Graphical abstract
Keywords: Fungi, Bacteria, Protista, Glycome, Carbohydrate, Glycan, Diversity, CSDB
Abstract
In this paper, we present the results of a systematic statistical analysis of the fungal glycome in comparison with the prokaryotic and protistal glycomes as described in the scientific literature and presented in the Carbohydrate Structure Database (CSDB). The monomeric and dimeric compositions of glycans, their non-carbohydrate modifications, glycosidic linkages, sizes of structures, branching degree and net charge are assessed. The obtained information can help elucidating carbohydrate molecular markers for various fungal classes which, in its turn, can be demanded for the development of diagnostic tools and carbohydrate-based vaccines against pathogenic fungi. It can also be useful for revealing specific glycosyltransferases active in a particular fungal species.
1. Introduction
The kingdom Fungi comprises a tremendous number of biological species that differ in their appearance, morphology, life strategies and ecology [1]. Fungi are virtually everywhere; according to estimations, the number of their species comes close to four million [2]. They can be found as high as in the stratosphere and as deep as on the ocean floor, in places as hot as deserts and as cold as Antarctic glaciers; some of them form indispensable relationships with plants and animals, and other spoil food supplies and provoke severe diseases in humans and other organisms [1], [3]. Due to the remarkable diversity in combination with a simple body structure, the accurate taxonomical classification of fungi is a challenging task, and the fungal taxonomy has been revised on several occasions [1], [2], [4]. Nevertheless, all these organisms have something in common: they are heterotrophic eukaryotes, and their cells are surrounded by the cell wall mainly composed of chitin [1].
Similar to prokaryotes, the cell wall both protects the fungal cell from the environment and provides it with the means of interactions with it. In general, the fungal cell wall comprises two layers: the relatively conserved inner layer, which serves as a skeleton and mainly consists of chitin and glucan, and the more variable outer layer, which includes glycoproteins bearing species-specific oligosaccharide moieties [5], [6]. Since cells of animals, including humans, have no cell walls, this part of fungal cells is an apparent candidate to become a drug target and a trigger of the immune response in higher organisms [5], [7], [8]. However, similar to bacteria, fungi have the ability to evade antimicrobial substances, as well as the protective immune system of a host organism, via cell wall modifications, including epitope masking [8], [9]. Thus, understanding the mechanisms of immune evasion and drug resistance of pathogenic fungi, as well as the response of ecologically significant species to various types of environmental stress, relies on the exact knowledge of the structure of the fungal cell wall under various conditions.
The Carbohydrate Structure Database (CSDB, http://csdb.glycoscience.ru) is a free curated repository of prokaryotic, fungal, protistal and plant glycans [10]. It stores structural, taxonomical, NMR spectroscopic, bibliographical and other data on glycans, glycopolymers, and glycoconjugates from organisms from these domains of life. Currently, CSDB is the only scientific database that provides a close-to-complete coverage on published carbohydrate structures from microorganisms (bacteria, fungi and protista) up to the year 2020. The completeness of coverage is one of the most important characteristics of a database because it means that if an empty set is returned to a search request, then the searched object does not exist (or, as in the case of CSDB, has not been published so far). Whereas the complete coverage on bacterial glycans in CSDB has been used in other studies and assessed statistically [11], [12], the expansion of CSDB to fungal glycans has been started relatively recently [13], and the completeness of coverage was achieved in 2021 [14]. Thus, the fungal glycome stored in CSDB has not been analyzed so far.
In this paper, we carry out the first systematic statistical analysis of the fungal glycome presented in the scientific literature, and compare its numerical metrics with these for other microorganisms from the domains of prokaryotes and protista.
2. Results and discussion
The CSDB Linear notation [15] (e.g. aDManp for α-d-mannopyranose) is used for text identifiers of mono- and oligomeric fragments throughout the text and in the figures in order to make the labels shorter. On first use, full names are provided where unobvious. The CSDB logic of identifying residues is utilized: a residue is generally an entity that is connected with other entities by bonds implying the elimination of water. For example, N-acetylglucosamine consists of two residues: 2-aminoglucose and acetic acid. Though imperfect from a biosynthetic viewpoint, this purely structural approach allows avoiding the combinatorial burst of building blocks which obscures the data cumulation.
2.1. General data
Currently, CSDB contains ca. 5900 fungal carbohydrate structures (ca. 20 % of all the structures stored in the database) from ca. 2900 publications. These structures are assigned to ca. 3650 organisms (ca. 25 % of all the organisms in the database). The coverage was reported as close to complete for fungi up to the year 2020 [14]. These data reflect the fullness of study of the fungal glycome per taxonomic class; they are calculated for fungal species from 15 defined classes, eight of which belong to the phylum Ascomycota (ca. 4600 structures), six – to Basidiomycota (ca. 1900 structures), and one – to Mucoromycota (ca. 150 structures) (see Fig. 1). There are also 67 and 40 structures from Ascomycota and Basidiomycota with the class unspecified, respectively, as well as few occurrences of glycan structures form other classes belonging to the phyla Ascomycota (Arthoniomycetes, Lichinomycetes, Mortierellomycetes, Orbiliomycetes, Pneumocystidomycetes), Basidiomycota (Agaricostilbomycetes, Cystobasidiomycetes, Dacrymycetes, Pucciniomycetes, Tritirachiomycetes), Mucoromycota (Umbelopsidomycetes), Blastocladiomycota (Blastocladiomycetes), Chytridiomycota (Chytridiomycetes, Monoblepharidomycetes), and Glomeromycota (Glomeromycetes). The full data on the current CSDB coverage are provided in Supplementary Table S1a.
Fig. 1.
Absolute abundance of fungal carbohydrate structures in CSDB per taxonomic class. The affiliation of the classes with the corresponding phyla is color-coded. See Supplementary Table S1b for the source data.
In the subsequent sections, the absolute class population (the number of distinct reported glycans from species belonging to a particular taxonomic class) is used as a normalization basis; thus, the relative occurrence corresponds to an average content of a certain structural feature in glycans from a particular class.
2.2. Monomers
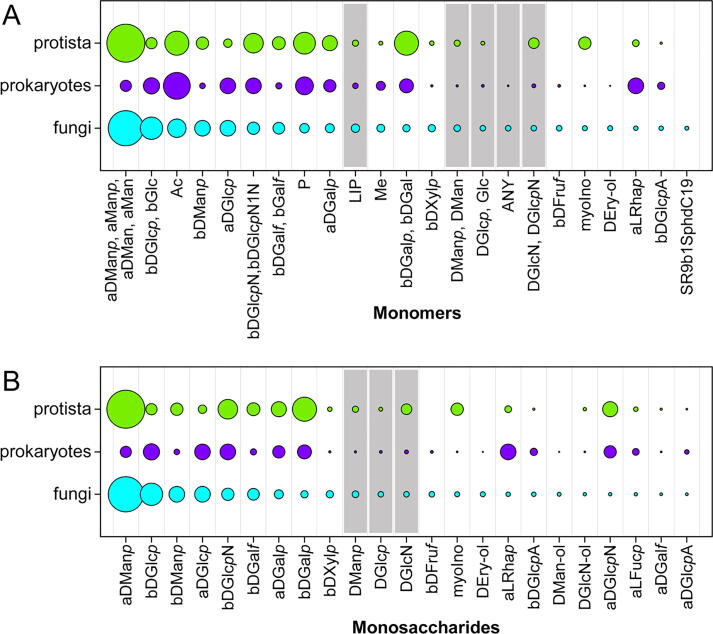
Fig. 2 shows the distribution of monomeric (A) and, in particular, monosaccharide (B) residues in fungal glycans as compared to those in prokaryotic and protistal ones. For clarity, only the most abundant residues present in more than 200 (Fig. 2A) or 100 (Fig. 2B) fungal structures are considered (thus, the plots can lack some other monomers that can be more frequent in bacteria and/or protista but virtually absent in fungi). According to the data stored in CSDB, mannose and glucose are the most frequent monomers in fungal carbohydrates, together with the acetic acid residue, which presumably originates from 2-acetamido-2-deoxy-glucopyranose residues. α-Mannose (aMan, aDMan, aManp, aDManp) is also the most frequent monomer in protistal glycans, as well as α- and β-d-galactose (aDGalp, bGalp, bDGalp, bGalf, bDGalf), 2-amino-2-deoxy-β-d-glucopyranose (bDGlcpN), acetic acid, and phosphoric acid, whereas in bacterial glycans, acetate is the most frequent residue. Other frequent bacterial monomers include bDGlcpN, α- and β-d-galactose, α- and β-d-glucose (Glc, DGlcp, aDGlcp, bGlc, bDGlcp), and α-l-rhamnopyranose (aLRhap) (Fig. 2A). The pseudo-residue 1,2-diamino-1,2-dideoxy-β-d-glucopyranose (bDGlcpN1N) originates from asparagine-linked root residues of N-glycans within glycoproteins, according to the way of recording N-glycosides in CSDB.
Fig. 2.
Distribution of all monomeric residues (A) and monosaccharides/alditols (B) in glycans from fungi, bacteria and protista (including unicellular algae). Only the residues found in more than 200 (A) or 100 (B) fungal structures are shown. Underdetermined entities, for which some configurations or the exact residue identity are not reported, are highlighted in grey. The bubble area is an average occurrence of a given residue per structure. See Supplementary Table S2a and S2b for the source data.
As for monosaccharide and alditol residues, α-d-mannopyranose (aDManp) is the most frequent monosaccharide found in fungi, as well as in protista, whereas in bacteria the highest frequency is observed for β-d-glucopyranose (bDGlcp), α-d-glucopyranose (aDGlcp), bDGlcpN, and aLRhap. Glucopyranose residues, which are also frequent in the fungal structures, possibly come from the skeletal layer of the fungal cell wall consisting of chitin and glucans (Fig. 2B). d-mannitol (DMan-ol) and 2-amino-2-deoxy-d-glucitol (DGlcN-ol) are probably analytical artifacts.
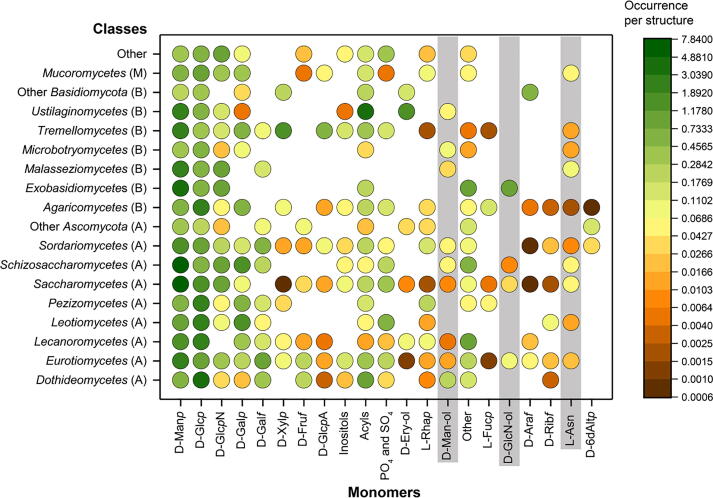
In accordance with the above-discussed abundance of all monomeric and purely monosaccharide components in fungal glycans, d-mannose and d-glucose are the most common monomers in the carbohydrate structures from species belonging to most of the classes stored in CSDB (Fig. 3). The only exception is Ustilaginomycetes, in which aliphatic acids are the most frequent monomers. In other classes, which are not defined separately in the plot, 2-amino-2-deoxy-d-glucopyranose is most frequent; being a building block of chitin, this residue is common for most of the fungal classes. For some classes, in which glycoproteins are more studied, l-asparagine is reported; it is an attachment site in N-glycoproteins.
Fig. 3.
Distribution of monomeric residues in fungal classes. The color of the bubbles corresponds to the occurrence of a given residue per structure in a given taxonomic class (see the logarithmic color scale on the right). Probable analytical or notation artifacts are highlighted in grey. Phyla for the classes are indicated in parentheses: (A), Ascomycota, (B), Basidiomycota, and (M), Mucoromycota. See Supplementary Table S3 for the source data.
Notably, glycans from the classes belonging to the phylum Basidiomycota contain no or rare d-fructofuranose, in contrast to the glycans from Ascomycota and Mucoromycota, whereas glycans from Mucoromycota, in their turn, contain no or rare d-galactofuranose (DGalf), d-xylopyranose (DXylp), inositols, d-erythritol (d-Ery-ol), l-fucopyranose (LFucp), d-arabinofuranose (DAraf), d-ribofuranose (DRibf), and D-6-deoxyaltropyranose (D-6dAltp). It should be noted that, according to Fig. 1, glycans from this phylum are significantly understudied in comparison with those from Ascomycota and Basidiomycota, and this can be the reason of some gaps in Fig. 3 and other figures.
d-Man-ol and DGlcN-ol are probably analytical artifacts, as well as l-Asn (this amino acid residue is an N-glycan attachment site in glycoproteins and is left as a part of glycan structures after the analytical processing and notation of N-glycoproteins). These residues are highlighted in grey in Fig. 3.
2.3. Unique building blocks
Revealing structural components unique for a certain taxonomic group is a potential basis for microorganism classification and possible antimicrobial therapy targeting. Upon preparation of the presented statistical data, animal carbohydrates have not been considered as a comparison basis to detect uniqueness, since they are not covered by CSDB. However, animal glycans and glycoconjugates, especially those from mammals, are known for their conservative composition limited by a few standard building blocks [11], [16]. Therefore, an atypical component identified as unique in accordance with the CSDB content is most likely unique in relation to all biota, including higher animals.
The monomeric residues unique for fungal glycans are mostly represented by carboxylic acids, such as 3,5-dihydroxy-decanoic acid, 2-hydroxy-stearic acid, 2R-hydroxy-octadec-3E-enoic acid, 18-hydroxy-oleic acid, murolic acid, protoconstipatic acid, 2,4,6,8,10,12,14,16,18-nonamethyl-5,9,13-trihydroxy-2E,6E,10E-icosatrienoic acid, 17R-hydroxy-oleic acid, 2R-hydroxy-hexadec-3E-enoic acid, and 2-heptyl-4,6-dihydroxybenzoic acid. There are also sordaricin (tetracyclic diterpenoid) and sphingoids (2S,3R,4E,8E)-9-methyl-4,8-sphingadienine-C18 and icosaphytosphingosine (presumably parts of sphingolipids). Of carbohydrate residues, β-d-altropyranose (bDAltp) and β-d-altruronic acid (bDAltpA), as well as β-3-deoxy-d-erythro-pentafuranose and 1,4-dideoxy-d-xylohexapyranose (carbon-linked 4-deoxy-d-xylohexapyranose in C-glycosides) are detected (Fig. 4).
Fig. 4.
Distribution of monomeric residues unique for fungi among all biota present in CSDB, per class. The color of the bubbles corresponds to the occurrence of a given residue per structure in a given taxonomic class (see the logarithmic color scale on the right). Phyla for the classes are indicated in parentheses: (A), Ascomycota, (B), Basidiomycota, and (M), Mucoromycota. 9-methyl-4,8-sphingadienine-C18 = (2S,3R,4E,8E)-9-methyl-4,8-sphingadienine-C18; nonamethyltrihydroxyicosatrienoic acid = 2,4,6,8,10,12,14,16,18-nonamethyl-5,9,13-trihydroxy-2E,6E,10E-icosatrienoic acid. See Supplementary Table S4 for the source data.
(2S,3R,4E,8E)-9-methyl-4,8-sphingadienine-C18 and 2-hydroxy-stearic acid are the monomers most common for all the fungal classes studied; they are found in glycolipids of various classes of the Acsomycota, Basidiomycota and Mucoromycota phyla. 3,5-Dihydroxy-decanoic acid is present in the carbohydrate conjugate structures from two fungal classes: Dothideomycetes and Sordariomycetes. Of note, in the former it is significantly more abundant than the other unique residues found in this class ((2S,3R,4E,8E)-9-methyl-4,8-sphingadienine-C18, 2R-hydroxy-hexadec-3E-enoic acid, sordaricin, and 20S-isocholesterol). Murolic / allo-murolic acid and protoconstipatic acid are found only in the structures from Lecanoromycetes; 20S-isocholesterol is specific for Dothideomycetes; 18-hydroxy-oleic acid, 17R-hydroxy-oleic acid and bDAltpA – for Saccharomycetes, 2-heptyl-4,6-dihydroxybenzoic acid – for Eurotiomycetes, whereas C-linked 4-deoxy-d-xylohexapyranose (according to the CSDB notation, it is 1,4-dideoxy-d-xylohexapyranose) and 2,4,6,8,10,12,14,16,18-nonamethyl-5,9,13-trihydroxy-2E,6E,10E-icosatrienoic acid are unique features of Sordariomycetes.
Sordariomycetes, Saccharomycetes, and Eurotiomycetes (Ascomycota) are characterized by the highest variety of monomeric residues: species of these classes contain 11, 8, and 8 out of 18 unique monomeric residues found in fungal glycans, respectively. In contrast, Tremellomycetes and Agaricomycetes (Basidiomycota) contain only two and three unique monomers, respectively, similarly to Mucoromycetes (Mucoromycota). Note that other fungal classes and residues, which are below the frequency threshold (see explanations to Supplementary Table S4) and thus are absent from the plot, can be significantly understudied in comparison with those present.
Fig. 5 shows the distribution of monosaccharides unique for fungal glycans from a cumulative viewpoint. Among them, there are several presumable analytical artifacts, such as d-glycero-d-manno-heptitol (DDmanHep-ol), and d-mannonic acid (DMan-onic). Among the other carbohydrate monomers unique for fungi, β-d-mannofuranose (bDManf) is found in the glycans from three classes (Sordariomycetes, Leotiomycetes, and Dothideomycetes); bDAltp, β-3-deoxy-d-erythro-pentofuranose (bD3deryPenf), bDAltpA, α-d-mannofuranose (aDManf), and α-D-6-deoxyallopyranose (aD6dAllp) are present in glycans from two classes each, whereas C-linked β-D-4-deoxy-xylohexopyranose (bD1,4dxylHexp), α-l-mannose (aLMan), l-xylopyranose (LXylp), and α-D6-sulphoquinovose (aDS6Qui) are characteristic for a single class each.
Fig. 5.
Distribution of monosaccharides unique for fungi. The height of the bars corresponds to the occurrence of a given monosaccharide per structure in a given taxonomic class and, cumulatively, in fungi. Phyla for the classes are indicated in parentheses: (A), Ascomycota, (B), Basidiomycota, and (M), Mucoromycota. The monosaccharides are sorted by total absolute abundance in fungal glycans (from 43 to 3 instances). See Supplementary Table S5 for the source data.
Sordariomycetes demonstrates the highest variety of unique monosaccharide components. bDAltp is the most frequent monosaccharide in the glycans from this class; they also contain relatively high amounts of bD3deryPenf and bD1,4dxylHexp and lower amounts of bDAltpA, bDManf, aLMan, and DLyx-ol. In contrast, glycans from Dothideomycetes, Lecanoromycetes, Microbotryomycetes, Mucoromycetes, and Umbelopsidomycetes are characterized by a single unique monosaccharide. In particular, structures from Mucoromycetes and Umbelopsidomycetes (Mucoromycota) contain aD6dAllp, which is a unique component of this phylum (Fig. 5). Note that comparisons of the occurrences are valid for the bars of the same color only, whereas the total height of a particular stacked bar demonstrates the abundance of monosaccharides in all the fungal structures stored in CSDB.
Unique disaccharides allow estimating specific glycosyltransferases active in organisms from a given taxon. These glycosyltransferases, in their turn, can be expressed in biotechnologically demanded bacteria for the enzymatic synthesis of immunogenic fungal glycans and the subsequent development of carbohydrate-based vaccines [17] against pathogenic fungi.
Distribution of unique disaccharides in fungal glycans is shown in Fig. 6. As in the case of monomeric residues, several of the dimers probably contain analytical artifacts, such as d-mannitol, d-glucitol and d-mannonic acid. The most common domain-specific dimer, which is found in the structures from species belonging to six fungal classes (Arthoniomycetes, Dothideomycetes, Eurotiomycetes, Lecanoromycetes, Sordariomycetes, and Malasseziomycetes), is bDGalf(1–6)aDManp. bDGalf(1–2)aDManp is found in glycans from species from five classes (Dothideomycetes, Eurotiomycetes, Leotiomycetes, Pezizomycetes, Sordariomycetes, all belonging to the phylum Ascomycota). On the contrary, bDXylp(1–3)aDManp is present only in glycans from species of the Tremellomycetes class. Glycans from species of the Agaricomycetes class are characterized by three unique dimers, bDManp(1–2)aDGalp, aDManp(1–6)aDGalp, and aLFucp(1–6)aDManp, which are not found in the structures from the other classes studied so far. aDGlcp(1–2)DEry-ol is present only in the glycans from Lecanoromycetes, and aLRhap(1–2)bDGalf – in those from Sordariomycetes.
Fig. 6.
Distribution of disaccharides unique for fungi. The height of the bars corresponds to the occurrence of a given disaccharide per structure in a given taxonomic class, and cumulatively, in fungi (a logarithmic scale is used for clarity). Phyla for the classes are indicated in parentheses: (A), Ascomycota, (B), Basidiomycota, and (M), Mucoromycota. The disaccharides are sorted by total absolute abundance in fungal glycans (from 299 to 8 instances). See Supplementary Table S6 for the source data.
From the viewpoint of the relative diversity of unique features in fungal classes, Eurotiomycetes contain ten of the disaccharides shown in Fig. 6, including one possible analytical artifact; Sordaryomycetes contain nine, including two possible analytical artifacts; and Agaricomycetes contain six, including one possible analytical artifact. Arthoniomycetes, Leotiomycetes, Lichinomycetes, Schizosaccharomycetes, Agaricostilbomycetes, and Malasseziomycetes contain only one unique dissacharide, according to the current CSDB coverage. Note that comparisons of the occurrences are valid for the bars of the same color only, whereas the total height of a particular stacked bar demonstrates the abundance of disaccharides in all the fungal structures stored in CSDB.
2.4. Modifications
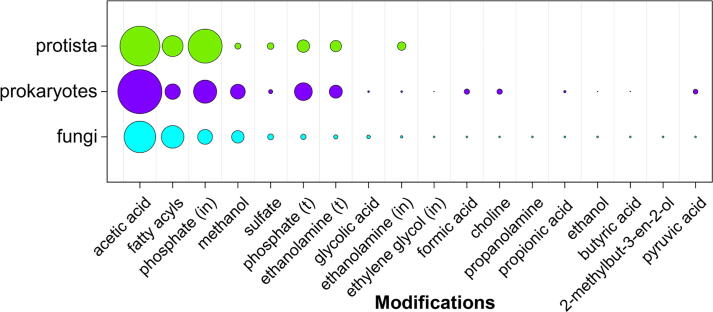
Various covalent modifications can modulate physicochemical and biological properties of natural glycans. Inline modifications are those substituted by monosaccharides or other residues, whereas terminal modifications of monosaccharides, especially O-linked acetates, can be present non-stoichiometrically (not in all the molecules, or not in all the repeating units of a polymer) without loss of the structure connectivity. Fig. 7 shows distribution of non-carbohydrate modifications found in glycans from fungi, prokaryotes and protista present in CSDB. Only the modifications most abundant in fungal structures are shown (glycans from prokaryotes and protista can contain other modifications, which are missing from fungal glycans and thus are absent from the plot).
Fig. 7.
Distribution of non-carbohydrate modifications in glycans from fungi, prokaryotes, and protista. Only the modification with greater than 10 occurrences in the fungal glycans are considered (including the “fatty acyls” superclass containing aliphatic acids, each with greater than 10 occurrences in fungi). (in) stands for inline location in a saccharide backbone; (t) stands for terminal location. The bubble area is an average occurrence of residues per structure. See Supplementary Table S7 for the source data.
The most abundant modification of monosacharides in all the three domains is acetylation, presumably due to the presence of the acetate moiety at N-glucosamine residues, which are among the most common structural elements of cell walls [18] (note that in CSDB, acetylated N-glucosamine residues are considered as two separate residues, glucosamine and acetic acid, linked via an amide bond). Among other frequent modifications of glycans from fungi, prokaryotes and protista are fatty acyls (including those from glycosylphosphatidylinositol anchors in eukaryotic species [19]) and phosphate groups (including those from phosphomannans in cell walls of yeasts [20], teichoic acids in cell walls of Gram-positive bacteria [21], lipid A as a part of lipopolysaccharides in outer membranes of Gram-negative bacteria [22], and glycosylphosphatidylinositol anchors in eukaryotic species [19]). Of the modifications unique for fungal structures, there are propanolamine and 2-methylbut-3-en-2-ol. Glycolic acid, ethylene glycol (inline), formic acid, choline, propionic acid, ethanol, butyric acid and pyruvic acid are found in structures from fungi and prokaryotes, but not from protista, possibly because the glycans from the latter are understudied in relation to the former.
2.5. Glycosidic linkages
Fig. 8, Fig. 9, and Fig. 10 show distribution of glycosidic linkages in glycans from fungi, prokaryotes, and protista, respectively. These data allow estimation of a glycosyltransferase pool required to cover a glycome of a certain taxonomic group. They can also be used for the evaluation of monosaccharide building blocks required for the automated synthesis of glycans of a certain taxonomic group [23].
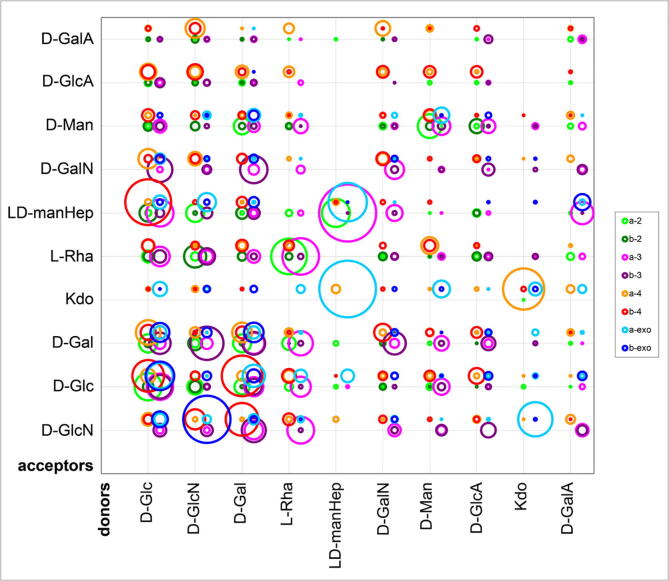
Fig. 8.
Distribution of glycosidic linkages in fungal carbohydrates and derivatives. The area of the circles corresponds to the occurrence of a given linkage per structure. The anomericity and position of the bond is color-coded. “Exo” means all positions above 4, assuming a linkage to an exocyclic tail. The acceptors d-Man-ol and d-GlcN-ol are probably analytical artifacts. Amino sugars include both N-acetylated and non-N-acetylated occurrences. The circle centers are slightly shifted to avoid overlaps. See Supplementary Table S8 for the source data.
Fig. 9.
Distribution of glycosidic linkages in bacterial and archaean carbohydrates and derivatives. The area of the circles corresponds to the occurrence of a given linkage per structure. The anomericity and position of the bond is color-coded. “Exo” means all positions above 4, assuming a linkage to an exocyclic tail. The acceptors d-Man-ol and d-GlcN-ol are probably analytical artifacts. Amino sugars include both N-acetylated and non-N-acetylated occurrences. ?-Gro (acceptor) stands for a residue of glycerol with an unknown absolute configuration (or unknown substitution position (1 or 3) in the case of d-glycerol); the data for d-Gro are not cumulated in this row (it occupies the 25th row in the sorted acceptor list). The circle centers are slightly shifted to avoid overlaps. See Supplementary Table S9 for the source data (the larger figure with 15 donor/acceptor residues is also provided).
Fig. 10.
Distribution of glycosidic linkages in protistal carbohydrates and derivatives. The area of the circles corresponds to the occurrence of a given linkage per structure. The anomericity and position of the bond is color-coded. “Exo” means all positions above 4, assuming a linkage to an exocyclic tail. Amino sugars include both N-acetylated and non-N-acetylated occurrencies. The acceptor d-GlcN-ol is probably an analytical artifact. The circle centers are slightly shifted to avoid overlaps. See Supplementary Table S10 for the source data.
In the figures, 10 of the most abundant counterparts in each domain are used to show the distribution. The donors (residues forming a linkage via their anomeric center; for more detailed explanation, please, refer to the CSDB Linear notation [15]) and acceptors are sorted independently in the abundance-decreasing order. In fungal glycans, dimannosides with α-1,2, α-1,3, and α-1,6 bonds are most frequent. They are followed by d-Man(β1-4)d-GlcN, diglucosides with β-1,3 and β-1,6 bonds, and d-GlcN(β1-4)d-GlcN (Fig. 8).
According to the CSDB content, bacterial glycans are significantly more diverse, as compared to fungal and protistal ones. The most frequent linkages include l-gro-d-manHep(α1-7)Kdo, l-gro-d-manHep(α1-3)l-gro-d-manHep, l-gro-d-manHep(α1-7)l-gro-d-manHep, d-Glc(β1-4)l-gro-d-manHep, d-GlcN(β1-6)d-GlcN, d-Gal(β1-4)d-Glc, and Kdo(α1-4)Kdo (l-gro-d-manHep = l-glycero-d-mannoheptose; Kdo = 3-deoxy-d-manno-oct-2-ulosonic acid).
As for glycans from protista, they most frequently contain dimannosides with α-1,2, α-1,3, and α-1,6 bonds, as well as d-Man(α1-4)d-GlcN, d-Man(β1-4)d-GlcN, d-GlcN(β1-4)d-GlcN, and d-Gal(β1-4)DMan. Thus, from the viewpoint of glycosidic bonds, fungal and protistal glycans demonstrate higher resemblance to each other than to bacterial glycans which corresponds to the closer positions of the eukaryotic domains in the phylogenetic tree of life [24].
2.6. Structure sizes
Distributions of sizes of glycan structures relative to the number of all residues and, in particular, monosaccharides for fungi, prokaryotes, and protista are provided in Fig. 11 and Fig. 12, respectively. Oligo- and polysaccharides are considered separately; in the latter case, the number of residues per regular repeating unit is used.
Fig. 11.
Distribution of structure sizes. The x-axis shows the number of residues per structure. Oligomers and regular repeating units are shown separately in the top and bottom plots, respectively (as oligo and poly). Logarithmic scales are used for better presentation. All residues are counted, including monovalent modifications. See Supplementary Table S11 for the source data.
Fig. 12.
Distribution of structure sizes. The x-axis shows the number of monosaccharides and alditols per structure. Oligomers and repeating units are shown separately in the top and bottom plots, respectively (as oligo and poly). Logarithmic scales are used for better presentation. See Supplementary Table S12 for the source data.
In the case of fungal oligomeric glycans and glycoconjugates, structures with two and three residues or one and two monosaccharides are most abundant. For bacterial oligomeric glycans, these numbers equals to three to six residues and two to four monosaccharides, respectively; for protistal oligomeric glycans – to four to six residues and three to seven monosaccharides, respectively (see Fig. 11 and Fig. 12).
In the case of fungal glycopolymers, structures with one to five residues or one to four monosaccharides are most abundant. For bacterial glycopolymers, the highest abundance is observed for structures with four to eight residues or four to five monosaccharides, whereas for protistal polymeric glycans – for structures with one and three residues or one and two monosaccharides, respectively (see Fig. 11 and Fig. 12).
Of note, whereas the maximal length of oligoglycans or repeating units reaches 26–80 residues in all the domains, larger structures are missing from CSDB, especially for protista where the total absolute coverage is low due to the lack of publications.
2.7. Branching degree and net charge
In this work, the branching degree, or antennarity, of a structure is defined as a ratio of the non-reducing termini count to the residue count. This parameter is identical to the branching index, as defined in [11], and shows how dendrite a given glycan molecule is. For glycopolymers, it shows the number of side chains per backbone repeating unit. The antennarity reflects the potential of a structure to carry non-reducing termini along the polymeric molecule. The immunogenicity of organisms is often related to glycoepitopes located at terminal positions of carbohydrate chains comprising the cell wall [17], [25], [26], [27].
Distribution of antennarity in fungal, prokaryotic and protistal structures present in CSDB is shown in Fig. 13. Glycans from all the three domains demonstrate a large peak at 0.3. In addition, fungal glycans also have a rather high peak at 0.5 (27.4 % structures) and a lower but wider peak at 0.7 (6.6 % structures). For glycans from prokaryotes and protista, there are also elevations in these areas, but they are less pronounced (14.9 % and 13.4 % at 0.5 and 1.7 % and 2.0 % at 0.7, respectively). Of note, bacterial glycans are characterized by a larger part of linear structures with zero antennarity (ca. 13.9 %), as compared to fungal and protistal ones (9.2 % and 6 %, respectively).
Fig. 13.
Distribution of antennarity (branching degree) in carbohydrate structures of fungi, prokaryotes and protista (including unicellular algae). See Supplementary Table S13 for the source data.
As for the net charge distribution (mean charge density, as defined in [11]), fungal glycans are characterized by a sharp narrow peak at 0.0 – 0.2) which encompasses ca. 79.5 % structures (79.1 % of them neutral) (Fig. 14). Glycans from prokaryotes and protista are also mostly neutral (41.3 % and 53.9 %, respectively), but there is a considerable part of the structures carrying negative charges from –0.6 to –0.2. This can be attributed to the higher abundance of uronic acids and phosphate groups in prokaryotes. In all the domains, structures with a positive net charge are rare, as most of the amino groups (the main source of a positive charge) are acetylated.
Fig. 14.
Net charge distribution in carbohydrate structures of fungi (A), prokaryotes (B) and protista (including unicellular algae) (C). Every bar corresponds to a range of net charges with a width of 0.2, including the lower (left) limit and excluding the upper (right) limit. Diamonds indicate the part of neutral structures in a given domain. See Supplementary Table S14 for the source data.
3. Conclusions
This paper presents the results of the systematic statistical analysis of the fungal glycome as compared to the prokaryotic and protistal glycomes presented in the scientific literature. The monomeric and dimeric compositions of glycans, their non-carbohydrate modifications, glycosidic linkages, sizes of structures, branching degrees and net charges are assessed. The obtained information on the monosaccharides unique for various fungal classes can help elucidating carbohydrate molecular markers for these classes, whereas unique disaccharides can be useful for determining specific glycosyltransferases active in particular fungal taxa. Such information can be demanded for the development of diagnostic tools and carbohydrate-based vaccines against pathogenic fungi. In addition, revealing structural components common and unique for certain taxonomic groups can be used as a potential basis for taxonomic classification of microorganisms.
4. Methods
The total abundance of structures per taxonomic ranks was obtained by using the Coverage statistics tool of CSDB (https://csdb.glycoscience.ru/database/core/covstat.html). The distribution of both unique and non-unique monomeric and dimeric fragments per taxonomic ranks was obtained by using the Fragment abundance tool of CSDB (https://csdb.glycoscience.ru/database/core/dimers.html). These built-in CSDB tools for the statistical analysis of glycomes were reported previously [12]. Distributions of glycosidic linkages, structure sizes, and branching degree were obtained by using dedicated SQL queries on a raw CSDB database, as imported from the reported source files [14]; the queries are referenced from the corresponding Supplementary Tables S8-S14 and copied in Supplementary Tables S15-S17. The charge distribution was generated by the dedicated PHP5 script from a previous work [14] run on a raw CSDB database.
All the data generated by the CSDB engine and the above queries and scripts were gathered in Microsoft Excel 2010 spreadsheets, manually checked for inconsistencies, normalized, and exported to OriginLab Origin Pro 2017 for visualization. The parameters inputted to the CSDB statistical tools are listed prior to data in Supplementary Table S1-S7. The manual operations used to refine the data (combining related entities, removing artifacts, filtering out inappropriate content, applying the occurrence cut-off, sorting, etc.) are listed in each supplementary table next to the input parameter summary.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This study was supported by the Russian Science Foundation, grant 18-14-00098-P.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2022.09.040.
Contributor Information
Philip V. Toukach, Email: netbox@toukach.ru.
Ksenia S. Egorova, Email: egorova-ks@ioc.ac.ru.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary Tables S1-S14 (the numbering corresponds to the figure numbers in the main text) include source data for the plots presented in the paper and description of formal procedures used to obtain them, including CSDB tool address, input parameters, and post-processing steps.
Supplementary Tables S15-S17 list SQL codes for queries used to obtain the data where no CSDB tool was available.
References
- 1.Naranjo-Ortiz M.A., Gabaldón T. Biol Rev Cambridge Philos Soc. 2019;94:2101. doi: 10.1111/brv.12550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lücking R., Aime M.C., Robbertse B., Miller A.N., Aoki T., Ariyawansa H.A., et al. Nat Microbiol. 2021;6:540. doi: 10.1038/s41564-021-00888-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Newbound M., McCarthy M.A., Lebel T. Landscape Urban Plann. 2010;96:138. [Google Scholar]
- 4.Lücking R., Aime M.C., Robbertse B., Miller A.N., Ariyawansa H.A., Aoki T., et al. IMA Fungus. 2020;11:14. doi: 10.1186/s43008-020-00033-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Latgé J.-P., Beauvais A. Curr Opin Microbiol. 2014;20:111. doi: 10.1016/j.mib.2014.05.009. [DOI] [PubMed] [Google Scholar]
- 6.Gow N.A.R., Latge J.-P., Munro C.A., Heitman J. Microbiol Spectrum. 2017;5:FUNK. doi: 10.1128/microbiolspec.funk-0035-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cortés J.C.G., Curto M.Á., Carvalho V.S.D., Pérez P., Ribas J.C. Biotechnol Adv. 2019;37 doi: 10.1016/j.biotechadv.2019.02.008. [DOI] [PubMed] [Google Scholar]
- 8.Lima S.L., Colombo A.L., de Almeida Junior J.N. Front Microbiol. 2019;10:2573. doi: 10.3389/fmicb.2019.02573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hopke A., Brown A.J.P., Hall R.A., Wheeler R.T. Trends Microbiol. 2018;26:284. doi: 10.1016/j.tim.2018.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toukach P.V., Egorova K.S. Nucleic Acids Res. 2016;44:D1229. doi: 10.1093/nar/gkv840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herget S., Toukach P.V., Ranzinger R., Hull W.E., Knirel Y.A., von der Lieth C.-W., et al. BMC Struct Biol. 2008;8:35. doi: 10.1186/1472-6807-8-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Egorova K.S., Kondakova A.N., Toukach P.V. Database. 2015 (2015):bav073. doi: 10.1093/database/bav073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Egorova K.S., Toukach P.V. Carbohydr Res. 2014;389:112. doi: 10.1016/j.carres.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 14.Toukach P.V., Egorova K.S. Sci Data. 2022;9:131. doi: 10.1038/s41597-022-01186-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toukach P.V., Egorova K.S. J Chem Inf Model. 2020;60:1276. doi: 10.1021/acs.jcim.9b00744. [DOI] [PubMed] [Google Scholar]
- 16.Adibekian A., Stallforth P., Hecht M.-L., Werz D.B., Gagneux P., Seeberger P.H. Chem Sci. 2011;2:337. [Google Scholar]
- 17.Astronomo R.D., Burton D.R. Nat Rev Drug Discovery. 2010;9:308. doi: 10.1038/nrd3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Varki A. Glycobiology. 2017;27:3. doi: 10.1093/glycob/cww086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kinoshita T., Fujita M. J Lipid Res. 2016;57:6. doi: 10.1194/jlr.R063313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masuoka J. Clin Microbiol Rev. 2004;17:281. doi: 10.1128/CMR.17.2.281-310.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown S., Santa Maria J.P., Walker S. Annu Rev Microbiol. 2013;67:313. doi: 10.1146/annurev-micro-092412-155620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rietschel E.T., Wollenweber H.-W., Zähringer U., Lüderitz O. Klin Wochenschr. 1982;60:705. doi: 10.1007/BF01716559. [DOI] [PubMed] [Google Scholar]
- 23.Pardo-Vargas A., Delbianco M., Seeberger P.H. Curr Opin Chem Biol. 2018;46:48. doi: 10.1016/j.cbpa.2018.04.007. [DOI] [PubMed] [Google Scholar]
- 24.Forterre P. Front Microbiol. 2015;6:717. doi: 10.3389/fmicb.2015.00717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Comstock L.E., Kasper D.L. Cell. 2006;126:847. doi: 10.1016/j.cell.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 26.Amon R., Reuven E.M., Leviatan Ben-Arye S., Padler-Karavani V. Carbohydr Res. 2014;389:115. doi: 10.1016/j.carres.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 27.Barreto-Bergter E., Figueiredo R.T. Front Cell Infect Microbiol. 2014;4:145. doi: 10.3389/fcimb.2014.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.