Abstract
The genomes of pathogenic Haemophilus influenzae strains are larger than that of Rd KW20 (Rd), the nonpathogenic laboratory strain whose genome has been sequenced. To identify potential virulence genes, we examined genes possessed by Int1, an invasive nonencapsulated isolate from a meningitis patient, but absent from Rd. Int1 was found to have a novel gene termed lav, predicted to encode a member of the AIDA-I/VirG/PerT family of virulence-associated autotransporters (ATs). Associated with lav are multiple repeats of the tetranucleotide GCAA, implicated in translational phase variation of surface molecules. Laterally acquired by H. influenzae, lav is restricted in distribution to a few pathogenic strains, including H. influenzae biotype aegyptius and Brazilian purpuric fever isolates. The DNA sequence of lav is surprisingly similar to that of a gene previously described for Neisseria meningitidis. Sequence comparisons suggest that lav was transferred relatively recently from Haemophilus to Neisseria, shortly before the divergence of N. meningitidis and Neisseria gonorrhoeae. Segments of lav predicted to encode passenger and β-domains differ sharply in G+C base content, supporting the idea that AT genes have evolved by fusing domains which originated in different genomes. Homology and base sequence comparisons suggest that a novel biotype aegyptius AT arose by swapping an unrelated sequence for the passenger domain of lav. The unusually mobile lav locus joins a growing list of genes transferred from H. influenzae to Neisseria. Frequent gene exchange suggests a common pool of hypervariable contingency genes and may help to explain the origin of invasiveness in certain respiratory pathogens.
Horizontal acquisition of virulence-determining genes has accelerated the evolution of bacterial pathogens (35). The recent availability of complete microbial genome sequences facilitates investigation of the history of virulence genes. Of particular interest are contingency loci, which encode phase-variable surface molecules involved in host tissue interactions (32). Contingency gene expression is commonly modulated by slipped-strand mispairing acting on simple sequence repeats within a control region. Although distantly related, the respiratory commensals and pathogens Haemophilus influenzae and Neisseria meningitidis each use tetranucleotide repeat sequences in translational phase variation of surface proteins (23, 37, 40). Such similarity prompts the question of whether repeat-regulated genes were present in a common ancestor, evolved independently, or had a recent common origin in laterally transferred DNA.
Several important human pathogens (Streptococcus pneumoniae, H. influenzae, and N. meningitidis) coexist in the respiratory flora of healthy individuals. Kroll et al., who first reported natural transfer of H. influenzae sequences to N. meningitidis, have suggested that genetic exchange between respiratory pathogens may spark the emergence of new invasive strains (25). They have identified three sequences, originally from Haemophilus and flagged by Haemophilus-specific uptake sequences (hUSs), which now reside in the meningococcal genome.
H. influenzae, a small, gram-negative bacterium, causes otitis media and bronchitis as well as invasive disease (meningitis and septicemia) (50). Invasive H. influenzae disease is usually caused by encapsulated strains of H. influenzae serotype b (Hib); near-universal immunization with conjugate Hib vaccine has largely eliminated Haemophilus meningitis from developed countries (3). Nonencapsulated (nontypeable) H. influenzae (NTHi), against which the Hib vaccine provides no protection, remains an important cause of respiratory infections (18). Although NTHi rarely causes invasive infection in immunocompetent hosts, sporadic exceptions warn of potential virulence. During the 1980s an outbreak of highly lethal septicemia among children in rural Brazil, termed Brazilian purpuric fever (BPF), was traced to a single NTHi clone related to H. influenzae biotype aegyptius strains previously associated only with conjunctivitis (7). In 1994, an NTHi strain (Int1; also called R2866) was isolated in the United States from the blood of a previously healthy, Hib-immunized child who developed meningitis (34).
H. influenzae is readily transformed with Haemophilus DNA; uptake requires the hUS, consisting of a conserved 9-bp core within an extended 29-bp consensus sequence. The H. influenzae strain Rd KW20 (hereafter referred to as Rd) genome, which has been sequenced (17), contains 1,465 hUSs. While most of these occur as single copies, 17% occur as pairs of inverted repeats in a stem-loop configuration. Stem-loop hUS pairs tend to occur at the ends of transcription units and may function as transcription terminators (44). At least one horizontally transferred island has been found inserted within a stem-loop hUS pair, suggesting that paired hUSs may play an additional role as potential targets for the insertion of virulence-associated genes, possibly mediated by phage integrases (10, 30).
N. meningitidis is a gram-negative bacterium phylogenetically distant from H. influenzae; N. meningitidis is classified in the β subdivision and H. influenzae is classified in the γ subdivision of the proteobacteria. In sub-Saharan Africa, N. meningitidis serogroup A causes epidemic meningitis, whereas in the developed world serogroups B and C cause endemic invasive disease (29, 47). Natural transformation is the primary means of genetic exchange among neisseriae; it is so frequent that lineage boundaries are blurred, requiring populations to be depicted as networks rather than as distinct clades (45). Transformation employs a neisseria-specific 10-bp uptake sequence (nUS) distinct from the hUS (14).
To identify NTHi pathogenicity genes, we initiated whole genome comparisons between Int1 and Rd, a nonpathogenic laboratory strain. Here we report the identification of lav, a candidate pathogenicity gene whose inferred translation product is homologous to virulence-associated autotransporters (AT) and whose DNA sequence is similar to NMB1527 (also called orf2 and nmrep3) and NMA1725 of N. meningitidis (24, 36, 37, 48). We present evidence that lav arose by fusion of segments from different organisms and has recently been transferred from Haemophilus to Neisseria.
MATERIALS AND METHODS
Bacteria.
H. influenzae was grown as previously described (30, 31); strains are described in Table 1.
TABLE 1.
Bacterial strainsa
| Strain | Source, disease, and/or description | Serotype; other relevant properties | lav insertb | Reference |
|---|---|---|---|---|
| Eagan (E1a) | CSF,c meningitis | Type b | − | 43 |
| Rd (R906) | Laboratory strain | Derived from type d | − | 9 |
| R539 | ATCC 9006 | Reference type a | − | |
| R538 | ATCC 9795 | Reference type b | − | |
| R540 | ATCC 9007 | Reference type c | − | |
| R541 | ATCC 9008 | Reference type d | − | |
| R542 | ATCC 8142 | Reference type e | − | |
| R543 | ATCC 9796 | Reference type f | − | |
| R1967 | ATCC 11116 | NTHi; reference biotype aegyptius | + | |
| R2140 (CDCF3031) | Blood, BPF | NTHi; biotype aegyptius | + | 7 |
| R2141 (CDCF3035) | Blood, BPF | BTHi; biotype aegyptius | + | 7 |
| R2866 (Int1) | Blood, meningitis | NTHi | + | 34 |
| R3001 | Bronchial isolate | NTHi | − | 31 |
| R3099 (602) | Chronic bronchitis | NTHi | − | 19 |
| R3100 (4564) | Chronic bronchitis | NTHi | − | 19 |
| R3101 (6173) | Chronic bronchitis | NTHi | − | 19 |
| R3151 (AAr160) | Tracheal aspirate | NTHi | + | 12 |
| R3157 (1128) | Otitis media | NTHi | + | 13 |
| R3169 | Invasive | NTHi | − | 33 |
| R3174 | Invasive | NTHi | − | 33 |
| R3179 (AAr73) | Nasopharynx | NTHi | − | 12 |
| R3237 | R2866 with TSTE insert in mutS | NTHi | + | —d |
| C2859 | CSF | NTHi | − | 31 |
| C2861 | CSF | NTHi | − | 31 |
| C2939 | Invasive | NTHi | − | 33 |
| C2965 | Invasive | NTHi | − | 33 |
H. influenzae, including laboratory strains and clinical isolates.
Presence (+) or absence (−) of lav-related insert in holB-tmk region.
CSF, cerebrospinal fluid.
T. Mutangadura, B. Williams, A. L. Smith, and M. Golomb, unpublished data.
Differential cloning of Int1 sequences.
An M13mp18 library was prepared from a partial Sau3A digest of Int1 DNA size fractionated to yield fragments between 0.75 and 1.5 kb. Bacterial clones containing recombinant plasmids were arrayed on nylon filters (1,200 colonies each). Replicate filters were probed at high stringency with digoxigenin-labeled Rd and Int1 genomic DNAs, using M13 clones from Pseudomonas aeruginosa, a G+C-rich organism as background controls. Clones hybridizing with Int1 but not with Rd were picked, and inserts were initially sequenced in one direction with a universal primer.
DNA isolation and PCR.
H. influenzae DNA isolation and long PCR were done as previously described (31). The holB-tmk region was amplified with forward primer 5′-CTTTAATCAGCACAGCATGATGCC, corresponding to H. influenzae Rd nucleotides 477248 to 477272, and reverse primer 5′-TGTTCAAGCTGATATTGAAAGTGC, corresponding to H. influenzae nucleotides 477396 to 477373 (17). DNA from BPF isolates R2140 and R2141 was amplified using the same reverse primer but with forward primer 5′-CACATTTTCAAACTGGCTTGAC.
DNA blotting.
The genomic blots, hybridization, procedures and stringent wash conditions used have been previously described (31). The lav probe was a gel-purified 1.4-kb PCR fragment made using Int1 lav internal primers 5′-GCTTTTGGCTGTTGATTACG and 5′-CCGCCCATTAAGCCAACGG, that was then digoxigenin labeled.
DNA sequencing.
PCR fragments were gel purified and sequenced directly or following subcloning into a phagemid vector; both strands were sequenced as previously described (31). Sequences were analyzed using BLASTX, and BLASTN (1, 2) and the GCG and Omiga packages.
Nucleotide sequence accession number.
The sequences reported in this paper have been deposited in GenBank and assigned accession no. AF385403 (lav) and AF385404 (las).
RESULTS
Identification of lav.
We constructed a bank of clones from the pathogenic NTH: strain Int1 and screened them by hybridization against whole genomic DNA from strain Int1 and, as a comparison, from nonpathogenic strain Rd. This differential screening was intended to identify genes specifically present in Int1 which potentially encode pathogenicity-related proteins. Of an initial 1,200 clones prepared from Int1 DNA, ∼10% were novel relative to Rd. Sequence 122 bore the 3′ end of an open reading frame (ORF) homologous to a family of AT, which includes VirG of Shigella flexneri (27), AIDA-I of Escherichia coli (4), VapA/nmrep2 of N. meningitidis (37), and PerT of Bordetella pertussis (15). These virulence factors are outer membrane proteins involved in adhesion, invasion, intercellular spread, or immune evasion (21, 28). Each consists of a carboxyl-terminal β-barrel domain, which forms a pore in the outer membrane; a linker peptide; a passenger effector domain, which is secreted through the pore; and an N-terminal signal sequence. The partial Int1 ORF was ∼90% identical in DNA sequence to orf2/nmrep3/NMB1525 of N. meningitidis strain MC58 (serogroup B), previously identified by its homology to other AT (24, 37).
Downstream from the presumed AT gene, sequence 122 contained a junction with 93% DNA homology to a gene in strain Rd, the 3′ end of H. influenzae gene HI 0456 (tmk), encoding thymidylate kinase. In Rd, the gene immediately downstream from tmk is H. influenzae gene HI 0455 (holB), which encodes the δ subunit of DNA polymerase III. The initiator codon of holB overlaps the terminator codon of tmk; a paired hUS in (+/−) inverted repeat configuration extends across the region of overlap (Fig. 1A).
FIG. 1.
(A) Arrangement of lav homologs in H. influenzae and N. meningitidis MC58 genomes. Neisserial genes are depicted as filled arrows (indicating orientation); H. influenzae-derived genes are depicted as open arrows. Hairpin symbols represent paired hUSs in stem-loop configuration. Relative to that in H. influenzae-Rd, the Int1 lav island is inserted within a stem-loop hUS pair, resulting in partial duplication of the hUS pair flanking the site of insertion within an Int1 ancestor. Fragments of H. influenzae holB and tmk in Neisseria are indicated in parentheses. (B) Repeat-containing regions of lav homologs in Int1 (hi lav), N. meningitidis MC58 (nm) (24, 37), N. gonorrhoeae, (ng) and biotype aegyptius (hae) reference strain R1967. (C) Inferred amino acid sequences of regions of Lav homologs, aligned with MEGALIGN (DNAStar). aa 1 to 40 are conserved sequences at the N terminus, including the predicted signal sequence cleavage site (arrow). aa 121 to 156 represent a less conserved region within the presumptive passenger domain, including a sequence bracketed by conserved cysteine residues (asterisks).
Rd-specific primers to the 5′ end of holB and the 3′ end of tmk were used to amplify Int1 genomic DNA by PCR. The predicted 125-bp fragment was obtained with Rd and a Hib (Eagan) template, but a 2.4-kb fragment was obtained with Int1, including 2.3 kb of novel sequence (Fig. 1A). The novel DNA is located within the (+/−) hUS pair at the holB-tmk junction, 18 bp of which are now found duplicated at either end of the island. The insert contains a single, 2,078-bp potential ORF termed lav (for like a VirG). The putative initiating ATG is in a different reading frame from the downstream coding region; 10 bp downstream are 19 copies of the tetranucleotide GCAA. A gain of one GCAA repeat creates a 2,082-bp ORF, potentially encoding a 693-amino-acid (aa) Lav polypeptide (pI = 9.3) with a 54-aa signal sequence. Lav is homologous at its carboxyl terminus to VirG and other AT.
Horizontal transfer of lav.
The complete DNA sequence of H. influenzae lav was 89% similar to orf2/nmrep3 of N. meningitidis MC58. The meningococcal gene is located in a region containing rfaF (encoding a heptosyl transferase involved in lipo-oligosaccharide biosynthesis) between a gene (orf1) whose inferred product is a small protein B homolog and a gene (dld) encoding a d-lactate dehydrogenase (4, 18, 48). N. meningitidis serotype B orf2 is part of a 2.45-kb island of similarity to H. influenzae (Fig. 1A). Four GCAA repeats follow the initiator codon of orf2/nmrep3, placing its translated product out of reading frame; loss of a GCAA would permit translation of a 678-aa protein. A homologous gene (NMA1725) 95% identical to N. meningitidis serotype B. orf2, but with three GCAA repeats, is found at the same site in the genome of N. meningitidis serogroup A strain Z2491 (36).
The unfinished Neisseria gonorrhoeae genome (http://www.genome.ou.edu) was searched for homology with N. meningitidis orf2. A gonococcal sequence 91% identical to N. meningitidis orf2 and 89% identical to H. influenzae lav was found on contig 140, at the same chromosomal site as in N. meningitidis. Endpoints of the island of homology with H. influenzae are identical—exactly 57 bp downstream from orf1 and 35 bp downstream from dld—in the N. meningitidis and N. gonorrhoeae genomes. N. gonorrhoeae orf2 is preceded by two GCAA repeats after the initiating codon (Fig. 1B), is also out of reading frame, and contains an in-frame stop codon and a frameshift mutation. Inferred proteins encoded by lav homologs contain highly conserved regions interspersed with divergent regions (Fig. 1C). The N-terminal region surrounding the signal sequence is highly conserved, as is the C-terminal end of the β-domain; the passenger domain contains two conserved cysteine residues which can potentially form a short loop of variable sequence (Fig. 1C).
Three considerations indicate that transfer from Haemophilus to Neisseria has occurred. (i) The neisserial island of similarity to H. influenzae includes a sequence upstream of orf2 which matches hUS consensus at all 29 bp and a paired (+/−) uptake sequence downstream of orf2 which matches consensus at all but 1 bp. These hUSs align with those in Int1. (ii) The N. gonorrhoeae island includes 116 bp from H. influenzae holB and 225 bp from H. influenzae tmk in the intergenic spaces flanking orf2. The N. meningitidis island also includes 116 bp from H. influenzae holB, but it has only 77 bp from H. influenzae tmk. The 77-bp tmk sequence aligns with 225 bp of the Rd sequence but with a 148-bp internal deletion. The presence in the meningococcal genome of nonfunctional bits of adjacent H. influenzae genes is highly suggestive of the direction of transfer. (iii) The G+C content of N. meningitidis serotype B orf2 is 40.0%, and that of N. gonorrhoeae is 39.7%. These values are much closer to the H. influenzae genomic average of 38.2% than to the neisserial average of 51.8%.
Recent transfer of lav from Haemophilus to Neisseria.
Genetic similarity suggests that the Haemophilus lav gene and the Neisseria orf2 gene diverged relatively recently, not long before orf2 genes diverged in Neisseria. As a more reliable indicator of genetic distance, we compared noncoding, intergenic DNA sequence, which was expected to be subject to fewer selective constraints than coding sequence. The 256-bp sequence immediately upstream of lav can be aligned for all four bacterial genomes (Fig. 2a). It includes 19 codons of holB, but the remaining 206 bp in H. influenzae (and all 256 bp in Neisseria) are noncoding. The H. influenzae sequence is 94.5% identical to that of N. gonorrhoeae and 93.8% identical to that of N. meningitidis, whereas sequences from the two neisserial species are 96.9% identical. The distance between the sequences for H. influenzae and Neisseria is approximately twice that between the sequences for neisserial species (Fig. 2b). Similar relative distances (but higher absolute divergences) were found for the lav region including the β-barrel plus the linker (Fig. 2b). In comparison, the DNA sequence of an evolutionarily conserved regulatory gene, rpoD (encoding the major sigma factor), is only 47% identical between H. influenzae and Neisseria, yet rpoD similarity between the two Neisseria sequences is 97% (Fig. 2b).
FIG. 2.
(a) Alignment (MEGALIGN) of DNA sequence 256 bp upstream of lav homologs. Strain designations are as described for Fig. 1, except that NM-B is MC58 and NM-A is Z2491. The initiating codon (complementary strand) of holB is indicated (left arrow), and the sequence includes 19 codons at the N terminus-encoding portion of of holB. The start of lav is also indicated (right arrow). An 18-bp sequence from an hUS is underlined; this sequence has been duplicated from a paired hUS at the tmk end of the Int1 lav island. (b) Phylograms of aligned sequences from the 256-bp upstream sequence (5′ to lav), from the β-barrel region (last 385 codons, including the predicted linker region), and from rpoD (17, 36, 48). Phylogenies were derived with the ClustalV program of MEGALIGN, using a combination of the unweighted pair group method using arithmetic averages algorithm and the neighbor-joining method (39, 46). (c) H. influenzae lav is anomalously similar to its N. meningitidis homologs relative to pairwise comparisons between housekeeping genes shared by H. influenzae and N. meningitidis. Linear regression of inferred amino acid sequence identity on DNA identity of 12 housekeeping genes (open squares) in N. meningitidis serotype B and H. influenzae Rd is shown. Filled circles (arrows) indicate Int1 lav compared to homologs in N. meningitidis serotype B and N. meningitidis serotype A, in ascending order of DNA similarity. Genes compared, in ascending order of DNA similarity, were tpiA, lig (lig-1 in N. meningitidis serotype B) dnaG, mutS, gyrA, rpoD, cysK, aroG, rpoB, recA, sucD, and ilvD (ilvD-1 in N. meningitidis serotype B). The sequences were compared using ClustalV (MEGALIGN).
The atypically high similarity of lav homologs, relative to that of other genes shared by Haemophilus and Neisseria, is evident in Fig. 2c, which shows a comparison of 12 pairs of evolutionarily conserved housekeeping genes shared by these species to provide a baseline. The mean DNA similarity between Haemophilus and Neisseria was 47.3% (standard deviation, 8.7%; range, 35.0 to 60.2%). The values for the lav homologs lie above the range of DNA similarity represented for these highly conserved genes. With respect to DNA similarity, the Lav amino acid sequence is more divergent than that of or typical housekeeping genes, presumably because lav is exposed to a different type of selection (Fig. 2c).
Chimeric origin of lav as evidence for domain shuffling.
A scan of G+C content across the lsi-dld region revealed additional detail (Fig. 3). The island of similarity between H. influenzae and neisserial chromosomes coincides with a dip in G+C content to a level approaching the H. influenzae genomic average; the profile is similar in detail for all three species, again indicating a recent common origin. G+C content is not uniform across lav but rather rises sharply at 850 ± 50 bp after the GCAA repeat region. Thus, Int1 lav is composed of two distinct segments, a 5′ segment of 283 codons averaging 35.1% G+C and a 3′ segment of 376 codons averaging 40.8% G+C.
FIG. 3.
G+C content of regions containing lav homologs, scanned with a sliding window of 100 bp, in 25-bp increments. hi, H. influenzae Int1; nm, N. meningitidis serotype A Z2491; ng, N. gonorrhoeae. The interval on the abscissa corresponds to bp 113 to 5147 of N. gonorrhoeae contig 140 aligned with N. meningitidis serotype A interval 1660274 to 1654800 (which has a deletion within tmk) by creating a 148-bp gap in the N. meningitidis serotype A sequence starting at bp 4033 and the entire Int1 sequence between tmk and holB.
The two segments defined by G+C content approximately coincide with predicted boundaries of the two major structural and functional domains of an AT. The closest homolog to Lav with functionally characterized domains is the 1,286-aa E. coli AIDA-I preprotein (31). Amino acid sequence homology begins near the processing site at AIDA-I Leu840 and includes the linker region and the 14-strand β-barrel, which begins at AIDA-I Ala1002. On the basis of alignment with AIDA-I, the β-domain of Int1 lav is predicted to extend from codon 385 to 659 codon and the linker is predicted to extend from codon ∼290 to codon 384. The low-G+C segment of lav (first 283 codons) thus corresponds roughly to the predicted passenger domain, and the high-G+C segment (last 376 codons) corresponds to the β-barrel domain plus linker.
The two segments of lav have evolved at markedly different rates. The role of selection in fixing mutations can be assessed by comparing silent-substitution rates (Ks = synonymous changes per 100 synonymous sites) to nonsilent-substitution rates (Ka = nonsynonymous changes per 100 nonsynonymous sites) (32); the Ks/Ka ratio is usually >10 for conserved housekeeping genes. Silent-substitution rates in the C terminus-encoding segment are consistent with recent common origin of these genes (Table 2). The Ks values derived from a comparison of Int1 and Neisseria are only 1.2- to 1.5-fold higher than those derived from a comparison of N. meningitidis and N. gonorrhoeae. However, nonsynonymous-substitution rates differ substantially between the segments encoding the N and C termini within each lineage. The Ks/Ka ratio is ≈4 for the segment encoding the C terminus, but it is ≤1 for the segment encoding the N terminus-segment (Table 2). These differences suggest that the two major segments of the gene are subject to different evolutionary constraints.
TABLE 2.
Substitution rates of domain-encoding regions of lav homologsa
| Pair | Lav N terminus
|
Lav C terminus
|
||||
|---|---|---|---|---|---|---|
| Ks | Ka | Ks/Ka | Ks | Ka | Ks/Ka | |
| N. meningitidis serotype A vs N. meningitidis serotype B | 0.167 (0.053) | 0.156 (0.028) | 1.07 | 0.093 (0.024) | 0.021 (0.008) | 4.36 |
| N. meningitidis serotype A vs N. gonorrhoeae | 0.096 (0.034) | 0.120 (0.024) | 0.80 | 0.201 (0.041) | 0.065 (0.014) | 3.08 |
| N. meningitidis serotype A vs Int1 | 0.157 (0.052) | 0.179 (0.030) | 0.88 | 0.246 (0.042) | 0.056 (0.011) | 4.36 |
| N. gonorrhoeae vs Int1 | 0.145 (0.044) | 0.162 (0.030) | 0.89 | 0.310 (0.046) | 0.075 (0.014) | 4.11 |
| Biotype aegyptius vs N. meningitidis serotype A | —b | — | — | 0.416 (0.065) | 0.095 (0.0.016) | 4.40 |
| Biotype aegyptius vs N. gonorrhoeae | — | — | — | 0.418 (0.063) | 0.096 (0.016) | 4.37 |
| Biotype aegyptius vs Int1 | — | — | — | 0.448 (0.070) | 0.100 (0.016) | 4.50 |
Putative domain-encoding DNA sequences of the lav gene (Int1), orf2 homologs from Neisseria, and biotype aegyptius las were aligned using Clustal W (GCG), and pairwise substitutions were analyzed using DIVERGE (GCG). N-terminal domain-encoding regions of lav homologs are the first 283 codons after the GCAA repeats; C-terminal domain-encoding regions are the remaining 376 codons. Values are given as means, with standard deviations in parentheses.
—, the biotype aegyptius las N-terminal domain-encoding region is too divergent to include in the alignment.
Distribution of lav.
Various H. influenzae strains and clinical isolates, including NTHi and representatives from each capsular serotype, were examined by PCR and genomic Southern analyses for the presence of an insert at the holB-tmk junction. The junction was amplified in 26 strains, including 8 from encapsulated lineages (Rd, Hib [Eagan], and reference type strains of serotypes a to f), 9 respiratory NTHi isolates, and 9 NTHi isolates from patients with invasive disease (Table 1). A 2.4-kb fragment containing a lav-related sequence was found at the holB-tmk site in six strains, representing just two clades of NTHi. Southern analysis confirmed the absence of a lav sequence in the other strains or at other chromosomal sites. Strains having lav-related islands included Int1 and two respiratory isolates previously identified as clonally related to Int1 (T. Mutangadura and M. Golomb, unpublished data), 1128 (isolated from a patient with otitis media) (13) and AAr160 (isolated from a tracheal aspirate) (12). Partial sequencing revealed lav genes at identical sites in both isolates. The sequence of the AAr160 lav island was identical to that of Int1 for 600 bp, starting at position 1 in Fig. 2a, with the exception of the number of GCAA repeats and a G→T polymorphism at Int1 position 382, which conservatively changes an L to an F within the lav-encoded signal sequence. A 2.4-kb island was found at the same chromosomal site in R1967, the reference biotype aegyptius strain, and in R2140 and R2141, clonally related biotype aegyptius strains isolated from patients with BPF (Table 3).
TABLE 3.
AT genes in holB-tmk island of NTHi genomes
| Strain | Source or associated disease | ORF type | No. of GCAA repeats | Nearest in-frame repeat no. |
|---|---|---|---|---|
| R2866 (Int1) | Meningitis | lav | 19 | 20 |
| R3157 (1128) | Otitis media | lav | 19 | 20 |
| R3151 (AAr160) | Tracheal aspirate | lav | 14 | 14 |
| R1967 (biotype aegyptius) | Conjunctivitis | las | 25 | 26 |
| R2140 | BPF (biotype aegyptius) | las | 11 | 11 |
| R2141 | BPF (biotype aegyptius) | las | 25 | 26 |
Domain shuffling and the emergence of a novel biotype aegyptius AT gene.
The holB-tmk island of R1967 (biotype aegyptius) had endpoints identical to those in Int1. The island contained a single potential ORF of 2,102 bp, at a position aligned with Int1 lav. Downstream from the predicted initiating ATG were 25 GCAA repeats, placing the coding sequence out of reading frame. Gain of a single repeat would allow translation of a 702-aa polypeptide with a predicted 61-aa signal polypeptide. The C-terminal β-domain of this polypeptide is strongly homologous to Lav. We refer to the putative biotype aegyptius gene at this site as las. Partial sequencing of the R2140 and R2141 islands (BPF) revealed las homologs (Table 3).
Like lav, biotype aegyptius las is a mosaic of low- and high-G+C segments (Fig. 4a). Although the region immediately upstream from biotype aegyptius las and the las β-domain are highly homologous (∼90%) to corresponding regions in lav (Fig. 2), the N terminus-encoding portion of the gene, following the GCAA repeats, has no significant homology to lav or to any sequence in the databanks at either the DNA or amino acid sequence level (Fig. 4b). Dispersed motifs within the inferred N-terminal region of Las suggest a distant common ancestry with H. influenzae Lav; thus, both proteins share the sequence SLWEPR(W/F)NS at corresponding sites (aa 222 to 230 in Int1 and aa. 226 to 234 in biotype aegyptius). The junction with the high-homology segment (Fig. 4a, a) is near the G+C transition point, which coincides with that in lav (∼850 bp or 283 codons). High amino acid sequence similarity starts at the position corresponding to lav codon 277 (biotype aegyptius codon 280). Evidently the lav and las genes diverged from an ancestral H. influenzae sequence by recombinational fusion of a novel passenger domain to the H. influenzae β-domain near the predicted passenger-linker boundary.
FIG. 4.
(a) G+C bias of biotype aegyptius las codons, computed using CodonPreference with a 25-codon window. The abscissa shows nucleotide position in las following the GCAA repeat region. The horizontal line indicates average H. influenzae codon usage. (b) Homology between H. influenzae lav and H. influenzae las, using a 10-bp window, as a function of nucleotide position.
DISCUSSION
Lateral acquisition of lav by NTHi.
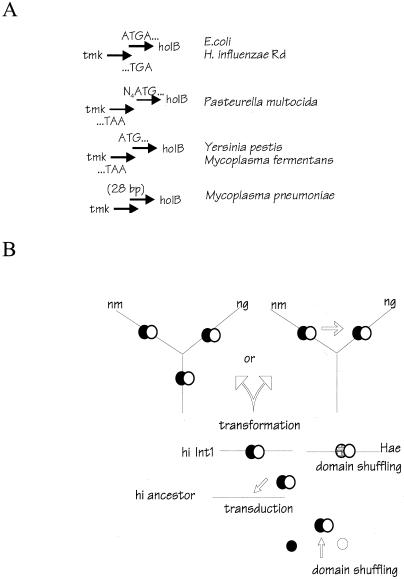
The restricted distribution of lav could be explained either by transfer from another species or by inheritance of the gene from an H. influenzae ancestor followed by loss within multiple lineages. Genomic comparisons favor lateral transfer. In Rd, holB and tmk overlap by 4 bp, with the terminating TGA codon of tmk overlapping the initiating ATG codon of holB. A diverse set of bacterial species have similar arrangements (Fig. 5A). Discussion In Pasteurella, sister genus to Haemophilus (38), the genes are contiguous but not overlapping (http://www.cbc.umn.edu/ResearchProjects/AGAC/Pm/pmhome.html). The tmk and holB genes are also contiguous in Buchnera, a gram-negative endocellular parasite of aphids that has a subset of the genes found in E. coli. Buchnera is believed to have diverged before the common ancestor of H. influenzae and E. coli appeared (41). The contiguity of holB and tmk in other gram-negative bacteria and even in Mycoplasma (phylogenetically closer to gram-positive bacteria) suggests that it is the ancestral arrangement for eubacteria. Thus, lav was most likely inserted into the chromosome of an Int1 ancestor.
FIG. 5.
(A) Arrangement of tmk and holB in various genomes. Nucleotides at the junction of the two genes are shown. In H. influenzae Rd and E. coli, the reading frame of holB is offset −2 relative to that of tmk; in all others shown, the relative reading frame of holB is offset −1. In P. multocida (http://www.cbc.umn.edu/ResearchProjects/AGAC/Pm/pmhome.html), the two genes are separated by 4 bp. GenBank accession numbers and sources are as follows: Rd, reference 17 E. coli, reference 5; P. multocida, contig 186, nucleotides 22421 to 23201; Yersinia pestis, accession no. AF065312 (11); Mycoplasma pneumoniae, accession no. AE000016 (22); and Mycoplasma fermentans, accession no. AF100324 (8). (B) Proposed scenario for origin of lav homologs in various organisms. The original form of the gene in H. influenzae is arbitrarily shown as Int1 lav but could have been either a lav or a las ancestor. nm, N. meningitidis; ng, N. gonorrhoeae; hi, H. influenzae; Hae, biotype aegyptius.
The present distribution of lav requires at least three interspecies transfers (Fig. 5B). The extrageneric origin of lav is supported by its limited distribution within Haemophilus, by G+C content analysis, and by analysis of junctions with adjoining genes. Although the lav segment does not differ in G+C content from the H. influenzae average, it is actually composed of two segments, one atypically low and one atypically high in G+C content. This suggests that the gene originated in another, unidentified, organism by fusion between segments from different species. The chimeric gene was then laterally acquired by H. influenzae-and inserted within a stem-loop hUS pair originally present in single copy in a chromosome resembling that of Rd. The stem-loop pair was partially duplicated during this integration, resulting in direct repeats flanking the lav island in an ancestor of Int1. Insertion of virulence-associated genes within hUS pairs has previously been noted for the tna cluster (30); the targeting of regions of high potential secondary structure suggests that paired hUSs, like tRNA genes, may serve as targets for phage integrases in Haemophilus.
Subsequently, the lav island with adjoining bits of holB and tmk was incorporated by Neisseria, most likely by transformation. Perhaps the fragment recombined within a region of weak, fortuitous homology, though none has been identified. (The two hUS pairs, located internally within the transferred fragment, presumably played no role in neisserial uptake of Haemophilus DNA. Their presence in Neisseria merely reflects the recent origin of this fragment in Haemophilus.) The strongest evidence indicating polarity of transfer is the presence of pseudogene fragments of the adjoining Haemophilus holB and tmk genes. (Functional, endogenous holB and tmk genes are located at different chromosomal sites in Neisseria.) Notably, the usual criteria for direction of transfer (G+C content, within-species distribution, and presence of signature sequences like the hUS) are less reliable and may be deceptive at times. For instance, the G+C content of the Lav-encoding part of the H. influenzae island is species typical not because of its origin within H. influenzae but because it is an artifact of averaging across two dissimilar segments. In general, detailed sequence analysis is needed to infer direction of lateral transfer.
The appearance of lav in both N. meningitidis and N. gonorrhoeae could be explained either by preexistence in a common ancestor or by acquisition by one lineage followed by homologous transformation into the other (Fig. 5B). A third possibility, independent acquisition by N. meningitidis and N. gonorrhoeae from two different H. influenzae lineages, is much less likely because the junction of neisserial sequence with the H. influenzae island is identical, to the base pair, in N. gonorrhoeae and N. meningitidis. The presence of lav (orf2) in all N. meningitidis strains surveyed to date (37) is consistent with its preexistence in an N. meningitidis ancestor, though it could also be explained by transmission across N. meningitidis lineages and strong selective advantage.
DNA sequence similarity attests that the meningococcus and the gonococcus have diverged relatively recently, although no time frame estimates have been published. Most genes are shared by both pathogens and are on average ≥98% identical (47, 49). If emergence of N. gonorrhoeae as a sexually transmitted pathogen required relatively high human population densities, divergence may have occurred as recently as the Neolithic period, 10,000 years ago. Supposing that lav preexisted in the common ancestor of N. gonorrhoeae and N. meningitidis, it must have emerged shortly before species divergence, since it is only slightly less similar between H. influenzae and Neisseria than between neisserial species. Once in Neisseria, lav evolved rapidly, especially within its passenger domain. Nonsilent- and silent-mutation rates are approximately equal within the passenger domains, suggesting either selective neutrality or (more likely) diversifying selection acting on variable regions interspersed within a more conserved framework, as suggested by the data presented in Fig. 1C.
In Haemophilus, phase variation by repeat slippage is associated with surface molecules exposed to immune surveillance (23, 51), and slippage rates increase with the number of repeats (20). The 19 GCAA repeats of Int1 lav and the 25 GCAA repeats of biotype aegyptius las should be relatively unstable in vivo, consistent with an actively expressed gene. We have detected phase variants ranging from 18 to 22 repeats in laboratory populations (data not shown). Although the Int1 lav variant described here is out of reading frame, in-frame variants would arise readily in natural populations and are represented by two clinical isolates (Table 3). Compared to Haemophilus orf2 genes, the N. meningitidis orf2 genes have fewer GCAA repeats; among 41 N. meningitidis isolates possessing orf2, the number of repeats ranged from 1 to 12 (37).
A mosaic origin for lav was inferred from a G+C content transition at the boundary of its presumed passenger domain with the linker and β-barrel domains. Similarly, the junction of nonhomology between lav and las coincides with the G+C transition and inferred domain boundaries of both genes. On the basis of quite different evidence (discordance between phylogenies based on individual domains), Loveless and Saier have proposed that AT proteins evolve by domain shuffling (28). A functionally novel AT can arise by linking a new passenger activity to a generic β-barrel pore. Our analysis provides independent evidence for the combinatorial origin and subsequent reshuffling of at least one AT protein.
Within the small sample of H. influenzae strains examined, lav was restricted to a few NTHi strains with unusual virulence potential. Although Int1 lacks the type b polysaccharide capsule which protects against lysis by human serum, it is more serum resistant than most NTHi strains (B. Williams and A. L. Smith, unpublished data). Int1, AAr160, and 1128 constitute a clade by various criteria, including the results of pulsed-field gel electrophoresis after restriction with rare-cutting enzymes (Mutangadura and Golomb, unpublished). All three strains have the phage HP2, otherwise present in only a small minority of NTHi strains (Williams and Smith, unpublished). The biotype aegyptius is associated with unusual virulence because it includes BPF isolates.
As biotype aegyptius strains and Int1 belong to different phylogenetic subgroups, it is unlikely that they inherited lav from a common ancestor. Rather, it is likely that the first H. influenzae clade to acquire the gene passed it to one or more other clades by transformation and homologous recombination within flanking DNA. Once a laterally transferred fragment has been acquired by a population of naturally transformable bacteria, it can readily be assimilated into the species by co-opting linked homologous sequence and uptake signals. Interstrain and interspecies transfer implies a shared selective advantage in certain host environments.
This is the second report of gene transfer from Haemophilus to Neisseria (25) and the first of a gene belonging to a family widely associated with virulence. Gene flow from Neisseria to Haemophilus has not yet been documented and may be rarer than gene flow in the opposite direction. Natural transformation is more frequent and promiscuous in Neisseria than in Haemophilus, and the requirement for the nUS can be bypassed experimentally (6). In contrast, the requirement of Haemophilus transformation for the hUS is stringent (42). Many genes in the N. meningitidis genome appear to be of external origin, as judged by anomalous G+C content; in contrast, the H. influenzae Rd genome contains fewer islands of unusual G+C content (16, 17, 36). One way to discover a sequence of external origin is to search a genome for heterologous uptake sequences. When Kroll et al. (25) searched the Rd genome with the 10-bp nUS, its only occurrence was within an ORF believed to have been transferred from Haemophilus to Neisseria. When the small Int1-specific library was searched with the nUS, a single hit was found. It consisted of two oppositely oriented nUS sequences in the −/+ configuration, separated by 71 bp, within a slightly longer region of dyad symmetry: TTTCAGACGGCATN67ATGCCGTCTGAAA (inverted repeat nUSs are underlined and N signifies any base). The probability that two nUSs will occur by chance within 100 bp of each other is <10−4. Inverse PCR identified a >2.5-kb island without significant homology to Rd (data not shown). The two nUSs are within a 1,431-bp ORF that is 65% identical in deduced amino acid sequence to the Vibrio cholerae ORF VC1769 product, a presumed methyltransferase (HsdM) subunit of a type I restriction endonuclease system (20). G+C content of this Int1 ORF is 46.1%, a value midway between H. influenzae and neisserial averages. The Vibrio ORF is 43% G+C and part of an island of anomalously low G+C content. Despite possession of a neisserial signature, this ORF did not match sequence entries for N. meningitidis serotype A, N. meningitidis serotype B, or N. gonorrhoeae. We conclude that the Int1 nUS-bearing ORF, although laterally derived, is unlikely to be from Neisseria.
Humans are the exclusive natural host of H. influenzae and the meningococcus. Virulence genes are likely to be recent specializations for competitive adaptation to a human host, and their most obvious sources are other human pathogens and commensals. Repeat-regulated genes such as lav are well equipped for horizontal mobility, since their regulatory system is self-contained. In contrast to selfish operons, which must be transferred as a unit (26), contingency genes are controlled without the need for accessory proteins or for extensive compatibility between regulatory elements from different species. The emergence of invasive NTHi strains, such as those causing BPF and meningitis in immunocompetent children, suggests that contingency gene transfer between dangerous respiratory pathogens may be an ongoing process.
ACKNOWLEDGMENTS
We are grateful to Robert Munson, Janet Gilsdorf, and Loek van Alphen for provision of bacterial strains, to Michael Calcutt for helpful discussions, and to Stephen Lory for advice and help in constructing the M13 library. We thank Arnie Kas, University of Washington, for an unpublished G+C scan program. Incompletely sequenced microbial genomes were accessed on the World Wide Web from the Gonococcal Sequencing Project at Oklahoma University and the Pasteurella multocida Sequencing Project at the University of Minnesota.
This work was supported by NIH grant 5RO1 HG01475 to Maynard Olson, University of Washington, by NIH grant AI 44002 to A.L.S., and by University of Missouri Research Board grants to M.G. and A.L.S.
REFERENCES
- 1.Altschul S F. A protein alignment scoring system sensitive at all evolutionary distances. J Mol Evol. 1993;36:290–300. doi: 10.1007/BF00160485. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Barbour M L. Conjugate vaccines and the carriage of Haemophilus influenzae type b: Emerg. Infect Dis. 1996;2:176–182. doi: 10.3201/eid0203.960303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benz I, Schmidt M A. AIDA-I, the adhesin involved in diffuse adherence of the diarrhoeagenic Escherichia coli strain 2787 (O126:H27), is synthesized via a precursor molecule. Mol Microbiol. 1992;6:1539–1546. doi: 10.1111/j.1365-2958.1992.tb00875.x. [DOI] [PubMed] [Google Scholar]
- 5.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G, Gregor J, Davis N W, Kirkpatrick H, Goeden M, Rose D, Mau R, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 6.Boyle-Vavra S, Seifert H S. Uptake-sequence-independent DNA transformation exists in Neisseria gonorrhoeae. Microbiology. 1996;142:2839–2845. doi: 10.1099/13500872-142-10-2839. [DOI] [PubMed] [Google Scholar]
- 7.Brenner D J, Mayer L W, Carlone G M, Harrison L H, Bibb W F, de Cunto Brandileone M C, Sottnek F O, Iriono K, Reeves M W, Swenson S J M, Birkness K A, Weyant R S, Berkley S F, Woods T C, Steigerwalt A G, Grimont P A D, McKinney R M, Fleming D W, Gheesling L L, Cooksey R C, Arko R J, Broome C V The Brazilian Purpuric Fever Study Group. Biochemical, genetic, and epidemiologic characterization of Haemophilus influenzae biogroup aegyptius (Haemophilus aegyptius) strains associated with Brazilian purpuric fever. J Clin Microbiol. 1988;26:1524–1534. doi: 10.1128/jcm.26.8.1524-1534.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calcutt M J, Kim M F, Karpas A B, Mühlradt P F, Wise K S. Differential posttranslational processing confers intraspecies variation of a major surface lipoprotein and a macrophage-activating lipopeptide of Mycoplasma fermentans. Infect Immun. 1999;67:760–771. doi: 10.1128/iai.67.2.760-771.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Catlin B W, Bendler J W, Goodgal S H. The type b capsulation locus of Haemophilus influenzae: map location and size. J Gen Microbiol. 1972;70:411–422. doi: 10.1099/00221287-70-3-411. [DOI] [PubMed] [Google Scholar]
- 10.Cheetham B F, Katz M E. A role for bacteriophages in the evolution and transfer of bacterial virulence determinants. Mol Microbiol. 1995;18:201–208. doi: 10.1111/j.1365-2958.1995.mmi_18020201.x. [DOI] [PubMed] [Google Scholar]
- 11.Chenal-Francisque V, Tourneux L, Carniel E, Christova P, de la Sierra I L, Barzu O, Gilles A M. The highly similar TMP kinases of Yersinia pestis and Escherichia coli differ markedly in their AZTMP phosphorylating activity. Eur J Biochem. 1999;265:112–119. doi: 10.1046/j.1432-1327.1999.00691.x. [DOI] [PubMed] [Google Scholar]
- 12.Clemans D L, Marrs C F, Patel M, Duncan M, Gilsdorf J R. Comparative analysis of Haemophilus influenzae hifA (pilin) genes. Infect Immun. 1998;66:656–663. doi: 10.1128/iai.66.2.656-663.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coleman T, Grass S, Munson R., Jr Molecular cloning, expression, and sequence of the pilin gene from nontypeable Haemophilus influenzae M37. Infect Immun. 1991;59:1716–1722. doi: 10.1128/iai.59.5.1716-1722.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elkins C, Thomas C E, Seifert H S, Sparling P F. Species-specific uptake of DNA by gonococci is mediated by a 10-base-pair sequence. J Bacteriol. 1991;173:3911–3913. doi: 10.1128/jb.173.12.3911-3913.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Everest P, Li J, Douce G, Charles I, De Azavedo J, Chatfield S, Dougan G, Roberts M. Role of the Bordetella pertussis P.69/pertactin protein and the P.69/pertactin RGD motif in the adherence to and invasion of mammalian cells. Microbiology. 1996;142:3261–3268. doi: 10.1099/13500872-142-11-3261. [DOI] [PubMed] [Google Scholar]
- 16.Faguy D M. The controlled chaos of shifty pathogens. Curr Biol. 2000;10:R498–R501. doi: 10.1016/s0960-9822(00)00558-3. [DOI] [PubMed] [Google Scholar]
- 17.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J-F, Dougherty B A, Merrick J M, McKenney K, Sutton G, FitzHugh W, Fields C, Gocayne J D, Scott J, Shirley R, Liu L-I, Glodek A, Kelley J M, Weidmann J F, Phillips C A, Spriggs T, Hedblom E, Cotton M D, Utterback T R, Hanna M C, Nguyen L D, Saudek D M, Brandon R C, Fine L D, Fritchman J L, Geoghagen N S M, Gnehm C L, McDonald L A, Small K V, Fraser C M, Smith H O, Venter J C. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 18.Foxwell A R, Kyd J M, Cripps A W. Nontypeable Haemophilus influenzae: pathogenesis and prevention. Microbiol Mol Biol Rev. 1998;62:294–308. doi: 10.1128/mmbr.62.2.294-308.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geluk F, Eijk P P, van Ham S M, Jansen H M, van Alphen L. The fimbria gene cluster of nonencapsulated Haemophilus influenzae. Infect Immun. 1998;66:406–417. doi: 10.1128/iai.66.2.406-417.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heidelberg J F, Eisen J A, Nelson W C, Clayton R A, Gwinn M L, Dodson R J, Haft D H, Hickey E K, Peterson J D, Umayam L, Gill S R, Nelson K E, Read T D, Tettelin H, Richardson D, Ermolaeva M D, Vamathevan J, Bass S, Qin H, Dragoi I, Sellers P, McDonald L, Utterback T, Fleishmann R D, Nierman W C, White O. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henderson I R, Navarro-Garcia F, Nataro J P. The great escape: structure and function of the autotransporter proteins. Trends Microbiol. 1998;6:370–378. doi: 10.1016/s0966-842x(98)01318-3. [DOI] [PubMed] [Google Scholar]
- 22.Himmelreich R, Hilbert H, Plagens H, Pirkl E, Li B C, Herrmann R. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 1996;24:4420–4449. doi: 10.1093/nar/24.22.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hood D W, Deadman M E, Jennings M P, Bisceric M, Fleischmann R D, Venter J C, Moxon E R. DNA repeats identify novel virulence genes in Haemophilus influenzae. Proc Natl Acad Sci USA. 1996;93:1121–1125. doi: 10.1073/pnas.93.20.11121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jennings M P, Bisercic M, Dunn K L R, Virji M, Martin A, Wilks K E, Richards J C, Moxon E R. Cloning and molecular analysis of the lsi1 (rfaF) gene of Neisseria meningitidis which encodes a heptosyl-2-transferase involved in LPS biosynthesis: evaluation of surface exposed carbohydrates in LPS mediated toxicity for human endothelial cells. Microb Pathog. 1995;19:391–407. doi: 10.1006/mpat.1995.0074. [DOI] [PubMed] [Google Scholar]
- 25.Kroll J S, Wilks K E, Farrant J L, Langford P R. Natural genetic exchange between Haemophilus and Neisseria: intergeneric transfer of chromosomal genes between major human pathogens. Proc Natl Acad Sci USA. 1998;95:12381–12385. doi: 10.1073/pnas.95.21.12381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lawrence J G, Roth J R. Selfish operons: horizontal transfer may drive the evolution of gene clusters. Genetics. 1996;143:1843–1860. doi: 10.1093/genetics/143.4.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lett M-C, Sasakawa C, Okada N, Sakai T, Makino S, Yamada M, Komatsu K, Yoshikawa M. virG, a plasmid-coded virulence gene of Shigella flexneri: identification of the virG protein and determination of the complete coding sequence. J Bacteriol. 1989;171:353–359. doi: 10.1128/jb.171.1.353-359.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loveless B J, Saier M H. A novel family of channel-forming, autotransporting, bacterial virulence factors. Mol Membr Biol. 1997;14:113–123. doi: 10.3109/09687689709048171. [DOI] [PubMed] [Google Scholar]
- 29.Maiden M C J, Feavers I M. Population genetics and global epidemiology of the human pathogen Neisseria meningitidis. In: Baumberg S, Young J P W, Wellington E M H, Saunders J R, editors. Population genetics of bacteria. Cambridge, United Kingdom: Cambridge University Press; 1995. pp. 269–293. [Google Scholar]
- 30.Martin K, Morlin G, Smith A, Nordyke A, Eisenstark A, Golomb M. The tryptophanase gene cluster of Haemophilus influenzae type b: evidence for horizontal gene transfer. J Bacteriol. 1998;180:107–118. doi: 10.1128/jb.180.1.107-118.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mhlanga-Mutangadura T, Morlin G, Smith A L, Eisenstark A, Golomb M. Evolution of the major pilus gene cluster of Haemophilus influenzae. J Bacteriol. 1998;180:4693–4703. doi: 10.1128/jb.180.17.4693-4703.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moxon E R, Rainey P B, Nowak M A, Lenski R E. Adaptive evolution of highly mutable loci in pathogenic bacteria. Curr Biol. 1994;4:24–33. doi: 10.1016/s0960-9822(00)00005-1. [DOI] [PubMed] [Google Scholar]
- 33.Mühlemann K, Balz M, Aebi S, Schopfer K. Molecular characteristics of Haemophilus influenzae causing invasive disease during the period of vaccination in Switzerland: analysis of strains isolated between 1986 and 1993. J Clin Microbiol. 1996;34:560–563. doi: 10.1128/jcm.34.3.560-563.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nizet V, Colina K F, Almquist J R, Rubens C E, Smith A L. A virulent nonencapsulated Haemophilus influenzae. J Infect Dis. 1996;173:180–186. doi: 10.1093/infdis/173.1.180. [DOI] [PubMed] [Google Scholar]
- 35.Ochman H. How Salmonella became a pathogen. Trends Microbiol. 1997;9:343–349. doi: 10.1016/S0966-842X(97)01099-8. [DOI] [PubMed] [Google Scholar]
- 36.Parkhill J, Achtman M, James K D, Bentley S D, Churcher C, Klee S R, Morelli G, Basham D, Brown D, Chillingworth T, Davies R M, Davis P, Devlin K, Feltwell T, Hamlin N, Holroyd S, Jagels K, Leather S, Moule S, Mungall K, Quail M A, Rajandream M A, Rutherford K M, Simmonds M, Skelton J, Whitehead S, Spratt B G, Barrell B G. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature. 2000;404:502–506. doi: 10.1038/35006655. [DOI] [PubMed] [Google Scholar]
- 37.Peak I R, Jennings M P, Hood D W, Moxon E R. Tetranucleotide repeats identify novel virulence determinant homologues in Neisseria meningitidis. Microb Pathog. 1999;26:13–23. doi: 10.1006/mpat.1998.0243. [DOI] [PubMed] [Google Scholar]
- 38.Pohl S. DNA relatedness in HPA. In: Kilian M, Frederiksen W, Biberstein E L, editors. Haemophilus, Actinobacillus, and Pasteurella. New York, N.Y: Academic Press; 1981. pp. 245–253. [Google Scholar]
- 39.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 40.Saunders N J, Jeffries A C, Peden J F, Hood D W, Tettelin H, Rappuoli R, Moxon E R. Repeat-associated phase variable genes in the complete genome sequence of Neisseria meningitidis strain MC58. Mol Microbiol. 2000;37:207–215. doi: 10.1046/j.1365-2958.2000.02000.x. [DOI] [PubMed] [Google Scholar]
- 41.Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature. 2000;407:81–86. doi: 10.1038/35024074. [DOI] [PubMed] [Google Scholar]
- 42.Sisco K L, Smith H O. Sequence-specific DNA uptake in Haemophilus transformation. Proc Natl Acad Sci USA. 1979;76:972–976. doi: 10.1073/pnas.76.2.972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith A L, Smith D H, Averill D R, Jr, Marino J, Moxon E R. Production of Haemophilus influenzae b meningitis in infant rats by intraperitoneal inoculation. Infect Immun. 1973;8:278–290. doi: 10.1128/iai.8.2.278-290.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith H O, Tomb J-F, Dougherty B A, Fleischmann R D, Venter J C. Frequency and distribution of DNA uptake signal sequences in the Haemophilus influenzae Rd genome. Science. 1995;269:538–540. doi: 10.1126/science.7542802. [DOI] [PubMed] [Google Scholar]
- 45.Smith J M, Smith N H, O'Rourke M, Spratt B G. How clonal are bacteria? Proc Natl Acad Sci USA. 1993;90:4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sneath P H A, Sokal R R. Numerical taxonomy. W. H. San Francisco, Calif: Freeman; 1973. [Google Scholar]
- 47.Spratt B G, Smith N H, Zhous J, O'Rourke M, Feil E. The population genetics of the pathogenic Neisseria. In: Baumberg S, Young J P W, Wellington E M H, Saunders J R, editors. Population genetics of bacteria. Cambridge, United Kingdom: University of Cambridge; 1995. pp. 269–160. [Google Scholar]
- 48.Tettelin H, Saunders N J, Heidelberg J, Jeffries A C, Nelson K E, Eisen J A, Ketchum K A, Hood D W, Peden J F, Dodson R J, Nelson W C, Gwinn M L, DeBoy R, Peterson J D, Hickey E K, Haft D H, Salzberg S L, White O, Fleischmann R D, Dougherty B A, Mason T, Ciecko A, Parksey D S, Blair E, Cittone H, Clark E B, Cotton M D, Utterback T R, Khouri H, Qin H, Vamathevan J, Gill J, Scarlato V, Masignani V, Pizza M, Grandi G, Sun L, Smith H O, Fraser C M, Moxon E R, Rappuoli R, Vente R J C. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science. 2000;287:1809–1815. doi: 10.1126/science.287.5459.1809. [DOI] [PubMed] [Google Scholar]
- 49.Tinsley C R, Nassif X. Analysis of the genetic differences between Neisseria meningitidis and Neisseria gonorrhoeae: two closely related bacteria expressing two different pathogenicities. Proc Natl Acad Sci USA. 1996;93:11109–11114. doi: 10.1073/pnas.93.20.11109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Turk D C. Clinical importance of Haemophilus influenzae—1981. In: Sell S H, Wright P F, editors. Haemophilus influenzae epidemiology, immunology, and prevention of disease. New York, N.Y: Elsevier/North-Holland Publishing Co.; 1982. pp. 3–9. [Google Scholar]
- 51.Weiser J N. The generation of diversity by Haemophilus influenzae. Trends Microbiol. 2000;8:433–435. doi: 10.1016/s0966-842x(00)01839-4. [DOI] [PubMed] [Google Scholar]