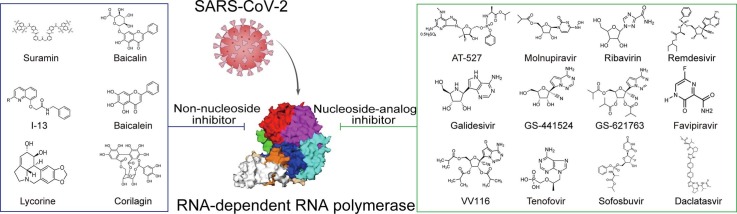
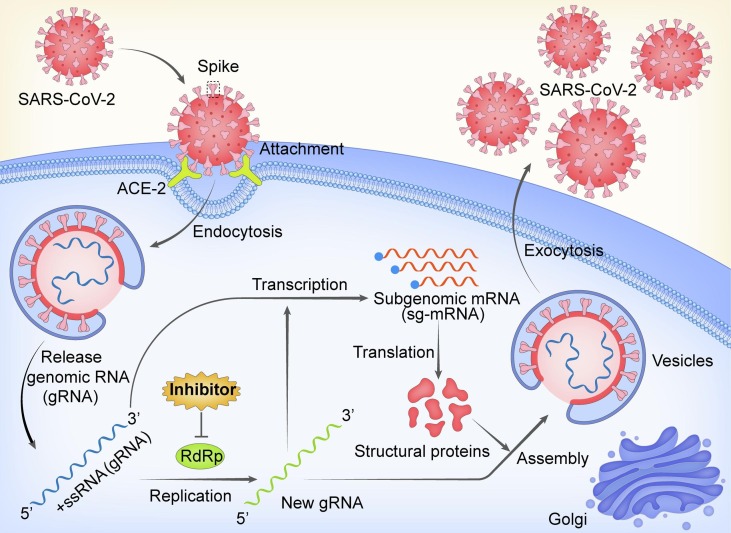
Graphical abstract
Keywords: COVID-19, SARS-CoV-2, Antiviral, RdRp, Inhibitors
Abbreviations: C-RTC, central replication-transcription complex; DENV, dengue virus; dpi, days post-infection; E-RTC, elongation replication-transcription complex; FDA, Food and Drug Administration; FPV, favipiravir; HBV, Hepatitis B virus; HCV, Hepatitis C virus; HIV, Human Immunodeficiency virus; IMPDH, inosine monophosphate dehydrogenase; MP, monophosphate; NAIs, nucleoside analog inhibitors; NHC, β-D-N4-hydroxycytidine; NiRAN, nidovirus RdRp-associated nucleotidyltransferase; NNIs, non-nucleoside inhibitors; nsp, nonstructural protein; NTP, nucleoside triphosphate; ORF, open reading frame; RBV, ribavirin; RdRp, RNA-dependent RNA polymerase; RTC, replication-transcription complex; RDV, remdesivir; SOF, sofosbuvir; TAF, tenofovir alafenamide; TDF, tenofovir disoproxil fumarate; UMP, uridine monophosphate; VOC, variant of concern; ZIKV, Zika virus
Abstract
The highly transmissible variants of SARS-CoV-2, the causative pathogen of the COVID-19 pandemic, bring new waves of infection worldwide. Identification of effective therapeutic drugs to combat the COVID-19 pandemic is an urgent global need. RNA-dependent RNA polymerase (RdRp), an essential enzyme for viral RNA replication, is the most promising target for antiviral drug research since it has no counterpart in human cells and shows the highest conservation across coronaviruses. This review summarizes recent progress in studies of RdRp inhibitors, focusing on interactions between these inhibitors and the enzyme complex, based on structural analysis, and their effectiveness. In addition, we propose new possible strategies to address the shortcomings of current inhibitors, which may guide the development of novel efficient inhibitors to combat COVID-19.
1. Introduction
At the end of 2019, a severe acute respiratory syndrome caused by a novel coronavirus (SARS-CoV-2) initially emerged in China and later spread rapidly around the world, causing a global pandemic COVID-19 [1]. As of September 2022, the World Health Organization (WHO) had recorded more than 611 million confirmed cases of COVID-19 globally and more than 6 million related deaths (https://covid19.who.int/).
SARS-CoV-2, the causative pathogen of COVID-19, is a member of betacoronavirus genus of the Coronaviridae family, which possesses a single-stranded, positive-sense RNA that shares ∼96% sequence identity with bat-origin RaTG13 CoV and ∼80% sequence identity with SARS-CoV. The 30-kb genome of this novel coronavirus contains 14 open reading frames (ORFs) that encode at least 27 proteins. The 3′ end of the genome encodes four typical structural proteins of coronaviruses, namely, spike (S), envelope (E), membrane (M), and nucleocapsid (N) protein, as well as eight accessory proteins, which have roles in viral pathogenesis and host immunity suppression. After being primed by the host serine protease TMPRSS2, the virus can enter host cells via attachment of its spike protein with the host cell angiotensin converting enzyme II (ACE2) receptor [2], [3]. In addition, the two leading open reading frames at the 5′ end, ORF 1a and ORF 1b, express two replication polyproteins, namely pp1a and pp1ab, which are further hydrolyzed into 16 nonstructural proteins (nsp1-16) [4]. Among those, nsp12, also known as RNA-dependent RNA polymerase (RdRp), is involved in directing viral genome replication and transcription [5] ( Fig. 1 ).
Fig. 1.
The life cycle of SARS-CoV-2 via RdRp. The RdRp is involved in virus genomic RNA (gRNA) and subgenomic mRNA (sg-mRNA) synthesis. Inhibitors targeting RdRp are promising candidates for antiviral therapy.
Different variants of SARS-CoV-2 are continually emerging, bringing new waves of infection worldwide. Following the D614G, Beta/Gamma, and Delta variants of concern (VOC), the Omicron variant is responsible for the largest wave pandemic at present [6]. All of these new variants contain mutations in the spike protein and are characterized by enhanced transmissibility or virulence, decreased sensitivity to current vaccines or neutralizing monoclonal antibodies (mAbs), and the ability to evade detection [7]. Given the concern over the immune escape by the variants of current vaccines, developing broad-spectrum and effective drugs against SARS-CoV-2 and its variants remains a priority strategy for treatment and prevention.
RdRp, a critical enzyme in the viral life cycle, is a prime target for developing effective antivirals against SARS-CoV-2 and many variants, because it is the most conserved protein across coronaviruses and lacks a human homolog. Therefore, we summarize the recent advances in the structural understanding of SARS-CoV-2 RdRp and the interaction mechanisms and effects of inhibitors targeting it, to help guide future research and drug discovery.
2. Function and structure of SARS-CoV-2 RdRp
After entry into the host cells, SARS-CoV-2 employs a so-called replication and transcription complex (RTC), assembled by nonstructural proteins, to synthesize the negative-strand template, the positive-strand genomic RNA and subgenomic mRNAs of virus [8]. The central component of the RTC is the highly conserved catalytic subunit nsp12 (RdRp), which is responsible for the synthesis of viral RNA. However, nsp12 alone displays extremely low enzymatic activity and needs to be stimulated with two accessory proteins (nsp7 and nsp8). Thus, the nsp12-nsp7-nsp8 subcomplex (known as holoenzyme RdRp, holo-RdRp) is defined as the minimal core component for mediating the synthesis of coronavirus RNA [9].
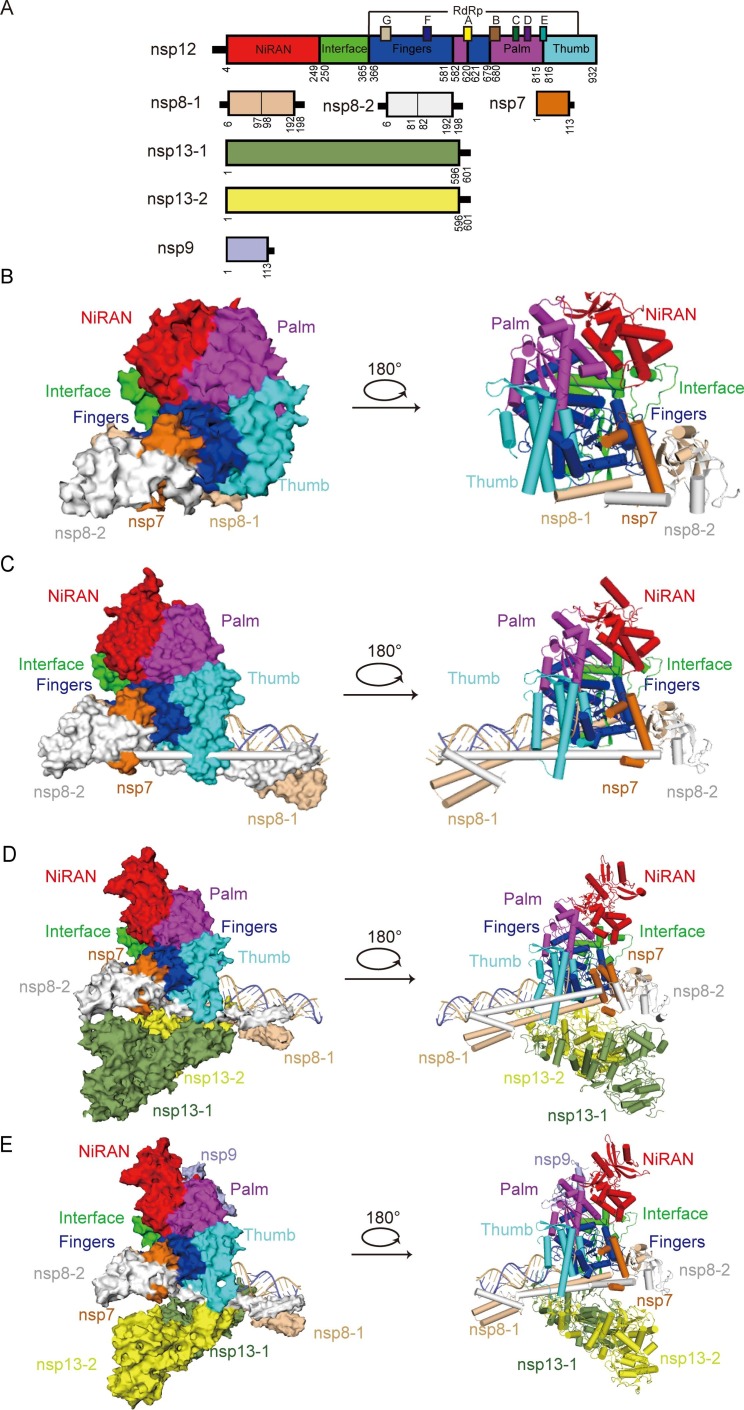
The structures of nsp12 in complex with template-primer RNA (central RTC, C-RTC), a mini RTC composed of helicase (nsp13) and C-RTC for the elongation of nucleic acid (elongation RTC, E-RTC), and an extended E-RTC composed of a single RNA-binding protein (nsp9) have been determined [8], [10] ( Fig. 2 A). Structural information about the SARS-CoV-2 core polymerase complex (nsp12-nsp7-nsp8) suggests that the nsp12 subunit contains an N-terminal NiRAN domain, an interface domain, a C-terminal RdRp domain, and a previously undefined β-hairpin motif. Among these, the RdRp domain adopts a typical right-hand configuration, composed of the palm, finger, and thumb subdomains, to form a catalytic chamber, in which the template-directed RNA synthesis takes place [11]. Within the core RdRp domain, the active site is subdivided into seven conserved motifs (A–G), mediating template-directed RNA synthesis [12]. The nsp7-nsp8 heterodimer binds the thumb subdomain of RdRp, and the second nsp8 (nsp8-2) subunit clamps the top region of the finger subdomain of RdRp, forming additional interactions with the interface domain ( Fig. 2 B). The partial double-stranded template-primer RNA is embraced by the palm-finger-thumb subdomains, and two nsp8 molecules promote the polymerase activity by stabilizing existing RNA through the interactions between positively charged residues of their long N-terminal helical extensions and the RNA backbone ( Fig. 2 C). Two nsp13 protomers, namely nsp13F (fingers) and nsp13T (thumb), bind to the plane region formed by the nsp12 and two nsp8 protomers ( Fig. 2 D). The conformation of nsp13F is conserved, which stabilizes the architecture of RTC. However, nsp13T, which channels the unpaired 5′ extension of the RNA template to the RdRp active site in E-RTC, moves down to the template-primer RNA clamped by nsp8-1 and nsp8-2 upon nsp9 binding, to facilitate the viral mRNA synthesis ( Fig. 2 E). The NiRAN domain of nsp12 at the back side of RdRp, which is absent in RNA viruses outside the order Nidovirales, possesses guanylyltransferase activity, catalyzing the formation of the cap core structure (GpppA). Meanwhile, nsp9 binds tightly to NiRAN, allowing the nsp9 N terminus to insert into the catalytic center of nsp12 NiRAN, which then inhibits the activity [13], [14].
Fig. 2.
Structure of SARS-CoV-2 replication-transcription complex (RTC) (A) The schematic diagram for the domain organization of each component in the RTC, containing nsp12, nsp7, two copies of nsp8 (nsp8-1 and nsp8-2), two copies of nsp13 (nsp13-1 and nsp13-2) and nsp9. The interdomain borders are labeled with residue numbers. In nsp12, the conserved sequence motifs A-G are depicted. The components and domains are colored as follows: nsp12 NiRAN, red; nsp12 Interface, green; nsp12 Fingers, blue; nsp12 Palm, magenta; nsp12 Thumb, cyan; nsp7, orange; nsp8-1, wheat; nsp8-2, white; nsp13-1, split-pea; nsp13-2, yellow; nsp9, light blue. The color coding corresponds to the figures throughout this manuscript. (B) Cryo-EM densities (left) and cartoon representations (right) of SARS-CoV-2 apo RdRp complex (nsp7-nsp82-nsp12) in different views (PDB ID:7BW4). (C) Cryo-EM densities (left) and cartoon representations (right) of SARS-CoV-2 polymerase in complexes with RNA (nsp7-nsp82-nsp12-RNA, central RTC) in different views (PDB ID:6YYT). The RNA is depicted with the template strand in slate and the primer/product strand in light orange. (D) Cryo-EM densities (left) and cartoon representations (right) of SARS-CoV-2 central RTC with nsp13 helicases (nsp7-nsp82-nsp12-nsp132-RNA, elongation RTC) in different views (PDB ID:6XEZ). (E) Cryo-EM densities (left) and cartoon representations (right) of SARS-CoV-2 elongation RTC with nsp9 (nsp7-nsp82-nsp12-nsp132-nsp9-RNA, extended E-RTC) in different views (PDB ID:7CYQ). Nsp9 binds to the catalytic center of the nsp12 (RdRp) NiRAN domain.
3. Inhibitors of RdRp
According to the structures and modes of action, inhibitors of RdRp fall into two main categories, one is nucleoside analog inhibitors acting at the substrate site, the other one is non-nucleoside inhibitors interacting with allosteric binding sites [15].
3.1. Nucleoside analog inhibitors (NAIs)
Nucleotide analogs are considered the most promising inhibitors of RdRp. NAIs are generally classified into three major classes: mutagenic nucleosides, obligate chain terminators and delayed chain terminators. To compete with the natural substrates of RdRp, NAIs require activation to the pharmacologically active triphosphate form (NTP) by host kinases, which lead them to be incorporated into the nascent viral RNA with the energy provided by the hydrolysis and release of the pyrophosphate group of the NTP. Once incorporated, NAIs then terminate RNA synthesis or induce mutations in progeny RNA. Since the pandemic broke out, many NAIs have been tested against SARS-CoV-2, and some structures of them have been solved to clarify their inhibition mechanisms.
3.1.1. Remdesivir
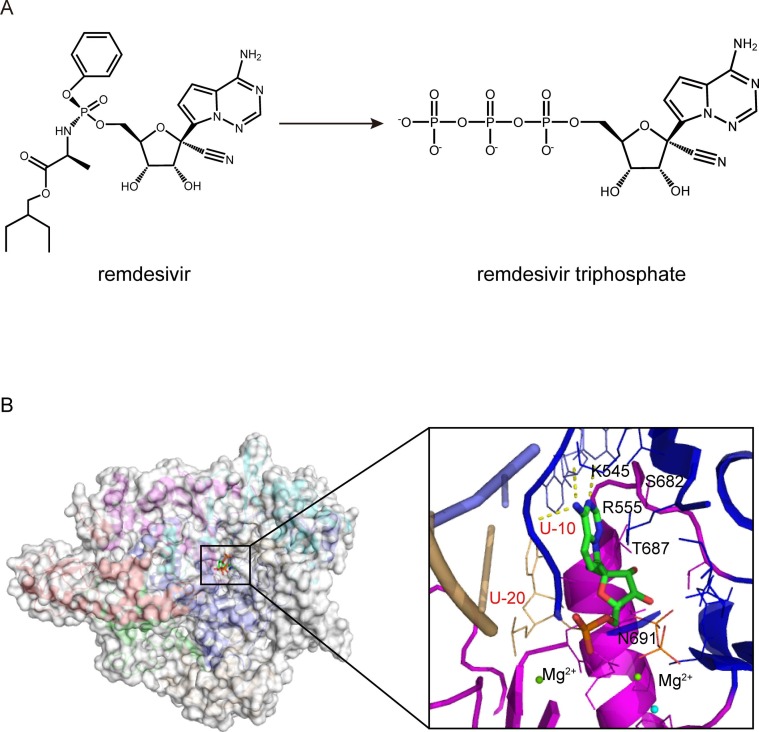
Remdesivir (RDV, formerly GS-5734) is a prodrug of a 1′-cyano-modified adenosine nucleotide analog, which was developed initially to combat Ebola virus in 2014 [16] ( Fig. 3 A). Nevertheless, the antiviral activity of RDV is considerably broad-spectrum and includes members of several virus families, including filoviruses (e.g., Ebola) and coronaviruses (e.g., SARS-CoV and MERS-CoV) [17], [18]. Upon cellular uptake, RDV is metabolized into the adenine metabolite and further processed into the monophosphate derivative (RDV-MP) and ultimately into the active nucleoside triphosphate (RDV-TP) derivative, which is the substrate competing with ATP for incorporation by the viral RdRp, thereby resulting in inhibiting viral RNA synthesis [19].
Fig. 3.
Cryo-EM structure of remdesivir bound to the RdRp complex of SARS-CoV-2(PDB ID:7BV2). (A) Chemical structures of remdesivir and remdesivir triphosphate. (B) Overall view of nsp12-nsp7-nsp8 in complex with template-primer RNA and remdesivir monophosphate (RDV-MP) and close-up view of the interactions between the SARS-CoV-2 RdRp active site and remdesivir. The potential polar contacts are shown as yellow dashed lines. The protein and RNA are shown in cartoons with the key residues involved in remdesivir interaction shown in lines and labeled in black and red, respectively. The RDV-MP is shown as a stick model.
RDV is the first candidate that has received much attention for COVID-19 therapy, demonstrating efficacy against SARS-CoV-2 and related coronaviruses both in vitro and in vivo animal models [20], [21], [22]. Apart from preclinical tests, the possible clinical use of RDV to fight against coronavirus has been supported. It reduced early-stage mortality and the need for high-flow oxygen supplementation as well as invasive mechanical ventilation among hospitalized COVID-19 patients [23]. In addition, an improvement in the primary endpoint was reported for patients receiving RDV, with a 31% faster time to recovery compared with the control, according to an interim analysis in the Adaptive COVID-19 Treatment Trial (NCT04280705) [24], [25]. To date, RDV is the first and only one FDA-authorized drug to treat COVID-19 patients in such severe emergency [26].
Furthermore, the structure of the RdRp-RNA-RDV complex has been solved [11], [13], [27] ( Fig. 3 B). RDV can be efficiently incorporated and allows subsequent catalysis. As an adenosine monophosphate analog, the RDV-MP molecule is incorporated into the primer strand and forms a base-stacking interaction with two hydrogen bonds with the uridine base from the template strand. In addition, RDV-MP interacts with residues K545 and R555 in motif F, which stabilize the incoming nucleotide in the correct position for catalysis [11]. Compared with other classical chain terminators, favorable reactivity for the RDV-TP over ATP and delayed chain termination of nascent viral RNA at the i + 3 position (where i corresponds to the position of the first incorporated RDV-MP) are observed to be the crucial features of the mechanism of inhibition with SARS-CoV-2 RdRp complexes [28], [29]. Structural analysis and enzymological characterization further suggested that the 1′-CN substituent of the incorporated RDV-MP will sterically clash with the sidechain of the S861 poised near the product strand backbone at the −4 position in the thumb domain of nsp12 upon further chain elongation, and probably lead to a significant distortion of the RNA position, hampering the translocation of RNA to the -4 position. Failure to translocate would arrest the RTC and lead to the termination of RNA synthesis unless the translocation impediment is repaired [27], [30]. Although the delayed termination induced by RDV is evident in the meta-stable complex, a recent study reveals that RDV can overcome the S861 roadblock under certain circumstances, allowing processive RNA synthesis by passing the corresponding “i + 3”-related SARS-CoV-2 nsp12 S861 site. In that case, RDV might become part of the RNA template or affect post-transcription events [31], [32]. Overall, although much has been learned about RDV for COVID-19 therapy and its possible side effects, there is still more to be learned.
3.1.2. Favipiravir
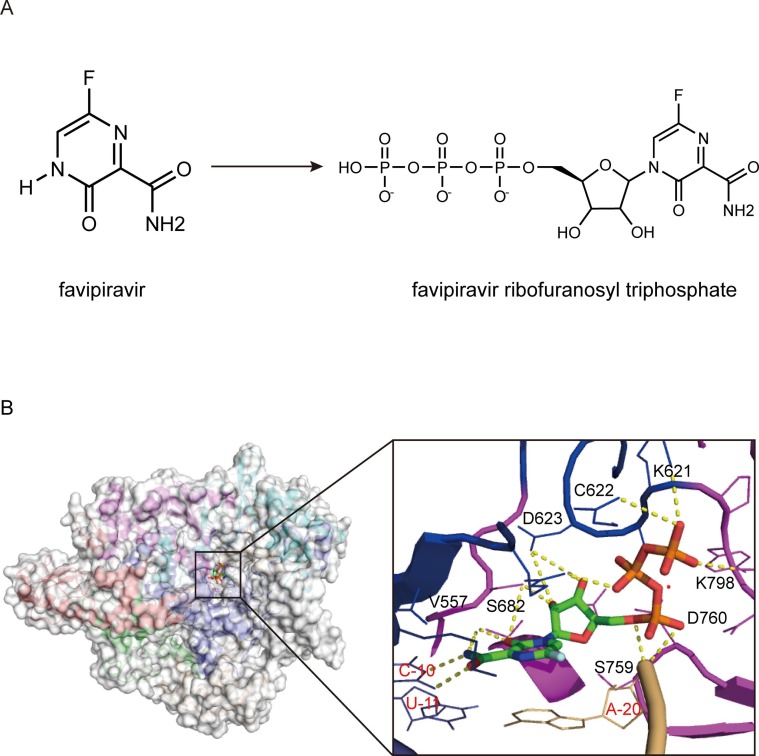
Favipiravir (FPV, T-705), a derivative of pyrazine carboxamide, was originally developed as an anti-influenza drug and is currently approved in Japan for treating influenza infection [33] ( Fig. 4 A). In addition to influenza, favipiravir has demonstrated antiviral activity against some other RNA viruses, including Ebola virus, flaviviruses, noroviruses, arenaviruses, foot-and-mouth disease virus, phleboviruses, hantaviruses, enteroviruses, Western equine encephalitis virus, rabies and respiratory syncytial virus [33].
Fig. 4.
Cryo-EM structure of favipiravir bound to the RdRp complex of SARS-CoV-2 (PDB ID:7CTT). (A) Chemical structures of favipiravir and favipiravir ribofuranosyl triphosphate (FPV-RTP). (B) Overall view of the FPV-RTP bound polymerase complex of SARS-CoV-2 and close-up views of recognition of FPV-RTP by SARS-CoV-2 polymerase complex. The potential polar contacts are shown as yellow dashed lines. The protein and RNA are shown in cartoons with the key residues involved in FPV-RTP interaction shown in lines and labeled in black and red, respectively. The FPV-RTP is shown as a stick model.
In COVID-19 clinical studies, FPV showed effectiveness in controlling disease progression, and its one-week clinical recovery rate of patients with moderate COVID-19 was 71% [34], [35]. After treating severe COVID-19 patients with FPV, the lymphocyte count can be improved [36]. Therefore, oral therapy with FPV has been recommended to tackle COVID-19 in several countries. For instance, in India, it is a prominent antiviral used in clinics for COVID-19 management and treatment [37].
Similar to RDV, FPV is a nucleotide analog inhibitor of RdRp. FPV (prodrug) can be metabolized in the body to be an active metabolite, namely favipiravir-ribofuranosyl-5′-triphosphate (FPV-RTP), via ribosylation and phosphorylation, which acts as a competitor to purine nucleosides. The FPV-RTP is weakly incorporated into the RNA primer strand and binds to the active site of the viral RdRp, transforming the binding pocket into a catalytically nonproductive conformation, and finally terminates replication of viral RNA at the site of incorporation [38]. FPV-RTP exerts its antiviral effect through lethal mutagenesis, where it is mis-incorporated into a nascent viral RNA by RdRp, and promiscuously paired to natural nucleotides cytosine (C) and uracil (U) ( Fig. 4 B). In the next cycle of RNA synthesis, the regions of the viral genome that have incorporated FPV-RTP will be prone to mutagenesis, and excess mutations will finally destroy the viruses [39], [40], [41]. In addition, FPV might be associated with slowed RNA synthesis and nonobligate chain termination.
Based on the risks of adverse reactions, including decreased red blood cell production in hematopoietic tissues, increased liver function parameters, teratogenicity and embryotoxicity to human offspring, the manufacturing, selling and clinical administration of FPV are still strictly limited, which need further exploration [42], [43], [44].
3.1.3. Molnupiravir
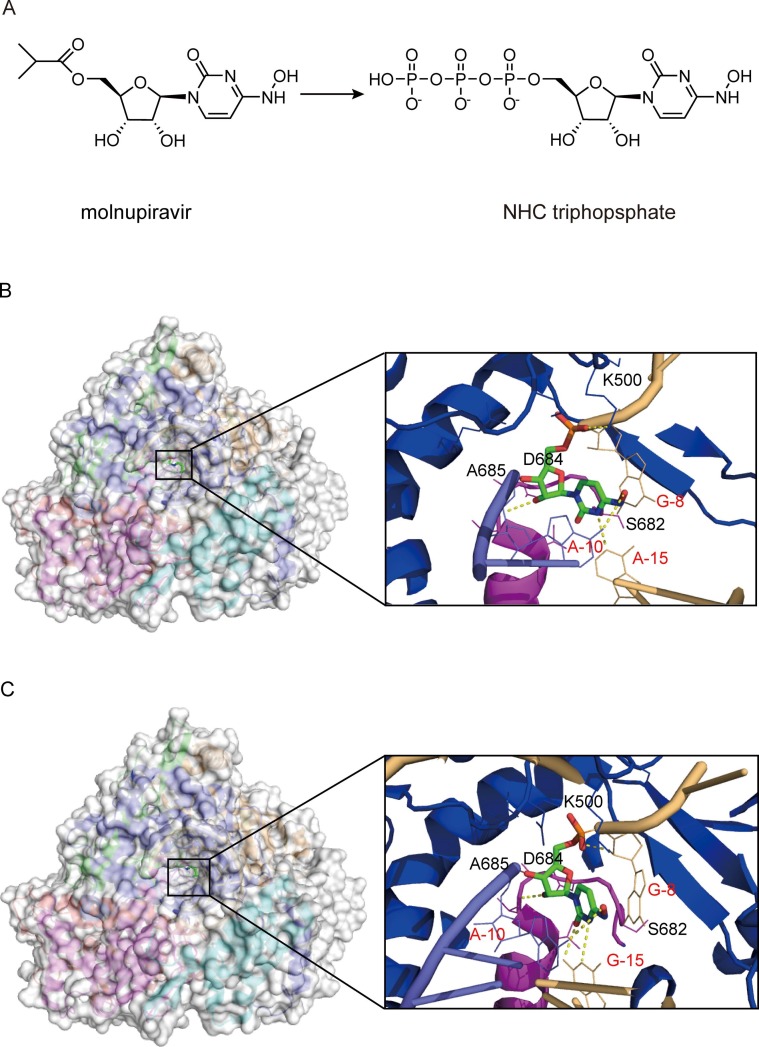
Molnupiravir (EIDD-2801, MK-4482) is an isopropyl ester prodrug of the ribonucleoside analog β-D-N4-hydroxycytidine (EIDD-1931 or NHC) [45] ( Fig. 5 A). Molnupiravir could efficiently hampered the replication of SARS-CoV-2 in human airway epithelial cell cultures and various animal-infected models [46], [47]. Moreover, molnupiravir is greater than 100-fold more active against SARS-CoV-2, compared with other drugs such as favipiravir. It could be also effective in treating patients with resistance to RDV [48], [49], [50]. Other available data from a positive interim analysis in a phase III study suggested that molnupiravir significantly reduced the risk of death in patients with mild to moderate COVID-19 by 50% compared with placebo [51]. Notably, most of the subjects were infected with mutant strains, reflecting the potential effectiveness of molnupiravir against new variants [52], [53]. Hence, molnupiravir was granted emergency use authorization by the Medicines and Healthcare products Regulatory Agency (MHRA) and the FDA for COVID-19 treatment [54].
Fig. 5.
Cryo-EM structure of NHC bound to the RdRp complex of SARS-CoV-2. (A) Chemical structures of molnupiravir and NHC triphosphate. (B and C) Overview of RdRp-RNA structure with an NHC monophosphate in the RNA template strand and close-up views of the interactions between the SARS-CoV-2 RdRp active site and NHC base paired to A(B) (PDB ID:7OZU) and base paired to G(C) (PDB ID:7OZV). The potential polar contacts are shown as yellow dashed lines. The protein and RNA are shown in cartoons with the key residues involved in NHC interaction shown in lines and labeled in black and red, respectively. The NHC are shown as stick models.
The antiviral mechanism of molnupiravir is similar to that of FPV. After oral administration of molnupiravir, the NHC appears rapidly in plasma, which is subsequently activated to triphosphate form in cells. After that, the NHC triphosphate is incorporated into viral RNA in place of cytosine or uracil by RdRp, causing mutations to break replication fidelity, ultimately resulting in an antiviral effect [46], [48], [55], [56]. Structures of the complexes containing mutagenesis products illustrated that the NHC embedded in the template can form stable base pairs with either G or A in the RdRp active site, explaining how the polymerase escapes proofreading and synthesizes mutated RNA [57], [58] ( Fig. 5 B and 5C). In particular, the cytidine base of this NHC forms an extra hydrogen bond with the guanine base from the template strand, which may explain the apparent higher potency of this NHC in inhibiting SARS-CoV-2 replication.
Nevertheless, it was reported that this NHC displays host mutational activity in an animal cell culture assay [49]. Due to the risks this suggests, it is essential to obtain a comprehensive understanding of the potential for side effects of molnupiravir through further studies.
3.1.4. Galidesivir
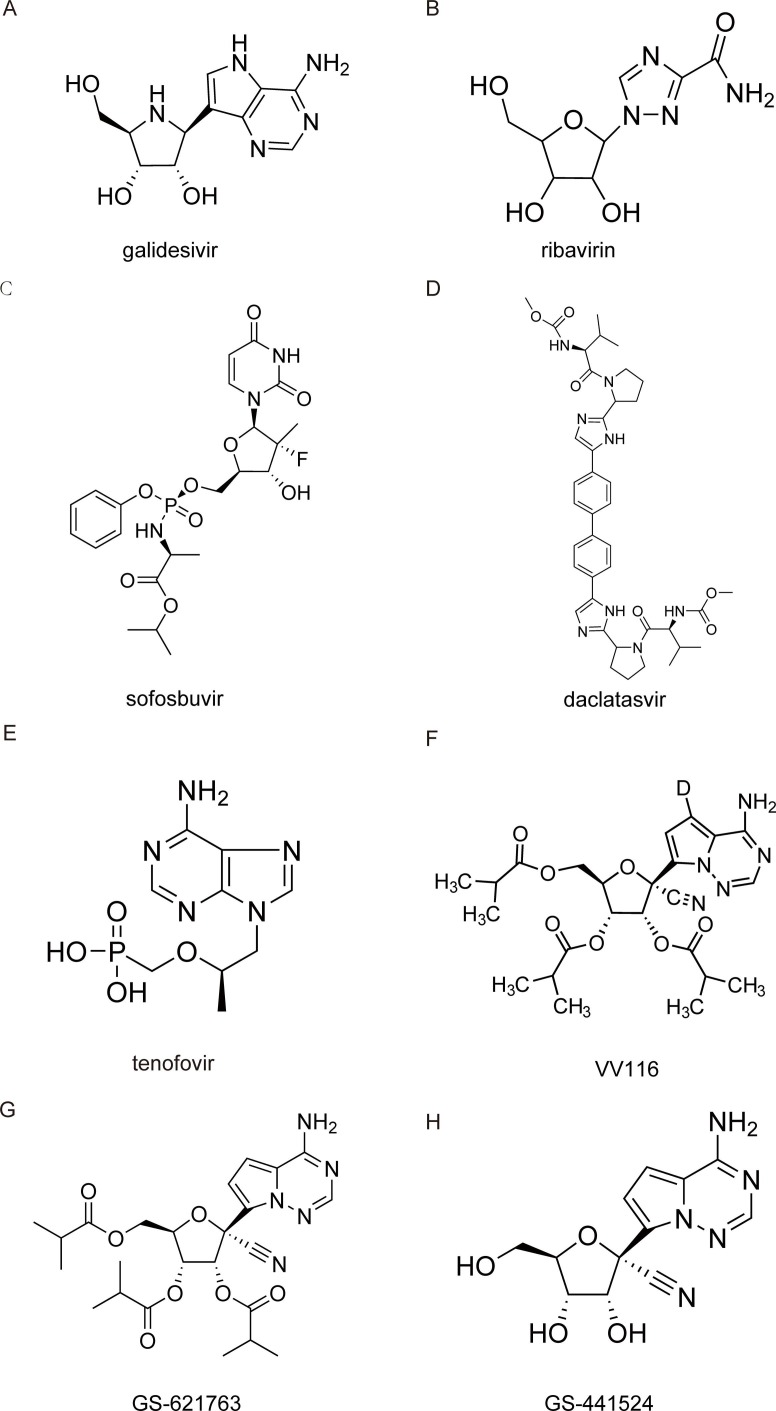
Galidesivir (BCX4430) is a novel synthetic adenosine analog designed as an inhibitor of RdRp ( Fig. 6 A). When the RdRp incorporates galidesivir triphosphate (BCX4430-TP), which prefers viral RNA polymerase rather than host polymerase, into nascent viral RNA strands, the structural change alters its electrostatic interaction, resulting in premature termination of the elongating RNA strand [59]. Computational modeling studies predicted that potential mechanism of galidesivir binding to SARS-CoV-2 RdRp is involves six hydrogen bonds and four hydrophobic interactions [60], [61].
Fig. 6.
Structures of nucleoside analog inhibitors.
In vitro and in animal models, galidesivir can inhibit RNA replication of many RNA viruses, including Ebola, Zika, Marburg, yellow fever and Rift Valley fever viruses [62], [63]. In SARS-CoV-2 infected Syrian golden hamster models, galidesivir reduced lung pathology when the treatment was initiated 24 h prior to infection, compared with untreated controls [64]. Based on these data, several clinical trials have been conducted to evaluate the efficiency of galidesivir against SARS-CoV-2.
3.1.5. Ribavirin
Discovered more than 40 years ago, ribavirin (RBV,1-β-d-ribofuranosyl-1,2,4-triazole-3-carboxamide) ( Fig. 6 B) is a guanosine analog with a broad-spectrum antiviral activity in both human cell lines and animal models [65].
RBV is converted by host kinases into RBV triphosphate (RBV-TP), which pairs with the cytidine triphosphate or uridine triphosphate in the RNA template. The ambiguous coding nature of its purine-mimicking nucleobase accounts for its antiviral effect through lethal mutagenesis [66]. In addition, RBV monophosphate is a potent inhibitor of the host inosine monophosphate dehydrogenase (IMPDH). IMPDH inhibition represses de novo synthesis of guanine nucleotides, thus affecting RNA polymerase fidelity through nucleotide pool imbalance. Alternatively, enhancing the induction of interferon-related genes is another proposed mechanism of RBV antiviral activity [67].
During the emergence of SARS-CoV-2, RBV was repurposed to treat COVID-19, and some promising results have been reported [68], [69]. An observational study has indicated that RBV is well tolerated in severe patients with severe COVID-19 and may benefit them by increasing the viral clearance, resulting in a higher survival rate [70]. Furthermore, RBV was employed in a Phase II trial in combination with interferon beta-1b and lopinavir-ritonavir, to treat patients with mild to moderate COVID-19. In that trial, the combination treatment group was superior in reducing the viral load compared with lopinavir-ritonavir alone. Moreover, a hastened clinical improvement with shorter duration of hospital stays was observed in the triple combination group [71].
Despite the potential efficacy in treating COVID-19, some side effects of RBV may be of concerned, primarily, hemolytic anemia. Furthermore, it cannot be prescribed to pregnant women and the elderly population because of fetal toxicity and reduction of calcium as well as magnesium levels in blood [72].
3.1.6. Sofosbuvir and daclatasvir
Sofosbuvir (GS‐7977; formerly PSI‐7977) ( Fig. 6 C) and daclatasvir ( Fig. 6 D) are considered safe and well-tolerated anti-HCV therapies. Sofosbuvir triphosphate competes with uridine to be incorporated by the HCV RNA polymerase (NS5B) into the elongating RNA strand, resulting in chain termination [73]. Daclatasvir inhibits HCV replication by binding to the N-terminus of nonstructural protein 5A (NS5A), affecting both viral RNA replication and virion assembly [74].
Based on the similarity of the replication mechanisms between HCV and coronavirus, it is hypothesized that sofosbuvir and daclatasvir might be potential options in the treatment of COVID-19, especially at the start of the disease and before the viral invasion into the lung parenchymal cells [75]. Indeed, both sofosbuvir and daclatasvir inhibit RNA synthesis in various cell lines via different mechanisms, and co-treatment of daclatasvir/sofosbuvir shows a cooperative antiviral effect on SARS-CoV-2 replication in respiratory cells [75]. An open-label, multi-center, and randomized clinical trial was conducted on 66 adults with moderate or severe COVID-19 in Iran, which showed that the addition of sofosbuvir and daclatasvir to standard care significantly reduced the duration of hospital stay compared with standard care alone [76].
Regarding the mechanism of antiviral activity, daclatasvir favors the unfolding of SARS-CoV-2 secondary RNA structures, whereas sofosbuvir triphosphate (SOF-TP) can be incorporated by the relatively low-fidelity SARS-CoV-2 RdRp, serving as an immediate polymerase reaction terminator [77], [78]. In addition, upon the incorporation of SOF-TP into RNA by the SARS-CoV-2 RdRp, sofosbuvir is removed at a lower rate by the SARS-CoV-2 exonuclease complex, relative to the natural nucleotide UMP, thus conferring a substantial level of resistance to excision [79].
Further studies are needed to confirm the efficacy, safety and mechanism of daclatasvir/sofosbuvir in combination with other drugs for prevention and treatment of COVID-19.
3.1.7. Tenofovir
Tenofovir is a broad-spectrum antiviral drug active against human immunodeficiency virus (HIV) and hepatitis B virus (HBV) infection, prodrug formulations of which are tenofovir disoproxil fumarate (TDF) and tenofovir alafenamide (TAF) [80] ( Fig. 6 E). As a nucleotide analog, it is reported that the active triphosphate form of the tenofovir diphosphate acts as an immediate chain terminator when incorporated during the synthesis of viral DNA [81], [80], [82].
Several in silico studies show that tenofovir can bind to SARS-CoV-2 RdRp tightly, with binding energies comparable to those of native nucleotides [61]. Polymerase extension experiments in vitro demonstrated that tenofovir diphosphate was incorporated by SARS-CoV-2 RdRp, where it terminated subsequent polymerase extension, providing a molecular basis for inhibition of the SARS-CoV-2 RdRp by this nucleotide analog [77]. Emtricitabine-tenofovir, a prescription medicine approved for HIV by the U.S. FDA, was assessed in the highly susceptible SARS-CoV-2 ferret infection model. Compared with the PBS-treated control group, the emtricitabine-tenofovir-treated group showed lower viral titers in nasal washes at eight days post-infection (dpi) than the PBS-treated group. In addition, it has been reported that TDF plus emtricitabine significantly decreased the SARS-CoV-2 viral burden in recently infected COVID-19 outpatients [83].
However, only a few in vitro studies on tenofovir/TDF/TAF efficacy have been described, and the results are contradictory, which may be due to the different experimental conditions [84], [85], [86]. Further studies should proceed to better evaluate the potential efficacy of this drug (alone or in combination with other antiviral drugs) for the prevention and early treatment of COVID-19.
3.1.8. VV116
Because RDV is an intravenously administered nucleotide prodrug and is therefore inconvenient for patients, VV116 (JT001), a new orally available RDV derivative, was developed ( Fig. 6 F). This prodrug was endowed with significantly improved oral absorption and a favorable tissue distribution profile, circumventing the liver-targeting issue of the phosphoramidate prodrugs [87], [88].
As a deuterated, tri-isobutyrate ester prodrug of the RDV parent nucleoside, VV116 is rapidly metabolized into the parent nucleoside (116-N1) in vivo. 116-N1 is intracellularly converted to the nucleoside triphosphate active form, which would interfere with the function of RdRp, thus inhibiting SARS-CoV-2 replication [89]. It has been demonstrated that VV116 showed potent activity against a panel of SARS-CoV-2 variants, including the Omicron variant [87], [90]. In addition, VV116 exhibited satisfactory safety and tolerability in healthy subjects in phase I studies [89]. VV116 has been approved for the treatment of COVID-19 in Uzbekistan and is currently being investigated in phase II/III clinical trials in patients with COVID-19.
3.1.9. GS-621763
GS-621763 ( Fig. 6 G), which demonstrated high oral bioavailability in two relevant animal species, including non-human primates [91], is designed for optimal delivery of RDV parent nucleoside GS-441524 into the systemic circulation. Oral GS-621763 is efficiently converted to plasma metabolite GS-441524 ( Fig. 6 H), and in the lungs to the triphosphate metabolite identical to that generated by RDV, demonstrating a consistent mechanism of activity, with blockade of SARS-CoV-2 replication by triggering delayed chain termination of the nascent viral RNA chain and template-dependent inhibition after incorporation into viral antigenomic RNA [30], [31]. Both GS-621763 and GS-441524 exhibited antiviral activity against SARS-CoV-2, including variants of concern (VOC) in lung cell lines and two different human primary lung cell culture systems that model the tissues targeted by SARS-CoV-2 in humans. Therapeutic GS-621763 administration reduced viral load and lung pathology and improved pulmonary function in a COVID-19 mouse model [92]. When dosed therapeutically against VOC P.1 (γ) in ferrets, oral GS-621763 completely blocked virus replication and prevented transmission to untreated contact animals [93]. With confirmed in vivo efficacy, GS-621763 may become a cornerstone in the first-line defense against future pandemic threats.
3.1.10. AT-527
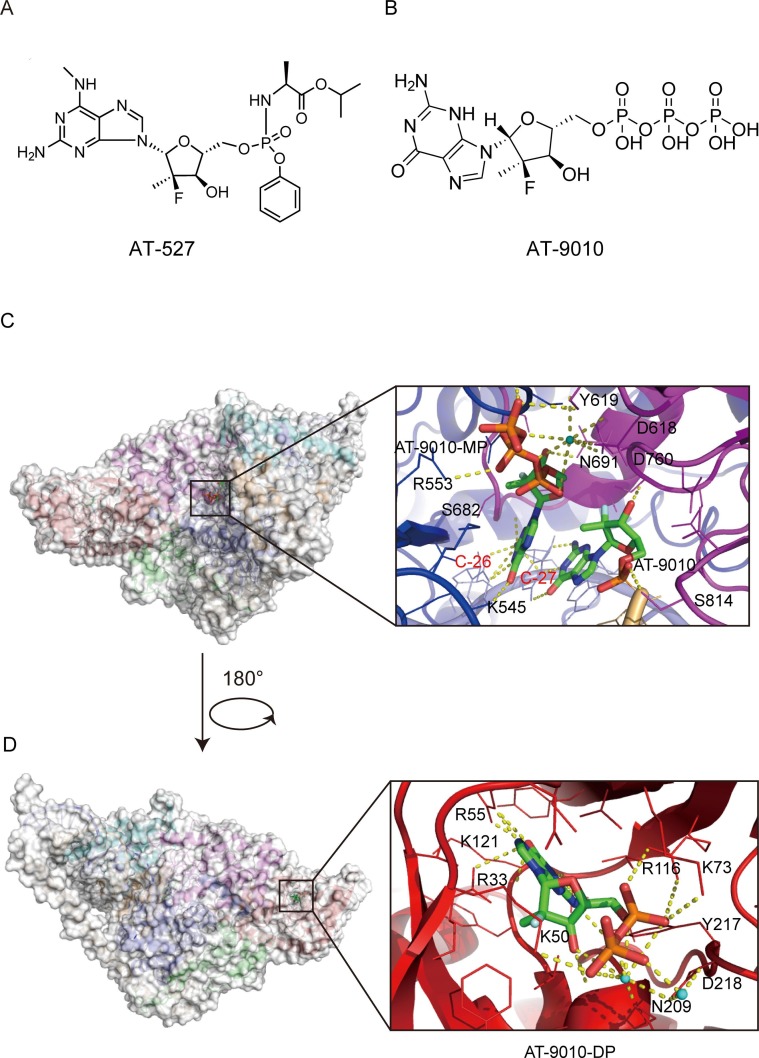
AT-527, an orally available prodrug of a guanosine nucleotide analog carrying a 2′-fluoro-2′C-methyl modified ribose ( Fig. 7 A), was shown to act as a potent broad-spectrum anti- coronavirus inhibitor in a variety of cell lines by targeting the RdRp activity [94]. After oral dosing, AT-527 is converted by cellular enzymes to the active triphosphate metabolite, AT-9010, which is structurally similar to that of sofosbuvir, with the only difference being that it has a guanine base in place of uracil [95] ( Fig. 7 B). AT-511, the free base of AT-527, also had potent antiviral activity when tested in vitro against several human coronaviruses, including SARS-CoV-2 [94]. AT-527 recently entered phase III clinical trials to treat COVID-19 [96].
Fig. 7.
Cryo-EM structure of AT-9010 bound to the RdRp complex of SARS-CoV-2 (PDB ID:7ED5) (A) Chemical structures of AT-527 and its active triphosphate form AT-9010. (B and C) Overall view of three AT-9010 molecules bound polymerase complex of SARS-CoV-2 and close up of RdRp catalytic site following AT-9010 incorporation(B) and binding of AT-9010 diphosphate (AT-9010-DP) in the NiRAN domain(C). In the RdRp active site, one AT-9010 monophosphate (AT-9010-MP) is incorporated at (+1) position, with an incoming AT-9010 occupying the (-1) position. In the NiRAN domain, AT-9010 is bound in its diphosphate form (AT-9010-DP). The potential polar contacts are shown as yellow dashed lines. The protein and RNA are shown in cartoons with the key residues involved in AT-9010 interaction shown in lines and labeled in black and red, respectively. AT-9010 are shown as stick models.
Ashleigh Shannon et al. reported a 2.98 Å cryo-EM structure of the SARS-CoV-2 nsp12-nsp7-nsp82-RNA complex, showing AT-9010 simultaneously bound to both NiRAN and RdRp active sites of nsp12 ( Fig. 7 C and 7D). AT-9010 is efficiently incorporated into viral RNA, causes immediate termination of RNA synthesis, and shows resistance to excision. In addition, AT-9010 outcompetes all native nucleotides for NiRAN binding, inhibiting its nucleotidyl transferase activity. The dual mechanism of action of AT-527 should attenuate the chance of resistance mutations and represents a promising research avenue against COVID-19 [97].
3.2. Non-nucleoside inhibitors (NNIs)
Due to the proofreading activity of nsp14 exonuclease, the potential of NAIs as antiviral drugs for COVID-19 treatment may be limited [98]. An alternative route to NAIs would be the discovery of NNIs, which generally show lower resistance barriers. NNIs also have several other advantages compared with NAIs, particularly their higher chemical diversity and likely per os administration. Therefore, the discovery of non-nucleoside analog-type drugs that target SARS-CoV-2 RdRp might be particularly advantageous.
Mechanistically, most NNIs change the conformation of RdRp by binding to allosteric sites on the surface of the polymerase and then affect the binding of the catalytically active site of RdRp to the substrates [99]. Some NNIs also exert anti-inflammatory, antioxidant, immunomodulatory and cardioprotective functions, with multifunctional therapeutic benefits in the clinical management of COVID-19 [100]. Herein, we outline the most promising NNIs of SARS-CoV-2 RdRp reported so far.
3.2.1. Suramin
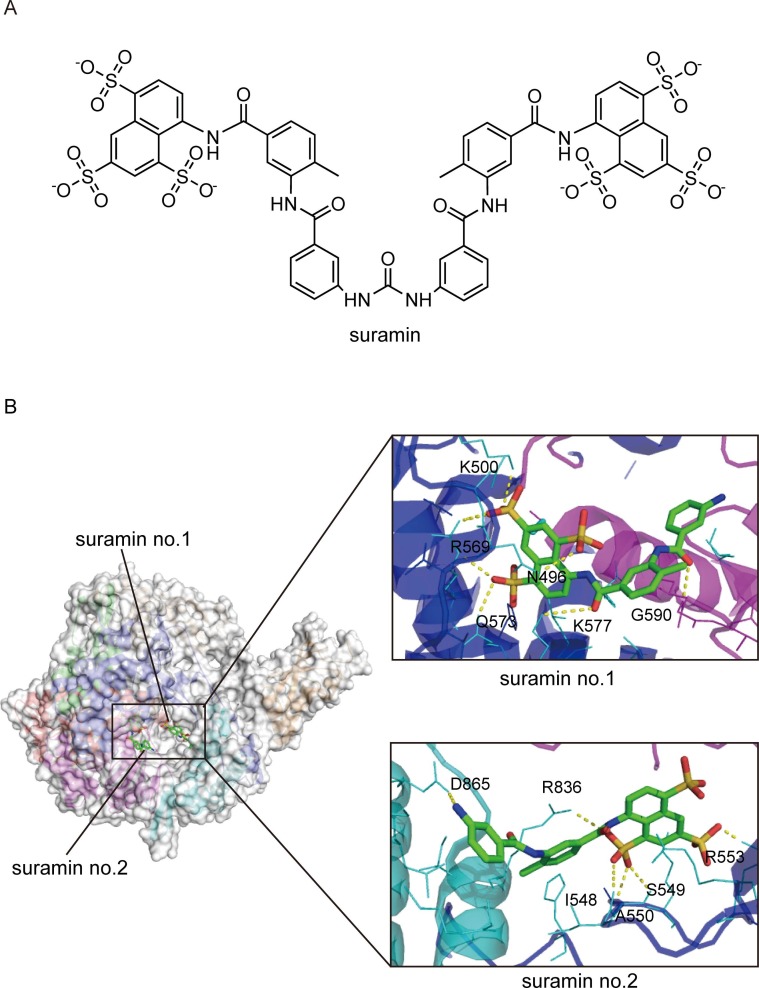
Suramin ( Fig. 8 A) is a century-old drug with a wide array of potential applications, from parasitic and viral diseases to cancer, snakebite, and autism. The antiviral activity of suramin has been known since the mid-20th century [101]. The viral inhibition mechanisms of suramin are diverse, including inhibition of viral attachment, viral entry, and release from host cells in part through interactions with viral capsid proteins [102], [103]. It has been recently demonstrated that suramin can inhibit SARS-CoV-2 infection and decrease the viral load by 2–3 logs in cell culture by preventing cellular entry of the virus. In a primary human airway epithelial cell culture model, suramin also inhibited the progression of infection [104].
Fig. 8.
Cryo-EM structure of suramin bound to the RdRp complex of SARS-CoV-2 (PDB ID:7D4F) (A) The chemical structure of suramin. (B) Overall view of the RdRp-suramin complex and close views of the interactions of the two suramin molecules with RdRp. Suramin no.1 directly blocks the binding of the RNA template strand, and suramin no.2 clashes with the RNA primer strand near the RdRp catalytic site. The potential polar contacts are shown as yellow dashed lines. The protein and RNA are shown in cartoons with the key residues involved in suramin interaction shown in lines and labeled. Suramin are shown as stick models.
Suramin is also a potent inhibitor of the SARS-CoV-2 RNA-dependent RNA polymerase. In terms of inhibition mechanism, suramin has two independent binding sites with SARS-CoV-2 RdRp ( Fig. 8 B). One of the sites (suramin no. 1) is located in a cavity formed by the N-termin us of the conserved motif G and motif B, directly blocking the binding of template RNA. The other site (suramin no. 2) is located in a cavity near the catalytically active site formed by the conservative motifs A, C, E, and F, thus blocking RNA primer binding by steric hindrance [105]. This unique binding pattern is consistent with suramin and its derivatives being at least 20-fold more potent than the triphosphate form of remdesivir (RDV-TP) in polymerase activity inhibition assays in vitro. In addition, suramin, as a polyanionic compound, may bind tightly to positively charged patches of RNA-binding proteins, thereby inhibiting RNA replication [106], [107]. In addition, it has been suggested that suramin, in combination with quinacrine, showed promising synergistic efficacy in inhibiting SARS-CoV-2 3CLpro [108]. These results support that suramin is an attractive drug candidate to combat SARS-CoV-2 infections by targeting multiple critical proteins in the viral life cycle.
However, suramin is also associated with unwanted side effects. The most frequent adverse reactions of suramin are nausea and vomiting for the treatment of onchocerciasis [109]. Clinical trials in cancer patients and HIV-infected patients treated with suramin documented other toxic effects such as proteinuria, liver toxicity, corneal damage, and adrenal insufficiency [110], [111]. As such, well-designed, properly controlled randomized trials and safety studies are essential for determining the feasibility of using suramin to treat COVID-19. Suramin analogs can also be developed to improve the potency and efficacy of the drug, as well as reduce its toxicity.
3.2.2. Quinoline derivatives
Quinoline, an important heterocyclic compound, is a scaffold used in the discovery of many new drug candidates [112]. It has low toxicity to humans on oral absorption and inhalation, with a logP value of 2.04 [113], [114]. Quinoline derivatives have a wide range of biological properties such as antibacterial, antifungal, anti-cancer, and antiviral activities [115], [116], [117].
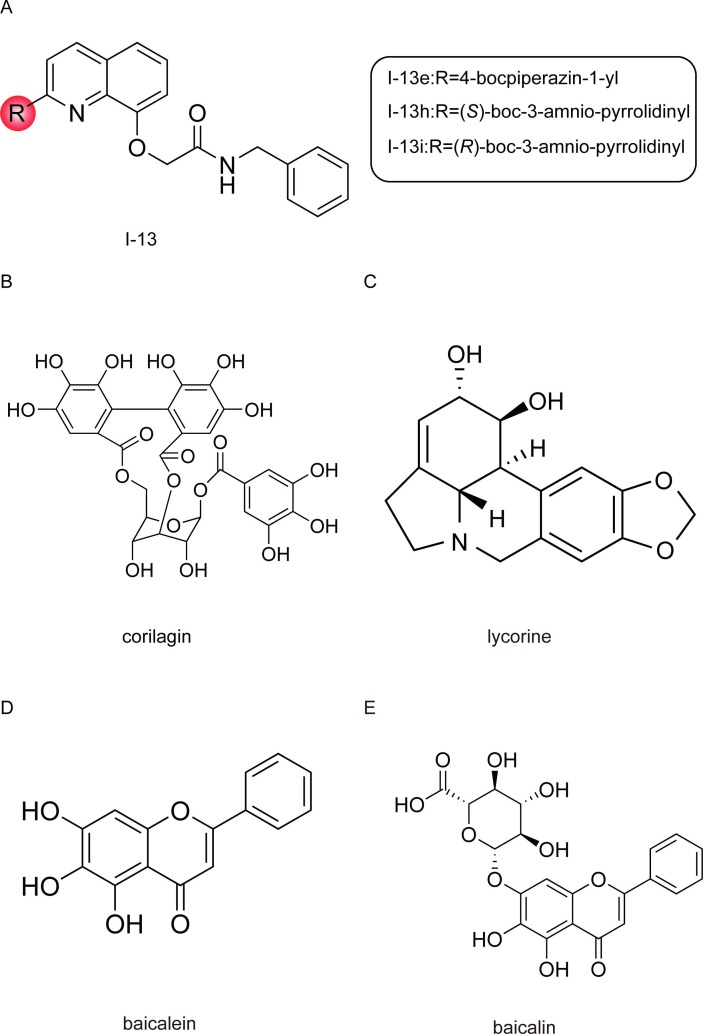
Three quinoline derivatives (I-13e, I-13 h, and I-13i) ( Fig. 9 A), which are previously reported to have significant activity against the influenza A virus RdRp, exhibit remarkable potency in inhibiting RNA synthesis by targeting SARS-CoV-2 RdRp and relatively low cytotoxicity in a cell-based assay, with therapeutic index (TI) (CC50/EC50) values of 65, 34, and 20, respectively [118]. Among these three compounds, I-13e showed the most potent inhibition of RNA synthesis and bypasses viral exonuclease activity concerns common with nucleotide inhibitors, thus holding the potential of being a drug candidate for the treatment of SARS-CoV-2. In addition, based on the activity assay, the C-2 group of the quinoline is crucial for antiviral activity, indicating that future work may focus on the modification of C-2 by introducing some large hydrophobic/hydrophilic groups [118].
Fig. 9.
Structures of non-nucleoside inhibitors.
3.2.3. Corilagin
Corilagin (RAI-S-37) ( Fig. 9 B), a gallotannin, is one of the primary active components of many traditional herbal plants. In the past few decades, corilagin was reported to exhibit anti-tumor, anti-inflammatory, and hepatoprotective activities [119].
Recently, it has been demonstrated that corilagin can effectively inhibit the polymerase activity in both cell-free and cell-based assays and potently inhibit SARS-CoV-2 infection. As expected, as a non-nucleoside inhibitor, corilagin fully resists the proofreading activity of SARS-CoV-2 nsp10-nsp14 exoribonuclease complex. Computational modeling predicts that corilagin binds to the palm domain of RdRp and prevents conformational changes required for nucleotide incorporation by RdRp. Importantly, the combination of corilagin with RDV exhibits additive activity against SARS-CoV-2 RdRp [120]. Specifically, corilagin did not show any toxicity in acute and sub-chronic toxicity studies. After oral administration of corilagin at higher dosages, no undesirable behavioral changes or weight loss were observed in mouse models, demonstrating that corilagin was well tolerated in vivo [121], [122]. These results suggest that corilagin holds promise of becoming an effective SARS-CoV-2 therapeutic.
3.2.4. Lycorine
Lycorine ( Fig. 9 C), a natural alkaloid extracted from the amaryllidaceae plant lycoris radiata, also a traditional Chinese medicinal herb, exhibits multiple pharmacological activities, including regulation of autophagy and the induction of cancer cell apoptosis, and has anti-inflammatory, anti-fungal, anti-malarial, anti-tumor and antiviral activities [123]. Lycorine was previously identified as a coronavirus inhibitor according to its potent inhibition of replication in diverse coronavirus, including SARS-CoV, HCoV-OC43, HCoV-NL63, MERS-CoV, and MHV-A59 in vitro [124], [125].
Lycorine exhibited dose-dependent inhibition of SARS-CoV-2 infection with a low nanomolar EC50 in infected cells without apparent cytotoxicity [126], [127]. A docking simulation showed that lycorine could interact with SARS-CoV-2 RdRp through three hydrogen bonds, and the predicted binding affinity of lycorine (-6.2 kcal/mol) was higher than that of RDV (-4.7 kcal/mol). In addition, lycorine directly inhibited MERS-CoV RdRp activity, and the inhibitory effect of lycorine on MERS-CoV RdRp activity was comparable with that of RDV. These results suggest that lycorine is a potent non-nucleoside direct-acting antiviral against emerging coronavirus infections and acts by inhibiting viral RdRp activity. Therefore, lycorine may be a candidate against the current COVID-19 pandemic [128].
3.2.5. Baicalein and baicalin
Baicalein and baicalin ( Fig. 9 D and 9E), the major bioactive phenolic flavonoid compounds extracted from the root of Scutellaria baicalensis Georgi (called Huang-Qin), are widely used in traditional Chinese medicine for the clinical treatment of hyperlipidemia, hypertension, atherosclerosis, dysentery, and inflammatory diseases [129]. After oral administration, baicalin is metabolized into baicalein in the intestine, and baicalein exerts a broad-spectrum antiviral effect, including against influenza virus, Zika virus (ZIKV), and dengue virus (DENV) [130], [131], [132]. Clinical laboratory assessments showed that single oral doses of 100–2800 mg of baicalein were safe and well tolerated by healthy subjects, and no serious accumulation of baicalein was observed in the multiple-dose (200–800 mg) study [133], [134].
Baicalein and baicalin are promising therapeutic candidates for COVID-19, exhibiting dose-dependent inhibitory effects against SARS-CoV-2 in Vero cells and Calu3 human lung cells, and baicalein is more potent compared to baicalin [135], [136]. In addition, baicalein inhibited the loss of body weight, reduced viral load, and relieved lung injury in mice infected with SARS-CoV-2 [137]. Further research showed that both compounds act as SARS-CoV-2 RdRp inhibitors, directly inhibiting the activity of the SARS-CoV-2 RdRp, which could give advantages to these two compounds as non-nucleoside analog SARS-CoV-2 RdRp inhibitors [138].
4. Discussion and perspectives
Following SARS-CoV in 2003 and MERS-CoV in 2012, SARS-CoV-2 is the third coronavirus that crossed the species barrier to cause severe diseases in humans in only 20 years, implying coronaviruses may pose further threats to human health in the future. On Nov. 25, 2021, about 23 months since the first reported case of COVID-19, a new SARS-CoV-2 variant of concern, Omicron, was reported and has split into a number of sub-lineages now. The Omicron variant hosted an unprecedented number of mutations in its spike gene, which reduced vaccine effectiveness, leading to an increased incidence of reinfections and breakthrough infections [139], [140], [141], [142]. It is a plausible strategy to develop broad-spectrum antivirals, together with vaccination programs to curb future transmissions.
In the life cycle of coronaviruses, RdRp is the prime broad-spectrum drug target due to its essential role in viral RNA synthesis, the lack of a host homolog, and high sequence and structural conservation among coronaviruses. Great efforts have been devoted to structural and functional studies of the RTC of SARS-CoV-2, to promote antiviral development against COVID-19 and other coronavirus-associated diseases. In this paper, all of the current knowledge on the inhibitors targeting RdRp of SARS-CoV-2, including nucleoside analog inhibitors and non-nucleoside inhibitors, is summarized.
It is clear that the most promising, broad-spectrum class of viral RdRp inhibitors is nucleoside analogs. However, NAIs are hindered by the proofreading activity of SARS-CoV-2 nsp14 exoribonuclease, which can excise erroneous mutagenic nucleotides incorporated by RdRp into the nascent viral RNA, thus augmenting the resistance of SARS-CoV-2 RdRp to NAIs [98], [143]. Therefore, to potentially inhibit SARS-CoV-2 replication, nsp14 exoribonuclease needs to be included in the screening assay for RdRp NAIs, and well-developed NAIs should be designed to either be less recognized by the exoribonuclease or be incorporated by RdRp at a rate exceeding exoribonuclease excision velocity. In addition, the rational design of new potent exonuclease inhibitors in further research represents another strategy to overcome this deficiency of NAIs against SARS-CoV-2 RdRp.
Compared with NAIs, which are vulnerable to exonuclease cleavage, NNIs have the potential to be highly potent antivirals against SARS-CoV-2. As we reviewed previously, some natural compounds and their derivatives have been repurposed as strong NNIs against SARS-CoV-2 RdRp. Besides, alkaloids [144], [145], plant polyphenols, including epigallocatechin-3-gallate (EGCG), silibinin, theaflavin [146], [147], [148], [149], and fungal derivative cordycepin [150] were also reported to establish strong and favorable binding interactions with the active site of SARS-CoV-2 RdRp by molecular dynamic simulation. It is recommended to test these natural products in the laboratory to determine their inhibitory efficacy, as such compounds may be promising alternative and complementary therapies for COVID-19 due to their diverse range of biological and therapeutic properties.
The continual emergence of mutations of SARS-CoV-2 is a cause of concern, as it may hinder the development of long-lasting effective RdRp inhibitors, in addition to promoting escape from vaccine-induced immune protection. Identifying potential hotspot residues contributing to antiviral resistance in SARS-CoV-2 is crucial for robust antiviral design and discovery. In addition, experience with other viral diseases suggests that combinations of inhibitors can provide a significant effect and prevent the evolution of drug-resistant virus mutants. Therefore, phytochemicals, in addition to other promising antivirals against SARS-CoV-2 RdRp, can be exploited for the development of a cocktail of inhibitors in the near future, which may have significant implications for alleviating the current global public health threat.
Author contributions
XX, XW, and YC conceived this review. YC, XL, WZ, WF, LY wrote the initial draft of the review, XW and XX revised and edited the manuscript.
CRediT authorship contribution statement
Xiaoying Xu: Conceptualization, Methodology, Supervision, Writing – review & editing, Project administration, Funding acquisition. Yuheng Chen: Investigation, Data curation, Writing – original draft. Xinyu Lu: Investigation, Data curation, Writing – original draft. Wanlin Zhang: Writing – original draft. Wenxiu Fang: Writing – original draft. Luping Yuan: Writing – original draft. Xiaoyan Wang: Conceptualization, Methodology, Supervision, Writing – review & editing, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This research was supported by the Zhejiang Provincial Natural Science Foundation of China (Grant No. LQ20C050002), the Research Project of Zhejiang Chinese Medical University(2019ZR09), National Natural Science Foundation of China (Grant No. 81973571), YUAN ZHI Outstanding Youth Research Foundation of ZCMU (2018) and YUAN ZHI Outstanding Doctor Research Foundation of ZCMU (2018). We also appreciate the support from the Public Research Platform of Academy of Chinese Medical Science, ZCMU.
References
- 1.Muralidar S., Ambi S.V., Sekaran S., Krishnan U.M. The emergence of COVID-19 as a global pandemic: Understanding the epidemiology, immune response and potential therapeutic targets of SARS-CoV-2. Biochimie. 2020;179:85–100. doi: 10.1016/j.biochi.2020.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu K., Pan X., Li L., Yu F., Zheng A., Du P., et al. Binding and molecular basis of the bat coronavirus RaTG13 virus to ACE2 in humans and other species. Cell. 2021;184(13):3438–3451.e10. doi: 10.1016/j.cell.2021.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.R. Arya, S. Kumari, B. Pandey, H. Mistry, S.C. Bihani, A. Das, et al., Structural insights into SARS-CoV-2 proteins, J. Mol. Biol. 433(2) (2021) 166725. [DOI] [PMC free article] [PubMed]
- 5.Jiang Y., Yin W., Xu H.E. RNA-dependent RNA polymerase: Structure, mechanism, and drug discovery for COVID-19. Biochem. Biophys. Res. Commun. 2021;538:47–53. doi: 10.1016/j.bbrc.2020.08.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Araf Y., Akter F., Tang Y.D., Fatemi R., Parvez M.S.A., Zheng C., et al. Omicron variant of SARS-CoV-2: Genomics, transmissibility, and responses to current COVID-19 vaccines. J. Med. Virol. 2022;94(5):1825–1832. doi: 10.1002/jmv.27588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao Y., Wang J., Jian F., Xiao T., Song W., Yisimayi A., et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature. 2022;602(7898):657–663. doi: 10.1038/s41586-021-04385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Q. Peng, R. Peng, B. Yuan, J. Zhao, M. Wang, X. Wang, et al., Structural and Biochemical Characterization of the nsp12-nsp7-nsp8 Core Polymerase Complex from SARS-CoV-2, Cell Rep. 31(11) (2020) 107774. [DOI] [PMC free article] [PubMed]
- 9.Subissi L., Posthuma C.C., Collet A., Zevenhoven-Dobbe J.C., Gorbalenya A.E., Decroly E., et al. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl. Acad. Sci. U. S. A. 2014;111(37) doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan L., Ge J., Zheng L., Zhang Y., Gao Y., Wang T., et al. Cryo-EM Structure of an Extended SARS-CoV-2 Replication and Transcription Complex Reveals an Intermediate State in Cap Synthesis. Cell. 2021;184(1):184–193.e10. doi: 10.1016/j.cell.2020.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yin W., Mao C., Luan X., Shen D.-D., Shen Q., Su H., et al. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368(6498):1499–1504. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368(6492):779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Q., Wu J., Wang H., Gao Y., Liu Q., Mu A., et al. Structural Basis for RNA Replication by the SARS-CoV-2 Polymerase. Cell. 2020;182(2):417–428.e13. doi: 10.1016/j.cell.2020.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hillen H.S., Kokic G., Farnung L., Dienemann C., Tegunov D., Cramer P. Structure of replicating SARS-CoV-2 polymerase. Nature. 2020;584(7819):154–156. doi: 10.1038/s41586-020-2368-8. [DOI] [PubMed] [Google Scholar]
- 15.Cannalire R., Cerchia C., Beccari A.R., Di Leva F.S., Summa V. Targeting SARS-CoV-2 Proteases and Polymerase for COVID-19 Treatment: State of the Art and Future Opportunities. J. Med. Chem. 2022;65(4):2716–2746. doi: 10.1021/acs.jmedchem.0c01140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santoro M.G., Carafoli E. Remdesivir: From Ebola to COVID-19. Biochem. Biophys. Res. Commun. 2021;538:145–150. doi: 10.1016/j.bbrc.2020.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown A.J., Won J.J., Graham R.L., Dinnon K.H., Sims A.C., Feng J.Y., et al. Broad spectrum antiviral remdesivir inhibits human endemic and zoonotic deltacoronaviruses with a highly divergent RNA dependent RNA polymerase. Antiviral Res. 2019;169 doi: 10.1016/j.antiviral.2019.104541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siegel D., Hui H.C., Doerffler E., Clarke M.O., Chun K., Zhang L., et al. Discovery and Synthesis of a Phosphoramidate Prodrug of a Pyrrolo[2,1-f][triazin-4-amino] Adenine C-Nucleoside (GS-5734) for the Treatment of Ebola and Emerging Viruses. J. Med. Chem. 2017;60(5):1648–1661. doi: 10.1021/acs.jmedchem.6b01594. [DOI] [PubMed] [Google Scholar]
- 19.Cardile A.P., Warren T.K., Martins K.A., Reisler R.B., Bavari S. Will There Be a Cure for Ebola? Annu. Rev. Pharmacol. Toxicol. 2017;57(1):329–348. doi: 10.1146/annurev-pharmtox-010716-105055. [DOI] [PubMed] [Google Scholar]
- 20.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30(3):269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Wit E., Feldmann F., Cronin J., Jordan R., Okumura A., Thomas T., et al. Prophylactic and therapeutic remdesivir (GS-5734) treatment in the rhesus macaque model of MERS-CoV infection. Proc. Natl. Acad. Sci. U. S. A. 2020;117(12):6771–6776. doi: 10.1073/pnas.1922083117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williamson B.N., Feldmann F., Schwarz B., Meade-White K., Porter D.P., Schulz J., et al. Clinical benefit of remdesivir in rhesus macaques infected with SARS-CoV-2. Nature. 2020;585(7824):273–276. doi: 10.1038/s41586-020-2423-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drożdżal S., Rosik J., Lechowicz K., Machaj F., Szostak B., Przybyciński J., et al. An update on drugs with therapeutic potential for SARS-CoV-2 (COVID-19) treatment. Drug Resist Updat. 2021;59 doi: 10.1016/j.drup.2021.100794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eastman R.T., Roth J.S., Brimacombe K.R., Simeonov A., Shen M., Patnaik S., et al. Remdesivir: A Review of Its Discovery and Development Leading to Emergency Use Authorization for Treatment of COVID-19. ACS Cent. Sci. 2020;6(5):672–683. doi: 10.1021/acscentsci.0c00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., et al. Remdesivir for the Treatment of Covid-19 - Final Report. N Engl J Med. 2020;383(19):1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lamb Y.N. Remdesivir: First Approval. Drugs. 2020;80(13):1355–1363. doi: 10.1007/s40265-020-01378-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kokic G., Hillen H.S., Tegunov D., Dienemann C., Seitz F., Schmitzova J., et al. Mechanism of SARS-CoV-2 polymerase stalling by remdesivir. Nat. Commun. 2021;12(1) doi: 10.1038/s41467-020-20542-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Agostini M.L., Andres E.L., Sims A.C., Graham R.L., Sheahan T.P., Lu X., et al. Coronavirus Susceptibility to the Antiviral Remdesivir (GS-5734) Is Mediated by the Viral Polymerase and the Proofreading Exoribonuclease. MBio. 2018;9(2) doi: 10.1128/mBio.00221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gordon C.J., Tchesnokov E.P., Feng J.Y., Porter D.P., Götte M. The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. J Biol Chem. 2020;295(15):4773–4779. doi: 10.1074/jbc.AC120.013056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gordon C.J., Tchesnokov E.P., Woolner E., Perry J.K., Feng J.Y., Porter D.P., et al. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J Biol Chem. 2020;295(20):6785–6797. doi: 10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tchesnokov E.P., Gordon C.J., Woolner E., Kocinkova D., Perry J.K., Feng J.Y., et al. Template-dependent inhibition of coronavirus RNA-dependent RNA polymerase by remdesivir reveals a second mechanism of action. J Biol Chem. 2020;295(47):16156–16165. doi: 10.1074/jbc.AC120.015720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.J. Wu, H. Wang, Q. Liu, R. Li, Y. Gao, X. Fang, et al., Remdesivir overcomes the S861 roadblock in SARS-CoV-2 polymerase elongation complex, Cell Rep. 37(4) (2021) 109882. [DOI] [PMC free article] [PubMed]
- 33.Shiraki K., Daikoku T. Favipiravir, an anti-influenza drug against life-threatening RNA virus infections. Pharmacol. Ther. 2020;209 doi: 10.1016/j.pharmthera.2020.107512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sreekanth Reddy O., Lai W.-F. Tackling COVID-19 Using Remdesivir and Favipiravir as Therapeutic Options. Chembiochem. 2021;22(6):939–948. doi: 10.1002/cbic.202000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen P.J., Chao C.M., Lai C.C. Clinical efficacy and safety of favipiravir in the treatment of COVID-19 patients. J Infect. 2021;82(5):186–230. doi: 10.1016/j.jinf.2020.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dauby N., Van Praet S., Vanhomwegen C., Veliziotis I., Konopnicki D., Roman A. Tolerability of favipiravir therapy in critically ill patients with COVID-19: A report of four cases. J. Med. Virol. 2021;93(2):689–691. doi: 10.1002/jmv.26488. [DOI] [PubMed] [Google Scholar]
- 37.Udwadia Z.F., Singh P., Barkate H., Patil S., Rangwala S., Pendse A., et al. Efficacy and safety of favipiravir, an oral RNA-dependent RNA polymerase inhibitor, in mild-to-moderate COVID-19: A randomized, comparative, open-label, multicenter, phase 3 clinical trial. Int J Infect Dis. 2021;103:62–71. doi: 10.1016/j.ijid.2020.11.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Naydenova K., Muir K.W., Wu L.F., Zhang Z., Coscia F., Peet M.J., et al. Structure of the SARS-CoV-2 RNA-dependent RNA polymerase in the presence of favipiravir-RTP. Proc. Natl. Acad. Sci. U. S. A. 2021;118(7) doi: 10.1073/pnas.2021946118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shannon A., Selisko B., Le N.T.T., Huchting J., Touret F., Piorkowski G., et al. Rapid incorporation of Favipiravir by the fast and permissive viral RNA polymerase complex results in SARS-CoV-2 lethal mutagenesis. Nat. Commun. 2020;11(1):4682. doi: 10.1038/s41467-020-18463-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peng Q., Peng R., Yuan B., Wang M., Zhao J., Fu L., et al. Structural Basis of SARS-CoV-2 Polymerase Inhibition by Favipiravir. Innovation (Cambridge (Mass.)) 2021;2(1) doi: 10.1016/j.xinn.2021.100080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hashemian S.M., Farhadi T., Velayati A.A. A review on favipiravir: the properties, function, and usefulness to treat COVID-19. Expert Rev Anti Infect Ther. 2021;19(8):1029–1037. doi: 10.1080/14787210.2021.1866545. [DOI] [PubMed] [Google Scholar]
- 42.Delang L., Abdelnabi R., Neyts J. Favipiravir as a potential countermeasure against neglected and emerging RNA viruses. Antiviral Res. 2018;153:85–94. doi: 10.1016/j.antiviral.2018.03.003. [DOI] [PubMed] [Google Scholar]
- 43.Nagata T., Lefor A.K., Hasegawa M., Ishii M. Favipiravir: a new medication for the Ebola virus disease pandemic. Disaster Med. Public Health Prep. 2015;9(1):79–81. doi: 10.1017/dmp.2014.151. [DOI] [PubMed] [Google Scholar]
- 44.Ghasemnejad Berenji M., Pashapour S. Favipiravir and COVID-19: A Simplified Summary. Drug Res. 2021;71(3):166–170. doi: 10.1055/a-1296-7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vicenti I., Zazzi M., Saladini F. SARS-CoV-2 RNA-dependent RNA polymerase as a therapeutic target for COVID-19. Expert Opin. Ther. Pat. 2021;31(4):325–337. doi: 10.1080/13543776.2021.1880568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pourkarim F., Pourtaghi-Anvarian S., Rezaee H. Molnupiravir: A new candidate for COVID-19 treatment. Pharmacol. Res. Perspect. 2022;10(1):e00909. doi: 10.1002/prp2.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wahl A., Gralinski L.E., Johnson C.E., Yao W., Kovarova M., Dinnon K.H., et al. SARS-CoV-2 infection is effectively treated and prevented by EIDD-2801. Nature. 2021;591(7850):451–457. doi: 10.1038/s41586-021-03312-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Urakova N., Kuznetsova V., Crossman D.K., Sokratian A., Guthrie D.B., Kolykhalov A.A., et al. β-d- -Hydroxycytidine Is a Potent Anti-alphavirus Compound That Induces a High Level of Mutations in the Viral Genome. J. Virol. 2018;92(3):e01965–e02017. doi: 10.1128/JVI.01965-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou S., Hill C.S., Sarkar S., Tse L.V., Woodburn B.M.D., Schinazi R.F., et al. β-d-N4-hydroxycytidine Inhibits SARS-CoV-2 Through Lethal Mutagenesis But Is Also Mutagenic To Mammalian Cells. J Infect Dis. 2021;224(3):415–419. doi: 10.1093/infdis/jiab247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Padhi A.K., Shukla R., Saudagar P., Tripathi T. High-throughput rational design of the remdesivir binding site in the RdRp of SARS-CoV-2: implications for potential resistance. IScience. 2021;24(1) doi: 10.1016/j.isci.2020.101992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jayk Bernal A., Gomes da Silva M.M., Musungaie D.B., Kovalchuk E., Gonzalez A., Delos Reyes V., et al. Molnupiravir for Oral Treatment of Covid-19 in Nonhospitalized Patients. N Engl J Med. 2022;386(6):509–520. doi: 10.1056/NEJMoa2116044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mahase E. Covid-19: Molnupiravir reduces risk of hospital admission or death by 50% in patients at risk, MSD reports. BMJ (Clinical Research ed.) 2021;375 doi: 10.1136/bmj.n2422. [DOI] [PubMed] [Google Scholar]
- 53.Singh A.K., Singh A., Singh R., Misra A. Molnupiravir in COVID-19: A systematic review of literature. Diabetes Metab. Syndr. 2021;15(6) doi: 10.1016/j.dsx.2021.102329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parums D.V. Editorial: Current Status of Oral Antiviral Drug Treatments for SARS-CoV-2 Infection in Non-Hospitalized Patients. Med Sci Monit. 2022;28:e935952. doi: 10.12659/MSM.935952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Painter W.P., Holman W., Bush J.A., Almazedi F., Malik H., Eraut N.C.J.E., et al. Human Safety, Tolerability, and Pharmacokinetics of Molnupiravir, a Novel Broad-Spectrum Oral Antiviral Agent with Activity Against SARS-CoV-2. Antimicrob. Agents Chemother. 2021;65(5) doi: 10.1128/AAC.02428-20. e02428-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Agostini M.L., Pruijssers A.J., Chappell J.D., Gribble J., Lu X., Andres E.L., et al. Small-Molecule Antiviral β-d- -Hydroxycytidine Inhibits a Proofreading-Intact Coronavirus with a High Genetic Barrier to Resistance. J. Virol. 2019;93(24) doi: 10.1128/JVI.01348-19. e01348-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gordon C.J., Tchesnokov E.P., Schinazi R.F., Götte M. Molnupiravir promotes SARS-CoV-2 mutagenesis via the RNA template. J Biol Chem. 2021;297(1) doi: 10.1016/j.jbc.2021.100770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kabinger F., Stiller C., Schmitzová J., Dienemann C., Kokic G., Hillen H.S., et al. Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis. Nat. Struct. Mol. Biol. 2021;28(9):740–746. doi: 10.1038/s41594-021-00651-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Warren T.K., Wells J., Panchal R.G., Stuthman K.S., Garza N.L., Van Tongeren S.A., et al. Protection against filovirus diseases by a novel broad-spectrum nucleoside analogue BCX4430. Nature. 2014;508(7496):402–405. doi: 10.1038/nature13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aftab S.O., Ghouri M.Z., Masood M.U., Haider Z., Khan Z., Ahmad A., et al. Analysis of SARS-CoV-2 RNA-dependent RNA polymerase as a potential therapeutic drug target using a computational approach. J. Transl. Med. 2020;18(1):275. doi: 10.1186/s12967-020-02439-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Elfiky A.A. Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): A molecular docking study. Life Sci. 2020;253 doi: 10.1016/j.lfs.2020.117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Julander J.G., Siddharthan V., Evans J., Taylor R., Tolbert K., Apuli C., et al. Efficacy of the broad-spectrum antiviral compound BCX4430 against Zika virus in cell culture and in a mouse model. Antiviral Res. 2017;137:14–22. doi: 10.1016/j.antiviral.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Taylor R., Kotian P., Warren T., Panchal R., Bavari S., Julander J., et al. BCX4430 - A broad-spectrum antiviral adenosine nucleoside analog under development for the treatment of Ebola virus disease. J Infect Public Health. 2016;9(3):220–226. doi: 10.1016/j.jiph.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Taylor R., Bowen R., Demarest J.F., DeSpirito M., Hartwig A., Bielefeldt-Ohmann H., et al. Activity of Galidesivir in a Hamster Model of SARS-CoV-2. Viruses. 2021;14(1) doi: 10.3390/v14010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sidwell R.W., Huffman J.H., Khare G.P., Allen L.B., Witkowski J.T., Robins R.K. Broad-spectrum antiviral activity of Virazole: 1-beta-D-ribofuranosyl-1,2,4-triazole-3-carboxamide. Science (New York, N.Y.) 1972;177(4050):705–706. doi: 10.1126/science.177.4050.705. [DOI] [PubMed] [Google Scholar]
- 66.Crotty S., Maag D., Arnold J.J., Zhong W., Lau J.Y., Hong Z., et al. The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat. Med. 2000;6(12):1375–1379. doi: 10.1038/82191. [DOI] [PubMed] [Google Scholar]
- 67.Ferron F., Subissi L., Silveira De Morais A.T., Le N.T.T., Sevajol M., Gluais L., et al. Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc. Natl. Acad. Sci. USA. 2018;115(2) doi: 10.1073/pnas.1718806115. E162–E71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Elfiky A.A. SARS-CoV-2 RNA dependent RNA polymerase (RdRp) targeting: an perspective. J. Biomol. Struct. Dyn. 2021;39(9):3204–3212. doi: 10.1080/07391102.2020.1761882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Elfiky A.A. Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sci. 2020;248 doi: 10.1016/j.lfs.2020.117477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xu Y., Li M., Zhou L., Liu D., He W., Liang W., et al. Ribavirin Treatment for Critically Ill COVID-19 Patients: An Observational Study. Infect. Drug Resist. 2021;14:5287–5291. doi: 10.2147/IDR.S330743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hung I.F.N., Lung K.C., Tso E.Y.K., Liu R., Chung T.W.H., Chu M.Y., et al. Triple combination of interferon beta-1b, lopinavir-ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. Lancet (London, England) 2020;395(10238):1695–1704. doi: 10.1016/S0140-6736(20)31042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brochot E., Castelain S., Duverlie G., Capron D., Nguyen-Khac E., François C. Ribavirin monitoring in chronic hepatitis C therapy: anaemia versus efficacy. Antivir. Ther. 2010;15(5):687–695. doi: 10.3851/IMP1609. [DOI] [PubMed] [Google Scholar]
- 73.Heo Y.A., Deeks E.D. Sofosbuvir/Velpatasvir/Voxilaprevir: A Review in Chronic Hepatitis C. Drugs. 2018;78(5):577–587. doi: 10.1007/s40265-018-0895-5. [DOI] [PubMed] [Google Scholar]
- 74.Smith M.A., Regal R.E., Mohammad R.A. Daclatasvir: A NS5A Replication Complex Inhibitor for Hepatitis C Infection. Ann Pharmacother. 2016;50(1):39–46. doi: 10.1177/1060028015610342. [DOI] [PubMed] [Google Scholar]
- 75.C.Q. Sacramento, N. Fintelman-Rodrigues, J.R. Temerozo, A.d.P.D. Da Silva, S.d.S.G. Dias, C.D.S. da Silva, et al., In vitro antiviral activity of the anti-HCV drugs daclatasvir and sofosbuvir against SARS-CoV-2, the aetiological agent of COVID-19, J Antimicrob Chemother.76(7) (2021) 1874–1885. [DOI] [PMC free article] [PubMed]
- 76.Sadeghi A., Ali Asgari A., Norouzi A., Kheiri Z., Anushirvani A., Montazeri M., et al. Sofosbuvir and daclatasvir compared with standard of care in the treatment of patients admitted to hospital with moderate or severe coronavirus infection (COVID-19): a randomized controlled trial. J. Antimicrob. Chemother. 2020;75(11):3379–3385. doi: 10.1093/jac/dkaa334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chien M., Anderson T.K., Jockusch S., Tao C., Li X., Kumar S., et al. Nucleotide Analogues as Inhibitors of SARS-CoV-2 Polymerase, a Key Drug Target for COVID-19. J. Proteome Res. 2020;19(11):4690–4697. doi: 10.1021/acs.jproteome.0c00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ahmad M., Dwivedy A., Mariadasse R., Tiwari S., Kar D., Jeyakanthan J., et al. Prediction of Small Molecule Inhibitors Targeting the Severe Acute Respiratory Syndrome Coronavirus-2 RNA-dependent RNA Polymerase. ACS Omega. 2020;5(29):18356–18366. doi: 10.1021/acsomega.0c02096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jockusch S., Tao C., Li X., Chien M., Kumar S., Morozova I., et al. Sofosbuvir terminated RNA is more resistant to SARS-CoV-2 proofreader than RNA terminated by Remdesivir. Sci. Rep. 2020;10(1):16577. doi: 10.1038/s41598-020-73641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.De Clercq E. Tenofovir alafenamide (TAF) as the successor of tenofovir disoproxil fumarate (TDF) Biochem. Pharmacol. 2016;119:1–7. doi: 10.1016/j.bcp.2016.04.015. [DOI] [PubMed] [Google Scholar]
- 81.Lou L. Advances in Nucleotide Antiviral Development from Scientific Discovery to Clinical Applications: Tenofovir Disoproxil Fumarate for Hepatitis B. J. Clin. Transl. Hepatol. 2013;1(1):33–38. doi: 10.14218/JCTH.2013.004XX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Feng J.Y., Ly J.K., Myrick F., Goodman D., White K.L., Svarovskaia E.S., et al. The triple combination of tenofovir, emtricitabine and efavirenz shows synergistic anti-HIV-1 activity in vitro: a mechanism of action study. Retrovirology. 2009;6:44. doi: 10.1186/1742-4690-6-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.J. J. Parienti, T. Prazuck, L. Peyro Saint Paul, A. Fournier, C. Valentin, S. Brucato, et al., Effect of Tenofovir Disoproxil Fumarate and Emtricitabine on nasopharyngeal SARS-CoV-2 viral load burden amongst outpatients with COVID-19: A pilot, randomized, open-label phase 2 trial, EClinicalMedicine. 38 (2021) 100993. [DOI] [PMC free article] [PubMed]
- 84.Feng J.Y., Du Pont V., Babusis D., Gordon C.J., Tchesnokov E.P., Perry J.K., et al. The Nucleoside/Nucleotide Analogs Tenofovir and Emtricitabine Are Inactive against SARS-CoV-2, Molecules (Basel. Switzerland) 2022;27(13):4212. doi: 10.3390/molecules27134212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Choy K.T., Wong A.Y.L., Kaewpreedee P., Sia S.F., Chen D., Hui K.P.Y., et al. Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antiviral Res. 2020;178 doi: 10.1016/j.antiviral.2020.104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.I. Zanella, D. Zizioli, F. Castelli, E. Quiros-Roldan, Tenofovir, Another Inexpensive, Well-Known and Widely Available Old Drug Repurposed for SARS-COV-2 Infection, Pharmaceuticals (Basel) 14(5) (2021) 454. [DOI] [PMC free article] [PubMed]
- 87.Xie Y., Yin W., Zhang Y., Shang W., Wang Z., Luan X., et al. Design and development of an oral remdesivir derivative VV116 against SARS-CoV-2. Cell Res. 2021;31(11):1212–1214. doi: 10.1038/s41422-021-00570-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li Y., Yang B., Quan Y., Li Z. Advancement of Prodrug Approaches for Nucleotide Antiviral Agents. Curr. Top. Med. Chem. 2021;21(32):2909–2927. doi: 10.2174/1568026621666210728094019. [DOI] [PubMed] [Google Scholar]
- 89.Qian H.J., Wang Y., Zhang M.Q., Xie Y.C., Wu Q.Q., Liang L.Y., et al. Safety, tolerability, and pharmacokinetics of VV116, an oral nucleoside analog against SARS-CoV-2, in Chinese healthy subjects. Acta Pharmacol. Sin. 2022;1–9 doi: 10.1038/s41401-022-00895-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shen Y., Ai J., Lin N., Zhang H., Li Y., Wang H., et al. An open, prospective cohort study of VV116 in Chinese participants infected with SARS-CoV-2 omicron variants. Emergi. Microbes Infect. 2022;11(1):1518–1523. doi: 10.1080/22221751.2022.2078230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mackman R.L., Hui H.C., Perron M., Murakami E., Palmiotti C., Lee G., et al. Prodrugs of a 1'-CN-4-Aza-7,9-dideazaadenosine -Nucleoside Leading to the Discovery of Remdesivir (GS-5734) as a Potent Inhibitor of Respiratory Syncytial Virus with Efficacy in the African Green Monkey Model of RSV. J. Med. Chem. 2021;64(8):5001–5017. doi: 10.1021/acs.jmedchem.1c00071. [DOI] [PubMed] [Google Scholar]
- 92.Schäfer A., Martinez D.R., Won J.J., Meganck R.M., Moreira F.R., Brown A.J., et al. Therapeutic treatment with an oral prodrug of the remdesivir parental nucleoside is protective against SARS-CoV-2 pathogenesis in mice. Sci. Transl. Med. 2022;14(643):eabm3410. doi: 10.1126/scitranslmed.abm3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cox R.M., Wolf J.D., Lieber C.M., Sourimant J., Lin M.J., Babusis D., et al. Oral prodrug of remdesivir parent GS-441524 is efficacious against SARS-CoV-2 in ferrets. Nat. Commun. 2021;12(1):6415. doi: 10.1038/s41467-021-26760-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Good S.S., Westover J., Jung K.H., Zhou X.-J., Moussa A., La Colla P., et al. AT-527, a Double Prodrug of a Guanosine Nucleotide Analog, Is a Potent Inhibitor of SARS-CoV-2 and a Promising Oral Antiviral for Treatment of COVID-19. Antimicrob. Agents Chemother. 2021;65(4) doi: 10.1128/AAC.02479-20. e02479–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.C.B. Dousson, Current and future use of nucleo(s)tide prodrugs in the treatment of hepatitis C virus infection, Antivir Chem Chemother. 26 (2018) 2040206618756430. [DOI] [PMC free article] [PubMed]
- 96.Han Y.J., Lee K.H., Yoon S., Nam S.W., Ryu S., Seong D., et al. Treatment of severe acute respiratory syndrome (SARS), Middle East respiratory syndrome (MERS), and coronavirus disease 2019 (COVID-19): a systematic review of in vitro, in vivo, and clinical trials. Theranostics. 2021;11(3):1207–1231. doi: 10.7150/thno.48342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shannon A., Fattorini V., Sama B., Selisko B., Feracci M., Falcou C., et al. A dual mechanism of action of AT-527 against SARS-CoV-2 polymerase. Nat. Commun. 2022;13(1):621. doi: 10.1038/s41467-022-28113-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ma Y., Wu L., Shaw N., Gao Y., Wang J., Sun Y., et al. Structural basis and functional analysis of the SARS coronavirus nsp14-nsp10 complex. Proc. Natl. Acad. Sci. U. S. A. 2015;112(30):9436–9441. doi: 10.1073/pnas.1508686112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sofia M.J., Chang W., Furman P.A., Mosley R.T., Ross B.S. Nucleoside, nucleotide, and non-nucleoside inhibitors of hepatitis C virus NS5B RNA-dependent RNA-polymerase. J. Med. Chem. 2012;55(6):2481–2531. doi: 10.1021/jm201384j. [DOI] [PubMed] [Google Scholar]
- 100.Naidu S.A.G., Mustafa G., Clemens R.A., Naidu A.S. Plant-Derived Natural Non-Nucleoside Analog Inhibitors (NNAIs) against Complex (nsp7/nsp8/nsp12) of SARS-CoV-2. J. Diet. Suppl. 2021:1–30. doi: 10.1080/19390211.2021.2006387. [DOI] [PubMed] [Google Scholar]
- 101.Wiedemar N., Hauser D.A., Mäser P. 100 Years of Suramin. Antimicrob. Agents Chemother. 2020;64(3) doi: 10.1128/AAC.01168-19. e01168-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Albulescu I.C., White-Scholten L., Tas A., Hoornweg T.E., Ferla S., Kovacikova K., et al. Suramin Inhibits Chikungunya Virus Replication by Interacting with Virions and Blocking the Early Steps of Infection. Viruses. 2020;12(3):314. doi: 10.3390/v12030314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Albulescu I.C., Kovacikova K., Tas A., Snijder E.J., van Hemert M.J. Suramin inhibits Zika virus replication by interfering with virus attachment and release of infectious particles. Antiviral Res. 2017;143:230–236. doi: 10.1016/j.antiviral.2017.04.016. [DOI] [PubMed] [Google Scholar]
- 104.Salgado-Benvindo C., Thaler M., Tas A., Ogando N.S., Bredenbeek P.J., Ninaber D.K., et al. Suramin Inhibits SARS-CoV-2 Infection in Cell Culture by Interfering with Early Steps of the Replication Cycle. Antimicrob. Agents Chemother. 2020;64(8) doi: 10.1128/AAC.00900-20. e00900-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yin W., Luan X., Li Z., Zhou Z., Wang Q., Gao M., et al. Structural basis for inhibition of the SARS-CoV-2 RNA polymerase by suramin. Nat. Struct. Mol. Biol. 2021;28(3):319–325. doi: 10.1038/s41594-021-00570-0. [DOI] [PubMed] [Google Scholar]
- 106.Bertolin A.P., Weissmann F., Zeng J., Posse V., Milligan J.C., Canal B., et al. Identifying SARS-CoV-2 antiviral compounds by screening for small molecule inhibitors of nsp12/7/8 RNA-dependent RNA polymerase. Biochem J. 2021;478(13):2425–2443. doi: 10.1042/BCJ20210200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wu K., Chong R.A., Yu Q., Bai J., Spratt D.E., Ching K., et al. Suramin inhibits cullin-RING E3 ubiquitin ligases. Proc. Natl. Acad. Sci. USA. 2016;113(14) doi: 10.1073/pnas.1601089113. E2011-E2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Eberle R.J., Olivier D.S., Amaral M.S., Gering I., Willbold D., Arni R.K., et al. The Repurposed Drugs Suramin and Quinacrine Cooperatively Inhibit SARS-CoV-2 3CL In Vitro. Viruses. 2021;13(5):873. doi: 10.3390/v13050873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Henß L., Beck S., Weidner T., Biedenkopf N., Sliva K., Weber C., et al. Suramin is a potent inhibitor of Chikungunya and Ebola virus cell entry. Virol J. 2016;13:149. doi: 10.1186/s12985-016-0607-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Stein C.A., LaRocca R.V., Thomas R., McAtee N., Myers C.E. Suramin: an anticancer drug with a unique mechanism of action. J Clin Oncol. 1989;7(4):499–508. doi: 10.1200/JCO.1989.7.4.499. [DOI] [PubMed] [Google Scholar]
- 111.L.D. Kaplan, P.R. Wolfe, P.A. Volberding, P. Feorino, J.A. Levy, D.I. Abrams, et al., Lack of response to suramin in patients with AIDS and AIDS-related complex, Am J Med. 82(3 Spec No) (1987) 615–620. [DOI] [PubMed]
- 112.Matada B.S., Pattanashettar R., Yernale N.G. A comprehensive review on the biological interest of quinoline and its derivatives. Biorg. Med. Chem. 2021;32 doi: 10.1016/j.bmc.2020.115973. [DOI] [PubMed] [Google Scholar]
- 113.De S., Aamna B., Sahu R., Parida S., Behera S.K., Dan A.K. Seeking heterocyclic scaffolds as antivirals against dengue virus. Eur. J. Med. Chem. 2022;240 doi: 10.1016/j.ejmech.2022.114576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Marella A., Tanwar O.P., Saha R., Ali M.R., Srivastava S., Akhter M., et al. Quinoline: A versatile heterocyclic. Saudi Pharm J. 2013;21(1):1–12. doi: 10.1016/j.jsps.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zheng X., Liang C., Wang L., Wang B., Liu Y., Feng S., et al. Discovery of Benzoazepinequinoline (BAQ) Derivatives as Novel, Potent, Orally Bioavailable Respiratory Syncytial Virus Fusion Inhibitors. J. Med. Chem. 2018;61(22):10228–10241. doi: 10.1021/acs.jmedchem.8b01394. [DOI] [PubMed] [Google Scholar]
- 116.Zhang G., Wang M., Zhao J., Wang Y., Zhu M., Wang J., et al. Design, synthesis and in vitro anti-influenza A virus evaluation of novel quinazoline derivatives containing S-acetamide and NH-acetamide moieties at C-4. Eur. J. Med. Chem. 2020;206 doi: 10.1016/j.ejmech.2020.112706. [DOI] [PubMed] [Google Scholar]
- 117.Wan Z., Hu D., Li P., Xie D., Gan X. Synthesis, Antiviral Bioactivity of Novel 4-Thioquinazoline Derivatives Containing Chalcone Moiety. Molecules (Basel, Switzerland) 2015;20(7):11861–11874. doi: 10.3390/molecules200711861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhao J., Zhang Y., Wang M., Liu Q., Lei X., Wu M., et al. Quinoline and Quinazoline Derivatives Inhibit Viral RNA Synthesis by SARS-CoV-2 RdRp. ACS Infect Dis. 2021;7(6):1535–1544. doi: 10.1021/acsinfecdis.1c00083. [DOI] [PubMed] [Google Scholar]
- 119.Li X., Deng Y., Zheng Z., Huang W., Chen L., Tong Q., et al. Corilagin, a promising medicinal herbal agent. Biomed. Pharmacother. 2018;99:43–50. doi: 10.1016/j.biopha.2018.01.030. [DOI] [PubMed] [Google Scholar]
- 120.Li Q., Yi D., Lei X., Zhao J., Zhang Y., Cui X., et al. Corilagin inhibits SARS-CoV-2 replication by targeting viral RNA-dependent RNA polymerase. Acta Pharmaceutica Sinica. B. 2021;11(6):1555–1567. doi: 10.1016/j.apsb.2021.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Reddy B.U., Mullick R., Kumar A., Sharma G., Bag P., Roy C.L., et al. A natural small molecule inhibitor corilagin blocks HCV replication and modulates oxidative stress to reduce liver damage. Antiviral Res. 2018;150:47–59. doi: 10.1016/j.antiviral.2017.12.004. [DOI] [PubMed] [Google Scholar]
- 122.Yang L.J., Chen R.H., Hamdoun S., Coghi P., Ng J.P.L., Zhang D.W., et al. Corilagin prevents SARS-CoV-2 infection by targeting RBD-ACE2 binding. Phytomedicine. 2021;87 doi: 10.1016/j.phymed.2021.153591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Xiao H., Xu X., Du L., Li X., Zhao H., Wang Z., et al. Lycorine and organ protection: Review of its potential effects and molecular mechanisms. Phytomedicine. 2022;104 doi: 10.1016/j.phymed.2022.154266. [DOI] [PubMed] [Google Scholar]
- 124.Shen L., Niu J., Wang C., Huang B., Wang W., Zhu N., et al. High-Throughput Screening and Identification of Potent Broad-Spectrum Inhibitors of Coronaviruses. J. Virol. 2019;93(12) doi: 10.1128/JVI.00023-19. e00023-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mani J.S., Johnson J.B., Steel J.C., Broszczak D.A., Neilsen P.M., Walsh K.B., et al. Natural product-derived phytochemicals as potential agents against coronaviruses: A review. Virus Res. 2020;284 doi: 10.1016/j.virusres.2020.197989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhang Y.N., Zhang Q.Y., Li X.D., Xiong J., Xiao S.Q., Wang Z., et al. Gemcitabine, lycorine and oxysophoridine inhibit novel coronavirus (SARS-CoV-2) in cell culture. Emerg. Microbes Infect. 2020;9(1):1170–1173. doi: 10.1080/22221751.2020.1772676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ren P.X., Shang W.J., Yin W.C., Ge H., Wang L., Zhang X.L., et al. A multi-targeting drug design strategy for identifying potent anti-SARS-CoV-2 inhibitors. Acta Pharmacol. Sin. 2022;43(2):483–493. doi: 10.1038/s41401-021-00668-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jin Y.H., Min J.S., Jeon S., Lee J., Kim S., Park T., et al. Lycorine, a non-nucleoside RNA dependent RNA polymerase inhibitor, as potential treatment for emerging coronavirus infections. Phytomedicine. 2021;86 doi: 10.1016/j.phymed.2020.153440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhao Q., Chen X.Y., Martin C. the golden herb from the garden of Chinese medicinal plants. Sci. Bull. (Beijing). 2016;61(18):1391–1398. doi: 10.1007/s11434-016-1136-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Low Z.X., OuYong B.M., Hassandarvish P., Poh C.L., Ramanathan B. Antiviral activity of silymarin and baicalein against dengue virus. Sci. Rep. 2021;11(1):21221. doi: 10.1038/s41598-021-98949-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Oo A., Teoh B.T., Sam S.S., Bakar S.A., Zandi K. Baicalein and baicalin as Zika virus inhibitors. Arch. Virol. 2019;164(2):585–593. doi: 10.1007/s00705-018-4083-4. [DOI] [PubMed] [Google Scholar]
- 132.Xu G., Dou J., Zhang L., Guo Q., Zhou C. Inhibitory effects of baicalein on the influenza virus in vivo is determined by baicalin in the serum. Biol. Pharm. Bull. 2010;33(2):238–243. doi: 10.1248/bpb.33.238. [DOI] [PubMed] [Google Scholar]
- 133.Li M., Shi A., Pang H., Xue W., Li Y., Cao G., et al. Safety, tolerability, and pharmacokinetics of a single ascending dose of baicalein chewable tablets in healthy subjects. J. Ethnopharmacol. 2014;156:210–215. doi: 10.1016/j.jep.2014.08.031. [DOI] [PubMed] [Google Scholar]
- 134.Pang H., Xue W., Shi A., Li M., Li Y., Cao G., et al. Multiple-Ascending-Dose Pharmacokinetics and Safety Evaluation of Baicalein Chewable Tablets in Healthy Chinese Volunteers. Clin. Drug Investig. 2016;36(9):713–724. doi: 10.1007/s40261-016-0418-7. [DOI] [PubMed] [Google Scholar]
- 135.Liu H., Ye F., Sun Q., Liang H., Li C., Li S., et al. extract and baicalein inhibit replication of SARS-CoV-2 and its 3C-like protease. J. Enzyme Inhib. Med. Chem. 2021;36(1):497–503. doi: 10.1080/14756366.2021.1873977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.S. Huang, Y.e. Liu, Y. Zhang, R. Zhang, C. Zhu, L. Fan, et al., Baicalein inhibits SARS-CoV-2/VSV replication with interfering mitochondrial oxidative phosphorylation in a mPTP dependent manner, Signal Transduction Targeted Ther. 5(1) (2020) 266. [DOI] [PMC free article] [PubMed]
- 137.Song J., Zhang L., Xu Y., Yang D., Zhang L., Yang S., et al. The comprehensive study on the therapeutic effects of baicalein for the treatment of COVID-19 in vivo and in vitro. Biochem. Pharmacol. 2021;183 doi: 10.1016/j.bcp.2020.114302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zandi K., Musall K., Oo A., Cao D., Liang B., Hassandarvish P., et al. Baicalein and Baicalin Inhibit SARS-CoV-2 RNA-Dependent-RNA Polymerase. Microorganisms. 2021;9(5):893. doi: 10.3390/microorganisms9050893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Callaway E. Omicron likely to weaken COVID vaccine protection. Nature. 2021;600(7889):367–368. doi: 10.1038/d41586-021-03672-3. [DOI] [PubMed] [Google Scholar]
- 140.Andrews N., Stowe J., Kirsebom F., Toffa S., Rickeard T., Gallagher E., et al. Covid-19 Vaccine Effectiveness against the Omicron (B.1.1.529) Variant. N. Engl. J. Med. 2022;386(16):1532–1546. doi: 10.1056/NEJMoa2119451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Karim S.S.A., Karim Q.A. Omicron SARS-CoV-2 variant: a new chapter in the COVID-19 pandemic. Lancet (London, England) 2021;398(10317):2126–2128. doi: 10.1016/S0140-6736(21)02758-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kuhlmann C., Mayer C.K., Claassen M., Maponga T., Burgers W.A., Keeton R., et al. Breakthrough infections with SARS-CoV-2 omicron despite mRNA vaccine booster dose. Lancet (London, England) 2022;399(10325):625–626. doi: 10.1016/S0140-6736(22)00090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhao J., Guo S., Yi D., Li Q., Ma L., Zhang Y., et al. A cell-based assay to discover inhibitors of SARS-CoV-2 RNA dependent RNA polymerase. Antiviral Res. 2021;190 doi: 10.1016/j.antiviral.2021.105078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.O.M. Ogunyemi, G.A. Gyebi, A.A. Elfiky, S.O. Afolabi, O.B. Ogunro, A.P. Adegunloye, et al., Alkaloids and flavonoids from African phytochemicals as potential inhibitors of SARS-Cov-2 RNA-dependent RNA polymerase: an perspective, Antivir. Chem. Chemother. 28 (2020) 2040206620984076. [DOI] [PMC free article] [PubMed]
- 145.Warowicka A., Nawrot R., Goździcka-Józefiak A. Antiviral activity of berberine. Arch. Virol. 2020;165(9):1935–1945. doi: 10.1007/s00705-020-04706-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Jin Y.H., Lee J., Jeon S., Kim S., Min J.S., Kwon S. Natural Polyphenols, 1,2,3,4,6-O-Pentagalloyglucose and Proanthocyanidins, as Broad-Spectrum Anticoronaviral Inhibitors Targeting Mpro and RdRp of SARS-CoV-2. Biomedicines. 2022;10(5):1170. doi: 10.3390/biomedicines10051170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Mhatre S., Srivastava T., Naik S., Patravale V. Antiviral activity of green tea and black tea polyphenols in prophylaxis and treatment of COVID-19: A review. Phytomedicine. 2021;85 doi: 10.1016/j.phymed.2020.153286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Singh S., Sk M.F., Sonawane A., Kar P., Sadhukhan S. Plant-derived natural polyphenols as potential antiviral drugs against SARS-CoV-2 RNA-dependent RNA polymerase (RdRp) inhibition: an analysis. J. Biomol. Struct. Dyn. 2021;39(16):6249–6264. doi: 10.1080/07391102.2020.1796810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hamdy R., Mostafa A., Abo Shama N.M., Soliman S.S.M., Fayed B. Comparative evaluation of flavonoids reveals the superiority and promising inhibition activity of silibinin against SARS-CoV-2. Phytother Res. 2022;36(7):2921–2939. doi: 10.1002/ptr.7486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Bibi S., Hasan M.M., Wang Y.B., Papadakos S.P., Yu H. Cordycepin as a Promising Inhibitor of SARS-CoV-2 RNA Dependent RNA Polymerase (RdRp) Curr. Med. Chem. 2022;29(1):152–162. doi: 10.2174/0929867328666210820114025. [DOI] [PubMed] [Google Scholar]