Abstract
Lethal heat stress generates oxidative stress in Saccharomyces cerevisiae, and anaerobic cells are several orders of magnitude more resistant than aerobic cells to a 50°C heat shock. Here we characterize the oxidative effects of this heat stress. The thermoprotective effect in anaerobic cells was not due to expression of HSP104 or any other heat shock gene, raising the possibility that the toxicity of lethal heat shock is due mainly to oxidative stress. Aerobic but not anaerobic heat stress caused elevated frequencies of forward mutations and interchromosomal DNA recombination. Oxidative DNA repair glycosylase-deficient strains under aerobic conditions showed a powerful induction of forward mutation frequencies compared to wild-type cells, which was completely abolished under anaerobiosis. We also investigated potential causes for this oxygen-dependent heat shock-induced genetic instability. Levels of sulfhydryl groups, dominated mainly by the high levels of the antioxidant glutathione (reduced form) and levels of vitamin E, decreased after aerobic heat stress but not after anaerobic heat stress. Aerobic heat stress also led to an increase in mitochondrial membrane disruption of several hundredfold, which was 100-fold reduced under anaerobic conditions.
All organisms have an optimal temperature range for growth. When the temperature rises above this optimal temperature, cellular growth ceases and toxicity ensues. For baker's yeast, Saccharomyces cerevisiae, this optimum is within the range of 25 to 35°C. Above 45°C, yeast cells are severely stressed and progressively die so that after 5 min at 50°C, more than 99% of growing nonadapted aerobic yeast cells have died. In the range of 35 to 37°C yeast cells are moderately stressed but continue to grow, developing a protective tolerance to higher lethal heat exposures. In these sublethal heat shock conditions, the cell responds by synthesizing a discrete subset of stress proteins via heat shock-dependent transcription pathways (heat shock, stress response, and yAP-1 elements) and concomitantly attains the increased capacity to resist higher lethal temperatures (38, 39, 46). Among the heat shock proteins in baker's yeast, only HSP104 has been conclusively associated with adaptive thermotolerance (37, 44, 45, 48). HSP70 is involved in adaptive thermotolerance only in the absence of HSP104 (49).
Furthermore, general inhibition of gene expression during heating does not block the acquisition of thermotolerance (5). In addition, the acquisition of thermotolerance is independent of mitotic cell cycle arrest and of the majority of the full spectrum of heat shock proteins (6). For instance o-phenanthroline, a cell cycle inhibitor, causes thermotolerance without induction of the characteristic pattern of heat shock proteins (6). Heat shock proteins are, however, thought to play a role during recovery from stressful conditions (33, 35). For example, Hsp104p is a multifunctional protein having known roles in solubilization of aggregated proteins following heat stress and maintenance of mRNA splicing during heat stress conditions (43, 44, 63).
The causative molecular events for heat killing are multifarious and poorly defined. Many investigations have focused on the endogenous defenses that cells summon in response to heat stress. Cellular thermotolerance is induced by many diverse factors, including prior exposure to a variety of sublethal stresses. The intracellular molecules that are associated with acquired thermotolerance include intracellular pH (H+), cyclic AMP, and intracellular concentrations of adenosine 5′-phosphosulfate 3′-phosphate and adenosine 5′-phosphosulfate (14–16, 18, 30). Adaptive thermotolerance in response to sublethal heat shock creates tolerance to unrelated agents (e.g., hydrogen peroxide and ethanol), suggesting that the molecular targets for each diverse stressor are general rather than specific. Proteins denature and become insoluble in response to a variety of stressful conditions, and denatured proteins and abortive protein synthesis can elicit thermotolerance (13, 22, 27). Many heat shock proteins are molecular chaperonins, refolding unraveled or aggregated proteins. It is not clear, however, whether protein denaturation is the primary cause of heat lethality or a consequence of some other initiating event. Lipid membranes are also similarly disrupted by increasing temperature (33, 34), and loss of bilayer integrity may contribute to heat toxicity, perhaps through an increase in proton permeability as proposed by Coote et al. (14).
We have previously shown that oxidative stress and antioxidant enzymes play a major role in heat induced cell death in yeast. Mutants deleted for the antioxidant genes catalase, superoxide dismutase, thioredoxin peroxidase, and cytochrome c peroxidase are more sensitive to the lethal effect of heat than isogenic wild-type cells (17, 36). Overexpression of catalase and superoxide dismutase genes cause an increase in thermotolerance. Anaerobic conditions cause a 500- to 20,000-fold increase in thermotolerance. By using an oxidation-dependent fluorescent molecular probe, a two- to threefold increase in fluorescence is found upon heating. These findings indicate that the toxic effect of lethal heat is mediated at least in part by reactive oxygen species (17). In support of this, cells respond to heat shock with the induction of the cytosolic catalase T gene in yeast (8, 39, 65) and superoxide dismutase in bacteria (7, 9).
Oxidative stress causes DNA damage. Spontaneous oxidative DNA lesions are formed by direct reaction of ⋅OH with base or deoxyribose components or as secondary breakdown products (19). Two N-glycosylases with broad substrate specificity for oxidized pyrimidines (Ntg1p and Ntg2p) and high sequence similarity to each other (41% identity, 63% similarity) have been found in S. cerevisiae (24). Ntg1p and Ntg2p have similar overlapping substrate specificities, which atypically includes formamidopyrimidine (FaPy) guanine and FaPy adenine lesions. Abasic sites are recognized by Ntg1p and Ntg2p, both of which have an intrinsic apurinic and apyrimidinic lyase activity (2, 21, 53). Deletion of either enzyme has been reported to result in a mild mutator phenotype, and in some strains a sensitivity to the oxidants H2O2 and menadione has been found in NTG1 deletion mutants (21) but not in others (57).
Purine oxidative base lesions include 8-oxoguanine, which can base pair with adenine during DNA synthesis, resulting in G:C→T:A transversion mutations. FaPy guanine and adenine lesions are formed by rupture of the imidazole ring following oxidative attack (reviewed in reference 19). In S. cerevisiae, the gene OGG1 removes 8-oxoguanine from DNA (25, 40).
Given the potential importance of oxidative stress during heat exposure, we have investigated some key oxidative stress determinants, including determination of heat toxicity and frequency of DNA mutations in oxidative glycosylase mutant cells, mitochondrial membrane lysis, and endogenous antioxidant concentrations following a 50°C exposure. By making use of the ability of yeast to grow either aerobically or anaerobically, we have determined the involvement of oxygen in these biological effects of lethal heat.
MATERIALS AND METHODS
Media and strains.
Media were prepared as described by Sherman et al. (54). Rich medium consisted of 1% yeast extract–2% Bacto Peptone supplemented with 0.8 mg of adenine liter and containing either 2% dextrose (YPAD) or 3% glycerol–1% ethanol (YPEG). Anaerobic media were prepared in Baxter 50-ml inoculation bottles with rubber bungs and crimped metal caps which were flushed with nitrogen. Inoculation of the bottles was achieved by insertion of a 19.5-gauge needle through the rubber bung. All anaerobic media were supplemented with 6 mg of ergosterol and 660 mg of Tween 80 per liter and contained 0.001% resazurin as an oxygen indicator.
All strains used in this study are described in Table 1. Strain JDY1 was made by transforming strain MCC123, obtained from Thorsness and Fox (59), with a TRP1 fragment disrupted with Salmonella hisG repeats flanking the URA3 gene. Selection for Ura+ colonies ensured integration of the hisG-URA3-hisG cassette (1). High spontaneous recombination of the hisG repeats results in loss of the URA3 gene, leaving the TRP1 gene disrupted by one hisG allele, an event which was selected for by growing the cells on minimal media containing 5-fluororotic acid (5-FOA). Colonies that were unable to grow on medium lacking tryptophan were isolated.
TABLE 1.
Strains used in this study
| Strain | Genotype | Reference or origin |
|---|---|---|
| Y433 | MATα ura3-52 leu2Δ98 ade2-101 his3Δ200 lys2801 | 51 |
| JDY47 | MATα ura3-52 leu2Δ98 ade2-101 his3Δ200 lys2-801 ogg1::hisG ntg1::hisG ntg2::URA3 | This study |
| W303-1a LEU2 | MATacan1-100 ade2-1 his3-11,15 trp1-1 ura3-1 | 48 |
| W303-1a Δhsp104 | MATacan1-100 ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 Δhsp104::LEU2 | 48 |
| S41 | MATa/α arg4-3/arg4-17 trp5-12/trp5-27 his1-1/his1-315 ade2-40/ade2-119 ilv1-92/ilv1-92 leu2-3,112/leu2-3,112 ura3-52/ura3-52 | 52 |
| RS112 | MATa/α ura3-52/ura3-52 leu2-3,112/leu2Δ98 trp5-27/TRP5 arg4-3/ARG4 ade2-40/ade2-101 ilv1-92/ILV1 HIS3::pRS6/his3Δ200LYS2/lys2-801 | 50 |
| AH22 | MATaleu2 his4 | 41 |
| YHT121 | MATaleu2 his4 gsh1::LEU2 | 41 |
| JDY1 | MATaura3-52 ade2-10 kar1-1 trp1::hisg [ρ+TRP1] | This study |
The glycosylase triple mutant (ntg1 ntg2 ogg1) was generated in strain Y433. Y433 was transformed with the disruption construct from pJD47 to create Y433 ogg1::hisG-URA3-hisG. pJD47 was created by PCR amplification of the OGG1 open reading frame (ORF), using primers containing BamHI and EcoRI tails: OGG1forward (5′-GGGGGATCCATGTCTTATAAATTCGGCAA-3′) and OGG1reverse (5′-GGGGAATTCCTAATCTATTTTTGCTTCTT-3′).
Restriction enzyme digestion with EcoRI and BamHI allowed cloning into the multiple cloning site of pUC19, creating pJD4. The blunt-end MscI restriction site located 71 bp into the OGG1 ORF was cut and ligated to the BglII linker, creating pJD5. The 3.85-kb hisG-URA3-hisG cassette was released from pNKY51 by digestion with BamHI/BgIII (1) and ligated to the pJD5 BglII site to create pJD47. Digestion with BamHI/EcoRI releases the 4.9-kb transforming fragment. Selection on 5-FOA plates enabled reuse of the URA3 marker and resulted in strain JDY23 (Y433 ogg1::hisG), which was verified by PCR.
Strain JDY23 was transformed with the disruption construct derived from pJD46. PJD46 was created by PCR amplification of a 786-bp PCR product containing a section of the NTG1 ORF, using a forward primer which contained an EcoRI restriction site and a reverse primer which was located 5′ to an endogenous HindIII restriction site: NTG1forward (5′-TGAGAATTCGACCGCTGGTAAAGACTGAA-3′) and NTG1reverse (5′-GCGCATCTACCCATTTCCAA-3′).
Digestion of the 786-bp PCR product with HindIII and EcoRI facilitated cloning into the pUC19 vector to create pJD40. Digestion of pJD40 with BglII allowed insertion of the 3.85-kb hisG-URA3-hisG cassette from pNKY51 by digestion with BamHI/BglII (1) into a site 383 bp into the NTG1 ORF to create pJD46. The disruptor construct was released by digestion of pJD46 with SalI/EcoRI. Selection on 5-FOA plates enabled reuse of the URA3 marker and resulted in strain JDY28 (Y433 ogg1::hisG ntg1::hisG), which was verified by PCR.
Strain JDY47 was generated from strain JDY28 by transformation with pJD61 containing the NTG2 disruption construct. pJD61 was created by PCR amplification of a 721-bp product containing a section of the NTG2 ORF, using a forward primer which contained an HindIII restriction sites and a reverse primer which was located 5′ to an endogenous EcoRI restriction site: NTG2forward (5′-AAAAAGCTTCTAGGAAGAGAAAGCATA-3′) and NTG2reverse (5′-GGTCCAACTCCAGGTAACGA-3′).
Digestion of the 721-bp PCR product with HindIII/EcoRI facilitated cloning into the pUC19 vector to create pJD48. Digestion of pJD48 with BsmFI released a 244-bp deletion from the ORF. Fill-in reaction and BglII linker ligation created pJD49. Digestion of pJD49 with BglII allowed insertion of a 1.1-kb BamHI fragmant containing the complete URA3 into the NTG2 ORF to create pJD61. Digestion of pJD61 with HindIII/EcoRI allowed release of the NTG2 deletion/disruption construct. Transformation with the NTG2 knockout construct into strain JDY28 created JDY47 (Y433 ogg1::hisG ntg1::hisG ntg2::URA3).
Aerobic lethal heat stress survival assay.
Strains were grown to log phase (5 × 106 cells/ml) in YPAD, washed in distilled water, and concentrated to 2 × 108 cells/ml. Aliquots of 110 μl were prepared in 0.6-ml PCR tubes for each time point and placed on ice. Lethal heating was performed at 50°C. Tubes were removed at set time points and immediately placed on ice. The aliquots were then diluted as needed and plated onto solid YPAD. Colonies were counted after 3 days of incubation at 30°C, and the percent viability was calculated with respect to unheated controls.
Anaerobic lethal heat stress survival assay.
Cultures were grown anaerobically to log phase (5 × 106 cells/ml) in YPAD and anaerobic YPAD. Sealed anaerobic cultures were opened inside an anaerobic glove box containing a nitrogen atmosphere with <1 ppm of oxygen. Cultures were sealed in 50-ml centrifuge tubes, removed from the glove box through the antechamber, and centrifuged at 3,000 rpm. Following centrifugation they were returned to the glove box, where they were washed in deoxygenated distilled water and concentrated to 2 × 108 cells/ml. Aliquots of 110 μl were dispensed into 0.6-ml tubes and sealed before the heat stress viability assay.
Reoxygenation experiments were performed with anaerobic cultures prepared as described above except that prior to lethal heat stress, the 0.6-ml tubes were opened to the atmosphere and vigorously mixed in a vortex mixer. Oxygen leakage was assessed by the redox-sensitive compound resazurin (Sigma Chemical Co.), which is colorless in reducing conditions and turns red after exposure to oxygen (61).
Deoxygenation experiments were performed using aerobic cultures that were resuspended inside the glove box in preequilibrated solutions. Heat stress and plating were performed inside the anaerobic atmosphere of the glove box, and plates were sealed in a large plastic bin to avoid dessication during colony growth.
Metabolic 35S labeling of protein synthesis.
W303-1a LEU2 and Δhsp104 LEU2 cells were grown aerobically and anaerobically in 50 ml of YPAD to 5 × 106 cells/ml. Each culture was halved, concentrated to 2 × 107 cells/ml, and resuspended in fresh YPAD. One half of each culture was shifted to 39°C for 15 min before the addition of 100 μCi of [35S]methionine (Tran35S-label; ICN Biochemicals); the other half was labeled at 23°C. Cells were labeled for 15 min before snap-freezing in liquid nitrogen. Anaerobic cultures were heat shocked and handled in an anaerobic glove box (<1 ppm of O2). Frozen cultures were thawed individually, washed in 50 mM HEPES buffer (pH 7.5) containing 1 mM phenylmethylsulfonyl fluoride and 1 mM EDTA, and resuspended to a final volume of 120 μl in a 1.5-ml microcentrifuge tube. An equal volume of acid-washed glass beads (425/600 mesh) was added, and samples were vigorously agitated in a vortex mixer for six periods of 30 s each, with incubation on ice between each period. After lysis of the cells, the tubes were centrifuged for 10 min at 4°C. The supernatant was transferred to a fresh 1.5-ml microcentrifuge tube and centrifuged for 10 minutes at 4°C. Crude extract was withdrawn using a Pasteur pipette, and a 10-μl aliquot was taken for protein determination. Proteins (40 μg/lane) were separated by polyacrylamide gel electrophoresis using a 5% stacking gel and an 8% running gel as described by Sambrook et al. (47).
Sulfhydryl analysis.
Total sulfhydryl content was determined by the method of Boyne and Ellman (10), using 5,5′-dithio-bis(2-nitrobenzoic acid) (DTNB). Four 50-ml anaerobic cultures were prepared by needle inoculation of strain RS112 (104 cells/ml) and were grown at 30°C to 107 cells/ml in anaerobic media. The culture bottles were transferred to an anaerobic glove box containing a nitrogen atmosphere with <1 ppm of oxygen. The cells were sealed in 50-ml tubes, removed from the glove box, and centrifuged at 3,000 × g for 10 min. Any cultures showing a color change to red, indicating oxygen leakage, were discarded from the experiment. Cells were returned to the glove box, which was then opened, and pooled before resuspension to a final concentration of 2 × 109 cells/ml. The pooled cell suspension was halved, and one half was removed from the glove box and exposed to atmospheric oxygen with vigorous shaking. The oxygenated cells were then transferred in 200-μl aliquots to 0.6-ml PCR tubes and heat stressed in a PCR thermal cycler for 0, 1, 2, and 3 min at 50°C. Samples were immediately placed on ice following heat treatment. Anaerobic samples were likewise dispensed into 0.6-ml PCR tubes within the glove box, the tubes were closed, and the cells were heat stressed as described above.
Following heat treatment, the cells were washed three times in cold 0.1 M sodium phosphate buffered to pH 7.0 and resuspended in 500 μl of the same buffer for each sample time point in a 15-ml plastic Falcon tube. Crude extract was prepared by glass bead lysis as described above. Each extract was diluted to 0.5 mg of total protein per ml and 200 μl of each sample was added to 800 μl of 0.1 M sodium phosphate buffered to pH 8.0 and 7 μl of Ellman reagent (0.01 M DTNB in 10 ml of sodium phosphate buffer [pH 7.0]). The samples were incubated on ice for 30 min before measurement of absorbance at 412 nm in a spectrophotometer. The extinction coefficient for DTNB, 13,600 M-1 cm-1, was used to determine the concentration of total sulfhydryl.
Measurement of gene conversion frequencies.
Prior to measurement of gene conversion frequencies, a fluctuation assay was performed on 10 cultures of diploid strain S41 (containing the alleles trp5-12 and trp5-27, suitable for scoring interchromosomal gene conversion events) (52, 68). The culture with the lowest spontaneous reversion frequency was used to seed cultures in further experiments.
Strain S41 (5 × 104 cells) was grown in liquid anaerobic YPAD to log phase (5 × 106 cells/ml). One half of the culture was used for anaerobic heat exposure in a PCR thermal cycler as described above for 0, 1, 2, and 3 min at 50°C; the other half was reoxygenated and subjected to aerobic heat exposure under the same conditions. Samples were plated on solid synthetic complete medium (SC) and on SC lacking tryptophan. Gene conversion frequencies were then measured by counting the number of Trp+ prototrophic colonies per viable cell.
Measurement of mitochondrial DNA release.
Prior to measurement of mitochondrial DNA release (58), a fluctuation assay was performed on 10 cultures of strain JDY1, and the culture with the lowest spontaneous Trp+ reversion frequency was selected for expansion in further experiments. Cultures were grown in liquid YPEG to 2 × 107 cells/ml. Cells were collected by centrifugation, and resuspended at 3 × 106 cells/ml in fresh YPEG for 6 h before assay to ensure that the cells were growing and to allow maximum mitochondrial number and mitochondrial membrane surface area. Experiments in the absence of oxygen were performed with liquid YPEG which was bubbled with N2 gas and allowed to equilibrate in the anaerobic glove box overnight. After 6 h of aerobic incubation in YPEG, cells were resuspended in the anaerobic medium within the glove box for 1 h prior to assay. Cells were heat exposed for 0, 1, 2, 3, 4, and 5 min at 50°C as described above. Cells were then plated for survivors and for TRP1 revertants.
Measurement of petite cell formation.
Strain RS112 (Table 1) was grown in YPAD to 107 cells/ml. Aerobic heat stress survival assays were performed for 0 and 5 min at 50°C. A total of 500 colonies were plated on 10 plates at each time point. After 3 days growth at 30°C, the number of small colonies (mean ± standard deviation [SD]) on each plate was determined for each time point and compared to the total colony number. Twenty random small colonies were replica plated to solid YPEG, and over 95% were [rho−]. Anaerobic heat stress was performed on the same cultures following introduction to the anaerobic chamber.
Vitamin E measurement.
Vitamin E was measured by high-pressure liquid chromatography (HPLC), using a Beckman ODS column. Strains AH22 and YHT121 (5 × 108 aerobically grown cells) were exposed to 50°C for 10 min and extracted with an excess of hexane. Vitamin E (α- and γ-tocopherol) was measured by UV detection V and quantified using purified vitamin E standards of known concentration.
Measurement of mutation frequency.
Forward mutation was measured at the CANI locus. Any mutation at the CANI locus resulting in disruption of function to the arginine permease leads to development of resistance to the drug canavanine (64). Cultures were halved, and aerobic lethal heat assays were performed on cells grown aerobically in YPAD for 66 h. Deoxygenated lethal heat stress assays were performed at the same time with cells resuspended in the anaerobic glove box as described above. Cells were plated on solid SC lacking arginine and on the same medium with l-canavanine (60 mg/liter). Forward mutation frequencies were measured by counting the number of canavanine-resistant colonies per viable cell.
RESULTS
Anaerobic thermotolerance is independent of HSP104.
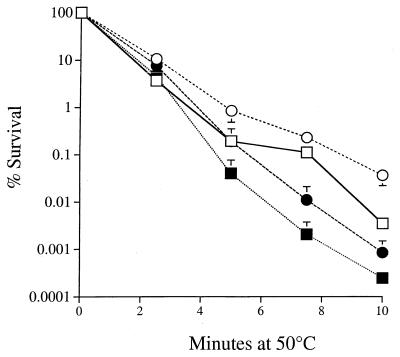
We have previously shown that growing cultures of S. cerevisiae are more resistant to heat stress in the absence of oxygen (17). Expression of HSP104 is primarily involved in the acquisition of thermotolerance (48). Thus, it was possible that the thermotolerance of anaerobically grown cells may be due to expression of HSP104 under these conditions. If this were true, then an hsp104 deletion mutant should not be thermotolerant under anaerobic conditions. To investigate this possibility, we performed heat survival assays on aerobic and anaerobic cultures of an hsp104 deletion mutant strain and the isogenic wild type (Fig. 1). After 10 min at 50°C, the growing cultures of both the wild type (W303-1a) and isogenic hsp104 deletion mutant exhibited sensitivities to lethal heat stress which were enhanced to the same extent in the presence of oxygen (Fig. 1). The hsp104 mutant was more sensitive to heat exposure than the wild type but was still able to develop the same degree of anaerobic thermotolerance as the wild type. Since the anaerobic cultures of the hsp104 deletion mutant did not become as resistant to heat as the anaerobic wild type, it can be inferred that the two processes of thermotolerance (anoxia and Hsp104p activity) are additive. In these experiments, the highest level of thermotolerance was attained in cells both exposed to low oxygen tension and containing a functional HSP104 gene. This indicates that the thermotolerance of anaerobic cells is independent of HSP104.
FIG. 1.
Anoxic thermoprotection is not dependent on HSP104. Cultures of W303-1a (wild type; circles) and Δhsp104 (squares) were grown in YPAD to 5 × 106 cells/ml. Anaerobic (open symbols) and aerobic (closed symbols) lethal heat stress assays were performed at 50°C for the indicated times. Data represent the average of three experiments ± SD.
HSP104 is not induced under anaerobic conditions.
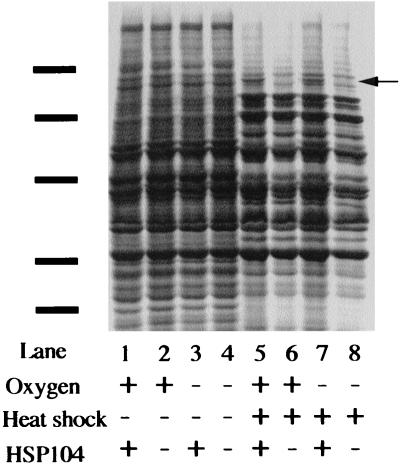
Anaerobic cells are heat tolerant. Thus, anaerobiosis may result in the induction of the heat shock response. The above experiment determined genetically that HSP104 does not play a role in the development of thermotolerance. We next wanted to determine on the basis of protein expression whether the heat shock response is induced in anaerobic cells. We examined the effect of anaerobiosis on total cellular protein synthesis in extracts obtained from cells grown anaerobically and reoxygenated at 23°C. We also compared the patterns of protein synthesis obtained in wild-type cells and hsp104 deletion mutant cells shifted to 39°C for 15 min (Fig. 2). The anaerobic pattern of protein synthesis at 23°C is indistinguishable from the pattern obtained from reoxygenated cells grown at 23°C. For comparison, heat-shocked cells show the induction of heat shock genes including HSP104, which is absent in the hsp104 deletion mutant. This indicates that the heat resistance of anaerobic cells is not due to overexpression of heat shock genes under anaerobic conditions.
FIG. 2.
Total protein synthesis was measured by [35S]methionine label. Anaerobic cells were labeled at 23 or 39°C for 15 min in the presence or absence of atmospheric oxygen. Solid bars indicate molecular weight markers (from the top, 107, 76, 52, 36.8, and 27.2 kDa). The arrow indicates the position of Hsp104p.
Oxidative heat stress increases the frequency of DNA released from mitochondria.
We determined the frequency of heat-induced membrane leakage in aerobic and anaerobic conditions. Since the mitochondrial membrane is impermeable to nucleic acids, release of mitochondrial DNA is a measure of mitochondrial membrane integrity (58). Strain JDY1 (Table 1) contains the TRP1 wild-type marker integrated into mitochondrial DNA (59). As the genetic code differs between genome and mitochondria, a genomic trp1 mutant containing such mitochondria is unable to grow on tryptophan omission medium. Trp+ reversions occur at a spontaneous frequency of approximately 1 to 5 per 106 cells in cultures grown in glycerol and ethanol as carbon sources (Table 2). These reversion events are due to leakage or destruction of the mitochondrial membranes, leading to escape of mitochondrial DNA and passage to the nucleus (58, 59). It has previously been shown that a nonlethal heat shock at 37°C caused a fivefold increase in mitochondrial membrane leakage (58).
TABLE 2.
Release of mitochondrial DNA by heat stress
| Min at 50°C | Oxygenated
|
Anaerobic
|
||
|---|---|---|---|---|
| Fold increasea in TRP1 reversion | Viability (%) | Fold increasea in TRP1 reversion | Viability (%) | |
| 0 | 1.0 | 100 | 1.0 | 100 |
| 1 | 26.0 ± 17.9 | 75 ± 28.4 | 2.1 ± 0.1 | 108 ± 3.5 |
| 2 | 93.1 ± 25.3 | 52 ± 16.3 | 3.0 ± 0.9 | 102 ± 16.5 |
| 3 | 211 ± 3.0 | 38 ± 3.1 | 7.0 ± 1.8 | 73.3 ± 10.3 |
| 4 | 214 ± 21.9 | 44 ± 6.4 | 9.6 ± 4.8 | 69.6 ± 12.3 |
| 5 | 687 ± 180 | 17 ± 5.2 | 5.1 ± 1.8 | 73.7 ± 16.0 |
Relative TRP1 reversions were averaged over two experiments. Similar results were obtained in two other experiments at different time points. Data are the means ± ranges.
After 5 min at 50°C, the aerated culture showed more than a 600-fold induction of the frequency of TRP1 reversion. This indicates that aerobic heat stress treatment caused an increase in ruptured mitochondrial membranes, releasing the mitochondrial genome containing the integrated TRP1 gene. Resuspension of aerobic cells in fresh medium equilibrated in anaerobic liquid YPEG and incubated in an atmosphere of <1 ppm of oxygen for 30 min prior to heat stress exposure resulted in only a 5-fold induction of TRP1 reversions, which is more than 100-fold lower than for the aerated culture (Table 2).
Lethal heat stress induces a petite phenotype.
The increases in Trp+ reversion in strain JDY1 led us to investigate the effect of lethal heat stress on the formation of petite [rho−] colonies. A loss of respiratory function leads to this phenotype, defined by the inability of the [rho−] cell to grow on nonfermentable carbon sources. Treatment of aerobic cultures with a 50°C aerobic heat stress resulted in over 300-fold increase in [rho−] cells after 5 min (from 2.4 ± 1.4 [mean ± SD] to 934 ± 90 petite cells/104 viable aerobic cells). Deoxygenation of the culture prior to heat stress reduced the frequency of petite cell formation by approximately 50% (3.9 ± 0.65 petite cells/104 viable anaerobic cells before heat stress; 532 ± 28 petite cells/104 anaerobic cells after heat stress), indicating that the presence of oxygen during a heat exposure can have major detrimental effects on mitochondrial function.
Total sulfhydryl content is decreased during lethal heat stress.
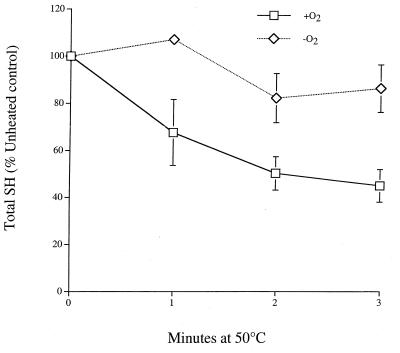
To further characterize the oxidizing effect of heat, total cellular reduced sulfhydryl content in growing cells of strain RS112 (Table 1) was measured using a thiol-specific reactant (Ellman reagent) (Fig. 3). Reoxygenated anaerobic cultures exposed to a 50°C heat stress lose 50 to 60% of their total thiol content within 3 min. Oxygenation alone did not measurably decrease total sulfhydryl content. Little or no decrease was observed in heat-stressed cells maintained in anaerobic conditions. The Ellman reagent will react with all thiol groups in the crude extract, the most abundant being the reduced form of glutathione (GSH). Glutathione is present in millimolar concentrations in the cytoplasm, and it is likely that the decrease in total reduced sulfhydryl content is due to GSH becoming oxidized. This is supported by our observation that precipitated proteins show no apparent decrease in thiol content when reacted with Ellman reagent following the same heat stress conditions (data not shown).
FIG. 3.
Anaerobic cultures (107 cells/ml) of RS112 were halved in an anaerobic glove box; one half was exposed to atmospheric oxygen immediately prior to a 50°C heat stress for the times indicated, while the other half was heat stressed in the absence of oxygen. Crude extract was obtained by glass bead lysis, and total sulfhydryl content was measured with Ellman reagent. Data represent the average of three experiments ± SD.
Vitamin E content is decreased in glutathione-deficient cells during heat stress.
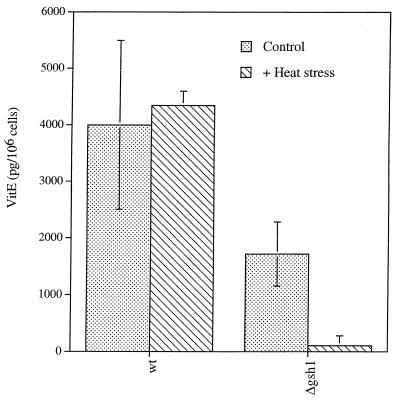
Vitamin E is an important lipid-soluble antioxidant which protects lipid membranes from the detrimental effects of lipid peroxidation by breaking free radical chain reactions occurring within membranes. Vitamin E is itself able to redox cycle, receiving electrons from the soluble antioxidants vitamin C and glutathione. Vitamin E was measured by HPLC in wild-type and glutathione-deficient cells. Glutathione-deficient cells had approximately 43% of the wild-type levels of vitamin E in unheated controls. Heat stress had no effect on vitamin E levels in wild-type cells; following heat stress, the vitamin E concentration was reduced to less than 6% of that of the unheated glutathione-deficient cells (Fig. 4).
FIG. 4.
Aerobic heat stress decreases vitamin E concentration in glutathione-deficient cells only. Vitamin E was measured in strain (wild type AH22 [wt]) and YHT121 (Δgsh1) cells following heat stress. Results are the means of three experiments ± SD.
Nuclear DNA is a target for heat-induced oxidative stress.
As heat stress was demonstrated to have oxidative damaging effects on mitochondria and both lipid and water-soluble antioxidants, we also investigated whether DNA might also be vulnerable to the oxidizing effects of heat exposure. DNA is sensitive to reactive oxygen attack, which can result in the formation of a variety of lesions, including double- and single-strand breaks, base modifications, and abasic sites (3). Oxidative DNA damage can lead to increases in mutation and DNA recombination frequencies. We measured nuclear DNA mutation frequencies and gene conversion frequencies following heat stress and found that both indicators of DNA damage are increased by heat exposure and that oxygen is involved in this process.
Gene conversion frequencies were determined in the diploid strain S41 by measuring the frequency of reversion of the trp5 marker. S41 contains two alleles, trp5-12 and trp5-27, which revert to Trp+ by gene conversion (68) at a frequency of approximately 1 per 105 cells. Lethal heat stress conditions leads to an increase in gene conversion frequency in aerated growing-phase cultures (Table 3). After 3 min at 50°C, the aerated culture showed an eightfold induction of gene conversion events, whereas there was less than a twofold induction under anaerobic conditions (Table 3). DNA recombination may repair oxidative DNA damage or is initiated in response to a particular DNA damage (e.g., double-or single-strand breaks).
TABLE 3.
Gene conversion frequencies are elevated by heat stress
| Min at 50°C | Fold increasea in TRP5 reversion
|
|
|---|---|---|
| Reoxygenated | Anaerobic | |
| 0 | 1.0 | 1.0 |
| 1 | 4.6 ± 3.9 | 1.9 ± 0.6 |
| 2 | 10.9 ± 6.9 | 1.9 ± 1.3 |
| 3 | 8.4 ± 3.8 | 1.4 ± 0.5 |
Mean of three experiments ± SD.
Forward mutations at the CAN1 locus were measured in strains Y433 and JDY47 (lacking all three oxidative DNA glycosylases NTG1, NTG2, and OGG1) after growth in YPAD for 66 h. Deletion of NTG1 and OGG1 alone resulted in smaller increases in heat-dependent mutation frequency compared to the oggIntgIntg2 triple mutant (data not shown). As the triple mutant showed the greatest increase in heat-induced mutation, this strain was used to determine the dependency of oxygen on this process.
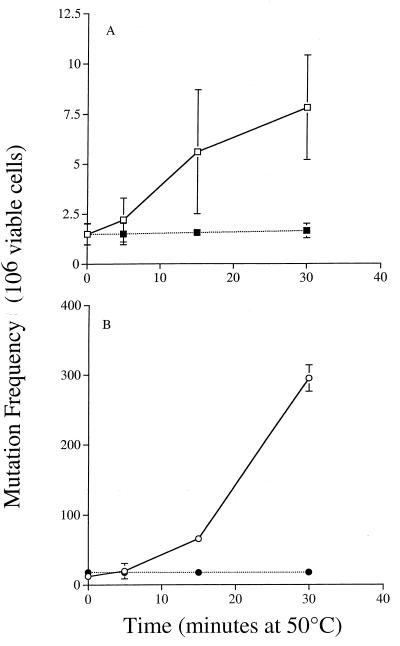
Mutation frequency increased approximately 5 fold in the wild type after 30 min at 50°C, while the glycosylase-deficient triple mutant displayed increases of almost 300-fold in mutation frequency after the same exposure compared to unheated controls (Fig. 5). Both wild-type and mutant cells showed no increase in mutation frequency when shifted to anaerobic conditions for heat stress. This suggests that heat stress generates specific oxidative base lesions which are normally recognized and repaired by the NTG1, NTG2, and OGG1 glycosylases. These DNA repair genes clearly are important in the maintenance of genetic integrity following oxidative heat stress, and repair of oxidative damage following heat stress may be their primary function.
FIG. 5.
Heat-induced mutations are dependent on oxygen and are repaired by the oxidative glycosylases Ntg1p, Ntg2p, and Ogg1p. Cultures of Y433 (wild type) (A) and (ntg1ntg2ogg1) (B) JDY47 were grown for 66 h in YPAD before being split. One half of each culture was assayed following heat stress for forward mutation at the CAN1 locus in the presence of oxygen (open symbols); the other half was assayed in the absence of oxygen after introduction to the anaerobic chamber (closed symbols). Results are the means of three experiments ± SD.
DISCUSSION
Hyperthermic conditions are stressful and lead to toxicity. The molecular events contributing to lethality are poorly defined, and previous investigators have focused mainly on adaptive endogenous thermotolerance. To further characterize the mechanism of heat-induced toxicity, we describe the consequences of a heat-associated oxidative stress on key intracellular macromolecules and antioxidants.
HSP104 is not involved in anaerobic thermotolerance.
The thermoprotective effect observed in anaerobic cells was not due to expression of HSP104 or other heat shock genes. Since anaerobic conditions did not cause thermotolerance by induction of heat shock proteins, it was possible that large differences in survival in heat stress anaerobic cultures are due to oxygen-dependent toxicity. If toxic oxygen species are developed during heat stress, then macromolecules within the cells should become oxidized only during aerobic heat stress. Thus, we compared the effects of aerobic and anaerobic lethal heat stresses on frequencies of interchromosomal DNA recombination and mutation, mitochondrial membrane integrity, formation of respiratory-deficient cells, and the soluble and nonsoluble antioxidant agents glutathione and vitamin E. Aerobic heat-stress caused effects on each of these endpoints, which were reduced or abolished by anaerobic conditions.
Increased recombination during heat exposure.
Specific oxidative DNA lesions and enzymatic DNA repair pathways have evolved to cope with the flux of oxidants which are generated during aerobic respiration as well as during times of increased radical production (31, 32). DNA strand breaks and increases in DNA recombination frequencies are observed in response to conditions that promote oxidative stress (3, 20, 66). Our finding that lethal heat induces an oxygen-dependent increase in the frequency of DNA recombination events agrees with the idea that the production of reactive oxygen species occurs in response to lethal heat shock. DNA recombination is stimulated in response to double- and single-stranded gaps caused by exposure to H2O2 (11, 42) and also during conditions where DNA replication is stalled, for example, when DNA polymerase encounters a blocking lesion (e.g., an abasic site or thymidine glycol lesion) (67; reviewed in reference 19). Oxidative DNA glycosylase enzymes repair heat-dependent mutations.
We also find that nuclear mutation frequencies increase in response to an aerobic heat stress and that the magnitude of this mutagenic effect is greatly increased in cells defective for repair of oxidatively modified base lesions. The specific glycosylase enzymes encoded by NTG1, NTG2, and OGG1 are responsible for repair of a variety of pyrimidine and purine base lesions, and when all three genes were deleted, the resultant strain was extremely susceptible to mutation following a heat exposure. The specificity of these glycosylases for oxygen radical-damaged bases argues for an involvement of oxygen radicals during heat-induced mutation. Indeed, when the mutation assays were performed on the same cells after introduction into a low-oxygen atmosphere (<1 ppm), the heat-induced mutations were abolished, indicating that the mutations are caused by oxygen radicals.
Oxidative stress during heat stress damages mitochondrial membranes.
To determine whether oxidative heat stress affects mitochondrial membranes, the escape of mitochondrial DNA via mitochondrial membrane leakage was assessed using aerobic cells of strain JDY1 growing in glycerol-ethanol (Table 2). Growth on nonfermentable carbon sources results in many small mitochondria compared to the few large and branched mitochondria observed in glucose-grown cells (62). During growth on nonfermentable carbon sources, the mitochondrial membrane surface area is larger (62) and hence the surface exposed to oxidative and thermal attack is greater. The results displayed in Table 2 demonstrate that oxygen deprivation immediately and substantially protects the mitochondrial membrane from rupture and release of mitochondrial DNA. Likewise, the formation of petite colonies was greatly increased by lethal heat exposure, and petite colony formation was suppressed by anaerobic conditions prior to heat stress. We speculate that ruptured mitochondria are one source of the oxidative stress associated with heat lethality. Leaky or broken mitochondria may release superoxide anions, which are generated by the electron transport chain and accumulate in the matrix lumen (4, 55, 60), into the cytoplasm. Release of superoxide anions from mitochondria might stimulate lipid peroxidation and lead to further mitochondrial lysis.
Nonenzymatic antioxidants are depleted by heat stress.
We have previously shown that enzymatic antioxidants such as superoxide dismutase, catalase, and cytochrome c peroxidase protect cells against lethal heat (17), and it has been reported that deletion of thioredoxin peroxidase sensitizes cells to lethal effects of heat stress (36). The primary nonenzymatic line of defense against oxidants is GSH (28, 56), which is found in millimolar concentrations in the cytoplasm and mitochondria. We demonstrate that reoxygenated anaerobic cells show a reduction in total cellular sulfhydryl content by 50% following a heat stress exposure, whereas the anaerobic cells are largely unaffected. The largest contribution to cellular sulfhydryl content is GSH. A loss of more than half of the intracellular GSH concentration within the first few minutes of lethal heat stress indicates that the cell has expended a tremendous concentration of reducing equivalents, indicative of a serious oxidative burden. The amount of GSH becoming oxidized during heat stress is further compounded when it is considered that GSH is constantly being renewed by glutathione reductase, which acts to maintain glutathione in a reduced form at the expense of cellular NADPH (26). Glutathione not only acts directly as an antioxidant but also maintains other important antioxidants, such as vitamin E and vitamin C, in the biologically active reduced form (12, 29). A 50% decrease in GSH concentration will directly affect the antioxidant potential of these other important reductants. Indeed, we observed that the cells deficient in GSH showed approximately 43% the total vitamin E content of the wild-type cells. After a heat stress exposure, the glutathione-deficient cells experienced an almost complete depletion of vitamin E that was not observed in wild-type cells, highlighting the importance of GSH as a thermoprotectant both within the cytoplasm and indirectly within the lipid membranes via vitamin E reduction. It is possible that the decrease in intracellular thiol reducing equivalents during heat stress may be due in part to the utilization of electrons from GSH to replenish the oxidized form of vitamin E.
Previous analysis of CHO cells depleted of glutathione by diethylmaleate demonstrated that a decreased GSH concentration causes cells to become thermosensitive (22). Mitochondrial GSH concentration is more important than cytoplasmic GSH levels for thermoprotection. Declining mitochondrial GSH concentrations coincide with the thermosensitization threshold response to the diethylmaleate concentration (23).
This study provides further support for an involvement of oxidative stress during heating. Several possible mechanisms can account for this. It is possible that iron and other Fenton reactive ions are released during thermal denaturation from sequestered intracellular sites or from metal-containing proteins such as iron-sulfur proteins. Alternatively, thermal damage to mitochondria may expose electron carrier components of the mitochondrial electron transport chain (e.g. ubisemiquinone) to potential electron donors in the cytoplasm, leading to uncontrolled redox cycling between electron carriers, cellular reductants, and oxygen. The superoxide anions produced could dismutate to hydrogen peroxide and diffuse throughout the cell, including the nucleus. Fenton reaction with iron or copper would produce the strongly oxidizing hydroxyl radical, which is capable of oxidizing most macromolecules in the cell, including, DNA, proteins, and lipids (12). Another possibility is that heat-induced protein denaturation may lower the normal level of cellular antioxidant enzymes, with the consequence of oxidative stress.
We have shown that heat stress combined with an oxygenated environment is sufficient to induced extensive damage to DNA, lipids, and cellular antioxidants. The repeated exposures to stressful temperatures that a unicellular organism such as S. cerevisiae is likely to encounter implicates thermal oxidative damages as relevant selection pressures during the evolution of antioxidant defenses and maintenance of genetic integrity.
ACKNOWLEDGMENTS
We thank Susan Lindquist, Edith Gralla, and Thomas Fox for supplying plasmids and strains. We thank the members of the our laboratory as well as Jim Anderson for analysis of vitamin E, and we thank Bruce Demple, Marianne Wessling-Resnick, and Dan Finley for valuable discussion.
This work was supported by grant CN-83 from the American Cancer Society and Research Career Development Award ES00299 from the National Institute of Environmental Health Sciences to R.H.S. J.F.D. was supported by NIH training grants 5-T32CA09078 and 2-T32ES07155.
REFERENCES
- 1.Alani E, Cao L, Kleckner N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics. 1987;116:541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alseth I, Eide L, Pirovano M, Rognes T, Seeberg E, Bjoras M. The Saccharomyces cerevisiae homologues of endonuclease III from Escherichia coli, Ntg1 and Ntg2, are both required for efficient repair of spontaneous and induced oxidative DNA damage in yeast. Mol Cell Biol. 1999;19:3779–3787. doi: 10.1128/mcb.19.5.3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ames B N, Shigenaga M K, Gold L S. DNA lesions, inducible DNA repair, and cell division: three key factors in mutagenesis and carcinogenesis. Environ Health Perspect. 1993;101:35–44. doi: 10.1289/ehp.93101s535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ames B N, Shigenaga M K, Hagen T M. Mitochondrial decay in aging. Biochim Biophys Acta. 1995;1271:165–170. doi: 10.1016/0925-4439(95)00024-x. [DOI] [PubMed] [Google Scholar]
- 5.Bader S B, Price B D, Mannheim-Rodman L A, Calderwood S K. Inhibition of heat shock gene expression does not block the development of thermotolerance. J Cell Physiol. 1992;151:56–62. doi: 10.1002/jcp.1041510110. [DOI] [PubMed] [Google Scholar]
- 6.Barnes C A, Johnston G C, Singer R A. Thermotolerance is independent of induction of the full spectrum of heat shock proteins and of cell cycle blockage in the yeast Saccharomyces cerevisiae. J Bacteriol. 1990;172:4352–4358. doi: 10.1128/jb.172.8.4352-4358.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Begonia G B, Salin M L. Elevation of superoxide dismutase in Halobacterium halobium by heat shock. J Bacteriol. 1991;173:5582–5584. doi: 10.1128/jb.173.17.5582-5584.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belazzi T, Wagner A, Wieser R, Schanz M, Adam G, Hartig A, Ruis H. Negative regulation of transcription of the Saccharomyces cerevisiae catalase T (CTT1) gene by cAMP is mediated by a positive control element. EMBO J. 1991;10:585–592. doi: 10.1002/j.1460-2075.1991.tb07985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benov L, Fridovich I. Superoxide dismutase protects against aerobic heat shock in Escherichia coli. J Bacteriol. 1995;177:3344–3346. doi: 10.1128/jb.177.11.3344-3346.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boyne A F, Ellman G L. A methodology for analysis of tissue sulfhydryl components. Anal Biochem. 1972;46:639–653. doi: 10.1016/0003-2697(72)90335-1. [DOI] [PubMed] [Google Scholar]
- 11.Brennan R J, Swoboda B E, Schiestl R H. Oxidative mutagens induce intrachromosomal recombination in yeast. Mutat Res. 1994;308:159–167. doi: 10.1016/0027-5107(94)90151-1. [DOI] [PubMed] [Google Scholar]
- 12.Buettner G R. The pecking order of free radicals and antioxidants: lipid peroxidation, alpha-tocopherol, and ascorbate. Arch Biochem Biophys. 1993;300:535–543. doi: 10.1006/abbi.1993.1074. [DOI] [PubMed] [Google Scholar]
- 13.Burgman P W, Kampinga H H, Konings A W. Possible role of localized protein denaturation in the mechanism of induction of thermotolerance by heat, sodium-arsenite and ethanol. Int J Hyperthermia. 1993;9:151–162. doi: 10.3109/02656739309061487. [DOI] [PubMed] [Google Scholar]
- 14.Coote P J, Cole M B, Jones M V. Induction of increased thermotolerance in Saccharomyces cerevisiae may be triggered by a mechanism involving intracellular pH. J Gen Microbiol. 1991;137:1701–1708. doi: 10.1099/00221287-137-7-1701. [DOI] [PubMed] [Google Scholar]
- 15.Coote P J, Jones M V, Edgar K, Cole M B. TPK gene products mediate cAMP-independent thermotolerance in Saccharomyces cerevisiae. J Gen Microbiol. 1992;138:2551–2557. doi: 10.1099/00221287-138-12-2551. [DOI] [PubMed] [Google Scholar]
- 16.Coote P J, Jones M V, Seymour I J, Rowe D L, Ferdinando D P, McArthur A J, Cole M B. Activity of the plasma membrane H(+)-ATPase is a key physiological determinant of thermotolerance in Saccharomyces cerevisiae. Microbiology. 1994;140:1881–1890. doi: 10.1099/13500872-140-8-1881. [DOI] [PubMed] [Google Scholar]
- 17.Davidson J F, Whyte B, Bissinger P H, Schiestl R H. Oxidative stress is involved in heat-induced cell death in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1996;93:5116–5121. doi: 10.1073/pnas.93.10.5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis M J, Coote P J, O'Byrne C P. Acid tolerance in Listeria monocytogenes: the adaptive acid tolerance response (ATR) and growth-phase-dependent acid resistance. Microbiology. 1996;142:2975–2982. doi: 10.1099/13500872-142-10-2975. [DOI] [PubMed] [Google Scholar]
- 19.Demple B, Harrison L. Repair of oxidative damage to DNA: enzymology and biology. Annu Rev Biochem. 1994;63:915–948. doi: 10.1146/annurev.bi.63.070194.004411. [DOI] [PubMed] [Google Scholar]
- 20.Dreher D, Junod A F. Role of oxygen free radicals in cancer development. Eur J Cancer. 1996;32A:30–38. doi: 10.1016/0959-8049(95)00531-5. [DOI] [PubMed] [Google Scholar]
- 21.Eide L, Bjoras M, Pirovano M, Alseth I, Berdal K G, Seeberg E. Base excision of oxidative purine and pyrimidine DNA damage in Saccharomyces cerevisiae by a DNA glycosylase with sequence similarity to endonuclease III from Escherichia coli. Proc Natl Acad Sci USA. 1996;93:10735–10740. doi: 10.1073/pnas.93.20.10735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freeman M L, Borrelli M J, Syed K, Senisterra G, Stafford D M, Lepock J R. Characterization of a signal generated by oxidation of protein thiols that activates the heat shock transcription factor. J Cell Physiol. 1995;164:356–366. doi: 10.1002/jcp.1041640216. [DOI] [PubMed] [Google Scholar]
- 23.Freeman M L, Meredith M J. Subcellular localization of glutathione and thermal sensitivity. Radiat Res. 1988;115:461–471. [PubMed] [Google Scholar]
- 24.Girard P M, Boiteux S. Repair of oxidized DNA bases in the yeast Saccharomyces cerevisiae. Biochimie. 1997;79:559–566. doi: 10.1016/s0300-9084(97)82004-4. [DOI] [PubMed] [Google Scholar]
- 25.Girard P M, Guibourt N, Boiteux S. The Ogg1 protein of Saccharomyces cerevisiae: a 7,8-dihydro-8- oxoguanine DNA glycosylase/AP lyase whose lysine 241 is a critical residue for catalytic activity. Nucleic Acids Res. 1997;25:3204–3211. doi: 10.1093/nar/25.16.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grant C M, Collinson L P, Roe J H, Dawes I W. Yeast glutathione reductase is required for protection against oxidative stress and is a target gene for yAP-1 transcriptional regulation. Mol Microbiol. 1996;21:171–179. doi: 10.1046/j.1365-2958.1996.6351340.x. [DOI] [PubMed] [Google Scholar]
- 27.Grant C M, Firoozan M, Tuite M F. Mistranslation induces the heat-shock response in the yeast Saccharomyces cerevisiae. Mol Microbiol. 1989;3:215–220. doi: 10.1111/j.1365-2958.1989.tb01810.x. [DOI] [PubMed] [Google Scholar]
- 28.Grant C M, MacIver F H, Dawes I W. Glutathione is an essential metabolite required for resistance to oxidative stress in the yeast Saccharomyces cerevisiae. Curr Genet. 1996;29:511–515. doi: 10.1007/BF02426954. [DOI] [PubMed] [Google Scholar]
- 29.Gumpricht E, Hildenbrandt G R, Scholz R W, Reddy C C. Glutathione-dependent protection against lipid peroxidation in sheep liver microsomes. Biochem Mol Biol Int. 1996;38:559–567. [PubMed] [Google Scholar]
- 30.Holyoak C D, Stratford M, McMullin Z, Cole M B, Crimmins K, Brown A J, Coote P J. Activity of the plasma membrane H(+)-ATPase and optimal glycolytic flux are required for rapid adaptation and growth of Saccharomyces cerevisiae in the presence of the weak-acid preservative sorbic acid. Appl Environ Microbiol. 1996;62:3158–3164. doi: 10.1128/aem.62.9.3158-3164.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu J J, Dubin N, Kurland D, Ma B L, Roush G C. The effects of hydrogen peroxide on DNA repair activities. Mutat Res. 1995;336:193–201. doi: 10.1016/0921-8777(94)00054-a. [DOI] [PubMed] [Google Scholar]
- 32.Hu J J, Roush G C, Berwick M, Dubin N, Mahabir S, Chandiramani M, Boorstein R. Effects of dietary supplementation of alpha-tocopherol on plasma glutathione and DNA repair activities. Cancer Epidemiol Biomarkers Prev. 1996;5:263–270. [PubMed] [Google Scholar]
- 33.Jozwiak Z. Involvement of heat shock proteins and cellular membranes in the development of thermotolerance. Arch Immunol Ther Exp. 1994;42:247–252. [PubMed] [Google Scholar]
- 34.Jozwiak Z, Leyko W. Role of membrane components in thermal injury of cells and development of thermotolerance. Int J Radiat Biol. 1992;62:743–756. doi: 10.1080/09553009214552701. [DOI] [PubMed] [Google Scholar]
- 35.Laszlo A. The thermoresistant state: protection from initial damage or better repair? Exp Cell Res. 1992;202:519–531. doi: 10.1016/0014-4827(92)90107-j. [DOI] [PubMed] [Google Scholar]
- 36.Lee S M, Park J W. Thermosensitive phenotype of yeast mutant lacking thioredoxin peroxidase. Arch Biochem Biophys. 1998;359:99–106. doi: 10.1006/abbi.1998.0896. [DOI] [PubMed] [Google Scholar]
- 37.Lindquist S, Kim G. Heat-shock protein 104 expression is sufficient for thermotolerance in yeast. Proc Natl Acad Sci USA. 1996;93:5301–5306. doi: 10.1073/pnas.93.11.5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mager W H, De Kruijff A J. Stress-induced transcriptional activation. Microbiol Rev. 1995;59:506–531. doi: 10.1128/mr.59.3.506-531.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marchler G, Schuller C, Adam G, Ruis H. A Saccharomyces cerevisiae UAS element controlled by protein kinase A activates transcription in response to a variety of stress conditions. EMBO J. 1993;12:1997–2003. doi: 10.1002/j.1460-2075.1993.tb05849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nash H M, Bruner S D, Scharer O D, Kawate T, Addona T A, Spooner E, Lane W S, Verdine G L. Cloning of a yeast 8-oxoguanine DNA glycosylase reveals the existence of a base-excision DNA-repair protein superfamily. Curr Biol. 1996;6:968–980. doi: 10.1016/s0960-9822(02)00641-3. [DOI] [PubMed] [Google Scholar]
- 41.Ohtake Y, Yabuuchi S. Molecular cloning of the gamma-glutamylcysteine synthetase gene of Saccharomyces cerevisiae. Yeast. 1991;7:953–961. doi: 10.1002/yea.320070907. [DOI] [PubMed] [Google Scholar]
- 42.Paques F, Haber J E. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parsell D A, Kowal A S, Lindquist S. Saccharomyces cerevisiae Hsp104 protein. Purification and characterization of ATP-induced structural changes. J Biol Chem. 1994;269:4480–4487. [PubMed] [Google Scholar]
- 44.Parsell D A, Kowal A S, Singer M A, Lindquist S. Protein disaggregation mediated by heat-shock protein Hsp104. Nature. 1994;372:475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- 45.Parsell D A, Sanchez Y, Stitzel J D, Lindquist S. Hsp104 is a highly conserved protein with two essential nucleotide-binding sites. Nature. 1991;353:270–273. doi: 10.1038/353270a0. [DOI] [PubMed] [Google Scholar]
- 46.Ruis H, Schuller C. Stress signaling in yeast. Bioessays. 1995;17:959–965. doi: 10.1002/bies.950171109. [DOI] [PubMed] [Google Scholar]
- 47.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 48.Sanchez Y, Lindquist S L. HSP104 required for induced thermotolerance. Science. 1990;248:1112–1115. doi: 10.1126/science.2188365. [DOI] [PubMed] [Google Scholar]
- 49.Sanchez Y, Parsell D A, Taulien J, Vogel J L, Craig E A, Lindquist S. Genetic evidence for a functional relationship between Hsp104 and Hsp70. J Bacteriol. 1993;175:6484–6491. doi: 10.1128/jb.175.20.6484-6491.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schiestl R H. Nonmutagenic carcinogens induce intrachromosomal recombination in yeast. Nature. 1989;337:285–288. doi: 10.1038/337285a0. [DOI] [PubMed] [Google Scholar]
- 51.Schiestl R H, Petes T D. Integration of DNA fragments by illegitimate recombination in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1991;88:7585–7589. doi: 10.1073/pnas.88.17.7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schiestl R H, Prakash S. RAD1, an excision repair gene of Saccharomyces cerevisiae, is also involved in recombination. Mol Cell Biol. 1988;8:3619–3626. doi: 10.1128/mcb.8.9.3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Senturker S, Auffret van der Kemp P, You H J, Doetsch P W, Dizdaroglu M, Boiteux S. Substrate specificities of the Ntg1 and Ntg2 proteins of Saccharomyces cerevisiae for oxidized DNA bases are not identical. Nucleic Acids Res. 1998;26:5270–5276. doi: 10.1093/nar/26.23.5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sherman F, Fink G R, Hicks J B. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- 55.Shigenaga M K, Hagen T M, Ames B N. Oxidative damage and mitochondrial decay in aging. Proc Natl Acad Sci USA. 1994;91:10771–10778. doi: 10.1073/pnas.91.23.10771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stephen D W, Jamieson D J. Glutathione is an important antioxidant molecule in the yeast Saccharomyces cerevisiae. FEMS Microbiol Lett. 1996;141:207–212. doi: 10.1111/j.1574-6968.1996.tb08386.x. [DOI] [PubMed] [Google Scholar]
- 57.Swanson R L, Morey N J, Doetsch P W, Jinks-Robertson S. Overlapping specificities of base excision repair, nucleotide excision repair, recombination, and translesion synthesis pathways for DNA base damage in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:2929–2935. doi: 10.1128/mcb.19.4.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thorsness P E, Fox T D. Escape of DNA from mitochondria to the nucleus in Saccharomyces cerevisiae. Nature. 1990;346:376–379. doi: 10.1038/346376a0. [DOI] [PubMed] [Google Scholar]
- 59.Thorsness P E, Fox T D. Nuclear mutations in Saccharomyces cerevisiae that affect the escape of DNA from mitochondria to the nucleus. Genetics. 1993;134:21–28. doi: 10.1093/genetics/134.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Turrens J F. Superoxide production by the mitochondrial respiratory chain. Biosci Rep. 1997;17:3–8. doi: 10.1023/a:1027374931887. [DOI] [PubMed] [Google Scholar]
- 61.Visser W, Scheffers W A, Batenburg-van der Vegte W H, van Dijken J P. Oxygen requirements of yeasts. Appl Environ Microbiol. 1990;56:3785–3792. doi: 10.1128/aem.56.12.3785-3792.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Visser W, van Spronsen E A, Nanninga N, Pronk J T, Gijs Kuenen J, van Dijken J P. Effects of growth conditions on mitochondrial morphology in Saccharomyces cerevisiae. Antonie Leeuwenhoek. 1995;67:243–253. doi: 10.1007/BF00873688. [DOI] [PubMed] [Google Scholar]
- 63.Vogel J L, Parsell D A, Lindquist S. Heat-shock proteins Hsp104 and Hsp70 reactivate mRNA splicing after heat inactivation. Curr Biol. 1995;5:306–317. doi: 10.1016/s0960-9822(95)00061-3. [DOI] [PubMed] [Google Scholar]
- 64.Whelan W L, Gocke E, Manney T R. The CAN1 locus of Saccharomyces cerevisiae: fine-structure analysis and forward mutation rates. Genetics. 1979;91:35–51. doi: 10.1093/genetics/91.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wieser R, Adam G, Wagner A, Schuller C, Marchler G, Ruis H, Krawiec Z, Bilinski T. Heat shock factor-independent heat control of transcription of the CTT1 gene encoding the cytosolic catalase T of Saccharomyces cerevisiae. J Biol Chem. 1991;266:12406–12411. [PubMed] [Google Scholar]
- 66.Wiseman H, Kaur H, Halliwell B. DNA damage and cancer: measurement and mechanism. Cancer Lett. 1995;93:113–120. doi: 10.1016/0304-3835(95)03792-U. [DOI] [PubMed] [Google Scholar]
- 67.Yap W Y, Kreuzer K N. Recombination hotspots in bacteriophage T4 are dependent on replication origins. Proc Natl Acad Sci USA. 1991;88:6043–6047. doi: 10.1073/pnas.88.14.6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zimmermann F K. Procedures used in the induction of mitotic recombination and mutation in the yeast Saccharomyces cerevisiae. Mutat Res. 1975;31:71–86. doi: 10.1016/0165-1161(75)90069-2. [DOI] [PubMed] [Google Scholar]