Abstract
Radial glial (RG) cells are the neural stem cells of the developing neocortex. Apical RG (aRG) cells can delaminate to generate basal RG (bRG) cells, a cell type associated with human brain expansion. Here, we report that aRG delamination is regulated by the post‐Golgi secretory pathway. Using in situ subcellular live imaging, we show that post‐Golgi transport of RAB6+ vesicles occurs toward the minus ends of microtubules and depends on dynein. We demonstrate that the apical determinant Crumbs3 (CRB3) is also transported by dynein. Double knockout of RAB6A/A' and RAB6B impairs apical localization of CRB3 and induces a retraction of aRG cell apical process, leading to delamination and ectopic division. These defects are phenocopied by knockout of the dynein activator LIS1. Overall, our results identify a RAB6‐dynein‐LIS1 complex for Golgi to apical surface transport in aRG cells, and highlights the role of this pathway in the maintenance of neuroepithelial integrity.
Keywords: cell polarity, dynein, neocortex development, polarized trafficking, RAB6
Subject Categories: Cell Adhesion, Polarity & Cytoskeleton; Membranes & Trafficking; Neuroscience
A RAB6‐Dynein‐LIS1 complex controls post‐Golgi apical transport of CRUMBS in neuronal progenitors. Impairment of this pathway alters apical junctions, causes delamination of these epithelial cells, and leads to the formation of basal progenitors.
Introduction
In the developing neocortex, all neurons derive from neural stem cells called radial glial (RG) progenitor cells(Paridaen & Huttner, 2014; Uzquiano et al, 2018). These highly elongated cells also serve as tracks for the migration of newborn neurons into the cortical plate. Two types of RG cells have been identified: apical RG (aRG) cells (also known as vRG cells), located in the ventricular zone (VZ), and basal RG cells (bRG cells, also known as oRG cells) located in the subventricular zone (Fietz et al, 2010; Hansen et al, 2010; Reillo et al, 2011; Fig 1A). aRG cells are common to all mammalian species while bRG cells, which originate from aRG cells, are rare in lissencephalic species such as mice but abundant in gyrencephalic species, including humans (Fernández et al, 2016; Florio et al, 2016; Penisson et al, 2019). aRG cells are tightly connected to each other by adherens junctions and form a pseudostratified epithelium lining the ventricle (Lee & Norden, 2013). They are highly polarized and display an apical process extending to the ventricular surface, and a long basal process, connecting to the pial surface (Fig 1A). Several studies have illustrated that apicobasal polarity is critical for the maintenance of aRG cells, and that its alteration can lead to aRG cell delamination from the neuroepithelium and to the generation of bRG‐like cells (Cappello et al, 2006; Itoh et al, 2013; Johnson et al, 2018; Narayanan et al, 2018; Tavano et al, 2018). In ferrets, the cell adhesion molecule cadherin 1 is downregulated at the critical period of bRG cell generation and its knockdown is sufficient to induce bRG cell generation(Martínez‐Martínez et al, 2016).
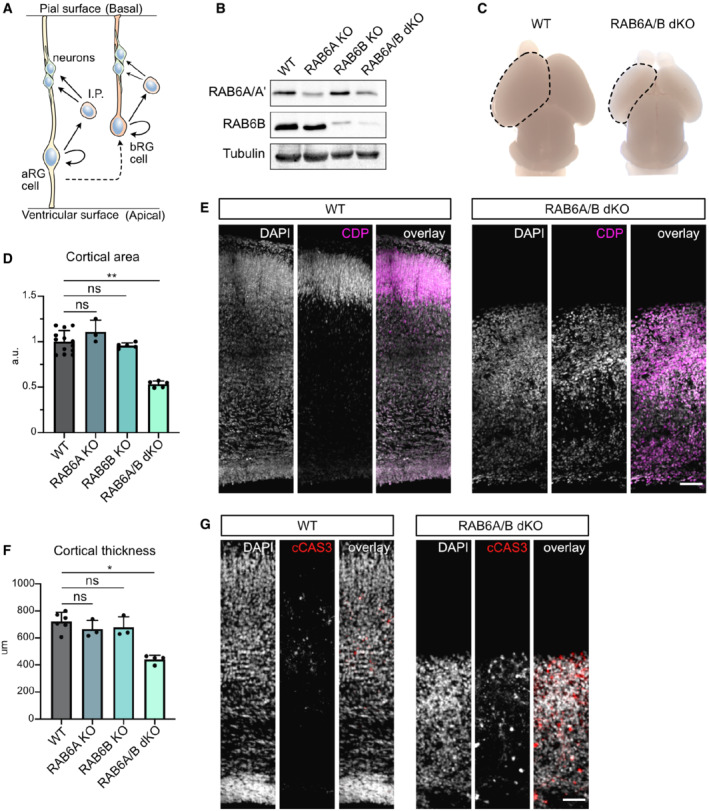
Figure 1. RAB6A/B double knockout causes microcephaly.
-
ASchematic representation of cortical neurogenesis. Apical radial glial (aRG) cells are epithelial cells and the main neuronal progenitors in mouse. Basal radial glial (bRG) cells are rare in mouse but are the most abundant progenitor population in human. They have delaminated from the neuroepithelium. I.P.: Intermediate Progenitor.
-
BWestern blot analysis of RAB6A/A' and RAB6B protein levels in WT, Emx1‐Cre; RAB6 loxP/loxP (RAB6A KO), RAB6B −/− (RAB6B KO) and Emx1‐Cre; RAB6A loxP/loxP ; RAB6B −/− (RAB6A/B dKO) E15.5 cortical extracts.
-
CP0 WT and RAB6A/B dKO brains. A cortical hemisphere is circled (dotted lines).
-
DCortical area in WT (N = 13 brains), RAB6A KO (N = 3 brains), RAB6B KO (N = 5 brains), and RAB6A/B dKO (N = 5 brains) at P0.
-
EWT and RAB6A/B dKO brains stained for layer II/III marker CDP at P0. Scale bar = 100 μm.
-
FCortical thickness (μm) in WT (N = 6 brains), RAB6A KO (N = 3 brains), RAB6B KO (N = 3 brains) and RAB6A/B dKO (N = 4 brains) at P0.
-
GImmunostaining for cleaved Caspase 3 (cCAS3) in WT and RAB6A/B dKO brains at P0. Scale bar = 50 μm.
Data information: (D, F) Kruskal–Wallis test with a Dunn post‐hoc test and Benjamini–Hochberg procedure. All error bars indicate SD.
Epithelial polarity is controlled by the PAR, Crumbs, and Scribble complexes which mutually interact to generate and maintain apical and basolateral domains. The Crumbs complex is composed of CRB, PALS1, and PATJ and is a major apical domain determinant (Bulgakova & Knust, 2009). In the mouse developing neocortex, knockout of CRB1 and CRB2 leads to an alteration of aRG cells apical junctions, while knockout of PALS1 causes severe polarity defects, apoptotic cell death, and microcephaly (Kim et al, 2010; Dudok et al, 2016). The establishment and maintenance of epithelial polarity also rely on polarized trafficking along the biosynthetic/secretory pathway. Newly synthesized transmembrane proteins are sorted in the Golgi apparatus/TGN (Trans‐Golgi Network) and are routed towards the apical or basolateral domains of epithelial cells, possibly transiting through endosomal compartments (Apodaca et al, 2012). In particular, the secretory pathway is essential for the apical targeting of newly synthesized CRB, the only transmembrane protein among the apical polarity complexes (Rodriguez‐Boulan & Macara, 2014).
RAB6 is a Golgi/TGN‐associated small GTPase which controls both anterograde and retrograde transport, from and toward the Golgi apparatus (Goud et al, 2018). Three RAB6 paralogs have been identified: ubiquitous RAB6A (and its splicing variant RAB6A'), RAB6B, predominantly expressed in the brain, and RAB6C, encoded by a primate‐specific retrogene and involved in cell cycle progression (Opdam et al, 2000; Goud et al, 2018). In non‐polarized cells, RAB6A is associated with most—if not all—post‐Golgi vesicles, irrespective of the transported cargo, suggesting that RAB6A is a general regulator of post‐Golgi trafficking (Fourriere et al, 2019). The exact role of RAB6B is poorly known but evidence exist that it acts redundantly with RAB6A in the secretory pathway (Homma et al, 2019). RAB6‐positive (RAB6+) secretory vesicles are transported to the cell surface by two plus end‐directed kinesins, KIF5B and KIF13B (Serra‐Marques et al, 2020). Retrograde transport toward the Golgi apparatus or the endoplasmic reticulum (ER) is driven by dynein (Matanis et al, 2002; Young et al, 2005). RAB6 recruits dynein and its partner dynactin through Bicaudal‐D (BicD) adaptor proteins, leading to dynein activation and processive movement along microtubules (Splinter et al, 2012; Mckenney et al, 2014; Schlager et al, 2014a; Huynh & Vale, 2017; Urnavicius et al, 2018). Dynein activity is further regulated by LIS1 (Elshenawy et al, 2020; Htet et al, 2020; Marzo et al, 2020), the dysfunction of which being the most common cause of human lissencephaly (Reiner et al, 1993; Marzo et al, 2020). LIS1 activates dynein, but can subsequently be released from an idling complex by RAB6 for processing movement (Yamada et al, 2013).
In polarized epithelial cells, the machinery controlling trafficking from the Golgi apparatus toward the apical surface was not clearly identified. Conflicting reports have involved both plus‐end directed and minus‐end directed microtubule motors (Tai et al, 1999; Noda et al, 2001; Jaulin et al, 2007; Bay et al, 2013; Aguilar‐Aragon et al, 2020). This is largely due to the limited ability to resolve vesicular transport and post‐Golgi trafficking events in polarized epithelial cells, because of the small size of these cells and to the thickness of epithelial tissues. Here, using a method for subcellular live imaging within embryonic brain slices, we show that apical transport of post‐Golgi RAB6+ vesicles is driven by dynein. RAB6A/B double KO leads to aRG cell delamination during interphase and to the formation of proliferating RG cells localized basally. LISbRG1 loss of function largely phenocopies RAB6A/B dKO, indicating that the RAB6‐dynein‐LIS1 apical trafficking pathway is required for preventing aRG cell delamination. Finally, we provide evidence that this pathway is critical for the apical transport of the major polarity determinant CRB3 in aRG cells.
Results
RAB6A/B double knockout causes microcephaly
To investigate the role of RAB6 during mouse neocortex development, we adopted a knockout approach. We confirmed RAB6A/A' and B expression in the developing brain, and observed that RAB6B expression strongly rises from E11.5, while RAB6A/A' levels remain constant (Fig EV1A). Because constitutive knockout of RAB6A (coding for the two isoforms RAB6A and RAB6A') leads to early developmental lethality (Shafaq‐Zadah et al, 2016), we previously generated a Cre‐inducible KO mouse model (Bardin et al, 2015). Dorsal cortex‐specific depletion of RAB6A, using the Emx1‐Cre driver, did not lead to any observable phenotype on neocortex development. To test for redundancy, we therefore generated a constitutive KO mouse for RAB6B, using Crispr‐Cas9. We obtained two lines, a 279 bp inversion affecting in exons 3 and 4, and a 1 bp deletion in exon 2, both leading to a premature stop codon. Both lines were viable and, as for conditional RAB6A KO, did not display any observable alterations of neocortex development. We therefore generated RAB6A/B double KO (RAB6A/B dKO) animals. Efficient protein depletion for RAB6A/A' and RAB6B in the embryonic cortex was verified by western blot (Fig 1B), residual RAB6A/A' signal in RAB6A/B dKO mice being likely due to the presence of non‐Cre expressing cells in the protein extract. Strikingly, RAB6A/B dKO mice were severely microcephalic. At P0, the cortical area and the cortical thickness of double mutant animals were reduced by half, while single KOs were unaffected (Fig 1C–F). Reduced brain size was likely the consequence of increased levels of apoptotic cell death observed in RAB6A/B dKO (Fig 1G). Neuronal positioning was also strongly affected, with layer II‐III neurons (CDP+) dispersed throughout the neocortex, suggesting impaired neuronal migration (Fig 1E). Therefore, loss of RAB6A/A' and RAB6B leads to microcephaly and altered neuronal positioning.
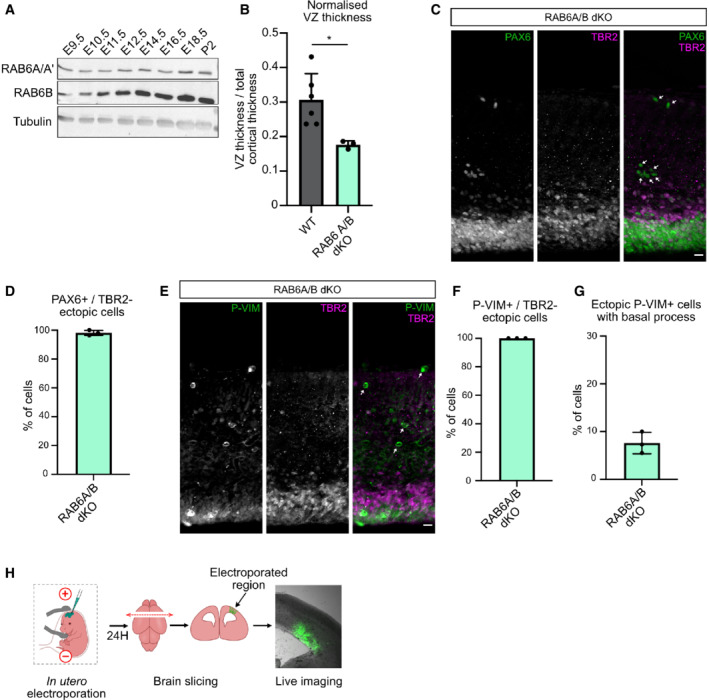
Figure EV1. RAB6 A/B dKO leads to ectopic TBR2‐negative basal progenitors.
-
ARAB6A/A' and RAB6B expression in the developing brain and at P2.
-
BVentricular zone (VZ) thickness normalized to total cortical thickness in N = 6 WT brains and N = 3 RAB6A/B dKO. Mann–Whitney U test, *P ≤ 0.05.
-
CPAX6 and TBR2 staining in RAB6A/B dKO E15.5 brains. Arrows indicate detached PAX6+/TBR2− cells. Scale bar = 25 μm.
-
DPercentage of ectopic PAX6+/TBR2− cells in RAB6A/B dKO E15.5 brains. RAB6A/B dKO: 470 cells from N = 3 brains.
-
EP‐VIM and TBR2 staining in RAB6A/B dKO E15.5 brains. Arrows indicate detached P‐VIM+/TBR2− cells. Scale bar = 25 μm.
-
FPercentage of ectopic P‐VIM+/TBR2− cells in RAB6A/B dKO E15.5 brains. RAB6A/B dKO: 99 cells from N = 3 brains.
-
GPercentage of ectopic P‐VIM+ cells that maintained a basal process in RAB6A/B dKO E15.5 brains. RAB6A/B dKO: 99 cells from N = 3 brains.
-
HSchematic representation of in utero electroporation and live imaging procedure in the mouse developing cortex.
Data information: All error bars indicate SD.
RAB6A/B dKO leads to aRG cell delamination during interphase
We next addressed the consequences of RAB6A/B dKO on the RG progenitor population. In E15.5 control as well as in single RAB6A and RAB6B KO brains, these epithelial cells were concentrated within the VZ. In RAB6A/B dKO, however, numerous RG cells could be observed above the VZ, suggesting delamination from the neuroepithelium (Fig 2A and B). Moreover, the size of the PAX6+ VZ was reduced, even when normalized to total cortical thickness, further indicating a loss of ventricular aRG cells (Fig EV1B). The presence of ectopic RG cells was confirmed by the strong increase in the fraction of mitotic RG cells located above the ventricular surface, positive for phospho‐Vimentin (p‐VIM), which specifically marks mitotic RG cells (Stahl et al, 2013; Vaid et al, 2018; Fig 2C and D). Both PAX6+ and P‐VIM+ ectopic cells were negative for the intermediate progenitor marker TBR2, indicating that differentiation was neither a cause nor a consequence of cell displacement (Fig EV1C–F). P‐VIM staining further revealed that basally located RG cells had lost their apical process and had therefore detached form the neuroepithelium. Notably, they had retracted their basal process, and as a consequence could not perform mitotic somal translocation (Ostrem et al, 2014) (Fig EV1G). Nevertheless, quantification of the mitotic index of PAX6+ RG cells indicated that ectopic RAB6A/B dKO RG cells proliferated at a normal rate (Fig 2E).
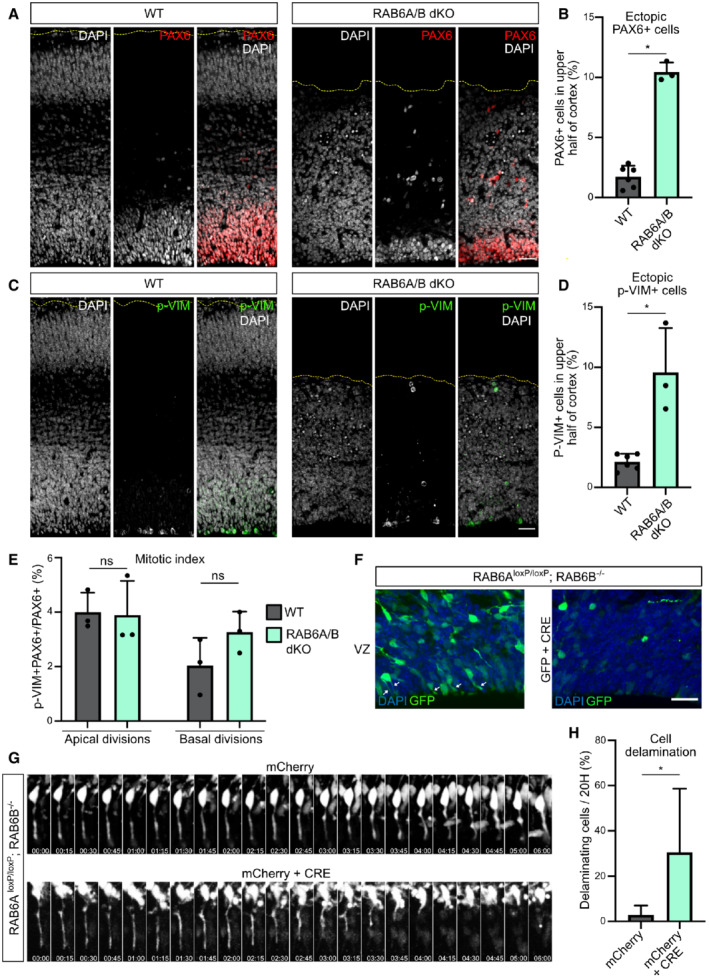
Figure 2. RAB6A/B dKO leads to aRG cell delamination during interphase.
-
APAX6 staining in WT and RAB6A/B dKO E15.5 brains. Scale bar = 50 μm.
-
BPercentage of PAX6+ cells located in the upper half of the cortex of WT and RAB6A/B dKO E15.5 brains. WT: 4282 cells from N = 6 brains. RAB6A/B dKO: 1,241 cells from N = 3 brains. Mann–Whitney U test.
-
CPhospho‐Vimentin (p‐Vim) staining in WT and RAB6A/B dKO E15.5 brains. Scale bar = 50 μm.
-
DPercentage p‐VIM+ cells dividing ectopically, in the upper half of the cortex of WT and RAB6A/B dKO E15.5 brains. WT: 1,713 cells from N = 6 brains. RAB6A/B dKO: 506 cells from N = 3 brains. Mann–Whitney U test.
-
EMitotic index (p‐VIM+ PAX6+ / PAX6+ cells) of RG cells dividing apically (at the ventricular surface) or basally (upper half) in WT and RAB6A/B dKO E15.5 brains. Apical divisions: N = 1,886 cells from 3 brains for WT and 1,511 cells from 3 brains for RAB6A/B dKO. Basal divisions: N = 643 cells from 3 brains for WT and 809 cells from 3 brains for RAB6A/B dKO. Mann–Whitney U test.
-
FElectroporation of RAB6A loxP/loxP ; RAB6B −/− E14.5 embryos with GFP (control) or GFP + CRE (RAB6A/B dKO) and fixation at E18.5. Localization of GFP+ cells in the ventricular zone (VZ). White arrows indicate apical processes. Scale bar = 25 μm.
-
GElectroporation of RAB6A loxP/loxP ; RAB6B −/− E14.5 embryos with mCherry (control) or mCherry + CRE (RAB6A/B dKO) and live imaging of delamination events at E17.5.
-
HApical endfoot detachment and retraction events during 20 h movies in mCherry or mCherry + CRE electroporated cells at E17.5. mCherry: N = 69 cells from 5 movies. mCherry + CRE: N = 52 cells from 4 movies. Fisher's exact test, *P ≤ 0.05.
Data information: All error bars indicate SD.
To investigate further whether these ectopic aRG cells had indeed delaminated from the neuroepithelium, we used in utero electroporation, which specifically targets the aRG cells and therefore allows to assess the position of these cells and their progeny over time (Fig EV1H). We electroporated a plasmid coding for the Cre recombinase, as well as GFP, into E14.5 RAB6A loxP/loxP ; RAB6B −/− brains, to deplete both RAB6A/A' and B specifically in the GFP‐expressing electroporated aRG cells. After 4 days in control GFP‐electroporated brains, numerous aRG cells could be observed connected to the ventricular surface by their apical processes (Fig 2F). In Cre‐expressing brains however, these cells were largely lost, suggesting that they had detached from the neuroepithelium (Fig 2F). To confirm that the presence of basally localized RG cells was indeed a consequence of apical process detachment during interphase, we live imaged aRG cells 3 days after Cre expression‐induced RAB6A/B dKO. While the majority of control cells maintained an apical attachment throughout 20 h‐long movies, a high proportion of Cre‐expressing RAB6A/B dKO RG cells were observed to detach from the neuroepithelium and retract their apical process toward the cell soma (Fig 2G and H; Movies EV1 and EV2). Together, these results indicate that double depletion of RAB6A/A' and B leads to the delamination of RG cells during interphase. These cells lose their elongated shape but maintain the expression of RG markers and continue to proliferate above the VZ.
Post‐Golgi apical trafficking occurs towards the minus ends of microtubules
To understand how RAB6 may be involved in the maintenance of aRG cell apical attachment to the ventricular surface, we investigated RAB6‐dependent post‐Golgi transport within the apical process. aRG cells are highly elongated cells and undergo interkinetic nuclear migration (INM), a process by which their nuclei translocate basally, before migrating back to the apical surface for mitosis (Hu et al, 2013; Baffet et al, 2015). As a consequence, the average distance between the Golgi apparatus, which follows the nucleus, and the apical surface, where the centrosome is located, is 17.84 μm, ranging from 0 to 46.81 μm, depending on the stage of INM (Fig 3A and B; Taverna et al, 2016).
Figure 3. Apical transport of RAB6A+ post‐Golgi vesicles is driven by dynein.
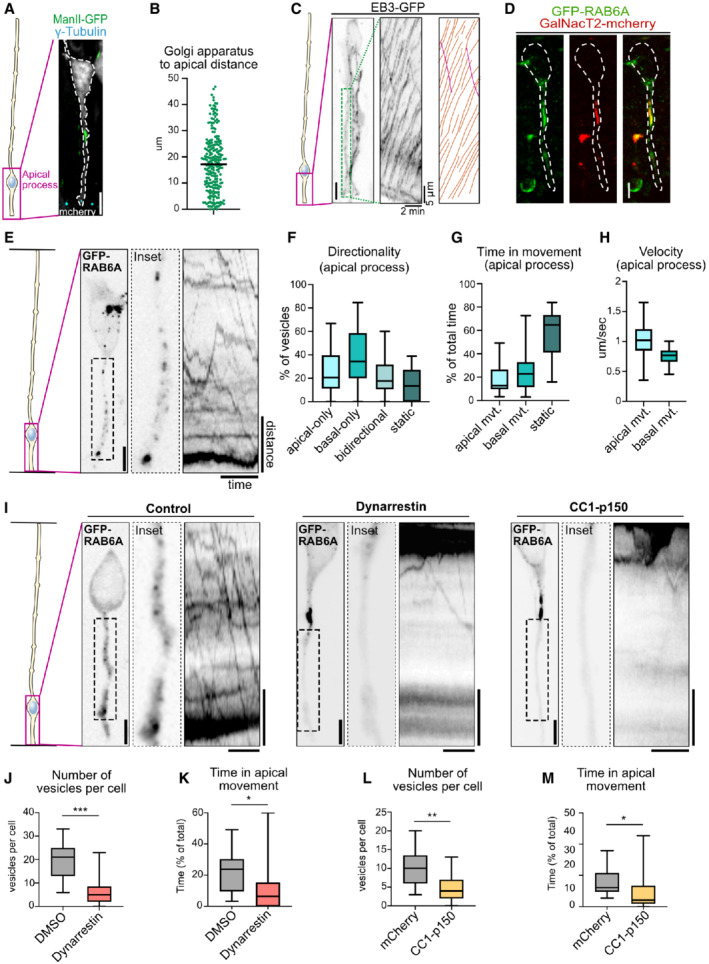
-
ALocalization of the Golgi apparatus (ManII‐GFP) and the centrosome (γ‐tubulin) in E15.5 mCherry‐electroporated radial glial cell. The Golgi apparatus is localized basally, away from the centrosome. Scale bar = 5 μm.
-
BAverage distance between the apical most part of the Golgi apparatus and the apical surface in aRG cells. N = 224 cells from three independent brains.
-
CLive imaging of EB3‐GFP in the apical process of an aRG cell at E15.5. Center: kymograph. Left: manual tracking of EB3 comets. Orange: basally growing. Pink: Apically growing. Scale bar = 5 μm.
-
DCo‐expression of GFP‐RAB6A and GalNacT2‐mCherry in E15.5 aRG cells reveals colocalization at the Golgi apparatus. Scale bar = 5 μm. Dashed line: cell outline.
-
ELive imaging of GFP‐RAB6A in aRG cells at E15.5 allows tracking of individual RAB6A+ vesicles in situ, from the basal Golgi apparatus toward the apical surface. Scale bar = 5 μm. Distance = 5 μm, time = 30 s.
-
FRAB6A+ vesicle directionality in apical processes of aRG cells over 1‐min movies.
-
GRelative time spent by RAB6A+ vesicles in apical, basal, or static phases.
-
HVelocity of apically and basally moving RAB6A+ vesicles.
-
II Live imaging of GFP‐RAB6A in control, dynarrestin‐treated, and CC1‐p150‐expressing aRG cells at E15.5. Scale bars = 5 μm. Distance = 5 μm, time = 30 s.
-
JNumber of RAB6A+ vesicles in the apical process of DMSO and dynarrestin‐treated mouse aRG cells.
-
KRelative time spent by RAB6A+ vesicles in apical movement phase, in DMSO and dynarrestin‐treated mouse aRG cells.
-
LNumber of RAB6A+ vesicles in the apical process of mCherry and CC1‐p150‐expressing aRG cells.
-
MRelative time spent by RAB6A+ vesicles in apical movement phase, in mCherry and CC1‐p150‐expressing aRG cells. DMSO treatment slightly affected RAB6A dynamics, as compared to mCherry control.
Data information: (F, G, H) N = 388 vesicles from 30 cells. (J, K, L, M) 216 vesicles from N = 11 cells for DMSO, 145 vesicles from N = 25 cells for dynarrestin, 173 vesicles from N = 17 cells for mCherry control, 71 vesicles from N = 15 cells for CC1‐p150. Mann–Whitney U test *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001. All boxplots: whiskers indicate min and max, boxes indicate 25th and 75th percentiles, and central band indicates the median.
To perform subcellular live imaging within thick organotypic brain slices, aRG cells are electroporated with fluorescent reporters in utero and, following 24 h of expression, brains are sliced and mounted for imaging on a CSU‐W1 spinning disk microscope equipped with a high working distance 100× objective (see methods) (Coquand et al, 2021). This approach allowed the visualization of growing microtubule plus ends in cells expressing the plus end tracking protein EB3 (Coquand et al, 2021). We confirmed our previous results, that is, the unipolar organization of the microtubule network with over 99% of plus ends growing in the basal direction, from the pericentrosomal apical surface (Fig 3C; Movie EV3). Notably, virtually no microtubules emanating from the Golgi area were observed to grow apically.
To visualize post‐Golgi transport vesicles, we electroporated aRG cells in utero with a GFP‐RAB6A expressing plasmid. The construct was expressed at low levels to avoid cytosolic accumulation, revealing a strong accumulation at the Golgi (Fig 3D). For live imaging, 3–5 planes were imaged to capture the entire apical process, leading to a temporal resolution of 600–1,000 ms. GFP‐RAB6A marked the Golgi apparatus, which sometimes appears fragmented as previously reported in these cells (Taverna et al, 2016), as well as small and dynamic vesicular structures that could often be observed budding from the Golgi (Fig EV2A; Movie EV4). Live imaging within the apical process (between the Golgi and the apical surface) revealed that RAB6A+ vesicles were bidirectional (Fig 3E; Movie EV5). Highly dynamic RAB6A+ vesicles could also be observed within the basal process (above the nucleus), where they also appeared highly dynamic (Fig EV2B; Movie EV6). In the apical process, manual tracking of individual RAB6A+ vesicles revealed that, throughout 1‐min movies, 39% displayed basal movement (toward the Golgi apparatus), 25% apical movement (toward the apical surface), 21% bidirectional movement, and 15% were static (Fig 3F). These vesicles spent 24% of their time moving in the basal direction, 18% moving in the apical direction, and 58% not moving (Fig 3G). Apically moving RAB6A+ vesicles moved faster than basally moving ones, in agreement with faster minus‐end transport reported in non‐polarized cells (Schlager et al, 2014b; Serra‐Marques et al, 2020; Fig 3H). Including pauses, RAB6A+ vesicles traveled on average 32.3 μm/min. They were often observed to disappear at the apical surface, suggesting apical fusion events, either with the plasma membrane or with another compartment (Fig EV2C; Movie EV7). Together, these results reveal that RAB6A+ vesicles traffic in a highly bidirectional manner between the perinuclear Golgi apparatus and the apical surface, which they reach following transport directed toward microtubule minus ends.
Figure EV2. RAB6A dynamics in aRG cells and dynarrestin validation.
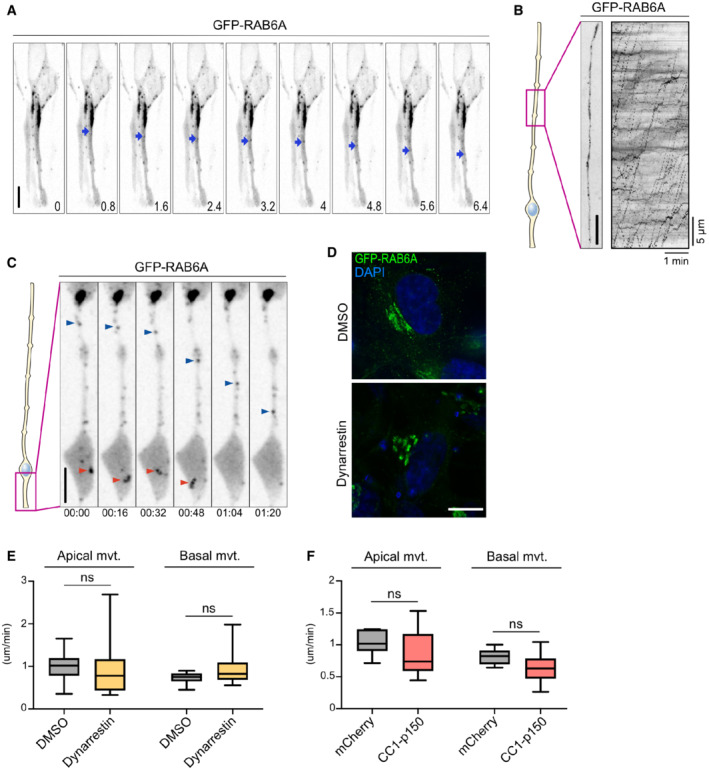
-
ALive imaging of GFP‐RAB6A in the apical process of an aRG cell at E15.5. At 0.8 s, a tubule is budding from the Golgi, leading to the formation of an apically moving vesicle. Blue arrowhead indicates RAB6A+ vesicle. Scale bar = 5 μm.
-
BLive imaging of GFP‐RAB6A in the basal process of an aRG cell at E15.5. Right: kymograph. Scale bar = 5 μm.
-
CLive imaging of GFP‐RAB6A in the apical process of an aRG cell at E15.5. Red arrowhead: a RAB6A+ vesicle can be seen disappearing in the endfoot, suggesting fusion with the apical membrane. Blue arrowhead: a RAB6A+ vesicle moving apically within the apical process. Scale bar = 10 μm.
-
DRPE‐1 cells transfected with GFP‐RAB6A to visualize the Golgi apparatus architecture, and treated for 4 h with 100 μM dynarrestin or DMSO. Scale bar = 10 μm.
-
EVelocity of apically and basally moving RAB6A vesicles within the apical process of DMSO and dynarrestin‐treated aRG cells. In all, 142 vesicles from N = 7 cells for DMSO, 74 vesicles from N = 18 cells for dynarrestin.
-
FVelocity of apically and basally moving RAB6A vesicles within the apical process of mcherry control and CC1‐p150‐expressing aRG cells. In all, 120 vesicles from N = 17 cells for mCherry control, 39 vesicles from N = 11 cells for CC1‐p150.
Data information: (E, F) Mann–Whitney U test. Boxplots whiskers indicate min and max, boxes indicate 25th and 75th percentiles, and central band indicates the median.
Apical transport of post‐Golgi RAB6A+ vesicles is driven by dynein
We next asked whether post‐Golgi apical transport of RAB6A+ vesicles was dependent on the minus end microtubule motor dynein. To test this, we treated brain slices with the dynein inhibitor dynarrestin, prior to live imaging (Höing et al, 2018). Because of its short stability, a new batch of dynarrestin was dissolved prior to each experiment, and validated in parallel for Golgi dispersal in RPE‐1 cells (Fig EV2D). Dynarrestin treatment in aRG cells led to a drastic inhibition of the trafficking of RAB6A+ vesicles into the apical process, as compared to dimethyl sulfoxide (DMSO)‐treated cells (Fig 3I; Movies EV8 and EV9). The total number of RAB6A+ vesicles observed within the apical process was severely reduced (Fig 3J). This result suggests that, in the absence of dynein activity, the balance between opposing motors was shifted toward kinesin‐dependent transport in the basal direction, leading to an emptying of the apical process. The vesicles that did manage to enter the apical process performed substantially less apically directed movements (Fig 3K). On the contrary, RAB6A+ vesicles in the cell soma and in the basal process remained highly mobile.
To confirm these results, we next overexpressed a truncated form of p150 Glued (CC1‐p150), which acts as a dominant negative for the dynactin complex (Tripathy et al, 2014). Expression of CC1‐p150 for 24 h in aRG cells led to a very similar outcome, impairing the localization of RAB6A+ vesicles into the apical process (Fig 3I and L; Movies EV8 and EV10). As for dynarrestin treatment, apical movement of RAB6A+ vesicles located within the apical process was markedly reduced (Fig 3M). Mobile RAB6A+ vesicles were still abundant in the soma and basal process. In both cells treated with dynarrestin or overexpressing CC1‐p150, the speed of RAB6A+ vesicles that were still moving was unaltered within the apical process (Fig EV2E and F). Together, these results indicate that post‐Golgi RAB6A+ vesicles travel toward the apical surface of aRG cells along a uniformly polarized microtubule network via dynein‐based transport.
Post‐Golgi apical transport of Crumbs is driven by dynein
Interphasic delamination is a consequence of destabilization of the adherens junctions, which are themselves dependent on properly established epithelial polarity. The transmembrane protein CRB is a major determinant of epithelial apical domain polarity and the only one to be transported along the secretory pathway. Accordingly, CRB3, the major Crumbs isoform expressed in mammalian epithelial cells (Margolis, 2018), and its partner PALS1 localize to the apical surface of aRG cells (Fig 4A). We therefore asked whether the RAB6‐dynein complex controls the apical transport of CRB3 in these cells. To distinguish between different trafficking pools—secretory and endolysosomal—we analyzed CRB3 trafficking using the RUSH system (Boncompain et al, 2012; Rodriguez‐Fraticelli et al, 2015) (Fig 4B). This assay allows for the retention of a cargo of interest in the ER and, upon addition of biotin, its release for trafficking along its secretory route. Following in utero electroporation, SBP‐CRB3‐GFP was efficiently retained in vivo within the ER and absent from the apical surface of aRG cells, indicating that endogenous biotin levels in mouse were not sufficient to induce its release (Fig 4C and D). To monitor SBP‐CRB3‐GFP trafficking, brain slices were incubated in the presence of biotin and fixed at different time points. At 20 min, CRB3 had arrived at the Golgi apparatus in most aRG cells (95.7 ± 5.2%), and by 60 min it strongly accumulated at the apical surface of over 90% of the cells (Fig 4C and D). In 40% of the cells, CRB3 was only detected at the apical surface, indicating that most of the protein pool had reached its final location (Fig 4C and E). We verified that the bright structure in which CRB3 was released was indeed the Golgi apparatus, by co‐expressing the Golgi‐resident enzyme GalNacT2. Upon biotin addition, but not before, a strong colocalization between CRB3 and GalNacT2 was indeed observed, confirming Golgi identity (Fig EV3A).
Figure 4. Post‐Golgi apical transport of Crumbs is driven by dynein.
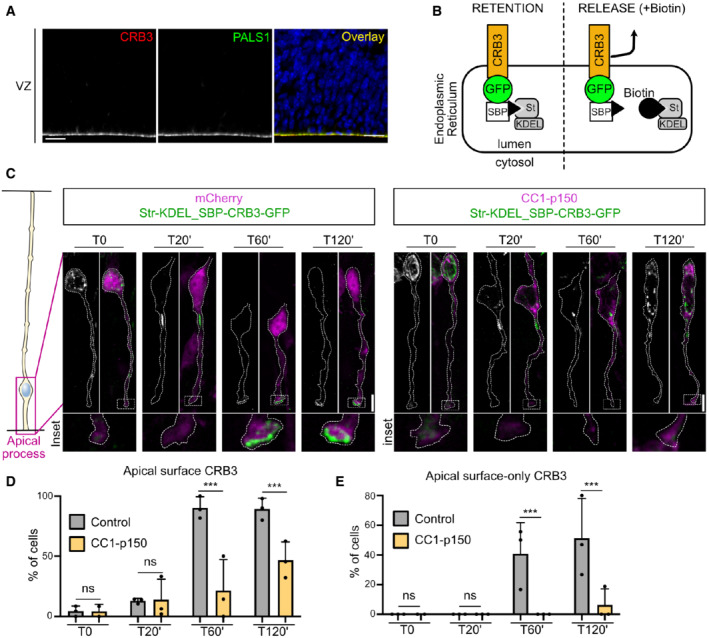
-
AImmuno‐staining for Crumbs3 (CRB3) and PALS1 in E15.5 embryonic cortex. Scale bar = 25 μm.
-
BSchematic representation of the RUSH system. CRB3 is retained in the endoplasmic reticulum (ER) until the addition of biotin, which releases it for trafficking. SBP: Streptavidin‐binding protein. St: Streptavidin.
-
CRUSH assay for CRB3‐GFP in control (mcherry) and dynactin‐inhibited radial glial cells (CC1‐p150‐dsRed), electroporated at E.15.5 and imaged at E16.5. Scale bar = 5 μm.
-
DCRB3 localization at the apical surface upon release.
-
EPercentage of cells with 100% of CRB3 signal at the apical surface upon release.
Data information: (D, E) mCherry (control): N = 3 brains per timepoint (361 cells total). CC1‐p150: N = 3 brains per timepoint (2 brains for T0) (268 cells total). Fisher's exact test and Benjamini–Hochberg procedure, ***P ≤ 0.001. All error bars indicate SD.
Figure EV3. CRB3 exits the Golgi within RAB6+ vesicles.
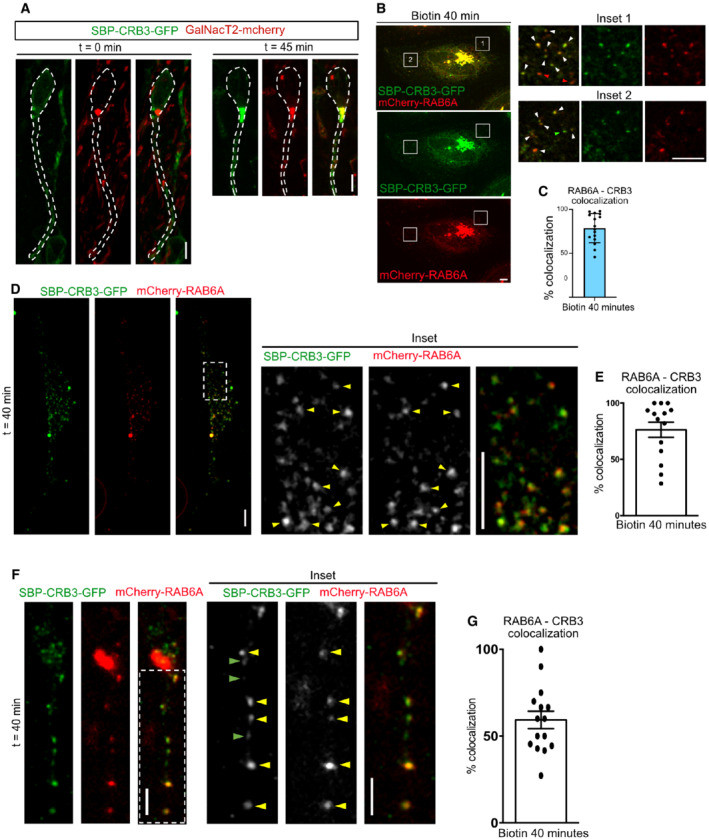
-
ASBP‐CRB3‐GFP and GalNacT2‐mCherry expression in aRG cells before and 45 min after addition of biotin. SBP‐CRB3‐GFP relocates from a diffuse perinuclear localization to the Golgi. Scale bar = 5 μm.
-
BSBP‐CRB3‐GFP and mCherry‐RAB6A localization in HeLa cells before and 40 min after addition of biotin. Scale bar = 5 μm. White arrowheads: colocalizing foci.
-
CQuantification of SBP‐CRB3‐GFP and mCherry‐RAB6A colocalization away from the Golgi apparatus 40 min after biotin addition. N = 16 cells from three independent experiments.
-
DSBP‐CRB3‐GFP and mCherry‐RAB6A localization in dissociated aRG cells cultivated in vitro, 40 min after addition of biotin. Scale bar = 5 μm. Yellow arrowheads: colocalizing foci.
-
EQuantification of SBP‐CRB3‐GFP and mCherry‐RAB6A colocalization away from the Golgi apparatus 40 min after biotin addition. N = 14 cells from three independent experiments.
-
FSBP‐CRB3‐GFP and mCherry‐RAB6A localization in aRG cells cultivated within brain slices, 40 min after addition of biotin. Scale bar = 5 μm. Yellow arrowheads: colocalizing foci.
-
GQuantification of SBP‐CRB3‐GFP and mCherry‐RAB6A colocalization away from the Golgi apparatus 40 min after biotin addition. N = 15 cells from three independent experiments.
Data information: All error bars indicate SD.
To test whether post‐Golgi transport of CRB3 toward the apical surface relies on dynein, we monitored SBP‐CRB3‐GFP trafficking in aRG cells expressing the CC1‐p150 dominant‐negative construct. As in control, 20 min after biotin treatment, CRB3 reached the Golgi apparatus (in 94.1 ± 3.6% of the cells), but at 60 min its trafficking toward the apical surface was severely affected (Fig 4C and D). By 120 min, it started to reach the apical surface, although exhibiting a twofold decrease compared to control cells. Moreover, almost no CC1‐expressing cell showed a localization of the total CRB3 pool at the apical surface, even after 120 min, as compared to half of control cells (Fig 4C and E). Therefore, post‐Golgi transport of Crumbs towards the apical surface of aRG cells is driven by the dynein–dynactin complex.
We and others have previously shown that post‐Golgi RAB6A+ vesicles contain a wide variety of cargoes (Grigoriev et al, 2007; Stehbens et al, 2014; Fourriere et al, 2019). We confirmed here that RAB6A+ vesicles also transport CRB3. HeLa cells expressing CRB3 in the RUSH system were imaged 30 min after biotin addition, when CRB3 has reached the Golgi apparatus and begun to exit it. At this timepoint, almost 80% of vesicles containing SBP‐CRB3‐GFP were positive for mcherry‐RAB6A, indicating that CRB3 largely exits the Golgi apparatus within RAB6A+ vesicles (Fig EV3B and C). To validate that RAB6A+ vesicles also transport CRB3 in aRG cells, we reproduced this experiment within in vitro cultivated mouse aRG cells (Fig EV3D and E). Finally, we confirmed these results in vivo, within in utero electroporated aRG cells (Fig EV3F and G).
LIS1 knockout leads to ectopically dividing progenitors
Because dynein apically transports RAB6+ vesicles containing CRB3, we next asked whether altered dynein would lead to aRG cell delamination, as observed in RAB6A/B dKO. To test this, we inactivated the dynein activator LIS1 in the mouse neocortex, using an inducible KO mouse model (Yingling et al, 2008). Emx1‐Cre; LIS1 loxP/loxP (LIS1 KO) were severely microcephalic, as previously described (Yingling et al, 2008). PAX6+ cells in E12.5 LIS1 KO were found dispersed throughout the entire tissue (Fig 5A). The majority of mitotic RG cells (PAX6+ p‐H3+) were localized basally, away from the apical surface where they are normally found, suggesting that they had delaminated (Fig 5A and B). Similarly, we observed a strong increase in the fraction of p‐VIM+ cells located above the VZ (Fig 5C and D). As in the RAB6A/B dKO, these ectopic pVIM+ cells were largely negative for the intermediate progenitor marker TBR2 (Fig EV4A and B), but had retracted their basal process (Fig EV4C). Therefore, inhibition of dynein through LIS1 loss of function largely phenocopies RAB6A/B dKO, suggesting that RAB6‐dynein‐LIS1‐dependent apical trafficking of CRB3 is required to prevent aRG cell delamination.
Figure 5. LIS1 knockout leads to ectopically dividing progenitors.
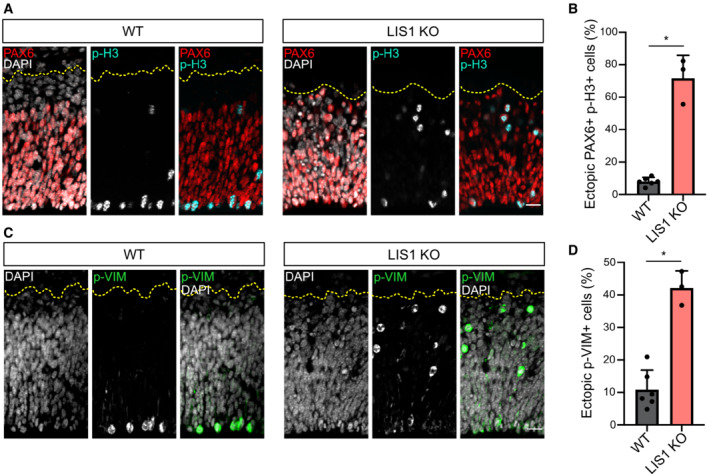
-
APAX6 and phospho‐Histone 3 (p‐H3) staining in WT and LIS1 KO E12.5 brains. Cortices were subdivided into five bins of equal size along the radial axis. Scale bar = 25 μm.
-
BPercentage of PH3+/PAX6+ cells located above the ventricular surface of WT and LIS1 KO E12.5 brains. WT: 1192 cells from N = 6 brains. LIS1 KO: 589 cells from N = 3 brains.
-
CPhospho‐Vimentin (p‐VIM) staining in WT and LIS1 KO E12.5 brains. Scale bar = 25 μm.
-
DPercentage p‐VIM+ cells dividing ectopically, away from the ventricular surface of WT and LIS1 KO E12.5 brains. WT: 1056 cells from N = 6 brains. LIS1 KO: 879 cells from N = 3 brains.
Data information: (B, D) Mann–Whitney U test, *P ≤ 0.05. All error bars indicate SD.
Figure EV4. LIS1 KO leads to ectopic TBR2‐negative basal progenitors.
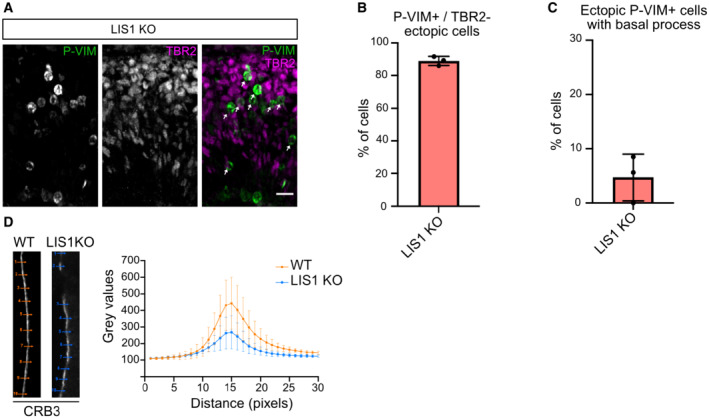
-
AP‐VIM and TBR2 staining in LIS1 KO E12.5 brains. Arrows indicate detached P‐VIM+/TBR2− cells. Scale bar = 25 μm.
-
BPercentage of ectopic P‐VIM+/TBR2− cells in LIS1 KO E12.5 brains. LIS1 KO: 301 cells from N = 3 brains. Error bars indicate SD.
-
CPercentage of ectopic P‐VIM+ cells that maintained a basal process in LIS1 KO E12.5 brains. LIS1 KO: 301 cells from N = 3 brains. Error bars indicate SD.
-
DCRB3 average apical signal intensity ± SEM in WT and LIS1 KO E12.5 brains. N = 3 brains per condition.
RAB6A/B and LIS1 are required of CRB localization and maintenance of adherens junctions
To confirm this model, we tested the consequence of LIS1 and RAB6A/B KO on the steady‐state levels of the Crumbs complex at the apical surface of aRG cells. LIS1 KO brains revealed altered apical localization of CRB3 and its partner PALS1 (Fig 6A). The CRB3 apical signal intensity was reduced, which we quantified using line scan fluorescent intensity measurements (Fig EV4D). Moreover, we observed the frequent appearance of patches that were completely devoid of CRB3 and PALS1. We quantified the number of empty patches along the ventricular surface, which were completely absent in control embryos but occurred at a frequency of 8.2 per mm in LIS1 KO embryos (Fig 6B). RAB6A/B dKO embryonic cortices also displayed an altered apical localization of CRB3 (Fig 6A). As observed in LIS1 KO brains, empty patches devoid of CRB3 and PALS1 occurred at a frequency of 4.3 per mm in RAB6A/B dKO (Fig 6B). On the other hand, single gene depletion of RAB6A or RAB6B had no effect.
Figure 6. RAB6A/B and LIS1 are required for Crumbs localization and junctional integrity.
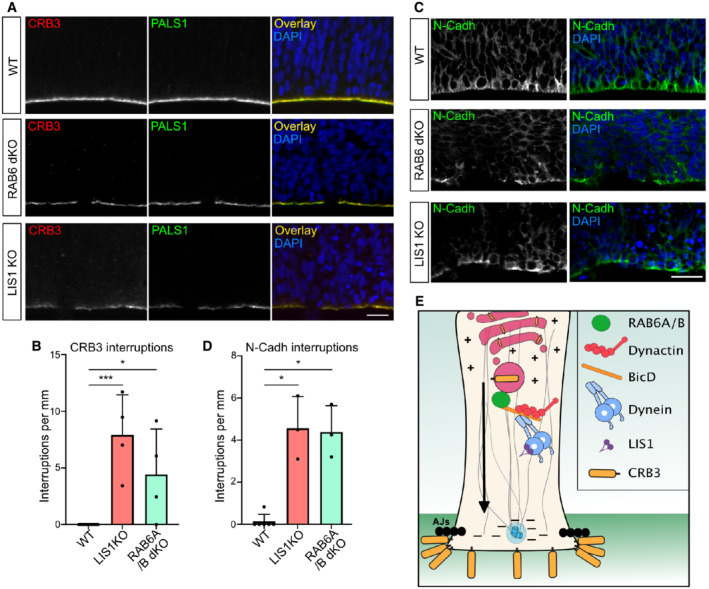
-
ACRB3 and PALS1 staining in WT, RAB6A/B dKO (E15.5), and LIS1 KO (E12.5) brains. Scale bar = 25 μm.
-
BQuantification of CRB3 staining interruptions along the ventricular boundary of WT (E12.5 and E15.5; N = 13 brains), LIS1 KO (E12.5, N = 4 brains), and RAB6A/B dKO (E15.5, N = 4 brains).
-
CN‐Cadherin staining in WT, RAB6A/B dKO (E15.5) and LIS1 KO (E12.5) brains. Scale bar = 25 μm.
-
DQuantification of N‐Cadh staining interruptions along the ventricular boundary of WT (E12.5 and E15.5; N = 6 brains), LIS1 KO (E12.5, N = 3 brains), and RAB6A/B dKO (E15.5, N = 3 brains).
-
EModel for post‐Golgi apical transport of CRB3 to the apical surface of aRG cells.
Data information: (B, D) Kruskal–Wallis test with a Dunn post‐hoc test and Benjamini–Hochberg procedure, *P ≤ 0.05, ***P ≤ 0.001. All error bars indicate SD.
Finally, to test whether the RAB6/dynein/LIS1 complex is required for proper integrity of the apical junctions, we stained embryonic brains for N‐Cadherin. These experiments revealed major junction defects, with abundant interruptions of the staining along the ventricular lining (Fig 6C and D). Even within regions positive for N‐Cadherin, the staining was highly abnormal. These results indicate that inhibition of RAB6 and LIS1 alters CRB3 localization and leads to a destabilization of the apical junctions and a delamination of the aRG cells (Fig 6E).
Discussion
A major finding of this study is that, in aRG cells, post‐Golgi apical trafficking occurs in the microtubule minus end direction, via the RAB6‐dynein‐LIS1 complex, and is required for the apical localization of the Crumbs complex (Fig 6E). As a consequence, genetic inactivation of RAB6A/B or LIS1 leads to CRB3 loss at the ventricular surface and a delamination of aRG cells, which maintain RG features, including fate and the ability to proliferate. We also establish aRG cells as a powerful epithelial model, enabling to resolve transport events in real time in situ.
Post‐Golgi transport is highly bidirectional in aRG cells
Dynein is largely viewed as a retrograde motor, driving trafficking toward the center of the cell. We show here that in epithelial cells, where microtubule minus ends concentrate apically, dynein controls exit from the Golgi apparatus and transport to the apical surface. We observed that apical transport is however highly bidirectional, with RAB6+ vesicles constantly alternating in the apical and basal directions. Therefore, rather than being transported in a strictly polarized manner, RAB6+ vesicles actively oscillate, increasing the chances of reaching and docking to the apical surface. In non‐polarized epithelial cells, although bi‐directional movement can be observed, the trafficking of post‐Golgi RAB6+ vesicles is largely unidirectional, moving toward the cell periphery in a kinesin‐dependent manner (Grigoriev et al, 2007; Miserey‐Lenkei et al, 2010; Serra‐Marques et al, 2020). The higher rate of minus end runs in aRG cells may point to a specific regulation of motors on RAB6+ vesicles upon epithelial polarization. Bicaudal family members, which recruit and activate dynein onto RAB6+ vesicles, are promising candidates for such regulation. Knockout of BICD2 in the mouse neocortex leads to the appearance of ectopically dividing progenitors, phenocopying LIS1 and RAB6A/B dKO, and suggesting apical polarity defects and delamination (Will et al, 2019). Transport in the minus end direction may be further biased by BICDR1, which is able to recruit two dynein molecules for faster movement, and induces strong accumulation of RAB6+ vesicles at the microtubule minus ends (Schlager et al, 2010, 2014b; Urnavicius et al, 2018).
The RAB6‐dynein‐LIS1 complex controls post‐Golgi apical transport of CRB3
Newly synthetized cargoes can traffic directly from the Golgi to the plasma membrane, though passage through intermediate recycling compartments was also proposed. We recently demonstrated that RAB6 acts as a general regulator of protein secretion and confirm here that CRB3 traffics within RAB6+ vesicles (Fourriere et al, 2019). Because RAB6+ vesicles directly fuse with the plasma membrane, via its docking factor ELKS (Grigoriev et al, 2007), we favor a model whereby CRB3 is directly transported from the Golgi to the apical surface. CRB is known to be further maintained apically through RAB11‐dependent and PLLP‐dependent recycling route, that leads to its final localization at tight junctions (Rodriguez‐Fraticelli et al, 2015; Aguilar‐Aragon et al, 2020). Retromer‐dependent transport back to the TGN was also described, indicating that the RAB6‐dynein‐LIS1 pathway we describe here may also play a role in CRB recycling (Pocha et al, 2011). Of note, RAB6+ vesicles were also abundant in the basal process of aRG cells, but the mechanism(s) for sorting of apical and basal post‐Golgi cargoes will require further investigation.
RAB6A and RA6B redundantly control polarized trafficking
We observed that, unlike double KO, single deletion of RAB6A or RAB6B did not affect brain development, indicating that they were largely acting redundantly. Such redundancy was previously observed in cultured neurons following shRNA‐mediated knockdown, as well as in MDCK cells where the very low levels of RAB6B are sufficient to compensate for RAB6A KO (Schlager et al, 2010, 2014b; Homma et al, 2019). We also show that RAB6A/A' and RAB6B act redundantly to control proper neuronal positioning, which may be caused by altered trafficking of adhesion molecules, including integrins (Shafaq‐Zadah et al, 2016).
Impaired apical post‐Golgi trafficking leads to aRG cell delamination
bRG cells are generated from aRG cells and their amplification is a hallmark of gyrencephaly. The expression of several factors is known to affect their production but the underlying mechanisms remain largely unclear (Stahl et al, 2013; Florio et al, 2015; Ju et al, 2016). aRG cells were proposed to detach due to mitotic spindle rotation, or downregulation of the adherens junctions (Konno et al, 2008; Ostrem et al, 2014; Martínez‐Martínez et al, 2016). Recent evidence has demonstrated that delamination can be associated with Golgi structure abnormalities, and that detached aRG cells can reintegrate into the epithelium at early developmental stages but not at later neurogenic states (Uzquiano et al, 2019; Fujita et al, 2020). Here, using live imaging, we demonstrate that altered post‐Golgi transport leads to a detachment of the apical process of aRG cells during interphase, and to the production of ectopically localized cells that maintain RG identity and proliferative capacity. These cells however appear to retract their basal process, potentially due to impaired integrin‐based transport to the basal end‐foot. We did not observe the appearance of folding patterns in KO brains, due to the presence of an apoptosis‐dependent microcephaly phenotype.
In conclusion, our results indicate that the maintenance of epithelial integrity during neocortex development relies on post‐Golgi transport to the apical surface of aRG cells. This pathway can control the balance between aRG cell maintenance and delamination, highlighting a site of action for factors that may participate in the generation of bRG cells.
Materials and Methods
Animal breeding and care
Animals
All experiments involving mice were carried out according to the recommendations of the European Community (2010/63/UE). The animals were bred and cared for in the Specific Pathogen Free Animal Facility of Institut Curie (agreement C 75‐05‐18). All animal procedures were approved by the ethics committee of the Institut Curie CEEA‐IC #118 and by French Ministry of Research (2016–002). Animals were housed at a temperature of 22°C, 50% humidity, and a 12/12 h light/dark cycle.
Mice
Generation of RAB6B knockout mice
The constitutive RAB6B knockout mice have been engineered using CRISPR/Cas9 technology to create a frame shift in the coding sequence. Two gRNA couples, respectively, targeting exons 2 and 4 (GGAAGACGTCTCTGATCACG and CCGAGACTCCACGGTGGCTG), and 3 and 4 (TGTACTTGGAAGACCGTACG and CAGCTACATCCGAGACTCCA) were selected. gRNAs and Cas9m RNA were prepared according to the online protocol from Feng Zhang, https://www.addgene.org/crispr/zhang/. Briefly, the forward and the reverse oligonucleotides specific for the selected gRNA sequences were annealed and cloned into px330 plasmid. To get Cas9 mRNA and gRNAs, an in vitro transcription was performed on Cas9 pCR2.1‐XL plasmid and gRNA plasmid using a T7 promoter, and the mMessage mMachine T7 ULTRA kit and MEGAshortscript T7 kit (Life Technologies), respectively. Cas9 mRNA and sgRNAs were then purified using the MEGAclear Kit (Thermo Fisher Scientific) and eluted in RNAse‐free water. The gRNAs and Cas9mRNA quality were evaluated on agarose gel.
Eight‐week‐old superovulated B6D2F1/J (C57BL/6J × DBA/2J) females from Charles River France were superovulated by intraperitoneal (i.p.) administration of 5 IU of Pregnant Mare Serum Gonadotropin followed by an additional i.p. injection of 5 IU Human Chorion Gonadotropin 48 h later. Superovulated females were mated to stud males of the same background. Zygotes were collected from the oviduct and were cultured in Cleave medium (Cook, K‐RVCL‐50) at 37°C under 5% CO2 until microinjection. An injection solution was prepared as following: Cas9 mRNA at 100 and 50 ng/μl for each gRNA in Brinster buffer (10 mM Tris–HCl pH 7.5; 0.25 mM EDTA) and passed through 0.22 μm pore size filter. Cytoplasmic microinjection was performed into mouse fertilized oocytes. Microinjected embryos were transferred into 0.5 dpc NMRI pseudo‐pregnant females with 12 zygotes per oviduct. Selected founders F0 carrying a 1 bp deletion in exon 2 and a 279 bp inversion, both leading to a premature STOP codon, were then backcrossed to C57BL6/N to segregate out undesired genetic events.
RAB6A/B dKO and LIS1 KO
RAB6A loxP/loxP mutant mice were previously generated and characterized (Bardin et al, 2015). RAB6A loxP/loxP mice were first crossed with RAB6B −/− mice to generate RAB6A loxP/loxP ; RAB6B −/− animals, which were viable and fertile. These animals were then crossed with Emx1‐Cre (JAX 005628) animals to generate Emx1; RAB6A loxP/loxP ; RAB6B −/− (RAB6A/B double knockout) animals. LIS1 conditional knockout mice (LIS1 −/−, also known as Pafah1b1‐loxP; Hirotsune et al, 1998) were crossed with Emx1‐Cre mice.
Expression constructs and antibodies
The following plasmids were used in this study: ManII‐GFP, CC1‐p150, Streptavidin‐KDEL SBP‐CRB3A‐GFP (Franck Perez); GFP‐RAB6A[54]; EB3‐GFP (gift from Matthieu Piel); mCherry2‐C1 vector (gift from Michael Davidson, Addgene plasmid #54563); Cre (gift from David Liu, Addgene plasmid #123133); pCAG‐Cre‐IRES2‐GFP vector (gift from Anjen Chenn, Addgene plasmid #26646); pCAG‐GFP vector (gift from Richard Vallee, Columbia University); TagRFP‐RAB6A (gift from Yuko Mimori‐Kiyosue, Riken Center, Japan).
Antibodies used in this study were mouse anti‐γTubulin (Sigma‐Aldrich, T5326), rat anti‐Crumbs3 (gift from André Le Bivic, Marseille), rabbit anti‐MPP5/PALS1 (Proteintech, 17710‐1‐AP), human anti‐GFP (recombinant antibody platform (Tab‐IP)—Institut Curie, A‐R‐H#11), rabbit anti‐Pax6 (Biolegend, B214847), goat anti‐phospho‐Histone 3 (Santa Cruz, SC‐12927), mouse anti‐phospho‐Vimentin (Abcam, 22651), CUX‐1 (Santa‐Cruz, discontinued), rabbit anti‐cleaved‐Caspase 3 (Cell Signaling, 9661S), rabbit anti‐RAB6A/A' (home‐made (Goud et al, 1990)), rabbit anti‐RAB6B (Proteintech, 10340‐1‐AP), human anti‐αTubulin (recombinant antibody platform (Tab‐IP)—Institut Curie, A‐R‐H#02). Secondary antibodies: donkey Alexa Fluor 488 anti‐mouse, anti‐rabbit, anti‐goat (Jackson laboratories 715‐545‐150, 711‐165‐152, 715‐605‐152), donkey Alexa Fluor 555 anti‐mouse, anti‐rabbit, anti‐goat (Jackson laboratories 715‐545‐150, 711‐165‐152, 715‐605‐152), donkey Alexa Fluor 647 anti‐mouse, anti‐rabbit, anti‐goat (Jackson laboratories 715‐545‐150, 711‐165‐152, 715‐605‐152).
Subcellular live imaging in mouse embryonic brain cortex slices
To record GFP‐RAB6A dynamics in radial glia in situ, we used the following approach. 24 h after the electroporation of E15.5 to E16.5 embryos, the pregnant mouse was sacrificed and the electroporated embryos recovered. Brains were dissected in artificial cerebrospinal fluid (ACSF) and 250‐μm thick coronal slices were prepared with a Leica VT1200S vibratome in ice‐cold ACSF. The slices were cultured on membrane filters over enriched medium (DMEM‐F12 containing B27, N2, 10 ng/ml FGF, 10 ng/ml EGF, 5% fetal bovine serum and 5% horse serum). After recovery in an incubator at 37°C, 5% CO2 for 2 h (or 48 h for human tissue to allow for construct expression), the filters were cut and carefully turned over on a 35 mm FluoroDish (WPI), to position the sample in direct contact with the glass, underneath the filter (to maintain the sample flat).
Live imaging was performed on a fully motorized spinning disk wide microscope driven by Metamorph software (Molecular Devices) and equipped with a Yokogawa CSU‐W1 scanner unit to increase the field of view and improve the resolution deep in the sample. The inverted microscope (Nikon Eclipse Ti2) was equipped with a high working distance (WD 0.3 mm) 100X SR HP Plan Apo 1.35 NA Silicon immersion (Nikon) and a Prime95B sCMOS camera (Photometrics). To maintain stable cell culture conditions (37°C, humidity, 5% CO2), time‐lapse imaging was performed on a STX stage top incubator (Tokai Hit). Z‐stacks of 3–5 μm range were taken on a Mad City Lab piezo stage (Nano Z500) with an interval of 1 μm. Maximum projections were generated from which kymographs were generated. Tracking and quantifications of GFP‐RAB6A+ vesicle dynamics were directly performed on the movies and the kymographs were used for validation and display purposes. Videos were mounted in Metamorph. Kymograph generation (KymographBuilder), Tracking of GFP‐RAB6A+ vesicles (manual tracking) as well as image modifications (brightness and contrast, background, gamma) were carried out on Fiji. Figures were assembled in Affinity Designer.
RUSH assay in situ
E15.5–E16.5 embryos were electroporated with a Streptavidin‐KDEL SBP‐CRB3‐GFP construct with or without CC1‐p150 for 24 h. Slicing and culture were performed as for subcellular live imaging experiments and sections were cultured in DMEM‐F12 medium supplemented with 4 nM avidin to prevent leakage due to circulating biotin. Biotin was added to the enriched medium (40 μM final) for the indicated period of time (37°C, 5% CO2) prior to paraformaldehyde fixation. Immunostaining against GFP was performed to amplify fluorescence (see immunostaining section) prior to mounting.
Statistical analysis
All the statistical analysis has been made using R 4.0.5. R Core Team (2021), R Foundation for Statistical Computing, Vienna, Austria (https://www.R‐project.org/). Due to the low sample sizes inherent to in vivo work, we conducted nonparametric analyses. Median comparisons between two conditions have been made with a Mann–Whitney U test (Figs 2B and D, 3J–M, 5B and D, and EV1B). When more than two conditions were compared, we used Kruskal–Wallis test with a Dunn post‐hoc test and Benjamini–Hochberg procedure to control the false discovery rate using the dunn.test package (Figs 1D and F, 2E, and 6B and D). These analyses have been made considering the animal as the statistical unit except for the Fig 3J–M. Embryos for a given condition come from different litters. For categorical data (Figs 2H and 4D and E) and data from Figs 3J–M and EV2E and F, we considered each cell as a statistical unit. Since the cells are electroporated in‐situ, we made the reasonable approximation that cells received their constructs independently and their properties are measured individually at the cell scale. We validated this hypothesis by repeating experiments in different independent animals to conclude that the effect was not due to cells coming from biased individuals due to an abnormal electroporation or an abnormal embryo. For categorial data, analysis has been made using Fisher's exact test (Fig 2H) accompanied with a Benjamini–Hochberg procedure to control the false discovery rate when more than two conditions were compared (Fig 4D and E). These categorical data are depicted as percentages for clarity. P values superior to 0.05 are considered as not significant. Due to the evident KO phenotypes, no blinding was performed.
Author contributions
Jean‐Baptiste Brault: Conceptualization; data curation; formal analysis; funding acquisition; writing – original draft; project administration. Sabine Bardin: Data curation; methodology. Marusa Lampic: Data curation. Jacopo A Carpentieri: Data curation. Laure Coquand: Data curation; methodology. Maxime Penisson: Data curation. Hugo Lachuer: Formal analysis. Guiliana Soraya Victoria: Data curation. Sarah Baloul: Data curation. Fatima El Marjou: Methodology. Gaelle Boncompain: Resources. Stephanie Miserey‐Lenkei: Resources. Richard Belvindrah: Data curation. Vincent Fraisier: Methodology. Fiona Francis: Resources. Franck Perez: Resources; funding acquisition; methodology. Bruno Goud: Conceptualization; formal analysis; writing – original draft; project administration; writing – review and editing. Alexandre D Baffet: Conceptualization; data curation; formal analysis; funding acquisition; investigation; visualization; methodology; writing – original draft; project administration; writing – review and editing.
Disclosure and competing interests statement
The authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
Movie EV1
Movie EV2
Movie EV3
Movie EV4
Movie EV5
Movie EV6
Movie EV7
Movie EV8
Movie EV9
Movie EV10
PDF+
Acknowledgements
The authors greatly acknowledge the Cell and Tissue Imaging (PICT‐IBiSA), Institut Curie, member of the French National Research Infrastructure France‐BioImaging (ANR10‐INBS‐04), and the Nikon BioImaging Center (Institut Curie, France). We greatly acknowledge the Recombinant Antibody Platform of the Institut Curie for the production of antibodies. We are grateful to Shinji Hirotsune for providing the Lis1 mouse line and embryos, and Yoann Saillour for aid generating Lis1 embryos. We thank D. Massey‐Harroche and A. Le Bivic for the anti‐Crumbs3 antibody. A.D.B. is an INSERM researcher. This work was supported by the ANR (ANR‐19‐CE13‐0002‐02), CNRS, Institut Curie, the Ville de Paris “Emergences” program, Labex CelTisPhyBio (11‐LBX‐0038), and PSL. F.F.'s lab was supported by the ANR‐16‐CE16‐0011‐03 and NEURON‐Full‐815‐006 STEM‐MCD grants.
EMBO reports (2022) 23: e54605
Contributor Information
Bruno Goud, Email: bruno.goud@curie.fr.
Alexandre D Baffet, Email: alexandre.baffet@curie.fr.
Data availability
No data that require deposition in a public database have been generated.
References
- Aguilar‐Aragon M, Fletcher G, Thompson BJ (2020) The cytoskeletal motor proteins dynein and MyoV direct apical transport of Crumbs. Dev Biol 459: 126–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apodaca G, Gallo LI, Bryant DM (2012) Role of membrane traffic in the generation of epithelial cell asymmetry. Nat Cell Biol 14: 1235–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baffet AD, Hu DJ, Vallee RB (2015) Cdk1 activates pre‐mitotic nuclear envelope dynein recruitment and apical nuclear migration in neural stem cells. Dev Cell 33: 703–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardin S, Miserey‐Lenkei S, Hurbain I, Garcia‐Castillo D, Raposo G, Goud B (2015) Phenotypic characterisation of RAB6A knockout mouse embryonic fibroblasts. Biol Cell 107: 427–439 [DOI] [PubMed] [Google Scholar]
- Bay AEP, Schreiner R, Mazzoni F, Carvajal‐Gonzalez JM, Gravotta D, Perret E, Mantaras GL, Zhu Y‐S, Rodriguez‐Boulan EJ (2013) The kinesin KIF16B mediates apical transcytosis of transferrin receptor in AP‐1B‐deficient epithelia. EMBO J 32: 2125–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boncompain G, Divoux S, Gareil N, de Forges H, Lescure A, Latreche L, Mercanti V, Jollivet F, Raposo G, Perez F (2012) Synchronization of secretory protein traffic in populations of cells. Nat Methods 9: 493–498 [DOI] [PubMed] [Google Scholar]
- Bulgakova NA, Knust E (2009) The Crumbs complex: From epithelial‐cell polarity to retinal degeneration. J Cell Sci 122: 2587–2596 [DOI] [PubMed] [Google Scholar]
- Cappello S, Attardo A, Wu X, Iwasato T, Itohara S, Wilsch‐Bräuninger M, Eilken HM, Rieger MA, Schroeder TT, Huttner WB et al (2006) The rho‐GTPase cdc42 regulates neural progenitor fate at the apical surface. Nat Neurosci 9: 1099–1107 [DOI] [PubMed] [Google Scholar]
- Coquand L, Victoria GS, Tata A, Carpentieri JA, Brault J‐B, Guimiot F, Fraisier V, Baffet AD (2021) CAMSAPs organize an acentrosomal microtubule network from basal varicosities in radial glial cells. J Cell Biol 220: e202003151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudok JJ, Murtaza M, Alves CH, Rashbass P, Wijnholds J (2016) Crumbs 2 prevents cortical abnormalities in mouse dorsal telencephalon. Neurosci Res 108: 12–23 [DOI] [PubMed] [Google Scholar]
- Elshenawy MM, Kusakci E, Volz S, Baumbach J, Bullock SL, Yildiz A (2020) Lis1 activates dynein motility by modulating its pairing with dynactin. Nat Cell Biol 22: 570–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández V, Llinares‐Benadero C, Borrell V (2016) Cerebral cortex expansion and folding: what have we learned? EMBO J 35: 1021–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fietz SA, Kelava I, Vogt J, Wilsch‐Bräuninger M, Stenzel D, Fish JL, Corbeil D, Riehn A, Distler W, Nitsch R et al (2010) OSVZ progenitors of human and ferret neocortex are epithelial‐like and expand by integrin signaling. Nat Neurosci 13: 690–699 [DOI] [PubMed] [Google Scholar]
- Florio M, Albert M, Taverna E, Namba T, Brandl H, Lewitus E, Haffner C, Sykes A, Wong FK, Peters J et al (2015) Human‐specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion. Science 347: 1465–1470 [DOI] [PubMed] [Google Scholar]
- Florio M, Borrell V, Huttner WB (2016) Human‐specific genomic signatures of neocortical expansion. Curr Opin Neurobiol 42: 33–44 [DOI] [PubMed] [Google Scholar]
- Fourriere L, Kasri A, Gareil N, Bardin S, Bousquet H, Pereira D, Perez F, Goud B, Boncompain G, Miserey‐Lenkei S (2019) RAB6 and microtubules restrict protein secretion to focal adhesions. J Cell Biol 218: 2215–2231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita I, Shitamukai A, Kusumoto F, Mase S, Suetsugu T, Omori A, Kato K, Abe T, Shioi G, Konno D et al (2020) Endfoot regeneration restricts radial glial state and prevents translocation into the outer subventricular zone in early mammalian brain development. Nat Cell Biol 22: 26–37 [DOI] [PubMed] [Google Scholar]
- Goud B, Zahraoui A, Tavitian A, Saraste J (1990) Small GTP‐binding protein associated with Golgi cisternae. Nature 345: 553–556 [DOI] [PubMed] [Google Scholar]
- Goud B, Liu S, Storrie B (2018) Rab proteins as major determinants of the Golgi complex structure. Small GTPases 9: 66–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriev I, Splinter D, Keijzer N, Wulf PS, Demmers J, Ohtsuka T, Modesti M, Maly IV, Grosveld F, Hoogenraad CC et al (2007) Rab6 regulates transport and targeting of exocytotic carriers. Dev Cell 13: 305–314 [DOI] [PubMed] [Google Scholar]
- Hansen DV, Lui JH, Parker PRL, Kriegstein AR (2010) Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature 464: 554–561 [DOI] [PubMed] [Google Scholar]
- Hirotsune S, Fleck MW, Gambello MJ, Bix GJ, Chen A, Clark GD, Ledbetter DH, McBain CJ, Wynshaw‐Boris A (1998) Graded reduction of Pafah1b1 (Lis1) activity results in neuronal migration defects and early embryonic lethality. Nat Genet 19: 333–339 [DOI] [PubMed] [Google Scholar]
- Höing S, Yeh T‐Y, Baumann M, Martinez NE, Habenberger P, Kremer L, Drexler HCA, Küchler P, Reinhardt P, Choidas A et al (2018) Dynarrestin, a novel inhibitor of cytoplasmic dynein. Cell Chem Biol 25: 357–369.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma Y, Kinoshita R, Kuchitsu Y, Wawro PS, Marubashi S, Oguchi ME, Ishida M, Fujita N, Fukuda M (2019) Comprehensive knockout analysis of the Rab family GTPases in epithelial cells. J Cell Biol 218: 2035–2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Htet ZM, Gillies JP, Baker RW, Leschziner AE, DeSantis ME, Reck‐Peterson SL (2020) LIS1 promotes the formation of activated cytoplasmic dynein‐1 complexes. Nat Cell Biol 22: 518–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu DJ‐K, Baffet AD, Nayak T, Akhmanova AS, Doye V, Vallee RB (2013) Dynein recruitment to nuclear pores activates apical nuclear migration and mitotic entry in brain progenitor cells. Cell 154: 1300–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh W, Vale RD (2017) Disease‐associated mutations in human BICD2 hyperactivate motility of dynein‐dynactin. J Cell Biol 216: 3051–3060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y, Moriyama Y, Hasegawa T, Endo TA, Toyoda T, Gotoh Y (2013) Scratch regulates neuronal migration onset via an epithelial‐mesenchymal transition–like mechanism. Nat Neurosci 16: 1–12 [DOI] [PubMed] [Google Scholar]
- Jaulin F, Xue X, Rodriguez‐Boulan E, Kreitzer G (2007) Polarization‐dependent selective transport to the apical membrane by KIF5B in MDCK cells. Dev Cell 13: 511–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MB, Sun X, Kodani A, Borges‐Monroy R, Girskis KM, Ryu SC, Wang PP, Patel K, Gonzalez DM, Woo YM et al (2018) Aspm knockout ferret reveals an evolutionary mechanism governing cerebral cortical size. Nature 556: 370–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju X‐C, Hou Q‐Q, Sheng A‐L, Wu K‐Y, Zhou Y, Jin Y, Wen T, Yang Z, Wang X, Luo Z‐G (2016) The hominoid‐specific gene TBC1D3 promotes generation of basal neural progenitors and induces cortical folding in mice. eLife 5: e18197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Lehtinen MK, Sessa A, Zappaterra MW, Cho S‐H, Gonzalez D, Boggan B, Austin CA, Wijnholds J, Gambello MJ et al (2010) The apical complex couples cell fate and cell survival to cerebral cortical development. Neuron 66: 69–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno D, Shioi G, Shitamukai A, Mori A, Kiyonari H, Miyata T, Matsuzaki F (2008) Neuroepithelial progenitors undergo LGN‐dependent planar divisions to maintain self‐renewability during mammalian neurogenesis. Nat Cell Biol 10: 93–101 [DOI] [PubMed] [Google Scholar]
- Lee HO, Norden C (2013) Mechanisms controlling arrangements and movements of nuclei in pseudostratified epithelia. Trends Cell Biol 23: 141–150 [DOI] [PubMed] [Google Scholar]
- Margolis B (2018) The Crumbs3 polarity protein. Cold Spring Harb Perspect Biol 10: a027961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez‐Martínez MÁ, Romero CDJ, Fernández V, Cárdenas A, Götz M, Borrell V (2016) A restricted period for formation of outer subventricular zone defined by Cdh1 and Trnp1 levels. Nat Commun 7: 11812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzo MG, Griswold JM, Markus SM (2020) Pac1/LIS1 stabilizes an uninhibited conformation of dynein to coordinate its localization and activity. Nat Cell Biol 22: 559–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matanis T, Akhmanova AS, Wulf P, Nery ED, Weide T, Stepanova T, Galjart N, Grosveld F, Goud B, de Zeeuw CI et al (2002) Bicaudal‐D regulates COPI‐independent Golgi–ER transport by recruiting the dynein–dynactin motor complex. Nat Cell Biol 4: 986–992 [DOI] [PubMed] [Google Scholar]
- Mckenney RJ, Huynh W, Tanenbaum ME, Bhabha G, Vale RD (2014) Activation of cytoplasmic dynein motility by dynactin‐cargo adapter complexes. Science 345: 337–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miserey‐Lenkei S, Chalancon G, Bardin S, Formstecher E, Goud B, Echard A (2010) Rab and actomyosin‐dependent fission of transport vesicles at the Golgi complex. Nat Cell Biol 12: 645–654 [DOI] [PubMed] [Google Scholar]
- Narayanan R, Pham L, Kerimoglu C, Watanabe T, Hernandez RC, Sokpor G, Ulmke PA, Kiszka KA, Tonchev AB, Rosenbusch J et al (2018) Chromatin remodeling BAF155 subunit regulates the genesis of basal progenitors in developing cortex. iScience 4: 109–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda Y, Okada Y, Saito N, Setou M, Xu Y, Zhang Z, Hirokawa N (2001) KIFC3, a microtubule minus end‐directed motor for the apical transport of annexin XIIIb‐associated triton‐insoluble membranes. J Cell Biol 155: 77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opdam FJ, Echard A, Croes HJ, van den Hurk JA, van de Vorstenbosch RA, Ginsel LA, Goud B, Fransen JA (2000) The small GTPase Rab6B, a novel Rab6 subfamily member, is cell‐type specifically expressed and localised to the Golgi apparatus. J Cell Sci 113: 2725–2735 [DOI] [PubMed] [Google Scholar]
- Ostrem BEL, Lui JH, Gertz CC, Kriegstein AR (2014) Control of outer radial glial stem cell mitosis in the human brain. Cell Rep 8: 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paridaen JT, Huttner WB (2014) Neurogenesis during development of the vertebrate central nervous system. EMBO Rep 15: 351–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penisson M, Ladewig J, Belvindrah R, Francis F (2019) Genes and mechanisms involved in the generation and amplification of basal radial glial cells. Front Cell Neurosci 13: 381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pocha SM, Wassmer T, Niehage C, Hoflack B, Knust E (2011) Retromer controls epithelial cell polarity by trafficking the apical determinant Crumbs. Curr Biol 21: 1111–1117 [DOI] [PubMed] [Google Scholar]
- Reillo I, Romero CDJ, García‐Cabezas MÁ, Borrell V (2011) A role for intermediate radial glia in the tangential expansion of the mammalian cerebral cortex. Cereb Cortex 21: 1674–1694 [DOI] [PubMed] [Google Scholar]
- Reiner O, Carrozzo R, Shen Y, Wehnert M, Faustinella F, Dobyns WB, Caskey CT, Ledbetter DH (1993) Isolation of a miller‐Dieker lissencephaly gene containing G protein beta‐subunit‐like repeats. Nature 364: 717–721 [DOI] [PubMed] [Google Scholar]
- Rodriguez‐Boulan E, Macara IG (2014) Organization and execution of the epithelial polarity programme. Nat Rev Mol Cell Biol 15: 225–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez‐Fraticelli AE, Bagwell J, Bosch‐Fortea M, Boncompain G, Reglero‐Real N, García‐León MJ, Andrés G, Toribio ML, Alonso MA, Millán J et al (2015) Developmental regulation of apical endocytosis controls epithelial patterning in vertebrate tubular organs. Nat Cell Biol 17: 241–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlager MA, Kapitein LC, Grigoriev I, Burzynski GM, Wulf PS, Keijzer N, de Graaff E, Fukuda M, Shepherd IT, Akhmanova AS et al (2010) Pericentrosomal targeting of Rab6 secretory vesicles by bicaudal‐D‐related protein 1 (BICDR‐1) regulates neuritogenesis. EMBO J 29: 1637–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlager MA, Hoang HT, Urnavicius L, Bullock SL, Carter AP (2014a) In vitro reconstitution of a highly processive recombinant human dynein complex. EMBO J 33: 1855–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlager MA, Serra‐Marques A, Grigoriev I, Gumy LF, da Silva ME, Wulf PS, Akhmanova AS, Hoogenraad CC (2014b) Bicaudal D family adaptor proteins control the velocity of dynein‐based movements. Cell Rep 8: 1248–1256 [DOI] [PubMed] [Google Scholar]
- Serra‐Marques A, Martin M, Katrukha EA, Grigoriev I, Peeters CA, Liu Q, Hooikaas PJ, Yao Y, Solianova V, Smal I et al (2020) Concerted action of kinesins KIF5B and KIF13B promotes efficient secretory vesicle transport to microtubule plus ends. eLife 9: e61302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafaq‐Zadah M, Gomes‐Santos CS, Bardin S, Maiuri P, Maurin M, Iranzo J, Gautreau A, Lamaze C, Caswell P, Goud B et al (2016) Persistent cell migration and adhesion rely on retrograde transport of β(1) integrin. Nat Cell Biol 18: 54–64 [DOI] [PubMed] [Google Scholar]
- Splinter D, Razafsky DS, Schlager MA, Serra‐Marques A, Grigoriev I, Demmers J, Keijzer N, Jiang K, Poser I, Hyman AA et al (2012) BICD2, dynactin, and LIS1 cooperate in regulating dynein recruitment to cellular structures. Mol Biol Cell 23: 4226–4241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl R, Walcher T, Romero CDJ, Pilz GA, Cappello S, Irmler M, Sanz‐Aquela JM, Beckers J, Blum R, Borrell V et al (2013) Trnp1 regulates expansion and folding of the mammalian cerebral cortex by control of radial glial fate. Cell 153: 535–549 [DOI] [PubMed] [Google Scholar]
- Stehbens SJ, Paszek M, Pemble H, Ettinger A, Gierke S, Wittmann T (2014) CLASPs link focal‐adhesion‐associated microtubule capture to localized exocytosis and adhesion site turnover. Nat Cell Biol 16: 561–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai AW, Chuang JZ, Bode C, Wolfrum U, Sung CH (1999) Rhodopsin's carboxy‐terminal cytoplasmic tail acts as a membrane receptor for cytoplasmic dynein by binding to the dynein light chain Tctex‐1. Cell 97: 877–887 [DOI] [PubMed] [Google Scholar]
- Tavano S, Taverna E, Kalebic N, Haffner C, Namba T, Dahl A, Wilsch‐Bräuninger M, Paridaen JTML, Huttner WB (2018) Insm1 induces neural progenitor delamination in developing neocortex via downregulation of the Adherens Junction Belt‐specific protein Plekha7. Neuron 97: 1299–1314.e8 [DOI] [PubMed] [Google Scholar]
- Taverna E, Mora‐Bermúdez F, Strzyz PJ, Florio M, Icha J, Haffner C, Norden C, Wilsch‐Bräuninger M, Huttner WB (2016) Non‐canonical features of the Golgi apparatus in bipolar epithelial neural stem cells. Sci Rep 6: 21206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathy SK, Weil SJ, Chen C, Anand P, Vallee RB, Gross SP (2014) Autoregulatory mechanism for dynactin control of processive and diffusive dynein transport. Nat Cell Biol 16: 1192–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnavicius L, Lau CK, Elshenawy MM, Morales‐Rios E, Motz C, Yildiz A, Carter AP (2018) Cryo‐EM shows how dynactin recruits two dyneins for faster movement. Nature 554: 202–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzquiano A, Ng IG, Nguyen L, Reiner O, Götz M, Matsuzaki F, Francis F (2018) Cortical progenitor biology: Key features mediating proliferation versus differentiation. J Neurochem 146: 500–525 [DOI] [PubMed] [Google Scholar]
- Uzquiano A, Cifuentes‐Diaz C, Jabali A, Romero DM, Houllier A, Dingli F, Maillard C, Boland A, Deleuze J‐F, Loew D et al (2019) Mutations in the heterotopia gene Eml1/EML1 severely disrupt the formation of primary cilia. Cell Rep 28: 1596–1611.e10 [DOI] [PubMed] [Google Scholar]
- Vaid S, Camp JG, Hersemann L, Oegema CE, Heninger A‐K, Winkler S, Brandl H, Sarov M, Treutlein B, Huttner WB et al (2018) A novel population of Hopx‐dependent basal radial glial cells in the developing mouse neocortex. Development 145: dev169276 [DOI] [PubMed] [Google Scholar]
- Will L, Portegies S, van Schelt J, van Luyk M, Jaarsma D, Hoogenraad CC (2019) Dynein activating adaptor BICD2 controls radial migration of upper‐layer cortical neurons in vivo . Acta Neuropathol Commun 7: 162–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M, Kumamoto K, Mikuni S, Arai Y, Kinjo M, Nagai T, Tsukasaki Y, Watanabe TM, Fukui M, Jin M et al (2013) Rab6a releases LIS1 from a dynein idling complex and activates dynein for retrograde movement. Nat Commun 4: 2033 [DOI] [PubMed] [Google Scholar]
- Yingling J, Youn YH, Darling D, Toyo‐oka K, Pramparo T, Hirotsune S, Wynshaw‐Boris A (2008) Neuroepithelial stem cell proliferation requires LIS1 for precise spindle orientation and symmetric division. Cell 132: 474–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J, Stauber T, Nery ED, Vernos I, Pepperkok R, Nilsson T (2005) Regulation of microtubule‐dependent recycling at the trans‐Golgi network by Rab6A and Rab6A'. Mol Biol Cell 16: 162–177 [DOI] [PMC free article] [PubMed] [Google Scholar]


