Abstract
Studies of germline polymorphisms as predictors of tumor response to anti-EGFR monocloncal antibody agents in metastatic colorectal cancer have reported inconsistent results. We performed a systematic review of studies from 1990 to September, 2015, followed by random-effects meta-analyses for polymorphisms examined in at least three studies. Of 87 studies, 40 passed criteria for systematic review and 23 for meta-analysis. The polymorphisms suitable for meta-analysis were: CCND1 (rs17852153), COX2 (rs20417), EGF (rs4444903), EGFR (rs712829, rs11543848, 3’UTR CA repeat), FCGR2A (rs1801274), FCGR3A (rs396991), IL8 (rs4073), KRAS (rs61764370), and VEGFA (rs3025039). Meta-analysis yielded nominal significance (at alpha=0.05) for rs4444903 and rs11543848, but showed no significant results after multiple testing correction; this was unchanged by sensitivity analyses to address subgroups, funnel-plot asymmetries, and study quality. This highlights a tendency for lack of replication in the face of initial positive results, and possibly the unsuitability of relying on tumor response as a surrogate marker in this setting.
Keywords: colorectal cancer, cetuximab, panitumumab, EGFR, polymorphism, meta-analysis
1. INTRODUCTION
Colorectal cancer (CRC) is a leading cause of cancer death, with a large fraction of patients developing advanced or metastatic disease.1 For most of these patients, systemic control of disease is paramount, and is now achievable via targeted therapies that directly inhibit molecular drivers of tumor proliferation. By far the most commonly used of these therapies are monoclonal antibodies to the epidermal growth factor receptor (EGFR), which include cetuximab and panitumumab. These drugs not only help to achieve systemic control in metastatic disease after other agents have failed, but also have a much-improved side-effect profile compared to traditional therapies such as irinotecan, oxaliplatin, and fluoropyrimidines.2
Because the majority of CRC patients show no response to anti-EGFR monoclonal antibody therapies, considerable efforts are underway to identify up-front the patients who will respond, so that the rest can be spared the time, expense and side effects of an ineffective treatment. One advance in this arena has been the recognition that tumor KRAS mutations are strongly associated with non-response to anti-EGFR drugs,3 since which KRAS testing has become routine. However, even after such testing, more than half of patients still show no response to anti-EGFR drugs4,5 indicating a pressing need for additional research. This ongoing effort has led to discovery and adoption of NRAS/HRAS testing in many jurisdictions,6,7 and has identified some well-studied candidate mutations in genes such as BRAF, PIK3CA, and PTEN whose effectiveness as predictive markers remains uncertain.5,8,9
Genetic alterations with potential as predictive biomarkers in this scenario may affect either tumor (somatic) or patient (germline) DNA. Alterations of somatic tumor DNA (i.e. “tumor gene mutations”) directly affect tumor cells, and can thus alter tumor characteristics such as growth rate, invasiveness, metastatic potential, and vulnerability to particular drugs. In contrast, alterations in germline DNA (i.e. “genetic polymorphisms”) directly affect patient cells and can thus influence patient factors such as drug bioavailability, kinetics, and metabolism, as well as host immune interactions and local tissue responses.
While many germline polymorphisms have been proposed as biomarkers in anti-EGFR monoclonal antibody treated CRC, studies have often yielded inconsistent results. This may be due to the lack of an underlying true association, heterogeneity of study population, or low power in smaller studies. For these reasons, we undertook a systematic review and meta-analysis to evaluate the association of these polymorphisms as putative clinical biomarkers of response to cetuximab/panitumumab therapy. We focused on response as an outcome because: (1) response was much more widely and uniformly reported among studies in this area, and (2) the majority of studies did not have control arms of patients not treated with cetuximab/panitumumab, rendering time-to-event outcomes incapable of distinguishing between prognostic and predictive associations.
2. MATERIALS AND METHODS
Systematic review methodology
Ovid MEDLINE was searched using a date-range of January, 1990 to September, 2015. The search string used was: “(exp Polymorphism, Genetic/ or polymorphism*.mp.) and (exp Colorectal Neoplasms/ or (colon or rectal or colorectal).mp.) and (neoplas* or tumor* or tumour* or cancer or carcinoma or adenocarcinoma).mp. and (cetuximab or panitumumab).mp.” Results were limited to English language publications and studies in human subjects. Methods used to identify additional papers included checking reference lists, communicating with researchers in the field, and using web search engines such as Google Scholar. The resulting studies were manually reviewed, with access of the full text articles as necessary to determine eligibility for systematic review. The inclusion criterion for the systematic review was that included studies had to evaluate one or more germline polymorphism as predictors of tumor response in colorectal cancer patients who were treated with an anti-epidermal growth factor receptor (anti-EGFR) monoclonal antibody (i.e. cetuximab or panitumumab). Exclusion criteria for the systematic review were as follows: review articles, case reports, studies with fewer than 25 patients, duplicate citations, repeat analyses of data published elsewhere, and presentations of subsets of data published elsewhere.
For eligible publications, the following data were abstracted: study authors, year of publication, characteristics of the patient population (country of recruitment or ethnicity, line of therapy, therapy during the study, total number of patients, intent of therapy, the KRAS status of included patients), the number and identity of tested polymorphisms, the reported outcomes, and whether sufficient data was reported to allow meta-analysis for each outcome. The study methodology was categorized as either prospective or retrospective (depending on method of cohort recruitment), with notation of whether the cohort was ascertained within a Phase II or randomized clinical trial. For studies with incomplete reporting of detailed results, attempts were made to contact study authors to obtain complete information. Authors were asked to provide either summary statistics or raw data from which summary statistics (e.g. counts) could be calculated.
Meta-Analysis Methodology
Polymorphisms identified in the systematic review were evaluated for their suitability for meta-analysis. Only polymorphisms with published results from at least three separate studies were included. Studies were thus included in the meta-analysis if they (1) reported data on an appropriate polymorphism and (2) reported data in sufficient detail to allow meta-analysis (i.e. counts of responders and non-responders with each genotype).
The study quality was evaluated using a schema based on the “Strengthening the Reporting of Genetic Association studies” (STREGA) framework10. Two independent reviewers read the full text of each article (and accessed supplementary published materials where needed), and assigned the study a score between 1 and 3 in each of 27 categories (see Appendix 1); results were totaled and averaged to produce a summary score for each article. Concordance was compared between the two reviewers, and then their scores were averaged to produce the final article score. The article scores were then plotted as a histogram, and equal-sized bins chosen to divide the articles into four categories (adequate, good, very good, excellent), as none fell into the poor category by qualitative analysis. A sensitivity analysis was then performed by evaluating each meta-analysis result, where possible, using only articles in the very good and excellent categories. Because response was the primary outcome for meta-analysis, included studies were also reviewed to determine the specific definition of response used.
For each polymorphism, alleles associated with improved outcomes in prior studies were recorded. These alleles were evaluated via meta-analysis using R (v2.15.1)11 and the metafor package (v1.7–0)12 under the assumptions of additive, dominant, and recessive genetic inheritance models. A random effects model was used, with evaluation of heterogeneity using the Higgins’ I2 summary statistic.13 The model with the lowest p-value for each polymorphism was retained, provided that the direction of association remained as previously proposed. In a few cases where certain studies had reported results only for particular genetic models, these models were preferred for meta-analysis to allow inclusion of the broadest range of data. Correction for multiple testing was performed using the Benjamini-Hochberg procedure to limit the false discovery rate to less than 5%. The potential for publication bias was evaluated using funnel plots.
For each polymorphism, we performed multiple sensitivity analyses that excluded: (1) studies showing up as unbalanced points in funnel plots; (2) KRAS unselected studies (i.e. including only studies with KRAS wild-type patients); (3) KRAS wild-type studies (i.e. including only studies with patients unselected by KRAS status).; (4) studies with only poor or adequate quality ratings; and (5) studies showing deviations from Hardy-Weinberg equilibrium (relevant to the FCGR polymorphisms only). A few sensitivity analyses did increase the strength of association for polymorphisms with borderline statistical significance, for instance to p=0.004 for EGF 61 in one sensitivity analysis; nonetheless, none retained significance at q < 0.05 after correcting for even the baseline level of multiple comparisons in this meta-analysis, let alone the additional tests involved in the sensitivity analysis.
3. RESULTS
Systematic review results
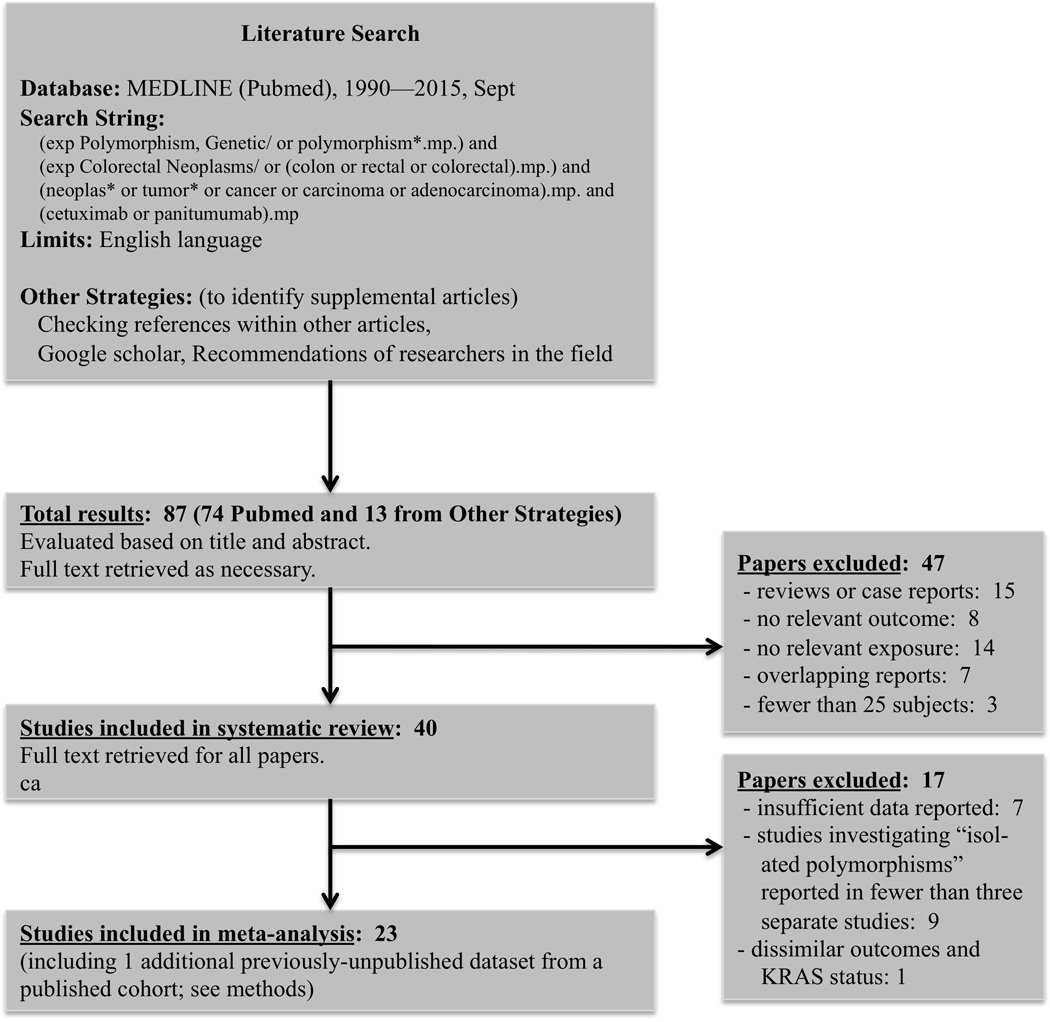
The systematic review yielded 87 studies, of which 47 failed to meet inclusion or exclusion criteria (Figure 1). Of the 40 remaining articles retained for systematic review, 23 were included in the subsequent meta-analysis. Of the 17 excluded studies, seven had insufficient data on covariate or response variables, nine involved polymorphisms that were assessed in fewer than three separate studies, and one14 was excluded for reporting only disease control rate in patients with KRAS mutant (rather than KRAS wild-type) tumors. Of those included in the meta-analysis, one study consisted of new data, published for the first time as part of this meta-analysis, on two of the polymorphisms (FCGR2A and FCGR3A), from analysis of samples from CO.17, a Phase III clinical trial of cetuximab versus best supportive care.2 These 23 studies were included for meta-analysis, and their characteristics are shown in Table 1.
Figure 1. Systematic review methodology and results.
Table 1. Summary of studies eligible for meta analysis, grouped by study methodology.
| Author | Year | Region | Study Methodology | N | Intent of Therapy | Other Chemotherapy | KRAS Status | Reported Outcomes |
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Saridaki 22 | 2014 | Belgium, France, USA | mixed | 559 | mixed | mixed | unselected | response, PFS, OS |
|
| ||||||||
| Loupakis 23 | 2014 | California | prospective cohort | 113 | mixed | irinotecan | wild-type | response, PFS, OS |
| Garm Spindler 24 | 2009 | Denmark | prospective cohort | 71 | salvage | irinotecan | unselected | response, PFS, OS |
| Graziano 25 | 2008 | Italy | prospective cohort | 110 | salvage | irinotecan | unselected | response, PFS, OS |
| Inoue 26 | 2014 | Japan | prospective cohort | 57 | salvage | mixed | wild-type | response, PFS, OS |
|
| ||||||||
| Sclafani 27 | 2014 | response, PFS, OS | prospective cohort within Phase II trial | 105 | adjuvant | capecitabine, oxaliplatin | wild-type | response, PFS, OS |
| Etienne-Grimaldi 28 | 2012 | France | prospective cohort within Phase II trial |
52 | first-line | irinotecan, folinic acid, UFT |
unselected | response, OS |
| Lurje 29 | 2008 | California | prospective cohort within Phase II trial | 130 | salvage | none | unselected | response, PFS, OS |
| Zhang 30 | 2010 | California | prospective cohort within Phase II trial | 65 | salvage | mixed | unselected | response, TTP, OS |
| Zhang 31 | 2011 | California | prospective cohort within Phase II trial | 111 | salvage | none | unselected | response, PFS, OS |
| Kjersem 32 | 2012 | Norway | prospective cohort within RCT | 180 | first-line | oxalplatin, folinic acid, UFT |
unselected | response, PFS, OS |
| Kjersem 19 | 2014 | Norway | prospective cohort within RCT | 504 | first-line | oxalplatin, folinic acid, UFT | unselected | response, PFS, OS |
|
| ||||||||
| Jonker 2 | 2015 | Canada & Australia |
prospective cohort within RCT | 138 | salvage | none | unselected | response, OS |
|
| ||||||||
| Hsieh 33 | 2012 | Taiwan | retrospective cohort | 118 | first-line | oxalplatin, folinic acid, UFT |
wild-type | response, PFS, OS |
| Negri 34 | 2014 | Italy | retrospective cohort | 86 | mixed | mixed | unselected | response, TTP, OS |
| Calemma 35 | 2012 | Italy | retrospective cohort | 50 | mixedC/P | mixed | wild-type | response, PFS, OS |
| Hu-Lieskovan 36 | 2011 | Europe | retrospective cohort | 130 | neoadjuvantLA | mixed | unselected | pathologic response |
| Bibeau 37 | 2009 | France | retrospective cohort | 69 | salvage | irinotecan | unselected | response, PFS, OS |
| Dahan 38 | 2011 | France | retrospective cohort | 58 | salvage | irinotecan | unselected | response, TTP, DSS |
| Park 39 | 2012 | Korean | retrospective cohort | 118 | salvage | mixed | unselected | response, PFS, OS |
| Paez 40 | 2010 | Caucasian | retrospective cohort | 104 | salvageC/P | mixed | unselected | response, PFS |
| Sebio 41 | 2013 | Spain | retrospective cohort | 100 | salvageC/P | mixed | wild-type | response |
| Geva 18 | 2015 | Europe (multicenter) |
retrospective cohort | 740 | salvage | mixed | wild-type | response, PFS, OS, DCR |
N = number participants; UFT = Tegafur/uracil; PFS = progression free survival; OS = overall survival; TTP = time to progression; DSS = disease free survival; LA = patients had locally advanced disease; C/P = patients received either cetuximab and panitumumab
Most studies (57%) used a prospective cohort design, with the remainder used a retrospective cohort design. Most studies (56%) gave anti-EGFR agents as a salvage therapy, while a few (22%) gave it as first-line or neoadjuvant therapy, and the remainder were mixed. KRAS status was unknown in most studies (70%) and limited to wild-type in the remainder. All studies reported response, and most also reported one or more survival outcomes. The geographic location of studies and their ethnic composition were diverse. While most studies investigated survival outcomes, incomplete reporting of these outcomes was common and was a major limitation to meta-analysis. For example, many studies reported full results (i.e. at least an effect size and measure of precision) only for selected polymorphisms, often those that were the most statistically significant. Included studies were subjected to quality review by two independent reviewers. These reviewers demonstrated good agreement with a Spearman correlation coefficient of 0.90, while the mean quality score was 1.96 (range 1.56 – 2.29), and thus the category cut-offs were chosen as: adequate (<1.8), good (1.8-<2.0), very good (2.0-<2.2), and excellent (≥2.2). The resulting quality ratings are also shown in Table 2, along with a summary of which studies investigated which of the polymorphisms that were studied via meta-analysis. The definitions of tumor response used are also shown in Table 2; most were variations of the RECIST criteria.15
Table 2. Matrix illustrating, for each study, which polymorphisms it investigated, its quality rating, and its stated definition of response.
| Study Reference | FCGR2A 131 R>H | FCGR3A 158 F>V | EGF 61 A>G | EGFR 497 R>K | KRAS Let-7 T>G | EGFR 3’UTR (CA)n S>L | EGFR −216 G>T | CCND1 870 A>G | VEGF 936 C>T | COX2 −765 G>C | IL-8 −251 T>A | Study Quality | Definition of Response |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Saridaki, 2014 | X | Good | Objective Resp. Rate | ||||||||||
| Loupakis, 2014 | X | Very Good | RECIST 1.0 | ||||||||||
| Garm Spindler, 2009 | X | Very Good | RECIST 1.0 | ||||||||||
| Graziano, 2008 | X | X | X | X | Good | RECIST 1.0 | |||||||
| Inoue, 2014 | X | X | X | Very Good | RECIST | ||||||||
| Sclafani, 2014 | X | X | Very Good | RECIST | |||||||||
| Etienne-Grimaldi, 2012 | X | X | X | X | X | Good | RECIST | ||||||
| Lurje, 2008 | X | X | X | X | X | X | X | X | X | Excellent | WHO criteria, modified | ||
| Zhang, 2010 | X | X | X | X | X | X | X | X | X | Adequate | RECIST | ||
| Zhang, 2011 | X | Adequate | WHO criteria, modified | ||||||||||
| Kjersem, 2012 | X | Very Good | RECIST | ||||||||||
| Kjersem, 2014 | X | X | Good | RECIST | |||||||||
| Dobrovic, 2015 | X | X | Excluded* | RECIST 1.0 | |||||||||
| Hsieh, 2012 | X | Good | RECIST | ||||||||||
| Negri, 2014 | X | X | Good | RECIST 1.1 | |||||||||
| Calemma, 2012 | X | X | Good | RECIST | |||||||||
| Hu-Lieskovan, 2011 | X | X | X | X | X | X | X | X | X | X | Good | Dworak grade | |
| Bibeau, 2009 | X | X | Good | RECIST 1.0 | |||||||||
| Dahan, 2011 | X | X | X | X | X | X | Good | RECIST, modified | |||||
| Park, 2012 | X | X | Very Good | RECIST 1.1 | |||||||||
| Paez, 2010 | X | X | Good | RECIST 1.0 | |||||||||
| Sebio, 2013 | X | Good | RECIST 1.1 | ||||||||||
| Geva, 2015 | X | X | Very Good | RECIST 1.0 or WHO |
Jonker, 2007 was excluded because no appropriate polymorphism-related manuscript was available, and because both quality reviewers were involved with the study.
Meta-analysis of tumor response
Eleven polymorphisms were suitable for meta-analysis (see Table 2). Although some polymorphisms were within genes clearly related to known mechanisms of drug activity (EGF, EGFR, FCGR, KRAS polymorphisms), others were relatively tangential (CCND1, COX2, IL8, VEGF polymorphisms). Most had some data reporting a putative functional consequence of carrying different polymorphic alleles. Among the relevant studies, both prospective and retrospective cohort study designs were well represented, and several were Phase II/III trials. The median number of patients per analysis was 110 (range 50 to 740).
For each polymorphism, we calculated a pooled effect with associated standard error, and Higgins’ I2 (see Table 3), and constructed funnel plots (not shown). Two polymorphisms showed pooled relative risks of response that differed significantly from 1.0 with alpha=0.05: the EGF A61G (rs4444903) and EGFR R497K (rs11543848) polymorphisms. However, neither result retained statistical significance after correction for multiple testing. Among the polymorphisms not associated with outcome were the FCGR2A H131R (rs1801274) and FCGR3A F158V (rs396991), which have garnered much interest and involved the largest number of studies among the polymorphisms reviewed.
Table 3. Meta-analysis results for polymorphisms with at least three studies.
Each polymorphism’s common name is listed along with the corresponding OMIM gene number and dbSNP polymorphism number. For each polymorphism, the results of meta-analysis are presented for the genetic model resulting in the lowest p-value. Data presented for each analysis include the pooled relative risk with corresponding confidence interval, number of studies contributing data to the analysis, Higgin’s I2, p-value, and false discovery rate q-value),
| Polymorphism | OMIM # | RS # | Test Allele (Model) | RR [95% CI] | N | Higgins’ I2 | p-val | q-val |
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| CCND1 870 A>G | 168461 | rs17852153 | A (Recessive) | 1.14 [0.64, 2.04] | 5 | 34.8% | 0.652 | 0.530 |
| COX2 −765 G>C | 600262 | rs20417 | C (Recessive) | 2.67 [0.69, 10.36] | 3 | 55.0% | 0.155 | 0.427 |
| EGF 61 A>G | 131530 | rs4444903 | G (Recessive) | 1.81 [1.08, 3.02] | 6 | 48.4% | 0.023 | 0.257 |
| EGFR −216 G>T | 131550 | rs712829 | T (Recessive) | 1.27 [0.78, 2.06] | 3 | 0.0% | 0.331 | 0.460 |
| EGFR 497 R>K | 131550 | rs11543848 | K (Recessive) | 1.52 [1.01, 2.31] | 6 | 0.0% | 0.047 | 0.460 |
| EGFR 3’UTR (CA)n S>L | 131550 | N/A | S (Recessive) | 1.23 [0.81, 1.85] | 7 | 45.4% | 0.334 | 0.259 |
| FCGR2A 131 R>H | 146790 | rs1801274 | R (Dominant)* | 1.09 [0.94, 1.27] | 15 | 0.0% | 0.251 | 0.275 |
| FCGR3A 158 F>V | 146740 | rs396991 | V (Recessive) | 1.03 [0.85, 1.24 ] | 15 | 0.0% | 0.781 | 0.781 |
| IL8 −251 T>A | 146930 | rs4073 | T (Recessive) | 1.29 [0.78, 2.13] | 3 | 0.0% | 0.324 | 0.530 |
| KRAS Let-7 T>G | 190070 | rs61764370 | G (Dominant) | 1.36 [0.70, 2.65] | 5 | 83.2% | 0.370 | 0.460 |
| VEGFA 936 C>T | 192240 | rs3025039 | T (Dominant) | 1.34 [0.81, 2.21] | 3 | 0.0% | 0.710 | 0.460 |
RR = relative risk; CI = confidence interval; A/C/G/T = represent respective oligonucleotides; S = the shorter number of CA repeats (with the other allele being L, the longer number of CA repeats); NS = previously non-significant; OS = significant association for overall survival; PFS = significant association for progression-free survival;
the recessive model had a slightly lower p-value for this polymorphism, but the dominant model was chosen to include the largest number of high-quality studies, not all of which reported sufficient data for analysis of all allele combinations
4. DISCUSSION
The present systematic review and meta-analysis of 23 eligible studies was able to evaluate pooled effects for 11 polymorphisms on tumor response in colorectal cancer patients treated with anti-EGFR monoclonal antibody therapies. Two polymorphisms demonstrated nominal statistical significance, but these associations were not robust to correction for multiple testing.
The published literature studying the association of germline polymorphisms with clinical outcome in anti-EGFR treated colorectal cancer patients presents certain challenges. Most prominently, published studies have historically been quite small, fewer than 150 patients. This has likely been due to logistic difficulties in assembling large patient cohorts from individual centers, the greater cost in dealing with large cohorts, and the fact that in the past, the number of centers offering anti-EGFR drugs and the number of patients receiving such agents were both small. This combination of multiple groups studying small samples increases the risk that any discovered statistically significant association is in fact a false positive. This is a central problem in all biomedical research, and is due in large part to factors beyond the control of individual researchers.16
One of the main tools available to address this challenge is meta-analysis, which allows evidence to be pooled across studies and can increase the precision of estimates as well as the statistical power to discover true associations, while also showing individual more extreme results in the context of all similar studies.17 In this context, it is expected that many associations showing statistical significance in particular studies will fail to find support after meta-analysis, and that this generally indicates the absence of a relevant underlying association. Indeed, this same trend has been observed in recent large, carefully-conducted studies on certain polymorphisms that failed to replicate significant associations observed in smaller prior studies.18,19 However, negative meta-analysis results must be taken with important caveats including study and patient heterogeneity, low minor allele frequencies (limiting meta-analysis study power), low response rates to anti-EGFR agents, and inappropriate use of response rate as a surrogate for survival in this context, trade-offs in statistical modeling, and poor reporting of outcomes in the published literature. We address each of these issues below.
Study Heterogeneity
An important limitation in this study was the heterogeneity of included study designs. While all studies enrolled patients with colorectal cancer (usually metastatic) and evaluated the association of polymorphisms with response to anti-EGFR therapy, there was room for substantial variation. Patients varied in whether they had prior surgery or chemotherapy, how many prior lines of chemotherapy might have been used, and which regimens had been previously tried. Studies also varied in whether patients were taking anti-EGFR drugs as monotherapy or along with other drugs, and also in the choice of any such drugs. The criteria employed for evaluating tumor response varied (see Table 2). The KRAS status of study patients (and indeed, whether KRAS testing was performed at all) also varied, with many studies of all patients regardless of KRAS status, and several others with only KRAS wild-type tumors. Finally, the country of recruitment and thus ethnicity distribution of patients varied considerably, with potential masking of significant associations that may be present only within particular ethnic groups. These differences represent a limitation that is difficult to fully resolve when the literature typically includes only three to six studies per polymorphism, which is too small a number for meta-regression.
However, as an alternate approach to address these sources of heterogeneity, along with the possibility that they may have led to false negative results, we performed multiple sensitivity analyses (see results section), some of which produced lower nominal p-values, but none of which were robust to correction for multiple testing, rendering them unlikely to be of great promise. Of course, it is difficult to entirely exclude the possibility a relevant, statistically significant result may be discoverable for one or more polymorphisms within some alternate subgroup of studies. However, an exhaustive search for such associations would also undoubtedly reveal many more false positives than true associations.
Statistical Modeling Tradeoffs
We chose the more conservative random effects model for meta-analysis rather than a fixed effects model. While this may reduce the study’s ability to identify true associations, the random effects model is most suited to situations with heterogeneity between study designs,20 which is certainly abundant in this case.
We corrected for multiple comparisons relating to two different factors: (1) the multiple polymorphisms being tested and (2) the multiple genetic models being tested per polymorphism. However, different genetic models do not represent independent statistical tests, since there is a strong correlation between the results of recessive, dominant, and recessive models for a given allele. Thus typical methods of correction, which assume independence of statistical tests, run the risk of being too conservative. We addressed this concern in two ways. First, we chose to limit the false discovery rate to q < 0.05 using the well-known Benjamini-Hochberg procedure. Second, realizing that even this procedure could yield overly stringent results for the stated false discovery rate in the setting of dependent tests, we performed a sensitivity analysis at a more liberal rate of q < 0.2, which still failed to show any significant results.
Finally, it could be argued that our approach to meta-analysis, which included separate tests for each of three genetic models, is less likely to find true associations due to the necessary correction for multiple testing. This argument would posit that had we instead chosen a single genetic model for each polymorphism based on prior publications, a significant association might have been found. To address this concern, we simulated perfect foreknowledge by picking the lowest p-value model for each polymorphism and correcting for multiple testing using only those 11 models. The conclusions were unchanged, with no model retaining significance at the chosen cutoff of q < 0.05. Even extending this approach of perfect foreknowledge further to allow selection of the lowest p-value model for each polymorphism including all sensitivity analyses, conclusions remained unchanged with no model retaining significance.
Limitations of Tumor Response as an Outcome
An important limitation of analyses of tumor response in cetuximab-treated colorectal carcinoma is the low rates of tumor response in the published literature. In CO.17, the study which originally demonstrated cetuximab efficacy in metastatic CRC, the response rate in the cetuximab treatment group was only 8%. This introduces a challenge with statistical power – even assuming that a beneficial effect exists for certain polymorphisms, this low rate would make it difficult to demonstrate. This issue is beginning to be addressed by new larger studies, as well as by the present meta-analysis.
However, an even more fundamental point relates to the hypothesized action of anti-EGFR agents in CRC. The low observed response rate may be a sign that the beneficial effects of cetuximab on survival (which is ultimately the most important outcome) are largely independent of tumor response. For example, if the mechanism was primarily inhibition of tumor growth (or anti-proliferation, given that EGFR acts on cellular growth) rather than inducing tumor shrinkage, then there may be a true benefit in the absence of any significant tumor response. It would thus be ideal to also perform a meta-analysis of patient survival in this clinical area. Unfortunately, our ability to do this was limited by inconsistent reporting of outcomes among reviewed studies, which is discussed below.
Inconsistent Outcome Reporting in Published Studies
Many reviewed studies reported survival outcomes, such as progression-free survival and overall survival, in addition to tumor response. Survival outcomes are of prime importance in evaluating oncology therapeutics, including anti-EGFR therapies, and would ideally be included in this study. Indeed, tumor response is often viewed as a surrogate for survival, which is the outcome of ultimate importance (at least for drug effectiveness; for efficacy, in contrast, tumor response may sometimes be the preferred outcome). Unfortunately, it was not possible to perform a meta-analysis of survival in the present study due to incomplete and/or irregular reporting of survival outcomes. Some studies did not investigate survival outcomes; many others did, but reported effect sizes and precisions for only selected analyses. Even when survival data was reported, its format was inconsistent, variably couched as hazard ratios, Kaplan-Meier plots, p-values for log-rank tests, or median survivals by patient group. Such problems are routine in the published survival literature, and represent a significant challenge to meta-analysis generally.21 This was particularly true as we also attempted to contact individual studies to request primary source data and/or re-analyzed data according to a single standard, which met with only marginal success. In the present study, meta-analysis was technically possible for selected polymorphisms (i.e. more than two studies reporting adequate survival outcomes for a given polymorphism); however, given the large proportion of relevant published results that would have been excluded due to inadequate reporting, the potential for biased and misleading results would be extreme. Consequently, the present study is necessarily limited to the more proximal outcome of tumor response, and a meta-analysis of survival outcomes must await more uniform reporting, more widespread sharing of unpublished data, or methodological advances that allow the incorporation of studies with incomplete data.
Conclusions
The present study represents the first systematic review and meta-analysis of germline polymorphisms as biomarkers of tumor response in CRC patients treated with anti-EGFR monoclonal antibody therapy. The resulting pooled analysis, which was possible for 11 of the reviewed polymorphisms, revealed no statistically significant associations after correction for multiple testing. Given the substantial heterogeneity in methodology among included studies, the relatively small numbers of analyzable studies for each polymorphism, and the inability to systematically analyze survival outcomes, this result cannot definitively exclude the possibility of a significant association for one of the included polymorphisms. Nonetheless, these findings were robust to multiple sensitivity analyses, and also parallel an observed trend in recent large, well-conducted studies in the area that have failed to replicate significant associations observed in smaller prior studies.18,19
Equally important in this study is how the results serve to highlight important issues for future research in this area. The present results argue for the use of potentially more fruitful approaches through the planning of larger studies, potentially within consortia to leverage the resources of multiple centers, and where possible in adopting an unbiased, genome-wide approach to biomarker discovery that will better facilitate data-sharing, patient-level meta-analysis, and validation of polymorphisms proposed by other groups. Finally, the current literature is quite variable in the reporting of survival outcomes, and it is crucial that future studies publish uniform data regarding all major clinical outcomes for all studied polymorphisms (at least as supplementary data), in order to minimize publication bias and facilitate aggregation of study results, which will be indispensable to future progress in this area.
Supplementary Material
9. APPENDIX 1. – Criteria used for quality review of included articles.
| Category | Criteria Summary |
|---|---|
| title and abstract | design in title or abstract; informative/balanced summary in abstract |
| intro - rationale | explain scientific background and rationale |
| intro - objectives | specific objectives; first report vs replication |
| meth - setting | setting, location, dates |
| meth - participants | eligibility criteria, sources, selection methods |
| meth - variables | outcomes, covariates, variants (standard nomenclature), ethnic confounding |
| meth - measurement | lab methods, source/storage of DNA, genotyping method, allele calling algorithm, error rates, call rates, laboratory identified |
| meth - size | how was study size arrived at |
| meth - Q vars | explain how quantitative variables were handled in analysis (choice of groupings) |
| Stats | describe all statistical methods, including confounding; software, version, options |
| any methods to examine subgroups and interactions | |
| how was missing data addressed | |
| loss to follow-up (cohort), matching (case/control) | |
| describe any sensitivity analyses | |
| state if HWE was considered and how | |
| describe any methods to address multiple comparisons or risk of false positives | |
| describe any methods used to address subject relatedness | |
| res - participants | numbers of individuals at each stage of study, reasons for non-participation; number successfully genotyped? |
| res - descriptive | participant characteristics, information on exposures and potential confounders; number of participants with missing data for variables of interest; follow-up time |
| res - outcome | outcomes by genotype over time (cohort); summary of outcomes by genotype (case-control) |
| res - main | unadjusted and (IA) adjusted estimates (which covariates?), precisions; report category boundaries if discretized; results of multiple comparisons adjustments |
| res - other | summarize results from all variants analyzed; (IA) how can more detailed results be accessed? |
| disc - results | summarize key results with relation to objectives |
| disc - limits | study limitations discussed (bias, imprecision, direction/magnitude of bias) |
| desc - interp | give cautious interpretation considering limitations, multiple testing, other studies |
| desc - general | discuss generalizability (external validity) |
| other - funds | give sources of funding and role of funders in present study and (IA) for original studies |
Footnotes
CONFLICT OF INTEREST
EKM: none
HL: Advisory board member for Merck KG, Bristol Myers Squibb Clinical Trial support, Merck Serono, and Bristol Myers Squibb
DJJ: none
DT: none
GM: Merck Serono honorarium
FG: none
JZ: Travel and research support Merck Serono
CK: Advisory board member for Amgen and Merck Serono
AD: Advisory board member for Amgen
CJO: none
GL: Advisory board member for Roche, Astra Zeneca, Novartis, Pfizer, and Merck Serono.
Contributor Information
Eric K Morgen, Dalla Lana School of Public Health, University of Toronto; Department of Pathobiology and Laboratory Medicine, University of Toronto, Toronto, Ontario, Canada.
Heinz-Josef Lenz, USC/Norris Comprehensive Cancer Center, Los Angeles, California..
Derek J Jonker, The Ottawa Hospital Research Institute, University of Ottawa, Ottawa, Ontario, Canada.
Dongsheng Tu, Canadian Cancer Trials Group, Queen’s University, Kingston, Canada.
Gerard Milano, Laboratoire d’Oncopharmacologie EA 3836, Centre Antoine Lacassagne, Nice, France..
Francesco Graziano, Division of Medical Oncology, Azienda “Ospedali Riuniti Marche Nord”, Pesaro, Italy..
John Zalcberg, Head, Cancer Research Program, School of Public Health and Preventive Medicine Faculty of Medicine, Monash University, Melbourne. Australia.
Christos S Karapetis, Flinders University and Flinders Medical Centre, Adelaide, Australia.
Alexander Dobrovic, Translational Genomics and Epigenomics Laboratory; Olivia Newton-John Cancer Research Institute, Heidelberg, Victoria, Australia; School of Cancer Medicine, La Trobe University, Bundoora, Victoria, Australia; Department of Pathology, University of Melbourne, Parkville, Victoria, Australia..
Chris J O’Callaghan, Canadian Cancer Trials Group, Queen’s University, Kingston, Ontario, Canada.
Geoffrey Liu, Dalla Lana School of Public Health, University of Toronto; Departments of Medicine and Medical Biophysics, University of Toronto, Toronto, Ontario, Canada.
REFERENCES
- 1.Donadon M, Ribero D, Morris-Stiff G, Abdalla EK, Vauthey J-N. New paradigm in the management of liver-only metastases from colorectal cancer. Gastrointest Cancer Res 2007; 1: 20–27. [PMC free article] [PubMed] [Google Scholar]
- 2.Jonker DJ, O’Callaghan CJ, Karapetis CS, Zalcberg JR, Tu D, Au H-J et al. Cetuximab for the treatment of colorectal cancer. N Engl J Medicine 2007; 357: 2040–2048. [DOI] [PubMed] [Google Scholar]
- 3.Bokemeyer C, Bondarenko I, Hartmann JT, de Braud F, Schuch G, Zubel A et al. Efficacy according to biomarker status of cetuximab plus FOLFOX-4 as first-line treatment for metastatic colorectal cancer: the OPUS study. Ann Oncol 2011; 22: 1535–1546. [DOI] [PubMed] [Google Scholar]
- 4.Soeda H, Shimodaira H, Watanabe M, Suzuki T, Gamoh M, Mori T et al. Clinical usefulness of KRAS, BRAF, and PIK3CA mutations as predictive markers of cetuximab efficacy in irinotecan- and oxaliplatin-refractory Japanese patients with metastatic colorectal cancer. Int J Clin Oncol 2013; 18: 670–677. [DOI] [PubMed] [Google Scholar]
- 5.Prognostic Custodio A. and predictive biomarkers for epidermal growth factor receptor-targeted therapy in colorectal cancer: beyond KRAS mutations. Crit Rev Oncol/Hematol 2013; 85: 45–81. [DOI] [PubMed] [Google Scholar]
- 6.Taniguchi H, Yamazaki K, Yoshino T, Muro K, Yatabe Y, Watanabe T et al. Japanese Society of Medical Oncology Clinical Guidelines: RAS (KRAS/NRAS) mutation testing in colorectal cancer patients. Cancer Sci 2015; 106: 324–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wong NACS, Gonzalez D, Salto-Tellez M, Butler R, Diaz-Cano SJ, Ilyas M et al. RAS testing of colorectal carcinoma—a guidance document from the Association of Clinical Pathologists Molecular Pathology and Diagnostics Group. J Clin Pathol 2014; 67: 751–757. [DOI] [PubMed] [Google Scholar]
- 8.Tol J, Dijkstra JR, Klomp M, Teerenstra S, Dommerholt M, Vink-Börger ME et al. Markers for EGFR pathway activation as predictor of outcome in metastatic colorectal cancer patients treated with or without cetuximab. Eur J Cancer 2010; 46: 1997–2009. [DOI] [PubMed] [Google Scholar]
- 9.Bokemeyer C, Van Cutsem E, Rougier P, Ciardiello F, Heeger S, Schlichting M et al. Addition of cetuximab to chemotherapy as first-line treatment for KRAS wild-type metastatic colorectal cancer: pooled analysis of the CRYSTAL and OPUS randomised clinical trials. Eur J Cancer 2012; 48: 1466–1475. [DOI] [PubMed] [Google Scholar]
- 10.Little J, Higgins JPT, Ioannidis JPA, Moher D, Gagnon F, von Elm E et al. STrengthening the REporting of Genetic Association Studies (STREGA)--an extension of the STROBE statement. Genet Epidemiol 2009; 33: 581–598. [DOI] [PubMed] [Google Scholar]
- 11.R Core Team. The R Project for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria, 2012. Available at: http://www.r-project.org/. [Google Scholar]
- 12.Viechtbauer W. Conducting meta-analyses in R with the metafor Package. J Stat Softw 2010; 36: 1–48. [Google Scholar]
- 13.Higgins JPT, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med 2002; 21: 1539–1558. [DOI] [PubMed] [Google Scholar]
- 14.Rodríguez J, Zarate R, Bandres E, Boni V, Hernández A, Sola JJ et al. Fc γ receptor polymorphisms as predictive markers of Cetuximab efficacy in epidermal growth factor receptor downstream-mutated metastatic colorectal cancer. Eur J Cancer 2012; 48: 1774–1780. [DOI] [PubMed] [Google Scholar]
- 15.Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubinstein L et al. New guidelines to evaluate the response to treatment in solid tumors. J Natl Cancer Inst 2000; 92: 205–216. Polymorphism meta-analysis for anti-EGFR monoclonal antibody response EK Morgen et al 540 The Pharmacogenomics Journal (2017), 535 – 542 © 2017 Macmillan Publishers Limited, part of Springer Nature.10655437 [Google Scholar]
- 16.Ioannidis JPA. Why most published research findings are false. PLoS Med 2005; 2: e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garg AX, Hackam D, Tonelli M. Systematic review and meta-analysis: when one study is just not enough. Clin J Am Soc Nephrol 2008; 3: 253–260. [DOI] [PubMed] [Google Scholar]
- 18.Geva R, Vecchione L, Kalogeras KT, Jensen BV, Lenz H-J, Yoshino T et al. FCGR polymorphisms and cetuximab efficacy in chemorefractory metastatic colorectal cancer: an international consortium study. Gut 2015; 64: 921–928. [DOI] [PubMed] [Google Scholar]
- 19.Kjersem JB, Skovlund E, Ikdahl T, Guren T, Kersten C, Dalsgaard AM et al. FCGR2A and FCGR3A polymorphisms and clinical outcome in metastatic colorectal cancer patients treated with first-line 5-fluorouracil/folinic acid and oxaliplatin +/− cetuximab. BMC Cancer 2014; 14: 340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Riley RD, Higgins JPT, Deeks JJ. Interpretation of random effects meta-analyses. BMJ 2011; 342: d549. [DOI] [PubMed] [Google Scholar]
- 21.Michiels S, Piedbois P, Burdett S, Syz N, Stewart L, Pignon J-P. Meta-analysis when only the median survival times are known: a comparison with individual patient data results. Int J Technol Assess Health Care 2005; 21: 119–125. [DOI] [PubMed] [Google Scholar]
- 22.Saridaki Z, Weidhaas JB, Lenz H-J, Laurent-Puig P, Jacobs B, De Schutter J et al. A let-7 microRNA-binding site polymorphism in KRAS predicts improved outcome in patients with metastatic colorectal cancer treated with salvage cetuximab/ panitumumab monotherapy. Clin Cancer Res 2014; 20: 4499–4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loupakis F, Antoniotti C, Cremolini C, Zhang W, Yang D, Wakatsuki T et al. Prospective study of EGFR intron 1 (CA)n repeats variants as predictors of benefit from cetuximab and irinotecan in chemo-refractory metastatic colorectal cancer (mCRC) patients. Pharmacogenomics J 2014; 14: 322–327. [DOI] [PubMed] [Google Scholar]
- 24.Garm Spindler K- L, Pallisgaard N, Rasmussen AA, Lindebjerg J, Andersen RF, Crüger D et al. The importance of KRAS mutations and EGF61A4G polymorphism to the effect of cetuximab and irinotecan in metastatic colorectal cancer. Ann Oncol 2009; 20: 879–884. [DOI] [PubMed] [Google Scholar]
- 25.Graziano F, Ruzzo A, Loupakis F, Canestrari E, Santini D, Catalano V et al. Pharmacogenetic profiling for cetuximab plus irinotecan therapy in patients with refractory advanced colorectal cancer. J Clin Oncol 2008; 26: 1427–1434. [DOI] [PubMed] [Google Scholar]
- 26.Inoue Y, Hazama S, Iwamoto S, Miyake Y, Matsuda C, Tsunedomi R et al. FcγR and EGFR Polymorphisms as Predictive Markers of Cetuximab Efficacy in Metastatic Colorectal Cancer. Mol Diagn Ther 2014; 18: 541–8. [DOI] [PubMed] [Google Scholar]
- 27.Sclafani F, Gonzalez de Castro D, Cunningham D, Hulkki Wilson S, Peckitt C, Capdevila J et al. FcγRIIa and FcγRIIIa polymorphisms and cetuximab benefit in the microscopic disease. Clin Cancer Res 2014; 20: 4511–4519. [DOI] [PubMed] [Google Scholar]
- 28.Etienne-Grimaldi M-C, Bennouna J, Formento J-L, Douillard J-Y, Francoual M, Hennebelle I et al. Multifactorial pharmacogenetic analysis in colorectal cancer patients receiving 5-fluorouracil-based therapy together with cetuximabirinotecan. Br J Clin Pharmacol 2012; 73: 776–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lurje G, Nagashima F, Zhang W, Yang D, Chang HM, Gordon MA et al. Polymorphisms in cyclooxygenase-2 and epidermal growth factor receptor are associated with progression-free survival independent of K-ras in metastatic colorectal cancer patients treated with single-agent cetuximab. Clin Cancer Res 2008; 14: 7884–7895. [DOI] [PubMed] [Google Scholar]
- 30.Zhang W, Azuma M, Lurje G, Gordon MA, Yang D, Pohl A et al. Molecular predictors of combination targeted therapies (cetuximab, bevacizumab) in irinotecanrefractory colorectal cancer (BOND-2 study). Anticancer Res 2010; 30: 4209–4217. [PubMed] [Google Scholar]
- 31.Zhang W, Labonte MJ, Lenz H-J. KRAS let-7 LCS6 SNP predicts cetuximab efficacy in KRASwt metastatic colorectal cancer patients: Does treatment combination partner matter? Ann Oncol 2011; 22: 484–485. [DOI] [PubMed] [Google Scholar]
- 32.Kjersem JB, Ikdahl T, Guren T, Skovlund E, Sorbye H, Hamfjord J et al. Let-7 miRNAbinding site polymorphism in the KRAS 3’UTR; colorectal cancer screening population prevalence and influence on clinical outcome in patients with metastatic colorectal cancer treated with 5-fluorouracil and oxaliplatin +/− cetuximab. BMC Cancer 2012; 12: 534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hsieh Y-Y, Tzeng C-H, Chen M-H, Chen P-M, Wang W-S. Epidermal growth factor receptor R521K polymorphism shows favorable outcomes in KRAS wild-type colorectal cancer patients treated with cetuximab-based chemotherapy. Cancer Sci 2012; 103: 791–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Negri FV, Musolino A, Naldi N, Bortesi B, Missale G, Laccabue D et al. Role of immunoglobulin G fragment C receptor polymorphism-mediated antibodydependant cellular cytotoxicity in colorectal cancer treated with cetuximab therapy. Pharmacogenomics J 2014; 14: 14–19. [DOI] [PubMed] [Google Scholar]
- 35.Calemma R, Ottaiano A, Trotta AM, Nasti G, Romano C, Napolitano M et al. Fc γ receptor IIIa polymorphisms in advanced colorectal cancer patients correlated with response to anti-EGFR antibodies and clinical outcome. J Transl Med 2012; 10: 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu-Lieskovan S, Vallbohmer D, Zhang W, Yang D, Pohl A, Labonte MJ et al. EGF61 polymorphism predicts complete pathologic response to cetuximab-based chemoradiation independent of KRAS status in locally advanced rectal cancer patients. Clin Cancer Res 2011; 17: 5161–5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bibeau F, Lopez-Crapez E, Di Fiore F, Thezenas S, Ychou M, Blanchard F et al. Impact of Fc{γ}RIIa-Fc{γ}RIIIa polymorphisms and KRAS mutations on the clinical outcome of patients with metastatic colorectal cancer treated with cetuximab plus irinotecan. J Clin Oncol 2009; 27: 1122–1129. [DOI] [PubMed] [Google Scholar]
- 38.Dahan L, Norguet E, Etienne-Grimaldi M-C, Formento J-L, Gasmi M, Nanni I et al. Pharmacogenetic profiling and cetuximab outcome in patients with advanced colorectal cancer. BMC Cancer 2011; 11: 496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park SJ, Hong YS, Lee J-L, Ryu M-H, Chang HM, Kim K et al. Genetic polymorphisms of FcγRIIa and FcγRIIIa are not predictive of clinical outcomes after cetuximab plus irinotecan chemotherapy in patients with metastatic colorectal cancer. Oncology 2012; 82: 83–89. [DOI] [PubMed] [Google Scholar]
- 40.Paez D, Paré L, Espinosa I, Salazar J, del Rio E, Barnadas A et al. Immunoglobulin G fragment C receptor polymorphisms and KRAS mutations: are they useful biomarkers of clinical outcome in advanced colorectal cancer treated with anti-EGFRbased therapy? Cancer Sci 2010; 101: 2048–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sebio A, Paré L, Páez D, Salazar J, González A, Sala N et al. The LCS6 polymorphism in the binding site of let-7 microRNA to the KRAS 3’-untranslated region: its role in the efficacy of anti-EGFR-based therapy in metastatic colorectal cancer patients. Pharmacogenet Genomics 2013; 23: 142–147. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.