Abstract
Expression of Bacillus subtilis aprE, encoding an extracellular alkaline protease, is positively regulated by phosphorylated DegU, the regulator of a two-component regulatory system, DegS-DegU. We found that the expression of an aprE′-′lacZ fusion was greatly reduced in a disruption mutant with a mutation of relA, which encodes the stringent factor RelA. The level of DegU in the relA mutant was similar to that in the wild-type cell. A relA degU double mutation did not result in a further decrease of the aprE′-′lacZ level found in a degU single mutant. The expression of the aprE′-′lacZ fusion in the relA mutant was stimulated by multicopy degR or the degU32(Hy) and degS200(Hy) mutations that cause the stabilization of phosphorylated DegU. Furthermore, the expression of sacB′-′lacZ, which is also dependent on phosphorylated DegU, was stimulated by the relA mutation, and this stimulation was not seen in the relA degU double mutant. These results show that RelA (or its product guanosine-3′,5′-bisdiphosphate [pp Gpp]) does not affect the phosphorylation of DegU and suggest that it participates in the expression of aprE and sacB through the regulation of DegU-dependent transcription.
Bacillus subtilis produces two major extracellular proteases, the alkaline and neutral proteases, which are produced after the cessation of exponential growth. The expression of the aprE gene, encoding the alkaline protease, is regulated at the level of transcription both by positive regulators including the two-component regulatory system DegS-DegU and by negative regulatory factors (5, 7, 12, 20). Among the positive factors, the DegS-DegU pair plays a central role, since disruption of either degS or degU results in a substantial decrease in aprE expression (3, 14), and obliterates the effects of the positive regulators (11, 15, 17, 18).
Microorganisms living in nature adapt to changing environments such as starvation, desiccation, osmotic stress, and temperature variations for survival. One such example of adaptation is the stringent response in which the synthesis of many high-molecular-weight components including stable RNA (rRNA and tRNA) becomes limited while gene expression of some others including biosynthesis genes of amino acids is activated (1). This response leads to adjustments of gene expression which are thought to be mediated by guanosine 3′,5′-bisdiphosphate (ppGpp) synthesized by the stringent factor RelA (1). ppGpp accumulation is provoked by many stress conditions including heat shock, oxidative stress, and deficiency of amino acids and carbon, nitrogen, and phosphate sources. Numerous studies have been performed on the Escherichia coli and Salmonella enterica serovar Typhimurium stringent response (1), and recently the Bacillus subtilis relA gene was cloned and studied by Wendrich and Marahiel (23).
B. subtilis is a gram-positive bacterium living in soil, where it may encounter severe fluctuations in the environment, including deficiency of nutrients and a variety of stresses. It may therefore be reasonable to assume that a number of adaptive responses have evolved to cope with such situations. One such example will be the secretion of proteases: they may be secreted from B. subtilis cells in response to limitation of the intracellular nitrogen source so that they can digest high-molecular-weight proteins present in the environments and provide the cells with amino acids. Since these enzymes are produced in considerable amounts, the production may be strictly controlled in response to the environmental conditions. These considerations prompted us to examine whether the stringent response is involved in aprE expression. In this paper we show that RelA exerts positive and negative regulation on aprE and sacB, respectively, in a DegU-dependent manner.
Involvement of relA in aprE expression.
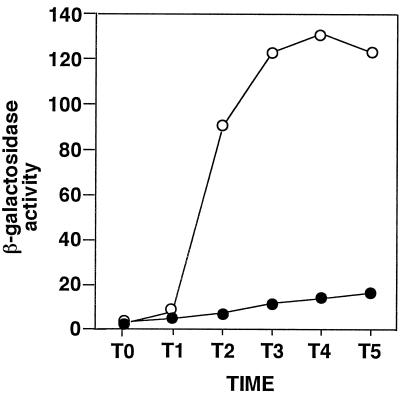
The relA gene, encoding a 734-amino-acid protein, is located at a region from nucleotides 2821998 to 2819794 on the B. subtilis chromosome (8). We disrupted the relA gene in strain YY102 (see the legend to Fig. 1) by insertion of tetracycline resistance gene (tet) (6) at the EcoT22I site (codon 333) by a double-crossover event. As shown in Fig. 1, the expression of aprE′-′lacZ was greatly reduced in the resultant relA333 mutant (HT1013) grown in Schaeffer medium. Disruption of the downstream gene yrvI (8) did not affect aprE′-′lacZ expression (data not shown), indicating that the above result was not due to a polar effect of relA disruption on the downstream gene. The level of ppGpp in strain HT1013 during vegetative and stationary-phase growth in Schaeffer medium was found to be about 1/20 the level found in the wild-type strain, CU741 (data not shown).
FIG. 1.
Effect of disruption of relA on aprE′-′lacZ expression. Cells grown overnight in Luria-Bertani medium were inoculated into Schaeffer sporulation medium (19) and harvested at hourly intervals after the cessation of logarithmic growth. β-Galactosidase activities are shown in Miller units. Numbers on the x axis represent the growth time in hours relative to the end of vegetative growth (T0). Symbols: ○, YY102 relA+, ●, HT1013 relA333. Strain YY102 (leuC7 trpC2 aprE::pSKK25) was constructed by insertion of a pUC18-derived, kanamycin resistance plasmid, pSKK25, carrying aprE′-′lacZ at the aprE locus of CU741 (22) by Campbell-type recombination. Strain HT1013 is described in the text.
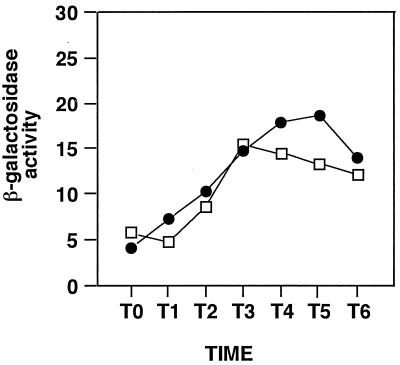
It has been demonstrated that the total level of aprE expression is the sum of degS-degU-dependent and -independent expression and that the former pathway accounts for most of the aprE′-′lacZ expression in nutritional medium (14). We next examined which of the two pathways leading to aprE expression described above is subject to RelA regulation. We constructed two strains carrying deletions in either degU or both degU and relA so that we could examine the effect of the relA333 mutation on DegU-independent aprE expression. As shown in Fig. 2, the β-galactosidase activities found in the two strains were almost the same. These results show that DegU-dependent aprE expression but not DegU-independent expression is subject to control by RelA.
FIG. 2.
Effect of relA mutation on aprE′-′lacZ expression in a degU-deficient mutant. The experimental conditions were the same as those described in the legend to Fig. 1. Numbers on the x axis represent the growth time in hours relative to the end of vegetative growth (T0). Colonies formed on a plate were inoculated into Luria-Bertani medium containing appropriate antibiotics (5 μg of chloramphenicol per ml and 10 μg of tetracycline per ml) and incubated overnight at 37°C. A 2-ml volume of the overnight culture was inoculated in 50 ml of Schaeffer sporulation medium without antibiotics, and samples were withdrawn at the times indicated. The data shown are those obtained in one of two sets of experiments performed under the same conditions and at the same time. β-Galactosidase activities are shown in Miller units. The parental strain B. subtilis CU741 did not show detectable β-galactosidase activity during the growth period. Symbols: ●, HT2113 (relA333 degU); □, YY309 (degU). Strain HT2113 (leuC7 trpC2 relA333 degU::cat aprE::pSKK25) was constructed by transformation of HT1013 with DNA from TT711 (leuC7 trpC2 degU::cat) (20). Strain YY309 (leuC7 trpC2 degU::cat aprE::pSKK25) was constructed by transformation of YY102 with DNA from TT711.
Effect of relA mutation on degU expression.
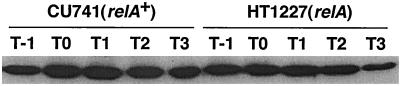
If the relA disruption caused a reduction in degU expression, the decrease in the expression of aprE in the relA-deficient mutant could be ascribed to a reduced level of DegU. To test this possibility, we examined the expression of degU by Western analysis of the relA strain. As shown in Fig. 3, the levels of DegU protein were similar in strain HT1013 and the wild-type strain throughout the time examined. We note that the highest level of DegU was attained after around T1 to T2, which is in contrast to the gradual decline of β-galactosidase activities derived from the degU′-′lacZ fusion after T0 (reference 17 and data not shown). These results indicate that the β-galactosidase activities derived from the degU′-′lacZ fusion do not reflect the actual level of DegU protein in the cell. This may be due either to the degradation of the chimeric β-galactosidase composed of the N-terminal DegU and E. coli β-galactosidase or to the instability of mRNA carrying the mRNA for the fusion protein.
FIG. 3.
Effect of relA disruption on degU expression as determined by Western analysis of DegU. B. subtilis HT1227 (leuC7 trpC2 relA333) was constructed by transformation of YY102 with DNA from HT1013. Numbers above the photograph indicate the growth time in hours relative to T0. DegU was purified by a previously described method (14), and the antibody against DegU was commercially prepared by Sawady Co. For Western blot analysis, cells collected from 10-ml cultures were resuspended in 1 ml of TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA) containing 1 mM phenylmethylsufonyl fluoride, and disrupted in a French press. Proteins were separated by electrophoresis in a 12.5% polyacrylamide gel, and DegU bands were detected by the DegU antibody using the BM Chemiluminescence Western blotting kit (Boehringer Mannheim Co.).
Enhanced expression of aprE by multicopy degR, degU32(Hy), and degS200(Hy) mutations in a relA background.
It has been demonstrated that multicopy degR or the degS200(Hy) mutation causes the stabilization of phosphorylated DegU (4, 5, 21). Similarly, the DegU protein encoded by the degU32(Hy) gene retains phosphate longer than the wild-type DegU protein does (4). The presence of these genetic traits results in enhanced expression of aprE′-′lacZ. Therefore, if the reduced expression of aprE′-′lacZ by the relA mutation is due to a defect in the phosphorylation process of DegU, the enhancing effects would not be observed in the relA background. In fact, the stimulatory effects of multicopy degR and the degU32(Hy) mutation, which require phosphorylation of DegU, are not observed in degS-deficient mutants (5, 15). A multicopy plasmid, pNC61, that we used for this experiment carries degR in a vector, pNC6 (15, 16). We found that the expression of aprE′-′lacZ in the relA cell (17% of the control level) was increased about 250-fold by the presence of pNC61 under conditions where the multicopy degR effect in the relA+ cell was 96-fold (Table 1, experiment 1). Likewise, both the degU32(Hy) and degS200(Hy) mutations caused the stimulation of aprE′-′lacZ expression in the relA cells to levels similar to those attained in the relA+ cells (Table 1, experiment 2). The differences in the magnitude of enhancement between multicopy degR and the degU32(Hy) or degS200(Hy) mutations may be due to the different modes of stabilization of phosphorylated DegU, although the precise mechanisms of stabilization by these genetic traits remain to be studied. These results show either that DegU is phosphorylated in part in the relA cell and stabilized by multicopy degR or the degU32(Hy) and degS200(Hy) mutations or that it is phosphorylated to a level similar to the wild-type level but does not show full activity due to a defect in some other process in the pathway of aprE expression.
TABLE 1.
Expression of aprE′-′lacZ in B. subtilis strains carrying either multicopy degR or degU32(Hy) and degS200(Hy) mutations
| Expt | Straina | Relevant genotype | β-Galactosidase activityb | Stimulationc |
|---|---|---|---|---|
| 1 | YY102(pNC6) | relA+ | 107 | 1 |
| YY102(pNC61) | relA+ multicopy degR | 10,330 | 96 | |
| HT1013(pNC6) | relA | 18 | 1 | |
| HT1013(pNC61) | relA multicopy degR | 4,650 | 258 | |
| 2 | YY102 | relA+ | 133 | 1 |
| YY202 | relA+degU32(Hy) | 9,380 | 70 | |
| YY299 | relA+degS200(Hy) | 7,260 | 54 | |
| HT1013 | relA | 20 | 1 | |
| HT1020 | relA degU32(Hy) | 1,020 | 51 | |
| HT1022 | relA degS200(Hy) | 820 | 41 |
Plasmids carried by the host strains are shown in parentheses. For strains YY102 and HT1013, refer to the legend to Fig. 1. Strains YY202 [trpC2 degU32(Hy) aprE::pSKK25] and YY299trpC2 [degS200(Hy) aprE::pSKK25] are the degU32(Hy) and degS200(Hy) derivatives, respectively, of YY102. Strains HT1020 [trpC2 degU32(Hy) relA::tet aprE::pSKK25] and HT1022 [trpC2 degS200(Hy) relA::tet aprE::pSKK25] are the degU32(Hy) and degS200(Hy) derivatives, respectively, of HT1013.
β-Galactosidase activities in Miller units were measured at T0 through T5 as shown in Fig. 1, and the values shown are the peak values observed during this period.
The values show the extent of stimulation caused by multicopy degR, degU32(Hy), and degS200(Hy) mutations relative to the control level, which was set as unity.
Effect of relA mutation on sacB′-′lacZ expression.
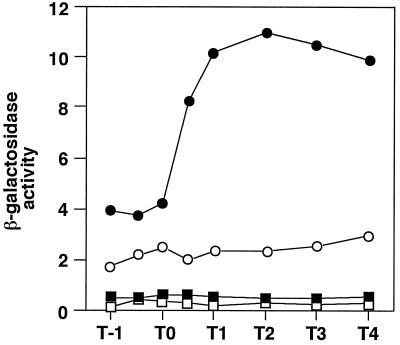
The expression of the sacB gene, encoding extracellular levansucrase, is also positively regulated by phosphorylated DegU. This was shown by the effect of a defect in levansucrase production by a strain carrying the degS42 mutation, which results in the production of an autophosphorylation-defective DegS protein, and by overproduction of the enzyme by strains carrying the degU32(Hy) or degS200(Hy) mutation or multicopy degR (10, 16). We therefore examined the effect of relA deficiency on sacB expression by using a sacB′-′lacZ translational fusion (9). In contrast to its negative effect on aprE expression, the relA mutation caused an enhancement of sacB′-′lacZ expression (Fig. 4). To test whether the enhanced expression of sacB in the relA-deficient mutant is still dependent on DegU, we introduced a degU null mutation into the relA mutant. As shown in Fig. 4, a degU deletion abolished the enhanced expression of sacB′-′lacZ caused by the relA mutation, and the resultant level was as low as that seen in the relA+ degU strain, indicating that the enhanced expression of sacB′-′lacZ in the relA mutant requires DegU.
FIG. 4.
Effect of relA and degU disruptions on sacB′-′lacZ expression. Cells were grown in SCHE medium (9) containing 2% sucrose, collected at the indicated times, and determined for β-galactosidase activity. Numbers on the x axis represent the growth time in hours relative to the end of vegetative growth (T0). The data shown are those obtained in one of two sets of experiments performed under the same conditions and at the same time. The host strain, B. subtilis CU741, did not show detectable β-Galactosidase activity during the growth period. β-Galactosidase activities are shown in Miller units. Symbols: ○, IN6052 (relA+); ●, IN7461 (relA333); □, IN6711 (relA+ degU); ■; IN7141 (relA333 degU). Strain IN6052 [leuC7 trpC2 amyE::(sacB′-′lacZ erm)] was constructed by transformation of CU741 with DNA from QB4624 (9). Strain IN7461 [leuC7 trpC2 relA::tet amyE::(sacB′-′lacZ erm)] was constructed by transformation of HT1227 with DNA from QB4624. Strains IN6711 and IN7141 were constructed by transformation of IN6052 and IN7461, respectively, with DNA from TT711.
In this work, we constructed a relA333 disruption mutation and found that it exerts different effects on the expression of the aprE and sacB genes, although they are in the same DegU regulon (13). From the findings that relA deficiency caused a reduction in aprE′-′lacZ expression but did not affect the intracellular level of DegU protein (Fig. 2), two possibilities are conceivable concerning the target site of RelA (or its reaction product ppGpp) in aprE expression. One is that the target is in a pathway leading to the phosphorylation of DegU. This possibility is, however, unlikely because of the following two results. First, the expression of aprE′-′lacZ was stimulated by multicopy degR or the degU32(Hy) and degS200(Hy) mutations in the relA cell (Table 1). Since the stimulation of aprE expression by these genetic traits is dependent on phosphorylated DegU (5, 15, 21), the results indicate that DegU is phosphorylated at least in part in the relA cells. Second, the expression of both the aprE and sacB genes is known to be regulated positively by phosphorylated DegU (4, 10, 21), but the effects of the relA mutation on these genes were found to be opposite (Fig. 1 and 4). The other possible target site may be in the transcription process, since it is generally accepted that ppGpp affects transcription through interaction with RNA polymerase (references 1 and 2 and references therein). When this notion is applied to the results obtained in this study, it follows that the phosphorylated DegU-dependent transcription is enhanced or inhibited by ppGpp for aprE and sacB, respectively. Here, phosphorylated DegU plays a major role, since a disruption of degU in a relA333 mutant resulted in complete inhibition of the expression of both genes (Fig. 2 and 4). It should be noted that ppGpp regulates numerous genes in positive and negative ways (1).
An entirely different notion is also possible, i.e., that there are two targets of RelA in the process of aprE or sacB expression. For sacB expression, for example, one target reduces phosphorylation of DegU while the other enhances some other process, with an overall effect being positive in the expression of sacB. This possibility, however, may be excluded, since if it were true, a degU knockout would affect only the DegU pathway and would not result in the complete loss of sacB′-′lacZ activity shown in Fig. 4. Therefore, we favor the hypothesis that RelA affects aprE and sacB expression at the level of transcription.
The differential regulation of aprE and sacB expression reported in this study is reminiscent of the opposite effect of high salt concentrations on aprE and sacB expression reported by Kunst and Rapoport (9). Although this seemed to imply a common mechanism between relA deficiency and high salt concentrations, addition of 1 M NaCl stimulated sacB′-′lacZ expression in both relA+ and relA strains, indicating that the high-salt effect is not mediated through RelA (data not shown).
Acknowledgments
We thank F. Ikebuchi, K. Nakata, and H. Ishida for technical assistance; Y. Sadaie for a bacterial strain; and M. Itaya for a plasmid. We also thank K. Ochi, Y. Ohashi, and N. Saito for ppGpp measurement.
This work was supported in part by a grant-in-aid for scientific research from the Ministry of Education, Science and Culture of Japan and by RIKEN Biodesign Research.
REFERENCES
- 1.Cashel M, Gentry D R, Hernandez V J, Vinella D. The stringent response. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella; cellular and molecular biology. 2nd ed. Washington, D.C.: American Society for Microbiology; 1996. pp. 1458–1496. [Google Scholar]
- 2.Choy H E. The study of guanosine 5′-diphosphate 3′diphosphate-mediated transcription regulation in vitro using a coupled transcription-translation system. J Biol Chem. 2000;275:6783–6789. doi: 10.1074/jbc.275.10.6783. [DOI] [PubMed] [Google Scholar]
- 3.Dahl M K, Msadek T, Kunst F, Rapoport G. Mutational analysis of the Bacillus subtilis DegU regulator and its phosphorylation by the DegS protein kinase. J Bacteriol. 1991;173:2539–2547. doi: 10.1128/jb.173.8.2539-2547.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dahl M K, Msadek T, Kunst F, Rapoport G. The phosphorylation state of the DegU response regulator acts as a molecular switch allowing either degradative enzyme synthesis or expression of genetic competence in Bacillus subtilis. J Biol Chem. 1992;267:14509–14514. [PubMed] [Google Scholar]
- 5.Henner D J, Ferrari E, Perego M, Hoch J A. Localization of Bacillus subtilis sacU(Hy) mutations to two linked genes with similarities to the conserved procaryotic family of two-component signaling systems. J Bacteriol. 1988;170:5102–5109. doi: 10.1128/jb.170.11.5102-5109.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Itaya M. Construction of a novel tetracycline resistance gene cassette useful as a marker on the Bacillus subtilis chromosome. Biosci Biotechnol Biochem. 1992;56:685–686. doi: 10.1271/bbb.56.685. [DOI] [PubMed] [Google Scholar]
- 7.Kunst F, Debarbouille M, Msadek T, Young M, Mauel C, Karamata D, Klier A, Rapoport G, Dedonder R. Deduced polypeptides encoded by the Bacillus subtilis sacU locus share homology with two-component sensor-regulator systems. J Bacteriol. 1988;170:5093–5101. doi: 10.1128/jb.170.11.5093-5101.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell S C, Bron S, Brouillet S, Bruschi C V, Caldwell B, Capuano V, Carter N M, Choi S K, Codani J J, Connerton I F, Danchin A, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–563. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 9.Kunst F, Rapoport G. Salt stress is an environmental signal affecting degradative enzyme synthesis in Bacillus subtilis. J Bacteriol. 1995;177:2403–2407. doi: 10.1128/jb.177.9.2403-2407.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lepesant J-A, Kunst F, Lepesant-Kejzlarova J, Dedonder R. Chromosomal location of mutations affecting sucrose metabolism in Bacillus subtilis. Mol Gen Genet. 1972;118:135–160. doi: 10.1007/BF00267084. [DOI] [PubMed] [Google Scholar]
- 11.Msadek T, Kunst F, Klier A, Rapoport G. DegS-DegU and ComP-ComA modulator-effector pairs control expression of the Bacillus subtilis pleiotropic regulatory gene degQ. J Bacteriol. 1991;173:2366–2377. doi: 10.1128/jb.173.7.2366-2377.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Msadek T, Kunst F, Rapoport G. Two-component regulatory systems. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C.: American Society for Microbiology; 1993. pp. 729–745. [Google Scholar]
- 13.Msadek T, Kunst F, Rapoport G. A signal transduction network in Bacillus subtilis includes the DegS/DegU and ComP/ComA two-component systems. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C.: American Society for Microbiology; 1995. pp. 447–471. [Google Scholar]
- 14.Mukai K, Kawata M, Tanaka T. Isolation and phosphorylation of the Bacillus subtilis degS and degU gene products. J Biol Chem. 1990;265:824–834. [PubMed] [Google Scholar]
- 15.Mukai K, Kawata-Mukai M, Tanaka T. Stabilization of phosphorylated Bacillus subtilis DegU by DegR. J Bacteriol. 1992;174:7954–7962. doi: 10.1128/jb.174.24.7954-7962.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagami Y, Tanaka T. Molecular cloning and nucleotide sequence of a DNA fragment from Bacillus natto that enhances production of extracellular proteases and levansucrase in Bacillus subtilis. J Bacteriol. 1986;166:20–28. doi: 10.1128/jb.166.1.20-28.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ogura M, Kawata-Mukai M, Itaya M, Takio K, Tanaka T. Multiple copies of the proB gene enhance degS-dependent extracellular protease production in Bacillus subtilis. J Bacteriol. 1994;176:5673–5680. doi: 10.1128/jb.176.18.5673-5680.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pang A S-H, Nathoo S, Wong S. Cloning and characterization of a pair of novel genes that regulates production of extracellular enzymes in Bacillus subtilis. J Bacteriol. 1991;173:46–54. doi: 10.1128/jb.173.1.46-54.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schaeffer P J, Millet J, Aubert J. Catabolite repression of bacterial sporulation. Proc Natl Acad Sci USA. 1965;54:704–711. doi: 10.1073/pnas.54.3.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanaka T, Kawata M. Cloning and characterization of Bacillus subtilis iep, which has positive and negative effects on production of extracellular proteases. J Bacteriol. 1988;170:3593–3600. doi: 10.1128/jb.170.8.3593-3600.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanaka T, Kawata M, Mukai K. Altered phosphorylation of Bacillus subtilis DegU caused by single amino acid changes in DegS. J Bacteriol. 1991;173:5507–5515. doi: 10.1128/jb.173.17.5507-5515.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ward J B, Jr, Zahler S A. Genetic studies of leucine biosynthesis in Bacillus subtilis. J Bacteriol. 1973;116:719–726. doi: 10.1128/jb.116.2.719-726.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wendrich T M, Marahiel M A. Cloning and characterization of a relA/spoT homologue from Bacillus subtilis. Mol Microbiol. 1997;26:65–79. doi: 10.1046/j.1365-2958.1997.5511919.x. [DOI] [PubMed] [Google Scholar]