Abstract
Background:
CD4+T cell responses to apolipoprotein B are well characterized in atherosclerotic mice and detectable in humans. CD4+T cells recognize antigenic peptides displayed on highly polymorphic Human Leukocyte Antigen-II. Immunogenicity of individual APOB peptides is largely unknown in humans. Only one HLA-II-restricted epitope was validated using the DRB1*07:01-APOB3036–3050 tetramer. We hypothesized that human APOB may contain discrete immunodominant CD4+T cell epitopes that trigger atherosclerosis-related autoimmune responses in donors with diverse HLA alleles.
Methods:
We selected twenty APOB-derived peptides (APOB20) from an in silico screen and experimentally validated binding to the most commonly occurring human HLA-II alleles. We optimized a restimulation-based workflow to evaluate antigenicity of multiple candidate peptides in HLA-typed donors. This included activation-induced marker (AIM) assay, intracellular cytokine staining (ICS), IFNУ–enzyme-linked immunospot (ELISpot) and Cytometric Bead Array (CBA). High-throughput sequencing delineated TCR clonalities of APOB-reactive CD4+T cells.
Results:
Using stringent positive, negative and crossover-stimulation controls, we confirmed specificity of expansion-based protocols to detect CD4+T cytokine responses to APOB20 pool. Ex vivo assessment of AIM+CD4+T cells revealed statistically significant autoimmune response to APOB20, but not to a ubiquitously-expressed negative control protein, actin. Resolution of CD4+T responses to the level of individual peptides using IFNУ-ELISpot, led to the discovery of six immunodominant epitopes (APOB6) that triggered robust CD4+T activation in most donors. APOB6-specific responding CD4+T cells were enriched in unique expanded TCR clonotypes and preferentially expressed memory markers. CBA analysis detected APOB6-induced secretion of both pro-inflammatory and regulatory cytokines. In clinical samples from patients with angiographically verified coronary artery disease (CAD), APOB6 stimulation induced higher activation and memory phenotypes, and augmented secretion of proinflammatory cytokines TNF and IFNУ, compared to patients with low CAD.
Conclusions:
Using three cohorts, each with ~20 donors, we discovered and validated six immunodominant, HLA-II-restricted APOB epitopes. Immune response to these APOB epitopes correlated with CAD severity.
Keywords: apolipoprotein B, immunodominant, HLA-II-restricted, TCR clonotypes, CAD
Graphical Abstract
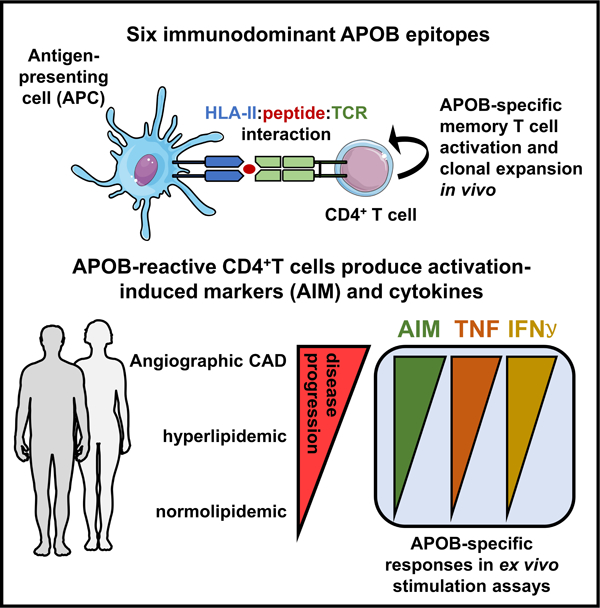
Introduction
Atherosclerosis, a pathophysiology that underlies many cardiovascular diseases, is an inflammatory disease of the artery wall with an obligatory secondary autoimmune component1–3. Negative selection of T cells in the thymus is incomplete and autoreactive CD4 and CD8 T cells are detectable in human blood4–6. Studies in transgenic mice that modelled tissue-specific expression patterns of endogenous antigens, demonstrated that autoreactive T cell responses against certain tissue-restricted self-antigens, unlike those targeting ubiquitously expressed proteins, are comparable to T cell responses to foreign antigen7, 8.
Recent studies have highlighted the involvement of an autoantigen-specific adaptive immune network in atherosclerosis3. Antigen-specific and MHC-II-restricted productive recall interactions between antigen-experienced CD4+T cells and antigen presenting cells (APCs) occur in the aortic walls of atherosclerotic mice9. Oligoclonal proliferation and expression of activation markers were observed in plaque-associated T cells, indicating that human T cells respond to unknown antigens in the vessel wall10, 11.
The best evidence for a relevant atherosclerosis antigen supports a major role for low-density lipoprotein (LDL) and its core protein Apolipoprotein B (APOB)12–16. Both humans and mice have circulating antibodies to native or modified LDL and APOB. While the antibodies against modified lipid moieties of LDL are predominantly of the immunoglobulin M (IgM) isotype, most antibodies to the protein component APOB are of the IgG isotype17–19. To produce IgG, B cells must undergo isotype switching, which requires help from activated CD4+ follicular helper T cells20. Thus, the existence of IgG antibodies to human APOB indicates that APOB-specific human CD4+T cells exist in vivo.
Peptide presentation on heterodimeric Human Leukocyte Antigen (HLA) Class II molecules (encoded by DP, DQ and DR alpha and beta chain genes)21 engages epitope-specific αβ T Cell Receptors (TCRs). Human HLA-II genes are highly polymorphic, with most numbers of allelic variants being reported for DP, DQ, DR beta chain (B) genes21. The αβTCR clonotypic repertoire is variable22 and predominantly consists of individual-specific private TCRs23. Based on immunogenicity studies in other autoimmune-related inflammatory diseases4, 24, 25, only specific TCR-HLA-II-epitope trimolecular combinations trigger CD4+T cell activation26. For this reason, it is important to resolve antigen-specific responses to single epitopes in HLA-typed donors and identify antigenic candidates that elicit strong CD4+T activation in a large fraction of individuals27.
Early attempts to elucidate the identities of human APOB-specific CD4+T cells involved characterization of in vitro stimulated splenocytes and T cell hybridomas generated from oxidized-LDL immunized human ApoB100 (huB100tg) transgenic mice28, 29. This study revealed preferential VDJ usage of antigen-specific mouse CD4+T cell clones. However, APOB-responsive human CD4+T cells remained unidentified. Using a DRB1*07:01 Class-II tetramer molecule, loaded with human APOB3036–3050, we previously provided the first evidence that APOB epitope-specific CD4+T cells exist in human peripheral blood mononuclear cells (PBMCs)1. Since an individual tetramer can screen only one HLA-II-peptide combination at a time, this study could not assess antigenicity of ABOB3036–3050 in the context of other HLA-II alleles or in comparison to other peptides.
Here, we systematically shortlisted 20 APOB epitopes from an in silico screen and HLA-II binding assays. We examined APOB-specific CD4+T responses using an optimized and integrated re-stimulation-based workflow that includes – (1) activation-induced marker (AIM) assay that facilitates detection and isolation of antigen-specific CD4+T cells based on the surface expression of antigen-induced markers, (2) enzyme-linked immune absorbent spot (ELISpot) assay for screening and deconvolution of responses at single epitope resolution, (3) flow-cytometry-based intracellular cytokine staining (ICS) assays for multiparametric assessment of cytokine producing epitope-specific T cells, and (4) cytometric bead array (CBA) to profile peptide-induced T helper cytokine secretion.
METHODS
Data Availability
All data, methods and essential research materials used for research and analysis have been provided. A detailed Material and Methods section is available in the Online Data Supplement. Additional data related to peptides and TCR sequences are detailed in Additional Files 1–3.
RESULTS
Broadly binding MHC Class-II restricted APOB peptides elicit antigen-specific responses in human CD4+T cells
The core HLA-II binding region of an epitope is a nonameric sequence. Because HLA-II molecules have an open binding pocket, a 15-mer sequence is typically analyzed (3 aa overhang on each side). The entire 4,563 aa long human APOB protein contains 4,549 possible 15-mer sequences. Those overlapping by >10 residues were removed because these sequences redundantly span the same nonameric core. Prediction tools available from the Immune Epitope Database (IEDB) were used to score the remaining 911 peptides for their ability to bind a reference set of the 27 most frequent and representative HLA DP, DQ and DR alleles (Table S1) which provide an estimated global coverage of >98% of individuals belonging to all the major races and ethnic groups30. Peptides predicted to bind most (≥75%) of the alleles, were experimentally verified in HLA-II binding assays31. For each peptide, the number of alleles that bound at <1000nM, was tabulated. The top 20 promiscuous binders (bind >50% of reference alleles) were selected for further study (Figure S1A, Table S2). This approach is supported by previous observations that the promiscuous HLA-II binders tend to be the most immunogenic27, 32. Homology analysis with BLAST and PEPMatch (Additional File 1) showed that these peptides did not share any significant sequence identity with other known epitopes in the IEDB database33. Using this APOB20 peptide pool, we optimized a set of re-stimulation-based assays (Table S3).
First, we established an in vitro expansion-based protocol (Figure S1B) that maximizes the sensitivity and reliability of the quantitation of autoantigen-specific CD4+T responses. Similar protocols have been used to resolve epitope-specific responses to self-antigens in other autoimmune-related diseases4. Expansion and restimulation of PBMCs with the APOB20 pool and multiparametric flow cytometry-based intracellular cytokine staining (ICS; antibodies in Table S4) detected significantly higher T helper cytokine responses in CD40L+CD4+T activated cells, as compared to unstimulated controls (Figure S1C). Most APOB-specific CD4+T cells produced Th1 cytokines, some produced IL-4 and others IL-17A. APOB20-induced IL-10 was detectable (Figure S1C) but rare (median frequency only 0.002% of CD4+T cells) (Figure S1D).
The efficacy and specificity of our expansion-based method was assessed in a crossover re-stimulation assay in which PBMCs were expanded with a peptide pool and then re-stimulated with either the cognate or a different pool. We utilized two sets of peptide pools – the experimental APOB20 pool and a positive control CEFX-II pool containing 68 epitopes from infectious agents, which are known to bind a broad range of HLA class-II alleles. PBMCs were expanded with either the APOB20 or the CEFX-II pool. Subsequent re-stimulation on day 14 with the cognate pool elicited positive responses in an IFNγ ELISpot assay (Figure 1A). Cross-over re-stimulation with the opposite pool yielded no responses. Unstimulated and PHA-L treated cells served as negative and positive controls, respectively. Average response magnitudes, expressed as spot forming cells (SFCs) per 106 PBMCs, were significantly higher for the cognate pool than to the opposite pool (Figure 1B). Specificity was further confirmed in an ICS assay for multiple CD4+T helper cytokines: TNF (Figure 1C), IFNγ (Figure 1D), IL-4 (Figure 1E) and IL-17A (Figure 1F). Significant induction of cytokine response over unstimulated background controls was detected with the cognate, but not with the non-cognate pool (Figure 1G–J). This shows that the expansion process boosts sensitive detection of an APOB-specific T cell response and is strictly antigen specific.
Figure 1: In vitro expanded antigen-specific T cells respond specifically to cognate peptides upon subsequent re-stimulation.
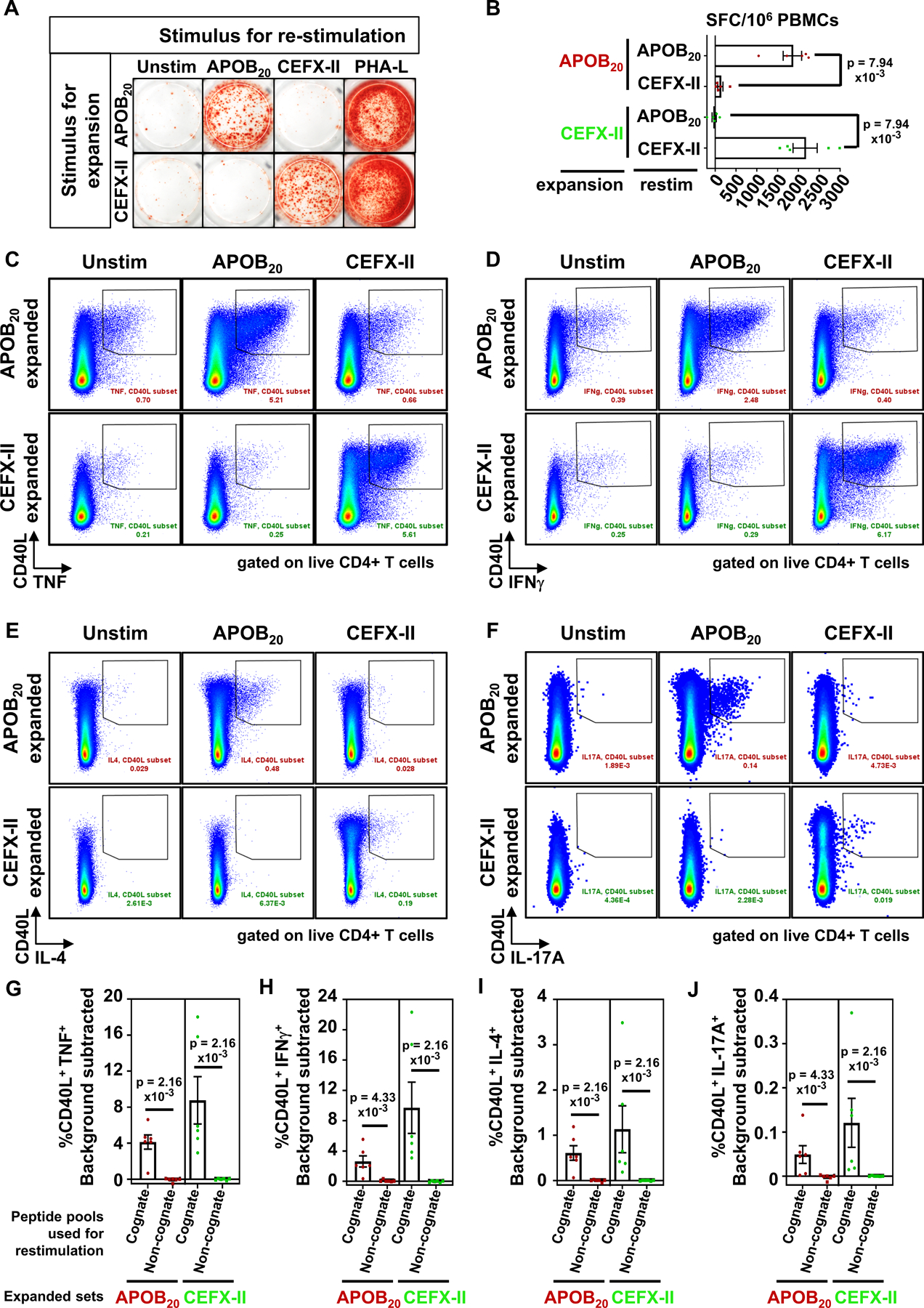
Human PBMCs were expanded for 14 days with the APOB20 or CEFX-II pool and then re-stimulated with the cognate or the opposite peptide pools. Antigen-induced T cell responses were monitored using IFNγ ELISpot assay (A and B) or FACS-based intracellular cytokine staining for TNF (C,G), IFNγ (D,H), IL-4 (E,I) and IL-17A (F,J) producing CD4+T cells. A) Representative images from an IFNγ ELISpot assay after 24h of re-stimulation. PHA-L: positive control. B) Numbers of IFNγ–secreting cells in stimulated sets minus the background signal in unstimulated controls, expressed as spot forming cells (SFCs) per 106 PBMCs. Five independent donors. C-F) Representative FACS plots gated on singlets, live, dump−, CD3+, CD8−, CD4+T cells, showing %CD40L+cytokine+ populations after 6h of re-stimulation. (F). G-J) Background subtracted frequencies of %CD40L+cytokine+ CD4+T cells in cognate or non-cognate pool-stimulated PBMCs. Six independent donors. Colored symbols represent data from individual donors. Bars show mean with standard error of mean (SEM). Statistical comparisons between cognate and non-cognate pools (B, G-J) were performed using Mann-Whitney test.
Significant autoreactive response to human APOB is detectable in the general population
Next, we optimized a 24h assay (Figure S2A–C) which is based on the surface expression of T cell activation-induced markers (AIM) and has been used in a range of studies to identify CD4+T cell responses against vaccine candidates and pathogen-derived epitopes34–37. It has been previously established that addition of anti-CD40 antibody during the stimulation phase inhibits CD40-CD40L interaction-induced internalization of CD40L and improves CD40L detection on the cell surface38. We found that the 24h activation regime, as opposed to 6h stimulation, improved the intensities of CD40L and CD69 surface expression and lowered the background in unstimulated controls (Figure S2C). This enhanced the signal to noise ratio of the antigen-specific CD4+T response (Figure S2C), allowing sensitive detection and accurate enumeration of low-frequency autoantigen-specific CD4+T cells in ex vivo assays. HLA-II restriction was verified using a pan HLA-II blocking antibody, which strongly inhibited APOB20-induced AIM marker upregulation (Figure S2D). Ex vivo stimulation of PBMCs from 21 donors (demographics in Table S5) with the APOB20 pool for 24h in the presence of anti-CD40 antibody significantly enhanced the frequencies of AIM+ (%CD40L+CD69+) CD4+ T cells in 100% of the donors tested (p<0.0001 versus unstimulated control, Figure 2A).
Figure 2: Activation-induced surface expression of CD40L and CD69 (AIM assay) allows sensitive detection of APOB-specific CD4+T cells in ex vivo stimulated PBMCs.
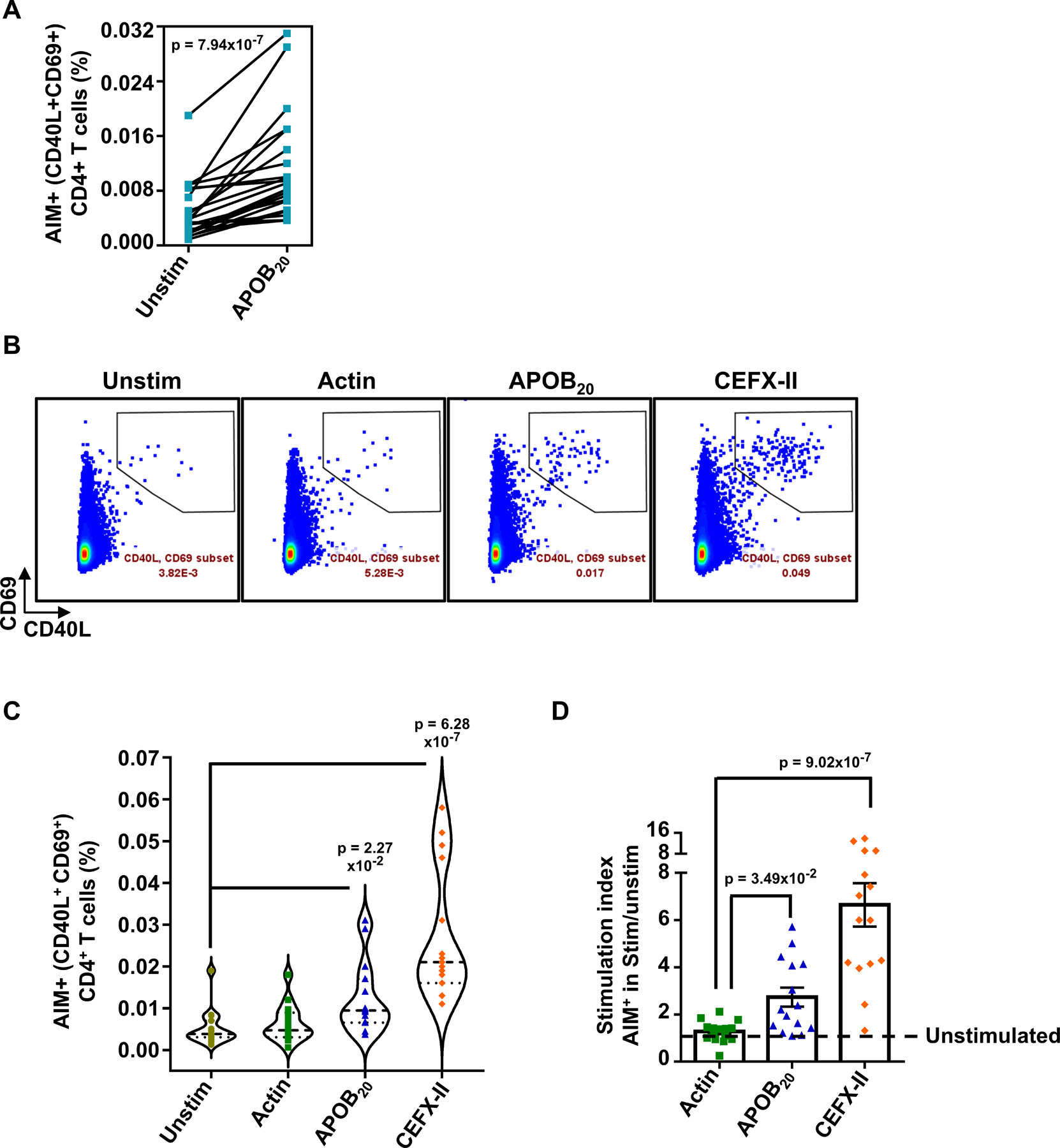
A) %CD40L+CD69+ (AIM+) CD4+T cells in paired sets of APOB20 stimulated and unstimulated PBMCs. 21 independent donors. B) Representative FACS plots and C) Violin plots showing median frequencies of %CD40L+CD69+ (AIM+) CD4+ T cells in unstimulated and 24h Actin pool or APOB20 pool or CEFX-II pool-stimulated PBMCs. D) Average stimulation indices (%CD40L+CD69+ CD4 T cells in stimulated sets over those in unstimulated control) for Actin, APOB20 and CEFX-II peptide pools. 15 independent donors. Colored symbols represent data from individual donors. Pairwise statistical comparisons (A) were performed with the Wilcoxon test. Statistical tests for mean response across different stimulation conditions and unstimulated (C) or actin (D) sets were performed using Kruskal-Wallis test with Dunn’s multiple comparison testing..
Next, we compared APOB-specific CD4+T responses with those triggered by the positive control CEFX-II pool and a negative control pool containing 92 peptides spanning human α-actin (Figure 2B–D). Actin is ubiquitously expressed and actin-specific T cells are known to undergo strong negative selection8. As compared to unstimulated controls (Mean %AIM+CD4+T = 0.005), stimulation with APOB20 (Mean = 0.012) and CEFX-II (Mean = 0.03) pools, but not with the actin pool (Mean = 0.006), resulted in significant increase in activated CD4+T cells (Figure 2B,C). Stimulation-induced fold increase, commonly expressed as stimulation index (SI = AIM+CD4+T cells in stimulated/unstimulated sets), was significantly higher for both APOB20 (Mean SI=2.73, p=0.0349 versus actin, p=0.0002 versus unstimulated) and CEFX-II (Mean SI=6.65, p<0.0001 versus actin or unstimulated) stimulations (Figure 2D). There was no statistically significant autoimmune response to actin (Mean SI=1.28, n.s.).
Discovery of immunodominant APOB-derived CD4 T cell epitopes
Not all peptides that bind HLA-II alleles exhibit comparable immunogenic potential. To resolve APOB-specific responses to single epitopes, we used the optimized IFNγ ELISpot assay (Figure 1A), because Th1 cytokine response to APOB peptides is more robust than other cytokines (Figure S1D) and is easily detectable in high-throughput screens. PBMCs from HLA-typed donors (demographics and HLA-II alleles in Tables S5, S6) were incubated with the APOB20 pool for 14 days with intermittent IL-2 feeding (schematic in Figure S3A). Subsequent re-stimulation with individual peptides resulted in a mosaic pattern of T cell responses of varying magnitudes that were associated with specific epitopes (Figure 3A). Restimulation with the APOB20 pool and with PHA-L served as positive controls, and unstimulated cells were negative controls. For each donor, APOB peptide-specific restimulation conditions were run in triplicate wells and the average number of spot-forming cells (SFCs) per 106 PBMCs was calculated. To determine responder frequencies, we defined a positive response based on established criteria set for detection of responses to other autoantigenic epitopes4: (1) ≥ 100 spot-forming cells per 106 PBMCs; (2) stimulation index ≥2 (3) p≤ 0.01 by Student’s t-test, mean response in stimulated vs unstimulated replicate wells. Using these criteria, six peptides (#2, 4, 5, 11, 12 and 17) were found to induce positive responses in ≥10 out of 19 donors tested (Figure 3B). The average magnitude of response elicited by each of these six epitopes (Figure 3C) was significantly higher than the average response to all the other epitope candidates (Figure 3D). Scrambled versions of these six epitopes (Figure S3B) elicited negligible responses (Figure S3C), confirming that only the original APOB peptides are recognized by human autoreactive T cells.
Figure 3: Deconvolution of T cell responses to APOB20 peptide pool identifies dominant antigenic sites in APOB.
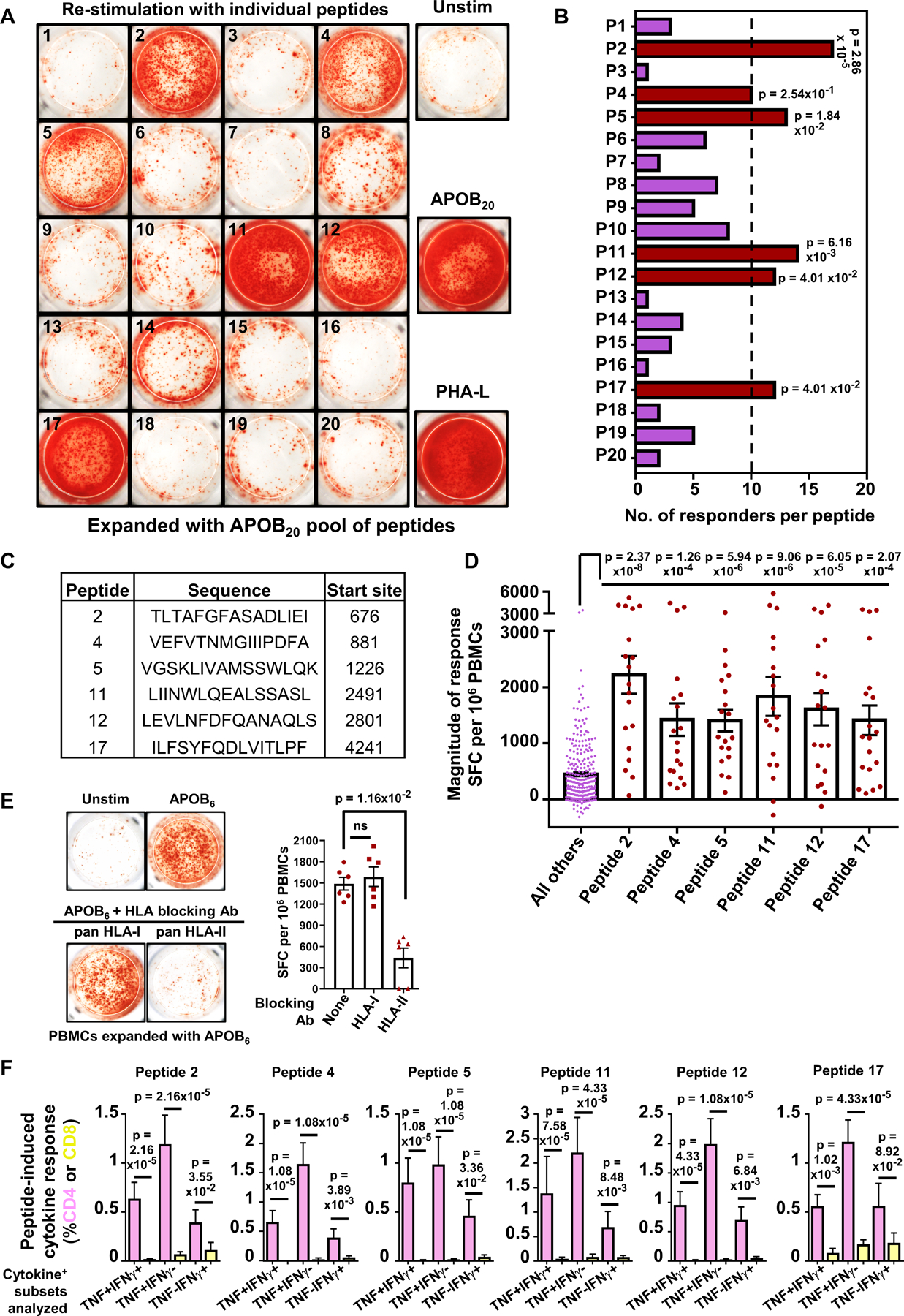
Human PBMCs were expanded with the APOB20 peptide pool and then re-stimulated with each peptide individually. Peptide-specific responses in PBMCs from 19 HLA-typed donors were evaluated using IFNγ ELISpot assay. A) Representative images from IFNγ ELISpot assay. PHA-L: positive control. B) Numbers of donors (X-axis) that respond to each peptide (Y-axis). C) Sequences and start sites of the dominant APOB epitopes. D) Average responses (spot forming cells, SFC) to individual peptides P2, 4, 5, 11, 12 or 17 and to the other 14 peptides (left, purple). E) Representative images of ELISpot wells and quantification showing IFNγ responses in PBMCs expanded and re-stimulated with a pool of dominant epitopes (APOB6), either alone or in the presence of pan-HLA-I or pan-HLA-II (DP+DQ+DR) blocking antibodies. Six independent donors. F) %TNF+IFNγ+, %TNF+IFNγ− and %TNF−IFNγ+ populations among CD4+ (pink) and CD8+ (yellow) T cell subsets are shown for each peptide. Ten independent donors. Colored symbols represent data from individual donors. Bars represent mean with SEM. Statistical tests for peptide-specific responder frequencies (B) were performed using Fisher’s exact test with FDR (false discovery rate) correction. Comparison of response magnitudes across peptides (D) or effect of blocking antibodies (E) were examined using Kruskal-Wallis test with Dunn’s multiple comparison testing. Statistical comparisons between cytokine responses in CD4+ vs CD8+ T cells (F) were performed using Mann-Whitney test. Red: dominant epitopes, purple: remaining peptides (B,D).
IFNγ responses to a combined pool of these HLA-II restricted dominant epitopes (APOB6) were inhibited by pan-HLA-II blocking antibodies, but remained unaffected by pan-HLA-I blocking (Figure 3E). Using HPLC-purified (>95% purity) peptides and flow cytometry-based ICS assays (schematic in Figure S4A), we validated that Th1 cytokine (TNF and IFNγ) responses to all six epitopes were mediated by CD4+T cells, with negligible CD8+T responses (Figure 3F, Figure S4B). None of the peptides induced any significant cell death in the expanded and restimulated T cells (Figure S4C). Furthermore, only cognate epitopes, and not an irrelevant peptide, triggered Th1 responses in activated CD4+T cells (Figure S4D).
Evaluation of ex vivo responses using the 24h AIM assay (Figure 2) revealed significant increase in %CD40L+CD69+CD4+T cells, as compared to unstimulated controls, in APOB6 and CEFX-II stimulated PBMCs, but not in response to the actin pool (Figure S4E).
Next, we used the RATE (Restrictor Analysis Tool for Epitopes) tool (IEDB.org) to computationally infer putative HLA-II associations of the dominant peptides (Additional File 2), based on observed positive responses (Figure 3B) and HLA-II allele expression in the screening cohort (Table S6)39, 40. As expected, these APOB peptides, which were shown to bind multiple HLA-II alleles in our binding assay (Figure S1A), did not exhibit significant positive association with any particular donor HLA-II allele.
To further examine dependency of APOB peptide immunogenicity on their measured binding affinities and expression of binder alleles in the responding donors, we constructed case-control scenarios of binder and non-binder peptide-HLA-II combinations. First, we studied the effect of pan-HLA-DR blockade on the immunogenicity of P2 in two donors (Figure S5A) whose alleles at DRB1 and DRB3/4/5 loci exhibited contrasting binding preferences for P2. A pan-HLA-DR blocking antibody inhibited responses to P2 in donor 1, who expressed HLA-DR alleles with strong binding affinities for P2. Immunogenicity of the same peptide in donor 2, expressing DQ and DP binder alleles and DR non-binder alleles, remained unaffected by HLA-DR blockade. Both donors expressed similar levels of HLA-DR molecules on B cells, the major antigen-presenting cells in peptide-expanded PBMCs.
In another strategy, we used single HLA-II transfected cell lines41 expressing either DPB1*05:01 or DRB3*02:02 (Figure S5B, left) and examined the immunogenicity of two contrasting sets of binder and non-binder APOB peptides (Figure S5B, right). We found that exogenous addition of either binder or non-binder peptide elicited responses in PBMCs, because they express a combination of all HLA-II alleles present in that donor. However, under conditions where peptide presentation was limited to only one HLA-II molecule (DPB1*05:01 or DRB3*02:02 cell lines), we found that the responses to each peptide were dependent on their binding preferences (Figure S5C).
In our deconvolution experiments (Figure 3B), 16 out of 19 donors identified at least 4 of the 6 dominant epitopes, with only one donor not responding to any of them (Figure S6A). Every donor expressed at least one binder allele for each of the six peptides (Figure S6B). Together these data (Figures S5, S6) suggested that although immunogenicity of dominant APOB epitopes relies on specific peptide-HLA-II binder combinations, a combined peptide pool of these six promiscuous dominant epitopes (APOB6) will bind most of the frequently occurring HLA-II alleles (Table S1) and allows evaluation of APOB-specific CD4+T responses in a range of donors, irrespective of their HLA-II allelic heterogeneity.
Dominant APOB epitopes activate oligoclonal populations of APOB-reactive CD4+ T cells
The TCR repertoire, diversity and clonality are important indicators of a T cell-mediated immunological reaction and reflect the immune status of an individual22, 42. To examine the clonotypic identities of autoreactive CD4+ T cells, we sequenced the hypervariable TCRβ CDR3 region using a DNA-based sequencing technique42 (Adaptive Biotechnologies), which have been used to profile clonal populations of other self-antigen specific T cells43. PBMCs from six HLA-typed donors (demographics and HLA-II alleles in Tables S5, S6) were expanded (14-days) and restimulated (24h) with the APOB6 pool. To isolate viable responder and non-responder CD4+T cells from the same pool of stimulated PBMCs (schematic in Figure S7A, gating strategy in Figure S7B), we used the AIM assay protocol (Figure 2). We assessed surface expression of the five most commonly used AIM markers – CD40L, CD69, CD25, OX-40 and 4–1BB, which maximized detection of APOB-induced activated (AIM+) CD4+ T cell repertoire, irrespective of their cytokine secretion potential. AIM− CD4+ T cells that did not express any of the activation markers tested were used as “non-responder” negative controls (gating strategy in Figure S7B). As a second control, unexpanded PBMCs (day 0 samples), sorted into naïve, central memory (TCM) and effector memory (TEM) CD4+T cells, were used (gating strategy in Figure S7C). This allowed tracing the APOB6-specific AIM+ T cell clones in naïve and memory T cell lineages. Genomic DNA, isolated from sorted AIM− and AIM+ Day 14 samples and naïve, TCM and TEM Day 0 samples from six donors, was used as the starting material for TCRVβ sequencing (schematic in Figure S7A) using an optimized two-step, amplification bias-controlled multiplex-PCR-based approach42, 44 (immunoSEQ Human TCRB Assay, Adaptive Biotechnologies).
As most T cells encode only one somatically rearranged TCR45, frequencies of CDR3 templates identified from genomic DNA strongly correlate with the relative abundances of the clonotypic T cells bearing that CDR3 sequence46. For our analysis, we focused on productive templates in which the nucleotide sequence of the CDR3 region is in-frame for protein translation and does not contain any stop codons. A comparison of total vs unique template counts (details in Table S7) detected in AIM+ and AIM− cells revealed an enrichment of a restricted set of unique rearrangements in the responding CD4+T cells (Figure 4A), indicating APOB-dependent selection of specific TCR clones. We determined 14,400 unique TCR rearrangements (Additional File 3) from a total of 149,065 productive TCRβ chain CDR3 templates identified in APOB-specific CD4+T cells that recognized these six immunodominant epitopes. Comparison of the repertoire characteristics of AIM− and AIM+ cells using diversity and clonality metrics47 (Figure 4B), confirmed that AIM+ populations are represented by fewer but more abundant unique clonotypes (higher average Simpson’s clonality). The APOB-AIM+CD4+T cells, and not the AIM−CD4+T cells, were significantly more oligoclonal than Day 0 subsets (Figure S7D), thereby validating the efficacy of the expansion and restimulation regime in detecting epitope specific oligoclonal CD4+T cells in a polyclonal pool of PBMCs. While expansion improves sensitivity, restimulation enforces specificity. Comparing the extent of similarity between APOB-specific AIM+ TCR repertoire and the different ex vivo subsets using Jaccard’s index47 (Figure 4C), revealed that the APOB-specific TCR repertoire exhibited some overlap with central and effector memory CD4+T cell TCRs, but not with naïve populations, suggesting that exposure to these epitopes and memory formation had occurred in vivo.
Figure 4: APOB-derived dominant epitopes trigger expansion and activation of an oligoclonal population of CD4+T cells in human PBMCs.
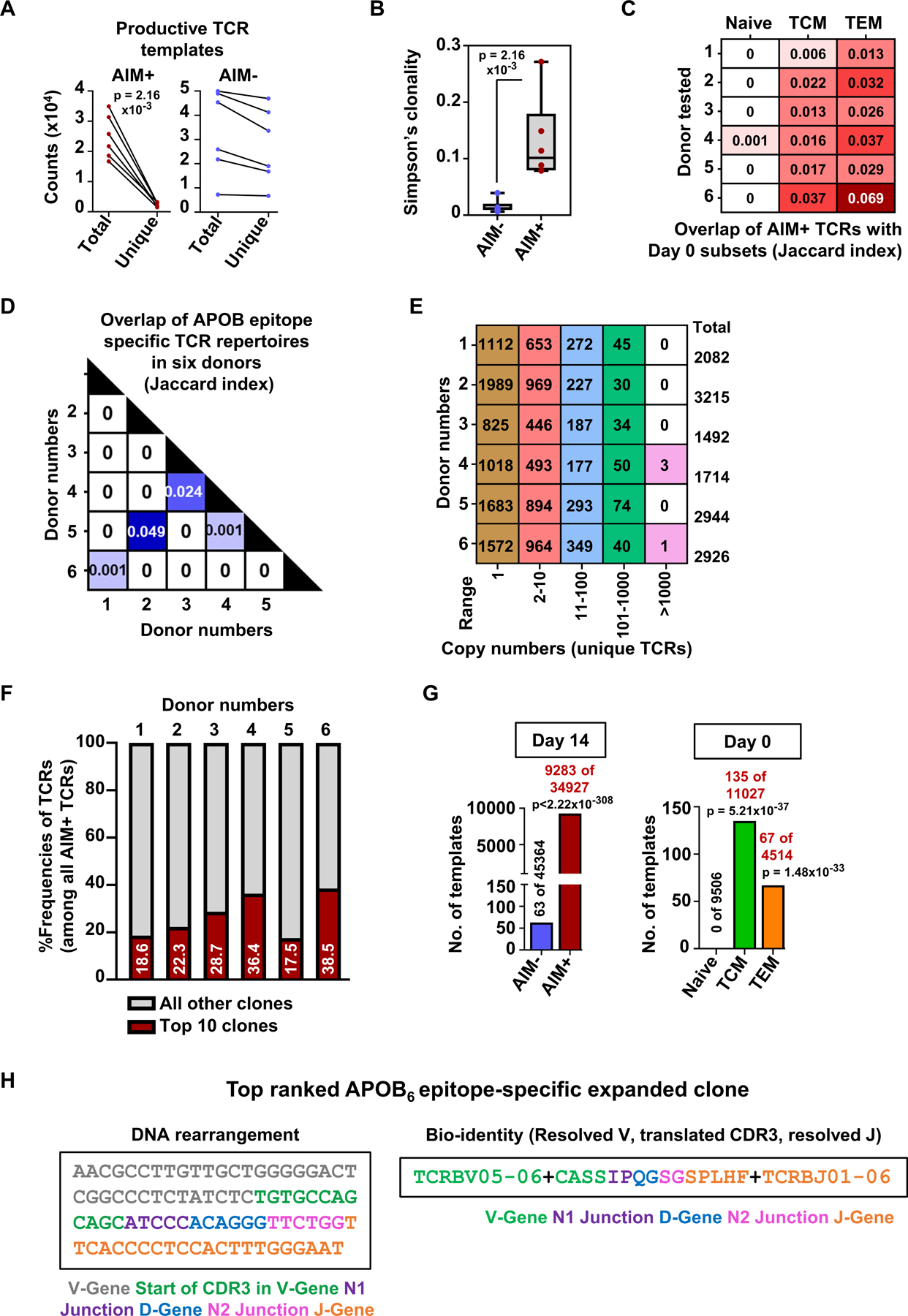
A) Counts of total and unique productive (in-frame for protein translation) TCR templates identified in control AIM− and APOB6-specific AIM+ CD4+T cells. Y-axis: counts x 104. B) Box plots showing Simpson’s clonality index measured for AIM− and AIM+ productive TCRβ sequences. C) Comparison of repertoire overlap between AIM+ productive TCRs and those from naïve or TCM or TEM subsets. Values denote the Jaccard index, a statistical measurement of similarity between sample sets. Rows represent data from individual donors. D) Matrix showing TCR repertoire overlap across donor-specific AIM+ productive TCRs E) Total numbers of unique productive TCR clones identified in AIM+ CD4+ T cells from individual donors (rows) and are present within a specific range of copy numbers (columns). F) Red bars and values showing the cumulative %frequencies of the top 10 most abundant rearrangements of all productive TCRs detected in AIM+ cells in each donor. Grey shaded areas represent the combined frequencies of all other clones. Six independent donors (A-F). G) Template copy numbers of the top-expanded clone (detected in Donor 6) in different sorted CD4+ T populations. Total numbers of productive TCR templates in AIM− and AIM+ subsets (Day 14) and in naïve, TCM and TEM lineages (Day 0) are also indicated. H) Nucleotide sequence and bio-identity of the top-ranked APOB6-specific TCR rearrangement. Details of the rearrangement are color coded by component. Colored symbols represent data from individual donors. Statistical comparisons between total and unique TCR counts (A) and Simpson’s clonality in AIM- and AIM+ populations (B) were performed using Mann-Whitney test. Statistical comparisons of specific TCR clonal frequencies in AIM− vs AIM+ cells and in naïve vs TCM or naïve vs TEM populations (G) were performed using Fisher’s exact test.
To assess whether shared TCR clones appear across AIM+ T cells from different donors, we examined repertoire overlap (Figure 4D) and performed Venn diagram analysis of the TCR rearrangements (Figure S7E). We found that most clones that were shared between two donors showed preferential expansion in one donor and were singletons (one copy) or rare (<10 copies) in the other donor. Only 6 clones, shared between donors 2 and 5, were present in >10 copies in both donors (Figure S7F). These data show that CD4+T responses to APOB6 consists predominantly private TCR clones restricted to individual donors. Analysis of the extent of APOB6 driven expansion of clonotypes within each donor showed that almost half of the AIM+ rearrangements were present at >1 copy number, with enrichments ranging from low (2–10) to moderate (11–100) to high (101–1000) (Figure 4E). In two donors, APOB-specific clones with >1000 copies were detected (Figure 4E). As the frequencies of the top 10 most abundant clones in each donor accounted for a significant fraction (frequencies 18–39%; average 27%) of all TCR templates (Figure 4F), we examined the sequence identities (Table S8) of these clones in greater detail. We verified their expansion over AIM− controls by examining clone-specific relative frequencies using Fisher’s exact test and odds ratio analysis (Table S8).
For some donors, we determined TCR sequences from CEFX-II stimulated PBMCs and compared the frequencies of top 10 APOB and top 10 CEFX-II TCRs (Table S9, Additional File 3) in APOB and CEFX-II-specific AIM+ subsets. Most TCRs were either highly enriched (having ~10-fold higher productive frequencies) or found exclusively in any one of the AIM+ groups. Only 3 of the top clones (found in donors 2 and 5) occurred at comparable frequencies in the two AIM+CD4+T subsets (Table S9). These findings suggest that some degree of TCR cross reactivity between APOB-specific CD4+T cells and some CEFX-II viral or bacterial antigen-specific CD4+T cells may exist in some donors. However, this finding is preliminary and requires a larger number of donors to become conclusive.
Statistical analysis of the most expanded APOB-AIM+ clone (Rank 1 in donor 6) using Fisher’s exact test confirmed significant APOB6-driven selection of this clone (frequencies in AIM− and AIM+ templates) and suggested the existence of an already expanded APOB6-specific memory T cell pool in vivo (frequencies in naïve vs TCM or naïve vs TEM templates) (Figure 4G). The nucleotide sequence of this TCR rearrangement and its bio-identity (translated CDR3 amino acid sequence and annotated V and J genes) are shown (Figure 4H).
APOB6-reactive CD4+T cells are enriched in antigen-experienced memory markers
Lineage-tracing of APOB-specific AIM+ TCR sequences in Figure 4 highlighted the existence of APOB6-responsive memory CD4+T cells in normal donors with no known conditions of heart disease. We examined this further in an ex vivo (without expansion) AIM assay of APOB6-stimulated PBMCs from 20 new donors enrolled in a validation cohort of HLA-typed healthy participants (demographic details in Table S10, donor HLA-II alleles in Table S11). The short stimulation regime (24h) avoids any potential non-specific skewing of phenotypes and therefore is most likely to reflect in vivo states. As before, responding and non-responding populations in peptide-stimulated PBMCs were defined using a sequential gating scheme (gating strategy in Figure S8A) to evaluate expressions of the T cell activation markers CD40L, CD69, CD25, OX-40 and 4–1BB. Analysis of multiple activation marker combinations helped us to capture all responding AIM+ CD4+T cells, irrespective of donor-to-donor heterogeneity in APOB6-induced AIM marker expressions (representative plots in Figure 5A). Comparison of naïve and memory (TCM+TEM) fractions within AIM−, AIM+ and all CD4+T cells (representative plots in Figure 5B) in APOB6 stimulated (Figure 5C) and CEFX-II stimulated (Figure S8B) PBMCs, revealed significantly higher fractions of memory cells and lower fractions of naïve cells in AIM+CD4+T cells, as compared to AIM− or all CD4+T cells. This confirmed that antigen-specific CD4+T cells that respond to dominant APOB epitopes are enriched in antigen-experienced memory T cell features, corroborating our previous conclusions from the lineage-tracing experiment in Figure 4.
Figure 5: Ex vivo activated APOB6-specific CD4+T cells are enriched in memory T cell markers.
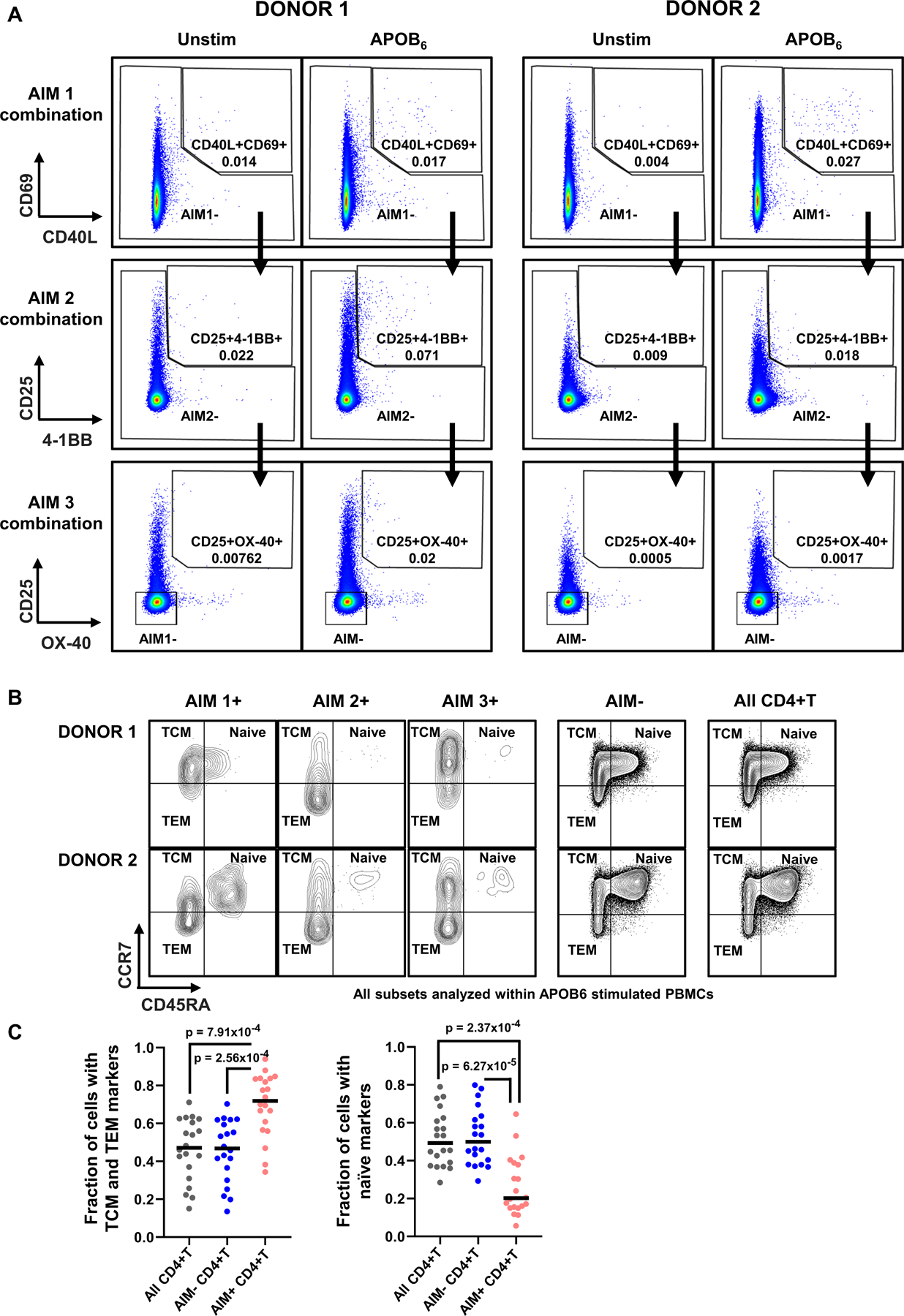
PBMCs were stimulated for 24h with the APOB6 peptide pool and surface expression of activation markers on CD4+T cells was assessed by flow cytometry. A) Representative FACS plots in two donors showing APOB6-induced expressions of three different combinations of T cell activation markers, as assessed in a sequential gating scheme. Top row: AIM1 %CD40L+CD69+; middle row: AIM2 %CD25+4–1BB+; bottom row: AIM3 %CD25+OX-40+. %AIM+CD4+T represent the sum of AIM1, AIM2 and AIM3 frequencies. B) Representative FACS plots in two donors showing CD45RA+CCR7+ naïve, CD45RA−CCR7+ central memory (TCM) and CD45RA−CCR7− effector memory (TEM) populations in AIM1+, AIM2+, AIM3+, AIM−, and all CD4+ T cells. C) Median frequencies of naïve and memory (TCM+TEM) fractions within AIM+, AIM− and all CD4+T subsets in APOB6 stimulated PBMCs. To calculate these fractions for the AIM+ subset, the numbers of cells expressing either naïve or memory markers were first counted within AIM1+, AIM2+ and AIM3+ CD4+T cells individually and then their sum was divided by the total number of AIM+ (AIM1+AIM2+AIM3) CD4+T cells in that donor. Twenty independent donors. Colored symbols represent data from individual donors. Statistical comparisons across different subsets (C) were performed using Kruskal-Wallis test with Dunn’s multiple comparison testing.
APOB6 triggers secretion of both proinflammatory and regulatory T helper cytokines
We used a commercially available (BD Biosciences) human Cytometric Bead Array (CBA) kit to examine the full spectrum of T helper cytokines (IL-2, TNF, IFNγ, IL-17A, IL-4, IL-10) secreted in response to APOB6 stimulation of PBMCs from donors in the validation cohort (Table S10). We used a short (24h) stimulation regime to avoid any phenotypic skewing that may occur in expansion-based methods. Ex vivo analysis also improved detection of IL-10 production, whose levels were very low in expansion-based ICS assays (Figure S1C,D).
As a negative control, we selected a contrasting set of APOB peptides (“neg peps”) that had exhibited low reactivity in our deconvolution experiment (Figure 3). These peptides contributed <5% to the cumulative response (Figure 6A) and had responder frequencies ≤10% (Figure 3B). As compared to the dominant APOB peptides (“pos peps”), responses to them were ~8-fold lower in a cellular proliferation assay (quantification and representative plots in Figure S9A) and ~3.5-fold lower in the 24h ex vivo AIM assay (Figure 6B, representative plots in Figure S9B).
Figure 6: Dominant APOB epitopes elicit diverse set of T helper cytokine responses in ex vivo stimulated PBMCs.
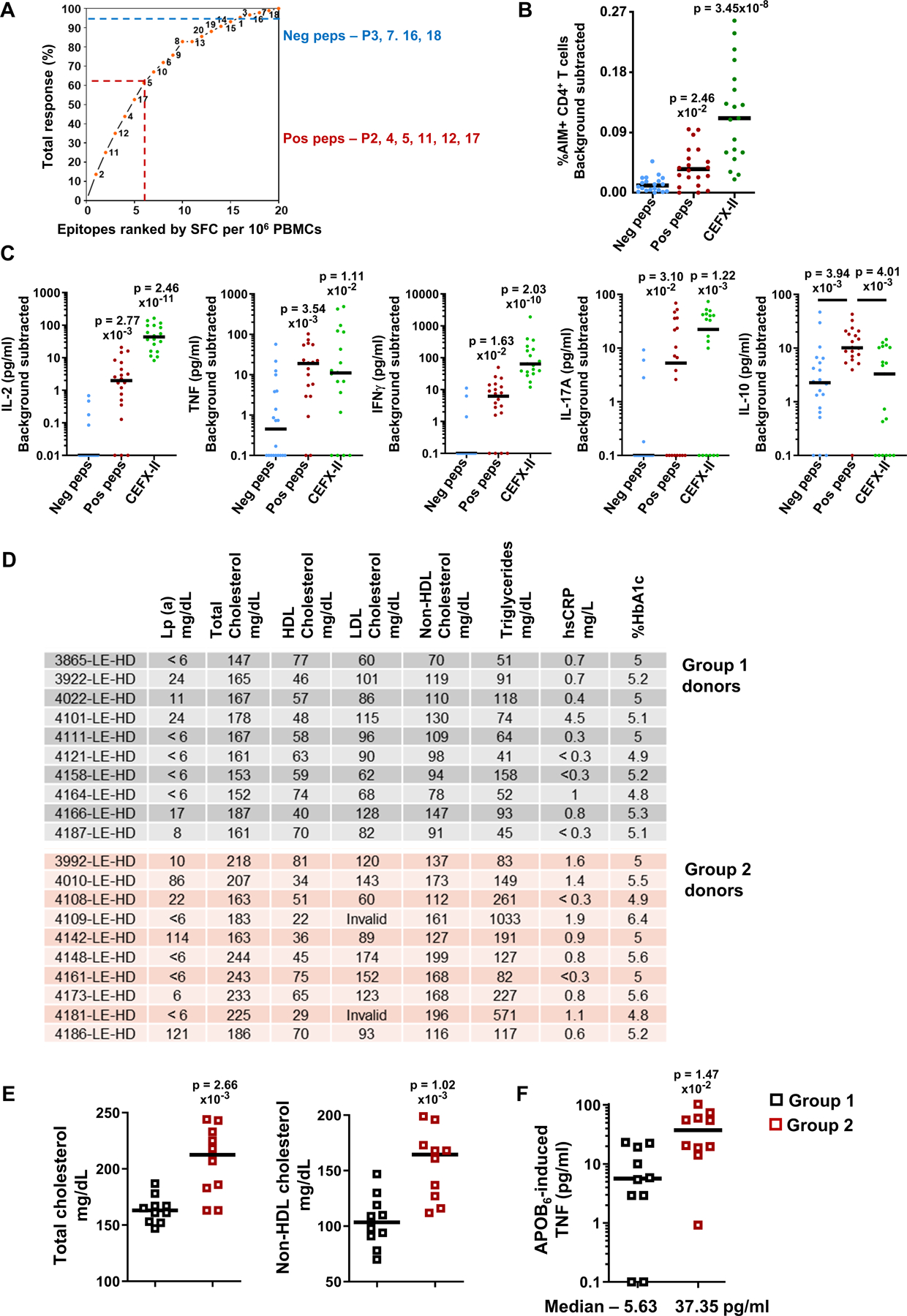
A) The 20 APOB peptides were ranked by their respective strength of elicited responses (from Figure 3). Ranked epitopes (X-axis) were plotted against their relative contributions to the cumulative aggregate response detected in all donors (Y-axis). Dominant “pos peps” APOB peptides P2,4,5,11,12,17 contributed >60% (dotted red line) to the cumulative response and had responder frequency >50%. APOB peptides P3,7,16,18 (neg peps) contributed <5% (dotted blue line) to the cumulative response and had responder frequencies ≤10%. B) Median frequencies of %AIM+CD4+T cells and C) Median secreted levels (pg/ml) of IL-2, TNF, IFNγ, IL-17A and IL-10, upon 24h stimulation with “neg” or “pos” APOB peptide pools or with CEFX-II pool. Twenty independent donors for APOB pools, nineteen for CEFX-II. D) Lipoprotein(a), lipid profile, hsCRP and HbA1c levels in the blood of group 1 and 2 donors on the day of sample collection. E) Median levels (mg/dL) of total (left) and non-HDL (right) cholesterol in blood. F) Median levels (pg/ml) of TNF secreted in response to 24h stimulation with APOB6 (pos peps) pool. Groups 1 and 2 each had ten independent donors. Y-axis (C, F) log10 transformed; data points with 0 or negative values were collapsed onto the minimum value on the scale. Colored symbols represent data from individual donors. Statistical comparisons across peptide pools (B, C) were performed using Kruskal-Wallis test with Dunn’s multiple comparison testing. Statistical tests between the two donors groups (E,F) were done using Mann-Whitney test.
We found that both the APOB6 and the CEFX-II pools induced significantly higher secretion of IL-2, TNF, IFNγ and IL-17A, as compared to the contrasting set of APOB peptides (Figure 6C). Unlike these cytokines, significantly higher induction of the regulatory cytokine IL-10 was triggered only by the dominant self-antigen-derived APOB peptides, but not by CEFX-II pool of bacterial and viral epitopes (Figure 6C). IL-4 was not detectable in our ex vivo assay.
APOB6 induces stronger proinflammatory responses in donors with abnormal or borderline risk levels of CVD-related lipids and lipoproteins in the blood
For all donors in our validation cohort, we ordered lab tests (at UCSD Center for Advanced Laboratory Medicine CALM) for multiple clinical parameters (Lipoprotein(a), lipid panel, comprehensive metabolic panel, hsCRP, HbA1c) on the day of sample collection (Table S12). We segregated the donors into two groups based on their lipid profile and Lp(a) levels. While none of the donors had any known conditions of heart disease, donors in group 2 exhibited abnormal or borderline risk levels of either Lp(a) [>30 mg/dL] or total cholesterol [>200 mg/dL] or HDL cholesterol [<40 mg/dL] or triglycerides [>200 mg/dL] (Figure 6D). These Group 2 donors, who had significantly higher levels of total and APOB-containing non-HDL cholesterol (Figure 6E), also secreted increased amounts of the proinflammatory cytokine TNF (Median 37.35 vs 5.63 pg/ml, Group 2 vs 1) upon ex vivo stimulation with APOB6 pool (Figure 6F). Levels of other APOB6-induced T helper cytokines did not differ significantly between the two groups (Figure S9C).
These data collectively suggest that the dominant epitopes in APOB represent suitable candidates to monitor autoreactive proinflammatory and regulatory responses in the general population.
CD4+T cell activation, memory marker expression and proinflammatory cytokine responses to APOB6 are heightened in CAD patients with higher disease burden
To examine APOB6-specific responses in patients with coronary artery disease (CAD), we analyzed clinical samples from the Coronary Assessment in Virginia (CAVA) cohort. We compared CD4+T responses to APOB6 in matched clinical samples (Table S13) from patients with high and low CAD, as assessed by angiographic disease burden, expressed as Gensini scores48. In the ex vivo (24h) AIM assay, stimulation of PBMCs with APOB6 pool triggered stronger CD4+T cell activation and higher expression of multiple AIM marker combinations (Figure 7A) in a patient with more severe CAD (Gensini 75), as compared to a patient with less severe disease (Gensini 0). Evaluation of APOB6-induced expression of all AIM combinations in 18 matched CAD patients (Table S13), detected ~3.5-fold higher frequencies of AIM+CD4+T cells in patients with higher Gensini scores (median %AIM+CD4+T 0.06 vs 0.21; Figure 7B).
Figure 7: Dominant APOB epitopes trigger increased CD4+T activation and augmented secretion of proinflammatory cytokines in patients with more severe CAD.
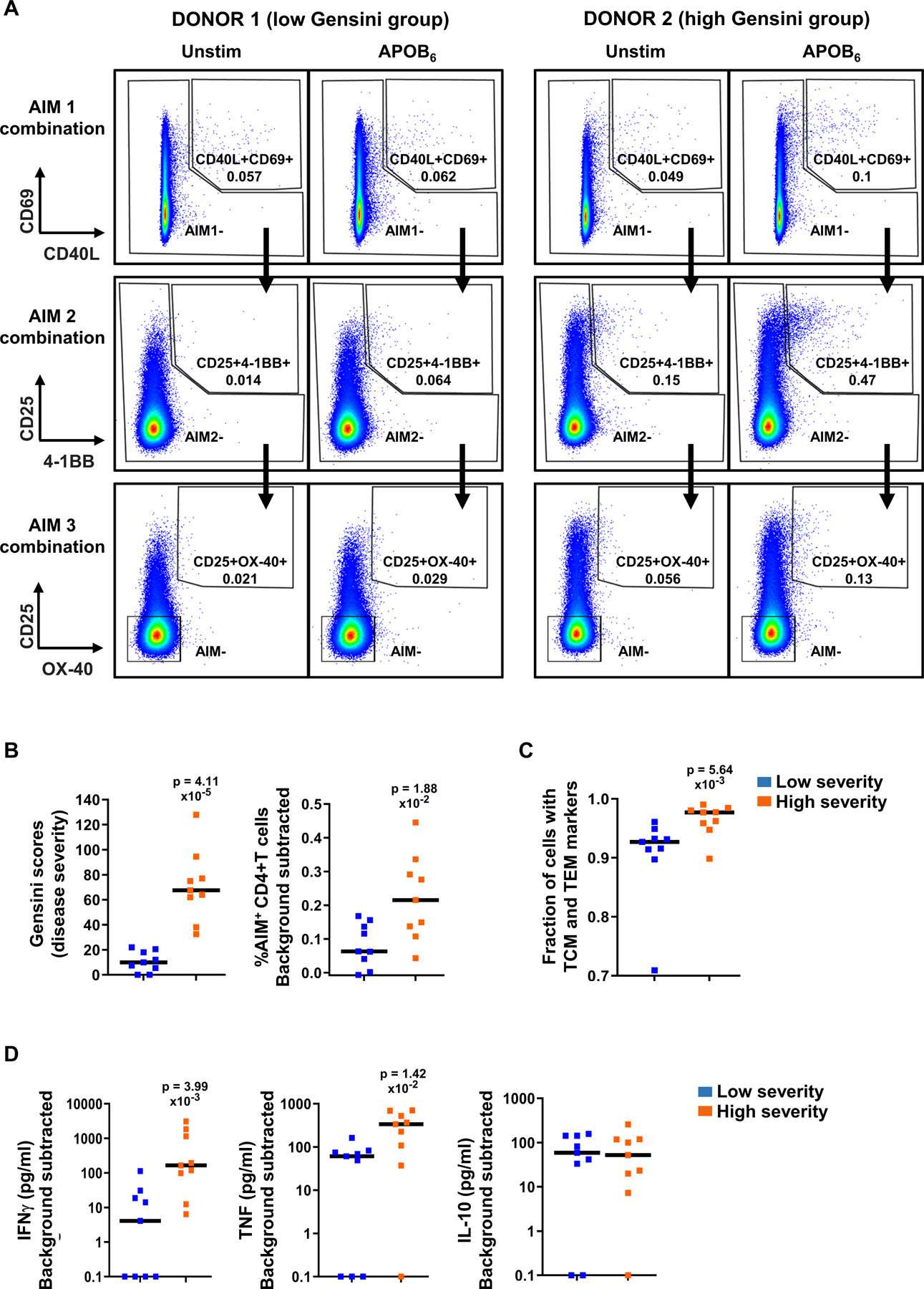
PBMCs from matched clinical samples from patients with low and high CAD severity were stimulated with APOB6 for 24h. CD4+T cell activation and naïve vs memory marker expression was examined using the AIM assay. Secreted T helper cytokines were profiled using CBA. A) Representative FACS plots showing frequencies of AIM+CD4+T cells (AIM1, 2 and 3 in the serial gating scheme) in unstimulated and APOB6 stimulated PBMCs. B) Median Gensini scores and frequencies of AIM+CD4+T cells (sum of %AIM1,2,3). C) Median frequencies of memory (TCM+TEM) cells in AIM+CD4+T cells, calculated as detailed in the legend of Figure 5C. D) Median secreted levels (pg/ml) of induced IFNγ, TNF and IL-10. Y-axis (D) log10 transformed; data points with 0 or negative values were collapsed onto the minimum value on the scale. Low and high severity groups each had nine independent donors. Colored symbols represent data from individual donors. Statistical comparisons between the two groups of patients (B-D) were performed using Mann-Whitney test.
APOB-specific AIM+CD4+T cells in this high CAD group exhibited greater skewing towards antigen-experienced phenotypes and contained significantly higher fractions of memory T cells (CD45RA−CCR7+ central memory and CD45RA−CCR7− effector memory) (Figure 7C), than the AIM+CD4+T cells in the low CAD group.
Next, we monitored cytokine responses to APOB6 in these patient samples. We detected significantly higher levels of secreted proinflammatory cytokines TNF (~5.5-fold; median 61.2 vs 335.3 pg/ml) and IFNγ (~40-fold; median 4.1 vs 164.6 pg/ml) in patients with more severe CAD than in the less severe group (Figure 7D). In contrast, APOB6-induced IL-10 levels were similar in both groups (median 59 vs 52 pg/ml, Figure 7D). Taken together, our data demonstrates that autoimmune CD4+T activation in response to dominant APOB epitopes and the resulting proinflammatory cytokines TNF and IFN-γ are highly informative, antigen-specific immune readouts and correlate positively with CAD.
In summary, we have systematically examined the antigenicity of an atherosclerosis-related autoantigen, the human APOB protein, and delineated the identities of all components of an immunodominant anti-APOB CD4+T trimolecular complex – the epitopes, epitope binding human HLA-II alleles and human autoreactive TCR clones that are activated by the APOB epitopes (Figure 8A). Stimulation with a combined pool of these six immunodominant peptides detected APOB-responsive memory CD4+T cells and triggered secretion of both proinflammatory and regulatory cytokines (Figure 8B) in normal donors with diverse HLA-II allelic background. In clinical samples, APOB6 stimulation detected heightened frequencies of activated CD4+T cells, increased skewing towards memory phenotype and augmented secretion of the proinflammatory cytokines TNF and IFNγ in patients with more severe disease (Figure 8C).
Figure 8: Broadly binding MHC Class-II-restricted immunodominant epitopes in human APOB allow phenotypic evaluation of atherosclerosis-related CD4+T responses and enable identification of APOB-specific autoreactive TCR clones.
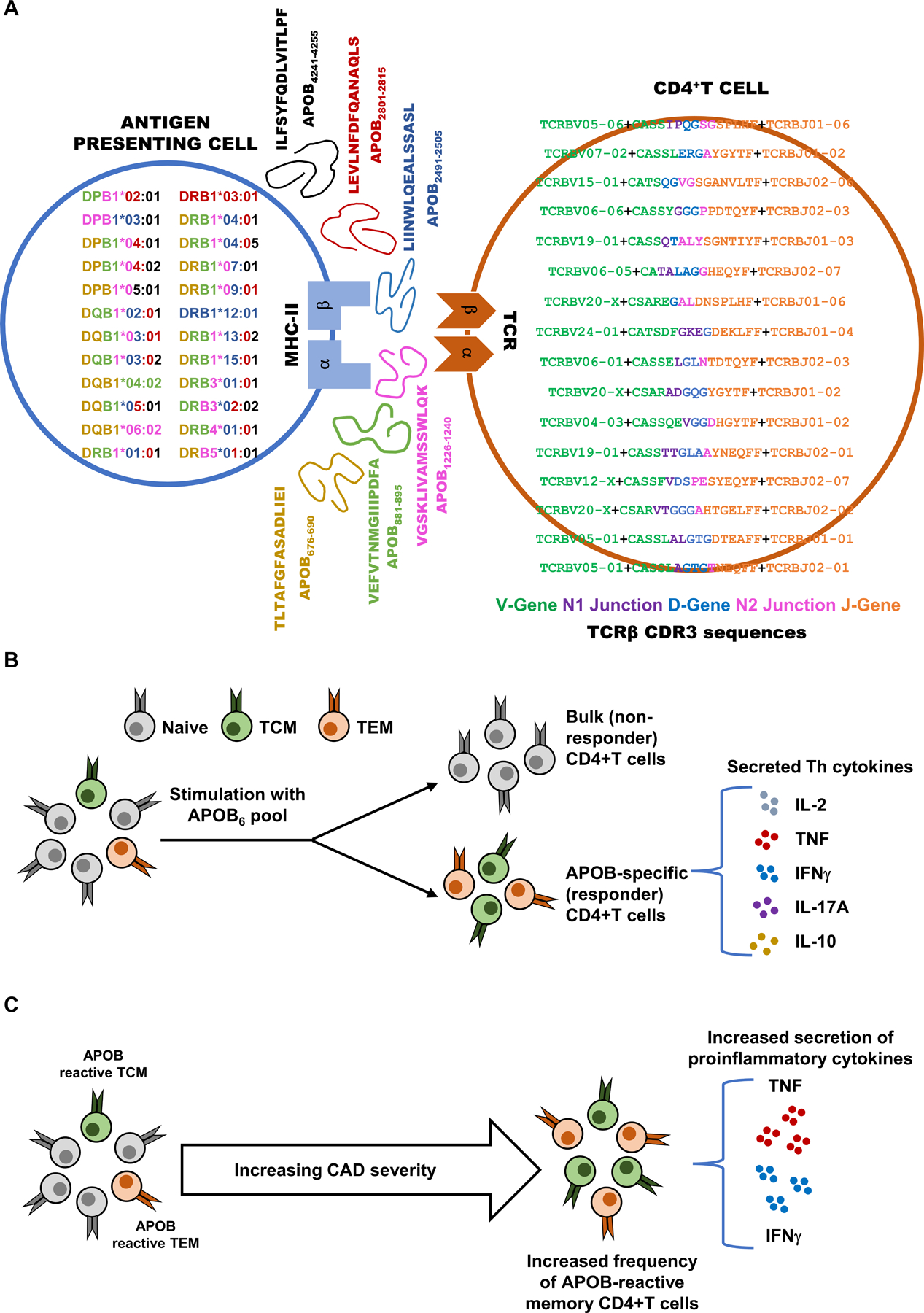
A) Human HLA-II binder alleles expressed in donor samples are shown in the antigen presenting cell (left). Each allele is color coded by the corresponding color of the APOB epitopes (middle, with sequence and position in APOB) that it binds. Bio-identities of clones whose AIM+ vs AIM− log10 odds ratios are > 1 and FDR corrected Fisher’s exact test p values < 10−200 are shown in the CD4+ T cell. Sequences are color coded by component. Resolved V gene green, translated CDR3 region (which includes the start of CDR3 encoded by the V gene green, N1 junction purple, D-gene blue, N2 junction pink, end of CDR3 in J gene orange) and resolved J-gene orange are shown. B) Stimulation of human PBMCs with a pool of dominant epitopes (APOB6) elicits memory CD4+T responses and triggers secretion of multiple T helper cytokines. C) Higher frequencies of antigen-experienced APOB6-reactive CD4+T cells and increased proinflammatory cytokine responses are observed in patients with more severe CAD.
DISCUSSION
Recent research has established a critical role of the immune system in shaping atherosclerosis3. Adaptive immune responses, defined by strict antigen-receptor specificity, represent precise targets for monitoring and understanding disease-specific immune responses. Recent single cell sequencing studies revealed the presence of T cells expressing activation and memory markers and oligoclonal populations of proliferated T cells in human plaques10, 11. However, the identities of the antigenic proteins and putative epitopes that trigger such atherosclerosis-related human T cell activation and proliferation remain unknown. Preclinical studies and clinical correlations of autoantibodies against apolipoprotein B, the LDL core protein, suggest APOB is a relevant atherosclerosis-related antigen12, 13. Two recent studies from our lab demonstrated the presence of APOB-specific CD4+T cells in human blood1, 2, suggesting that human autoreactive T cells respond to APOB. However, immunodominant HLA-II-restricted CD4+T cell epitopes in APOB have not been reported.
We developed a restimulation-based workflow that allowed sensitive detection of statistically significant APOB-specific CD4+T cell responses in the general population. The presence of epitope-specific autoreactive T cells in blood of healthy people, sometimes at frequencies similar to those in diseased individuals, have also been reported in other autoimmune-related conditions5, 6, 24, 49. We mapped six immunodominant APOB epitopes in HLA-typed donors and delineated TCRs of the autoreactive CD4+T clones using high-throughput sequencing strategies. In our screening cohort, we observed stronger responses in PBMCs from some subjects compared to others, across all six peptides. The validation cohort showed that these stronger responses to APOB6 were preferentially seen in hyperlipidemic subjects. The clinical cohort demonstrated that the response was elevated even further in subjects with severe CAD.
APOB-specific responding T cells were enriched in memory phenotypes. In comparison to a contrasting set of poorly immunogenic APOB peptides, the dominant epitopes (APOB6) triggered strong CD4+T activation and robust secretion of both pro-inflammatory and regulatory T helper cytokines. APOB6-dependent CD4+T activation and memory marker expression in the responding cells were higher in PBMCs from CAD patients with high Gensini scores as compared to those with low-Gensini controls. TNF and IFN-γ were more abundantly secreted in CAD cases compared to controls. The integrated workflow described here represents an optimized strategy to interrogate the dynamic behavior (fluctuations in frequencies and phenotypes, expansion and proliferation of specific public or private TCRs) of autoreactive T cells under homeostatic and diseased conditions.
Immuno-profiling of CDR3 sequences have been employed to track disease-specific autoreactive T cell clones in longitudinal samples, case-control studies and across different immune compartments in various autoimmune conditions50–52. Unique disease-related clones that can discriminate between autoimmune disorders have been identified through TCRβ repertoire analysis of peripheral blood samples from patients of systemic lupus erythematosus or rheumatoid arthritis and from healthy controls53. Preexisting T cells undergo recall responses upon sensitization with relevant autoantigens and exhibit disease-associated fluctuations in frequencies, memory phenotypes, cytokine secretion and tissue distribution52, 54. Identification of their TCR sequences has improved our understanding of epitope-specific autoreactive responses55 and has opened up new avenues for improved disease prognosis53, 56 and targeted interventions57. Here, we expand this paradigm to human atherosclerosis, the pathophysiology driving CAD.
In conclusion, this study lays the foundation for understanding the constitutive CD4+T cell response to APOB in humans. For the first time, the molecular triad is resolved at the sequence level: human MHC-II, human TCRβ CDR3 sequences and six immunodominant human APOB peptides that engage these MHC and TCR molecules. Proinflammatory antigen-experienced autoimmune responses to these dominant APOB epitopes escalate in clinical atherosclerosis. Knowledge of these epitopes can form the basis for future strategies for antigen-specific immune risk assessment or tolerogenic immunotherapies like those currently being explored in type 1 diabetes58 and celiac disease59.
Supplementary Material
Novelty and Significance.
What is known?
CD4+T cells are critical modulators of human atherosclerosis, yet the epitopes they recognize are largely unknown.
Apolipoprotein B is a clinically relevant atherosclerosis-related autoantigen.
APOB-specific CD4+T cells are detectable in human blood.
What new information does this article contribute?
Six immunodominant epitopes in human APOB: TLTAFGFASADLIEI, VEFVTNMGIIIPDFA, VGSKLIVAMSSWLQK, LIINWLQEALSSASL, LEVLNFDFQANAQLS and ILFSYFQDLVITLPF, that bind multiple HLA-II alleles, trigger robust CD4+T cell activation and oligoclonal proliferation in a majority of donors.
By sequencing the TCRβ CDR3, we define the full triad: HLA-II, peptide and autoreactive TCRs for these 6 immunodominant epitopes.
Collectively these six peptides detect APOB-specific CD4+T activation, enrichment of memory markers in responding cells and induction of proinflammatory and regulatory cytokine responses in healthy donors expressing diverse HLA alleles.
Severe coronary artery disease in humans is associated with heightened activation of CD4+T cells, skewed expression of antigen-experienced phenotypes, and increased secretion of proinflammatory cytokines in response to these dominant APOB epitopes.
We find that a highly significant autoreactive CD4+T response to human APOB peptides is already detectable in the general population and is increased in people with known lipid risk factors. An integrated restimulation-based workflow resolved individual epitope-specific CD4+T responses in a range of donors with heterogeneous HLA-II allelic background, and allowed mapping and validation of dominant CD4+T cell activating epitopes and determination of unique TCRβ sequences of autoreactive APOB-specific CD4+T cells. The six immunodominant epitopes discovered in this study likely represent the major drivers of an APOB-specific autoimmune response. A combined pool of these peptides enabled comprehensive assessment of the frequencies and phenotypes of APOB-reactive CD4+T cells and peptide-induced T helper cytokine responses in more than sixty donors distributed into three cohorts – a screening and a validation cohort of healthy participants and a clinical (CAVA) cohort of matched CAD patients with low and high disease severity.
Acknowledgments
We thank members of the clinical core and Flow Cytometry core at LJI. We thank members at clinical and CATH labs at UCSD and UVA. We sincerely thank all participants in the study.
Sources of Funding
The authors’ work is funded by The National Institutes of Health, HL145241 and HL136275 to K.L., and “The Tullie and Rickey Families SPARK Awards for Innovations in Immunology at La Jolla Institute” to P.R.
Nonstandard Abbreviations and Acronyms
- AIM
activation-induced marker
- APC
antigen-presenting cell
- APOB
apolipoprotein B
- BLAST
basic local alignment search tool
- CBA
cytometric bead array
- CAD
coronary artery disease
- CD
cluster of differentiation
- CDR3β
complementarity-determining region 3 of the beta chain
- ELISpot
enzyme-linked immune absorbent spot
- FACS
fluorescence-activated cell sorting
- HLA
human leukocyte antigen
- ICS
intracellular cytokine staining
- IEDB
immune epitope database
- IFNG
interferon gamma
- Ig
immunoglobulin
- IL
interleukin
- LDL
low-density lipoprotein
- MHC
major histocompatibility complex
- PBMC
peripheral blood mononuclear cell
- TCR
T-cell receptor
- TNF
tumor necrosis factor
Footnotes
Disclosures
K.L. is a founder of Atherovax, Inc. K.L. and P.R. are named as co-inventors on pending patent applications held by La Jolla Institute for Immunology relating to TCR-based cardiovascular diagnostics and therapeutics.
Expanded Materials and Methods
Online Figures S1–S9
Online legends for data sets in Additional files 1–3
REFERENCES
- 1.Kimura T, Kobiyama K, Winkels H, Tse K, Miller J, Vassallo M, et al. Regulatory cd4(+) t cells recognize mhc-ii-restricted peptide epitopes of apolipoprotein b. Circulation 2018;138:1130–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wolf D, Gerhardt T, Winkels H, Michel NA, Pramod AB, Ghosheh Y, et al. Pathogenic autoimmunity in atherosclerosis evolves from initially protective apolipoprotein b100-reactive cd4(+) t-regulatory cells. Circulation 2020;142:1279–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roy P, Orecchioni M, Ley K. How the immune system shapes atherosclerosis: Roles of innate and adaptive immunity. Nat Rev Immunol 2021 [DOI] [PMC free article] [PubMed]
- 4.Sulzer D, Alcalay RN, Garretti F, Cote L, Kanter E, Agin-Liebes J, et al. T cells from patients with parkinson’s disease recognize alpha-synuclein peptides. Nature 2017;546:656–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gonzalez-Duque S, Azoury ME, Colli ML, Afonso G, Turatsinze JV, Nigi L, et al. Conventional and neo-antigenic peptides presented by beta cells are targeted by circulating naive cd8+ t cells in type 1 diabetic and healthy donors. Cell Metab 2018;28:946–960 e946 [DOI] [PubMed] [Google Scholar]
- 6.Zou J, Hannier S, Cairns LS, Barker RN, Rees AJ, Turner AN, et al. Healthy individuals have goodpasture autoantigen-reactive t cells. J Am Soc Nephrol 2008;19:396–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Legoux FP, Lim JB, Cauley AW, Dikiy S, Ertelt J, Mariani TJ, et al. Cd4+ t cell tolerance to tissue-restricted self antigens is mediated by antigen-specific regulatory t cells rather than deletion. Immunity 2015;43:896–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malhotra D, Linehan JL, Dileepan T, Lee YJ, Purtha WE, Lu JV, et al. Tolerance is established in polyclonal cd4(+) t cells by distinct mechanisms, according to self-peptide expression patterns. Nat Immunol 2016;17:187–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koltsova EK, Garcia Z, Chodaczek G, Landau M, McArdle S, Scott SR, et al. Dynamic t cell-apc interactions sustain chronic inflammation in atherosclerosis. J Clin Invest 2012;122:3114–3126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernandez DM, Rahman AH, Fernandez NF, Chudnovskiy A, Amir ED, Amadori L, et al. Single-cell immune landscape of human atherosclerotic plaques. Nat Med 2019;25:1576–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Depuydt MAC, Prange KHM, Slenders L, Ord T, Elbersen D, Boltjes A, et al. Microanatomy of the human atherosclerotic plaque by single-cell transcriptomics. Circ Res 2020;127:1437–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marchini T, Hansen S, Wolf D. Apob-specific cd4(+) t cells in mouse and human atherosclerosis. Cells 2021;10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nilsson J, Bjorkbacka H, Fredrikson GN. Apolipoprotein b100 autoimmunity and atherosclerosis - disease mechanisms and therapeutic potential. Curr Opin Lipidol 2012;23:422–428 [DOI] [PubMed] [Google Scholar]
- 14.Fredrikson GN, Hedblad B, Berglund G, Alm R, Ares M, Cercek B, et al. Identification of immune responses against aldehyde-modified peptide sequences in apob associated with cardiovascular disease. Arterioscler Thromb Vasc Biol 2003;23:872–878 [DOI] [PubMed] [Google Scholar]
- 15.Frostegard J, Wu R, Giscombe R, Holm G, Lefvert AK, Nilsson J. Induction of t-cell activation by oxidized low density lipoprotein. Arterioscler Thromb 1992;12:461–467 [DOI] [PubMed] [Google Scholar]
- 16.Gistera A, Klement ML, Polyzos KA, Mailer RKW, Duhlin A, Karlsson MCI, et al. Low-density lipoprotein-reactive t cells regulate plasma cholesterol levels and development of atherosclerosis in humanized hypercholesterolemic mice. Circulation 2018;138:2513–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prasad A, Clopton P, Ayers C, Khera A, de Lemos JA, Witztum JL, et al. Relationship of autoantibodies to mda-ldl and apob-immune complexes to sex, ethnicity, subclinical atherosclerosis, and cardiovascular events. Arterioscler Thromb Vasc Biol 2017;37:1213–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsiantoulas D, Diehl CJ, Witztum JL, Binder CJ. B cells and humoral immunity in atherosclerosis. Circ Res 2014;114:1743–1756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsimikas S, Miyanohara A, Hartvigsen K, Merki E, Shaw PX, Chou MY, et al. Human oxidation-specific antibodies reduce foam cell formation and atherosclerosis progression. J Am Coll Cardiol 2011;58:1715–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crotty S A brief history of t cell help to b cells. Nat Rev Immunol 2015;15:185–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robinson J, Barker DJ, Georgiou X, Cooper MA, Flicek P, Marsh SGE. Ipd-imgt/hla database. Nucleic Acids Res 2020;48:D948–D955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Attaf M, Huseby E, Sewell AK. Alphabeta t cell receptors as predictors of health and disease. Cell Mol Immunol 2015;12:391–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turner SJ, Doherty PC, McCluskey J, Rossjohn J. Structural determinants of t-cell receptor bias in immunity. Nat Rev Immunol 2006;6:883–894 [DOI] [PubMed] [Google Scholar]
- 24.Ota K, Matsui M, Milford EL, Mackin GA, Weiner HL, Hafler DA. T-cell recognition of an immunodominant myelin basic protein epitope in multiple sclerosis. Nature 1990;346:183–187 [DOI] [PubMed] [Google Scholar]
- 25.Purcell AW, Sechi S, DiLorenzo TP. The evolving landscape of autoantigen discovery and characterization in type 1 diabetes. Diabetes 2019;68:879–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Provenzano M, Panelli MC, Mocellin S, Bracci L, Sais G, Stroncek DF, et al. Mhc-peptide specificity and t-cell epitope mapping: Where immunotherapy starts. Trends Mol Med 2006;12:465–472 [DOI] [PubMed] [Google Scholar]
- 27.Tian Y, da Silva Antunes R, Sidney J, Lindestam Arlehamn CS, Grifoni A, Dhanda SK, et al. A review on t cell epitopes identified using prediction and cell-mediated immune models for mycobacterium tuberculosis and bordetella pertussis. Front Immunol 2018;9:2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hermansson A, Ketelhuth DF, Strodthoff D, Wurm M, Hansson EM, Nicoletti A, et al. Inhibition of t cell response to native low-density lipoprotein reduces atherosclerosis. J Exp Med 2010;207:1081–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gistera A, Hermansson A, Strodthoff D, Klement ML, Hedin U, Fredrikson GN, et al. Vaccination against t-cell epitopes of native apob100 reduces vascular inflammation and disease in a humanized mouse model of atherosclerosis. J Intern Med 2017;281:383–397 [DOI] [PubMed] [Google Scholar]
- 30.Greenbaum J, Sidney J, Chung J, Brander C, Peters B, Sette A. Functional classification of class ii human leukocyte antigen (hla) molecules reveals seven different supertypes and a surprising degree of repertoire sharing across supertypes. Immunogenetics 2011;63:325–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sidney J, Southwood S, Moore C, Oseroff C, Pinilla C, Grey HM, et al. Measurement of mhc/peptide interactions by gel filtration or monoclonal antibody capture. Curr Protoc Immunol 2013;Chapter 18:Unit 18 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paul S, Grifoni A, Peters B, Sette A. Major histocompatibility complex binding, eluted ligands, and immunogenicity: Benchmark testing and predictions. Front Immunol 2019;10:3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vita R, Mahajan S, Overton JA, Dhanda SK, Martini S, Cantrell JR, et al. The immune epitope database (iedb): 2018 update. Nucleic Acids Res 2019;47:D339–D343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.da Silva Antunes R, Babor M, Carpenter C, Khalil N, Cortese M, Mentzer AJ, et al. Th1/th17 polarization persists following whole-cell pertussis vaccination despite repeated acellular boosters. J Clin Invest 2018;128:3853–3865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dan JM, Havenar-Daughton C, Kendric K, Al-Kolla R, Kaushik K, Rosales SL, et al. Recurrent group a streptococcus tonsillitis is an immunosusceptibility disease involving antibody deficiency and aberrant tfh cells. Sci Transl Med 2019;11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Moderbacher CR, et al. Targets of t cell responses to sars-cov-2 coronavirus in humans with covid-19 disease and unexposed individuals. Cell 2020;181:1489–1501 e1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mateus J, Grifoni A, Tarke A, Sidney J, Ramirez SI, Dan JM, et al. Selective and cross-reactive sars-cov-2 t cell epitopes in unexposed humans. Science 2020;370:89–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frentsch M, Arbach O, Kirchhoff D, Moewes B, Worm M, Rothe M, et al. Direct access to cd4+ t cells specific for defined antigens according to cd154 expression. Nat Med 2005;11:1118–1124 [DOI] [PubMed] [Google Scholar]
- 39.Paul S, Arlehamn CSL, Schulten V, Westernberg L, Sidney J, Peters B, et al. Experimental validation of the rate tool for inferring hla restrictions of t cell epitopes. BMC Immunol 2017;18:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paul S, Dillon MBC, Arlehamn CSL, Huang H, Davis MM, McKinney DM, et al. A population response analysis approach to assign class ii hla-epitope restrictions. J Immunol 2015;194:6164–6176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McKinney DM, Southwood S, Hinz D, Oseroff C, Arlehamn CS, Schulten V, et al. A strategy to determine hla class ii restriction broadly covering the dr, dp, and dq allelic variants most commonly expressed in the general population. Immunogenetics 2013;65:357–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robins H Immunosequencing: Applications of immune repertoire deep sequencing. Curr Opin Immunol 2013;25:646–652 [DOI] [PubMed] [Google Scholar]
- 43.Latorre D, Kallweit U, Armentani E, Foglierini M, Mele F, Cassotta A, et al. T cells in patients with narcolepsy target self-antigens of hypocretin neurons. Nature 2018;562:63–68 [DOI] [PubMed] [Google Scholar]
- 44.Carlson CS, Emerson RO, Sherwood AM, Desmarais C, Chung MW, Parsons JM, et al. Using synthetic templates to design an unbiased multiplex pcr assay. Nat Commun 2013;4:2680. [DOI] [PubMed] [Google Scholar]
- 45.Brady BL, Steinel NC, Bassing CH. Antigen receptor allelic exclusion: An update and reappraisal. J Immunol 2010;185:3801–3808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosati E, Dowds CM, Liaskou E, Henriksen EKK, Karlsen TH, Franke A. Overview of methodologies for t-cell receptor repertoire analysis. BMC Biotechnol 2017;17:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chiffelle J, Genolet R, Perez MA, Coukos G, Zoete V, Harari A. T-cell repertoire analysis and metrics of diversity and clonality. Curr Opin Biotechnol 2020;65:284–295 [DOI] [PubMed] [Google Scholar]
- 48.Gensini GG. A more meaningful scoring system for determining the severity of coronary heart disease. Am J Cardiol 1983;51:606. [DOI] [PubMed] [Google Scholar]
- 49.Lindestam Arlehamn CS, Pham J, Alcalay RN, Frazier A, Shorr E, Carpenter C, et al. Widespread tau-specific cd4 t cell reactivity in the general population. J Immunol 2019;203:84–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goebels N, Hofstetter H, Schmidt S, Brunner C, Wekerle H, Hohlfeld R. Repertoire dynamics of autoreactive t cells in multiple sclerosis patients and healthy subjects: Epitope spreading versus clonal persistence. Brain 2000;123 Pt 3:508–518 [DOI] [PubMed] [Google Scholar]
- 51.Musters A, Klarenbeek PL, Doorenspleet ME, Balzaretti G, Esveldt REE, van Schaik BDC, et al. In rheumatoid arthritis, synovitis at different inflammatory sites is dominated by shared but patient-specific t cell clones. J Immunol 2018;201:417–422 [DOI] [PubMed] [Google Scholar]
- 52.Risnes LF, Christophersen A, Dahal-Koirala S, Neumann RS, Sandve GK, Sarna VK, et al. Disease-driving cd4+ t cell clonotypes persist for decades in celiac disease. J Clin Invest 2018;128:2642–2650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu X, Zhang W, Zhao M, Fu L, Liu L, Wu J, et al. T cell receptor beta repertoires as novel diagnostic markers for systemic lupus erythematosus and rheumatoid arthritis. Ann Rheum Dis 2019;78:1070–1078 [DOI] [PubMed] [Google Scholar]
- 54.Ogura H, Preston-Hurlburt P, Perdigoto AL, Amodio M, Krishnaswamy S, Clark P, et al. Identification and analysis of islet antigen-specific cd8(+) t cells with t cell libraries. J Immunol 2018;201:1662–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Petersen J, Montserrat V, Mujico JR, Loh KL, Beringer DX, van Lummel M, et al. T-cell receptor recognition of hla-dq2-gliadin complexes associated with celiac disease. Nat Struct Mol Biol 2014;21:480–488 [DOI] [PubMed] [Google Scholar]
- 56.Di Sante G, Tolusso B, Fedele AL, Gremese E, Alivernini S, Nicolo C, et al. Collagen specific t-cell repertoire and hla-dr alleles: Biomarkers of active refractory rheumatoid arthritis. EBioMedicine 2015;2:2037–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ostrov DA, Alkanani A, McDaniel KA, Case S, Baschal EE, Pyle L, et al. Methyldopa blocks mhc class ii binding to disease-specific antigens in autoimmune diabetes. J Clin Invest 2018;128:1888–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nikolic T, Zwaginga JJ, Uitbeijerse BS, Woittiez NJ, de Koning EJ, Aanstoot HJ, et al. Safety and feasibility of intradermal injection with tolerogenic dendritic cells pulsed with proinsulin peptide-for type 1 diabetes. Lancet Diabetes Endocrinol 2020;8:470–472 [DOI] [PubMed] [Google Scholar]
- 59.Hardy MY, Goel G, Russell AK, Chen Yi Mei SLG, Brown GJE, Wang S, et al. A sensitive whole blood assay detects antigen-stimulated cytokine release from cd4+ t cells and facilitates immunomonitoring in a phase 2 clinical trial of nexvax2 in coeliac disease. Front Immunol 2021;12:661622. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data, methods and essential research materials used for research and analysis have been provided. A detailed Material and Methods section is available in the Online Data Supplement. Additional data related to peptides and TCR sequences are detailed in Additional Files 1–3.


