Abstract
The exploration of advanced probes for cancer diagnosis and treatment is of high importance in fundamental research and clinical practice. In comparison with the traditional “always‐on” probes, the emerging activatable probes enjoy advantages in promoted accuracy for tumor theranostics by specifically releasing or activating fluorophores at the targeting sites. The main designing principle for these probes is to incorporate responsive groups that can specifically react with the biomarkers (e. g., enzymes) involved in tumorigenesis and progression, realizing the controlled activation in tumors. In this review, we summarize the latest advances in the molecular design and biomedical application of enzyme‐responsive organic fluorescent probes. Particularly, the fluorophores can be endowed with ability of generating reactive oxygen species (ROS) to afford the photosensitizers, highlighting the potential of these probes in simultaneous tumor imaging and therapy with rational design. We hope that this review could inspire more research interests in the development of tumor‐targeting theranostic probes for advanced biological studies.
Keywords: enzyme-activatable probes, functional groups, peptide-responsive, photodynamic therapy, tumor fluorescent imaging
Activatable probes for tumor imaging and therapy enjoy advantages in comparison to traditional “always‐on” probes such as improved accuracy by specifically releasing or activating fluorophores at the targeting sites. The authors summarize recent advances in the molecular design and biomedical applications of enzyme‐activatable fluorescent probes for tumor theranostics reported over the past five years.

1. Introduction
Cancer is one of the main public health risks threatening human health and survival. According to the latest global cancer burden data, there were 19.29 million new cases and 9.96 million deaths worldwide reported in 2020. [1] At present, more than 90 % of cancer‐related deaths are caused by metastatic diseases and the related complications. [2] Therefore, developing effective tumor diagnostic and therapeutic platforms, especially for detection and intervention of cancer at early stage for reducing recurrence risk, has become urgently needed. [3] Among a variety of advanced techniques in preclinical and clinical implementation,[ 4 , 5 , 6 , 7 , 8 , 9 ] monitoring and analyzing the tumor biomarkers, including antigens, [10] enzymes [11] and reactive oxygen species (ROS), [12] has become highly important for early detection of tumors.
Currently, non‐ionizing fluorescence imaging has been used to monitor complex pathological processes at the molecular and cellular levels, owing to its inherent advantages of high temporal and spatial resolution with good sensitivity. [13] Depending on their signal output characters, the organic fluorescent probes can be generally categorized into two types: “always‐on” and “activatable/responsive” probes. [14] The fluorescent signal of the former is always present, leading to visualization of the target with a limited signal‐to‐noise ratio. Responsive probes, by contrast, are not fluorescent before being activated to turn on by a specific biological target, resulting in a relatively higher contrast at the targeted site. The molecular structure of responsive fluorescent probe is mainly composed of fluorophore, linker and recognition group. When the fluorescent probes meet the targeting substrates (e. g., tumor biomarkers), weak bonding, chemical reaction, or a combination of both happens between them. As a result, the fluorescent signal of the probe is altered accordingly to indicate the presence of biomarkers. [15]
By rational molecular design, the organic fluorophores can be endowed with the capability of generating ROS under light irradiation to afford photosensitizers (PSs). ROS are highly active and toxic substances that react rapidly with subcellular components, leading to tumor cell death. [16] In the past decades, photodynamic therapy (PDT) with the aid of PSs has attracted increasing attention, enjoying non‐invasive damage to tumor, minimal damage to normal organs and negligible multidrug resistance.[ 17 , 18 , 19 , 20 , 21 ] In general, the therapeutic effect of PDT on tumors involves tumor cell death caused by ROS, tumor‐associated vascular injury, and the initiation of specific immune responses against tumor cells. [22] Despite the rapid development of PDT, traditional photosensitizers may still cause toxic side effects in normal cells during PDT due to the lack of targeting and specificity. Referring to the targeted transport and responsive release of anticancer prodrugs, [23] the exploration of biomarker‐activatable PSs can help solve the problem of poor specificity in tumor PDT.
Among tumor biomarkers, enzymes have been of special interest to researchers. As a result of excessive expression of the enzymes associated with tumors,[ 11 , 14 ] the activation of fluorophores or PSs can be restricted to the local tumor lesions while they remain silent in normal tissues. According to the working mechanism, there are two main types of enzyme‐activatable organic fluorescent probes: one is based on the enzyme digestion response of peptides serving as linker or coating in the probes, the other is based on the biochemical reaction response of functional groups on fluorescent molecules. The fluorescence of the former is usually quenched by an inhibitor or due to the conformation of assembler before reacting with the targets. After cleavage of the peptide linker by an enzyme, its fluorescence and ROS generation ability are restored. [24] The molecular structures of the latter with reactive chemical functional groups only change in the presence of target enzyme, thus creating PSs at the target site with lighted‐up fluorescence. [25]
In recent years, enzyme‐activatable probes have emerged in abundance because of their great advantages in precise imaging and therapy.[ 11 , 26 ] A few reviews have summarized the promising progress of disease‐related enzyme‐responsive probes.[ 15 , 27 ] However, very few reviews focus on enzyme‐activatable organic fluorescent probes associated with a therapeutic function. Herein, we provide a tutorial review of recent advances in enzyme‐activatable theranostic probes. In this article, we summarize the core chemical structures reported in the last five years on which the construction of enzyme‐responsive probes depends. The first part consists in an introduction to fluorescence imaging, photodynamic therapy, and design strategies of enzyme‐activatable probes. In the second part, the peptide‐based enzyme‐responsive probes for tumor fluorescence imaging and PDT are described. In the third part, we introduce newly developed enzyme‐responsive fluorescent probes based on biochemical reactions (e. g., redox and hydrolysis reactions) of functional groups on molecules. Finally, the summary and outlook in this emerging field are presented, to highlight the opportunities in exploring advanced activatable theranostic probes for precise diagnosis and treatment of cancer. We hope that this review will provide guidance for the structural design of new generation enzyme‐based responsive fluorescent probes for disease theranostics.
2. Enzyme‐Responsive Probes Based on Peptides
It is well known that certain peptidases, such as matrix metalloproteinases (MMPs), [28] caspases, [29] fibroblast activating protein (FAP), [30] cathepsins, [31] and furin, [32] are overexpressed in different tumor cells (Table 1). This provides a basis for researchers to design enzyme‐cleavable probes based on corresponding peptides. These overexpressed enzymes cut off specific peptide sequences, leading to the release of active substances (e. g., fluorophores, photosensitizers and drugs) for enhanced fluorescence signals and therapeutic effects. In general, the fluorescence features of fluorophores used for these probes fall into two categories: aggregation‐caused quenching (ACQ) [33] and aggregation‐induced emission (AIE). [34] Traditional fluorophores are usually relying on ACQ, and the probes constructed according to this principle show negligible or weak fluorescence in the assembly (aggregation) state, and strong fluorescence after the responsive disassembly. On the contrary, the probes constructed from AIE fluorophores are often designed to promote the molecular aggregation in response to enzymes, thus exhibiting enhanced fluorescence and PDT. The significantly enhanced fluorescence signal could further improve the signal‐to‐noise ratio of the AIE probes in imaging. And it is worth mentioning that AIE probes often have the advantages of photobleaching resistance and high fluorescence quantum yield, which promote the imaging time and resolution. In addition to the above‐mentioned probes with fluorescence “turn‐on” signature by altering the aggregation state of fluorophores, another main class of responsive probes is constructed by linking a fluorophore to a quencher. The quencher is detached to release fluorescence only upon the cleavage of linker by enzymes.
Table 1.
Peptide‐based enzyme responsive probes.
|
Enzyme |
Targeting ligands |
Response mechanism |
Overexpressed cell type |
Ref. |
|---|---|---|---|---|
|
MMP‐2 |
GPLGIAGQ peptide; gelatin |
Aggregation state change; quencher removal |
Glioblastoma multiforme, colorectal adenocarcinoma, breast, pancreatic and ovarian cancer cells, etc. |
[38, 39] [40] |
|
MMP‐9 |
PLGVRG peptide; gelatin |
Aggregation state change; quencher removal |
Colon, melanoma, breast, lung, and ovarian cancer cells, etc. |
[38, 39] |
|
MMP‐14 |
SGRIGFLRTA peptide |
Quencher removal |
Ovarian, lung, breast, bladder, and endometrial cancer cells, etc. |
[38] |
|
Caspase‐3 |
DEVD peptide |
Aggregation state change; Quencher removal |
Hepatocellular carcinoma, etc. |
[42, 43] |
|
FAP‐α |
KGPGPNQC peptide; GPA peptide |
Accumulation of fluorophores; quencher removal |
Breast, ovarian, lung, colorectal, gastric, pancreatic cancer and cutaneous melanoma, etc. |
[47] |
|
Cathepsin B |
GFLG peptide |
Quencher removal |
Ovarian and intestinal cancer cells, etc. |
[50, 51] |
|
Furin |
RVRR peptide |
Aggregation state change |
Rhabdomyosarcoma and breast cancer, etc. |
[52, 53] |
2.1. Fluorescent Probes Responsive to MMPs
Matrix metalloproteinases (MMPs) represent the most prominent proteases related to tumorigenesis. [35] MMPs degrade various protein components in the extracellular matrix (ECM) for promoting tumor invasion and metastasis. In general, MMPs are overexpressed in several tumors including glioblastoma multiforme, colorectal adenocarcinoma, breast cancers, pancreatic cancers and ovarian cancers. The ischemia and hypoxia induced by tumor growth further promote their expression. Studies have shown that the active MMP‐2 activity detected in colorectal cancer is, on average, 10 times higher than in normal mucosa. [36] Therefore, probes can be designed to response to protein decomposition mediated by MMPs. According to the literature, most of the reported probes are based on the substrate sequences of MMP‐2 and MMP‐9. [37] Since MMPs are abundant in the tumor interstitial space, probes fused with the specific substrate peptides can be designed to control the release of diagnostic and therapeutic agents within the tumor microenvironment.
As described herein, MMPs with an elevated expression in tumor ECM are crucial in tumor progression. Therefore, making use of MMP‐cleavable sequences can readily achieve the release of PSs or fluorophores. For example, Frochot and co‐workers reported novel photodynamic molecular beacons (PMBs) [38] activated by MMP‐2 and/or MMP‐9 and/or MMP‐14. These PMBs are basically constructed from a photosensitizer and a singlet oxygen (1O2) quencher which are linked by the MMP‐cleavable peptide linker. Based on the fact that peptide SGRIGFLRTA can be cut by multiple MMPs, an MMP‐cleavable peptide ligand (H‐GRIGFLRTAKGG‐OH) was synthesized as the linker. Before being activated by MMPs, the PS in PMBs was quenched by the quencher due to the fluorescence resonance energy transfer (FRET) effect. The ability of PS to generate fluorescence and ROS by photoexcitation was restored only after the cleavage of linker by overexpressed MMPs at the tumor site. The graphical abstract (Figure 1) of this work illustrates a common model of enzyme‐activable fluorescent probes.
Figure 1.
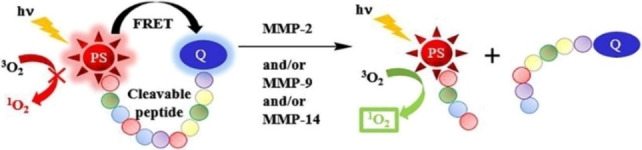
Demonstration of the working mechanism for an enzyme‐activable probe. Reproduced with permission from Ref. [38]. Copyright 2018, Elsevier.
More specifically, the peptide sequence of GPLGIAGQ can be cleaved into GPLG and IAGQ by MMP‐2. [37] In addition to cutting peptide sequences, MMP‐2 and MMP‐9 can also degrade gelatin at high temperatures (above 40 °C). [39] So it is an important strategy for releasing and activating PSs by degrading the gelatin nanoparticles (NPs) loading with PSs. A gelatin‐based nanoreactor (MCGPD∼RGD NPs) is reported to realize enhanced chemotherapy and PDT/PTT‐combined cancer therapy (Figure 2). [40] In this nano‐system, the Arg‐Gly‐Asp (RGD) peptides provide targeting ability and ensure sufficient accumulation of NPs at the tumor site. Under 808 nm laser irradiation, the overexpressed MMP‐2 and heat generated by polydopamine (PDA) coating work together to trigger gelatin degradation, leading to release of chlorin‐e6 (Ce6), metformin (Met), and doxorubicin (DOX) from the MCGPD‐RGD NPs. Metformin alleviates tumor hypoxia and promotes O2‐dependent PDT based on Ce6 (which can excite oxygen to form 1O2 under light irradiation). Besides, the heat generated by the PDA coating directly kills tumor cells, affording photothermal therapy (PTT) and amplifying the effect of chemotherapy (by DOX) for tumor growth inhibition.
Figure 2.
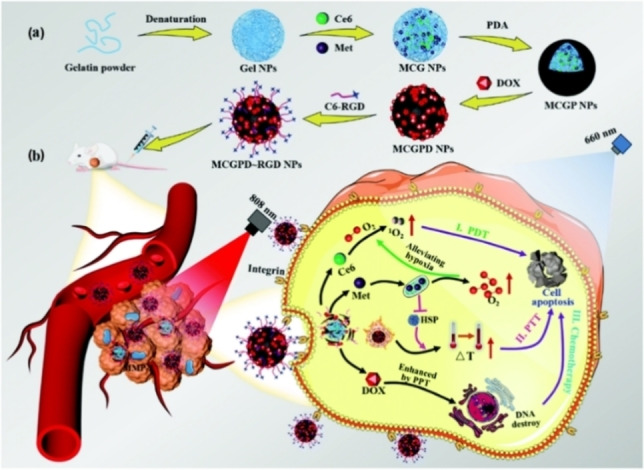
Schematic illustration for (a) the synthesis and (b) the antitumor mechanism of MCGPD‐RGD NPs. Reproduced with permission from Ref. [40]. Copyright 2021 The Royal Society of Chemistry.
2.2. Fluorescent Probes Responsive to Caspases
Caspases, cysteinyl‐aspartate‐specific proteinases, are closely related to eukaryotic cell apoptosis that can trigger a chain reaction of enzymes, leading to degradation of chromosomal DNA and cell disintegration. [29] Caspases are a family of proteases, including multiple subfamilies. The subfamily caspase‐3, including caspase‐3, −6, and −7, plays the role of apoptosis executor. [29a] Caspase‐3 is overexpressed in cancer cells such as those of oral tongue squamous cell carcinoma (∼30‐fold higher than in normal tissue), [29d] providing useful information about tumor status.
Caspase‐3/7 selectively cut the DEVD protein sequence, making DEVD‐based peptides cleavable linkers to construct the responsive probes. [41] Because the DEVD peptides are water‐soluble and the fluorophores are more hydrophobic, the solubility of DEVD‐containing probes is altered after being cut by enzymes in the aqueous environment. For example, AIE fluorophores in the probe will aggregate due to their poor water solubility after releasing the hydrophilic DEVD peptides, which will show obvious fluorescence “turn‐on” signature.
Meade and co‐workers reported a caspase‐activatable fluorescent‐magnetic resonance bimodal probe (CP1) by combining tetraphenylethylene (TPE) with a Gd‐chelate (Figure 3). [42] TPE is a typical aggregation‐induced emission luminogen (AIEgen). In the presence of caspase‐3/7, the DEVD peptide was cleaved and the remaining fragments containing TPE self‐assemble into nanoparticles, triggering the AIE effect to turn on fluorescence. Time‐lapse fluorescence imaging of apoptotic HeLa cells (Figure 3c) indicated that CP1 had a good potential in monitoring elevated caspase‐3/7 activity during apoptosis. The shortwave (∼480 nm) fluorescence of TPE cannot penetrate the tissue, but deep tissue imaging was achieved by the Gd‐chelate combined with magnetic resonance (MR) imaging.
Figure 3.
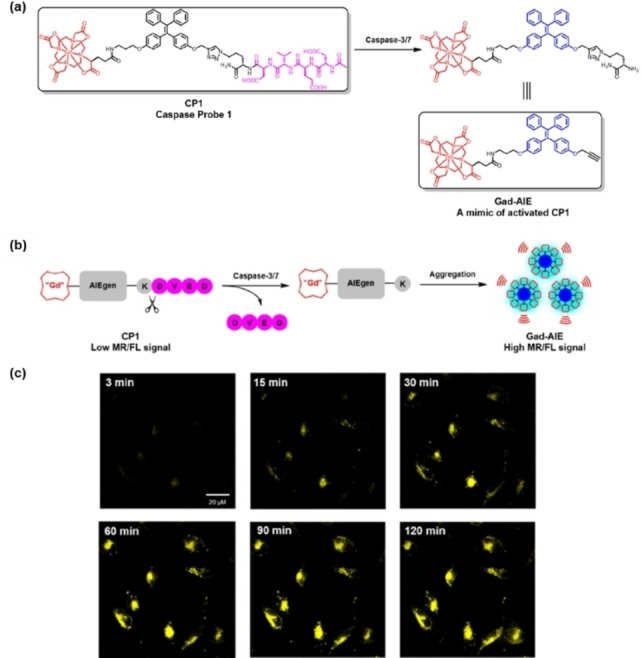
(a) Chemical structures of CP1 and Gad‐AIE; (b) sensing mechanism of CP1; (c) time‐lapse fluorescence imaging of apoptotic HeLa cells incubated with CP1 (50 μm) prior to apoptosis induction with sodium tetradecyl sulfate (STS, 1 μm). Reproduced with permission from Ref. [42]. Copyright 2019 American Chemical Society.
Depth growth in fluorescence imaging is largely dependent on the development of near‐infrared (NIR) fluorescent probes (fluorescence emission wavelength at 700–1700 nm). In 2019, Wang and co‐workers developed a tumor‐selective cascade activatable self‐detained system (TCASS) within an imaging probe (1) containing a cyanine dye (Cy) to offer NIR fluorescence for cancer imaging (Figure 4). [43] The probe 1 was designed to consist of (i) a recognition motif, a peptide that selectively binds to tumor cells with overexpressed X‐linked inhibitor of apoptosis protein (XIAP); (ii) an enzymatically degradable linker, a peptide substrate capable of being specifically cleaved by caspase‐3; (iii) a self‐assembly motif, a hydrophobic peptide derived from amyloid‐β (Aβ) to facilitate molecular self‐assembly; and (iv) a functional molecule, a cyanine dye (Cy) or a chemotherapeutics doxorubicin (DOX). After internalization into cancer cells, the recognition motif bound to XIAP and activated the caspase‐3/7 pathway. Subsequently, the activated caspase‐3/7 cleaved the DEVD peptide in 1. The residual fragments then rapidly self‐assembled to form fibrous β‐sheet superstructures, leading to the enrichment and enhanced retention of Cy in tumors with enhanced imaging effect. The fluorescence imaging of H460 tumor‐bearing mice indicates that the tumor fluorescence signal after injection of 1 is much stronger than that of mice injected with a non‐targeted control probe (2) or a non‐assembled control probe (3). Even 5 days after injection of 1, the fluorescence signal of the tumors was still clearly observed. It was also demonstrated that this system had a long‐term therapeutic effect when the dye was replaced with DOX.
Figure 4.
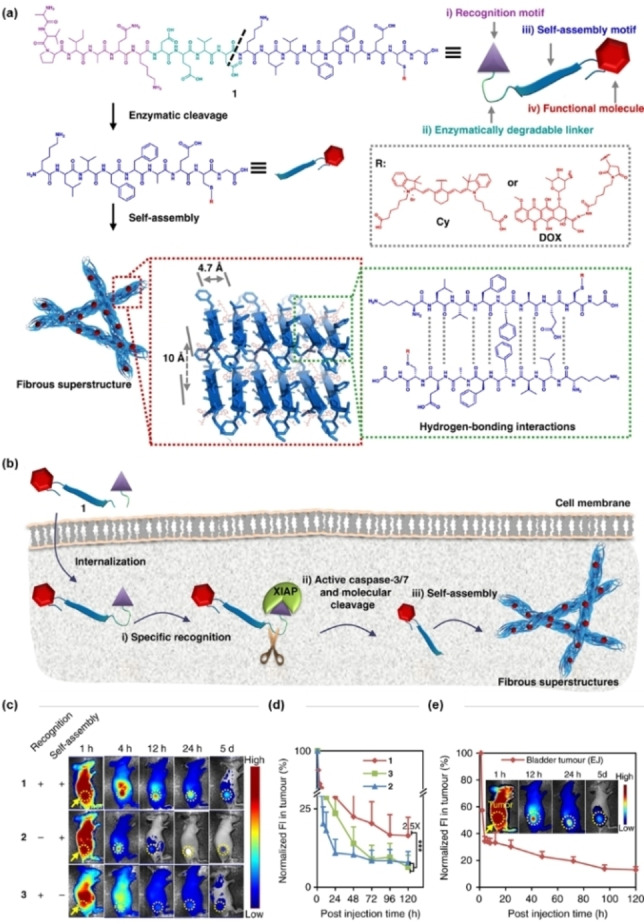
(a) Molecular design and chemical structure of the tumor‐selective cascade activatable self‐detained system; (b) schematic illustration of the working mechanism of probe 1; (c) representative NIR fluorescence images of probes 1, 2 and 3 on H460 tumor‐bearing mice after intravenous injection. Images were acquired at 1, 4, 12, 24 h and 5 days post injection. Probes 2 and 3 are the control probes. The probe concentration used in the in vivo experiment was 14 mg kg−1; (d) normalized fluorescence intensity in tumors in c; (e) NIR fluorescence images and normalized fluorescence intensity of tumor on tumor‐bearing mice after IV injection. Reproduced with permission from Ref. [43]. Copyright 2019 Springer Nature.
2.3. Fluorescent Probes Responsive to FAP‐α
Fibroblasts, the main component of tumor stroma, interact with cancer cells to activate and become cancer‐associated fibroblasts (CAFs). [44] Compared with fibroblasts in normal tissues, CAFs have corresponding changes in phenotype, activity and function, and thus play a supporting role in the occurrence and development of tumors. Fibroblast activation protein‐α (FAP‐α) is an antigen molecule specifically expressed on the surface of CAFs, belonging to the serine protease family, which can enhance the degradation of tumor stroma and promote tumor growth and invasion. [45] Studies have shown that FAP‐α is selectively expressed on the surface of stromal fibroblasts in more than 90 % of epithelial malignancies, including breast, ovarian, lung, colorectal, gastric, pancreatic cancer and cutaneous melanoma, etc. [30] Therefore, FAP‐α as a target for tumor targeted diagnosis and treatment has been attracting much attention.
Previous studies have explored FAP‐α activable fluorescent probes based on the chromophore‐quencher model. For example, Cheng and co‐workers developed ANPFAP that linked a NIR dye Cy5.5 and a quencher QSY21 by a FAP‐α‐specific peptide sequence (KGPGPNQC). [46] After the cleavage of the KGPGPNQC sequence by overexpressed FAP‐α, the FRET effect was lifted between Cy5.5 and QSY21, and the fluorescence of the dye was turned on to provide a high contrast fluorescence imaging in U87MG tumor models. Recently reported FAP‐α‐responsive fluorescence probes are mainly designed to target tumors by FAP‐α‐cleavable peptides, improving the imaging through the accumulation of fluorophore at the target site. In 2019, Wang and co‐workers constructed a NIR fluorescent probe based on a peptide that specifically responds to FAP‐α. [47] The probe (Molecule 1) consists of a tailoring motif, a self‐assembly motif, and two hydrophilic motifs (Figure 5). The Cy dye is attached to the self‐assembly motif. On the surface of CAFs, the peptide sequence GPAX (X designates any amino acid) on the tailoring motif was specifically cleaved by FAP‐α, leading to self‐assembly of the residue fragments to form β‐sheet nanofibers. These nanofibers containing the dyes can accumulate around the tumor with a long retention time. Experimental results showed that the fluorescence signal of Molecule 1 was 5.5 times stronger than that of the control probe (Molecule 4) that could not aggregate. The selective assembly of this probe resulted in a ∼4‐fold and ∼5‐fold higher fluorescent signal of tumors in comparison to that of the liver and kidney, respectively. Due to the enhanced imaging capability, this probe allowed detection of small tumors approximately 2 mm in diameter.
Figure 5.
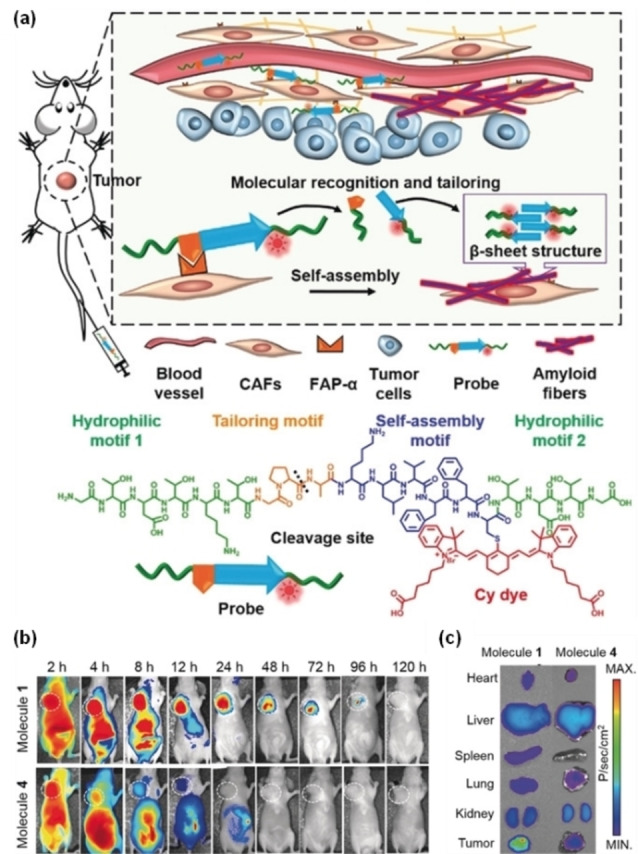
(a) Schematic illustration of the self‐assembly process and chemical structure of the probe with four motifs; (b) the time‐dependent NIR fluorescent images of mice bearing CAFs from 2–120 h after intravenous administration of Molecule 1 and Molecule 4; (c) NIR fluorescence distribution in heart, liver, spleen, lung, kidney, and tumor of Molecules 1 and 4 at 48 h post intravenous injection at a dose of 14 mg kg−1. Molecule 1 is the probe, and Molecule 4 is the control probe. Reproduced with permission from Ref. [47]. Copyright 2019 Wiley‐VCH.
2.4. Fluorescent Probes Responsive to Cathepsins
Cathepsins belong to the cysteine family, and they are active in reducing and slightly acidic environments (e. g., lysosomes). [48] Similar to matrix metalloproteinases, they also affect extracellular matrix degradation and promote tumor proliferation, invasion, angiogenesis and metastasis. There are data suggesting that the relative amount of cathepsin B in melanoma is at least three times greater than that in normal mouse tissue. [49] Due to their overexpression in multiple tumors (ovarian cancer, lung cancer and intestinal cancer, etc.), [31] cathepsins are considered as potential biomarkers for clinical tumor diagnosis and evaluation.
In 2017, Yoon and co‐workers reported a cathepsin B (CTSB)‐activated fluorescent probe (CyA−P−CyB) for the in vivo imaging and PDT of tumors (Figure 6). [50] The probe (CyA−P−CyB) consisted of two cyanine moieties (CyA and CyB) linked by a peptide Gly‐Phe‐Leu‐Gly (GFLG) that can be selectively cleaved by cathepsin B. In the probe, CyA acted as a fluorescent donor and CyB acted as a quencher. The initial probe CyA−P−CyB did not emit fluorescence because of the FRET from CyA to CyB, but it showed profound fluorescent signal (at 720 nm) when the peptide linker was cleaved by CTSB. Notably, the probe also exhibited effective PDT since a PDT reagent (Cy−S−Ph−NH2) derived from CyB is released after the cleavage of peptide linker. This PDT reagent could kill tumor cells with overexpression of CTSB by generating ROS under 808 nm laser irradiation. Thus, the probe (CyA−P−CyB) was successfully used in precise imaging and therapy of CTSB‐overexpressing tumors.
Figure 6.
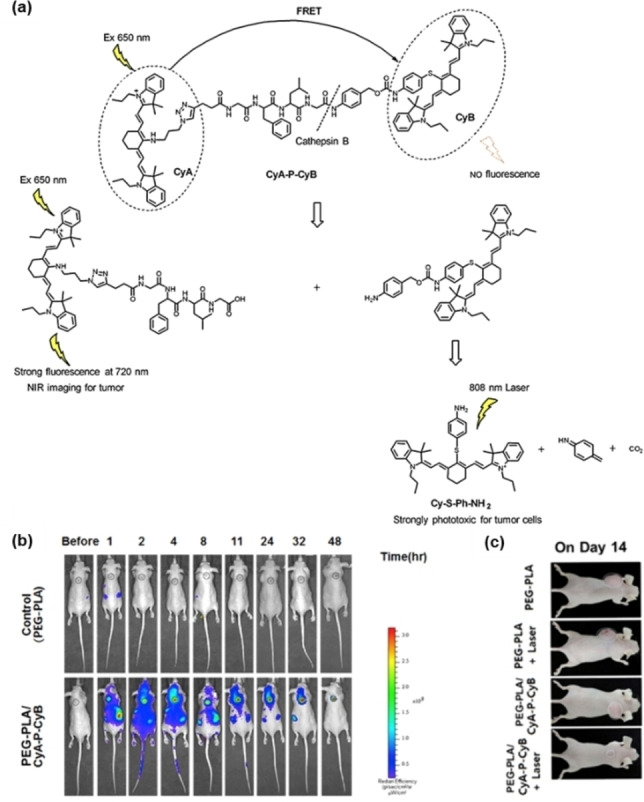
(a) Illustration for the responsive mechanism of probe CyA−P−CyB; (b) NIR images of tumor‐bearing mice were collected over 48 h after the intravenous injections of PEG‐PLA/CyA−P−CyB; (c) photos of mice bearing H640 tumors on day 14 after administration of the indicated treatments. Reproduced with permission from Ref. [50]. Copyright 2017, Elsevier.
Cancer treatment is always faced with the challenge of improving treatment efficiency and reducing side effects. In order to overcome these challenges, Wang and co‐workers designed a multifunctional polymer prodrug micelle. [51] The micelles were prepared by self‐assembly of the amphiphilic copolymer and Ce6. After cellular uptake, the GFLG peptides in the amphiphilic copolymer were degraded by cathepsin B. This process further triggered the disassembly of the micelles to release free Ce6, followed by augmented generation of singlet oxygen under laser irradiation at 660 nm. Based on the CTSB‐targeted release of PSs, accurate imaging and PDT of the tumors was realized.
2.5. Fluorescent Probes Responsive to Furin
Furin protease is a preprotein invertase, located in the trans‐Golgi network, which can be activated in an acidic environment. Furin protease can cleave precursor proteins with a specific sequence to produce mature proteins with biological activity. Studies have shown that the furin protease is generally overexpressed (∼3‐ to 5‐fold) in head and neck squamous cell carcinoma, breast cancer, rhabdomyosarcoma, and other tumors. [32]
Researchers have designed some responsive fluorescent probes to detect the furin enzyme overexpressed by cancer cells. For example, Li and co‐workers successfully designed and synthesized a chimeric peptide probe RVRR‐FFF‐NBD (C‐3) that can recognize the overexpression of furin. [52] Due to the amphiphilic property of chimeric peptide RVRR‐FFF‐NBD, it can self‐assemble into nano‐micelles. When the probe (C‐3) was endocytosed by furin‐overexpressing cells, the RVRR sequence was enzymatically hydrolyzed to release the fluorescent fragment FFF‐NBD that emitted green fluorescence (Figure 7). The cancer cells with different expression levels of furin were then identified through the varied intracellular fluorescence intensities. In 2017, Liang's group reported a dual AIE fluorescent probe Ac‐Arg‐Val‐Arg‐Arg‐Cys(StBu)‐Lys(TPE)‐CBT based on the RVRR peptide. [53] In their design, a AIE fluorogen (TPE) was introduced at the side chain of lysine. When the ingestion by furin‐overexpressing tumor cells happened, the ethylene disulfide was reduced. As a result, intermolecular condensation was triggered to form dimers and oligomers that rapidly self‐assembled into nanoparticles in situ. Due to the aggregation‐induced emission (AIE) effect of TPE, the fluorescence signal from internalized probe was significantly enhanced.
Figure 7.
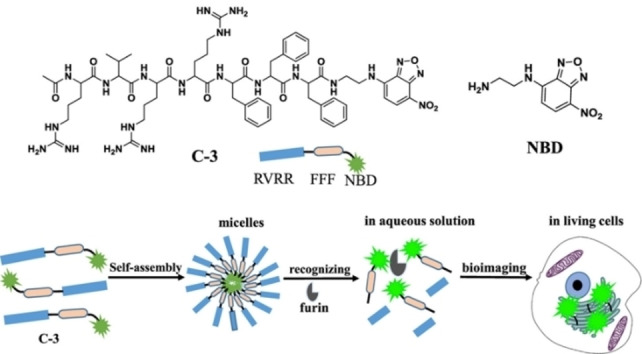
Schematic illustration of the self‐assembly process and furin detection using the probe C‐3. Reproduced with permission from Ref. [52]. Copyright 2019, American Chemical Society.
3. Enzyme‐Responsive Probes Based on Functional Groups
In addition to the peptide‐cleavable enzymes in tumors, there are another type of overexpressed enzymes that catalyze biochemical reactions involving redox and hydrolysis reactions (e. g., alkaline phosphatase, [54] quinone oxidoreductase‐1, [55] azoreductase, [56] etc., Table 2). Using these overexpressed enzymes as biomarkers, some new enzyme‐responsive probes have been designed based on the characteristic functional chemical groups of these enzyme‐catalyzed substrates. After the functional groups in probes react with these enzymes, the solubility and/or the molecular structure of the fluorophores change. As a result, the fluorescence emission and ROS generation ability are thus promoted. In addition to the above‐mentioned “aggregation state change” and “quencher removal” response mechanisms, the molecular energy levels change by tuning the distribution of π‐electrons in the conjugated system of fluorophores upon chemical reaction. In this section, we summarize representative enzyme‐responsive organic fluorescent probes based on reactive functional groups reported over the past give years, hoping to provide guidance for the molecular design of new theranostic probes.
Table 2.
Enzyme‐responsive probes based on functional groups.
|
Enzyme |
Functional groups |
Reactions |
Overexpressed cell type |
Ref. |
|---|---|---|---|---|
|
Alkaline phosphatase |
Phosphate ester |
Phosphate hydrolysis |
Cervical, colon, and liver cancer cells, etc. |
[58,59] [60] |
|
NAD(P)H: quinone oxidoreductase‐1 |
Quinone |
Quinone reduced to hydroquinone |
Ovarian and colon cancer cells, etc. |
[62,63] |
|
Azoreductase |
Azo bonds |
Azo reduced to amine |
Breast and ovarian cancer cells, etc. |
[65,66] |
|
Tyrosinase |
Catechol or monophenol |
Phenol oxidized to quinone |
Melanoma and liver cancer cells, etc. |
[69] |
|
Nitroreductase |
Nitro group |
Nitro group reduced to amino group |
Breast and cervical cancer cells, etc. |
[73,74] |
|
Carboxylesterase |
Carboxyl ester; amide |
Hydrolysis |
Rhabdomyosarcoma and breast cancer, etc. |
[76,77] [78,79] |
3.1. Fluorescent Probes Responsive to Alkaline Phosphatases
Alkaline phosphatase (ALP) is a subfamily of phosphatases that catalyze the hydrolysis of phosphate fractions of proteins, nucleic acids, and other biomolecules. [57] Up‐regulated expression (∼5‐fold) of ALP is related to the occurrence and development of diseases such as prostate cancer and cervical cancer. [54]
For accurate detection of ALP, Zhang and co‐workers constructed a fluorescent probe (NALP). [58] Intact NALP is non‐fluorescent because the phosphate group blocks the intramolecular charge transfer (ICT). Only after the phosphate was hydrolyzed by the overexpressed ALP, the structure of NALP changed to form a semi‐cyanine fluorophore with strong fluorescence. On the contrary, other non‐specific enzymes and biological species did not affect the fluorescence of NALP, indicating its high selectivity to APL. In addition to cells, the NALP probe could be also used for monitoring the ALP activity in tumor tissues in nude mice. Hou and co‐workers constructed a novel NIR fluorescent probe (APT) via integrating a self‐immolative unit and a fluorophore with the phosphate group as recognition group. [59] The APT probe exhibited a high sensitivity for ALP with a detection limit of 0.89 U/L. In addition, acetaminophen‐induced alkaline phosphatase upregulation was observed in living zebrafish.
In biomedical research and clinical applications, the long‐term dynamic observation of cancer cells is usually limited by the aggregation‐caused quenching (ACQ) effect of the traditional fluorescent probes. To address this challenge, Yoon's group reported an amphiphilic AIEgen of DQM‐ALP for ALP imaging in a recent study. [60] DQM‐ALP was composed of a quinoline‐malononitrile core and a phosphate group, in which the core was a hydrophobic AIE module and the phosphate group acted as the ALP reaction site to offer the overall hydrophilicity (Figure 8). After the reaction with alkaline phosphatase, the resulting DQM‐OH rapidly aggregated, resulting in enhanced fluorescence. Depending on this working mechanism, DQM‐ALP could distinguish normal tissue from tumor tissue by fluorescence imaging. The photophysical properties of DQM‐ALP remained unchanged under light irradiation at 350 mW cm−2 for at least 2 h. Good photostability ensured that it could be observed for a long time. This allowed for the analysis of spatially heterogeneous ALP activity with a high spatial resolution on a two‐photon imaging platform, creatively providing the recognition of cancer‐related submillimeter tumorigenicity.
Figure 8.
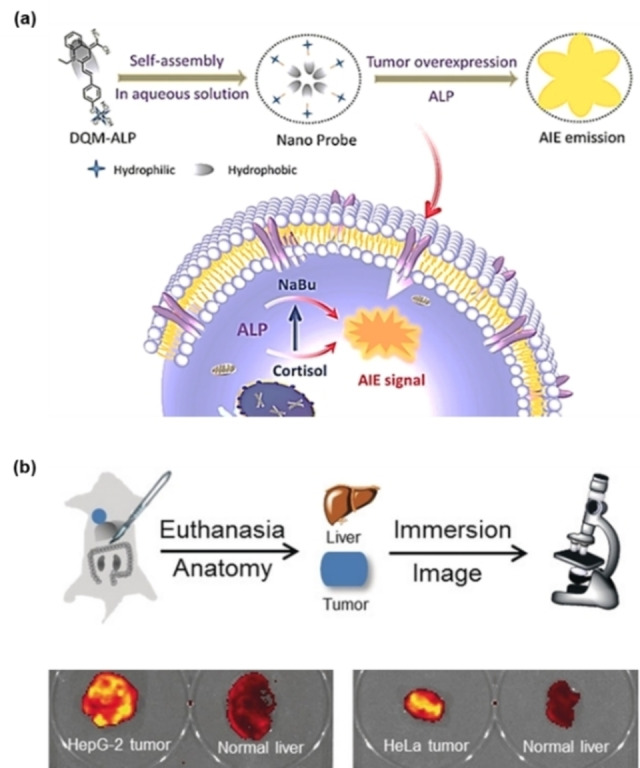
(a) Schematic illustration of the assembly process of DQM‐ALP; (b) fluorescence imaging of tumors and normal tissues after immersion in DQM‐ALP solution. Reproduced with permission from Ref. [60]. Copyright 2020, Wiley‐VCH.
3.2. Fluorescent Probes Responsive to NAD(P)H: Quinone Oxidoreductase‐1
NAD(P)H: quinone oxidoreductase‐1 (NQO1) is a flavin protein that is widely distributed in organs such as the liver, kidney, and gastrointestinal tract. [55] This enzyme can catalyze the two‐electron reduction of quinone and its derivatives, protecting cells from redox cycles and oxidative stress. The expression level of NQO1 is usually 20‐ to 50‐fold higher in tumor cells (e. g., liver, colon, breast, lung, and kidney cancer cells) compared with normal tissue cells. Therefore, NQO1 is considered as an important target for anticancer drug design and tumor diagnosis. [61]
In 2017, McCarley's group reported a cyanine dye‐based near‐infrared fluorescent probe Q3STCy for NQO1 recognition. [62] This probe exhibited a low background fluorescence before reaction with NQO1 and achieved detection of NQO1 with a high signal‐to‐noise ratio upon releasing hydroquinone. Due to the high selectivity and rapid response, Q3STCy can detect metastatic tumors with a diameter of 0.5 mm. Zhao and co‐workers constructed a nano‐probe (Nano‐NQO1) [63] for quantitative detection of NQO1 by using a polymer micelle assembly strategy. Nano‐NQO1 was assembled from an amphiphilic copolymer (mPEG‐DSPE) and a NQO1‐responsive molecule (BOD‐NQO1). The core molecule BOD‐NQO1 was made of a BODIPY‐based fluorophore, an NQO1 recognition moiety (quinone) and a spontaneously cleavable linker (Figure 9). Compared with the direct use of molecule BOD‐NQO1, Nano‐NQO1 formed by encapsuling BOD‐NQO1 with the polymer had a good solubility and biocompatibility. Since the fluorescence changed from green to red after the probe reacted with NQO1, ratiometric cell imaging was realized to distinguish NQO1‐positive cancer cells (HT‐29 cells) from NQO1‐negative cells (HT‐596 cells).
Figure 9.
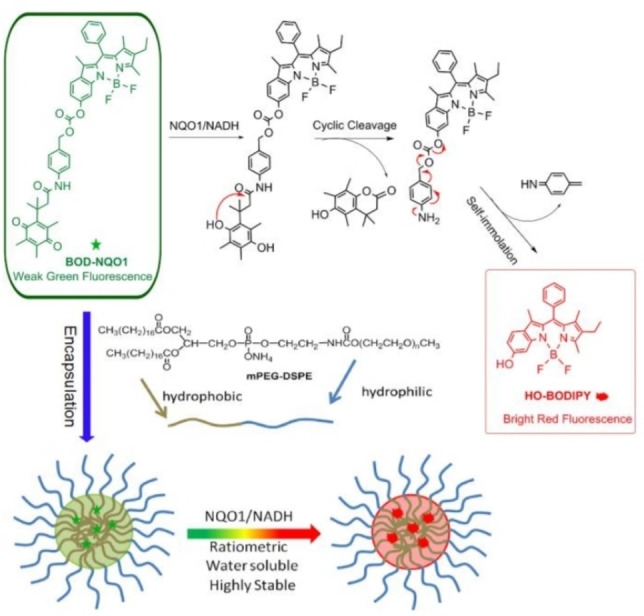
Schematic representation of the reaction process of BOD‐NQO1 and polymeric micelle assembly of nanoprobes with ratiometric response to NQO1. Reproduced with permission from Ref. [63]. Copyright 2017, Elsevier.
3.3. Fluorescent Probes Responsive to Azoreductases
Hypoxia is one of the most prominent features of solid tumors, and its degree is closely related to the local concentration of reductive substances such as azoreductase (AzoR). [56] AzoR can catalyze the reductive cleavage of azo bonds in the structure of azobenzene group. This specific reductive reaction provides a strategy to construct hypoxia sensitive probes using azobenzene as the bioactive linker. At present, azoreductase‐specific fluorescent probes have shown promising prospects in biological imaging and drug delivery in the hypoxic tumors. For instance, Nagano's group prepared a series of FRET‐based fluorescent probes for the sensing of azoreductase. [64] These non‐emissive probes were constructed by linking a cyanine dye and quencher (Black Hole Quencher‐3) with an azo bond. Upon activation by azoreductase under a hypoxic environment, strong fluorescence was released from these probes with increasing intensity in 10 min. The NIR fluorescence imaging of cancer cells under normoxic and hypoxic conditions demonstrated that these probes specifically exhibited fluorescence under hypoxic condition, indicating their selectivity to hypoxic tumors.
In 2019, Wu and co‐workers developed an intelligent probe (NR‐azo) for precise imaging and treating solid tumors. [65] NR‐azo could respond specifically to tumor hypoxia, releasing the fluorescent NR‐NH2 and active anticancer drugs under the action of azoreductase. The activated fluorophores were used to detect and image tumor hypoxia, while the released therapeutic drug can eliminate tumors. The approach of this work thus provides a strategy for designing chromophore‐drug combinational probes for hypoxic tumor theranostics. Following a similar strategy, Shi and co‐workers constructed an azoreductase‐responsive probe (AP−N=N−CY) for targeting the colon. [66] As shown in Figure 10, the precursor podophyllotoxin derivative (AdP), a common anticancer prodrug, was linked to a NIR fluorophore (Cy) via an activated azobenzene group to afford AP−N=N−CY. This probe was targeted to the site of intestinal lesions and was activated in situ by the local overexpressed azoreductase, facilitating its therapeutic role.
Figure 10.
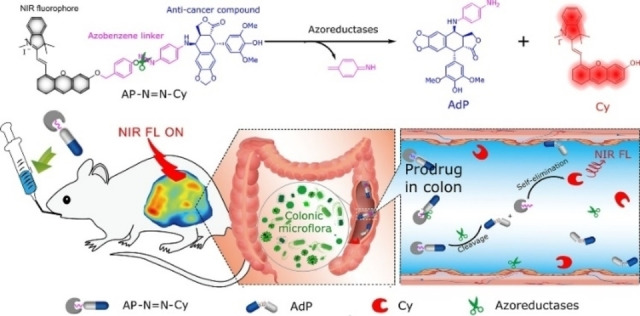
Schematic illustration of probe AP−N=N−CY in response to azoreductase. Reproduced with permission from Ref. [66]. Copyright 2020, American Chemical Society.
3.4. Fluorescent Probes Responsive to Tyrosinases
Tyrosinase (TYR) is a copper‐containing enzyme which can convert catechol or monophenol derivatives in organisms to the corresponding quinones. [67] Since TYR plays a vital role in the biosynthesis of melanin and is overexpressed (∼10‐fold) in malignant melanoma, [68] this enzyme is considered as a potential biomarker for melanoma diagnosis.
In 2018, Liang and co‐workers developed a tyrosinase‐activated fluorescent probe (MB1) under the guidance of theoretical thermodynamic analysis. [69] The authors designed and synthesized MB1 by coupling 4‐(2‐aminoethyl) phenol (recognition group) to methyl blue (MB) with a rationally modified urea bond. As a response to the overexpressed TYR, the recognition group was interrupted for releasing MB (Figure 11). TYR could effectively activate the probe, even at a very low TYR concentration in a short incubation time. In addition to melanoma‐specific imaging, MB1 demonstrated its potential in PDT to kill melanoma cells, owing to the ability of MB in generating singlet oxygen upon light irradiation. Similarly, Wu and co‐workers constructed an NBR‐AP probe by using phenol and an urea bond as the recognition group. [70] This probe produced strong fluorescence in living cells and tumors through TYR‐mediated phenol oxidation and urea bond hydrolysis. More importantly, the NBR‐AP probe could image cancer metastases in mice.
Figure 11.
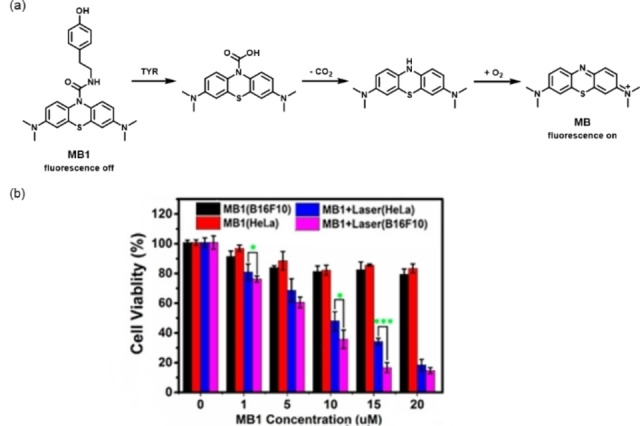
(a) Proposed reaction mechanism of the TYR‐catalyzed release of MB from MB1; (b) toxicity of MB1 to B16F10 cells and HeLa cells with and without laser irradiation. Reproduced with permission from Ref. [69]. Copyright 2018, American Chemical Society.
3.5. Fluorescent Probes Responsive to Nitroreductases
Nitroreductase (NTR) is another overexpressed reductase in the hypoxic tumor micro‐environment which can catalyze the reduction of nitro to amino groups in biomolecules. [71] It has been confirmed that the expression level of NTR is positively correlated with the degree of hypoxia in solid tumors. [72] Therefore, monitoring NTR activity is of great significance to the cancer screening at its early stage and the evaluation of tumor hypoxia and prognosis.
In 2019, Peng and co‐workers reported an NTR‐responsive probe (ICy−N). [73] Following reaction with NTR, nitro reduction and subsequent self‐combustion exposed phenolic hydroxyl groups, releasing ICy−OH with a NIR fluorescent (716 nm) signal (Figure 12). The released ICy−OH served as an effective PS for tumor ablation. The iodine was introduced into the fluorophore (ICy−OH) to increase the yield of singlet oxygen, which can be attributed to improved intersystem crossing through the “heavy atom” effect. During PDT, the activated ICy−N showed high phototoxicity (IC50=0.63 μm) under 660 nm light irradiation. Notably, this probe could be specifically anchored to mitochondria for promoting apoptosis (Figure 12b). Based on these facts, it has shown a good capacity in inhibiting tumor growth.
Figure 12.
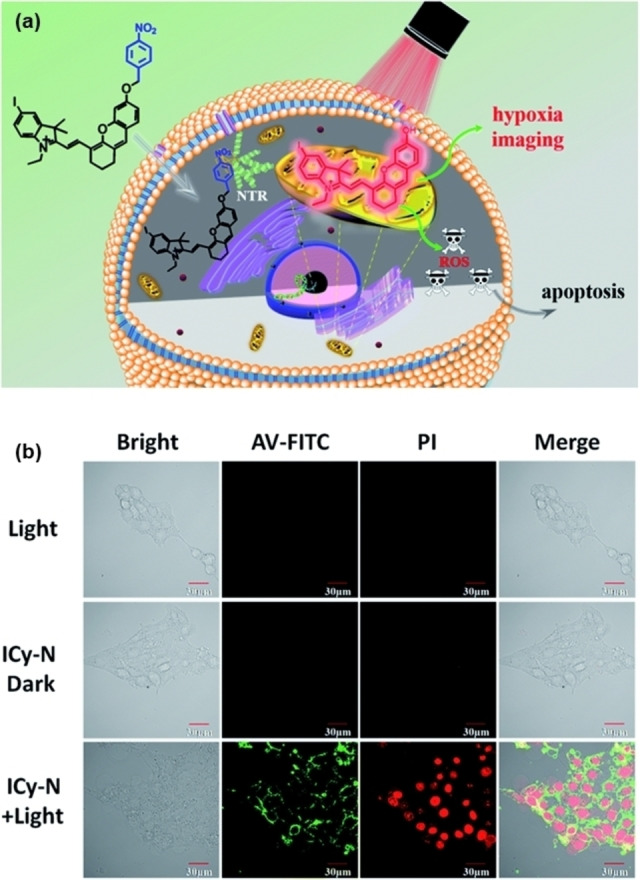
(a) Schematic illustration of the response process of ICy−N for NTR; (b) ICy−N induced apoptosis assays of 4T1 cells. Confocal microscopy imaging of annexin V‐FITC and PI stained 4T1 cells after different treatments. The green channel of annexin V‐FITC was excited at 488 nm and collected at 500–550 nm; the red channel of PI was excited at 561 nm and collected at 590–640 nm. Reproduced with permission from Ref. [73]. Copyright 2019, The Royal Society of Chemistry.
Recently, Wu and co‐workers developed an NTR‐activated AIEgen (Q‐NO2) for the localization of breast cancer metastasis. [74] The probe Q‐NO2 consists of a chromophore and a fluorescence quencher (nitrobenzene). In addition to quenching fluorescence, nitrobenzene also acts as a responsive group for NTR. Under the catalytic action of NTR, the nitro group of the probe Q‐NO2 was rapidly reduced to an amine group. The self‐elimination reaction was then carried out to generate a hydroxyl group with a strong electron‐donating effect. Thus, the quenching effect was eliminated, and the output of AIE fluorescence and photoacoustic signal was restored. The in vivo NTR activity tracking experiments showed that the probe Q‐NO2 could monitor not only the orthotopic breast tumors but also the continuous metastasis to lymph nodes and lungs.
3.6. Fluorescent Probes Responsive to Carboxylesterases
Carboxylesterases (CEs) are polymeric proteins belonging to the esterase family. They are mainly distributed in cytoplasm, mitochondria, and endoplasmic reticulum of cells. Through catalyzing the hydrolysis of carboxylester, amide, and thioester bonds, CEs are widely involved in the biotransformation of endogenous and exogenous substances, playing an important role in detoxification and metabolism. It has been reported that CEs are overexpressed (∼3‐fold) in a variety of tumor cells, especially in liver cancer cells. [75]
In 2020, Cai and co‐workers developed a carboxylesterase‐activatable PS based on perylene monoimide (PMI). [76] The tetrachloroperylene monoimide PS (P1) was assembled with folate (FA)‐modified albumins to afford nanoclusters (FHP) with an average diameter of ∼100 nm. The chemical structure of P1 contained two ester bonds, which were specifically hydrolyzed by the overexpressed CEs in tumor cells. Under the catalytic hydrolysis reaction of CEs, P1 was turned into carboxylic hydrolysate (P2) with the increasing molecular polarity and water solubility. Once P1 was hydrolyzed, FHP decomposed into ultra‐small nanoparticles (about 10 nm), facilitating the deeper penetration in tumor. Furthermore, the generation of P2 and the disassembly of FHP resulted in augmented fluorescence intensity (∼8 times) and singlet oxygen generation (∼4 times), to promote in‐situ fluorescence imaging and PDT of HepG2 liver cancer cells. Through enzyme‐triggered probe disassembly, FHP achieved significant tumor growth suppression in vivo with minimal side effects by activatable PDT in deep tissues.
The targeting ability of the single targeting probes can be restricted by the heterogeneous expression of the biomarker receptors in the complex tumor microenvironment. To address this issue, modifying the probes with more targeting groups to target two or more tumor biomarkers may lead to a higher accuracy. For example, Chen and co‐workers reported a fluorescence probe (NIR‐CBT) based on biotin and fluorophore Cy5.5 for simultaneously targeting biotin receptors and CEs. [77] NIR‐CBT self‐assembled into nanoparticles through a reduction condensation reaction, resulting in quenched fluorescence. After NIR‐CBT‐NP targeted to HepG2 cells by biotin, the overexpressed CEs cleaved the ester bond to restore the fluorescence of Cy5.5. Thus, the precise imaging of HepG2 tumors was realized by NIR‐CBT‐NP.
In 2021, Yu and co‐workers developed a NIR two‐photon fluorescent probe (DCM−Cl−CE) by screening a series of recognition moieties of substrates. [78] The DCM fluorophore was used as the reporter, while the dimethyl carbamate acted as the specific recognition group. This DCM−Cl−CE possessed outstanding sensitivity with a low detection limit of 0.013 U mL−1 and excellent selectivity toward CEs over the interference of Butyrylcholinesterase/Acetylcholinesterase. The CE activity was then effectively evaluated in orthotopic HepG2 tumor‐bearing mice under different conditions and 3D imaging in vivo. To afford the cancer‐killing ability, Zhang and colleagues developed a CE‐triggered multifunctional prodrug BDP−L−CPT (Figure 13) by the conjugation of a boron dipyrromethene (BDP)‐based PS to the active site of the chemotherapeutic drug camptothecin (CPT). [79] The activity of CPT was effectively shielded in the prodrug by chemical modification. The phenyl benzoate linker in BDP−L−CPT was selectively cleaved by endogenous carboxylesterase overexpressed in cancer cells, followed by self‐immolation to release free CPT. Therefore, the prodrug BDP−L−CPT exhibited excellent cytotoxicity against HepG2 and HeLa tumor cells by combining PDT and chemotherapy. The IC50 values were dropping to 0.11 μm for HepG2 cells and 0.13 μm for HeLa cells, which were significantly lower than those of BDP−COOH (0.13 and 0.15 μm, respectively) with only PDT effect. The remarkable combination antitumor efficacy of the prodrug was also obtained in ICR mouse H22 xenografts.
Figure 13.
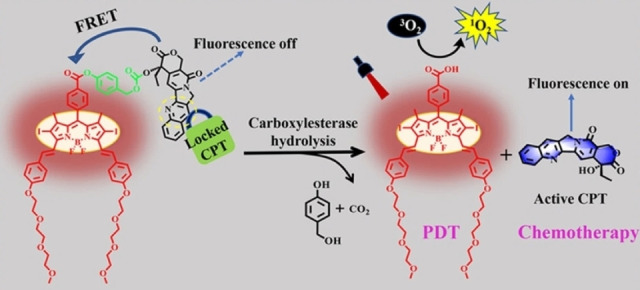
Schematic illustration of proposed drug release and action mechanism for the probe BDP−L−CPT. Reproduced with permission from Ref. [79]. Copyright 2022, American Chemical Society.
4. Summary and Outlook
Enzyme‐activatable fluorescent probes have been widely involved in the visualization and intervention of pathological processes owing to their unique advantages including good biocompatibility, tumor‐specificity, easy cellular metabolism, and high sensitivity. In this review, we have systematically summarized the design strategies, molecular structures, and biomedical applications of enzyme‐activatable organic fluorescent probes reported during the past five years. Although there has been substantial development of enzyme‐activatable fluorescent probes for the diagnosis and treatment of tumors in recent years, some challenges still need to be addressed in future.
The effective information obtained from in vivo imaging is still limited owing to the insufficient depth of tissue penetration in fluorescence imaging. Developing enzyme‐activatable probes by using fluorophores in the second near‐infrared region (1000 to 1700 nm) will be a feasible way to solve this problem. Enzyme‐responsive probes with multi‐mode imaging function in combination with magnetic resonance imaging will also be good weapons for the diagnosis of tumors in complex organisms.
Most of these probes that have a PDT function require abundant oxygen to produce singlet oxygen. However, most solid tumors are in hypoxic condition, limiting their therapeutic effectiveness. Using type‐I PSs to construct theranostic probes can be a promising strategy to overcome the tumor hypoxia via an oxygen‐independent PDT pathway.
The current applications of enzyme‐activatable fluorescent probes are relatively simple, and most of them are only used for imaging and treatment of cell and mouse models.
Therefore, in order to promote the clinical translation of these probes, interdisciplinary translational research with input from chemists, biologists, and medical personnel should be carried out to explore their potential in clinical uses.
Conflict of interest
The authors declare no conflict of interest.
5.
Biographical Information
Song‐Bo Lu received his B.Sc. degree from the Southern University of Science and Technology in 2017, and his M.Sc. degree from Harbin Institute of Technology in 2019. He is now pursuing his Ph.D. degree at the Department of Biomedical Engineering of Southern University of Science and Technology. His current research focuses on the development of organic photosensitizers for tumor theranostics.

Biographical Information
Luyao Wei received her Ph.D. degree from the Tsinghua‐Berkeley Shenzhen Institute of Tsinghua University in 2021. In 2022, she joined the Southern University of Science and Technology as a Postdoc in the Department of Biomedical Engineering. Her research interest is to explore optical imaging probes based on ultrasmall up‐conversion nanoparticles for biomedical applications.

Biographical Information
Wenjing He received her Ph.D. degree from the University of Science and Technology of China in 2021. She is currently working as a postdoctoral researcher in the Department of Biomedical Engineering at Southern University of Science and Technology. Her current research interest is focused on the design and synthesis of functional photosensitizers for biomedical applications.

Biographical Information
Kai Li received his Ph.D. degree in the Department of Chemical and Biomolecular Engineering from the National University of Singapore in 2011. He then joined the Institute of Materials Research and Engineering (A*STAR, Singapore) as a research scientist. He was a visiting scholar in the Molecular Imaging Program at Stanford from 2015 to 2017, working on stem cell tracking for translational research. In 2018, he joined the Southern University of Science and Technology as an Associate Professor in the Department of Biomedical Engineering. His research interest is the exploration of conjugated polymer/oligomer‐based optical probes for biomedical applications such as in vivo imaging.

Acknowledgements
The authors are grateful to the Ministry of Science and Technology of China (2020YFA0908900), the National Natural Science Foundation of China (U21 A2097, 31870991), the Department of Science and Technology of Guangdong Province (2022B1212010003, 2019ZT08Y191, 2019QN01Y640), the Shenzhen Key Laboratory of Smart Healthcare Engineering (ZDSYS20200811144003009), and the Shenzhen Science and Technology Program (JSGG20200225151916021, KQTD20190929172743294) for financial support.
S.-B. Lu, L. Wei, W. He, Z.-Y. Bi, Y. Qian, J. Wang, H. Lei, K. Li, ChemistryOpen 2022, 11, e202200137.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1. Sung H., Ferlay J., Siegel R. L., Laversanne M., Soerjomataram I., Jemal A., Bray F., Ca-Cancer J. Clin. 2021, 70, 7–30. [DOI] [PubMed] [Google Scholar]
- 2. Gao L., Zhang C., Gao D., Liu H., Yu X., Lai J., Wang F., Lin J., Liu Z., Theranostics 2016, 6, 627–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cardoso F., Senkus-Konefka E., Fallowfield L., Costa A., Castiglione M., Ann. Oncol. 2010, 21, v15-v19. [DOI] [PubMed] [Google Scholar]
- 4. Luger N. M., Mach D. B., Sevcik M. A., Mantyh P. W., J. Pain Symptom Manag. 2005, 29, 32–46. [DOI] [PubMed] [Google Scholar]
- 5. Yang K., Yang G., Chen L., Cheng L., Wang L., Ge C., Liu Z., Biomaterials 2015, 38, 1–9. [DOI] [PubMed] [Google Scholar]
- 6. Gaikwad H. K., Tsvirkun D., Ben-Nun Y., Merquiol E., Popovtzer R., Blum G., Nano Lett. 2018, 18, 1582–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Teng Z., Wang R., Zhou Y., Kolios M., Wang Y., Zhang N., Wang Z., Zheng Y., Lu G., Biomaterials 2017, 134, 43–50. [DOI] [PubMed] [Google Scholar]
- 8. Guo B., Feng Z., Hu D., Xu S., Middha E., Pan Y., Liu C., Zheng H., Qian J., Sheng Z., Liu B., Adv. Mater. 2019, 31, 1902504. [DOI] [PubMed] [Google Scholar]
- 9. Dzutsev A., Badger J. H., Perez-Chanona E., Roy S., Salcedo R., Smith C. K., Trinchieri G., Annu. Rev. Immunol. 2017, 35, 199–228. [DOI] [PubMed] [Google Scholar]
- 10.
- 10a. Benchimol S., Fuks A., Jothy S., Beauchemin N., Shirota K., Stanners C. P., Cell 1989, 57, 327–334; [DOI] [PubMed] [Google Scholar]
- 10b. Grunnet M., Sorensen J. B., Lung Cancer 2012, 76, 138–143. [DOI] [PubMed] [Google Scholar]
- 11.
- 11a. Sharma A., Kim E.-J., Shi H., Lee J. Y., Chung B. G., Kim J. S., Biomaterials 2018, 155, 145–151; [DOI] [PubMed] [Google Scholar]
- 11b. Kawatani M., Yamamoto K., Yamada D., Kamiya M., Miyakawa J., Miyama Y., Kojima R., Morikawa T., Kume H., Urano Y., J. Am. Chem. Soc. 2019, 141, 10409–10416; [DOI] [PubMed] [Google Scholar]
- 11c. Miao Q., Yeo D. C., Wiraja C., Zhang J., Ning X., Xu C., Pu K., Angew. Chem. Int. Ed. 2018, 57, 1256–1260; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2018, 130, 1270–1274. [Google Scholar]
- 12.
- 12a. Zhu H., Zhang Z., Long S., Du J., Fan J., Peng X., Nat. Protoc. 2018, 13, 2348–2361; [DOI] [PubMed] [Google Scholar]
- 12b. Li H., Yao Q., Fan J., Du J., Wang J., Peng X., Biosens. Bioelectron. 2017, 94, 536–543. [DOI] [PubMed] [Google Scholar]
- 13.
- 13a. Sedgwick A. C., Wu L., Han H.-H., Bull S. D., He X.-P., James T. D., Sessler J. L., Tang B. Z., Tian H., Yoon J., Chem. Soc. Rev. 2018, 47, 8842–8880; [DOI] [PubMed] [Google Scholar]
- 13b. Wang C., Qiao Q., Chi W., Chen J., Liu W., Tan D., McKechnie S., Lyu D., Jiang X.-F., Zhou W., Xu N., Zhang Q., Xu Z., Liu X., Angew. Chem. Int. Ed. 2020, 59, 10160–10172; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2020, 132, 10246–10258; [Google Scholar]
- 13c. Park S.-H., Kwon N., Lee J.-H., Yoon J., Shin I., Chem. Soc. Rev. 2020, 49, 143–179; [DOI] [PubMed] [Google Scholar]
- 13d. Li Y., Zha M., Kang T., Li C., Wu X., Wang S., Lu S.-B., Lee Y.-S., Wu Y.-R., Ni J.-S., Li K., Small 2022, 18, 2105362; [DOI] [PubMed] [Google Scholar]
- 13e. Li Y., Zhou H., Bi R., Li X., Zha M., Yang Y., Ni J.-S., Liew W. H., Olivo M., Yao K., Liu J., Chen H., Li K., J. Mater. Chem. B 2021, 9, 9951–9960; [DOI] [PubMed] [Google Scholar]
- 13f. Li Y., Zha M., Yang G., Wang S., Ni J.-S., Li K., Chem. Eur. J. 2021, 27, 13085–13091; [DOI] [PubMed] [Google Scholar]
- 13g. Li Y., Li Z., Hu D., Wang S., Zha M., Lu S.-B., Sheng Z., Li K., Chem. Commun. 2021, 57, 6420–6423; [DOI] [PubMed] [Google Scholar]
- 13h. Yang G., Ni J.-S., Li Y., Zha M., Tu Y., Li K., Angew. Chem. Int. Ed. 2021, 60, 5386–5393; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2021, 133, 5446–5453; [Google Scholar]
- 13i. Zhu W., Kang M., Wu Q., Zhang Z., Wu Y., Li C., Li K., Wang L., Wang D., Tang B. Z., Adv. Funct. Mater. 2021, 31, 202007026; [Google Scholar]
- 13j. Ma Y.-L., Sun C., Li Z., Wang Z., Wei J., Cheng Q., Zheng L.-S., Chang X.-Y., Li K., Wang R., Jiang W., CCS Chem. 2021, 3, 2143–2155; [Google Scholar]
- 13k. Li Y., Hu D., Sheng Z., Min T., Zha M., Ni J.-S., Zheng H., Li K., Biomaterials 2021, 264, 120365. [DOI] [PubMed] [Google Scholar]
- 14.
- 14a. Mochida A., Ogata F., Nagaya T., Choyke P. L., Kobayashi H., Bioorg. Med. Chem. 2018, 26, 925–930; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14b. Ni J.-S., Li Y., Yue W., Liu B., Li K., Theranostics 2020, 10, 1923–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.
- 15a. Lovell J. F., Liu T. W. B., Chen J., Zheng G., Chem. Rev. 2010, 110, 2839–2857; [DOI] [PubMed] [Google Scholar]
- 15b. Qin H., Ding Y., Mujeeb A., Zhao Y., Nie G., Mol. Pharmacol. 2017, 92, 219–231; [DOI] [PubMed] [Google Scholar]
- 15c. Li H., Kim D., Yao Q., Ge H., Chung J., Fan J., Wang J., Peng X., Yoon J., Angew. Chem. Int. Ed. 2021, 60, 17268–17289; [DOI] [PubMed] [Google Scholar]
- 15d. Hu F., Yuan Y., Mao D., Wu W., Liu B., Biomaterials 2017, 144, 53–59. [DOI] [PubMed] [Google Scholar]
- 16.
- 16a. Feng Z., Guo J., Liu X., Song H., Zhang C., Huang P., Dong A., Kong D., Wang W., Biomaterials 2020, 255, 120210; [DOI] [PubMed] [Google Scholar]
- 16b. Liu Y., Pan Y., Cao W., Xia F., Liu B., Niu J., Alfranca G., Sun X., Ma L., de la Fuente J. M., Song J., Ni J., Cui D., Theranostics 2019, 9, 6867–6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hu D., Sheng Z., Gao G., Siu F., Liu C., Wan Q., Gong P., Zheng H., Ma Y., Cai L., Biomaterials 2016, 93, 10–19. [DOI] [PubMed] [Google Scholar]
- 18. Fan W., Shen B., Bu W., Zheng X., He Q., Cui Z., Ni D., Zhao K., Zhang S., Shi J., Biomaterials 2015, 69, 89–98. [DOI] [PubMed] [Google Scholar]
- 19. Huang C., Zheng J., Ma D., Liu N., Zhu C., Li J., Yang R., J. Mater. Chem. B 2018, 6, 6424–6430. [DOI] [PubMed] [Google Scholar]
- 20. Kim M. M., Darafsheh A., Photochem. Photobiol. 2020, 96, 280–294. [DOI] [PubMed] [Google Scholar]
- 21.
- 21a. Dolmans D. E. J. G. J., Fukumura D., Jain R. K., Nat. Rev. Cancer 2003, 3, 380–387; [DOI] [PubMed] [Google Scholar]
- 21b. Jiao X., Li Y., Niu J., Xie X., Wang X., Tang B., Anal. Chem. 2018, 90, 533–555. [DOI] [PubMed] [Google Scholar]
- 22.
- 22a. Hwang H. S., Shin H., Han J., Na K., J. Pharm. Investig. 2018, 48, 143–151; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22b. Yang G., Lu S.-B., Li C., Chen F., Ni J.-S., Zha M., Li Y., Gao J., Kang T., Liu C., Li K., Chem. Sci. 2021, 12, 14773–14780; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22c. Zha M., Yang G., Li Y., Zhang C., Li B., Li K., Adv. Healthcare Mater. 2021, 10, 2101066. [DOI] [PubMed] [Google Scholar]
- 23. Cao Z., Li W., Liu R., Li X., Li H., Liu L., Chen Y., Lv C., Liu Y., Biomed. Pharmacother. 2019, 118, 109340. [DOI] [PubMed] [Google Scholar]
- 24. Sun W., Li M., Fan J., Peng X., Acc. Chem. Res. 2019, 52, 2818–2831. [DOI] [PubMed] [Google Scholar]
- 25.
- 25a. Chi W., Chen J., Liu W., Wang C., Qi Q., Qiao Q., Tan T. M., Xiong K., Liu X., Kang K., Chang Y.-T., Xu Z., Liu X., J. Am. Chem. Soc. 2020, 142, 6777–6785; [DOI] [PubMed] [Google Scholar]
- 25b. Lee M. H., Kim J. S., Sessler J. L., Chem. Soc. Rev. 2015, 44, 4185–4191; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25c. Yuan L., Lin W., Zheng K., Zhu S., Acc. Chem. Res. 2013, 46, 1462–1473; [DOI] [PubMed] [Google Scholar]
- 25d. Tu Y., Liu J., Zhang H., Peng Q., Lam J. W. Y., Tang B. Z., Angew. Chem. Int. Ed. 2019, 58, 14911–14914; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2019, 131, 15053–15056. [Google Scholar]
- 26.
- 26a. Chiba M., Ichikawa Y., Kamiya M., Komatsu T., Ueno T., Hanaoka K., Nagano T., Lange N., Urano Y., Angew. Chem. Int. Ed. 2017, 56, 10418–10422; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2017, 129, 10554–10558; [Google Scholar]
- 26b. He S., Li J., Lyu Y., Huang J., Pu K., J. Am. Chem. Soc. 2020, 142, 7075–7082; [DOI] [PubMed] [Google Scholar]
- 26c. Lee H. W., Lim C. S., Choi H., Cho M. K., Noh C.-K., Lee K., Shin S. J., Kim H. M., Anal. Chem. 2019, 91, 14705–14711; [DOI] [PubMed] [Google Scholar]
- 26d. Jung H. S., Lee J.-H., Kim K., Koo S., Verwilst P., Sessler J. L., Kang C., Kim J. S., J. Am. Chem. Soc. 2017, 139, 9972–9978; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26e. Li H., Yao Q., Xu F., Li Y., Kim D., Chung J., Baek G., Wu X., Hillman P. F., Lee E. Y., Ge H., Fan J., Wang J., Nam S.-J., Peng X., Yoon J., Angew. Chem. Int. Ed. 2020, 59, 10186–10195; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2020, 132, 10272–10281; [Google Scholar]
- 26f. Li H., Yao Q., Sun W., Shao K., Lu Y., Chung J., Kim D., Fan J., Long S., Du J., Li Y., Wang J., Yoon J., Peng X., J. Am. Chem. Soc. 2020, 142, 6381–6389; [DOI] [PubMed] [Google Scholar]
- 26g. Zhou X., Li H., Shi C., Xu F., Zhang Z., Yao Q., Ma H., Sun W., Shao K., Du J., Long S., Fan J., Wang J., Peng X., Biomaterials 2020, 253, 120089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.
- 27a. Zhang J., Chai X., He X.-P., Kim H.-J., Yoon J., Tian H., Chem. Soc. Rev. 2019, 48, 683–722; [DOI] [PubMed] [Google Scholar]
- 27b. Liu H.-W., Chen L., Xu C., Li Z., Zhang H., Zhang X.-B., Tan W., Chem. Soc. Rev. 2018, 47, 7140–7180; [DOI] [PubMed] [Google Scholar]
- 27c. Wu X., Wang R., Kwon N., Ma H., Yoon J., Chem. Soc. Rev. 2022, 51, 450–463. [DOI] [PubMed] [Google Scholar]
- 28. Zhang Z., Yang J., Lu J., Lin J., Zeng S., Luo Q., J. Biomed. Opt. 2008, 13, 011006. [DOI] [PubMed] [Google Scholar]
- 29.
- 29a. Nakagawara A., Nakamura Y., Ikeda H., Hiwasa T., Kuida K., Su M. S., Zhao H., Cnaan A., Sakiyama S., Cancer Res. 1997, 57, 4578–4584; [PubMed] [Google Scholar]
- 29b. Persad R., Liu C., Wu T.-T., Houlihan P. S., Hamilton S. R., Diehl A. M., Rashid A., Mod. Pathol. 2004, 17, 861–867; [DOI] [PubMed] [Google Scholar]
- 29c. Stefflova K., Chen J., Marotta D., Li H., Zheng G., J. Med. Chem. 2006, 49, 3850–3856; [DOI] [PubMed] [Google Scholar]
- 29d. Liu P.-F., Hu Y.-C., Kang B.-H., Tseng Y.-K., Wu P.-C., Liang C.-C., Hou Y.-Y., Fu T.-Y., Liou H.-H., Hsieh I−C., Ger L.-P., Shu C.-W., PLoS ONE 2017, 12, e0180620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu R., Li H., Liu L., Yu J., Ren X., Cancer Biol. Ther. 2012, 13, 123–129. [DOI] [PubMed] [Google Scholar]
- 31.
- 31a. Jedeszko C., Sloane B. F., Biol. Chem. 2004, 385, 1017–1027; [DOI] [PubMed] [Google Scholar]
- 31b. Blum G., von Degenfeld G., Merchant M. J., Blau H. M., Bogyo M., Nat. Chem. Biol. 2007, 3, 668–677. [DOI] [PubMed] [Google Scholar]
- 32.
- 32a. Thomas G., Nat. Rev. Mol. Cell Biol. 2002, 3, 753–766; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32b. Jia H., Ding D., Hu J., Dai J., Yang J., Li G., Lou X., Xia F., Adv. Mater. 2021, 2104615. [DOI] [PubMed] [Google Scholar]
- 33.J. B. Birks, Photophysics of aromatic molecules, Wiley, Hoboken, NJ, USA 1970.
- 34. Luo J., Xie Z., Lam J. W. Y., Cheng L., Chen H., Qiu C., Kwok H. S., Zhan X., Liu Y., Zhu D., Tang B. Z., Chem. Commun. 2001, 1740–1741. [DOI] [PubMed] [Google Scholar]
- 35. Kessenbrock K., Plaks V., Werb Z., Cell 2010, 141, 52–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.
- 36a. Giambernardi T. A., Grant G. M., Taylor G. P., Hay R. J., Maher V. M., McCormick J. J., Klebe R. J., Matrix Biol. 1998, 16, 483–496; [DOI] [PubMed] [Google Scholar]
- 36b. Murnane M. J., Cai J., Shuja S., McAneny D., Klepeis V., Willett J. B., Int. J. Cancer 2009, 125, 2893–2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.
- 37a. Bremer C., Bredow S., Mahmood U., Weissleder R., Tung C. H., Radiology 2001, 221, 523–529; [DOI] [PubMed] [Google Scholar]
- 37b. Van Valckenborgh E., Mincher D., Di Salvo A., Van Riet I., Young L., Van Camp B., Vanderkerken K., Leukemia 2005, 19, 1628–1633; [DOI] [PubMed] [Google Scholar]
- 37c. Kline T., Torgov M. Y., Mendelsohn B. A., Cerveny C. G., Senter P. D., Mol. Pharm. 2004, 1, 9–22. [DOI] [PubMed] [Google Scholar]
- 38. Stallivieri A., Colombeau L., Devy J., Etique N., Chaintreuil C., Myrzakhmetov B., Achard M., Baros F., Arnoux P., Vanderesse R., Frochot C., Bioorg. Med. Chem. 2018, 26, 688–702. [DOI] [PubMed] [Google Scholar]
- 39. Bu L.-L., Wang H.-Q., Pan Y., Chen L., Wu H., Wu X., Zhao C., Rao L., Liu B., Sun Z.-J., J. Nanobiotechnol. 2021, 19, 379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Meng X., Song J., Lei Y., Zhang X., Chen Z., Lu Z., Zhang L., Wang Z., Biomater. Sci. 2021, 9, 7456–7470. [DOI] [PubMed] [Google Scholar]
- 41.
- 41a. Chen J., Stefflova K., Niedre M. J., Wilson B. C., Chance B., Glickson J. D., Zheng G., J. Am. Chem. Soc. 2004, 126, 11450–11451; [DOI] [PubMed] [Google Scholar]
- 41b. Li S.-Y., Cheng H., Xie B.-R., Qiu W.-X., Song L.-L., Zhuo R.-X., Zhang X.-Z., Biomaterials 2016, 104, 297–309. [DOI] [PubMed] [Google Scholar]
- 42. Li H., Parigi G., Luchinat C., Meade T. J., J. Am. Chem. Soc. 2019, 141, 6224–6233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. An H.-W., Li L.-L., Wang Y., Wang Z., Hou D., Lin Y.-X., Qiao S.-L., Wang M.-D., Yang C., Cong Y., Ma Y., Zhao X.-X., Cai Q., Chen W.-T., Lu C.-Q., Xu W., Wang H., Zhao Y., Nat. Commun. 2019, 10, 4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mueller M. M., Fusenig N. E., Nat. Rev. Cancer 2004, 4, 839–849. [DOI] [PubMed] [Google Scholar]
- 45.
- 45a. Orimo A., Gupta P. B., Sgroi D. C., Arenzana-Seisdedos F., Delaunay T., Naeem R., Carey V. J., Richardson A. L., Weinberg R. A., Cell 2005, 121, 335–348; [DOI] [PubMed] [Google Scholar]
- 45b. Chen W.-T., Kelly T., Cancer Metastasis Rev. 2003, 22, 259–269. [DOI] [PubMed] [Google Scholar]
- 46. Li J., Chen K., Liu H., Cheng K., Yang M., Zhang J., Cheng J. D., Zhang Y., Cheng Z., Bioconjugate Chem. 2012, 23, 1704–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhao X.-X., Li L.-L., Zhao Y., An H.-W., Cai Q., Lang J.-Y., Han X.-X., Peng B., Fei Y., Liu H., Qin H., Nie G., Wang H., Angew. Chem. Int. Ed. 2019, 58, 15287–15294; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2019, 131, 15431–15438. [Google Scholar]
- 48. Verbovsek U., Van Noorden C. J., Lah T. T., Semin. Cancer Biol. 2015, 35, 71–84. [DOI] [PubMed] [Google Scholar]
- 49.
- 49a. Anja P., Anahid J., Janko K., Semin. Cancer Biol. 2018, 53, 168–177;30063965 [Google Scholar]
- 49b. Qian F., Bajkowski A. S., Steiner D. F., Chan S. J., Frankfater A., Cancer Res. 1989, 49, 4870–4875. [PubMed] [Google Scholar]
- 50. Chen X., Lee D., Yu S., Kim G., Lee S., Cho Y., Jeong H., Nam K. T., Yoon J., Biomaterials 2017, 122, 130–140. [DOI] [PubMed] [Google Scholar]
- 51. Hu C., Zhuang W., Yu T., Chen L., Liang Z., Li G., Wang Y., J. Mater. Chem. B 2020, 8, 5267–5279. [DOI] [PubMed] [Google Scholar]
- 52. Li X., Cao C., Wei P., Xu M., Liu Z., Liu L., Zhong Y., Li R., Zhou Y., Yi T., ACS Appl. Mater. Interfaces 2019, 11, 12327–12334. [DOI] [PubMed] [Google Scholar]
- 53. Liu X., Liang G., Chem. Commun. 2017, 53, 1037–1040. [DOI] [PubMed] [Google Scholar]
- 54.
- 54a. Wolf P. L., Clin J., Lab. Anal. 1994, 8, 172–176; [DOI] [PubMed] [Google Scholar]
- 54b. Rao S. R., Snaith A. E., Marino D., Cheng X., Lwin S. T., Orriss I. R., Hamdy F. C., Edwards C. M., Br. J. Cancer 2017, 116, 227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Belinsky M., Jaiswal A. K., Cancer Metastasis Rev. 1993, 12, 103–117. [DOI] [PubMed] [Google Scholar]
- 56.
- 56a. Verwilst P., Han J., Lee J., Mun S., Kang H.-G., Kim J. S., Biomaterials 2017, 115, 104–114; [DOI] [PubMed] [Google Scholar]
- 56b. Tian Y., Li Y., Jiang W.-L., Zhou D.-Y., Fei J., Li C.-Y., Anal. Chem. 2019, 91, 10901–10907. [DOI] [PubMed] [Google Scholar]
- 57. Coleman J. E., Annu. Rev. Bioph. Biom. 1992, 21, 441–483. [DOI] [PubMed] [Google Scholar]
- 58. Liu H.-W., Hu X.-X., Zhu L., Li K., Rong Q., Yuan L., Zhang X.-B., Tan W., Talanta 2017, 175, 421–426. [DOI] [PubMed] [Google Scholar]
- 59. Zhang X., Chen X., Liu K., Zhang Y., Gao G., Huang X., Hou S., Anal. Chim. Acta 2020, 1094, 113–121. [DOI] [PubMed] [Google Scholar]
- 60. Li H., Yao Q., Xu F., Li Y., Kim D., Chung J., Baek G., Wu X., Hillman P. F., Lee E. Y., Ge H., Fan J., Wang J., Nam S.-J., Peng X., Yoon J., Angew. Chem. Int. Ed. 2020, 59, 10186–10195; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2020, 132, 10272–10281. [Google Scholar]
- 61.
- 61a. Zhang C., Zhai B.-B., Peng T., Zhong Z., Xu L., Zhang Q.-Z., Li L.-Y., Yi L., Xi Z., Dyes Pigm. 2017, 143, 245–251; [Google Scholar]
- 61b. Kwon N., Cho M. K., Park S. J., Kim D., Nam S.-J., Cui L., Kim H. M., Yoon J., Chem. Commun. 2017, 53, 525–528. [DOI] [PubMed] [Google Scholar]
- 62. Shen Z., Prasai B., Nakamura Y., Kobayashi H., Jackson M. S., McCarley R. L., ACS Chem. Biol. 2017, 12, 1121–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Fei Q., Zhou L., Wang F., Shi B., Li C., Wang R., Zhao C., Dyes Pigm. 2017, 136, 846–851. [Google Scholar]
- 64. Kiyose K., Hanaoka K., Oushiki D., Nakamura T., Kajimura M., Suematsu M., Nishimatsu H., Yamane T., Terai T., Hirata Y., J. Am. Chem. Soc. 2010, 132, 15846–15848. [DOI] [PubMed] [Google Scholar]
- 65. Huang J., Wu Y., Zeng F., Wu S., Theranostics 2019, 9, 7313–7324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhao X.-B., Ha W., Gao K., Shi Y.-P., Anal. Chem. 2020, 92, 9039–9047. [DOI] [PubMed] [Google Scholar]
- 67. Rolff M., Schottenheim J., Decker H., Tuczek F., Chem. Soc. Rev. 2011, 40, 4077–4098. [DOI] [PubMed] [Google Scholar]
- 68. Osella-Abate S., Savoia P., Quaglino P., Fierro M. T., Leporati C., Ortoncelli M., Bernengo M. G., Br. J. Cancer 2003, 89, 1457–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Li Z., Wang Y.-F., Zeng C., Hu L., Liang X.-J., Anal. Chem. 2018, 90, 3666–3669. [DOI] [PubMed] [Google Scholar]
- 70. Zhan C., Cheng J., Li B., Huang S., Zeng F., Wu S., Anal. Chem. 2018, 90, 8807–8815. [DOI] [PubMed] [Google Scholar]
- 71.
- 71a. Vaupel P., Thews O., Hoeckel M., Med. Oncol. 2001, 18, 243–259; [DOI] [PubMed] [Google Scholar]
- 71b. Liu Y., Teng L., Chen L., Ma H., Liu H.-W., Zhang X.-B., Chem. Sci. 2018, 9, 5347–5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kumar R., Kim E.-J., Han J., Lee H., Shin W. S., Kim H. M., Bhuniya S., Kim J. S., Hong K. S., Biomaterials 2016, 104, 119–128. [DOI] [PubMed] [Google Scholar]
- 73. Xu F., Li H., Yao Q., Ge H., Fan J., Sun W., Wang J., Peng X., Chem. Sci. 2019, 10, 10586–10594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ouyang J., Sun L., Zeng Z., Zeng C., Zeng F., Wu S., Angew. Chem. Int. Ed. 2020, 59, 10111–10121; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2020, 132, 10197–10207. [Google Scholar]
- 75.
- 75a. Reichl P., Mikulits W., Oncol. Rep. 2016, 36, 613–625; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75b. Xu G., Zhang W., Ma M. K., McLeod H. L., Clin. Cancer Res. 2002, 8, 2605–2611. [PubMed] [Google Scholar]
- 76. Cai Y., Ni D., Cheng W., Ji C., Wang Y., Müllen K., Su Z., Liu Y., Chen C., Yin M., Angew. Chem. Int. Ed. 2020, 59, 14014–14018; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2020, 132, 14118–14122. [Google Scholar]
- 77. Chen P., Kuang W., Zheng Z., Yang S., Liu Y., Su L., Zhao K., Liang G., Theranostics 2019, 9, 7359–7369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wu X., Wang R., Qi S., Kwon N., Han J., Kim H., Li H., Yu F., Yoon J., Angew. Chem. Int. Ed. 2021, 60, 15418–15425. [DOI] [PubMed] [Google Scholar]
- 79. Zhang H.-X., Lin H.-H., Su D., Yang D.-C., Liu J.-Y., Mol. Pharmaceutics 2022, 19, 630–641. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


