Abstract
Background and Objectives:
Vector-borne bacterial diseases represent a substantial public health burden and rodents have been recognized as important reservoir hosts for many zoonotic pathogens. This study investigates bacterial pathogens in a small mammal community of the southwestern United States of America.
Methods:
A total of 473 samples from 13 wild rodent and 1 lagomorph species were tested for pathogens of public health significance: Bartonella, Brucella, Yersinia, Borrelia, Rickettsia spp., and Anaplasma phagocytophilum.
Results:
Three animals were positive for Yersinia pestis, and one Sylvilagus audubonii had a novel Borrelia sp. of the relapsing fever group. No Brucella, Rickettsia, or A. phagocytophilum infections were detected. Bartonella prevalence ranged between 0% and 87.5% by animal species, with 74.3% in the predominant Neotoma micropus and 78% in the second most abundant N. albigula. The mean duration of Bartonella bacteremia in mark-recaptured N. micropus and N. albigula was 4.4 months, ranging from <1 to 18 months, and differed among Bartonella genogroups. Phylogenetic analysis of the Bartonella citrate synthase gene (gltA) revealed 9 genogroups and 13 subgroups. Seven genogroups clustered with known or previously reported Bartonella species and strains while two were distant enough to represent new Bartonella species. We report, for the first time, the detection of Bartonella alsatica in North America in Sylvilagus audubonii and expand the known host range of Bartonella washoensis to include Otospermophilus variegatus.
Interpretation and Conclusion:
This work broadens our knowledge of the hosts and geographic range of bacterial pathogens that could guide future surveillance efforts and improves our understanding of the dynamics of Bartonella infection in wild small mammals.
Keywords: woodrat, bacteria, Bartonella, small mammals, vector-borne pathogens
Introduction
Rodents are important reservoir hosts for many zoonotic pathogens, with ∼10.7% of rodent species being hosts to 85 zoonotic pathogens (Bordes et al. 2015, Han et al. 2016). Despite the ubiquity of rodents and the diversity, distribution, and epidemiological significance of their pathogens, rodent-borne diseases are still greatly underinvestigated (Kosoy et al. 2015). Several bacterial pathogen genera of epidemiological significance to humans have been previously detected in rodents, including Yersinia, Bartonella, Brucella, Borrelia, Anaplasma, and Rickettsia.
The study area in North Central New Mexico, United States of America is a natural focus of plague caused by flea-borne Yersinia pestis. Although plague is a rare disease, about half of the human cases in the United States of America are found in New Mexico (Plague 2019). A previous study targeted the plague pathogen in rodent fleas and specific Y. pestis antibodies in animal sera (Kosoy et al. 2017). In the current study, we were interested in detection of Y. pestis DNA in blood.
Bartonella species from wild rodents, including Bartonella washoensis in ground squirrels (Kosoy et al. 2003) or Bartonella vinsonii subsp. arupensis in deer mice (Welch et al. 1999), have been implicated as potential public health threats. Bartonella species were previously cultured from Neotoma woodrats from New Mexico and detected in their fleas (Morway et al. 2008). We hypothesized that molecular methods would allow us to reveal Bartonella species that are hard to culture and thus could have been missed in prior studies, altering estimates of infection prevalence and diversity. Furthermore, we analyzed Bartonella prevalence across species, predicted variation in the probability of infection in one highly sampled species, and assessed temporal patterns of infection in serially sampled individuals.
First isolated from Neotoma woodrats in Utah in 1957 (Stoenner and Lackman 1957), Brucella neotomae was thought to be limited to woodrats (Moreno 2014) until its isolation from cerebrospinal fluid of two men with neurobrucellosis in Costa Rica in 2008 and 2011 (Suarez-Esquivel et al. 2017). We tested our samples to determine if woodrats in New Mexico could harbor Brucella neotomae or other Brucella species.
Finally, we included tick-borne pathogens Borrelia spp., Rickettsia spp., and Anaplasma phagocytophilum because of their public health significance and recent reports of increased incidence in the United States of America (Rosenberg et al. 2018, Binder and Armstrong 2019). A. phagocytophilum was previously reported from rodents in California, Connecticut, and Minnesota (Atif 2016). Transmission of A. phagocytophilum between Neotoma mexicana woodrats in Colorado was maintained by Ixodes spinipalpis ticks (Zeidner et al. 2000). Borrelia burgdorferi was detected in rodents from California (Brown and Lane 1994, Brown et al. 2006) and rabbits from Texas (Burgess and Windberg 1989). The emerging pathogen Rickettsia felis and the agent of murine typhus R. typhi were reported from wildlife and their fleas in New Mexico and California (Williams et al. 1992, Stevenson et al. 2005).
In this study, we investigated the presence of multiple pathogens in a natural community of small mammals in New Mexico to gain a more comprehensive view of the complex epidemiological system there and to broaden our understanding of pathogen/host interactions and the implications for human health.
Materials and Methods
Sample collection and DNA extraction
We tested 473 whole blood samples from 13 rodent and 1 lagomorph species (Table 1) captured in the Eldorado subdivision of Santa Fe County, New Mexico, United States of America from November 2002 to July 2004 as part of a markrecapture surveillance study (Morway et al. 2008). Animals were captured at 89 trap stations selected by identification of freshly occupied woodrat den. Each trapping station had a small Sherman trap (2 × 2.5 × 6.5″) for small mouse-size rodents, a large Sherman trap (3 × 3.5 × 9″) for rat-size rodents, and a Tomahawk trap (4 × 4 × 10″) for squirrel-size rodents and rabbits. Traps were baited with oats, peanut butter, and molasses, set in the afternoon, and checked the following morning. Captured animals were anesthetized with a mixture of isoflurane and oxygen and marked individually with ear tag or/and subcutaneous transponder (AVID, Folsom, LA). A retroorbital bleed was performed; blood was collected with heparinized microhematocrit capillary tubes and kept on dry ice until placed at −80°C in the laboratory until processing. All animal handling procedures were approved by the Center for Disease and Control’s Division of Vector-Borne Diseases Institutional Animal Care and Use Committee, protocol number 06–008. We extracted DNA using the KingFisher Flex Purification System and the associated MagMAX Pathogen RNA/DNA Kit (ThermoFisher, Waltham, MA) according to the manufacturer’s protocols.
Table 1.
Prevalence of Bartonella Species in a Small Mammal Community in the Eldorado Subdivision of Santa Fe County, Northern New Mexico, United States of America in 2002–2004
| Family | Common name | Latin name | Tested samples | Percent of total number of samples | Bartonella positive | Bartonella prevalence [95% CI] |
|---|---|---|---|---|---|---|
|
| ||||||
| Cricetidae | White-throated woodrat | Neotoma albigula | 50 | 10.6 | 39 | 78 [64.8–87.2] |
| Southern Plains woodrat | Neotoma micropus | 272 | 57.5 | 202 | 74.3 [68.8–79.1] | |
| Northern grasshopper mouse | Onychomys leucogaster | 8 | 1.7 | 7 | 87.5 [52.9–97.8] | |
| White-footed mouse | Peromyscus leucopus | 25 | 5.3 | 12 | 48 [30–66.5] | |
| Deer mouse | Peromyscus maniculatus | 14 | 3 | 3 | 21.4 [7.6–47.6] | |
| Pinyon mouse | Peromyscus truei | 12 | 2.5 | 2 | 16.7 [4.7–44.8] | |
| Western harvest mouse | Reithrodontomys megalotis | 2 | 0.4 | 1 | 50 [9.5–90.5] | |
| Heteromyidae | Ord’s kangaroo rat | Dipodomys ordii | 43 | 9.1 | 30 | 69.8 [54.9–81.4] |
| Banner-tailed kangaroo rat | Dipodomys spectabilis | 2 | 0.4 | 0 | 0 [0–65.8] | |
| Silky pocket mouse | Perognathus flavus | 1 | 0.2 | 0 | 0 [0–79.3] | |
| Leporidae | Desert cottontail | Sylvilagus audubonii | 10 | 2.1 | 7 | 70 [39.7–89.2] |
| Muridae | House mouse | Mus musculus | 1 | 0.2 | 0 | 0 [0–79.3] |
| Sciuridae | Rock squirrel | Otospermophilus variegatus | 27 | 5.7 | 10 | 37 [21.5–55.8] |
| Spotted ground squirrel | Xerospermophilus spilosoma | 6 | 1.3 | 0 | 0 [0–39] | |
| Total | 473 | 313 | 66.2 [61.8–70.3] | |||
Pathogen detection, sequencing, and phylogenetic analysis
Initial detection of Bartonella, Brucella, and Yersinia DNA was implemented using a multiplex quantitative real-time PCR (qPCR) protocol targeting the Bartonella transfer mRNA gene (ssrA), Brucella insertion sequence (IS711), and Yersinia peptidoglycan-associated lipoprotein (pal). In addition to the qPCR analysis, all samples were tested for Bartonella species by conventional PCR for the 16S-23SrRNAgene intergenic transcribed spacer (ITS) region; ssrA and/or ITS-positive samples were tested by nested PCR for the citrate synthase gene (gltA). To reduce risk of false positives, samples were considered Bartonella positive only if they tested positive for two out of three targets (ssrA, ITS, gltA) and were successfully sequenced. We used gltA sequences for phylogenetic analysis as they clearly distinguish Bartonella species and subspecies (La Scola et al. 2003), and gltA is the most widely used marker for Bartonella genotyping (Kosoy et al. 2018). Yersinia qPCR-positive samples were confirmed by conventional PCR for the plasmogen activator gene pla (Bai et al. 2017). A multiplex qPCR protocol was used for detection of p44 and msp4 genes from A. phagocytophilum and fliD and the 18S rRNA gene from Borrelia burgdorferi and Borrelia spp. Testing for Rickettsia species was conducted by qPCR targeting the gltA gene. All primers, probes, and conditions are given in Supplementary Table S1.
Positive (Bartonella doshiae, Brucella melitensis, Y. pseudotuberculosis, Borrelia burgdorferi, A. phagocytophilum, R. felis) and negative (deionized water) controls were used in all respective reactions to evaluate the presence of bacterial DNA and to detect potential contamination, respectively. PCR reactions were considered positive if they had cycle threshold value Ct <40 and characteristic amplification plots.
Conventional PCR products were separated by 1.5% gel electrophoresis and visualized by Biotium GelGreen stain (Biotium, Hayward, CA).Positive PCR products were purified using the QIA quick PCR Purification Kit (QIAGEN,Valencia, CA) according to the manufacturer’s protocols and sequenced in both directions with the same primers on an Applied Bio-systems Model 3130 Genetic Analyzer (Applied Biosystems, Foster City, CA). We assembled forward and reverse sequences using the SeqMan Pro program in Lasergene v12 (DNASTAR, Madison, WI). All sequences were aligned with MAFFT v7.187 (Katoh and Standley 2013), trimmed to equal lengths with Gblocks v0.91b(Castresana 2000),and compared with other Bartonella strains from rodents, rodent ectoparasites, and known Bartonella species. A neighbor-joining tree was produced with MEGA v7.0.26 (Kumar et al. 2016) from the 351bp alignment of 348 gltA sequences using B. tamiae as the outgroup. We used the Tamura-Nei model (Tamura and Nei1993) of sequence evolution for calculating branch lengths and tested branch support using 1000 bootstrap replicates. Clades of Bartonella genogroups were designated according to the standards of La Scola et al. (2003), wherein gltA sequences with <96% homology with another Bartonella species may be defined as novel.
Statistical analysis
Prevalence was estimated by species across all monthly sampling time points when rodents were captured using the number of infected animals out of the total sampled. For the most frequently sampled species, N. micropus, we estimated prevalence at monthly sampling time points between November 2002 and October 2003. Confidence intervals for prevalence were calculated with Wilson score intervals (Wilson 1927). Differences in Bartonella prevalence between species were analyzed using a chi-square test and logistic regression considering prevalence as a binomial variable, using Dipodomys ordii as the index species. Host specificity of Bartonella genogroups was examined by plotting the relative abundance of each genogroup (i.e., the percent of all infections attributed to a genogroup) by host species and host family.
Variation in Bartonella prevalence in the sampled N. micropus population over time was analyzed using binomial logistic regression with days since the first monthly sampling point (November 2002) as the independent variable. Individual-level variation in the probability of Bartonella infection was analyzed using generalized linear mixed modeling. Previous studies have indicated that Bartonella prevalence in rodent populations can vary over time, by sex, and by animal weight (Kosoy et al. 2004, Morway et al. 2008, Bai et al. 2011). We included data on these variables as well as the presence and number of fleas recorded on each sampled N. micropus individual. All factors were included as fixed effects in a global model, with individual ear tag numbers as a random effect to account for multiple sampling of some individuals (n = 36). We also included an interaction term between individual sex and weight in the global model to account for potential differences in the change in the probability of infection with weight between males and females, a pattern that has not been evaluated in past studies (Kosoy et al. 2004, Morway et al. 2008, Bai et al. 2011). Model selection was performed on the global model that included all fixed and random effects and interactions. The best model was chosen based on the lowest Akaike information criterion with a correction (AICc) for finite sample sizes (Burnham and Anderson 2004). Goodness of fit for the best model was assessed using the area under the receiver operating characteristic curve (AUC) (Hosmer and Lemeshow 2000). All statistical analyses were performed in R v3.6.1 (Team RC 2013). For all tests, α = 0.05 was chosen as the threshold level for statistical significance.
Results
The tested animals represented 14 species of 5 families and 2 orders, Rodentia (97.9%) and Lagomorpha (2.1%). The most abundant were N. micropus (57.5%), followed by N. albigula (10.6%) and D. ordii (9.1%) (Table 1). Among N. micropus, more adult females than adult males were present in the population at all times, with an average ratio of 1.71:1.
One N. micropus, one N. albigula, and one Peromyscus truei tested positive for Y. pestis. One Sylvilagus sudubonii tested positive for novel Borrelia spp. of relapsing fever group and work continues to characterize it. All samples tested negative for A. phagocytophilum, Brucella, and Rickettsia species.
Patterns of Bartonella infection status
In contrast with the rare occurrence of other pathogens, 66% of samples tested positive for Bartonella species (Table 1). Since host species carry predominately their own Bartonella genogroups and any sharing of genogroups among species makes up a small proportion of the total infections within each host species, we decided to look at the trends in prevalence over time for N. micropus only, instead of those in the community, particularly because N. micropus constitutes the vast majority of samples taken. Sampling from other host species was too sparse to analyze prevalence over time, so we summarized prevalence over the whole sampling period (Table 1). There was significant variation in prevalence among species in the community (χ2 = 72.4, df = 13, p<0.001). According to binomial regression analysis and considering D. ordii as the index species (at 70% prevalence), Otospermophilus variegatus, P. maniculatus, and P. truei had significantly lower prevalence than D. ordii. Prevalence in P. leucopus was lower than D. ordii, but this difference was only marginally significant (p<0.1). Bartonella prevalence in the remaining sampled species was not significantly different compared with D. ordii (Table 1).
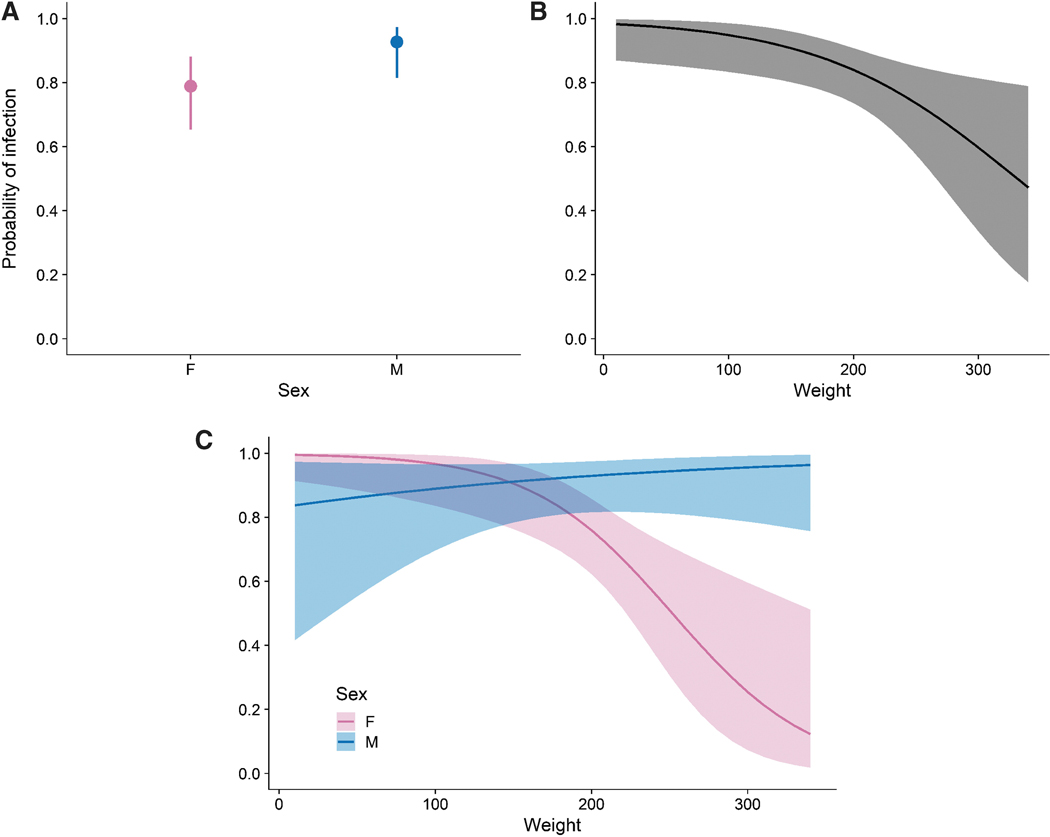
Analysis of prevalence variation over time makes it clear that Bartonella infection is enzootic at high prevalence in N. micropus (Fig. 1). Prevalence over the period of sampling increased slightly from 71% to 79%, however, this change was not significant (F = 0.13, df = 1, p = 0.73). We also looked at other factors that could predict Bartonella infection status at the time of capture in sampled N. micropus individuals, including date of capture, sex, age, body weight, whether fleas were present, the count of fleas, an interaction term between sex and body weight, and a random effect for individual ear tag numbers. The best model according to AICc was the model that included sex (F = 8.8, df = 1, p = 0.0034), weight (F = 3.5, df = 1, p = 0.062), an interaction between sex and weight (F = 10.6, df = 1, p = 0.0011), and a random effect of ear tags (Supplementary Table S3). With AUC = 0.89, the data demonstrate a good fit to the model. Averaging over weight, males were more likely to be infected than females (Fig. 2A). Combining the sexes, the probability of infection declines with body weight (Fig. 2B). However, with the inclusion of the significant interaction term between sex and weight, the model indicates that the probability of Bartonella infection decreases with weight for females while the probability of infection is constant or slightly increasing with weight for males (Fig. 2C); females were significantly less likely to be infected than males only if they weighed over 200g.
FIG. 1.
Bartonella prevalence in Neotoma micropus over time. Circles show estimated prevalence and gray outline shows 95% confidence intervals. The dashed line is the predicted fit for the binomial regression. Numbers above show the number of animals sampled.
FIG. 2.
Modeled probability of Bartonella infection in sampled N. micropus individuals including terms for sex, weight, an interaction between sex and weight, and a random effect for individual ear tag. Panels show the predicted probability of infection accounting for the effect of sex, averaging over weight (A); the effect of weight, with sexes combined (B); and the interaction of sex and weight (C).
Bartonella species diversity
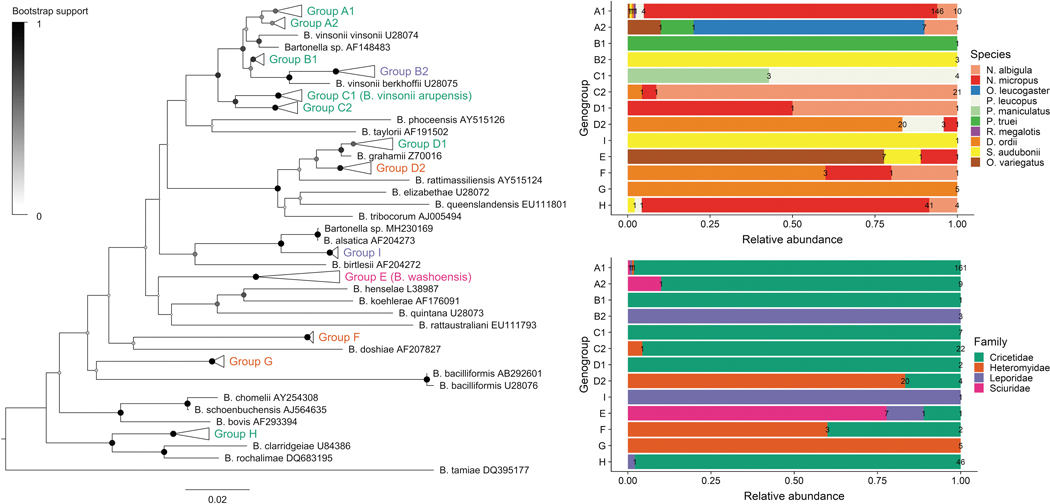
Based on sequence similarity, 296 gltA sequences were clustered into nine phylogenetic genogroups (A–I) and 13 subgroups (Fig. 1). Groups A–C all cluster within the Bartonella vinsonii species complex. The net average genetic distances between groups ranged from 6.7% to 13.8%, considering groups A–C together as a clade. The net average genetic distances between subgroups A–C and between D1 and D2 ranged from 2.2% to 3.2%. We uploaded GenBank sequences obtained from new phylogenetic groups and from new hosts in previously reported groups (Supplementary Table S2).
Genogroup A includes subgroups A1 and A2 and clusters with Bartonella vinsonii subsp. vinsonii with 97.5% homology (Table 2). Group A1 is more abundant (95.3%) than group A2 (4.7%), but both present strong host specificity. Group A1 contains sequences obtained primarily from N. micropus (89%) and N. albigula (6.1%) and includes a previously reported Bartonella strain from N. albigula (Rubio et al. 2014). In addition, we detected this ecotype in P. leucopus (2.4%) and S. audubonii, O. variegatus, R. megalotis, and D. ordii (0.6% each). Group A2 consists of sequences obtained from Onychomys leucogaster (70%) and two previously published sequences from O. leucogaster (Bai et al. 2007). We also detected this ecotype in N. albigula, P. truei, and O. variegatus (10% each).
Table 2.
Phylogenetic Relationships Between Detected Bartonella Genogroups, Known Bartonella Species, and Reference Sequences from GenBank
| Genogroup | Related Bartonella species | Primary hosts in current study | Closest matching GenBank record |
|---|---|---|---|
|
| |||
| A1 | B. vinsonii subsp. vinsonii | N. micropus, N. albigula | KJ719287: N. albigula, Mexico |
| A2 | B. vinsonii subsp. vinsonii | O. leucogaster | DQ357610: O. leucogaster, Kansas, United States of America |
| DQ357613: O. leucogaster, Kansas, United States of America | |||
| B1 | B. vinsonii subsp. berkhoffii | P. truei | AF148489: O. leucogaster, New Mexico, United States of America |
| B2 | B. vinsonii subsp. berkhoffii | S. audubonii | AF148486: S. audubonii, New Mexico, United States of America |
| C1 | B. vinsonii subsp. arupensis | P. leucopus, P. maniculatus | FJ946836: dog, Thailand |
| FJ946844: dog, Thailand | |||
| C2 | B. vinsonii subsp. arupensis | N. albigula | AF148487: N. albigula, New Mexico, United States of America |
| AF148491: N. albigula, New Mexico, United States of America | |||
| AF148493: N. albigula, New Mexico, United States of America | |||
| D1 | B. grahamii | N. micropus, N. albigula | KJ175044: Myodes rutilus, China |
| KX549996: Apodemus agrarius, China | |||
| D2 | B. grahamii | D. ordii | KJ719293: D. spectabilis, Mexico |
| KJ719295: D. merriami, Mexico | |||
| E | B. washoensis | O. variegatus | AF470616: O. beecheyi, Nevada, United States of America |
| AY071858: O. beecheyi, Nevada, United States of America | |||
| FJ719016: human, California, United States of America | |||
| F | B. doshiae | D. ordii | |
| G | B. bacilliformis | D. ordii | KJ719297: D. merriami, Mexico |
| KJ719298: D. merriami, Mexico | |||
| H | B. rochalimae | N. micropus, N. albigula | EU549693: Orchopeas sexdentatus (flea), N. micropus (host), New Mexico, United States of America |
| I | B. alsatica | S. audubonii | AF204273: Oryctolagus cuniculus, France |
Within genogroup B, subgroups B1 and B2 cluster with Bartonella vinsonii subsp. berkhoffii with 96.9% homology. Group B1 includes one P. truei sequence and a previously published sequence from O. leucogaster. Group B2 contains sequences from S. audubonii and a previously reported sequence from S. audubonii. Genogroup C clusters closely with Bartonella vinsonii subsp. arupensis with 98.7% homology. Group C1 includes sequences from P. leucopus (57%) and P. maniculatus (43%) and groups with previously detected sequences from stray dogs in Thailand (Bai et al. 2010). Group C2 contains sequences from N. albigula (87.5%), N. micropus (4.2%), and D. ordii (4.2%) and clusters with sequences previously reported from N. albigula (Fig. 3).
FIG. 3.
Phylogenetic tree and host range of Bartonella genogroups. The neighbor-joining tree was produced from a 351 bp alignment of the gltA gene. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 pseudoreplicates) are shown as colored circles at each branch. Evolutionary distances were computed using the Tamura-Nei method and are in the units of the number of base substitutions per site. Relative abundance and counts of sequences from each genogroup are summarized to the right of the tree based on the host species and the host family from which the sequence was obtained.
Group D1 includes one N. albigula and one N. micropus sequence, as well as previously detected sequences from Myodes rutilus (Li et al. 2015) and Apodemus agrarius in China. Group D2 is more distant from Bartonella grahamii (97.2% homology) compared with D1 (98.9%). Sequences from D. ordii comprise 83.3% of group D2, 12.5% from P. leucopus, and 4.2% from N. micropus. Group D2 also contains sequences previously reported from Dipodomys spp. from Mexico (Rubio et al. 2014).
Genogroup E includes sequences obtained from O. variegatus (77.8%), S. audubonii (11.1%), and N. micropus (11.1%) and clusters with Bartonella washoensis from O. beecheyi (Kosoy et al. 2003) and a human patient from California (Probert et al. 2009). Genogroup F includes sequences from D. ordii, N. micropus, and N. albigula and clusters with Bartonella doshiae (89.6% homology), but the bootstrap support for this relationship was only 30%. Genogroup G contains sequences from D. ordii and clustered with two previously published sequences from D. merriami (Rubio et al. 2014). The closest Bartonella species to genogroup G was Bartonella bacilliformis with 87.7% homology, but the bootstrap support for this relationship was only 24%.
Sequences in the second most abundant genogroup H were from N. micropus (87.2%), N. albigula (8.5%), S. audubonii (2.1%), and P. leucopus (2.1%). The group clustered with a previously reported sequence from the flea Orchopeas sexdentatus collected from N. micropus. The closest species to this group was Bartonella rochalimae with 95.1% homology. Genogroup I included one sequence from S. audubonii and presented 97.4% homology to Bartonella alsatica isolated from the blood of European rabbits (Heller et al. 1999).
Across all host species, the most numerous genogroup was A1 found in 54.5% of all tested samples, followed by groups H, D2, and C2, found in 15.6%, 8%, and 7.6% of samples, accordingly. Bartonella genogroups generally showed patterns of specificity at the levels of host species and families (Figs. 3 and 4; Table 2). Genogroups A1, A2, B1, C1, C2, D1, and H were primarily associated with species in the Cricetidae family and genogroups D2, F, and G were associated with D. ordii in the Heteromyidae family. Genogroups B2 and I were associated with S. audubonii and E was mainly found in O. variegatus.
FIG. 4.
Relative abundance of Bartonella genogroups A–I in host species and families.
Bartonella infection in recaptured Neotoma woodrats
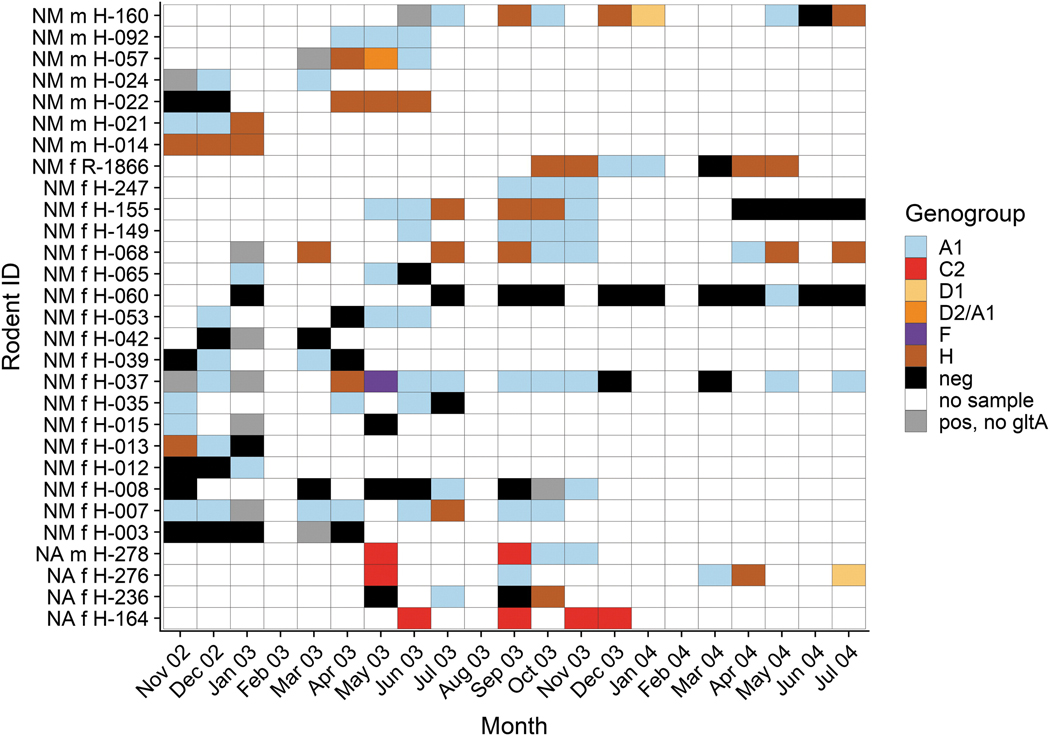
A total of 29 Neotoma spp. woodrats were recaptured more than three times over the course of the study and most recaptured animals were captured at the same trap stations or one nearby. The likelihood of recapture was not significantly different between sexes compared with their frequency in the population at large, either for N. micropus (χ2 = 2.6, p = 0.11), or for N. albigula (χ2 = 0, p = 1). Most (25/29) individuals were positive at multiple sampling time points and in many cases the same Bartonella genogroup was detected across months of sampling (Fig. 5).
FIG. 5.
Resampling history, infection course, and genotypic characterization of sequentially recovered Bartonella samples from 29 woodrats (NM = N. micropus and NA = N. albigula) captured three and more times during 21 months of the study. The genogroups of Bartonella recovered from bacteremic woodrats at a sample month are shown with different colors. The classification ‘‘pos, no gltA’’ means that the sample was positive by ITS and real-time ssrA tests, but no gltA sequence was obtained to determine the Bartonella genogroup present. ITS, intergenic transcribed spacer.
We examined the duration of Bartonella infections in all recaptured animals. Infection durations for any Bartonella genogroup is the longest duration that an individual could have been considered positive, including months when no sample was taken but not including months when the individual tested negative. Individuals showed a broad variation in infection durations with a range <1 to 18 months with a median infection duration of 3 months and a mean of 4.4 months (Fig. 6). A large proportion of individuals showed an infection duration of <1 month, meaning the individual only tested positive at a single sampling time point within a month, but at no time after. To determine infection durations for Bartonella genogroups, each infection timeline was split across different genogroups for an individual (Fig. 6). Mean durations for A1, C2, and H were 1.8, 3.3, and 1.0 months, respectively.
FIG. 6.
Infection duration for any Bartonella genogroup (left panel) and for separate genogroups (right panel). Individual points for infection durations of each genogroup in an individual are shown as open circles, the median is a thick black line on the box plot, and the mean is an open diamond. Numbers above the box plots show the counts of infections for each genogroup. Some individuals had multiple infections for the same genogroup over their timeline, for example, they were infected with A1, then H, then A1 again.
Discussion
In this study, we performed surveillance for several bacterial pathogens of public health concern in a community of rodents in New Mexico. While we successfully detected Y. pestis, Borrelia sp., and Bartonella spp. in species within the community, we were unable to detect the presence of Brucella, Rickettsia, or A. phagocytophilum DNA in any samples. These results should not however be interpreted as a true absence of these bacteria in the community, since a lack of sensitivity in our assays may have missed some positive samples. We acknowledge that multiplex assays can be less sensitive than uniplex assays, especially to any target pathogen present at low abundance in the same specimen as another, highly abundant target pathogen. We may therefore have missed coinfecting bacteria in some specimens infected with Bartonella, so the actual prevalence of the other pathogens targeted in our study could be higher than reported here and should be investigated further.
We detected Y. pestis DNA in blood from N. micropus, N. albigula, and P. truei. A prior study in New Mexico found fleas from N. micropus positive for Y. pestis DNA and detected antibodies to Y. pestis antigen in six species: N. micropus, O. leucogaster, P. leucopus, P. maniculatus, P. truei, O. variegatus, and S. audubonii (Kosoy et al. 2017). Small-scale die-offs in woodrats are suggested to support the maintenance of plague in the active southwestern United States of America focus (Kosoy et al. 2017). Our results indicate that the presence of Y. pestis in rodent communities can be detected in rodent blood as part of a broad pathogen surveillance program. Future studies will be needed to elucidate the long-term dynamics and enzootic maintenance of Y. pestis among diverse rodent communities.
Overall, we determined that Bartonella infection is common in many small mammal species in our study area in New Mexico. High variation in Bartonella prevalence among host species and host specificity of Bartonella genogroups makes it more feasible to compare prevalence at the host species and genus levels, rather than in the whole small mammal community. Host specificity of Bartonella strains in rodents has been described in other studies (Kosoy et al. 1997, 2000, Jardine et al. 2005, Bai et al. 2007, 2011, Rubio et al. 2014). This host specificity is likely due to historical adaptation of Bartonella bacteria to a particular host species or a set of related species. Accordingly, we found that variants within genogroups tend to infect certain genera and families of hosts more frequently than others within this community (Fig. 1).
As predicted, Bartonella prevalence determined by molecular methods in N. micropus and N. albigula in this study was higher than the previous findings in the same area (Morway et al. 2008), as the molecular methods allowed us to find genogroups that are potentially hard to culture, such as the Bartonella rochalimae-like genogroup H (Harms et al. 2017). Other studies found Bartonella prevalence of 36.7% and 50% in N. albigula and N. micropus woodrats, respectively, in the United States of America (Bai et al. 2009) and 75% in N. albigula in Mexico (Rubio et al. 2014).
Bartonella prevalence was lower in November 2002 through March 2003, during the prereproductive period of N. micropus, which breeds in the early spring and produces one litter (Braun and Mares 1989). A similar pattern was registered in another study in New Mexico that also found an increase in flea loads during the early reproductive period (Morway et al. 2008), which may also explain the slight increase in Bartonella prevalence over time. The sex bias in the likelihood of Bartonella infection in N. micropus (Fig. 4A), also noted in the prior study (Morway et al. 2008), could be explained by the social organization and promiscuity of this species (Braun and Mares 1989, Conditt and Ribble 1997, Suchecki et al. 2004, Baxter et al. 2009), with mature male home ranges overlapping an average of three female home ranges, thereby increasing the number of potential contacts with infected fleas on a female mate or in a female’s roost. In addition, elevated testosterone levels can have immunosuppressive effects that could lead to higher susceptibility of infection (Marriott and Huet-Hudson 2006) and may also affect male social behavior that can lead to higher pathogen exposure (Grear et al. 2009). These immunological factors can also account for the decrease in Bartonella prevalence with increasing weight in female but not male N. micropus (Fig. 4C).
Similar patterns were previously reported in cotton rats (Kosoy et al. 2004) and prairie dogs (Bai et al. 2008), although these studies did not examine the effect of weight on prevalence in males and females separately. The patterns in duration of infection indicate that Bartonella infection is likely enzootic in this population of Neotoma woodrats and that individuals can be serially infected. Many individuals are consistently positive when sampled and can carry infection of the same Bartonella genogroup for many months. Prior laboratory studies indicate that the length of infection may differ between Bartonella species (Telfer et al. 2007). Additionally, previous studies of serially sampled cotton rats (Sigmodon hispidus) have shown similar patterns of consistent infection of Bartonella genotypes across multiple weeks of sampling (Kosoy et al. 2004, Bai et al. 2011). However, it is unclear from these past studies and in the present study, whether these patterns of infection are due to chronic, persistent infection within individuals that go dormant and occasionally reactivate; if rodent individuals are clearing and acquiring new infections between sampling periods; or if the patterns reflect both processes occurring simultaneously. Future studies may consider removing arthropod vectors from the community (Jardine et al. 2006) to ascertain the infectious period of natural Bartonella infections in these rodents in the absence of reinfection.
Bartonella prevalence in Peromyscus mice varied among the species and averaged 33.3%. Another study found an average Bartonella prevalence of 44.2% in Peromyscus mice in the United States of America (Bai et al. 2009). Bartonella prevalence in P. leucopus in our study fits into the wide range of prevalence for the species by culturing methods: 0–40% in Georgia, 6.3–76% in North Carolina, 23.2% in the western United States of America (Kosoy et al. 1997, Bai et al. 2009); and by molecular methods: 5–10% in Minnesota and Wisconsin (Hofmeister et al. 1998) and 50% in Mexico (Rubio et al. 2014). Bartonella prevalence in P. maniculatus and P. truei was lower than in prior studies demonstrating 47– 82% prevalence in other areas (Bai et al. 2009, 2011, Rubio et al. 2014, Ziedins et al. 2016). Bartonella prevalence in O. leucogaster in New Mexico fits into the 25–90% prevalence range, whereas prevalence in D. ordii is higher than in previous studies from Mexico and the United States of America (Bai et al. 2007, 2009, Rubio et al. 2014).
Detection of Bartonella infection in S. audubonii and in Sylvilagus rabbits is the first to our knowledge. A study in central California found no Bartonella spp. in Sylvilagus bachmani riparius and their ticks Haemaphysalis leporispalustris (Schmitz et al. 2014). Additionally, this is first molecular detection of Bartonella infection in O. variegatus, although Bartonella has been reported in the congener O. beecheyi in California (Osikowicz et al. 2016, Ziedins et al. 2016). For D. spectabilis, P. flavus, M. musculus, and X. spilosoma more samples will need to be collected to confirm if Bartonella is truly absent in these species.
Phylogenetic analysis of gltA sequences revealed diverse Bartonella species and subspecies in this small mammal community in New Mexico. The subgroups A–C match the criterion used for the separation of Bartonella species set out by La Scola et al. (2003), wherein >96% homology is used to cluster a gltA sequence within a Bartonella species, and therefore qualify as subspecies or ecotypes of Bartonella vinsonii and subgroups D as ecotypes of Bartonella grahamii. Whereas Bartonella vinsonii subsp. vinsonii and Bartonella vinsonii subsp. arupensis were previously reported in rodents, Bartonella vinsonii subsp. berkhoffii is usually associated with Caniformia (Kosoy and Goodrich 2018). Both Bartonella vinsonii subsp. arupensis and berkhoffii are recognized as human pathogens (Welch et al. 1999, Roux et al. 2000, Fenollar et al. 2005, Myint et al. 2011, Breitschwerdt et al. 2019). Bartonella grahamii is known in voles, mice, rats, and lagomorphs from Europe, Asia, and North America (Ellis et al. 1999, Jardine et al. 2005, Inoue et al. 2009, Rubio et al. 2014, Rao et al. 2015). Using a separate phylogenetic tree of Bartonella grahamii sequences (Fig. 7), we determined that genogroup D1 with Neotoma woodrat sequences clusters within a clade that contains Bartonella grahamii sequences associated with voles and mice in North America, Europe, and China and is separate from a clade containing sequences from Apodemus mice in Asia, whereas group D2 with sequences from D. ordii is distinct from both clades and clusters with the previously published sequences from Dipodomys kangaroo rats from Mexico (Rubio et al. 2014). Bartonella grahamii is a zoonotic pathogen detected in patients with ocular infections and cat scratch disease (Kerkhoff et al. 1999, Serratrice et al. 2003, Oksi et al. 2013).
FIG. 7.
Phylogenetic of B. grahamii and genogroups D1 and D2. The neighbor-joining tree was produced from a 351bp alignment of 41 gltA sequences. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown as colored circles at each branch. Evolutionary distances were computed using the Tamura-Nei method and are in the units of the number of base substitutions per site. DO = Dipodomys ordii; NM = N. micropus; PL = Peromyscus leucopus.
Detection of Bartonella washoensis in O. variegatus, S. audubonii, and N. micropus expands the range of rodent hosts of this pathogen. First isolated from a patient with fever and myocarditis in Nevada, Bartonella washoensis was later isolated from ground squirrels, likely a reservoir and a source of the infection (Kosoy et al. 2003). Bartonella washoensis was implicated in a case of human meningitis in California (Probert et al. 2009) and isolated from a dog with mitral valve endocarditis (Chomel et al. 2003).
Genogroups F and G likely represent new Bartonella species based on their phylogenetic distance from the other genogroups and known Bartonella species, however, further studies are needed to fully characterize them at additional genetic loci. Group H clustered with Bartonella rochalimae but was distant enough to represent an independent species. Bartonella rochalimae was isolated from an American woman after visiting Peru (Eremeeva et al. 2007). The pathogen has been reported mainly from Caniformia animals (Kosoy and Goodrich 2018). However, Bartonella rochalimae-like isolates have also been isolated from rodents (mainly rats) worldwide (Lin et al. 2008, Gundi et al. 2012, Buffet et al. 2013).
The close phylogenetic association of genogroup I with Bartonella alsatica represents the first molecular detection of this agent outside of Europe to our knowledge and broadens its host range to include S. audubonii. Bartonella alsatica was first isolated from the blood of asymptomatic wild European rabbits O. cuniculus in France (Heller et al. 1999) and was considered apathogenic until the transmission of Bartonella alsatica was reported in humans with culture-negative endocarditis in association with wild European rabbits (Raoult et al. 2006, Angelakis et al. 2008, Jeanclaude et al. 2009). While it is possible that Bartonella alsatica was inadvertently introduced to the United States with the European rabbit and/or its fleas, we consider this scenario unlikely. The lower homology of genogroup I to Bartonella alsatica (97.4%) at the gltA locus makes it more likely that the bacterial lineages infecting European O. cuniculus and American S. audubonii evolved from a common ancestor when these genera diverged from one another millions of years ago. Future studies should attempt detection and phylogenetic characterization of Bartonella alsatica strains from a broader diversity of lagomorphs to explore any biogeographical patterns, as has been done for Bartonella washoensis (Inoue et al. 2011) and Bartonella grahamii (Inoue et al. 2009).
Conclusion
In conclusion, we determined that Bartonella infection is common in many small mammal species in New Mexico, discovered high genetic diversity of Bartonella species there, and provided evidence of host-specific associations between Bartonella genogroups and their hosts. We uncovered two novel Bartonella species and updated the classification of Bartonella grahamii phylogenetic groups. We also report the first molecular detection of Bartonella alsatica outside of Europe and in a new host S. audubonii and broaden the known host ranges of other Bartonella species. As most known species of Bartonella are recognized as zoonotic pathogens, it is important to classify pathogen diversity and potential threats to human populations. Paired with the detection of Y. pestis and Borrelia sp., this study provides important data for assessing the occurrence of bacterial pathogens in wild rodent populations.
Supplementary Material
Acknowledgments
The authors are grateful to Allen Richards and Ju Jiang for providing DNA of Anaplasma phagocytophilum and Rickettsia felis and to Andrias Hojgaard for Borrelia spp. DNA. The authors thank the reviewers for careful revision of the article, thoughtful comments, and constructive suggestions. Special thanks should be given to the staff of the Steven B. Thacker CDC Library for their invaluable assistance in obtaining scientific materials.
Funding Information
The authors received no specific funding for this work.
Footnotes
Author Disclosure Statement
No conflicting financial interests exist.
References
- Angelakis E, Lepidi H, Canel A, Rispal P, et al. Human case of Bartonella alsatica lymphadenitis. Emerg Infect Dis 2008; 14:1951–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atif FA. Alpha proteobacteria of genus Anaplasma (Rick-ettsiales: Anaplasmataceae): Epidemiology and characteristics of Anaplasma species related to veterinary and public health importance. Parasitology 2016; 143:659–685. [DOI] [PubMed] [Google Scholar]
- Bai Y, Calisher CH, Kosoy MY, Root JJ, et al. Persistent infection or successive reinfection of deer mice with Bartonella vinsonii subsp. arupensis. Appl Environ Microbiol 2011; 77: 1728–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Kosoy M, Calisher C, Cully J Jr., et al. Effects of rodent community diversity and composition on prevalence of an endemic bacterial pathogen-Bartonella. 2009; 10: 3–11. [Google Scholar]
- Bai Y, Kosoy MY, Boonmar S, Sawatwong P, et al. Enrichment culture and molecular identification of diverse Bartonella species in stray dogs. Vet Microbiol 2010; 146:314–319. [DOI] [PubMed] [Google Scholar]
- Bai Y, Kosoy MY, Cully JF, Bala T, et al. Acquisition of nonspecific Bartonella strains by the northern grasshopper mouse (Onychomys leucogaster). FEMS Microbiol Ecol 2007; 61: 438–448. [DOI] [PubMed] [Google Scholar]
- Bai Y, Kosoy MY, Ray C, Brinkerhoff RJ, et al. Temporal and spatial patterns of Bartonella infection in black-tailed prairie dogs (Cynomys ludovicianus). Microb Ecol 2008; 56:373– 382. [DOI] [PubMed] [Google Scholar]
- Bai Y, Urushadze L, Osikowicz L, McKee C, et al. Molecular survey of bacterial zoonotic agents in bats from the country of Georgia (Caucasus). PLoS One 2017; 12:e0171175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter BD, Mendez-Harclerode FM, Fulhorst CF, Bradley RD. A molecular examination of relatedness, multiple paternity, and cohabitation of the Southern Plains woodrat (Neotoma micropus). J Mammal 2009; 90:819–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder AM, Armstrong PA. Increase in reports of tick-borne rickettsial diseases in the United States. Am J Nurs 2019; 119:20–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordes F, Blasdell K, Morand S.Transmission ecology of rodent-borne diseases: New frontiers. Integr Zool 2015; 10:424–435. [DOI] [PubMed] [Google Scholar]
- Braun J, Mares M. Neotoma micropus. J Mammalian Species 1989; 330:1–9. [Google Scholar]
- Breitschwerdt EB, Maggi RG, Quach C, Bradley JM. Bartonella spp. bloodstream infection in a Canadian family. Vector Borne Zoonotic Dis 2019; 19:234–241. [DOI] [PubMed] [Google Scholar]
- Brown RN, Lane RS. Natural and experimental Borrelia burgdorferi infections in woodrats and deer mice from California. J Wildl Dis 1994; 30:389–398. [DOI] [PubMed] [Google Scholar]
- Brown RN, Peot MA, Lane RS. Sylvatic maintenance of Borrelia burgdorferi (Spirochaetales) in Northern California: Untangling the web of transmission. J Med Entomol 2006; 43:743–751. [DOI] [PubMed] [Google Scholar]
- Buffet JP, Kosoy M, Vayssier-Taussat M. Natural history of Bartonella-infecting rodents in light of new knowledge on genomics, diversity and evolution. Future Microbiol 2013; 8: 1117–1128. [DOI] [PubMed] [Google Scholar]
- Burgess EC, Windberg LA. Borrelia sp. infection in coyotes, black-tailed jack rabbits and desert cottontails in southern Texas. J Wildl Dis 1989; 25:47–51. [DOI] [PubMed] [Google Scholar]
- Burnham KP, Anderson DR. Multimodel inference: Understanding AIC and BIC in model selection. Sociol Methods Res 2004; 33:261–304. [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 2000; 17:540–552. [DOI] [PubMed] [Google Scholar]
- Chomel BB, Wey AC, Kasten RW. Isolation of Bartonella washoensis from a dog with mitral valve endocarditis. J Clin Microbiol 2003; 41:5327–5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conditt SA, Ribble DO. Social organization of Neotoma micropus, the southern plains woodrat. Am Midl Nat 1997:290–297. [Google Scholar]
- Ellis BA, Regnery RL, Beati L, Bacellar F, et al. Rats of the genus Rattus are reservoir hosts for pathogenic Bartonella species: An old world origin for a new world disease? J Infect Dis 1999; 180:220–224. [DOI] [PubMed] [Google Scholar]
- Eremeeva ME, Gerns HL, Lydy SL, Goo JS, et al. Bacteremia, fever, and splenomegaly caused by a newly recognized Bartonella species. N Engl J Med 2007; 356:2381–2387. [DOI] [PubMed] [Google Scholar]
- Fenollar F, Sire S, Raoult D. Bartonella vinsonii subsp. arupensis as an agent of blood culture-negative endocarditis in a human. J Clin Microbiol 2005; 43:945–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grear DA, Perkins SE, Hudson PJ. Does elevated testosterone result in increased exposure and transmission of parasites? Ecol Lett 2009; 12:528–537. [DOI] [PubMed] [Google Scholar]
- Gundi VA, Billeter SA, Rood MP, Kosoy MY. Bartonella spp. in rats and zoonoses, Los Angeles, California, USA. Emerg Infect Dis 2012; 18:631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han BA, Kramer AM, Drake JM. Global patterns of zoonotic disease in mammals. Trends Parasitol 2016; 32:565–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms A, Segers FH, Quebatte M, Mistl C, et al. Evolutionary dynamics of pathoadaptation revealed by three independent acquisitions of the VirB/D4 Type IV secretion system in Bartonella. Genome Biol Evol 2017; 9:761–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller R, Kubina M, Mariet P, Riegel P, et al. Bartonella alsatica sp. nov., a new Bartonella species isolated from the blood of wild rabbits. Int J Syst Bacteriol 1999; 49 Pt 1:283–288. [DOI] [PubMed] [Google Scholar]
- Hofmeister EK, Kolbert CP, Abdulkarim AS, Magera JM, et al. Cosegregation of a novel Bartonella species with Borrelia burgdorferi and Babesia microti in Peromyscus leucopus. J Infect Dis 1998; 177:409–416. [DOI] [PubMed] [Google Scholar]
- Hosmer DW, Lemeshow S. Applied Logistic Regression, Second Edition. New York: Wiley and Sons, Inc., 2000. [Google Scholar]
- Inoue K, Kabeya H, Hagiya K, Kosoy MY, et al. Multi-locus sequence analysis reveals host specific association between Bartonella washoensis and squirrels. Vet Microbiol 2011; 148:60–65. [DOI] [PubMed] [Google Scholar]
- Inoue K, Kabeya H, Kosoy MY, Bai Y, et al. Evolutional and geographical relationships of Bartonella grahamii isolates from wild rodents by multi-locus sequencing analysis. Microb Ecol 2009; 57:534–541. [DOI] [PubMed] [Google Scholar]
- Jardine C, Appleyard G, Kosoy MY, McColl D, et al. Rodentassociated Bartonella in Saskatchewan, Canada. Vector Borne Zoonotic Dis 2005; 5:402–409. [DOI] [PubMed] [Google Scholar]
- Jardine C, Waldner C, Wobeser G, Leighton FA. Effect of experimental ectoparasite control on Bartonella infections in wild Richardson’s ground squirrels. J Wildl Dis 2006; 42: 750–758. [DOI] [PubMed] [Google Scholar]
- Jeanclaude D, Godmer P, Leveiller D, Pouedras P, et al. Bartonella alsatica endocarditis in a French patient in close contact with rabbits. Clin Microbiol Infect 2009; 15 Suppl 2: 110–111. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol 2013; 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerkhoff FT, Bergmans AM, van Der Zee A, Rothova A. Demonstration of Bartonella grahamii DNA in ocular fluids of a patient with neuroretinitis. J Clin Microbiol 1999; 37:4034–4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosoy M, Goodrich I. Comparative ecology of Bartonella and Brucella infections in wild carnivores. Front Vet Sci 2018; 5: 322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosoy M, Khlyap L, Cosson JF, Morand S. Aboriginal and invasive rats of genus Rattus as hosts of infectious agents. Vector Borne Zoonotic Dis 2015; 15:3–12. [DOI] [PubMed] [Google Scholar]
- Kosoy M, Mandel E, Green D, Marston E, et al. Prospective studies of Bartonella of rodents. Part I. Demographic and temporal patterns in population dynamics. Vector Borne Zoonotic Dis 2004; 4:285–295. [DOI] [PubMed] [Google Scholar]
- Kosoy M, McKee C, Albayrak L, Fofanov Y. Genotyping of Bartonella bacteria and their animal hosts: Current status and perspectives. Parasitology 2018; 145:543–562. [DOI] [PubMed] [Google Scholar]
- Kosoy M, Murray M, Gilmore RD Jr., Bai Y, et al. Bartonella strains from ground squirrels are identical to Bartonella washoensis isolated from a human patient. J Clin Microbiol 2003; 41:645–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosoy M, Reynolds P, BaiY, Sheff K, et al. Small-scale die-offs in woodrats support long-term maintenance of plague in the U.S. Southwest. Vector Borne Zoonotic Dis 2017; 17:635–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosoy MY, Regnery RL, Tzianabos T, Marston EL, et al. Distribution, diversity, and host specificity of Bartonella in rodents from the Southeastern United States. Am J Trop Med Hyg 1997; 57:578–588. [DOI] [PubMed] [Google Scholar]
- Kosoy MY, Saito EK, Green D, Marston EL, et al. Experimental evidence of host specificity of Bartonella infection in rodents. Comp Immunol Microbiol Infect Dis 2000; 23:221–238. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 2016; 33:1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Scola B, Zeaiter Z, Khamis A, Raoult D. Gene-sequencebased criteria for species definition in bacteriology: The Bartonella paradigm. Trends Microbiol 2003; 11:318–321. [DOI] [PubMed] [Google Scholar]
- Li DM, Hou Y, Song XP, Fu YQ, et al. High prevalence and genetic heterogeneity of rodent-borne Bartonella species on Heixiazi Island, China. Appl Environ Microbiol 2015; 81: 7981–7992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JW, Chen CY, Chen WC, Chomel BB, et al. Isolation of Bartonella species from rodents in Taiwan including a strain closely related to ‘Bartonella rochalimae’ from Rattus norvegicus. J Med Microbiol 2008; 57:1496–1501. [DOI] [PubMed] [Google Scholar]
- Marriott I, Huet-Hudson YM. Sexual dimorphism in innate immune responses to infectious organisms. Immunol Res 2006; 34:177–192. [DOI] [PubMed] [Google Scholar]
- Moreno E. Retrospective and prospective perspectives on zoonotic brucellosis. Front Microbiol 2014; 5:213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morway C, Kosoy M, Eisen R, Montenieri J, et al. A longitudinal study of Bartonella infection in populations of woodrats and their fleas. J Vector Ecol 2008; 33:353–364. [DOI] [PubMed] [Google Scholar]
- Myint KS, Gibbons RV, Iverson J, Shrestha SK, et al. Serological response to Bartonella species in febrile patients from Nepal. Trans R Soc Trop Med Hyg 2011; 105:740–742. [DOI] [PubMed] [Google Scholar]
- New Mexico Department of Health. Plague. 2019. Available at https://nmhealth.org/about/erd/ideb/zdp/plg/
- Oksi J, Rantala S, Kilpinen S, Silvennoinen R, et al. Cat scratch disease caused by Bartonella grahamii in an immunocompromised patient. J Clin Microbiol 2013; 51:2781–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osikowicz LM, Billeter SA, Rizzo MF, Rood MP, et al. Distribution and diversity of Bartonella washoensis strains in ground squirrels from California and their potential link to human cases. Vector Borne Zoonotic Dis 2016; 16:683–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probert W, Louie JK, Tucker JR, Longoria R, et al. Meningitis due to a ‘‘Bartonella washoensis’’-like human pathogen. J Clin Microbiol 2009; 47:2332–2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao HX, Yu J, Guo P, Ma YC, et al. Bartonella species detected in the plateau pikas (Ochotona curzoiae) from Qinghai Plateau in China. Biomed Environ Sci 2015; 28:674–678. [DOI] [PubMed] [Google Scholar]
- Raoult D, Roblot F, Rolain JM, Besnier JM, et al. First isolation of Bartonella alsatica from a valve of a patient with endocarditis. J Clin Microbiol 2006; 44:278–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg R, Lindsey NP, Fischer M, Gregory CJ, et al. Vital signs: Trends in reported vectorborne disease cases—United States and territories, 2004–2016. MMWR Morb Mortal Wkly Rep 2018; 67:496–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux V, Eykyn SJ, Wyllie S, Raoult D. Bartonella vinsonii subsp. berkhoffii as an agent of afebrile blood culturenegative endocarditis in a human. J Clin Microbiol 2000; 38: 1698–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio AV, Avila-Flores R, Osikowicz LM, Bai Y, et al. Prevalence and genetic diversity of Bartonella strains in rodents from northwestern Mexico. Vector Borne Zoonotic Dis 2014; 14:838–845. [DOI] [PubMed] [Google Scholar]
- Schmitz KM, Foley JE, Kasten RW, Chomel BB, et al. Prevalence of vector-borne bacterial pathogens in riparian brush rabbits (Sylvilagus bachmani riparius) and their ticks. J Wildl Dis 2014; 50:369–373. [DOI] [PubMed] [Google Scholar]
- Serratrice J, Rolain JM, Granel B, Ene N, et al. [Bilateral retinal artery branch occlusions revealing Bartonella grahamii infection]. Rev Med Interne 2003; 24:629–630 (Article in French). [DOI] [PubMed] [Google Scholar]
- Stevenson HL, Labruna MB, Montenieri JA, Kosoy MY, et al. Detection of Rickettsia felis in a New World flea species, Anomiopsyllus nudata (Siphonaptera: Ctenophthalmidae). J Med Entomol 2005; 42:163–167. [DOI] [PubMed] [Google Scholar]
- Stoenner H, Lackman D. A new species of Brucella isolated from the desert wood rat, Neotoma lepida Thomas. Am J Vet Res 1957; 18:947. [PubMed] [Google Scholar]
- Suarez-Esquivel M, Ruiz-Villalobos N, Jimenez-Rojas C, Barquero-Calvo E, et al. Brucella neotomae infection in humans, Costa Rica. Emerg Infect Dis 2017; 23:997–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchecki J, Ruthven D, Fulhorst C, Bradley R. Natural history of the southern plains woodrat Neotoma micropus (Rodentia: Muridae) from southern Texas. Texas J Sci 2004; 56:131–140. [Google Scholar]
- Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 1993; 10:512–526. [DOI] [PubMed] [Google Scholar]
- Team RC. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. 2013. Available at http://www.R-project.org. [Google Scholar]
- Telfer S, Begon M, Bennett M, Bown KJ, et al. Contrasting dynamics of Bartonella spp. in cyclic field vole populations: The impact of vector and host dynamics. Parasitology 2007; 134:413–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch DF, Carroll KC, Hofmeister EK, Persing DH, et al. Isolation of a new subspecies, Bartonella vinsonii subsp. arupensis, from a cattle rancher: Identity with isolates found in conjunction with Borrelia burgdorferi and Babesia microti among naturally infected mice. J Clin Microbiol 1999; 37: 2598–2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SG, Sacci J, Schriefer M, Andersen E, et al. Typhus and typhuslike rickettsiae associated with opossums and their fleas in Los Angeles County, California. J Clin Microbiol 1992; 30:1758–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson EB. Probable inference, the law of succession, and statistical inference. J Am Stat Assoc 1927; 22:209–212. [Google Scholar]
- Zeidner NS, Burkot TR, Massung R, Nicholson WL, et al. Transmission of the agent of human granulocytic ehrlichiosis by Ixodes spinipalpis ticks: Evidence of an enzootic cycle of dual infection with Borrelia burgdorferi in Northern Colorado. J Infect Dis 2000; 182:616–619. [DOI] [PubMed] [Google Scholar]
- Ziedins AC, Chomel BB, Kasten RW, Kjemtrup AM, et al. Molecular epidemiology of Bartonella species isolated from ground squirrels and other rodents in northern California. Epidemiol Infect 2016; 144:1837–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.