Abstract
In triple negative breast cancer (TNBC) treatment, early prediction of pathological complete response (PCR) from chemotherapy before surgical operations is crucial for optimal treatment planning. We propose a novel deep learning-based system to predict PCR to neoadjuvant chemotherapy for TNBC patients with multi-stained histopathology images of serial tissue sections. By first performing tumor cell detection and recognition in a cell detection module, we produce a set of feature maps that capture cell type, shape, and location information. Next, a newly designed spatial attention module integrates such feature maps with original pathology images in multiple stains for enhanced PCR prediction in a dedicated prediction module. We compare it with baseline models that either use a single-stained slide or have no spatial attention module in place. Our proposed system yields 78.3% and 87.5% of accuracy for patch-, and patient-level PCR prediction, respectively, outperforming all other baseline models. Additionally, the heatmaps generated from the spatial attention module can help pathologists in targeting tissue regions important for disease assessment. Our system presents high efficiency and effectiveness and improves interpretability, making it highly promising for immediate clinical and translational impact.
Keywords: Breast cancer, Convolutional neural network, Pathological complete response, Serial pathology images, Spatial attention
1. Introduction
In triple negative breast cancer (TNBC) treatment, pathological complete response (PCR) to neoadjuvant chemotherapy (NAC) is defined as the lack of all signs of cancer, especially the absence of cancer cells in pathology images of tissue samples dissected during surgery. It plays an important role in treatment planning and assessment [1,2]. Patients with negative PCR tend to have longer event-free survival and overall survival [1]. Therefore, the accurate prediction of patient PCR to neoadjuvant chemotherapy can significantly help enhance clinical treatment planning by avoiding unnecessary chemotherapy treatment for some patient cohorts. Accurate PCR prediction has a significant clinical impact as it not only reduces adverse chemotherapy effects on patient life quality but also makes it possible to go for other alternative regimes in this treatment window before surgery. However, a precise PCR prediction remains a challenging and unsolved problem.
Early studies have been conducted to predict PCR with ultrasound [3], CT/PET [4], or MRI [5,6]. However, they all have limited accuracy, thus not feasible to be deployed into clinical settings. To our best knowledge, only one study has utilized pathology images for this prediction task, which has limited success [7]. The development of a PCR prediction system using patient histopathology images is indeed conceptually rationale and promising, as histopathology images are the direct imaging source for PCR review. Furthermore, two important biomarkers, Ki-67 and phosphohistone-H3 (PHH3) characterizing tumor cell proliferation circle, are known to have a strong relationship with PCR [8,9]. However, these essential biomarkers from immunohistochemistry (IHC) images have not been jointly studied with spatially aligned tumor phenotypic information from adjacent Hematoxylin and Eosin (H&E) stained slides. Due to the absence of necessary technology development, integrated use of pathophysiological biomarkers with pathology structure features for PCR prediction remains unexplored by far. Recently, deep learning has been rapidly developed in the machine learning research field [10]. This technology has achieved groundbreaking milestones not only in conventional computer science problems [10,11], but also in a large variety of biomedical studies [12–14]. However, deep learning based studies on PCR prediction in breast cancer so far only used radiology images [15,16], limiting the system prediction accuracy and interpretability. Overall, a robust, effective, and interpretable deep learning system for PCR prediction using histopathology images is still in its primitive stage.
To address this unmet clinical need, we have developed a deep learning system that predicts PCR to neoadjuvant chemotherapy in TNBC patients with integrated use of histopathology images of serial tissue sections in multiple stains. The novelty and contribution of this work are threefold. 1) Biomarkers and pathology features integration: Instead of using single stained histopathology images, pathology images of serial tissue sections in three PCR relevant stains, including H&E, Ki-67, and PHH3, are jointly utilized in our proposed system, providing complementary molecular and pathology information. 2) Multi-task: Our proposed system detects and classifies cells before PCR prediction. Therefore, key information, such as cell type, shape, spatial organization, and the cell proliferation cycle status is provided to the PCR prediction module. This process emulates the reviewing process pathologists follow in clinical settings, making the system more rationale and interpretable. 3) Spatial attention mechanism: We have designed a novel spatial attention module that informs the PCR prediction module of tissue spatial importance map. Additionally, the spatial attention module produces heatmaps that make pathologists more informed about the machine-based PCR prediction process, significantly improving the system’s interpretability.
2. Methodology
2.1. Image Registration
To enable a joint use of serial histopathology images in multiple stains, we follow a two-step process for pathology image registration [17]. First, the global structure transformations are estimated with low-resolution image representations that can readily fit machine memory for processing. Each whole slide image is scaled down by 16 times. The serial Ki-67 and PHH3 IHC slides are matched to the corresponding reference H&E slide by both rigid and non-rigid registration sequentially. These two registration steps at the low image resolution result in transformations that restore global tissue rotation, translation, scale, and local tissue deformation. Second, low-resolution transformations are mapped to and aggregated at the high image resolution level. Each reference H&E slide is partitioned into a set of 8, 000 × 8, 000 image regions of interest at the high image resolution. The mapped and aggregated transformations are applied to H&E image regions. After mapping and interpolation, the registered image regions are extracted from the serial Ki-67 and PHH3 slides at the high resolution. Additionally, these initial registered Ki-67 and PHH3 images are subject to a second round of rigid registration for final matched image triplets. We present the schema of the registration working pipeline in Fig. 1.
Fig. 1.
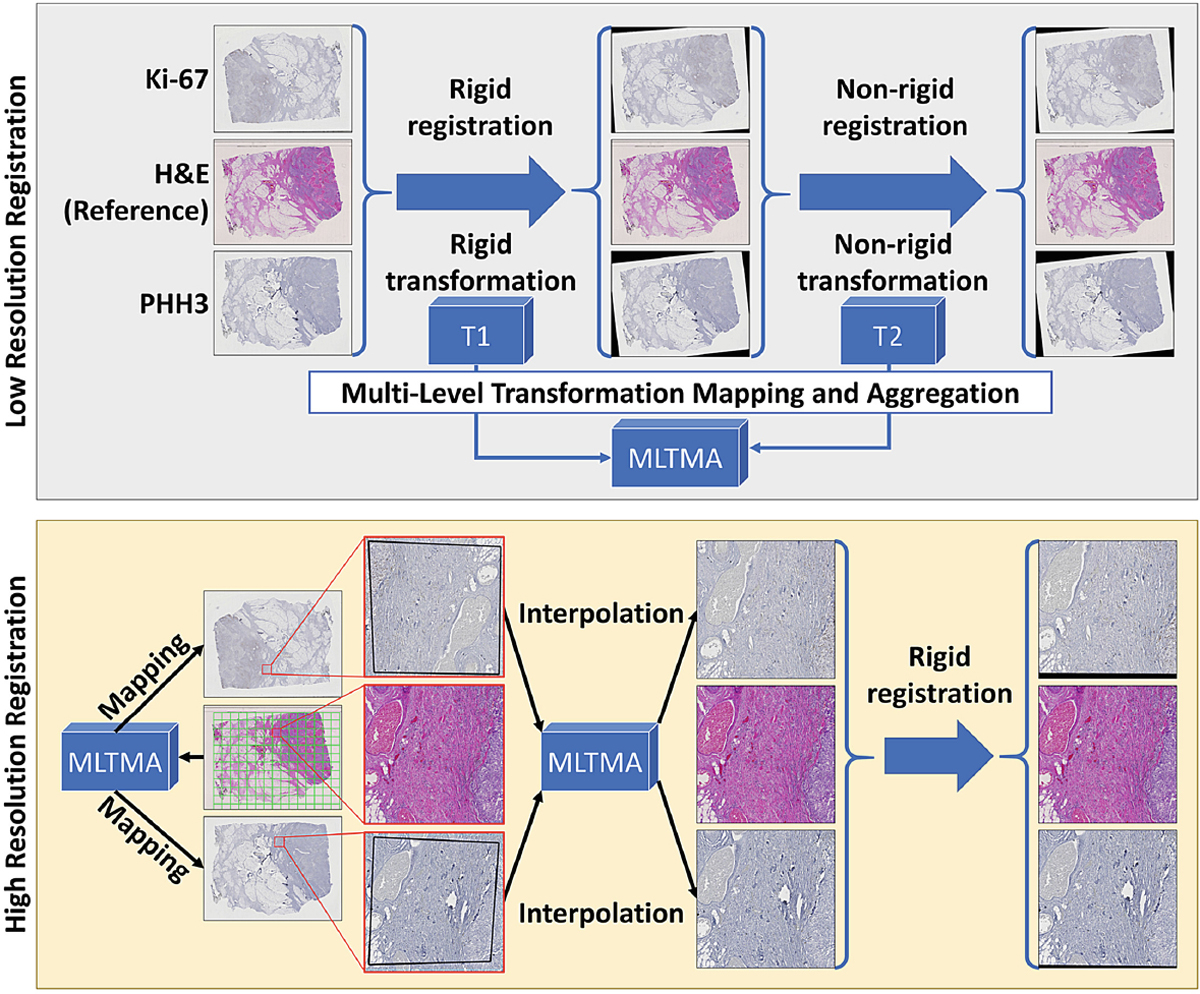
Schema of registration working pipeline.
2.2. System Architecture
Presented in Fig. 2, our proposed spatial attention-based deep learning PCR prediction system consists of three primary modules: Cell Detection Module (CDM), Spatial Attention Module (SAM), and Prediction Module (PM).
Fig. 2.
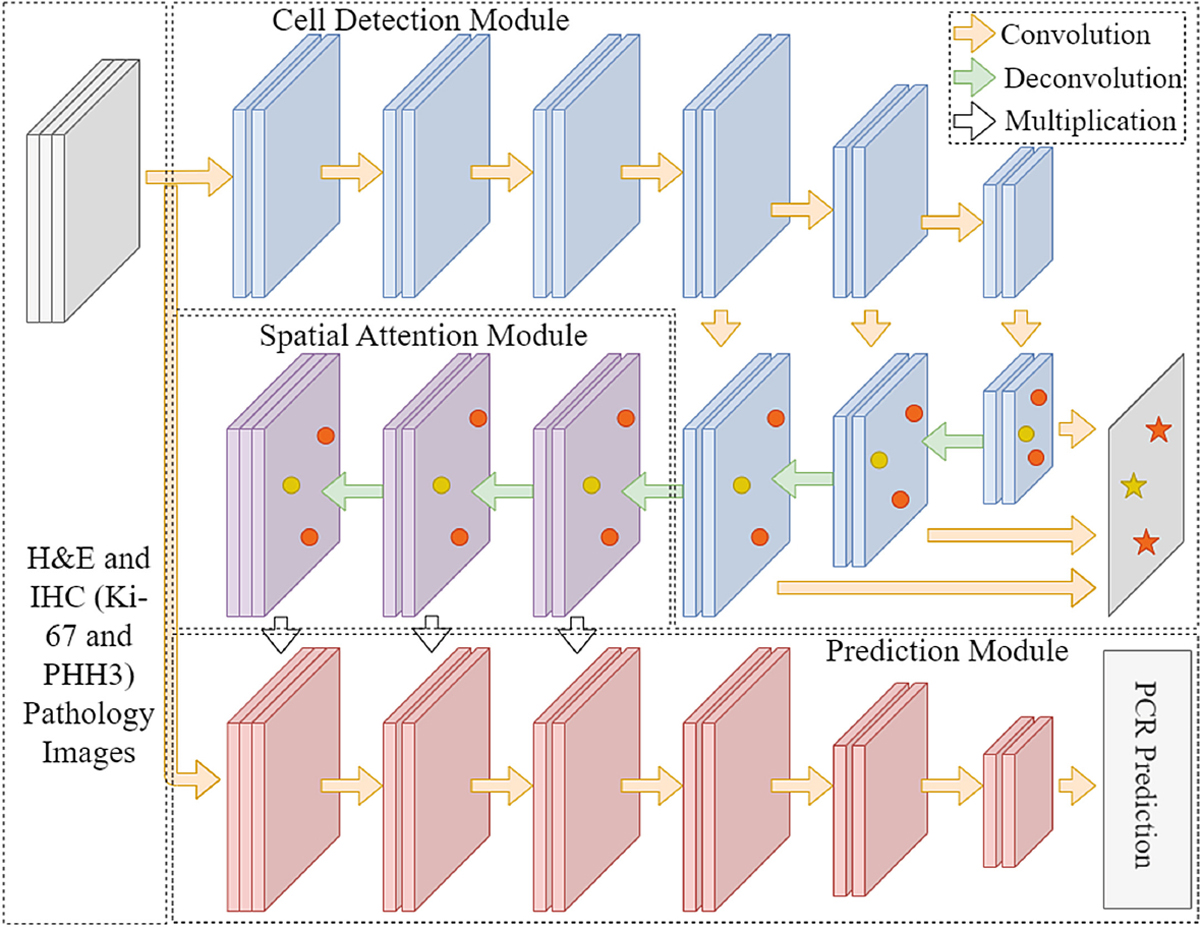
Architecture schema of our proposed multi-task deep learning system leveraging multi-stained histopathology images of serial tissue sections.
Cell Detection Module:
CDM is designed to recognize cell locations and types in H&E stained histopathology images. The high-level feature maps produced by CDM capture information on cell location, spatial organization, intensity, and types. Such critical information is provided to the next module to form spatial attention. We choose YOLO version 4 as the CDM backbone [18]. YOLO, as the first one-stage detection system, compared with two-stage detection systems, is more efficient in the global feature map generation [19]. With numerous modifications to the architecture, especially the feature-pyramid-like detector header for enhanced spatial feature extractions from different scales, YOLOv4 is improved to accommodate object variations in size [20]. Thus, we use YOLO version 4 for cell detection and classification in CDM.
Spatial Attention Module:
SAM consists of three blocks. In each block, there are sequentially two convolutional layers and one deconvolutional layer for upsampling the input feature maps. Informed of such information on cell location, spatial organization, intensity, and class label derived from the detection module, SAM generates spatial attention maps with each pixel value ranging from 0 to 1. These attention maps highlight tissue areas which the system is guided to pay special attention to. SAM includes three blocks, each producing a spatial attention map. The resulting spatial attention maps are respectively multiplied with the original pathology images and the intermediate results from the prediction module in a pixel-wise manner. The essence of the SAM is that it produces a spatial attention map that guides the following prediction module to dynamically emphasize important tissue areas essential to the final PCR prediction.
Prediction Module:
PM is used for making the final prediction from spatial attention-guided serial histopathology images in multiple stains. Three different pathology images (H&E, Ki-67, and PHH3) multiplied with spatial attention maps are set as the input of the prediction module. Taking into account the effectiveness and model complexity, we use VGG-16 as the backbone of the prediction module because of its simplicity and efficiency [21]. While the final prediction target in this study is PCR, it is natural to extend this architecture to predict other clinical outcomes, such as residual cancer burden or overall survival.
2.3. Dataset and Training
Our data are pre-NAC biopsies, which were collected before neoadjuvant therapy. We define PCR as having no evidence of residual invasive carcinoma in both the breast tissue and regional lymph nodes with the Residual Cancer Burden (RCB) value being zero. Non-PCR covers varying levels in response with evidence of residual invasive carcinoma. Note that RCB value is calculated based on the lymph nodes and Primary Tumor Bed. A total of 75 NAC treated TNBC cases are collected before neoadjuvant therapy from Dekalb Medical Center in Emory University Healthcare. Of these patients, 43 had PCR and 32 patients had residual disease (i.e. non-PCR). Formalin-fixed paraffin-embedded serial section samples for this study are obtained with information on clinical outcomes. The serial sections are H&E stained and immunohistochemically stained for Ki-67 and PHH3.
Our model training process has two steps. First, we train the Cell Detection Module with 868 40x H&E pathology image regions for tumor cell and TIL (Tumor-infiltrating lymphocytes) detection. Bounding boxes for cells of interest (53,314 tumor cells and 20,966 TILs) are labeled and classified by expert pathologists. Non-overlapped 416 × 416 image patches are cropped from these image regions for training. With the optimizer stochastic gradient descent (SGD) and the loss function for YOLOv4 [18], CDM is trained for 200 epochs with the learning rate of 0.001 using one NVIDIA V100 GPU. When the CDM is fully trained, the CDM is frozen in the later training process to avoid the overwhelming computation burden. For PCR prediction training, we use 1, 038 40x registered H&E, Ki-67, and PHH3 pathology image regions of size 8, 000 × 8, 000 in pixel. Note this data set is independent from the detection dataset for the CDM training. Typical registered pathology image triplets are presented in Fig. 3. The training set includes 455 regional images from 35 randomly selected patients, while 583 regional images from the remaining 40 patients are allocated for the testing dataset. To facilitate model training, we further partition these registered pathology image regions into non-overlapped image patches of 416 × 416 in size, generating 41,157 image patches in the training set and 46,029 in the testing set, respectively. With trained CDM fixed, SGD as the optimizer, learning rate as 0.001 for 100 epochs, SAM and PM are further trained with the cross-entropy loss for the PCR prediction.
Fig. 3.
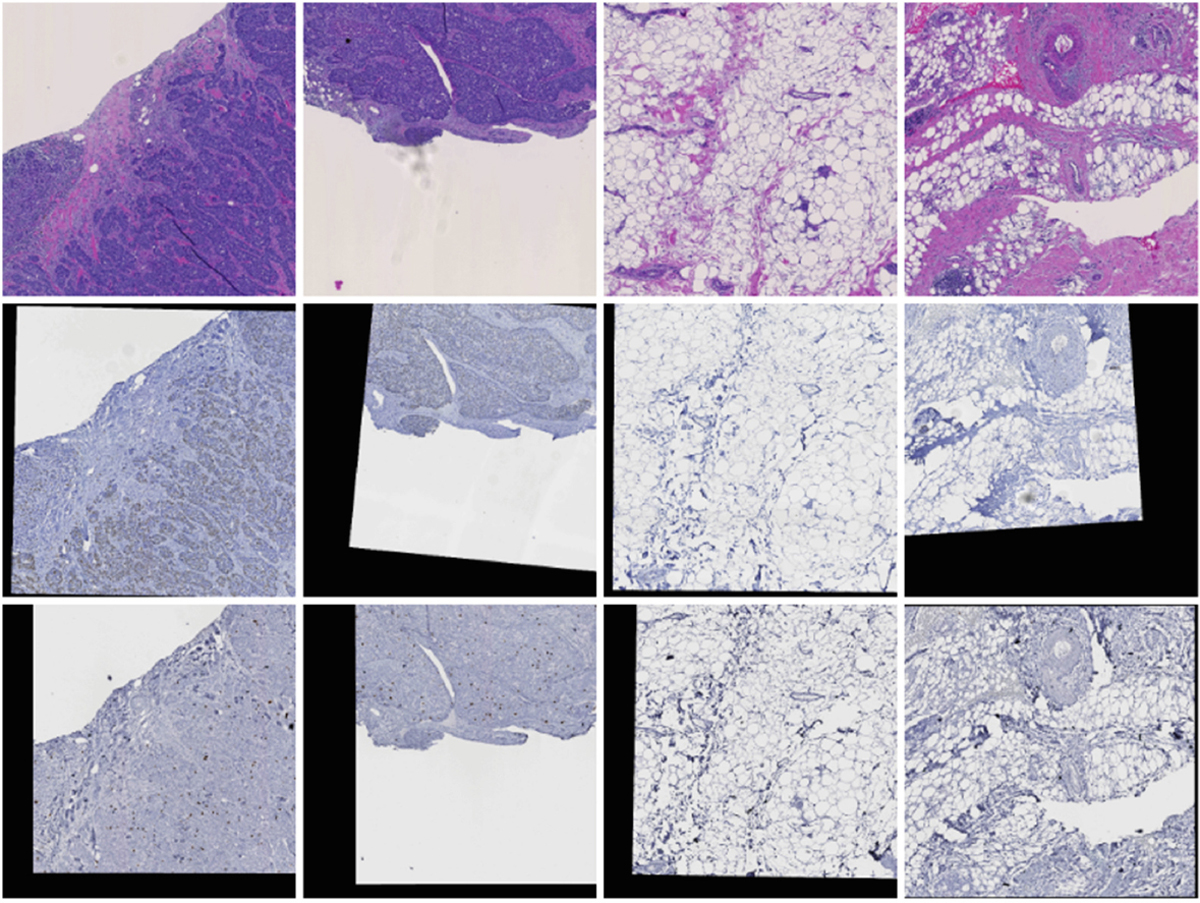
Four sample cases of registered pathology images in (Top) H&E, (Middle) Ki67, and (Bottom) PHH3 stain.
3. Experiments and Results
Our two main contributions to the PCR prediction system architecture are 1) the integrative use of H&E and IHC biomarker images of adjacent tissue sections and 2) spatial attention-based prediction. To evaluate the effectiveness of these new modules, we have designed three other baseline systems for comparison. The first baseline system only takes H&E histopathology images as input and has only a prediction module. With the first system serving as the building block, the second model upgrades its input and processes the multi-stained input images. By contrast, the third system for comparison takes H&E images, but it is equipped with the spatial attention mechanism that leverages information from the detection model to guide the prediction module. In other words, “single stain” systems are only provided with H&E stained pathology images without adjacent Ki-67 and PHH3 images. “Prediction only” systems only have a prediction module without the cell detection module and spatial attention module. Finally, “spatial attention” systems retain the same pre-trained detection module for fair comparisons. All models are fully trained to their best performance with the same training and testing configuration.
In Table 1, we present detailed performance comparisons between our proposed system and other baseline models based on prediction accuracy, area under the curve (AUC), sensitivity, specificity, and balanced accuracy (BA). The baseline system taking only H&E stained images and including only the prediction module performs the worst, with 0.507 and 0.622 for accuracy and AUC respectively at the image patch level. Information from images of adjacent tissue sections in multiple stains can improve the prediction performance of the baseline system to 0.635 and 0.644 for accuracy and AUC respectively. When the baseline system includes the spatial attention module integrating the detection with the prediction module, the resulting system performance is significantly improved with accuracy higher than 70%. Our best proposed model, by including both the multi-stain and spatial attention modules, achieves the best accuracy of 0.783 and AUC of 0.803, respectively. Although our proposed model is not the best by sensitivity, it outperforms others by BA. BA is considered as a more general metric as it is the arithmetic average of sensitivity and specificity. With the progressively improved performance demonstrated in Table 1, it is manifested that our proposed two innovative modules help improve the PCR prediction performance significantly.
Table 1.
Comparison of PCR prediction performance of progressively improved deep learning models at the image patch level by metrics of accuracy, AUC, sensitivity, specificity, and balanced accuracy (BA).
| Single stain + Prediction only | Multi-stain + Prediction only | Single stain + SAM prediction | Multi-stain + SAM prediction | |
|---|---|---|---|---|
|
| ||||
| Accuracy | 0.507 | 0.635 | 0.709 | 0.783 |
| AUC | 0.622 | 0.644 | 0.762 | 0.803 |
| Sensitivity | 0.726 | 0.625 | 0.765 | 0.701 |
| Specificity | 0.467 | 0.612 | 0.664 | 0.829 |
| BA | 0.596 | 0.619 | 0.715 | 0.765 |
Additionally, we present in Table 2 PCR prediction performance at different levels. As described in the dataset section, our testing image patches in 416×416 pixels are cropped from 583 8, 000 × 8, 000 image regions of 40 TNBC patients. The PCR prediction at the image region level is determined by the max voting of the patch level results from the region, while the prediction at the patient level is determined by the max voting of the region level results from that patient. As shown in Table 2, the prediction performance of our proposed model is superior to that of other baseline models at all levels, achieving 78.3%, 83.7%, and 87.5% accuracy for patch, region, and patient-level prediction, respectively.
Table 2.
Prediction performance comparison at patch, region, and patient-level with the number of corrected predictions, total cases, and accuracy.
| Single stain + Prediction only | Multi-stain + Prediction only | Single stain + SAM prediction | Multi-stain + SAM prediction | |
|---|---|---|---|---|
|
| ||||
| Patient level | 18/40 (45.0%) | 27/40 (67.5%) | 32/40 (80.0%) | 35/40 (87.5%) |
| Region level | 289/583 (49.6%) | 387/583 (66.4%) | 446/583 (76.5%) | 488/583 (83.7%) |
| Patch level | 23325/46029 (50.7%) | 29248/46029 (63.5%) | 32674/46029 (70.9%) | 36059/46029 (78.3%) |
4. Discussion
To our best knowledge, only one study so far aims at predicting PCR to neoadjuvant chemotherapy with histopathology images [7]. By univariate and multivariate analysis with 15 manually designed image features, the best performance achieved by this method was 4.46 by the metric of odds ratio [7]. By comparison, our proposed multi-stain and multi-task deep learning system achieves an odds ratio of 13.3 and 51 at the patch and patient-level, respectively.
Our model is the first work on predicting PCR with multi-stained histopathology images of serial tissue sections. The existing systems use only pathology images of tissue biopsies stained by a single stain, leading to limited accuracy. They also lack full automation for the prediction. As tissue-derived molecular data do not preserve high resolution spatial information in most cases, it has always been a challenge to integrate such information with histopathology features within the same tissue space. As a result, it is ideal to spatially map multiple molecular signatures to the same histology space, enabling multi-modal microscopy integrative analysis for better clinical prediction power. When it comes to PCR prediction, both conventional H&E and IHC biomarker pathology images play an important role. It has been reported that aggressive TNBCs promote cell proliferation with a faster cell cycling kinetics and enhances cell cycle progression [22]. This enhanced cell cycle kinetics can be captured by Ki67 and PHH3 stains in adjacent tissue sections. As a result, we design a PCR prediction system that combines multi-stained serial slides to improve both prediction accuracy and robustness. The superior performance of our novel multi-stain spatial attention system indicates that an integrated analysis of multi-stained serial tissue sections can substantially improve the PCR prediction for NAC TNBC patients.
Our system is also the first work on predicting PCR from pathology images using deep learning techniques. Specifically, we develop the spatial attention module for guided PCR prediction with knowledge from cell detection. Representative spatial attention maps from the spatial attention module are presented in Fig. 4. After the spatial attention module, important areas suggested for spatial attention are highlighted in red. Such attention guidance mechanism effectively directs the following prediction module for enhanced PCR prediction. In the meanwhile, human pathologists may benefit from such interpretable information for treatment planning in clinical practice too.
Fig. 4.
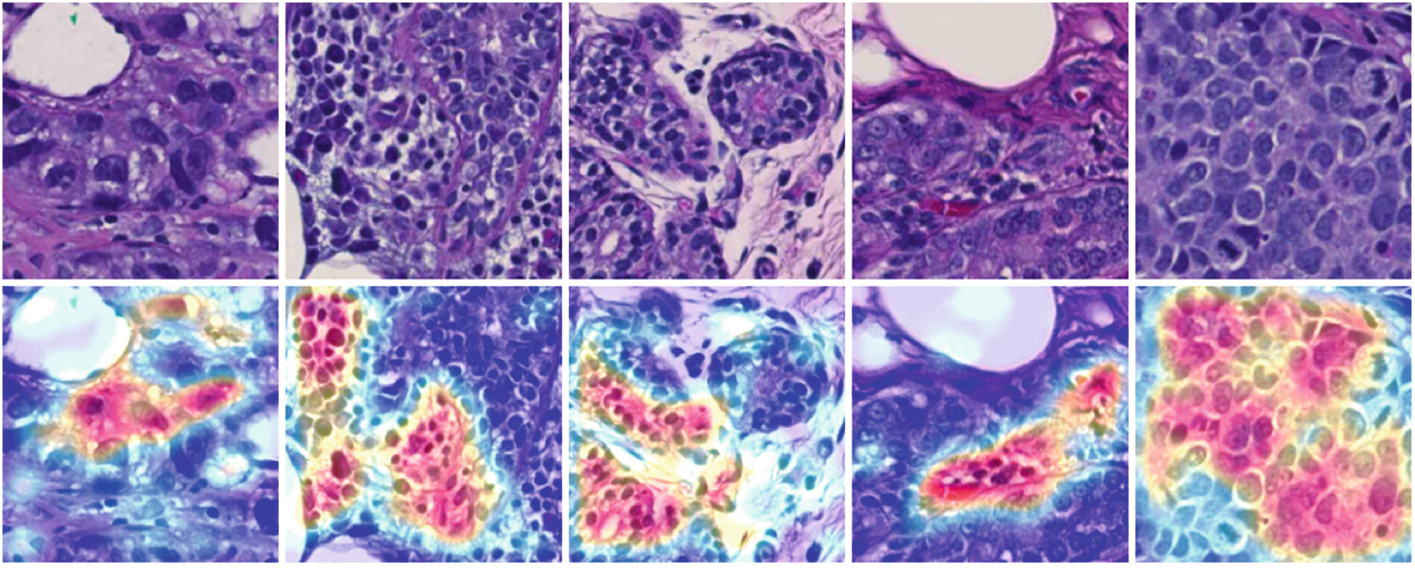
Typical spatial attention maps from the spatial attention module.
For future work, first, we will explore incorporating non-imaging features to the PCR prediction system, as several non-imaging clinical variables are proven to be correlated with the PCR. Second, since our current system requires images in three stains to be well registered, we will study improving the reliability of the system for unregistered multi-stained images. Last, besides PCR, we will expand the prediction to additional clinical outcomes such as recurrence and overall survival.
5. Conclusion
In this paper, we present a novel deep learning-based system for the prediction of PCR to neoadjuvant chemotherapy for TNBC patients. By integrating detection and prediction tasks with a new spatial attention mechanism, our proposed system can capture and jointly use information from histopathology images of adjacent tissue sections in three stains (i.e. H&E, Ki-67, and PHH3). Our comparative study with three baseline models demonstrates progressively improved prediction performance of our system. The prediction effectiveness and high interpretability enabled by the spatial attention mechanism suggest its promising clinical potential for enhancing treatment planning in clinical settings.
Funding.
This research was supported by National Institute of Health (U01CA 242936), National Science Foundation (ACI 1443054, IIS 1350885), and CNPq.
References
- 1.Cortazar P, et al. : Pathological complete response and long-term clinical benefit in breast cancer: the CTNeoBC pooled analysis. Lancet 384(9938), 164–172 (2014) [DOI] [PubMed] [Google Scholar]
- 2.Ring AE, Smith IE, Ashley S, Fulford LG, Lakhani SR: Oestrogen receptor status, pathological complete response and prognosis in patients receiving neoadjuvant chemotherapy for early breast cancer. Br. J. Cancer 91(12), 2012–2017 (2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Evans A, et al. : Prediction of pathological complete response to neoadjuvant chemotherapy for primary breast cancer comparing interim ultrasound, shear wave elastography and MRI. Eur. J. Ultrasound 39(04), 422–431 (2018) [DOI] [PubMed] [Google Scholar]
- 4.van Stiphout RGPM, et al. : Development and external validation of a predictive model for pathological complete response of rectal cancer patients including sequential PET-CT imaging. Radiother. Oncol. 98(1), 126–133 (2011) [DOI] [PubMed] [Google Scholar]
- 5.Gollub MJ, et al. : Dynamic contrast enhanced-MRI for the detection of pathological complete response to neoadjuvant chemotherapy for locally advanced rectal cancer. Eur. Radiol. 22(4), 821–831 (2012) [DOI] [PubMed] [Google Scholar]
- 6.Yu N, Leung VWY, Meterissian S: MRI performance in detecting PCR after neoadjuvant chemotherapy by molecular subtype of breast cancer. World J. Surg. 43(9), 2254–2261 (2019) [DOI] [PubMed] [Google Scholar]
- 7.Raza Ali H, et al. : Computational pathology of pre-treatment biopsies identifies lymphocyte density as a predictor of response to neoadjuvant chemotherapy in breast cancer. Breast Cancer Res. 18(1), 1–11 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tőkés T, et al. : Expression of cell cycle markers is predictive of the response to primary systemic therapy of locally advanced breast cancer. Virchows Arch. 468(6), 675–686 (2016). 10.1007/s00428-016-1925-x [DOI] [PubMed] [Google Scholar]
- 9.Penault-Llorca F, Radosevic-Robin N: Ki67 assessment in breast cancer: an update. Pathology 49(2), 166–171 (2017) [DOI] [PubMed] [Google Scholar]
- 10.LeCun Y, Bengio Y, Hinton G: Deep learning. Nature 521(7553), 436–444 (2015) [DOI] [PubMed] [Google Scholar]
- 11.Deng L, Dong Yu.: Deep learning: methods and applications. Found. Trends Signal Process. 7(3–4), 197–387 (2014) [Google Scholar]
- 12.Cheplygina V, de Bruijne M, Pluim JPW: Not-so-supervised: a survey of semi-supervised, multi-instance, and transfer learning in medical image analysis. Med. Image Anal. 54, 280–296 (2019) [DOI] [PubMed] [Google Scholar]
- 13.Shen D, Guorong W, Suk H-I: Deep learning in medical image analysis. Annu. Rev. Biomed. Eng 19, 221–248 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Litjens G, et al. : A survey on deep learning in medical image analysis. Med. Image Anal. 42, 60–88 (2017) [DOI] [PubMed] [Google Scholar]
- 15.Yu-Hong Q, Zhu H-T, Cao K, Li X-T, Ye M, Sun Y-S: Prediction of pathological complete response to neoadjuvant chemotherapy in breast cancer using a deep learning (DL) method. Thoracic Cancer 11(3), 651–658 (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cui Y, et al. : Radiomics analysis of multiparametric MRI for prediction of pathological complete response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer. Eur. Radiol. 29(3), 1211–1220 (2019) [DOI] [PubMed] [Google Scholar]
- 17.Rossetti B, Wang F, Zhang P, Teodoro G, Brat D, Kong J: Dynamic registration for gigapixel serial whole slide images. In: IEEE International Symposium on Biomedical Imaging: From Nano to Macro (ISBI), pp. 424–428 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bochkovskiy A, Wang C-Y, Liao H-YM: YOLOv4: optimal speed and accuracy of object detection. arXiv preprint arXiv:2004.10934 (2020) [Google Scholar]
- 19.Redmon J, Divvala S, Girshick R, Farhadi A: You only look once: unified, real-time object detection. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), June 2016 [Google Scholar]
- 20.Lin T-Y, Dollár P, Girshick R, He K, Hariharan B, Belongie S: Feature pyramid networks for object detection. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 2117–2125 (2017) [Google Scholar]
- 21.Simonyan K, Zisserman A: Very deep convolutional networks for large-scale image recognition. In: International Conference on Learning Representations (2015) [Google Scholar]
- 22.Pannu V, et al. : HSET overexpression fuels tumor progression via centrosome clustering-independent mechanisms in breast cancer patients. Oncotarget 6(8), 6076 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]


