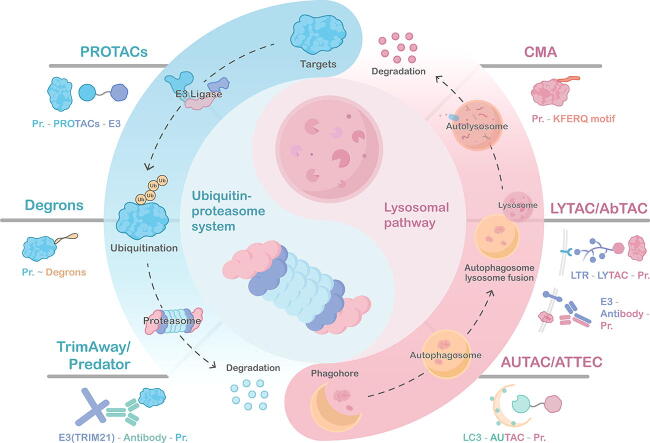
Graphical abstract
Abstract
In the eukaryotic cellular milieu, proteins are continuously synthesized and degraded effectively via endogenous protein degradation machineries such as the ubiquitin–proteasome and lysosome pathways. By reengineering and repurposing these natural protein regulatory mechanisms, the targeted protein degradation (TPD) strategies are presenting biologists with powerful tools to manipulate the abundance of proteins of interest directly, precisely, and reversibly at the post-translational level. In recent years, TPD is gaining massive attention and is recognized as a paradigm shift both in basic research, application-oriented synthetic biology, and pioneering clinical work. In this review, we summarize the updated information, especially the engineering efforts and developmental route, of current state-of-the-art TPD technology such as Trim-Away, LYTACs, and AUTACs. Besides, the general design principle, benefits, problems, and opportunities to be addressed were further analyzed, with the aim of providing guidelines for exploration, discovery, and further application of novel TPD tools in the future.
1. Introduction
The expression of genes is precisely regulated in mammalian cells so that the timing, location, and abundance of protein meet the needs of growth, development, and reproduction of cells, which is of great significance for living cells to sense and process surrounding signals and adapt to the environment with high precision [1], [2]. Gene expression regulation techniques, including loss-of-function methods and gain-of-function methods, enable scientists to manipulate gene networks that control cellular functions and or physiological states, which open possibilities for uncovering the complex landscape of living cells and engineering artificial cell functionalities. Loss-of-function gene expression regulation techniques that interrupted gene expression and corresponding cellular functions can act at particular levels (DNA, RNA, and protein level) [3]. Gene knockout techniques based on homologous recombination have been widely used to disrupt protein functions at the DNA level by inactivating genes that are responsible for a protein product [4]. In recent years, gene editing techniques, as exemplified by the zinc finger nuclease, TALE nuclease, and CRISPR/Cas9, have emerged as powerful tools for gene expression regulation due to their simplicity, convenience, and high efficiency [5], [6], [7]. Techniques that act at the RNA level, such as RNA interference (RNAi) techniques, can be utilized to manipulate the expression of the target gene by affecting the stability of the target mRNA or interfering with its translation process [8]. Additionally, engineered site-specific transcriptional regulators and synthetic promoters, e.g. The TetON/OFF system, provide tools for controllable, reversible transcriptional initialization or repression of the target gene [9]. However, these techniques described above focus on preventing the synthesis of protein, the reduction of the protein abundance of target genes in cells largely depends on the turnover of existing protein products, which provides cells with enough time to initiate compensatory adaptation and secondary side effects that could mask or alter corresponding loss-of-function phenotypes of the gene of interest. Furthermore, these techniques that indirectly manipulate the protein cannot effectively change the abundance of the long-lived proteins [10].
Recently, the reengineering and repurposing of natural protein regulatory mechanisms have bred a new area of what is termed targeted protein degradation (TPD), which is rapidly gaining massive attention. The TPD techniques harness the power of natural protein degradation systems such as ubiquitin–proteasome system (UPS) and lysosomal system and re-orient them to directly degrade the protein of interest (POI), enabling the direct depletion and removal of the target protein with high selectivity and efficiency. These emerging techniques, such as well-known UPS-based PROTACs, molecular glues, and Trim-Away, and most recently, lysosomal system-based LYTACs, AbTAC, AUTAC, and ATTEC techniques offer biologists with powerful tools to manipulate the abundance of POI directly, accurately, and reversibly at a post-translational level.
TPD techniques are recognized as a paradigm shift not only in basic research but also in pioneering clinical work. Compared to traditional small-molecule drugs (small-molecule inhibitors) that based on the “occupation-driven” pharmacological principles, the TPD techniques represent a brand-new “event-driven” mode that degrades protein directly with fewer restrictions, e.g. requiring clear active sites or necessary ligand-binding pocket on pathological protein targets, allowing precise depletion of many targets that previously recognized as the “undruggable”. Additionally, the TPD technique can rapidly deplete the target protein using high-efficiency endogenous protein degradation machinery within minutes to hours, instead of the days or weeks that are required for the manipulation of mRNA or genomic DNA in nucleic-acid therapeutics such as CRISPR/Cas9 and RNAi techniques. For those pathogenic proteins that have similar spatial structure as the normal protein, TPD-based drugs can selectively bind and induce the precise degradation of pathogenic variants without affecting normal proteins expression and function, while CRISPR and RNAi techniques might affect the functioning of all variants of target proteins simultaneously, leading to unexpected side effects or cytotoxicity [11].
In this review, we summarize the updated information, particularly the functional parts, engineering efforts, and developmental route, of current cutting-edge TPD technology such as Trim-Away, LYTACs, and AUTACs. Besides, the general design principle, benefits, problems, and opportunities to be addressed were further analyzed, with the aim of providing guidelines for future discovery and further application of novel TPD tools.
1.1. Natural protein regulation machinery - ubiquitin–proteasome system and lysosomal pathway
There are two main ways of intracellular protein degradation in eukaryotic cells: the Ubiquitin-Proteasome System (UPS), and the lysosomal pathway [12], [13] (Fig. 1). The ubiquitin–proteasome system is the major proteolytic pathway responsible for about 80 % selective degradation of endogenous protein in eukaryotic cells [14]. In the UPS process, misplaced folded proteins, denatured proteins, and mutant proteins were recognized, ubiquitinated, and sent to degradation by 26 s proteasome [15], [16], [17] (Fig. 1A). The detailed mechanisms and functions of different types of ubiquitination signals have been discussed in several reviews [18], [19], which we won’t discuss further in this review. In general, K11/K48-linked polyubiquitination chains serve as the degradation signal in the second step that ubiquitinated proteins are recognized and rapidly degraded by 26 s proteasome [14], [20] (Fig. 1A). Notably, the specificity of UPS is largely dependent on the selectivity recognition between the E3 ligase and the corresponding target protein. There are over 600 encoded E3 ligases in human genomes and can be divided into three main families according to the functional domain and the ubiquitin-transferring manner: RING family, HECT family, and RBR family [21].
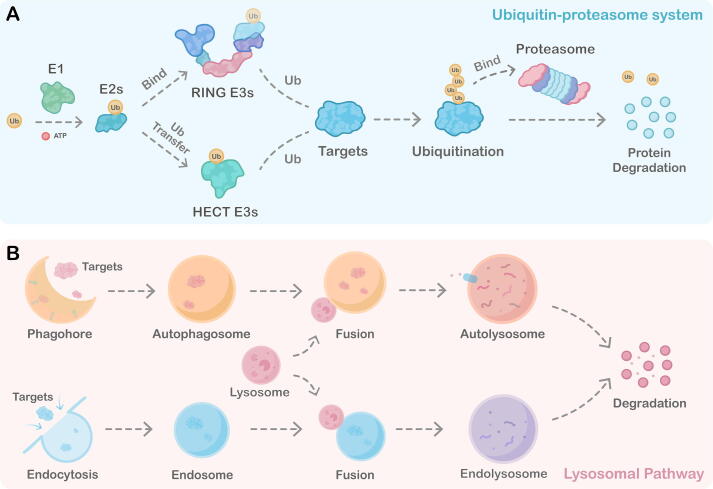
Fig. 1.
Two main protein quality control machinery in mammalian cells: the ubiquitin–proteasome system and the Lysosomal system. A) Schematic representation of the ubiquitin–proteasome system. The ubiquitin–proteasome pathway is a multi-enzyme cascade process. Firstly, E1 ubiquitin-activating enzyme activates ubiquitin molecules via consuming ATP. The ubiquitin is then transferred from E1 to E2 ubiquitin-conjugating enzyme. With target protein specifically recognized by the E3 ligase, E3s recruit the E2 and therefore transfers the ubiquitin to the protein or ubiquitin-tagged protein in the following rounds. The polyubiquitin chain-labeled target protein is then degraded by the 26 s proteasome. B) Schematic representation of the lysosomal system. The lysosomal system includes the endosome-lysosomal pathway and autophagy-lysosome pathway. The autophagy pathway begins with an isolated membrane structure called the phagophore. This phagophore expands, and swallows up substrates in cells, including proteins and other bio-/macro-molecules, then isolates them in autophagosomes. The loaded autophagosome will mature and fuse with the lysosome, leading to the final degradation of the cargoes. In the endosomal-lysosomal pathway, targets such as aggregates and proteins are engulfed in the endosome via endocytosis, and the endosome is formed. Then the early endosome is transformed into the late endosome and fused with the lysosome to form an endolysosome. The cargoes in the endolysosome finally degrade by the acid environment and hydrolases that lysosome carries. Therefore, both the endosomal-lysosomal pathway and autophagy-lysosome pathway can degrade target cargoes.
The RING (Really interesting new genes)-finger family is characterized by a canonical E2-binding RING domain, which can act as a bridge to directly transfer activated ubiquitin from E2 to the target protein, while the E3s themselves do not interact with ubiquitin directly. The RING-finger family E3s contains cIAP, TRIM5α, APC/C, etc. [22]. Among them, Cullin-RING ligase (CRL, shown in Fig. 1A is the Skp, Cullin, F-box-containing complex, or SCF complex for short, a representative number of CRL) is the largest class of multi-subunit RING finger E3 ligases.
The HECT (homologous to E6-AP C-terminus) E3s have a conserved HECT domain, and its cysteine (Cys) residue can form a thioester bond with E2-bound ubiquitin molecules, thus transferring ubiquitin molecules from E2 to E3s, and then HECT E3s itself presents ubiquitin to the substrates (Fig. 1A).
The RBR (RING-in-between RING) E3 ligase is a newly discovered ubiquitin ligase in recent years, and some of its members have special ubiquitin transfer activity. As a quick example, RBR E3 ligase consists of the RING1 domain, BRcat (benign-catalytic) domain, and C-terminal Rcat (required for catalysis) domain [23]. The detailed ubiquitin-transfer mechanism is regarded as the hybrid of RING and HECT [24].
In recent years, another intracellular protein degradation pathway, the lysosomal pathway, which is independent of the proteasome, has gradually become the focus of novel TPD techniques. The lysosome is a kind of acidic organelle, which receives, degrades, and recycles substances from the plasma membrane or cytoplasm through endocytosis, phagocytosis, and autophagy. Additionally, the lysosomal pathway is capable of digesting the local cytoplasm or organelles of the cell. Regulating the level of intracellular substances through lysosomes is of great significance to maintain normal metabolic activities. In mammalian cells, lysosomal pathways mainly include the endosomal-lysosomal pathway and autophagy-lysosome pathway, aiming to eliminate exogenous or endogenous targets, respectively (Fig. 1B). At present, these two pathways have been used to degrade pathogenic proteins associated with various diseases, such as cancer, Huntington’s disease, etc [25].
In mammalian cells, both UPS and lysosomal pathways are critical in the maintenance of cellular homeostasis while they differ mechanically and functionally [12], [13]. UPS effectively degrades short-lived, soluble unfolded, or misfolded proteins and peptides, while the autophagy-lysosome system is responsible for the elimination of long-lived proteins, insoluble protein aggregates, and dysfunctional organelles, such as degenerated mitochondria. Nevertheless, owing to their unique but intrinsic capability of controlling protein abundance with high selectivity and efficiency, as a “lesson from nature”, re-purposing and re-engineering of these two pathways to directly remove protein of interest bred the concept of targeted protein degradation, which essentially open a new chapter for both basic research and clinical applications.
2. UPS-based protein degradation methods
2.1. Re-engineering of E3 ligase
Since E3 ubiquitin ligase provides the specificity and selectivity for target protein ubiquitination, engineering of E3s, commonly by modifying substrate-binding domain or fusing a targeting module to E3, might re-orient the targeting specificity of E3 ligase, enabling the selective and rapid degradation of protein of interest via ubiquitin–proteasome systems.
In 2000, Zhou et al. re-directed E3 ligase to the target protein pRB in yeast and human osteosarcoma cells by fusing Cdc4p, the F-box protein in E3 ligase SCF complex which selectively binds to targets, with pRB-binding E7N, and demonstrated that target protein was effectively degraded by the new E3 ligase. Their results prove that the UPS in mammalian cells can be utilized and re-directed for specific degradation of a given protein [26]. These E3s engineering techniques that fuse different targeting modules to modify SCF complex F-box protein β-TrCP to redirect UPS were further applied to the degradation of various targets, such as β-catenin [27], [28], [29], CyclinA/CDK2 [30], and many other functional proteins. Additionally, the loss-of-function phenotypes mediated by E3s were verified at the cellular level and in vivo compared with classic gene knockdown techniques, e.g., siRNA.
Moreover, the engineered E3s proposed by Cong et al. is capable of degrading β-catenin in the cytoplasm while maintaining normal β-catenin located on the membrane [31], indicating that this technique has the potential to distinguish proteins with different subcellular localization. Engineered β-TrCP established by Su et al. demonstrated the linker between the F-box and β-TrCP should remain instead of directly fusing the targeting module(APCbc4), otherwise, it would affect the interaction between F-box protein and Skp1, indicating that this linker may be related to the normal folding and function of E3 enzyme protein [28].
Notably, except for invalidated off-target concerns, these works illustrate the power of the new mindset that E3s can be engineered, functionally re-directed, and used as a gene expression knockdown tool that functions specifically at the protein level in basic research.
2.2. Degrons
Degron is a specific amino acid sequence that can be recognized by intracellular proteases, leading to the degradation and clearance of degron-tagged proteins [32]. Most of the degron exist in the DNA sequence that encodes the target protein, and a few are added to the target protein during or after translation. So far, there have been many TPD techniques based on the mechanism of degron. Apart from constitutional degron that is knocked into the gene of interest to destabilize target protein, which has been discussed in many reviews [33], [34], [35], other degrons, based on their controllable manner, can be divided into four types: phospho-degron, small molecule-induced degron, temperature-induced degron, and light-induced degron.
Phospho-degron is a phosphorylated motif that can specifically interact with appropriate F-box proteins. It is formed in protein turnover processes where protein kinases mediated the phosphorylation of target protein [36], [37]. So far, two crystal structures of phosphor-degrons that bind to F-box protein have been reported: β-catenin phospho-degrons (DpSGIHpS) bind to the surface of WD40 repeats in WD40 through arginine residues [38]. Phosphate degrader (LPpTPP) originated from cyclin E, and can specifically interact with yeast protein CDC4, which is homologous with human F-box protein Fbw7 [38], [39].
Small molecule-induced degron is a class of inducible degrons controlled by chemical molecules. At present, this widely-used degron toolbox includes AID (auxin-inducible degron) [40], DD (destabilization domain) [41], LID (ligand-induced degradation) [42], TIPI (tobacco etch virus protease-induced protein inactivation) [43], deGradFP (degrade green fluorescent protein) [44], SMASh (small molecule-assisted shutoff) [45], etc.
As a quick example, The key element of the AID technique is auxin (indole-3-acetic acid) which is capable of recruiting transport inhibitor response 1 (TIR1), an F-box protein, and binding AID peptides consisting of 44 amino acids [40]. AID was firstly constructed and utilized as a degron in yeast and mammalian cell lines, allowing auxin-induced degradation of target protein CenP-h, as demonstrated by Kohei Nishimura and Masato Kanemaki[46]. Then, the AID technique was further applied to species including Caenorhabditis elegans [47], Drosophila melanogaster [48], etc.
SMASh techniques require target protein fused with a SMASh tag that consists of three parts: an NS3 cleavage site, an NS3 protease from hepatitis C virus (HCV), and a degron [45]. In the presence of NS3 protease inhibitor (asunaprevir), the cleavage activity of NS3 protease is inhibited, and degron-tagged target protein will therefore be degraded through currently unknown pathways [49]. It is noteworthy that SMASh allows asunaprevir (NS3 protease inhibitor)-triggered degradation of target protein while removing the prior modification of SMASh-tag under normal conditions, which largely attenuates side-effect brought by degron modification.
Temperature-induced degron mainly utilizes the temperature sensitivity of a dihydrofolate reductase (DHFR) mutant cloned from mice, enabling a temperature-activated degradation of the target protein. For example, Suzuki et al. constructed a temperature-sensitive degron in Saccharomyces cerevisiae, and demonstrated the abundance of the target protein can be easily controlled by changing the temperature of the incubator [50].
Light-induced degron includes the LOV (light-oxygen-voltage) regulation system cloned from Arabidopsis thaliana and the ODC (ornithine decarboxylase) system derived from mice, enabling controllable protein degradation in a spatiotemporal manner. At present, Light-induced degrons have been widely used in different organisms such as c-elegans [51] and Danio rerio [52].
2.3. Tag-specific degrader
Tag-specific degrader focuses on targeting protein pre-fused with TAG, so it can rapidly induce targeted degradation of POI without the need to specify the targets. In this case, the protein degradation molecules are optimized to bind the protein tags, thereby removing the target protein fusion conjugates. The main tags widely used are HaloTag [53],dTag systems [54] and IKZF3 [58].
HaloTag (33KDa) is a common self-labeling tag, which combines covalently with chlorinated alkanes [53]. So far, the HaloTag degrader of E3 ligase VHL and IAP has been well-described, which are active in the low nanomole range.
The dTAG technique includes a FKBPF36V protein tag fused to the target protein and a bivalent degradant molecule (dTAG) [54]. DTAG selectively binds to FKBP at one site and E3 ligase CRBN at another site to induce the degradation of the fusion protein structure. DTAG method has been used many times to validate target proteins in different disease models [55], [56], [57]. Compared with the HaloTag system, dTAG has a catalytic effect and has been proven to be effective in both in vivo and in vitro applications.
IKZF3 peptidic degrader is another degradation tag for IMiD-induced degradation [58]. At the boundary of the zinc finger-2 domain of IKZF3, the researchers found a 25-amino-acid degrader (3 kDa), which can be fused with a POI for IMiD-induced degradation. It is less likely for this degrader to interfere with the natural function of the target protein due to its smaller size. Another advantage of IMIDS is its high bioavailability. It can work in the central nervous system (CNS), which is the only ligand-induced system known to be suitable for regulating brain proteins. So far, it has been reported that this degradation only degrades ∼ 50 % of the tagged proteins, which may need further optimization.
2.4. Chimeric molecules linking E3 enzyme to the substrate: PROTACs
PROTACs (Proteolysis Targeting Chimeras) technique utilized small molecule chimeras that bind substrate protein and E3 ubiquitin ligase simultaneously, leading to the formation of ternary complex and therefore mediating the ubiquitination and degradation of protein targets (Fig. 2) [7], [59], [60]. The conventional PROTACs molecules are composed of three parts: an E3s-recruiting ligand, a POI-targeting warhead, and a flexible linker that links two ligands.
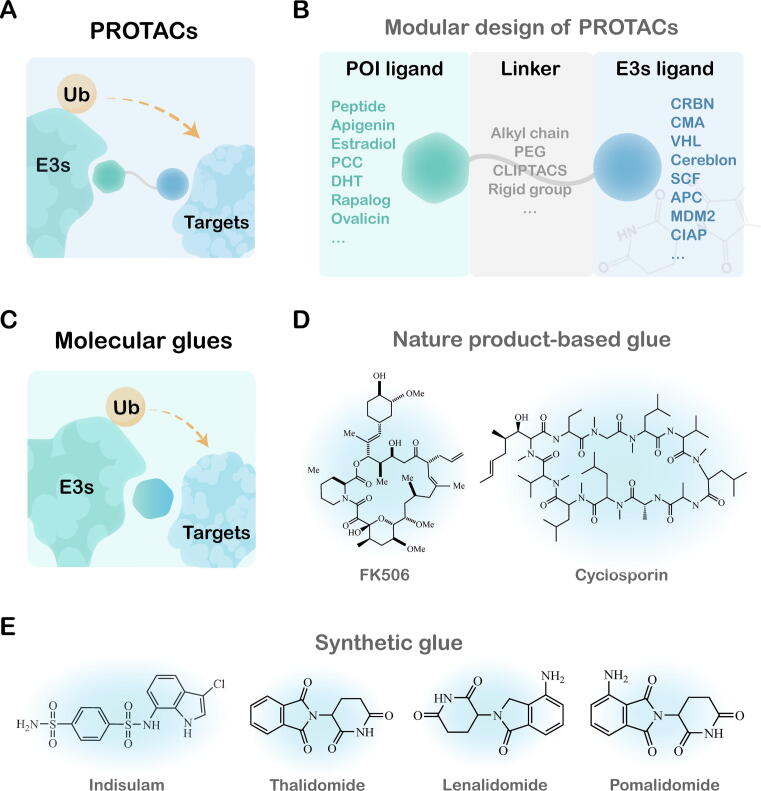
Fig. 2.
Schematic representation and structure of PROTAC and Molecular Glue. A) Schematic representation of PROTAC. Bifunctional PROTAC contains a ligand targeting E3 ubiquitin ligase, a linker, and a warhead targeting POI. B) Modular design of PROTACs. C) Schematic representation of molecular glue. Molecular glue induces the protein–protein interaction between POI and E3 ubiquitin ligase by binding to E3 ubiquitin ligase or POI. D) Chemical structures of nature product-based glue. E) Chemical structures of synthetic glue.
Geldanamycin-estradiol hybrids were the precursor of the PROTACs. Previous studies have found that Geldanamucin not only inhibits the proliferation of cancer cells but also degrades many non-specific protein targets, such as ER (estrogen receptor), HER2 (human epidermal growth factor receptor-2), tumor suppressor protein p53, Raf-1, etc., which leads to the strong cytotoxicity. To address this issue, researchers linked Geldanamucin with estradiol (targeting ER) or testosterone (targeting AR) to improve its selectivity and specificity, which successfully degraded ER and AR. These results also demonstrate that chimeric is capable of mediating the specific degradation of protein targets [60].
2.4.1. First-generation PROTACs molecule: The origin of PROTACs
First-generation PROTACs, firstly proposed by Sakamoto in 2001, was based on peptide E3-recruiting ligand. The author designed a chimeric molecule named Protac-1 (Proteolysis-targeting chimeric molecule 1), which was composed of ovalicin (OVA) and phosphopeptide derived from IkBα (IPP). Phosphopeptide acts as the E3s-recruiting ligand recognized by β-TrCP in the E3 ligase SCF complex, while the OVA binds to the MetAP2 (methionine aminopeptidase-2), serving as the POI-targeting warhead. Thus, Protac-1 recruits MetAP2 to the E3s and induces the degradation of MetAP2 via the ubiquitin–proteasome system [61]. Furthermore, the OVA was replaced by estradiol and DHT, and the new Protac-2 and Protac-3 molecules enabled the targeting and degradation of ER and AR, respectively [62]. This peptide-based mode opens new possibilities for the depletion of target proteins. However, it suffers from stability and cell permeability issues.
2.4.2. Second-generation PROTACs molecules: full-small molecule PROTACs based on small molecule E3 recruitment module
To improve cell permeability, the second-generation PROTACs replaced the E3-recruiting ligand with small molecules in 2008 [63], [64]. Owing to the replaceable E3-recruiting ligand and POI-targeting warhead, PROTACs can theoretically degrade any protein, including those recognized as “undruggable” targets, if their ligands are known or identified. Nowadays, PROTACs techniques have successfully targeted methionine aminopeptidase 2, AR, cellular retinoic acid binding protein, ER, Tau-related proteins, kinases, etc., showing good prospects for fundamental and applied life sciences [62], [56], [57], [58], [59] (see Table 1). Currently, PROTACs are generating immense scientific, clinical and commercial interest, while PROTACs-based medicines are hopefully on the verge of fruition. For example, two oral PROTAC (ARV-110, ARV-471) from Arvinas company, are currently in phase II clinical trials, aiming to treat metastatic prostate cancer and breast cancer resistant to androgen deprivation therapy. The deficiency is that its high molecular weight leads to poor water solubility, oral bioavailability, and membrane permeability [60]. Recent products indicate that PROTACs are expected to significantly influence drug discovery in the coming years (Table 1).
Table 1.
Worldwide PROTAC projects in the clinical stage (Data before June 2022).
| Degrader | Target | Company | E3 ligase | Highest phase | Ref. |
|---|---|---|---|---|---|
| ARV-110 | AR | Arvinas | CRBN | Phase VII | [65] |
| ARV-471 | ER | Arvinas; Pizer | CRBN | Phase VII | [66] |
| CFT7455 | IKZF3, IKZF1 | C4 Therapeutics | CRBN | Phase VII | [67] |
| AC0682 | ER α | Accutar Biotech | CRBN | Phase I | [68] |
| AC0176 | AR | Accutar Biotech | Undisclosed | Phase I | [68] |
| ARV-766 | AR | Arvinas | Undisclosed | Phase I | [69] |
| BGB-16673 | BTK | Beigene | CRBN | Phase I | [70] |
| CC-94676 | AR | BMS | CRBN | Phase I | [71] |
| DT2216 | BCL-XL | Dialectic Therapeutics; University of Florida | VHL | Phase I | [72] |
| FHD-609 | BRD9 | Foghorn Therapeutics | Undisclosed | Phase I | [73] |
| HP518 | AR | Hinova | Undisclosed | Phase I | [74] |
| KPG-818 | IKZF 1/3 | Kangpu | CRBN | Phase Ib/2a | [75] |
| KPG-121 | IKZF 1/3; CK 1a | Kangpu | CRBN | Phase I | [76] |
| KT-333 | STAT3 | Kymera | Undisclosed | Phase I | [77] |
| KT-413 | IRK4 | Kymera | CRBN | Phase I | [78] |
| KT-474 | IRK4 | Kymera; Sanofi | Undisclosed | Phase I | [79] |
| NX-2127 | BTK | Nurix Therapeutics | CRBN | Phase I | [80] |
| NX-5948 | BTK | Nurix Therapeutics | CRBN | Phase I | [81] |
| AC0676 | BTK | Accutar Biotech | Undisclosed | IND-Enabling | [68] |
| CFT8634 | BRD9 | C4 Therapeutics | CRBN | IND-Enabling | [82] |
| CFT8919 | EGFR-L858R | C4 Therapeutics | CRBN | IND-Enabling | [83] |
| CG001419 | TRK | Cullgen | CRBN | IND-Enabling | [84] |
2.5. Molecular glue
Molecular glue is a range of small molecules that mainly induces or stabilizes the PPI between an E3 ligase and target protein, which leads to protein degradation without the need for a binding pocket on target protein (Fig. 2) [85], [86]. The earliest discovery of molecular glue originated from the study of biological effects by S.Schreiber of Harvard University in 1991. Cyclosporin A (CsA) and tacrolimus (FK506) “stick” their respective target proteins to calcineurin respectively, resulting in the inhibition of the target, cyclophilin and FKBP, which is called the earliest molecular glue [88]. Reported molecular glue degraders also include synthetic glue such as thalidomide analogues and aryl sulfonamides [89]. They do not depend on the ligand pocket of the target protein but utilize the protein interaction interface between the receptor and the target protein to reprogram the selectivity of ubiquitin ligase and drive multiple rounds of ubiquitination. Compared with chimeric molecules PROTAC, molecular glue has lower molecular weight, simpler chemical structure, smaller steric interference, and better properties, which is in accordance with the pharmacological “rule of 5”. However, molecular glue has several shortcomings, for example, it cannot be designed or identified through large-scale screening of components like PROTAC, which limits its broad application [87].
2.6. Trim-Away techniques
In 2017, a novel protein degradation technique, termed Trim-Away, was proposed. In this approach, antibodies are introduced into the cell by microinjection or electroporation, enabling high specific recognition of antigens (target proteins) and the formation of an antigen–antibody complex. Furthermore, E3 ligase TRIM21 which can recognize the Fc region of antibody is utilized to bind to antigen–antibody complex and therefore mediate the antibody-dependent intracellular neutralization (ADIN) process, which leads to the ubiquitination of the complex and subsequent degradation via 26 s proteasome, achieving specific recognition and depletion of the target protein [90], [91] (Fig. 3). The Trim-Away technique not only requires no prior modification of target protein as AID and deGradFP do, but also minimalizes off-target effects that CRISPR, PROTACs, and RNAi technology have using antigen–antibody specific interaction to recognize target protein [7], [10], [92]. The latest research shows that the Trim-Away technique can also be applied in vivo using zebrafish [93]. However, the introduction of antibodies requires microinjection, electroporation, and other complicated techniques, which inevitably affect the physiological state of cells and restrict further clinical application.
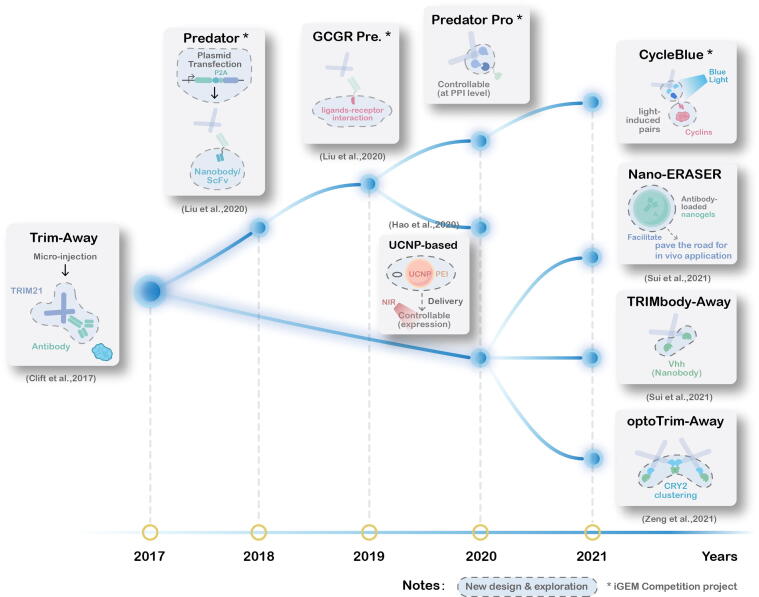
Fig. 3.
Targeted protein degradation techniques originate from Trim-Away techniques. The map shows the latest developments in Trim-Away technology since 2017. Dotted line: new design & exploration. *: iGEM Competition project.
In 2018, Liu et al developed a novel TPD technique, the Predator technique, based on molecular mechanisms of Trim-Away [94]. The Predator technique first replaced the microinjection method used in Trim-Away techniques with a constitutional expression strategy, enabling an easy-to-use, low-cost, and genetically-encoded TPD technique. Additionally, they utilized a modular chimeric antibody consisting of the E3s-recruiting module (e.g., Fc region that recruits TRIM21) and the targeting module (e.g., nanobodies, scFv, peptide aptamers, or any peptide-based targeting module), to mimic the function of the antibody in Trim-Away. This programmable, modular design of Predator is advantageous for achieving control over any protein if only the corresponding targeting module has been identified (Fig. 3) [94]. In 2019, a novel GCGR Predator which introduces ligand-receptor interaction to the Predator technique was proposed and validated, this new Predator enables the control over hyperglycemia by degrading GCGR (hepatic glucagon receptor) [94]. From an engineering prospect, there are still many shortcomings in Predator techniques, for example, it is impossible to directly control the whole degradation process after Predator plasmids are introduced to cells. Therefore, Liu and members of the NUDT_CHINA iGEM team developed Predator Pro, which is directly initiated and regulated by exogenous signals, e.g., small molecule rapamycin, as an effective extension of the current synthetic biology toolbox for protein abundance control [95]. In 2021, the NUDT_CHINA team further constructed a blue light-induced protein degradation system, CycleBlue, achieving the Spatio-temporal control of cell cycle via regulation of cyclins (Fig. 3) [96]. Although continuous effort has been put into Predator technique, several fundamental problems, such as the off-target effect, has not been addressed properly.
In addition, the Nano-ERASER technique developed by Xu et al aimed to simplify the delivery of antibodies, as microinjection or electroporation was inconvenient and required specialized instruments and techniques. They successfully degraded the coatomer protein complex ζ1 of cancer cells, killing them while sparing normal cells, which opens the window for in vivo application of Trim-Away [97]. Chen et al. also replaced full-size mAbs with a smaller nanobody (VHH, ∼15 kDa), and constructed a novel method named TRIMBody-Away by fusing nanobody and RBCC motifs of TRIM21 to increase tissue penetration [98]. In 2020, Zeng et al constructed opto-Trim-Away using CRY2, an optogenetic tool for light-induced dimerization, to realize the light-controlled Trim-Away system (Fig. 3) [99].
In addition to technological advances, Trim-Away has been used extensively to regulate the expression of target genes or proteins in fundamental research (as summarized in Table 2). As a quick example, RIF1-gene knockout experiments were carried out by introducing anti-RIF1 antibodies into oocytes electroporation, achieving a quick decrease in the RIF1 protein level, the author utilized this approach to explore the function of RIF1 in epigenetic abnormalities of zygotes [100].
Table 2.
Applications of Trim-Away Technology.
| Targets | Target Module | Projects | Ref. |
|---|---|---|---|
| RIF1 | Monoclonal, rabbit anti-RIF1 Antibody | Elevated RIF1 participates in the epigenetic abnormalities of zygotes by regulating histone modifications on MuERV-L in obese mice | [100] |
| TRF1 | Monoclonal, rat anti-TRF1 Antibody | TRF1 Depletion Reveals Mutual Regulation Between Telomeres, Kinetochores, and Inner Centromeres in Mouse Oocytes. | [101] |
| BCR/ABL | Monoclonal, rabbit anti-BCR/ABL antibody | Intracellular delivery of anti-BCR/ABL antibody by PLGA nanoparticles suppresses the oncogenesis of chronic myeloid leukemia cells. | [102] |
| Cops3 | Monoclonal, rabbit anti-COPS3 antibody | The COP9 signalosome subunit 3 is necessary for early embryo survival by way of a stable protein deposit in mouse oocytes | [103] |
| STIP1 | Monoclonal, mouse anti-STIP1 antibody | Intracellular targeting of STIP1 inhibits human cancer cell line growth | [104] |
| Rec8 | Monoclonal, rabbit anti-Rec8 antibody | Deprotection of centromeric cohesin at meiosis II requires APC/C activity but not kinetochore tension. | [105] |
| Pericentrin | Monoclonal, mouse anti-Pericentrin antibody | Two mechanisms drive pronuclear migration in mouse zygotes | [106] |
| RBD (SARS-COV 2) | full-length sACE2 fused to Fc | Targeted intracellular degradation of SARS-CoV-2 via computationally optimized peptide fusions. | [107] |
| TDP43 | Monoclonal, E6 mouse IgG2A anti-RRM1 TDP43 antibody | Monoclonal full-length antibody against TAR DNA binding protein 43 reduces related proteinopathy in neurons. | [108] |
| Nup133, POM121, NDC1 |
Mouse anti-Nup133, Rabbit anti-Nup133, Rabbit anti-POM121, Mouse anti-NDC1, |
G4C2 Repeat RNA Initiates a POM121-Mediated Reduction in Specific Nucleoporins in C9orf72 ALS/FTD. | [109] |
| CyK4 | Monoclonal, mouse anti-CYK4 antibody | Symmetry breaking in hydrodynamic forces drives meiotic spindle rotation in mammalian oocytes | [110] |
| BTG4 |
Monoclonal, rabbit anti-BTG4 antibody | Characterization of zygotic genome activation-dependent maternal mRNA clearance in mouse | [111] |
| RCC1 | Polyclonal, goat anti-RCC1 antibody | Ran GTP is essential for MI spindle assembly and function both in humans and mice | [112] |
| PKM2 | Monoclonal, rabbit anti-PKM2 antibody | Glucose metabolism distinguishes TE from ICM fate during mammalian embryogenesis | [113] |
| Tead4 |
Monoclonal, mouse anti-Tead4 antibody | A framework for TRIM21-mediated protein depletion in early mouse embryos | [114] |
| CENPF | Monoclonal, sheep anti-CENPF antibody | Loss of CENPF leads to developmental failure in mouse embryos | [115] |
| SNAP23 |
Monoclonal, mouse anti-SNAP23 antibody | SNAP23 is required for constitutive and regulated exocytosis in mouse oocytes | [116] |
| MHTT/wtHTT | Monoclonal, mouse anti-mHTT antibody | Targeting the HTT levels offer systematic, mechanism-driven routes towards curing HD | [117] |
3. Targeted protein degradation based on lysosomal pathway
Although many UPS-based TPD studies show favorable degradation efficiency in vivo and in vitro, it has limited effect on the degradation of macromolecular proteins, aggregates, extracellular proteins, and organelles. However, many extracellular proteins and cell membrane proteins are also closely related to the occurrence and development of cancer, autoimmune diseases and other diseases [118]. Therefore, establishing targeted degradation techniques that can target these substances is of great importance for basic research or clinical applications.
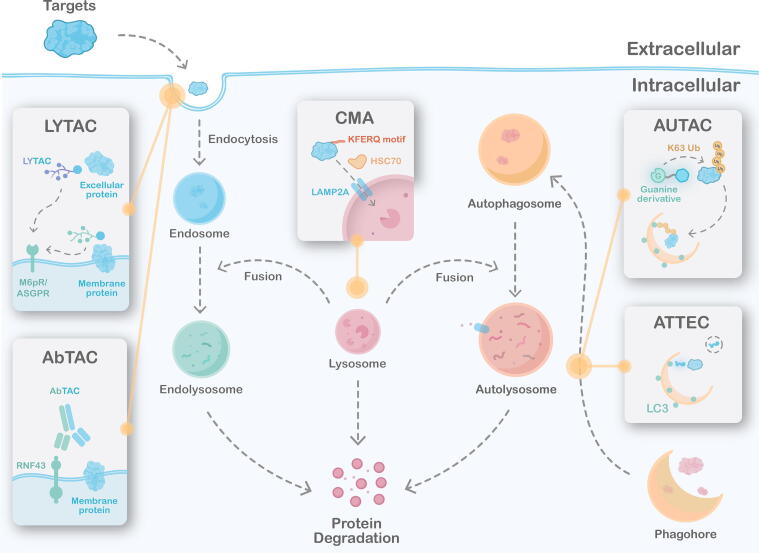
In recent years, the lysosomal pathway, a proteasome-independent intracellular protein degradation pathway, has been widely used to develop TPD methods. There are two pathways leading to lysosome, endosomal-lysosome pathway and autophagy-lysosome pathway (Fig. 1), capable of clearing substances in cells, selectively removing protein aggregates, pathogens, and redundant or damaged organelles. At present, these two pathways have been used by several techniques to enable targeted protein degradation [25] (Fig. 4).
Fig. 4.
Targeted protein degradation techniques based on the lysosomal pathway. LYTAC conjugates a glycan tag, mannose-6-phosphonate (M6Pn), to serine or lysine residues on antibodies, which induces receptor (M6Pr/ASGPR)-mediated internalization and subsequent degradation of target proteins. AbTAC degrades membrane protein through a similar mechanism that is mediated by a chimeric antibody and RNF43. CMA-based degrader uses a POI-binding sequence to bind with the target protein, which is then transported to the lysosome for degradation. AUTAC consists of three parts: a degradation tag based on guanine derivative, a linker, and a warhead specificity binds to the target. The AUTAC molecule triggers the K63-linked polyubiquitination and subsequent lysosome-mediated degradation. ATTEC is a compound capable of binding to not only LC3 but also POI. In the autophagy-lysosome pathway, target protein, as well as ATTEC and associated lipidated LC3, are incorporated into double-membrane autophagosomes. After fusion with lysosomes, the encapsulated substances can be degraded.
3.1. Lysosomal targeted chimeric system based on endosomal-lysosomal pathway
Bertozzi et al. developed LYTAC, which is a new technology to induce the degradation of extracellular or membrane proteins through the endosomal-lysosome pathway [119]. LYTAC is a bifunctional chimera, which binds to the extracellular domain of the target and the cell-surface lysosome-targeting receptors (LTRs) to form a ternary complex, leading to protein phagocytosis and degradation, while LTR is then shuttled back to the membrane to repeat the cycle. Cationic independent mannose 6-phosphate receptor (CI-M6PR) is a representative receptor in the LTR family. Coutinho et al designed an LYTAC with CI-M6PR, which successfully led to the transport and degradation of the target protein [120], such as EGFR [119], PD-L1, etc.
Although the expression of CI-MPR in cells is prevalent, the expression of some LTRs is, however, tissue-specific, enabling tissue-specific degradation of target proteins. For example, ASGPR is a liver-specific LTR [121], [122]. The ASGPR-based LYTAC molecule comprises a fusion of the antibody with N-acetyl galactosamine (GalNAc), which targets ASGPR [123]. These studies showed the broad application potential of LYTAC in extracellular and transmembrane proteins. To achieve more tissue-specific targeting, it is necessary to screen other specific LTRs. Another research recently published by the Bertozzi team extends the concept of targeting cell surface molecules to sialoglycans [124] and showed that removing sialoglycan from cancer cells with antibody-sialidase conjugates can improve the anti-tumor immune response of mice by a Siglec-E-dependent mechanism.
It should be noted that in comparison with POI inhibition, LYTAC directly degrades proteins, thereby preventing the potential activation of other downstream pathways that may be induced by inhibitors. Furthermore, this strategy prevents compensation and cellular adaptation since they are more efficient than genetic techniques like CRISPR-Cas9. However, the degradation process induced by LYTAC often lasts several days, which can be reduced to a few minutes if UPS is used.
3.2. Targeted protein degradation techniques based on autophagy-lysosome pathway
Autophagy is an important process in eukaryotic cells for macromolecular substances and organelles turnover through lysosomes under the regulation of autophagy-related genes. Briefly, damaged macromolecules or organelles are wrapped by autophagosomes (bilayer autophagy vesicles) and engulfed by lysosomes for degradation and recycling. There are three main types of autophagy: macroautophagy, commonly called “autophagy”, in which the substrate protein is encapsulated by autophagosomes and then engulfed by the lysosome for degradation; microautophagy, which is a non-selective lysosomal degradative process characterized by direct engulfment of cytoplasm and mediates invagination of cytoplasmic cargo[125]; CMA (Chaperone-mediated autophagy), mediated by molecular chaperones like Heat shock cognate protein 70 (Hsc7), is a highly selective autophagy process, which can only degrade certain proteins but not organelles (Fig. 4) [126].
3.2.1. CMA-based protein degradation system
CMA mainly recognizes and degrades soluble cytoplasmic protein substrates with KFERQ sequence, namely CTM (CMA targeting motif) [127]. Previous studies have fused CPP, CTM and PBD (Protein binding domain) to form a chimeric protein, which can specifically degrade DAPK1 (Death associated protein kinase 1) and PSD-95 (Postsynaptic Density Protein-95) in primary neurons [128]. In addition, Zhou et al. also synthesized a peptide that can induce the degradation of CDK5 through the specific binding of TAT-CDK5-CTM to CDK5 [129]. Experiments have shown that the peptide can not only prevent the death of neurons but also reduce the cerebral infarction of mice with arterial occlusion. This CMA-based protein degradation provides a new method for regulating endogenous protein abundance. Xu's team discovered the regulatory factor HIP1R can interact with PD-L1 directly and transport it to lysosome for subsequent degradation. They also found that the interaction between HIP1R and PD-L1 depends on the structural motif of PD-L1 and the signal peptide targeting to lysosome. Then, the team firstly designed a peptide containing a PD-L1 binding sequence from HIP1R and a CMA sorting tag with sequence KFERQQKILDQRFFE, which can significantly reduce PD-L1 [130]. The CMA provides a powerful tool and a new strategy for scientific research and treatment of diseases caused by protein misfolding. However, there are still some shortcomings: (1) the design of peptides requires a domain with high affinity to the target protein; (2) the related research and therapeutic potential of CMA is still limited by the low efficiency of transmembrane peptide delivery.
3.2.2. AUTAC-based protein degradation system
After the formation of phagophore, cells require further multi-step degradation processes for autophagy. After that, soluble LC3-I (microtubule-associated protein light chain3-I) is conjugated to lipids to transform into LC3-II and then combines with phagophore to form autophagosomes [127]. After the target is modified by K63-linked polyubiquitin, it is recognized and captured by autophagosomes through the surface receptor LC3-I. Finally, autophagosomes combine with lysosomes to form autolysosomes, and the cargoes inside are degraded.
In early 2019, Arimoto et al. constructed an AUTAC (autophagy-targeting chimera) molecule that uses autophagy-lysosome pathway to regulate endogenous protein levels [131]. AUTAC molecules contain small ligands of the target protein and guanine derivatives as degradation tags, which can trigger K63 polyubiquitin [131]. Liang et al. showed a new AUTAC, which degrades protein BRD4 by targeting LC3 and exhibits good anti-proliferative activity in many tumor cells [132]. However, the mechanism of selective autophagy process is extremely complex and the effect on the whole cell is still unclear, so its stability is poor and further study is necessary. In addition, the mechanism of ubiquitination of K63 induced by guanine derivatives, the miss effect of AUTAC technology, and the functionality in vivo need further investigation.
3.2.3. ATTEC-based protein degradation system
ATTEC (autophagosome-tethering compound) functions through interacting with POI and LC3 [133], which is a more direct strategy to utilize autophagy to degrade targets. It was found that ATTEC, obtained by a high-throughput screening strategy, can interact with both LC3 and mHTT (major mutant HTT protein), and can rescue HD (Huntington’s disease) phenotypes [134]. Fu et al. developed ld-ATTECS to remove lipid droplets (LD), which may help treat chronic diseases such as obesity and cardiovascular disease [135]. It is one of the few examples of directed degradation of non-protein targets. In the mouse model, ld-ATTECs treatment reduced body weight, liver weight, liver LDS, and serum TAG and cholesterol levels [135].
Compared with PROTAC and LYTAC, ATTEC can manipulate protein levels more effectively due to its small size. However, it is still not clear about the detail of the interface between the POI and LC3 induced by ATTEC, and requires further study.
3.2.4. AUTOTAC-based protein degradation system
Recently, many autophagy-lysosome pathway-based degraders, such as AUTAC and ATTEC, have been developed. However, ATTEC targets mHTT or lipid droplets directly at autophagosomes, while AUTAC uses S-guanosine, which depends on the ubiquitination of the target. It is necessary to develop a universal TPD platform independent of ubiquitin and proteasome. Ji et al. constructed the AUTOTAC, which consists of a module interacting with the ZZ domain of p62 and a POI targeting module [136]. The AUTOTAC promoted the oligomerization and activation of p62 and degraded POI through autophagy-lysosome pathway, mediating the targeted degradation of individual proteins and aggregates. Ji et.al. proved that AUTOTAC can effectively remove misfolded tau protein in mouse model expressing human pathological tau mutants. In contrast, proteasome-based techniques, such as PROTAC and molecular glue, are usually ineffective when dealing with misfolded proteins. Additionally, AUTOTAC can also effectively remove a variety of oncoproteins, such as the degradation of AR [136].
4. Conclusion
Over the past two decades, the world has witnessed the rapid development of TPD technology. Techniques based on UPS, e.g., PROTACs, molecular glue, Trim-Away, Predator techniques, achieve rapid, direct manipulation of target protein, enabling a more specific and direct knock-down of genes of interest in basic research, while providing a novel toolkit for applied life science such as biotechnology or synthetic biology. Molecular glue, especially thalidomide derivatives have approved by FDA in clinical usage and achieved good therapeutic effects. In recent years, CMA, LYTAC, ATTEC and AUTAC techniques based on lysosomal pathways provide us with a new road towards the manipulation of target proteins, further expanding the scope and toolbox of TPD. However, compared with UPS, the detailed mechanism and cost of lysosomal pathway-based techniques are still not fully understood, Take AbTAC technique as a quick example, it is not clear whether the “hijacking” lysosomal pathway affects the functioning or physiological state of cells (Table 3). Therefore, from the views of both basic research and clinical application, TPD techniques/drugs based on the lysosomal pathway still have a long way to go [10], [59], [137], [138]. Additionally, from the prospect of biotechnology, it should be noted that TPD techniques with low-cost, convenience, high-efficiency and high-specificity have not been widely used, or even developed, yet. There is a pressing need to further modify the existing TPD techniques or develop novel TPD techniques. In considering the development of novel TPD techniques, the main exploration direction can be roughly divided into the following parts:
-
1)
The mining and discovery of novel functional E3 ligase. We need to continuously explore, identify and screen the new E3 ligase, as well as identify their ligand, catalytic activity, stability, tissue-specific expression profile and protein structure. Expanding and mining existing E3s family libraries to identify new functional proteins or domains that can be used in TPD techniques [139], and then combined with re-design and re-engineering of new E3 ligase, PROTACs and other techniques, to explore the possibility of using novel or engineering E3s in UPS. If the mechanism is clear, proteins involved in deubiquitylation and SUMOylation can also be harnessed to extend the scope of TPD, adding novel, useful tools to the TPD toolbox.
-
2)
Exploration of targeting module. The key to TPD techniques is the targeting module which the selectivity and specificity of system rely on. Identifying, characterizing, quantifying and screening a new targeting module-target pairs, and constructing a database for standardized targeting module or likewise functional module (such as nanobodies, as Wilton et al’s sdAb-DB [140]; scFv; aptamers; etc.), would be advantageous for modular design and engineering of TPD techniques. In addition, new targeting modules can also be designed, evolved and characterized using techniques from directed evolution, quantitative biology and synthetic biology.
-
3)
Iterative refinement of the TPD techniques: Following rational design principles in synthetic biology, modular design, iterative optimization and mathematical modeling can be utilized to identify the optimized combination of targeting module, E3s/E3s-recruiting module and linker between two modules. Additionally, the scope of the targeted ubiquitin degradation system could be extended to the recognition and degradation of specific post-translational modifications such as phosphorylation, and palmitoylation, or applied to subcellular localized proteins, e.g., nuclear proteins, a subunit of protein complex, isoform or mutant of proteins, etc.
-
4)
Controllable TPD system. Controllable elements used in synthetic biology such as tissue-specific promoter, chemical molecules/light/microwave/magnetic field-induced promoter or PPI (such as protein dimerization/dissociation) pairs can be coupled with TPD techniques, enabling a controllable or context-dependent (such as cancers, diseases, etc.) TPD with spatiotemporal precision. Adding more control layers for
-
5)
Delivery of the TPD system. From the clinical application point of view, commonly-used vectors in gene therapy, such as retrovirus vectors, and adenovirus vector can be used to transport the genetically-encoded TPD system to the body, their compatibility and transfer efficiency should be further validated and discussed. In addition, from the prospect of basic research, transfection methods such as cationic carriers, liposomes, nanoparticles and other materials can encapsulate DNA, but how they can be coupled with the TPD system with more should be investigated further.
Table 3.
Advantages and limitations of the different TPD technology-based degradation systems.
| Degradation pathways | Degradation system | Advantages | Limitations | Highest phase | Ref. |
|---|---|---|---|---|---|
| TPD via ubiquitin–proteasome | PROTAC |
In vivo; do not require tight binding; improved selectivity and efficiency |
High molecular weight (800 kDa) and high surface area; low solubility; poor cell permeability and low oral bioavailability; safety concerns |
Phase II | [7], [59], [60] |
| Molecular glue | Good pharmacology; specific to ligase and target |
lack of rational design; poor in general substrate selectivity |
Approved | [141] | |
| AID | Controllable protein degradation can be achieved through the addition time of Auxin. | Complicated experimental design; unclear activity of TIR1 in non-plant cells |
Exploratory | [40] | |
| SMASh | FDA-approved HCV drug | Not suitable for studying biological process with fast kinetics; needs modification of SMASh system | Exploratory | [45] | |
| Trim-Away | Improved selectivity and efficiency | Poor cell membrane permeability; need introduction of antibody with complicated instruments |
Exploratory | [90], [91] | |
| TPD via endosome-lysosomal pathway | LYTAC | Independent of ubiquitination-proteasome degradation | Difficult to determine the optimal linking site; requires an antibody to maintain its characteristics; usually takes a few days | Exploratory | [118] |
| TPD via autophagy lysosome pathway | CMA-based degrader | Faster degradation rate, better reversibility; dose-dependence; stronger specificity; easy design strategy | Poor cellular membrane permeability | Exploratory | [128] |
| AUTAC | Not only can degrade cytoplasmic proteins, but also achieve fragmented organelle degradation | Lack of detailed mechanisms |
Exploratory | [127] | |
| ATTEC | Low molecular weight, good transmembrane activity, and better pharmacokinetics | High molecular design costs; low versatility | Exploratory | [133] | |
| AUTOTAC | Independent of ubiquitin on POI | Slow degradation rate | Exploratory | [136] |
Although the development and broad application of TPD technology are still in its infancy, by virtue of its advantages such as direct manipulation of target protein, high specificity, and rapid time of actin. It is no doubt that TPD is bound to open a new era in the field of basic research and drug discovery.
5. Funding information
This work was supported by a grant from the National Natural Science Foundation of China (No. 82003646), the National Natural Science Foundation of China (No. 31870855), and the Natural Science Foundation of Hunan Province, China (No. 2020JJ5680).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Contributor Information
Chuanyang Liu, Email: liuchuanyang13@nudt.edu.cn.
Lingyun Zhu, Email: lingyunzhu@nudt.edu.cn.
References
- 1.Cameron D.E., Bashor C.J., Collins J.J. A brief history of synthetic biology. Nat Rev Microbiol. 2014;12(5):381–390. doi: 10.1038/nrmicro3239. [DOI] [PubMed] [Google Scholar]
- 2.Xie M., Fussenegger M. Mammalian designer cells: Engineering principles and biomedical applications. Biotechnol J. 2015;10(7):10051018. doi: 10.1002/biot.201400642. [DOI] [PubMed] [Google Scholar]
- 3.Schneekloth J.S., Jr F.F.N., Koldobskiy M. Chemical genetic control of protein levels: selective in vivo targeted degradation. J Am Chem Soc. 2004;126(12):3748–3754. doi: 10.1021/ja039025z. [DOI] [PubMed] [Google Scholar]
- 4.Capecchi M.R. Altering the genome by homologous recombination. Science. 1989;244(4910):1288–1292. doi: 10.1126/science.2660260. [DOI] [PubMed] [Google Scholar]
- 5.Doudna J.A., Emmanuelle C. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346(6213):1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 6.Eder J.H., Paul L. Springer International Publishing. Springer Int Publishing. 2015;232:1–20. [Google Scholar]
- 7.Martinez-Lage M., Puig-Serra P., Menendez P., et al. CRISPR/Cas9 for Cancer Therapy: Hopes and Challenges. Biomedicines. 2018;6(4) doi: 10.3390/biomedicines6040105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elbashir S.M., Harborth J., Lendeckel W., et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411(6836):494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 9.Xie M., Fussenegger M. Designing cell function: assembly of synthetic gene circuits for cell biology applications. Nat Rev Mol Cell Bio. 2018;19(8):507–525. doi: 10.1038/s41580-018-0024-z. [DOI] [PubMed] [Google Scholar]
- 10.Clift D., McEwan W.A., Labzin L.I., et al. A Method for the Acute and Rapid Degradation of Endogenous Proteins. Cell. 2017;171(7):1692–1706 e1618. doi: 10.1016/j.cell.2017.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu T., Yoon H., Xiong Y. Targeted protein degradation as a powerful research tool in basic biology and drug target discovery. Nat Struct Mol Biol. 2020;27(7):605–614. doi: 10.1038/s41594-020-0438-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ding W.X., Yin X.M. Sorting, recognition and activation of the misfolded protein degradation pathways through macroautophagy and the proteasome. Autophagy. 2008;4(2):141–150. doi: 10.4161/auto.5190. [DOI] [PubMed] [Google Scholar]
- 13.Bennett E.J., Bence N.F., Jayakumar R. Global impairment of the ubiquitin-proteasome system by nuclear or cytoplasmic protein aggregates precedes inclusion body formation. Mol Cell. 2005;17(3):351–365. doi: 10.1016/j.molcel.2004.12.021. [DOI] [PubMed] [Google Scholar]
- 14.Hochstrasser M. Ubiquitin, proteasomes, and the regulation of intracellular protein degradation. Curr Opin Cell Biol. 1995;7(2):215–223. doi: 10.1016/0955-0674(95)80031-x. [DOI] [PubMed] [Google Scholar]
- 15.Goldberg A.L. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426(6968):895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- 16.Chan N.C., Salazar A.M., Pham A.H. Broad activation of the ubiquitin-proteasome system by Parkin is critical for mitophagy. Hum Mol Genet. 2011;20(9):1726–1737. doi: 10.1093/hmg/ddr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hao J., Shen R., Li Y. Cancer-testis antigen HCA587/MAGE-C2 interacts with BS69 and promotes its degradation in the ubiquitin-proteasome pathway. Biochem Biophys Res Commun. 2014;449(4):386–391. doi: 10.1016/j.bbrc.2014.05.078. [DOI] [PubMed] [Google Scholar]
- 18.Mulder M.P.C., Witting K.F., Ovaa Huib. Cracking the Ubiquitin Code: The Ubiquitin Toolbox. Curr Issues Mol Biol. 2020:1–20. doi: 10.21775/cimb.037.001. [DOI] [PubMed] [Google Scholar]
- 19.Yang Q., Zhao J., Chen D., et al. E3 ubiquitin ligases: styles, structures and functions. Mol Biomed. 2021;2(1):23. doi: 10.1186/s43556-021-00043-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flick K., Raasi S., Zhang H. A ubiquitin-interacting motif protects polyubiquitinated Met4 from degradation by the 26S proteasome. Nat Cell Biol. 2006;8(5):509–515. doi: 10.1038/ncb1402. [DOI] [PubMed] [Google Scholar]
- 21.Morreale F.E., Walden H. Types of Ubiquitin Ligases. Cell. 2016;165(1):248–248 e241. doi: 10.1016/j.cell.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 22.Bertrand M.J., Lippens S., Staes A. cIAP1/2 are direct E3 ligases conjugating diverse types of ubiquitin chains to receptor interacting proteins kinases 1 to 4 (RIP1-4) PLoS ONE. 2011;6(9) doi: 10.1371/journal.pone.0022356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spratt D.E., Walden H., Shaw G.S. RBR E3 ubiquitin ligases: new structures, new insights, new questions. Biochem J. 2014;458(3):421–437. doi: 10.1042/BJ20140006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dove K.K., Stieglitz B., Duncan E.D., et al. Molecular insights into RBR E3 ligase ubiquitin transfer mechanisms. EMBO Rep. 2016;17(8):1221–1235. doi: 10.15252/embr.201642641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ding Y., Fei Y.Y., Lu B.X. Emerging new concepts of degrader technologies. Trends Pharmacol Sci. 2020;41(7):464–474. doi: 10.1016/j.tips.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou P., Bogacki R., Mcreynolds L. Harnessing the ubiquitination machinery to target the degradation of specific cellular proteins. Mol Cell. 2000;6(3):751–756. doi: 10.1016/s1097-2765(00)00074-5. [DOI] [PubMed] [Google Scholar]
- 27.Liu J., Stevens J., Matsunami N. Targeted degradation of β-catenin by chimeric F-box fusion proteins. Biochem Biophys Res Commun. 2004;313(4):1023–1029. doi: 10.1016/j.bbrc.2003.12.035. [DOI] [PubMed] [Google Scholar]
- 28.Su Y., Ishikawa S., Kojima M. Eradication of pathogenic -catenin by Skp1/Cullin/F box ubiquitination machinery. Proc Natl Acad Sci. 2003;100(22):12729–12734. doi: 10.1073/pnas.2133261100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cong F., Zhang J., Pao W. A protein knockdown strategy to study the function of β-catenin in tumorigenesis. BMC Mol Biol. 2003;4(1):10. doi: 10.1186/1471-2199-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen W. Proteasome-Mediated Destruction of the Cyclin A/Cyclin-Dependent Kinase 2 Complex Suppresses Tumor Cell Growth in Vitro and in Vivo. Cancer Res. 2004;64(11):3949–3957. doi: 10.1158/0008-5472.CAN-03-3906. [DOI] [PubMed] [Google Scholar]
- 31.Chan T.A., Wang Z., Dang L.H. Targeted inactivation of CTNNB1 reveals unexpected effects of β-catenin mutation. Proc Natl Acad Sci. 2002;99(1):8265–8270. doi: 10.1073/pnas.082240999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Melvin A.T., Woss G.S., Park J.H. Measuring activ ity in the ubiquitin-proteasome system: From large scale discoveries to single cells analysis. Cell Biochem Biophys. 2013;67(1):75–89. doi: 10.1007/s12013-013-9621-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen R.P., Gaynor A.S., Chen W. Synthetic biology approaches for targeted protein degradation. Biotechnol Adv. 2019;107446 doi: 10.1016/j.biotechadv.2019.107446. [DOI] [PubMed] [Google Scholar]
- 34.Chassin H., Müller M., Tigges M., et al. A modular degron library for synthetic circuits in mammalian cells. Nat Commun. 2019;10(1):2013. doi: 10.1038/s41467-019-09974-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schapira M., Calabrese M.F., Bullock A.N., et al. Targeted protein degradation: expanding the toolbox. Nat Rev Drug Discov. 2019:1–15. doi: 10.1038/s41573-019-0047-y. [DOI] [PubMed] [Google Scholar]
- 36.Koepp D.M., Schaefer L.K., Ye X., et al. Phosphorylation-dependent ubiquitylation of cyclin E by SCFFbw7 ubiquitin ligase. Science. 2001;294:173–177. doi: 10.1126/science.1065203. [DOI] [PubMed] [Google Scholar]
- 37.Nash P., Tang X., Orlicky S., et al. Multisite phosphorylation of a Cdk inhibitor sets a threshold for the onset of DNA replication. Nature. 2001;414:514–521. doi: 10.1038/35107009. [DOI] [PubMed] [Google Scholar]
- 38.Wu G., Xu G., Schulman B.A., et al. Structure of a beta-TrCP1-Skp1-beta-catenin complex: Destruction motif binding and lysine specificity of the SCF(beta-TrCP1) ubiquitin ligase. Mol Cell Biol. 2003;11:1445–1456. doi: 10.1016/s1097-2765(03)00234-x. [DOI] [PubMed] [Google Scholar]
- 39.Strohmaier H., Spruck C.H., Kaiser P., et al. Human F-box protein hCdc4 targets cyclin E for proteolysis and is mutated in a breast cancer cell line. Nature. 2001;413:316–322. doi: 10.1038/35095076. [DOI] [PubMed] [Google Scholar]
- 40.Tan X., Calderon-Villalobos L.I.A., Sharon M. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446(7136):640–645. doi: 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- 41.Banaszynski L.A., Chen L.-C., Maynard-Smith L.A., et al. A Rapid, Reversible, and Tunable Method to Regulate Protein Function in Living Cells Using Synthetic Small Molecules. Cell. 2006;126(5):995–1004. doi: 10.1016/j.cell.2006.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Richman S.A., Wang Liang-Chuan, Khire Uday R., et al. Ligand-Induced Degradation of a CAR Permits Reversible Remote Control of CAR T Cell Activity In Vitro and In Vivo. Mol Ther J Am Soc Gene Ther. 2020;28(7):1600–1613. doi: 10.1016/j.ymthe.2020.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taxis C., Stier G., Spadaccini R., et al. Efficient protein depletion by genetically controlled deprotection of a dormant N-degron. Mol Syst Biol. 2009;5(1):267. doi: 10.1038/msb.2009.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Caussinus E., Affolter M. deGradFP: A System to Knockdown GFP-Tagged Proteins [J] Methods Mol Biol. 2016;1478 doi: 10.1007/978-1-4939-6371-3_9. [DOI] [PubMed] [Google Scholar]
- 45.Chung H.K., Jacobs C.L., Huo Y.W. Tunable and reversible drug control of protein production via a self-excising degron. Nat Chem Biol. 2015;11(9):713–720. doi: 10.1038/nchembio.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nishimura K., Fukagawa T., Takisawa H. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods. 2009;6(12):917–922. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- 47.Zhang L., Ward J.D., Cheng Z. The auxin-inducible degradation (AID) system enables versatile conditional protein depletion in C. elegans. Development. 2015;142(24):4374–4384. doi: 10.1242/dev.129635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trost M., Blattner A.C., Lehner C.F.J.F. Regulated protein depletion by the auxin-inducible degradation system in Drosophila melanogaster. Fly. 2016;10(1):35–46. doi: 10.1080/19336934.2016.1168552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verma R., Mohl D., Deshaies R.J. Harnessing the Power of Proteolysis for Targeted Protein Inactivation. Mol Cell. 2020;77(3):446–460. doi: 10.1016/j.molcel.2020.01.010. [DOI] [PubMed] [Google Scholar]
- 50.Suzuki T, Varshavsky A. Degradation signals in the lysine-asparagine sequence space. 0261-4189 (Print)). [DOI] [PMC free article] [PubMed]
- 51.Hermann A, Liewald JF, Gottschalk A. A photosensitive degron enables acute light-induced protein degradation in the nervous system. 1879-0445 (Electronic)). [DOI] [PubMed]
- 52.Bonger K.M., Rakhit R., Payumo A.Y. General method for regulating protein stability with light. ACS Chem Biol. 2014;9(1):111–115. doi: 10.1021/cb400755b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Los G.V., Encell L.P., McDougall M.G., et al. HaloTag: A Novel Protein Labeling Technology for Cell Imaging and Protein Analysis. ACS Chem Biol. 2008;3(6):373–382. doi: 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- 54.Nabet B., Roberts J.M., Buckley D.L. The dTAG system forimmediate and target-specific protein degradation. Nat Chem Biol. 2018;14(5):431–441. doi: 10.1038/s41589-018-0021-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Erb M.A., Scott T.G., Li B.E., et al. Transcription control by the ENL YEATS domain in acute leukaemia. Nature. 2017;543(7644):270–274. doi: 10.1038/nature21688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brunetti L., Gundry M.C., Sorcini D., et al. Mutant NPM1 Maintains the Leukemic State through HOX Expression. Cancer Cell. 2018;34(3):499–512.e499. doi: 10.1016/j.ccell.2018.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weintraub A.S., Li C.H., Zamudio A.V., et al. YY1 Is a Structural Regulator of Enhancer-Promoter Loops. Cell. 2017;171(7):1573–1588.e1528. doi: 10.1016/j.cell.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koduri V. Peptidic degron for IMiD-induced degradation of heterologous proteins. Proc Natl Acad Sci USA. 2019;116:2539–2544. doi: 10.1073/pnas.1818109116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ottis P., Crews C.M. Proteolysis-Targeting Chimeras: Induced Protein Degradation as a Therapeutic Strategy. ACS Chem Biol. 2017;12(4):892–898. doi: 10.1021/acschembio.6b01068. [DOI] [PubMed] [Google Scholar]
- 60.Zou Y., Ma D., Wang Y. The PROTAC technology in drug development. Cell Biochem Funct. 2019 doi: 10.1002/cbf.3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sakamoto K.M., Kim K.B., Kumagai A. Protacs: Chimeric molecules that target proteins to the Skp1-Cullin-F box complex for ubiquitination and degradation. Proc Natl Acad Sci. 2001;98(15):8554–8559. doi: 10.1073/pnas.141230798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sakamoto K.M. Development of Protacs to Target Cancer-promoting Proteins for Ubiquitination and Degradation. Mol Cell Proteomics. 2003;2(12):1350–1358. doi: 10.1074/mcp.T300009-MCP200. [DOI] [PubMed] [Google Scholar]
- 63.Vassilev L.T. In Vivo Activation of the p53 Pathway by Small-Molecule Antagonists of MDM2. Science. 2004;303(5659):844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 64.Schneekloth A.R., Pucheault M., Tae H.S. Targeted intracellular protein degradation induced by a small molecule: En route to chemical proteomics. Bioorg Med Chem Lett. 2008;18(22):5904–5908. doi: 10.1016/j.bmcl.2008.07.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gao X., Iii H., Vuky J., et al. Phase 1/2 study of ARV-110, an androgen receptor (AR) PROTAC degrader, in metastatic castration-resistant prostate cancer (mCRPC) J Clin Oncol. 2022;40:17. [Google Scholar]
- 66.Hamilton E., Schott A., Nanda R., et al. ARV-471, an estrogen receptor (ER) PROTACdegrader, combined with palbociclib in advanced ER+/human epidermal growth factor receptor 2–negative (HER2-) breast cancer: Phase 1b cohort (part C) of a phase 1/2 study. J Clin Oncol. 2022;40 TPS1120-TPS1120. [Google Scholar]
- 67.Lonial S., Richard S., Matous J., et al. Pharmacokinetic (PK) profile of a novel IKZF1/3 degrader, CFT7455, enables significant potency advantage over other IKZF1/3 degraders in models of multiple myeloma (MM) and the results of the initial treatment cohort from a first-in-human (FIH) phase 1/2 study of CFT7455 in MM. Cancer Res. 2022;82 Abstract nr CT186. [Google Scholar]
- 68.Accutar Biotech Pipeline [2022.07]. https://www.accutarbio.com/workflow/.
- 69.Arvinas pipeline [2022.07]. https://www.arvinas.com/pipeline-programs/pipeline.
- 70.Tam C., Cheah Chan, Stevens D., et al. P686: a phase 1 first in-human study of BGB-16673, a Bruton tyrosine kinase protein degrader, in patients (PTS) with B-cell malignancies (trial in progress) HemaSphere. 2022;6:582–583. [Google Scholar]
- 71.BMS pipeline [2022.07]. https://www.bmsscience.com/.
- 72.Jaiswal A., Williamson E., Hromas R., et al. Degradation of Bcl-xL by DT2216 is lethal to T-cell acute lymphoblastic leukemia. Cancer Res. 2022;82 Abstract nr 5654. [Google Scholar]
- 73.Flagship Pioneering Pipeline [2022.07]. https://www.flagshippioneering.com.
- 74.Hinova pipeline [2022.07]. http://www.hinovapharma.com/en/chanpinxianguan.html.
- 75.Chuansheng Ge, Baisong Liao, Lei Zhang. KPG-818, a novel cereblon modulator, inhibits hematological malignancies in preclinical models. In: Proceedings of the Annual Meeting of the American Association for Cancer Research 2020; 2020 Apr 27-28 and Jun 22-24. Philadelphia (PA): AACR. Cancer Res 2020, 2020, 80(16 Suppl): Abstract nr 6367.
- 76.Chuansheng Ge, Lei Zhang, Baisong Liao. KPG-121, a novel CRBN modulator, potently inhibits growth of metastatic castration resistant prostate cancer as a single agent or in combination with androgen receptor signaling inhibitors both in vitro and in vivo. In: Proceedings of the Annual Meeting of the American Association for Cancer Research 2020; 2020 Apr 27-28 and Jun 22-24. Philadelphia (PA): AACR. Cancer Res 2020, 2020, 80(16 Suppl): Abstract nr 5327.
- 77.Starodub A., Gollerkeri A., De Savi C., et al. Phase 1 study of KT-333, a targeted protein degrader, in patients with relapsed or refractory lymphomas, large granular lymphocytic leukemia, and solid tumors. J Clin Oncol. 2022;40 TPS3171-TPS3171. [Google Scholar]
- 78.Stevens D., Ewesuedo R., McDonald A., et al. Phase 1 study of KT-413, a targeted protein degrader, in adult patients with relapsed or refractory B-cell non-Hodgkin lymphoma. J Clin Oncol. 2022;40 TPS3170-TPS3170. [Google Scholar]
- 79.Kymera pipeline [2022.07]. https://www.kymeratx.com/pipeline/.
- 80.Mato A., Danilov A., Patel M., et al. A first-in-human phase 1 trial of NX-2127, a first-in-class oral BTK degrader with IMiD-like activity, in patients with relapsed and refractory B-cell malignancies. J Clin Oncol. 2022;40 TPS7581-TPS7581. [Google Scholar]
- 81.Kim Linton G., Collins D.-S., et al. P650: a first-in-human phase 1 trial of NX-5948, an oral BTK degrader, in patients with relapsed and refractory B-cell malignancies. HemaSphere. 2022;6:548–549. [Google Scholar]
- 82.Jackson K., Agafonov R., Carlson M., et al. The discovery and characterization of CFT8634: A potent and selective degrader of BRD9 for the treatment of SMARCB1-perturbed cancers. Cancer Res. 2022;82 Abstract nr ND09. [Google Scholar]
- 83.C4 Therapeutics Pipeline [2022.07]. https://c4therapeutics.com/pipeline.
- 84.Cullgen Pipeline [2022.07]. https://www.cullgen.com/.
- 85.Stanton B.Z., Chory E.J., Crabtree G.R. Chemically induced proximity in biology and medicine. Science. 2018;359:eaao5902. doi: 10.1126/science.aao5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gerry C.J., Schreiber S.L. Unifying principles of bifunctional, proximity-inducing small molecules. Nat Chem Biol. 2020;16:369–378. doi: 10.1038/s41589-020-0469-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stoeckli E.T. Protocadherins: not just neuron glue, more too! Dev Cell. 2014;30:643–644. doi: 10.1016/j.devcel.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 88.Schreiber S.L. Chemistry and biology of the immunophilins and their immunosuppressive ligands. Science. 1991;251:283–287. doi: 10.1126/science.1702904. [DOI] [PubMed] [Google Scholar]
- 89.Chamberlain P.P., Cathers B.E. Cereblon modulators: Low molecular weight inducers of protein degradation. Drug Discov Today Technol. 2019;31:29–34. doi: 10.1016/j.ddtec.2019.02.004. [DOI] [PubMed] [Google Scholar]
- 90.Vaysburd M., Watkinson R.E., Cooper H. Intracellular antibody receptor TRIM21 prevents fatal viral infection. Proc Natl Acad Sci. 2013;110(30):12397–12401. doi: 10.1073/pnas.1301918110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Keeble A.H., Khan Z., Forster A. TRIM21 is an IgG receptor that is structurally, thermodynamically, and kinetically conserved. Proc Natl Acad Sci U S A. 2008;105(16):6045–6050. doi: 10.1073/pnas.0800159105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Neklesa T.K., Winkler J.D., Crews C.M. Targeted protein degradation by PROTACs. Pharmacol Ther. 2017;174:138–144. doi: 10.1016/j.pharmthera.2017.02.027. [DOI] [PubMed] [Google Scholar]
- 93.Chen X., Liu M., Lou H. Degradation of endogenous proteins and generation of a null-like phenotype in zebrafish using Trim-Away technology. Genome Biol. 2019;20(1) doi: 10.1186/s13059-019-1624-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liu C., Kuang J., Qiu X., et al. Cold Spring Harbor Laboratory; 2020. Predator: A novel method for targeted protein degradation. [Google Scholar]
- 95.2020 iGEM NUDT_CHINA Results [2022.07]. https://2020.igem.org/Team:NUDT_CHINA/Results.
- 96.2021 iGEM NUDT_CHINA Results [2022.07]. https://2021.igem.org/Team:NUDT_CHINA/Results.
- 97.Sui B., Wang M., Cheng C., et al. Nanogel-Facilitated Protein Intracellular Specific Degradation through Trim-Away. Adv Funct Mater. 2021;31:2010556. doi: 10.1002/adfm.202010556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chen G., Kong Y., Li Y., et al. A Promising Intracellular Protein-Degradation Strategy: TRIMbody-Away Technique Based on Nanobody Fragment. Biomolecules. 2021;11(10) doi: 10.3390/biom11101512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zeng J., Santos A.F., Mukadam A.S., et al. Target-induced clustering activates Trim-Away of pathogens and proteins. Nat Struct Mol Biol. 2021;28(3):278–289. doi: 10.1038/s41594-021-00560-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Huang J., Gaizhen R.u., Sun L., et al. Elevated RIF1 participates in the epigenetic abnormalities of zygotes by regulating histone modifications on MuERV-L in obese mice. Mol Med. 2022;28:17. doi: 10.1186/s10020-022-00446-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jeon H.-J., Oh J.S. TRF1 Depletion Reveals Mutual Regulation Between Telomeres, Kinetochores, and Inner Centromeres in Mouse Oocytes. Front Cell Dev Biol. 2021;9 doi: 10.3389/fcell.2021.749116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jiang G., Huang Z., Yuan Y., et al. Intracellular delivery of anti-BCR/ABL antibody by PLGA nanoparticles suppresses the oncogenesis of chronic myeloid leukemia cells. J Hematol Oncol. 2021;14 doi: 10.1186/s13045-021-01150-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Israel S., Drexler H., Fuellen G., et al. The COP9 signalosome subunit 3 is necessary for early embryo survival by way of a stable protein deposit in mouse oocytes. Mol Hum Reprod. 2021;27 doi: 10.1093/molehr/gaab048. [DOI] [PubMed] [Google Scholar]
- 104.Lin C.-Y., Chen S.-H., Tsai C.-L., et al. Intracellular targeting of STIP1 inhibits human cancer cell line growth. Transl Cancer Res. 2021;10:1313–1323. doi: 10.21037/tcr-20-3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mengoli V., Jonak K., Lyzak O., et al. Deprotection of centromeric cohesin at meiosis II requires APC/C activity but not kinetochore tension. EMBO J. 2021;40 doi: 10.15252/embj.2020106812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Scheffler K., Uraji J., Jentoft I.a., et al. Two mechanisms drive pronuclear migration in mouse zygotes. Nat Commun. 2021;12:841. doi: 10.1038/s41467-021-21020-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chatterjee P., Ponnapati M., Kramme C., et al. Targeted intracellular degradation of SARS-CoV-2 via computationally optimized peptide fusions. Commun Biol. 2020;3 doi: 10.1038/s42003-020-01470-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pozzi S., Codron P., Soucy G., et al. Monoclonal full-length antibody against TAR DNA binding protein 43 reduces related proteinopathy in neurons. JCI Insight. 2020;5 doi: 10.1172/jci.insight.140420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Coyne A., Zaepfel B., Hayes L., et al. G4C2 Repeat RNA Initiates a POM121-Mediated Reduction in Specific Nucleoporins in C9orf72 ALS/FTD. Neuron. 2020;107 doi: 10.1016/j.neuron.2020.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang H., Li Y., Yang J., et al. Symmetry breaking in hydrodynamic forces drives meiotic spindle rotation in mammalian oocytes. Sci Adv. 2020;6:eaaz5004. doi: 10.1126/sciadv.aaz5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sha Q.-Q., Zhu Y., Li S., et al. Characterization of zygotic genome activation-dependent maternal mRNA clearance in mouse. Nucleic Acids Res. 2019;48 doi: 10.1093/nar/gkz1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Drutovic D., Duan X., Li R., et al. Ran GTP and importin β regulate meiosis I spindle assembly and function in mouse oocytes. EMBO J. 2019;39 doi: 10.15252/embj.2019101689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Fangtao Chi, Mark Sharpley, Raghavendra Nagaraj, et al. Glucose metabolism distinguishes TE from ICM fate during mammalian embryogenesis [M]. 2019. [DOI] [PMC free article] [PubMed]
- 114.Israel S., Casser E., Drexler H., et al. A framework for TRIM21-mediated protein depletion in early mouse embryos: Recapitulation of Tead4 null phenotype over three days. BMC Genomics. 2019;20 doi: 10.1186/s12864-019-6106-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jie Z., Wang X.-Y., Han Z., et al. Loss of CENPF leads to developmental failure in mouse embryos. Cell Cycle. 2019;18:1–16. doi: 10.1080/15384101.2019.1661173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mehlmann L., Uliasz T., Lowther K. SNAP23 is required for constitutive and regulated exocytosis in mouse oocytes. Biol Reprod. 2019;101 doi: 10.1093/biolre/ioz106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jarosińska O., Rüdiger S. Molecular Strategies to Target Protein Aggregation in Huntington’s Disease. Front Mol Biosci. 2021;8 doi: 10.3389/fmolb.2021.769184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Uhlén M., Fagerberg L., Proteomics H.B.M. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 119.Banik S.M., Pedram K., Wisnovsky S. Lysosome-targeting chimaeras for degradation of extracellular proteins. Nature. 2020;584(7820):291–297. doi: 10.1038/s41586-020-2545-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Coutinho M.F., Prata M.J., Alves S. A shortcut to the lysosome: the mannose-6-phosphate-independent pathway. Mol Genet Metab. 2012;107(3):257–266. doi: 10.1016/j.ymgme.2012.07.012. [DOI] [PubMed] [Google Scholar]
- 121.Park E.I., Mi Y., Unverzagt H.-J., Baenziger J.U. The asialoglycoprotein receptor clears glycoconjugates terminating with sialic acidα2,6GalNAc. PNAS. 2005;102(47):17125–17129. doi: 10.1073/pnas.0508537102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zimmermann T.S., Karsten V., Chan A., et al. Clinical Proof of Concept for a Novel Hepatocyte-Targeting GalNAc-siRNA Conjugate. Mol Ther. 2017;25(1):71–78. doi: 10.1016/j.ymthe.2016.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ahn G., Banik S.M., Miller C.L., et al. LYTACs that engage the asialoglycoprotein receptor for targeted protein degradation. Nat Chem Biol. 2021;17(9):937–946. doi: 10.1038/s41589-021-00770-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gray M.A., Stanczak M.A., Mantuano N.R., et al. Targeted glycan degradation potentiates the anticancer immune response in vivo. Nat Chem Biol. 2020;16(12):1376–1384. doi: 10.1038/s41589-020-0622-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Schuck S. Microautophagy – distinct molecular mechanisms handle cargoes of many sizes. J Cell Sci. 2020;133(17) doi: 10.1242/jcs.246322. [DOI] [PubMed] [Google Scholar]
- 126.Pei J., Wang G., Feng L., et al. Targeting Lysosomal Degradation Pathways: New Strategies and Techniques for Drug Discovery. J Med Chem. 2021;64(7):3493–3507. doi: 10.1021/acs.jmedchem.0c01689. [DOI] [PubMed] [Google Scholar]
- 127.Sinha R.A., Singh B.K., Yen P.M. Reciprocal crosstalk between autophagic and endocrine signaling in metabolic homeostasis. Endocr Rev. 2017;38(1):69–102. doi: 10.1210/er.2016-1103. [DOI] [PubMed] [Google Scholar]
- 128.Fan X.L., Jin W.Y., Lu J. Rapid and reversible knockdown of endogenous proteins by peptide- directed lysosomal degradation. Nat Neurosci. 2014;17(3):471–480. doi: 10.1038/nn.3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhou Y.-F., Wang J., Deng M.-F., et al. The Peptide-Directed Lysosomal Degradation of CDK5 Exerts Therapeutic Effects against Stroke. Aging Dis. 2019;10(5):1140–1145. doi: 10.14336/AD.2018.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wang H., Yao H., Li C. HIP1R targets PD-L1 to lysosomal degradation to alter T cell–mediated cytotoxicity. Nat Chem Biol. 2019;15:42–50. doi: 10.1038/s41589-018-0161-x. [DOI] [PubMed] [Google Scholar]
- 131.Takahashi D., Moriyama J., Nakamura T. AUTACs: cargo-specific degraders using selective autophagy. Mol Cell. 2019;76(5):797–810.e710. doi: 10.1016/j.molcel.2019.09.009. [DOI] [PubMed] [Google Scholar]
- 132.Pei J., Pan X., Wang A., et al. Developing potent LC3-targeting AUTAC tools for protein degradation with selective autophagy. Chem Commun (Camb) 2021;57(97):13194–13197. doi: 10.1039/d1cc04661f. [DOI] [PubMed] [Google Scholar]
- 133.Li Z.Y., Wang C., Wang Z.Y. Allele-selective lowering of mutant HTT protein by HTT-LC3 linker compounds. Nature. 2019;575(7781):203–209. doi: 10.1038/s41586-019-1722-1. [DOI] [PubMed] [Google Scholar]
- 134.Tabrizi S.J., Flower M.D., Ross C.A. Huntington disease: new insights into molecular pathogenesis and therapeutic opportunities. Nat Rev Neurol. 2020;16(10):529–546. doi: 10.1038/s41582-020-0389-4. [DOI] [PubMed] [Google Scholar]
- 135.Fu Y., Chen N., Wang Z., et al. Degradation of lipid droplets by chimeric autophagy-tethering compounds. Cell Res. 2021;31(9):965–979. doi: 10.1038/s41422-021-00532-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ji C.H., Kim H.Y., Lee M.J., et al. The AUTOTAC chemical biology platform for targeted protein degradation via the autophagy-lysosome system. Nat Commun. 2022;13(1):904. doi: 10.1038/s41467-022-28520-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zeng L.R., Vega-Sanchez M.E., Zhu T. Ubiquitination-mediated protein degradation and modification: an emerging theme in plant-microbe interactions. Cell Res. 2006;16(5):413–426. doi: 10.1038/sj.cr.7310053. [DOI] [PubMed] [Google Scholar]
- 138.Zheng J., Tan C., Xue P. Proteolysis targeting peptide (PROTAP) strategy for protein ubiquitination and degradation. Biochem Biophys Res Commun. 2016;470(4):936–940. doi: 10.1016/j.bbrc.2016.01.158. [DOI] [PubMed] [Google Scholar]
- 139.Kramer L.T., Zhang X. Expanding the landscape of E3 ligases for targeted protein degradation. Curr Res Chem Biol. 2022;2 [Google Scholar]
- 140.Wilton E.E., Opyr M.P., Kailasam S., et al. sdAb-DB: The Single Domain Antibody Database. Acs Synth Biol. 2018;7(11):2480–2484. doi: 10.1021/acssynbio.8b00407. [DOI] [PubMed] [Google Scholar]
- 141.Surka C., Jin L., Mbong N. CC-90009, a novelcereblon E3 ligase modulator, targets acute myeloid leukemia blasts andleukemia stem cells. Blood. 2021;137(5):661–677. doi: 10.1182/blood.2020008676. [DOI] [PMC free article] [PubMed] [Google Scholar]