Abstract
Transforming growth factor beta receptor Ⅱ (TGFBR2), a core member of the transforming growth factor-β (TGF-β) signaling pathway. To date, chicken TGFBR2 (cTGFBR2) genomic structure has not been fully explored. Here, the complete sequences of cTGFBR2 transcript isoforms were determined by 5′ and 3′ rapid amplification of cDNA ends (5′ & 3′ RACE) and reverse transcription polymerase chain reaction (RT-PCR); the tissue expression profiling of cTGFBR2 transcript isoforms was performed using quantitative real-time polymerase chain reaction (qRT-PCR). The results showed that cTGFBR2 gene produced 3 transcript isoforms though alternative transcription initiation, splicing, and polyadenylation, which were designated as cTGFBR2-1, cTGFBR2-2, and cTGFBR2-3, respectively. These 3 cTGFBR2 transcript isoforms encoded 3 protein isoforms: cTGFBR2-1, cTGFBR2-2, and cTGFBR2-3. Duplication analysis revealed that, unlike other animal species, cTGFBR2 gene harbored a 5.5-kb intragenic tandem duplication. Tissue expression profiling in the 4-wk-old Arbor Acres (AA) broiler chickens showed that cTGFBR2-1 was ubiquitously expressed, with high expression in abdominal fat, subcutaneous fat, lung, gizzard, and muscle; cTGFBR2-2 was highly expressed in heart, kidney, gizzard, and muscle; cTGFBR2-3 was weakly expressed in all the tested chicken tissues. Tissue expression profiling in the 7-wk-old broiler chickens of the fat and lean lines of Northeast Agricultural University broiler lines divergently selected for abdominal fat content (NEAUHLF) showed that cTGFBR2-1 was significantly differentially expressed in all the tested tissues except heart, cTGFBR2-2 was significantly differentially expressed in all the tested tissues except subcutaneous fat and liver, and cTGFBR2-3 was significantly differentially expressed in all the tested tissues between the lean and fat lines. Intriguingly, in the fat line, the 3 cTGFBR2 transcript isoforms were expressed to varying degrees in all the 3 tested fat tissues, while in the lean line, only cTGFBR2-1 was expressed in all the 3 tested fat tissues. This is the first report of intragenic tandem duplication within TGFBR2 gene. Our findings pave the way for further studies on the functions and regulation of cTGFBR2 gene.
Key words: chicken, TGFBR2, transcript isoform, intragenic tandem duplication, gene expression
INTRODUCTION
Transforming growth factor-β (TGF-β) signaling pathway regulates a variety of biological processes throughout embryonic development and postnatal life, as well as during the presentation of human disease (Lönn et al., 2009; Moustakas and Heldin, 2009; Pardali et al., 2010; Xu et al., 2012). There are 3 TGF-β forms (TGF-β1, 2, and 3) and 3 TGF-β receptors (TGFBR1, 2, and 3) (Vander Ark et al., 2018). TGFBR2 plays an important role in the TGF-β signaling pathway. Upon ligand binding, TGFBR2 forms a signaling complex with TGFBR1 and activates TGFBR1 via phosphorylation. The activated TGFBR1 in turn phosphorylates its downstream receptor-regulated SMADs (R-SMADs, SMAD2/3). Subsequently, the activated R-SMADs bind to SMAD4, migrate to the nucleus, and regulate TGF-β target gene transcription.
TGFBR2 serves as an initial regulator of the TGF-β signaling pathway and is involved in many cellular processes (Moustakas and Heldin, 2009). Loss or reduction of its expression can lead to uncontrolled cell growth (Yuan et al., 2020). Defects by TGFBR2 knockout involve many phylogenetic and major organ systems, including developmental defects of the kidney, skeleton, and lymphatic network (James et al., 2013; Peters et al., 2017; Dumbrava et al., 2021). During the development of chicken embryos, TGFBR2 shows dynamically and frequently overlapping expression patterns in numerous embryonic cell layers and structures, and is involved in chicken-specific developmental processes including somitogenesis, cardiogenesis, and vasculogenesis (Cooley et al., 2014).
TGFBR2 genomic structure has been intensively studied in mammals. Both human and mouse TGFBR2 genes are composed of 8 exons and 7 introns. Due to alternative splicing, TGFBR2 gene generates 2 transcript isoforms in humans and mice, which encode 2 protein isoforms, named as TβRII and TβRII-B, respectively (Hirai and Fijita, 1996; Krishnaveni et al., 2006). Recently, a novel human TGFBR2 transcript isoform named as TβRII-SE was discovered, which encodes a soluble protein (Bertolio et al., 2021). To date, chicken TGFBR2 (cTGFBR2) genomic structure has not been fully explored, and whether the cTGFBR2 gene encodes multiple transcript and protein isoforms is unclear. In the present study, we for the first time characterized cTGFBR2 transcript isoforms, dissected the cTGFBR2 genomic structure and identified a 5.5-kb intragenic tandem duplication.
MATERIALS AND METHODS
Experimental Animals and Tissue Collection
Our animal experiments were conducted according to the guidelines for the care and use of experimental animals established by the Ministry of Science and Technology of the People's Republic of China (approval no. 2006-398) and were approved by the Laboratory Animal Management Committee of Northeast Agricultural University (Harbin, Heilongjiang, PRC). All experimental chickens were fed under the same environmental conditions with free intake of food and water. Abdominal fat, subcutaneous fat, gizzard fat, heart, liver, spleen, lung, kidney, gizzard, proventriculus, pancreas, and muscle tissue were collected from 4-wk-old Arbor Acres (AA) broiler chickens. All of the above tissues except the lung were collected from 7-wk-old broiler chickens of fat and lean lines of Northeast Agricultural University broiler lines divergently selected for abdominal fat content (NEAUHLF) (24th generation). All samples were washed with a solution containing 0.75% NaCl, snap frozen in liquid nitrogen, and stored at −80°C until RNA isolation.
RNA Isolation and Reverse Transcription
The frozen tissue samples were ground into powder in liquid nitrogen by using a mortar and pestle. Total RNAs were isolated using Trizol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. RNA quality was assessed by denaturing formaldehyde agarose gel electrophoresis and quantified by using a spectrophotometer. The extracted RNA was reverse transcribed using the PrimeScriptTM RT reagent Kit (Takara, Dalian, China) according to the manufacturer's protocol.
5′ and 3′ Rapid Amplification of cDNA Ends
The 5′ and 3′ rapid amplification of cDNA ends (5′ & 3′ RACE) of cTGFBR2 was performed using the SMARTer RACE 5′/3′ Kit (Takara, Dalian, China) according to the manufacturer's protocol. The gene-specific primers for the 5′ and 3′ RACE were designed by Primer 6.0 based on cTGFBR2 mRNA (NM_205428.1). The primer sequences are listed in Table 1. The RACE products were separated by agarose gel electrophoresis and recovered using DNA Gel Recovery Kit (Axygen, Union City, CA) following the manufacturer's protocol. All recovered products were cloned using pEASY blunt simple cloning vector (TransGen, Beijing, China) according to the manufacturer's protocol, and transformed into Escherichia coli (E. coli). Single colonies were picked for PCR identification and sent to Genewiz biological company (Suzhou, China) for Sanger sequencing.
Table 1.
The primers used in the present study.
| Type | Primer name | Primer sequence |
|---|---|---|
| RACE primer | 5′RACE-GSP1 | R: ACAGCGAGATGTCATTTCCCAGA |
| 5′RACE-GSP2 | F: TTTCCCAGAGGACCAAAGC | |
| 3′RACE-GSP1 | F: CTCCATGGCTTTGGTCCTCTG | |
| 3′RACE-GSP2 | F: CTCGCTGTAATGGTGTTGG | |
| RT-PCR primer | TGFBR2-1-F/TGFBR2-1-R | F: TGCAGCGCCGAAGTGAAGTTTTC |
| R: TACATCTCCCTGCCCAGAGCCAC | ||
| TGFBR2-1-F/ TGFBR2-2-R | F: TGCAGCGCCGAAGTGAAGTTTTC | |
| R: GAACAGAGTCAGGCTGTGGTATG | ||
| TGFBR2-1-F/ TGFBR2-3-R | F: TGCAGCGCCGAAGTGAAGTTTTC | |
| R: GTCCGTAAAGACCACTCAACATA | ||
| TGFBR2-2-F/ TGFBR2-1-R | F: GGGGAGAAGGAACTACTGTAAGA | |
| R: TACATCTCCCTGCCCAGAGCCAC | ||
| TGFBR2-2-F/ TGFBR2-2-R | F: GGGGAGAAGGAACTACTGTAAGA | |
| R: GAACAGAGTCAGGCTGTGGTATG | ||
| TGFBR2-2-F/ TGFBR2-3-R | F: GGGGAGAAGGAACTACTGTAAGA | |
| R: GTCCGTAAAGACCACTCAACATA | ||
| PCR primer | Genome- F/R | F: AGAAGGATGATGGGGGACTGA |
| R: TGGTTTGAGCACGTTGTTGC | ||
| qRT-PCR primer | cTGFBR2-total-F/R | F: CTAAGAGACAGAGGGCGACC |
| R: ACAGCTTCTCCCTGAGAGCT | ||
| cTGFBR2-1-F/R | F: TAGAAGCAAGGAAAATGG | |
| R: CTGGCTTATGGGGATCAA | ||
| cTGFBR2-3-F/R | F: TTGGTCCTCTGGGAAATG | |
| R: AGATGGGCTTTGTAGTGC | ||
| TBP-F/R | F: GCGTTTTGCTGCTGTTATTATGAG | |
| R: TCCTTGCTGCCAGTCTGGAC |
Abbreviations: TGFBR2, transforming growth factor beta receptor Ⅱ; TBP, TATA-box binding protein.
Reverse Transcription Polymerase Chain Reaction and Genomic PCR
Reverse transcription polymerase chain reaction (RT-PCR) was used to identify cTGFBR2 transcript isoforms on the cDNAs generated from various chicken tissues. The primers are shown in Table 1. Thermal cycling was performed as follows: 5 min for the initial denaturation at 95°C, followed by 40 cycles of 15 s at 95°C, 15 s at 60°C, and 180 s at 72°C, and ending with a final extension time of 10 min at 72°C in a Master cycler nexus GSX1 (Eppendorf, Hamburg, Germany). Genomic PCR was used to detect the intragenic tandem duplication of cTGFBR2 gene on blood genomic DNAs extracted from 4 different chicken breeds (Northeast Agricultural University F2 Resource Line, NEAUHLF, AA broiler chickens, and Lindian chickens), which were stored in our laboratory. The primers are shown in Table 1. The forward primer was located in exon 3 of cTGFBR2 gene and the reverse primer was located in exon 2′. If there is an intragenic tandem duplication comprising exons 2 and 3 and intron 2 in the cTGFBR2 gene, genomic PCR amplification with this primer pair generates a 2,000-bp product, otherwise, no PCR products would be obtained. Thermal cycling was performed as follows: 5 min for the initial denaturation at 95°C, followed by 40 cycles of 30 s at 95°C, 30 s at 63°C, 150 s at 72°C, and ending with a final extension time of 10 min at 72°C in a Master cycler nexus GSX1 (Eppendorf, Hamburg, Germany). PCR products were purified, cloned into pEASY blunt simple cloning vector (TransGen, Beijing, China), and sent to Genewiz biological company (Suzhou, China) for Sanger sequencing.
Quantitative Real-Time Polymerase Chain Reaction
The relative expression of cTGFBR2-1, cTGFBR2-2, and cTGFBR2-3 were determined by quantitative real-time polymerase chain reaction (qRT-PCR) using FastStart Universal SYBR Green Master [Rox] (Roche, Basel, Switzerland). The primers are shown in Table 1. The expression levels of cTGFBR2-1 and cTGFBR2-3 were detected with primer pairs cTGFBR2-1-F/R and cTGFBR2-3-F/R, respectively. The expression level of cTGFBR2-2 was calculated by the total expression of cTGFBR2-1 plus cTGFBR2-2 detected with the primer pair cTGFBR2-total-F/R minus the cTGFBR2-1 expression detected with the primer pair cTGFBR2-1-F/R. The qRT-PCR was performed on ABI Prism 7500 sequence detection system (Applied Biosystems, Foster City, CA). The cycling conditions were used as follows: 30 s at 95°C, followed by 40 cycles of 5 s at 95°C and 30 s at 60°C. TATA-box binding protein (TBP) was used as an internal reference, and the relative gene expression was calculated using the 2−ΔCt method.
Western Blotting Analysis
The frozen tissue samples were ground into powder in liquid nitrogen by using a mortar and pestle and lysed using RIPA buffer containing 1% PMSF (Beyotime, Shanghai, China). Proteins were separated by 12% SDS-PAGE and transferred onto nitrocellulose membranes (Millipore, Bedford, MA). Then the membranes were blocked for 120 min and incubated overnight at 4°C with primary antibodies against TGFBR2 (1:1,000, Santa, Dallas, Texas) and β-actin (1:1,000, Beyotime, Shanghai, China). After the blots were rinsed with PBST 3 times, they were incubated with HRP-conjugated anti-mouse secondary antibody (1:3,000, Beyotime, Shanghai, China) for 60 min at room temperature. The blots were observed with an ECL Plus detection kit (Beyotime, Shanghai, China).
Bioinformatics Analysis
cTGFBR2 genomic sequence was downloaded from the National Center for Biotechnology Information (NCBI) website (https://www.ncbi.nlm.nih.gov/nuccore/NC_052533.1?report=fasta&from=39709134&to=39773208). Intragenic tandem duplication analysis was performed using the DNASTAR software. Open reading frame (ORF) analysis and sequence identity analysis were performed using the DNAMAN software. Sequence alignment was performed using the basic local alignment search tool (BLAST) (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Protein domain analysis was performed using SMART (http://smart.embl-heidelberg.de/). Polyadenylation signal (PAS) analysis was performed using the ITB tools (http://itbtools.ba.itb.cnr.it/utrscan).
Statistical Analysis
Results are expressed as means ± SD. Comparisons between groups were performed using the multiple t tests. Statistical differences were considered significant when P < 0.05.
RESULTS
Identification of the 5′ and 3′ Terminal Sequences of cTGFBR2 mRNA
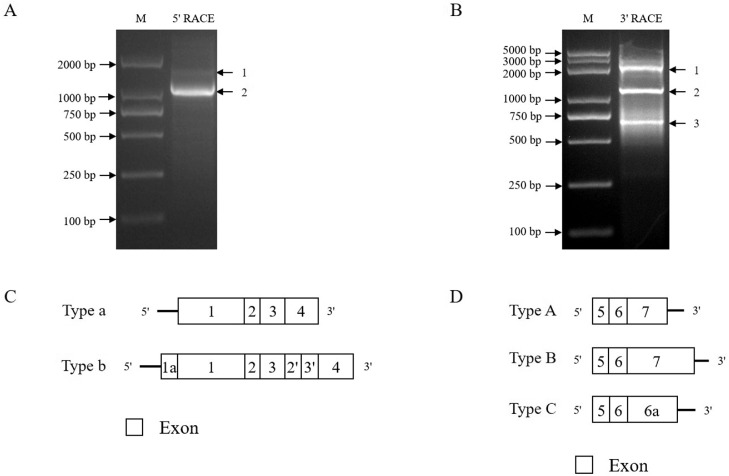
To obtain the full-length cTGFBR2 mRNA, we performed the 5′ and 3′ RACE on the pooled cDNA samples. The 5′ RACE and 3′ RACE PCR products were analyzed by electrophoresis on 2% agarose gels. The results showed that there were 2 different-sized 5′ RACE PCR products (Figure 1A) and 3 different-sized 3′ RACE PCR products (Figure 1B). All 5′ and 3′ RACE PCR products were purified, cloned and sequenced. A total of 36 recombinant plasmids containing the 5′ RACE PCR products were sequenced. The sequencing analysis identified 2 distinct 5′-terminal sequences of cTGFBR2: 1,493 (11 colonies) and 1,918 bp (25 colonies) in size, which were designated as Type a and Type b, respectively (Figure 1C). A total of 82 recombinant plasmids containing the 3′ RACE PCR products were sequenced. The sequencing analysis identified 3 distinct 3′-terminal sequences of cTGFBR2: 913 (39 colonies), 2,560 (26 colonies), and 1,640 bp (17 colonies), which were designated as Type A, Type B, and Type C, respectively (Figure 1D).
Figure 1.
Identification of the 5′ and 3′ terminal regions of cTGFBR2 transcript isoforms. (A) 5′ RACE analysis of cTGFBR2 transcript isoforms. M, marker. (B) 3′ RACE analysis of cTGFBR2 transcript isoforms. M, marker. (C) Schematic structure of the 5′-terminal region of cTGFBR2 transcript isoforms. (D) Schematic structure of the 3′-terminal regions of cTGFBR2 transcript isoforms. Exons are numbered inside the boxes.
Identification of cTGFBR2 Transcript Isoforms
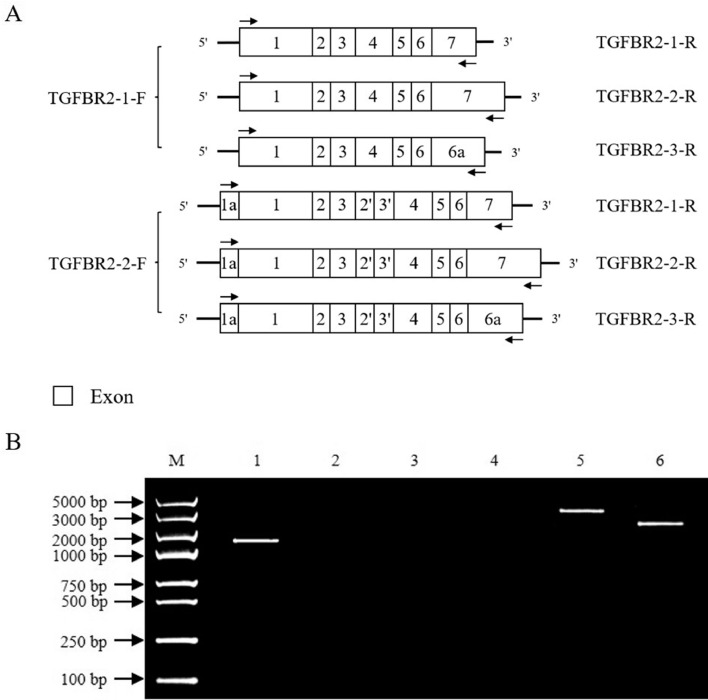
Based on the 5′ and 3′ RACE analysis results, we speculated that the cTGFBR2 gene may generates at most 6 transcript isoforms (Figure 2A). To identify cTGFBR2 transcript isoforms, we designed the transcript isoform-specific primer sets for the 6 possible transcript isoforms (Table 1 and Figure 2A) and performed RT-PCR on the pooled cDNA. The expected RT-PCR product sizes for 6 possible cTGFBR2 transcript isoforms were 1,953, 2,904, 2,360, 2,028, 3,321, and 2,777 bp, respectively. The RT-PCR products were analyzed by electrophoresis on 2% agarose gels. The RT-PCR with primer sets TGFBR2-1-F/TGFBR2-1-R, TGFBR2-2-F/TGFBR2-2-R, and TGFBR2-2-F/TGFBR2-3-R yielded the expected RT-PCR products, but RT-PCR with other 3 primer sets (TGFBR2-1-F/TGFBR2-2-R, TGFBR2-1-F/TGFBR2-3-R, and TGFBR2-2-F/TGFBR2-1-R) yielded no RT-PCR products (Figure 2B). The 3 expected RT-PCR products were cloned and a total of 180 recombinant plasmids were sequenced. The sequencing results showed, as expected, the sizes of these 3 RT-PCR products were 1,953 (52 colonies), 3,321 (49 colonies), and 2,777 bp (79 colonies), respectively. Collectively, these results suggest that the cTGFBR2 gene produces 3 transcript isoforms, which were designated as cTGFBR2-1, cTGFBR2-2, and cTGFBR2-3. Their nucleotide sequences were deposited in GenBank database under accession numbers ON164837, ON164838, and ON164839, respectively.
Figure 2.
Identification of cTGFBR2 transcript isoforms. (A) Schematic structure of 6 possible cTGFBR2 transcript isoforms and schematic location of the 6 transcript isoform-specific primer sets used for the identification of cTGFBR2 transcript isoforms. (B) RT-PCR identification of cTGFBR2 transcript isoforms. RT-PCR was performed on pooled cDNA to identify cTGFBR2 transcript isoforms with the 6 transcript-specific primer sets. RT-PCR products were analyzed by 2% agarose gel electrophoresis. M, marker. Lane 1, RT-PCR with primer set TGFBR2-1-F and TGFBR2-1-R; Lane 2, RT-PCR with primer set TGFBR2-1-F and TGFBR2-2-R; Lane 3, RT-PCR with primer set TGFBR2-1-F and TGFBR2-3-R; Lane 4, RT-PCR with primer set TGFBR2-2-F and TGFBR2-1-R; Lane 5, RT-PCR with primer set TGFBR2-2-F and TGFBR2-2-R; Lane 6, RT-PCR with primer set TGFBR2-2-F and TGFBR2-3-R.
There are 2 cTGFBR2 transcript sequences (NM_205428.2 and XM_015281321.4) in GenBank database. Sequence identity analysis using DNAMAN software showed that cTGFBR2-1 shared 57.88 and 50.36% nucleotide identities, respectively, with the 2 reported cTGFBR2 transcript sequences; cTGFBR2-2 shared 86.08 and 97.18% nucleotide identities, respectively, and cTGFBR2-3 shared 46.71 and 57.88% nucleotide identities, respectively. The 5′ UTR sizes of the 3 cTGFBR2 transcript isoforms were 140, 223, and 223 bp, respectively. The 3′ UTR sizes of the 3 cTGFBR2 transcript isoforms were 535, 2,196, and 1,413 bp, respectively. The coding region sequence of cTGFBR2-1 and cTGFBR2-2 were completely identical to that of NM_205428.2 and XM_015281321.4, respectively. The cTGFBR2-1 contained an additional 23 nucleotides in 5′ UTR and lacked 1,663 nucleotides in 3′ UTR, compared with the NM_205428.2. The cTGFBR2-2 contained an additional 106 nucleotides in 5′ UTR and lacked 2 nucleotides in 3′ UTR, compared with the XM_015281321.4. The cTGFBR2-3 contained an additional 83 nucleotides in 5′ UTR, and an additional 878 nucleotides in 3′ UTR, compared with the cTGFBR2-1. The cTGFBR2-3 possessed the same 5′ UTR as cTGFBR2-2 but lacked 783 nucleotides in 3′ UTR relative to the cTGFBR2-2.
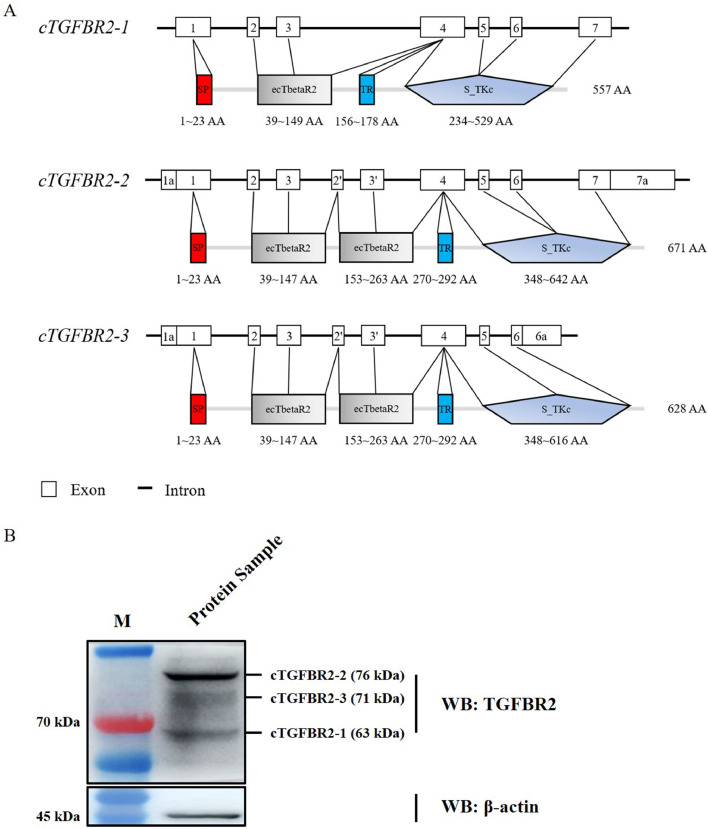
ORF analysis using DNAMAN software showed that these 3 identified cTGFBR2 transcript isoforms could encode 3 protein isoforms: cTGFBR2-1 (557 amino acids), cTGFBR2-2 (671 amino acids), and cTGFBR2-3 (628 amino acids; Figure 3A). Comparison of amino acid sequences of cTGFBR2-1, cTGFBR2-2, cTGFBR2-3 with the published chicken TGFBR2 (cTGFBR2) revealed that, cTGFBR2-1 shared 100% identity with NP_990759.1 (encoded by NM_205428.2), cTGFBR2-2 shared 100% identity with XP_015136807.1 (encoded by XM_015281321.4), but cTGFBR2-3 shared 74.52 and 91.51% identity with NP_990759.1 and XP_015136807.1, respectively. By comparison, cTGFBR2-3 contained an additional 114 amino acids at its N-terminal, compared with the cTGFBR2-1, while lacked the C-terminal 43 amino acids, compared with the cTGFBR2-1 and cTGFBR2-2 (Figure S1).
Figure 3.
Structural analysis and detection of cTGFBR2 protein isoforms. (A) The domains of cTGFBR2 isoforms were predicted using SMART (http://smart.embl-heidelberg.de/). SP, signal peptide; ecTbetaR2, Transforming growth factor beta receptor 2 ectodomain; TR, Transmembrane region; S_TKc, Serine/threonine-protein kinases. (B) Western blotting analysis of cTGFBR2 protein isoform expression in the pooled tissue protein lysate (abdominal fat, heart, liver, kidney, gizzard, and muscle). The pooled tissue protein lysate was generated from the 4-wk-old AA broilers and immunoblotted with the indicated antibodies.
The protein structural domain analysis using the SMART database showed that, similar to human and mouse TGFBR2, the 3 cTGFBR2 isoforms consisted of 4 primary structural domains: the signal peptide (SP), transforming growth factor beta receptor 2 ectodomain (ecTbetaR2), transmembrane region (TR), and serine/threonine-protein kinases (S_TKc; Figure 3A). By comparison, cTGFBR2-2 and cTGFBR2-3 have 1 more ecTbetaR2 domain than cTGFBR2-1 (Figure 3A). To confirm the presence of these 3 cTGFBR2 isoforms, we performed western blotting using TGFBR2 monoclonal antibody (sc-17799, Santa, Dallas, Texas) to detect cTGFBR2 in the pooled protein sample consisting of abdominal fat, heart, liver, kidney, gizzard, muscle in equal proportions. Western blotting detected the predicted 63, 76, and 71 kDa protein bands of cTGFBR2 isoforms (Figure 3B), suggesting that cTGFBR2 gene indeed encodes 3 protein isoforms.
Characterization of cTGFBR2 Genomic Structure
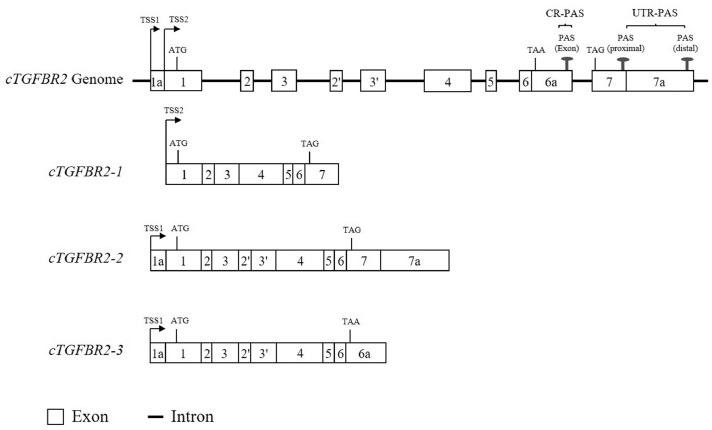
To determine the genomic structure of the cTGFBR2 gene, we aligned these 3 cTGFBR2 transcript isoform sequences against chicken genome sequence (GRCg7b) using the BLAST at the NCBI. As expected, these 3 cTGFBR2 transcript isoforms were mapped to the cTGFBR2 locus on chromosome 2 in the Gallus gallus genome database (GRCg7b). The cTGFBR2 gene spanned a region of approximately 64-kb (Chr.2: 39,709,028-39,773,208). The nucleotide lengths of each exon and intron of cTGFBR2 gene are as follows: exon 1a/1 (83/222 bp), intron 1 (26,763 bp), exon 2 (151 bp), intron 2 (1,517 bp), exon 3 (191 bp), intron 3 (1,747 bp), exon 2′ (151 bp), intron 4 (1,622 bp), exon 3′ (191 bp), intron 5 (11,718 bp), exon 4 (800 bp), intron 6 (4,245 bp), exon 5 (142 bp), intron 7 (8,713 bp), exons 6/6a (128/1,464 bp), intron 8 (1,955 bp), and exons 7/7a (715/1,661 bp; Figure 4). The cTGFBR2-1 consisted of 7 exons (exons 1-7), cTGFBR2-2 consisted of 9 exons (exon 1a/1, exons 2-6, exons 2′ and 3′, exons 7/7a), and cTGFBR2-3 consisted of 8 exons (exon 1a/1, exons 2-5, exons 2′ and 3′, exons 6/6a; Figure 4). All the intron/exon splice junctions followed the GT-AG rule.
Figure 4.
Schematic structure of cTGFBR2 genomic and transcript isoforms. Abbreviations: ATG, start codon; TAA and TAG, stop codon; TSS, transcription start site; PAS, polyadenylation signal; CR-PAS, coding region PAS; UTR-PAS, untranslated region PAS.
Furthermore, BLAST analysis revealed that the cTGFBR2 gene had 2 alternative transcription start sites (TSS1 and TSS2), which were 82 bp apart. The cTGFBR2-2 and cTGFBR2-3 shared TSS1 and cTGFBR2-1 employed TSS2 (Figure 4). These 3 cTGFBR2 transcript isoforms shared the same start codon (ATG) on exon 1, the cTGFBR2-1 and cTGFBR2-2 shared the same stop codon (TAG) located in exon 7, whereas cTGFBR2-3 positioned its stop codon (TAA) in exon 6a (Figure 4). Sequence analysis showed that cTGFBR2 gene had 3 alternative 3′ UTRs (535, 2,196, and 1,413 bp, respectively), indicating that the cTGFBR2 gene undergoes alternative polyadenylation (APA). We used ITB tools to identify the PASes for the 3 alternative 3′ UTRs. The results showed that as expected, cTGFBR2 gene had 3 PASes (AATAAA), one of the PASes, located in exon 6a, was 6 bp upstream of the poly (A) site of cTGFBR2-3 (Figure 4). The other 2 PASes, located in the exons 7 and 7a, were 9 and 16 bp, respectively, upstream of the poly (A) sites of cTGFBR2-1 and cTGFBR2-2 (Figure 4). The PAS in exon 6a was a coding region PAS (CR-PAS), which led to the production of the novel protein isoform cTGFBR2-3. The other 2 PASes were the untranslated region PAS (UTR-PAS), and did not affect the coding capacity of cTGFBR2, but caused the different 3′ UTR lengths between cTGFBR2-1 and cTGFBR2-2.
Intragenic Duplication of cTGFBR2 Gene
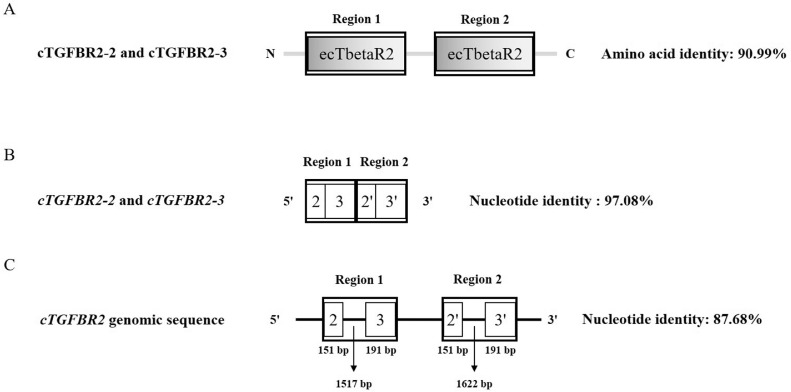
The structural domain analysis showed that the cTGFBR2-1 protein isoform had 1 ecTbetaR2 domain, while both cTGFBR2-2 and cTGFBR2-3 protein isoforms had 2 tandem ecTbetaR2 domains, which had high amino acid sequence identity (90.99%; Figure 5A). Using DNAMAN software, we analyzed the mRNA sequences coding the 2 tandem ecTbetaR2 domains in cTGFBR2-2 and cTGFBR2-3. The results showed that the coding sequences for the 2 tandem ecTbetaR2 domains (324 and 330 bp, respectively) shared 97.08% nucleotide identity (Figure 5B).
Figure 5.
Schematic of cTGFBR2 intragenic duplication. (A) Schematic of cTGFBR2 with a tandem repeat of 2 ecTbetaR2 domains. (B) Schematic of cTGFBR2 transcript isoforms with a tandem duplication of exons 2 and 3. (C) Schematic of cTGFBR2 genomic intragenic duplication. The black square represents the region of intragenic duplication.
Protein domain repeats derive from the intragenic tandem duplication (Björklund et al., 2010). To verify the presence of intragenic tandem duplication in the cTGFBR2 gene locus, we performed duplication analysis of the cTGFBR2 genomic sequence (Chr.2: 39,709,028-39,773,208, 64-kb) using DNASTAR software. The results displayed that the cTGFBR2 genomic sequence had an intragenic tandem duplication of 5.5-kb (Chr.2: 39,736,096-39,741,665). Duplication region 1 (Chr.2: 39,736,096-39,737,954) was 1,859 bp in length and consisted of exons 2 and 3, and intron 2. Duplication region 2 (Chr.2: 39,739,702-39,741,665) was 1,964 bp in length and consisted of exons 2′ and 3′, and intron 4. These 2 intragenic duplication regions shared 87.68% nucleotide identity (Figure 5C), and the nucleotide identities between exon 2 and exon 2′, exon 3 and exon 3′, intron 2 and intron 4, were 93.38, 100.00, and 85.71%, respectively. Duplication region 1 encoded the first ecTbetaR2 domain of cTGFBR2-2 and cTGFBR2-3, and duplication region 2 encoded the second ecTbetaR2 domain of cTGFBR2-2 and cTGFBR2-3.
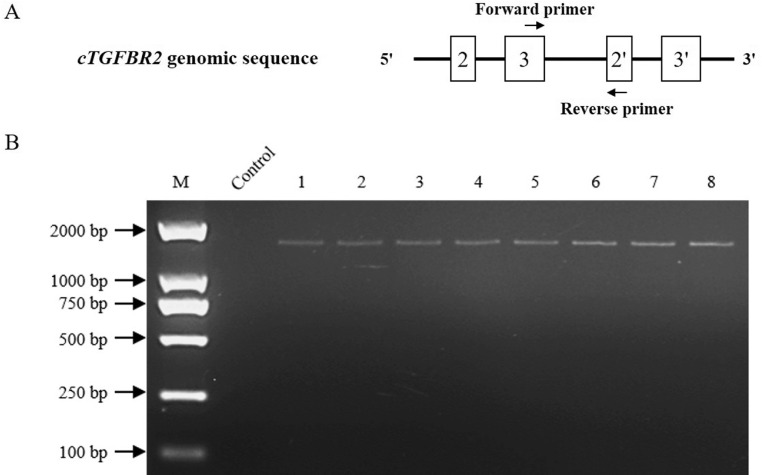
To test whether this cTGFBR2 intragenic tandem duplication is commonly present in chickens, we performed the duplication-specific genomic PCR to detect the intragenic tandem duplication in 4 chicken breeds: Northeast Agricultural University F2 Resource Line, NEAUHLF, AA broiler chickens, and Lindian chickens (a local Chinese chicken breed). The genomic PCR results showed that a single band of the expected size (2,000-bp) was amplified using the duplication-specific primer pair Genome-F/R in all the 4 tested chicken breeds (Figures 6A and 6B), and Sanger sequencing further confirmed that cTGFBR2 gene harbored the intragenic tandem duplication in all the tested chickens. To test whether this intragenic tandem duplication is chicken-specific, we performed bioinformatics analysis of TGFBR2 genomic, mRNA, and protein sequences in various animal species including goose, duck, horse, penguin, eagle, human, and mouse. The bioinformatics analysis showed that there was no intragenic tandem duplication in TGFBR2 genomic and mRNA sequences, and no tandem domain repeats in TGFBR2 protein in these tested animal species. Taken together, these results suggest that the tandem domain repeat and intragenic tandem duplication of the TGFBR2 gene is chicken-specific.
Figure 6.
PCR identification of cTGFBR2 intragenic duplication. (A) Schematic locations of PCR primers used for the identification of cTGFBR2 intragenic duplication. (B) Genomic PCR identification of the intragenic duplication of cTGFBR2 in various chicken breeds. M, marker; Control, water as the negative control; Lanes 1 and 2, Northeast Agricultural University F2 Resource Line; Lanes 3 and 4, Lindian chicken breed; Lanes 5 and 6, NEAUHLF; Lanes 7 and 8, AA broiler chicken.
Tissue Expression Patterns of cTGFBR2 Transcript Isoforms
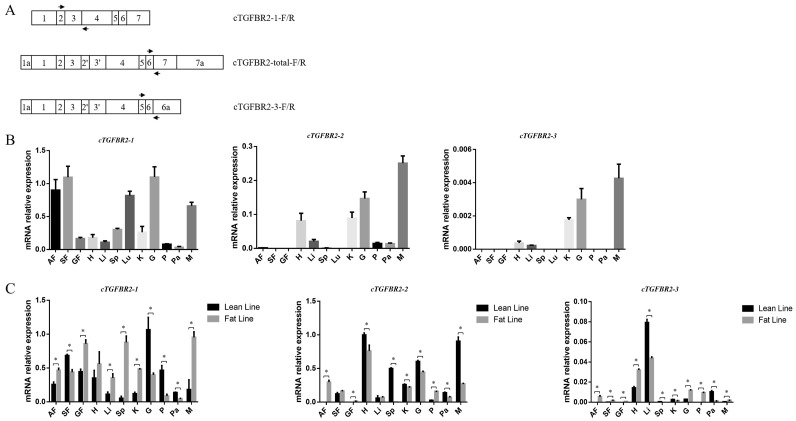
To understand the functions of these 3 cTGFBR2 transcript isoforms, we performed tissue expression profiling of the 3 cTGFBR2 transcript isoforms in various tissues of the 4-wk-old AA broiler chickens using qRT-PCR with the primer pairs (Table 1, Figure 7A). The results showed that the cTGFBR2-1 was ubiquitously expressed in all the tested chicken tissues, with high expression in abdominal fat, subcutaneous fat, lung, gizzard, and muscle, but low expression in gizzard fat, heart, liver, spleen, kidney, proventriculus, and pancreas. The cTGFBR2-2 was highly expressed in the heart, kidney, gizzard, and muscle, whereas low or no expression was detected in abdominal fat, subcutaneous fat, gizzard fat, liver, spleen, lung, proventriculus, and pancreas. The cTGFBR2-3 was weakly expressed in all the tested tissues, with no expression in abdominal fat, subcutaneous fat, gizzard fat, spleen, lung, proventriculus, and pancreas (Figure 7B). These results suggest that the 3 cTGFBR2 transcript isoforms may play different roles in different tissues and organs.
Figure 7.
Tissue expression profiling of cTGFBR2 transcript isoforms in various chicken tissues. (A) Schematic locations of qRT-PCR primers used in the tissue expression profiling of cTGFBR2 transcript isoforms. (B) Expression profiling of the 3 cTGFBR2 transcript isoforms in various tissues of the 4-wk-old AA chickens. (C) Expression comparison of the 3 cTGFBR2 transcript isoforms in various tissues of fat and lean lines of NEAUHLF. Abbreviations: AF, abdominal fat; SF, subcutaneous fat; GF, gizzard fat; H, heart; Li, liver; Sp, spleen; Lu, lung; K, kidney; G, gizzard; P, proventriculus; Pa, pancreas; M, muscle.
To confirm the tissue expression profiling results, we also performed tissue expression profiling of the 3 cTGFBR2 transcript isoforms in the 7-wk-old chickens of NEAUHLF. In both lean and fat lines, only cTGFBR2-1 was ubiquitously expressed in all the tested tissues (Figure 7C). Comparison of gene expression between the lean and fat lines revealed that cTGFBR2-1 was significantly differentially expressed in all the tested tissues except the heart, and cTGFBR2-2 was significantly differentially expressed in all the tested tissues except the subcutaneous fat and liver, and cTGFBR2-3 was significantly differentially expressed in all the tested tissues (Figure 7C, P < 0.05).
It is worth noting that, in the fat line, the 3 cTGFBR2 transcript isoforms were expressed to varying degrees in all the 3 tested fat tissues (Figure 7C). However, in the lean line, only cTGFBR2-1 was expressed in all the 3 tested fat tissues, cTGFBR2-2 and cTGFBR2-3 were only expressed in subcutaneous fat (Figure 7C). These results suggest that the expression of the 3 cTGFBR2 transcript isoforms is differentially regulated in adipose tissues at different anatomic sites.
DISCUSSION
In the present study, we demonstrated that cTGFBR2 gene produces 3 transcript isoforms (cTGFBR2-1, cTGFBR2-2, cTGFBR2-3) though alternative transcription initiation, splicing, and polyadenylation and these transcript isoforms encode 3 cTGFBR2 protein isoforms (cTGFBR2-1, cTGFBR2-2, cTGFBR2-3). Among these 3 cTGFBR2 transcript isoforms, cTGFBR2-3 was a novel cTGFBR2 transcript isoform, which encodes a novel cTGFBR2 protein isoform.
Intragenic duplication plays an important role in the evolution, inheritance and variation of life, and involves gene expression, transcription regulation, chromosome construction and physiological metabolism (Brahmachari et al., 1995; Saier Jr, 2016; Nava Rodrigues et al., 2019; Xu et al., 2022). For example, a recent study showed the crest phenotype in domestic chicken is caused by a 197 bp duplication in the intron of HOXC10 (Li et al., 2021). Many eukaryotic proteins contain tandem domain repeats (Light et al., 2012), which are often caused by intragenic duplication (Björklund et al., 2010). In the present study, we demonstrated that the intragenic duplication resulted in a tandem repeat of 2 tandem ecTbetaR2 domains in cTGFBR2. Protein domain repeats have 2 modes of evolution: 1) the conservative mode, whereby repeats occur before species differentiation and thus the same repeat structure is maintained within different species (Hughes, 2000); 2) the concerted mode, whereby repeats are homogenized within species by independent duplication after species differentiation (Thomas et al., 1997). In the present study, we demonstrated that the intragenic tandem duplication of the TGFBR2 gene is chicken-specific. We reason that intragenic tandem duplication of cTGFBR2 gene occurs after species differentiation, and has evolved in a concerted fashion in chickens.
It has been shown that there are marked species differences in TGF-β signaling pathway (Kruithof et al., 2012). For example, chickens had significantly higher TGF-β signaling activity in lower jaw skeleton than ducks (Smith et al., 2022). Chickens and mice exhibited distinct TGF-β ligand expression patterns during the epithelial to mesenchymal cell transformation (Azhar et al., 2003; Molin et al., 2003) and had contradictory requirements for the TGF-β ligands and receptors during the endocardial-to-mesenchymal cell transformation (Jiao et al., 2006; Yamagishi et al., 2012). In the present study, we provided evidence that TGFBR2, the core receptor of the TGF-β signaling pathway, differs in structure between chickens and other species. Our finding might partially explain the differences in TGF-β signaling pathway activity between chickens and other species.
APA is one of the important post-transcriptional mechanisms and plays an important regulatory role in many biological processes (Nourse et al., 2020). PAS is divided into CR-PAS and UTR-PAS, of which UTR-PAS is the most common (Chen et al., 2017). UTR-PAS generates various transcript isoforms with different 3′ UTR lengths (Jambhekar and Derisi, 2007). CR-PAS causes genes to encode different C-terminal protein isoforms, and some of the isoforms may lack functional domains (Tian and Manley, 2017). In the present study, we for the first time demonstrated that cTGFBR2 gene undergoes APA and that cTGFBR2 gene possesses both UTR-PAS and CR-PAS. Similarly, chicken growth hormone receptor (GHR) gene has been shown to have both CR-PAS and UTR-PAS (Lau et al., 2007). TGFBR2 is known to play vital roles in multiple developmental processes. Considering the importance of APA in posttranscriptional regulation, it is essential to explore the roles and molecular mechanisms of APA in cTGFBR2.
Tissue expression profiling analysis showed that the cTGFBR2-1 was ubiquitously expressed in all the tested tissues of the 4-wk-old AA broiler chickens and the 7-wk-old broiler chickens of NEAUHAL, which was similar to the tissue expression pattern of human TGFBR2 (Fagerberg et al., 2014), suggesting that cTGFBR2-1 and human TGFBR2 may have similar functions in tissue development. In the present study, our results showed that cTGFBR2-1 in 10 tested tissues, cTGFBR2-2 in 9 tested tissues, and cTGFBR2-3 in all the tested tissues, displayed significantly differential expression levels between the fat and lean lines of NEAUHAL, suggesting that these 3 cTGFBR2 transcript isoforms may contribute to various trait differences between the fat and lean lines of NEAUHAL.
Intriguingly, in the present study, we found that the expression of these cTGFBR2 transcript isoforms in the abdominal fat and gizzard fat was significantly higher in the fat line than in the lean line (Figure 7C). It has been known that activation of TGF-β signaling pathway inhibits adipogenesis, and the expression of adipogenesis-related genes such as PPARγ (Feng and Derynck, 2005; Hill, 2009; Li and Wu, 2020). TGFBR2 knockdown in hASCs cells significantly increased the expression of PPARγ and C/EBPα and promoted adipogenesis (Kim et al., 2009). Our previous study found that PPARγ mRNA expression in chicken abdominal fat was significantly higher in the fat line than in the lean line at 7-wk-old broiler chickens of NEAUHAL (Sun et al., 2014). Based on our previous study, and the inhibitory role of TGF-β signaling pathway in mammalian adipogenesis and adipogenesis-related genes expression, we supposed that the expression of the 3 cTGFBR2 transcript isoforms in abdominal fat would be lower in the fat line than in the lean line. However, our expression results contradicted this supposition. This unexpected result may be explained by 2 reasons. First, it has been shown that TGFBR2 gene expression was promoted by activation of PI3K/Akt signaling pathway (Budi et al., 2015), and that PI3K/Akt pathway activity was higher in obese individuals than in normal individuals (Chi et al., 2017). Hence, PI3K/Akt signaling pathway activity in the abdominal fat may be higher in the fat line than in the lean line, leading to increased expression of cTGFBR2 transcript isoforms in the fat line relative to the lean line. Second, adipose tissue TGFBR2 expression may be differentially regulated between chickens and mammals.
In conclusion, we defined cTGFBR2 genomic structure, characterized cTGFBR2 transcript and protein isoforms, and identified an intragenic tandem duplication within cTGFBR2. To the best of our knowledge, this is the first report of the intragenic tandem duplication within TGFBR2 gene. Our findings pave the way for further investigating the roles and regulation of cTGFBR2 gene.
ACKNOWLEDGMENTS
This work was supported by National 973 Project of China (No.2009CB941604), China National Natural Science Foundation Grant (No.31872346), and the China Agriculture Research System of MOF and MARA (No. CARS-41).
Disclosures
We declare no conflicts of interest.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.psj.2022.102169.
Appendix. Supplementary materials
Figure S1. The alignment of protein sequences of the 3 cTGFBR2 protein isoforms (cTGFBR2-1, cTGFBR2-1, and cTGFBR2-3). The red highlighting indicates the amino acid sequences of the signal peptide in 3 protein isoforms. The gray highlighting indicates the amino acid sequences of the unique ecTbetaR2 in cTGFBR2-1 and the first ecTbetaR2 in cTGFBR2-2 and cTGFBR2-3. The yellow highlighting indicates the amino acid sequences of the second ecTbetaR2 in cTGFBR2-2 and cTGFBR2-3. The blue highlighting indicates the amino acid sequences of the transmembrane region in 3 cTGFBR2 protein isoforms. The green highlighting indicates the amino acid sequences of the serine/threonine-protein kinases in 3 cTGFBR2 protein isoforms. The black square indicates the amino acid sequence differences among 3 protein isoforms (cTGFBR2-1, cTGFBR2-2, and cTGFBR2-3).
REFERENCES
- Azhar M., Schultz J.E.J., Grupp I., Dorn G.W., Meneton P., Molin D.G.M., Gittenberger-de Groot A.C., Doetschman T. Transforming growth factor beta in cardiovascular development and function. Cytokine. Growth. Factor. Rev. 2003;14:391–407. doi: 10.1016/s1359-6101(03)00044-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolio M.S., La Colla A., Carrea A., Romo A., Canziani G., Echarte S.M., Campisano S., Barletta G.P., Monzon A.M., Rodriguez T.M., Chisari A.N., Dewey R.A. A novel splice variant of human TGF-β type II receptor encodes a soluble protein and its Fc-Tagged version prevents liver fibrosis in vivo. Front. Cell. Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.690397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björklund A.K., Light S., Sagit R., Elofsson A. Nebulin: a study of protein repeat evolution. J. Mol. Biol. 2010;402:38–51. doi: 10.1016/j.jmb.2010.07.011. [DOI] [PubMed] [Google Scholar]
- Brahmachari S.K., Meera G., Sarkar P.S., Balagurumoorthy P., Tripathi J., Raghavan S., Shaligram U., Pataskar S. Simple repetitive sequences in the genome: structure and functional significance. Electrophoresis. 1995;16:1705–1714. doi: 10.1002/elps.11501601283. [DOI] [PubMed] [Google Scholar]
- Budi E.H., Muthusamy B.P., Derynck R. The insulin response integrates increased TGF-β signaling through Akt-induced enhancement of cell surface delivery of TGF-β receptors. Sci. Signal. 2015;8:ra96. doi: 10.1126/scisignal.aaa9432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Jia Q., Song Y., Fu H., Wei G., Ni T. Alternative polyadenylation: methods, findings, and impacts. Genom. Proteom. Bioinf. 2017;15:287–300. doi: 10.1016/j.gpb.2017.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi Y., Li J., Li N., Chen Z., Ma L., Peng W., Pan X., Li M., Yu W., He X., Geng B., Cui Q., Liu Y., Yang J. FAM3A enhances adipogenesis of 3T3-L1 preadipocytes via activation of ATP-P2 receptor-Akt signaling pathway. Oncotarget. 2017;8:45862–45873. doi: 10.18632/oncotarget.17578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooley J.R., Yatskievych T.A., Antin P.B. Embryonic expression of the transforming growth factor beta ligand and receptor genes in chicken. Dev. Dyn. 2014;243:497–508. doi: 10.1002/dvdy.24085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumbrava M.G., Lacanlale J.L., Rowan C.J., Rosenblum N.D. Transforming growth factor beta signaling functions during mammalian kidney development. Pediatr. Nephrol. 2021;36:1663–1672. doi: 10.1007/s00467-020-04739-5. [DOI] [PubMed] [Google Scholar]
- Fagerberg L., Hallstrom B.M., Oksvold P., Kampf C., Djureinovic D., Odeberg J., Habuka M., Tahmasebpoor S., Danielsson A., Edlund K., Asplund A., Sjostedt E., Lundberg E., Szigyarto C.A., Skogs M., Takanen J.O., Berling H., Tegel H., Mulder J., Nilsson P., Schwenk J.M., Lindskog C., Danielsson F., Mardinoglu A., Sivertsson Å., von Feilitzen K., Forsberg M., Zwahlen M., Olsson I., Navani S., Huss M., Nielsen J., Ponten F., Uhlén M. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteomics. 2014;13:397–406. doi: 10.1074/mcp.M113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X.H., Derynck R. Specificity and versatility in TGF-β signaling through smads. Annu. Rev. Cell. Dev. Biol. 2005;21:659–693. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- Hill C.S. Nucleocytoplasmic shuttling of smad proteins. Cell. Res. 2009;19:36–46. doi: 10.1038/cr.2008.325. [DOI] [PubMed] [Google Scholar]
- Hirai R., Fijita T. A human transforming growth factor-β type II receptor that contains an insertion in the extracellular domain. Exp. Cell. Res. 1996;223:135–141. doi: 10.1006/excr.1996.0066. [DOI] [PubMed] [Google Scholar]
- Hughes A.L. Modes of evolution in the protease and kringle domains of the plasminogen-prothrombin family. Mol. Phylogenet. Evol. 2000;14:469–478. doi: 10.1006/mpev.1999.0685. [DOI] [PubMed] [Google Scholar]
- Jambhekar A., Derisi J.L. Cis-acting determinants of asymmetric, cytoplasmic RNA transport. RNA. 2007;13:625–642. doi: 10.1261/rna.262607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James J.M., Nalbandian A., Mukouyama Y.S. TGFβ signaling is required for sprouting lymphangiogenesis during lymphatic network development in the skin. Development. 2013;140:3903–3914. doi: 10.1242/dev.095026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao K., Langworthy M., Batts L., Brown C.B., Moses H.L., Baldwin H.S. Tgfβ signaling is required for atrioventricular cushion mesenchyme remodeling during in vivo cardiac development. Development. 2006;133:4585–4593. doi: 10.1242/dev.02597. [DOI] [PubMed] [Google Scholar]
- Kim Y.J., Hwang S.J., Bae Y.C., Jung J.S. MiR-21 regulates adipogenic differentiation through the modulation of TGF-β signaling in mesenchymal stem cells derived from human adipose tissue. Stem. Cells. 2009;27:3093–3102. doi: 10.1002/stem.235. [DOI] [PubMed] [Google Scholar]
- Krishnaveni M.S., Hansen J.L., Seeger W., Morty R.E., Sheikh S.P., Eickelberg O. Constitutive homo- and hetero-oligomerization of TβRII-B, an alternatively spliced variant of the mouse TGF-β type II receptor. Biochem. Biophys. Res. Commun. 2006;351:651–657. doi: 10.1016/j.bbrc.2006.10.083. [DOI] [PubMed] [Google Scholar]
- Kruithof B.P.T., Duim S.N., Moerkamp A.T., Goumans M.J. TGFβ and BMP signaling in cardiac cushion formation: lessons from mice and chicken. Differentiation. 2012;84:89–102. doi: 10.1016/j.diff.2012.04.003. [DOI] [PubMed] [Google Scholar]
- Lau J.S., Yip C.W., Law K.M., Leung F.C. Cloning and characterization of chicken growth hormone binding protein (cGHBP) Domest. Anim. Endocrin. 2007;33:107–121. doi: 10.1016/j.domaniend.2006.04.012. [DOI] [PubMed] [Google Scholar]
- Li J., Lee M.O., Davis B.W., Wu P., Hsieh Li S.M., Chuong C.M., Andersson L. The crest phenotype in domestic chicken is caused by a 197 bp duplication in the intron of HOXC10. G3 (Bethesda) 2021;11:jkaa048. doi: 10.1093/g3journal/jkaa048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.N., Wu J.F. TGF-β/SMAD signaling regulation of mesenchymal stem cells in adipocyte commitment. Stem. Cell. Res. Ther. 2020;11:41. doi: 10.1186/s13287-020-1552-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light S., Sagit R., Ithychanda S.S., Qin J., Elofsson A. The evolution of filamin-a protein domain repeat perspective. J. Struct. Biol. 2012;179:289–298. doi: 10.1016/j.jsb.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lönn P., Morén A., Raja E., Dahl M., Moustakas A. Regulating the stability of TGFβ receptors and smads. Cell. Res. 2009;19:21–35. doi: 10.1038/cr.2008.308. [DOI] [PubMed] [Google Scholar]
- Molin D.G., Bartram U., Van der Heiden K., Van Iperen L., Speer C.P., Hierck B.P., Poelmann R.E., Gittenberger-de-Groot A.C. Expression patterns of Tgfβ1-3 associate with myocardialisation of the outflow tract and the development of the epicardium and the fibrous heart skeleton. Dev. Dyn. 2003;227:431–444. doi: 10.1002/dvdy.10314. [DOI] [PubMed] [Google Scholar]
- Moustakas A., Heldin C.H. The regulation of TGFβ signal transduction. Development. 2009;136:3699–3714. doi: 10.1242/dev.030338. [DOI] [PubMed] [Google Scholar]
- Nava Rodrigues D., Casiraghi N., Romanel A., Crespo M., Miranda S., Rescigno P., Figueiredo I., Riisnaes R., Carreira S., Sumanasuriya S., Gasperini P., Sharp A., Mateo J., Makay A., McNair C., Schiewer M., Knudsen K., Boysen G., Demichelis F., de Bono J.S. RB1 heterogeneity in advanced metastatic castration-resistant prostate cancer. Clin. Cancer. Res. 2019;25:687–697. doi: 10.1158/1078-0432.CCR-18-2068. [DOI] [PubMed] [Google Scholar]
- Nourse J., Spada S., Danckwardt S. Emerging roles of RNA 3′-end cleavage and polyadenylation in pathogenesis, diagnosis and therapy of human disorders. Biomolecules. 2020;10:915. doi: 10.3390/biom10060915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardali E., Goumans M.J., ten Dijke P. Signaling by members of the TGF-β family in vascular morphogenesis and disease. Trends. Cell. Biol. 2010;20:556–567. doi: 10.1016/j.tcb.2010.06.006. [DOI] [PubMed] [Google Scholar]
- Peters S.B., Wang Y., Serra R. Tgfbr2 is required in Osterix expressing cells for postnatal skeletal development. Bone. 2017;97:54–64. doi: 10.1016/j.bone.2016.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier M.H., Jr Transport protein evolution deduced from analysis of sequence, topology and structure. Curr. Opin. Struct. Biol. 2016;38:9–17. doi: 10.1016/j.sbi.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S.S., Chu D., Qu T., Aggleton J.A., Schneider R.A. Species-specific sensitivity to TGFβ signaling and changes to the Mmp13 promoter underlie avian jaw development and evolution. eLife. 2022;11:e66005. doi: 10.7554/eLife.66005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y.N., Gao Y., Qiao S.P., Wang S.Z., Duan K., Wang Y.X., Li H., Wang N. Epigenetic DNA methylation in the promoters of peroxisome proliferator-activated receptor γ in chicken lines divergently selected for fatness. J. Anim. Sci. 2014;92:48–53. doi: 10.2527/jas.2013-6962. [DOI] [PubMed] [Google Scholar]
- Thomas G.H., Newbern E.C., Korte C.C., Bales M.A., Muse S.V., Clark A.G., Kiehart D.P. Intragenic duplication and divergence in the spectrin superfamily of proteins. Mol. Biol. Evol. 1997;14:1285–1295. doi: 10.1093/oxfordjournals.molbev.a025738. [DOI] [PubMed] [Google Scholar]
- Tian B., Manley J.L. Alternative polyadenylation of mRNA precursors. Nat. Rev. Mol. Cell. Biol. 2017;18:18–30. doi: 10.1038/nrm.2016.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Ark A., Cao J., Li X. TGF-β receptors: in and beyond TGF-β signaling. Cell. Signal. 2018;52:112–120. doi: 10.1016/j.cellsig.2018.09.002. [DOI] [PubMed] [Google Scholar]
- Xu F., Aref-Eshghi E., Wu J., Schubert J., Wertheim G., Bhatti T., Pogoriler J., Patel M., Cao K., Long A., Fan Z., Denenberg E.H., Fanning E.A., Wilmoth D.M., Luo M., Conlin L.K., Dain A.S., Zelley K., Baldino S., Balamuth N., MacFarland S., Li M.M., Zhong Y. A novel TP53 tandem duplication in a child with Li-Fraumeni syndrome. Cold. Spring. Harb. Mol. Case. Stud. 2022;8 doi: 10.1101/mcs.a006181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P., Liu J., Derynck R. Post-translational regulation of TGF-β receptor and smad signaling. FEBS. Lett. 2012;586:1871–1884. doi: 10.1016/j.febslet.2012.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi T., Ando K., Nakamura H., Nakajima Y. Expression of the Tgfβ2 gene during chick embryogenesis. Anat. Rec. (Hoboken). 2012;295:257–267. doi: 10.1002/ar.22400. [DOI] [PubMed] [Google Scholar]
- Yuan J., Yi K., Yang L. TGFBR2 regulates hedgehog pathway and cervical cancer cell proliferation and migration by mediating SMAD4. J. Proteome. Res. 2020;19:3377–3385. doi: 10.1021/acs.jproteome.0c00239. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. The alignment of protein sequences of the 3 cTGFBR2 protein isoforms (cTGFBR2-1, cTGFBR2-1, and cTGFBR2-3). The red highlighting indicates the amino acid sequences of the signal peptide in 3 protein isoforms. The gray highlighting indicates the amino acid sequences of the unique ecTbetaR2 in cTGFBR2-1 and the first ecTbetaR2 in cTGFBR2-2 and cTGFBR2-3. The yellow highlighting indicates the amino acid sequences of the second ecTbetaR2 in cTGFBR2-2 and cTGFBR2-3. The blue highlighting indicates the amino acid sequences of the transmembrane region in 3 cTGFBR2 protein isoforms. The green highlighting indicates the amino acid sequences of the serine/threonine-protein kinases in 3 cTGFBR2 protein isoforms. The black square indicates the amino acid sequence differences among 3 protein isoforms (cTGFBR2-1, cTGFBR2-2, and cTGFBR2-3).