Abstract
Human mesenchymal stem cells (MSCs) are isolated from various adult and perinatal tissues. Although mesenchymal stem cells from multiple sources exhibit similar morphology and cell surface markers, they differ in their properties. In this study, we determined that the expression of integrin alpha 6 (ITGA6) and ITGA6 antisense RNA (ITGA6-AS1) correlates with the proliferation, cell size, and differentiation potential. The expression of ITGA6 was inversely correlated with ITGA6-AS1 in MSCs. The expression of ITGA6 was higher, but ITGA6-AS1 was lower in MSCs from cord placenta junction, cord tissue, and Wharton’s jelly. In contrast, ITGA6 expression was lower, while ITGA6-AS1 was higher in MSCs from the placenta. The bioinformatic analysis showed that ITGA6 genomic DNA transcribes ITGA6-AS1 from the reverse strand, overlapping ITGA6 exon-2. Additionally, we identify several putative promoters (P1-P10) of ITGA6. ITGA6-P10 is CG rich and contains CGI. EMBOSS Cpgplot software revealed a CGI length of 180 bp that extends from nucleotide 125 to 304 of the P10 sequence. We suggest that the post-transcriptional regulation of the ITGA6 in mesenchymal stem cells is controlled by the ITGA6-AS1, which could be a critical factor responsible for the heterogeneity in function and cell fate of human MSCs. These results may provide further impetus for investigations to unravel the mechanisms of ITGA6 regulation that could help maintain or improve the properties of mesenchymal stem cells.
Keywords: integrin alpha 6, integrin alpha 6-antisense 1, lncRNA, mesenchymal stem cells, self-renewal, promoters
Introduction
Human mesenchymal stem cells (MSCs) display promising therapeutic properties including propensity to differentiate into multilineage, the ability to home to the site of damage, induce immunomodulatory and anti-inflammatory responses, and are heterogeneous cells that provide neuroprotection (Beeravolu et al., 2016; Beeravolu et al., 2017; Brown et al., 2021). Although MSCs from multiple sources exhibit similar morphology and cell surface markers, they display heterogeneity in features depending upon the source, method of isolation, culture media, and mechanical cues (Kolf et al., 2007; Chen et al., 2015; Elahi et al., 2016; Costa et al., 2021). The culture conditions, intracellular matrix (ICM), and extracellular matrix (ECM) of MSCs can influence genetic-epigenetic inheritance (Maurer, 2011; Sullivan et al., 2014; Assis-Ribas et al., 2018; Matta et al., 2019). Genetic-epigenetic can also influence the MSC’s fate; differentiation into progenitors and terminally differentiated cells (Teven et al., 2011; Cakouros and Gronthos, 2020; Nieto-Nicolau et al., 2020).
The long non-coding RNA (lncRNA) plays an important role in the post-transcriptional regulation of genes. They are the host of microRNAs (miRNAs) produced from non-coding RNAs forming an imperfect stem-loop secondary structure (Ballantyne et al., 2016; He et al., 2019; Zhang et al., 2019; Sun et al., 2021). lncRNA inactivates or destabilizes mRNA by pairing at a specific complementarity sequence or generating a miRNA that targets the mRNA (Zealy et al., 2018; Zhang et al., 2019; Ma et al., 2021; Statello et al., 2021). Antisense RNAs are lncRNAs subtype (Sun et al., 2021) that can generate diverse transcripts, play multifunctional roles including embryonic pluripotency, differentiation and development, are widespread in humans and other eukaryotes (Villegas and Zaphiropoulos, 2015). lncRNAs are also reported to be functional regulators of MSCs (Ju et al., 2019; Li et al., 2019; Xie et al., 2019).
An important group of proteins, the integrin family that includes integrins alpha (ITGA), and beta proteins, play a critical role in the MSCs fate. They are cell surface adhesion receptors that support signaling across the plasma membrane in both ICM and ECM and mediate the intracellular signals response to the ECM (Hynes, 2002). Further, integrins activate a wide range of signaling pathways and are considered the main factor involved in regulating cell growth and mobility, cellular shape, survival, and differentiation associated with ECM interaction (Akiyama, 1996; Yamada, 1997; Chen and Sheppard, 2007; Schwartz, 2010; Becerra-Bayona et al., 2018; Peterson and Koval, 2021). Thus, integrins are a prime link between the MSCs’ ICM and ECM.
Among the ITGAs, ITGA6, also known as CD49f, is a transmembrane glycoprotein adhesion receptor protein which show higher expression in bone marrow (BM) MSCs (Nieto-Nicolau et al., 2020). Downregulation of ITGA6 impaired the cell proliferation and migration of BM-MSCs via the protein kinase B (AKT) pathway and the cell cycle inhibitor proteins p53 and p21 (Nieto-Nicolau et al., 2020). Additionally, overexpression of ITGA6 was found to be associated with an osteoporotic vertebral fracture in elderly women (Jales Neto et al., 2020), bone metastasis and ductal carcinoma (Martin and Jiang, 2014), colon cancer-initiating cells (Haraguchi et al., 2013), cervical squamous cell carcinoma (Hou et al., 2015), and in the invasion, metastasis and poor prognosis in human gallbladder carcinoma (Zhang et al., 2016).
Our study showed a correlation between the expression of ITGA6 and ITGA6 antisense RNA1 (AS1), also known as AC078883.3, is inversely proportional to the properties of MSCs isolated from various sources. The ITGA6 genomic DNA expresses ITGA6-AS1 from the reverse strand. Our bioinformatic analysis identified several putative promoters (P1-P10) of ITGA6, and ITGA6-P10 is CG-rich and contains CGI of 180 bp that extends from nucleotide 125 to 304. Our results and bioinformatic analysis indicate that post-transcriptional regulation of the ITGA6 in MSCs may be controlled by the ITGA6-AS1, which could be a critical factor responsible for the heterogeneity in the cell function and fate of human MSCs. These results may provide further impetus for investigations to unravel the mechanisms of ITGA6 regulation that could help maintain or improve the properties of MSCs.
Materials and methods
Human mesenchymal stem cell cultures
In this study, we used well-characterized MSCs isolated from six sources, BM, chorion (CH), cord placenta junction (CPJ), cord tissue (CT), Wharton’s jelly (WJ), and placenta (PC) as described previously (Beeravolu et al., 2016; Beeravolu et al., 2017). All cells were grown in a growth medium (GM) containing DMEM nutrient mix F12 medium (DMEM/F12; Life Technologies, Carlsbad, CA, United States), supplemented with 10% fetal bovine serum (FBS; VWR, Radnor, PA, United States), and 5.6% of antibiotic solution (0.1% gentamicin, 0.2% streptomycin, and 0.12% penicillin) (Sigma Aldrich, St Louis, MO, United States) in a humidified 5% CO2 atmosphere at 37 °C. All cells were grown to 70% confluency at passage 6 (P6).
Determination of Cell size, Doubling Time, and Colony Forming Efficiency of MSCs.
MSC size was measured by using microscopy and ImageJ. The doubling time (DT) and colony-forming efficiency of MSCs were determined as described previously (Beeravolu et al., 2016; Beeravolu et al., 2017).
RNA Extraction and Analysis of ITGA6 and ITGA6-AS1 Expression.
Total RNA was extracted from 70% confluent cells using the GeneJET RNA purification kit (ThermoFisher Scientific) following the manufacturer’s instructions. RNA was treated with DNase and incubated at 37 °C for 30 min in the thermocycler (Bio-Rad, Hercules, CA, United States). cDNA was synthesized using iScript kit (Bio-Rad), and RT-qPCR was performed by using SsoAdvanced universal SYBR Green Supermix Kit (Bio-Rad) on CFX96 Real-Time System (Bio-Rad). A 10 µL reaction was used, which included 5 µL SYBR green, 3 µL of distilled water, 0.5 µL of forward primer, 0.5 µL of reverse primer, and 1 µL of 1:10 diluted cDNA. Each reaction was exposed to the following conditions: 98 °C for 10 min, followed by 30 s of 98°C, 20 s of 60°C, and 30 s of 72 C for 44 cycles in 96-well optical reaction plates (Bio-Rad). Human gene GAPDH was used to normalize fold gene expression. Primer sets for ITGA6, ITGA6-AS1, and GAPDH are listed in Supplementary Table S1. The 2−∆∆Cq method was used to analyze relative gene expression (fold change) data obtained by real-time quantitative PCR (Schmittgen and Livak, 2008).
Bioinformatic analysis and genomic databases
The URLs of genomics databases and bioinformatics tools used in this study are shown in Supplementary Table S2. The genomic features of ITGA6 and ITGA6-AS1 loci are searched using online public genomics databases NCBI-Gene, UCSC Genome Browser, Ensembl Genome Browser, EMBL-EBI, and Eukaryotic promoter database. EMBOSS Matcher was used to identify local similarities in two input sequences. EMBOSS Needle was used for optimal global sequence alignment and EMBOSS Cpgplot to locate and plot CpG islands (CGI) in nucleotide sequence(s). The miRBase search tool to discover mature microRNA (miR) sequences from long non-coding RNA ITGA6 antisense RNA 1 (ITGA6-AS1), NCBI Reference Sequence: NR_1,57,573.1. Identification of the miR recognition elements (MREs) in the ITGA6 mRNA, NM001079818.3, was performed by RNA22 v2 miR target detection tool to find miR that binds to the target ITGA6 mRNA MRE. The UCSC genome browser “get DNA” and “Blat” tools were used to retrieve and analyze sequences in the forward or reverse strands and recover the identified sequences from earlier versions to the updated GRCh38/hg38 version.
Statistical analysis
The statistical tests, t-test, and one-way ANOVA were performed using Excel Data Analysis Toolpack and GraphPad Prism 7.01 (GraphPad Software, Inc., La Jolla, CA, United States) for various parameters. A significant difference was assessed at p < 0.05.
Results
Properties of mesenchymal stem cells from various sources
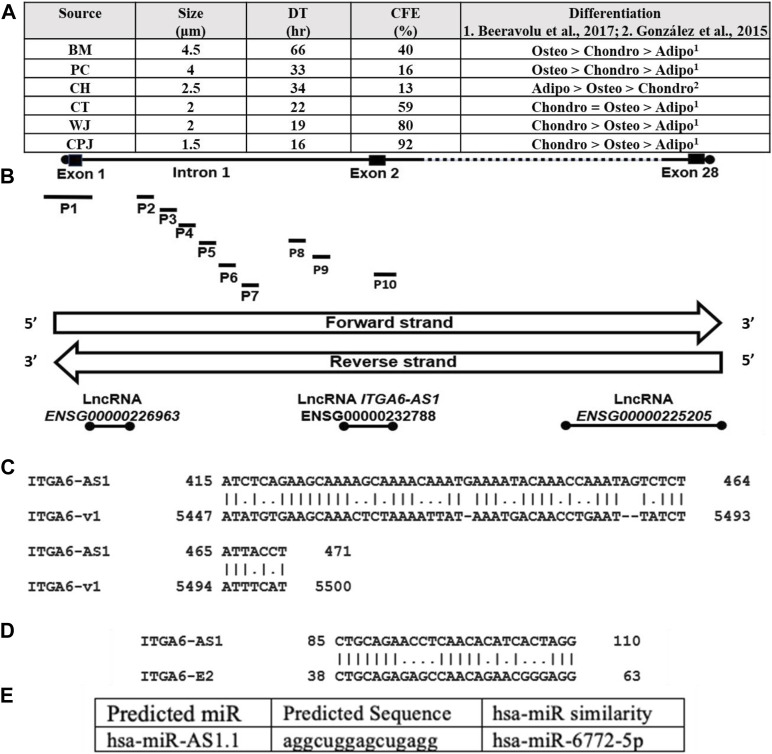
MSCs isolated from various sources expressed specific cell surface markers including CD29, CD44, CD73, CD90, CD105 as determined by flow cytometry (Brown et al., 2019). All MSCs (P6) were cultured to 70% confluency and analyzed for various properties (Figure 1A). A cell size comparison showed that MSCs from BM, PC, CH, CT, WJ, and CPJ had sizes of 4.5, 4, 2.5, 2, 2, and 1.5 μm, respectively. BM MSCs had the largest, and CPJ had the smallest cell size. The growth rate determined based on the DT correlated with the size of MSCs. The BM cells had the highest, and CPJ had the lowest DT. We also analyzed the clonogenicity of MSCs from various sources. The results showed that MSCs had CFE of 13, 16, 40, 59, 80, and 92% from CH, PC, BM, CT, WJ, and CPJ, respectively. CPJ MSCs had the highest, and CH had the lowest CFE. We have previously shown that MSCs from BM and PC favored differentiation toward the osteogenic lineage more but less toward adipogenic lineage (Beeravolu et al., 2017). MSCs from CPJ and WJ showed increased differentiation towards chondrogenic lineage. CT MSC had a similar propensity to differentiate between chondrogenic and osteogenic lineages. CPJ, WJ and CT had a lower differentiation potential toward the adipogenic lineage. CH MSCs displayed increased adipogenic differentiation but decreased chondrogenic differentiation potential (González et al., 2015). CPJ and WJ displayed increased differentiation towards the chondrogenic lineage. CH MSCs had the lowest clonogenicity potential. BM and PC MSCs with the greater tendency to differentiate into osteogenic lineage were larger in cell size and doubling time. CPJ cells had the smallest cell size, doubling time, and largest CFE compared to the MSCs from other sources. Our published results showed that ITGA6 expression varied significantly among the MSCs from BM, CH, PC, CT, WJ, and CPJ (Brown et al., 2019). ITGA6 expression was highest in CPJ and lowest in both BM and PC. Our results showed a clear correlation between the ITGA6 expression and the properties of MSCs.
FIGURE 1.
(A) Characteristics including cell size, doubling time, CFE, and differentiation potential of human MSCs from six sources, BM. PC, CH, CT, WJ, and CPJ. The ITGA6 is mapped at chr2:172,427,336–172,506,459 forward strand. (B) ITGA6 genomic DNA hosts three lncRNA loci and ten promoters (P1 to P10) and the lncRNA are coded by the reverse strand (Data from NCBI-Gene and Ensembl databases). (C) The ITGA6-AS1 partial complementarity with 3′UTR of ITGA6 mRNA type 1/variant 1 (v1). (D) ITGA6-AS1 showed 65.4% alignment to 26 bp of ITGA6 exon 2 (E2). (E) Predicted miR sequence hsa-miR-AS1.1, for details, see Supplementary Table S6.
Bioinformatic analysis of integrins alpha 6 gene
A search of the public databases showed eight genes of the human integrin alpha gene family hosting 18 members. In addition, several lncRNA, including three antisense and multiple alternative promoters (Supplementary Table S3). These genes code for exons ranging from 26 to 32, producing 1 to 17 transcripts. These transcripts were generated by promoters ranging from 1–10. ITGA6 has the most putative promoters. Each of the genes also expressed 0 to 3 lncRNA. One of the lncRNA of ITGA2, ITGA6, and ITGA9 was antisense RNA, ITGA2-AS1, ITGA6-AS1, and ITGA9-AS1, and they are likely to be involved in regulating the respective genes. The identity of 10 promoters of ITGA6 is shown in Supplementary Table S4. One of ITGA6 promoters overlaps ITGA6-AS1 and ITGA6 exon2 sequences (Figure 1B). The ITGA6 (NCBI Gene ID: 3,655) occupies a region of 79,124 bps on the forward strand of the short arm of chromosome two mapped to cytogenetic location 2q31.1 at GRCh38/hg38 genomic coordinates: chr2:172,427,336–172,506,459 and generate six mRNA types/variants. The global alignment of two ITGA6 mRNA variants, ITGA6 mRNA variant one versus the other five mRNA variants, showed alignment similarities in the range of 91.1%–99.2% (Supplementary Table S5). ITGA6 mRNA variant one was selected for further bioinformatics analysis in the current study. Thus, we investigated the potential association of ITGA6-AS1 with ITGA6 expression, considering the critical roles of ITGA6 in the determination of MSCs types.
Potential involvement of integrins alpha 6-antisense 1 in the regulation of integrins alpha 6
Since some of the studies have shown that ITGA6 is regulated by ITGA6-AS1 (Song et al., 2021), we investigated the sequence ITGA6-AS1 transcript pairing with ITGA6 transcript variant 1. The ITGA6-AS1 was coded by the opposite strand of DNA coding for the ITGA6 exon-2 (Figure 1B). Local alignment analysis by EMBOSS Matcher of ITGA6-AS1 NR_157,573.1 transcript of 442 bp and ITGA6 transcript type 1 (NM_001079818.3) of 5,686 bp showed partial complementarity with 64.9% similarity along 57 bp (Figure 1C). The identified partial complementarity mapped at positions 5,447 to 5,500 in the 3′untranslated region (UTR) of ITGA6 mRNA type 1/variant 1 (v1). Furthermore, the ITGA6-AS1 showed 65.4% alignment to 26 bp of ITGA6 exon 2 (E2) (Figure 1D).
Knowing the lncRNA loci are sources of microRNA (Sun et al., 2021), we explored the presence of microRNA in lncRNA ITGA6-AS1 transcript by using miRbase, human (Homo sapiens) option tool. Four miRNA sequences identified were referred to by us as human (hsa)-miR-AS1.1, hsa-miR-AS1.2, hsa-miR-AS1.3, and hsa-miR-AS1.4 that showed similarities to hsa-miR-6772–5p, hsa-miR-7109–3p, hsa-miR-6797–5p, and hsa-miR-1911–5p respectively (Figure 1E, and Supplementary Table S6). The in-silico analysis by RNA22 v2 tool for microRNA recognition elements (MRE) in the target transcript showed that miR hsa-miR-AS1.2 could not target ITGA6 MRE and thus cannot interfere with ITGA6 transcription. The other three identified miR sequences target ITGA6 MRE of ITGA6 transcript at ITGA6 3′-UTR and ITGA6 exon 1 (Supplementary Table S7). The low p-values, 6.18E-2–1.21E-1, represent a greater chance of the ITGA6 transcript containing a valid ITGA6 MRE.
Genomic context of integrins alpha 6 exon 2 promoter
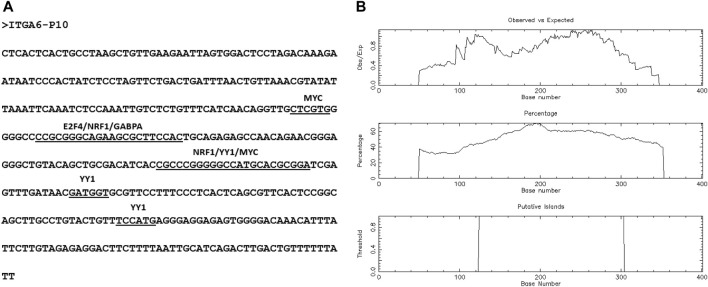
Ensembl database showed ten ITGA6 alternative putative promoters from P1 to P10 (Supplementary Table S4). Two of 10 ITGA6 promoters, P1 and P10, were mapped at ITGA6 exon-1 and exon-2, respectively, and eight promoters, P2 to P9 mapped in the ITGA6 intron-1 (Figure 1B). The divergent location of ITGA6 exon two and ITGA6-AS1, overlapping promoter ITGA6 exon-2 (P10) sequences, suggests P10 may possess bidirectional activity. Consistent with this observation, the bioinformatics analysis demonstrated P10 has the features and properties of a bidirectional promoter (Yang and Elnitski, 2008; Seila et al., 2009; Orekhova and Rubtsov, 2013; Al-Obaide et al., 2021). In addition to the divergent configuration of ITGA6-AS1 and P10, the 402-nucleotide sequence of P10 lacked a TATA box motif and enriched in binding sites for several transcription factors GABPA, MYC, E2F4, NRF1, YY1 found in bidirectional promoters (Figure 2A). Additionally, bioinformatics analysis demonstrated that the ITGA6-P10 is CG rich and contains CGI. EMBOSS Cpgplot software revealed a CGI length of 180 bp in the P10 sequence that extends from nucleotide 125 to 304 along P10 sequence (Figure 2B).
FIGURE 2.
The predicted features of P10, bidirectional promoter. (A) P10 sequence shows transcription factors binding sites. (B) Identification of CpG Island in the P10 sequence. The following options were searched: Window size, 100; minimum sequence length, 150; minimum Obs/Exp CpG, >0.6; %C + %G, >50.00%.
Quantification of expression of integrin alpha 6 and integrin alpha 6-antisense 1 expression in mesenchymal stem cells
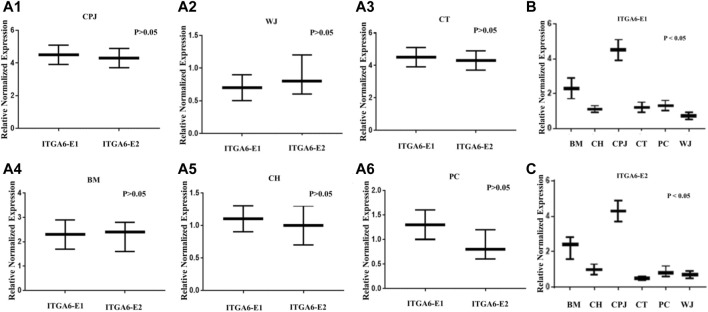
To validate the bioinformatic findings and significance that might be influencing the properties of MSCs, we analyzed the expression of both ITGA6 and ITGA6-AS1 by RT-qPCR. The results depicted in Figures 3A1–A6 show no significant difference between the expression of ITGA6-E1 and ITGA6-E2 in the same tested MSCs. However, there was a statistically significant difference in the expressions of ITGA6-E1 in the samples of MSCs from various sources (Figure 3B). A similar trend in the expression of ITGA6-E2 was observed among various MSCs (Figure 3C). The relative normalized expression values for ITGA6-E1 and ITGA6-E2 showed that CPJ-MSCs had a 2 to 5-fold increase in expression levels over other MSCs.
FIGURE 3.
The expression of ITGA6 in the MSCs. (A1–A6) The t-test showed no statistically significant difference in expressions of ITGA6-E1 and ITGA6-E2 in the same MSC, p > 0.05. (B–C) A one-way ANOVA revealed a statistically significant difference in the expressions of ITGA6-E1 and ITGA6-E2, respectively in the six MSCs, p < 0.05.
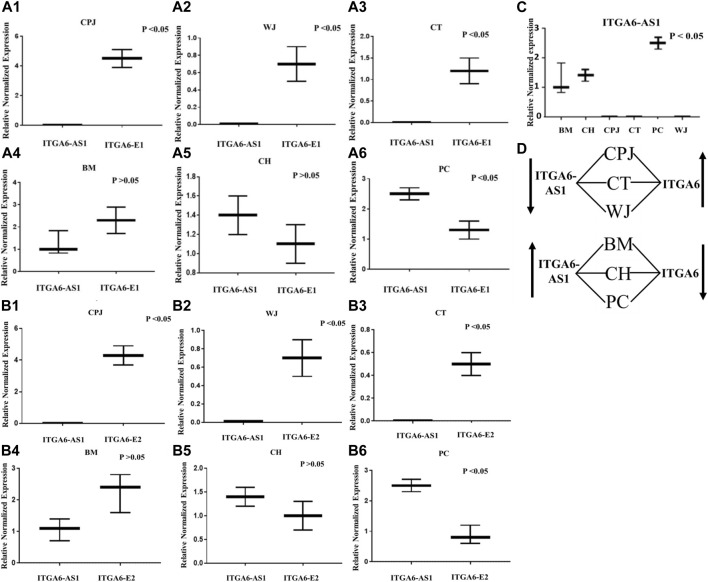
We then compare the expression of ITGA6 and ITGAS6-AS1 in the MSCs from all six sources. The results reported in Figure 4 show that the expression of ITGA6 was inversely proportional to the ITGA6-AS1. The primer set directed against ITGA6-E1 indicated significantly higher expression of ITGA6 but lower expression of ITGA6-AS1 in MSCs from CPJ, WJ, and CT (Figures 4A1–A3). In contrast, the difference in the expression of ITGA6 and ITGA6-AS1 was insignificance in the MSCs from BM and CH (Figures 4A4–A5). On the other hand, the expression of ITGA6-AS1 was significantly higher than ITGA6 in PC MSCs (Figure 4A6). A similar trend was noted when the primer set against ITGA6-E2 was used (Figures 4B1–B6). The expression of ITGA6-AS1 was significantly higher in MSCs from BM, CH, and PC when compared to CPJ CT and WJ. The highest expression of the antisense RNA was observed in PC MSCs (Figure 4C). There was a clear pattern in the expression of ITGA6 and ITGA6-AS1 in the MSC samples, as summarized in Figure 4D. When ITGA6-AS1 was high, expression of ITGA6 was low, suggesting its potential role in the regulation of ITGA6 in MSCs. Based on the bioinformatics analysis, we proposed that P10 is involved in the expression of ITGA6-AS1 that overlaps ITGA6 exon two sequence. However, further studies are warranted to define the function of P10 unambiguously.
FIGURE 4.
The expression of ITGA6-E1 and ITGA6-AS1 in MSCs. (A1–A6) Expression of ITGA6-E1 and ITGA6-AS1 in the same CPJ, WJ, CT, BM, CH and PC, respectively. (B1–B6) Expressions of ITGA6-E2 and ITGA6-AS1 in the same CPJ, WJ, CT, BM, CH and PC, respectively. (C) A one-way ANOVA revealed a statistically significant difference in ITGA6-AS1 expressions in the tested MSCs, p < 0.05. (D) Sketch showing correlation of ITGA6-AS1 and ITGA6 expressions in MSCs.
Discussion
ITGA6 is one of the many cell surface markers commonly found in over 30 types of stem cells (Krebsbach and Villa-Diaz, 2017). While ITGA6’s role in a broad range of stem cell populations has been linked to maintaining the self-renewal of pluripotent stem cells and breast and glioblastoma cancer stem cells, its function in the multipotent MSCs is not well understood.
Our study of the six human MSCs provided evidence of a potential correlation between the ITGA6 and its antisense RNA, ITGA6-AS1, and MSCs characteristics for the first time. Elevated expression of ITGA6 in BM MSCs from different donors was correlated with high clonogenicity, migration, differentiation, low doubling time, and proliferation (Nieto-Nicolau et al., 2020; (Lee et al., 2009; Yu et al., 2012; Nystedt et al., 2013). However, our studies showed low ITGA6 expression, poor clonogenicity, and higher doubling time in BM MSCs. On the other hand, we found significantly higher clonogenicity and ITGA6 expression and low doubling time in MSCs derived from human umbilical cord tissues (CT, CPJ, and WJ).
RNA generated from lncRNA has been shown to be a widespread phenomenon in regulating genes in eukaryotes and humans (Villegas and Zaphiropoulos, 2015). The plausible explanation for the ITGA6-AS1 regulatory role originates from the transcript’s sequence features. Antisense RNAs are lncRNAs subtypes that can alter the stability and translation of cytoplasmic mRNAs (Statello et al., 2021; Sun et al., 2021). Also, lncRNAs are host for microRNA (miRNA). The miRNA can be produced from non-coding RNA transcripts (Ballantyne et al., 2016; He et al., 2019; Zhang et al., 2019). The three identified miR sequences from ITGA6-AS1 transcript in this study can potentially target MREs in the ITGA6 transcript at ITGA6 3′-UTR, and ITGA6 exon 1. Previous studies reported miRNAs could target 5′ and 3′ UTRs as well as exons (Broughton and Pasquinelli, 2016; Plotnikova et al., 2019). The UTRs at the 3′ end of mRNA transcripts play a crucial role in gene expression and contain important sequences that influence the fate of mRNA (Matoulkova et al., 2012; Mayr, 2019). The regulatory mechanism of miRNAs functions through inactivating the target mRNA by the silencing complex (RISC) (Tafrihi and Hasheminasab, 2019; Al-Obaide et al., 2021). Further, in this study, we showed putative partial binding of ITGA6-AS1 to the complementary region of the ITGA6 at exon two and 3′UTR, such partial binding may have a consequential post-transcriptional influence on the ITGA6 mRNA and cause mRNA stabilization or destabilization (He et al., 2019). Although the reported data in this study suggest ITGA6-AS1 is involved in the ITGA6 regulation, more detailed studies are required to explore the ITGA6-AS1 role in ITGA6 expression in MSCs.
Additional research could be conducted to explore the inverse correlation in the expression of ITGA6 and ITGA6-AS1. The NCBI-Nucleotide and Ensembl databases showed ITGA6 has several different splice variants; quantifying these variants may indicate transcription preference of ITGA6 transcript type correlated with ITGA6-AS1. Overexpression or inhibition analysis of ITGA6-AS1 should be performed to assess its effect on ITGA6 expression. It is also of interest to investigate the interaction between ITGA6-AS1 and lncRNA–ITGA6 mRNA transcripts by RNA-RNA pulldown assay. Further work is warranted to show the potential roles of ITGA6 bidirectional promoter (P10) usage in the expression of ITGA6-AS1 and ITGA6 in the MSCs. Our results also detected intrinsic elements in the putative ITGA6 that generate microRNAs from the ITGA6-AS1, which could impart epigenetic regulation of ITGA6. A clear understanding of ITGA6-AS1 regulation of the gene could provide novel ways to improve the therapeutic potential of MSCs.
Acknowledgments
The study was also supported by the OU-WB Institute for Stem Cell and Regenerative Medicine (ISCRM), Oakland University, Rochester, MI, Ascension–Providence Hospital, Southfield, MI, and Michigan Head and Spine Institute, Southfield, MI. We also acknowledge support from Oakland University for the Provost Graduate (AI, CB) and Undergraduate (MM) Research Awards. We thank Ibtisam Ismael Al-Obaidi for technical assistance.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
RC conceived, initiated, and supervised the study. MA conceived the study, searched, collected, curated datasets, and performed data analysis and figures. AI, MM, CB, MC, and PA performed cell culture and RT-qPCR, collected data, and prepared the figures. MC provided suggestions and participated in writing the manuscript. MA and RC wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by National Institutes of Health R16 NS129748-01 grant to G. Rasul Chaudhry.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.968228/full#supplementary-material
References
- Akiyama S. K. (1996). Integrins in cell adhesion and signaling. Hum. Cell. 9, 181–186. [PubMed] [Google Scholar]
- Al-Obaide M. A. I., Al O., Vasylyeva T. L. (2021). Unexplored regulatory sequences of divergently paired GLA and HNRNPH2 loci pertinent to Fabry disease in human kidney and skin cells: Presence of an active bidirectional promoter. Exp. Ther. Med. 21, 154. 10.3892/etm.2020.9586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assis-Ribas T., Forni M. F., Winnischofer S. M. B., Sogayar M. C., Trombetta-Lima M. (2018). Extracellular matrix dynamics during mesenchymal stem cells differentiation. Dev. Biol. 437, 63–74. 10.1016/j.ydbio.2018.03.002 [DOI] [PubMed] [Google Scholar]
- Ballantyne M. D., McDonald R. A., Baker A. H. (2016). lncRNA/MicroRNA interactions in the vasculature. Clin. Pharmacol. Ther. 99, 494–501. 10.1002/cpt.355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becerra-Bayona S. M., Guiza-Arguello V. R., Russell B., Höök M., Hahn M. S. (2018). Influence of collagen-based integrin α(1) and α(2) mediated signaling on human mesenchymal stem cell osteogenesis in three dimensional contexts. J. Biomed. Mat. Res. A 106, 2594–2604. 10.1002/jbm.a.36451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeravolu N., Khan I., McKee C., Dinda S., Thibodeau B., Wilson G., et al. (2016). Isolation and comparative analysis of potential stem/progenitor cells from different regions of human umbilical cord. Stem Cell. Res. 16, 696–711. 10.1016/j.scr.2016.04.010 [DOI] [PubMed] [Google Scholar]
- Beeravolu N., McKee C., Alamri A., Mikhael S., Brown C., Perez-Cruet M., et al. (2017). Isolation and characterization of mesenchymal stromal cells from human umbilical cord and fetal placenta. J. Vis. Exp. 122, 55224–55293. 10.3791/55224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton J. P., Pasquinelli A. E. (2016). A tale of two sequences: microRNA-target chimeric reads. Genet. Sel. Evol. 48, 31. 10.1186/s12711-016-0209-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C., McKee C., Bakshi S., Walker K., Hakman E., Halassy S., et al. (2019). Mesenchymal stem cells: Cell therapy and regeneration potential. J. Tissue Eng. Regen. Med. 13, 1738–1755. 10.1002/term.2914 [DOI] [PubMed] [Google Scholar]
- Brown C., McKee C., Halassy S., Kojan S., Feinstein D. L., Chaudhry G. R. (2021). Neural stem cells derived from primitive mesenchymal stem cells reversed disease symptoms and promoted neurogenesis in an experimental autoimmune encephalomyelitis mouse model of multiple sclerosis. Stem Cell. Res. Ther. 12, 499. 10.1186/s13287-021-02563-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cakouros D., Gronthos S. (2020). Epigenetic regulators of mesenchymal stem/stromal cell lineage determination. Curr. Osteoporos. Rep. 18, 597–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Sheppard D. (2007). Identification and molecular characterization of multiple phenotypes in integrin knockout mice. Methods Enzymol. 426, 291–305. 10.1016/S0076-6879(07)26013-6 [DOI] [PubMed] [Google Scholar]
- Chen J. Y., Mou X. Z., Du X. C., Xiang C. (2015). Comparative analysis of biological characteristics of adult mesenchymal stem cells with different tissue origins. Asian pac. J. Trop. Med. 8, 739–746. 10.1016/j.apjtm.2015.07.022 [DOI] [PubMed] [Google Scholar]
- Costa L. A., Eiro N., Fraile M., Gonzalez L. O., Saá J., Garcia-Portabella P., et al. (2021). Functional heterogeneity of mesenchymal stem cells from natural niches to culture conditions: Implications for further clinical uses. Cell. Mol. Life Sci. 78, 447–467. 10.1007/s00018-020-03600-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elahi K. C., Klein G., Avci-Adali M., Sievert K. D., MacNeil S., Aicher W. K. (2016). Human mesenchymal stromal cells from different sources diverge in their expression of cell surface proteins and display distinct differentiation patterns. Stem Cells Int. 2016, 5646384. 10.1155/2016/5646384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González P. L., Carvajal C., Cuenca J., Alcayaga-Miranda F., Figueroa F. E., Bartolucci J., et al. (2015). Chorion mesenchymal stem cells show superior differentiation, immunosuppressive, and angiogenic potentials in comparison with haploidentical maternal placental cells. Stem Cells Transl. Med. 4, 1109–1121. 10.5966/sctm.2015-0022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraguchi N., Ishii H., Mimori K., Ohta K., Uemura M., Nishimura J., et al. (2013). CD49f-positive cell population efficiently enriches colon cancer-initiating cells. Int. J. Oncol. 43, 425–430. 10.3892/ijo.2013.1955 [DOI] [PubMed] [Google Scholar]
- He R. Z., Luo D. X., Mo Y. Y. (2019). Emerging roles of lncRNAs in the post-transcriptional regulation in cancer. Genes. Dis. 6, 6–15. 10.1016/j.gendis.2019.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou T., Zhang W., Tong C., Kazobinka G., Huang X., Huang Y., et al. (2015). Putative stem cell markers in cervical squamous cell carcinoma are correlated with poor clinical outcome. BMC Cancer 15, 785. 10.1186/s12885-015-1826-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes R. O. (2002). Integrins: Bidirectional, allosteric signaling machines. Cell. 110, 673–687. 10.1016/s0092-8674(02)00971-6 [DOI] [PubMed] [Google Scholar]
- Jales Neto L. H., Wicik Z., Torres G. H. F., Takayama L., Caparbo V. F., Lopes N. H. M., et al. (2020). Overexpression of SNTG2, TRAF3IP2, and ITGA6 transcripts is associated with osteoporotic vertebral fracture in elderly women from community. Mol. Genet. Genomic Med. 8, e1391. 10.1002/mgg3.1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju C., Liu R., Zhang Y. W., Zhang Y., Zhou R., Sun J., et al. (2019). Mesenchymal stem cell-associated lncRNA in osteogenic differentiation. Biomed. Pharmacother. 115, 108912. 10.1016/j.biopha.2019.108912 [DOI] [PubMed] [Google Scholar]
- Kolf C. M., Cho E., Tuan R. S. (2007). Mesenchymal stromal cells. Biology of adult mesenchymal stem cells: Regulation of niche, self-renewal and differentiation. Arthritis Res. Ther. 9, 204. 10.1186/ar2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebsbach P. H., Villa-Diaz L. G. (2017). The role of integrin α6 (CD49f) in stem cells: More than a conserved biomarker. Stem Cells Dev. 26, 1090–1099. 10.1089/scd.2016.0319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R. H., Seo M. J., Pulin A. A., Gregory C. A., Ylostalo J., Prockop D. J. (2009). The CD34-like protein PODXL and alpha6-integrin (CD49f) identify early progenitor MSCs with increased clonogenicity and migration to infarcted heart in mice. Blood 113, 816–826. 10.1182/blood-2007-12-128702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Han H., Song S., Fan G., Xu H., Zhou W., et al. (2019). HOXC10 regulates osteogenesis of mesenchymal stromal cells through interaction with its natural antisense transcript lncHOXC-AS3. Stem Cells 37, 247–256. 10.1002/stem.2925 [DOI] [PubMed] [Google Scholar]
- Ma W., Liu Y., Ma H., Ren Z., Yan J. (2021). Cis-acting: A pattern of lncRNAs for gene regulation in induced pluripotent stem cells from patients with down syndrome determined by integrative analysis of lncRNA and mRNA profiling data. Exp. Ther. Med. 22, 701. 10.3892/etm.2021.10133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin T. A., Jiang W. G. (2014). Evaluation of the expression of stem cell markers in human breast cancer reveals a correlation with clinical progression and metastatic disease in ductal carcinoma. Oncol. Rep. 31, 262–272. 10.3892/or.2013.2813 [DOI] [PubMed] [Google Scholar]
- Matoulkova E., Michalova E., Vojtesek B., Hrstka R. (2012). The role of the 3' untranslated region in post-transcriptional regulation of protein expression in mammalian cells. RNA Biol. 9, 563–576. 10.4161/rna.20231 [DOI] [PubMed] [Google Scholar]
- Matta C., Boocock D. J., Fellows C. R., Miosge N., Dixon J. E., Liddell S., et al. (2019). Molecular phenotyping of the surfaceome of migratory chondroprogenitors and mesenchymal stem cells using biotinylation, glycocapture and quantitative LC-MS/MS proteomic analysis. Sci. Rep. 9, 9018. 10.1038/s41598-019-44957-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer M. H. (2011). Proteomic definitions of mesenchymal stem cells. Stem Cells Int. 2011, 704256. 10.4061/2011/704256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C. (2019). What are 3' UTRs doing? Cold Spring Harb. Perspect. Biol. 11, a034728. 10.1101/cshperspect.a034728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto-Nicolau N., de la Torre R. M., Fariñas O., Savio A., Vilarrodona A., Casaroli-Marano R. P. (2020). Extrinsic modulation of integrin α6 and progenitor cell behavior in mesenchymal stem cells. Stem Cell. Res. 47, 101899. 10.1016/j.scr.2020.101899 [DOI] [PubMed] [Google Scholar]
- Nystedt J., Anderson H., Tikkanen J., Pietilä M., Hirvonen T., Takalo R., et al. (2013). Cell surface structures influence lung clearance rate of systemically infused mesenchymal stromal cells. Stem Cells 31, 317–326. 10.1002/stem.1271 [DOI] [PubMed] [Google Scholar]
- Orekhova A. S., Rubtsov P. M. (2013). Bidirectional promoters in the transcription of mammalian genomes. Biochemistry. 78, 335–341. 10.1134/S0006297913040020 [DOI] [PubMed] [Google Scholar]
- Peterson R. J., Koval M. (2021). Above the matrix: Functional roles for apically localized integrins. Front. Cell. Dev. Biol. 9, 699407. 10.3389/fcell.2021.699407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotnikova O., Baranova A., Skoblov M. (2019). Comprehensive analysis of human microRNA-mRNA interactome. Front. Genet. 10, 933. 10.3389/fgene.2019.00933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen T. D., Livak K. J. (2008). Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108. 10.1038/nprot.2008.73 [DOI] [PubMed] [Google Scholar]
- Schwartz M. A. (2010). Integrins and extracellular matrix in mechanotransduction. Cold Spring Harb. Perspect. Biol. 2, a005066. 10.1101/cshperspect.a005066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seila A. C., Core L. J., Lis J. T., Sharp P. A. (2009). Divergent transcription: A new feature of active promoters. Cell. Cycle 8, 2557–2564. 10.4161/cc.8.16.9305 [DOI] [PubMed] [Google Scholar]
- Song S., Zhang J., Su Q., Zhang W., Jiang Y., Fan G., et al. (2021). Downregulation of ITGA6 confers to the invasion of multiple myeloma and promotes progression to plasma cell leukaemia. Br. J. Cancer 124, 1843–1853. 10.1038/s41416-021-01362-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Statello L., Guo C. J., Chen L. L., Huarte M. (2021). Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell. Biol. 22, 96–118. 10.1038/s41580-020-00315-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan K. E., Quinn K. P., Tang K. M., Georgakoudi I., Black L. D. (2014). Extracellular matrix remodeling following myocardial infarction influences the therapeutic potential of mesenchymal stem cells. Stem Cell. Res. Ther. 5, 14. 10.1186/scrt403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q., Song Y. J., Prasanth K. V. (2021). One locus with two roles: microRNA-independent functions of microRNA-host-gene locus-encoded long noncoding RNAs. Wiley Interdiscip. Rev. RNA 12, e1625. 10.1002/wrna.1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tafrihi M., Hasheminasab E. (2019). MiRNAs: Biology, biogenesis, their web-based tools, and databases. Microrna 8, 4–27. 10.2174/2211536607666180827111633 [DOI] [PubMed] [Google Scholar]
- Teven C. M., Liu X., Hu N., Tang N., Kim S. H., Huang E., et al. (2011). Epigenetic regulation of mesenchymal stem cells: A focus on osteogenic and adipogenic differentiation. Stem Cells Int., 201371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villegas V. E., Zaphiropoulos P. G. (2015). Neighboring gene regulation by antisense long non-coding RNAs. Int. J. Mol. Sci. 16, 3251–3266. 10.3390/ijms16023251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z. Y., Wang P., Wu Y. F., Shen H. Y. (2019). Long non-coding RNA: The functional regulator of mesenchymal stem cells. World J. Stem Cells 11, 167–179. 10.4252/wjsc.v11.i3.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K. M. (1997). Integrin signaling. Matrix Biol. 16, 137–141. 10.1016/s0945-053x(97)90001-9 [DOI] [PubMed] [Google Scholar]
- Yang M. Q., Elnitski L. L. (2008). Diversity of core promoter elements comprising human bidirectional promoters. BMC Genomics 9, S3. 10.1186/1471-2164-9-S2-S3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K. R., Yang S. R., Jung J. W., Kim H., Ko K., Han D. W., et al. (2012). CD49f enhances multipotency and maintains stemness through the direct regulation of OCT4 and SOX2. Stem Cells 30, 876–887. 10.1002/stem.1052 [DOI] [PubMed] [Google Scholar]
- Zealy R. W., Fomin M., Davila S., Makowsky D., Thigpen H., McDowell C. H., et al. (2018). Long noncoding RNA complementarity and target transcripts abundance. Biochim. Biophys. Acta. Gene Regul. Mech. 1861, 224–234. 10.1016/j.bbagrm.2018.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D. H., Yang Z. L., Zhou E. X., Miao X. Y., Zou Q., Li J. H., et al. (2016). Overexpression of Thy1 and ITGA6 is associated with invasion, metastasis and poor prognosis in human gallbladder carcinoma. Oncol. Lett. 12, 5136–5144. 10.3892/ol.2016.5341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Wang W., Zhu W., Dong J., Cheng Y., Yin Z., et al. (2019). Mechanisms and functions of long non-coding RNAs at multiple regulatory levels. Int. J. Mol. Sci. 20, E5573. 10.3390/ijms20225573 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.