ABSTRACT
The gut microbiota plays a fundamental role in human nutrition and metabolism and may have direct implications for type 2 diabetes and associated preconditions. An improved understanding of relations between human gut microbiota and glucose metabolism could lead to novel opportunities for type 2 diabetes prevention, but human observational studies reporting on such findings have not been extensively reviewed. Here, we review the literature on associations between gut microbiota and markers and stages of glucose dysregulation and insulin resistance in healthy adults and in adults with metabolic disease and risk factors. We present the current evidence for identified key bacteria and their potential roles in glucose metabolism independent of overweight, obesity, and metabolic drugs. We provide support for SCFAs mediating such effects and discuss the role of diet, as well as metabolites derived from diet and gut microbiota interactions. From 5983 initially identified PubMed records, 45 original studies were eligible and reviewed. α Diversity and 45 bacterial taxa were associated with selected outcomes. Six taxa were most frequently associated with glucose metabolism: Akkermansia muciniphila, Bifidobacterium longum, Clostridium leptum group, Faecalibacterium prausnitzii, and Faecalibacterium (inversely associated) and Dorea (directly associated). For Dorea and A. muciniphila, associations were independent of metabolic drugs and body measures. For A. muciniphila and F. prausnitzii, limited evidence supported SCFA mediation of potential effects on glucose metabolism. We conclude that observational studies applying metagenomics sequencing to identify species-level relations are warranted, as are studies accounting for confounding factors and investigating SCFA and postprandial glucose metabolism. Such advances in the field will, together with mechanistic and prospective studies and investigations into diet–gut microbiota interactions, have the potential to bring critical insight into roles of gut microbiota and microbial metabolites in human glucose metabolism and to contribute toward the development of novel prevention strategies for type 2 diabetes, including precision nutrition.
Keywords: glucose metabolism, gut microbiota, humans, insulin resistance, prediabetes, short-chain fatty acids, microbial metabolites, diet–gut microbiota interactions, type 2 diabetes prevention, precision nutrition
Introduction
Mounting evidence has linked the gut microbiota to health status in humans. It has become evident that gut microbes play a fundamental role in human nutrition and metabolism (1, 2) and that alterations in microbiota composition, diversity, and function may have direct implications for metabolic derangements including type 2 diabetes and associated preconditions (1, 3). This insight has led to extensive research to identify microbial taxa and functions which could dictate or be targets for preventative actions and treatment, including dietary strategies (4–6). At present, we recognize that specific dietary components, such as certain dietary fibers, can increase the abundance of specific gut bacteria (7, 8) or result in differential health responses determined by specific bacteria (9). Yet, it is unknown which gut bacteria, and if bacteria determined a priori, can be utilized for precision prevention in the general population or in different risk groups.
Type 2 diabetes is a global epidemic and prevalence is projected to increase (10, 11). Human observational studies show significant differences in the composition and function of the gut microbiota between healthy individuals and people with prevalent type 2 diabetes (3, 12, 13). However, these observations could partly or entirely be driven by antidiabetic medication, drugs for common metabolic comorbidities (14, 15), overweight or obesity (16, 17), and other confounding factors. Causal evidence for the role of gut microbiota in the etiology of type 2 diabetes is mostly limited to animal studies and it remains unclear in what specific processes of the pathogenesis gut microbiota may be involved (3).
Perturbed glucose metabolism plays a central role in type 2 diabetes development. Interactions between the gut microbiota and diet, especially dietary fibers, are closely linked with glucose regulation (18–22), potentially through mechanisms involving microbial metabolites including, but not limited to, SCFAs and succinate (13, 23–25). Novel insights into relations between gut microbiota, microbial metabolites, and dysregulations in glucose metabolism can contribute to the understanding of the role of gut microbiota in type 2 diabetes development and lead to novel opportunities for prevention. Yet, despite the increasing number of human observational studies in this field, this literature has not been extensively reviewed.
The aim of this scoping review was to identify and summarize the evidence for gut bacteria most frequently and consistently associating with markers and stages of glucose dysregulation and insulin resistance in adults with and without metabolic disease and common metabolic risk factors. We assessed if there was evidence supporting a role for these bacteria in glucose metabolism independent of overweight, obesity, and metabolic drugs and the potential mediation by SCFAs and succinate. Finally, we identified limitations in the current literature and provide future directives that could contribute toward the development of novel microbiota-based prevention strategies for type 2 diabetes, including precision nutrition.
Methods
A scoping review was conducted through PubMed literature searches to capture studies reporting on associations between fecal bacteria at different taxonomic levels, as well as α diversity, and diabetes risk outcomes related to blood glucose and insulin. Glucose-related outcomes included fasting and postprandial glucose, glycated hemoglobin (HbA1c), and presence of prediabetes as opposed to normoglycemia. Insulin-related outcomes included fasting and postprandial insulin concentrations and insulin resistance, e.g., HOMA-IR, insulin sensitivity measured with euglycemic clamp, and presence of insulin resistance.
The included literature was identified and reviewed based on defined search strategies, criteria, and research questions defined using the PICo (population or problem, interest, and context) framework (26, 27) and the Preferred Reporting Items for Systematic Reviews and Meta-Analyses extension for Scoping Reviews (PRISMA-ScR) checklist (Supplemental Figure 1, Supplemental Table 1). No protocol was registered or published a priori. PubMed-specific title and abstract words (TiAb) and Medical Subject Headings (MeSH) search terms were used for title and abstract words and study index categories, respectively (Supplemental Tables 2–4). Three literature searches acted complementarily: 1 initial search (22 November, 2019) and 2 searches specified to capture the most recently published literature (14 June, 2021) and to expand the review on associations with postprandial glucose and insulin (21 December, 2021), respectively (Supplemental Tables 2–4). Duplicate articles were removed using the Amsterdam Efficient Deduplication (AED) method (28). The gut microbiota–related search terms were general to allow for identification of any relevant associations for fecal bacteria regardless of prior knowledge. A broad range of study designs were eligible, including intervention studies from which baseline associations were extracted. For quality control purposes, studies with populations or groups (for group-based comparisons) with ≤5 participants were excluded (Supplemental Table 1). Studies were also excluded if populations had undergone recent treatments that could influence the gut microbiota (e.g., antibiotic or antidiarrheal treatment) or presented with extreme bowel habits or gastrointestinal symptoms. Other exclusion criteria were non-English publications and studies with populations with diseases other than type 2 diabetes or cardiovascular disease or traits that were considered nonrepresentative of the general population, e.g., professional athletes and pregnant females.
Records were first assessed based on titles and abstracts using Rayyan QCRI (29); each record was evaluated by 2 individuals in a blinded manner and conflicting assessments were resolved upon discussion or assessment of the full text. Eligible full texts were read by ≥1 individuals. Each included study was charted through an independent process and a piloted form and extracted results were limited to statistically significant results and, if applicable, results that remained statistically significant after adjusting for multiple comparisons (Supplemental Table 5). Adjustments, including adjustments for diet, sex, and age, were noted for each included article, as were whether studies reported on SCFAs, SCFA-associated enzymes, other metabolites, or incretins. Results were only included if there were ≥2 studies supporting an inverse/direct association with either glucose- or insulin-related outcomes. Any studies reporting on contradictory findings were also included. For example, if 2 studies found direct associations between a bacterium and glucose outcomes, and 1 study found an inverse association, all 3 studies were reviewed and reported. Taxa were reported using the name recognized by NCBI Taxonomy (30) at time of submission, bacteria reported under different synonyms were merged, and any strain-level results were reported as species. Notably, results for “Clostridium leptum” were reported as “C. leptum group” when group-specific qPCR primers had been used, because the C. leptum group (also known as Clostridial cluster IV) includes other species and is predominated by Faecalibacterium prausnitzii, not C. leptum (31).
Charted data on study characteristics were summarized and visualized as a bar plot. Taxonomic trees were generated using PhyloT version 2 (32) and visualized and annotated using iTOL version 6 (33). Venn diagrams were created using tools available at http://bioinformatics.psb.ugent.be.
Results
Characteristics of reviewed studies
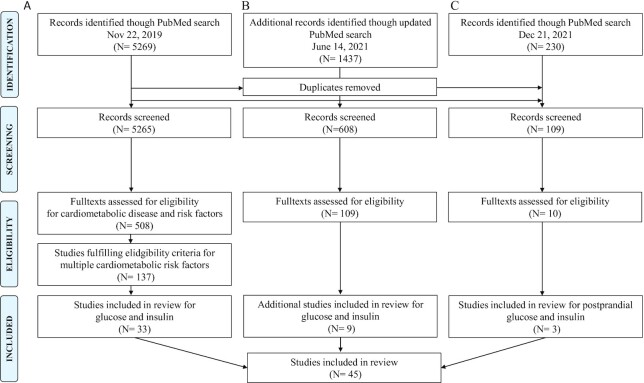
A total of 45 studies out of 5982 initially identified records were eligible and included in this review (Figure 1, Supplemental Table 5). This included 20 cross-sectional studies, 15 case–control studies, and 10 intervention-based designs, many with study populations ranging from 100 to 2000 participants (Figure 2) and the majority published in the past 5 y (n = 33). Most studies (n = 40) included females and males and had 1 study location, but 2 studies included populations from different countries (34, 35) and 1 study included a discovery cohort and a validation cohort based in the same country (Sweden) (36). Asian (n = 21) and European (n = 15) populations were overrepresented. Most studies (n = 28) had applied 16S ribosomal RNA (rRNA) gene amplification and sequencing to measure the gut microbiota, fewer had used metagenomic shotgun sequencing (n = 11) or qPCR (n = 6). Only 3 studies had investigated postprandial glucose and insulin response in relation to gut microbiota (18, 37, 38), 1 of which also reported on associations with fasting glucose and insulin concentrations (37) (Supplemental Figure 2B).
FIGURE 1.
Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) flow diagram summarizing the process of screening and identification of the literature in the scoping review. (A) Original literature search. (B) Updated literature search specified to capture recent studies reporting on associations with glucose and insulin and differences across glycemic and insulinemic groups. (C) Literature search specified for postprandial glucose and insulin.
FIGURE 2.
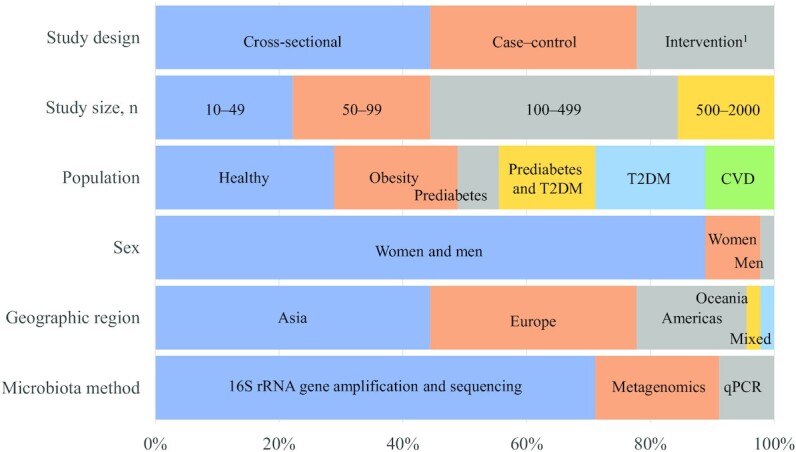
Bar plot of main characteristics of reviewed studies (n = 45). Data are presented as percentages of studies belonging to the different categories of study design, study size, type of population with regards to metabolic disease and obesity, sex, geographical country of population, and main type of methods measuring the gut microbiota. 1Only baseline results from intervention studies were included. CVD, cardiovascular disease; rRNA, ribosomal RNA; T2DM, type 2 diabetes mellitus.
Bacteria associated with glucose metabolism outcomes and their taxonomic relations
In total, 45 taxa (phyla, orders, families, genera, and species) and α diversity were identified to be related to the studied outcomes (Tables 1 and 2, Supplemental Figure 2A). Out of these, 40 taxa were associated with glucose and 17 were associated with insulin. Most taxa were presented as species. Based on the totality of the studies included, we found that F. prausnitzii was inversely associated with both fasting (39–41) and postprandial insulin (37, 38) and HOMA-IR (42). Enterobacteriaceae had a direct association with fasting (35, 43) and postprandial glucose (18) and Haemophilus parainfluenzae was inversely associated with fasting glucose and HbA1c (40), prediabetes (44), and postprandial glucose (37) (Tables 1 and 2, Supplemental Table 5). [Ruminococcus] gnavus had a higher abundance in groups with prediabetes (45) and glucose intolerance (36) and was elevated with postprandial glucose (37).
TABLE 1.
Summary of reported associations between bacteria on different taxonomic levels and glucose-related outcomes1
| Gut microbiota | Glucose2 | Postprandial glucose | |||
|---|---|---|---|---|---|
| Unadjusted association | Adjusted for body measures3 | Adjusted for metabolic drugs4 | SCFAs or enzymes measured | ||
| Phyla | |||||
| Bacteroidetes | ↓ (48, 88) | ||||
| Firmicutes | ↓ (48, 60) | ↓ (39) | (60) | ||
| ↑ (89) | ↑ (39) | ||||
| Synergistetes | ↑ (90) | ↑ (51) | ↑ (51) | ||
| Order | |||||
| Eubacteriales5 | ↓ (45) | ↓ (41, 45) | |||
| ↑ (39) | |||||
| Families | |||||
| Christensenellaceae | ↓ (73) | ↓ (45) | ↓ (45) | (73) | |
| Enterobacteriaceae | ↑ (43) | ↑ (35) | (35, 43) | ↑ (18) | |
| Lachnospiraceae | ↓ (39, 45) | ||||
| ↑ (34) | ↑ (34) | ||||
| Oscillospiraceae6 | ↓ (43, 67) | ↓ (45) | (43) | ||
| ↑ (35) | ↑ (34) | ↑ (34) | (35) | ||
| Veillonellaceae | ↓ (35) | (35) | |||
| ↑ (43) | ↑ (34) | (43) | |||
| Genera | |||||
| Akkermansia | ↓ (53) | ↓ (52) | |||
| Alistipes | ↓ (61, 91) | ||||
| Anaerostipes | ↑ (73, 92) | (73) | |||
| Bacteroides | ↓ (92) | ↓ (45) | ↓ (45) | ||
| ↑ (34) | ↑ (34) | ||||
| Blautia | ↓ (93) | ↓ (45) | |||
| ↑ (34) | ↑ (34, 39, 45) | ||||
| Clostridium | ↓ (39, 45) | ||||
| ↑ (39) | |||||
| Dorea | ↑ (34, 39, 78) | ↑ (34, 39, 45, 78) | |||
| Escherichia-Shigella | ↑ (65, 87) | ||||
| Faecalibacterium | ↓ (61–65) | ↓ (34, 39) | ↓ (34) | ||
| ↑ (34) | ↑ (34) | ||||
| Lactobacillus | ↑ (53) | ↑ (34, 59) | ↑ (34, 59) | ||
| Megasphaera | ↑ (53, 92) | ↑ (34) | ↑ (34) | ||
| Parasutterella | ↓ (90, 92) | ||||
| Prevotella | ↓ (62) | ↓ (34) | ↓ (34) | ||
| ↑ (34) | ↑ (34) | ||||
| Ruminococcus | ↓ (61, 91) | ↓ (45) | ↓ (45) | ||
| Slackia | ↑ (90) | ↑ (34) | ↑ (34) | ||
| Streptococcus | ↑ (34, 45) | ↑ (34, 45) | |||
| Species | |||||
| Akkermansia muciniphila | ↓ (46, 48, 49) | ↓ (45, 47) | ↓ (45, 47) | (46) | |
| Bifidobacterium longum | ↓ (74, 75) | ↓ (36, 39) | ↓ (36, 39) | (36) | |
| Blautia coccoides group | ↓ (83) | ↓ (59) | (83) | ||
| Blautia obeum | ↓ (36) | ↓ (36) | (36) | ||
| ↑ (40) | ↑ (34) | ↑ (34) | |||
| Blautia wexlerae | ↑ (34, 36) | ↑ (34, 36) | (36) | ||
| Clostridium leptum group7 | ↓ (42, 48, 61) | ↓ (59) | ↓ (59) | ||
| Clostridium leptum7 | ↑ (94) | ||||
| Dialister invisus | ↓ (40) | ↓ (44) | ↓ (44) | ||
| Escherichia coli | ↑ (42) | ↑ (44) | ↑ (44) | ||
| Enterocloster bolteae | ↑ (75) | ↑ (36, 39) | ↑ (36, 39) | (36) | |
| Faecalibacterium prausnitzii | ↓ (40, 42) | ↓ (36, 44) | ↓ (36, 39, 44) | (36) | |
| ↑ (66) | ↑ (34, 45) | ↑ (34, 45) | |||
| Haemophilus parainfluenzae | ↓ (40) | ↓ (44) | ↓ (37) | ||
| Intestinibacter bartlettii | ↓ (75) | ↓ (39) | ↓ (39) | ||
| Megasphaera elsdenii | ↑ (34, 44) | ↑ (34, 44) | |||
| [Ruminococcus] gnavus | ↑ (36) | ↑ (36, 45) | (36) | ↑ (37) | |
| Streptococcus salivarius | ↑ (75) | ↑ (44) | |||
| Diversity | |||||
| α Diversity | ↓ (35) | ↓ (51) | ↓ (45, 51) | (35) | |
| ↑ (73) | (73) | ||||
Associations with postprandial glucose concentrations and remaining glucose outcomes are shown separately and further divided according to whether estimations were unadjusted or accounting for body measures and/or metabolic drugs and whether SCFAs or genes encoding for enzymes in SCFA biosynthesis were analyzed. ↓ and ↑ refer to inverse and direct associations, respectively. HbA1c, glycated hemoglobin.
Glucose includes the studied outcomes fasting and nonfasting glucose measures, HbA1c, and comparisons between groups of individuals with or without prediabetes.
Body measures refers to BMI, body weight, body fat or body fat percentage, waist circumference, or waist-to-hip ratio.
Metabolic drugs include drugs such as antidiabetic drugs and drugs lowering blood pressure or blood lipids.
Previously Clostridiales.
Previously Ruminococcaceae.
Articles reporting on C. leptum were investigated for methodology and divided into C. leptum group when using group-level primers and qPCR and C. leptum when based on universal primers, 16S ribosomal RNA gene sequencing, and databases. C. leptum group is predominated by the species F. prausnitzii and these results were merged with those reported as Clostridium cluster IV (synonym).
TABLE 2.
Summary of reported associations between bacteria on different taxonomic levels and insulin-related outcomes1
| Gut microbiota | Insulin2 | Postprandial insulin | |||
|---|---|---|---|---|---|
| Unadjusted association | Adjusted for body measures3 | Adjusted for metabolic drugs4 | SCFAs or enzymes measured | ||
| Phyla | |||||
| Bacteroidetes | ↓ (48, 89) | ||||
| ↑ (60) | (60) | ||||
| Firmicutes | ↓ (48) | ↓ (41, 60) | ↓ (39, 41) | (60) | |
| ↑ (39) | |||||
| Proteobacteria | ↓ (41) | ↓ (41) | |||
| ↑ (89) | ↑ (60) | (60) | |||
| Order | |||||
| Eubacteriales5 | ↓ (39, 41) | ||||
| ↑ (39) | |||||
| Families | |||||
| Clostridiaceae | ↓ (45, 78, 95) | ↓ (45, 78) | |||
| Lachnospiraceae | ↓ (39, 41, 45) | ||||
| ↑ (41) | |||||
| Oscillospiraceae6 | ↓ (60) | ↓ (45) | (60) | ||
| Genera | |||||
| Bifidobacterium | ↓ (48) | ↓ (58) | ↓ (58) | ||
| Blautia | ↑ (39, 45) | ||||
| Clostridium | ↓ (78) | ↓ (39, 45, 78) | |||
| ↑ (39) | |||||
| Prevotella | ↑ (48, 87) | ||||
| Species | |||||
| Akkermansia muciniphila | ↓ (46, 48, 50) | ↓ (47) | ↓ (47) | (46) | |
| Bacteroides intestinalis | ↓ (40) | ↓ (41) | |||
| Bifidobacterium longum | ↓ (58) | ↓ (39) | ↓ (39) | ||
| Blautia coccoides group | ↓ (58) | ↓ (58, 59) | |||
| Clostridium leptum group | ↓ (42, 48) | ↓ (59) | ↓ (58, 59) | ||
| Faecalibacterium prausnitzii | ↓ (40, 42) | ↓ (41) | ↓ (39, 41) | ↓ (37, 38) | |
Associations with postprandial insulin concentrations and remaining insulin outcomes are shown separately and further divided according to whether estimations were unadjusted or accounting for body measures and/or metabolic drugs and whether SCFAs or genes encoding for enzymes in SCFA biosynthesis were analyzed. ↓ and ↑ refer to inverse and direct associations, respectively.
Insulin includes the studied outcomes fasting insulin, HOMA-IR, and measures of insulin resistance/sensitivity and comparisons between groups of individuals with or without insulin resistance.
Body measures refers to BMI, body weight, body fat or body fat percentage, waist circumference, or waist-to-hip ratio.
Metabolic drugs include drugs such as antidiabetic drugs and drugs lowering blood pressure or blood lipids.
Previsously Clostridiales.
Previously Ruminococcaceae.
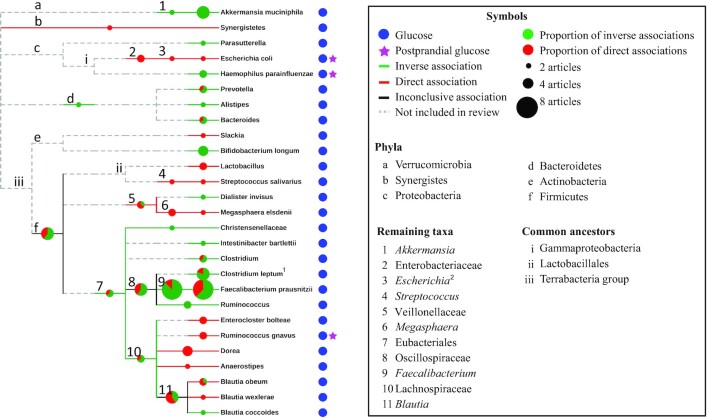
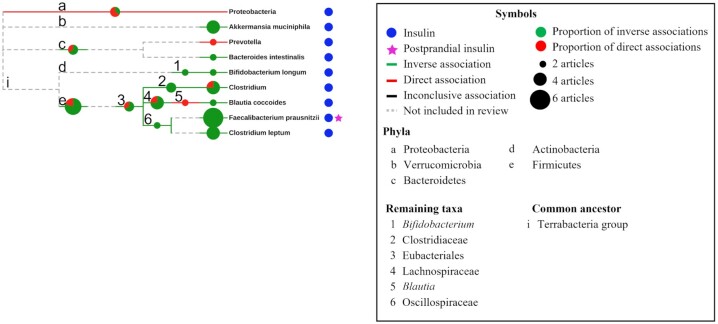
The taxonomic relations between bacteria were investigated in relation to our findings (Figures 3 and 4). Overall, there were consistent inverse associations with glucose and insulin for taxa belonging to the Oscillospiraceae family (formerly known as Ruminococcaceae) including Faecalibacterium, F. prausnitzii, and the C. leptum group. Inverse associations were also found with insulin for Bifidobacterium and Bifidobacterium longum (Figure 4). For glucose, direct associations were observed with Enterobacteriaceae and its taxa Escherichia-Shigella and Escherichia coli, Megasphaera and Megasphaera elsdenii, and with Streptococcus and Streptococcus salivarius (Figure 3). The Bacteroidetes phylum and genera (Prevotella, Bacteroides, and Alistipes) as well as Akkermansia and Akkermansia muciniphila were inversely associated with glucose. Results were typically more consistent for genera and species than for phyla and orders.
FIGURE 3.
Taxonomic trees for fecal bacteria associating with glucose-related outcomes and differences across glycemic groups. The 40 bacterial taxa associating with glucose belonged to 5 phyla and organized into 27 leaves. Meaning of symbols and colors are explained in the accompanying box. Pie charts show the proportion of studies that reported on inverse and direct associations, respectively. The size of the pie chart is proportional to the number of studies and studies that reported on both an inverse and a direct association for any given taxon were counted twice (supporting both the inverse and the direct association). 1Clostridium leptum refers to results both for the C. leptum group, where all studies showed inverse associations, and for C. leptum with unknown specificity, showing a direct association with insulin. 2Escherichia refers to results for Escherichia-Shigella.
FIGURE 4.
Taxonomic trees for fecal bacteria associating with insulin-related outcomes and differences across insulinemic groups. The 17 bacterial taxa associating with insulin belonged to 5 phyla and organized into 9 leaves. Meaning of symbols and colors are explained in the accompanying box. Pie charts show the proportion of studies that reported on inverse and direct associations, respectively. The size of the pie chart is proportional to the number of studies and studies that reported on both an inverse and a direct association for any given taxon were counted twice (supporting both the inverse and the direct association).
Evidence for the role of key gut bacteria in glucose regulation
The most consistent and inverse associations with glucose- and/or insulin-related outcomes were found for A. muciniphila, B. longum, Faecalibacterium, the C. leptum group, and F. prausnitzii (Tables 1 and 2, Figures 3 and 4). Dorea was consistently elevated with prediabetes and positively associated with glucose concentrations. The findings for these bacteria are discussed in greater detail in what follows. Special attention was given to taxonomic relations and associations with SCFAs and microbial genes encoding for enzymes involved in SCFA biosynthesis. We also report on evidence that supports or contradicts the potential role of these taxa in glucose metabolism independent of body weight, body composition, and metabolic drugs.
A. muciniphila and Akkermansia
The abundance of A. muciniphila was inversely associated with fasting glucose and HbA1c (45–49) and with insulin concentrations and insulin resistance (46–48, 50) and was lower in treatment-naïve individuals with prediabetes than in normoglycemic controls (45, 51) (Tables 1 and 2, Figures 3 and 4). Inverse associations were also found for the genus level (52, 53), where the association between Akkermansia and fasting glucose withstood statistical correction for metabolic disease diagnosis, use of metabolic drugs, as well as sex, age, and lifestyle factors (52) (Table 1, Figure 3). Similarly, several studies found the relation between A. muciniphila and blood glucose and insulin to be independent of BMI (45, 47), diabetes diagnosis and treatment (45), as well as sex and age (46, 47, 50) (Supplemental Table 5). In 2 studies, bimodal distributions of A. muciniphila were identified, where individuals with the lower A. muciniphila abundance had higher glycemic and insulinemic measures (46, 47), including an elevated glucose response after an oral-glucose-tolerance test (46). Another study found that patients with refractory diabetes, defined as a persistent elevation in HbA1c despite glucose-lowering medications, had lower A. muciniphila abundance than individuals with type 2 diabetes and lower HbA1c levels (49).
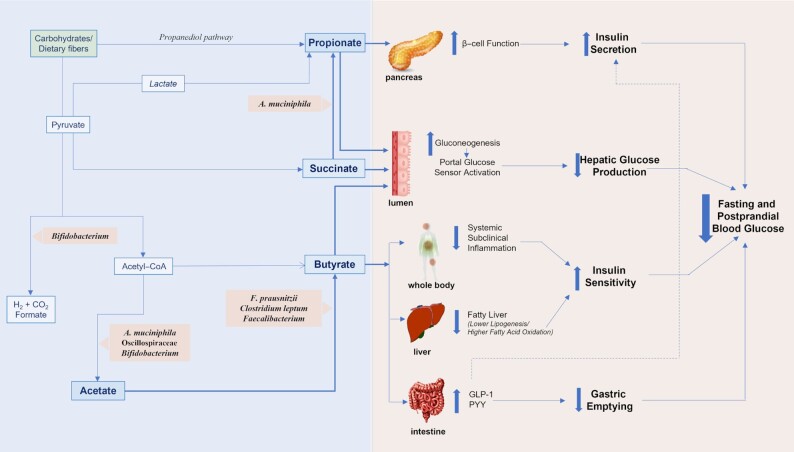
A. muciniphila is a mucin-degrading bacterium known to produce acetate and propionate (54), 2 SCFAs which have been suggested to affect β-cell function and insulin secretion (55–57) (Figure 5). In the study by Dao et al. (46), A. muciniphila was correlated with circulating acetate, consistent with the proposed mechanisms. Neither propionate nor butyrate were measured, nor were SCFAs in feces.
FIGURE 5.
Proposed mechanisms of how SCFA-producing bacteria and SCFAs affect fasting and postprandial glucose metabolism in humans in the context of dietary fiber and based on present knowledge. SCFA-producing bacteria included in this review are shown. A. muciniphila, Akkermansia muciniphila; F. prausnitzii, Faecalibacterium prausnitzii; GLP-1, glucagon-like peptide-1; PYY, peptide YY.
F. prausnitzii, C. leptum group, Faecalibacterium, and Oscillospiraceae
We found consistent evidence for reduced abundances of F. prausnitzii (37–42), C. leptum group (42, 48, 58, 59), and Oscillospiraceae (45, 60) with elevated insulin concentrations and insulin resistance (Table 2, Figure 4). This included 2 studies that reported on postprandial insulin responses and inverse associations with F. prausnitzii (37, 38). Several studies had excluded participants taking antidiabetic medications (39, 45, 59) and other metabolic drugs (59) and 1 study found null associations with metformin, a common antidiabetic drug (41) (Supplemental Table 5). There was limited and overall inconclusive evidence for potential bias of overweight and obesity.
Consistent inverse associations were also observed for glucose with the C. leptum group (42, 48, 59, 61) and Faecalibacterium (39, 61–65), with the exception of 1 study with inconclusive results for Faecalibacterium and prediabetes (34) (Table 1, Figure 3). Five studies, which had applied metagenomics sequencing (36, 39, 40, 44) or qPCR (42), showed consistent inverse associations between F. prausnitzii and fasting glucose, HbA1c, and prediabetes. In contrast, 3 studies using 16S rRNA gene amplification and sequencing (with lower specificity for F. prausnitzii) found direct associations with fasting glucose and prediabetes (34, 45, 66). Inconsistent results were also found for Oscillospiraceae but could not be attributed to differences in methodology (34, 35, 43, 45, 67) (Supplemental Table 5). There was no conclusive evidence regarding the role of adiposity and metabolic drugs in the associations with glucose for the C. leptum group, F. prausnitzii, Faecalibacterium, or Oscillospiraceae.
None of the studies for F. prausnitzii, the C. leptum group, Oscillospiraceae, or Faecalibacterium had measured butyrate, a major fermentation product for these taxa (e.g., after dietary fiber degradation (68)) and an SCFA which has been shown to affect fasting and postprandial glucose metabolism through various mechanisms (23, 57, 69–72) (Figure 5). However, Wu et al. (36) found that the lower abundance of F. prausnitzii in individuals with prediabetes was accompanied by a reduction of bacterial genes linked to butyrate biosynthesis. Some Oscillospiraceae taxa produce acetate, and 2 studies found a direct association between Oscillospiraceae and circulating acetate (60, 73). Plasma acetate was furthermore inversely correlated to HbA1c and was lower in individuals with obesity and insulin resistance than in insulin-sensitive controls with obesity (60).
B. longum and Bifidobacterium
The abundance of B. longum was frequently found to be lower with elevated glucose (36, 39, 74, 75) and insulin measures (39, 58) (Tables 1 and 2, Figures 3 and 4). Several studies reported on relations that were independent of BMI, body fat, age, and sex (36, 39), and dietary intake (39, 58), but not for BMI and dietary intake combined (58) (Supplemental Table 5). Similar results were found on the genus level. Bifidobacterium was inversely associated with insulin and insulin resistance in participants with obesity in 2 studies (48, 58), where 1 of the studies had adjusted for dietary intake and for BMI and excluded participants taking any medications (58) (Table 2, Figure 4). Two studies reporting on B. longum (39) and Bifidobacterium (48) had excluded participants based on probiotic consumption (e.g., B. longum) before enrollment.
Notably, there were inverse associations between baseline B. longum and 2-h glucose measures at a 4-y follow-up time point as well as the change in glucose concentrations over time, in a nested case–control study where B. longum had correlated with fasting glucose at baseline (75).
Bifidobacteria mainly produce acetate (and formate) during fermentation (76, 77) (Figure 5). No study reporting on Bifidobacterium or B. longum had measured acetate or other SCFAs.
Dorea
Dorea was consistently and positively associated with glucose measures, possibly independently of metabolic drugs as shown in all 4 studies (34, 39, 45, 78) (Table 1, Figure 3). Fasting glucose was positively correlated with Dorea in 3 studies (39, 45, 78), a correlation which remained statistically significant after adjustment for age, body fat, and diet in females with obesity (39), and was not associated with BMI in a population with overweight and obesity (78) (Supplemental Table 5). Two studies showed elevated concentrations in subjects with prediabetes in comparisons with normoglycemic controls, matched or adjusted for age and sex (34, 45) and for waist-to-hip ratio and inflammatory markers (34). In 1 of these studies, Dorea was only elevated in Danish but not Indian individuals with prediabetes (34). However, 2 Dorea species (D. formicigenerans and D. longicatena) were elevated in the Indian group of individuals with prediabetes which had comparable weight and waist-to-hip ratio with the normoglycemic controls. Three of the 4 studies included Danish populations (34, 39, 45).
None of the studies measured acetate, one of the main end products of fermentation for Dorea.
Discussion
In this scoping review we identified α diversity and 45 taxa, from phyla to species, that were associated with the selected glucose metabolism outcomes. Among these, A. muciniphila, B. longum, Faecalibacterium, the C. leptum group, and F. prausnitzii were most consistently inversely associated with glucose and/or insulin, whereas Dorea was directly associated with glucose. These taxa represent key candidates for future investigations and development of type 2 diabetes prevention strategies. Moreover, there was some evidence to suggest that associations may be independent of body measures, metabolic drugs, and other covariates (e.g., age, sex, and diet), particularly for A. muciniphila and Dorea, as well as potential mediation by SCFAs for A. muciniphila and F. prausnitzii.
Our findings should be placed in context with the strengths and limitations of the reviewed literature. Of the 45 eligible studies, the majority were published in the past 5 y, many with moderate to large study populations (100–2000) and mixed-sex study populations. Study design, glucose metabolism outcomes, and the presence of metabolic disease and risk factors varied. There was a lack of metagenomics sequencing, postprandial measures of glucose and insulin, and SCFA analysis; study populations were predominantly Asian or European; and there is a need for procedures accounting for confounding factors. Importantly, evidence from prospective studies and well-controlled intervention trials in this field is warranted to infer causality. Owing to the broad nature of this scoping review and for feasibility purposes, null, rare, or statistically nonsignificant associations were not included, neither were associations for co-occurring bacteria or groups of bacteria sharing functional traits (e.g., butyrate-producing bacteria). For the same reason, although we assessed whether studies had accounted for diet when reporting gut microbiota–glucose metabolism associations, we did not assess response to dietary trials. We regard work on gut microbiota–diet interactions and evidence toward using diet to alter the abundance and activity of specific (key) bacteria as crucial and complementary to the current literature and the present review.
Some microbial taxa belonging to the same lineage had similar relations with glucose metabolism outcomes. This can reflect shared attributes among bacteria within a taxon but also result from differences in sequencing depth across studies. In other words, associations for taxa at higher taxonomic ranks (including Akkermansia and Faecalibacterium) may to a large extent reflect associations for 1 or few prominent taxa at lower ranks (e.g., A. muciniphila and F. prausnitzii). Studying relations at the species level, and also strain and clade level, using metagenomic sequencing is warranted to elucidate relations for gut microbiota with glucose metabolism in humans and to distinguish between similarities and unique characteristics for bacteria within a given taxon. This includes investigating whether F. prausnitzii or other bacteria drive the associations for the C. leptum group. Metagenomic sequencing can also provide information on bacterial function and metabolic capabilities and thus provide insight on functional gene redundancy independent of taxonomic origin as well as on potential underlying mechanisms. It is also important to acknowledge that methodological steps such as DNA isolation and extraction can affect gut microbiota analysis and contribute to differences in results between studies and that method harmonization is needed (79–82).
SCFA production remains one of the best-characterized metabolic attributes of human gut bacteria and SCFAs are commonly proposed to underlie observed relations between gut microbiota and health outcomes in humans. Despite this, only 5 studies in the current literature reported on SCFAs (43, 46, 60, 73, 83), 2 reported on genes that encode for enzymes involved in SCFA biosynthesis (35, 36), and no study reported on succinate. Yet, taken together, results support the proposed mechanism: direct associations for A. muciniphila and Oscillospiraceae with circulating acetate (46, 60, 73), a coinciding reduction of F. prausnitzii and bacterial genes related to butyrate production with prediabetes (36), an inverse association between plasma acetate and insulin resistance (60), and inverse associations for total organic acid content, acetate, and propionate with HbA1c. The exception is the direct association between Dorea and glucose concentrations and the acetate-producing capabilities of Dorea. It is well established that SCFA concentrations measured in feces and blood poorly reflect SCFA production in the gut but are meaningful indicators of the locally and systemically available SCFAs that could exhibit different biological effects (84). Ongoing research is investigating SCFA concentrations, and differences in SCFA production across individuals, after the consumption of different dietary fibers (6, 8, 85) and whether supplementation of butyrate or butyrate-producing bacteria can prevent or delay development of type 2 diabetes (13). There are also investigations into cross-feedings and co-occurrence of bacteria, the flux of SCFAs, and shifts in the gut microbiota metabolism with type 2 diabetes (1, 57). This development will contribute complementary and critical insight to the role of SCFAs in glucose regulation as well as to the complex relations between human gut bacteria and potentially toward novel preventative strategies for diabetes. Importantly, SCFA production may only represent a fraction of the mechanisms through which gut microbiota may affect glucose metabolism and effects may also be mediated by microbial metabolites such as trimethylamine N-oxide (TMAO), indole-3-propionic acid (IPA), imidazole propionate (ImP), and bile acids, and be contingent on diet (86).
Lastly, Bifidobacterium, A. muciniphila, Faecalibacterium, and F. prausnitzii have consistently been found at lower abundances in individuals with prevalent type 2 diabetes than in individuals with normal glucose regulation (12), thus supporting a potential role of these bacteria in contrasting the progression toward type 2 diabetes. The inverse associations between baseline B. longum and type 2 diabetes incidence and the change in blood glucose over time (75) are also promising in terms of prevention but require further investigation. Mechanistic studies are required to disentangle the roles of individual bacteria in type 2 diabetes etiology and whether gut bacteria are involved in specific steps of diabetes development. In the present literature we found little support for a gradual shift toward the type 2 diabetic state for taxa reported in this review (36, 41, 44, 51, 53, 75, 87), with the exception of Akkermansia (53) and a shift in the overall microbial composition toward impaired glucose tolerance and untreated type 2 diabetes (36). Future studies may also evaluate the potential to use baseline A. muciniphila abundance for screening and stratification purposes for type 2 diabetes prevention, based on the bimodal abundance reported (46, 47).
Conclusions
In this scoping review we identified and extensively reviewed 45 original studies and identified A. muciniphila, B. longum, Faecalibacterium, the C. leptum group, F. prausnitzii, and Dorea as key bacteria for future investigation. For some of these bacteria, there was evidence to suggest SCFA mediation as well as associations independent of overweight, obesity, and metabolic drugs, and all, except Dorea, have previously been consistently and inversely associated with prevalent type 2 diabetes. Yet, several limitations in the present literature must be addressed in future studies, including an increased application of metagenomic sequencing with harmonized sample preparation procedures, increasing diverse representation in study populations, taking measures to account for confounding factors, and increasing investigations into postprandial glucose metabolism and mediation by SCFAs and other microbial metabolites. In addition, prospective studies, well-controlled intervention trials, and studies investigating the role of diet–gut microbiota interactions are needed to infer causality. Taken together, this will have the potential to bring critical insight into roles of gut microbiota and microbial metabolites in human glucose metabolism and to contribute toward the development of novel prevention strategies for type 2 diabetes, including precision nutrition.
Supplementary Material
Acknowledgements
The authors’ responsibilities were as follows—MSAP-B, RL, GR, SÅ, and YH: designed the research; MSAP-B, SÅ, YH, GC, and CV: conducted the research; MSAP-B, GC, CV, and JD: analyzed the data; MSAP-B and RL: wrote the paper; RL: had primary responsibility for the final content; and all authors: read and approved the final manuscript.
Notes
Author disclosures: The authors report no conflicts of interest
This work is part of the Food4GutMarKIT project, supported by FORMAS grant 2019-04639 (to RL). MSAP-B is supported by FORMAS grant 2017-02003 under the umbrella of the European Joint Programming Initiative “A Healthy Diet for a Healthy Life” (JPI HDHL) and of the ERA-NET Cofund ERA-HDHL (GA No. 727565 of the EU Horizon 2020 Research and Innovation Programme). RL is supported by FORMAS research grant Dnr 2020-01113 and by Swedish Research Council grant 2019-01264. The funders did not have a role in the design, implementation, analysis, or interpretation in the present scoping review.
Supplemental Figures 1 and 2 and Supplemental Tables 1–5 are available from the “Supplementary data” link in the online posting of the article and from the same link in the online table of contents at https://academic.oup.com/ajcn/.
GR and RL contributed equally to this work as senior investigators.
Contributor Information
Marie S A Palmnäs-Bédard, Department of Biology and Biological Engineering, Division of Food and Nutrition Science, Chalmers University of Technology, Gothenburg, Sweden.
Giuseppina Costabile, Department of Clinical Medicine and Surgery, University of Naples Federico II, Naples, Italy; Task Force on Microbiome Studies, University of Naples Federico II, Naples, Italy.
Claudia Vetrani, Department of Clinical Medicine and Surgery, University of Naples Federico II, Naples, Italy.
Sebastian Åberg, Department of Biology and Biological Engineering, Division of Food and Nutrition Science, Chalmers University of Technology, Gothenburg, Sweden.
Yommine Hjalmarsson, Department of Communication and Learning in Science, Chalmers University of Technology, Gothenburg, Sweden.
Johan Dicksved, Department of Animal Nutrition and Management, Swedish University of Agricultural Sciences, Uppsala, Sweden.
Gabriele Riccardi, Department of Clinical Medicine and Surgery, University of Naples Federico II, Naples, Italy; Task Force on Microbiome Studies, University of Naples Federico II, Naples, Italy.
Rikard Landberg, Department of Biology and Biological Engineering, Division of Food and Nutrition Science, Chalmers University of Technology, Gothenburg, Sweden; Department of Public Health and Clinical Medicine, Umeå University, Umeå, Sweden.
Data availability
Data described in the article, code book, and analytic code will be made available upon request pending application and all data used in this article are derived from the charted data present in Supplemental Table 5.
References
- 1. Daisley BA, Koenig D, Engelbrecht K, Doney L, Hards K, Al KFet al. Emerging connections between gut microbiome bioenergetics and chronic metabolic diseases. Cell Rep. 2021;37(10):110087. [DOI] [PubMed] [Google Scholar]
- 2. Fan Y, Pedersen O. Gut microbiota in human metabolic health and disease. Nat Rev Microbiol. 2021;19(1):55–71. [DOI] [PubMed] [Google Scholar]
- 3. Koh A, Bäckhed F. From association to causality: the role of the gut microbiota and its functional products on host metabolism. Mol Cell. 2020;78(4):584–96. [DOI] [PubMed] [Google Scholar]
- 4. Brunkwall L, Orho-Melander M. The gut microbiome as a target for prevention and treatment of hyperglycaemia in type 2 diabetes: from current human evidence to future possibilities. Diabetologia. 2017;60(6):943–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhang X, Cai X, Zheng X. Gut microbiome-oriented therapy for metabolic diseases: challenges and opportunities towards clinical translation. Trends Pharmacol Sci. 2021;42(12):984–7. [DOI] [PubMed] [Google Scholar]
- 6. Rivellese A. Short-chain fatty acids in plasma after intake of fermentable cereal fibres- an extended postprandial study [Internet]. 2022. Clinicaltrials.gov identifier: NCT05443828. [Accessed 2022 Jul 6]. Available from: http://www.clinicaltrials.gov/ct2/show/NCT05443828.
- 7. Deehan EC, Yang C, Perez-Muñoz ME, Nguyen NK, Cheng CC, Triador Let al. Precision microbiome modulation with discrete dietary fiber structures directs short-chain fatty acid production. Cell Host Microbe. 2020;27(3):389–404.e6. [DOI] [PubMed] [Google Scholar]
- 8. So D, Whelan K, Rossi M, Morrison M, Holtmann G, Kelly JTet al. Dietary fiber intervention on gut microbiota composition in healthy adults: a systematic review and meta-analysis. Am J Clin Nutr. 2018;107(6):965–83. [DOI] [PubMed] [Google Scholar]
- 9. Christensen L, Sørensen CV, Wøhlk FU, Kjølbæk L, Astrup A, Sanz Yet al. Microbial enterotypes beyond genus level: Bacteroides species as a predictive biomarker for weight change upon controlled intervention with arabinoxylan oligosaccharides in overweight subjects. Gut Microbes. 2020;12(1):1847627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Saeedi P, Petersohn I, Salpea P, Malanda B, Karuranga S, Unwin Net al. Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045: results from the International Diabetes Federation Diabetes Atlas 9th edition. Diabetes Res Clin Pract. 2019;157:107843. [DOI] [PubMed] [Google Scholar]
- 11. Ge K. The transition of Chinese dietary guidelines and food guide pagoda. Asia Pac J Clin Nutr. 2011;20(3):439–46. [PubMed] [Google Scholar]
- 12. Gurung M, Li Z, You H, Rodrigues R, Jump DB, Morgun Aet al. Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine. 2020;51:102590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Arora T, Tremaroli V. Therapeutic potential of butyrate for treatment of type 2 diabetes. Front Endocrinol (Lausanne). 2021;12:761834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vieira-Silva S, Falony G, Belda E, Nielsen T, Aron-Wisnewsky J, Chakaroun Ret al. Statin therapy is associated with lower prevalence of gut microbiota dysbiosis. Nature. 2020;581(7808):310–15. [DOI] [PubMed] [Google Scholar]
- 15. Wu H, Esteve E, Tremaroli V, Khan MT, Caesar R, Manneras-Holm Let al. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat Med. 2017;23(7):850–8. [DOI] [PubMed] [Google Scholar]
- 16. Safari S, Abdoli M, Amini M, Aminorroaya A, Feizi A. A 16-year prospective cohort study to evaluate effects of long-term fluctuations in obesity indices of prediabetics on the incidence of future diabetes. Sci Rep. 2021;11(1):11635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Goossens GH. The metabolic phenotype in obesity: fat mass, body fat distribution, and adipose tissue function. Obes Facts. 2017;10(3):207–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zeevi D, Korem T, Zmora N, Israeli D, Rothschild D, Weinberger Aet al. Personalized nutrition by prediction of glycemic responses. Cell. 2015;163(5):1079–94. [DOI] [PubMed] [Google Scholar]
- 19. Berry SE, Valdes AM, Drew DA, Asnicar F, Mazidi M, Wolf Jet al. Human postprandial responses to food and potential for precision nutrition. Nat Med. 2020;26(6):964–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hjorth MF, Christensen L, Kjølbæk L, Larsen LH, Roager HM, Kiilerich Pet al. Pretreatment Prevotella-to-Bacteroides ratio and markers of glucose metabolism as prognostic markers for dietary weight loss maintenance. Eur J Clin Nutr. 2020;74(2):338–47. [DOI] [PubMed] [Google Scholar]
- 21. Korem T, Zeevi D, Zmora N, Weissbrod O, Bar N, Lotan-Pompan Met al. Bread affects clinical parameters and induces gut microbiome-associated personal glycemic responses. Cell Metab. 2017;25(6):1243–53.e5. [DOI] [PubMed] [Google Scholar]
- 22. Kovatcheva-Datchary P, Nilsson A, Akrami R, Lee YS, De Vadder F, Arora Tet al. Dietary fiber-induced improvement in glucose metabolism is associated with increased abundance of Prevotella. Cell Metab. 2015;22(6):971–82. [DOI] [PubMed] [Google Scholar]
- 23. De Vadder F, Kovatcheva-Datchary P, Goncalves D, Vinera J, Zitoun C, Duchampt Aet al. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell. 2014;156(1–2):84–96. [DOI] [PubMed] [Google Scholar]
- 24. De Vadder F, Kovatcheva-Datchary P, Zitoun C, Duchampt A, Backhed F, Mithieux G. Microbiota-produced succinate improves glucose homeostasis via intestinal gluconeogenesis. Cell Metab. 2016;24(1):151–7. [DOI] [PubMed] [Google Scholar]
- 25. Yamashita H. Biological function of acetic acid–improvement in obesity and glucose tolerance by acetic acid in type 2 diabetic rats. Crit Rev Food Sci Nutr. 2016;56(sup1):S171–5. [DOI] [PubMed] [Google Scholar]
- 26. Richardson WS, Wilson MC, Nishikawa J, Hayward RS. The well-built clinical question: a key to evidence-based decisions. ACP J Club. 1995;123(3):A12–13. [PubMed] [Google Scholar]
- 27. Miller SA, Forrest JL. Enhancing your practice through evidence-based decision making: PICO, learning how to ask good questions. J Evid Based Dent Pract. 2001;1(2):136–41. [Google Scholar]
- 28. Otten R, Vries RD, Schoonmade L. Amsterdam Efficient Deduplication (AED) method. 2019. Available from: 10.5281/ZENODO.3582927. [DOI]
- 29. Ouzzani M, Hammady H, Fedorowicz Z, Elmagarmid A. Rayyan—a web and mobile app for systematic reviews. Syst Rev. 2016;5(1):210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schoch CL, Ciufo S, Domrachev M, Hotton CL, Kannan S, Khovanskaya Ret al. NCBI Taxonomy: a comprehensive update on curation, resources and tools. Database. 2020:baaa062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lay C, Dore J, Rigottier-Gois L. Separation of bacteria of the Clostridium leptum subgroup from the human colonic microbiota by fluorescence-activated cell sorting or group-specific PCR using 16S rRNA gene oligonucleotides. FEMS Microbiol Ecol. 2007;60(3):513–20. [DOI] [PubMed] [Google Scholar]
- 32. PhyloT v2: a phylogenetic tree generator, based on NCBI or GTD taxonomy [Internet]. 2022. [cited 14 February, 2022]. Ivica Letunic, Heidelberg, Germany. Available from: https://phylot.biobyte.de/. [Google Scholar]
- 33. Letunic I, Bork P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49(W1):W293–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pinna NK, Anjana RM, Saxena S, Dutta A, Gnanaprakash V, Rameshkumar Get al. Trans-ethnic gut microbial signatures of prediabetic subjects from India and Denmark. Genome Med. 2021;13(1):36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fei N, Bernabé BP, Lie L, Baghdan D, Bedu-Addo K, Plange-Rhule Jet al. The human microbiota is associated with cardiometabolic risk across the epidemiologic transition. PLoS One. 2019;14(7):e0215262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wu H, Tremaroli V, Schmidt C, Lundqvist A, Olsson LM, Krämer Met al. The gut microbiota in prediabetes and diabetes: a population-based cross-sectional study. Cell Metab. 2020;32(3):379–90.e3. [DOI] [PubMed] [Google Scholar]
- 37. Asnicar F, Berry SE, Valdes AM, Nguyen LH, Piccinno G, Drew DAet al. Microbiome connections with host metabolism and habitual diet from 1,098 deeply phenotyped individuals. Nat Med. 2021;27(2):321–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Xu J, Lian F, Zhao L, Zhao Y, Chen X, Zhang Xet al. Structural modulation of gut microbiota during alleviation of type 2 diabetes with a Chinese herbal formula. ISME J. 2015;9(3):552–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Brahe LK, Le Chatelier E, Prifti E, Pons N, Kennedy S, Hansen Tet al. Specific gut microbiota features and metabolic markers in postmenopausal women with obesity. Nutr Diabetes. 2015;5(6):e159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu R, Hong J, Xu X, Feng Q, Zhang D, Gu Yet al. Gut microbiome and serum metabolome alterations in obesity and after weight-loss intervention. Nat Med. 2017;23(7):859–68. [DOI] [PubMed] [Google Scholar]
- 41. Karlsson FH, Tremaroli V, Nookaew I, Bergström G, Behre CJ, Fagerberg Bet al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498(7452):99–103. [DOI] [PubMed] [Google Scholar]
- 42. Furet J-P, Kong L-C, Tap J, Poitou C, Basdevant A, Bouillot J-Let al. Differential adaptation of human gut microbiota to bariatric surgery–induced weight loss: links with metabolic and low-grade inflammation markers. Diabetes. 2010;59(12):3049–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gonai M, Shigehisa A, Kigawa I, Kurasaki K, Chonan O, Matsuki Tet al. Galacto-oligosaccharides ameliorate dysbiotic Bifidobacteriaceae decline in Japanese patients with type 2 diabetes. Benef Microbes. 2017;8(5):705–16. [DOI] [PubMed] [Google Scholar]
- 44. Zhong H, Ren H, Lu Y, Fang C, Hou G, Yang Zet al. Distinct gut metagenomics and metaproteomics signatures in prediabetics and treatment-naive type 2 diabetics. EBioMedicine. 2019;47:373–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Allin KH, Tremaroli V, Caesar R, Jensen BAH, Damgaard MTF, Bahl MIet al. Aberrant intestinal microbiota in individuals with prediabetes. Diabetologia. 2018;61(4):810–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dao MC, Everard A, Aron-Wisnewsky J, Sokolovska N, Prifti E, Verger EOet al. Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity: relationship with gut microbiome richness and ecology. Gut. 2016;65(3):426–36. [DOI] [PubMed] [Google Scholar]
- 47. Mitsou EK, Detopoulou M, Kakali A, Fragopoulou E, Nomikos T, Antonopoulou Set al. Mining possible associations of faecal A . muciniphila colonisation patterns with host adiposity and cardiometabolic markers in an adult population. Benef Microbes. 2019;10(7):741–9. [DOI] [PubMed] [Google Scholar]
- 48. Tabasi M, Eybpoosh S, Sadeghpour Heravi F, Siadat SD, Mousavian G, Elyasinia Fet al. Gut microbiota and serum biomarker analyses in obese patients diagnosed with diabetes and hypothyroid disorder. Metab Syndr Relat Disord. 2021;19(3):144–51. [DOI] [PubMed] [Google Scholar]
- 49. Shih C-T, Yeh Y-T, Lin C-C, Yang L-Y, Chiang C-P. Akkermansia muciniphila is negatively correlated with hemoglobin A1c in refractory diabetes. Microorganisms. 2020;8(9):1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yassour M, Lim MY, Yun HS, Tickle TL, Sung J, Song Y-Met al. Sub-clinical detection of gut microbial biomarkers of obesity and type 2 diabetes. Genome Med. 2016;8(1):17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nuli R, Cai J, Kadeer A, Zhang Y, Mohemaiti P. Integrative analysis toward different glucose tolerance-related gut microbiota and diet. Front Endocrinol. 2019;10:295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhong X, Harrington JM, Millar SR, Perry IJ, O'Toole PW, Phillips CM. Gut microbiota associations with metabolic health and obesity status in older adults. Nutrients. 2020;12(8):2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gaike AH, Paul D, Bhute S, Dhotre DP, Pande P, Upadhyaya Set al. The gut microbial diversity of newly diagnosed diabetics but not of prediabetics is significantly different from that of healthy nondiabetics. mSystems. 2020;5(2):e00578–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Derrien M, Vaughan EE, Plugge CM, de Vos WM. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol. 2004;54(5):1469–76. [DOI] [PubMed] [Google Scholar]
- 55. Chambers ES, Byrne CS, Morrison DJ, Murphy KG, Preston T, Tedford Cet al. Dietary supplementation with inulin-propionate ester or inulin improves insulin sensitivity in adults with overweight and obesity with distinct effects on the gut microbiota, plasma metabolome and systemic inflammatory responses: a randomised cross-over trial. Gut. 2019;68(8):1430–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tilg H, Cani PD, Mayer EA. Gut microbiome and liver diseases. Gut. 2016;65(12):2035–44. [DOI] [PubMed] [Google Scholar]
- 57. Blaak EE, Canfora EE, Theis S, Frost G, Groen AK, Mithieux Get al. Short chain fatty acids in human gut and metabolic health. Benef Microbes. 2020;11(5):411–55. [DOI] [PubMed] [Google Scholar]
- 58. Teixeira TFS, Grześkowiak ŁM, Salminen S, Laitinen K, Bressan J, do Carmo Gouveia Peluzio M. Faecal levels of Bifidobacterium and Clostridium coccoides but not plasma lipopolysaccharide are inversely related to insulin and HOMA index in women. Clin Nutr. 2013;32(6):1017–22. [DOI] [PubMed] [Google Scholar]
- 59. Chen P-C, Chien Y-W, Yang S-C. The alteration of gut microbiota in newly diagnosed type 2 diabetic patients. Nutrition. 2019;63–64:51–6. [DOI] [PubMed] [Google Scholar]
- 60. Moreno-Navarrete JM, Serino M, Blasco-Baque V, Azalbert V, Barton RH, Cardellini Met al. Gut microbiota interacts with markers of adipose tissue browning, insulin action and plasma acetate in morbid obesity. Mol Nutr Food Res. 2018;62(3):201700721. [DOI] [PubMed] [Google Scholar]
- 61. Zeng Q, Li D, He Y, Li Y, Yang Z, Zhao Xet al. Discrepant gut microbiota markers for the classification of obesity-related metabolic abnormalities. Sci Rep. 2019;9(1):13424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tao S, Li L, Li L, Liu Y, Ren Q, Shi Met al. Understanding the gut–kidney axis among biopsy-proven diabetic nephropathy, type 2 diabetes mellitus and healthy controls: an analysis of the gut microbiota composition. Acta Diabetol. 2019;56(5):581–92. [DOI] [PubMed] [Google Scholar]
- 63. Aron-Wisnewsky J, Prifti E, Belda E, Ichou F, Kayser BD, Dao MCet al. Major microbiota dysbiosis in severe obesity: fate after bariatric surgery. Gut. 2019;68(1):70–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Salamon D, Sroka-Oleksiak A, Kapusta P, Szopa M, Mrozińska S, Ludwig-Słomczyńska AHet al. Characteristics of gut microbiota in adult patients with type 1 and type 2 diabetes based on next-generation sequencing of the 16S rRNA gene fragment. Pol Arch Intern Med. 2018;128(6):336–43. [DOI] [PubMed] [Google Scholar]
- 65. Gao R, Zhu C, Li H, Yin M, Pan C, Huang Let al. Dysbiosis signatures of gut microbiota along the sequence from healthy, young patients to those with overweight and obesity. Obesity (Silver Spring). 2018;26(2):351–61. [DOI] [PubMed] [Google Scholar]
- 66. Feng J, Tang H, Li M, Pang X, Wang L, Zhang Met al. The abundance of fecal Faecalibacterium prausnitzii in relation to obesity and gender in Chinese adults. Arch Microbiol. 2014;196(1):73–7. [DOI] [PubMed] [Google Scholar]
- 67. Leite AZ, Rodrigues NC, Gonzaga MI, Paiolo JCC, de Souza CA, Stefanutto NAVet al. Detection of increased plasma interleukin-6 levels and prevalence of Prevotella copri and Bacteroides vulgatus in the feces of type 2 diabetes patients. Front Immunol. 2017;8:1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Louis P, Flint HJ. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett. 2009;294(1):1–8. [DOI] [PubMed] [Google Scholar]
- 69. Chen J, Vitetta L. Gut microbiota metabolites in NAFLD pathogenesis and therapeutic implications. Int J Mol Sci. 2020;21(15):5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bouter K, Bakker GJ, Levin E, Hartstra AV, Kootte RS, Udayappan SDet al. Differential metabolic effects of oral butyrate treatment in lean versus metabolic syndrome subjects. Clin Transl Gastroenterol. 2018;9(5):e155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Canfora EE, Jocken JW, Blaak EE. Short-chain fatty acids in control of body weight and insulin sensitivity. Nat Rev Endocrinol. 2015;11(10):577–91. [DOI] [PubMed] [Google Scholar]
- 72. Zhao L, Zhang F, Ding X, Wu G, Lam Y, Wang Xet al. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science. 2018;359(6380):1151–6. [DOI] [PubMed] [Google Scholar]
- 73. Org E, Blum Y, Kasela S, Mehrabian M, Kuusisto J, Kangas AJet al. Relationships between gut microbiota, plasma metabolites, and metabolic syndrome traits in the METSIM cohort. Genome Biol. 2017;18(1):70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Haro C, Garcia-Carpintero S, Alcala-Diaz JF, Gomez-Delgado F, Delgado-Lista J, Perez-Martinez Pet al. The gut microbial community in metabolic syndrome patients is modified by diet. J Nutr Biochem. 2016;27:27–31. [DOI] [PubMed] [Google Scholar]
- 75. Wang L, Yu X, Xu X, Ming J, Wang Z, Gao Bet al. The fecal microbiota is already altered in normoglycemic individuals who go on to have type 2 diabetes. Front Cell Infect Microbiol. 2021;11:598672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Tsukuda N, Yahagi K, Hara T, Watanabe Y, Matsumoto H, Mori Het al. Key bacterial taxa and metabolic pathways affecting gut short-chain fatty acid profiles in early life. ISME J. 2021;15(9):2574–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Moens F, Verce M, De Vuyst L. Lactate- and acetate-based cross-feeding interactions between selected strains of lactobacilli, bifidobacteria and colon bacteria in the presence of inulin-type fructans. Int J Food Microbiol. 2017;241:225–36. [DOI] [PubMed] [Google Scholar]
- 78. Naderpoor N, Mousa A, Gomez-Arango LF, Barrett HL, Dekker Nitert M, de Courten B. Faecal microbiota are related to insulin sensitivity and secretion in overweight or obese adults. J Clin Med. 2019;8(4):452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Fiedorova K, Radvansky M, Nemcova E, Grombirikova H, Bosak J, Cernochova Met al. The impact of DNA extraction methods on stool bacterial and fungal microbiota community recovery. Front Microbiol. 2019;10:821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Greathouse KL, Sinha R, Vogtmann E. DNA extraction for human microbiome studies: the issue of standardization. Genome Biol. 2019;20(1):212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Lim MY, Song E-J, Kim SH, Lee J, Nam Y-D. Comparison of DNA extraction methods for human gut microbial community profiling. Syst Appl Microbiol. 2018;41(2):151–7. [DOI] [PubMed] [Google Scholar]
- 82. Yang F, Sun J, Luo H, Ren H, Zhou H, Lin Yet al. Assessment of fecal DNA extraction protocols for metagenomic studies. Gigascience. 2020;9(7):giaa071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Yamashiro K, Tanaka R, Urabe T, Ueno Y, Yamashiro Y, Nomoto Ket al. Gut dysbiosis is associated with metabolism and systemic inflammation in patients with ischemic stroke. PLoS One. 2017;12(2):e0171521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Müller M, González Hernández MA, Goossens GH, Reijnders D, Holst JJ, Jocken JWEet al. Circulating but not faecal short-chain fatty acids are related to insulin sensitivity, lipolysis and GLP-1 concentrations in humans. Sci Rep. 2019;9(1):12515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Amalia R, Noer ER, Muniroh M, Afifah DN, Kumoro AC, Pramono A. The effect of fibre intervention on serum and faecal short-chain fatty acids in human with overweight or obesity: a systematic review of human intervention studies. J Biomed Transl Res. 2022;8(1):38–44. [Google Scholar]
- 86. Zhu T, Goodarzi MO. Metabolites linking the gut microbiome with risk for type 2 diabetes. Curr Nutr Rep. 2020;9(2):83–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Diener C, Reyes-Escogido ML, Jimenez-Ceja LM, Matus M, Gomez-Navarro CM, Chu NDet al. Progressive shifts in the gut microbiome reflect prediabetes and diabetes development in a treatment-naive Mexican cohort. Front Endocrinol. 2020;11:602326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Emoto T, Yamashita T, Sasaki N, Hirota Y, Hayashi T, So Aet al. Analysis of gut microbiota in coronary artery disease patients: a possible link between gut microbiota and coronary artery disease. J Atheroscler Thromb. 2016;23(8):908–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Sanchez-Alcoholado L, Castellano-Castillo D, Jordán-Martínez L, Moreno-Indias I, Cardila-Cruz P, Elena Det al. Role of gut microbiota on cardio-metabolic parameters and immunity in coronary artery disease patients with and without type-2 diabetes mellitus. Front Microbiol. 2017;8:1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Yang J, Summanen PH, Henning SM, Hsu M, Lam H, Huang Jet al. Xylooligosaccharide supplementation alters gut bacteria in both healthy and prediabetic adults: a pilot study. Front Physiol. 2015;6:216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Li N, Wang X, Sun C, Wu X, Lu M, Si Yet al. Change of intestinal microbiota in cerebral ischemic stroke patients. BMC Microbiol. 2019;19(1):191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Li H, Liu B, Song J, An Z, Zeng X, Li Jet al. Characteristics of gut microbiota in patients with hypertension and/or hyperlipidemia: a cross-sectional study on rural residents in Xinxiang County, Henan Province. Microorganisms. 2019;7(10):399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Inoue R, Ohue-Kitano R, Tsukahara T, Tanaka M, Masuda S, Inoue Tet al. Prediction of functional profiles of gut microbiota from 16S rRNA metagenomic data provides a more robust evaluation of gut dysbiosis occurring in Japanese type 2 diabetic patients. J Clin Biochem Nutr. 2017;61(3):217–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Zupancic ML, Cantarel BL, Liu Z, Drabek EF, Ryan KA, Cirimotich Set al. Analysis of the gut microbiota in the Old Order Amish and its relation to the metabolic syndrome. PLoS One. 2012;7(8):e43052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Zhou W, Sailani MR, Contrepois K, Zhou Y, Ahadi S, Leopold SRet al. Longitudinal multi-omics of host–microbe dynamics in prediabetes. Nature. 2019;569(7758):663–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data described in the article, code book, and analytic code will be made available upon request pending application and all data used in this article are derived from the charted data present in Supplemental Table 5.