Abstract
Introduction:
CLN3 Batten disease is a rare pediatric neurodegenerative lysosomal disorder caused by biallelic disease-associated variants in CLN3. Despite decades of intense research, specific biofluid biomarkers of disease status have not been reported, hindering clinical development of therapies. Thus, we sought to determine whether individuals with CLN3 Batten disease have elevated levels of specific metabolites in blood.
Methods:
We performed an exhaustive metabolomic screen using serum samples from a novel minipig model of CLN3 Batten disease and validated findings in CLN3 pig serum and CSF and Cln3 mouse serum. We further validate biomarker candidates with a retrospective analysis of plasma and CSF samples collected from participants in a natural history study. Plasma samples were evaluated from 22 phenotyped individuals with CLN3 disease, 15 heterozygous carriers, and 6 non-affected non-carriers (NANC).
Results:
CLN3 pig serum samples from 4 ages exhibited large elevations in 4 glycerophosphodiester species: glycerophosphoinositol (GPI), glycerophosphoethanolamine (GPE), glycerophosphocholine (GPC), and glycerophosphoserine (GPS). GPI and GPE exhibited the largest elevations, with similar elevations found in CLN3 pig CSF and Cln3 mouse serum. In plasma samples from individuals with CLN3 disease, glycerophosphoethanolamine and glycerophosphoinositol were significantly elevated with glycerophosphoinositol exhibiting the clearest separation (mean 0.1338 vs 0.04401 nmol/mL for non-affected non-carriers). Glycerophosphoinositol demonstrated excellent sensitivity and specificity as a biomarker, with a receiver operating characteristic area under the curve of 0.9848 (P = .0003).
Conclusions:
GPE and GPI could have utility as biomarkers of CLN3 disease status. GPI, in particular, shows consistent elevations across a diverse cohort of individuals with CLN3. This raises the potential to use these biomarkers as a blood-based diagnostic test or as an efficacy measure for disease-modifying therapies.
Keywords: Biomarkers, neuronal ceroid-lipofuscinoses, metabolomics, lipid metabolism
Introduction
CLN3 Batten disease (CLN3 disease) is the most common form of neuronal ceroid lipofuscinosis, a group of rare pediatric lysosomal storage disorders with neurocentric pathology. Each of the 14 forms of the disease are caused by mutations in a different gene and has slightly different but overlapping cellular and clinical manifestations. The CLN3 form is characterized by vision loss in childhood with protracted progression that eventually involves seizures, dementia, motor dysfunction, and premature death in the second to third decade of life. 1 The disease is caused by biallelic mutations in the CLN3 gene that are believed to result in a loss of CLN3 function. 2 However, despite decades of intense research, the primary molecular function of the CLN3 protein remains poorly understood. Likewise, little is known regarding upstream disease etiology, and reliable, specific biomarkers of disease status have not been reported. 3 With several potential therapies in clinical trials, sensitive, minimally invasive biomarkers could facilitate a faster path to regulatory approval.
Biomarkers for CLN3 disease could potentially reflect pathological changes in a wide range of pathways and processes. While substrates or products of a potential catalytic or transport function for CLN3 would perhaps be the most sensitive and specific biomarkers, these have not been reported. Instead, most investigations have focused on species that are presumably products of downstream pathology, including lysosomal proteins, inflammatory molecules, and products of neurodegeneration.1,4-10 While interesting patterns have been noted, existing biomarker candidates are limited in their specificity for CLN3 disease, their sensitivity for detecting early changes, and their potential utility for monitoring early treatment responses.
To gain insights into CLN3 function and potential biomarkers, we performed an exhaustive metabolomic screen using serum samples collected from a novel minipig model of CLN3 disease. Several mouse models of CLN3 disease have been developed, but none faithfully recapitulate all key hallmarks of the disease such as severely reduced lifespan, blindness, and seizures. Likewise, the small body size of rodents is accompanied by key differences in metabolic pathways as compared to humans and other large mammals, limiting their utility for biomarker screening. We generated a minipig model of CLN3 disease that recapitulates the most common CLN3 mutation observed in patients, 11 a 966 base pair (often referenced as ~1 KB) deletion in exons 7 and 8 (c.461-280_677+382del; p.[Gly154Alafs*29, Val155_Gly264del], referred to here as CLN3Δex7-8) that results in a premature stop codon in exon nine.
Materials and Methods
Animal ethics statement
Yucatan miniature pigs (mini pigs) were maintained at Precigen Exemplar under an approved IACUC protocol. Wild type and homozygous Cln3Δex7-8 (Jackson Laboratory # 017895), 12 Cln1R151X (Jackson Laboratory # 026197), 13 and Cln6nclf (Jackson Laboratory # 003605) 14 mutant mice on C57BL/6J backgrounds were used for all studies. All mice were housed in an AAALAC International-accredited facility under identical conditions with Institutional Animal Care and Use Committee (IACUC) approval (US Department of Agriculture [USDA] license 46-R-0009; protocol #178-02-24D, Sanford Research, Sioux Falls, SD, USA).
CLN3Δex7-8 mini pig generation
Transgenic CLN3Δex7-8 mini pigs were generated as previously described.15,16 Briefly, wild type fetal Yucatan mini pig fibroblasts were transduced with a recombinant AAV1 carrying a CLN3Δex7-8-Neo targeting vector spanning exon 6 through intron 9 and lacking a ~1 KB region encompassing exons 7 and 8 (recapitulating the most common mutation in CLN3 disease). After antibiotic selection, PCR-positive clones were treated with a recombinant AAV1 carrying a cre recombinase expression cassette to remove the integrated selection cassette. Recombined clones were screened by Southern blot and sequencing to identify those with on-target integrations. Nuclear transfer and embryo transfer were performed at Precigen Exemplar Genetics (Germantown, MA, USA). Abdominal ultrasound was used to check recipient animals for pregnancy after day 21 and throughout gestation.
Pig and mouse biofluid collection
Pig serum collection
Animals were anesthetized with xylazine (TKX) and isoflurane (1%-2%) and a 16G needle was inserted into the right ventricle of the heart. A 20-cc syringe was attached to the needle and approximately 20 mL of blood was obtained. The syringe was emptied into 2 10 mL Monojet™ blood collection tubes via a Saf-T Holder™ transfer device. The blood collection tubes were set aside for 30 minutes at room temperature to allow the sample to clot. Then the tubes were then centrifuged at 3100 × g for 10 minutes at room temperature to retrieve serum. Serum was collected into multiple 2 mL polypropylene screw-top tubes and stored at −80°C.
Pig CSF collection
Animals were anesthetized with xylazine (TKX) and isoflurane (1%-2%) and a region approximately 1 to 2 cm away from the nuchal crest was punctured with an 18-gage, 6-inch spinal needle. The needle was inserted 3 to 5 inches deep depending on age and size of the animal. Once the dura matter was reached, a 12-cc syringe was attached to the needle and ~10 mL of CSF was drawn. The CSF was then transferred into multiple 2 mL polypropylene screw-top tubes and stored at −80°C.
Human ethics statement
Individuals with CLN3 disease were evaluated as part of a Natural History study approved by the Eunice Kennedy Shriver National Institute of Child Health and Human Development Institutional Review Board (NCT03307304). The primary inclusion criteria included the individuals having at least 1 disease-associated variant in CLN3, and clinical symptoms suggestive of CLN3-related disorders. Written informed consent and assent were provided by parents or guardians and participants older than 7 years of age, respectively. Extensive phenotyping was completed, and related phenotypic data used in the analyses here were previously published. Participants from the natural history study have a consistent individual identifier (SP. . .) across publications 4 to facilitate data comparisons.
Human plasma collection
Plasma samples were collected via venipuncture into sodium heparin tubes, following an overnight fast. Samples were transported at room temperature and processed and aliquoted for storage at −80°C within 1 hour of collection. We used the same process to collect samples from unaffected parents and siblings of individuals with CLN3 disease for use as carrier and non-affected non-carrier (NANC) controls.
Human CSF collection
CSF samples were collected under sedation using standard procedures. Following collection, CSF samples were transported at room temperature and aliquoted and frozen at −80°C within 1 hour of collection. Samples with observable blood contamination were centrifuged at 3400 rpm for 10 minutes prior to collection of supernatant for storage. NANC CSF samples were sourced from Precision Medicine (Carlsbad, CA).
Quanterix Simoa™ Neurology 4-plex assay
Pig serum samples were analyzed using a Simoa™ Human Neurology 4-plex kit and a Simoa™ HD-1 analyzer. Samples were run in singlet at a 1:4 dilution in a blinded manner with plate locations randomized.
Metabolomics screening
Metabolon (Morrisville, NC, USA) performed metabolomic analyses.
Sample accessioning
Samples were shipped overnight to Metabolon on dry ice. Samples were inventoried following receipt and immediately stored at −80°C until processed.
Sample preparation
Proteins were precipitated with methanol while vigorously shaking for 2 minutes (Glen Mills GenoGrinder 2000), and were then centrifuged. The resulting supernatant was subsequently divided into 5 fractions: 2 were utilized for 2 separate reversed phase (RP)/UPLC-MS/MS with positive ion electrospray ionization (ESI) methods, 1 was utilized for RP/UPLC-MS/MS with negative ion ESI, 1 was utilized for HILIC/UPLC-MS/MS with negative ion ESI, and 1 sample was set aside as a backup. Organic solvent was removed with a TurboVap® (Zymark).
Quality assurance
Experimental samples were analyzed alongside several controls: a pooled matrix sample which served as a technical replicate; water samples as blanks; and a mixture of QC standards that were selected for their ability to not interfere with measurement of endogenous compounds. Technical variability was assessed by calculating the median relative standard deviation (RSD) for the standards within each sample. Process variability was assessed by calculating the median RSD for all endogenous molecules (ie, non-instrument standards) present in all of the pooled matrix samples. Experimental samples were randomized and QC samples were positioned evenly throughout the experimental injections.
Ultrahigh performance liquid chromatography-tandem mass spectroscopy
Assays utilized a Waters ACQUITY ultra-performance liquid chromatography (UPLC) and a Thermo Scientific Q-Exactive high resolution/accurate mass spectrometer coupled with a heated electrospray ionization (HESI-II) source and an Orbitrap mass analyzer (operated at 35 000 mass resolution). Sample extracts were dried and then reconstituted in solvents amenable to each of the 4 aforementioned methods. The first aliquot was analyzed using acidic positive ion conditions that were optimized for hydrophilic compounds. The extract was gradient-eluted from a C18 column (Waters UPLC BEH C18-2.1 × 100 mm, 1.7 µm) using water and methanol with 0.05% perfluoropentanoic acid (PFPA) and 0.1% formic acid (FA). The second aliquot was analyzed using acidic positive ion conditions optimized for hydrophobic compounds. With this method, the extract was gradient-eluted from the same C18 column using methanol, acetonitrile, water, 0.05% PFPA, and 0.01% FA (operated at an overall higher organic content). The third aliquot was analyzed with basic negative ion-optimized conditions using a dedicated C18 column. These extracts were gradient-eluted using methanol and water with 6.5 mM Ammonium Bicarbonate at pH 8. The last aliquot was analyzed with negative ionization after elution from a HILIC column (Waters UPLC BEH Amide 2.1 × 150 mm, 1.7 µm) with a gradient containing water and acetonitrile with 10 mM Ammonium Formate, pH 10.8. MS analysis alternated between MS and data-dependent MSn scans with dynamic exclusion. Scan ranges varied slightly between methods but covered 70 to 1000 m/z.
Bioinformatics
Informatics consisted of 3 major software components for: (1) data extraction and peak-identification, (2) data processing tools for compound identification and QC, and (3) visualization and information interpretation tools. The hardware used for these tools included the LAN backbone, and a database server with Oracle 10.2.0.1 Enterprise Edition.
Data extraction and compound identification/quantification
Raw data were extracted, peaks were identified and QC was processed using the above tools. Compounds were identified by comparison to an existing library of purified standards or frequently encountered unknown entities. This library is maintained based on data for authenticated standards including mass to charge ratio (m/z), the retention time/index (RI), and chromatographic data (including spectral data). Compound identifications were based on retention index, mass, and the MS/MS forward and reverse scores between the standards and the experimental data. Over 3300 commercially available standards were acquired for determination of their analytical characteristics. Metabolite levels were quantified using area-under-the-curve.
Targeted glycerophosphodiester analysis
Targeted UPLC-MRM/MS analyses of GPI, GPE, GPC, and GPS were conducted by Creative Proteomics (Shirley, NY, USA).
Sample preparation
Samples were shipped overnight on dry ice to the vendor. Thirty μL of each sample was mixed with 90 μL of acetonitrile-2% formic acid. The mixture was vortexed for 1 minute at 3000 rpm and then sonicated in an ice-water bath for 2 minutes, followed by centrifugal clarification at 21 000 rpm at 5°C for 10 minutes. Fifty microliter of the clear supernatant was mixed with 100 μL of 50% acetonitrile.
Standards of GPI, GPE, and GPC were used to prepare stock solutions in 60% acetonitrile. This solution was used to make serially diluted standard solutions in the range of 0.00005 to 40 μM.
LC-MS analysis
UPLC-MRM/MS
Ten μL aliquots of each standard solution and each sample solution were injected to run LC-MRM/MS on a Waters Acuity UPLC coupled to a Sciex QTRAP 6500 Plus mass spectrometer with a positive-ion (+) detection for quantitation of GPC and GPS and with negative-ion (−) detection for quantitation of GPI and GPE, in 2 rounds of LC injections.
An amide UPLC column (2.1 × 100 mm, 1.7 μm) was used for LC separation, with the use of an ammonium formate solution (A) and an acetonitrile-0.01% formic acid solution (B) as the mobile phase for gradient elution at 35°C and 0.3 mL/min, with an efficient gradient of 80% to 20% B in 8 minutes.
Concentrations of individual metabolites were calculated by constructing linear-regression curves of the metabolites (except GPS, for which a standard was not available) with peak areas versus concentrations of standard solutions and then interpolating the calibration curves with the peak areas measured from injections of sample solutions in an appropriate concentration range for each compound. Peak areas were used for relative quantitation of GPS.
Statistics
Samples were blinded for metabolomics and targeted glycerophosphodiester analyses. Formal power analyses were not performed; sample sizes were selected based on prior experience with a mouse model of CLN3 disease. 12 N’s for each experiment were determined based upon sample availability, results observed in prior studies, and experience with similar study designs and animal models.
Statistical tests used are described in the figure legends for each experiment. No outlier animals or data points were excluded. The metabolite volcano plot utilized an two-tailed homoscedastic t-test that was not corrected for multiple comparisons. Longitudinal comparisons utilized two-way ANOVAs with two-tailed Sidak’s multiple comparisons post-hoc tests. Two-group single time-point comparisons utilized one-tailed t-tests based on hypotheses on directionality of change derived from prior experiments. Human glycerophosphodiesters comparisons utilized Brown-Forsythe one-way ANOVAs with Dunnett’s multiple comparisons post-hoc tests due to non-Gaussian distribution. UBDRS correlations were examined with Pearson correlation. Statistical calculations were performed using GraphPad Prism 9.2.0 (San Diego, CA, USA).
Study Approval
Yucatan miniature pigs (mini pigs) were generated and maintained at Precigen Exemplar under an approved IACUC protocol. Mice were housed in an AAALAC International-accredited facility under Institutional Animal Care and Use Committee (IACUC) approval. Samples from individuals with CLN3 disease were collected as part of a Natural History study approved by the Eunice Kennedy Shriver National Institute of Child Health and Human Development Institutional Review Board (NCT03307304). Written informed consent and assent were provided by parents or guardians and participants older than 7 years of age, respectively, prior to participation.
Results
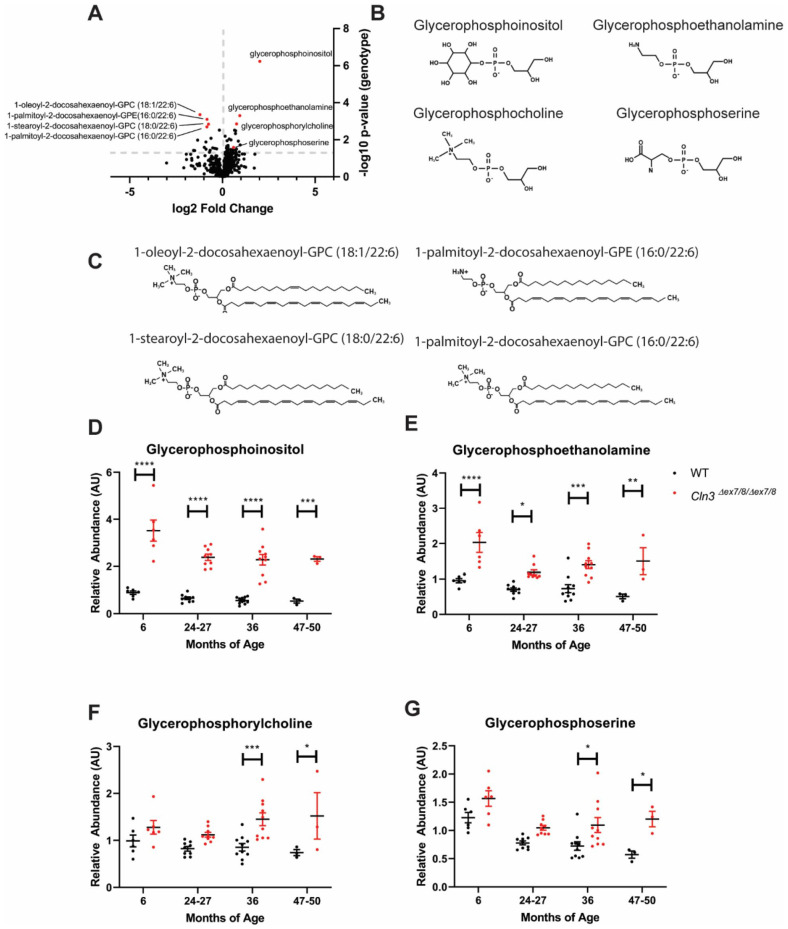
CLN3Δex7-8 pigs were generated with somatic cell nuclear transfer of Yucatan minipig fibroblast nuclei in which the endogenous CLN3 gene was replaced with a targeting construct that recapitulates the CLN3Δex7-8 mutation. 15 CLN3Δex7-8 pigs exhibit progressive brain pathology beginning by 2 months of age, retinal dysfunction by 14 months of age, and gait abnormalities by 15 months of age. 15 We examined common blood-based markers of neurodegeneration and neuroinflammation as potential biomarkers, but observed only mild phenotypes that weren’t evident until 36 months of age (Supplemental Figure 1). To identify novel biomarkers with enhanced potential for monitoring disease status across the lifespan, we quantified relative levels of 350 lipids and 451 small molecule metabolites in serum samples from male and female homozygous CLN3Δex7-8 and wild type pigs at approximately 6, 24 to 27, 36, and 47 to 50 months of age. At 36 months of age, 38 species were significantly upregulated in CLN3Δex7-8 samples (Figure 1A). Strikingly, 4 of the most significantly upregulated species were glycerophosphodiesters with a high degree of structural similarity, consisting of a glycerol backbone with unoccupied sn1 and sn2 positions and a phosphodiester-linked R group at the sn3 position (Figure 1B). Of these, glycerophosphoinositol (GPI) exhibited by far the greatest fold change, with more minor elevations in glycerophosphoserine (GPS), glycerophosphocholine (GPC), and glycerophosphoethanolamine (GPE). Several species were also downregulated in CLN3Δex7-8 samples, most notably 4 phospholipids with docosahexaenoyl groups at the sn2 position (Figure 1A and C).
Figure 1.
GPI, GPE, GPC, and GPS are elevated in CLN3Δex7-8 mini pigs. Relative levels of 350 lipids and 451 small molecule metabolites were quantified in serum samples from male and female homozygous CLN3Δex7-8 and wild type pigs at 6 (n = 6/group), 24 to 27 (n = 9/group), 36 (n = 10/group), and 48 (n = 3/group) months of age: (A) at 36 months of age, 38 species were significantly upregulated in CLN3Δex7-8 samples including 4 glycerophosphodiesters (red points on graph) and 4 docosahexaenoyl-conjugated phospholipids (red points on graph), (B) chemical structures of glycerophosphodiesters significantly upregulated in CLN3Δex7-8 pigs, (C) chemical structures of phospholipids significantly downregulated in CLN3Δex7-8 pigs, and (D-G) longitudinal plots of glycerophosphodiester levels in wild type and CLN3Δex7-8 pigs. GPI and GPE are significantly elevated at 6, 24 to 27, 36, and 47 to 50 months of age, while GPC and GPS are significantly elevated 36 and 47 to 50 months only. Uncorrected two-tailed homoscedastic t-test in (A). Two-way ANOVA with two-tailed Sidak’s multiple comparisons test in (D) to (G).
*P < .05. **P < .005. ***P < .0005. ****P < .0001.
Longitudinal patterns were remarkably stable over time for each of the 4 glycerophosphodiesters, with significant elevations at each of the 4 ages for GPI and GPE (Figure 1D-G). The phospholipid species exhibited similar trends, but with lower fold changes and greater variability. Thus, we focused on the glycerophosphodiesters for subsequent investigations. Serum elevations for each of the 4 species were confirmed using an orthogonal semi-quantitative UHPLC-MS/MS platform (Supplemental Figure 2) developed with individual analyte standards for GPI, GPE, and GPC. Cerebrospinal fluid (CSF) samples from 36-month-old CLN3Δex7-8 and wild type pigs were also analyzed using the same platform, finding significant elevations in the CLN3Δex7-8 samples for GPI, but not GPE, GPC, or GPS (Supplemental Figure 3). We also quantified levels of each of the 4 species in serum samples from male and female homozygous Cln3Δex7-812 and wild type mice at 6, 12, 18, and 24 months of age, again finding elevations for GPI and GPE, albeit with more variable longitudinal patterns (Supplemental Figure 4). To examine whether these changes were specific to CLN3 disease, we examined serum samples from mouse models for 2 other genetic forms of Batten disease. None of the glycerophosphodiesters were elevated in serum samples from mouse models of CLN1 13 or CLN6 14 Batten disease (Supplemental Figures 5 and 6).
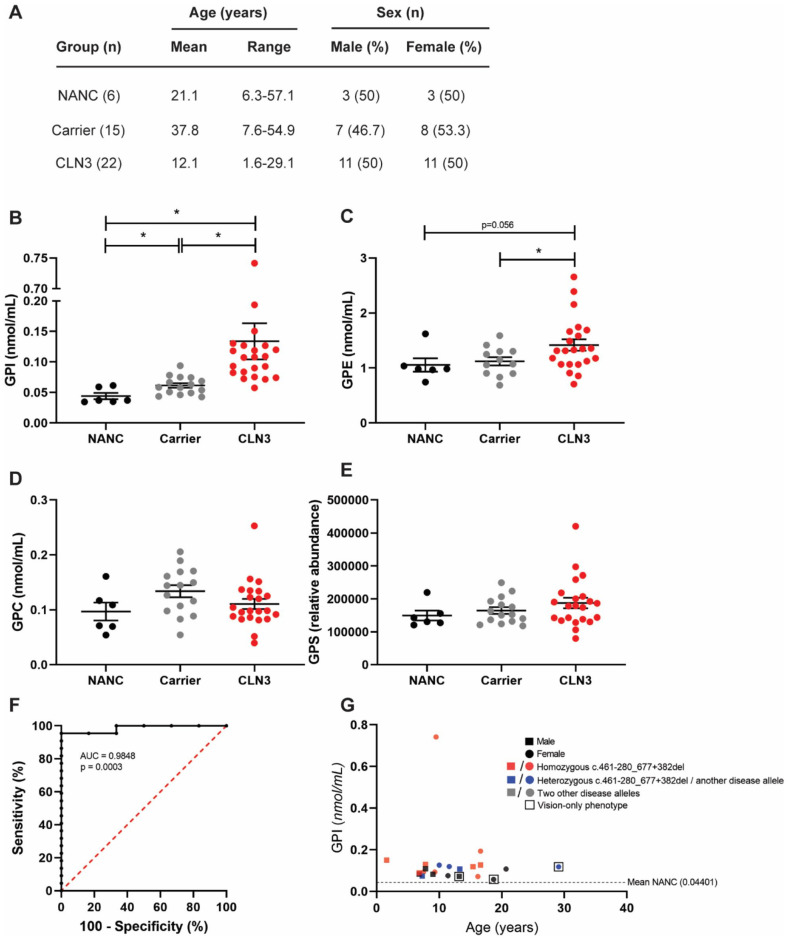
Collectively, the patterns observed across species and time points suggested that the glycerophosphodiesters species may be closely linked to CLN3 disease etiology and could thus be useful as clinical biomarkers of disease status. To investigate this, we examined levels of each of the species in plasma samples from 22 phenotyped individuals with CLN3 disease, 4 15 heterozygous carriers, and 6 non-affected non-carriers (NANC). The individuals with CLN3 covered a wide range of genotypes, ages, and phenotypes (Figure 2A and Supplemental Table 1). 4 While plasma GPS and GPC were unchanged across the 3 groups, plasma GPE and GPI were significantly elevated in the CLN3 samples, with GPI exhibiting the clearest separation (Figure 2B-D). Plasma GPI demonstrated excellent sensitivity and specificity as a biomarker, with a receiver operating characteristic (ROC) area under the curve (AUC) of 0.9848 (P = .0003, Figure 2G) when comparing CLN3 and NANC. We also examined levels of the 4 glycerophosphodiesters in CSF samples from a subset of 8 of the individuals with CLN3 disease, finding similar, statistically significant elevations in GPI levels (Supplemental Figure 7).
Figure 2.
GPI and GPE are elevated in plasma samples from individuals with CLN3 disease. GPI, GPE, GPC, and GPS levels were quantified in plasma samples from individuals with CLN3 disease, heterozygous carriers, and non-affected non-carriers (NANC): (A) demographics for each group in the cohort, (B) GPI levels were elevated in both carriers and individuals with CLN3 disease, with higher levels in the CLN3 group, (C) GPE levels were higher in individuals with CLN3 as compared to carriers and trended toward higher levels as compared to NANC. No significant differences were noted for GPC (D) or GPS (E), (G) ROC curve for CLN3 versus NANC (AUC = 0.9848, P = .0003). (G) GPI levels (y-axis) as a function of age (x-axis) for individuals with CLN3 disease. Males are represented by square points and females are represented by circles. Samples from individuals with homozygous c.461-280_677+382del mutations are shown in red; compound heterozygotes with a single c.461-280_677+382del mutation are shown in blue. Samples from individuals with other classes of mutations are shown in gray. Samples from individuals with vision-only phenotypes are overlayed with a clear box. One-way ANOVA test with one-tailed Dunnett’s multiple comparisons test for (B) to (E). Wilson/Brown 95% confidence interval for (F).
*P < .05.
CLN3 affected individuals exhibited a wide range of plasma GPI levels ranging from 0.05749 to 0.7417 nmol/mL with a mean of 0.1338 nmol/mL (SD = 0.1392 nmol/mL). While the entirety of this range is substantially elevated over the mean for NANC (0.04401 nmol/mL, SD = 0.01599 nmol/mL), we were interested in potential relationships between GPI levels in CLN3 affected individuals and other demographic and phenotypic data points. However, the data did not reveal any clear patterns. GPI levels in the CLN3 group were not significantly impacted by sex, age, or genotype, and did not correlate significantly with clinical severity as measured by United Batten Disease Rating Scale (UBDRS) scores (Supplemental Figure 8). 17 The cohort also included 3 patients with rare vision-only phenotypes, 18 and GPI levels for these patients were intermediate among the broader cohort.
Surprisingly, GPI was also elevated to intermediate levels in heterozygous carriers with a mean of 0.06150 nmol/mL (SD = 0.01261). These levels are statistically different from both individuals with CLN3 and NANC. Since CLN3 carriers are apparently free from any observable CLN3 disease process and would thus not be expected to exhibit changes in markers related to the neuroinflammation, neurodegeneration, and neuronal dysfunction, these intermediate levels strongly suggest that GPI could be closely linked to upstream CLN3 disease etiology or even to the function of CLN3.
Discussion
As demonstrated, GPE and GPI could have utility as biomarkers of CLN3 disease status. GPI, in particular, shows consistent elevations across a diverse cohort of individuals with CLN3, as well as in multiple animal models of the disease. GPI offers several advantages over generic biomarkers of neurodegeneration and neuroinflammation. Namely, GPI is elevated early and throughout the lifespan in CLN3 animal models, even prior to the onset of overt neurodegeneration. Similarly, even the youngest affected individual (1.6 year of age) in the cohort analyzed here exhibited pronounced GPI elevations in both plasma and CSF. Such early and consistent elevations have not been observed for other biomarkers of CLN3 disease. Additionally, our observation that heterozygous carriers exhibit intermediate serum GPI elevations suggests a dose-response relationship wherein GPI levels are closely linked to levels of functional CLN3 protein. The studies presented here were limited to a translational scope, and more work will be required to elucidate the mechanisms by which CLN3 deficiency results in GPI elevation. A comprehensive analysis of related phospholipid species could help deduce a more complete picture of the deficiencies leading to glycerophosphodiester elevations in CLN3 disease.
One intriguing possibility is that CLN3 has a direct influence on metabolic pathways that generate, consume, or transport glycerophosphodiesters. Structural similarity has been noted between CLN3 and major facilitator superfamily transporters, 2 1 member of which is a transporter of GPI in yeast. 19 Thus, GPI and GPE elevations in individuals with heterozygous and homozygous CLN3 mutations could be a key insight leading to a long-sought primary molecular function for CLN3.
Conclusion
These results raise the potential to use these biomarkers as a blood-based diagnostic test (eg, a newborn screen) or as an efficacy measure for disease-modifying therapies. Therapies that restore CLN3 function, such as the CLN3 gene therapy currently in clinical trials 20 , may reduce GPI in various tissues and biofluids. CSF levels, in particular, may be valuable for monitoring the efficacy of CNS-directed therapies. Additionally, combining GPI or GPE with biomarkers that correlate with clinical severity, such as neurofilament light, may provide for a comprehensive assessment of molecular and clinical disease status.
Supplemental Material
Supplemental material, sj-docx-1-bmi-10.1177_11772719221107765 for Glycerophosphoinositol is Elevated in Blood Samples From CLN3Δex7-8 pigs, Cln3Δex7-8 Mice, and CLN3-Affected Individuals by Jon J Brudvig, Vicki J Swier, Tyler B Johnson, Jacob C Cain, Melissa Pratt, Mitch Rechtzigel, Hannah Leppert, An N Dang Do, Forbes D Porter and Jill M Weimer in Biomarker Insights
Supplemental material, sj-docx-2-bmi-10.1177_11772719221107765 for Glycerophosphoinositol is Elevated in Blood Samples From CLN3Δex7-8 pigs, Cln3Δex7-8 Mice, and CLN3-Affected Individuals by Jon J Brudvig, Vicki J Swier, Tyler B Johnson, Jacob C Cain, Melissa Pratt, Mitch Rechtzigel, Hannah Leppert, An N Dang Do, Forbes D Porter and Jill M Weimer in Biomarker Insights
Supplemental material, sj-xlsx-3-bmi-10.1177_11772719221107765 for Glycerophosphoinositol is Elevated in Blood Samples From CLN3Δex7-8 pigs, Cln3Δex7-8 Mice, and CLN3-Affected Individuals by Jon J Brudvig, Vicki J Swier, Tyler B Johnson, Jacob C Cain, Melissa Pratt, Mitch Rechtzigel, Hannah Leppert, An N Dang Do, Forbes D Porter and Jill M Weimer in Biomarker Insights
Footnotes
The author(s) declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: J.J.B., T.B.J., J.C.C., and J.M.W. are employees of Amicus Therapeutics, Inc. and hold equity in the company in the form of stock-based compensation. A.D.D. and F.D.P. have Collaborative Research and Development Agreements with Amicus Therapeutics, Inc.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was funded by a grant to J.J.B. from the Forebatten Foundation. Funding for the CLN3 natural history study was provided by an NIH Clinical Center Bench-to-Bedside award and the NICHD intramural research program (ZIA HD008989). We thank study participants and families for their invaluable contributions.
Author Contributions: J.J.B., T.B.J., J.C.C., A.D.D., F.D.P., and J.M.W. conceived the study; J.J.B., V.J.S., M.P., and M.R. performed the experiments; J.J.B. analyzed the data; J.J.B. wrote the manuscript with editorial input from T.B.J., J.C.C., A.D.D., F.D.P., and J.M.W.; J.J.B. and H.L. generated the figures; A.D.D. and F.D.P. implemented and conducted the natural history study.
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Johnson TB, Cain JT, White KA, Ramirez-Montealegre D, Pearce DA, Weimer JM. Therapeutic landscape for Batten disease: current treatments and future prospects. Nat Rev Neurol. 2019;15:161-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mirza M, Vainshtein A, DiRonza A, et al. The CLN3 gene and protein: what we know. Mol Genet Genomic Med. 2019;7:e859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brudvig JJ, Weimer JM. On the cusp of cures: breakthroughs in Batten disease research. Curr Opin Neurobiol. 2022;72:48-54. [DOI] [PubMed] [Google Scholar]
- 4. Dang Do AN, Sinaii N, Masvekar RR, et al. Neurofilament light chain levels correlate with clinical measures in CLN3 disease. Genet Med Off J Am Coll Med Genet. 2020;23:751-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Timm D, Cain JT, Geraets RD, et al. Searching for novel biomarkers using a mouse model of CLN3-Batten disease. PLoS One. 2018;13:e0201470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sleat DE, Tannous A, Sohar I, et al. Proteomic analysis of brain and cerebrospinal fluid from the three major forms of neuronal ceroid lipofuscinosis reveals potential biomarkers. J Proteome Res. 2017;16:3787-3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sleat DE, Wiseman JA, El-Banna M, et al. Analysis of brain and cerebrospinal fluid from mouse models of the three major forms of neuronal ceroid lipofuscinosis reveals changes in the lysosomal proteome. Mol Cell Proteomics. 2019;18:2244-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Iwan K, Clayton R, Mills P, et al. Urine proteomics analysis of patients with neuronal ceroid lipofuscinoses. iScience. 2021;24:102020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Soldati C, Lopez-Fabuel I, Wanderlingh LG, et al. Repurposing of tamoxifen ameliorates CLN3 and CLN7 disease phenotype. EMBO Mol Med. 2021;13:e13742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lebrun AH, Moll-Khosrawi P, Pohl S, et al. Analysis of potential biomarkers and modifier genes affecting the clinical course of CLN3 disease. Mol Med. 2011;17:1253-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Munroe PB, Mitchison HM, O’Rawe AM, et al. Spectrum of mutations in the Batten disease gene, CLN3. Am J Hum Genet. 1997;61:310-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cotman SL. Cln3 deltaex7/8 knock-in mice with the common JNCL mutation exhibit progressive neurologic disease that begins before birth. Hum Mol Genet. 2002;11:2709-2721. [DOI] [PubMed] [Google Scholar]
- 13. Miller JN, Kovács AD, Pearce DA. The novel Cln1(R151X) mouse model of infantile neuronal ceroid lipofuscinosis (INCL) for testing nonsense suppression therapy. Hum Mol Genet. 2014;24:185-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bronson RT, Donahue LR, Johnson KR, Tanner A, Lane PW, Faust JR. Neuronal ceroid lipofuscinosis (NCLF), a new disorder of the mouse linked to chromosome 9. Am J Med Genet. 1998;77:289-297. [DOI] [PubMed] [Google Scholar]
- 15. Johnson TB, Sturdevant DA, White KA, et al. Characterization of a novel porcine model of CLN3-Batten disease. Mol Genet Metab. 2019;126:S81. [Google Scholar]
- 16. White KA, Swier VJ, Cain JT, et al. A porcine model of neurofibromatosis type 1 that mimics the human disease. JCI Insight. 2018;3:e120402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Marshall FJ, de Blieck EA, Mink JW, et al. A clinical rating scale for Batten disease: reliable and relevant for clinical trials. Neurology. 2005;65:275-279. [DOI] [PubMed] [Google Scholar]
- 18. Ku CA, Hull S, Arno G, et al. Detailed clinical phenotype and molecular genetic findings in CLN3-associated isolated retinal degeneration. JAMA Opthalmol. 2017;135:749-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Patton-Vogt JL, Henry SA. GIT1, a gene encoding a novel transporter for glycerophosphoinositol in Saccharomyces cerevisiae. Genetics. 1998;149:1707-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. de los Reyes E, Aylward S, Islam M, et al. An open-label, phase 1/2a, AAV9-CLN3 gene transfer clinical trial for juvenile neuronal ceroid lipofuscinosis. Paper presented at: 17th Annual WORLD Symposium; February 7-12, 2021; Orlando, FL. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-bmi-10.1177_11772719221107765 for Glycerophosphoinositol is Elevated in Blood Samples From CLN3Δex7-8 pigs, Cln3Δex7-8 Mice, and CLN3-Affected Individuals by Jon J Brudvig, Vicki J Swier, Tyler B Johnson, Jacob C Cain, Melissa Pratt, Mitch Rechtzigel, Hannah Leppert, An N Dang Do, Forbes D Porter and Jill M Weimer in Biomarker Insights
Supplemental material, sj-docx-2-bmi-10.1177_11772719221107765 for Glycerophosphoinositol is Elevated in Blood Samples From CLN3Δex7-8 pigs, Cln3Δex7-8 Mice, and CLN3-Affected Individuals by Jon J Brudvig, Vicki J Swier, Tyler B Johnson, Jacob C Cain, Melissa Pratt, Mitch Rechtzigel, Hannah Leppert, An N Dang Do, Forbes D Porter and Jill M Weimer in Biomarker Insights
Supplemental material, sj-xlsx-3-bmi-10.1177_11772719221107765 for Glycerophosphoinositol is Elevated in Blood Samples From CLN3Δex7-8 pigs, Cln3Δex7-8 Mice, and CLN3-Affected Individuals by Jon J Brudvig, Vicki J Swier, Tyler B Johnson, Jacob C Cain, Melissa Pratt, Mitch Rechtzigel, Hannah Leppert, An N Dang Do, Forbes D Porter and Jill M Weimer in Biomarker Insights