Abstract
Mutation in the senataxin (SETX) gene causes an autosomal dominant neuromuscular disorder, amyotrophic lateral sclerosis 4 (ALS4), characterized by degeneration of motor neurons, muscle weakness and atrophy. SETX is an RNA-DNA helicase that mediates resolution of co-transcriptional RNA:DNA hybrids (R-loops). The process of R-loop resolution is essential for the normal functioning of cells, including neurons. The molecular basis of ALS4 pathogenesis and the mechanism of R-loop resolution are unclear. We report that the zinc finger protein ZPR1 binds to RNA:DNA hybrids, recruits SETX onto R-loops and is critical for R-loop resolution. ZPR1 deficiency disrupts the integrity of R-loop resolution complexes containing SETX and causes increased R-loop accumulation throughout gene transcription. We uncover that SETX is a downstream target of ZPR1 and that overexpression of ZPR1 can rescue R-loop resolution complexe assembly in SETX-deficient cells but not vice versa. To uncover the mechanism of R-loop resolution, we examined the function of SETX-ZPR1 complexes using two genetic motor neuron disease models with altered R-loop resolution. Notably, chronic low levels of SETX-ZPR1 complexes onto R-loops result in a decrease of R-loop resolution activity causing an increase in R-loop levels in spinal muscular atrophy. ZPR1 overexpression increases recruitment of SETX onto R-loops, decreases R-loops and rescues the spinal muscular atrophy phenotype in motor neurons and patient cells. Strikingly, interaction of SETX with ZPR1 is disrupted in ALS4 patients that have heterozygous SETX (L389S) mutation. ZPR1 fails to recruit the mutant SETX homodimer but recruits the heterodimer with partially disrupted interaction between SETX and ZPR1. Interestingly, disruption of SETX-ZPR1 complexes causes increase in R-loop resolution activity leading to fewer R-loops in ALS4. Modulation of ZPR1 levels regulates R-loop accumulation and rescues the pathogenic R-loop phenotype in ALS4 patient cells. These findings originate a new concept, ‘opposite alterations in a cell biological activity (R-loop resolution) result in similar pathogenesis (neurodegeneration) in different genetic motor neuron disorders’. We propose that ZPR1 collaborates with SETX and may function as a molecular brake to regulate SETX-dependent R-loop resolution activity critical for the normal functioning of motor neurons.
Keywords: amyotrophic lateral sclerosis 4, R-loops, ZPR1, spinal muscular atrophy, DNA damage response
Mutations in the senataxin (SETX) gene cause ALS4, a juvenile form of ALS. Kannan et al. provide insights into the molecular basis of ALS4 pathogenesis by showing that SETX mutations disrupt the interaction between SETX and the zinc finger protein ZPR1, interfering with the resolution of RNA-DNA hybrids (R-loops).
Introduction
Amyotrophic lateral sclerosis 4 (ALS4) is an autosomal dominant neuromuscular disorder caused by mutation in the senataxin (SETX) gene. ALS4 is classified as a juvenile form of ALS and characterized by chronic degeneration of upper and lower motor neurons, distal muscle weakness and atrophy.1–3 SETX is an RNA-DNA helicase involved in the resolution of RNA:DNA hybrids (R-loops) formed during transcription.4 R-loops consist of three nucleic acids strands, nascent RNA hybridized to the transcribing DNA strand (RNA:DNA hybrid) and a complementary DNA strand. R-loops play important roles in physiological processes, including immunoglobin (Ig) class switching, gene expression, DNA repair and genome instability.5–7 Defects in R-loop metabolism are associated with human diseases such as cancer and neurodegenerative disorders.8–11 Thus, precise regulation of R-loop resolution is critical for the normal functioning and survival of the cell.12
The molecular mechanism of R-loop resolution is not well understood. Many factors have been identified that modulate R-loop levels or interact with R-loops, but their specific biochemical contribution to R-loop metabolism remains to be validated.13–15 One of the key factors is SETX, an ATP-dependent RNA-DNA helicase that unwinds RNA:DNA hybrids and contributes to R-loop resolution.16–19 Other critical factors that modulate RNA:DNA hybrid resolution during RNA polymerase II (RNAPII)-dependent gene transcription include XRN2, a 5′-3′-exonuclease that promotes SETX-dependent resolution of R-loops at G-rich transcription pause sites.16 RNA helicase A, also known as DHX9, increases R-loop formation in cells with splicing defects20 and enhances transcription termination by suppressing R-loop accumulation.14 SETX forms complexes with RNAPII and survival motor neuron (SMN) proteins and these protein complexes are involved in mRNA biogenesis that includes transcription, splicing and R-loop resolution.21–23 SMN directly interacts with RNAPII, and the disruption of RNAPII and SMN interactions by mutations in RNAPII causes defects in transcription termination.24 Mutation of the SMN1 gene causes spinal muscular atrophy (SMA).25 Chronic SMN deficiency causes downregulation of SETX, resulting in R-loop accumulation and DNA damage that lead to genomic instability and neurodegeneration in SMA.26
In this study, we identify that the zinc finger protein ZPR1 forms complexes with SETX and R-loops. ZPR1 is evolutionarily conserved and is essential for cell viability in yeast and mammals.27–30 ZPR1 interacts with SMN and is required for SMN translocation from the cytoplasm to the nucleus.31 ZPR1 also interacts with RNAPII and is part of SMN-RNAPII complexes.32 ZPR1 deficiency causes neurodegeneration and contributes to respiratory distress associated with SMA pathogenesis.33–36 ZPR1 is a protective modifier, it upregulates expression of SMN and rescues SMA in mice.32 However, the physiological function of ZPR1 is unknown.
Here, we show that ZPR1 binds to RNA:DNA hybrids, recruits SETX onto R-loops and is critical for the resolution of RNA:DNA hybrids. To unravel the molecular mechanism of R-loop resolution, we investigated the role of ZPR1-SETX complexes using two disease models with altered R-loop metabolism: SMA with increased R-loops and ALS4 with decreased R-loops. In SMA, chronic low levels of ZPR1-SETX complexes impair the efficiency of R-loop resolution resulting in R-loop accumulation. In ALS4, interaction of SETX with ZPR1 is disrupted in patients that have heterozygous SETX (L389S) mutation. ZPR1 fails to recruit mutant SETX homodimer but recruits heterodimer with partially disrupted interaction between SETX and ZPR1. Disruption of ZPR1-SETX complexes results in ZPR1-dependent gain in R-loop resolution activity leading to fewer R-loops in ALS4. Modulation of ZPR1 levels regulates SETX abundance, assembly and R-loop resolution activity of R-loop resolution complexes (RLRC), and rescues pathogenic R-loop phenotype in ALS4 patient cells. These data suggest that ZPR1 tethers to RNA:DNA hybrids, recruits SETX onto R-loops and may function to regulate SETX-dependent R-loop resolution activity. Together, these findings originate a novel concept, opposite alterations in R-loop resolution activity result in similar pathogenesis, motor neuron degeneration, in different genetic motor neuron disorders, SMA and ALS4. We propose that ZPR1’s potential to modulate R-loop levels could be exploited therapeutically in different disease conditions.
Materials and methods
Mice
SMA [Smn−/+; SMN2+/+; SMNΔ7+/+] mice37 on FVB/N background and wild-type FVB/N were purchased from the Jackson Laboratory (mouse line #4299) and maintained in our laboratory. SMA carrier mice were crossed with TFZP (Flag-Zpr1−/+) mice on pure FVB/N background to generate Z-SMA carrier mice with Flag-Zpr1−/+ [Smn−/+; SMN2+/+; SMNΔ7+/+; Flag-Zpr1−/+] as described in a recent study.32 Z-SMA carrier mice were bred to generate SMA [Smn−/+; SMN2+/+; SMNΔ7+/+] and Z-SMA [Smn−/−; SMN2+/+; SMNΔ7+/+; Flag-Zpr1−/+] littermates.32 All animals were housed in a facility accredited by the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC). All animal experimental and biochemical procedures were approved by the Institutional Animal Care and Use Committee (IACUC) and by the Institutional Biosafety Committee (IBC) of the Texas Tech University Health Sciences Center El Paso. Animals were treated humanely, and euthanasia was performed using methods approved by the American Veterinary Medical Association (AVMA).
ALS4 and SMA patient primary fibroblasts and mammalian cell culture
Human control (Normal #1, #2 and #5) and ALS4 patient primary fibroblasts (ALS4 #3, #4 and #6) were received from Dr Kenneth H. Fischbeck, NINDS and grown in Dulbecco’s modified Eagle medium (DMEM) with foetal bovine serum (FBS), 100 units/ml penicillin and 100 µg/ml streptomycin.38 Human control fibroblasts cell line, WI-38 and primary fibroblasts derived from SMA type I patients, GM03813 and GM09677 were cultured in DMEM supplemented with 10% FBS.31 Cells were maintained at 37°C in a humidified atmosphere containing 5% CO2. Cell lysates were prepared from cultured cells using Triton lysis buffer (TLB) for immunoprecipitation (IP) and immunoblot analysis.39 SMA patient cells were cultured on glass coverslips (1 × 105 cells/well) in six well plates and transfected with phrGFPIIc (GFP) or phrGFPIIc-FlagZPR1 (ZPR1-GFP) (1 µg/well) using Lipofectamine® 2000 and incubated for 30 h.26,31 COS-7 cells were transfected with pDEST53 containing GFP-SETX (1-667, wild-type) (a gift from Dr Stephen C. West, The Francis Crick Institute, London, UK) or GFP-SETX (1-667, L389S) cDNA generated by site-directed mutagenesis kit (Agilent Technologies) and harvested 30 h post-transfection. ALS4 patient cells were infected with adenovirus expressing recombinant green fluorescent protein (GFP) (Ad5-GFP) and ZPR1-GFP (Ad5-ZPR1-GFP) at 100 MOI (multiplicity of infection) for 48 h. Cells were either harvested for preparing cell lysate or fixed post-treatment for immunoblot and immunofluorescence analyses, respectively.
Primary spinal cord neuron culture
Mice spinal cords were dissected from 7-day-old normal, SMA and Z-SMA mice were cut into small pieces and explants cultured and differentiated in vitro for 12–14 days in serum-free neurobasal medium supplemented with B-27 (1×), 25 mM glucose, 25 mM KCl, 2 mM glutamine and penicillin/streptomycin using eight-well chamber microscope slides coated with laminin and poly-d-lysine (Corning).26,32,40 The morphology and identity of the spinal cord neurons were established by staining with specific markers, including neuron-specific β-tubulin-III, choline acetyltransferase (ChAT) and homeobox Hlxb9 (Hb9).26,40 Neurons were either fixed (4% paraformaldehyde, PFA) for immunofluorescence analysis or harvested for extraction of protein for immunoblot analysis.
Knockdown of SETX and ZPR1 expression
HeLa cells were cultured in MEM/EBSS supplemented with 10% FBS, 100 units/ml penicillin, 100 µg/ml streptomycin and 1 mM sodium pyruvate and maintained at 37°C in a humidified atmosphere containing 5% CO2. ALS4 and HeLa cells were plated on to a six well plate (1 × 105 cells/well) with glass coverslips for immunofluorescence or in petri dishes for immunoprecipitation and immunoblot analyses. Cells were mock transfected (Control) or transfected with either antisense oligonucleotides (100 nM) against human ZPR1 (As-ZPR1) 5′-CATGGCCACCACGCGCAATT-3′ and scrambled sequence oligonucleotide 5′-CACGGCTACCTCGCACAAGT-3′ (scrambled) or silencer select siRNA (ID #s22952) against human Senataxin (SETX) (Ambion, Life Technology Corp) and scramble siRNA (Dharmacon) using Lipofectamine®2000. Cells were harvested 30 h post-transfection and examined by immunoprecipitation, immunoblot and immunofluorescence analyses. HEK-293 and COS-7 cells were grown in DMEM medium with 10% FBS. To examine the effect of SETX-deficiency on ZPR1 interaction with R-loops, HEK-293 cells were double-transfected by first transfecting with phrGFPIIc (GFP) or phrGFPIIc-FlagZPR1 (ZPR1-GFP) (1.0 µg/well) using Lipofectamine. Plasmid transfected cells were re-transfected after 24 h with siSETX to knockdown SETX. HeLa cells were infected with Ad-h-SETX (Vector Biolabs) at 100 MOI and transfected with As-ZPR1 at 24 h post-infection.26 Cells were harvested after 30 h post-second transfection for immunoprecipitation and immunoblot analyses.26
Immunoblot analysis
Protein extracts for immunoblot analysis were prepared from Control, SMA (GM03813 and GM09677), ALS4 patient cells (ALS4 #3 and ALS4 #4), SMA patient cells and COS-7 cells transfected with phrGFPIIc (GFP) or phrGFPIIc-FlagZPR1 (ZPR1-GFP), HeLa and ALS4 patient cells transfected with As-ZPR1 or scramble oligos, HeLa cells transfected with siSETX or scramble siRNA and HEK-293 cells transfected with phrGFPIIc (GFP) or phrGFPIIc-FlagZPR1 (ZPR1-GFP) and siSETX. Cell lysates were prepared using TLB (20 mM Tris-HCl, pH 7.4, 137 mM NaCl, 1% Triton-X100, 2 mM EDTA, 10% glycerol, 25 mM β-glycerophosphate, pH 7.4, 2 mM sodium pyrophosphate) and freshly added 1 mM sodium orthovanadate, 1 mM PMSF, 1× protease inhibitor cocktail (Millipore Sigma).28 Specific proteins were detected by automated capillary western blot system, Wes System (ProteinSimple), which utilizes capillary based electrophoretic separation and detection of proteins using antibodies as described previously.26 Signal intensity (area) of the protein was normalized to the peak area of loading control α-tubulin. The following primary antibodies were used for immunoblot analysis, SMN (610647, clone 8) from BD Biosciences, ZPR1 (Clone LG-C61),32,35 α-tubulin (#T8203) from Sigma-Aldrich, γH2AX (phospho Ser139) (ab26350), p-DNA-PKcs (Ser2056) (ab18192) and SETX (ab220827), total-DNA-PKcs (ab53701) from Abcam, GFP (A11122) from ThermoFisher and α-tubulin (T8203) from Millipore Sigma. Data analysis and quantitation of protein levels were performed using Compass Software (ProteinSimple).26,32 The relative levels of proteins [mean ± standard error of the mean (SEM)] normalized to tubulin, are presented.
Immunofluorescence analysis
Human fibroblasts (control and derived from ALS4 and SMA patients) cultured on glass coverslips were washed with PBS and fixed in pre-chilled methanol (−20°C) for 5 min followed by 2 min in pre-chilled acetone at −20°C.31 Cells were also fixed with 4% PFA as described in other methods sections. PFA fixed cells were washed with PBS, permeabilized by incubation with 0.1% Triton X100 for 5 min and washed 3× with PBS. Cells were blocked with 3% bovine serum albumin (BSA) in PBS with 0.5% Tween 20 (PBS-T) for 30 min at room temperature.26,32 Cells were double-labelled by sequential incubation with primary antibody against ZPR1 (clone LG1)31 or SMN (clone 8) or RNA:DNA hybrids (R-loops) (S9.6 antibody purified from hybridoma HB-8730, ATCC)41 for 1 h, washed 3× with PBS-T for 5 min each and incubated with secondary antibody Alexa488-conjugated anti-mouse IgG, washed 3× with PBS-T for 5 min each and followed by staining with second primary antibody against p80 Coilin (rabbit polyclonal, NBP2-15939, Novus Biologicals and mouse monoclonal; Clone 56; BD Biosciences; 612074) or DNA damage response (DDR) markers, γH2AX (rabbit NB100-79967, Novas Biologicals and mouse ab26350, Abcam), 53BP1 (4937, Cell Signaling), or SETX (ab220827, Abcam), p-DNA-PKcs (Ser2056) (ab18192), anti-β-tubulin class-III neuron-specific antibody (clone TUJ1, MAB1195, R & D Systems) for 1 h, washed 3× with PBS-T for 5 min, incubated with secondary antibody Alexa594-conjugated anti-rabbit IgG, washed 3× with PBS-T for 5 min. For ribonuclease H (RNase H) enzyme treatment, control and As-ZPR1 transfected cells were extracted with CSK buffer containing 0.1% Triton X-100 for 5 min followed by washing with CSK buffer without Triton-X100. Cells were incubated in RNase H digestion buffer [20 mM HEPES, pH 8.0, 50 mM KCl, 4 mM MgCl2, 1 mM 1, 4-dithiothreitol (DTT), 50 μg/ml BSA, ∼10 U RNase H/ml] for 20 min at 37°C.26,32 After RNase H treatment cells were washed with PBS and fixed in 4% PFA and double stained for R-loops and Coilin. Processed coverslips were mounted on microscope slides with mounting medium containing DAPI (Vectashield) and edges were sealed with a coat of clear nail polish. Stained cells were examined using confocal immunofluorescence microscope equipped with acousto-optical beam splitter (AOBS) and ultraviolet (405 nm), visible (488 nm, 594 nm) and infrared (633 nm) lasers (Leica-TCS-SP5). Quantification of IF intensity of R-loops accumulated in the nucleus of cells was performed using NIH ImageJ software.
Real-time quantitative PCR analysis
Total RNA was isolated from control HeLa cells or cells transfected with As-ZPR1 and scrambled sequence using RNeasy Mini Kit (Qiagen). Total RNA (100 ng) per sample was reverse transcribed using SuperScript VILO cDNA synthesis Kit. Real-time quantitative PCR (RT-qPCR) amplification was performed using Power SYBR™ Green PCR Master Mix. Relative mRNA levels normalized to GAPDH were calculated using the 2−ΔΔCT method.26,32,35,42 The primer sequences were as follows: human GAPDH primers: Forward (5′- ATAGGCGAGATCCCTCCAA-3′) and reverse (5′-TGAAGACGCCAGTGGAC-3′); SETX primers, forward 5′-CTTCATCCTCGGACATTTGAG-3′; and reverse 5′-TTAATAATGGCACCACGCTTC-3′; and human ZPR1 primers hZPR1-Jxn-E9/E10_F2 (5′-CGGACCAATGAGGTGAAATCTGGAGG-3′) and reverse hZPR1-Jxn-E11/E12_R3 (5′-GGATTTTTGGTCACCAGTTC-3′).
Immunoprecipitation
Whole cell protein extracts were prepared using 1×TLB (20 mM Tris-HCl, pH 7.4, 137 mM NaCl, 1% Triton-X100, 2 mM EDTA, 10% glycerol, 25 mM β-glycerophosphate, pH 7.4, 2 mM sodium pyrophosphate) with freshly added protease inhibitor cocktail (Millipore Sigma). Total protein in cell lysates was determined using Bradford’s method using Coomassie protein assay reagent (ThermoScientific).43 Cell lysates were prepared and diluted to 10 mg protein/ml. Cell lysates were treated with ribonucleases, RNase T1 (5 U/ml) (AM2283), RNase III (50 U/ml) (AM2290), RNase H (50 U/ml) (AM2293), RNase A (20 μg/ml) (Thermo Scientific), DNase I (50 μg/ml) (DN-25, Sigma) incubated at 37°C for 30 min before immunoprecipitations. Enzymatic reactions were stopped by adding 0.5 M EDTA.44 Antibodies (3 μg) to ZPR1 (clone LG-C61), R-loops (S9.6), and SETX (ab220827, Abcam) were prebound to 30 μl of protein A–agarose beads for 2 h at 4°C. Immunoprecipitations were performed using cell lysate 300 μl (10 mg/ml) diluted to 1 ml with TLB and incubated with beads only or beads bound to antibodies (3 μg) and incubated for 3 h at 4°C with rotation. Beads were then washed with TLB three times for 10 min each. Proteins of immune complexes bound to beads were then denatured by boiling for 5 min in Laemmli sample buffer (30 μl) with β-mercaptoethanol.43 After cooling at room temperature, beads were centrifuged at high speed for 5 min and 5 μl of supernatant was used for IB analysis using automated capillary Wes System. Quantitation of protein levels were performed by measuring signal intensity (area) of proteins of interest in control and samples with various treatments (knockdown, adenoviral infection etc.) normalized to background and loading control (α-tubulin) and plotted relative to signal intensity (area) of the control sample.
Glutathione S-transferase pulldown assay
Total cell lysates from HeLa cells were prepared using TLB with PI cocktail. Recombinant glutathione S-transferase (GST) and GST-ZPR1 fusion proteins were produced in bacteria (BL21) using PGEX-5x-2 (GST) and PGEX-5x-2/GST-hZPR1 vectors and purified using glutathione agarose beads spin columns (Pierce® GST Spin Purification Kit).28 GST and GST-ZPR1 fusion protein (5 μg) were bound to glutathione-agarose beads for 2 h, at 4°C. The beads were then incubated with cell lysate for 3 h, at 4°C with rotation, washed with TLB three times for 10 min each. Protein complexes bound to beads were eluted and denatured by boiling for 5 min in Laemmli sample buffer (30 μl) with β-mercaptoethanol.28,43 After cooling at room temperature, beads were centrifuged at high speed for 5 min and 5 μl of supernatant was used for immunoblot analysis using automated capillary Wes System.
DNA-RNA immunoprecipitation and R-loop mapping
DNA-RNA immunoprecipitation (DRIP) was performed using the S9.6 antibody against RNA:DNA hybrids.16,45 The monoclonal antibody against RNA:DNA hybrids was purified from S9.6 hybridoma cell line, HB-8730, ATCC.41 Control and transfected cells, As-ZPR1, siSETX, were collected by trypsinization and washed with PBS. Genomic DNA (gDNA) with R-loops was extracted using genomic DNA purification kit with nuclear lysis (Pure-link genomic DNA isolation kit, ThermoFisher). Isolated gDNA was sonicated for 20 min with 15 s pulses and 50 s rest in between pulses and the tubes were kept on ice to shear the DNA into fragments of an average size of 500 bp in length. DRIP was performed by adding fragmented DNA/RNA treated with or without RNase H to S9.6 antibody or anti-mouse IgG (negative control) pre-bound to magnetic protein A beads and incubated for overnight at 4°C with constant rotation.24,46 Beads were then washed and treated with Proteinase K incubated at 62°C for 2 h and 95°C for 10 min. Precipitated DNA was then purified using PCR purification kit before proceeding to qPCR. To map R-loop accumulation throughout the gene transcription, we used input and immuno-precipitated DNA, and SYBR green-based quantitative real-time PCR with primer pairs covering different regions (5′-UTR to 3′-UTR) of human β-actin (ACTB) and GAPDH genes.24 RNA:DNA hybrid enrichment (R-loop accumulation) during transcription of ACTB and GAPDH was calculated relative to input DNA.
Dot-blot analysis: R-loop quantification
Genomic DNA was isolated from cultured cells using PureLink genomic DNA kit (ThermoFisher). Genomic DNA (1.0 μg/well) spotted on Biodyne B nylon membrane presoaked in phosphate-buffered saline (PBS) (ThermoFisher Scientific) using a 96 well Bio-Dot microfiltration apparatus (Biorad). DNA on the membrane was UV-crosslinked (120 mJ/cm2) using Spectrolinker XL-1500 (Spectronics Corporation). Membrane was blocked in 5% non-fat dried milk prepared in PBS-T (0.1% Tween-20) for 1 h followed by incubation with mouse S9.6 antibody (1.0 mg/ml) (1:1000 dilution) for overnight at 4°C.38,47 Blots were washed 3× with PBS-T for 5 min, incubated with secondary antibody, goat anti-mouse HRP (1:5000 dilution) for 1 h at room temperature, washed 3× with PBS-T for 5 min and then developed using Pierce ECL Western Blotting Substrate kit and chemiluminescence digital images captured using Image Quant LAS 4000. NIH ImageJ software was used for densitometric quantification of blots.
Statistical analysis
The quantitative analysis of continuously distributed data is presented as scattered plots or box-and-whisker and violin plots with quantitative elements, including median with interquartile interval, minimum and maximum ranges. Statistical analysis performed using either one-way analysis of variance (ANOVA) or Student’s t-test (unpaired, two-tailed) with GraphPad Prism (version 5.0 d). The value P = 0.05 or less was considered significant. In all experiments with cells, ‘n’ represents the number of times an experiment was performed. A minimum of n = 3 was used in all the experiments, unless otherwise specified in some experiments.
Data availability
The data that support the findings and conclusions of this study are included in this research article and in the Supplementary material, and are also available from the corresponding author upon reasonable request.
Results
ZPR1 forms endogenous complexes with SETX
ZPR1 is an evolutionarily conserved and ubiquitously expressed protein in eukaryotes and is essential for cell viability.27,28,30 However, the biochemical and physiological functions of ZPR1 are unknown. Here, we began by investigating its interaction with other proteins. We previously showed that several proteins co-immunoprecipitate (co-IP) with ZPR1 from 35S-methionine-labelled cell lysates31 Three ZPR1 interacting proteins were identified, the epidermal growth factor receptor (EGFR),48 eukaryotic translation elongation factor-1A (eEF1A)28 and SMN.31 A few other co-immunoprecipitating proteins, including a prominent protein band at the molecular weight (MW) ∼300 kDa, remain to be identified (Supplementary Fig. 1).31
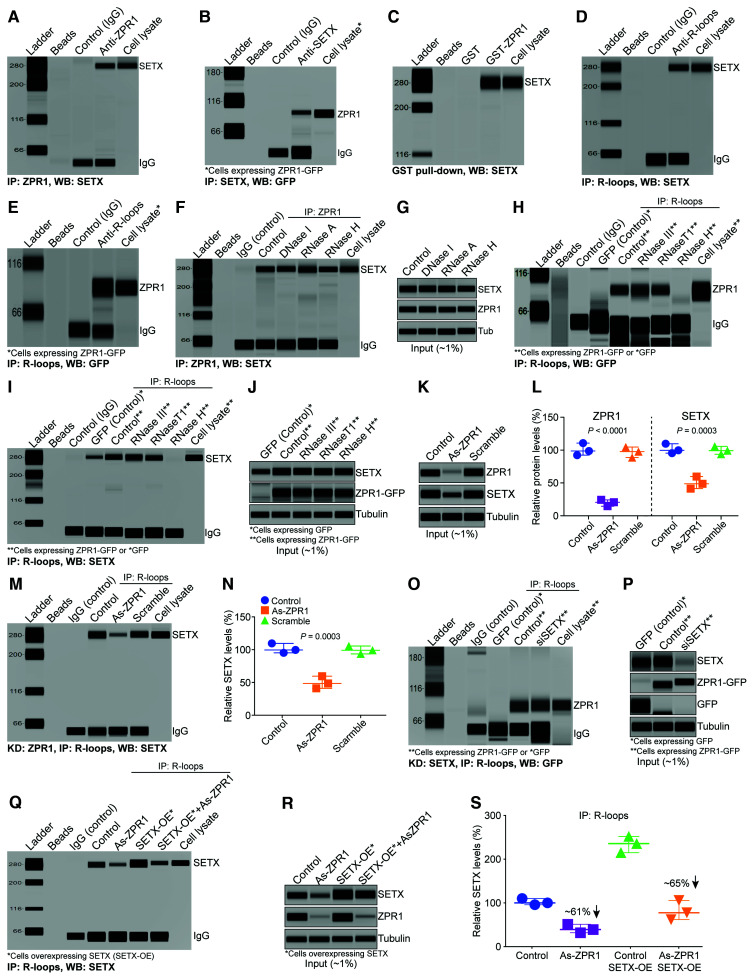
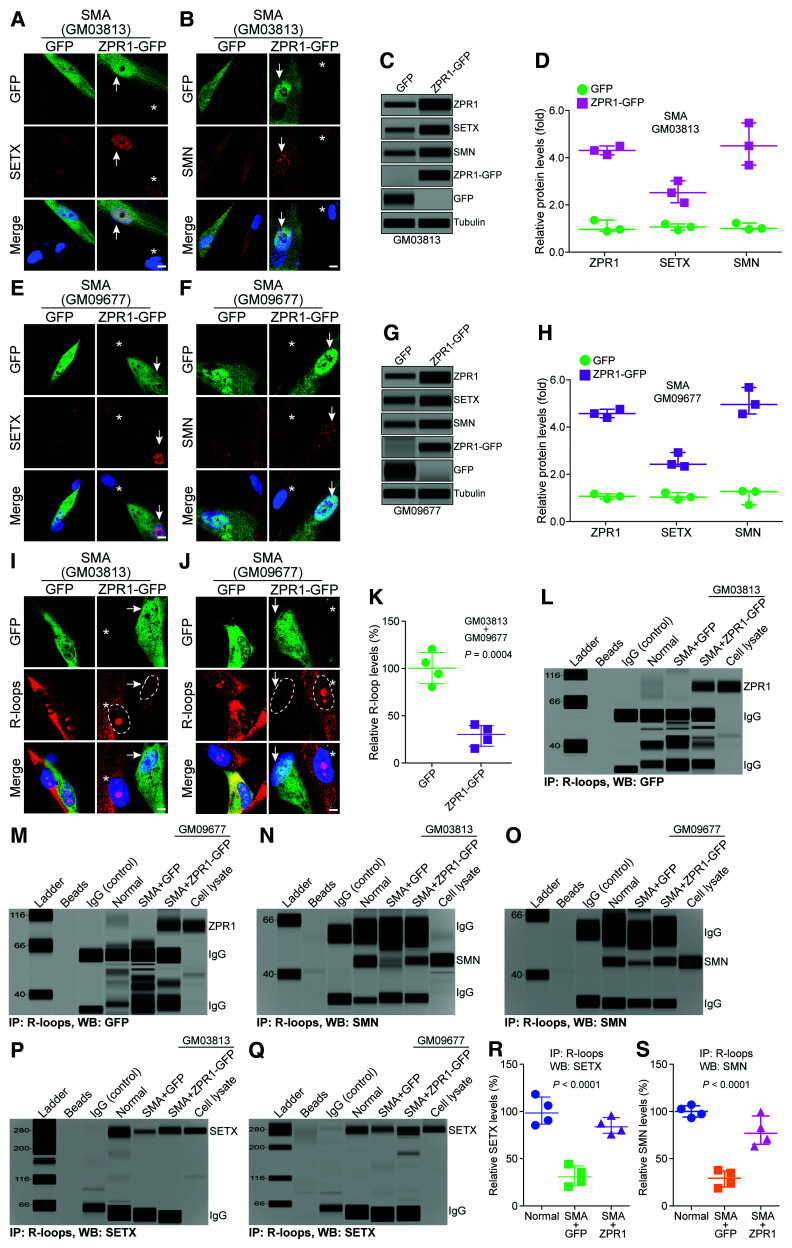
ZPR1 interacts and co-localizes with SMN,31 which interacts and co-localizes with SETX (∼300 kDa) in subnuclear bodies,21,24,26 raising the possibility that the 300 kDa MW protein might be SETX. To test this possibility, we examined the interaction of endogenous ZPR1 with SETX by immunoprecipitation followed by automated western blot analysis. We found that SETX co-IP with ZPR1 from HeLa cell lysates (Fig. 1A). Conversely, immunoprecipitation with SETX antibody shows that ZPR1 co-IP with SETX (Fig. 1B). For the SETX IP, we used COS7 cells expressing ZPR1-GFP because ZPR1 MW is ∼52–54 kDa and migrates with heavy chain IgG, making it difficult to detect in western blot analysis, whereas ZPR1-GFP MW is ∼80 kDa, runs above the IgG band and allows its unequivocal detection. Further, GST pulldown assay using purified recombinant GST-ZPR1 protein shows that ZPR1 can efficiently pulldown SETX from cell lysates (Fig. 1C). These data suggest that ZPR1 forms complexes with SETX in vivo.
Figure 1.
ZPR1 interacts with SETX and R-loops and facilitates SETX recruitment onto R-loops. (A–C) ZPR1 and SETX physically interact and form complexes with R-loops. Immunoprecipitations (IPs) and GST pulldowns were examined by capillary-based automated western blot system. (A) Immunoprecipitation of ZPR1 with antibody against ZPR1 (IP: ZPR1) from HeLa cell lysate followed by western blot (WB) analysis using antibody against SETX (WB: SETX) shows SETX binds in vivo with ZPR1. (B) Immunoprecipitation of SETX with antibody against SETX (IP: SETX) from COS-7 cells expressing recombinant ZPR1-GFP* followed by western blot with antibody against GFP (WB: GFP) to detect fusion protein ZPR1-GFP. (C) GST pulldown assay shows recombinant GST-ZPR1 fusion protein pulls down SETX from HeLa cell lysate. (D and E) ZPR1 interacts in vivo with R-loops and is part of SETX containing RLRCs. Immunoprecipitation of R-loops (IP: R-loops) were performed using monoclonal antibody (S9.6) against RNA:DNA hybrids from HeLa cell lysate followed by western blot analysis. Immunoprecipitation with S9.6 antibody shows co-immunoprecipitation (Co-IP) of (D) SETX and (E) ZPR1 with R-loops. (F) Effect of DNase I, RNase A and RNase H on the binding of ZPR1 with SETX. (G) Western blot analysis of input of proteins in the cell lysate (∼1%) used for IPs. (H and I) Effect of RNA III, RNase T1 and RNase H on the binding of (H) ZPR1 with R-loops and (I) SETX with R-loops. (J) Western blot analysis of input of proteins, SETX, ZPR1, tubulin, in the cell lysates treated with ribonucleases. (K) Immunoblots of ZPR1, SETX and tubulin from cell lysates, Control, As-ZPR1 and Scramble. (L) Quantitation of changes in ZPR1 and SETX protein levels with ZPR1 knockdown are shown as a scatter plot with median and range (min, median, max). ZPR1: Control (92.36, 98.65, 110.7), As-ZPR1 (14.65, 20.65, 24.21), Scramble (88.69, 97.65, 104.7); SETX: Control (94.32, 98.65, 108.7), As-ZPR1 (40.32, 47.65, 58.64), Scramble (92.65, 98.21, 104.70). Quantitative (mean ± SEM, n = 3) and statistical (ANOVA) analyses of immunoblots show KD of ZPR1 levels to (19.84 ± 2.79%, P < 0.0001) causes decrease in SETX levels to (48.87 ± 5.32%, P = 0.0003) compared to Control and Scramble. (M) Immunoprecipitation of R-loops with S9.6 antibody shows ZPR1 knockdown (As-ZPR1) causes a decrease in SETX binding with R-loops. (N) Quantitation of changes in SETX protein levels co-IP with R-loops with ZPR1 knockdown is presented as a scatter plot with median and range (min, median, max). Control (94.32, 98.65, 108.70), As-ZPR1 (40.32, 47.65, 58.64), Scramble (92.65, 98.21, 104.70). Quantitation (mean ± SEM, n = 3, ANOVA) of SETX co-immunoprecipitation with R-loops shows that SETX binding with R-loops is reduced to (25.08 ± 2.53%, P = 0.0003) by ZPR1 knockdown. (O) SETX knockdown (siSETX) does not affect binding of ZPR1 with R-loops as assessed by immunoprecipitation of R-loops in HEK-293 cells overexpressing either GFP or ZPR1-GFP. (P) Immunoblots of ZPR1-GFP, SETX, GFP and tubulin from cell lysates used for IP. (Q–S) Effect of SETX overexpression on SETX binding with R-loops in ZPR1-deficient cells. (Q) Immunoprecipitation of R-loops followed by western blot analysis for SETX. (R) Immunoblot analysis of input proteins, SETX, ZPR1 and tubulin. (S) Quantitation of SETX protein levels co-immunoprecipitated with R-loops in Control and treated cells are presented as a scatter plot with median and range (min, median, max). Control (94.32, 98.65, 108.70); As-ZPR1 (30.32, 37.65, 49.64); SETX OE (213.50, 234.00, 250.40); SETX OE + As-ZPR1 (60.48, 76.24, 104.20). Quantification of SETX levels immunoprecipitated with R-loop antibody, As-ZPR1 (39.20 ± 5.63%, P = 0.0010) compared to control (100.50 ± 4.24%) and As-ZPR1 + SETX-OE (80.31 ± 12.78%, P = 0.0008) compared to Control + SETX-OE (232.60 ± 10.69%). All full-length blots are included in Supplementary Fig. 3.
ZPR1 forms endogenous complexes with R-loops
SETX binds to RNA:DNA hybrids and is an RNA-DNA helicase. ZPR1 contains two zinc fingers that may have affinity for binding to nucleic acids. To test whether ZPR1 binds to RNA:DNA hybrids, we examined the binding of ZPR1 with labelled DNA and RNA:DNA hybrids using electrophoretic-mobility shift assay in vitro (Supplementary Fig. 2A–C) and found that this was indeed the case. We also tested ZPR1 affinity for single-stranded (ssRNA) and double-stranded (dsRNA). These in vitro data show that ZPR1 has low binding affinity for ssRNA and did not bind to dsRNA (Supplementary Fig. 2D and E). To test the interaction of ZPR1 with R-loops in vivo, we used an antibody against RNA:DNA hybrids (S9.6) used for the detection and IP of R-loops.41,49 Immunoprecipitation of R-loops followed by WB showed that SETX co-immunoprecipitates with R-loops (Fig. 1D). Notably, we found that ZPR1 also co-immunoprecipitates with R-loops (Fig. 1E). To test the specificity of ZPR1 interaction with SETX and R-loops, we examined the effect of ribonucleases treatment on in vivo interactions. We found that the treatment of cell lysates with DNase I, RNase A and RNase H before immunoprecipitation did not affect the binding of ZPR1 with SETX (Fig. 1F and G and Supplementary Fig. 3A) suggesting that the binding of ZPR1 with SETX is independent of RNA and DNA. Notably, treatments with RNase T1 (digest ssRNA), RNase III (digest dsRNA) and RNase H (digest RNA:DNA hybrids) show that only RNase H abolishes the binding of ZPR1 (Fig. 1H) and SETX (Fig. 1I and J and Supplementary Fig. 3B) with R-loops. These data suggest that ZPR1 and SETX specifically bind to RNA:DNA hybrids and demonstrate the specificity of S9.6 antibody for the IP of RNA:DNA hybrid (R-loop) complexes with ZPR1 and SETX. Together, these data suggest that ZPR1 and SETX interact and may form endogenous complexes with R-loops.
ZPR1 is critical for SETX binding with R-loops
The interaction between ZPR1 and SETX and their association with R-loops suggest that they may collaborate to regulate R-loop resolution. To gain mechanistic insight into this process, we examined the effect of ZPR1 knockdown on SETX binding with R-loops. ZPR1 knockdown in HeLa cells causes decrease in ZPR1 levels to ∼20% and SETX levels to ∼49% compared to control and scramble treated cells (Fig. 1K and L and full-length blots in Supplementary Fig. 3C). Immunoprecipitation of R-loops using S9.6 antibody shows that ZPR1-deficiency causes marked decrease in the binding of SETX with R-loops in vivo (Fig. 1M). Quantitation shows that SETX co-immunoprecipitation decreased to ∼25% in ZPR1-deficient cells (Fig. 1N). This observation raised a question; is the decrease in SETX co-IP with R-loops is because of SETX downregulation in ZPR1-deficient cells? To address this, we analysed quantitative data and found that ZPR1 knockdown (∼80%) causes ∼50% reduction in SETX levels (Fig. 1K and L). About 50-fold higher amount of SETX protein with reference to ∼1% input (Fig. 1K) was used for immunoprecipitation (Fig. 1M). These data show that the amount of SETX (∼50%) present in the cell lysate (As-ZPR1) is ∼2.5-fold higher than ZPR1 (∼20%) for immunoprecipitation and is not limiting compared to ZPR1 levels. Thus, the decrease in co-IP of SETX with R-loops is not because of the downregulation of SETX in ZPR1-deficient cells but due to acute loss of ZPR1 suggesting that ZPR1 may be critical for binding of SETX with R-loops. Notably, knockdown of SETX did not affect the binding of ZPR1 to R-loops (Fig. 1O and P and full-length blots in Supplementary Fig. 3D). To further test whether ZPR1 is critical for SETX binding to R-loops, we examined the effect of SETX overexpression on the rescue of SETX binding with R-loops in ZPR1-deficient cells (Fig. 1Q and R and Supplementary Fig. 3E). The quantitative data show that SETX binding was decreased by ∼61% in ZPR1-deficient cells (As-ZPR1) and ∼65% in ZPR1-deficient cells with SETX overexpression (As-ZPR1 + SETX-OE), suggesting that SETX overexpression did not rescue SETX binding with R-loops in ZPR1-deficient cells (Fig. 1S). These data suggest that ZPR1 is required for in vivo binding of SETX with R-loops and support the idea that ZPR1 may recruit SETX onto R-loops.
Together, these data suggest that ZPR1 recruits SETX onto R-loops and is critical for in vivo assembly of the core complex of proteins and nucleic acids, SETX-ZPR1-RNA:DNA hybrids. We call this the ‘R-loop resolution complex (RLRC)’ to illustrate its critical role in resolving RNA:DNA hybrids formed during transcription.
ZPR1-deficiency causes downregulation of SETX and accumulation of R-loops
To test whether ZPR1 contributes to the physiological function of SETX in R-loop resolution, we investigated the effect of ZPR1 deficiency on SETX cellular distribution and R-loop accumulation. SETX co-localizes with SMN in nuclear gems.26 ZPR1 is required for SMN and p80 coilin accumulation in subnuclear bodies, including gems and Cajal bodies (CBs).27 We found that ZPR1 co-localizes with SETX and is required for the accumulation of SETX in subnuclear bodies, including gems and Cajal bodies in HeLa and WI-38 cells Control HeLa cells show co-localization of SETX with ZPR1 (Fig. 2A), SMN (Fig. 2B) and coilin (Fig. 2C) in subnuclear bodies (low magnification multi-cell images are shown in Supplementary Fig. 4). Quantification of SETX co-localization show the highest co-localization with ZPR1 (79.60 ± 3.03%) compared to SMN (54.01 ± 4.51%) (gems) and coilin (47.00 ± 4.46%) (Cajal bodies) (Fig. 2D). Quantitative and immunofluorescence analyses of SETX co-localization in WI-38 cells show similar trend and the highest SETX co-localization with ZPR1 (84.61 ± 2.12%) (low magnification multi-cell images are shown in Supplementary Fig. 5 and higher mag images and quantification in Supplementary Fig. 6). These data show that the majority of SETX+ nuclear bodies co-localize with ZPR1 and suggest functional collaboration between ZPR1+ and SETX+ nuclear bodies. Notably, knockdown of ZPR1 (As-ZPR1) causes disruption of SETX+, SMN+ (gems) and coilin+ (Cajal bodies) and decreases the staining intensity of SETX compared to control and scramble treated cells (Fig. 2A–C). These data suggest that ZPR1 is required for SETX localization in gems and Cajal bodies, and indicate that ZPR1 deficiency may cause defects in SETX function.
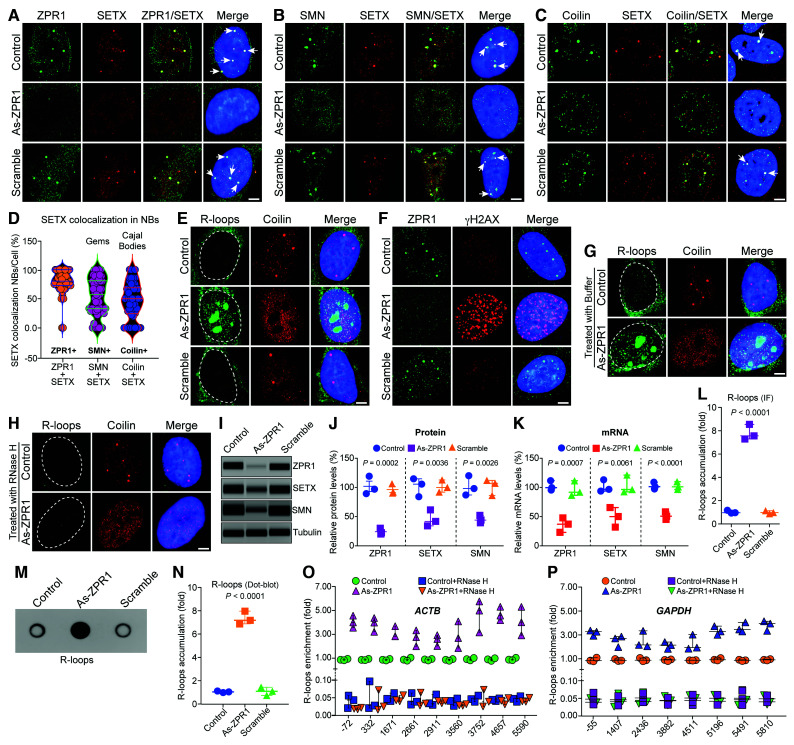
Figure 2.
ZPR1 co-localizes with SETX in nuclear bodies and its deficiency causes disruption of gems and Cajal bodies, downregulation of SETX and accumulation of R-loops. HeLa cells (Control) or transfected with 100 nM antisense oligonucleotides against human ZPR1 (As-ZPR1) or scrambled sequence oligo (Scramble) were fixed and stained with antibodies for immunofluorescence (IF) analysis. (A) Control and scramble oligo treated HeLa cells show ZPR1 (green) and SETX (red) co-localize in subnuclear foci (arrows) and knockdown of ZPR1 (As-ZPR1) causes disruption of SETX+ foci and shows decrease in staining of SETX (red). (B) SETX (red) co-localizes with SMN (green) in nuclear gems (arrows) in control cells. As-ZPR1 causes disruption of SETX+ (red) foci and SMN+ (green) gems. (C) SETX (red) co-localizes with Coilin (green) in Cajal bodies (CBs) (arrows) in control cells. As-ZPR1 causes disruption of SETX+ foci and Cajal bodies. (D) Quantification of SETX co-localization in subnuclear bodies (NBs)/cell (%) is shown as a violin plot with median and interquartile range (Q1, median, Q3) (50 cells/group). SETX co-localization with ZPR1: ZPR1 + SETX (72.73, 80.00, 100.00), SMN: SMN + SETX (32.89, 57.14, 80.83) and coilin: Coilin + SETX (24.31, 50.00. 68.75). Quantification of SETX co-localization (mean ± SEM, n = 50 cells/group) show the highest co-localization with ZPR1 (79.60 ± 3.03%) compared to SMN (54.01 ± 4.51%) (gems) and Coilin (47.00 ± 4.46%) (Cajal bodies). (E) Accumulation of RNA:DNA hybrids (R-loops) in ZPR1-deficient cells detected by monoclonal antibody (S9.6). As-ZPR1 causes accumulation of R-loops (green) and disruption of Cajal bodies (Coilin) (red). (F) ZPR1 (green) deficiency causes accumulation of γH2AX foci, a marker for DNA damage (red). (G and H) Specificity of R-loops detection by S9.6 antibody established by digestion of R-loops with the RNase H enzyme. Cells transfected with As-ZPR1 were permeabilized and treated with (G) buffer only and (H) RNase H enzyme for 20 min at room temperature, washed and fixed with 4% PFA. R-loops (green), coilin (red), nuclei (blue). Dotted circular lines indicate nuclei. Scale bar = 5.0 μm. (I–K) ZPR1 knockdown causes downregulation of SETX. (I) Immunoblots (IBs) of ZPR1, SETX, SMN and tubulin from cell lysates of Control, As-ZPR1 and Scramble transfected HeLa cells. Full-length blots are included in Supplementary Fig. 7C. (J) Quantitation of changes in ZPR1, SETX, and SMN protein levels with ZPR1 knockdown are shown as a scatter plot with median and range (min, median, max). ZPR1: Control (87.69, 95.35, 117.60), As-ZPR1 (18.36, 20.63, 28.32), Scramble (88.69, 90.32, 104.70); SETX: Control (82.79, 104.50, 113.70), As-ZPR1 (32.65, 40.36, 60.21), Scramble (90.65, 98.63, 111.70); SMN: Control (85.32, 96.32, 118.40), As-ZPR1 (37.65, 42.32, 50.32), Scramble (84.36, 105.40, 110.30). Quantitative (mean ± SEM, n = 3) and statistical analysis (ANOVA) show knockdown of ZPR1 levels to (22.44 ± 3.01%, P = 0.0002) decreases SETX levels to (44.41 ± 8.20%, P = 0.0036) and SMN levels to (43.43 ± 3.69%, P = 0.0026) compared to Control and Scramble. (K) Quantitation of changes in ZPR1, SETX, and SMN mRNA levels with ZPR1 knockdown are shown as a scatter plot with median and range. ZPR1: Control (93.70, 97.90, 109.90), As-ZPR1 (21.07, 35.09, 45.98), Scramble (84.65, 90.02, 109.70); SETX: Control (92.50, 95.50, 112.00), As-ZPR1 (30.50, 48.60, 64.17), Scramble (91.20, 95.20, 120.00); SMN: Control (95.70, 99.60. 107.20), As-ZPR1 (43.60, 49.00, 56.40), Scramble (94.87, 99.30, 108.00). Knockdown of ZPR1 mRNA expression to (34.05 ± 7.21%, P = 0.0007) causes downregulation of SETX mRNA expression to (47.76 ± 9.72%, P = 0.0061) and SMN mRNA to (49.67 ± 3.71%, P < 0.0001) compared to Control and Scramble. (L) Quantitative analysis of nuclear R-loop immunofluorescence intensity with NIH ImageJ software show ZPR1-deficient cells (As-ZPR1) accumulate R-loops (7.80 ± 0.37-fold, P < 0.0001) compared to Control and Scramble cells. R-loops nuclear intensity levels were quantified from three experiments (30 cells/group). Quantitative analysis of R-loop levels is shown as a scatter plot with median and range. Control (0.94, 0.97, 1.17), As-ZPR1 (7.29, 7.56, 8.54), Scramble (0.88, 0.98, 1.13). (M) Dot-blot analysis of R-loops using S9.6 antibody and genomic DNA isolated from control, As-ZPR1 and Scramble treated cells. (N) Densitometric quantitative analysis of R-loop levels in dot-blot shown as a scatter plot with median and range. Control (0.93, 0.99, 1.06), As-ZPR1 (6.81, 7.13, 7.91), Scramble (0.68, 1.04, 1.39). Quantitation of R-loop levels in dot-blot shows ZPR1-deficient cells (As-ZPR1) accumulate R-loops (7.28 ± 0.32-fold, P < 0.0001) compared to control and scramble cells. (O and P) Quantitative mapping of R-loop accumulation throughout transcription of the β-Actin (ACTB) and GAPDH genes. DRIP was performed using S9.6 antibody and genomic DNA prepared from control, control + RNase H, ZPR1-deficient (As-ZPR1) and As-ZPR1 + RNase H treated HeLa cells. DRIP and input DNA were used for qPCR analysis using specific primers pairs to amplify different regions of R-loop accumulation during transcription of the ACTB gene in (O) control, control + RNase H, As-ZPR1 and As-ZPR1 + RNase H, and (P) the GAPDH gene in control, control + RNase H, As-ZPR1 and As-ZPR1 + RNase H. Quantitative analysis (mean ± SEM, n = 3) shows ZPR1-deficiency causes ∼4-5-fold R-loop accumulation throughout transcription, including transcription start. Loss of R-loops with RNase H treatment shows specificity of DRIP analysis.
SETX is an ATP-dependent helicase required for unwinding and resolution of RNA:DNA hybrids formed during transcription.50 To test whether disruption of SETX+ bodies upon ZPR1 knockdown correlates with altered R-loop resolution, we examined the effect of ZPR1-deficiency on R-loops using an antibody against RNA:DNA hybrids (S9.6) that detects R-loops. Control cells show low levels of R-loops in the nucleus compared to cytoplasm, where they are associated with mitochondrial transcription51 (Fig. 2E). Notably, knockdown of ZPR1 (As-ZPR1) causes marked accumulation of R-loops in the nucleus (Fig. 2E). Staining of ZPR1-deficient cells with an antibody against γH2AX, a marker for DNA damage, shows accumulation of γH2AX foci in the nucleus (Fig. 2F), which indicates that ZPR1-deficieny may cause R-loop-mediated DNA damage.6 To test the specificity of the S9.6 antibody for R-loop detection, we treated control and transfected (As-ZPR1) cells with permeabilization buffer without (Fig. 2G) or with RNase H that digests R-loops (Fig. 2H). Control and ZPR1-deficient cells (As-ZPR1) cells show that R-loops can be resolved by digestion of RNA using exogenous RNase H, establishing the specificity of R-loop detection and accumulation (Fig. 2H). Further, complementation of ZPR1-deficient cells (As-ZPR1) with mouse ZPR1-GFP causes decrease in R-loops and rescues DNA damage accumulation (Supplementary Fig. 7A and B). Together, these results establish that ZPR1 deficiency negatively impacts on R-loop resolution.
The reduced staining of SETX in ZPR1-deficient cells (Fig. 2A–C) indicates that ZPR1 may influence SETX levels. Quantitative analysis of immunoblots shows that knockdown of ZPR1 levels to ∼22% (P = 0.0002) causes decrease in SETX ∼55% (P = 0.0036) and SMN ∼56% (P = 0.0026) protein levels (Fig. 2I and J and Supplementary Fig. 7C) compared to control and scramble treated cells. Analysis of mRNA levels shows that ZPR1 knockdown to ∼34% (P = 0.0007) causes downregulation of SETX mRNA to ∼47% (P = 0.0061) and SMN mRNA to ∼50% (P < 0.0001) compared to control and scramble (Fig. 2K). These data suggest that SETX may be a downstream target of ZPR1. Quantitation of nuclear R-loops revealed by immunostaining (Fig. 2E) shows ZPR1-deficiency (Fig. 2L) causes marked increase (∼7.80-fold, P < 0.0001) in R-loop accumulation. We noted discrepancies in the literature about the specificity of S9.6 antibody regarding absolute quantitation of R-loops in vivo, including its affinity towards AT-rich dsRNA, which is ∼5-fold lower than RNA:DNA hybrids, but may contribute to quantitation using the immunofluorescence method.52–54 However, our study requires measurements of relative changes in R-loop levels under different experimental conditions. Nevertheless, to eliminate any contribution by dsRNA and to get better insight into the relative changes in R-loop levels, we used dot-blot method utilizing purified genomic DNA.38,47 Quantification of R-loops immunoblots (Fig. 2M) shows similar increase (∼7.3-fold, P < 0.0001) in ZPR1-deficient cells (As-ZPR1) (Fig. 2N). Comparison of R-loop levels measured with the immunofluorescence (∼7.80-fold) and dot-bolt (∼7.3-fold) methods show slightly lower value (0.5-fold) for the dot-blot method suggesting that the statistically significant relative increase in R-loop accumulation is not an artefact of immunofluorescence measurement and is caused by ZPR1 deficiency.
To further investigate the role of ZPR1 in R-loop resolution, we mapped the accumulation of R-loops during transcription of beta-actin (ACTB) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) genes using DRIP with and without RNase H treatment followed by real-time qPCR.24,46 Analysis of ACTB gene transcripts shows that ZPR1 deficiency (As-ZPR1) causes ∼4-5-fold R-loop accumulation throughout the transcription (Fig. 2O). Similarly, GAPDH gene transcripts show that ZPR1 deficiency (As-ZPR1) causes ∼3-4-fold R-loop accumulation throughout the transcription (Fig. 2P). Together, these data suggest that ZPR1 is critical for R-loop resolution.
SETX-deficiency causes disruption of ZPR1+ NBs, gems and Cajal bodies, and accumulation of R-loops
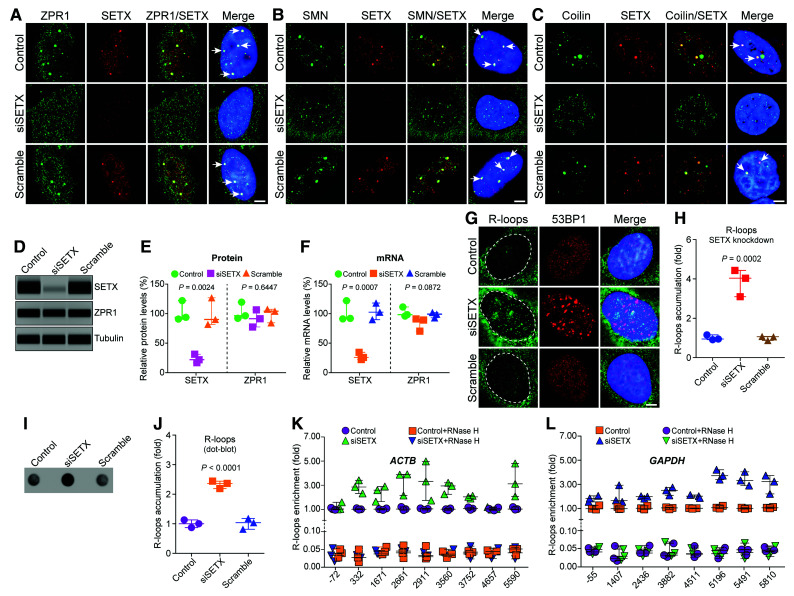
The disruption of SETX NBs and the downregulation of SETX in ZPR1-deficient cells suggest that ZPR1 may regulate SETX abundance and raise the question of whether SETX might also influence the integrity of NBs and ZPR1 levels. Knockdown of SETX (siSETX) causes disruption of ZPR1+, SMN+ (gems) and coilin+ (Cajal bodies) and results in mislocalization of ZPR1 in the nucleoplasm, fragmentation of gems and CBs in smaller nuclear foci compared to control and scramble treated cells (Fig. 3A–C). In contrast to ZPR1 (Fig. 2I–K), knockdown of SETX (siSETX) and reduction of its levels to ∼22% (P = 0.0024) did not significantly decrease ZPR1 levels, which remained ∼90% (P = 0.6447) (Fig. 3D and E and Supplementary Fig. 7D). This result was supported by mRNA analysis that shows knockdown of SETX expression to ∼26% (P = 0.0007) did not significantly alter ZPR1 mRNA expression ∼82% (P = 0.0872) compared to control and scramble (Fig. 3F). However, SETX knockdown causes disruption of ZPR1+ subnuclear bodies and redistributes ZPR1 in the nucleoplasm (Fig. 3A). Disruption of NBs and mis-localization of ZPR1 in SETX-deficient cells suggest that SETX-deficiency may influence ZPR1-dependent R-loop resolution. SETX-deficiency causes accumulation of R-loops and 53BP1 foci, a marker of DNA damage, in the nucleus, effects similar to the ones caused by ZPR1-deficiency (Fig. 3G and see Fig. 2E for comparison). Quantitation of nuclear R-loops revealed by immunostaining (Fig. 3G) shows SETX-deficiency causes ∼3.85-fold R-loop accumulation (Fig. 3H), which is lower than ZPR1-deficiency (∼7.80-fold) (Fig. 2L). Dot-blot analysis of R-loops (Fig. 3I) and quantitation (Fig. 3J) also show increase in accumulation of R-loops (∼2.4-fold, P < 0.0001) in SETX-deficient cells (siSETX).
Figure 3.
SETX deficiency causes disruption of ZPR1 positive nuvlear bodies, gems and Cajal bodies and accumulation of R-loops during transcription. HeLa cells, untransfected (Control) or transfected with 100 nM siRNA against SETX (siSETX) and siRNA with scrambled sequence (Scramble), were fixed and stained with antibodies for immunofluorescence (IF) analysis. (A) Control and Scramble oligo treated HeLa cells show ZPR1 (green) and SETX (red) co-localize in subnuclear foci (arrows) and knockdown of SETX (siSETX) causes disruption of SETX+ and ZPR1+ foci and results in mislocalization of ZPR1 (green) in the nucleoplasm. (B) SETX (red) co-localizes with SMN (green) in nuclear gems (arrows) in control cells. Knockdown of SETX (siSETX) causes disruption of SMN+ foci (gems) resulting many smaller gem-like foci. (C) SETX (red) co-localizes with coilin (green) in Cajal bodies (CBs) (arrows) in control cells. Knockdown of SETX (siSETX) causes disruption of Cajal bodies and result in several small coilin+ foci. (D) Immunoblots of SETX, ZPR1 and tubulin from cell lysates of Control, siSETX and Scramble transfected HeLa cells. Full-length blots are included in Supplementary Fig. 7D. (E) Quantitation of changes in SETX and ZPR1 protein levels with SETX knockdown (siSETX) are shown as a scatter plot with median and range (min, median, max). SETX: Control (89.32, 92.39, 120.70), siSETX (15.32, 20.35, 29.63), Scramble (80.32, 88.62, 125.60); ZPR1: Control (90.32, 94.65, 117.70), siSETX (75.63, 89.65, 104.60), Scramble (82.69, 102.40, 105.60). Quantitation shows knockdown of SETX (siSETX) levels to (21.77 ± 4.19%, P = 0.0024) did not significantly change ZPR1 levels (89.97 ± 8.37%, P = 0.6447) compared to Control and Scramble. (F) Quantitation of changes in SETX and ZPR1 mRNA levels with SETX knockdown (siSETX) are shown as a scatter plot with median and range (min, median, max). SETX: Control (89.53, 90.31, 120.40), siSETX (20.55, 24.12, 32.72), Scramble (88.26, 100.80, 116.40); ZPR1: Control (94.23, 96.48, 109.50), siSETX (68.64, 87.54, 89.33), Scramble (91.10, 97.36, 100.20). Knockdown of SETX mRNA expression to (25.80 ± 3.61%, P = 0.0007) does not significantly alter ZPR1 mRNA expression (81.83 ± 6.61%, P = 0.0872) compared to Control and Scramble. (G) SETX-deficiency (siSETX) causes accumulation of R-loops (green) and 53BP1 foci (red), a marker for DNA damage. (H) Quantitative analysis of nuclear R-loop immunofluorescence intensity with NIH ImageJ software is shown as a scatter plot with median and range (min, median, max). Control (0.91, 0.95, 1.15), siSETX (3.09, 4.03, 4.43), Scramble (0.87, 1.06, 1.07). Quantitation of nuclear R-loops shows SETX-deficient cells (siSETX) accumulate R-loops (3.85 ± 0.39-fold, P = 0.0002) compared to Control and Scramble cells. R-loops nuclear intensity levels were quantified from three experiments (30 cells/group). (I) Dot-blot analysis of R-loops using S9.6 antibody and genomic DNA isolated from Control, siSETX and Scramble treated cells. (J) Densitometric quantitative analysis of R-loop levels in dot-blot shown as a scatter plot with median with range (min, median, max). Control (0.87, 1.0, 1.13), siSETX (2.19. 2.36, 2.44), Scramble (0.81, 1.03, 1.18). SETX-deficient cells (siSETX) accumulate higher R-loops (2.33 ± 0.07-fold, P < 0.0001) compared to control and scramble cells. (K and L) Quantitative mapping of R-loop accumulation throughout transcription of the β-Actin (ACTB) and GAPDH genes. DRIP was performed using S9.6 antibody and genomic DNA prepared from control, control + RNase H, SETX-deficient (siSETX) and siSETX + RNase H treated HeLa cells. DRIP and input DNA were used for qPCR analysis using specific primers pairs to amplify different regions of R-loop accumulation during transcription of the ACTB gene in (K) control, control + RNase H, siSETX and siSETX + RNase H (L) the GAPDH gene in control, control + RNase H, As-ZPR1 and siSETX + RNase H. Quantitative analysis (mean ± SEM, n = 3) of SETX-deficiency shows R-loop accumulation (∼2-3-fold) throughout transcription except at the start of transcription compared to control. Loss of R-loops with RNase H treatment shows specificity of DRIP analysis.
Analysis of R-loops during transcription of ACTB and GAPDH genes using DRIP with and without RNase H treatment followed by real-time qPCR shows that SETX-deficiency (siSETX) causes ∼2-3-fold (ACTB) and ∼1-3-fold (GAPDH) R-loop accumulation throughout the transcription (Fig. 3I and J). Comparison of the effect of ZPR1 and SETX deficiencies shows that ZPR1-deficiency results in higher ∼4-fold accumulation of R-loops around the start of transcription (−72, 5′-UTR) and (−55, 5′-UTR) regions compared to SETX-deficiency in ACTB and GAPDH genes, respectively (Fig. 2O and P and Fig. 3I and J). These data suggest ZPR1 may be critical for initiating R-loop resolution at the start of transcription.
Together, these data demonstrate that SETX-dependent mis-localization of ZPR1 may be a cause of R-loop accumulation in SETX-deficient cells and indicate a functional contribution of SETX in ZPR1-dependent resolution of R-loops.
Chronic low levels of ZPR1 impair assembly of R-loop resolution complexes in SMA
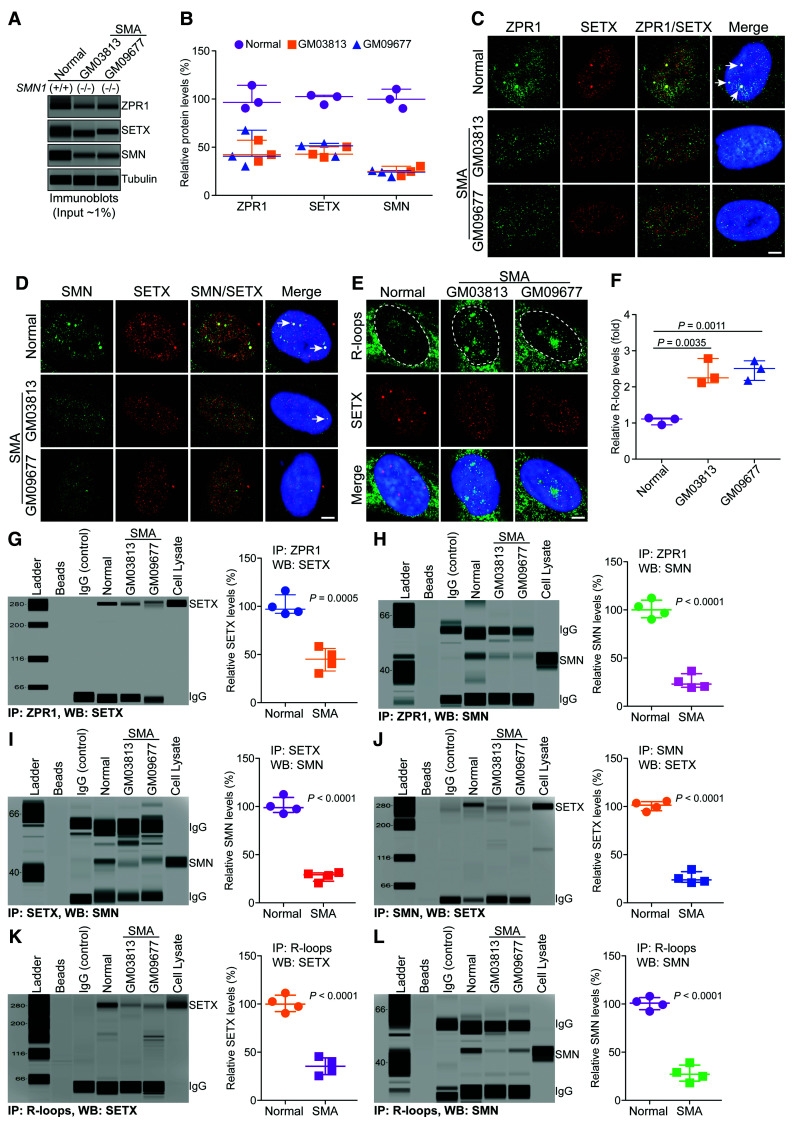
SMA is a motor neuron disorder caused by mutations in the SMN1 gene that result in low levels of SMN and neurodegeneration.25,36 Chronic SMN deficiency causes accumulation of pathogenic R-loops and DNA damage leading to genomic instability and neurodegeneration in SMA.24,26,55 However, the molecular mechanism of R-loop accumulation in SMA is unclear. SMN forms in vivo complexes with SETX.21 Interestingly, ZPR1 is downregulated in SMA patients.31,33,56 It is possible that ZPR1 deficiency may contribute to R-loop accumulation associated with SMA pathogenesis. To test this possibility, we examined in vivo binding of SETX and SMN with R-loops using fibroblasts derived from two SMA patients. Quantitative analysis shows low levels (∼45%) of ZPR1 in SMA patient fibroblasts compared to non-SMA (normal) fibroblasts (Fig. 4A and B). SETX levels were also decreased ∼46% in SMA (Fig. 4A and B and Supplementary Fig. 8A). These data suggest that chronic low levels of ZPR1 in SMA correlate with SETX downregulation, which is consistent with data in Fig. 2 and supports the idea that SETX may be a downstream target of ZPR1. Co-localization of ZPR1 with SETX (Fig. 4C), and co-localization of SMN with SETX (Fig. 4D) in subnuclear foci is disrupted in SMA compared to normal cells, indicating that the disruption of subnuclear bodies and mislocalization of core components of RLRC may be a cause of R-loop accumulation. Analysis of cells stained with an antibody against R-loops (Fig. 4E) and quantitation of nuclear R-loop immunofluorescence intensity shows ∼2.5-fold higher R-loop accumulation in SMA patient cells compared to control cells (Fig. 4F). Dot-blot analysis of R-loops and quantitation also show increase in R-loop accumulation in SMA, GM03813 (∼2.4-fold, P = 0.0093) and GM09677 (∼3.2-fold, P = 0.0064) compared to normal cells (Supplementary Fig. 8B and C).
Figure 4.
Chronic low levels of ZPR1 impair assembly of RLRC in SMA. Cultured WI-38 (Normal) and primary fibroblast derived from SMA type I patients, GM03813 and GM09677 (SMA) that have homozygous deletion of the SMN1 gene, were used for immunofluorescence, immunoprecipitation and immunoblot analyses. (A) Representative capillary-blot images of proteins are shown (full-length blots are included in Supplementary Fig. 8). (B) Comparison of protein levels between normal and SMA patient cells (GM03813 and GM09677) is shown as a scatter plot with median and range (min, median, max). ZPR1: Normal (90.58, 96.54, 114.30), GM03813 (35.42, 42.36, 57.32), GM09677 (30.25, 40.65, 67.65); SETX: Normal (94.21, 102.60, 104.00), GM03813 (39.50, 42.81, 50.49), GM09677 (40.42, 51.39, 54.05); SMN: Normal (90.32, 99.87, 110.30), GM03813 (20.31, 24.81, 30.29), GM09677 (19.31, 24.20, 25.68). Quantitative analysis (mean ± SEM, n = 3, t-test, unpaired) show SMN1 mutation results in the low levels SMN in GM03813 (25.14 ± 2.88%, P = 0.0003) and GM09677 (23.06 ± 1.92%, P = 0.0002) SMA patient cells compared to normal cells. Chronic SMN-deficiency is known to cause splicing defects and alter expression of many genes. Analysis of core components of RLRC shows ZPR1 levels decreased to (45.03 ± 6.46%, P = 0.0045) in GM03813 and (46.18 ± 11.15%, P = 0.0148) in GM09677. SETX levels decreased to (44.27 ± 3.25%, P = 0.0002) in GM03813 and (48.62 ± 4.17%, P = 0.0006) in GM09677 compared to control. (C–E) Representative images are presented for double-labelled immunostainings that show chronic SMN-deficiency causes disruption of subnuclear bodies and mislocalization of RLRC core proteins, ZPR1 and SETX of RLRC: (C) ZPR1 (green) co-localizes with SETX (red) in subnuclear foci in normal (arrows) but is mislocalized in SMA. (D) SMN (green) co-localizes with SETX (red) in subnuclear foci in normal but is mislocalized in SMA. (E) Accumulation of R-loops (green) and disruption of SETX (red) subnuclear bodies in SMA patients compared normal cells. Nuclei were stained with DAPI (blue). Scale bar = 5.0 μm. Dotted circular lines indicate nuclei. (F) Comparison of nuclear R-loop intensity between normal and SMA (GM03813, GM09677) patient cells is shown as a scatter plot with median and range. Normal (0.95, 1.11, 1.14), GM03813 (2.11, 2.25, 2.78), GM09677 (2.17, 2.51, 2.72). Quantitative (mean ± SEM, n = 3) and statistical (unpaired t-test) analysis of R-loop intensity in the nucleus show increase in R-loop accumulation in SMA, GM03813 (2.38 ± 0.20-fold, P = 0.0035) and GM09677 (2.46 ± 0.15-fold, P = 0.0011) compared to normal cells. R-loops nuclear intensity levels were quantified from three experiments (30 cells/group). Data from dot-blot and quantitative analysis of R-loop is included in Supplementary Fig. 8B and C. Chronic ZPR1-deficiency results in decrease of ZPR1 complexes with SETX and SMN in SMA compared to normal cells. Immunoprecipitation of ZPR1 shows decrease in co-IP of (G) SETX and (H) SMN from SMA (GM03813, GM09677) compared to normal cells. Comparison of SETX and SMN co-IP levels with ZPR1 in Normal and SMA patient cells is presented as a scatter plot with median and range. SETX: Normal (92.30, 97.01, 116.7), SMA (30.48, 45.21, 58.40); SMN: Normal (90.23, 100.10, 112.30) and SMA (19.40, 23.06, 36.59). Quantitation (mean ± SEM, n = 4) and statistical (unpaired t-test) analysis shows decreased SETX (44.83 ± 6.02%, P = 0.0005) and SMN (25.53 ± 3.91%, P < 0.0001) co-IP with ZPR1 in SMA cells compared to normal cells. (I) Immunoprecipitation of SETX shows decrease in co-IP of SMN from SMA compared to normal cells. Quantitation of SMN co-IP with SETX in SMA compared to normal cells is shown as a scatter plot with median and range. Normal (92.40, 98.69, 112.40) and SMA (20.52, 29.20, 31.40). Quantitation shows decrease in SMN (27.58 ± 2.43%, P < 0.0001) co-IP levels with SETX in SMA cells compared to normal. (J) Immunoprecipitation of SMN shows decrease in co-IP of SETX from SMA compared to normal cells. Quantitation of SETX co-IP with SMN in SMA compared to normal cells is shown as a scatter plot with median and range. Normal (94.30, 101.30, 105.40) and SMA (20.98, 23.87, 105.40). Quantitation shows decrease in SETX (25.84 ± 3.07%, P < 0.0001) co-IP levels with SMN in SMA cells compared to normal. (K and L) Comparison of accumulation of SETX and SMN on R-loops and assembly of RLRC between normal and SMA cells. (K) Immunoprecipitation of R-loops shows decreased association of SETX with RNA:DNA hybrids in SMA compared to normal cells. Quantitation of SETX (35.40 ± 4.53%, P < 0.0001) co-IP levels with R-loops in normal and SMA cells is presented as a scatter plot with median and range. SETX: Normal (90.30, 99.87, 111.30) and SMA (25.49, 35.36, 45.39). (L) Immunoprecipitation of R-loops shows decreased association of SMN with RNA:DNA hybrids in SMA compared to normal cells. Quantitation of SMN (27.83 ± 4.36%, P < 0.0001) co-IP levels with R-loops in normal and SMA cells is presented as a scatter plot with median and range. Normal (92.30, 100.80, 107.90) and SMA (18.20, 27.03, 39.07).
To test whether R-loop accumulation might be because of defective assembly of RLRC core proteins onto R-loops, we examined RLRC assembly in SMA. Analysis of ZPR1 endogenous complexes shows decreased co-immunoprecipitation of SETX (∼45%, P = 0.0005) (Fig. 4G) and SMN (∼26%, P < 0.0001) (Fig. 4H) in SMA. Further, immunoprecipitations of SETX show decrease in co-IP levels of SMN (∼28%, P < 0.0001) (Fig. 4I) and immunoprecipitation of SMN show decrease in co-IP levels of SETX (∼26%, P < 0.0001) (Fig. 4J) suggesting low levels of endogenous SETX-SMN complexes in SMA (Fig. 4I and J). These data thus reveal decreased levels of ZPR1-SETX, ZPR1-SMN and SETX-SMN complexes or interactions that are critical for R-loop resolution. Immunoprecipitation of R-loops shows marked decrease in in vivo association of SETX (∼35%, P < 0.0001) (Fig. 4K) and SMN (∼28%, P < 0.0001) (Fig. 4L) with R-loops. Altogether, these results suggest that ZPR1 downregulation causes defects in the assembly of RLRC, which may lead to inefficient R-loop resolution and result in R-loop accumulation in SMA.
ZPR1 rescues defective RLRC assembly and prevents pathogenic R-loop accumulation in SMA
To test the hypothesis that ZPR1 is central to R-loop resolution, we performed the rescue experiment with ZPR1 overexpression in SMA patient cells. Control cells expressing GFP did not show any change in the levels or cellular localization of SETX and SMN (GFP panels). Notably, ZPR1 overexpression increased SETX and SMN accumulation in the nucleus of GM03813 (Fig. 5A and B) and GM09677 (Fig. 5E and F) cells expressing ZPR1-GFP (arrows) compared to cells without expression (non-transfected, indicated by asterisks). Increase in ZPR1 (∼4-fold) expression increases the levels of SETX (∼2.5-fold) and SMN (∼4.5) in GM03813 cells (Fig. 5C and D and Supplementary Fig. 9A) and in GM09677 cells, SETX (∼2.5-fold) and SMN (∼5.0-fold) (Fig. 5G and H and Supplementary Fig. 9B). These data suggest that modulation in ZPR1 levels alters the expression of SETX and SMN and support the idea that these critical proteins are downstream targets of ZPR1. Next, we tested whether increase in ZPR1, SETX and SMN levels would reduce R-loop accumulation. We found that both SMA cell lines, GM03813 and GM09677, overexpressing ZPR1-GFP (arrows) show decrease in R-loop accumulation compared to non-transfected controls (asterisks) (Fig. 5I and J). ZPR1 overexpression causes ∼71% reduction in the accumulation of R-loops in SMA patient cells (Fig. 5K).
Figure 5.
ZPR1 rescues assembly of RLRC and averts accumulation of pathogenic R-loops in SMA. SMA patient primary fibroblast cell lines, GM03813 (A, B and I) and GM09677 (E, F and J) were transfected with phrGFP (GFP) or phrZPR1-GFP (ZPR1-GFP), fixed and stained with antibodies against SETX, SMN and R-loops for immunofluorescence or cell lysates were prepared for immunoprecipitation and immunoblot analyses. Ectopic ZPR1 expression elevates levels of SETX and SMN in SMA cells. (A and E) SETX (red) and ZPR1-GFP (green), and (B and F) SMN (red) and ZPR1-GFP (green). Nuclei were stained with DAPI (blue). Scale bar = 5.0 μm. Arrows show transfected cells and asterisks indicate non-transfected cells. Immunoblots show ZPR1 overexpression increases SETX and SMN levels in SMA patient cells (C) GM03813 and (G) GM09677 (full-length blots, Supplementary Fig. 9). (D) Quantitation of changes in protein levels with ZPR1 overexpression in SMA cells (GM03813) is shown as a scatter plot with median and range (min, median, max). ZPR1: GM03813 + GFP (0.81, 0.89, 1.28), GM03813 + ZPR1-GFP (4.05, 4.23, 4.42); SETX: GM03813 + GFP (0.86, 1.00, 1.12), GM03813 + ZPR1-GFP (2.02, 2.45, 2.95); SMN: GM03813 + GFP (0.89, 0.93, 1.16), GM03813 + ZPR1-GFP (3.61, 4.43, 5.40). The comparison of different protein levels (mean ± SEM, n = 3) in GM03813 + ZPR1-GFP cells show increase in levels of ZPR1 to (4.23 ± 0.10-fold, P < 0.0001), SETX (2.47 ± 0.26-fold, P = 0.0060), and SMN (4.48 ± 0.51-fold, P = 0.0026) compared to control GM03813 + GFP cells. (H) Quantitation of changes in protein levels with ZPR1 overexpression in SMA cells (GM09677) is shown as a scatter plot with median and range. ZPR1: GM09677 + GFP (0.89, 1.00, 1.10), GM09677 + ZPR1-GFP (4.33, 4.50, 4.69); SETX: GM09677 + GFP (0.87, 0.96, 1.15), GM09677 + ZPR1-GFP (2.27, 2.35, 2.85); SMN: GM09677 + GFP (0.64, 1.20, 1.22), GM09677 + ZPR1-GFP (4.49, 4.89, 5.61). The comparison of different protein levels (mean ± SEM, n = 3) in GM09677 + ZPR1-GFP cells show increase in levels of ZPR1 (4.50 ± 0.10-fold, P < 0.0001), SETX (2.49 ± 0.18-fold, P = 0.0017) and SMN (4.99 ± 0.32-fold, P = 0.0005) compared to control GM09677 + GFP cells. ZPR1 complementation decreases R-loop accumulation in SMA patient cells. (I) GM03813, R-loops (red) and ZPR1-GFP (green). (J) GM09677, R-loops (red) and ZPR1-GFP (green). Arrows show transfected ZPR1-GFP+ cells with decreased R-loops, asterisks indicate non-transfected cells with R-loop in the nucleus (dotted circular lines). Control cells GFP− or GFP+ show GFP expression did not alter accumulation of R-loops. ZPR1 complementation decreases R-loop accumulation in ZPR1-GFP + SMA patient cells. (K) Quantitation of nuclear R-loop immunofluorescence intensity in SMA + GFP and SMA + ZPR1-GFP patient cells is shown as a scatter plot with median and range. SMA + GFP (80.54, 100.30, 120.60), SMA + ZPR1-GFP (15.40, 30.28, 40.87). Quantitative (mean ± SEM, n = 4) shows R-loop accumulation in SMA cells (GM03813 + GM09677) decreased to (29.21 ± 5.69%, P = 0.0004) in ZPR1-GFP+ compared to control cells. R-loops nuclear intensity levels were quantified from three experiments (30 cells/group). (L–S) ZPR1 rescues defects in the assembly of core RLRC proteins, ZPR1 and SETX with R-loops in SMA patient cells. Immunoprecipitation using S9.6 antibody against R-loops show association of ZPR1-GFP with R-loops in (L) SMA GM03813 + ZPR1-GFP and (M) SMA GM09677 + ZPR1-GFP cells. (N and O) Immunoprecipitations of R-loops from GM03813 + ZPR1-GFP and GM09677 + ZPR1-GFP show increased association of SMN with R-loops compared to SMA + GFP cells. (P and Q) Immunoprecipitations of R-loops from GM03813 + ZPR1-GFP and GM09677 + ZPR1-GFP show increased association of SETX with R-loops compared to SMA + GFP cells. (R) Quantitation of SETX co-IP with R-loops in Normal, SMA + GFP, and SMA + ZPR1-GFP are presented as a scatter plot with median and range. Normal (85.44, 98.43, 118.30), SMA + GFP (20.56, 30.87, 44.34), SMA + ZPR1-GFP (75.94, 83.80, 95.63). Quantitation and comparison of SETX in vivo association with R-loops between Normal, SMA + GFP and SMA + ZPR1-GFP shows that ZPR1 increases SETX binding with R-loops or accumulation in RLRC from 31.66 ± 5.30% (SMA + GFP) to 84.79 ± 4.31% (P < 0.0001) (SMA + ZPR1-GFP). (S) Quantitation of SMN co-IP with R-loops between Normal, SMA + GFP, and SMA + ZPR1-GFP are presented as a scatter plot with median and range. Normal (93.27, 100.20, 107.30), SMA + GFP (18.78, 29.45, 37.84), SMA + ZPR1-GFP (63.35, 76.94, 99.65). Quantitation and comparison of SMN in vivo association with R-loops between Normal, SMA + GFP and SMA + ZPR1-GFP shows that ZPR1 increases SMN binding with R-loops or accumulation in RLRC from 28.88 ± 4.5% (SMA + GFP) to 79.22 ± 7.9% (P = 0.0015) (SMA + ZPR1-GFP).
The decrease in R-loop levels in ZPR1 overexpressing SMA patient cells suggests that ZPR1 might rescue the assembly of RLRC and improve the efficiency of R-loop resolution. Analysis of R-loop IPs shows that ZPR1-GFP co-immunoprecipitates in patient cells GM03813 (Fig. 5L) and GM09677 (Fig. 5M). We have shown previously that ZPR1-GFP retains its biological activity and rescues viability of ZPR1-null cells.28 ZPR1 overexpression increases SMN co-immunoprecipitation with R-loops in SMA patient cell lines GM03813 (Fig. 5N) and GM09677 (Fig. 5O). Notably, ZPR1 also increases in vivo association of SETX with R-loops in ZPR1 complemented compared to control SMA patient cells, GM03813 (Fig. 5P) and GM09677 (Fig. 5Q). IP of R-loops from SMA cells (SMA + GFP) shows low levels (∼32%) of SETX co-IP compared to normal cells. R-loop IP of ZPR1 complemented cells (SMA + ZPR1-GFP) show higher levels (∼85%) of SETX co-IP, ∼2.5-fold increase, compared to control (SMA + GFP) cells (Fig. 5R). Similarly, immunoprecipitation of R-loops from SMA + GFP shows low levels of SMN (∼29%) co-IP and SMA + ZPR1-GFP cells show higher levels of SMN (∼80%) co-IP, ∼2.75-fold increase, compared to control (SMA + GFP) cells (Fig. 5S). Together, these data show that ZPR1 overexpression recruits more SETX and SMN, and improves the assembly of RLRC, which enhances R-loop resolution and rescues the pathogenic R-loop phenotype in SMA patient cells as shown in Fig. 5I–K. These data support our hypothesis that ZPR1 is critical for recruiting SETX and may be central to the process of R-loop resolution.
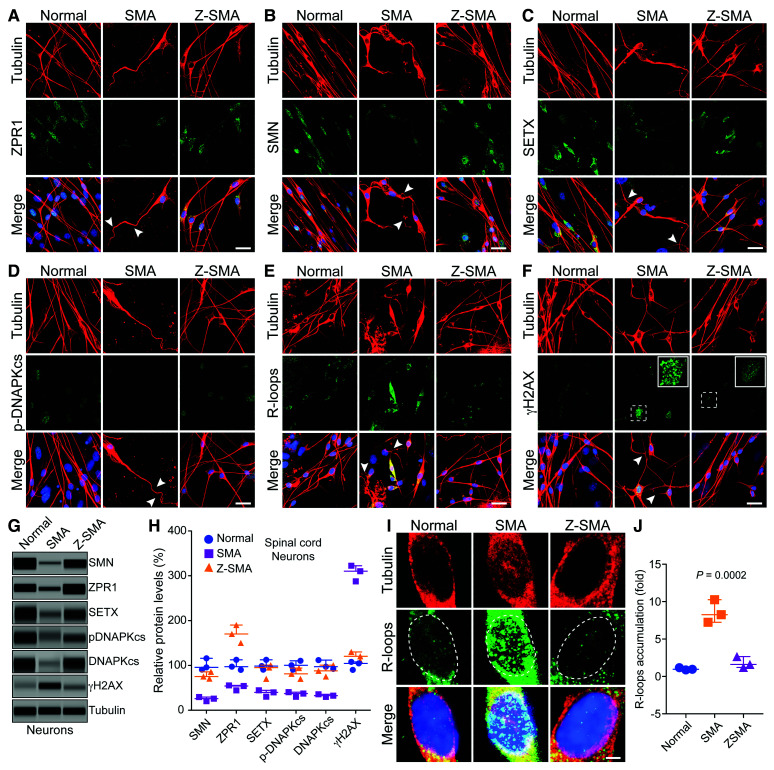
ZPR1 overexpression in vivo rescues R-loop mediated DNA damage and prevents degeneration of motor neurons in SMA
SMA is a neurodegenerative disorder characterized by degeneration of spinal cord motor neurons. To test another important aspect of our hypothesis as to whether ZPR1 rescues molecular defects in vivo and specifically in SMA spinal cord motor neurons, we used primary cultured spinal cord neurons derived from 7-day-old normal, SMA and transgenic Z-SMA mice (SMA mice overexpress Flag-Zpr1 under the control of mouse Rosa26 promoter).32 Combined deficiency of SETX and DNA-activated protein kinase catalytic subunit (DNA-PKcs), critical for non-homologous end joining (NHEJ)-mediated DNA repair in neurons, causes R-loop accumulation and DNA damage, and inefficient DNA damage repair leading to genomic instability and motor neuron degeneration in SMA.26 To test whether ZPR1 overexpression will restore SETX and DNA-PKcs levels and reduce DNA damage in motor neurons, we examined and compared primary spinal cord neurons cultured from Normal, SMA and Z-SMA mice using immunofluorescence and immunoblot analyses (Fig. 6). We established a method for culture of primary spinal cord neurons from postnatal mice, which stain positive for known motor neuron markers, including ChAT and Hlxb9 (Hb9), suggesting that cultured spinal cord neurons may retain characteristics of motor neurons.26,32,33,40 Comparison of morphology of cultured primary neurons stained with neuron-specific β-tubulin-III shows that neurons from Z-SMA mice were healthy and were rescued of axonal defects such as retraction, bending, folding of axons (shown with arrowheads) compared to neurons from SMA mice (Fig. 6A–F). Staining of neurons for ZPR1 shows increased ZPR1 levels in neurons from Z-SMA mice (Fig. 6A) that is supported by immunoblot quantitative analysis (Fig. 6G, H and Supplementary Fig. 10). Notably, increase in ZPR1 levels results in increased staining and levels of SMN, SETX and p-DNA-PKcs (Fig. 6B-D, G and H). The increase in SETX levels (∼2.3-fold) suggests that there might be a decrease in the accumulation of R-loops, a concept supported by quantitative analysis of nuclear R-loops, which shows marked decrease (∼4.7-fold) in R-loop levels in Z-SMA compared to SMA neurons (Fig. 6E, I and J). Increase in total DNA-PKcs (∼2.69-fold) and activated p-DNA-PKcs (∼2.28-fold) levels suggest an improvement in the efficiency of DNA repair and reduction in DNA damage, further supported by immunofluorescence and immunoblot analyses of γH2AX, a DNA damage marker, which show ∼2.6-fold decrease in γH2AX levels (Fig. 6F–H). These data suggest that ZPR1 overexpression restores levels of SETX and DNA-PKcs, critical for R-loop resolution and DNA damage repair respectively, and rescues R-loop-mediated DNA damage overall preventing degeneration of SMA neurons.
Figure 6.
ZPR1 overexpression in vivo rescues DNA damage associated with R-loop accumulation and prevents degeneration of motor neurons in SMA. Primary spinal cord neurons were cultured from 7-day-old normal, SMA and Z-SMA (SMA mice with ZPR1 overexpression under the control of mouse Rosa26 promoter) mice. Neurons were differentiated in vitro for 12 days and stained with antibodies against neuron-specific β-tubulin-III (red), SMN, SETX, p-DNA-PKcs, R-loops and γH2AX, and immunofluorescence was examined by confocal microscopy. (A–F) Axonal defects include retraction, bending, folding of axons (arrowheads) that indicate degeneration of SMN-deficient neurons. (A) Staining of neurons with ZPR1 (green) and β-tubulin (red), (B) staining of neurons with SMN (green) and β-tubulin (red), (C) SETX (green) and β-tubulin (red), (D) p-DNA-PKcs (green) and β-tubulin (red), (E) R-loops (green) and β-tubulin (red) and (F) γH2AX (green) and β-tubulin (red). Insets show higher magnification of punctate staining of γH2AX foci indicating DNA damage. Z-SMA neurons with in vivo ZPR1 overexpression show rescue of degenerative features. Nuclei were stained with DAPI (blue). Scale bar = 25 μm. (G) Immunoblot analysis of cultured primary spinal cord motor neurons from Normal, SMA and Z-SMA mice for detecting changes in levels of ZPR1, SMN, SETX, DNA-PKcs, p-DNA-PKcs, and DNA damage marker, γH2AX (full-length blots, Supplementary Fig. 10). (H) Quantitation of protein levels in motor neurons from Normal, SMA, and Z-SMA mice is shown as a scatter plot with median and range (min, median, max). SMN: Normal (90.54, 95.64, 115.90), SMA (20.36, 25.87, 29.54), Z-SMA (70.65, 75.32, 85.65); ZPR1: Normal (90.21, 97.25, 112.50), SMA (44.25, 54.21, 55.47), Z-SMA (150.20, 170.30, 190.30); SETX: Normal (90.65, 98.54, 112.50), SMA (30.25, 39.87, 44.56), Z-SMA (70.25, 81.25, 94.21); p-DNA-PKcs: Normal (90.54, 100.30, 110.30), SMA (30.21, 37.32, 40.25), Z-SMA (70.25, 81.25, 94.21); DNA-PKcs: Normal (92.36, 97.25, 112.00), SMA (29.87, 32.65, 35.68), Z-SMA (75.32, 87.98, 100.70); γH2AX: Normal (90.25, 104,50, 108.0), SMA (287.5, 310.30, 322.60), Z-SMA (110.20, 120.40, 130.30). Statistical analysis (ANOVA) of immunoblot data (mean ± SEM, n = 3 mice/group) from spinal cord neurons shows increase in ZPR1 levels (1.70 ± 0.11-fold, P = 0.0001) results in increase of SMN levels to (77.21 ± 4.43%, P = 0.0002), SETX (88.73 ± 9.36%, P = 0.0012), p-DNA-PKcs (81.90 ± 6.92%, P = 0.0037) and total DNA-PKcs (87.98 ± 7.31%, P = 0.0018) leading to a marked decrease in γH2AX levels from 306.80 ± 10.26% to 120.30 ± 5.77% (P = 0.0002). (I) Higher magnification images of nuclei of neurons stained with antibody against R-loops (green) and β-tubulin (red) from Normal, SMA and Z-SMA spinal cord neurons (dotted lines show nuclei). (J) Quantitation of nuclear immunofluorescence R-loop intensity in Normal, SMA, and Z-SMA spinal cord neurons is shown as a scatter plot with median and range. Normal (0.87, 0.95, 1.17), SMA (7.2, 8.25, 10.25), ZSMA (1.20, 1.60, 2.65). Quantitative analysis of nuclear R-loop immunofluorescence shows marked reduction in R-loop accumulation (1.82 ± 0.43-fold) in Z-SMA compared to SMA (8.58 ± 0.88)-fold (P = 0.0023, t-test), and compared to Normal (1.04 ± 0.09-fold) (P = 0.0002, ANOVA) neurons.
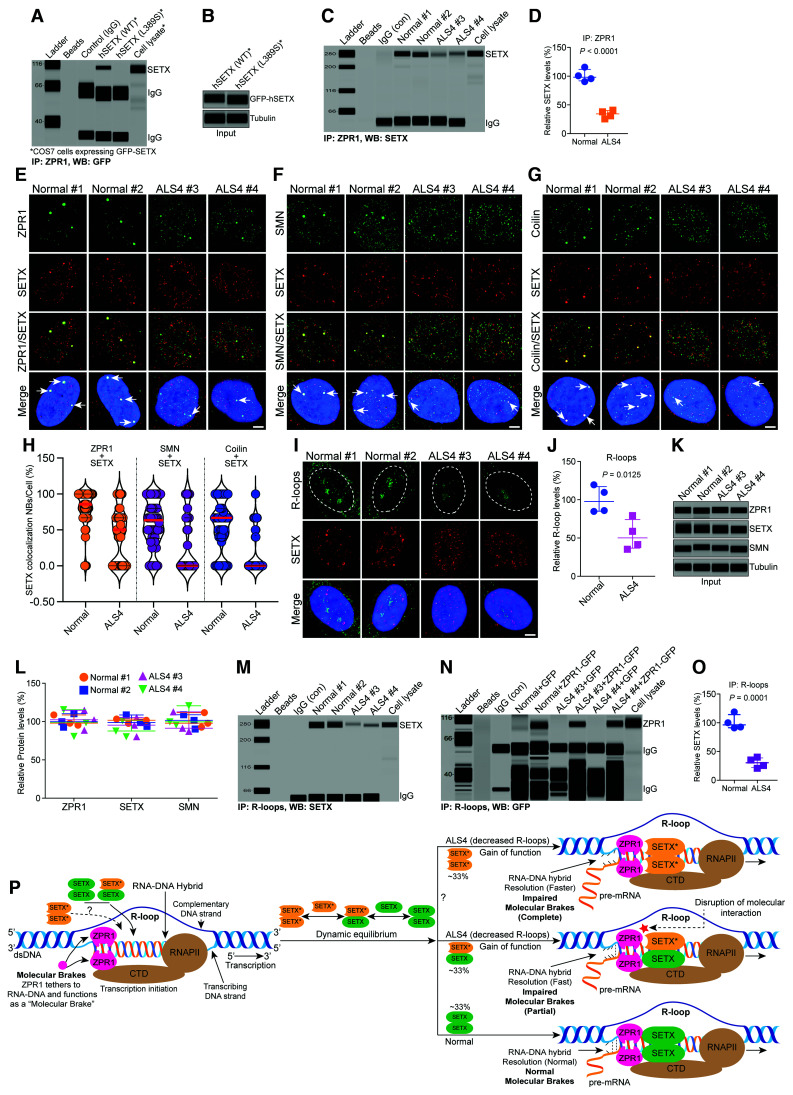
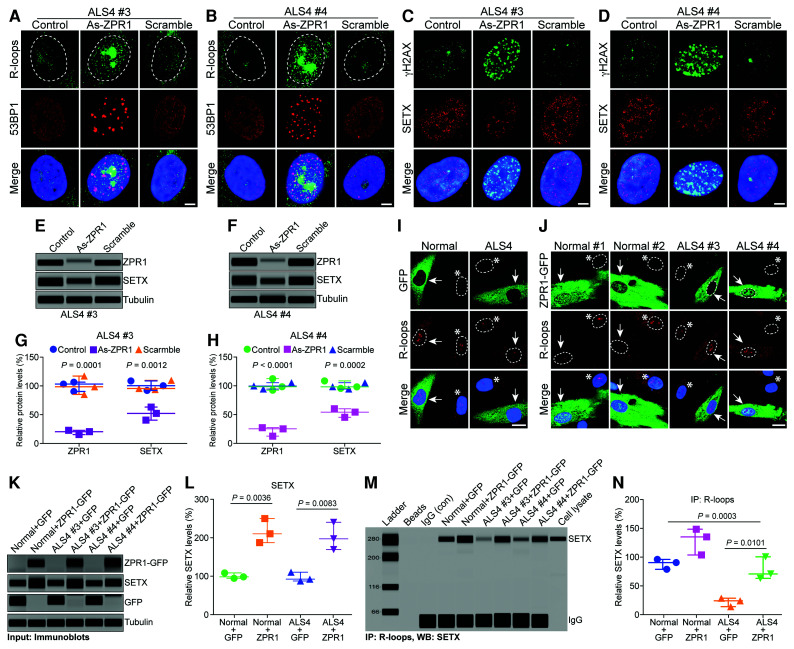
Interaction of SETX with ZPR1 is disrupted in ALS4 patients
To further test our hypothesis that ZPR1 and SETX collaborate to regulate R-loop metabolism, we thought that disruption of ZPR1 interaction with SETX might provide additional insight into the function of ZPR1-SETX complexes. The NH2-terminal of SETX is involved in protein-protein interaction.57 We anticipated that mutations reported in the NH2-terminal of SETX might disrupt its interaction with ZPR1. Mutational analysis using recombinant SETX protein revealed that SETX mutation L389S, which causes ALS4,3 disrupts interaction of SETX with ZPR1 (Fig. 7A–B and Supplementary Fig. 11A). Based on these data, we anticipated that the interaction between SETX and ZPR1 might be disrupted in ALS4.
Figure 7.
Interaction of SETX with ZPR1 is disrupted in ALS4 patients and ZPR1 fails to recruit mutant SETX onto R-loops in ALS4. Mutational analysis shows that SETX L389S mutation, which causes autosomal dominant ALS4, disrupts interaction of SETX with ZPR1. (A) COS7 cells were transfected with plasmids pDEST53 expressing GFP-hSETX (1–667) (WT) and GFP-hSETX (1–667) (L389S). Immunoprecipitation was performed using anti-ZPR1 antibody followed by western blot with anti-GFP to detect GFP-hSETX. (B) Immunoblots of cell lysate from cells expressing GFP-hSETX (WT) and GFP-hSETX (L389S). To gain insight into the contribution of disruption of SETX-ZPR1 complexes in the pathogenesis of ALS4, we used fibroblasts derived from normal subjects and ALS4 patients that have heterozygous SETX mutation L389S (SETX+/L389S). Cultured control, Normal #1 and Normal #2, and ALS4 cell lines, ALS4 #3 and ALS4 #4 were used for immunofluorescence, immunoblot and immunoprecipitation analyses. (C) Interaction of SETX with ZPR1 is disrupted in ALS4 patients. Immunoprecipitation of ZPR1 shows decrease in co-immunoprecipitation of SETX from ALS4 patients compared to control (Normal) fibroblast. (D) Quantitation of SETX co-IP with ZPR1 in Normal and ALS4 patient cells is presented as a scatter plot with median and range (min, median, max). Normal (90.54, 98.21, 115.70), ALS4 (25.54, 34.44, 40.87). Quantitation of SETX levels (mean ± SEM, n = 4) in ZPR1 immunoprecipitation shows that the levels of SETX binding with ZPR1 are reduced to (33.82 ± 3.45%, P < 0.0001) in ALS4 compared to control cells. Representative images with highest co-localization are presented for double-labelled immunostainings: (E) Control cells (Normal #1 and Normal #2) stained with ZPR1 (green) and SETX (red) show ZPR1 and SETX co-localize in subnuclear bodies (arrows). In contrast, SETX mutation causes partial disruption of subnuclear bodies and mislocalization of ZPR1 and SETX in ALS4 patient cells. (F) Cells stained with SMN (green) and SETX (red) show SETX co-localizes with SMN in nuclear gems bodies (arrows) in cells from Normal subjects. The SETX mutation causes partial disruption of SMN+ gems and results in smaller SMN+ foci in ALS4 patient cells. (G) Cells stained with coilin (green) and SETX (red) show SETX co-localizes with coilin in Cajal bodies (arrows) in Normal cells. Notably, SETX mutation causes disruption of CBs and results in smaller CBs in ALS4 compared to Normal cells. (H) Quantification of SETX co-localization in subnuclear bodies (NBs)/cell (%) in normal and ALS4 patient cells is shown as a violin plot with median and interquartile range (Q1, median, Q3) (50 cells/group). In normal cells, SETX co-localization with ZPR1: ZPR1 + SETX (66.67, 85.71, 100.00), SMN: SMN + SETX (38.33, 63.33, 80.00) and coilin: Coilin + SETX (38.33, 66.67. 80.00). In ALS4 cells, SETX co-localization with ZPR1: ZPR1 + SETX (0.0, 50.00, 66.67), SMN: SMN + SETX (0.0, 0.0, 66.67) and coilin: Coilin + SETX (0.0, 0.0. 42.50). Quantification of SETX co-localization in normal cells (mean ± SEM, n = 50 cells/group) shows higher co-localization with ZPR1 (80.04 ± 3.64%) compared to SMN (57.87 ± 4.60%) (gems) and Coilin (55.06 ± 4.99%) (Cajal bodies). SETX co-localization in ALS4 cells also show higher co-localization with ZPR1 (41.88 ± 5.21%) compared to SMN (32.50 ± 5.82%) and Coilin (17.60 ± 4.17%). The overall SETX co-localization is reduced by ∼48% (ZPR1, P < 0.0001), ∼44% (SMN, P = 0.0009) and ∼68% (Coilin, P < 0.0001) in ALS4 compared to normal cells. (I) Immunofluorescence analysis of R-loops (green) and SETX (red) shows reduced intensity of R-loops in ALS4 compared to normal cells. Nuclei were stained with DAPI (blue). Scale bar = 5.0 μm. (J) Quantitation of R-loop nuclear immunofluorescence intensity in Normal and ALS4 patient cells is shown as a scatter plot with median and range. Normal (84.76, 97.82, 119.60), ALS4 (35.28, 50.17, 79.25). Quantitative (mean ± SEM, n = 4) and statistical (unpaired t-test) analysis of R-loop nuclear immunofluorescence intensity in cells show fewer R-loops (53.72 ± 9.85%, P = 0.0125) in ALS4 patient cells (ALS4 #3 + ALS4 #4) compared to control (Normal #1 + Normal #2) cells. Quantification of R-loop levels using dot-blot analysis is included in Supplementary Fig. 12. (K) Immunoblot analysis of proteins ZPR1, SETX, SMN and tubulin in Normal #1 and Normal #2, and ALS4 #3 and ALS4 #4, patient cells. (L) Quantitation of protein levels in Normal and ALS4 patient cells is shown as a scatter plot with median and range (min, median, max). ZPR1: Normal #1 (95.36, 97.98, 108.7), Normal #2 (92.65, 99.85, 110.30), and ALS4 #3 (88.65, 102.40, 114.90), ALS4 #4 (88.65, 102.40, 114.9); SETX: Normal #1 (97.65, 101.70, 103.30), Normal #2 (93.75, 98.73, 108.7), and ALS4 #3 (80.32, 95.32, 105.40), ALS4 #4 (80.25, 87.69, 101.30); SMN: Normal #1 (92.65, 97.85, 111.70), Normal #2 (90.35, 101.50, 109.00), and ALS4 #3 (87.21, 91.25, 112.70), ALS4 #4 (95.64, 99.65, 120.70). Quantitative analysis of protein levels (mean ± SEM, n = 3) shows SETX mutation did not significantly change the levels of ZPR1 (98.27 ± 10.10%, P = 0.8252), SMN (97.04 ± 7.89%, P = 0.9080) and SETX (93.67 ± 7.27%, P = 0.3903) in ALS4 compared to normal cells. (M) SETX fails to associate in vivo with R-loops in ALS4. IP of R-loops shows marked decrease in SETX co-immunoprecipitation in ALS4 compared to control suggesting that ZPR1 fails to recruit mutant SETX to R-loops. (N) Immunoprecipitations of R-loops from Normal and ALS4 subjects infected with Ad5-GFP and Ad5-ZPR1-GFP show that the binding of ZPR1 with R-loops is unaffected under ALS4 disease conditions and supports the idea that ZPR1 interaction with SETX is critical for the recruitment of SETX onto R-loops. (O) Quantitation of SETX co-IP with R-loops in Normal and ALS4 patient cells is presented as a scatter plot with median and range. Normal (91.34, 96.62, 119.2), ALS4 (20.76, 30.49, 40.32). Quantitative and statistical analysis of SETX co-IP (mean ± SEM, n = 4) with R-loop immunoprecipitation shows that SETX binding with R-loops reduced to (30.51 ± 4.43%, P = 0.0001) in ALS4 compared to control cells. (P) Graphical illustration of the molecular mechanism of R-loop resolution in ALS4 patients that have SETX L389S mutation. ZPR1 tethers to RNA:DNA hybrid and may function as a ‘molecular brake’ to control the speed of R-loop resolution by regulating translocase/helicase activity of SETX during transcription. SETX mutation alters dynamic equilibrium of SETX dimers and causes disruption of SETX-ZPR1 complexes that may result in partial impairment of the molecular brake leading to faster resolution (gain-of-function) and fewer R-loops in ALS4. All full-length blots are included in Supplementary Fig. 11.
To test our hypothesis, we examined interaction of SETX with ZPR1 using fibroblasts derived from ALS4 patients that have the SETX L389S mutation.38 We found that SETX co-immunoprecipitation with ZPR1 was reduced in experiments with fibroblasts isolated from three different ALS4 patients compared to fibroblasts isolated from three normal subjects (Fig. 7C and Supplementary Fig. 12G). Quantitative analysis of SETX co-IP with ZPR1 shows a marked reduction in the amount of SETX immunoprecipitation; ∼34% (P < 0.0001) with ZPR1 in ALS4 compared to controls (Normal) fibroblasts (Fig. 7D). These data indicate partial loss of SETX-ZPR1 complexes. This is likely because ALS4 is an autosomal dominant disease caused by heterozygous mutation, SETX+/L389S, in the SETX gene. These data indicate that mutation in SETX disrupts ZPR1-SETX complexes in ALS4.
Mislocalization of SETX and ZPR1 in ALS4 patient cells
The observation that SETX-ZPR1 complexes are disrupted in ALS4 suggests that the mutation in SETX might also affect cellular localization and alter levels of SETX and ZPR1. We examined cellular distribution and levels of ZPR1 and SETX in ALS4 patient cells. Control (Normal) cells show ZPR1 co-localizes with SETX (Fig. 7E). In contrast, ALS4 patient cells show decreased co-localization of ZPR1 and SETX in subnuclear bodies. In addition, the size of ZPR1 and SETX subnuclear bodies is reduced in ALS4 cells (Fig. 7E). SETX also co-localizes with SMN and coilin in control cells from normal subjects (Fig. 7F and G). Notably, SETX co-localization with SMN+ (gems) and coilin+ (CBs) is also markedly reduced in ALS4 patient cells (Fig. 7F and G). These data show that mutation in SETX results in mislocalization or redistribution of ZPR1, SETX, SMN and coilin in the nucleoplasm of ALS4 patient cells (Supplementary Fig. 13). Additional data from a third set of control and ALS4 cell lines is included in Supplementary Fig. 12A–C. Quantification of SETX co-localization in normal cells (mean ± SEM) shows higher co-localization with ZPR1 (80.04 ± 3.64%) compared to SMN (57.87 ± 4.60%) (gems) and Coilin (55.06 ± 4.99%) (CBs) (Fig. 7H), which is consistent with data from HeLa (Fig. 2D) and WI-38 (Supplementary Fig. 4D) cells. Notably, SETX co-localization in ALS4 patient cells also show higher co-localization with ZPR1 (41.88 ± 5.21%) compared to SMN (32.50 ± 5.82%) and Coilin (17.60 ± 4.17%) but the overall SETX co-localization is reduced by ∼48% (ZPR1), ∼44% (SMN) and ∼68% (coilin) in ALS4 compared to normal cells. Cellular mislocalization of ZPR1, SMN and coilin and disruption of SETX interaction with ZPR1 indicate the possibility of altered R-loop resolution in ALS4. Immunofluorescence and dot-blot quantifications show R-loop levels were reduced to (∼54%) and (∼42%), respectively, in ALS4 patient cells compared to control cells (Fig. 7I–J and Supplementary Fig. 12D–F). These data are consistent with recently published findings, which hypothesized that mutation in SETX may be a cause of gain-of-function in SETX-dependent R-loop resolution activity leading to fewer R-loops in ALS4.38,58 Notably, comparison of ZPR1, SETX and SMN protein as well as mRNA levels did not show any significant difference between control and ALS4 patient cells (Fig. 7K–L and Supplementary Figs 11B and 12I). These are intriguing findings, which suggest that disruption of SETX-ZPR1 complexes may be the cause of gain-of-function in R-loop resolution activity leading to fewer R-loops in ALS4.
Mutation in SETX decreases in vivo association with R-loops in ALS4
It is possible that the interaction of ZPR1 with SETX is critical for the recruitment of SETX onto R-loops. Therefore, the disruption of ZPR1 interaction with SETX in ALS4 may impair recruitment of mutant SETX (SETX*) onto R-loops. To test this, we examined the effect of disruption of SETX-ZPR1 complexes on the binding of SETX with R-loops using ALS4 patient fibroblasts. As anticipated, we found that SETX co-immunoprecipitation with R-loops was markedly reduced from ALS4 patient fibroblasts (SETX+/L389S) compared to fibroblasts isolated from normal subjects (SETX+/+) (Fig. 7M and Supplementary Fig. 12H). To determine whether the disruption of ZPR1-SETX interaction also affects ZPR1 binding with R-loops, we performed IP of R-loops followed by immunoblot analysis of ZPR1. We found that in vivo association of ZPR1 with R-loops was unaffected in ALS4 patient cells (Fig. 7N). These data show that disruption of SETX interaction with ZPR1 results in decreased association of SETX with R-loops in vivo and support the idea that interaction of ZPR1 with SETX is critical for ZPR1 to recruit SETX onto R-loops.
Quantitative analysis of SETX co-IP with R-loops shows a marked reduction in the amount of SETX immunoprecipitation; ∼31% with R-loops in ALS4 compared to controls fibroblasts (Fig. 7O). These data show unanticipated low levels of SETX co-immunoprecipitation, while SETX levels are unchanged in ALS4 compared to control fibroblasts (Fig. 7K and L). However, theoretically 50% of the produced SETX protein would be mutant SETX* in ALS4 patients (SETX+/L389S). In addition, SETX forms a homodimer and SETX mutation L389S does not affect its dimerization.57 These findings suggest that SETX may form three types of dimers, SETX-SETX, SETX-SETX* and SETX*-SETX*, with ∼33.3% contribution of each to total SETX pool in ALS4 (see graphical illustration in Fig. 7P). Notably, ZPR1 does not self-dimerize and does not form homodimers.30 Thus, the observation that mutation in SETX disrupts its interaction with ZPR1 suggests that SETX and ZPR1 may form two types of complexes with 1:1 and 1:0.5 stoichiometry, namely ZPR1-SETX-SETX-ZPR1 (normal) and ZPR1-SETX-SETX* (ALS4), respectively. It is possible that SETX heterodimers containing mutant SETX* (SETX-SETX*) may have reduced efficiency of recruitment by ZPR1 onto R-loops, which is supported by decreased SETX binding (∼31–34%) with ZPR1 and R-loops in ALS4. Our data show that ZPR1 tethers to RNA:DNA hybrids, recruits SETX onto R-loops and required for R-loop resolution. Based on current and published findings, we hypothesize that ZPR1 may regulate the activity of SETX by controlling the speed of R-loop resolution by acting as some kind of a brake. Therefore, we propose to refer to ZPR1 as a ‘molecular brake’ to regulate SETX-dependent RLRC activity (Fig. 7P, graphical model of hypothesis). Mutation in SETX disrupts its binding with ZPR1, which may cause partial impairment of the molecular brake resulting in higher activity of R-loop resolution (gain-of-function) leading to fewer R-loops in ALS4 (Fig. 7P). Together, these data suggest functional collaboration between SETX and ZPR1 in regulating R-loop resolution activity.
Modulation of ZPR1 levels regulates R-loop accumulation and rescues pathogenic R-loop phenotype in ALS4 patient cells
Comparison of data from SMA and ALS4 fibroblasts show similar decrease (∼65%) in SETX association with R-loops but contrasting levels of R-loop accumulation in SMA (high) and ALS4 (low). These data raise the question of whether ALS4 patient cells have normal biochemical potential to accumulate R-loops. To address this, we examined the effect of ZPR1 knockdown on R-loop accumulation. We found that ZPR1 knockdown causes R-loop accumulation in both ALS4 fibroblasts lines (As-ZPR1) compared control and scramble oligo treated ALS4 fibroblasts (Fig. 8A and B). Thus, ALS4 cells retain the biochemical potential and the molecular machinery for R-loop accumulation. We also found that ZPR1-deficient ALS4 fibroblasts accumulated 53BP1 (Fig. 8A and B) and γH2AX foci (Fig. 8C and D), markers for DNA damage and double strand breaks. Quantitative analysis of ZPR1 knockdown shows ∼50% downregulation of SETX in both ALS4 cell lines suggesting that downregulation of mutant SETX* may also contribute to R-loop accumulation in ZPR1-deficient cells (Fig. 8E–H and Supplementary Fig. 14A and B). Thus, R-loop accumulation in ALS4 cells triggers similar downstream molecular events, which compromise genomic integrity5, with those observed (Fig. 2) and reported in control and SMA cells.26
Figure 8.
Modulation of ZPR1 levels regulates R-loop accumulation and rescues pathogenic R-loop phenotype in ALS4 patient cells. (A–D) Knockdown of ZPR1 in ALS4 patient cells increases R-loop accumulation and causes activation of DNA damage response. ALS4 cells were untransfected (Control) or transfected with (100 nM) antisense oligos (As-ZPR1) or scrambled sequence oligo (Scramble) (100 nM). Representative images are presented for double-labelled immunostainings: (A) ALS4 #3 patient cells stained for R-loops (green) and 53BP1 (red) and (B) ALS4 #4 patient cells stained for R-loops (green) and 53BP1 (red) show marked increase in R-loop accumulation in ZPR1-deficient ALS4 patient cells (As-ZPR1) compared to control and scramble oligo treated cells, which causes DNA damage response and accumulation of 53BP1. ZPR1-deficiency causes downregulation of SETX and DNA damage. (C) ALS4 #3 patient cells stained for SETX (red) and γH2AX (green) and (D) ALS4 #4 patient cells stained for SETX (red) and γH2AX (green) show marked increase γH2AX foci in the nucleus of ZPR1-deficient ALS4 patient cells (As-ZPR1) compared to control and scramble oligo treated cells. Scale bar = 5.0 μm. Quantitation (mean ± SEM, n = 3) of immunoblots from ALS4 #3 (E, G) and ALS4 #4 (F and H) control, As-ZPR1 and scramble samples show KD of ZPR1 to (19.69 ± 2.13%, P = 0.0001) in ALS4 #3 and (21.7 ± 4.614%, P < 0.0001) in ALS4 #4. ZPR1 KD decreases SETX level to (52.06 ± 6.55%, P = 0.0012) in ALS4 #3 and (53.28 ± 4.33%, P = 0.0002) in ALS4 #4. (G) Quantitation of changes in ZPR1 and SETX levels with ZPR1 knockdown in ALS4 #3 patient cells is shown as a scatter plot with median and range (min, median, max). ZPR1: ALS4 #3 Control (90.32, 103.30, 106.60), ALS4 #3 – As-ZPR1 (15.64, 20.54, 22.89), ALS4 #3 – Scramble (85.24, 98.54, 117.3); SETX: ALS4 #3 Control (92.21, 100.30, 108.60), ALS4 #3 – As-ZPR1 (40.56, 52.36, 63.25), ALS4 #3 – Scramble (93.68, 95.47, 109.60). (H) Quantitation of changes in ZPR1 and SETX levels with ZPR1 knockdown in ALS4 #4 patient cells is shown as a scatter plot with median and range. ZPR1: ALS4 #4 Control (92.31, 98.32, 112.40), ALS4 #4 – As-ZPR1 (12.54, 25.31, 27.25), ALS4 #4 – Scramble (94.32, 99.65, 105.3); SETX: ALS4 #4 Control (95.45, 98.24, 108.6), ALS4 #4 – As-ZPR1 (45.32, 54.32, 60.21), ALS4 #4 – Scramble (94.32, 98.32, 105.3). (I and J) Overexpression of ZPR1 in ALS4 patient cells rescues disease phenotype and improves accumulation of R-loops. (I) Control experiment with Normal and ALS4 patient cells infected with adenovirus Ad5-GFP (100 MOI) expressing GFP (green) and stained with antibody against R-loops (red) did not show any change in the normal levels of R-loop fluorescence intensity in the nucleus. (J) Normal and ALS4 cells infected with Ad5-ZPR1-GFP expressing ZPR1-GFP (green) and stained for R-loops (red) show that ZPR1 overexpression decreases R-loop accumulation in the Normal (arrows) and rescues R-loop accumulation in ALS4 patient cells (arrows). Nuclei were stained with DAPI (blue). Scale bar = 20 μm. Arrows show infected cells and asterisks indicate non-infected cells. Dotted circular lines indicate nuclei. (K–N) ZPR1 overexpression improves in vivo association of SETX with R-loops in ALS4 patient cells. (K) Immunoblots of ZPR1-GFP, SETX, GFP and tubulin proteins in Normal and ALS4 patient cells overexpressing GFP and ZPR1-GFP. (L) Quantitation of SETX levels in Normal and ALS4 patient cells overexpressing GFP and ZPR1-GFP is presented as a scatter plot with median and range. Normal-GFP (94.65, 98.36, 108.70); Normal-ZPR1-GFP (187.40, 210.40, 250.00); ALS4-GFP (87.65, 92.35, 110.30); ALS4-ZPR1-GFP (169.40, 197.30, 240.30). (M) Immunoprecipitation of R-loops from Normal and ALS4 cells overexpressing GFP and ZPR1-GFP shows increase in in vivo binding of SETX with R-loops. (N) Quantitation of SETX protein levels co-IP with R-loops in Normal and ALS4 patient cells overexpressing GFP and ZPR1-GFP is presented as a scatter plot with median and range. Normal-GFP (90.76, 102.50, 107.90); Normal-ZPR1-GFP (115.60, 147.00, 160.30); ALS4-GFP (25.98, 36.10, 40.53); ALS4-ZPR1-GFP (74.98, 82.76, 112.40). Quantitation and comparison of SETX levels in co-IP with R-loops show increase in SETX binding with R-loops (90.03 ± 11.39%) compared to (34.24 ± 4.30%, P = 0.0101) in ALS4 expressing ZPR1-GFP and GFP, respectively. All full-length blots are included in Supplementary Fig. 14. Enlarged images for (I) and (J) are included in Supplementary Figs 15 and 16.
Further, we examined the effect of ZPR1 overexpression on R-loop accumulation in ALS4 patient cells using adenoviral infection Ad-GFP (GFP) and Ad-ZPR1-GFP (ZPR1-GFP). Control experiment did not show any change in R-loop staining of cells expressing GFP (green, arrows) compared to cells that were not infected (asterisks) in normal and ALS4 fibroblasts (Fig. 8I, see enlarged images in Supplementary Fig. 15). Notably, normal subject-derived fibroblasts expressing ZPR1-GFP (arrows) show marked decrease in R-loop staining compared to non-infected cells (asterisks) (Fig. 8J, see enlarged images in Supplementary Fig. 16). These data suggest that ZPR1 has the potential to accelerate R-loop resolution under normal conditions. Interestingly, overexpression of ZPR1 in ALS4 patient fibroblasts causes increase in accumulation of R-loops (arrows) compared to non-infected cells (asterisks) (Fig. 8J) suggesting that ZPR1 can rescue cellular phenotype associated with ALS4 pathogenesis. To gain insight into the mechanism of rescue of pathogenic ALS4 phenotype, we examined the effect of ZPR1 overexpression on levels of SETX and its in vivo association with R-loops. We found ∼2.0-fold increase in SETX levels in normal and ALS4 patient cells, supporting the idea of SETX being a downstream target of ZPR1 (Fig. 8K–L and Supplementary Fig. 14C). Further, immunoprecipitation of R-loops from normal and ALS4 cells overexpressing ZPR1 shows increase in in vivo binding of SETX with R-loops compared to control cells (Fig. 8M). Quantitation of SETX levels co-IP with R-loops shows significant improvement in the binding of SETX from (34.24 ± 4.30%) to (90.03 ± 11.39%, P = 0.0101) in ALS4 cells with ZPR1 overexpression (ALS4 + ZPR1-GFP) compared to control (ALS4 + GFP) cells (Fig. 8N). These data suggest that ZPR1 has the potential to regulate SETX levels under normal and ALS4 conditions and rescues pathogenic R-loop phenotype in ALS4 patient cells. Together, these findings provide insight into the function of ZPR1-SETX complexes in R-loop resolution and indicate the disruption of ZPR1-SETX complexes as the molecular basis for ALS4 pathogenesis.
Discussion
In this study, we provide insight into the molecular basis of R-loop resolution and the pathogenesis of ALS4. We uncovered the critical role of SETX-ZPR1 complexes in R-loop resolution and identify the putative function of ZPR1 to regulate SETX-dependent R-loop resolution activity. Overall, findings of this study allowed to delineate the molecular mechanism of R-loop resolution under the normal and ALS4 conditions (Supplementary Video 1). We demonstrate that ZPR1 forms endogenous complexes with SETX and R-loops. ZPR1 is critical for recruiting SETX onto R-loops and is required for the resolution of RNA:DNA hybrids formed during transcription. The observation that SETX interacts with ZPR1 and shows highest co-localization (∼80%) with ZPR1+ nuclear bodies compared to SMN+ (gems) (∼55%) and coilin+ (Cajal bodies) (∼50%), indicates that these proteins may function jointly. Moreover, because ZPR1 is required for the recruitment of SETX onto R-loops, ZPR1 may be involved in the physiological functions of SETX. Our data demonstrate that low levels of SETX-ZPR1 complexes result in the loss-of-function leading to an increase in R-loop levels and motor neuron degeneration in SMA. Interestingly, SETX mutation (L389S) disrupts SETX-ZPR1 complexes, which results in gain-of-function leading to fewer R-loops and motor neuron degeneration in ALS4. Together, these findings originate a novel concept in the field of cell and molecular biology, opposite alterations in a cell biological activity (R-loop resolution) result in similar pathogenesis (neurodegeneration) in different genetic motor neuron disorders.
Findings of this study unravel the key steps that may be a part of the molecular mechanism of R-loop resolution under normal and ALS4 disease conditions. Our data demonstrate that following key steps may contribute to R-loop resolution: (i) ZPR1 tethers to RNA:DNA hybrids; (ii) recruits SETX onto R-loops; and (iii) regulates SETX-dependent RLRC activity. Based on current and published findings, we propose that ZPR1 may regulate the activity of SETX by controlling the speed of R-loop resolution. In this scenario, ZPR1 may function as a ‘molecular brake’ to modulate the speed of SETX-dependent RLRC activity. Mutation in SETX disrupts its interaction with ZPR1, which may cause partial impairment of the molecular brake, resulting in higher activity of R-loop resolution (gain-of-function) leading to fewer R-loops in ALS4 compared to control. The graphical illustration of our hypothesis and the mechanism of R-loop resolution in mammalian cells is shown in a video clip with the animation and narration of the molecular mechanisms of R-loop resolution under the normal and ALS4 disease conditions (Supplementary Video 1).
ZPR1 is essential for cell viability, however, the physiological function of ZPR1 that is critical for cell survival is unknown.27,28 Here we show that ZPR1 deficiency causes downregulation of SETX, suggesting that SETX is likely a downstream target of ZPR1. Accumulation of R-loops throughout gene transcription in ZPR1-deficient cells suggests that ZPR1-deficiency may primarily impair R-loop resolution while transcription continues and R-loop-mediated DNA damage causes genomic instability leading to neurodegeneration and cell death.27,59 These findings suggest that ZPR1 is critical for resolution of co-transcriptional R-loops and contributes to one of the fundamental cellular processes essential for cell viability.
Diversion from the normal R-loop levels is linked to neurodegenerative disorders such as ALS4 (low R-loops) and SMA (high R-loops). It is intriguing that contrasting levels of R-loops result in a common phenotype of motor neuron degeneration in two genetic diseases, ALS4 and SMA, caused by mutations in the SETX and SMN1 genes, respectively3,25 Our data demonstrate that ALS4 and SMA patient cells have a similar defect, which is ∼67–70% decrease in SETX association with R-loops but opposite cellular phenotype with low and high levels of R-loops, respectively. Comparison of SMA and ALS4 patient cells data reveals key molecular differences that may explain these contrasting as well as fascinating cellular phenotypes.
In SMA, expression of ZPR1 and SETX are downregulated, which results in low levels of ZPR1-SETX complexes. Downregulation of ZPR1 and SETX is likely the consequence of global splicing defects caused by SMN deficiency in SMA.60 In addition, defects in splicing can alter transcription because of interdependence between transcription and splicing and may contribute to R-loop accumulation.20,61–63 The decreased levels of ZPR1-SETX complexes impair the efficiency of R-loop resolution (loss-of-function) and result in higher levels of R-loops in SMA. This is supported by the observations that the knockdown of either ZPR1 or SETX results in low levels of ZPR1-SETX complexes and causes R-loop accumulation. Interestingly, cells derived from ataxia oculomotor apraxia 2 (AOA2) patients that have homozygous SETX mutation (N2037D), located in the helicase domain, show increased accumulation of R-loops suggesting that the likely loss of helicase activity (loss-of-function) results in inefficient resolution of R-loops,64 which is consistent with our observations of increased R-loop accumulation in SETX-deficient HeLa cells and SMA cells with chronic low levels of SETX.
Interestingly, the levels of ZPR1 and SETX are not altered in ALS4. However, mutation (L389S) in SETX abolishes its interaction with ZPR1, which impairs ZPR1 ability to recruit mutant SETX* to R-loops resulting in decreased levels of SETX onto R-loops but results in fewer R-loops in ALS4. In addition, the observation that the mutation in SETX disrupts interaction with ZPR1 but does not affect ZPR1 binding with R-loops in ALS4 suggesting that ZPR1 can bind to R-loops independently of SETX and supports the idea that ZPR1 binds first and then recruits SETX to R-loops. Therefore, the disruption of ZPR1-SETX complexes may be the cause of increase in R-loop resolution activity leading to fewer R-loops in ALS4. The next question, how disruption of ZPR1 and SETX interaction results in the gain-of-function is fascinating. The fact that only one SETX allele is mutated in autosomal dominant ALS43 and the findings that wild-type SETX can self-dimerize (SETX-SETX) and form dimers with mutant SETX* (SETX-SETX*)57 suggest that ZPR1 can form two types of complexes, ZPR1-SETX-SETX-ZPR1 (normal) and ZPR1-SETX-SETX* (ALS4). Therefore, ZPR1 might be able to recruit SETX-SETX* heterodimer onto R-loops but the partial disruption of molecular interaction between ZPR1 and SETX* (ZPR1-SETX-SETX*) may impair ZPR1’s ability to collaborate with and regulate mutant SETX* associated helicase/translocase activity. Therefore, disruption of ZPR1 and SETX interaction could result in uncontrolled (higher) SETX-dependent R-loop resolution activity in ALS4 (see graphical illustration in Fig. 7P). We propose that ZPR1 collaborates with SETX and functions as a molecular brake to regulate SETX-dependent R-loop resolution activity. It is possible that ZPR1-SETX-SETX* (ALS4) complexes possess hyper-activity compared to ZPR1-SETX-SETX-ZPR1 (normal) complexes because of the partial impairment of the molecular brake resulting in faster R-loop resolution leading to fewer R-loops in ALS4 compared to normal. Our additional data further support the proposed model and also demonstrate that SETX is a downstream target of ZPR1. Knockdown of ZPR1 causes downregulation of SETX expression, which result in accumulation of R-loops in ALS4 patient cells, similar to SMA cells, suggesting that ZPR1-dependent activity is also critical for R-loop resolution in ALS4 cells. Notably, overexpression of ZPR1 increases SETX levels, improves recruitment of SETX onto R-loops and rescues pathogenic R-loop phenotype in ALS4 patient cells. These data suggest that (i) increasing the ratio of normal ZPR1-SETX-SETX-ZPR1 versus mutant ZPR1-SETX-SETX* (ALS4) complexes can rescue pathogenic R-loop phenotype in ALS4; and (ii) ZPR1 may represent a potential molecular target that could be exploited therapeutically. These data suggest that ZPR1 may be a potential regulator of SETX-dependent R-loop resolution activity. It is also possible that other factors associated with R-loops, including RNA and DNA helicases such as DHX9, DDX5 and BLM or Sgs1 (orthologue of human Bloom’s syndrome helicase BLM), and factors such as XRN2, PRMT5, FANCD2 (Fanconi Anemia Complementation Group2) may be part of the RLRC and may contribute directly or indirectly to R-loop metabolism and mRNA biogenesis.14,20,65–68
Our study also provides insight into the mechanism of predominant degeneration of motor neurons in patients with ALS4 and SMA. Our data demonstrate that ZPR1 regulates expression of SETX levels. In SMA, chronic low levels of ZPR1 may contribute to downregulation of SETX. Decrease in ZPR1-SETX complexes results in R-loop accumulation that causes DNA double-strand breaks (DSBs).26,55 SMA motor neurons express low levels of DNA-PKcs, which is required for NHEJ-mediated DNA repair, the primary DSBs repair mechanism available in post-mitotic neurons. Deficiency of DNA-PKcs may impair DNA repair in motor neurons leading to genomic instability and neurodegeneration in SMA.26 Increase in ZPR1 levels resulted in two-pronged improvement in SMA. ZPR1 overexpression restores SETX levels and improves assembly and activity of RLRC, reducing R-loop accumulation in SMA. Furthermore, ZPR1 increases DNA-PKcs levels and rescues DNA damage in neurons, preventing neurodegeneration in SMA. These data suggest that genomic instability may be the cause of selective degeneration of motor neurons in SMA.
ALS4 is caused by heterozygous mutation in the SETX gene and characterized by motor neuron degeneration and neuromuscular weakness. However, the molecular basis of pathogenesis caused by the mutant SETX protein was unclear. We provide insight into the molecular events altered by mutant SETX, which contribute to pathogenic low levels of R-loops causing neurodegeneration in ALS4. Our data demonstrate that disruption of SETX interaction with ZPR1 caused by the SETX mutation (L389S) may be a cause of increase in R-loop resolution activity leading to fewer R-loops in ALS4. Low levels of R-loops are shown to decrease the expression of BMP and activin membrane bound inhibitor (BAMBI), a negative regulator of transforming growth factor-β (TGF-β) pathway, in cells derived from ALS4 patients.38 BAMBI binds to TGF-β receptor and blocks the interaction of TGF-β with receptors, and negatively regulating signalling.69 The TGF-β pathway plays an important role in survival and axon guidance of motor neurons70,71 and in the pathogenesis of ALS.72–74 Therefore, mutant SETX-mediated decrease in R-loop levels and R-loop-dependent downregulation of BAMBI may cause pathogenic increase in TGF-β signalling and contribute to motor neuron degeneration in ALS4.38 In addition, mislocalization of nuclear TAR DNA binding protein (TARDBP or TDP-43) in the cytoplasm of ALS4 patient spinal cord motor neurons may also contribute to neurodegeneration in ALS4 through a common pathogenic mechanism involved in ALS caused by mutations in TDP-43.75,76
Currently, there is no treatment available for a growing number of incurable disorders caused by defects in R-loop metabolism. The observation that ZPR1 can regulate R-loop accumulation and rescue pathogenic R-loop phenotypes in SMA and ALS4 patient cells suggest that ZPR1’s ability to modulate R-loop levels could be exploited therapeutically to develop treatments for diseases with altered R-loop metabolism.
Supplementary Material
Acknowledgements
We thank Dr Kenneth H. Fischbeck and Dr Christopher Grunseich, Neurogenetics Branch, National Institutes of Health for the generous gift of fibroblasts derived from normal subjects and ALS4 patients, and for comments on the manuscript. We thank Dr Stephen C. West, The Francis Crick Institute, London, UK, for the generous gift of plasmids with recombinant GFP-SETX (WT) and GFP-SETX (1-667).
Abbreviations
- ALS4
amyotrophic lateral sclerosis 4
- DNA-PKcs
DNA-activated protein kinase-catalytic subunit
- DRIP
DNA-RNA immunoprecipitation
- GST =
glutathione S-transferase
- RLRC =
R-loop resolution complexes
- SMA =
spinal muscular atrophy
- SMN =
survival motor neuron
Contributor Information
Annapoorna Kannan, Center of Emphasis in Neurosciences, Texas Tech University Health Sciences Center El Paso, El Paso, TX 79905, USA; Department of Molecular and Translational Medicine, Paul L. Foster School of Medicine, Texas Tech University Health Sciences Center El Paso, El Paso, TX 79905, USA.
Juliana Cuartas, Center of Emphasis in Neurosciences, Texas Tech University Health Sciences Center El Paso, El Paso, TX 79905, USA; Department of Molecular and Translational Medicine, Paul L. Foster School of Medicine, Texas Tech University Health Sciences Center El Paso, El Paso, TX 79905, USA.
Pratik Gangwani, Automated Driving Compute System Architecture, GM Global Technical Center - Sloan Engineering Center, Warren, MI 48092, USA.
Dana Branzei, The FIRC Institute of Molecular Oncology Foundation, IFOM Foundation, Milan 20139, Italy; Istituto di Genetica Molecolare, Consiglio Nazionale delle Ricerche (IGM-CNR), 27100 Pavia, Italy.
Laxman Gangwani, Center of Emphasis in Neurosciences, Texas Tech University Health Sciences Center El Paso, El Paso, TX 79905, USA; Department of Molecular and Translational Medicine, Paul L. Foster School of Medicine, Texas Tech University Health Sciences Center El Paso, El Paso, TX 79905, USA.
Funding
This study was supported by research grants to L.G. from the US National Institutes of Health (R01NS064224 and R01NS115834), and Cure SMA (GAN 2019), and a research grant to D.B. from the Italian Association for Cancer Research (AIRC) IG 23710). JC is a research mentee supported by NIH diversity supplement to L.G. (R01NS115834-01A1S1).
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- 1.Chance PF, Rabin BA, Ryan SG, et al. Linkage of the gene for an autosomal dominant form of juvenile amyotrophic lateral sclerosis to chromosome 9q34. Am J Hum Genet 1998;62:633–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rabin BA, Griffin JW, Crain BJ, Scavina M, Chance PF, Cornblath DR. Autosomal dominant juvenile amyotrophic lateral sclerosis. Brain 1999;122(Pt 8):1539–1550. [DOI] [PubMed] [Google Scholar]
- 3.Chen Y-Z, Bennett CL, Huynh HM, et al. DNA/RNA helicase gene mutations in a form of juvenile amyotrophic lateral sclerosis (ALS4). Am J Hum Genet 2004;74:1128–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Groh M, Albulescu LO, Cristini A, Gromak N. Senataxin: Genome Guardian at the Interface of Transcription and Neurodegeneration. J Mol Biol 2017;429:3181–3195. [DOI] [PubMed] [Google Scholar]
- 5.Crossley MP, Bocek M, Cimprich KA. R-loops as cellular regulators and genomic threats. Mol Cell 2019;73:398–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costantino L, Koshland D. Genome-wide map of r-loop-induced damage reveals how a subset of R-loops contributes to genomic instability. Mol Cell 2018;71:487–497.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Skourti-Stathaki K, Proudfoot NJ. A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes Dev 2014;28:1384–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wells JP, White J, Stirling PC. R loops and their composite cancer connections. Trends Cancer 2019;5:619–631. [DOI] [PubMed] [Google Scholar]
- 9.Richard P, Manley JL. R loops and links to human disease. J Mol Biol 2017;429:3168–3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia-Muse T, Aguilera A. R loops: from physiological to pathological roles. Cell 2019;179:604–618. [DOI] [PubMed] [Google Scholar]
- 11.Sollier J, Cimprich KA. Breaking bad: R-loops and genome integrity. Trends Cell Biol 2015;25:514–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allison DF, Wang GG. R-loops: formation, function, and relevance to cell stress. Cell Stress 2019;3:38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhatia V, Herrera-Moyano E, Aguilera A, Gomez-Gonzalez B. The role of replication-associated repair factors on R-loops. Genes (Basel) 2017;8:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cristini A, Groh M, Kristiansen MS, Gromak N. RNA/DNA hybrid interactome identifies DXH9 as a molecular player in transcriptional termination and R-loop-associated DNA damage. Cell Rep 2018;23:1891–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang IX, Grunseich C, Fox J, et al. Human proteins that interact with RNA/DNA hybrids. Genome Res 2018;28:1405–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skourti-Stathaki K, Proudfoot NJ, Gromak N. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol Cell 2011;42:794–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cohen S, Puget N, Lin Y-L, et al. Senataxin resolves RNA:DNA hybrids forming at DNA double-strand breaks to prevent translocations. Nat Commun 2018;9:533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mischo HE, Gómez-González B, Grzechnik P, et al. Yeast Sen1 helicase protects the genome from transcription-associated instability. Mol Cell 2011;41:21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rawal CC, Zardoni L, Di Terlizzi M, et al. Senataxin ortholog Sen1 limits DNA:RNA hybrid accumulation at DNA double-strand breaks to control end resection and repair fidelity. Cell Rep 2020;31:107603. [DOI] [PubMed] [Google Scholar]
- 20.Chakraborty P, Huang JTJ, Hiom K. DHX9 helicase promotes R-loop formation in cells with impaired RNA splicing. Nat Commun 2018;9:4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suraweera A, Lim Y, Woods R, et al. Functional role for senataxin, defective in ataxia oculomotor apraxia type 2, in transcriptional regulation. Hum Mol Genet 2009;18:3384–3396. [DOI] [PubMed] [Google Scholar]
- 22.Ursic D, Chinchilla K, Finkel JS, Culbertson MR. Multiple protein/protein and protein/RNA interactions suggest roles for yeast DNA/RNA helicase Sen1p in transcription, transcription-coupled DNA repair and RNA processing. Nucleic Acids Res 2004;32:2441–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuce O, West SC. Senataxin, defective in the neurodegenerative disorder ataxia with oculomotor apraxia 2, lies at the interface of transcription and the DNA damage response. Mol Cell Biol 2013;33:406–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao DY, Gish G, Braunschweig U, et al. SMN and symmetric arginine dimethylation of RNA polymerase II C-terminal domain control termination. Nature 2016;529(7584):48–53. [DOI] [PubMed] [Google Scholar]
- 25.Lefebvre S, Burglen L, Reboullet S, et al. Identification and characterization of a spinal muscular atrophy- determining gene. Cell 1995;80:155–165. [DOI] [PubMed] [Google Scholar]
- 26.Kannan A, Bhatia K, Branzei D, Gangwani L. Combined deficiency of Senataxin and DNA-PKcs causes DNA damage accumulation and neurodegeneration in spinal muscular atrophy. Nucleic Acids Res 2018;46:8326–8346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gangwani L, Flavell RA, Davis RJ. ZPR1 is essential for survival and is required for localization of the survival motor neurons (SMN) protein to Cajal bodies. Mol Cell Biol 2005;25:2744–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gangwani L, Mikrut M, Galcheva-Gargova Z, Davis RJ. Interaction of ZPR1 with translation elongation factor-1alpha in proliferating cells. J Cell Biol 1998;143:1471–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galcheva-Gargova Z, Gangwani L, Konstantinov KN, et al. The cytoplasmic zinc finger protein ZPR1 accumulates in the nucleolus of proliferating cells. Mol Biol Cell 1998;9:2963–2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mishra AK, Gangwani L, Davis RJ, Lambright DG. Structural insights into the interaction of the evolutionarily conserved ZPR1 domain tandem with eukaryotic EF1A, receptors, and SMN complexes. Proc Natl Acad Sci U S A 2007;104:13930–13935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gangwani L, Mikrut M, Theroux S, Sharma M, Davis RJ. Spinal muscular atrophy disrupts the interaction of ZPR1 with the SMN protein. Nat Cell Biol 2001;3:376–383. [DOI] [PubMed] [Google Scholar]
- 32.Kannan A, Jiang X, He L, Ahmad S, Gangwani L. ZPR1 prevents R-loop accumulation, upregulates SMN2 expression and rescues spinal muscular atrophy. Brain 2020;143:69–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ahmad S, Wang Y, Shaik GM, Burghes AH, Gangwani L. The zinc finger protein ZPR1 is a potential modifier of spinal muscular atrophy. Hum Mol Genet 2012;21:2745–2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doran B, Gherbesi N, Hendricks G, Flavell RA, Davis RJ, Gangwani L. Deficiency of the zinc finger protein ZPR1 causes neurodegeneration. Proc Natl Acad Sci U S A 2006;103:7471–7475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Genabai NK, Kannan A, Ahmad S, Jiang X, Bhatia K, Gangwani L. Deregulation of ZPR1 causes respiratory failure in spinal muscular atrophy. Sci Rep 2017;7:8295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ahmad S, Bhatia K, Kannan A, Gangwani L. Molecular mechanisms of neurodegeneration in spinal muscular atrophy. J Exp Neurosci 2016;10:39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Le TT, Pham LT, Butchbach ME, et al. SMNDelta7, the major product of the centromeric survival motor neuron (SMN2) gene, extends survival in mice with spinal muscular atrophy and associates with full-length SMN. Hum Mol Genet 2005;14:845–857. [DOI] [PubMed] [Google Scholar]
- 38.Grunseich C, Wang IX, Watts JA, et al. Senataxin mutation reveals how R-loops promote transcription by blocking DNA nethylation at gene promoters. Mol Cell 2018;69:426–437.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gangwani L. Deficiency of the zinc finger protein ZPR1 causes defects in transcription and cell cycle progression. J Biol Chem 2006;281:40330–40340. [DOI] [PubMed] [Google Scholar]
- 40.Genabai NK, Ahmad S, Zhang Z, Jiang X, Gabaldon CA, Gangwani L. Genetic inhibition of JNK3 ameliorates spinal muscular atrophy. Hum Mol Genet 2015;24:6986–7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boguslawski SJ, Smith DE, Michalak MA, et al. Characterization of monoclonal antibody to DNA·RNA and its application to immunodetection of hybrids. J Immunol Methods 1986;89:123–130. [DOI] [PubMed] [Google Scholar]
- 42.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001;25:402–408. [DOI] [PubMed] [Google Scholar]
- 43.Gangwani L, Khurana JP, Maheshwari SC. Inhibition of chloroplast protein phosphorylation by cAMP in Lemna paucicostata 6746. Phytochemistry 1996;41:49–54. [Google Scholar]
- 44.Ausubel FM, Brent R, Kingston RE, et al., eds. Enzymatic Manipulation of DNA and RNA: Ribonucleases. Current Protocols in Molecular Biology. John Wiley & Sons, Inc.; 2006. [Google Scholar]
- 45.Halasz L, Karanyi Z, Boros-Olah B, et al. RNA-DNA hybrid (R-loop) immunoprecipitation mapping: an analytical workflow to evaluate inherent biases. Genome Res 2017;27:1063–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Makharashvili N, Arora S, Yin Y, et al. Sae2/CtIP prevents R-loop accumulation in eukaryotic cells. Elife 2018;7:e42733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sollier J, Stork CT, Garcia-Rubio ML, Paulsen RD, Aguilera A, Cimprich KA. Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Mol Cell 2014;56:777–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Galcheva-Gargova Z, Konstantinov KN, Wu IH, Klier FG, Barrett T, Davis RJ. Binding of zinc finger protein ZPR1 to the epidermal growth factor receptor. Science 1996;272(5269):1797–1802. [DOI] [PubMed] [Google Scholar]
- 49.Stork CT, Bocek M, Crossley MP, et al. Co-transcriptional R-loops are the main cause of estrogen-induced DNA damage. Elife 2016;5:e17548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bennett CL, La Spada AR. Senataxin, A Novel Helicase at the Interface of RNA Transcriptome Regulation and Neurobiology: From Normal Function to Pathological Roles in Motor Neuron Disease and Cerebellar Degeneration. Adv Neurobiol 2018;20:265–281. [DOI] [PubMed] [Google Scholar]
- 51.Brown TA, Tkachuk AN, Clayton DA. Native R-loops persist throughout the mouse mitochondrial DNA genome. J Biol Chem 2008;283:36743–36751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Konig F, Schubert T, Langst G. The monoclonal S9.6 antibody exhibits highly variable binding affinities towards different R-loop sequences. PLoS One 2017;12:e0178875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Phillips DD, Garboczi DN, Singh K, Hu Z, Leppla SH, Leysath CE. The sub-nanomolar binding of DNA-RNA hybrids by the single-chain Fv fragment of antibody S9.6. J Mol Recognit 2013;26:376–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smolka JA, Sanz LA, Hartono SR, Chédin F. Recognition of RNA by the S9.6 antibody creates pervasive artifacts when imaging RNA:DNA hybrids. J Cell Biol 2021;220:e202004079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jangi M, Fleet C, Cullen P, et al. SMN deficiency in severe models of spinal muscular atrophy causes widespread intron retention and DNA damage. Proc Natl Acad Sci U S A 2017;114:E2347–E2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Helmken C, Hofmann Y, Schoenen F, et al. Evidence for a modifying pathway in SMA discordant families: reduced SMN level decreases the amount of its interacting partners and Htra2-beta1. Hum Genet 2003;114:11–21. [DOI] [PubMed] [Google Scholar]
- 57.Bennett CL, Chen Y, Vignali M, et al. Protein interaction analysis of senataxin and the ALS4 L389S mutant yields insights into senataxin post-translational modification and uncovers mutant-specific binding with a brain cytoplasmic RNA-encoded peptide. PLoS One 2013;8:e78837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fogel BL, Cho E, Wahnich A, et al. Mutation of senataxin alters disease-specific transcriptional networks in patients with ataxia with oculomotor apraxia type 2. Hum Mol Genet 2014;23:4758–4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jiang X, Kannan A, Gangwani L. ZPR1-Dependent Neurodegeneration Is Mediated by the JNK Signaling Pathway. J Exp Neurosci 2019;13:1179069519867915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Z, Lotti F, Dittmar K, et al. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell 2008;133:585–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Merkhofer EC, Hu P, Johnson TL. Introduction to cotranscriptional RNA splicing. Methods Mol Biol 2014;1126:83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Paulsen RD, Soni DV, Wollman R, et al. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol Cell 2009;35:228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kornblihtt AR, de la Mata M, Fededa JP, Munoz MJ, Nogues G. Multiple links between transcription and splicing. RNA 2004;10:1489–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Becherel OJ, Sun J, Yeo AJ, et al. A new model to study neurodegeneration in ataxia oculomotor apraxia type 2. Hum Mol Genet 2015;24:5759–5774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chang EY-C, Novoa CA, Aristizabal MJ, et al. RECQ-like helicases Sgs1 and BLM regulate R-loop–associated genome instability. J Cell Biol 2017;216:3991–4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Madireddy A, Kosiyatrakul ST, Boisvert RA, et al. FANCD2 Facilitates replication through common fragile sites. Mol Cell 2016;64:388–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liang Z, Liang F, Teng Y, et al. Binding of FANCI-FANCD2 complex to RNA and R-loops stimulates robust FANCD2 monoubiquitination. Cell Rep 2019;26:564–572.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Villarreal OD, Mersaoui SY, Yu Z, Masson JY, Richard S. Genome-wide R-loop analysis defines unique roles for DDX5, XRN2, and PRMT5 in DNA/RNA hybrid resolution. Life Sci Alliance 2020;3:e202000762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Onichtchouk D, Chen Y-G, Dosch R, et al. Silencing of TGF-β signalling by the pseudoreceptor BAMBI. Nature 1999;401(6752):480–485. [DOI] [PubMed] [Google Scholar]
- 70.Colavita A, Krishna S, Zheng H, Padgett RW, Culotti JG. Pioneer axon guidance by UNC-129, a C. elegans TGF-beta. Science 1998;281(5377):706–709. [DOI] [PubMed] [Google Scholar]
- 71.Martinou J-C, Le Van Thai A, Valette A, Weber MJ. Transforming growth factor β1 is a potent survival factor for rat embryo motoneurons in culture. Brain Res Dev Brain Res 1990;52(1-2):175–181. [DOI] [PubMed] [Google Scholar]
- 72.Endo F, Komine O, Fujimori-Tonou N, et al. Astrocyte-derived TGF- β1 accelerates disease progression in ALS mice by interfering with the neuroprotective functions of microglia and T cells. Cell Rep 2015;11:592–604. [DOI] [PubMed] [Google Scholar]
- 73.Houi K, Kobayashi T, Kato S, Mochio S, Inoue K. Increased plasma TGF-β1 in patients with amyotrophic lateral sclerosis. Acta Neurol Scand 2002;106:299–301. [DOI] [PubMed] [Google Scholar]
- 74.Phatnani HP, Guarnieri P, Friedman BA, et al. Intricate interplay between astrocytes and motor neurons in ALS. Proc Natl Acad Sci U S A 2013;110:E756–E765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bennett CL, Dastidar SG, Ling S-C, et al. Senataxin mutations elicit motor neuron degeneration phenotypes and yield TDP-43 mislocalization in ALS4 mice and human patients. Acta Neuropathol 2018;136:425–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Corcia P, Couratier P, Blasco H, et al. Genetics of amyotrophic lateral sclerosis. Rev Neurol 2017;173:254–262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings and conclusions of this study are included in this research article and in the Supplementary material, and are also available from the corresponding author upon reasonable request.