed that OxyR binds and regulates the promoter of the fur gene, one of the sites identified by our search (23). Here we examine OxyR binding to the sites upstream of the dsbG, fhuF, nmpC, and ybaL genes and to sites within the yfdI and ybbW open reading frames (ORFs).
TABLE 2.
Positions with the highest information content
| Screen | Site | Information content (bits) | Genomic position | Gene |
|---|---|---|---|---|
| Initial | 1 | 26.0 | 890054 | Upstream of grxA |
| 2 | 23.1 | 637851 | Upstream of dsbG | |
| 3 | 19.5 | 638089 | Upstream of ahpC | |
| 4 | 19.1 | 4131337 | Upstream of katG | |
| 5 | 16.0 | 2467337 | In yfdI | |
| 6 | 14.5 | 710103 | Upstream of fur | |
| 7 | 13.9 | 537879 | In ybbW | |
| 8 | 13.9 | 4156029 | Upstream of oxyR | |
| 9 | 13.8 | 2698016 | Downstream of yfhL | |
| 10 | 13.7 | 1103699 | In csgA | |
| 11 | 13.5 | 77803 | In yabM | |
| 12 | 13.3 | 576117 | Upstream of nmpC | |
| 13 | 13.2 | 1524021 | Upstream of ansP | |
| 14 | 13.1 | 3266461 | In yhaC | |
| 15 | 12.7 | 3406233 | In panF | |
| 16 | 12.4 | 1485303 | In ydcF | |
| 17 | 12.4 | 4071146 | In yihU | |
| 18 | 12.3 | 848228 | Upstream of dps | |
| 19 | 12.2 | 502543 | Upstream of ybaL | |
| 20 | 11.8 | 4603357 | Upstream of fhuF-2 | |
| 25 | 11.2 | 3643862 | gorA | |
| Improved | 1 | 29.3 | 637851 | Upstream of dsbG |
| 2 | 27.0 | 890054 | Upstream of grxA | |
| 3 | 26.0 | 710103 | Upstream of fur | |
| 4 | 23.0 | 2467337 | In yfdI | |
| 5 | 21.8 | 638089 | Upstream of ahpC | |
| 6 | 19.1 | 4603357 | Upstream of fhuF-2 | |
| 7 | 18.3 | 4603273 | Upstream of fhuF-1 | |
| 8 | 18.0 | 3666966 | Upstream of yhjA | |
| 9 | 17.6 | 4131337 | Upstream of katG | |
| 10 | 17.4 | 1762663 | Upstream of sufA | |
| 11 | 17.3 | 1475504 | Upstream of ynbA | |
| 12 | 17.3 | 2069355 | Upstream of flu | |
| 13 | 16.4 | 2167805 | In gatR | |
| 14 | 16.0 | 3266461 | In yhaC | |
| 15 | 15.8 | 3272480 | In yhaU | |
| 16 | 15.8 | 1211333 | Upstream of elbA | |
| 17 | 15.5 | 2698016 | Downstream of yfhL | |
| 18 | 15.5 | 1596260 | Upstream of ydeK | |
| 19 | 15.3 | 4156029 | Upstream of oxyR | |
| 20 | 15.1 | 1118367 | Upstream of yceA | |
| 38 | 13.1 | 848228 | Upstream of dps | |
| 77 | 11.4 | 2716640 | Upstream of trxC | |
| 114 | 10.3 | 497211 | Upstream of hemH | |
| 356 | 7.4 | 3643862 | Upstream of gorA |
OxyR binding upstream of dsbG.
The computer calculation predicted an OxyR binding site centered at position 637851, 54 bp upstream of the dsbG start codon and 238 bp away from the center of a known OxyR binding site proximal to the divergently transcribed ahpC gene (Fig. 1). The predicted site has an information content of 23.1 bits, which is higher than that of all known OxyR sites except the one in the grxA promoter region (26.0 bits). The dsbG site is homologous to the OxyR binding site detected upstream of the putative Salmonella enterica serovar Typhimurium dsbG gene (18) and previously designated Salmonella orf (19). To verify OxyR binding to this E. coli site, we carried out DNase I footprinting assays. We found that oxidized (Fig. 2A) but not reduced (data not shown) OxyR binds to the predicted site with an affinity that is comparable to that for the site in the ahpC promoter.
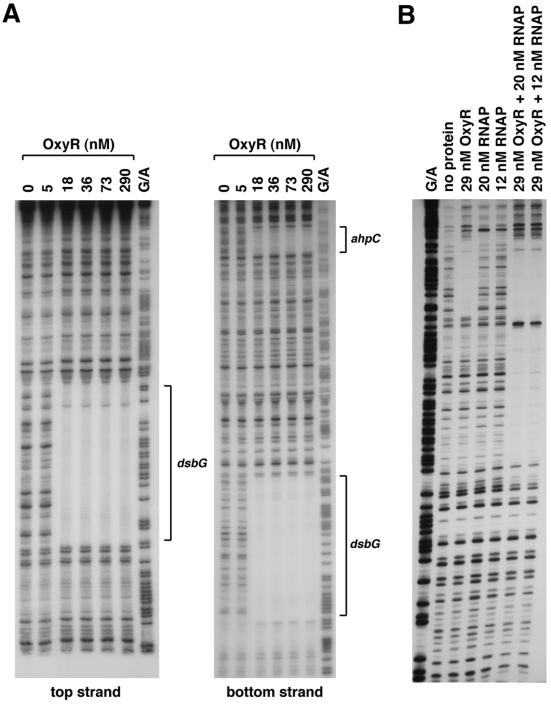
FIG. 2.
DNase I footprinting assays of oxidized OxyR binding to the top and bottom strands of the dsbG promoter in the absence of RNA polymerase (A) and to the top strand in the presence of RNA polymerase (B). The regions protected by OxyR are indicated by the brackets in panel A. All samples were run in parallel with Maxam-Gilbert G/A sequencing ladders. (A) For OxyR binding to the top strand relative to the dsbG promoter, the 230-bp EcoRI-BamHI fragment of pGSO123 was labeled with 32P at the EcoRI site. For OxyR binding to the bottom strand relative to the dsbG promoter, the 32P-labeled primer 710 and unlabeled primer 709 were used to PCR amplify a 430-bp fragment containing both the ahpC and dsbG promoter sequences. (B) For OxyR and RNA polymerase binding to the top strand relative to the dsbG promoter, a 250-bp fragment was PCR amplified from pGSO124 using primers 710 and 726. The amplified fragment was digested with EcoRI, labeled with 32P, and then digested with SmaI.
OxyR binding upstream of dsbG activates ahpCF expression.
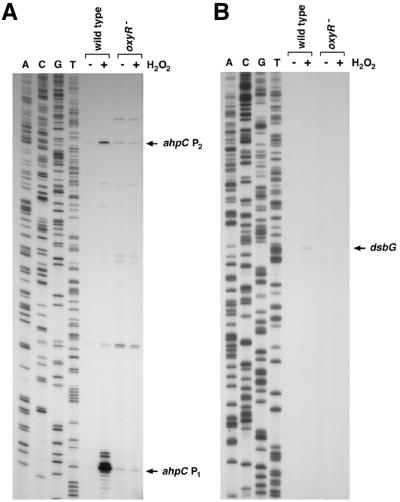
For all previously known OxyR-activated genes, OxyR binds at a site directly upstream of the −35 region of a target gene (19). Thus, the proximity of the OxyR binding site to the dsbG start codon led us to speculate that OxyR binding to this site is required for OxyR activation of dsbG expression in response to oxidative stress. However, multiple primer extension experiments failed to detect a dsbG transcription start in the vicinity of the OxyR binding site. To further elucidate the role of OxyR binding to the site upstream of dsbG, we examined RNA polymerase binding in the presence of OxyR. Previous studies have shown that oxidized OxyR binding leads to RNA polymerase recruitment to the promoter (6). As shown in Fig. 2B, RNA polymerase did not bind to the promoter fragment in the absence of other proteins. RNA polymerase binding was observed in the presence of OxyR. Surprisingly, the binding was to sequences upstream rather than downstream of the OxyR binding site relative to the dsbG start codon. This result suggested that OxyR binding to the site adjacent to dsbG was functioning to activate expression of a transcript encoding ahpC. Indeed, Northern blotting (data not shown) and primer extension assays (Fig. 3) of wild-type and ΔoxyR mutant cells grown in rich medium showed that hydrogen peroxide treatment led to the OxyR-dependent induction of two ahpC mRNAs, one transcript initiating at position 638144 and a second initiating at position 637916. Primer extension assays carried out with a variety of primers throughout the entire ahpC-dsbG inter-ORF region led to the detection of a hydrogen peroxide-induced transcript encoding dsbG initiating at position 638023. This start suggests that there is an approximately 100-bp overlap between the longer ahpC transcript and the dsbG mRNA. Whole-genome expression experiments also show the presence of overlapping ahpC and dsbG mRNAs (C. Rosenow, unpublished data), but the reason for the overlapping transcripts has not been elucidated.
FIG. 3.
Primer extension assays of ahpC and dsbG expression in wild-type and ΔoxyR mutant strains grown in LB medium. Exponential-phase cultures were split into two aliquots: one aliquot was left untreated, and the other was treated with 1 mM hydrogen peroxide. The cells were harvested after 10 min, total RNA was isolated, and primer extension assays were carried out with primer 709 specific to ahpC and primer 610 specific to dsbG. The neighboring sequencing reactions were carried out with the same primers.
OxyR binding to the fhuF promoter.
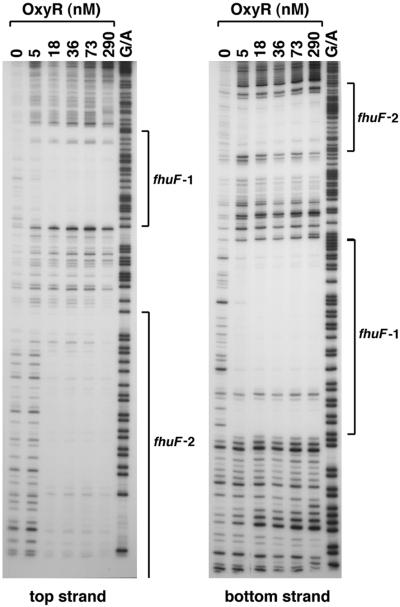
The computer search indicated an OxyR binding site centered at coordinate 4603357, 126 bases upstream of the start codon of the fhuF gene (Fig. 4). This site has an information content of 11.8 bits, which is lower than that of all the known OxyR binding sites except the one in the gorA promoter (11.2 bits). However, given our previous findings that OxyR regulates the expression of the iron metabolism regulator Fur, possible OxyR regulation of a putative iron reductase was of interest. Thus, we carried out DNase I footprinting experiments to test for OxyR binding to the promoter region of fhuF. As shown in Fig. 5, the site predicted by the computational search (fhuF-2) indeed is protected from DNase I digestion by the oxidized OxyR protein. Interestingly, we observed another oxidized OxyR binding site (fhuF-1) of slightly higher affinity centered at coordinate 4603273, 42 bases upstream of the fhuF start codon. Since the second site had an information content of 4.8 bits, OxyR binding to this position was an indication that the initial binding site model could be improved.
FIG. 5.
DNase I footprinting assays of oxidized OxyR binding to the top and bottom strands of the fhuF promoter. The regions protected by OxyR on both strands are indicated by the brackets. The 240-bp BamHI-EcoRI fragment of pGSO129 was labeled with 32P at either the BamHI site (top strand) or the EcoRI site (bottom strand). The samples were run in parallel with Maxam-Gilbert G/A sequencing ladders.
The fhuF gene has been shown previously to be repressed by the Fur transcription factor (16), and a Fur binding site in the fhuF promoter region has been suggested previously (8). As we have done for OxyR, we have built a Fur DNA binding model and scanned the entire E. coli genome (K. A. Lewis, B. Doan, M. Zheng, G. Storz, and T. D. Schneider, unpublished data). One interesting prediction of the Fur binding model was the presence of two clusters of Fur binding sites that overlapped the OxyR binding sites in the fhuF promoter. DNase I footprinting experiments with purified Fur protein (K. A. Lewis, B. Doan, M. Zheng, G. Storz, and T. D. Schneider, unpublished data) verified this prediction and showed that there are two regions of high- and low-affinity Fur binding, overlapping with the fhuF-2 and fhuF-1 OxyR binding sites, respectively (indicated in Fig. 4).
OxyR and Fur repression of fhuF.
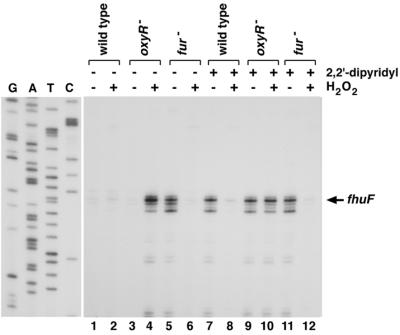
To determine whether fhuF is regulated by OxyR, the wild-type and the ΔoxyR and Δfur mutant strains were grown in the absence and presence of the iron chelator 2,2′-dipyridyl. We then examined fhuF expression without and with hydrogen peroxide treatment by primer extension assays (Fig. 6). These assays showed that, in the absence of oxidative stress, fhuF mRNA levels are strongly repressed by Fur under iron-rich (compare lanes 1 and 3 with lane 5) but not iron-poor (compare lanes 7, 9, and 11) conditions. This repression is in agreement with the strong Fur regulation reported by Stojiljkovic et al. (16). Treatment with hydrogen peroxide led to fhuF repression in the wild-type and the Δfur mutant strains, but not the ΔoxyR mutant strain, under both iron-rich and iron-depleted conditions (compare lanes 2, 6, 8, and 12 with lanes 4 and 10). These results show that OxyR represses fhuF expression under conditions of oxidative stress. The derepression of fhuF observed when the ΔoxyR strain grown under iron-rich conditions was treated with hydrogen peroxide may be due to reduced Fur binding upon oxidative damage to iron-loaded Fur.
FIG. 6.
Primer extension assays of fhuF expression in wild-type, ΔoxyR, and Δfur strains grown in LB medium without (lanes 1 to 6) and with (lanes 7 to 12) 1 mM 2,2′-dipyridyl. Exponential-phase cultures were split into two aliquots: one aliquot was left untreated, and the other was treated with 1 mM hydrogen peroxide. The cells were harvested after 10 min, total RNA was isolated, and primer extension assays were carried out with primer 706 specific to fhuF. The neighboring sequencing reactions were carried out with the same primer.
The start of the fhuF message was mapped to two adjacent A residues located 30 bases upstream of the ATG start codon, situated in the middle of the high-affinity OxyR binding site (Fig. 6). Based on sequence analysis, Müller et al. (8) predicted that the Fur-dependent transcription start at position 4603305 was located close to the high-affinity Fur site 123 bp upstream of the ATG codon. In our primer extension assay, we see very little expression from this predicted upstream start. It is possible that this predicted promoter is more active under other growth conditions. Regardless, given the two overlapping OxyR and Fur binding sites, it is clear that fhuF expression is tightly regulated in response to both oxidative stress and intracellular iron levels.
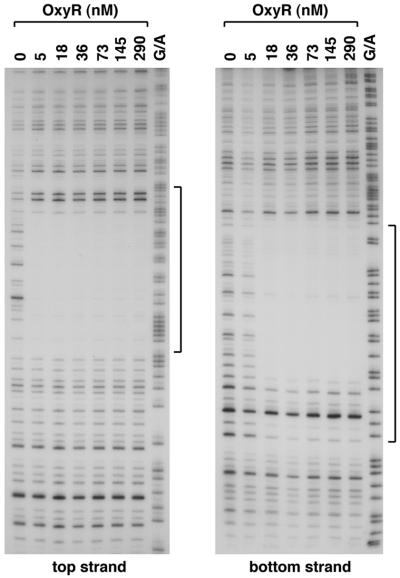
OxyR binding to the yfdI gene.
In addition to predicting OxyR binding sites upstream of the fur (23), dsbG, and fhuF genes, our computer model predicted OxyR sites centered 9 bp upstream of the nmpC and 81 bp upstream of the ybaL start codons as well as sites within ORFs. To test whether OxyR bound to these sites, we carried out DNase I footprinting experiments. We did not observe strong binding to the nmpC and ybaL promoter regions or to a fragment carrying the predicted site within the ybbW ORF (data not shown). However, we did observe strong binding by oxidized OxyR (Fig. 7 and 8) and weak binding by reduced OxyR (data not shown) to the predicted site within the yfdI ORF. The role of OxyR binding to this site is not clear. We carried out primer extension assays using five different primers but failed to detect the start of an OxyR-regulated transcript on either strand within the vicinity of the binding site. Microarray experiments showed that the expression of a transcript encoded within yfdI is repressed upon treatment with hydrogen peroxide, but this repression is observed in both a wild-type strain and an ΔoxyR mutant strain (24).
FIG. 8.
DNase I footprinting assays of oxidized OxyR binding to the top and bottom strands of the yfdI gene. The regions protected by OxyR on both strands are indicated by the brackets. The 180-bp BamHI-EcoRI fragment of pGSO130 was labeled with 32P at either the BamHI site (top strand) or the EcoRI site (bottom strand). The samples were run in parallel with Maxam-Gilbert G/A sequencing ladders.
Improved search for new OxyR binding sites.
Seven previously identified sites (oxyR, katG, ahpC, dps, gorA, mom, and grxA), five OxyR binding sites identified in the initial computational search (fur, dsbG, fhuF-1, fhuF-2, and yfdI), two sites identified by our DNA microarray studies (sufA and hemH) (24), and two sites identified in other studies (flu and trxC) (9; M. Zheng and G. Storz, unpublished data) were used to generate a new model to search the complete E. coli genome. A direct comparison of the initial and the improved models is given online (http://www.lecb.ncifcrf.gov/∼toms/paper/zheng.storz2001/), and the 20 sites with the highest information content (ranging from 15.1 to 29.3 bits) based on the second model are listed in Table 2. Although the overall compositions of the two models were similar, a few differences were noted: additional bases were accommodated at some positions (such as a G at position 5 relative to the center of the binding site), and there was some focusing on specific residues at other positions (such as on A at position 4). Interestingly, using the second model, four of the known OxyR binding sites, at the dps, trxC, hemH, and gor promoters, were no longer among the 20 sites with the highest information content. The gorA gene does not show strong OxyR regulation (7), but dps, trxC, and hemH are clearly induced by OxyR in response to oxidative stress (9, 17, 24). It also is noteworthy that several of the top 20 sites identified by the improved search are within ORFs.
DISCUSSION
Use of a computational approach based on information theory to identify OxyR binding sites.
As the complete sequences of more and more genomes become available, the extraction of biological information by computational analysis of sequence data is quickly becoming an indispensable tool in biological research. While searches for gene homology are routine, searches for DNA binding sites are less common, in part because the sequences recognized by a DNA binding protein are short and often vary considerably from a simple consensus sequence. A quantitative model based on information theory was constructed for describing the DNA recognition sequences for the OxyR transcription factor. This model was used to search for additional OxyR target sequences that were then experimentally tested for OxyR binding in vitro and OxyR-dependent expression in vivo.
Our computational search for OxyR binding sites allowed us to identify several new OxyR target genes. However, our findings also underscore the fact that computational searches are limited by the models used for the searches. Our initial model was based on nine OxyR binding sites. We tested for OxyR binding to six of the sites predicted to have high information content by the initial model; however, only three of these sites were found to be bound by OxyR in DNase I footprinting assays. In addition, our initial search indicated a low information content (4.7 bits) for the fhuF-1 site found to be bound with high affinity. Three bases present in the fhuF-1 binding sequence were not present in the nine sites used to construct the initial model. Thus, a penalty was given to this site. Larger data sets should allow significant improvements in the model, and in our improved search based on 16 binding sites, the fhuF-1 site was found to have 18.3 bits of information. It will be interesting to determine whether a higher percentage of the sites predicted by the improved model experimentally will be found to be OxyR binding sites.
With the improved model, two documented OxyR binding sites are not among the 100 sites with the highest information content. Possibly, OxyR actually binds more than 100 sites in the genome. The OxyR concentration in the cell has not been determined, but the first possibility would require OxyR to be in fair abundance. Alternatively, further improvements in our OxyR binding site model are needed. It is conceivable that there are subclasses of OxyR binding sites such that OxyR binding would be better represented by two or more models. For example, a model based solely on sites from which OxyR activates transcription might be stronger in predicting other sites where OxyR binds as an activator. Differences in the mode of OxyR regulation in the context of other transcription factors also may necessitate differences in the OxyR binding site at some genes. Additional microarray experiments to identify genomic DNA fragments bound to OxyR and to further examine the transcriptional profiles in different oxyR mutant strains should allow us to determine the total number of OxyR-regulated genes and to further refine our OxyR binding site model.
Identification of OxyR-regulated genes.
Our current search has led to the identification of a new OxyR-activated gene, dsbG, and a new OxyR-repressed gene, fhuF. The periplasmic DsbG protein has been shown to be a chaperone and disulfide bond isomerase involved in the correct folding of disulfide-containing proteins (2, 14). Periplasmic disulfide bond formation is a highly controlled process involving a series of Dsb proteins (5). The rate of disulfide bond formation is likely to increase upon exposure to hydrogen peroxide. Thus, the induction of disulfide bond isomerases may be critical in a defense against peroxide stress. In this context, it would be interesting to test whether expression of other E. coli Dsb proteins is modulated by oxidative stress. The FhuF protein has been suggested previously to be a ferric ion reductase that reduces siderophore-bound ferric ion upon import into the cytoplasm (8). Repression of fhuF transcription by OxyR may lead to decreased levels of FhuF protein which in turn would slow iron uptake, thus providing another mechanism for the cell to minimize the escalation of oxidative damage by the Fenton reaction.
Noncanonical modes of regulation by OxyR.
For most of the OxyR-activated genes, OxyR binding sites are located upstream of the −35 region (19). It is likely that OxyR binding to these sequences allows for RNA polymerase recruitment to the promoters. Our discovery of a binding site proximal to the dsbG gene led us to suggest that OxyR binding to this site would lead to OxyR regulation of dsbG. Unexpectedly, we found that OxyR binding to the dsbG-proximal site regulates the expression of the divergent ahpC transcript, while OxyR binding to the ahpC-proximal site regulates expression of the dsbG gene. These findings illustrate how predictions about promoters and regulation solely based on sequence analysis may be misleading and reinforce the need to experimentally test computer-based predictions. The respective starts of the dsbG and ahpC transcripts indicate that the two transcripts overlap by over 100 nucleotides. The reason for the overlap is not known, but pairing between transcripts may allow for an additional level of regulation. Whole-genome expression experiments indicate that more than 150 E. coli RNAs overlap with transcripts on the opposing strand (C. Rosenow, unpublished data). Thus, the overlap between the 5′ ends of the dsbG and ahpC mRNAs may represent a more widespread phenomenon.
Only three OxyR-repressed genes were known previously. OxyR represses its own expression, as well as transcription of the Mu phage mom and the E. coli flu genes. Our current data show that OxyR also represses the expression of fhuF. In contrast to the other repressed genes, however, OxyR binds to two separate sites in the fhuF promoter region. Since the higher-affinity fhuF-1 binding site covers the +1, −10, and −35 sequences of the fhuF promoter, we suggest that OxyR represses fhuF expression by blocking RNA polymerase binding to the promoter. The reason for OxyR binding to the lower-affinity fhuF-2 site is less obvious. It is likely that OxyR binding to fhuF-2 also contributes to repression. In addition, the presence of two OxyR sites may be required to allow for coordinate regulation by both the OxyR and Fur transcription factors.
Unexpectedly, our studies showed that OxyR binds to sequences within the coding region of yfdI. This finding raises questions as to the role of OxyR binding to coding sequences and whether other transcription factors bind to coding sequences. Whole-genome expression experiments suggest that there is a surprisingly high amount of transcription from the noncoding strand of E. coli (13). Thus, it is conceivable that OxyR is regulating the expression of a transcript encoded by the strand opposite yfdI. Alternatively, OxyR may be acting at a distance to regulate neighboring genes or has a structural role in maintaining the proper chromosome architecture. Together, our studies emphasize that OxyR has functions beyond recruiting RNA polymerase to promoters juxtaposed to OxyR binding sites. However, mutational studies to selectively eliminate OxyR binding to individual sites in the yfdI gene and the ahpC, dsbG, and fhuF promoter regions are necessary to fully delineate the roles of OxyR at each of the new binding sites.
ACKNOWLEDGMENTS
We appreciate the editorial comments of J. Imlay and R. LaRossa.
This work was supported by the intramural programs of the National Institute of Child Health and Human Development and the National Cancer Institute and a fellowship from the American Cancer Society (M.Z.).
REFERENCES
- 1.Andersen C L, Matthey-Dupraz A, Missiakas D, Raina S. A new Escherichia coli gene, dsbG, encodes a periplasmic protein involved in disulphide bond formation, required for recycling DsbA/DsbB and DsbC redox proteins. Mol Microbiol. 1997;26:121–132. doi: 10.1046/j.1365-2958.1997.5581925.x. [DOI] [PubMed] [Google Scholar]
- 2.Bessette P H, Cotto J J, Gilbert H F, Georgiou G. In vivo and in vitro function of the Escherichia coli periplasmic cysteine oxidoreductase DsbG. J Biol Chem. 1999;274:7784–7792. doi: 10.1074/jbc.274.12.7784. [DOI] [PubMed] [Google Scholar]
- 3.Blattner F R, Plunkett G, Bloch C A, Perna N T, Burland V, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 4.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 5.Debarbieux L, Beckwith J. Electron avenue: pathways of disulfide bond formation and isomerization. Cell. 1999;99:117–119. doi: 10.1016/s0092-8674(00)81642-6. [DOI] [PubMed] [Google Scholar]
- 6.Kullik I, Toledano M B, Tartaglia L A, Storz G. Mutational analysis of the redox-sensitive transcriptional regulator OxyR: regions important for oxidation and transcriptional activation. J Bacteriol. 1995;177:1275–1284. doi: 10.1128/jb.177.5.1275-1284.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michán C, Manchado M, Dorado G, Pueyo C. In vivo transcription of the Escherichia coli oxyR regulon as a function of growth phase and in response to oxidative stress. J Bacteriol. 1999;181:2759–2764. doi: 10.1128/jb.181.9.2759-2764.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Müller K, Matzanke B F, Schünemann V, Trautwein A X, Hantke K. FhuF, an iron-regulated protein of Escherichia coli with a new type of [2Fe-2S] center. Eur J Biochem. 1998;258:1001–1008. doi: 10.1046/j.1432-1327.1998.2581001.x. [DOI] [PubMed] [Google Scholar]
- 9.Ritz D, Patel H, Doan B, Zheng M, Åslund F, Storz G, Beckwith J. Thioredoxin 2 is involved in the oxidative stress response in Escherichia coli. J Biol Chem. 2000;275:2505–2512. doi: 10.1074/jbc.275.4.2505. [DOI] [PubMed] [Google Scholar]
- 10.Schneider T D. Reading of DNA sequence logos: prediction of major groove binding by information theory. Methods Enzymol. 1996;274:445–455. doi: 10.1016/s0076-6879(96)74036-3. [DOI] [PubMed] [Google Scholar]
- 11.Schneider T D. Information content of individual genetic sequences. J Theor Biol. 1997;189:427–441. doi: 10.1006/jtbi.1997.0540. [DOI] [PubMed] [Google Scholar]
- 12.Schneider T D. Sequence walkers: a graphical method to display how binding proteins interact with DNA or RNA sequences. Nucleic Acids Res. 1997;25:4408–4415. doi: 10.1093/nar/25.21.4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Selinger D W, Cheung K J, Mei R, Johansson E M, Richmond C S, Blattner F R, Lockhart D J, Church G M. RNA expression analysis using a 30 base pair resolution Escherichia coli genome array. Nat Biotechnol. 2000;18:1262–1268. doi: 10.1038/82367. [DOI] [PubMed] [Google Scholar]
- 14.Shao F, Bader M W, Jakob U, Bardwell J C A. DsbG, a protein disulfide isomerase with chaperone activity. J Biol Chem. 2000;275:13349–13352. doi: 10.1074/jbc.275.18.13349. [DOI] [PubMed] [Google Scholar]
- 15.Silhavy T J, Berman M L, Enquist L W. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1984. [Google Scholar]
- 16.Stojiljkovic I, Bäumler A J, Hantke K. Fur regulon in Gram-negative bacteria: identification and characterization of new iron-regulated Escherichia coli genes by a fur titration assay. J Mol Biol. 1994;236:531–545. doi: 10.1006/jmbi.1994.1163. [DOI] [PubMed] [Google Scholar]
- 17.Storz G, Zheng M. Oxidative stress. In: Storz G, Hengge-Aronis R, editors. Bacterial stress responses. Washington, D.C.: ASM Press; 2000. pp. 47–59. [Google Scholar]
- 18.Tartaglia L A, Storz G, Ames B N. Identification and molecular analysis of oxyR-regulated promoters important for the bacterial adaptation to oxidative stress. J Mol Biol. 1989;210:709–719. doi: 10.1016/0022-2836(89)90104-6. [DOI] [PubMed] [Google Scholar]
- 19.Toledano M B, Kullik I, Trinh F, Baird P T, Schneider T D, Storz G. Redox-dependent shift of OxyR-DNA contacts along an extended DNA-binding site: a mechanism for differential promoter selection. Cell. 1994;78:897–909. doi: 10.1016/s0092-8674(94)90702-1. [DOI] [PubMed] [Google Scholar]
- 20.van Straaten M, Missiakas D, Raina S, Darby N J. The functional properties of DsbG, a thiol-disulfide oxidoreductase from the periplasm of Escherichia coli. FEBS Lett. 1998;428:255–258. doi: 10.1016/s0014-5793(98)00539-0. [DOI] [PubMed] [Google Scholar]
- 21.Zhang A, Altuvia S, Tiwari A, Argaman L, Hengge-Aronis R, Storz G. The OxyS regulatory RNA represses rpoS translation and binds the Hfq (HF-I) protein. EMBO J. 1998;17:6061–6068. doi: 10.1093/emboj/17.20.6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng M, Åslund F, Storz G. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science. 1998;279:1718–1721. doi: 10.1126/science.279.5357.1718. [DOI] [PubMed] [Google Scholar]
- 23.Zheng M, Doan B, Schneider T D, Storz G. OxyR and SoxRS regulation of fur. J Bacteriol. 1999;181:4639–4643. doi: 10.1128/jb.181.15.4639-4643.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng M, Wang X, Templeton L J, Smulski D R, LaRossa R A, Storz G. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J Bacteriol. 2001;183:4562–4570. doi: 10.1128/JB.183.15.4562-4570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]