Abstract
Ambystoma mexicanum (axolotl) embryos and juveniles have been used as model organisms for developmental and regenerative research for many years. This neotenic aquatic species maintains the unique capability to regenerate most, if not all, of its tissues well into adulthood. With large externally developing embryos, axolotls were one of the original model species for developmental biology. However, increased access to, and use of, organisms with sequenced and annotated genomes, such as Xenopus laevis and tropicalis and Danio rerio, reduced the prevalence of axolotls as models in embryogenesis studies. Recent sequencing of the large axolotl genome opens up new possibilities for defining the recipes that drive the formation and regeneration of tissues like the limbs and spinal cord. However, to decode the large A. mexicanum genome will take a herculean effort, community resources, and the development of novel techniques. Here, we provide an updated axolotl-staging chart ranging from one-cell stage to immature adult, paired with a perspective on both historical and current axolotl research that spans from their use in early studies of development to the recent cutting-edge research, employment of transgenesis, high-resolution imaging, and study of mechanisms deployed in regeneration.
Keywords: axolotl, Ambystoma mexicanum, embryonic development, staging, regeneration
1 |. INTRODUCTION
Ambystoma mexicanum, the Mexican salamander commonly known as the axolotl, is an endangered amphibian with amazing regenerative capabilities. Although adult animals range in size, axolotls typically grow up to 20–35 cm in length from head to tail and live an aquatic lifestyle.1 As natural predators, axolotls are capable of eating many organisms that cross their path. However, due to deterioration of their natural ecosystem, axolotls currently hold a critically endangered status.2,3
Unlike most other amphibious salamanders, axolotls are neotenic animals that retain juvenile traits throughout their lifetime.4 Their close relatives, like the tiger salamander (Ambystoma tigrinum), undergo metamorphosis as they mature, losing the fringed gills and caudal fin that make axolotls so distinctive.5 Their neoteny is possible because axolotls lack thyroid-stimulating hormone, a precursor to thyroxine: the necessary component to kick-start metamorphosis. In fact, an injection of iodine or thyroxine can be used to stimulate the transition, but the results are stochastic and regeneration capacity can be reduced in some cases.6,7 Demircan et al. identified reductions in some tissues’ regenerative capacities and complete inhibition in others after thyroid hormone treatment.4 In contrast, Monaghan et al. found that while body size had no effect on regeneration, thyroxine-induced metamorphosis reduced regeneration rates by twofold and produced forelimb and digit abnormalities.7 Further research is needed to pinpoint the regenerative outcome of induced metamorphosis in axolotl and understand the mechanisms of growth regulation during regeneration.
A current gap in axolotl research is the application of new, precise tools for understanding cells at the individual and collective levels, and studies of gene and protein expression and function across development and regeneration. These experiments are key to support comparative developmental studies and identify conserved and divergent regulatory modules controlling axolotl developmental and regenerative programs. So much can be done with these animals, such as gain and loss of function,8–10 CRISPR-mediated transgenesis,10,11 single-cell characterization,12–16 live imaging,17–20 transplants and cell lineage tracing,20–27 and the use of ex vivo explants28 to compare the developmental processes of this unique salamander to the robust body of avian, frog, zebrafish, and mouse developmental research. For example, axolotl gills can regenerate, yet they are one of three modes of respiration at the organism’s disposal, and their full role remains unknown.29
Axolotls have the potential to become more prevalent developmental models, similar to Xenopus, but the continuity of tools and resources is lacking. Specifically, there are few stock resource centers for ease of stocking, housing, and sharing transgenic animals. However, the NIH-funded Ambystoma Genetic Stock Center (AGSC) has created Sal-Site (https://ambystoma.uky.edu/quick-links/sal-site) to provide information and access to resources for investigators.30,31 Recent work characterizing mutant laboratory axolotls identified that all individuals from the AGSC contain small portions of the tiger salamander (Ambystoma tigrinum) genome, as there appears to be genetic cross-contamination.32 These findings provoke the questions: how similar are laboratory axolotls to those in the wild, and how might discoveries differ based on genomic differences? Other tools like cross-species tools for gain and loss of function, validated antibodies that recognize axolotl proteins, and interspecies comparison of transcriptional networks would greatly improve knowledge acquisition capabilities.
2 |. NATURAL HISTORY OF THE AXOLOTL
Axolotls have been used as research organisms for over 150 years, and their vast potential stems from a humble beginning.33 Native axolotls originated in Lake Texcoco, thriving along the lake’s banks, and later, the canals of the Aztec city-state of Xochimilco. The Spaniards took notice of the Aztec people’s fondness for eating this aquatic species in the mid-16th century.34 Then, during the Spanish conquest and colonial rule, Xochimilco expanded rapidly, to the detriment of axolotls. The 19th century saw the first scientific interest in this species, and in 1863, six axolotls were transported from Mexico to the Jardin Des Plantes in Paris. These original six individuals propagated the majority of the present-day research axolotl population. As a result, many axolotls used in research have high genetic similarity, with an inbreeding coefficient of 35%, which substantially exceeds the threshold of 12% that indicates breeding between first cousins and concerns ecologists and geneticists greatly.35 This inbreeding decreases the viability of axolotls as a genetic model and potentially increases their disease susceptibility. Despite this challenge, axolotls spread as laboratory animals due to the ease of year-round breeding in captivity and naturally-occurring developmental mutations.36 However, axolotls are near extinction in their native Mexico, begetting a lack of genetic diversity in wild axolotls as well. Therefore, without remediation, science may lose secrets hidden in the axolotl’s genetic diversity on both fronts.37 Despite these challenges, the axolotl makes up for these genetic pitfalls with many redeeming qualities.
3 |. BACK TO THE FUTURE: AXOLOTLS AS MODELS FOR EMBRYONIC DEVELOPMENT
Amphibian embryos are an excellent model system for the study of cell and developmental biology due to their large sizes, resiliency, and large clutch sizes. As the major limb and spinal cord regenerative model system, axolotls are well studied in juvenile and larval stages;10,38 however, we lack detailed information about the molecular mechanisms that drive their early developmental processes. While axolotls retain their juvenile characteristics into adulthood, their development proceeds through distinct stages (Figure 1).39,40 In contrast to many other aquatic research models, axolotls utilize internal fertilization, externally laying single one-cell embryos over the course of several hours.41 Morphologically, early axolotl development appears to proceed similarly to other aquatic amphibians like Xenopus laevis or Xenopus tropicalis. For example, axolotls develop more rapidly at warmer temperatures, as noted previously.39,40 At their preferred cooler temperatures (17–18°C), axolotls progress through rapid cleavage stages (Figure 1, cleavage-blastula, 0–24 hours post laying, HPL), stereotypical gastrulation (Figure 1, gastrula, 24–54 hours HPL) and neurulation (Figure 1, neurula, 54–72 hours HPL). Neurulation is followed by tailbud and tadpole stages (prehatched and hatched, 72–340 hours + HPL) where organogenesis and growth occur. After hatching, limb morphogenesis (characterized in detail by the literature42) occurs and animals continue to grow and mature (Figure 1, larva-adult, or juvenile adult, 2–18 months post-fertilization). However, axolotl development proceeds at a slower rate during early development than Xenopus, and the animals are not sexually mature until they are approximately 1 year of age or older.40,43,44
FIGURE 1.
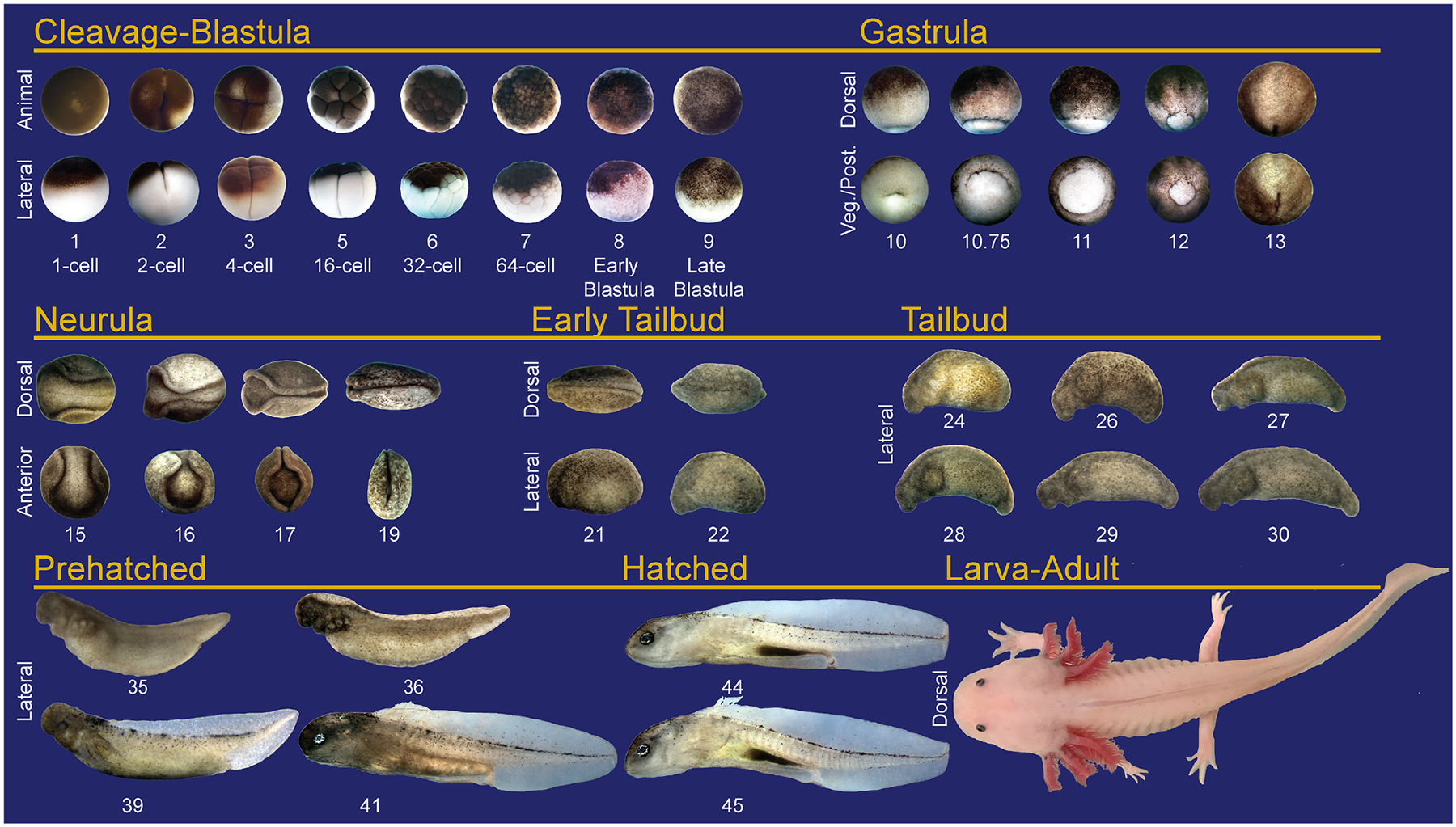
Staging series for Ambystoma mexicanum from fertilization through maturation. Stages were grouped into eight different categories: cleavage-blastula (0–24 hours post laying, HPL), gastrula (24–54 hours HPL), neurula (54–72 hours HPL), early tailbud (72–83 hours HPL), tailbud (83–122 hours HPL), prehatched (122–342 + HPL), hatched (342 hours to 1–3 months postlaying/fertilization), and larva-adult. Hatched larva to sexually mature adult can take up to 18 months depending on tank density. At cleavage/blastula stages, both animal and lateral views are shown. Similar to Xenopus laevis and tropicalis, Ambystoma embryos have pigment differentials on the animal and vegetal poles, the animal poles being dark and the vegetal poles light colored and filled with yolk even in leucistic animals as shown. At gastrula stages, we show both dorsal and posterior views. As gastrulation proceeds, the blastopore closes and the animals begin to neurulate. At neurula stages, we show dorsal and anterior views. After neural tube closure, the embryos begin axis elongation throughout the early tailbud and tailbud stages. At these stages, we show lateral views. The gill arches become visible during tailbud stages and further develop in the prehatch tadpoles (lateral view). At these stages, gills become more pronounced and eyes gain pigment and become visible. Prehatch embryos remain in their thick jelly coats in natural settings. In hatched tadpoles, the dorsal fin becomes more transparent, the gills branch and grow outward (lateral view). We show a dorsal view of a larva-adult. This animal is approximately 1 year old, but is not yet fully grown or sexually mature
A current gap exists in axolotl research in the study of embryogenesis at the cell and tissue levels and the comparative understanding of the epigenetic, transcriptomic, and proteomic changes that occur in early stages and primary formation compared to regeneration. A regulatory map of the processes that control primary development during embryogenesis in axolotl would be informative in its own right but would also inform regeneration studies in larvae, juveniles, and adult animals. It is difficult to ascertain which developmental or regenerative mechanisms are conserved with other vertebrates, including in “distantly” related fish, avians, and rodents, or even with more closely related amphibians such as X. laevis and tropicalis with the current gaps in knowledge. For the most fruitful research outcomes, it is important to know where these animals come from (primary development) in order to understand where they are going (regeneration).
Quantitative three-dimensional imaging and reconstruction of axolotl neurula embryos has provided some detail of the mechanical forces and changes that occur at the earliest developmental stages.45,46 Additional work focused on the role of ion transport during neurulation.47,48 However, more work is needed to validate structural and molecular similarities between axolotl development and other vertebrates. Without validated antibodies and molecular stains to spatiotemporally mark specific tissue derivatives in axolotl embryos, characterization and comparative analyses of cell and tissue specification and differentiation timing will be difficult. Although there is a single study of transcriptomic changes during embryogenesis in axolotl,49 the work was published prior to the axolotl genome sequencing and should be reassessed with this recent knowledge. Further, scientists are creating additional molecular tools for spatiotemporal profiling of gene and protein expression, but more research is needed to understand the molecular mechanisms driving development in these early stages.
Early embryonic fate specification in axolotls is somewhat varied from their closely related frog embryos. Axolotl ectoderm forms neural tissue in the absence of an organizer signal28 whereas in frog embryos, ectoderm forms non-neural ectoderm or epidermis in the absence of an organizer.50 However, neural induction and continued development requires additional instructive signaling in both organisms.28,51
The study of neural development is often paired with analyses of neural crest cell formation and migration, as the two tissues arise from the same ectodermal germ layer. Axolotls were a well-used model for the first wave of neural crest research that lasted roughly from the 1920s to 1950s, in which several scientists observed the migration and differentiation of this stem-like cell population through trunk and cranial neural crest transplantation experiments. Newth’s 1954 study supported the idea that neural crest cells are specified but not determined at the neurula stage.52 Delving further into the realm of understanding these migratory cells, Epperlein and Löfberg discovered that trunk neural crest-derived melanophores and xanthophores create the pigment pattern in axolotls.53 Cell tracing studies have also been performed on axolotl trunk neural crest cells and have shown that axolotl neural crest cells have different potentials depending on where they originate in the anterior–posterior axis, similar to chicken embryos.22,24,54 In addition, CRISPR-mediated genome editing and morpholino knockdowns have been used successfully in axolotl embryos, suggesting that it is a tractable model for functional studies.10,55–58 Moury and Jacobson identified that competence to become neural crest cells is not limited to the neural folds. They also proposed that epidermis-neural plate signaling and forces generated by local interactions between the two tissues induce neural crest cells, thus supporting a hypothesis of mixed intrinsic and extrinsic factors controlling neural crest cell development in axolotl embryos.58 These experiments established the nature of neural crest cells and began to answer questions on the timing of neural crest specification and migration. Between the 1960s and 2000s, the shift toward X. laevis as a developmental model and abundance of research in the avian neural crest contributed to a short-age of publications in this field.
The brink of the 21st century saw a resurgence in axolotl developmental research, picking up from the surge of neural crest cell research in urodele amphibians from the 1920s to 1950s.59 This second wave of neural crest cell research in axolotls resumed in the early 2000s, as new techniques gave scientists the ability to reconsider classic questions in neural crest development. This era also upheld the importance of comparative studies and centered the axolotl in the field of evolution of development (“evo-devo”). Epperlein et al. took advantage of lipophilic dye injections and the novel neural crest cell marker, AP-2, to define the order of neural crest cell migratory streams in axolotl and the potential of neural crest cells to change their migration path when displaced off course.22 Scanning electron microscopy (SEM) identified detailed information on neural crest cell migratory stream routes and stream assembly, as well as cell shape and orientation across stages.59 Comparing axolotl neural crest cell migration to other vertebrates identified that axolotls exhibit earlier neural crest cell migration than newts, but retain the distinct migratory streams common to amphibians.59 Epperlein et al. demonstrated the unique migratory patterns and timing of neural crest cell development in axolotls.53 Explant experiments showed intrinsic neural crest cell patterning and segmental migration, but the support of extrinsic signaling from the neighboring epidermis was necessary to maintain distinct streams.60 These two waves of axolotl neural crest cell research provide a strong basis for future axolotl studies, and it is clear this model organism still has much untapped potential.
4 |. AXOLOTLS AS MODELS FOR EVOLUTION OF DEVELOPMENT (EVO-DEVO)
While lamprey and hydra are renowned model organisms for studies of evolutionarily conserved and divergent developmental and regeneration mechanisms, axolotls carve out their own niche in the field of evo-devo studies.59 As developmental models, X. laevis and D. rerio are currently more prominent than the axolotl, but the newly sequenced axolotl genome paired with epigenetic and transcriptomic analyses provide traction to re-invigorate the use of axolotl in both comparative and mechanistic studies of conserved and divergent evolutionary processes.61–63 Rationale for the use of zebrafish and frog embryos over axolotls in developmental studies may include the ease of in vitro fertilization and jelly coat removal in frogs64,65 and genetic tractability in zebrafish,66 but axolotls provide a unique evolutionary niche. Frogs and fish develop faster and can be more resilient to cleavage-stage manipulation than axolotls. As a result, many laboratory techniques have been created or adapted for Xenopus, including in situ hybridization, immunohistochemistry, transgenic methods, and expression cloning.67–69 However, the axolotl reigns supreme as a vertebrate regenerative model, and techniques pioneered in other aquatic models can, and have been, readily be adapted for use in axolotl studies.
Their large genome, neoteny, ability to regenerate, and lack of a genome-wide duplication make axolotls the perfect tetrapod models that fill a unique evolutionary niche compared to other vertebrate models.62,63,70–72 To date, multiple studies have used axolotls as a comparative model of development. Axolotls are unique in their lack of Pax3, which drives neural crest and mesoderm development in multiple vertebrate species.62 Its absence suggests that there are some differences in the molecular mechanisms driving complex developmental processes across species. However, multiple studies identified developmental similarities as well. Analysis of pelvic development in axolotl and lungfish identified conserved mechanisms of chondrogenesis and musculogenesis between the lobe-finned fish and the tetrapod.73 In addition, studies of axolotl dentition identified a conserved ecto-endodermal boundary as the potential mediator of tooth development across multiple species.26 Further, loss of function studies identified that Bapx1 is necessary for jaw joint formation in multiple vertebrates, including axolotls.74 The current body of evo-devo research using axolotl as a bridge animal have identified similarities and differences from genomic to morphological scales. Access to newly developing tools and an annotated genome will advance these studies tremendously in the future.
5 |. THE ULTIMATE REGENERATOR
Although multiple animals have limited capabilities to regenerate, the axolotl has devised unique, extensive, and elegant methods of regenerating multiple tissues (Table 1).111–113 Here, we will focus on a subset of regeneration studies that span both historical and current work. Neoteny places axolotls at a unique intersection of developmental and regenerative research potential. They exhibit scarless wound healing and a lower cancer incidence,6,87 and they are an experimentally accessible organism for cell plasticity studies.3,107 They also develop quite slowly compared to other aquatic organisms such as zebrafish and frogs, lending them to experimental embryology studies, including cell fate tracing methods.21,60,114–119
TABLE 1.
Summary of tissues and structures that regenerate in axolotls after injury
| Tissue | Type of injury | Regeneration (Y/N) | References |
|---|---|---|---|
| Ectoderm derived | |||
| Brain | Incision to left dorsal pallium | Yes, regenerate multiple original neuron populations, but connectivity anomalies do occur | 75–78 |
| Lens | Lensectomy | Yes* (lost 2 weeks after hatching) | 79,80 |
| Retina | Retinectomy | Yes | 81–83 |
| Spinal cord | Amputation and spinal cord transection | Yes | 20,84–86 |
| Skin | Full thickness excisional (FTE) wounding with 4 mm biopsy punches | Yes | 87–90 |
| Sensory hair cells | Laser ablation | Yes | 91–93 |
| Mesoderm derived | |||
| Heart | Ventricular resection and cryoinjury | Yes | 94–98 |
| Ovary | Partial ovariectomy | Yes | 99 |
| Skeletal muscle | Limb amputation | Yes | 16,100,101 |
| Endoderm derived | |||
| Gills | Partial amputation | Yes | 29 |
| Lung | Partial amputation | Yes | 102 |
| Derived from multiple germ layers | |||
| Jaw/tooth | Dentectomy | Yes, teeth are nerve dependent, lower jaw is independent | 103 |
| Limb | Amputation | Yes | 12,13,15,38,42,104–106 |
| Tail | Amputation | Yes | 10,27,84,91,104,107–110 |
Most tissues within the axolotl regenerate in some capacity, making the species highly valuable for such studies. However, the axolotl does not necessarily employ the same mode of regeneration across tissues (Table 1). For example, dentectomy studies show that tooth regeneration is a nerve-dependent process while the lower jaw can regenerate without innervation.103 Also, axolotls can only regenerate their lens within the first 2 weeks of life, contrasting with other tissues’ extended regenerative capacity.79 Apoptotic tissue degradation is a precursor to the regenerative process following axolotl limb injuries, and an axolotl must remove injured cells and reduce immune cell counts to a specific balance to avoid unwanted damage.120 There are multiple in-depth recent reviews on the subject of axolotl regeneration,112,113,121,122 but the current body of research suggests that salamanders respond to injury signals in a way that is distinct from mammals. One of the secrets to axolotl limb and tail regeneration lies in its ability to form a blastema after injury. The blastema is a region of dedifferentiated cells that forms from underlying tissues at the site of injury.13,21,38,119,123–125 This transient structure interacts with the wound epidermis in similar ways to mesodermal–epidermal interactions seen during embryonic development.55,88 However, dedifferentiation and blastemal formation are not sufficient to drive the formation of a fully functional and patterned limb or tail after amputation. Although not an exhaustive list, signaling from immune cells, nerve cells, the inflammation response, and whole organism proliferation responses also occur and are important for regeneration after injury.14,15,102,126,127
While the body of research on axolotls and other salamanders has uncovered many details of their regenerative potential, the mechanistic basis of neoteny remains largely unknown. However, there may be developmental origins linking the neotenic state of axolotls with their exemplary regeneration capacity.128,129 Recent work has identified that many tissues maintain populations of stem-like cells, allowing for growth, wound healing, and regeneration. Embryonic stem-like cells, including neural crest cells, may be a key to a subset of axolotl regenerative capabilities.130–133 Understanding these mechanisms is crucial to bridge the gap in knowledge between axolotl developmental and regenerative programs.
6 |. FUTURE OF AMBYSTOMA AS A RESEARCH ORGANISM
New tools broaden the horizons for comparative and functional research in axolotls even further (Table 2). Obtaining the sequenced axolotl genome in 2018 and multiple bulk and single-cell transcriptomic atlases of developing and regenerating embryos and tissues have provided a baseline for functional studies, such as the analysis of genes and proteins that aid in regeneration. Specifically, single-cell sequencing atlases have been created from lineage-specific12,14–16 and unbiased13 samples during forelimb regeneration. By performing single-cell characterization of changes in gene expression during multiple stages of axolotl limb regeneration, previous studies identified that connective tissue cells revert to embryonic profiles,12 there are mitochondrial-specific genes supporting regeneration,16 and that this process is paired with changes in the immune response in lineage-specific and unbiased cell populations.13,14 In addition to transcriptomic analyses, recent characterization of the chromatin landscape using the assay for transposase-accessible chromatin using sequencing (ATAC-seq) in regenerating axolotl limbs has added complexity to the story by identifying changes during the eight stages of regeneration.139
TABLE 2.
Summary of recent developments in molecular tools in axolotl research
| Tool | Use | References |
|---|---|---|
| Sequenced genome | Key to study of sequences that regulate development, aid regeneration, and so on | 61–63 |
| Foamy virus | Gene transfer method used for regeneration studies | 134 |
| Baculo virus | Gene transfer and gene overexpression | 135 |
| Vesicular stomatitis virus | Gene transfer and neural cell labeling | 136 |
| Retroviruses | Infection in vivo and in vitro to target specific cell types in regeneration | 137 |
| Germline transgenic strains | Cell tracing and mutagenesis during development and regeneration | 20,23,134,137 |
| Click chemistry | Lung injury in axolotl salamanders induces an organ-wide proliferation response | 20,102,110 |
| Single-cell sequencing | Single-cell transcriptomic datasets from multiple germ layers and cell types in regenerating axolotl tissues | 12,14–16,138 |
| CRISPR-mediated genomic mutation | Implementation of genomic deletion via CRISPR/Cas9-mediated genome editing in axolotl | 10,25,57 |
A wide topic for future axolotl research is the investigation of the unique programming axolotls hold that makes their scarless wound healing, lower incidence of cancer, and regenerative capabilities currently unattainable in other vertebrates. Current research shows that regeneration is not as simple as rebooting embryonic programming; rather, it appears as though wound healing and regeneration responses are much more complex than simple dedifferentiation and redifferentiation, and depend on the type of cells, stage of regeneration, environment, and tissue-type. The process of dedifferentiation is necessary for regeneration of certain tissues, but it does not provide a complete picture of the process. Dedifferentiated cells can cross over to new cell lineages, and single-cell atlases support the dedifferentiation concept in specific tissues.12,138 However, the limits of this fate-switching are yet to be completely tested, and further work is needed to confirm this process across species.27,100 A related gap in knowledge is the exploration of the axolotl immune system in the regeneration of different tissues, although there is strong groundwork laid for future studies in limb, spinal cord, craniofacial, and heart tissues, among others.14,15,97,140 A decline in the immune system is correlated with aging and lack of regeneration in other vertebrates,141 and axolotls must maintain their immune systems in a careful balance to regenerate injured tissues successfully. As most regeneration studies have been performed in immature axolotls, it will be interesting to see how future studies determine whether the same reduction in immune response occurs in mature animals.
Even with the identification of novel genes and cell types during regeneration, the field still lacks many of the tools that would allow for fast analysis of functional relevance to define mechanisms, identify gene regulatory networks, and link the similarities and differences between development and regeneration in axolotls. However, with the founding of the International Society for Regenerative Biology (https://internationalsocietyforregenerativebiology.org/), the creation of AxoBase, a new online axolotl resource (https://www.axobase.org/), the NIH-funded support for Sal-Site (https://ambystoma.uky.edu/quick-links/sal-site) and annual Salamander Meetings bringing together researchers from across the globe, the future of research using the axolotl as a model organism is promising.
7 |. POP CULTURE ICONS
Secondary to their obvious importance in discovering the secrets of vertebrate regeneration and to the potential discoveries of new developmental pathways, these anomalous salamanders are pop culture icons. Axolotls are conspicuously adorable and hold a high status in the modern world. They are incorporated into video games, cartoons, and social media posts. We would be remiss to omit the charismatic draw of this animal in society. In Japan, the axolotl is known as the wooper looper/rooper (https://www.caudata.org), made popular by a commercial marketing campaign that was then followed by the creation of an axolotl named Wooper in Pokémon cartoons and video games. Further, other popular video games such as Fortnite and Minecraft introduced axolotls as passive characters in 2021,142 and Build-A-Bear created a buildable axolotl plush toy. On social media outlets like Twitter, axolotls are used for scientific communication and public engagement (e.g., #ChonkTheAxolotl), but are also popular in avatars and art. Most recently, the axolotl has been featured on the 50 peso bill from the Bank of Mexico as a representative of ancient Mexico. These animals provide the scientific community with answers while they provide the world with joy.
8 |. EXPERIMENTAL PROCEDURES
8.1 |. Animal Husbandry
All use of axolotl adults and embryos was performed in accordance with the UC Davis approved IACUC protocol #21448. Axolotls were bred in house and embryos were collected for fixation at multiple stages in preparation for imaging. Embryos were fixed in 4% paraformaldehyde solution in 2% phosphate buffer for 1 hour and then were either dehydrated step-wise into 100% methanol for storage or were imaged immediately in 1× TBS (500 mM Tris-HCl pH 7.4, 1.5 M NaCl, and 10 mM CaCl2) containing 0.1% Triton X-100 (TBST+ Ca2+).
8.2 |. Imaging
All whole mount embryos (Figure 1) were imaged using a Zeiss Microscopy Camera Axiocam 208 color mounted to a Zeiss Stemi 305 dissecting microscope. Zen Blue software was used for processing.
ACKNOWLEDGMENTS
The authors would like to thank the Rogers Lab at UC Davis for their efforts in caring for the axolotl colony. Special thanks to Mr. Connor Christensen for his assistance with rearing and collecting axolotl embryos and to the UC Davis PREP Program, who supported Mr. Christensen. The authors dedicate this article to #ChonkTheAxolotl and her siblings, who have brought unending joy to our lab and to the scientific community.
REFERENCES
- 1.Riquelme-Guzm an C, Schuez M, Böhm A, et al. Postembryonic development and aging of the appendicular skeleton in Ambystoma mexicanum. Dev Dyn. 2022;251(6):1015–1034. 10.1002/dvdy.407 [DOI] [PubMed] [Google Scholar]
- 2.IUCN. IUCN SSC Amphibian Specialist Group. 2020. Ambystoma mexicanum. The IUCN Red List of Threatened Species 2020: e.T1095A53947343
- 3.Voss SR, Woodcock MR, Zambrano L. A tale of two axolotls. Bioscience. 2015;65(12):1134–1140. doi: 10.1093/biosci/biv153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Demircan T, Ilhan AE, Ayturk N, Yildirim B, Ozturk G, Keskin I. A histological atlas of the tissues and organs of neotenic and metamorphosed axolotl. Acta Histochem. 2016; 118(7):746–759. doi: 10.1016/j.acthis.2016.07.006 [DOI] [PubMed] [Google Scholar]
- 5.Ryan ME, Johnson JR, Fitzpatrick BM. Invasive hybrid tiger salamander genotypes impact native amphibians. Proc Natl Acad Sci U S A. 2009;106(27):11166–11171. doi: 10.1073/pnas.0902252106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Demircan T, Ovezmyradov G, Yıldırım B, et al. Experimentally induced metamorphosis in highly regenerative axolotl (ambystoma mexicanum) under constant diet restructures microbiota. Sci Rep. 2018;8(1):10974. doi: 10.1038/s41598-018-29373-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Monaghan JR, Stier AC, Michonneau F, et al. Experimentally induced metamorphosis in axolotls reduces regenerative rate and fidelity. Regeneration (Oxf). 2014;1(1):2–14. doi: 10.1002/reg2.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fei JF, Schuez M, Knapp D, Taniguchi Y, Drechsel DN, Tanaka EM. Efficient gene knockin in axolotl and its use to test the role of satellite cells in limb regeneration. Proc Natl Acad Sci U S A. 2017;114(47):12501–12506. doi: 10.1073/pnas.1706855114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lou WP-K, Wang L, Long C, Liu L, Fei J-F. Direct Gene Knock-out of Axolotl Spinal Cord Neural Stem Cells via Electroporation of CAS9 Protein-gRNA Complexes. Journal of Visualized Experiments. 2019;(149). 10.3791/59850 [DOI] [PubMed] [Google Scholar]
- 10.Fei JF, Schuez M, Tazaki A, Taniguchi Y, Roensch K, Tanaka EM. CRISPR-mediated genomic deletion of Sox2 in the axolotl shows a requirement in spinal cord neural stem cell amplification during tail regeneration. Stem Cell Rep. 2014;3(3):444–459. doi: 10.1016/j.stemcr.2014.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanor LD, Flowers GP, Crews CM. Multiplex CRISPR/Cas screen in regenerating haploid limbs of chimeric Axolotls. eLife. 2020;9. 10.7554/elife.48511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerber T, Murawala P, Knapp D, et al. Single-cell analysis uncovers convergence of cell identities during axolotl limb regeneration. Science. 2018;362(6413). 10.1126/science.aaq0681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leigh ND, Dunlap GS, Johnson K, et al. Transcriptomic landscape of the blastema niche in regenerating adult axolotl limbs at single-cell resolution. Nat Commun. 2018;9(1):5153. doi: 10.1038/s41467-018-07604-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodgers AK, Smith JJ, Voss SR. Identification of immune and non-immune cells in regenerating axolotl limbs by single-cell sequencing. Exp Cell Res. 2020;394(2):112149. doi: 10.1016/j.yexcr.2020.112149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Wei X, Zhou L, et al. Dynamic cell transition and immune response landscapes of axolotl limb regeneration revealed by single-cell analysis. Protein Cell. 2021;12(1):57–66. doi: 10.1007/s13238-020-00763-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qin T, Fan CM, Wang TZ, et al. Single-cell RNA-seq reveals novel mitochondria-related musculoskeletal cell populations during adult axolotl limb regeneration process. Cell Death Differ. 2021;28(3):1110–1125. doi: 10.1038/s41418-020-00640-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ozkucur N, Epperlein HH, Funk RH. Ion imaging during axolotl tail regeneration in vivo. Dev Dyn. 2010;239(7):2048–2057. doi: 10.1002/dvdy.22323 [DOI] [PubMed] [Google Scholar]
- 18.Masselink W, Tanaka EM. Toward whole tissue imaging of axolotl regeneration. Dev Dyn. 2021;250(6):800–806. doi: 10.1002/dvdy.282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Currie JD, Kawaguchi A, Traspas RM, Schuez M, Chara O, Tanaka EM. Live imaging of axolotl digit regeneration reveals spatiotemporal choreography of diverse connective tissue progenitor pools. Dev Cell. 2016;39(4):411–423. doi: 10.1016/j.devcel.2016.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duerr TJ, Jeon EK, Wells KM, et al. A constitutively expressed fluorescent ubiquitination-based cell-cycle indicator (FUCCI) in axolotls for studying tissue regeneration. Development. 2022;149(6). 10.1242/dev.199637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muneoka K, Fox WF, Bryant SV. Cellular contribution from dermis and cartilage to the regenerating limb blastema in axolotls. Dev Biol. 1986;116(1):256–260. doi: 10.1016/0012-1606 [DOI] [PubMed] [Google Scholar]
- 22.Epperlein H, Meulemans D, Bronner-Fraser M, Steinbeisser H, Selleck MA. Analysis of cranial neural crest migratory pathways in axolotl using cell markers and transplantation. Development. 2000;127(12):2751–2761. [DOI] [PubMed] [Google Scholar]
- 23.Sobkow L, Epperlein HH, Herklotz S, Straube WL, Tanaka EM. A germline GFP transgenic axolotl and its use to track cell fate: dual origin of the fin mesenchyme during development and the fate of blood cells during regeneration. Dev Biol. 2006;290(2):386–397. doi: 10.1016/j.ydbio.2005.11.037 [DOI] [PubMed] [Google Scholar]
- 24.Epperlein HH, Selleck MA, Meulemans D, et al. Migratory patterns and developmental potential of trunk neural crest cells in the axolotl embryo. Dev Dyn. 2007;236(2):389–403. doi: 10.1002/dvdy.21039 [DOI] [PubMed] [Google Scholar]
- 25.Flowers GP, Sanor LD, Crews CM. Lineage tracing of genome-edited alleles reveals high fidelity axolotl limb regeneration. eLife. 2017;6. 10.7554/elife.25726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soukup V, Tazaki A, Yamazaki Y, et al. Oral and palatal dentition of axolotl arises from a common tooth-competent zone along the Ecto-endodermal boundary. Front Cell Dev Biol. 2020;8:622308. doi: 10.3389/fcell.2020.622308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Echeverri K, Tanaka EM. Ectoderm to mesoderm lineage switching during axolotl tail regeneration. Science. 2002; 298(5600):1993–1996. doi: 10.1126/science.1077804 [DOI] [PubMed] [Google Scholar]
- 28.Hurtado C, De Robertis EM. Neural induction in the absence of organizer in salamanders is mediated by MAPK. Dev Biol. 2007;307(2):282–289. doi: 10.1016/j.ydbio.2007.04.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saito N, Nishimura K, Makanae A, Satoh A. Fgf- and bmp-signaling regulate gill regeneration in Ambystoma mexicanum. Dev Biol. 2019;452(2):104–113. doi: 10.1016/j.ydbio.2019.04.011 [DOI] [PubMed] [Google Scholar]
- 30.Baddar NW, Woodcock MR, Khatri S, Kump DK, Voss SR. Sal-site: research resources for the Mexican axolotl. Methods Mol Biol. 2015;1290:321–336. doi: 10.1007/978-1-4939-2495-0_25 [DOI] [PubMed] [Google Scholar]
- 31.Smith JJ, Putta S, Walker JA, et al. Sal-site: integrating new and existing ambystomatid salamander research and informational resources. BMC Genomics. 2005;6:181. doi: 10.1186/1471-2164-6-181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Woodcock MR, Vaughn-Wolfe J, Elias A, et al. Identification of mutant genes and Introgressed Tiger salamander DNA in the laboratory axolotl, Ambystoma mexicanum. Sci Rep. 2017; 7(1):6. doi: 10.1038/s41598-017-00059-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reiss C, Olsson L, Hossfeld U. The history of the oldest self-sustaining laboratory animal: 150 years of axolotl research. J Exp Zool B Mol Dev Evol. 2015;324(5):393–404. doi: 10.1002/jez.b.22617 [DOI] [PubMed] [Google Scholar]
- 34.Malacinski GM. The Mexican axolotl, Ambystoma mexicanum: its biology and developmental genetics, and its autonomous cell-lethal genes. Am Zool. 1978;18(2):195–206. doi: 10.1093/icb/18.2.195 [DOI] [Google Scholar]
- 35.Vance E Biology’s beloved amphibian - the axolotl - is racing towards extinction. Nature. 2017;551(7680):286–289. doi: 10.1038/d41586-017-05921-w [DOI] [PubMed] [Google Scholar]
- 36.Smith SC, Bashir NS, Armstrong JB. Redneck, a new mutant of the axolotl (Ambystoma mexicanum) likely affects the development of cranial neural crest. Int J Dev Biol. 2001;45: 685–688. [PubMed] [Google Scholar]
- 37.Vance E The axolotl paradox. Nature. 2017;551(16):286–289. [DOI] [PubMed] [Google Scholar]
- 38.McCusker C, Bryant SV, Gardiner DM. The axolotl limb blastema: cellular and molecular mechanisms driving blastema formation and limb regeneration in tetrapods. Regeneration (Oxf). 2015;2(2):54–71. doi: 10.1002/reg2.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bordzilovskaya NP, Dettlaff TA, Duhon ST. Developmental-stage series of Axolotl embryos. In: Armstrong JB, Malacinski GM, ed. Developmental Biology of the Axolotl. New York, NY: Oxford University Press; 1989:201–219. [Google Scholar]
- 40.Schreckenberg GM, Jacobson AG. Normal stages of development of the axolotl Ambystoma mexicanum. Dev Biol. 1975; 42(2):391–400. doi: 10.1016/0012-1606(75)90343-7 [DOI] [PubMed] [Google Scholar]
- 41.Khattak S, Murawala P, Andreas H, et al. Optimized axolotl (Ambystoma mexicanum) husbandry, breeding, metamorphosis, transgenesis and tamoxifen-mediated recombination. Nat Protoc. 2014;9(3):529–540. doi: 10.1038/nprot.2014.040 [DOI] [PubMed] [Google Scholar]
- 42.Nye HL, Cameron JA, Chernoff EA, Stocum DL. Extending the table of stages of normal development of the axolotl: limb development. Dev Dyn. 2003;226(3):555–560. doi: 10.1002/dvdy.10237 [DOI] [PubMed] [Google Scholar]
- 43.Semlitsch RD. Reproductive strategy of a facultatively paedomorphic salamander Ambystoma talpoideum. Oecologia. 1985; 65(3):305–313. doi: 10.1007/BF00378903 [DOI] [PubMed] [Google Scholar]
- 44.Kakebeen AD, Wills AE. More than just a bandage: Closing the gap between injury and appendage regeneration. Frontiers in Physiology. 2019;10. 10.3389/fphys.2019.00081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Veldhuis JH, Brodland GW, Wiebe CJ, Bootsma GJ. Multi-view robotic microscope reveals the in-plane kinematics of amphibian neurulation. Ann Biomed Eng. 2005;33(6):821–828. doi: 10.1007/s10439-005-3309-2 [DOI] [PubMed] [Google Scholar]
- 46.Bootsma GJ, Brodland GW. Automated 3-D reconstruction of the surface of live early-stage amphibian embryos. IEEE Trans Biomed Eng. 2005;52(8):1407–1414. doi: 10.1109/TBME.2005.851500 [DOI] [PubMed] [Google Scholar]
- 47.Shi R, Borgens RB. Embryonic neuroepithelial sodium transport, the resulting physiological potential, and cranial development. Dev Biol. 1994;165(1):105–116. doi: 10.1006/dbio.1994.1238 [DOI] [PubMed] [Google Scholar]
- 48.Shi R, Borgens RB. Three-dimensional gradients of voltage during development of the nervous system as invisible coordinates for the establishment of embryonic pattern. Dev Dyn. 1995;202(2):101–114. doi: 10.1002/aja.1002020202 [DOI] [PubMed] [Google Scholar]
- 49.Jiang P, Nelson JD, Leng N, et al. Analysis of embryonic development in the unsequenced axolotl: waves of transcriptomic upheaval and stability. Dev Biol. 2017;426(2):143–154. doi: 10.1016/j.ydbio.2016.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sato SM, Sargent TD. Development of neural inducing capacity in dissociated Xenopus embryos. Dev Biol. 1989;134(1): 263–266. [DOI] [PubMed] [Google Scholar]
- 51.Launay C, Fromentoux V, Shi DL, Boucaut JC. A truncated FGF receptor blocks neural induction by endogenous Xenopus inducers. Development. 1996;122(3):869–880. [DOI] [PubMed] [Google Scholar]
- 52.Newth DR. Determination in the cranial neural crest of the axolotl. Development. 1954;2(2):101–105. doi: 10.1242/dev.2.2.101 [DOI] [Google Scholar]
- 53.Epperlein HH, Löfberg J. Xanthophores in chromatophore groups of the premigratory neural crest initiate the pigment pattern of the axolotl larva. Wilehm Roux Arch Dev Biol. 1984; 193(6):357–369. doi: 10.1007/BF00848226 [DOI] [PubMed] [Google Scholar]
- 54.Simoes-Costa M, Bronner ME. Reprogramming of avian neural crest axial identity and cell fate. Science. 2016;352(6293): 1570–1573. doi: 10.1126/science.aaf2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schnapp E, Kragl M, Rubin L, Tanaka EM. Hedgehog signaling controls dorsoventral patterning, blastema cell proliferation and cartilage induction during axolotl tail regeneration. Development. 2005;132(14):3243–3253. doi: 10.1242/dev.01906 [DOI] [PubMed] [Google Scholar]
- 56.Flowers GP, Timberlake AT, McLean KC, Monaghan JR, Crews CM. Highly efficient targeted mutagenesis in axolotl using Cas9 RNA-guided nuclease. Development. 2014;141(10): 2165–2171. doi: 10.1242/dev.105072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Flowers GP, Crews CM. Generating and identifying axolotls with targeted mutations using Cas9 RNA-guided nuclease. Methods Mol Biol. 2015;1290:279–295. doi: 10.1007/978-1-4939-2495-0_22 [DOI] [PubMed] [Google Scholar]
- 58.Moury JD, Jacobson AG. The origins of neural crest cells in the axolotl. Dev Biol. 1990;141(2):243–253. [DOI] [PubMed] [Google Scholar]
- 59.Falck P, Hanken J, Olsson L. Cranial neural crest emergence and migration in the Mexican axolotl (Ambystoma mexicanum). Fortschr Zool. 2002;105(3):195–202. doi: 10.1078/0944-2006-00079 [DOI] [PubMed] [Google Scholar]
- 60.Cerny R, Meulemans D, Berger J, et al. Combined intrinsic and extrinsic influences pattern cranial neural crest migration and pharyngeal arch morphogenesis in axolotl. Dev Biol. 2004; 266(2):252–269. doi: 10.1016/j.ydbio.2003.09.039 [DOI] [PubMed] [Google Scholar]
- 61.Schloissnig S, Kawaguchi A, Nowoshilow S, et al. The giant axolotl genome uncovers the evolution, scaling, and transcriptional control of complex gene loci. Proceedings of the National Academy of Sciences. 2021;118(15). 10.1073/pnas.2017176118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nowoshilow S, Schloissnig S, Fei JF, et al. The axolotl genome and the evolution of key tissue formation regulators. Nature. 2018;554(7690):50–55. doi: 10.1038/nature25458 [DOI] [PubMed] [Google Scholar]
- 63.Smith JJ, Timoshevskaya N, Timoshevskiy VA, Keinath MC, Hardy D, Voss SR. A chromosome-scale assembly of the axolotl genome. Genome Res. 2019;29(2):317–324. doi: 10.1101/gr.241901.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sive HL, Grainger RM, Harland RM. Xenopus laevis in vitro fertilization and natural mating methods. CSH Protoc. 2007; 2007:prot4737. doi: 10.1101/pdb.prot4737 [DOI] [PubMed] [Google Scholar]
- 65.Sive HL, Grainger RM, Harland RM. Dejellying Xenopus laevis embryos. CSH Protoc. 2007;2007:prot4731. doi: 10.1101/pdb.prot4731 [DOI] [PubMed] [Google Scholar]
- 66.Briggs JP. The zebrafish: a new model organism for integrative physiology. Am J Physiol Regul Integr Comp Physiol. 2002; 282(1):R3–R9. doi: 10.1152/ajpregu.00589.2001 [DOI] [PubMed] [Google Scholar]
- 67.Hemmati-Brivanlou A, Frank D, Bolce ME, Brown BD, Sive HL, Harland RM. Localization of specific mRNAs in Xenopus embryos by whole-mount in situ hybridization. Development. 1990;110(2):325–330. [DOI] [PubMed] [Google Scholar]
- 68.Sive HL, Grainger RM, Harland RM. Baskets for In Situ Hybridization and Immunohistochemistry: Figure 1. Cold Spring Harbor Protocols. 2007;2007(8):pdb.prot4777. 10.1101/pdb.prot4777 [DOI] [PubMed] [Google Scholar]
- 69.Smith WC, Harland RM. Expression cloning of noggin, a new dorsalizing factor localized to the Spemann organizer in Xenopus embryos. Cell. 1992;70(5):829–840. doi: 10.1016/0092-8674 [DOI] [PubMed] [Google Scholar]
- 70.Amemiya CT, Alfoldi J, Lee AP, et al. The African coelacanth genome provides insights into tetrapod evolution. Nature. 2013;496(7445):311–316. doi: 10.1038/nature12027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Smith JJ, Kuraku S, Holt C, et al. Sequencing of the sea lamprey (Petromyzon marinus) genome provides insights into vertebrate evolution. Nat Genet. 2013;45(4):415–421. doi: 10.1038/ng.2568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Smith JJ, Timoshevskaya N, Ye CX, et al. The sea lamprey germline genome provides insights into programmed genome rearrangement and vertebrate evolution (vol 50, pg 270, 2018). Nat Genet. 2018;50(11):1617. doi: 10.1038/s41588-018-0199-4 [DOI] [PubMed] [Google Scholar]
- 73.Boisvert CA, Joss JM, Ahlberg PE. Comparative pelvic development of the axolotl (Ambystoma mexicanum) and the Australian lungfish (Neoceratodus forsteri): conservation and innovation across the fish-tetrapod transition. Evodevo. 2013; 4(1):3. doi: 10.1186/2041-9139-4-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lukas P, Olsson L. Bapx1 is required for jaw joint development in amphibians. Evol Dev. 2018;20(6):192–206. doi: 10.1111/ede.12267 [DOI] [PubMed] [Google Scholar]
- 75.Kirsche K, Kirsche W. Regenerative processes in the telencephalon of Ambystoma mexicanum. J Hirnforsch. 1964;7:421–436. [PubMed] [Google Scholar]
- 76.Kirsche K, Kirsche W. Experimental study on the influence of olfactory nerve regeneration on forebrain regeneration of Ambystoma mexicanum. J Hirnforsch. 1964;7(3):315–333. [PubMed] [Google Scholar]
- 77.Amamoto R, Huerta VGL, Takahashi E, Dai G, Grant AK, Fu Z, Arlotta P. Adult axolotls can regenerate original neuronal diversity in response to brain injury. eLife. 2016;5. 10.7554/elife.13998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Burr HS. Regeneration in the brain of amblystoma. I. the regeneration of the forebrain. J Comp Neurol. 1916;26:203–211. doi: 10.1002/cne.900260203 [DOI] [Google Scholar]
- 79.Sousounis K, Athippozhy AT, Voss SR, Tsonis PA. Plasticity for axolotl lens regeneration is associated with age-related changes in gene expression. Regeneration (Oxf). 2014;1(3):47–57. doi: 10.1002/reg2.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Toro E Die Regeneration der Linse in fruhen Entwicklungsstadien bei Amblystoma mexicanum. Wilhelm Roux Arch Entwickl Mech Org. 1932;126(1):185–206. doi: 10.1007/BF00586380 [DOI] [PubMed] [Google Scholar]
- 81.Grigoryan E, Markitantova Y. Cellular and Molecular Pre-conditions for Retinal Pigment Epithelium (RPE) Natural Reprogramming during Retinal Regeneration in Urodela. Biomedicines. 2016;4(4):28. 10.3390/biomedicines4040028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Svistunov SA, Mitashov VI. Proliferative activity of the pigment epithelium and regenerating retinal cells in Ambystoma mexicanum. Ontogenez. 1983;14(6):597–606. [PubMed] [Google Scholar]
- 83.Yew DT. Eye enucleation and regeneration of neural retina in axolotl larvae (Ambystoma mexicanum). Anat Anz. 1985; 158(3):217–229. [PubMed] [Google Scholar]
- 84.Tazaki A, Tanaka EM, Fei JF. Salamander spinal cord regeneration: the ultimate positive control in vertebrate spinal cord regeneration. Dev Biol. 2017;432(1):63–71. doi: 10.1016/j.ydbio.2017.09.034 [DOI] [PubMed] [Google Scholar]
- 85.Walker SE, Sabin KZ, Gearhart MD, Yamamoto K, Echeverri K. Regulation of stem cell identity by miR-200a during spinal cord regeneration. Development. 2022;149(3). 10.1242/dev.200033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Baddar N, Dwaraka VB, Ponomareva LV, Thorson JS, Voss SR. Chemical genetics of regeneration: contrasting temporal effects of CoCl2 on axolotl tail regeneration. Dev Dyn. 2021;250(6):852–865. doi: 10.1002/dvdy.294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Seifert AW, Monaghan JR, Voss SR, Maden M. Skin regeneration in adult axolotls: a blueprint for scar-free healing in vertebrates. PLoS One. 2012;7(4):e32875. doi: 10.1371/journal.pone.0032875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tsai S Inhibition of wound epidermis formation via full skin flap surgery during axolotl limb regeneration. Journal of Visualized Experiments. 2020;(160). 10.3791/61522 [DOI] [PubMed] [Google Scholar]
- 89.Slack MJ. Morphogenetic properties of the skin in axolotl limb regeneration. J Embryol Exp Morphol. 1980;58:265–288. [PubMed] [Google Scholar]
- 90.Wigmore P, Holder N. The effect of replacing different regions of limb skin with head skin on regeneration in the axolotl. J Embryol Exp Morphol. 1986;98:237–249. [PubMed] [Google Scholar]
- 91.Jones JE, Corwin JT. Replacement of lateral line sensory organs during tail regeneration in salamanders: identification of progenitor cells and analysis of leukocyte activity. J Neurosci. 1993;13(3):1022–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jones JE, Corwin JT. Regeneration of sensory cells after laser ablation in the lateral line system: hair cell lineage and macrophage behavior revealed by time-lapse video microscopy. J Neurosci. 1996;16(2):649–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Balak KJ, Corwin JT, Jones JE. Regenerated hair cells can originate from supporting cell progeny: evidence from photo-toxicity and laser ablation experiments in the lateral line system. J Neurosci. 1990;10(8):2502–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cano-Martinez A, Vargas-Gonzalez A, Guarner-Lans V, Prado-Zayago E, Leon-Oleda M, Nieto-Lima B. Functional and structural regeneration in the axolotl heart (Ambystoma mexicanum) after partial ventricular amputation. Arch Cardiol Mex. 2010;80(2):79–86. [PubMed] [Google Scholar]
- 95.Cutie S, Huang GN. Vertebrate cardiac regeneration: evolutionary and developmental perspectives. Cell Regen. 2021; 10(1):6. doi: 10.1186/s13619-020-00068-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yamamoto S, Kashimoto R, Furukawa S, Sakamoto H, Satoh A. Comparing nerve-mediated FGF signalling in the early initiation phase of organ regeneration across mutliple amphibian species. J Exp Zool B Mol Dev Evol. 2021;336(7): 529–539. doi: 10.1002/jez.b.23093 [DOI] [PubMed] [Google Scholar]
- 97.Pedersen K, Rasmussen RK, Dittrich A, Lauridsen H. Cardiac regeneration in the axolotl is unaffected by alterations in leukocyte numbers induced by lipopolysaccharide and prednisolone. BMC Res Notes. 2021;14(1):157. doi: 10.1186/s13104-021-05574-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dittrich A, Lauridsen H. Cryo-injury induced heart regeneration in the axolotl and echocardiography and unbiased quantitative histology to evaluate regenerative progression. Journal of Visualized Experiments. 2021;(171). 10.3791/61966 [DOI] [PubMed] [Google Scholar]
- 99.Erler P, Sweeney A, Monaghan JR. Regulation of injury-induced ovarian regeneration by activation of Oogonial stem cells. Stem Cells. 2017;35(1):236–247. doi: 10.1002/stem.2504 [DOI] [PubMed] [Google Scholar]
- 100.Sandoval-Guzman T, Wang H, Khattak S, et al. Fundamental differences in dedifferentiation and stem cell recruitment during skeletal muscle regeneration in two salamander species. Cell Stem Cell. 2014;14(2):174–187. doi: 10.1016/j.stem.2013.11.007 [DOI] [PubMed] [Google Scholar]
- 101.Wu CH, Huang TY, Chen BS, Chiou LL, Lee HS. Long-duration muscle dedifferentiation during limb regeneration in axolotls. PLoS One. 2015;10(2):e0116068. doi: 10.1371/journal.pone.0116068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jensen TB, Giunta P, Schultz NG, et al. Lung injury in axolotl salamanders induces an organ-wide proliferation response. Dev Dyn. 2021;250(6):866–879. doi: 10.1002/dvdy.315 [DOI] [PubMed] [Google Scholar]
- 103.Makanae A, Tajika Y, Nishimura K, Saito N, Tanaka JI, Satoh A. Neural regulation in tooth regeneration of Ambystoma mexicanum. Sci Rep. 2020;10(1):9323. doi: 10.1038/s41598-020-66142-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Spallanzani L, Maty M. Pre-1801 Imprint Collection (Library of Congress). An Essay on Animal Reproductions London: T. Becket and P.A. de Hondt; 1769:86. [Google Scholar]
- 105.Butler EG. X-radiation and regeneration in Amblystoma. Science. 1931;74(1908):100–101. doi: 10.1126/science.74.1908.100 [DOI] [PubMed] [Google Scholar]
- 106.Joven A, Elewa A, Simon A. Model systems for regeneration: salamanders. Development. 2019;146(14). 10.1242/dev.167700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tanaka EM. Cell differentiation and cell fate during urodele tail and limb regeneration. Curr Opin Genet Dev. 2003;13(5): 497–501. doi: 10.1016/j.gde.2003.08.003 [DOI] [PubMed] [Google Scholar]
- 108.Jaques R The inhibitory action of thiouracil on the melano-genesis of the regenerating tail of axolotl. Experientia. 1950; 6(4):148–149. doi: 10.1007/BF02153096 [DOI] [PubMed] [Google Scholar]
- 109.Al Haj Baddar NW, Chithrala A, Voss SR. Amputation-induced reactive oxygen species signaling is required for axolotl tail regeneration. Dev Dyn. 2019;248(2):189–196. doi: 10.1002/dvdy.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Voss SR, Ponomareva LV, Dwaraka VB, et al. HDAC regulates transcription at the outset of axolotl tail regeneration. Sci Rep. 2019;9(1):6751. doi: 10.1038/s41598-019-43230-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Durant F, Whited JL. Finding solutions for fibrosis: understanding the innate mechanisms used by super-regenerator vertebrates to combat scarring. Adv Sci (Weinh). 2021;8(15): e2100407. doi: 10.1002/advs.202100407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.McCusker C, Monaghan J, Whited J. Salamander models for elucidating mechanisms of developmental biology, evolution, and regeneration: part one. Dev Dyn. 2021;250(6):750–752. doi: 10.1002/dvdy.358 [DOI] [PubMed] [Google Scholar]
- 113.Dwaraka VB, Voss SR. Towards comparative analyses of salamander limb regeneration. J Exp Zool B Mol Dev Evol. 2021; 336(2):129–144. doi: 10.1002/jez.b.22902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rollman-Dinsmore C, Bryant SV. The distribution of marked dermal cells from small localized implants in limb regenerates. Dev Biol. 1984;106(2):275–281. doi: 10.1016/0012-1606 [DOI] [PubMed] [Google Scholar]
- 115.Gillespie LL, Armstrong JB. Induction of triploid and gynogenetic diploid axolotls (Ambystoma-Mexicanum) by hydrostatic-pressure. J Exp Zool. 1979;210(1):117–121. doi: 10.1002/jez.1402100112 [DOI] [Google Scholar]
- 116.Muneoka K, Holler-Dinsmore GV, Bryant SV. A quantitative analysis of regeneration from chimaeric limb stumps in the axolotl. J Embryol Exp Morphol. 1985;90:1–12. [PubMed] [Google Scholar]
- 117.Chardin H, Vilain C, Charlemagne J. Characterization of axolotl heavy and light immunoglobulin chains by monoclonal antibodies. Hybridoma. 1987;6(6):627–635. doi: 10.1089/hyb.1987.6.627 [DOI] [PubMed] [Google Scholar]
- 118.Pescitelli MJ, Stocum DL. The origin of skeletal structures during intercalary regeneration of larval Ambystoma limbs. Dev Biol. 1980;79(2):255–275. doi: 10.1016/0012-1606 [DOI] [PubMed] [Google Scholar]
- 119.Echeverri K, Clarke JD, Tanaka EM. In vivo imaging indicates muscle fiber dedifferentiation is a major contributor to the regenerating tail blastema. Dev Biol. 2001;236(1):151–164. doi: 10.1006/dbio.2001.0312 [DOI] [PubMed] [Google Scholar]
- 120.Bucan V, Peck C-T, Nasser I, Liebsch C, Vogt PM, Strauß S. Identification of axolotl BH3-only proteins and expression in axolotl organs and apoptotic limb regeneration tissue. Biol Open. 2018;7(8):bio036293. doi: 10.1242/bio.036293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tanaka EM. Regeneration. Cell. 2003;113(5):559–562. 10.1016/s0092-8674(03)00395-7 [DOI] [PubMed] [Google Scholar]
- 122.Vieira WA, Wells KM, McCusker CD. Advancements to the axolotl model for regeneration and aging. Gerontology. 2020; 66(3):212–222. doi: 10.1159/000504294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Demircan T Dissecting the molecular signature of spinal cord regeneration in the axolotl model. Cureus. 2020;12(2):e7014. doi: 10.7759/cureus.7014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Demircan T, Hacibektasoglu H, Sibai M, et al. Preclinical molecular signatures of spinal cord functional restoration: optimizing the metamorphic axolotl (Ambystoma mexicanum) model in regenerative medicine. OMICS. 2020;24(6):370–378. doi: 10.1089/omi.2020.0024 [DOI] [PubMed] [Google Scholar]
- 125.Gardiner DM, Muneoka K, Bryant SV. The migration of dermal cells during blastema formation in axolotls. Dev Biol. 1986;118(2):488–493. doi: 10.1016/0012-1606 [DOI] [PubMed] [Google Scholar]
- 126.Wells KM, Kelley K, Baumel M, Vieira WA, McCusker CD. Neural control of growth and size in the axolotl limb regenerate. eLife. 2021;10. 10.7554/elife.68584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Cura Costa E, Otsuki L, Rodrigo Albors A, Tanaka EM, Chara O. Spatiotemporal control of cell cycle acceleration during axolotl spinal cord regeneration. eLife. 2021;10. 10.7554/elife.55665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Shimokawa T, Yasutaka S, Kominami R, Shinohara H. Lmx-1b and Wnt-7a expression in axolotl limb during development and regeneration. Okajimas Folia Anat Jpn. 2013;89(4):119–124. doi: 10.2535/ofaj.89.119 [DOI] [PubMed] [Google Scholar]
- 129.Voss SR, Epperlein HH, Tanaka EM. Ambystoma mexicanum, the axolotl: a versatile amphibian model for regeneration, development, and evolution studies. Cold Spring Harb Protoc. 2009;2009(8):emo128. doi: 10.1101/pdb.emo128 [DOI] [PubMed] [Google Scholar]
- 130.Kruger GM, Mosher JT, Bixby S, Joseph N, Iwashita T, Morrison SJ. Neural crest stem cells persist in the adult gut but undergo changes in self-renewal, neuronal subtype potential, and factor responsiveness. Neuron. 2002;35(4):657–669. doi: 10.1016/s0896-6273(02)00827-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zeuner MT, Didenko NN, Humphries D, et al. Isolation and characterization of neural crest-derived stem cells from adult ovine palatal tissue. Front Cell Dev Biol. 2018;6:39. doi: 10.3389/fcell.2018.00039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mehrotra P, Tseropoulos G, Bronner ME, Andreadis ST. Adult tissue-derived neural crest-like stem cells: sources, regulatory networks, and translational potential. Stem Cells Transl Med. 2020;9(3):328–341. doi: 10.1002/sctm.19-0173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Moghadasi Boroujeni S, Koontz A, Tseropoulos G, et al. Neural crest stem cells from human epidermis of aged donors maintain their multipotency in vitro and in vivo. Sci Rep. 2019;9(1):9750. doi: 10.1038/s41598-019-46140-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Khattak S, Sandoval-Guzman T, Stanke N, Protze S, Tanaka EM, Lindemann D. Foamy virus for efficient gene transfer in regeneration studies. BMC Dev Biol. 2013;13:17. doi: 10.1186/1471-213X-13-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Oliveira CR, Lemaitre R, Murawala P, Tazaki A, Drechsel DN, Tanaka EM. Pseudotyped baculovirus is an effective gene expression tool for studying molecular function during axolotl limb regeneration. Dev Biol. 2018;433(2):262–275. doi: 10.1016/j.ydbio.2017.10.008 [DOI] [PubMed] [Google Scholar]
- 136.Mundell NA, Beier KT, Pan YA, et al. Vesicular stomatitis virus enables gene transfer and transsynaptic tracing in a wide range of organisms. J Comp Neurol. 2015;523(11):1639–1663. doi: 10.1002/cne.23761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Whited JL, Tsai SL, Beier KT, et al. Pseudotyped retroviruses for infecting axolotl in vivo and in vitro. Development. 2013; 140(5):1137–1146. doi: 10.1242/dev.087734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lin TY, Gerber T, Taniguchi-Sugiura Y, et al. Fibroblast dedifferentiation as a determinant of successful regeneration. Dev Cell. 2021;56(10):1541–1551 e6. doi: 10.1016/j.devcel.2021.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wei X, Li H, Guo Y, et al. An ATAC-seq dataset uncovers the regulatory landscape during axolotl limb regeneration. Front Cell Dev Biol. 2021;9:651145. doi: 10.3389/fcell.2021.651145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Charbonneau AM, Astrom P, Salo T, Roy S, Tran SD. Axolotls’ and mices’ oral-maxillofacial trephining wounds heal differently. Cells Tissues Organs. 2021;210(4):260–274. doi: 10.1159/000518036 [DOI] [PubMed] [Google Scholar]
- 141.Weyand CM, Goronzy JJ. Aging of the immune system. Mechanisms and therapeutic targets. Ann Am Thorac Soc. 2016;13-(Suppl 5):S422–S428. doi: 10.1513/AnnalsATS.201602-095AW [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Needleman SE, Rodriguez S. Videogames ‘Fortnite,’ ‘Minecraft’ catapult smiley salamander to global fame. Wall Street J. 2022. https://www.wsj.com/articles/videogames-fortnite-minecraft-catapult-smiley-salamander-to-global-fame-11646666221 [Google Scholar]


