Abstract
Aims
Osteosarcoma (OS) is the most common primary malignant bone sarcoma among children and adolescents. Treatment is based on neo-adjuvant and adjuvant chemotherapy, using the standard drugs cisplatin, methotrexate, doxorubicin, and ifosfamide (IFO). Due to the high capacity of tumor resistance, the current work aimed to analyze genes related to cycle control and cell differentiation in OS cells sensitive to and with induced resistance to IFO. This was to assess whether the differentiated expression of these genes may affect resistance to the drug IFO used in OS treatment, and thus establish possible biomarkers of disease progression.
Materials and methods
In this work, the treatment-sensitive OS U2OS lineage was used, and the same lineage was submitted to the process of induction of IFO resistance. These cells were evaluated by MTT, migration and proliferation assays and submitted to gene expression analysis.
Key findings
The results demonstrate that after induction of resistance to IFO, resistant U2OS cells show a more aggressive tumor behavior, with greater capacity for cell migration, proliferation, and invasion compared to sensitive cells. Gene analysis indicates that resistance-induced cells have differentiated expression of the genes EPB41L3, GADD45A, IER3, OXCT1, UBE2L6, UBE2A ALPL, and EFNB2. Our results suggest new perspectives on possible resistance biomarkers, especially the genes EFNB2 and EPB41L3, given that these genes have rarely been studied their expression linked to osteosarcoma. They show how the resistance induction model can be useful for studies on tumor cell behavior.
Keywords: Osteosarcoma, Ifosfamide, Resistance-induced
Highlights
-
•
The U2OS cell line with induced resistance demonstrated greater capacity for cell migration and proliferation.
-
•
With the resistance induction to Ifosfamide, the EPB41L3, IER3, OXCT1 and EFNB2 genes were differently expressed.
-
•
EPB41L3, IER3, OXCT1 genes have potential to develop new tests in the search for biomarkers to detect aggressive tumors.
1. Introduction
Osteosarcoma (OS) is one of the most common types of primary bone tumors and most often affects the epiphysis region of long bones [1]. Approximately 20–25% of patients present metastases at diagnosis, a factor that implies worse survival of these patients, among whom only 1 in 4 patients has a 5-year disease-free survival. Overall, about 30% of metastatic tumors do not respond to chemotherapy [2].
The most commonly used anticancer agents are doxorubicin (DOX), ifosfamide (IFO), cisplatin (CIS), and high doses of methotrexate (MTX) [3,4]. The drug IFO is an alkylating agent that, due to its phosphoric mustard metabolites, intercalates in the double strand of DNA, generating cross-links in the N-7 region of the guanine base, thus leading to cellular apoptosis [5,6].
Drug resistance can be defined as intrinsic resistance, when tumor cells already have mutations in genes responsible for anti-apoptotic activities, drug efflux, cell migration, and other activities [7], or acquired resistance that occurs due to the selective process generated by drug treatment. These two forms of resistance can occur in OS [8],9.
In this context, the current study aimed to induce resistance to IFO in the U2OS lineage of human osteosarcoma to assess how these cells behave toward the drug IFO and to analyze the gene expression of EFNB2, EPB41L3, GADD45A, ALPL, OXCT1, IER3, UBE2A, and UBE2L6 involved in tumor resistance, in both sensitive and resistant cells. EPB41L3 is a tumor suppressor that inhibits cell proliferation and promotes apoptosis, in addition to encoding a protein that assists in the organization of the actin cytoskeleton; GADD45A is responsible for cell cycle control and stimulates DNA repair by excision of bases; IER3 influences the activation of the ERK pathway, which is responsible for cell proliferation, survival, and differentiation; OXCT1 plays a role in the metabolism of ketone bodies and is used by tumor cells as a source of ATP; the genes UBE2L6 and UBE2A are part of the family of enzymes that assist in the ubiquitination process, and they are responsible for protein degradation mediated by proteasomes and which is highly important in tumorigenesis; in this case the E2 group of enzymes are related to tumor progression involving mechanisms of DNA repair in tumor cells, apoptosis, and signaling of oncogenic pathways, among others; ALPL is a gene that plays a fundamental role in the formation of osteocytes and mineralization of the bone matrix; and, finally, EFNB2 regulates axon orientation, angiogenesis, and epithelial cell migration. In this context, it is of great importance to understand how dysregulation in the expression of these genes may be related to the degree of tumor aggressiveness and the process of resistance to the drug IFO.
In this work, we demonstrated the process of inducing resistance to IFO and the expression of genes that are differentially expressed in the absence and presence of the drug, suggesting that IFO leads to a process of selective pressure on resistant cells; when it is removed from the culture, the cells return to being sensitive to the drug. Furthermore, the genes involved in the process of cell cycle regulation and differentiation present a difference between cells that have been induced to be resistant to IFO and cells that are sensitive.
2. Materials and methods
2.1. Cell culture and MTT assay
Human osteosarcoma U2OS lineage cells (BCRJ code:0304/ATCC: HTB-96) were cultured with DMEM high glucose medium (Sigma-Aldrich) supplemented with 10% fetal bovine serum (FBS). The cells were maintained at 37˚C and 5% CO2 and cell culture was performed in the absence of antibiotics. The alkylating agent IFO >98% (Sigma-Aldrich®) C2H95Cl2N2O2P, with a molecular weight of 261.09 g/mol, was solubilized with DMSO and kept in stock solution of 1 mM divided into aliquots of approximately 10 ml, protected from light at a temperature of −20o C.
Before starting the IFO resistance induction protocol, an initial IC50 was performed to determine the percentage of cellular metabolic activity in an IFO concentration gradient (5.0 μM; 10 μM; 15 μM; 20 μM; 30 μM and 40 μM); this assay was performed using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay, which measures the mitochondrial activity of the cells [9].
The initial IC50 value was obtained from a nonlinear regression calculation.
For the MTT assay, a total of 1 × 104 IFO-treated cells were used per well in 96 plates, with a final volume at 150 μM (cells with DMEM medium and 10% FBS). Positive control consisted of cells and DMEM medium with 10% FBS, without treatment, and negative control consisted of only cells with medium and dimethyl sulfoxide (DMSO). After adding the IFO concentrations, the plate was kept in the chamber under the same culture conditions for 24 h, and then all the medium was removed and 90 μM of medium plus 10 μM of MTT (4 mg/mL) were added. The plate was incubated again for another 4 h with the addition of 60 μM of DMSO for 30 min and then read on a microplate spectrophotometer (BioTek Eon Microplate Spectrophotometer S N 265483) at an absorbance of 570 nm.
2.2. Induced resistance
The resistance induction protocol was performed [10]. When cells reached confluence between 70% and 80%, they received 20% of the concentration of 25 μM IFO and were incubated for 24 h under the same culture conditions. They were then washed with phosphate-buffered saline 1X (PBS), and when they reached the same confluence as the previous step, they received 20% of the initial IC50 value and the whole process was repeated three times. Cells were treated with 20%, 40%, 60%, 80%, and 100% of the 25 μM value. After four months of resistance induction, the U2OS R+ lineage was obtained.
A new MTT assay, as mentioned above, was performed to verify the sensitivity of the U2OS lineage after resistance induction (R+) compared to the sensitive U2OS lineage (S). In this new MTT assay, the IFO concentrations used were 10 μM, 20 μM, 30 μM, 40 μM, 50 μM, and 60 μM. The U2OS R+ lineage was maintained in culture with 35 μM of the drug, and some of the cells that went through the resistance induction protocol began to be cultivated in the absence of the drug (U2OS R-).
2.3. Migration assay
For the migration test, a total of 2.5 × 105 cells per well were used. After reaching 100% confluence, three parallel lines were made with the aid of a 200 μL tip; then the culture medium was removed and washed once with PBS [11]. Culture medium was added to each well without the addition of FBS, with the exception of the positive control, which had the addition of 2% FBS.
The cells were incubated at 37° C, and the measurements were photo-documented at previously established time intervals, starting from the moment after the incision of the lines (time 0 h), followed by intervals of 24, 48, and 72 h after the incision, always at the same previously marked points and corresponding to the location with the absence of cells at time 0 h. The number of cells was counted using the ImageJ® software, where the cell migration capacity was measured in relation to the groups of sensitive cells (S) and resistant cells (U2OS R+).
2.4. Survival test without membership
Cells were seeded in a quantity of 2.5 × 105 cells per well containing LB Agar covering the entire surface of a 6-well plate [12]. The cells were maintained in culture for 6 days in a humidified chamber with an atmosphere of 5% CO2 at 37°C, with changes of DMEM medium supplemented with 10% fetal bovine serum every 48 h. At the end of the culture time, the cells were subjected to an MTT test to assess cell viability.
2.5. Proliferation assay
A total of 2.5 × 105 cells per well were seeded in 6-well plates for re-counting. The values obtained at 0 h and 48 h were analyzed. The cells were trypsinized, washed with PBS, counted, and PDT was calculated following the formula PDT=(T2−T1) × log_10 2 ÷ log_10(N2/N1), where T corresponds to the assay time and N the number of cells counted [13].
2.6. RNA extraction, cDNA synthesis, and real-time qPCR
To evaluate the gene expression profile, the qPCR of the groups U2OS R -; U2OS R+, and U2OS S was performed. Total RNA from U2OS S, U2OS R+, and U2OS R - cells was extracted using the TRIzol method (Invitrogen, Osaka, Japan). Integrity and quantification were determined using qubit (Invitrogen) and nanodrop (Thermo Scientific Nanodrop 2000) and the ratios 260/280 and 260/230 in the range of approximately 2.0 were considered.
To remove contaminating genomic DNA, the preparation (10–100 μg) was subsequently incubated with 10 units of RQ1 RNase-free DNASE I (Promega, M6101). RNA at a concentration of 1 μg/μL was used for cDNA synthesis using the High-Capacity cDNA Reverse Transcription Kit (ThermoFisher USA), according to the manufacturer's manual. Therefore, a sensitive and reproducible quantitative PCR based on SyBr green using the StepOnePlus Real time PCR System@ (Applied Biosystems) was developed. The analysis was performed using the 2−ꙙCq method [14]. All samples were normalized using endogenous control for GAPDH. Table 1 shows the sequence of resistance genes used. All reactions were performed in biological and experimental triplicate.
Table 1.
Primer (10 mM) sequences used for detection of the resistance genes in S, R+ and R-cell groups.
| Ref. No. | Gene | Sequence 5’ – 3′ Product length (bp) |
|---|---|---|
| 262967769(fwd) | EPB41L3 | 5′- AGT GAG TTC CGC TTT GCA CCA AAC-3' (fwd) 24 |
| 262967770(rev) | 5′- AAA TGC ATC TCT GCT TCT GCT GGC -3' (rev); 24 | |
| 262967775(fwd) | GADD45A | 5′-CAT GTT CGT CAT GGG TGT GAA CCA- 3′(fwd), 24 |
| 262967776(rev) | 5′-AGT GAT GGC ATG GAC TGT GGT CAT-3′(rev). 24 | |
| 260377267(fwd) | IER3 | 5′-TCT TTC TGC TGC TCA CCA TCG TCT-3′(fwd), 24 |
| 260377268(rev) | 5′-GCT CCG AAG TCA GAT TAA AGGGCT-3′(rev), 24 | |
| 262967781(fwd) | OXCT1 | 5′-GACAGTGGATGACGTACAGAAG-3′(fwd) 22 |
| 2629677782(rev) | 5′CACGCAGCCTGGTACAAATA-3′(rev) 20 | |
| 262967771(fwd) | UBE2L6 | 5′-AGT ATC CGT TCA AGC CTC CCA TGA-3′(fwd), 24 |
| 262967772(rev) | 5′-AAG GCT TCC AGT TCT CAC TGC TGA-3′(rev); 24 | |
| 262967789(fwd) | UBE2A | 5′GAACAAAGCTGGCGTGATTG3′(fwd), 20 |
| 262967790(rev) | 5′AGGAGTAGGGAGGTGAC AA 3′(rev), 20 | |
| 262967785(fwd) | EFNB2 | 5′CTTTCCCAGAGGACACCTAATG3(fwd), 21 |
| 262967786(rev) | 5′GTGCTGTGCTTCAGTCAATTC 3′(rev) 22 | |
| 262967767(fwd) | GAPDH | 5′- CAT GTT CGT CAT GGG TGT GAA CCA -3' (fwd); 24 |
| 262967768(rev) | 5′- AGT GAT GGC ATG GAC TGT GGT CAT -3' (rev), 24 |
2.7. Data analysis
Statistical analyses were performed using the GraphPad Prism 8.0.1 program. The IC50 value was obtained by means of a non-linear regression, and the differences between the groups of cells were compared using two-way ANOVA and the Tukey post-hoc test. Gene expression results were analyzed with one-way ANOVA. Proliferation assay was obtained by unpaired t-test.
The statistical difference in the gene expression of S cells in relation to R+ and R-cells was represented by p < 0.0001 (****). The significance value was determined from p < 0.05.
3. Results
3.1. Initial IC50 and induced resistance
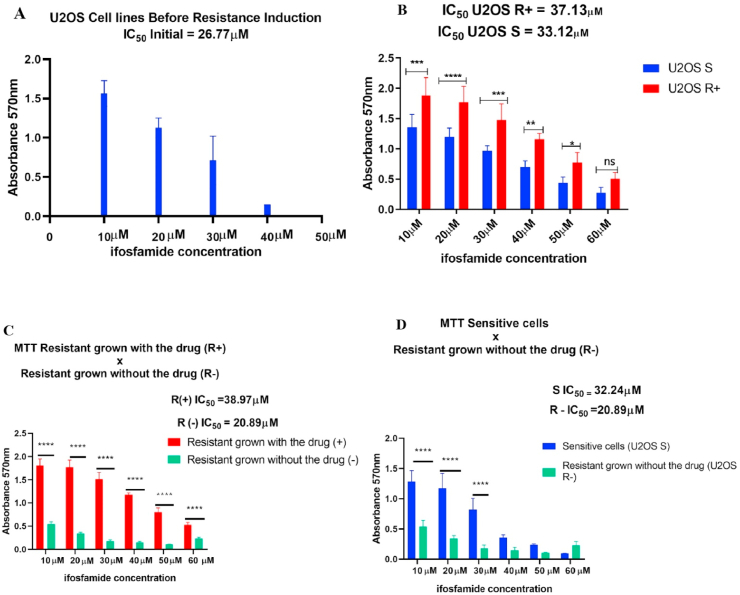
To obtain the U2OS lineage that was resistant to IFO, an initial MTT assay was performed, where the concentration of IFO used in the resistance induction protocol was determined, as shown in Fig. 1A. The IC50 from the U2OS lineage before induction was 26.77 μM. Subsequently, a new MTT was performed in cells that were induced to resistance to IFO (U2OS R+) and also in sensitive cells (U2OS S).
Fig. 1.
Induction of resistance to IFO. In A the MTT assay demonstrated that 26.77 μM of IFO inhibited approximately 50% of osteosarcoma U2OS cells. B indicates that after the resistance induction protocol, the IFO value necessary to inhibit 50% of the cells became 37.13 μM. In addition, the U2OS R+ group showed a significant difference in the rate of metabolic activity in relation to the U2OS S cells with *p < 0.05 and p < 0.0001 (****) at different drug concentrations. C- Significant difference between U2OS R+ and U2OS R-cells at all drug concentrations with p < 0.0001 (****). D – U2OS R- and U2OS S cells with significant differences in concentrations of 10 μM, 20 μM, and 30 μM (****); however, there was no difference in the higher concentrations of IFO, indicating the sensitivity to treatment in the two cell groups.
The IC50 of the U2OS R+ group was 37.13 μM, while in the S group the IC50 was 33.12 μM, as shown in Fig. 1 B. After confirming the resistance in U2OS R+ cells, these cells were cultivated in the absence of the drug (U2OS R-) to verify that, even without IFO stimulation, these cells remained resistant. In a new MTT, the U2OS R-group presented an IC50 of 20.89 μM; the U2OS R+ group had an IC50 of 38.97 μM, and the U2OS S group an IC50 of 32.24 μM (Fig. 1C and 1D). These results indicate that although the process of inducing resistance occurred as expected, this resistance only persists when cells are maintained in culture in the presence of the IFO drug, while when resistance is induced and subsequently cultured without the drug, these cells again become sensitive to IFO treatment.
3.2. Migration assay
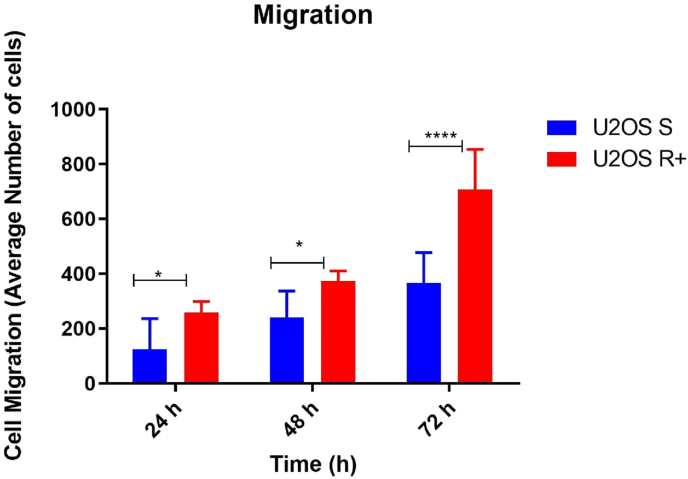
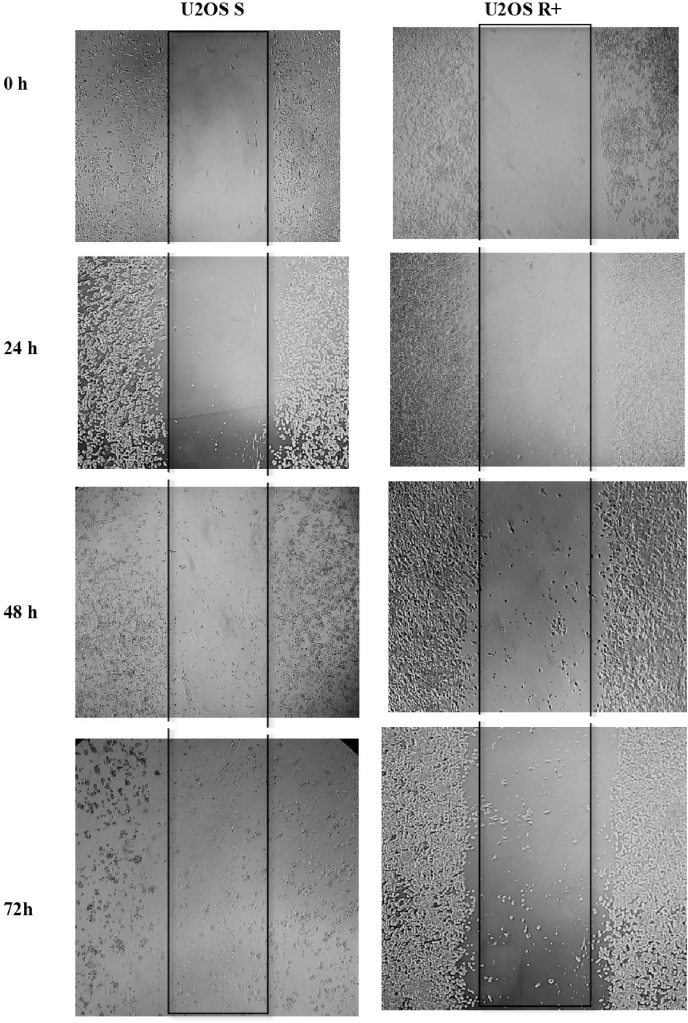
After 24 h, there was a statistical difference between U2OS R+ and U2OS S cells (*), and the same pattern of statistical difference occurred at 48 h and at 72 h. The U2OS R+ group continued to show a greater migration capacity p < 0.0001 (****), showing that the U2OS R+ cell group had a higher migration capacity compared to the U2OS S cell groups (Fig. 2). In Fig. 3, it is possible to observe the cell migration in vitro at the initial time (0 h), 24 h, 48 h, and 72 h respectively.
Fig. 2.
Migration of the U2OS S and U2OS R+ lineages. The x-axis corresponds to the time in hours and the y-axis is the average of the number of cells that migrated. At 24 and 48 h, U2OS R+ cells had greater migration capacity compared to U2OS S cells (*), and at 72 h the difference in migration between the sensitive and resistant groups was of p < 0.0001 (****).
Fig. 3.
In vitro cell migration of U2OS S and U2OS R+ groups at 0 h, 24 h, 48 h, and 72 h intervals.
3.3. Survival test without adherence
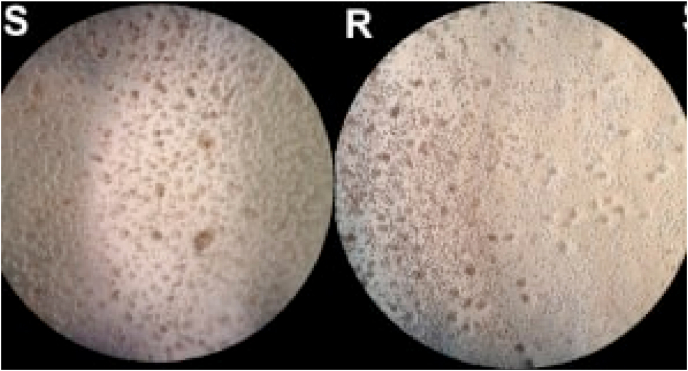
After six days of cultivation, counting performed using the Neubauer chamber and the MTT assay demonstrated that the number of cells increased compared to the beginning, the mean U2OS R+ cells to 3.36 × 105 and the U2OS S cells to 3.86 × 105 cells at the end of the experiment. This suggests that the IFO-sensitive and resistant osteosarcoma U2OS lineages have the ability to survive without plaque adherence and remain capable of cell proliferation. Fig. 4 shows the adherence of U2OS S cells and U2OS R+ cells, revealing their degradation potential.
Fig. 4.
Non-adherence survival test. Analysis of cells not adhered to the LB agar-coated bottom plate. U2OS S (S) represents the group of sensitive cells, U2OS R+ (R) the group of resistant cells. Optical microscopy view. 10x magnification.
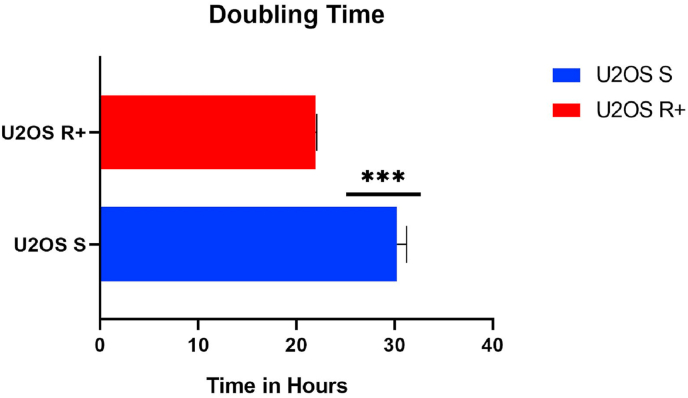
3.4. Proliferation assay
The group of U2OS R+ cells showed a shorter proliferation time compared to U2OS S cells, with P value ≤ 0.0001 (***), as indicated in Fig. 5. The proliferation capacity of the U2OS R+ group was on average 21 h, while the proliferation of U2OS S occurred in an average time of 30 h.
Fig. 5.
Cell proliferation assay – Doubling Time, indicating that U2OS S cells have an average time of 30 h to proliferate, while U2OS R+ cells have a shorter proliferation time, corresponding to 21 h.
3.5. Gene expression profile
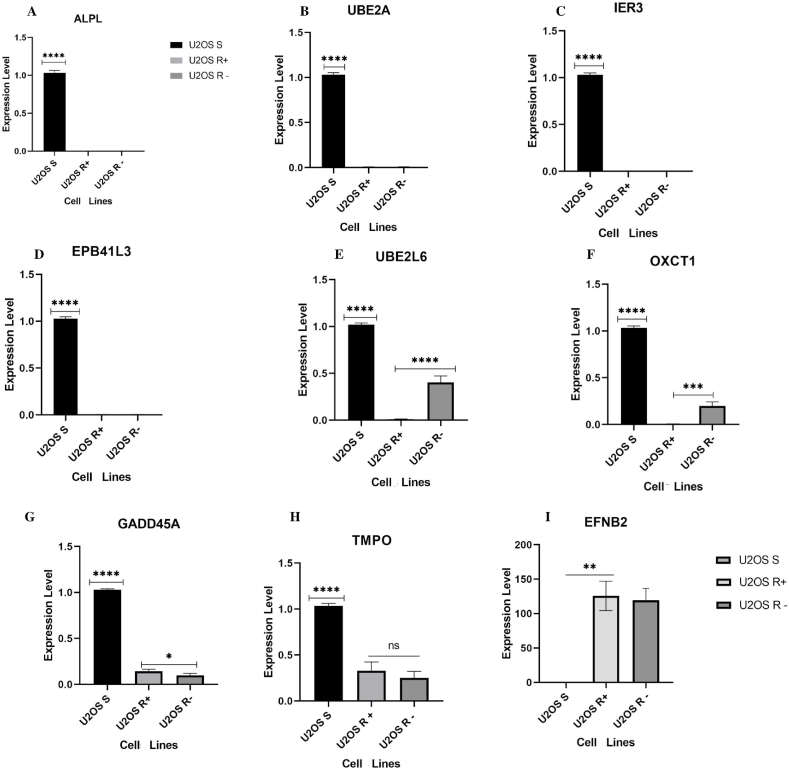
To assess whether the U2OS R+ group has different levels of expression of resistance genes compared to the U2OS S group, a qPCR was performed with the genes EFNB2; ALPL; TMPO; UBE2L6; UBE2A; EPB41L3; GADD45A; OXCT1, and IER3. We also evaluated whether the resistance-induced cells maintained the same gene expression profile when the IFO stimulus (U2OS R-) was removed.
The U2OS S lineage presented the genes ALPL, UBE2A, IER3 and EPB41L3 up-regulated in comparison with U2OS R+ and U2OS R-cells (p < 0.0001). In contrast, these same genes were down-regulated in the U2OS R+ and U2OS R-groups, and there was no significant difference between them, as shown in Fig. 6A–D.
Fig. 6.
Expression of tumor resistance genes in human osteosarcoma U2OS lineage groups. Expression levels were evaluated according to the mean of experimental and biological triplicates. The black bar indicates group U2OS S; the light gray bar U2OS R+ and the dark gray bar U2OS R-. The x-axis represents the group of cells and the y-axis the expression level. Graphs A – D represent the genes that were up-regulated in lineage U2OS S compared to groups U2OS R+ and U2OS R- (****p < 0.0001). Graph E represents the expression of UBE2L6 up-regulated in U2OS S (****p < 0.0001), and U2OS R-had greater expression (****p < 0.0001) compared to U2OS R+. Graph F – OXCT1 with high expression in U2OS S (****p < 0.0001) and U2OS R- (***p = 0.0004) with greater expression in relation to U2OS R+. Graph G – GADD45A up-regulated in U2OS S (****P < 0.0001), and in group U2OS R+ the expression was higher than in U2OS R- (*p = 0.0442). Graph H shows high expression of TMPO in U2OS S (****P < 0.0001) and there was no significant difference in expression between groups U2OS R+ and U2OS R-; Graph I – EFNB2 up-regulated in U2OS R+ and U2OS R- (****p < 0.0001) in comparison to group U2OS S, which was down-regulated. The results were validated using one-way ANOVA.
As regards gene UBE2L6, there was a high expression in the U2OS S group in relation to the other groups of resistant cells (p < 0.0001). When compared to the groups of resistant cells, it was observed that the U2OS R-group continued to show a high expression of UBE2L6 when compared to U2OS R+ (p < 0.0001). The same level of expression between the cell groups was repeated with OXCT1, as shown in Fig. 6E–F.
Analysis of GADD45A showed a high expression in the U2OS S group (p < 0.0001), although cell groups U2OS R+ and U2OS R-presented a lower expression of this gene when compared to U2OS S; group U2OS R+ presented a significant difference (p = 0.0442) in relation to U2OS R-, indicating that in resistance-induced cells that are later cultivated without the drug that is responsible for generating selective pressure, GADD45A starts to have its expression down-regulated (Fig. 6G).
In the same way, TMPO was up-regulated in U2OS S and, although it presented expression in the U2OS R+ and U2OS R-groups, there was not any significant difference between them (Fig. 6H). Finally, we had a different result from the expression levels presented up until now: the expression of EFNB2 was down-regulated in U2OS S and gained an up-regulated profile in U2OS R+ and U2OS R-cells, where its expression was approximately 150 times greater in these cell groups than in U2OS, a finding that can be seen in Fig. 6I.
4. Discussion
In the current study, the induction of resistance in cells of the U2OS lineage using IFO was demonstrated. However, MTT assays showed that in the absence of the drug, resistant cells regained sensitivity to the treatment. These results suggest that the cultivation of resistant cells without the drug can generate a re-sensitization of the cells, classifying the previously observed resistance as temporal or reversible [15]. There are not many data in the literature to explain this change in the behavior of resistant cells in the absence and presence of the drug in osteosarcoma studies, so this work may provide new data that lead to gaining a better understanding of the complexity of tumor resistance in this type of cancer.
By evaluating the IC50 of the U2OS lineage at the beginning of the tests and the IC50 obtained later in the new MTT assay, it was possible to observe an increase in the dose of IFO that maintains 50% of the cells with metabolic activity. This result suggests that U2OS S cells show tumor heterogeneity and intrinsic resistance to IFO, which is probably due to the fact that groups of tumor cells are composed of multiple clones of tumor cells that compete with each other for selective advantage [16]. Drug-sensitive tumor cell populations may have a small subpopulation of treatment-resistant cells, evidencing once again the heterogeneity within a cancer cell population [15]. Another hypothesis for the increase and variation in resistance of the U2OS S lineage suggests that it would be due to the phenomenon of phenotypic plasticity, which can occur spontaneously in tumor cells [17]. According to He et al. (2011), the J82 lineage of drug-sensitive bladder cancer was able to develop a highly tumorigenic phenotype without the presence of selective pressures, that is, without drug treatment. This phenomenon is called phenotypic plasticity and the same was observed in vivo, corroborating the data described in this work.
Even with this variation in drug resistance observed in the U2OS S lineage, migration, proliferation, and survival-free assays demonstrate that the U2OS R+ lineage has a more aggressive and resistant tumor phenotype. This result was expected, since OS cells with induced resistance have a greater capacity for migration and invasion compared to parental cells [18,19]. In this study, it was observed that both sensitive and resistant groups formed cell aggregates in the adherence-free survival assay. OS cells with metastatic potential in vivo form cell aggregates that become indispensable for their transport via the bloodstream or the lymphatic system, until they establish themselves in a new anatomical site [20].
The expression profile was similar in U2OS R+ and U2OS R-cells, with no significant difference between them, but the expression levels in the U2OS S group were significantly different in relation to the other groups.
Ephrin B2 (EFNB2) is variably expressed in tumor cells, and its blockage has been seen to inhibit angiogenesis in solid tumors. Over-expression of ephrin B2 has also been correlated with a worse response to chemotherapy and radiotherapy. As it is an important mediator of the invasion, migration and angiogenesis of tumor cells, which are characteristic of metastatic cells, its use as a critical biomarker in prognosis can be observed [21].
Ephrins exert their biological functions by bonding with Eph receptors, which include the biggest family of tyrosine kinase receptors. As well as signaling via receptor, the ephrins can act independently from the Eph receptor by means of reverse signaling, when the cytoplasmic domain of ephrin is phosphorylated, resulting in the activation of cell signaling. Both reverse signaling and signaling that depends on the receptor have been implicated in the development and progression of cancer [22].
Little is known about the expression of this gene in OS. The results obtained have demonstrated a greater capacity for migration and proliferation in U2OS R+, indicating more aggressive tumor behavior. The high level of expression of the R+ and R-lineages in comparison with the S lineage corroborates the data found in the literature.
Bruheim et al. (2009) demonstrated that OS xenografts with an unsatisfactory response to IFO treatment showed a low expression of GADD45A, relating this fact to tumor resistance to IFO. These data corroborate the current study, where only sensitive cells showed positive expression of GADD45A. This gene is responsible for regulating the cell cycle and apoptotic processes in response to physiological and environmental stress, which in OS and other tumors usually show methylation in the CpG region due to epigenetic mechanisms [23,24]. Tumors that lack GADD45A expression have greater angiogenesis and cell migration activity [25].
Corroborating this information, the present study showed that U2OS R+ cells, in addition to not showing gene expression of GADD45A, also demonstrated resistance to IFO and a high capacity for cell migration and proliferation when compared to U2OS S cells. However, the U2OS S group showed upregulation of GADD45A and a less aggressive tumor profile.
Genes such as ALPL and EPB41L3 were downregulated in IFO-resistant OS xenografts [26], and this profile was also repeated in the current study, where U2OS R+ cells showed resistance to IFO and low expression of these genes, while U2OS S cells sensitive to IFO had high levels of expression. Levels of ALPL in blood plasma are often associated with primary bone lesions and, because of this, it has been shown to be a potential biomarker for the follow-up of OS [27]. Elevated levels of ALPL were related to low disease survival [27]; however, in the current study, low ALPL expression was reported in the most sensitive lineage, while the resistant lineage showed high levels of this gene. This reinforces the idea that further studies are needed in these IFO-sensitive and resistant lineages, involving the related pathways, evaluating the genes and how they interact with each other.
The gene EPB41L3 may have a dual role in OS, because despite being found in high levels in OS cells, as represented in the U2OS S group, when this gene is downregulated it leads to the process of mesenchymal epithelium, generating metastasis and a more aggressive tumor [28], corroborating the tests performed on U2OS R+ cells.
In cases of esophageal squamous cell carcinoma (ESCC), EPB41L3 undergoes the methylation process and its expression is reduced, and thus the progression of the cell cycle occurs, demonstrating that this gene plays an important role in tumor suppression [29].
In the U2OS S and U2OS R+ lineages, EPB41L3 can be evaluated later as a target for the follow-up of the disease and for characterization of a more aggressive and drug-resistant tumor, since this gene showed differentiated expression between the sensitive and resistant lineages of OS.
Classes of genes responsible for ubiquitination, such as UBE2L6 and UBE2A [30], in addition to the IER3 gene, which is regulated in response to cellular stress and its influence on tumor progression [31,32], and the OXCT1 gene that participates in the metabolic process [33], are not well described in OS cells. All these genes were down-regulated in U2OS R+ and U2OS R-cells and up-regulated in U2OS S, but it was reported that the UBE2A gene is up-regulated in OS lineages and, when silenced, reduces the process of cell migration, invasion, and proliferation [34]. In the same way, the IER3 gene is usually up-regulated in more aggressive cancers, such as in some lineages of lung cancer [32], and in pancreatic cancer [31].
In OS xenografts, UBE2A and IER3 were up-regulated in IFO-resistant samples, which could explain why the U2OS S group has intrinsic resistance to IFO, as indicated in the metabolic activity tests, while the U2OS R+ group, despite having the UBE2A gene down-regulated, showed resistance to IFO, as was also observed for UBE2L6 and IER3. In the case of cervical cancer, the low expression of IER3 is related to the development of the tumor [35], but there are still not many data about this gene in OS. Based on its expression profile in different tumors, this gene becomes a strong candidate for further studies to better elucidate the role of IER3 in induced resistance to IFO in OS.
Finally, when analyzing the OXCT1 gene responsible for the synthesis and degradation of ketone bodies, participating in tumor progression and intercellular signaling [33], the U2OS S lineage showed high expression of this gene, while after resistance induction this gene was suppressed. Deletions in this gene that generate changes in the synthesis and degradation pathways of ketone bodies are characteristic of OS [36].
In breast cancer cells, the high expression of OXCT1 induces cell growth and metastasis, characterizing this gene as a metabolic oncogene [37]. In our study, the sensitive lineage, despite presenting positive regulation of this gene, presented less aggressive tumor behavior, compared to the lineage that was induced to resistance and showed low levels of expression of OXCT1, indicating that the process of metabolizing ketone bodies becomes inactive, but other metabolizing processes that lead to resistance may be involved [38].
A key point demonstrated in our work is that after the induction of resistance to IFO, when these cells are cultured with and without the drug, the expression profile of the analyzed genes remains similar, with no significant difference. However, when cultured in the absence of the drug after the resistance induction protocol, represented by the group of U2OS R-cells, these cells regain sensitivity to IFO. There are probably other pathways that influence this behavior, and, in addition, the current study evaluated the mRNA level, and it is clear that tests at the protein level would be necessary to assess whether in fact the translation into proteins corresponds to the expression levels of the analyzed genes.
5. Conclusion
When we induced resistance to IFO in the U2OS lineage, the cells presented more aggressive tumor behavior, in addition to deregulation in the expression levels of genes associated with resistance.
Among the results reported here, the behavior of resistant cells in the absence of the drug drew attention, because when subjected to the IFO sensitivity assay, these cells showed a significant reduction in resistance. However, the expression levels of U2OS R-remained similar to those of U2OS R+ cells. This result brings new data about the expression of the genes EPB41L3, IER3, OXCT1, and EFNB2 in the U2OS lineage, showing how they may play an important role in the development of resistance. Not many studies have evaluated these genes with this specific lineage. Therefore, to better evaluate how they affect the resistance and aggressiveness of the U2OS lineage, functional studies are needed with these genes as targets.
Author contribution to study
Maria Tereza de Oliveira Rodrigues.: Investigation, Writing - Original Draft, formal analysis, Visualization, Conceptualization and Methodology. Lucas Pereira da Silva: Investigation, Formal analysis, Writing - Original Draft, Visualization, Conceptualization and Methodology. Robert Edward Pogue: Conceptualization, Methodology. Juliana Lott de Carvalho: Conceptualization, Methodology. Andrea Barreto Motoyama: Conceptualization, Methodology, Resources - provided the lineage used. Thuany de Alencar e Silva: Formal analysis, Methodology. Hilana dos Santos Sena Brunel: Resources, support in cell culture. Maria Fátima Grossi de Sá: Resources, Writing - Review & Editing. Rosângela Vieira de Andrade: Project administration, Supervision, Resources, Methodology, Writing - Review & Editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by grants from the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brasil (CAPES) (88887.613109/2021-00), Universidade Católica de Brasília (88887.200777/2018-00 – UCB), Conselho nacional de Desenvolvimento científico e tecnológico (CNPq), and Fundação de Apoio a Pesquisa (FAP-DF - 193.001.533/2016). We would like to thank the company Bio inova for the partnership and availability of the laboratory for the tests carried out.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2022.101357.
Contributor Information
Maria Tereza de Oliveira Rodrigues, Email: Mariaatereza96@gmail.com.
Lucas Pereira da Silva, Email: biomedicolucasilva@gmail.com.
Robert Edward Pogue, Email: repogue@gmail.com.
Juliana Lott de Carvalho, Email: juliana.lott@unb.br.
Andrea Barretto Motoyama, Email: andreabm@unb.br.
Thuany de Alencar e Silva, Email: thuanyalencar@gmail.com.
Hilana dos Santos Sena Brunel, Email: lanasena@gmail.com.
Maria Fátima Grossi de Sá, Email: fatima.grossi@embrapa.br.
Rosângela Vieira de Andrade, Email: rosangelav@pos.ucb.br.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
I have shared the link to my data at the attach file
References
- 1.Van Driel M., Van Leeuwen J.P.T.M. Cancer and bone: a complex complex. Arch. Biochem. Biophys. 2014;561:159–166. doi: 10.1016/j.abb.2014.07.013. [DOI] [PubMed] [Google Scholar]
- 2.Meazza C., Scanagatta P. Metastatic osteosarcoma: a challenging multidisciplinary treatment. Expert Rev. Anticancer Ther. 2016;16:543–556. doi: 10.1586/14737140.2016.1168697. [DOI] [PubMed] [Google Scholar]
- 3.Moreno F., Cacciavillano W., Cipolla M., Coirini M., Streitenberger P., López Martí J., Palladino M., Morici M., Onoratelli M., Drago G., Schifino A., Cores M., Rose A., Jotomliansky J., Varel M., García Lombardi M. Vol. 64. 2017. (Childhood Osteosarcoma: Incidence and Survival in Argentina. Report from the National Pediatric Cancer Registry, ROHA Network 2000–2013, Pediatr. Blood Cancer). [DOI] [PubMed] [Google Scholar]
- 4.Varshney J., Scott M., Largaespada D., Subramanian S. Understanding the osteosarcoma pathobiology: a comparative oncology approach. Vet. Sci. 2016;3:3. doi: 10.3390/vetsci3010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sarbay H., Demir Ü.F., Yılmaz G., Atay A.A., Malbora B. Ifosfamide induced encephalopathy in a child with osteosarcoma. J. Oncol. Pharm. Pract. 2020 doi: 10.1177/1078155220963545. [DOI] [PubMed] [Google Scholar]
- 6.Ilyas S., Tabasum R., Iftikhar A., Nazir M., Hussain A., Hussain A., Ali M.S., Saleem F., Saleem U., Froeyen M., Abdullah I., Mirza M.U., Ahmad S. Effect of Berberis vulgaris L. root extract on ifosfamide-induced in vivo toxicity and in vitro cytotoxicity. Sci. Rep. 2021;11:1–14. doi: 10.1038/s41598-020-80579-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He H., Ni J., Huang J. Molecular mechanisms of chemoresistance in osteosarcoma. Oncol. Lett. 2014;7:1352–1362. doi: 10.3892/ol.2014.1935. (review) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lilienthal I., Herold N. Targeting molecular mechanisms underlying treatment efficacy and resistance in osteosarcoma: a review of current and future strategies. Int. J. Mol. Sci. 2020;21:1–56. doi: 10.3390/ijms21186885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mobarak H., Fathi E., Farahzadi R., Zarghami N., Javanmardi S. L-carnitine significantly decreased aging of rat adipose tissue-derived mesenchymal stem cells. Vet. Res. Commun. 2017;41:41–47. doi: 10.1007/s11259-016-9670-9. [DOI] [PubMed] [Google Scholar]
- 10.Wang J., Li G. Mechanisms of methotrexate resistance in osteosarcoma cell lines and strategies for overcoming this resistance. Oncol. Lett. 2015;9:940–944. doi: 10.3892/ol.2014.2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pijuan J., Barceló C., Moreno D.F., Maiques O., Sisó P., Marti R.M., Macià A., Panosa A. In vitro cell migration, invasion, and adhesion assays: from cell imaging to data analysis. Front. Cell Dev. Biol. 2019;7:1–16. doi: 10.3389/fcell.2019.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao W., Wu D., Wang Y., Wang Z., Zou C., Dai Y., Ng C., Teoh J.Y., Chan F.L. Development of a novel and economical agar-based non-adherent three-dimensional culture method for enrichment of cancer stem-like cells. Stem Cell Res. Ther. 2018:1–13. doi: 10.1186/s13287-018-0987-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martins T.M. da M., de Paula A.C.C., Gomes D.A., Goes A.M. Alkaline phosphatase expression/activity and multilineage differentiation potential are the differences between fibroblasts and orbital fat-derived stem cells – a study in animal serum-free culture conditions. Stem Cell Rev. Reports. 2014;10:697–711. doi: 10.1007/s12015-014-9529-9. [DOI] [PubMed] [Google Scholar]
- 14.Nolan T., Hands R.E., Bustin S.A. Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 2006;1:1559–1582. doi: 10.1038/nprot.2006.236. [DOI] [PubMed] [Google Scholar]
- 15.Sharma1 * Sreenath V., Lee1 Diana Y., Li1 Bihua, Quinlan1 Margaret P., Takahashi1 Fumiyuki, Maheswaran1 Shyamala, McDermott1 Ultan, Azizian1 Nancy, Lee Zou1, Fischbach1 Michael A., Wong2 Kwok-Kin, Brandstetter2 Kathleyn, Ben Wittner1, Ramaswamy1 Sridhar, Ma A chromatin-mediated reversible drug tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027.A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schiavone K., Garnier D., Heymann M.F., Heymann D. The heterogeneity of osteosarcoma: the role played by cancer stem cells. Adv. Exp. Med. Biol. 2019;1139:187–200. doi: 10.1007/978-3-030-14366-4_11. [DOI] [PubMed] [Google Scholar]
- 17.He K., Xu T., Goldkorn A. Cancer cells cyclically lose and regain a drug-resistant highly-tumorigenic phenotype in culture and in tumor xenografts. Mol. Cancer Therapeut. 2011;10:938–948. doi: 10.1161/CIRCULATIONAHA.110.956839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng A., Xu J., He P., Chen Y., Fang R., Shao X. Decrease in stathmin expression by arsenic trioxide inhibits the proliferation and invasion of osteosarcoma cells via the MAPK signal pathway. Oncol. Lett. 2017;14:1333–1340. doi: 10.3892/ol.2017.6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qiu E., Gao Y., Zhang B., Xia T., Zhang Z., Shang G. Upregulation of cell division cycle 20 in cisplatin resistance-induced epithelial-mesenchymal transition in osteosarcoma cells. Am. J. Transl. Res. 2020;12:1309–1318. [PMC free article] [PubMed] [Google Scholar]
- 20.Lewis V.O., Devarajan E., Cardó-Vila M., Thomas D.G., Kleinerman E.S., Marchiò S., Sidman R.L., Pasqualini R., Arap W. BMTP-11 is active in preclinical models of human osteosarcoma and a candidate targeted drug for clinical translation. Proc. Natl. Acad. Sci. U.S.A. 2017;114:8065–8070. doi: 10.1073/pnas.1704173114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oweida A., Bhatia S., Hirsch K., Calame D., Griego A., Keysar S., Pitts T., Sharma J., Eckhardt G., Jimeno A., Wang X.J., Parkash G., Califano J., Karam S.D. Ephrin-B2 overexpression predicts for poor prognosis and response to therapy in solid tumors. Mol. Carcinog. 2017;56:1189–1196. doi: 10.1002/mc.22574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kullander K., Klein R. Mechanisms and functions of Eph and ephrin signalling. Nat. Rev. Mol. Cell Biol. 2002;3:475–486. doi: 10.1038/nrm856. [DOI] [PubMed] [Google Scholar]
- 23.Al-Romaih K., Somers G.R., Bayani J., Hughes S., Prasad M., Cutz J.C., Xue H., Zielenska M., Wang Y., Squire J.A. Modulation by decitabine of gene expression and growth of osteosarcoma U2OS cells in vitro and in xenografts: identification of apoptotic genes as targets for demethylation. Cancer Cell Int. 2007;7:1–16. doi: 10.1186/1475-2867-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang F., wang Liu W., Chen H., Zhu J., hua Huang A., Zhou F., Gan Y., hua Zhang Y., Ma L. Carfilzomib inhibits the growth of lung adenocarcinoma via upregulation of Gadd45a expression. J. Zhejiang Univ. - Sci. B. 2020;21:64–76. doi: 10.1631/jzus.B1900551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang F., Zhang W., Li D., Zhan Q. Gadd45a suppresses tumor angiogenesis via inhibition of the mTOR/STAT3 protein pathway. J. Biol. Chem. 2013;288:6552–6560. doi: 10.1074/jbc.M112.418335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bruheim S., Xi Y., Ju J., Fodstad O. Gene expression profiles classify human osteosarcoma xenografts according to sensitivity to doxorubicin, cisplatin, and ifosfamide. Clin. Cancer Res. 2009;15:7161–7169. doi: 10.1158/1078-0432.CCR-08-2816. [DOI] [PubMed] [Google Scholar]
- 27.Kim S.H., Shin K.H., Moon S.H., Jang J., Kim H.S., Suh J.S., Yang W.I. Reassessment of alkaline phosphatase as serum tumor marker with high specificity in osteosarcoma. Cancer Med. 2017;6:1311–1322. doi: 10.1002/cam4.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yuan X., Piao L., Wang L., Han X., Tong L., Shao S., Xu X., Zhuang M., Liu Z. Vol. 12. 2020. pp. 1–15. (Erythrocyte Membrane Protein Band 4 . 1-like 3 Inhibits Osteosarcoma Cell Invasion through Regulation of Snai1-Induced Epithelial-To- Mesenchymal Transition). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeng R., Liu Y., Jiang Z.J., Huang J.P., Wang Y., Li X.F., Bin Xiong W., Wu X.C., Zhang J.R., Wang Q.E., Zheng Y.F. EPB41L3 is a potential tumor suppressor gene and prognostic indicator in esophageal squamous cell carcinoma. Int. J. Oncol. 2018;52:1443–1454. doi: 10.3892/ijo.2018.4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hosseini S.M., Okoye I., Chaleshtari M.G., Hazhirkarzar B., Mohamadnejad J., Azizi G., Hojjat-Farsangi M., Mohammadi H., Shotorbani S.S., Jadidi-Niaragh F. E2 ubiquitin-conjugating enzymes in cancer: implications for immunotherapeutic interventions. Clin. Chim. Acta. 2019;498:126–134. doi: 10.1016/j.cca.2019.08.020. [DOI] [PubMed] [Google Scholar]
- 31.Molejon M.I., Iovanna J.L. IER3 in pancreatic carcinogenesis. Oncotarget. 2015;6:15712–15713. doi: 10.18632/oncotarget.4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Z., Yu H.S., Yao R.Y., Qiu W.S., Yue L., Sui A.H., Liu X.P., Liu S.H. Construction and expression of an eukaryotic expression vector containing the IER3 gene. Asian Pac. J. Cancer Prev. APJCP. 2013;14:507–510. doi: 10.7314/APJCP.2013.14.1.507. [DOI] [PubMed] [Google Scholar]
- 33.Zhang S., Xie C. The role of OXCT1 in the pathogenesis of cancer as a rate-limiting enzyme of ketone body metabolism. Life Sci. 2017;183:110–115. doi: 10.1016/j.lfs.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y., Leng H., Chen H., Wang L., Jiang N., Huo X., Yu B. Knockdown of UBE2T inhibits osteosarcoma cell proliferation, migration, and invasion by suppressing the PI3K/Akt signaling pathway. Oncol. Res. 2016;24:361–369. doi: 10.3727/096504016X14685034103310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jin H., Suh D.S., Kim T.H., Yeom J.H., Lee K., Bae J. IER3 is a crucial mediator of TAp73β-induced apoptosis in cervical cancer and confers etoposide sensitivity. Sci. Rep. 2015;5:1–11. doi: 10.1038/srep08367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Du X., Yang J., Yang D., Tian W., Zhu Z. The genetic basis for inactivation of Wnt pathway in human osteosarcoma. BMC Cancer. 2014;14:1–10. doi: 10.1186/1471-2407-14-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martinez-Outschoorn U.E., Lin Z., Whitaker-Menezes D., Howell A., Sotgia F., Lisanti M.P. Ketone body utilization drives tumor growth and metastasis. Cell Cycle. 2012;11:3964–3971. doi: 10.4161/cc.22137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wenjing Du M.D., Jiang Peng, Mancuso Anthony, Stonestrom Aaron, Brewer X.Y., Minn Andy J., Mak Tak W., Wu Mian. TAp73 enhances the pentose phosphate pathway and supports cell proliferation. Nat. Cell Biol. 2013;15:991–1000. doi: 10.1038/ncb2789.TAp73. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
I have shared the link to my data at the attach file