Abstract
Five histone deacetylase genes (HDA1, RPD3, HOS1, HOS2, and HOS3) have been cloned from Candida albicans and characterized. Sequence analysis and comparison with 17 additional deacetylases resulted in a phylogenetic tree composed of three major groups. Transcription of the deacetylases HDA1 and RPD3 is down-regulated in the opaque phase of the white-opaque transition in strain WO-1. HOS3 is selectively transcribed as a 2.5-kb transcript in the white phase and as a less-abundant 2.3-kb transcript in the opaque phase. HDA1 and RPD3 were independently deleted in strain WO-1, and both switching between the white and opaque phases and the downstream regulation of phase-specific genes were analyzed. Deletion of HDA1 resulted in an increase in the frequency of switching from the white phase to the opaque phase, but had no effect on the frequency of switching from the opaque phase to the white phase. Deletion of RPD3 resulted in an increase in the frequency of switching in both directions. Deletion of HDA1 resulted in reduced white-phase-specific expression of the EFG1 3.2-kb transcript, but had no significant effect on white-phase-specific expression of WH11 or opaque-phase-specific expression of OP4, SAP1, and SAP3. Deletion of RPD3 resulted in reduced opaque-phase-specific expression of OP4, SAP1, and SAP3 and a slight reduction of white-phase-specific expression of WH11 and 3.2-kb EFG1. Deletion of neither HDA1 nor RPD3 affected the high level of white-phase expression and the low level of opaque-phase expression of the MADS box protein gene MCM1, which has been implicated in the regulation of opaque-phase-specific gene expression. In addition, there was no effect on the phase-regulated levels of expression of the other deacetylase genes. These results demonstrate that the two deacetylase genes HDA1 and RPD3 play distinct roles in the suppression of switching, that the two play distinct and selective roles in the regulation of phase-specific genes, and that the deacetylases are in turn regulated by switching.
Most strains of Candida albicans and related species switch, or can be induced to switch, between two or more general phenotypes distinguishable by colony morphology (14, 41, 42, 44, 48). Switching regulates a number of phenotypic characteristics, including putative virulence traits, and for that reason, it has been considered a higher-order virulence factor (42). Switching has been demonstrated to occur at sites of commensalism (21) and infection (48–50) and to occur at higher frequencies in strains causing infections than in commensal strains (21, 24, 61). Therefore, it has been proposed that switching provides colonizing populations with phenotypic variants for rapid adaptation in response to environmental challenges, such as drug therapy and the immune response, as well as changes in host physiology, the competing microflora, and anatomical locale (34, 45).
By using the simple white-opaque transition in C. albicans strain WO-1 (42) as a model system, it has been demonstrated that switching involves the transcription of a number of phase-specific genes (3, 13, 22, 32, 33, 46, 53–55) and that the differential expression of these genes is accomplished through phase-regulated transcription factors (30, 46, 47, 51, 56), leading to a model in which a reversible, spontaneous change at a master switch locus leads to the downstream activation and deactivation of unlinked phase-regulated genes (46, 47). Since the cis-acting regulatory sequences of coordinately expressed genes differ, models have been developed that accommodate different DNA binding proteins (46). However, in spite of recent progress in functionally characterizing the promoters of phase-regulated genes, little is known about the actual switch event or how genes are coordinately regulated in the white-opaque transition. Because switching occurs spontaneously and reversibly at relatively high frequencies (10−4 to >10−2), it had been suggested (37, 44) that it either represents a spontaneous and reversible reorganization of DNA at a master switch locus, which occurs in bacterial switching systems (15), or a reversible alteration in the state of chromatin at a master switch locus, which occurs in yeast when genes are placed at sites adjacent to subtelomeric regions (17, 36). Because the “silent information regulator” gene SIR2 plays a role in gene repression in the latter mechanism, it was suggested that a C. albicans homolog of SIR2 may play a role in the regulation of switching at a master switch locus in C. albicans (37, 44). Perez-Martin et al. (35) subsequently demonstrated that deletion of a C. albicans SIR2 homolog in strain CAI4 did in fact result in up-regulation of the switching system in that strain, which is analogous to the first switching system described in C. albicans strain 3153A (41, 54). In addition to the “silent information regulator” (Sir) proteins, other classes of proteins are involved in the repression or silencing of developmentally regulated genes in eukaryotes. One such class of genes, the histone deacetylases, has been demonstrated to regulate chromatin structure through selective histone deacetylation, which in turn affects chromatin folding and interactions between DNA and DNA-binding proteins (2, 63). The role of the deacetylases in gene regulation has been demonstrated by generating mutants with loss of function or with dominant-negative effects in both Saccharomyces cerevisiae (25, 39, 58) and human cell lines (18, 20).
Recently, we tested whether the specific deacetylase inhibitor trichostatin A (10, 65) affected the white-opaque transition (26). We found that the inhibitor caused a selective increase in the frequency of switching in the white-to-opaque transition, but had no effect on the frequency of switching in the opaque-to-white transition, suggesting that deacetylation through a trichostatin-sensitive deacetylase selectively suppresses switching in one direction (26). Since the deacetylase Hda1p is highly sensitive to trichostatin A (7), we deleted the HDA1 gene and obtained a mutant phenotype similar to that of trichostatin A-treated cells, supporting the conclusion that deacetylation through Hda1p suppresses switching selectively in the white-to-opaque direction (26). In addition to playing a role in switching, deacetylation may also play a role in the regulation of phase-specific gene transcription. We therefore cloned five of the major C. albicans histone deacetylase genes with homology to the genes that encode the five known histone deacetylases in S. cerevisiae (HDA1, RPD3, HOS1, HOS2, and HOS3) and analyzed their deduced protein sequences and expression patterns in the white-opaque transition. We have also generated, in addition to the HDA1 deletion mutant, a deletion mutant of RPD3. We present evidence that the expression of the deacetylase genes is affected by switching and that, while HDA1 plays a selective role in suppressing the basic switch from white to opaque, but not from opaque to white, RPD3 plays a role in suppressing the basic switch events in both directions. In addition, both HDA1 and RPD3 play indirect but distinct roles in regulating the levels of expression of select phase-specific genes.
MATERIALS AND METHODS
Maintenance of stock cultures.
C. albicans strain WO-1 (42) was periodically cloned from a frozen stock culture on agar containing modified Lee's medium (4). Strains TS3.3, a ura3 auxotroph (55), and TUI7, a URA3 prototrophic derivative of TS3.3, were maintained on agar containing modified Lee's medium with or without 0.01 mM uridine, respectively. Both the HDA1 and RPD3 homozygous mutant strains were maintained on agar containing modified Lee's medium. The genotypes of all strains used in this study are described in Table 1.
TABLE 1.
Genotypes of strains used in this study
| Strain | Parent | Relevant genotype | Source or reference |
|---|---|---|---|
| WO-1 | Wild type | 41 | |
| TS3.3 | Red3/6 | ade2/ade2, Δura3::ADE2/Δura3::ADE2 | 54 |
| HDhe20 | TS3.3 | ade2/ade2, Δura3::ADE2/Δura3::ADE2, Δhda1::CAT-URA3-CAT/HDA1 | This study |
| HDheF21 | HDhe20 | ade2/ade2, Δura3::ADE2/Δura3::ADE2, Δhda1::CAT/HDA1 | This study |
| HDho15 | HDheF21 | ade2/ade2, Δura3::ADE2/Δura3::ADE2, Δhda1::CAT/Δhda1::hisG-URA3-hisG | This study |
| HDho19 | HDheF21 | ade2/ade2, Δura3::ADE2/Δura3::ADE2, Δhda1::CAT/Δhda1::hisG-URA3-hisG | This study |
| RPhe2 | TS3.3 | ade2/ade2, Δura3::ADE2/Δura3::ADE2, Δrpd3::hisG-URA3-hisG/RPD3 | This study |
| RPheF4 | RPhe2 | ade2/ade2, Δura3::ADE2/Δura3::ADE2, Δrpd3::hisG/RPD3 | This study |
| RPho19 | RPheF4 | ade2/ade2, Δura3::ADE2/Δura3::ADE2, Δrpd3::hisG/Δrpd3::hisG-URA3-hisG | This study |
| RPho3 | RPheF4 | ade2/ade2, Δura3::ADE2/Δura3::ADE2, Δrpd3::hisG/Δrpd3::hisG-URA3-hisG | This study |
| TU17 | TS3.3 | ade2/ade2, URA3/Δura3::ADE2 | This study |
Cloning the deacetylase genes HDA1 and RPD3.
Based on short DNA sequences reported in the Stanford C. albicans genome database (www-sequence.stanford.edu), two sets of primers, DeACT5′/DeACT3′ and RPD5′/RPD3′ (Table 2), were designed to amplify by PCR DNA fragments containing HDA1 and RPD3 sequences, respectively, with genomic DNA of strain WO-1 as a template and with Taq polymerase. The PCR-derived fragments were then used to screen a λEMBL3A genomic library of C. albicans strain WO-1 (53). Of approximately 100,000 plaques screened for the HDA1 fragment or for the RPD3 fragment, 30 to 40 putative clones were identified. Ten putative clones of each gene were purified by a secondary screen and analyzed by Southern blot hybridization with the respective probe to assess purity. Based on these preliminary results, λDA3.1 and λRP2.1 were chosen to represent CaHDA1 and CaRPD3, respectively.
TABLE 2.
Primers used in this study
| Primer | Sequence |
|---|---|
| DeACT5′ | 5′-GAT TGG ATC AGC AAT ATT TAC C-3′ |
| DeACT3′ | 5′-GAT GTG ATT GGA GCT TGT CA-3′ |
| FANHDA5′ | 5′-GCG TCG CGA ATG TCG ACT GGT CAA GAA GAA-3′ |
| FANHDA3′ | 5′-GCG CCG CGG TAA GTA GCG ATT CCA GAT-3′ |
| RPD5′ | 5′-TCA TCA TAG AAC TCT CCA TCA-3′ |
| RPD3′ | 5′-AAG TGG TGA TCG ATT AGG ACC-3′ |
| FANRPD5′ | 5′-ATT GGG CCC ATG TAT CAA GAA TTA CCA TTT G-3′ |
| FANRPD3′ | 5′-ATT GGG CCC CAC ATT ATT ATT TAA TTT ATC-3′ |
| HOS1-5′ | 5′-CCT GGG CCC ATG GCA ACA AAG AGA-3′ |
| HOS1-3′ | 5′-ATA GGG CCC TAG AGA CAA TAA ATG-3′ |
| HOS2-5′ | 5′-CCT GGG CCC ATG ACG ATA CTG ATA-3′ |
| HOS2-3′ | 5′-ATA GGG CCC AGT CAT TAG TTC TCC-3′ |
| HOS3-5′ | 5′-CCT GGG CCC ATG TCG ACT TCA AAA-3′ |
| HOS3-3′ | 5′-ATA GGG CCC TAT ATT GGA TAC CTT-3′ |
| SAP3-5′ | 5′-CCT TCT CTA AAA TTA TGG AT-3′ |
| SAP3-3′ | 5′-TTG ATT TCA CCT TGG GGA CC-3′ |
| OP4NDEIF | 5′-GG CAT ATG AAG TTT TCA CAA GCC-3′ |
| OP4SPHIR | 5′-GGT GGT TGC TCT TCC GCA CTA ATA AAG TTT TCT TTT-3′ |
| FANEFG15′ | 5′-GCG TCG CGA ATG TCA ACG TAT TCT ATA-3′ |
| FANEFG13′ | 5′-GCG CCG CGG CTT TTC TTC TTT GGC AAC AGT CGT-3′ |
| RL5′ | 5′-TAG AAC CAT TGA CAG ACG-3′ |
| RL3′ | 5′-GTG GAG AAT TAT CAT CGC-3′ |
| McMORF5 | 5′-GAA TTA TCA GTA TTG ACA GGT ACT CAA GTG-3′ |
| CMORF3 | 5′-AAT GTG ATG AGC ATG ACC TGC AGC ACC-3′ |
| Tupbamf | 5′-TAA CGG ATC CTT ATG TCC ATG TAT CCC CAA-3′ |
| Tuppstr | 5′-CAA CTG CAG CTC ACA TAT ATA TAT ACT ACA CAC TTA-3′ |
To characterize HDA1, λDA3.1 was digested with SalI, and a 2.1-kb fragment that hybridized with the respective probe was subcloned into pGEM3Z to generate pDA14.1. The SalI DNA fragment was sequenced in both directions with an ABI model 373A automatic sequencing system and fluorescent Big Dye termination chemistry (PE-ABI Inc., Foster City, Calif.). In addition, approximately 1 kb of the 5′ upstream sequence and 0.5 kb of the 3′ downstream sequence of the SalI fragment were determined with λDA3.1 as a template. To characterize RPD3, a 4-kb sequence of λRP2.1 was directly sequenced with the same protocol applied to pDA14.1. Then, by using the specific primer pair FANRPD5′/FANRPD3′ (Table 2), the full-length RPD3 open reading frame (ORF) was amplified by PCR and subcloned into pGEM5Z between end-repaired SacI sites to generate pGRP16/5.
Cloning the deacetylase genes HOS1, HOS2, and HOS3.
Homologs to S. cerevisiae HOS1, HOS2, and HOS3 were identified in the Stanford C. albicans genome database and then isolated by PCR with the 5′ and 3′ primers HOS1-5′/HOS1-3′, HOS2-5′/HOS2-3′, and HOS3-5′/HOS3-3′, respectively (Table 2), and with strain WO-1 DNA as a template. The HOS1, HOS2, and HOS3 DNA fragments encompassing the entire ORFs were subcloned into pGEM-T easy vector to derive pC65.5, pC85.2, and pC58.14, respectively. Approximately 500 to 600 bp of both the 5′ and 3′ ends of each of the three ORFs were initially analyzed to confirm the identity of each deacetylase gene.
Analysis of protein sequences and construction of a phylogenetic tree.
Alignment of multiple protein sequences was performed with the Clustal W/Jalview multiple alignment editor, version 4 (60). Pairwise comparisons between protein sequences were performed with the PROTDIST program of the PHYLIP package, version 3.57C (http://evolution.genetics.washington.edu/Phyl.p.html). The unrooted dendrograms were generated by using the FITCH program of the PHYLIP package (the Fitch-Margoliash least-squares distance method) (11). The data set was subjected to a bootstrap analysis (1,000 replicates) by sequential use of the SECBOOT, PROTDIST, and Consensus programs. Genetic distances were derived by the Day'hoff PAM matrix algorithm (8).
Construction of homozygous HDA1 deletion mutants.
Two different deletion cassettes were constructed, each spanning the essential deacetylation motifs. To generate the heterozygote, a chloramphenicol acetyltransferase (CAT)-URA3-CAT-based cassette (55) was constructed. The pDA14.1 plasmid containing the 2.1-kb SalI fragment of HDA1 (Fig. 1A) was digested with BclI, deleting 1,147 bp of the HDA1 ORF, end repaired with T4 DNA polymerase, and dephosphorylated with shrimp alkaline phosphatase (SAP). The linearized vector was purified by Tris-borate-EDTA (TBE)-agarose gel electrophoresis and ligated with the 3.5-kb CAT-URA3-CAT cassette from the plasmid pCUC (55). The CAT-URA3-CAT cassette was isolated by digesting pCUC with BamHI, followed by end repair with T4 DNA polymerase. The resulting plasmid, pA48.1, contained the deleted version of HDA1 (Fig. 1A). pA48.1 was digested with SalI, and 25 μg was used to transform the ura3− strain TS3.3 (Table 1). Recovered transformants were tested for heterozygosity by digestion of genomic DNA with XbaI and analyzed by Southern blot hybridization with the HDA1 ORF. One confirmed heterozygous clone was subjected to 5-fluoroorotic acid (5-FOA) treatment in order to induce “pop-outs” of the URA3 gene. To generate the homozygote, a hisG-URA3-hisG-based cassette (12) was constructed. The BclI-BclI deletion fragment of plasmid pDA14.1 was isolated, end repaired with T4 DNA polymerase, dephosphorylated with SAP, and subcloned at the EcoRV site of pGEM5Z to generate pC25.1 (Fig. 1A). pC25.1 was digested with EcoRV and BglII to delete 702 bp of DNA, end repaired with T4 DNA polymerase, dephosphorylated with SAP, and ligated with the hisG-URA3-hisG cassette from plasmid pMB7 (55). The hisG-URA3-hisG cassette was prepared by digesting PMB7 with SalI and BglII and end repaired with T4 DNA polymerase. The resultant disruption cassette, pC88.10 (Fig. 1A), was then digested with ApaI and SacI. This DNA fragment could only target the functional HDA1 allele. It was used to transform the selected heterozygous ura3− strain. Transformants, obtained on selection plates, were tested for homozygosity by Southern analysis as described earlier.
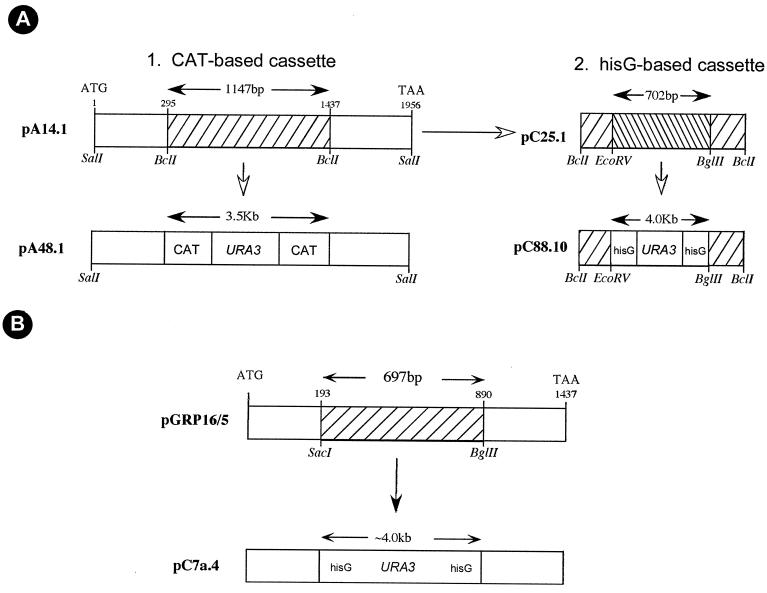
FIG. 1.
Maps of the deletion cassettes of HDA1 and RPD3. (A) HDA1 deletion cassettes. Panel 1 shows plasmid pDA14.1, which contains the entire HDA1 ORF subcloned as a SalI-SalI fragment. The ATG start codon is at 1 bp, and the TAA stop codon is at 1,956 bp. The plasmid pA48.1 was derived by replacing 1,147 bp of the HDA1 ORF with 3.5 kb of the CAT-URA-CAT cassette. Panel 2 shows plasmid pC88.10, generated by subcloning 1,147 bp of the BclI-BclI fragment into another plasmid to derive pC25.1, followed by substitution of a 702-bp EcoRV-BglII region of the HDA1 ORF with 4.0 kb of the hisG-URA3-hisG cassette. (B) RPD3 deletion cassette. The plasmid pGRP16/5 contained the entire RPD3 ORF derived by PCR. The ATG start-codon is at 1 bp, and the TAA stop codon is at 1,437 bp. The plasmid pC7a.4 was derived by deleting 697 bp of the RPD3 ORF and replacing it with the 4.0-kb hisG-URA3-hisG cassette.
Construction of homozygous RPD3 deletion mutants.
The hisG-URA3-hisG cassette was used to delete both alleles of RPD3. To construct the disruption cassette, pGRP16/5 was digested with SacI and BglII to delete 697 bp. The 4.0-kb hisG-URA3-hisG cassette was then inserted in its place to generate the plasmid pC7a.4 (Fig. 1B). The RPD3 disruption cassette was prepared by digesting pC7a.4 with ApaI and NsiI and used in both the first and second rounds of transformation to obtain first the heterozygous strain and second the homozygous strain. Prior to the second round of transformation, the selected heterozygous clone was subjected to the 5-FOA “pop-out” protocol to derive ura3 auxotrophy. The heterozygous and homozygous strains were confirmed by probing Southern blots of PstI-digested DNA with the RPD3 ORF.
Northern and Southern blot analyses.
Northern analyses were performed according to methods previously described (54, 55). To ensure that the growth conditions for the compared cell preparations were similar, cells of each cell type were removed from agar cultures and grown to the stationary phase in liquid culture. White-phase cells were then diluted to 106 and opaque-phase cells were diluted to 5 × 105 in fresh growth medium and grown to mid-log phase (9 × 106 and 5 × 106 per ml, respectively) prior to harvesting. Total RNA was extracted with the RNeasy mini kit according to manufacturer's specifications (Qiagen, Inc., Santa Clarita, Calif.). The fold differences in transcript levels were measured according to methods recently described (55). The hybridization probes for WH11, SAP1, EFG1, and OP4 contained the full-length ORFs, derived either by PCR with specific primers (Table 2) or by digesting them from the respective plasmids containing the cognate DNA inserts (32, 33, 53, 55). The probe for SAP3 was derived by PCR with the proximal activation sequence pAS3 as a template and the specific primers SAP3-5′ and SAP3-3′ (Table 2). Probes for HDA1 encompassed either the full-length ORF or the 702-bp EcoRV-BglII deletion fragment from pC25.1, as described earlier. Probes for RPD3 encompassed either the full-length ORF or the 697-bp SacI-BglII deletion fragment from pGRP16/5, as described earlier. The probes for HOS1, HOS2, HOS3, MCM1, and TUP1 encompassed the full-length ORFs obtained by PCR with the primer pairs previously described (Table 2). Southern analyses were performed according to methods previously described (33, 52, 55). Specific restriction enzymes and probes are described in Results.
RESULTS
Cloning and characterization of the five histone deacetylase genes.
A search of the C. albicans genome database for homology at the amino acid level initially revealed five genes homologous to each of the S. cerevisiae histone deacetylase genes HDA1, RPD3, HOS1, HOS2, and HOS3. HDA1 and RPD3 were cloned by probing a lambda genomic library of strain WO-1 (42) with PCR-generated DNA fragments based on nucleotide sequences reported in the Stanford C. albicans genome database. The three HOS genes were amplified from WO-1 genomic DNA by PCR with primers based on reported nucleotide sequences. The cloned HDA1, RPD3, HOS1, HOS2, and HOS3 homologs contained ORFs encoding proteins with sizes of 653, 478, 392, 454, and 713 amino acids, respectively (Fig. 2).
FIG. 2.
Sequence comparison of the five C. albicans histone deacetylases with S. cerevisiae Hda1p and Rpd3p. The sequences were aligned with the Clustal W multiple alignment editor (60). The rectangular unshaded box in the center represents the highly conserved deacetylation motif. The gray shaded area, including the unshaded rectangular box, contains amino acid residues conserved to various degrees in the deacetylases. The identical residues among all deacetylases are denoted by stars, while conservative replacements of amino acid residues are denoted by either two stacked solid circles (based on similar functional groups) or one solid circle (based on similar effects on secondary structure). Conserved histidines are presented in boldface. The accession numbers for HDA1 and RPD3 in GenBank are AF377894 and AF377895, respectively. Other accession numbers are as follows: ScHda1p, Saccharomyces cerevisiae Z71297; SCRpd3p, S. cerevisiae P32561. Ca, Candida albicans.
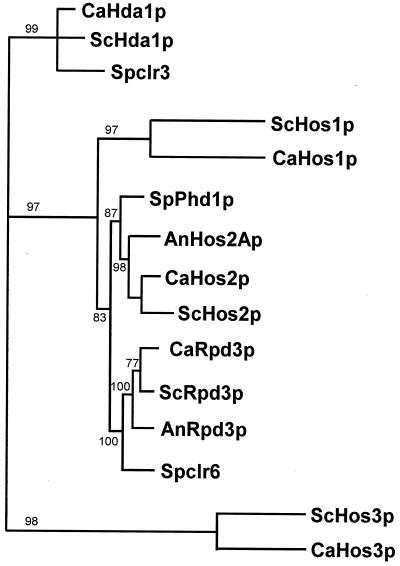
To test whether the five C. albicans histone deacetylase proteins represented distinct members of the histone deacetylase superfamily, alignment of these proteins and 12 additional fungal histone deacetylases was performed with the Clustal W multiple alignment protocol described in Materials and Methods. The alignment of approximately 330 amino acids included the nine sequence blocks that identify histone deacetylase subtypes (29). The phylogenetic analysis (Fig. 3) revealed that the five C. albicans deacetylases grouped into three distinct classes: the Hda1 class (C. albicans Hda1p), the Rpd3 class (C. albicans Rpd3p, HOS1p, and HOS2p), and the Hos3 class (C. albicans HOS3p). In every class, the C. albicans deacetylases were most similar to the corresponding S. cerevisiae deacetylase (Fig. 3).
FIG. 3.
Phylogenetic analysis of fungal histone deacetylases. The protein sequences of 15 different histone deacetylases from various fungi were aligned by using the Clustal W/Jalview multiple alignment editor, version 4, and subjected to phylogenetic analysis with PHYLIP software, version 3.57. The data set was bootstrapped, and the genetic distances were derived by the Dayhoff PAM matrix algorithm (8). The results of the bootstrap analysis (1,000 replicates) are shown either above or below the branches. All of the bootstrap values were above 77%, suggesting that the nodes are significant and reflect the correct phylogeny. Proteins from different fungal species are indicated by two-letter prefixes: Ca, Candida albicans; An, Aspergillus nidulans; Sc, Saccharomyces cerevisiae; and Sp, Schizosaccharomyces pombe. Accession numbers for the published sequences are as follows: ScHda1p, Z71297; Spclr3, AFO64207; ScHos1p, Z49219; SpPhd1p, BAA23598; AnHos2Ap, AF164342; ScHos2p, X91837; ScRpd3p, P32561; AnRpd3p, AF163862; Spclr6, AFO64206; ScHos3p, 1143503.
Further comparison of the five C. albicans histone deacetylase proteins with S. cerevisiae Hda1p and ScRpd3p revealed that a 200-amino-acid stretch in the middle of all seven contained highly conserved amino acid residues (Fig. 2). In particular, a highly conserved functional deacetylation motif of 53 to 63 amino acid residues resided in the middle of this stretch (see box in Fig. 2). Pairwise comparison of the seven proteins revealed four highly conserved histidines (presented in boldface in Fig. 2) organized in pairs and separated by roughly the same number of amino acids (5, 10, 18, 26, 65). Comparison of the deacetylation motifs of the five C. albicans deacetylases and 17 additional deacetylases revealed 12 identical amino acid residues and overall a high level of conservation (data not shown).
Expression of the deacetylase genes in the white-opaque transition.
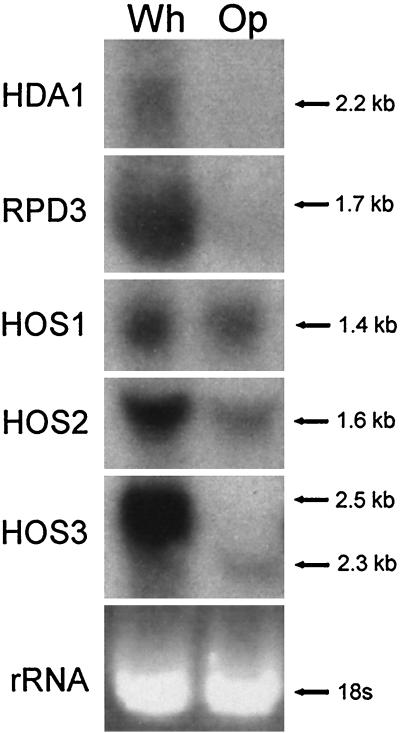
Northern analysis revealed that expression of the deacetylases differed between white- and opaque-phase cells. The transcript levels of HDA1 and RPD3 were significantly lower in the opaque phase (Fig. 4). The transcript levels of HOS1 and HOS2 were slightly lower in the opaque phase (Fig. 4). In the case of HOS3, both the levels and the molecular size of the transcripts differed between the two phases. In the white phase, the molecular size of the transcript was 2.5 kb, and in the opaque phase, the molecular size of the less-abundant transcript was 2.3 kb. Northern analysis with two independent RNA samples revealed similar results.
FIG. 4.
Northern blot analysis of mRNA expression of the five cloned histone deacetylase genes of C. albicans strain WO-1 in the white (Wh) and opaque (Op) phases. Blots were probed with the full-length ORF of each of the five deacetylase genes. The ethidium bromide-stained 18S rRNA band is presented at the bottom of the hybridization patterns to assess loading. Molecular sizes of the bands are presented to the right of each blot. Note that HOS3 is expressed as an abundant 2.5-kb message in the white phase and a far-less-abundant 2.3-kb transcript in the opaque phase.
Creating deletion mutants of HDA1 and RPD3.
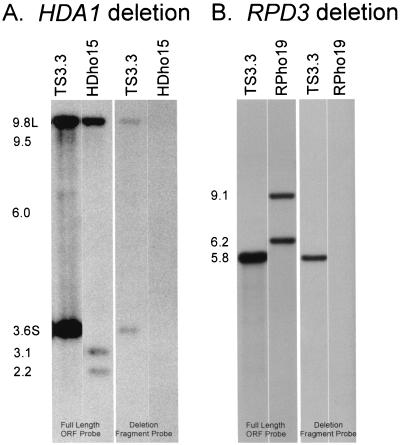
To create deletion mutants of HDA1 and RPD3, we employed the “urablast” gene knockout strategy (12) in strain WO-1 (42). To create the HDA1 deletion mutant, the ura3− strain TS3.3 was first transformed with the CAT-URA3-CAT-based HDA1 disruption cassette from pA48.1 (Fig. 1A). Of 40 transformants, 3 proved to be heterozygous by Southern analysis with the full-length HDA1 ORF and the EcoRV-BglII deletion fragment of the probe. The Southern blot of the ura3− derivative strain TS3.3 contained two bands at 9.8 and 3.6 kb, representing the “large” (L) and “small” (S) alleles of HDA1 (Fig. 5A). The ura3− heterozygote HdheF21 was transformed with the hisG-URA3-hisG-based HDA1 deletion cassette from pC88.10 (Fig. 1A). In one transformation experiment, 30 transformants were obtained, of which 1 proved to be homozygous, and in a second transformation experiment, 40 transformants were obtained, 7 of which proved to be homozygous. For further analysis, the homozygous transformant HDho15 was selected from the first transformation, and HDho11 and HDho19 were selected from the second. When Southern blots were probed with the deleted HDA1 fragment, the parental strain TS3.3 exhibited the two allelic bands at 9.8 and 3.6 kb, and the homozygous strain HDho15 exhibited neither band, confirming that both alleles of HDA1 were deleted in HDho15 (Fig. 5A). A similar analysis proved that both alleles of HDA1 were deleted in HDho19 and HDho11 (data not shown).
FIG. 5.
Southern blot analysis of null mutants of HDA1 and RPD3. (A) Analysis of the Hda1− mutant. Approximately 3 μg (each) of total genomic DNA from the ura− derivative TS3.3 and the homozygous deletion mutant HDho15 was individually digested with XboI and subjected to Southern blotting. Duplicate blots were hybridized with either the full-length HDA1 ORF probe or the EcoRV-BglII deletion fragment. (B) Analysis of the Rpd3− mutant. Approximately 3 μg (each) of total genomic DNA from TS3.3 and the homozygous deletion mutant RPho19 was individually digested with PstI and subjected to Southern blotting. Duplicate blots were hybridized with either the full-length RPD3 ORF probe or the SacI-BglII deletion fragment. The molecular sizes of the expected fragments are shown to the left of the panels.
To create the RPD3 deletion mutant, one urablast cassette based on hisG-URA3-hisG was used to knock out both alleles. TS3.3 was transformed with the disruption cassette from pC7a.4 (Fig. 1B). Of 60 transformants, 44 (73%) proved to be heterozygous for RPD3. When Southern blots of PstI-digested TS3.3 DNA were probed with the RPD3 ORF, a single band was identified at 5.8 kb (Fig. 5B). When the URA− heterozygote strain RPheF4 was retransformed with the pC7a.4 cassette, 40 transformants were obtained. Six proved to be homozygous disruptants. Two transformants, RPho19 and RPho3, were chosen for further analysis. The 5.8-kb band was missing in Southern blots of RPho19 (Fig. 5B) and RPho3 (data not shown) probed with the RPD3 ORF. Both contained the 6.2- and 9.1-kb molecular size bands. When probed with the deleted RPD3 fragment, the parent exhibited a 5.8-kb band (Fig. 5B), while RPho19 (Fig. 5B) and RPho3 (data not shown) exhibited no bands, confirming that both alleles of RPD3 were deleted in the latter strains.
Effects of HDA1 deletion on switching and phase-specific gene expression.
We recently demonstrated that treatment with the deacetylase inhibitor trichostatin A (TSA) or deletion of the most TSA-sensitive gene, HDA1, had a selective effect on switching (26). While both TSA-treated cells and cells of the two independent mutant strains HDho15 and HDho19 switched from the opaque to the white phenotype at frequencies comparable to that of untreated wild-type cells (∼10−3), both TSA-treated and mutant cells switched from the white- to the opaque-phase phenotype at frequencies more than an order of magnitude greater than that of wild-type cells (∼3 × 10−2) (26). These results demonstrated that although the deletion of HDA1 selectively increased the frequency of switching in the white-to-opaque direction, it had no effect on the unique signature morphology of opaque-phase cells.
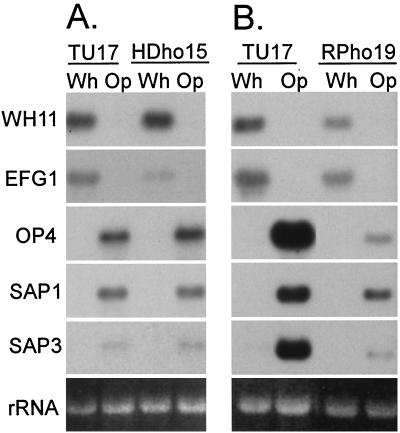
To assess the effects of deleting HDA1 on phase-specific gene expression, Northern blots of total cellular RNA of white- and opaque-phase cells of the parent strain TU17 and the deletion strains HDho15 (Fig. 6A) and HDho19 (data not shown) were probed with the white-phase-specific genes WH11 (53) and EFG1 (55) and the opaque-phase-specific genes OP4 (32), SAP1 (33), and SAP3 (22, 62). Deletion of HDA1 had little effect on the basic developmental regulation of most of the phase-regulated genes tested (Fig. 6A). It also had no significant effect on the level of expression of WH11 in the white phase or on the level of expression of OP4, SAP1, or SAP3 in the opaque phase (Fig. 6A). Deletion did, however, reduce the level of the EFG1 transcript in the white phase fivefold. Similar results were obtained in a repeat experiment in which RNA was extracted from independent growth cultures of TU17 and both mutant strains HDho15 and HDho19.
FIG. 6.
(A) Northern blots of white (Wh)- and opaque (Op)-phase cells of TU17 and HDho15 were probed with the phase-specific genes WH11, OP4, EFG1, SAP1, and SAP3. (B) Northern blots of white- and opaque-phase cells of TU17 and RPho19 were probed with the phase-specific genes WH11, OP4, SAP1, and SAP3. In each case, the same blot was stripped and probed with the five genes. The ethidium bromide-stained 18S rRNA band is presented at the bottom of each set of hybridization patterns to assess loading. Repeats of each experiment gave similar results.
When opaque-phase cell cultures are shifted from 25°C to 42°C, they switch en masse to the white phase (32, 38, 42, 53, 55). The average cell commits to the white-phase phenotype between 3 and 4 h, concomitant with the second cell doubling and a switch in phase-specific gene expression. At the time of phenotypic commitment, the expression of WH11 is activated and the capacity to induce OP4 and SAP1 expression by shifting cells from 37°C back to 25°C is lost (32, 42, 53, 55). To test whether HDA1 plays a role in this transition, opaque-phase cells of the control strain TU17 and the homozygous deletion mutant HDho15 were shifted from 25°C to 42°C. After 1 h (prior to phenotypic commitment) and 7 h (after phenotypic commitment), the samples were shifted back to 25°C. After 1 h at 42°C, both TU17 and HDho15 cells contained no detectable OP4 message. A shift to 25°C after 1 h at 42°C induced reexpression of OP4 in both TU17 and HDho15. After 7 h at 42°C, TU17 cells contained no OP4 message, and HDho15 cells contained a negligible level (data not shown). A subsequent shift to 25°C did not reactivate OP4 expression in either strain (data not shown), demonstrating that the normal switch in OP4 regulation had occurred. After 1 h at 42°C, both TU17 and HDho15 cells contained no detectable WH11 transcript, and a subsequent shift to 25°C did not up-regulate WH11 expression in either strain (data not shown). After 7 h at 42°C, both TU17 and HDho15 cells expressed a low level of WH11 transcript, and a shift to 25°C resulted in up-regulation of WH11 in both strains (data not shown), demonstrating that the switch in WH11 regulation had occurred in the mutant. These results demonstrate that deletion of HDA1 has no discernible effect on the basic activation and deactivation of phase-specific genes at the point of phenotypic commitment.
The effect of RPD3 deletion on switching and phase-specific gene expression.
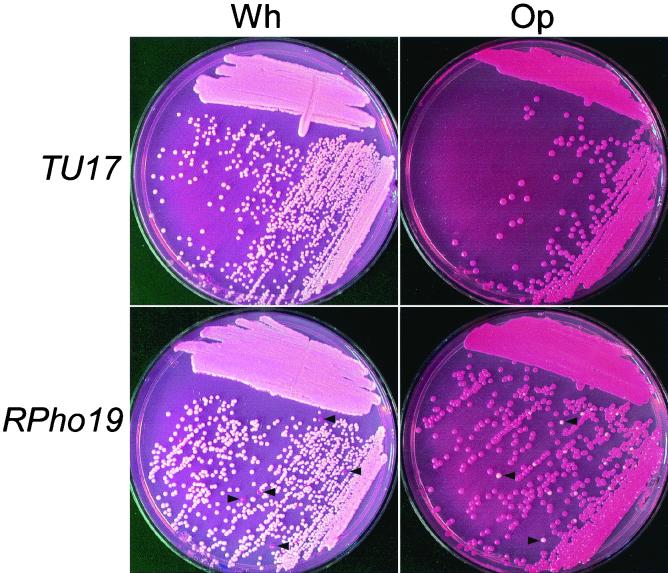
Deletion of RPD3 also had an effect on the frequency of switching, but the effect in this case was on switching in both directions. While the frequency of opaque-phase CFU in 5-day-old white-phase colonies of the parental strain TU17 was 4 × 10−4, those of the homozygous deletion mutants RPho19 and RPho3 were 1 × 10−2 and 2 × 10−2, respectively, representing 25- and 50-fold increases, respectively (Table 3). And while the frequency of white-phase CFU in 5-day-old opaque-phase colonies of the parental strain TU17 was 6 × 10−4, those of the homozygous deletion mutants RPho19 and RPho3 were 5 × 10−2 and 10−2, representing 83- and 17-fold increases, respectively (Table 3). The increases in the frequencies of the transition from white to opaque and opaque to white in RPD3 deletion mutants are evident in cultures in which cells from white and opaque colonies are streaked across nutrient agar (Fig. 7). The same differences were also evident in the frequency of sectored colonies (Table 3). Scanning electron micrographs of white- and opaque-phase cells revealed that the RPD3 deletion had no effect on either the smooth-surfaced, round budding phenotype of white-phase cells or the pimpled, elongate phenotype of opaque-phase cells (data not shown).
TABLE 3.
Effects of deletion of putative deacetylase gene RPD3 on frequency of switching in the white-opaque transitiona
| Initial phenotype of strain | Genotype | No. of colonies
(frequency [CFU])
|
Fold increase over TU17 | No. of original-phase colonies with opposite-phase sectors (frequency [CFU]) | ||
|---|---|---|---|---|---|---|
| Total | White phase | Opaque phase | ||||
| White | ||||||
| TU17 | RPD3/RPD3 | 4,633 | 4,631 | 2 (4 × 10−4) | 1 (2 × 10−4) | |
| RPho19 | rpd3/rpd3 | 2,104 | 2,073 | 31 (1 × 10−2) | 25 | 66 (3 × 10−2) |
| RPho27 | rpd3/rpd3 | 3,776 | 3,720 | 56 (2 × 10−2) | 50 | 15 (4 × 10−3) |
| RPho19+27 | rpd3/rpd3 | 5,880 | 5,793 | 87 (1 × 10−2) | 25 | 71 (1 × 10−2) |
| Opaque | ||||||
| TU17 | RPD3/RPD3 | 3,466 | 2 (6 × 10−4) | 3,464 | 0 (<3 × 10−4) | |
| RPho19 | rpd3/rpd3 | 1,662 | 139 (5 × 10−2) | 2,773 | 83 | 32 (1 × 10−2) |
| RPho27 | rpd3/rpd3 | 2,398 | 24 (1 × 10−2) | 2,374 | 16 | 8 (3 × 10−3) |
| RPho19+27 | rpd3/rpd3 | 4,060 | 163 (3 × 10−2) | 5,147 | 50 | 40 (1 × 10−2) |
Cells from white- or opaque-phase colonies were plated and incubated at 25°C for 5 days, and then the proportions of colony phenotypes (white, opaque, or sectored) were counted.
FIG. 7.
Streaks of cells from white (Wh)- and opaque (Op)-phase colonies of strain TU17 and the RPD3 deletion mutant RPho19. Arrowheads denote opaque colonies and white colonies resulting from elevated frequencies of switching in the white and opaque phases, respectively, of RPho19.
To assess the effect of deleting RPD3 on phase-specific gene expression, Northern blots of total cellular RNA of white- and opaque-phase cells of the parent strain TU17 and the deletion mutant RPho19 were probed with the white-phase-specific genes WH11 (53) and EFG1 (55) and the opaque-phase-specific genes OP4 (32), SAP1 (33), and SAP3 (22, 62). Deletion of RPD3 had no effect on the developmental regulation of any of the phase-specific genes (Fig. 6B). It had a small effect on the transcript levels of the white-phase-specific genes WH11 and EFG1 and the opaque-phase-specific gene SAP1, all of which were approximately twofold lower (Fig. 6B). Deletion of RPD3 caused 10- to 15-fold decreases in the transcript levels of the two opaque-phase-specific genes OP4 and SAP3 (Fig. 6B). Similar results were obtained in a repeat experiment in which RNA was extracted from independent growth cultures of TU17 and two deletion strains, RPho19 and RPho3 (data not shown).
The expression of MCM1 and TUP1 in HDA1 and RPD3 deletion mutants.
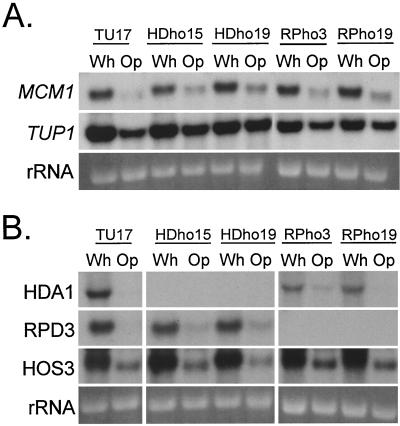
The opaque-phase-specific genes OP4 (30) and SAP3 (46; S. Lockhart and D. R. Soll, unpublished observations) contain MADS box protein consensus binding sites associated with the cis-acting activation sequences in their promoters, suggesting that a MADS box protein is involved in the regulation of these opaque-phase-specific genes. The MADS box binding sequences exhibited greatest homology to the MCM1 binding site in S. cerevisiae (30). We therefore cloned MCM1 by PCR using a sequence identified in the Stanford C. albicans database. Homology to the S. cerevisiae MCM1 gene was confirmed by partial sequencing. The cloned sequence was then used to probe Northern blots of the parental strain TU17, the HDA1 deletion mutants HDho15 and HDho19, and the RPD3 deletion mutants RPho3 and RPho19. We first found that the MCM1 transcript was high in white-phase cells and low in opaque-phase cells in parental strain TU17 (Fig. 8A). Neither deletion of HDA1 nor deletion of RPD3 affected the levels of expression in the white or opaque phase (Fig. 8A). The same blot was probed with TUP1, a transcription factor gene (6) expressed in white- and opaque-phase cells, but at slightly lower levels in the latter (R. Zhao, S. R. Lockhart, and D. R. Soll, unpublished data). Neither deletion of HDA1 nor deletion of RPD3 affected levels of expression in the white or opaque phase (Fig. 8A).
FIG. 8.
(A) Northern blot analysis of the expression of the MADS box protein gene MCM1 and the constitutively expressed transcription factor gene TUP1 in the white and opaque phases of the parent strain TU17, the HDA1 deletion mutants HDho15 and HDho19, and the RPD3 deletion mutants RPho3 and RPho19. The ethidium bromide-stained 18S rRNA band is shown at the bottom of each lane to assess loading. (B) Northern blot analysis of the expression of other deacetylase genes in the HDA1 mutants HDho15 and HDho19 and in the RPD3 mutants RPho3 and RPho19. The ethidium bromide-stained 18S rRNA band is shown at the bottom of each lane to assess loading.
Expression of the deacetylases in the HDA1 and RPD3 deletion mutants.
The deletion mutants HDA1 and RPD3 showed no hybridization bands with the respective probes, demonstrating that these strains were devoid of the respective deacetylase mRNAs (Fig. 8B). The deletion of HDA1 had no effect on the transcript levels of RPD3 and HOS3 (Fig. 8B) or HOS1 and HOS2 (data not shown) in either the white or opaque phase. Likewise, deletion of RPD3 had no effect on the transcript levels of HDA1 and HOS3 (Fig. 8B) or HOS1 and HOS2 (data not shown).
DISCUSSION
The molecular mechanisms involved in the downstream regulation of phase-specific genes in the process of high-frequency phenotypic switching have begun to emerge in recent years. In the white-opaque transition, which has been used as an experimental model system to study these mechanisms (46, 47), it has been demonstrated that phase-regulated genes are activated through the interaction of phase-specific trans-acting factors with specific cis-acting sequences in the promoters of these genes (30, 51, 55, 56). Because the cis-acting sequences in coordinately activated genes of the same phase differ, the simplest model of regulation involving a common trans-acting DNA-binding protein has been excluded (46), and alternative more complex models have been entertained (46). The emerging complexity of the circuitry that regulates the expression of phase-specific genes in the white-opaque transition should have been expected, given the complexity of the molecular mechanisms regulating eukaryotic gene expression in general. Indeed, in the regulation of a gene, it has become clear that in addition to activators and repressors, chromatin-modulating machinery is recruited that is involved in establishing transcriptionally active or inactive states (57). Among the classes of proteins involved in chromatin modifications, the deacetylases have been demonstrated to function in the repression of gene loci through the selective deacetylation of histones H3 and H4 (2, 63). To test whether deacetylases also play a role in switching, we initially performed an experiment with the deacetylase inhibitor TSA and found that it caused a dramatic and selective increase in the frequency of switching in the white-to-opaque direction, but had no effect on switching in the opaque-to-white direction (26). Since TSA preferentially inhibits the major deacetylase Hda1p (7), we deleted the gene coding for this protein in C. albicans strain WO-1 and found that the mutant phenotype was similar to that of TSA-treated cells (26). Switching was selectively upregulated in the white-to-opaque direction only, suggesting that deacetylation through Hda1p functioned either at a “master switch locus” to suppress the event or at the site of an activator of the switch event (26). Here we have extended these studies by identifying the members of the deacetylase family in C. albicans, examining the effects of switching on expression, testing whether HDA1 plays a role in phase-specific gene expression, and testing whether a second major deacetylase, RPD3, also plays a role in switching and phase-specific gene expression.
The histone deacetylases in C. albicans.
We cloned five histone deacetylases from C. albicans with homology to the five deacetylases in S. cerevisiae. By generating phylogenetic trees based on homology comparisons between the amino acid sequences of the five cloned C. albicans deacetylase genes, the five S. cerevisiae deacetylase genes and deacetylase genes from additional fungi, three groups were identified. One contained C. albicans Hda1p; the second contained C. albicans Hos1p, Hos2p, and Rpd3p; and the third contained C. albicans Hos3p. In each group, the deacetylase with the highest homology to each C. albicans deacetylase was the S. cerevisiae homolog. A comparison of the deacetylation motif of 22 deacetylases that included the five C. albicans deacetylases revealed 12 identical amino acid residues and very high levels of homology overall. By using a more rigorous search of the protein database, a sixth C. albicans deacetylase was recently identified in the Stanford C. albicans genome database with homology to Rpd3p (data not shown). We are in the process of characterizing the role of this new member of the histone deacetylase family by gene deletion and dominant-negative mutation strategies.
Both HDA1 and RPD3 play roles in the switching process.
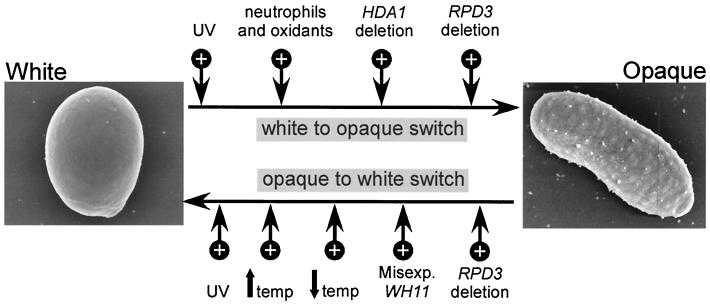
We had previously demonstrated the selective role of HDA1 in suppressing switching in the white-to-opaque direction (26), so we considered the possibility that a second deacetylase might selectively suppress switching in the opaque-to-white direction. We were therefore surprised to find that deletion of RPD3 resulted in an increase in the frequency of switching in both the white-to-opaque and opaque-to-white directions. The observation that deletion of either deacetylase results in an increase in switching in the white-to-opaque direction, but only deletion of RPD3 results in an increase in switching in the opaque-to-white direction, suggests the mechanisms for switching in the two directions are distinct. Other observations support this conclusion. First, an increase in temperature from 25 to >37°C leads to the mass conversion of opaque to white, but has no apparent effect on switching in the white-to-opaque direction (32, 38, 42, 53, 55). A decrease in temperature also selectively stimulates switching in the opaque-to-white direction only (42). Second, Kolotila and Diamond (27) demonstrated that leukocytes and oxidants selectively stimulated switching in the white-to-opaque direction, like TSA treatment and deletion of HDA1 (26). Third, misexpression of the white-phase-specific gene WH11 selectively stimulates switching in the opaque-to-white direction, but not the white-to-opaque direction (28). Finally, UV treatment stimulates switching in both the white-to-opaque direction and the opaque-to-white direction (32), as we have demonstrated here for RPD3 deletion. The effects of these different treatments on the frequencies of switching in the two directions are summarized in Fig. 9. It should be noted that although the characteristics of switching in the alternate directions differ, we cannot rule out the likely possibility that the reversible switch event occurs at a single locus.
FIG. 9.
Diagram of the different treatments, environmental conditions, and deacetylase mutations that cause increases (+) in the frequency of switching in the white-to-opaque and opaque-to-white directions. UV treatment (31) and treatment with neutrophils and oxidants (27) are shown. An up arrow shows a shift to high temperature (temp) (32, 38, 42, 51), and a down arrow shows a shift to low temperature (42). misexp., misexpression of WH11 in the opaque phase (28).
Since deacetylases play a role in the repression of gene expression (20, 40, 59, 64), we must first entertain the possibility that Hda1p and Rpd3p suppress switching by removing acetyl groups from histone 3 and/or 4 at the actual site of the switch event. In this case, a switch event would be affected by increased or altered acetylation patterns through one of the many histone acetyltransferases in an activation complex at the master switch locus. However, we must also entertain the alternate possibility that one or both histone deacetylases function to suppress one or more trans-activators of switching, which act directly upon the master switch locus. In this case, deletion of the deacetylase leads to up-regulation of the activator and, in turn, to up-regulation of the primary switch event.
The roles of HDA1 and RPD3 in the regulation of phase-specific gene expression and other deacetylases.
The histone deacetylases and SIR2, which was recently demonstrated to have NAD-dependent histone deacetylase activity (23), play roles in the expression of large numbers of different genes in S. cerevisiae. Recently, Bernstein et al. (5) performed a bioinformatic analysis of genes up-regulated at least 1.5-fold in rpd3, hda1, and sir2 mutants. They found that while the gene coding for Hda1p plays a more prominent role in regulating carbon metabolite and carbohydrate transport and utilization, RPD3 plays a role in cell cycle progression, and SIR2 plays a role in amino acid biosynthesis. There were also overlapping roles of the three genes in these three functional categories. In the more limited study of gene expression that we performed, we found no instance in which deletion of a deacetylase resulted in up-regulation of a phase-specific gene (Table 4). We did, however, observe down-regulation. In the case of the HDA1 deletion, white-phase expression of EFG1 was down-regulated, but there was no significant effect on the white-phase-specific expression of WH11 or HOS3 or the opaque-phase-specific expression of OP4, SAP1, and SAP3 (Table 4). There was also no effect on the white-phase-enriched expression of MCM1 (Table 4). In the case of RPD3 deletion, there was a small decrease in the expression of the white-phase genes WH11 and EFG1, as well as a more dramatic decrease in the opaque-phase genes OP4, SAP1, and SAP3 (Table 4). As in the case of the HDA1 deletion, there was no effect on the phase-regulated expression of MCM1 (Table 4). Neither deletion of HDA1 nor deletion of RPD3 affected the phase-regulated expression of the four other deacetylases (Table 4).
TABLE 4.
Summary of the effects of the HDA1 and RPD3 deletions on gene expression during the white-opaque transition
| Gene type | Expression
levela
|
|||||
|---|---|---|---|---|---|---|
| White
phase
|
Opaque phase
|
|||||
| TU17 | HDho15 | RPho19 | TU17 | HDho15 | RPho19 | |
| White-phase specific | ||||||
| WH11 | ++++ | ++++ | +++ | − | − | − |
| EFG1 (3.2 kb) | ++++ | ++(↓) | +++ | − | − | − |
| HOS3 (2.5 kb) | ++++ | ++++ | ++++ | − | − | − |
| Opaque-phase specific | ||||||
| OP4 | − | − | − | ++++ | ++++ | ++(↓) |
| SAP1 | − | − | − | ++++ | ++++ | ++(↓) |
| SAP3 | − | − | − | ++++ | ++++ | +(↓) |
| MCM1 | ++++ | ++++ | ++++ | + | + | + |
| HDA1 | ++++ | − | +++ | + | + | + |
| RPD3 | ++++ | ++++ | − | + | + | + |
| HOS1 | ++++ | ++++ | ++++ | +++ | ++ | ++ |
| HOS2 | ++++ | ++++ | ++++ | ++ | ++ | ++ |
| HOS3 (2.5 kb) | ++++ | ++++ | ++++ | − | − | − |
| HOS3 (2.3 kb) | − | − | − | ++++ | ++++ | ++++ |
| Constitutive | ||||||
| TUP1 | ++++ | ++++ | ++++ | +++ | +++ | +++ |
++++, full expression; +++, slightly reduced expression; ++, very reduced expression; +, extremely reduced expression; −, no expression. A down arrow in parentheses represents significant down-regulation.
Our observation that the deletion of HDA1 or RPD3 led only to down-regulation, not up-regulation, of select phase-specific genes is interesting, but not unique. Deletion of RPD3 in S. cerevisiae leads to increased repression of reporter gene expression at the mating type loci, telomeres, and ribosomal DNA (9, 20, 39), all loci under the regulation of Sir proteins (9, 17, 44). Increased silencing at these loci results presumably from up-regulation of silencing genes usually suppressed by the activity of the deacetylases. We assume that the down-regulation of select phase-specific genes in the deacetylase deletion mutants of C. albicans is effected by similar mechanisms.
Conclusion.
We have presented evidence that the relationship between high-frequency phenotypic switching and the deacetylases is complex. First, both HDA1 and RPD3 play roles in the suppression of switching: the former in the selective suppression of switching in the white-to-opaque direction and the latter in both the white-to-opaque and opaque-to-white directions. The results do not distinguish in either case whether suppression is mediated by deacetylation directly at the site of the switch event or indirectly at the site of a gene that encodes an activator of the switch event. Second, switching affects the expression of deacetylases. For four of the five deacetylases, expression is higher in the white phase. Third, deletion of HDA1 or RPD3 in no case results in up-regulation of a phase-specific or phase-enriched gene, and in select cases, it results in down-regulation. Our results therefore demonstrate that the deacetylases play distinct roles not only in the suppression of switching, but also in the activation of select phase-regulated genes, in the latter case presumably through the down-regulation of suppressor genes.
ACKNOWLEDGMENTS
We thank C. Pujol and S. Lachke for assistance with portions of this work. We also thank S. Lockhart for the MCM1 probe.
The major portion of this research was supported by NIH grant AI2392 to D.R.S. The research performed by A. J. S. Klar was sponsored by the National Cancer Institute, U.S. Department of Health and Human Services.
REFERENCES
- 1.Anderson J M, Soll D R. The unique phenotype of opaque cells in the white-opaque transition in Candida albicans. J Bacteriol. 1987;169:5579–5588. doi: 10.1128/jb.169.12.5579-5588.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ayer D E. Histone deacetylases: transcriptional repression with SINers and NuRDs. Trends Cell Biol. 1999;9:193–198. doi: 10.1016/s0962-8924(99)01536-6. [DOI] [PubMed] [Google Scholar]
- 3.Balan I, Alareo A-M, Raymond M. The Candida albicans CDR3gene codes for an opaque-phase ABC transporter. J Bacteriol. 1997;179:7210–7218. doi: 10.1128/jb.179.23.7210-7218.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bedell G W, Soll D R. The effects of low concentrations of zinc on the growth and dimorphism of Candida albicans: evidence for zinc-resistant and -sensitive pathways for mycelium formation. Infect Immun. 1979;26:348–354. doi: 10.1128/iai.26.1.348-354.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernstein B E, Tong J K, Schreiber S L. Genome wide studies of histone deacetylase function in yeast. Proc Natl Acad Sci USA. 2000;97:13708–13712. doi: 10.1073/pnas.250477697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun B R, Johnson A D. Control of filament function in Candida albicans by the transcription repressor TUP1. Science. 1997;77:105–109. doi: 10.1126/science.277.5322.105. [DOI] [PubMed] [Google Scholar]
- 7.Carmen A A, Rundlett S E, Grunstein M. HDA1 and HDA3are components of a yeast histone deacetylase (HDA) complex. J Biol Chem. 1996;271:15837–15844. doi: 10.1074/jbc.271.26.15837. [DOI] [PubMed] [Google Scholar]
- 8.Dayhoff M O. Atlas of protein sequence and structure. Washington, D.C.: National Biomedical Research Foundation; 1979. [Google Scholar]
- 9.De Rubertis F, Kadosh D, Henchoz S, Pauli D, Reuter G, Struhl K, Spierer P. The histone deacetylase RPD3 counteracts genomic silencing in Drosophilaand yeast. Nature. 1996;384:589–591. doi: 10.1038/384589a0. [DOI] [PubMed] [Google Scholar]
- 10.Ekwall K, Olsson T, Turner B M, Cranston G, Allshire R C. Transient inhibition of histone deacetylation alters the structural and functional imprint at fission yeast centromeres. Cell. 1997;91:1021–1032. doi: 10.1016/s0092-8674(00)80492-4. [DOI] [PubMed] [Google Scholar]
- 11.Fitch W M, Margoliash E. Construction of phylogenetic trees. Science. 1967;155:279–284. doi: 10.1126/science.155.3760.279. [DOI] [PubMed] [Google Scholar]
- 12.Fonzi W A, Irwin M Y. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franz R, Michel S, Morschhauser J. A fourth gene from the Candida albicansCDR family of ABC transporters. Gene. 1998;220:91–98. doi: 10.1016/s0378-1119(98)00412-0. [DOI] [PubMed] [Google Scholar]
- 14.Gil C, Pomes R, Nombela C. A complementation analysis by parasexual recombination of Candida albicansmorphological mutants. J Gen Microbiol. 1988;134:1587–1595. doi: 10.1099/00221287-134-6-1587. [DOI] [PubMed] [Google Scholar]
- 15.Glasgow A C, Hughes K T, Simon M I. Bacterial DNA inversion systems. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C.: American Society for Microbiology; 1989. pp. 637–659. [Google Scholar]
- 16.Gottlieb S, Esposito S. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell. 1989;56:771–776. doi: 10.1016/0092-8674(89)90681-8. [DOI] [PubMed] [Google Scholar]
- 17.Gottschling D E, Aparacio O M, Billington B L, Zakian V A. Position effect at S. cerevisiaetelomeres: reversible repression of Pol H transcription. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- 18.Grozinger C M, Hassig C A, Schreiber S L. Three proteins define a class of human histone deacetylases related to yeast Hda1p. Proc Natl Acad Sci USA. 1999;96:4868–4873. doi: 10.1073/pnas.96.9.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guarente L. Sir2 links chromatin silencing, metabolism, and aging. Genes Dev. 2000;14:1021–1026. [PubMed] [Google Scholar]
- 20.Hassig C A, Tong J K, Fleischer T C, Owa T, Grable P G, Ayer D E, Schreiber S L. A role for histone deacetylase activity in HDAC1-mediated transcriptional repression. Proc Natl Acad Sci USA. 1998;95:3519–3524. doi: 10.1073/pnas.95.7.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hellstein J, Vawter-Hugart H, Fotos P, Schmid J, Soll D R. Genetic similarity and phenotypic diversity of commensal and pathogenic strains of Candida albicansisolated from the oral cavity. J Clin Microbiol. 1993;31:3190–3199. doi: 10.1128/jcm.31.12.3190-3199.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hube B, Monod M, Schofield D, Brown A, Gow N. Expression of seven members of the gene family encoding aspartyl proteinases in Candida albicans. Mol Microbiol. 1994;14:87–99. doi: 10.1111/j.1365-2958.1994.tb01269.x. [DOI] [PubMed] [Google Scholar]
- 23.Imai S-I, Armstrong C M, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 24.Jones S, White G, Hunter P R. Increased phenotype switching in strains of Candida albicansassociated with invasive infections. J Clin Microbiol. 1994;32:2869–2870. doi: 10.1128/jcm.32.11.2869-2870.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kadosh D, Struhl K. Histone deacetylase activity of Rpd3 is important for transcriptional repression in vivo. Genes Dev. 1998;12:797–805. doi: 10.1101/gad.12.6.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klar, A. J. S., T. Srikantha, and D. R. Soll. A histone deacetylation inhibitor and mutant promote colony-type switching of the human pathogen Candida albicans. Genetics, in press. [DOI] [PMC free article] [PubMed]
- 27.Kolotila M P, Diamond R D. Effects of neutrophils and in vitro oxidants on survival and phenotypic switching of Candida albicansWO-1. Infect Immun. 1990;58:1174–1179. doi: 10.1128/iai.58.5.1174-1179.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kvaal C A, Srikantha T, Soll D R. Misexpression of the white-phase-specific gene WH11 in the opaque phase of Candida albicansaffects switching and virulence. Infect Immun. 1997;65:4468–4475. doi: 10.1128/iai.65.11.4468-4475.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leipe D D, Landsman D. Histone deacetylases, acetoin utilization proteins and acetylpolyamine amidohydrolases are members of an ancient protein superfamily. Nucleic Acids Res. 1997;25:3693–3697. doi: 10.1093/nar/25.18.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lockhart S R, Nguyen M, Srikantha T, Soll D R. A MADS box protein consensus binding site is necessary and sufficient for activation of the opaque-phase-specific gene OP4 of Candida albicans. J Bacteriol. 1998;180:6607–6616. doi: 10.1128/jb.180.24.6607-6616.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morrow B, Anderson J, Wilson E, Soll D R. Bidirectional stimulation of the white-opaque transition of Candida albicansby ultraviolet irradiation. J Gen Microbiol. 1989;135:1201–1208. doi: 10.1099/00221287-135-5-1201. [DOI] [PubMed] [Google Scholar]
- 32.Morrow B, Srikantha T, Anderson J, Soll D R. Coordinate regulation of two opaque-phase-specific genes during white-opaque switching in Candida albicans. Infect Immun. 1993;61:1823–1828. doi: 10.1128/iai.61.5.1823-1828.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morrow B, Srikantha T, Soll D R. Transcription of the gene for a pepsinogen, PEP1, is regulated by white-opaque switching in Candida albicans. Mol Cell Biol. 1992;12:2997–3005. doi: 10.1128/mcb.12.7.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Odds F C. Switch of phenotype as an escape mechanism of the intruder. Mycoses. 1997;40:S9–S12. doi: 10.1111/j.1439-0507.1997.tb00556.x. [DOI] [PubMed] [Google Scholar]
- 35.Perez-Martin J, Uria J A, Johnson A D. Phenotypic switching in Candida albicans is controlled by a SIR2gene. EMBO J. 1999;18:2580–2592. doi: 10.1093/emboj/18.9.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pilus L, Rine J. Epigenetic inheritance of transcriptional states of S. cerevisiae. Cell. 1989;59:637–647. doi: 10.1016/0092-8674(89)90009-3. [DOI] [PubMed] [Google Scholar]
- 37.Ramsey H, Morrow B, Soll D R. An increase in switching frequency correlates with an increase in recombination of the ribosomal chromosomes of Candida albicansstrain 3153A. Microbiology. 1994;140:1525–1531. doi: 10.1099/13500872-140-7-1525. [DOI] [PubMed] [Google Scholar]
- 38.Rikkerink E H A, Magee B B, Magee P T. Opaque-white phenotypic transition: a programmed morphological transition in Candida albicans. J Bacteriol. 1988;170:895–899. doi: 10.1128/jb.170.2.895-899.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rundlett S E, Carmen A A, Kobayashi R, Bavykin S, Turner B M, Grunstein M. HDA1 and RPD3are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc Natl Acad Sci USA. 1996;93:14503–14508. doi: 10.1073/pnas.93.25.14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rundlett S E, Carmen A A, Suka N, Turner B M, Grunstein M. Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature. 1998;392:831–836. doi: 10.1038/33952. [DOI] [PubMed] [Google Scholar]
- 41.Slutsky B, Buffo J J, Soll D R. High frequency switching of colony morphology in Candida albicans. Science. 1985;230:666–669. doi: 10.1126/science.3901258. [DOI] [PubMed] [Google Scholar]
- 42.Slutsky B, Staebell M, Anderson J, Risen L, Pfaller M, Soll D R. “White-opaque transition”: a second high-frequency switching system in Candida albicans. J Bacteriol. 1987;169:189–197. doi: 10.1128/jb.169.1.189-197.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith J S, Caputo E, Boeke J D. A genetic screen for ribosomal DNA silencing defects identifies multiple DNA replication and chromatin-modulating factors. Mol Cell Biol. 1999;19:3184–3197. doi: 10.1128/mcb.19.4.3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soll D R. High-frequency switching in Candida albicans. Clin Microbiol Rev. 1992;5:183–203. doi: 10.1128/cmr.5.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soll D R. New fungal strategies. Edinburgh, Scotland: Churchill Livingstone; 1992. pp. 156–172. [Google Scholar]
- 46.Soll, D. R. The molecular biology of switching in Candida. In R. Cihlar and R. Calderone (ed.), Fungal pathogenesis: principles and clinical application, in press. Marcel Dekker, Inc., New York, N.Y.
- 47.Soll, D. R. Phenotypic switching. In R. Calderone (ed.), Candida and candidiasis, in press. American Society for Microbiology, Washington, D.C.
- 48.Soll D R, Galask R, Isley S, Rao T V G, Stone D, Hicks J, Schmid J, Mac K, Hanna C. Switching of Candida albicansduring successive episodes of recurrent vaginitis. J Clin Microbiol. 1989;27:681–690. doi: 10.1128/jcm.27.4.681-690.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Soll D R, Langtimm C J, McDowell J, Hicks J, Galask R. High-frequency switching in Candidastrains isolated from vaginitis patients. J Clin Microbiol. 1987;25:1611–1622. doi: 10.1128/jcm.25.9.1611-1622.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Soll D R, Staebell M, Langtimm C J, Pfaller M, Hicks J, Rao T V G. Multiple Candidastrains in the course of a single systemic infection. J Clin Microbiol. 1988;26:1448–1459. doi: 10.1128/jcm.26.8.1448-1459.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Srikantha T, Chandrasekhar A, Soll D R. Functional analysis of the promoter of the phase-specific WH11 gene of Candida albicans. Mol Cell Biol. 1995;15:1797–1805. doi: 10.1128/mcb.15.3.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Srikantha T, Klapach A, Lorenz W W, Tsai L K, Laughlin L A, Gorman J A, Soll D R. The sea pansy Renilla reniformis luciferase serves as a sensitive bioluminescent reporter for differential gene expression in Candida albicans. J Bacteriol. 1996;178:121–129. doi: 10.1128/jb.178.1.121-129.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Srikantha T, Soll D R. A white-specific gene in the white-opaque switching system of Candida albicans. Gene. 1993;131:53–60. doi: 10.1016/0378-1119(93)90668-s. [DOI] [PubMed] [Google Scholar]
- 54.Srikantha T, Tsai L, Daniels K, Enger L, Highley K, Soll D R. The two-component hybrid kinase regulator CaNIK1 of Candida albicans. Microbiology. 1998;144:2715–2729. doi: 10.1099/00221287-144-10-2715. [DOI] [PubMed] [Google Scholar]
- 55.Srikantha T, Tsai L K, Daniels K, Soll D R. EFGI null mutants of Candida albicanscan switch, but cannot express the complete phenotype of white-phase budding cells. J Bacteriol. 2000;182:1580–1591. doi: 10.1128/jb.182.6.1580-1591.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Srikantha T, Tsai L K, Soll D R. The WH11 gene of Candida albicansis regulated in two distinct developmental programs through the same transcription activation sequences. J Bacteriol. 1997;179:3837–3844. doi: 10.1128/jb.179.12.3837-3844.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Struhl K. Fundamentally different logic of gene regulation in eukaryotes and prokaryotes. Cell. 1999;98:1–4. doi: 10.1016/S0092-8674(00)80599-1. [DOI] [PubMed] [Google Scholar]
- 58.Suka N, Carmen A A, Rundlett S, Grunstein M. The regulation of gene activity by histones and the histone deacetylase RPD3. Cold Spring Harbor Symp Quant Biol. 1998;63:391–399. doi: 10.1101/sqb.1998.63.391. [DOI] [PubMed] [Google Scholar]
- 59.Sun Z-W, Hampsey M. A general requirement for the Sin3-Rpd3 histone deacetylase complex in regulating silencing in Saccharomyces cerevisiae. Genetics. 1999;152:921–932. doi: 10.1093/genetics/152.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. The CLUSTAL:X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vargas K, Messer S A, Pfaller M, Lockhart S R, Stapleton J T, Soll D R. Elevated phenotypic switching and drug resistance of Candida albicansfrom human immunodeficiency virus-positive individuals prior to first thrush episode. J Clin Microbiol. 2000;38:3595–3607. doi: 10.1128/jcm.38.10.3595-3607.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.White T C, Miyasaki S H, Agabian N. Three distinct secreted aspartyl proteinases in Candida albicans. J Bacteriol. 1993;175:6126–6133. doi: 10.1128/jb.175.19.6126-6133.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Workman J L, Kingston R E. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 64.Wu M, Newcomb L, Heideman W. Regulation of gene expression by glucose in Saccharomyces cerevisiae: a role for ADA2 and ADA3/NGG1. J Bacteriol. 1999;181:4755–4760. doi: 10.1128/jb.181.16.4755-4760.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yoshida M, Horinouchi S, Beppu T. Trichostatin A and trapoxin: novel chemical probes for the role of histone acetylation in chromatin structure and function. Bioessays. 1995;17:423–430. doi: 10.1002/bies.950170510. [DOI] [PubMed] [Google Scholar]