Abstract
Background
Multiple mutation (MM) within a single gene has recently been reported as a mechanism involved in carcinogenesis. The present study investigated the clinical significance of MMs in hepatocellular carcinoma (HCC).
Methods
Two hundred twenty-three surgically resected HCCs were subjected to gene expression profiling and whole-exome sequencing.
Results
MMs in individual genes were detected in 178 samples (MM tumors: 79.8%). The remaining samples all carried a single mutation (SM tumors: 20.2%). Recurrence-free survival in the MM group was significantly worse in comparison to the SM group (P = 0.012). A Cox proportional hazard analysis revealed that MM tumor was an independent predictor for worse a prognosis (hazard ratio, 1.72; 95% confidence interval, 1.01–3.17; P = 0.045). MMs were frequently observed across in various genes, especially MUC16 (15% of samples had at least one mutation in the gene) and CTNNB1 (14%). Although the MUC16 mRNA expression of MUC16 wild-type and MUC16 SM tumors did not differ to a statistically significant extent, the expression in MUC16 MM tumors was significantly enhanced in comparison to MUC16 SM tumors (P < 0.001). In MUC16, MMs were associated with viral hepatitis, higher tumor marker levels and vascular invasion. The MUC16 MMs group showed significantly worse recurrence-free survival in comparison to the MUC16 SM group (P = 0.022), while no significant difference was observed between the MUC16 SM group and the MUC16 wild-type group (P = 0.324).
Conclusions
MM was a relatively common event that may occur selectively in specific oncogenes and is involved in aggressive malignant behavior.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12885-022-10143-z.
Keywords: Hepatocellular carcinoma, Multiple mutations, Hypermutation, MUC16
Introduction
Hepatocellular carcinoma (HCC) is the third leading cause of cancer-related death worldwide [1]. Advances in next-generation sequencing have enabled the examination of cancer genomes and led to the discovery of driver alterations. In HCC research, these advances have enabled the processing of the HCC genome, and somatic mutations, structural alterations, HBV integration [2], RNA editing and retrotransposon changes [3] have been identified. Somatic mutation in the TERT promoter has been identified as the most frequent alteration (approximately 60%) in HCC [4]. In coding regions, whole-exome sequencing detected frequent mutations in candidate driver genes, such as TP53 (31%), ARID1A (28.2%), CTNNB1 (18.8%), MTDH (14.7%), AXIN1 (14.2%), CDKN2A (11.7%), and ARID2 (10.9%) [3]. These comprehensive genomic analyses have already identified many pathways as potential therapeutic targets. However, most of the genetic alterations identified were shown to occur with low frequency. Thus, these findings indicate that HCC might be a disease for which the development of molecular targeted treatment is challenging.
Multiple mutations (MMs) in the same oncogene have been newly characterized as one of the mechanisms for the promotion of carcinogenesis [5]. Although tumour suppressor genes (TSGs) are known to be affected by multiple loss-of-function mutations [6, 7], no study has investigated MMs arising in the same oncogene during cancer initiation and development in a structured and consistent way. Saito et al. found that MMs in PIK3CA, a representative oncogene, resulted in increased activation of downstream signaling and dependency on the mutated genes as well as an increased sensitivity to specific inhibitors. Furthermore, MMs were reported to be functionally weak, infrequent mutations, preferentially in cis, suggesting that they act in combination to increase oncogenicity [5]. MMs in the same oncogene were reported to be more frequent than expected, with 9% of pan-cancer samples with mutations in oncogenes harboring MMs. These findings indicated that oncogenic MMs are a relatively common driver event, suggesting an underlying mechanism of clonal selection of suboptimal mutations with a low frequency. The previous report was a pan-cancer analysis to overview the landscape of MMs in the same oncogene. Therefore, the frequency and clinical significance of MMs in individual cancer types remain unclear.
Here, we performed comprehensive genetic profiling of HCCs using whole-exome sequencing (WES) and gene expression profiling (GEP) analysis in a large Japanese population. To overview MMs in HCCs and assess the clinical relevance of MMs in HCC patients, we investigated the accumulation of MMs in each gene and the association between MMs and clinicopathological information.
Methods
Ethics statement
To investigate the biological characteristics of cancer and diathesis of each patient with cancer, the Shizuoka Cancer Center started Project High-tech Omics-based Patient Evaluation (HOPE) in 2014 [8]. Project HOPE was designed according to the “Ethical Guidelines for Human Genome and Genetic Analysis Research” revised in 2013 [8]. Written consent was obtained from all patients participating in Project HOPE. The present study used the data from Project HOPE and was approved by the Institutional Review Board of Shizuoka Cancer Center (approval no. 25–33). The study protocol conforms to the ethical guidelines of the Declaration of Helsinki.
Patient selection and study design
From January 2014 to March 2019, 223 surgically resected HCCs were analyzed in Project HOPE. Written informed consent was obtained from all participants. All tumor tissues were pathologically diagnosed as HCC. Tumor tissue samples were dissected from fresh surgical specimens. The surrounding normal tissue was also obtained whenever possible. In addition, peripheral blood was collected as a control for WES. DNA was extracted from tissue samples using a QIAamp DNA Blood MINI Kit (Qiagen, Venlo, The Netherlands). For RNA analysis, tissue samples were submerged in RNAlater solution (Thermo Fisher Scientific), minced and stored at 4 °C overnight before RNA extraction. To validate our findings in other cohort, mutation profiles were extracted from the public database in the TCGA project [9].
Treatment strategy for HCCs
The details of the surgical strategy and procedure have been previously reported [10]. No preoperative or postoperative adjuvant therapy was performed. Patients underwent physical examinations and blood testing every three months postoperatively. Serial CT or liver ultrasonography was performed in each patient every three to six months. When HCC recurrence was detected, the most appropriate therapy was applied after considering the patient’s liver function and tumor factors. Therapy options included repeat hepatectomy, transcatheter arterial chemoembolization, radiofrequency ablation, or sorafenib.
WES analysis of HCC tissues using next-generation sequencing
WES analysis was performed as previously described [11, 12]. Briefly, DNA was subjected to WES on an Ion Proton System (Thermo Fisher Scientific). Torrent Suite software (ver. 4.4; Thermo Fisher Scientific) was used to convert binary raw data into sequence reads that were mapped to the reference human genome (UCSC, hg19). The mapping results were stored as BAM files. Two BAM files uploaded to the Ion Reporter system were analyzed simultaneously. For this analysis, AmpliSeq exome tumor-normal pair workflow (ver. 4.4, Thermo Fisher Scientific) with a Custom Hotspot file was used, and this Custom Hotspot file specifies the somatic and pathogenic mutations registered in COSMIC and ClinVar. The sequence data derived from blood samples were used as matched controls. Mutations fulfilling at least one of the following criteria were discarded as false positive: (1) quality score < 60, (2) depth of coverage < 20, (3) variant read observed in one strand only, (4) clipped sequence length < 100 (avg_clipped_length < 100), (5) variant located on either sequence end (avg_pos_as_fraction < 0.05), and (6) mutation matches one on an in-house false-positive list. Parameters specified in criteria (4) and (5) were calculated by bam-readcount with option “-q 1” (ver. 0.8.0) (https://github.com/genome/bam-readcount).
GEP using DNA microarray analysis
GEP analysis was performed as previously described [11, 13]. Total RNA was extracted using an miRNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. RNA samples with an RNA integrity number of greater than or equal to 6 were used for DNA microarray analysis. Labeled samples were hybridized to a SurePrint G3 Human Gene Expression 8 × 60 K v2 Microarray (Agilent Technologies, Santa Clara, CA, USA). The fold change between tumor and normal tissues from the same patient was calculated from the normalized values.
Clinicopathological variables
Data on clinicopathological characteristics were collected from a prospectively maintained HCC database at Shizuoka Cancer Center Hospital. Tumor size was measured at its largest diameter.
Construction of a catalogue of cancer-related genes
The classification for oncogenes and TSGs were obtained from COSMIC Cancer Gene Census [14], and OncoKB Cancer Gene List [15] as previously described, in our analysis pipeline Shizuoka Multi-omics Analysis Protocol [16].
CCLE cell line data
The mutation call data (depmap_19Q1_mutation_calls_v2.csv) for 1,601 cell lines and drug-sensitivity data (v17.3_fitted_dose_response.xlsx) for 1,065 cell lines were obtained from the DepMap (https://depmap.org/portal/), CCLE (https://portals.broadinstitute.org/ccle/) and Genomics of Drug Sensitivity in Cancer (GDSC; https://www.cancerrxgene.org/) databases.
Statistical analyses
Continuous variables were expressed as the median with the interquartile range (IQR) and compared using the Mann–Whitney U test or an analysis of variance. AFP and PIVKAII were compared by log-transformation in base 10. A univariate analysis for categorical variables was performed by the chi-square test and Fisher’s exact probability test. For comparisons of three or more groups, p-values were adjusted using the Bonferroni method. The thresholds generally accepted in clinical settings were employed as the cut-off value of continuous variables for statistical processing. In the liver damage classification [17], the ICG R15 value is classified into three groups by cut-offs at 15% and 40%. Taking this into account, a cut-off value of 20% was used to define the 2 groups in the present study. For AFP and PIVKAII, 200 ng/mL [18] and 100 mAU/mL [19] were used in our study, in reference to previous reports. The overall survival (OS) and relapse-free survival (RFS) were calculated using the Kaplan–Meier method, and the log-rank test was used to evaluate the statistical significance of the differences. A Cox proportional hazard regression analysis was used for the multivariate prognostic analysis. All statistical analyses were performed using the JMP software package, version 14.0 for Mac (SAS Institute Inc., Cary, NC, USA). A P-value lower than 0.050 was considered statistically significant.
Results
Tumor samples and patients
In total, 223 HCC tissues were analyzed. The median patient age was 71 years old (IQR: 65–77 years). The present cohort consisted of 182 males (82%) and 41 females (18%). Small tumors tended to be excluded from Project HOPE, since the removal of tumor tissue samples in patients with small tumors would make their pathological diagnosis difficult. The study included cases in which resection was performed for recurrence or remnant after previous treatment; 4 cases of transcatheter arterial embolization for rupture (2%), 4 cases of transcatheter arterial chemoembolization (TACE, 2%), 4 cases of raiofrequency ablation (RFA, 2%), 4 cases of RFA with TACE (2%), and 1 case of proton beam treatment (0%) were included. The median tumor size was 35 mm (range: 24–70 mm). The median follow-up period was 34.1 months; the 3-year OS was 81.5% and the median RFS after surgery was 27.6 months.
Overview of MMs in individual genes
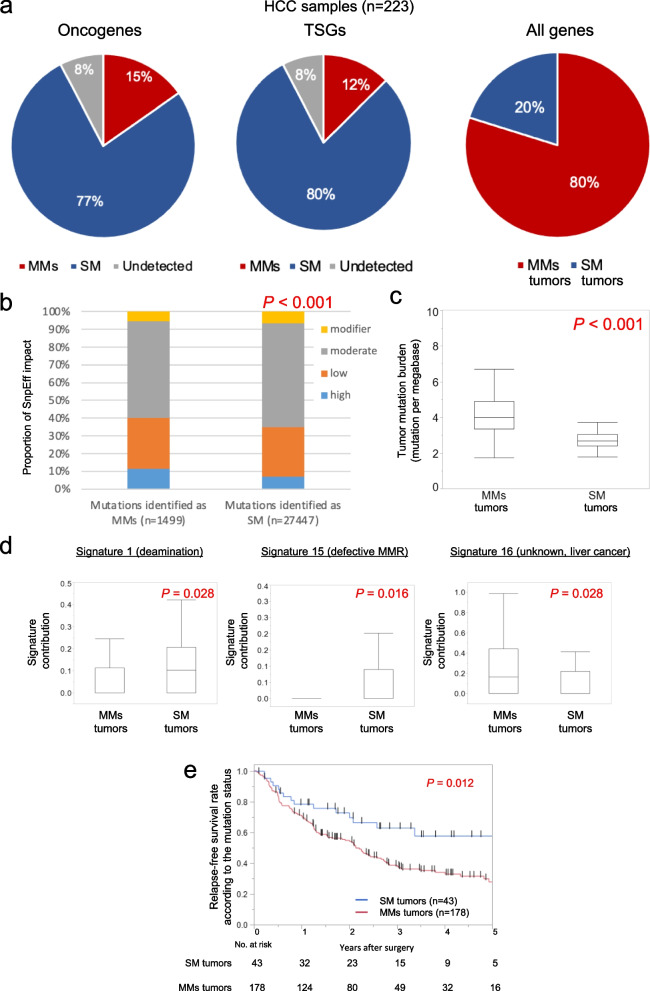
In the 223 samples, MMs within a oncogene were identified in 35 (15%) samples and MMs within a TSG were identified in 29 (12%) samples. For all genes, MMs within individual genes was identified in 178 samples (79.8%, MM tumors). All the remaining samples carried single mutation (20.2%, SM tumors, Fig. 1a). To compare the impact of genomic variant annotations and functional effect between mutations identified as SM and mutations identified as MMs, genomic variant were classified by SnpEffs into four levels in accordance with the variety of alterations as follows: high: nonsense mutation, frame-shift mutation and splice site mutation; moderate: missense mutation and in-frame indel; low: synonymous mutation; and modifier: untranslated region mutation. Mutations identified as MMs showed a higher fraction of high impact mutations than mutatios identified as SM; a larger impact on the protein structure caused by amino acid alterations were found in mutations identified as MMs than in mutations identified as SM (Fig. 1b). Furthermore, we evaluated the correlation between tumor mutation burden (TMB) and mutational signatures of the COSMIC database and MMs using deconstructSigs [20]. Supplementary Fig. 1 shows MMs, TMB, and signature contributions in samples with mutation count of > 50. The TMB was significantly higher in MMs tumors than that in SM tumors (Fig. 1c). As shown in Fig. 1d, three signature scores were significantly varied betweem MMs tumors and SM tumors. To assess the clinical impact on the presence of MMs in HCC, we performed prognostic analysis according to the presence of MMs. The RFS was significantly worse in the group with MMs tumors than in the group with SM tumors (P = 0.012, Fig. 1e). To consider the potential confounding of TMB with MMs, the prognostic analysis included the TMB. The distribution of TMB is shown in Supplementary Fig. 2. The cut-off value was set to 6.65 as 95% tile. The Cox proportional hazard analysis for RFS of all 223 patients who underwent resection identified MMs as an independent predictor for prognosis (hazard ratio, 1.72; 95% confidence interval, 1.01–3.17; P = 0.045) and showed that microvascular invasion (P < 0.001) was an independent factor that predicted poor survival (Table 1). No significant prognostic effect was found in the TMB.
Fig. 1.
Fraction of multiple mutations in individual genes. a Percentages of HCCs with single mutation (SM) and multiple mutations (MMs) according to the classification for oncogenes and tumor suppressor genes (TSGs). b Comparison of the impact of genomic variant annotations and functional effect by SnpEff. c A comparison of TMB between MMs tumors and SM tumors. The TMB was significantly higher in MMs tumors than that in SM tumors. d Comparisons of signature contributions between MMs tumors and SM tumors. Three signature scores were significantly varied betweem MMs tumors and SM tumors. e Prognostic analysis according to the presence of MMs. The RFS was significantly worse in the group with MMs tumors than in the group with SM tumors (P = 0.012, log-rank test)
Table 1.
Univariate and multivariate analyses for relapse-free survival after hepatectomy
| Variable | Univariate | Multivariate | |||||
|---|---|---|---|---|---|---|---|
| HR | 95% CI | P-value † | HR | 95% CI | P-value ‡ | ||
| Genetic signatures | |||||||
| Tumor mutation burden | |||||||
| Hyper | 146 | 1.41 | 0.97–2.10 | 0.079 | |||
| Hypo | 77 | ||||||
| Multiple mutations | |||||||
| Present | 178 | 1.97 | 1.19–3.53 | 0.012 | 1.72 | 1.01–3.17 | 0.045 |
| Absent | 45 | ||||||
| Clinicopathological factors | |||||||
| Sex | |||||||
| Male | 182 | 1.22 | 0.78–2.01 | 0.402 | |||
| Female | 41 | ||||||
| Age, years | |||||||
| ≥ 70 | 114 | 0.97 | 0.69–1.39 | 0.893 | |||
| < 70 | 109 | ||||||
| HBV or HCV | |||||||
| Negative | 112 | 0.92 | 0.65–1.31 | 0.655 | |||
| Positive | 111 | ||||||
| ICG-R15, % | |||||||
| ≥ 20 | 13 | 1.30 | 0.64–2.36 | 0.424 | |||
| < 20 | 210 | ||||||
| AFP, ng/ml | |||||||
| ≥ 200 | 48 | 1.40 | 0.91–2.09 | 0.106 | |||
| < 200 | 171 | ||||||
| PIVKA-II, mAU/ml | |||||||
| ≥ 100 | 121 | 1.84 | 1.27–2.69 | 0.001 | 1.34 | 0.88–2.06 | 0.177 |
| < 100 | 95 | ||||||
| Tumor size, mm | |||||||
| ≥ 30 | 132 | 1.68 | 1.16–2.46 | 0.006 | 1.23 | 0.84–1.96 | 0.257 |
| < 30 | 91 | ||||||
| Macrovascular invasion | |||||||
| Positive | 15 | 2.43 | 1.23–4.32 | 0.004 | 1.72 | 0.86–3.10 | 0.119 |
| Negative | 208 | ||||||
| Microvascular invasion | |||||||
| Positive | 75 | 2.26 | 1.58–3.22 | < 0.001 | 1.96 | 1.36–2.83 | < 0.001 |
| Negative | 148 | ||||||
| Liver damage | |||||||
| Liver cirrhosis | 88 | 1.36 | 0.95–1.94 | 0.085 | |||
| No cirrhosis | 135 | ||||||
Significant values are in bold
HR Hazard ratio, CI Confidence interval, HBV Hepatitis B virus, HCV Hepatitis C virus, ICG Indocyanine green, RFS Relapse-free survival, AFP Alpha fetoprotein
†Kaplan–Meier method. Significance was determined by the log-rank test
‡Multivariate survival analysis was performed using Cox’s proportional hazard model
Frequent MMs in a variety of oncogenes in HCC
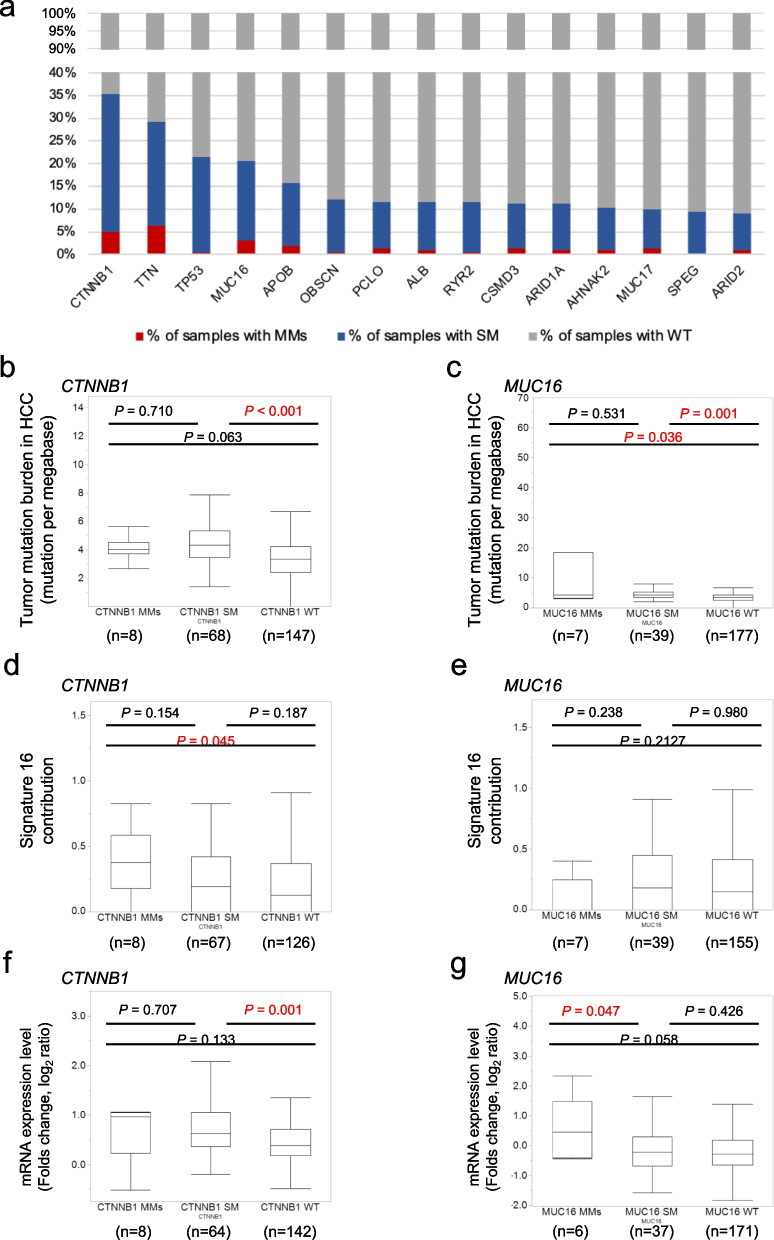
Figure 2a shows the percentages of samples with wild type, single mutations, and multiple mutations for 14 genes with ≥ 20 mutated samples in the present cohort (n = 223). MMs were frequently observed across a wide variety of genes; we found that 5% or more of the mutated samples carried MMs across 26 genes, particularly in MUC16 (15% of samples with mutation in the gene) and CTNNB1 (14%), suggesting that MMs within individual oncogenes is a relatively common phenomenon. We decided to focus on CTNNB1 and MUC16, which are recognized as being associated with cancer, for a further investigation. To assess the confounding of TMB with MMs, correlations between MMs in CTNNB1 (Fig. 2b) and MUC16 (Fig. 2c) and TMB were investigated. In both genes, significant differences in the TMB were found between samples with SM and samples with the wild-type gene (CTNNB1, P < 0.001; MUC16, P = 0.001), but no significant differences were found between SM and MMs (CTNNB1, P = 0.710, MUC16, P = 0.531). These findings suggest that MMs are not just a reflection of TMB but that there is selection pressure to accumulate mutations in specific genes. Therefore, we evaluated the mutational pattern of MMs in the genes. Using deconstructSigs [20], mutational signatures of the COSMIC database were investigated. The liver-cancer-specific signature 16 [21] was significantly higher in samples with MMs in CTNNB1 than samples with the wild-type CTNNB1, although no significant differences in the signature score between samples with SM in CTNNB1 and samples with the wild-type CTNNB1 (Fig. 2d). No significant differences in the signature 16 score between samples with MMs in MUC16 and samples with the wild-type MUC16 and between samples with SM in MUC16 and samples with the wild-type MUC16 were confirmed (Fig. 2e). The details of all mutations identified in MUC16, including protein change and DNA change are presented in Supplementary Table 1. The distribution of mutations and fraction of MMs for each position in CTNNB1 and MUC16 are shown in Supplementary Fig. 3. In CTNNB1, most mutations were located in major hotspots of exon 3. No significant difference of the frequency was observed between CTNNB1 SM tumors and CTNNB1 MMs tumors. In MUC16, mutations frequently located at exon 3 and there was no significant difference in the frequency between MUC16 SM and MUC16 MMs tumors. We investigated the allelic configuration of MMs by phasing from WES reads, which revealed that most MMs (83%) in CTNNB1 were present in cis. While all of the MMs in MUC16 was not located in a same amplicon, therefore the allelic configuration of MMs in MUC16 could not be investigated in the present study. Next, we investigated the impact of MMs on gene expression in CTNNB1 (Fig. 2f) and MUC16 (Fig. 2g). The gene expression raw data, including expression in tumors, expression in non-tumor areas as normal tissue, and fold-changes, in all cases are shown in Supplementary Table 2. In MUC16, MMs had larger alterations of gene expression; although there was no significant difference of expression between samples with wild-type MUC16 and samples with SM in MUC16, the expression in samples with MMs in MUC16 was significantly enhanced compared with samples with SM in MUC16 (P = 0.047). These results suggest that individually suboptimal mutations can confer enhanced oncogenic potential in combination as MMs. Based on the findings, for further investigation, we focused on MUC16 as a candidate oncogene to validate the impact of MMs.
Fig. 2.
Frequent MMs in a variety of oncogenes in HCC. a The percentages of samples with wild type, single mutation, and multiple mutations for 14 genes with 20 or more mutated samples in the present cohort (n = 223). Correlations between MMs in CTNNB1 (b) and MUC16 (c) and TMB were found between samples with SM and samples with the wild-type gene (CTNNB1, P < 0.001; MUC16, P = 0.001, Mann–Whitney U test) but not between SM and MMs (CTNNB1, P = 0.710, MUC16, P = 0.531, Mann–Whitney U test). The signature 16 was significantly higher in samples with MMs in CTNNB1 than samples with the wild-type CTNNB1, although no significant differences in the signature score between samples with SM in CTNNB1 and samples with the wild-type CTNNB1 (d). No significant differences in the signature 16 score according tot the mutational status in MUC16 (e). The impact of MMs on gene expression in CTNNB1 (f) and MUC16 (g)
Functional relevance of MMs in oncogenes
To assess the impact of MMs on phenotypes in cancer cell lines, an analysis of drug sensitivity screens in Cancer Cell Line Encyclopedia (CCLE) cell lines [22] was performed. Box plots (Supplementary Fig. 4) show sensitivity to regorafenib for 27 CCLE liver cancer cell lines, according to MUC16 mutational status. The results revealed that cells harboring in MMs in MUC16 exhibited a tendency of higher sensitivity to regorafenib than those with no or single MUC16 mutations, pointing to the potential value of MMs as a predictive marker for targeted therapies.
Clinical outcomes and MMs in MUC16
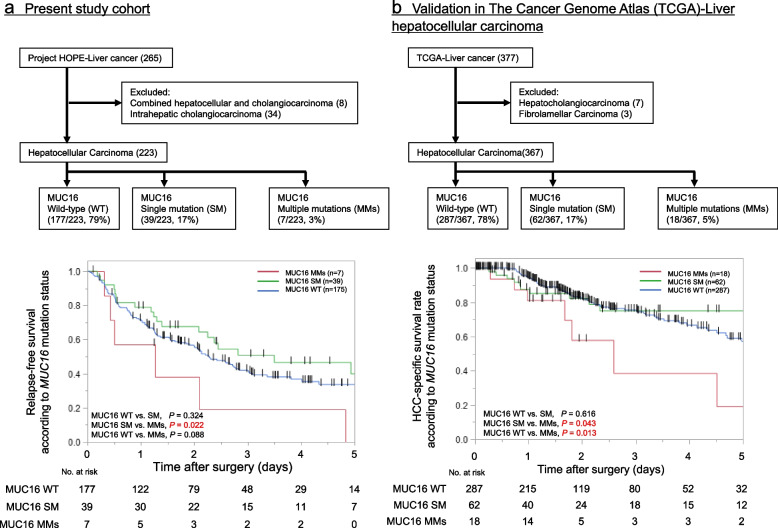
To assess the clinical impact of MMs in individual oncogenes, the clinicopathological factors according to mutational status in MUC16 were investigated (Table 2). MMs in MUC16 were associated with hepatitis C (P < 0.001), increased PIVKAII levels (P = 0.032) and macro vascular invasion (P = 0.041). Patient RFS was significantly worse in the group with MUC16 MMs than in the group with MUC16 SM (P = 0.022), although there was no significant difference between the group with MUC16 SM and the group with wild-type MUC16 (P = 0.324, Fig. 3a). Using TCGA data sets, we checked HCC-specific survival according to mutational status in MUC16 (Fig. 3b). No significant differences in Kaplan–Meier survival curves were observed between the group with MUC16 SM and the group with wild-type MUC16 (P = 0.616). Patient HCC-specific survival was significantly worse in the group with MUC16 MMs than in the group with MUC16 SM (P = 0.043) and in the group with wild-type MUC16 (P = 0.013).
Table 2.
Clinicopathological factors according to the number of MUC16 mutations
| Variable | Wild-type | Single mutation | Multiple mutations | P-value |
|---|---|---|---|---|
| N = 177 | N = 39 | N = 7 | ||
| Gender, N, (%) | ||||
| Male | 145 (82%) | 31 (79%) | 6 (86%) | < 0.001 |
| Female | 32 (18%) | 8 (21%) | 1 (14%) | |
| Age, years, (IQR) | 70 (64–76) | 74 (68–78) | 65 (64–73) | 0.022 |
| HBV or HCV, N, (%) | ||||
| HCV | 53 (30%) | 11 (28%) | 5 (71%) | < 0.001 |
| HBV | 34 (19%) | 8 (21%) | 0 (0%) | |
| NBNC | 90 (51%) | 20 (51%) | 2 (29%) | |
| ICG-R15, %, (IQR) | 9.5 (6.1–13.7) | 10.7 (7.5–15.7) | 13.4 (5.9–15.1) | 0.445 |
| AFP, ng/ml, (IQR) | 9.2 (3.7–144.1) | 5.2 (2.3–24.2) | 641.4 (7.4–883.7) | 0.107 |
| PIVKAII, mAU/ml, (IQR) | 116.0 (32.6–1495.0) | 414.0 (24.8–3537.5) | 5670.0 (160.0–37,800.0) | 0.032 |
| Tumor size, mm, (IQR) | 35 (22–65) | 36 (25–76) | 63 (33–78) | |
| Macrovascular invasion, N, (%) | ||||
| Positive | 12 (7%) | 1 (3%) | 2 (29%) | 0.041 |
| Microvascular invasion, N, (%) | ||||
| Positive | 58 (33%) | 15 (38%) | 2 (29%) | 0.761 |
Continuous variables expressed as median and interquartile range (IQR)
Significant values are in bold
HBV Hepatitis B virus, HCV Hepatitis C virus, ICG Indocyanine green, AFP Alpha fetoprotein, NBNC Non B non C
Fig. 3.
Prognostic impact of MMs in MUC16 and its validation in TCGA. The frequency of SM and MMs in MUC16 was 17% and 3% in our study and 17% and 5% in TCGA. a Patient RFS was significantly worse in the group with MUC16 MMs than in the group with MUC16 SM (P = 0.022), although there was no significant difference between the group with MUC16 SM and the group with wild-type MUC16 in our cohort (P = 0.324, log-rank test). b In TCGA data sets, no significant differences in HCC-specific survival were observed between the group with MUC16 SM and the group with wild-type MUC16 (P = 0.616). Patient HCC-specific survival was significantly worse in the group with MUC16 MMs than in the group with MUC16 SM (P = 0.043) and in the group with wild-type MUC16 (P = 0.013)
Discussion
In this study, we performed comprehensive WES for 223 HCCs. The results revealed that a high frequency of mutations in genes did not always correspond to a high frequency of MMs. Instead, MMs were revealed to accumulate selectively in specific genes. The signature of mutations identified as MMs, including mutation type and functional impact, also varied compared with that of mutations identified as SM. Furthermore, the GEP results implied that MMs had a greater impact on gene expression than SMs in some oncogenes, a trend was identified that MMs led to additional up-regulation (gain-of-function) of gene expression. Consequently, MMs are not just a reflection of mutation burden but occur in specific genes and pathways and thus contribute to carcinogenesis and/or acquisition of malignant potential in HCC.
The overall landscape of MMs was recently reported through a pan-cancer analysis of 60,954 cancer samples [5]. The study identified that oncogenic MMs were a relatively common driver event and thus MMs provide a novel underlying mechanism for cancer development. These observations reinforce the idea that MMs in the same oncogene cooperate to potentiate tumor-promoting activity. These findings also indicate the potential usefulness of MMs as a biomarker and a target for molecular-targeted therapy. However, the signature and clinical relevance of MMs in HCCs had remained unclear. In the present study, detailed clinical information led to the validation of the clinical significance of MMs in HCCs, and the present results identified MMs as an independent predictor for prognosis in HCC. To date, there has been no report on the clinical relevance of MMs, and therefore the present study provides further evidence of the prognostic impact of MMs.
The clinical impact of MMs in some genes in a sample led us to assess the clinical impact of MMs in individual oncogenes. Aberrant activation of WNT/β-catenin signaling is a driving molecular event in a wide range of tumors, including HCCs [23]. Somatic missense mutations in exon 3 of CTNNB1 are frequently reported in HCCs (10.0%–32.8% in genome-wide sequencing studies) [3, 24]. Consistent with previous reports, we found that mutations in CTNNB1 were frequently identified in HCC and MMs in CTNNB1 were also frequently found in 14% (11/79) of samples with at least one mutation. However, a significant prognostic difference between SM and MMs in CTNNB1 was not identified (data not shown). The conflicting prognostic impact of mutated CTNNB1 due to the bilateral nature was reported. HCC cases with the existence of an interaction between WNT activation and TGF-beta activation show poor survival, whereas HCCs harboring mutant CTNNB1 show generally favorable prognosis [25]. Therefore, for further investigation, we focused on MUC16, in which MMs were identified in 15.2% (7/46) of mutated samples, as a candidate oncogene to validate the impact of MMs.
MUC16 encodes a protein also known as ovarian carcinoma antigen CA125, which is clinically recommended as a screening biomarker for ovarian cancer. Although CA125 is not a general biomarker for HCC, high preoperative serum CA125 levels have been reported to serve as an independent prognostic factor for the OS and RFS in HCC [26]. MUC16 is an important membrane protein that maintains the normal cellular function and is involved in cancer development. Knockdown of MUC16 in HCC cell lines revealed that MUC16 plays a suppressive role in migration and invasion [26]. Furthermore, it has been reported that MUC16 promotes proliferation and invasion via activation of the JAK2/STAT3 pathway in cervical cancer [27]. Thus, alterations in MUC16 enhance the tumor invasive potential. In contrast, an increase in CA125 in serum was associated with an increase in malignancy and mortality, suggesting that mutations in MUC16 may affect the structural stability of MUC16, causing the tumor-derived protein to shed from the surface of hepatocarcinoma cells. Although the differences between SMs and MMs require further investigation, this may explain the association between mutations in MUC16 and high tumor marker levels and vascular invasion. The present results demonstrated that the presence of MMs in MUC16 was associated with higher tumor markers and vascular invasion. Patient RFS was significantly worse in the group with MMs than in the group with SM. The findings support the idea that MMs in the same oncogene cooperate to potentiate tumor-promoting activity.
The findings that the accumulation of MMs in specific oncogenes led us to assess the impact of MMs on phenotypes in cancer cell lines. Analysis of drug sensitivity screens in CCLE cell lines [22] revealed that cells harboring in MMs in MUC16 exhibited a higher sensitivity to regorafenib than those with no or single MUC16 mutations, indicating the potential value of MMs as a predictive marker for targeted therapies. Similarly, a previous study [5] reported that cells harboring MMs in PIK3CA exhibited a higher sensitivity to PI3K inhibitors compared with those with no or single PIK3CA mutations, suggesting that MMs may be useful as predictive markers for targeted therapies. The correlation between mutational status in an oncogene and the sensitivity to the targeted drug for the molecule should be further explored in future studies.
During recent years, new immune-modulatory agents have been introduced for HCC treatment, eventually leading to the clinical breakthrough of immune checkpoint inhibitors (ICIs) targeting programmed death-1 (PD-1), programmed death-ligand 1 (PD-L1), or cytotoxic T lymphocyte antigen-4 (CTLA-4) [28]. The TMB, which correlates with MMs, has received increasing attention owing to its potential to estimate the efficacy of the response to ICIs. The present results indicated that MMs were associated with the TMB to some extent; however, MMs were not just a reflection of mutation burden but occurred in specific genes and pathways through a selection mechanism to contribute to carcinogenesis and/or acquisition of malignant potential. These findings support a potential association between MMs and sensitivity to ICIs. Although we cannot draw any conclusions from these results, it may be worthwhile to examine MMs and the sensitivity to ICIs for future perspectives.
This study had several limitations. First, the number of MMs in each specific gene was relatively small, and therefore investigation of the clinical significance of MMs in each gene could not be conducted because of under power statistics. Ideally, the clinical significance of MMs in specific candidate genes will be investigated to establish a biomarker for targeted therapy. Second, the previous study [5] reported that the proportion of MMs in cis was particularly high (86%) in oncogenes with MMs. Consistent with the previous reports, most MMs (83%) in CTNNB1 were present in cis in the present study. However, all of the MMs in MUC16 was not located in a same amplicon, therefore the allelic configuration of MMs in MUC16 could not be investigated.
Conclusions
MMs are a relatively common event that selectively occurs in specific oncogenes and is involved in aggressive malignant behavior. This is the study investigating the clinical significance of MMs in patients with HCC and provide important insights into the development of personalized treatment strategies for HCCs.
Supplementary Information
Additional file 1: Supplementary Fig. 1. Mutation signature in the sample with multiple mutations. Supplementary Fig. 2. The distribution of TMB in HCCs. Supplementary Fig. 3. The mutational mapping of MMs in CTNNB1 and MUC16. Supplementary Fig. 4. Drug sensitivity screens in Cancer Cell Line Encyclopedia.
Additional file 2: Supplementary Table 1. Mutations identified in MUC16.
Additional file 3: Supplementary Table 2. Gene expression raw data.
Acknowledgements
Not applicable
Abbreviations
- CCLE
Cancer Cell Line Encyclopedia
- CTLA-4
Cytotoxic T lymphocyte antigen‐4
- GEP
Gene expression profiling
- HOPE
High-tech Omics-based Patient Evaluation
- ICIs
Immune checkpoint inhibitors
- MMs
Multiple mutations
- PD-1
Programmed death-1
- PD-L1
Programmed death-ligand 1
- OS
Overall survival
- RFS
Relapse-free survival
- WES
Whole-exome sequencing
Authors’ contributions
Authorship will be based on the Recommendations of the International Committee of Medical Journal Editors (ICMJE, www.icmje.org). TI has drafted the work; YO and KO have substantively revised the manuscript; KU, TS, TI, YY, RA, KO, Shimpei Otsuka, Sumiko Ohnami, TN, KH, TS, KU, and KY have made substantial contributions to the design of the study; KY, KO, TI, and YO have made substantial contributions to the conception and design of the study and have substantively revised the manuscript. All authors have read and approved the final manuscript. All authors have agreed both to be personally accountable for the author’s own contributions and to ensure that questions related to the accuracy or integrity of any part of the work, even ones in which the author was not personally involved, are appropriately investigated, resolved, and the resolution documented in the literature.
Funding
Not applicable.
Availability of data and materials
Data for this study is confidential patient information regulated by the IRB of the institution. The datasets generated and/or analyzed during the current study are available in the National Bioscience Database Center repository (accession no. hum0127; https://humandbs.biosciencedbc.jp/en/).
Declarations
Ethics approval and consent to participate
We used archived tissue samples for this study that were obtained with patient consent at Shizuoka Cancer Center. Written informed consent was obtained from all participants. The study was approved by the ethics committee of Shizuoka Cancer Center (approval no. 25–33). The study protocol conforms to the ethics guidelines of the 1975 Declaration of Helsinki.
Consent for publication
Not applicable.
Competing interests
The authors have no conflicts of interest to declare.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Neuveut C, Wei Y, Buendia MA. Mechanisms of HBV-related hepatocarcinogenesis. J Hepatol. 2010;52(4):594–604. doi: 10.1016/j.jhep.2009.10.033. [DOI] [PubMed] [Google Scholar]
- 3.Shibata T, Aburatani H. Exploration of liver cancer genomes. Nat Rev Gastroenterol Hepatol. 2014;11(6):340–349. doi: 10.1038/nrgastro.2014.6. [DOI] [PubMed] [Google Scholar]
- 4.Nault JC, Mallet M, Pilati C, Calderaro J, Bioulac-Sage P, Laurent C, Laurent A, Cherqui D, Balabaud C, Zucman-Rossi J. High frequency of telomerase reverse-transcriptase promoter somatic mutations in hepatocellular carcinoma and preneoplastic lesions. Nat Commun. 2013;4:2218. doi: 10.1038/ncomms3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saito Y, Koya J, Araki M, Kogure Y, Shingaki S, Tabata M, McClure MB, Yoshifuji K, Matsumoto S, Isaka Y, et al. Landscape and function of multiple mutations within individual oncogenes. Nature. 2020;582(7810):95–99. doi: 10.1038/s41586-020-2175-2. [DOI] [PubMed] [Google Scholar]
- 6.Garraway LA, Lander ES. Lessons from the cancer genome. Cell. 2013;153(1):17–37. doi: 10.1016/j.cell.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 7.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamaguchi K, Urakami K, Ohshima K, Mochizuki T, Akiyama Y, Uesaka K, Nakajima T, Takahashi M, Tamai S, Kusuhara M. Implementation of individualized medicine for cancer patients by multiomics-based analyses-the Project HOPE. Biomed Research. 2014;35(6):407–412. doi: 10.2220/biomedres.35.407. [DOI] [PubMed] [Google Scholar]
- 9.Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, Kovatich AJ, Benz CC, Levine DA, Lee AV, et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018;173(2):400–416 e411. doi: 10.1016/j.cell.2018.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okamura Y, Ito T, Sugiura T, Mori K, Uesaka K. Anatomic versus nonanatomic hepatectomy for a solitary hepatocellular carcinoma : a case-controlled study with propensity score matching. J Gastrointest Surg. 2014;18(11):1994–2002. [DOI] [PubMed]
- 11.Hatakeyama K, Ohshima K, Nagashima T, Ohnami S, Ohnami S, Serizawa M, Shimoda Y, Maruyama K, Akiyama Y, Urakami K, et al. Molecular profiling and sequential somatic mutation shift in hypermutator tumours harbouring POLE mutations. Sci Rep. 2018;8(1):8700. doi: 10.1038/s41598-018-26967-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagashima T, Shimoda Y, Tanabe T, Naruoka A, Saito J, Serizawa M, Ohshima K, Urakami K, Ohnami S, Ohnami S, et al. Optimizing an ion semiconductor sequencing data analysis method to identify somatic mutations in the genomes of cancer cells in clinical tissue samples. Biomed Res. 2016;37(6):359–366. doi: 10.2220/biomedres.37.359. [DOI] [PubMed] [Google Scholar]
- 13.Ohshima K, Hatakeyama K, Nagashima T, Watanabe Y, Kanto K, Doi Y, Ide T, Shimoda Y, Tanabe T, Ohnami S, et al. Integrated analysis of gene expression and copy number identified potential cancer driver genes with amplification-dependent overexpression in 1,454 solid tumors. Sci Rep. 2017;7(1):641. doi: 10.1038/s41598-017-00219-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sondka Z, Bamford S, Cole CG, Ward SA, Dunham I, Forbes SA. The COSMIC Cancer Gene Census: describing genetic dysfunction across all human cancers. Nat Rev Cancer. 2018;18(11):696–705. doi: 10.1038/s41568-018-0060-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, Rudolph JE, Yaeger R, Soumerai T, Nissan MH et al. OncoKB: a precision oncology knowledge base. JCO Precis Oncol. 2017;2017:1–16. [DOI] [PMC free article] [PubMed]
- 16.Nagashima T, Yamaguchi K, Urakami K, Shimoda Y, Ohnami S, Ohshima K, Tanabe T, Naruoka A, Kamada F, Serizawa M, et al. Japanese version of the Cancer Genome Atlas, JCGA, established using fresh frozen tumors obtained from 5143 cancer patients. Cancer Sci. 2020;111(2):687–699. doi: 10.1111/cas.14290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Omagari K, Ohba K, Kadokawa Y, Hazama H, Masuda J, Kinoshita H, Matsuo I, Ohnita K, Mizuta Y, Hayashida K, et al. Comparison of the grade evaluated by “Liver damage” of Liver Cancer Study Group of Japan and Child-Pugh classification in patients with hepatocellular carcinoma. Hepatol Res. 2006;34(4):266–72. [DOI] [PubMed]
- 18.Zhang J, Chen G, Zhang P, Zhang J, Li X, Gan D, Cao X, Han M, Du H, Ye Y. The threshold of alpha-fetoprotein (AFP) for the diagnosis of hepatocellular carcinoma: a systematic review and meta-analysis. PLoS ONE. 2020;15(2):e0228857. doi: 10.1371/journal.pone.0228857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kitai S, Kudo M, Izumi N, Kaneko S, Ku Y, Kokudo N, Sakamoto M, Takayama T, Nakashima O, Kadoya M, et al. Validation of three staging systems for hepatocellular carcinoma (JIS score, biomarker-combined JIS score and BCLC system) in 4,649 cases from a Japanese nationwide survey. Dig Dis. 2014;32(6):717–724. doi: 10.1159/000368008. [DOI] [PubMed] [Google Scholar]
- 20.Rosenthal R, McGranahan N, Herrero J, Taylor BS, Swanton C. DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol. 2016;17:31. doi: 10.1186/s13059-016-0893-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Letouzé E, Shinde J, Renault V, Couchy G, Blanc J-F, Tubacher E, Bayard Q, Bacq D, Meyer V, Semhoun J et al. Mutational signatures reveal the dynamic interplay of risk factors and cellular processes during liver tumorigenesis. Nat Commun. 2017;8(1):1–13. [DOI] [PMC free article] [PubMed]
- 22.Ghandi M, Huang FW, Jané-Valbuena J, Kryukov GV, Lo CC, McDonald ER, 3rd, Barretina J, Gelfand ET, Bielski CM, Li H, et al. Next-generation characterization of the cancer cell line encyclopedia. Nature. 2019;569(7757):503–508. doi: 10.1038/s41586-019-1186-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Polakis P. Wnt signaling and cancer. Genes Dev. 2000;14(15):1837–1851. doi: 10.1101/gad.14.15.1837. [DOI] [PubMed] [Google Scholar]
- 24.de La Coste A, Romagnolo B, Billuart P, Renard CA, Buendia MA, Soubrane O, Fabre M, Chelly J, Beldjord C, Kahn A, et al. Somatic mutations of the beta-catenin gene are frequent in mouse and human hepatocellular carcinomas. Proc Natl Acad Sci USA. 1998;95(15):8847–8851. doi: 10.1073/pnas.95.15.8847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoshida Y, Nijman SM, Kobayashi M, Chan JA, Brunet JP, Chiang DY, Villanueva A, Newell P, Ikeda K, Hashimoto M, et al. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Can Res. 2009;69(18):7385–7392. doi: 10.1158/0008-5472.CAN-09-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang Y, Huang X, Zeng J, Lin J. Knockdown of MUC16 (CA125) enhances the migration and invasion of hepatocellular carcinoma cells. Front Oncol. 2021;11:667669. doi: 10.3389/fonc.2021.667669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen H, Guo M, Wang L, Cui X. MUC16 facilitates cervical cancer progression via JAK2/STAT3 phosphorylation-mediated cyclooxygenase-2 expression. Genes Genomics. 2020;42(2):127–133. doi: 10.1007/s13258-019-00885-9. [DOI] [PubMed] [Google Scholar]
- 28.Herbst RS, Baas P, Kim DW, Felip E, Pérez-Gracia JL, Han JY, Molina J, Kim JH, Arvis CD, Ahn MJ, et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet (London, England) 2016;387(10027):1540–1550. doi: 10.1016/S0140-6736(15)01281-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplementary Fig. 1. Mutation signature in the sample with multiple mutations. Supplementary Fig. 2. The distribution of TMB in HCCs. Supplementary Fig. 3. The mutational mapping of MMs in CTNNB1 and MUC16. Supplementary Fig. 4. Drug sensitivity screens in Cancer Cell Line Encyclopedia.
Additional file 2: Supplementary Table 1. Mutations identified in MUC16.
Additional file 3: Supplementary Table 2. Gene expression raw data.
Data Availability Statement
Data for this study is confidential patient information regulated by the IRB of the institution. The datasets generated and/or analyzed during the current study are available in the National Bioscience Database Center repository (accession no. hum0127; https://humandbs.biosciencedbc.jp/en/).