Abstract
AR signalling pathway reactivation plays a key role in the development of castration-resistant prostate cancer (CRPC). High-mobility group protein B1 (HMGB1) is an important factor involved in the occurrence and development of a variety of tumours by regulating gene transcription. In the present study, the association between HMGB1 and prostate cancer (PCa) and the effects of HMGB1 on androgen receptor (AR) transcription and signalling pathway reactivation in PCa cells in vitro and in vivo were evaluated. A bioinformatics method was used to determine the mRNA expression level of HMGB1 in PCa specimens and its correlation with the mRNA expression of AR. Immunohistochemical staining was used to detect the expression of these proteins in clinical PCa samples. Reporter gene and ChIP assays were performed to determine the activity of AR and the effect of HMGB1 on the ability of AR to bind to the promoters of prostate specific antigen and transmembrane protease, serine 2. A bioluminescence resonance energy transfer assay was employed to observe the direct interaction between HMGB1 and AR protein. Additionally, a castrated nude mouse xenograft tumour model was established to verify the effect of HMGB1 on PCa. The results revealed that HMGB1 expression was significantly increased in PCa specimens, which may have a strong correlation with AR expression. Moreover, HMGB1 could reactivate the AR signalling pathway, directly interact with AR, and promote the development of CRPC in an androgen-independent manner. The results of the present study indicated that HMGB1 promoted the development of CRPC by interacting with AR, which inferred that decreasing the expression of HMGB1 may be a potential effective method for CRPC prevention and treatment.
Keywords: CRPC, AR, HMGB1, bioinformatics method
Introduction
In 2020, prostate cancer (PCa) became the most frequently diagnosed cancer in 112 countries and the leading cause of cancer-related mortality in 48 countries among men (1). In recent decades, researchers have found that the activation of androgen receptor (AR) signalling plays a crucial role in the development of PCa (2). Thus, androgen deprivation therapy (ADT) is an important and standard treatment for PCa. In China, the majority of patients with PCa have progressed to an advanced stage at the time of diagnosis, and ADT has been adopted as the first-line treatment when seeking treatment (3). However, the effect of ADT on prolonging the survival time of patients with PCa is not satisfactory. Upon ADT, almost all patients will further develop castration-resistant prostate cancer (CRPC), with a mean survival time of only 14.5 months (4,5). Hence, there is an urgent need to explore the specific mechanism of CRPC and ameliorate the prognosis of patients with CRPC.
Most patients with CRPC exhibit a continuous increase in prostate-specific antigen (PSA) levels despite the use of ADT and testosterone levels. The increase in PSA levels indicates that intratumoural androgen biosynthesis is functional in CRPC cells (6). Intratumoural androgen is considered the key factor for the continued activation of AR in CRPC. After binding with intratumoural androgen, the inactive AR protein changes its conformation, translocates from the cytoplasm to the nucleus, and forms AR dimers (7). Furthermore, the AR dimer is able to bind to androgen response elements (AREs), such as the promoter or enhancer regions of downstream genes (7), which promote the development of PCa. Although new hormonal agents (e.g., enzalutamide or abiraterone) have been shown to prolong the overall survival time of patients with CRPC by blocking intratumoural androgen biosynthesis and activity, most of the patients will develop resistance to hormonal agents in only several months (8,9). The reincrease in PSA in drug-resistant patients with CRPC suggests that the AR signalling pathway is reactivated in androgen-independent conditions (6,9). A previous study by the authors confirmed that the AR protein could still be dimerized and bind to ARE as well as promote the transcription of its downstream genes under androgen deprivation conditions, which conferred CRPC cell growth. By contrast, the dimerization and activity of AR as well as the growth of the tumour cells were inhibited after mutation of the dimerization sites (F23, 27A/L26A and A596T/S597T) (10). Therefore, the AR protein still plays a key role in the reactivation of the AR signalling pathway even under androgen deprivation conditions. It is important to identify target genes that can interact with AR proteins and regulate the ability of AR proteins to bind to AREs.
High-mobility group protein B1 (HMGB1), which plays an important role in the activation of transcription factor signalling pathways, has been identified to be involved in the occurrence and development of a variety of tumours, including non-small-cell lung, cervical, and colorectal cancer (11). Moreover, an increasing number of studies have demonstrated that the overexpression of HMGB1 is related to the poor prognosis of PCa and CRPC (12,13). Although these studies initially demonstrated that HMGB1 was associated with the progression, clinical pathological characteristics, and poor prognosis of PCa, the molecular mechanism by which HMGB1 promotes the development of CRPC remains unclear. A previous study suggested that HMGB1 enhances oestrogen receptor (ER) dimer binding to the oestrogen response element (ERE) and then enhances the transcription of its downstream genes (14). Notably, the protein structure domain and active form (dimerization) of ER were similar to those of AR (15), which implied that the activation of ER and AR may be regulated by the same regulatory factors. Additionally, some studies have indicated that estrone sulfate and dehydroepiandrosterone sulfate could enhance the transcriptional activity of ER and AR simultaneously (16). RO48-8071 was demonstrated to inhibit the transcriptional activity of ER as well as AR (17). Hence, it is surmised that HMGB1 may interact with AR protein, promote the ability of AR protein to bind to ARE, and ultimately promote the development of CRPC.
To the best of our knowledge, there are no other studies reporting that HMGB1 protein could regulate AR transactivating activity, which is also well known as AR transcriptional activity, in PCa. The aim of the present study was to investigate the association between HMGB1 and/or AR expression and the pathological features of patients with PCa to verify the regulatory effect and mechanisms of HMGB1 on AR transactivating activity in order to provide a theoretical basis for applying novel targeted drugs for CRPC treatment.
Materials and methods
Bioinformatics analysis
The Oncomine (https://www.oncomine.org/resource/login.html), GEPIA (http://gepia.cancer-pku.cn/index.html), and DepMap Portal (https://depmap.org/portal/interactive/) databases were used to analyse the gene expression levels of HMGB1 and AR in the prostate gland, PCa tissues, androgen-dependent PCa cells (LNCaP and MDAPCA2B), and CRPC cells (VCaP and 22RV1). The data retrieved from Oncomine originated from GSE6099 (18), GSE68907 (19), GSE6919 (20) and a study of Vanaja et al (21). Furthermore, the correlation between HMGB1 mRNA expression and AR mRNA expression was analysed using the GEPIA database.
Specimens
The present study was approved by the Ethics Committee of the School of Nursing, Jilin University (Changchun, China). Paraffin-embedded specimens were collected from 2001–2009 at the Prostate Diseases Prevention and Treatment Research Centre of Jilin University (Changchun, China). All patients were diagnosed during a PCa screening study for high-risk males. The patients were informed and agreed to participate in the PCa-relative study. The inclusion criteria were as follows: i) Male and over 50 years old; ii) the level of serum PSA exceeded 4 pg/ml or an existing prostate tumor suggested by imaging; iii) initially diagnosed with PCa; and iv) the pathological diagnoses were determined as prostate adenocarcinoma by an experienced urological pathologist working at the Second Hospital of Jilin University. The exclusion criteria were as follows: i) Individual history of other malignant tumors; ii) the patients had previously undergone radical prostatectomy or other treatments, such as hormone treatment or radiotherapy (22). Five patients with benign prostate hyperplasia (BPH) were included as control subjects. The clinicopathological features of the patients included are presented in Table I. The clinicopathological features of prostatic needle biopsy samples are shown in Table II.
Table I.
Clinicopathological features of patients with PCa.
| Clinical features | No. of cases (%) |
|---|---|
| Age, years | |
| <73 | 25 (49.02) |
| ≥73 | 26 (50.98) |
| Pathological grade | |
| BPH | 5 (9.80) |
| GS=6 | 10 (19.61) |
| GS=7 | 10 (19.61) |
| GS=8 | 11 (21.57) |
| GS=9 | 12 (23.53) |
| GS=10 | 3 (5.88) |
| PSA level, pg/ml | |
| <54.3 | 25 (49.02) |
| ≥54.3 | 26 (50.98) |
PCa, prostate cancer; BPH, benign prostate hyperplasia; GS, Gleason score; PSA, prostate specific antigen.
Table II.
Clinicopathological features of prostatic needle biopsy samples.
| Samples | Case no. | Mean age (range) | Mean PSA level (range) |
|---|---|---|---|
| BPH | 5 | 75 (67–84) | 19.3 (9.1-29.44) |
| GS=6 | 10 | 77.4 (63–83) | 33.24 (4-96.8) |
| GS=7 | 10 | 68.4 (56–82) | 83.95 (9.4-300) |
| GS=8 | 11 | 70.9 (64–84) | 82.96 (28-254.3) |
| GS=9 | 12 | 73.25 (54–90) | 75.48 (3-126.1) |
| GS=10 | 3 | 75 (70–78) | 29.9 (1.7-49) |
PSA, prostate specific antigen; BPH, benign prostate hyperplasia; GS, Gleason score.
Immunohistochemistry
Immunohistochemical (IHC) staining was performed as previously described (23). Briefly, after being fixed in 10% formalin at room temperature for 24 h, PCa samples or mouse tumour samples were embedded in paraffin and cut into 3-µm-thick sections. After deparaffinization by xylol and rehydration by a descending alcohol series, the sections were soaked in EDTA retrieval buffers and heated in a microwave oven. Nonspecifc binding was blocked by 5% bovine serum albumin (product code AR1006; Boster Biological Technology, Inc.) at room temperature for 20 min. Subsequently, the histological sections were stained with rabbit anti-AR antibody (product code ab74272; 1:200; Abcam) and anti-HMGB1 antibody (product code A00066-1; 1:200; Boster Biological Technology, Inc.) at 4°C overnight. Goat anti-rabbit IgG conjugated with horseradish peroxidase (cat. no. SA00001-2; 1:200; ProteinTech Group, Inc.) was used as the secondary antibody, and this staining procedure was carried out at 37°C for 30 min. Reactive products were visualized with 3,3′-diaminobenzidene (Boster Biological Technology, Inc.) as the chromogen, and the sections were counterstained with 0.1% haematoxylin (Boster Biological Technology, Inc.) at room temperature for 2 min. Histological images were captured under a light microscope (Olympus Corporation) with an objective magnification of ×200 or ×400. The positive cell density was assessed using Image-Pro Plus 6.0 software (Media Cybernetics, Inc.), and the results are presented as the mean optical density (MOD) values (22).
PCa cell lines and reagents
PC3 (ATCC no. CRL-1435), C4-2 (ATCC no. CRL-3314), VCaP (ATCC no. CRL-2876), and DU145 (ATCC no. HTB-81) cells were obtained from the American Type Culture Collection. LNCaP95 cells were provided from the College of Life Science of Jilin University (Changchun, China). LNCaP (cat. no. CL-0143) and 22RV1 (cat. no. CL-0004) cells were obtained from the Procell Life Science & Technology Co., Ltd. These cells were cultured in RPMI-1640 medium (HyClone; Cytiva) supplemented with 10% foetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) in a humidified atmosphere with 5% CO2 at 37°C.
Lentiviral vector plasmid pLVX-Puro (cat. no. 632164; Clontech; Takara Bio USA, Inc.) was used to construct pLVX-HMGB1 plasmid and pLVX-shRNA1 (cat. no. 632177; Clontech; Takara Bio USA, Inc.) was used to construct PLVX-shHMGB1, PLVX-shAR, and PLVX-shScramble plasmids. The interference sequences were as follows: shHMGB1, 5′-GGACAAGGCCCGTTATGAA-3′; shAR, 5′-TAGTGCAATCATTTCTGCTGGC-3′; and shScramble, 5′-GTATAAGTCAACTGTTGAC-3′. The lentiviruses used in this study were packaged using a 4th generation lentiviral packaging system (cat. no. 631275; Clontech; Takara Bio USA, Inc.) according to the manufacturer's instructions. Briefly, after mixing the lentiviral vector plasmids with premixed complex including five lentiviral packaging plasmids and transfection reagent, the nanoparticle complex solution was added into Lenti-X 293T cells (cat. no. 632180; Clontech; Takara Bio USA, Inc.). After 24 and 48 h, the media was collected and concentrated using the Lenti-X Concentrator (cat. no. 631231; Clontech; Takara Bio USA, Inc.) according to the manufacturer's instructions. The concentration medium was used to detect the titer of the virus using a Lenti-X qRT-PCR Titration Kit (cat. no. 631235; Clontech; Takara Bio USA, Inc.) according to the manufacturer's instructions, and then stored at −80°C.
To generate LNCaP-neo and LNCaP-HMGB1 cells, LNCaP cells were infected with 30 MOI of pLVX-HMGB1 containing the HMGB1 expression gene or the empty vector pLVX (the final titer of the virus was 7.5×105 IFU/ml). The media were changed 48 h later. For stable transfection, the cells were selected in medium containing puromycin (2 µM) and maintained in medium containing puromycin (1 µM). Puromycin selection was continued for 1 week prior to harvesting cells for downstream analysis, or 2 weeks prior to further knock down AR. To detect the effect of HMGB1 on AR transcriptional activity, LNCaP-HMGB1 cells were further infected with 30 MOI of pLVX-shAR containing AR shRNA or pLVX-shScramble (the final titer of the virus was 7.5×105 IFU/ml). To generate 22RV1-shScramble and 22RV1-shHMGB1 cells, 22RV1 cells were infected with 30 MOI of pLVX-shHMGB1 containing HMGB1 shRNA or pLVX-shScramble (the final titer of the virus was 7.5×105 IFU/ml).
Cell Counting Kit-8 (CCK-8) assay
LNCaP-neo, LNCaP-HMGB1, 22RV1-shScramble and 22RV1-shHMGB1 cells were seeded into 96-well plates (1,000 cells/well) and cultured in RPMI-1640 medium (HyClone; Cytiva) supplemented with 10% charcoal-stripped FBS (Thermo Fisher Scientific, Inc.). Subsequently, 48 and 96 h later, CCK-8 reagent (10 µl/well; BIOSS) was added to each well. Following culture at 37°C for 1.5 h, the absorbance of each well was measured at 450 nm using a microplate reader (Cytation 5; BioTek Instruments, Inc.).
Reporter gene assay
The reporter gene assay was performed according to the manufacturer's instructions. Briefly, LNCaP-neo, LNCaP-HMGB1, 22RV1-shScramble and 22RV1-shHMGB1 cells were transiently transfected with an ARE-luciferase reporter plasmid using Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific, Inc.). The ARE-luciferase reporter plasmid was provided by Dr Robert Matusik at Vanderbilt University School of Medicine (Nashville, USA) as indicated in a previous study by the authors (24). After incubating with the transfection mixture at 37°C for 3 h, the cells were divided equally into 24-well plates and cultured in charcoal-stripped medium for 96 h. Subsequently, the cells were lysed in reporter lysis buffer (Promega Corporation), and the luciferase activity was determined using the Luciferase Assay System (Promega Corporation) and normalized based on the protein concentrations for each sample (24).
Reverse transcription-quantitative PCR (RT-qPCR)
LNCaP-neo, LNCaP-HMGB1, 22RV1-shScramble and 22RV1-shHMGB1 cells were seeded into 6-well plates at a density of 3×105 cells/well. After being treated with charcoal-stripped medium for 96 h, the total RNA was extracted using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.), and first-strand cDNA synthesis was performed using a cDNA synthesis kit (cat. no. AT311-02; TransGen Biotech Co., Ltd.) according to the manufacturer's instructions. Real-time PCR was used to detect the transcription levels of PSA and TMPRSS2 using the SYBR Green qPCR kit (cat. no. AQ132; TransGen Biotech Co., Ltd.) according to the manufacturer's instructions, and GAPDH was used as an internal control gene. The following primers were used: TMPRSS2 forward, 5′-ACACACCGATTCTCGTCCT-3′ and reverse, 5′-TGGCCTACTCTGGAAGTTCA-3′; PSA forward, 5′-CGATTCTTCAGGAGGCTCAT-3′ and reverse, 5′-GCTGCCCACTGCATCAG-3′; and GAPDH forward, 5′-GAAGGTGAAGGTCGGAGTC-3′ and reverse, 5′-GAAGATGGTGATGGGATTTC-3′. PCR was performed at 94°C for 30 sec, followed by 45 cycles of amplification at 94°C for 5 sec, 51°C for 15 sec and 72°C for 10 sec using ABI-Q3 (Thermo Fisher Scientific, Inc.). mRNA expression levels were quantified using 2−ΔΔCq method (25).
Western blotting
Soluble proteins were extracted from LNCaP, 22RV1, and PC3 cells treated by the indicated manner using RIPA lysis (Beyotime Institute of Biotechnology). The protein concentrations were measured by a Coomassie Plus (Bradford) Assay Kit (Thermo Fisher Scientific, Inc.). Proteins, at a concentration of 15 µg, were then subjected to each lane of a 4–15% Precast Tris-Glycine Gel (TransGen Biotech Co., Ltd.). Prestained standards (TransGen Biotech Co., Ltd.) were used as molecular weight markers. Separated proteins were electrophoretically transferred onto polyvinylidene difluoride (PVDF) membranes (iBlot system; Invitrogen; Thermo Fisher Scientific, Inc.). The membranes were then blocked with 5% skim milk for 1 h at room temperature, and incubated with primary antibodies, including anti-AR (product code ab74272; 1:100; Abcam), anti-HMGB1 (product code A00066-1; 1:1,000; Boster Biological Technology, Inc.), and anti-GAPDH (product code ab245355; 1:5,000; Abcam), at room temperature for 2 h. Subsequently, following incubation with secondary-HRP antibodies (cat. no. SA00001-2; 1:20,000; ProteinTech Group, Inc.) at room temperature for 1 h, the protein levels were detected with an ECL plus kit and developed via an imaging densitometer (both from Clinx Science Instruments Co., Ltd.). The assay was repeated three times independently.
Bioluminescence resonance energy transfer (BRET) assay
To further detect the interaction between HMGB1 and AR protein, PC3 cells were co-transfected with an HMGB1-RLuc BRET fusion plasmid (Donor) and an AR-Turbo BRET fusion plasmid (Receptor). After being cultured in 10% charcoal-stripped medium for 48 h, the cells were detached with 5 mM EDTA in PBS and resuspended in PBS with 1% sucrose. Subsequently, the cells were counted and seeded in triplicate into a 96-well white-wall microplate at 1×105 cells/well. Freshly prepared coelenterazine (Nanolight Technologies; Prolume Ltd.) in water was added to the cells at a final concentration of 25 µM. BRET readings at 528 and 635 nm were obtained immediately with a Synergy 2 microplate reader (BioTek Instruments, Inc.). The BRET ratio was calculated by subtracting the ratio of the 635-nm emission and the 528-nm emission obtained from cells coexpressing the HMGB1-RLuc and AR-Turbo fusion proteins from the background BRET ratio resulting from cells expressing the RLuc fusion protein alone in the same experiment: BRET ratio=(emission at 635 nm)/(emission at 528 nm)-(emission at 635 nm RLuc only)/(emission at 528 nm RLuc only) (10).
Chromatin immunoprecipitation (ChIP) assay
To illustrate the effect of HMGB1 on the reactivation of the AR signalling pathway, a Magnetic Bead ChIP assay kit (cat. no. 26157; Thermo Fisher Scientific, Inc.) was used with an AR antibody (product code ab108341; Abcam) to determine the effect of HMGB1 on AR binding events at known AREs of the PSA and TMPRSS2 genes. The ChIP assay was performed according to the manufacturer's instructions. Briefly, after being treated with charcoal-stripped medium for 48 h, the cells were incubated with 1% formaldehyde at room temperature for 10 min. The nuclear DNA-protein complexes of LNCaP-neo, LNCaP-HMGB1, 22RV1-shScramble, and 22RV1-shHMGB1 cells were extracted using Lysis Buffer (200 µl for each IP reaction) of the kit, and then were centrifuged at 9,000 × g for 5 min (4°C). Subsequently, the nuclei was sonicated to produce 200–500 bp fragments. The chromatin extract was incubated with antibodies against AR (2 µg for each IP reaction), 10 µl RNA Polymerase II (Thermo Fisher Scientific, Inc.), or 2 µl IgG (Thermo Fisher Scientific, Inc.) overnight at 4°C. A uniform suspension of 20 µl containing Protein A/G Magnetic Beads was added to each IP reaction and incubated for 2 h at 4°C with mixing. Following incubation, the beads were collected and washed three times with IP Wash Buffer 1. After immunoprecipitation, the IP Elution Buffer was used to elute the protein-chromatin complex from the beads at 65°C for 30 min, and the complex was then treated with proteinase K at 65°C for 1.5 h. Finally, the purified DNA was analysed using RT-qPCR.
In vivo effect of HMGB1 on the development of PCa
A total number of 20 male BALB/c nu/nu mice (age, seven weeks old; weight, 15–20 g) were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. The animal experiments were approved by the Animal Ethics Committee of Basic Medical College of Jilin University (Changchun, China), complied with the recommendations of the ARRIVE guidelines, and were carried out in accordance with the Good Laboratory Practice Regulations. Approximately 0.1 ml of LNCaP-neo or LNCaP-HMGB1 cells was mixed with 50% Matrigel and subcutaneously implanted into the right flank region of BALB/c nu/nu mice at a dosage of 4×106 cells/mouse. When the tumour volume reached 100 mm3, the mice were subjected to sham surgery (intact group) or castration (castration group). The tumour volume was calculated as follows: V=L × W2/2, where L and W represent the length and width of the tumour, respectively (26). The weight and tumour volume were measured weekly. When the volume of tumours in the intact group or castration group reached 2,000 mm3, all mice were euthanized by cervical dislocation under 2–3% isoflurane anaesthesia, and tumour samples were weighed and collected.
ELISA
After mice were anaesthetized with isoflurane (a concentration of 2 to 3% for induction and 1.5 to 2% for maintenance), mouse serum was collected and used for PSA detection. The level of PSA was determined by an ELISA kit (cat. no. MM-0421H1; Meimian; http://www.mmbio.cn/) according to the manufacturer's instructions. Finally, the absorbance was detected at 450 nm using a microplate reader (BioTek Instruments, Inc.).
Statistical analysis
Statistical analysis was performed with multiple comparisons using one-factor analysis of variance (ANOVA) followed by Dunnett's post hoc test or Bonferroni's correction test, and the differences among two dependent samples were analysed using an unpaired Student's t-test. The linear relationships between two continuous variables were analyzed by Pearson's correlation analysis. Spearman's correlation analysis was used to determine the correlations between rank variables and continuous variables. All data were analysed with the statistical software SPSS 17.0 (SPSS, Inc.), and the results are expressed as the means ± SD. The results were repeated three times independently. P<0.05 was considered to indicate a statistically significant difference.
Results
Association of AR and HMGB1 mRNA levels in PCa
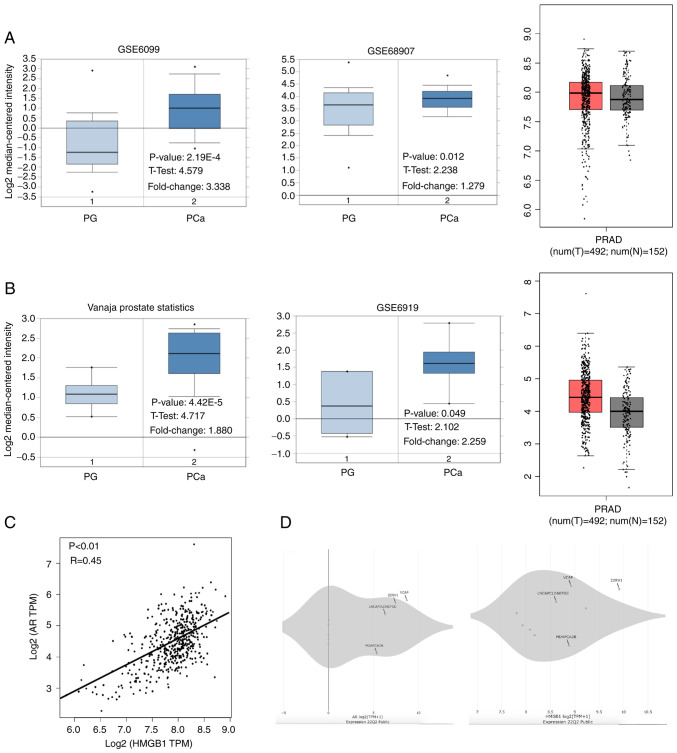
The associations between the mRNA levels of AR and HMGB1 in PCa were analysed using the Oncomine and GEPIA databases. As shown in Fig. 1A and B, the mRNA levels of AR and HMGB1 were increased in PCa tissues compared with normal prostate glands (P<0.05). Interestingly, a significant correlation between AR and HMGB1 mRNA expression in prostate tissues was found using Pearson analysis in the GEPIA database (R=0.45, P<0.01; Fig. 1C). Additionally, the data showed that the mRNA level of AR and HMGB1 was increased in CPRC cells (VCaP and 22RV1) compared with androgen-dependent PCa cells (LNCaP and MDAPCA2B) (Fig. 1D). The data also revealed that the expression levels of AR protein and HMGB1 protein were higher in 22RV1 cells than that in LNCaP cells (Fig. S1). LNCaP cells were employed to overexpress HMGB1 because of its low expression of HMGB1 and androgen-dependent feature. Moreover, although HMGB1 was upregulated in both 22RV1 and LNCaP95 cells compared with the other PCa cell lines, it was determined that 22RV1 cells exhibited a higher HMGB1 expression than that in LNCaP95 cells, indicating that 22RV1 cells may be more suitable for silencing HMGB1 in the subsequent experiments (Fig. S1).
Figure 1.
Roles of HMGB1 mRNA in the development of PCa and the correlation with the expression of AR mRNA, analysed using bioinformatics. (A) The levels of HMGB1 mRNA in normal PG and PCa were analysed using the Oncomine and GEPIA databases. (B) The levels of AR mRNA in normal PG and PCa were analysed using the Oncomine and GEPIA databases. (C) The correlation between the expression of HMGB1 mRNA and AR mRNA was detected by the GEPIA database. (D) The levels of AR mRNA and HMGB1 mRNA in VCaP, 22RV1, LNCaP, and MDAPCA2B cells were analysed using the DepMap Portal database. HMGB1, high-mobility group protein B1; PCa, prostate cancer; AR, androgen receptor; PG, prostate gland; PRAD, prostate adenocarcinoma.
Association of AR and HMGB1 proteins in human prostate tissues
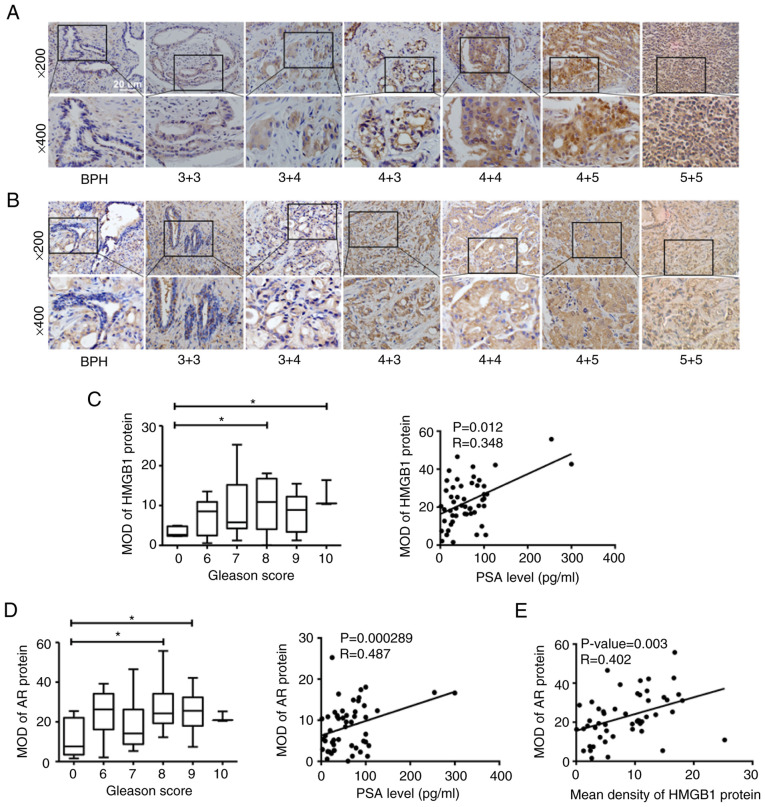
In this study, IHC staining was carried out to detect the protein expression of HMGB1 and AR in human prostate needle biopsy tissue specimens, including BPH and PCa. Positive staining of HMGB1 and AR in various pathologic categories is presented in Fig. 2A and B. By contrast to a few disseminated cells in BPH samples, cytoplasmic brown staining for HMGB1 and AR protein was widely observed in PCa tissues. For PCa specimens, the protein expression levels of HMGB1 and AR gradually increased in the epithelial cytoplasm of PCa cells according to the increasing PSA level (HMGB1: r=0.348, P=0.012; AR: r=0.487, P=0.000289) as well as the Gleason Score (GS) (Fig. 2C and D). These results indicated that both HMGB1 and AR are closely associated with the development of PCa. Moreover, the data showed a significant positive correlation between HMGB1 and AR expression in PCa tissues (r=0.402, P=0.003; Fig. 2E). The correlations of AR and HMGB1 expression with Gleason score, PSA and age are also presented in Table III.
Figure 2.
Expression levels of HMGB1 and AR proteins in the specimens of patients with PCa detected by IHC staining. (A) The expression levels of HMGB1 protein in PCa specimens with different Gleason scores. (B) The expression levels of AR protein in PCa specimens with different Gleason scores. (C) The association between HMGB1 protein expression in the specimens with Gleason scores and PSA levels. (D) The association between AR protein expression in the specimens and Gleason scores and PSA levels. (E) The correlation between HMGB1 protein expression and AR protein expression in the specimens of the patients. *P<0.05. HMGB1, high-mobility group protein B1; AR, androgen receptor; PCa, prostate cancer; IHC, immunohistochemical; MOD, mean optical density.
Table III.
Correlation of AR and HMGB1 expression with Gleason score, PSA and age.
| Prostate tumor | ||||
|---|---|---|---|---|
|
|
||||
| Variable | Values | Gleason scorea | PSAb | Ageb |
| MOD of AR Protein | R | 0.259 | 0.487b | −0.018 |
| P-value | 0.067 | 0.000 | 0.9 | |
| MOD of HMGB1 Protein | R | 0.298a | 0.348b | −0.182 |
| P-value | 0.033 | 0.012 | 0.202 | |
Spearman's test with significance, P<0.05;
Person's test with significance, P<0.05 and P<0.01. AR, androgen receptor; HMGB1, high-mobility group protein B1; PSA, prostate specific antigen; MOD, mean optical density.
HMGB1 promotes the proliferation of PCa cells
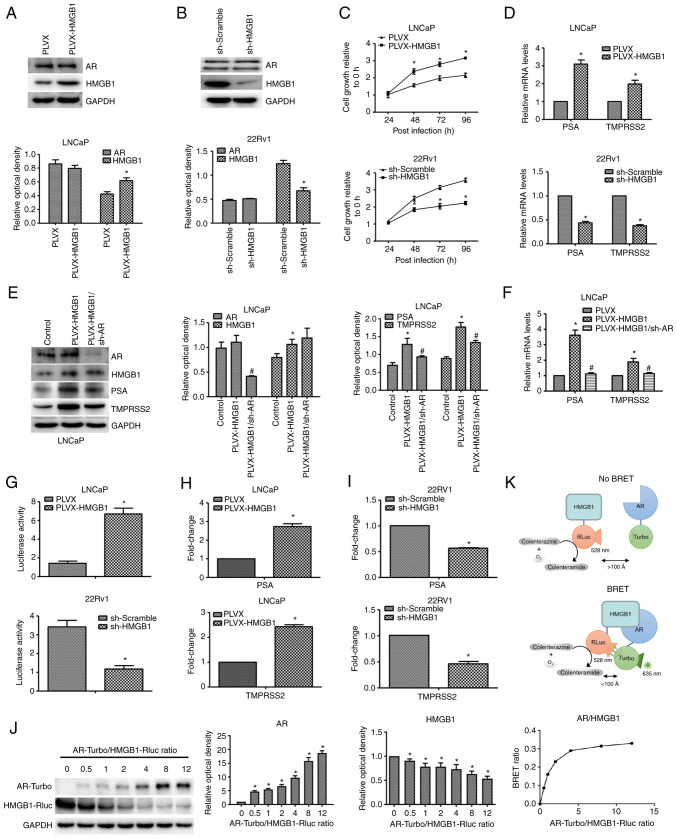
Following transfection with HMGB1 or shHMGB1 using the pLVX plasmid, the expression of HMGB1 protein was successfully overexpressed or silenced in LNCaP or 22RV1 cells (Fig. 3A and B), respectively. The effect of HMGB1 expression on LNCaP and 22RV1 cells was examined using a CCK-8 assay under charcoal-stripped conditions. As revealed in Fig. 3C, HMGB1 overexpression significantly promoted the growth ability of LNCaP cells, while silencing of HMGB1 inhibited the viability of 22RV1 cells. However, no significant effect of HMGB1 on AR protein expression was detected (Fig. 3A and B).
Figure 3.
HMGB1 protein activates the AR signalling pathway by directly interacting with AR protein in PCa in vitro. (A) Following transfection with exogenous HMGB1, the expression levels of AR and HMGB1 in LNCaP were detected by western blotting. *P<0.05 compared with pLVX or sh-Scramble. (B) Following transfection with sh-HMGB1, the expression levels of AR and HMGB1 in 22RV1 cells were detected by western blotting. *P<0.05 compared with pLVX or sh-Scramble. (C) The growth abilities of LNCaP-neo, LNCaP-HMGB1, 22RV1-shScramble, and 22RV1-shHMGB1 cells were detected by CCK-8 assay. *P<0.05 compared with the 0-h group. (D) The effects of HMGB1 on the transcription levels of PSA and TMPRSS2 in LNCaP-neo, LNCaP-HMGB1, 22RV1-shScramble, and 22RV1-shHMGB1 cells were examined by RT-qPCR. *P<0.05 compared with pLVX or sh-Scramble. (E) After treatment with pLVX-shAR, AR, HMGB1, PSA, and TMPRSS2 protein expression levels in LNCaP-neo, LNCaP-HMGB1 and LNCaP-HMGB1/shAR cells were detected by western blotting. *P<0.05 compared with the control group; #P<0.05 compared with the pLVX-HMGB1 group. (F) The effects of HMGB1 and AR on the transcription levels of PSA and TMPRSS2 in LNCaP-neo, LNCaP-HMGB1 and LNCaP-HMGB1/shAR cells were examined by RT-qPCR. *P<0.05 compared with the pLVX group; #P<0.05 compared with the pLVX-HMGB1 group. (G) Following transfection with exogenous HMGB1 or sh-HMGB1, the transactivating activity of AR in LNCaP-neo, LNCaP-HMGB1, 22RV1-shScramble, and 22RV1-shHMGB1 cells was determined by a reporter gene assay. *P<0.05 compared with the pLVX group. (H and I) ChIP assay followed by RT-qPCR was used to detect the effect of HMGB1 on the ability of AR to bind to the promoters of PSA and TMPRSS2 in LNCaP and 22RV1 cells. *P<0.05 compared with the pLVX group or sh-Scramble group. (J) AR-Turbo and HMGB1-Rluc, the BRET fusion constructs, were cotransfected into PC3 cells, and the BRET signal was measured after the addition of the coelenterazine substrate. Western blotting revealed the fold changes of the expression levels of the fusion proteins. *P<0.05 compared with the AR-Turbo/HMGB1-Rluc ratio=0 group. (K) A schematic of the principle of the BRET assay. HMGB1, high-mobility group protein B1; AR, androgen receptor; PCa, prostate cancer; PSA, prostate specific antigen; TMPRSS2, transmembrane protease, serine 2; RT-qPCR, reverse transcription-quantitative PCR; ChIP, chromatin immunoprecipitation; BRET, bioluminescence resonance energy transfer.
HMGB1 promotes the androgen-independent transactivating activity of AR
As demonstrated in Fig. 3D, overexpression of HMGB1 significantly increased the levels of PSA and TMPRSS2 mRNA in LNCaP-HMGB1 cells under charcoal-stripped conditions, which indicated that HMGB1 promotes the transactivating activity of AR. Moreover, the increasing levels of PSA and TMPRSS2 mRNA and protein were almost completely diminished by AR knockdown (Fig. 3E and F). Furthermore, overexpression of HMGB1 significantly promoted the luciferase activity of ARE in LNCaP-HMGB1 cells under charcoal-stripped conditions, while the luciferase activity of ARE was inhibited after silencing of HMGB1 in 22RV1 cells (Fig. 3G). These results indicated that HMGB1 promoted the androgen-independent transactivating activity of AR by increasing the recruitment of AR to ARE-luc.
HMGB1 promotes the binding of AR to the promoter/enhancer of AR downstream genes
To explore the underlying molecular mechanism of HMGB1 on androgen-independent reactivation of the AR signalling pathway, a ChIP assay was used to detect the binding levels of the promoter/enhancer of AR downstream genes. As shown in Fig. 3H, the overexpression of HMGB1 in LNCaP cells significantly upregulated the binding levels of the promoter/enhancer of PSA and TMPRSS2 in the ChIP assay, indicating that HMGB1 could strengthen the ability of AR to bind with the promoter/enhancer of its downstream genes PSA and TMPRSS2 under androgen-free conditions. These results were further confirmed by the data which revealed that silencing of HMGB1 in 22RV1 cells significantly downregulated the binding levels of the promoter/enhancer of PSA and TMPRSS2 in the ChIP assay (Fig. 3I).
HMGB1 initiates the reactivation of the AR signalling pathway by directly interacting with the AR protein
To further demonstrate the way in which HMGB1 initiates the reactivation of the AR signalling pathway, a BRET assay was performed to identify the interaction between AR and HMGB1 proteins. The BRET saturation curve for the combination of AR/HMGB1 fusion proteins in PC3 cells is presented in Fig. 3J. The BRET ratios increased hyperbolically and rapidly saturated with the increase in the ratio of energy acceptor to energy donor, indicating specific protein interactions (10). The BRET data revealed the ability of HMGB1 protein to interact with AR protein. The mechanisms of the BRET assay are shown in Fig. 3K.
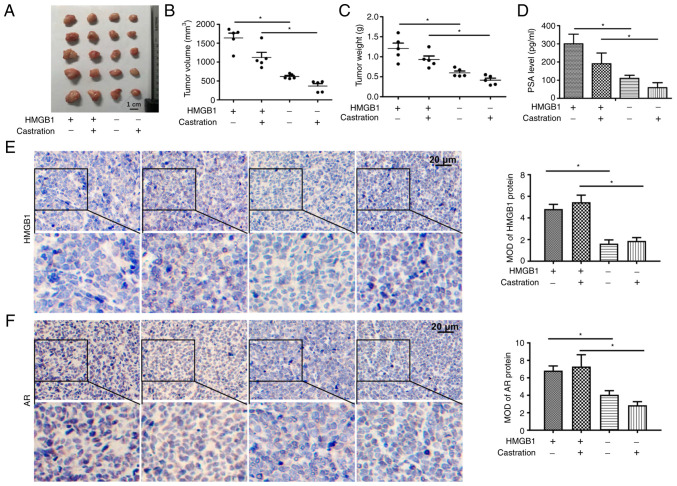
HMGB1 promotes the development of PCa in vivo
To verify whether overexpression of HMGB1 could promote the development of PCa, a LNCaP xenograft model was developed. The results revealed that overexpression of HMGB1 significantly increased the volume and weight of prostate tumours (Fig. 4A-C). Furthermore, the serum PSA level of mice exhibited the same tendency as the growth of the tumours (Fig. 4D). The data from IHC demonstrated that the expression of HMGB1 in tumour tissue was significantly increased in the two groups injected with LNCaP-HMGB1 cells (Fig. 4E). As shown in Fig. 4F, overexpression of HMGB1 significantly increased AR protein expression in the cell nuclei of the tumour tissues in both the intact and castration groups. These results suggested that HMGB1 promoted the development of PCa in an androgen-independent manner.
Figure 4.
HMGB1 promotes the development of prostate cancer in vivo. (A-C) HMGB1 significantly increased the volume and weight of tumours in a LNCaP xenograft model. (D) The effect of HMGB1 on the level of PSA in mouse serum was examined by ELISA. (E) HMGB1 protein expression in mouse tumour tissues detected by IHC. (F) AR protein expression in mouse tumour tissues detected by IHC. *P<0.05. HMGB1, high-mobility group protein B1; PSA, prostate specific antigen; IHC, immunohistochemical; AR, androgen receptor; MOD, mean optical density.
Discussion
The present study is the first, to the best of our knowledge, to reveal the association between HMGB1 and AR protein in CRPC cells and PCa patients. Using bioinformatics analysis and IHC, it was demonstrated that the expression levels of AR and HMGB1 were not only associated to the malignant progression of PCa but also correlated with each other. By overexpressing HMGB1 in LNCaP cells and silencing HMGB1 in 22RV1 cells, and performing a ChIP assay, it was further revealed that the ability of AR protein to bind to ARE is mediated by HMGB1 protein upon androgen deprivation. Notably, using BRET, the interaction effect between HMGB1 protein and AR protein, was directly identified, indicating the requirement of HMGB1 protein for important AR protein functions in CRPC.
The finding in the present study, on the correlation between HMGB1 expression and the malignant progression of PCa patients is in accordance with a previous study (27). Wang et al determined that the expression of HMGB1 protein was induced by enzalutamide in mouse CRPC tissues (28). These findings provide clinical and experimental evidence for the hypothesis proposed by the authors, that HMGB1 may be a promising target for CRPC treatment. Furthermore, it was revealed that the expression levels of HMGB1 in PCa samples were significantly correlated with the levels of AR. Together with the consideration of the similarity between the structure of ER and AR (15) as well as the effect of HMGB1 on ER signalling pathway activation (29), it was hypothesized that HMGB1 protein contributed to CRPC development by interacting with AR protein and then reactivating the AR signalling pathway.
A previous study revealed that HMGB1 knockdown inhibited the development of enzalutamide-induced CRPC in a mouse model (28). This is supported by the results of the present study showing that HMGB1 knockdown inhibited the growth of androgen-independent 22RV1 cells and promoted the growth of androgen-dependent LNCaP cells without the existence of androgen. Moreover, the in vivo data showed that overexpression of the HMGB1 protein attenuated the treatment effect of ADT on LNCaP-bearing mice. This indicated that HMGB1 could promote the transformation of PCa into CRPC in the presence of ADT. Thus, it is suggested that targeting HMGB1 may be a potential strategy for CRPC prevention. Interestingly, it was determined that the expression of AR was not regulated by HMGB1 protein in vitro, whereas the expression levels were significantly associated in the tissues of patients PCa. A plausible explanation is that correlation in expression does not signify that AR expression is directly regulated by HMGB1, and HMGB1 may only promote AR activity instead of expression in vitro. The in vivo results that HMGB1 overexpression promoted AR protein expression in the cell nucleus of tumour tissues provided a further explanation. It is well known that AR protein is predominantly located in the cytoplasm. Only after a nuclear translocation process can the activity of AR protein be activated with or without the presence of androgens (7,30). From this perspective, the in vivo data of the present study indicated that HMGB1 could activate AR activity by promoting the nuclear translocation process of AR. However, the detailed mechanism requires further investigation. Furthermore, it is suggested that this may also be due to the short regulation time of HMGB1 on AR in vitro. This short time is only enough for HMGB1 to promote AR activity instead of expression. However, this assumption requires more experiments for verification. Although the conclusion that HMGB1 protein could regulate the development of CRPC was in accordance with a previous study, a different mechanism of the effect of HMGB1 protein on CRPC was also identified in the present study.
Based on the data that HMGB1 protein could induce androgen-independent increases in the mRNA expression levels of PSA and TMPRSS2 mRNA, two AR downstream genes, in an AR-dependent manner, it is suggested that the mechanism may be related to the regulation of AR transactivating activity to its downstream genes. Interestingly, the results of the reporter gene assay revealed that HMGB1 could enhance AR transactivating activity without the existence of androgen, which confirmed the aforementioned hypothesis. Therefore, the potential mechanism of these results is continually being explored. To date, studies of AR transactivating activity in CRPC have focused mostly on its ability to bind to ARE. Selenium suppresses the binding of AR to the ARE and then inhibits the AR signalling pathway (31). Collak et al determined that Yes associated protein 1 (YAP1) and AR could form a complex and ultimately bind to the ARE of PSA (32). Interestingly, it was determined that HMGB1 promoted the ability of AR binding to the AREs of PSA and TMPRSS2 genes in a ChIP assay, which directly identified the promoting effect of HMGB1 protein on AR protein activity.
AR activity is affected not only by ligand binding but also by homodimerization and by interactions with cofactors (7). Over 170 cofactors of AR have been found to bind to AR and influence the transactivating activity of AR at low levels of androgen (6). The data of BRET performed in the present study, indicated that HMGB1 protein could be a cofactor and directly interact with AR protein through the formation of an HMGB1/AR complex in an androgen-independent manner in CRPC. This, together with the finding that the binding of AR protein to ARE was promoted by HMGB1 protein, demonstrated that the HMGB1/AR complex could contribute to the activation of the AR signalling pathway, which may be the key mechanism by which the HMGB1 protein promotes the malignant progression of PCa. Thus, disrupting AR/HMGB1 interactions may be an effective strategy for CRPC prevention as well as treatment.
In conclusion, the present study revealed the effects of HMGB1 on the development of CRPC in vitro and in vivo and described its mechanisms. The results demonstrated that HMGB1 promoted the development of CRPC by interacting with AR. Targeting HMGB1 may be a novel and effective treatment strategy for CPRC prevention and treatment.
Supplementary Material
Acknowledgements
Not applicable.
Glossary
Abbreviations
- CRPC
castration-resistant prostate cancer
- AR
androgen receptor
- AREs
androgen response elements
- HMGB1
high-mobility group protein B1
- PCa
prostate cancer
- ADT
androgen deprivation therapy
- PSA
prostate specific antigen
- ER
estrogen receptor
- ERE
estrogen response element
- IHC
immunohistochemical
- MOD
mean optical density
- BRET
bioluminescence resonance energy transfer
- ChIP
Chromatin immunoprecipitation
- BPH
benign prostate hyperplasia
Funding Statement
The present study was supported by the National Natural Science Foundation of China Project (grant no. 81602228) and Jilin Province Development and Reform Commission (grant no. 2019C050-5).
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
DX, JC, and LZ conceived and designed the experiments. JC and TW performed the experiments including data collection and data analysis. ZY and KH analyzed the data. JC and TW wrote the original manuscript. ZY and YY contributed to revising the manuscript for intellectual content and language editing. LZ and KH supervised the study and were involved in project management. LZ and JC confirm the authenticity of the raw data. All authors read and approved the final manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of the School of Nursing, Jilin University (Changchun, China), and the animal experiments were approved by the Animal Ethics Committee of Basic Medical College of Jilin University (Changchun, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing financial interests or personal relationships that could have influenced the research reported in the present study.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Glina S, Rivero MA, Morales A, Morgentaler A. Studies on prostatic cancer I. The effect of castration, of estrogen and of androgen injection on serum phosphatases in metastatic carcinoma of the prostate by Charles Huggins and Clarence V. Hodges. J Sex Med. 2010;7:640–644. doi: 10.1111/j.1743-6109.2009.01680.x. [DOI] [PubMed] [Google Scholar]
- 3.Guan W, Li F, Zhao Z, Zhang Z, Hu J, Zhang Y. Tumor-associated macrophage promotes the survival of cancer cells upon docetaxel chemotherapy via the CSF1/CSF1R-CXCL12/CXCR4 axis in castration-resistant prostate cancer. Genes (Basel) 2021;12:773. doi: 10.3390/genes12050773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kluetz PG, Ning YM, Maher VE, Zhang L, Tang S, Ghosh D, Aziz R, Palmby T, Pfuma E, Zirkelbach JF, et al. Abiraterone acetate in combination with prednisone for the treatment of patients with metastatic castration-resistant prostate cancer: U.S. Food and drug administration drug approval summary. Clin Cancer Res. 2013;19:6650–6656. doi: 10.1158/1078-0432.CCR-13-2134. [DOI] [PubMed] [Google Scholar]
- 5.Scher HI, Fizazi K, Saad F, Taplin ME, Sternberg CN, Miller K, de Wit R, Mulders P, Chi KN, Shore ND, et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med. 2012;367:1187–1197. doi: 10.1056/NEJMoa1207506. [DOI] [PubMed] [Google Scholar]
- 6.Egan A, Dong Y, Zhang H, Qi Y, Balk SP, Sartor O. Castration-resistant prostate cancer: Adaptive responses in the androgen axis. Cancer Treat Rev. 2014;40:426–433. doi: 10.1016/j.ctrv.2013.09.011. [DOI] [PubMed] [Google Scholar]
- 7.van Royen ME, van Cappellen WA, de Vos C, Houtsmuller AB, Trapman J. Stepwise androgen receptor dimerization. J Cell Sci. 2012;125:1970–1979. doi: 10.1242/jcs.096792. [DOI] [PubMed] [Google Scholar]
- 8.Tran C, Ouk S, Clegg NJ, Chen Y, Watson PA, Arora V, Wongvipat J, Smith-Jones PM, Yoo D, Kwon A, et al. Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science. 2009;324:787–790. doi: 10.1126/science.1168175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ryan CJ, Smith MR, Fizazi K, Saad F, Mulders PF, Sternberg CN, Miller K, Logothetis CJ, Shore ND, Small EJ, et al. Abiraterone acetate plus prednisone versus placebo plus prednisone in chemotherapy-naive men with metastatic castration-resistant prostate cancer (COU-AA-302): Final overall survival analysis of a randomised, double-blind, placebo-controlled phase 3 study. Lancet Oncol. 2015;16:152–160. doi: 10.1016/S1470-2045(14)71205-7. [DOI] [PubMed] [Google Scholar]
- 10.Xu D, Zhan Y, Qi Y, Cao B, Bai S, Xu W, Gambhir SS, Lee P, Sartor O, Flemington EK, et al. Androgen receptor splice variants dimerize to transactivate target genes. Cancer Res. 2015;75:3663–3671. doi: 10.1158/0008-5472.CAN-15-0381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xue J, Suarez JS, Minaai M, Li S, Gaudino G, Pass HI, Carbone M, Yang H. HMGB1 as a therapeutic target in disease. J Cell Physiol. 2021;236:3406–3419. doi: 10.1002/jcp.30125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jung AR, Kim GE, Kim MY, Ha US, Hong SH, Lee JY, Kim SW, Park YH. HMGB1 promotes tumor progression and invasion through HMGB1/TNFR1/NF-κB axis in castration-resistant prostate cancer. Am J Cancer Res. 2021;11:2215–2227. [PMC free article] [PubMed] [Google Scholar]
- 13.Chou YE, Yang PJ, Lin CY, Chen YY, Chiang WL, Lin PX, Huang ZY, Huang M, Ho YC, Yang SF. The impact of HMGB1 polymorphisms on prostate cancer progression and clinicopathological characteristics. Int J Environ Res Public Health. 2020;17:7247. doi: 10.3390/ijerph17197247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Das D, Peterson RC, Scovell WM. High mobility group B proteins facilitate strong estrogen receptor binding to classical and half-site estrogen response elements and relax binding selectivity. Mol Endocrinol. 2004;18:2616–2632. doi: 10.1210/me.2004-0125. [DOI] [PubMed] [Google Scholar]
- 15.Powell E, Xu W. Intermolecular interactions identify ligand-selective activity of estrogen receptor alpha/beta dimers. Proc Natl Acad Sci USA. 2008;105:19012–19017. doi: 10.1073/pnas.0807274105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bjerregaard-Olesen C, Ghisari M, Kjeldsen LS, Wielsøe M, Bonefeld-Jørgensen EC. Estrone sulfate and dehydroepiandrosterone sulfate: Transactivation of the estrogen and androgen receptor. Steroids. 2016;105:50–58. doi: 10.1016/j.steroids.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 17.Mafuvadze B, Liang YY, Hyder SM. Cholesterol synthesis inhibitor RO 48-8071 suppresses transcriptional activity of human estrogen and androgen receptor. Oncol Rep. 2014;32:1727–1733. doi: 10.3892/or.2014.3332. [DOI] [PubMed] [Google Scholar]
- 18.Tomlins SA, Mehra R, Rhodes DR, Cao X, Wang L, Dhanasekaran SM, Kalyana-Sundaram S, Wei JT, Rubin MA, Pienta KJ, et al. Integrative molecular concept modeling of prostate cancer progression. Nat Genet. 2007;39:41–51. doi: 10.1038/ng1935. [DOI] [PubMed] [Google Scholar]
- 19.Singh D, Febbo PG, Ross K, Jackson DG, Manola J, Ladd C, Tamayo P, Renshaw AA, D'Amico AV, Richie JP, et al. Gene expression correlates of clinical prostate cancer behavior. Cancer Cell. 2002;1:203–209. doi: 10.1016/S1535-6108(02)00030-2. [DOI] [PubMed] [Google Scholar]
- 20.Yu YP, Landsittel D, Jing L, Nelson J, Ren B, Liu L, McDonald C, Thomas R, Dhir R, Finkelstein S, et al. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol. 2004;22:2790–2799. doi: 10.1200/JCO.2004.05.158. [DOI] [PubMed] [Google Scholar]
- 21.Vanaja DK, Cheville JC, Iturria SJ, Young CY. Transcriptional silencing of zinc finger protein 185 identified by expression profiling is associated with prostate cancer progression. Cancer Res. 2003;63:3877–3882. [PubMed] [Google Scholar]
- 22.Tian Y, Zhao L, Zhang H, Liu X, Zhao L, Zhao X, Li Y, Li J. AKR1C3 overexpression may serve as a promising biomarker for prostate cancer progression. Diagn Pathol. 2014;9:42. doi: 10.1186/1746-1596-9-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J, Xia Y, Peng Y, Wu S, Liu W, Zhang H, Wang T, Yang Z, Zhao S, Zhao L. Analysis of the association between KIN17 expression and the clinical features/prognosis of epithelial ovarian cancer, and the effects of KIN17 in SKOV3 cells. Oncol Lett. 2021;21:475. doi: 10.3892/ol.2021.12736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Y, Liu T, Hu C, Xia H, Liu W, Chen J, Wu S, Jiang Y, Xu Y, Liu W, Zhao L. Ferroptosis inducer erastin downregulates androgen receptor and its splice variants in castration-resistant prostate cancer. Oncol Rep. 2021;45:25. doi: 10.3892/or.2021.7976. [DOI] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Chen J, Zhao S, Tan W, Wang T, Wu S, Wang C, Jiang Y, Zhou T, Zhang Z, Zhao L. Attenuated Salmonella carrying plasmid co-expressing HPV16 L1 and siRNA-E6 for cervical cancer therapy. Sci Rep. 2021;11:20083. doi: 10.1038/s41598-021-99425-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li T, Gui Y, Yuan T, Liao G, Bian C, Jiang Q, Huang S, Liu B, Wu D. Overexpression of high mobility group box 1 with poor prognosis in patients after radical prostatectomy. BJU Int. 2012;110:E1125–E1130. doi: 10.1111/j.1464-410X.2012.11277.x. [DOI] [PubMed] [Google Scholar]
- 28.Wang C, Peng G, Huang H, Liu F, Kong DP, Dong KQ, Dai LH, Zhou Z, Wang KJ, Yang J, et al. Blocking the feedback loop between neuroendocrine differentiation and macrophages improves the therapeutic effects of enzalutamide (MDV3100) on prostate cancer. Clin Cancer Res. 2018;24:708–723. doi: 10.1158/1078-0432.CCR-17-2446. [DOI] [PubMed] [Google Scholar]
- 29.Ogawa S, Inoue S, Watanabe T, Hiroi H, Orimo A, Hosoi T, Ouchi Y, Muramatsu M. The complete primary structure of human estrogen receptor beta (hER beta) and its heterodimerization with ER alpha in vivo and in vitro. Biochem Biophys Res Commun. 1998;243:122–126. doi: 10.1006/bbrc.1997.7893. [DOI] [PubMed] [Google Scholar]
- 30.Cao B, Qi Y, Zhang G, Xu D, Zhan Y, Alvarez X, Guo Z, Fu X, Plymate SR, Sartor O, et al. Androgen receptor splice variants activating the full-length receptor in mediating resistance to androgen-directed therapy. Oncotarget. 2014;5:1646–1656. doi: 10.18632/oncotarget.1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dong Y, Lee SO, Zhang H, Marshall J, Gao AC, Ip C. Prostate specific antigen expression is down-regulated by selenium through disruption of androgen receptor signaling. Cancer Res. 2004;64:19–22. doi: 10.1158/0008-5472.CAN-03-2789. [DOI] [PubMed] [Google Scholar]
- 32.Collak FK, Demir U, Sagir F. YAP1 is involved in tumorigenic properties of prostate cancer cells. Pathol Oncol Res. 2020;26:867–876. doi: 10.1007/s12253-019-00634-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.