Abstract
In Streptococcus pneumoniae, a fermentative aerotolerant and catalase-deficient human pathogen, oxidases with molecular oxygen as substrate are important for virulence and for competence. The signal-transducing two-component systems CiaRH and ComDE mediate the response to oxygen, culminating in competence. In this work we show that the two-component MicAB system, whose MicB kinase carries a PAS domain, is also involved in competence repression under oxygen limitation. Autophosphorylation of recombinant MicB and phosphotransfer to recombinant MicA have been demonstrated. Mutational analysis and in vitro assays showed that the C-terminal part of the protein and residue L100 in the N-terminal cap of its PAS domain are both crucial for autokinase activity in vitro. Although no insertion mutation in micA was obtained, expression of the mutated allele micA59DA did not change bacterial growth and overcame competence repression under microaerobiosis. This was related to a strong instability of MicA59DA-PO4 in vitro. Thus, mutations which either reduced the stability of MicA-PO4 or abolished kinase activity in MicB were related to competence derepression under microaerobiosis, suggesting that MicA-PO4 is involved in competence repression when oxygen becomes limiting. The micAB genes are flanked by mutY and orfC. MutY is an adenine glycosylase involved in the repair of oxidized pyrimidines. OrfC shows the features of a metal binding protein. We did not obtain insertion mutation in orfC, suggesting its requirement for growth. It is proposed that MicAB, with its PAS motif, may belong to a set of functions important in the protection of the cell against oxidative stress, including the control of competence.
In the catalase-negative pathogen Streptococcus pneumoniae, which has essentially fermentative metabolism, oxygen limitation in a microaerobic atmosphere abolishes developmental competence. Nox, an NADH oxidase that produces water by reducing O2 as it recycles NADH, has been shown to contribute to competence regulation by oxygen (3, 8). Studies of oxygen-independent mutant strains demonstrated the involvement of the two-component systems (TCSs) CiaRH and ComDE in this regulation (7, 8). To characterize in more detail the regulatory network facilitating bacterial adaptation to oxygen availability, we searched for amino acid sequences corresponding to motifs putatively involved in O2 and redox sensing, in the publicly available pneumococcal genome sequence (http://www.tigr.org).
The PAS domain may perceive cell energetic status by sensing oxygen, redox potential, ligands, proton motive force, and light (22; for review, see reference 25). PAS domains have been found in bacterial, archaeal, and eukaryotic proteins. Redox sensing and the corresponding signal transduction via two-component systems carrying PAS domains is one strategy used by bacteria and archaea for adaptation to variations in ambient oxygen concentration (4). PAS domains are frequently found upstream from the kinase transmitter domain. A heme-containing domain in a sensor histidine kinase from Sinorhizobium meliloti directly detects oxygen (11); however, in most cases little is known about the primary signals triggering the kinase activity. The presence of a PAS motif in the essential two-component HK02-RR02 system was recently described for S. pneumoniae (15), but its role in competence was not elucidated. The HK02-RR02 TCS has been detected in virulent strains of S. pneumoniae, in which it was designated 492HK-RR. Insertional mutagenesis of the kinase did not impair growth in mouse lungs (26). By genome analysis we found this protein to be the only one containing a PAS domain in S. pneumoniae and have established its involvement in competence regulation by oxygen. The kinase has therefore been designated MicB for its role under microaerobiosis, and its cognate response regulator was named MicA. Mutational and biochemical studies allowed us to demonstrate that MicB autophosphorylation requires the L100 residue of PAS and that the D59 residue of MicA is involved in the stability of the phosphorylated form of the response regulator. Genetic dissection and biochemical analysis suggest that MicA-PO4 functions upstream of ComDE to repress competence when oxygen is limiting.
MATERIALS AND METHODS
Bacterial strains, growth conditions, antibiotic susceptibility, and S. pneumoniae transformation.
The bacterial strains used in this study are listed in Table 1. Escherichia coli was grown and induced by 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) under standard conditions (23). S. pneumoniae was grown, transformed, and tested for competence development under aerobic or microaerobic conditions as described previously (7). Antibiotic susceptibility to erythromycin and lincomycin was determined on plates containing various concentrations of these antibiotics. Plates were incubated for 12 h at 37°C under aerobic and microaerobic conditions. For strain screening, antibiotics were used at the following concentrations; for E. coli, 100 μg of ampicillin ml−1 and 50 μg of spectinomycin ml−1; for S. pneumoniae, 2 μg of erythromycin ml−1, 2 μg of rifampin ml−1, 40 μg of kanamycin ml−1, and 10 μg of spectinomycin ml−1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Phenotypea | Source or reference |
|---|---|---|---|
| Strain | |||
| S. pneumoniae | |||
| Cp1015 | Rx derivate; str1 hexA | Smr | 19 |
| Cp6602 | comE38KE micB::km | Kanr | This work |
| Cp7002 | micB::km | Kanr | This work |
| Cp7003 | micB100LR | This work | |
| Cp7009 | micA59DA | This work | |
| Cp7039 | micA59DA comE::km | Kanr | This work |
| Cp7089 | micA59DA comA::ery | Eryr | This work |
| E. coli | |||
| DH5α | supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Bethesda Research Laboratories | |
| TOPI0 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL endA nupG | Invitrogen | |
| Plasmid | |||
| pAM239 | Derived from pBR322; ColE1 origin; lacZα selection | Sptr | 10 |
| pFS16 | 0.68-kb HindIII amplicon from Cp1015 containing the 16S gene inserted into pAM239 | Sptr | 7 |
| pPFMD52 | 1.92-kb KpnI/HindIII amplicon (primers FMD52 and Rmic) from pPT12 inserted into pWSK29 | Ampr | This work |
| pPFMD59 | 1.90-kb KpnI/HindIII amplicon (primers FMD59 and Rmic) from pPT12 inserted into pWSK29 | Ampr | This work |
| pPFMD105 | 1.0-kb BamHI/HindIII amplicon (primers FMD105 and Rmic) from pPT12 inserted into pWSK29 | Ampr | This work |
| pPFMG128 | 1.0-kb BamHI/HindIII amplicon (primers FMG128 and Rmic) from pPT12 inserted into pWSK29 | Ampr | This work |
| pPFML100 | 1.0-kb BamHI/HindIII amplicon (primers FML100 and Rmic) from pPT12 inserted into pWSK29 | Ampr | This work |
| pPFMN120 | 1.0-kb BamHI/HindIII amplicon (primers FMN120 and Rmic) from pPT12 inserted into pWSK29 | Ampr | This work |
| pPJ1 | pUC derivative containing an HincII fragment carrying the kanamycin resistance gene | Ampr Kanr | 21 |
| pPMAE | 0.7-kb BamHI/EcoRI amplicon (primers FMAE and RMAE) from pPT12 containing the micA gene inserted into pTrcHis2A | Ampr | This work |
| pPMAE59 | 0.7-kb BamHI/EcoRI amplicon (primers FMAE and RMAE) from Cp7009 containing the micA59DA gene inserted into pTrcHis2A | Ampr | This work |
| pPMAI | 0.4-kb EcoRI/HindIII amplicon (primers FMAI and RMAI) from pPT12 containing an internal fragment of micA inserted into pAM239 | Sptr | This work |
| pPMBE1 | 1.23-kb BamHI/EcoRI amplicon (primers FMBE and RMBE) from pPT12 containing the micB gene inserted into pTrcHis2A | Ampr | This work |
| pPMBE100 | 1.23-kb BamHI/EcoRI amplicon (primers FMBE and RMBE) from Cp7003 containing the micB100LR gene inserted into pTrcHis2A | Ampr | This work |
| pPMBEN | 0.88-kb BamHI/SalI DNA fragment from pPMBE1 containing the N terminal of micB∗ inserted into pTrcHis2A | Ampr | This work |
| pPMCI | 0.5-kb EcoRI/HindIII amplicon (primers ForfCin and RorfCin) from pPT14 containing an internal fragment of orfC inserted into pAM239 | Sptr | This work |
| pPT12 | 2.1-kb KpnI/HindIII amplicon (primers Fmic and Rmic) from Cp1015 containing the micAB genes inserted into pWSK29 | Ampr | This work |
| pPT12-Km | pPT12 micB::km | Ampr Kanr | This work |
| pPT14 | 0.98-kb XbaI/XhoI amplicon (primers ForfC and RorfC) from Cp1015 containing the orfC gene inserted into pWSK29 | Ampr | This work |
| pPT14-Km | pPT14 orfC::km | Ampr Kanr | This work |
| pPT18 | 2.9-kb XbaI-PstI fragment from Cp1015 amplified by PCR containing the comCDE operon, inserted into pWSK29 | Ampr | 7 |
| pPT18-KCl | pPT18 comE::km | Ampr Kanr | This work |
| pTrcHis2A | pUC-derived vector for the production and purification of recombinant proteins in E. coli | Ampr | Invitrogen |
| pWSK29 | Cloning vector derived from pBlueScript | Ampr | 29 |
| pXF208 | Plasmid containing an internal fragment of comA | Emr | 6 |
Amp, ampicillin; Ery, erythromycin; Kan, kanamycin; Spt, spectinomycin.
DNA manipulation, plasmid construction, and sequencing.
Standard recombinant DNA techniques were used, as described by Sambrook et al. (23). Restriction enzyme digestions were conducted according to the manufacturers' instructions, and digestion products were separated by electrophoresis in agarose gels in Tris-borate-EDTA buffer (23). All PCRs were performed in a GeneAmp PCR System 9600 (Perkin-Elmer Cetus). The cloning vectors were pWSK29 and pAM239 (Table 1). The nucleotide sequence of all the constructs was established by dye-terminator cycle sequencing with an automated 373 DNA Sequencer (Perkin-Elmer Applied Systems).
Cloning the micAB-orfC genes.
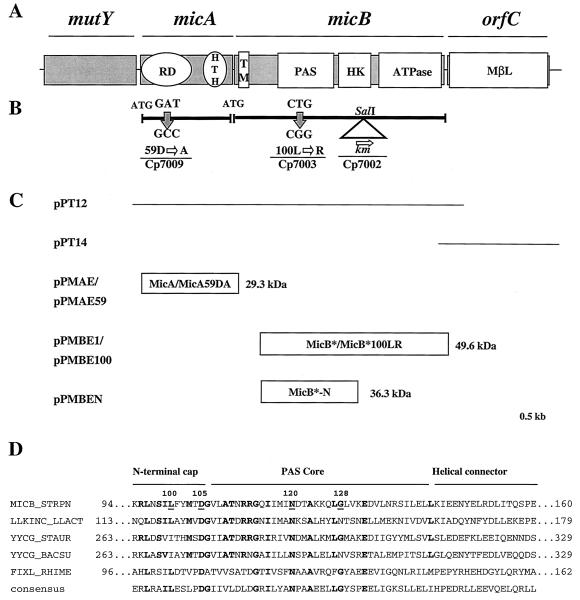
The nucleotide sequence of the open reading frame (ORF) containing the PAS domain (Fig. 1A) was obtained from the serotype 4 strain of S. pneumoniae, the genome of which has been published (see The Institute for Genomic Research website at http://www.tigr.org). An ORF was identified and micB was designated, flanked by micA and orfC (Fig1A). Primers Fmic (5′-CGATATGGTACCGAATTACCCACTTGCCAAACCC) and Rmic (5′-CACACAAGCTTCTAGTCTTCTACTTCATCCTCCCATA) were designed to amplify by PCR the micAB genes from DNA of Cp1015. The resulting amplicon was digested with KpnI and HindIII and cloned to give pPT12 (Fig. 1C and Table 1). For orfC, primers FmicC (5′-GGCGCGTCTAGACCAAGAGTGAATACGGCAAGGG) and RmicC (5′-CGCTCGCTCGAGGCTCCCTTTTTTAATGGTAACACC) were used to amplify orfC from Cp1015 DNA, and the PCR product, digested with XbaI and XhoI, was cloned to give pPT14 (Fig. 1C and Table 1). The complete nucleotide sequence of micAB-orfC from strain Cp1015 is available in GenBank (accession number AF219111).
FIG. 1.
Organization of the mutY-micAB-orfC locus with the conserved modules in MicA and MicB and strategy for producing the recombinant proteins. (A) Gene organization with significant conserved motifs in MicA, MicB, and OrfC. RD, receiver domain; HTH, helix turn helix; TM, transmembrane domain; HK, histidine kinase; MβL, metallo-β-lactamase. (B) Site-specific mutagenesis of MicA and MicB. (C) Cloning strategy for the nucleotide sequence determination of micAB and orfC and the expression of recombinant proteins in E. coli (the sizes of the recombinant proteins were calculated with the addition of 2.5 kDa for the His tag). (D) Sequence alignment of candidate PAS domains in MicB orthologues from B. subtilis (YYCG Bacsu, GenBank no. D78193), L. lactis (L1KINC Llact, GenBank no. AF178425), S. aureus (YYCG Staur, GenBank no. AF136709), FixL from S. meliloti (FIXL Rhime, GenBank no. J03174), and the PAS consensus sequence (http://smart.embl-heidelberg.de/). Identical residues are in bold. Residues chosen as mutagenesis targets are underlined.
Insertional mutagenesis.
For gene disruption two strategies were set up. Either the kanamycin-resistance cassette, aphA-3, with a candidate transcription terminator at the 5′ extreme (ΔG° = −26.5 kcal/mol) from pPJ1 (21), was inserted into relevant genes, previously cloned in pWSK29 (29), or internal amplicons of the relevant genes were cloned into pAM239 (10) to be used for plasmid insertion mutagenesis. The wild-type recipient Cp1015 was transformed by the mutagenic plasmids (10 μg/ml), and, accordingly, kanamycin or spectinomycin selection was applied to select for allelic exchange (kanamycin) or plasmid insertion (spectinomycin). Transformation efficiency of the recipient strain, Cp1015, was determined using chromosomal DNA (1 μg/ml) from the rifampin-resistant Cp1016 strain. Routinely the transformant recovery for Rifr transformants was >2% of the total population. For micB mutagenesis the km cassette was inserted into the SalI site of micB in pPT12 to generate pPT12-Km (Table 1). Transformation of strain Cp1015 with pPT12-Km yielded 1% Kmr transformants. The putative recombinants were checked by PCR with primers Fmic and Rmic, and one recombinant clone was retained and designated strain Cp7002 (micB::km). For micA mutagenesis, an internal amplicon was obtained with primers FmaI (5′-CATCACGAATTCTTATTATTCTGGATTTGATGCTTCC) and RmaI (5′-GGTCACAAGCTTGCAAGTGTTCGCGCGTGATGACTTG) and inserted between the EcoRI and HindIII sites of pAM239 to obtain pPMAI (Table 1). Strain Cp1015 was transformed with pPMAI, and spectinomycin selection was used. Mutagenesis of orfC was attempted with pPT14-Km containing orfC::km (Table 1) and pPMCI containing the amplicon obtained with primers ForfC in 5′-GGGCATGGATCCGCTGAAATTAACCGTAAGCCAG and RorfC in 5′-AGCGCCGAATTCCGATTTCCTAGCGTCCGAAT inserted between the EcoRI and HindIII sites of pAM239 (Table 1). Strain Cp1015 was transformed with pPMCI, and spectinomycin selection was applied accordingly.
Point mutagenesis.
Missense mutations were obtained by PCR amplification of the relevant genes using mutagenic primers carrying a single restriction site to facilitate the selection of recombinant clones after transformation, as described elsewhere (3). The resulting amplicons were cloned into pWSK29, and the resulting plasmids were used to transform the wild-type strain, Cp1015. Recombinant clones were screened by restriction analysis of the amplicons obtained with primers Fmic and Rmic. The mutations were confirmed by DNA sequencing using primers FMBT (5′-TTGTGACCCTCTTATTACTG) for micB and RMAT (5′-CAACTCACGATTGGAG) for micA. We obtained 2 to 4% recombinants by this procedure. Mutagenesis of residues L100 to R, D105 to A, N120 to I, and G128 to Q in MicB was achieved using the mutagenic primers FML100 (5′-TGGATCCGAGGCTAAATAGTATCCGGTTTTATATGACAG), FMD105 (5′-GCCGGATCCTGTTTTATATGACAGCTGGGGTTCTTGCGAC), FMN120 (5′-CCGGATCCAGATTATCATGATTATCGATACAGCCAAGAAG) and FMG128 (5′-CCGGGATCCAAGAAGCAACTGCAGTTGGTTAAGGAAGATG), respectively, and the complementary primer Rmic. The resulting plasmids were pPFML100, pPFMD105, pPFN120, and pPFMG128, respectively. Mutagenesis of residues D52 to Q and D59 to A in MicA was achieved with the mutagenic primers FMD52 (5′-CGATATGGTACCGATATTATTATTCTGCAGTTGATGCTTCCAGAA) and FMD59 (5′-CGATATGGTACCATGCTTCCAGAAATTGCCGGTTTAGAAGTTGCT), respectively, and the complementary Rmic oligonucleotide. This gave pPFMD52 and pPFMD59, respectively (Table 1).
Northern blot analysis.
Total RNA preparation, Northern blots, and probe labeling were carried out as described elsewhere (7). For studies of the micAB region, we used freshly prepared RNA that had not been kept frozen. Specific mRNA species were detected by hybridization with the following 32P-labeled probes: a 540-bp comE fragment from pPT18 (GenBank sequence U33315; position, 2450 to 2990), a 730-bp micB fragment from pPT12 (GenBank sequence AF219111; position, 1100 to 1830); a 490-bp orfC fragment from pPmicCin (GenBank sequence AF219111; position, 2342 to 2832); and a 650-bp 16S rRNA fragment from pP16S (GenBank sequence X58312; position, 166 to 816) from Cp1015.
Overproduction and isolation of His6-tagged MicA and MicB proteins in E. coli.
The DNA fragment encoding the cytoplasmic region of MicB, with the sequence encoding the first 38 N-terminal amino acids deleted, designated MicB*, was obtained by PCR amplification using primers FMBE (5′-GGTGAGGATCCGCGTGATAATATTCAGTTGAAGCAAGTCAAT) and RMBE (5′-GCGCGAATTCTTGTCTTCTACTTCATCCTCCCATACTTCTTC) containing EcoRI and BamHI restriction sites, respectively, facilitating insertion into the expression vector pTrcHis2A (Invitrogen) to give pPMBE1. To delete the C-terminal domain of MicB, which carries the kinase and the ATPase module, a SalI-EcoRI DNA fragment from pPMBE1 was inserted into pTrcHis2A to give pPMBEN. Primers FMAE (5′-GGCACGGATCCGATGAAAAAAATACTAATTGTAGATGATGAG) and RMAE (5′-CGCGAATTCTTAGCATTATTTCTCATGTAATACCCTACACC), containing EcoRI and BamHI restriction sites, respectively, were used to clone micA for insertion into pTrcHis2A to give pPMAE. For MicB*100LR and MicA59DA, the DNA targets for amplification were obtained from S. pneumoniae Cp7003 and Cp7009, respectively (Table 1). The corresponding amplicons were inserted into pTrcHis2A as described for MicB∗ to give pPMBE100 and pPMAE59, respectively.
E. coli TOP10 cells carrying the relevant plasmid were induced with 1 mM IPTG and lysed under denaturing conditions, according to Invitrogen's recommendations. The lysate was loaded onto a 3-ml Ni-nitrilotriacetic acid column. The column was washed with 0.5 M NaCl and 10 mM imidazole in 20 mM sodium phosphate buffer (pH 6). Refolding was achieved in a linear 7 to 0 M urea gradient in 20 mM sodium phosphate buffer (pH 6.0) containing 0.5 M NaCl. Proteins were eluted in 0.5 M imidazole and 0.5 M NaCl in 20 mM sodium phosphate buffer (pH 6.0). The samples were dialyzed against 50 mM Tris (pH 7.2), 1 mM EDTA, 100 mM KCl, 1 mM dithiothreitol (DTT) and 30% glycerol (vol/vol). Aliquots were frozen and stored at −80°C. Protein concentration was determined as described by Bradford (5). The mass of the recombinant proteins was estimated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using mid-range protein molecular mass markers (Promega) and was compared to their sizes predicted from the length of the coding fragments with the addition of 2.5 kDa from His tags.
Protein phosphorylation assays.
Proteins were phosphorylated in vitro by incubation with [γ-32P]ATP (Amersham Pharmacia Biotech) at 37°C for 10 min and analyzed by electrophoresis in 12% polyacrylamide gels after the addition of 100 μl of 2× loading buffer (pH 8.0; 0.2 M Tris, 50 mM EDTA, 0.1 M DTT, 8% SDS, 40% glycerol, 0.1% bromophenol blue). The 32P-labeled bands were detected by autoradiography, and the signals were quantified by densitometry using a Personal Densitometer SI (Molecular Dynamics) with the ImageQuant version 3.0 Fast Scan software package. As a control, samples were incubated with [α-32P]ATP in the same conditions as the assays. For MicB* autophosphorylation, 5 μg (100 pmol) of protein was incubated at 23°C in 100 μl of phosphorylation buffer (pH 7.5; 50 mM Tris buffer, 0.1 μM [γ-32P]ATP, 200 μCi/μmol, 5 mM DTT, 50 mM KCl, 0.1 mM EDTA, 10% [vol/vol] glycerol). Aliquots were removed at intervals, kept at 4°C, and analyzed as described above. For phosphotransfer from MicB*-PO4 to MicA and to MicA59DA, MicB* phosphorylation reaction was allowed for 4 min, 300 pmol (9 μg) of the wild-type and mutated MicA were added to the assay mixture, and 16-μl aliquots were removed at intervals, transferred at 4°C, and analyzed as described above. The negative control was a sample in which MicB∗ was omitted. To investigate the stability of MicA-PO4 and MicA59DA-PO4, isotopic dilution of [γ-32P]ATP in the reaction mixture was achieved by adding unlabeled ATP to a final concentration of 10 mM. Aliquots were removed at intervals and analyzed as previously described. MicA-PO4 was treated with acid or alkali by incubation with 0.1 N HCl or 0.1 N NaOH for 20 min at 23°C. Samples were analyzed as described above.
RESULTS
Cloning and nucleotide analysis of the micAB-orfC locus from the RX strain Cp1015.
We searched for proteins that might be involved in bacterial adaptation to oxygen by screening the published genome of S. pneumoniae (http://www.tigr.org), using the WU-BLAST (version 2.0) program and the PAS motif of the FixL protein of S. meliloti as a probe (12). A single ORF containing the PAS motif was found in contig 4189. According to further functional characterization, this ORF was named micB. ORFs flanking micB were designated micA and orfC. The micAB genes are located close to the antimutator, mutY (2, 16, 24) (Fig. 1A).
As a prerequisite to mutational analysis, the nucleotide sequence of micAB and orfC from the reference strain Cp1015 cloned in plasmids pPT12 and pPT14, respectively, has been established (Fig. 1C and Materials and Methods). The deduced amino acid sequences (GenBank accession number AF219111) were identical with the sequence published by Lange et al. (15; GenBank accession number AJ006392) except for the conservative V44 to I change and the R270 to C missense mutation in the kinase domain of MicB.
Localized mutagenesis of micAB-orfC and phenotypic characterization of the mutant strains.
The physiological role of micAB (492RR-492HK; HK02-RR02) and orfC has been assessed in vegetative growth, antibiotic susceptibility, and competence development in response to oxygen availability by using a mutational strategy.
The plasmids pPMAI, pPT12-Km, pPT14-Km, and pPMCI (Table 1) were used in the transformation of strain Cp1015 to obtain insertion mutations in micA, micB, and orfC, respectively. Recombinants were obtained only with pPT12-Km, resulting in strain Cp7002 carrying a micB insertion mutation (Fig. 1). For orfC mutagenesis, plasmids pPT14-Km and pPMCI (Table 1) were used to transform Cp1015 (see Materials and Methods). No recombinants were obtained in three independent experiments with each of these plasmids as donor DNA. This suggests that orfC cannot be disrupted and is probably required for in vitro growth in the Cp1015 genetic background. Indeed, Northern blotting of total RNA with an orfC-specific DNA probe allowed verification of the presence of orfC mRNA in all the mutant strains studied (data not shown). Virulent strains of S. pneumoniae deleted for orfC were selectively impaired in growth in vivo (26). Functional OrfC is likely required for S. pneumoniae growth in specific conditions, depending on the genetic background. We investigated further the function of MicA and MicB by introducing missense mutations at critical sites.
The conserved N120 and G128 residues in the PAS core and L100 and D105 in the N-terminal cap (see Fig. 1B and D) (22, 25) were chosen as targets for MicB mutagenesis for technical reasons inherent to the mutagenesis strategy (see Materials and Methods). Strain Cp1015 was transformed with the mutagenic plasmids pPFML100, pPFMG128, pPFMD105, and pPFMN120 to introduce the mutations into the resident chromosome by allelic exchange (Table 1 and Materials and Methods). Cultures were plated without selection, and for each transformation 100 colonies were checked. The presence of the mutation vas verified by restriction analysis of the relevant amplicons (see Materials and Methods). Transformants were obtained following transformation with pPFML100 only, giving Cp7003 carrying the micB100LR mutation.
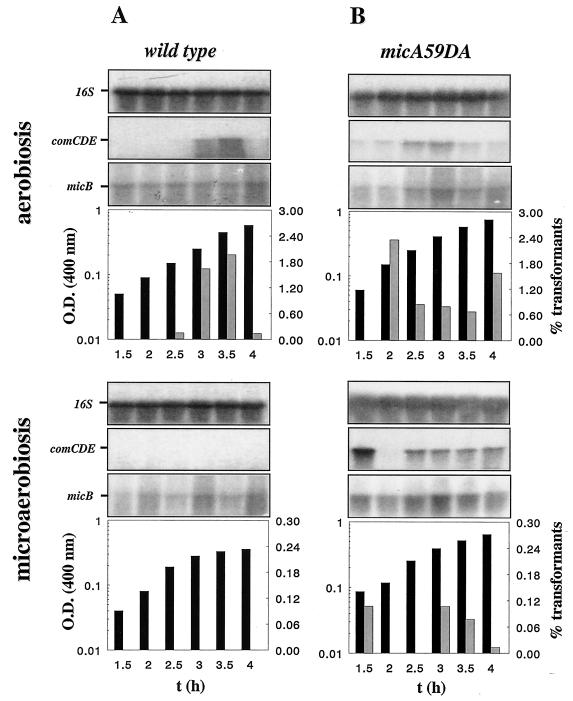
In MicA, we mutated putative phosphorylation sites (see Materials and Methods). Plasmids pPFMD52 and pPFMD59, carrying the mutated alleles micA52DQ and micA59DA, respectively (Table 1), were used to transform Cp1015. Recombinant clones were obtained only with pPFMD59. Alignment of the sequence of MicA with that of the well studied CheY of E. coli showed that the D59 residue corresponds to residue D64 of CheY. In CheY, D64 is part of a very rigid “γ-turn loop,” which has not been mutated (28). In S. pneumoniae, mutation of this loop did not diminish bacterial growth and one clone carrying the micA59DA mutation gave strain Cp7009 (Table 1 and Materials and Methods).
Mutant strains Cp7002, Cp7003, and Cp7009 had growth characteristics similar to those of the wild type. They were further analyzed for developmental competence and susceptibility to erythromycin and lincomycin. None of these mutations affected the level of resistance of the bacteria to these antibiotics (data not shown). This finding was in contrast with the observed hypersensitivity to macrolide and lincosamide antibiotics under aerobiosis in yycFG temperature-conditional mutants of Staphylococcus aureus (17).
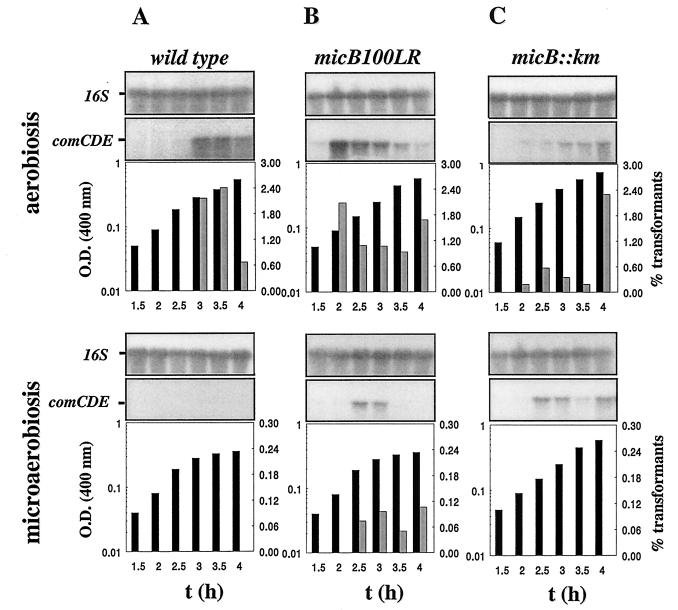
In competence tests for both strains Cp7003 (micB100LR) and Cp7009 (micA59DA), comCDE mRNA (7) was detected in cultures grown under microaerobic conditions, and transformants were obtained (Fig. 2 and 3). In contrast, the micB::km mutant strain, Cp7002, was not transformable under microaerobiosis despite the presence of significant amounts of comCDE mRNA in these bacteria (Fig. 2C). MicA59DA and MicB100LR therefore allow comCDE transcription and bacterial transformability, while the micB::km mutation dissociated comCDE transcript levels and bacterial transformability under oxygen limitation, leading to transformant recovery below the detection level (Fig. 3B). This possibly reveals a strong developmental checkpoint after comCDE transcription. In the comE38KE genetic background, in which the ComE38KE hyperactive response regulator is expressed (7), the micB::km mutation was silent because in cultures grown microaerobically Cp6600 (comE38KE) and Cp6602 (comE38KE, micB::km) gave 20 and 15% Rifr transformants, respectively, in the same experiment. Thus, comE38KE is epistatic to micB::km. This suggests posttranscriptional ComE regulation by MicB.
FIG. 2.
Effect of the micB100LR and micB::km mutations on competence control by O2. Strains Cp1015 (A), Cp7003 (B), and Cp7002 (C) were grown under aerobiosis or microaerobiosis. Aliquots were withdrawn at 30-min intervals and analyzed for change in biomass by determinations of the optical density (OD) at 400 nm (black bars), transformability (grey bars), or by Northern blotting. For Northern blotting, a DNA probe specific for comE (see Materials and Methods) was used to detect comCDE transcripts. We used 16S rRNA as a qualitative and quantitative internal control. Quantitative densitometry of 16S rRNA gave less than 25% variability between samples taken throughout the period of growth for a given culture. The experiment was repeated with independent cultures to test reproducibility. The columns are aligned with the corresponding signals in the Northern blots.
FIG. 3.
Effect of the micA59DA mutation on competence control by O2. Strains Cp7009 and Cp1015 were grown under aerobiosis or microaerobiosis. Aliquots of cultures were withdrawn at 30-min intervals and analyzed as described in the legend to Fig. 1. For Northern blotting, probes specific for comE and micB (see Materials and Methods) were used to detect comCDE and micAB RNA, respectively. The signal for 16S rRNA differed by less than 24% between wells. The experiment was repeated with independent cultures to check reproducibility.
To investigate further this point, production of competence stimulating factor (CSF) by micB::km mutants and their response to the synthetic competence stimulating peptide (CSP) has been addressed. Media conditioned by bacteria from strain Cp7002 grown microaerobically during 2 and 4 h contained a CSF activity equivalent to that of media conditioned by the wild-type strain Cp1015 grown in the same conditions, as measured by complementation of strain Cp1008 defective for CSF production. Indeed, these media were 103-fold less active than those produced by the oxygen-independent strain Cp6600 grown microaerobically. Taken as controls, media conditioned by aerobically growing bacteria from these three strains showed similar activity with regard to Cp1008 activation. This indicates that CSF concentrations in cultures of strains Cp7002 and Cp1015 growing under oxygen limitation did not reach the threshold level to trigger competence development. It has been verified that in the CSP-induced competence regime, strain Cp7002 (micB::km) showed the same activation pattern as strain Cp1015 with a threshold CSP requirement at 1 ng/ml, indicating that expression of the micB::km mutation specifically affects developmental competence but not competence triggered by addition of extra CSP to the culture. Despite a significant amount of comCDE mRNA in bacteria from strain Cp7002 grown microaerobically, its CSF production remained under the threshold level for competence activation, indicating a posttranscriptional control by full-size MicB.
We investigated further the relationship between micA and the competence autoregulation network encoded by comCDE and comAB. The response regulator ComE and the ComAB system facilitating the maturation and export of the competence-stimulating peptide are essential for competence development (for review, see reference 13). Genetic dissection showed that alleles comA::ery and comE::km, both of which abolished competence, were epistatic to micA59DA because no transformants were obtained in bacteria from microaerobic cultures of strains Cp7089 (comA::ery miA59DA) and Cp7039 (comE::km micA59DA), respectively. Thus, competence control by micA occurred upstream from the CSP-ComDE circuit. It should be noticed that mutation micA59DA did not significantly change the cellular levels of micB mRNA during competence development (Fig. 3B).
In order to obtain more precise insight into the mechanism of competence regulation by MicAB, biochemical analysis of the recombinant MicA and MicB and their corresponding mutated proteins has been undertaken.
Biochemical characterization of the two-component MicAB system and impact of the mutations which alter competence regulation on phosphorylation and phosphotransfer.
Analysis of the primary sequences of MicA and MicB strongly suggested that they form a two-component signaling system. Signal transduction systems involving phosphotransfer have been extensively described from genome analysis of S. pneumoniae. However, to date, no biochemical evidence has been obtained of autokinase activity and phosphotransfer in these bacteria.
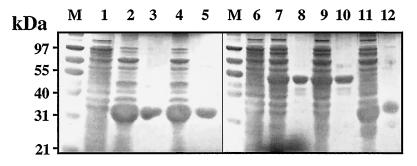
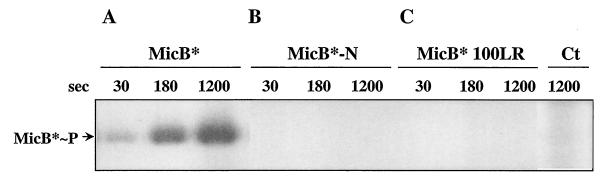
We expressed genes encoding recombinant proteins with a (His)6 tag fused to the C terminus in E. coli. The full-size MicA and MicA59DA products, as well as MicB*, with the transmembrane domains deleted (to facilitate expression in E. coli), MicB*100LR, and truncated MicB*-N were purified according to procedures described in Materials and Methods (Fig. 1C and 4). MicB* was autophosphorylated in the presence of [γ-32P]ATP (Fig. 5A). Autophosphorylation was abolished in the truncated product, MicB*-N (Fig. 5B). This demonstrates that MicB* autokinase activity is in the C-terminal part of the protein as suggested by computer-assisted prediction (Fig. 1A). MicB*100LR displayed no autokinase activity (Fig. 5C), consistent with residue L100 in the N-terminal cap of PAS being required for MicB* autokinase activity.
FIG. 4.
Purification of MicA and MicB* recombinant proteins from E. coli extracts. Cultures of strain TOP10 of E. coli producing the recombinant pneumococcal proteins (see Fig. 1) were induced by IPTG and lysed, and the recombinant proteins were purified as described in Materials and Methods. The proteins were analyzed by SDS-PAGE, and the gels were stained with Coomassie blue. Lane M, molecular mass standard; lanes 1 and 6, total protein from IPTG-induced cultures of E. coli TOP10 containing pTrcHis2A; lanes 2, 4, 7, 9, and 11, total protein from IPTG-induced bacteria containing pPMAE, pPMAE59, pPMBE1, pPMBE100, and pPMBEN, respectively; lanes 3, 5, 8, 10, and 12, purified MicA, MicA59DA, MicB*, MicB*100LR, and MicB*-N, respectively.
FIG. 5.
MicB* autokinase activity lies in the C-terminal part of the protein and is under the control of PAS. MicB*, MicB*-N, and MicB*100LR were autophosphosphorylated with [γ-32P]ATP at 23°C in 48 μl of phosphorylation buffer (see Materials and Methods) containing 12 pmol of purified protein. At intervals, 16-μl aliquots were mixed with 2× loading buffer and run on SDS-12% PAGE gels for analysis as described in Materials and Methods. (C) Control in which [α-32P]/ATP replaced [γ-32P]/ATP in the reaction mixture containing MicB*.
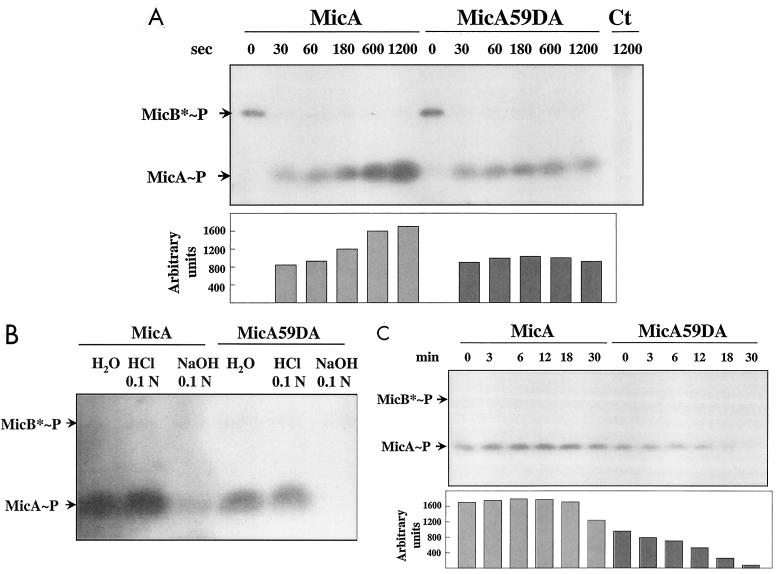
We also analyzed phosphotransfer from MicB*-PO4 to MicA. Both wild-type MicA and mutant MicA59DA were rapidly phosphorylated by MicB*-PO4 because MicB-PO4 was no longer detected after incubation for 30 s in the presence of MicA. The amount of MicA-PO4 increased with time from 30 to 1,200 s whereas the amount of MicA59DA-PO4 remained constant (Fig. 6A). Both phosphorylated proteins were labile in alkali and stable in acid (Fig. 6B), which indicates that an aspartate residue was likely phosphorylated in both (1, 11). However, MicA59DA-PO4 had a half-life of 12 min whereas MicA-PO4 had a half-life of over 20 min (Fig. 6C), indicating that the phosphorylated form of MicA59DA was unstable due to the missense mutation. It is possible that the phosphorylated residue in MicA59DA-PO4 is labile. Alternatively, the MicA59DA mutation may induce conformational changes in the protein resulting in higher suceptibility to phosphatases as described for other response regulators (14, 30).
FIG. 6.
Phosphotransfer from MicB-PO4 to MicA and MicA59DA and properties of the phosphorylated proteins. (A) The MicA and MicA59DA proteins (9 μg corresponding to 300 pmol) were added to the MicB* phosphorylation mixture at 240 s, and aliquots were analyzed at intervals. Ct, control sample containing MicA in the absence of MicB*. (B) MicA-PO4 and MicA59DA-PO4 were incubated with 0.1 N HCl or 0.1 N NaOH for 20 min at 23°C, and proteins were analyzed and compared to the control incubated with H2O. (C) MicA-PO4 and MicA59DA-PO4 were incubated at 23°C in phosphorylation buffer supplemented with 10 mM ATP. Aliquots were analyzed at intervals. Proteins were subjected to electrophoresis in 12% polyacrylamide gels. Signals on autoradiographs were quantified by densitometry (see Materials and Methods) and are expressed in arbitrary units. The bars are aligned with the corresponding signals.
DISCUSSION
In S. pneumoniae, oxygen limitation induces competence repression, thereby abolishing gene transfer by transformation. Previous work has provided evidence for an O2 signaling pathway encoded by nox, ciaRH, and comCDE (3, 7, 8). We provide here evidence that the autokinase MicB, which has a PAS motif, is involved in this regulation. Autophosphorylation of MicB and phosphotransfer to the response regulator, MicA, demonstrated that they form a signal transduction system. Mutations which abolish the kinase activity of MicB or lower the stability of MicA-PO4 both relieve competence repression under microaerobiosis. Thus, one function for the kinase MicB and the essential response regulator MicA is the perception of signals produced in response to oxygen deprivation and their transduction, culminating in competence repression.
Recombinant MicB was autophosphorylated with [γ-32P]ATP, and phosphotransfer to MicA was observed in vitro, providing the first biochemical evidence for signaling via phosphotransfer in S. pneumoniae. Mutational and biochemical analysis showed that residue L100 in the N-terminal cap of the PAS domain in recombinant MicB was important for C-terminal kinase activity in vitro. Although the N-terminal cap of PAS is the less conserved part in this motif (22, 25), residue L100 in cap is important for the kinase activity of MicB in vitro and for competence repression under microaerobiosis in vivo.
Insertion mutations in the gene encoding the response regulator, MicA, were not obtained in any genetic background (15, 26; this work), which suggests that this protein is essential in S. pneumoniae. Instability of recombinant MicA59DA-PO4 compared to MicA-PO4 was related to competence expression under oxygen limitation in mutant strains expressing the micA59DA allele but did not significantly affect growth. Taken as a whole, these data show that mutations which impact the level of phosphorylated MicA, either by abolishing the kinase activity of MicB or by lowering the stability of MicA-PO4 in vitro, allow accumulation of comCDE mRNA in bacteria grown under microaerobiosis. This indicates that the essential response regulator MicA represses competence when phosphorylated by MicB under oxygen limitation. The TCS CiaRH is a strong repressor of competence development both in aerobic- and microaerobic-grown cultures (7), and the NADH oxidase belongs to the competence signaling network (8). MicAB might exert its control through CiaRH regulation or via a complementary pathway. The signal recognized by the PAS domain controlling MicB phosphorylation remains to be characterized, as well as the MicA targets.
Oxic growth is required for induction of competence but not for transformation of competent bacteria. Indeed, when cultures were incubated under oxygen limitation, bacteria already committed to competence (competent or precompetent) remained transformable in contrast to noncompetent bacteria, which grew without expressing any competence and therefore did not transform. Occasionally, stored cultures from strain Cp1015 grown aerobically at low pH (19) contain some competent bacteria which account for residual transformation observed under oxygen limitation (3 and data not shown). Although the oxygen-dependent checkpoint for competence development is not yet characterized, the contribution of the NADH oxidase, Nox, which has O2 as substrate, has been demonstrated. Loss of function mutation in nox results in early and limited competence expression in cultures growing aerobically (3–8).
For the purpose of this study we focused our screening of mutant strains transformable under oxygen limitation and showing similar growth characteristics to the wild type. However, some mutated alleles of micA and micB were not tolerated in pneumococcus whatever the oxygen status of the culture. This identifies residues in MicA and also in MicB which are probably crucial for bacterial growth.
The micAB genes are flanked by mutY and orfC. OrfC is important for the growth of strains 23F and 0100993 in vivo (26) and Cp1015 in vitro, regardless of ambient oxygen concentration. OrfC shows the characteristic signature of a metal binding protein (15) and sequence similarity to YycJ of S. aureus, Lactococcus lactis and Bacillus subtilis (9, 17, 20). In B. subtilis, insertion mutation in yycJ is silent (9), and this issue has not been addressed for the yycJ of S. aureus (17). MutY of S. pneumoniae was recently identified by computer-aided studies and by complementation of a mutY mutant of E. coli (24). This antimutator gene in E. coli and S. pneumoniae encodes an adenine glycosylase specific for A/G miss-match pairs. The major in vivo substrate for this enzyme is probably the adenine from A/7,8-dihydro-8-oxoguanine, a product of oxidative damage to DNA (2, 16, 18). The MutY of E. coli has the highly conserved stretch of four cysteines, Cys-X6-Cys-X2-Cys-X5-Cys, that coordinates the (4Fe-4S)2+ cluster loop (FCL). These (4Fe-4S)2+ clusters have been extensively described for proteins that sense the redox status of the cells (27). MutY of S. pneumoniae has no FCL domain (24) and lies near micAB, a signaling system involved in the cellular response to oxygen.
In conclusion, this work provides a functional characterization of the essential signal-transducing two-component MicAB system. Phosphotransfer through MicAB represses competence under oxygen limitation. Although competence development during growth is not essential, competence regulation by MicAB might indicate nodes that are shared between the regulatory networks controlling essential biological processes and competence development. MicAB orthologues have been found in the transformable B. subtilis (9) and in bacteria in which transformation has never been described, such as L. lactis (20), and the pathogen S. aureus (17). When investigated by mutational analysis, the response regulator was shown to be essential for bacteria growth and involved in cell division and membrane integrity. Corresponding mutant strains exhibited a more pronounced phenotype in aerobic cultures (9, 17). It is proposed as a hypothesis that MicAB and its orthologues are involved in the bacterial adaptation to oxygen, whatever the mechanism specifically used by the different species. In S. pneumoniae, competence expression requires high oxygen levels (7) and the NADH oxidase which has O2 as a second substrate (3, 8). MicAB with its PAS signature contributes to competence regulation. Utilization of soluble DNA may have the advantage of making it possible to correct oxidative damage to chromosomal DNA, in addition to other repair enzymes such as MutY. Work is in progress to determine the role of MicAB in the relationship between MutY and competence regulation by oxygen in this pathogen.
ACKNOWLEDGMENTS
This work was supported by Université Paul Sabatier Toulouse and Rhône-Poulenc Rorer, France. J.R.E. was supported by an RPR postdoctoral fellowship.
We thank Franck Pasta for discussion. We are grateful to Delphine Dos Santos, Saliha Mimar, and Suzanne Eychenne for technical assistance. We thank the Technical Department of Institut Louis Bugnard/INSERM, Toulouse, for providing access to certain pieces of apparatus. We thank the Institute for Genomic Research website at http: //www.tigr.org for the S. pneumoniae genome sequence.
REFERENCES
- 1.Appleby J L, Bourret R B. Activation of CheY mutant D57N by phosphorylation at an alternative site, Ser-56. Mol Microbiol. 1999;34:915–925. doi: 10.1046/j.1365-2958.1999.01653.x. [DOI] [PubMed] [Google Scholar]
- 2.Au K G, Cabrera M, Miller J H, Modrich P. Escherichia coli mutY gene product is required for specific A-G/C-G mismatch correction. Proc Natl Acad Sci USA. 1988;85:9163–9166. doi: 10.1073/pnas.85.23.9163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Auzat I, Chapuy-Regaud S, Le Bras G, Dos Santos D, Ogunniyi A D, Le Thomas I, Garel J-R, Paton J C, Trombe M-C. The NADH oxidase of Streptococcus pneumoniae: its involvement in competence and virulence. Mol Microbiol. 1999;34:1018–1028. doi: 10.1046/j.1365-2958.1999.01663.x. [DOI] [PubMed] [Google Scholar]
- 4.Bauer C E, Elsen S, Bird T H. Mechanisms for redox control of gene expression. Annu Rev Microbiol. 1999;53:495–523. doi: 10.1146/annurev.micro.53.1.495. [DOI] [PubMed] [Google Scholar]
- 5.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 6.Chandler M S, Morrison D A. Competence for genetic transformation in Streptococcus pneumoniae: molecular cloning of com, a competence control locus. J Bacteriol. 1987;169:2005–2011. doi: 10.1128/jb.169.5.2005-2011.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Echenique J R, Chapuy-Regaud S, Trombe M-C. Competence regulation by oxygen in Streptococcus pneumoniae: involvement of ciaRH and comCDE. Mol Microbiol. 2000;36:688–696. doi: 10.1046/j.1365-2958.2000.01891.x. [DOI] [PubMed] [Google Scholar]
- 8.Echenique J R, Trombe M-C. Competence modulation by the NADH oxidase of Streptococcus pneumoniae involves signal transduction. J Bacteriol. 2001;183:768–772. doi: 10.1128/JB.183.2.768-772.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fabret C, Hoch J A. A two-component signal transduction system essential for growth of Bacillus subtilis: implications for anti-infective therapy. J Bacteriol. 1998;180:6375–6383. doi: 10.1128/jb.180.23.6375-6383.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gil D. Elaboration et caracterisation d'un nouveau type de vecteur de clonage a nombre de copies regulable. Thèse de l'Université. Toulouse, France: Université Paul Sabatier; 1990. [Google Scholar]
- 11.Gilles-Gonzalez M A, Ditta G S, Helinski D R. A haemoprotein with kinase activity encoded by the oxygen sensor of Rhizobium meliloti. Nature. 1991;350:170–172. doi: 10.1038/350170a0. [DOI] [PubMed] [Google Scholar]
- 12.Gong W, Hao B, Mansy S S, Gonzalez G, Gilles-Gonzalez M A, Chan M K. Structure of a biological oxygen sensor: a new mechanism for heme-driven signal transduction. Proc Natl Acad Sci USA. 1998;95:15177–15182. doi: 10.1073/pnas.95.26.15177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Havarstein L S. Identification of a competence regulon in Streptococcus pneumoniae by genomic analysis. Trends Microbiol. 1998;6:297–299. doi: 10.1016/s0966-842x(98)01328-6. [DOI] [PubMed] [Google Scholar]
- 14.Hess J F, Oosawa K, Kaplan N, Simon M I. Phosphorylation of three proteins in the signaling pathway of bacterial chemotaxis. Cell. 1988;53:79–87. doi: 10.1016/0092-8674(88)90489-8. [DOI] [PubMed] [Google Scholar]
- 15.Lange R, Wagner C, de Saizieu A, Flint N, Molnos J, Stieger M, Caspers P, Kamber M, Keck W, Amrein K E. Domain organization and molecular characterization of 13 two-component systems identified by genome sequencing of Streptococcus pneumoniae. Gene. 1999;237:223–234. doi: 10.1016/s0378-1119(99)00266-8. [DOI] [PubMed] [Google Scholar]
- 16.Lu A L, Cuipa M J, Ip M S, Shanabruch W G. Specific A/G-to-C.G mismatch repair in Salmonella typhimurium LT2 requires the mutB gene product. J Bacteriol. 1990;172:1232–1240. doi: 10.1128/jb.172.3.1232-1240.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin P K, Li T, Sun D, Biek D P, Schmid M B. Role in cell permeability of an essential two-component system in Staphylococcus aureus. J Bacteriol. 1999;181:3666–3673. doi: 10.1128/jb.181.12.3666-3673.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michaels M L, Tchou J, Grollman A P, Miller J H. A repair system for 8-oxo-7,8-dihydrodeoxyguanine. Biochemistry. 1992;31:10964–10968. doi: 10.1021/bi00160a004. [DOI] [PubMed] [Google Scholar]
- 19.Morrison D A, Trombe M-C, Hayden M K, Waszak G A, Chen J D. Isolation of transformation-deficient Streptococcus pneumoniae mutants defective in control of competence, using insertion-duplication mutagenesis with the erythromycin resistance determinant of pAM beta 1. J Bacteriol. 1984;159:870–876. doi: 10.1128/jb.159.3.870-876.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Connell-Motherway M, van Sinderen D, Morel-Deville F, Fitzgerald G F, Ehrlich S D, Morel P. Six putative two-component regulatory systems isolated from Lactococcus lactis subsp. cremoris MG1363. Microbiology. 2000;146:935–947. doi: 10.1099/00221287-146-4-935. [DOI] [PubMed] [Google Scholar]
- 21.Peeters B P, de Boer J H, Bron S, Venema G. Structural plasmid instability in Bacillus subtilis: effect of direct and inverted repeats. Mol Gen Genet. 1988;212:450–458. doi: 10.1007/BF00330849. [DOI] [PubMed] [Google Scholar]
- 22.Pellequer J L, Wager-Smith K A, Kay S A, Getzoff E D. Photoactive yellow protein: a structural prototype for the three-dimensional fold of the PAS domain superfamily. Proc Natl Acad Sci USA. 1998;95:5884–5890. doi: 10.1073/pnas.95.11.5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 24.Samrakandi M M, Pasta F. Hyperrecombination in Streptococcus pneumoniae depends on an atypical mutY homologue. J Bacteriol. 2000;182:3353–3360. doi: 10.1128/jb.182.12.3353-3360.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taylor B L, Zhulin I B. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev. 1999;63:479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Throup J P, Koretke K K, Bryant A P, Ingraham K A, Chalker A F, Ge Y, Marra A, Wallis N G, Brown J R, Holmes D J, Rosenberg M, Burnham M K. A genomic analysis of two-component signal transduction in Streptococcus pneumoniae. Mol Microbiol. 2000;35:566–576. doi: 10.1046/j.1365-2958.2000.01725.x. [DOI] [PubMed] [Google Scholar]
- 27.Unden G, Schirawski J. The oxygen-responsive transcriptional regulator FNR of Escherichia coli: the search for signals and reactions. Mol Microbiol. 1997;25:205–210. doi: 10.1046/j.1365-2958.1997.4731841.x. [DOI] [PubMed] [Google Scholar]
- 28.Volz K. Structural conservation in the CheY superfamily. Biochemistry. 1993;32:11741–11753. doi: 10.1021/bi00095a001. [DOI] [PubMed] [Google Scholar]
- 29.Wang R F, Kushner S R. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- 30.Weiss V, Magasanik B. Phosphorylation of nitrogen regulator I (NRI) of Escherichia coli. Proc Natl Acad Sci USA. 1988;85:8919–8923. doi: 10.1073/pnas.85.23.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]