Bacteria are highly adaptable organisms capable of growth on countless carbon and nitrogen sources and of occupying an inexhaustible variety of ecological niches. Any particular bacterium possesses a subset of these capabilities that are encoded by a repertoire of genes normally kept unexpressed unless called upon. The key to adaptability in bacteria is their capacity to express only those genes for enzymes and pathways that they need for maximal growth in the environment in which they find themselves. This is achieved by their ability to recognize the composition of their environment by sensing signals emanating from it.
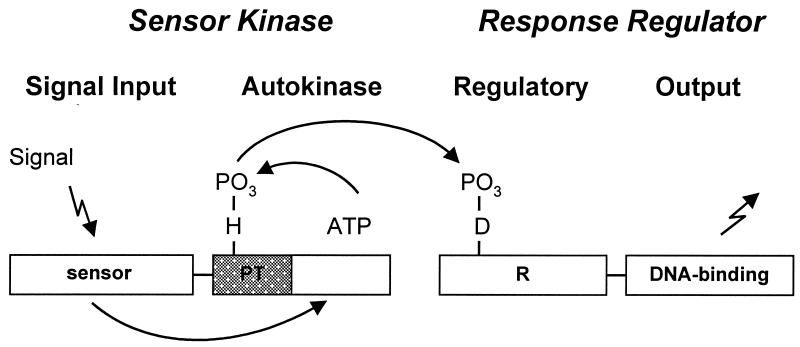
One of the major mechanisms of signal recognition leading to specific gene expression is the two-component system and its more-complex variant, the phosphorelay (Fig. 1). Two-component systems consist of a signal recognition sensor kinase that autophosphorylates on a histidine, usually in response to the presence of a signal, and a response regulator transcription factor that activates or represses gene expression when phosphorylated by the sensor kinase to which it is mated. Thus, sensor kinases and response regulators come in pairs; the sensor kinase detects signals and the response regulator carries out the action that the presence of the signal engenders.
FIG. 1.
Domain structure of two-component systems. Sensor kinases generally consist of a signal input domain coupled to an autokinase domain which can be divided into a histidine phosphotransferase subdomain (PT) and an ATP-binding subdomain. Detection of the stimulus by the sensor kinase induces the hydrolysis of ATP and phosphorylation of the histidine. Response regulators have a regulatory domain (R) that controls the activity of the output domain. Transfer of the phosphoryl group from the histidine of the sensor kinase to the aspartate of the regulatory domain reverses its activity on the output domain.
Two-component systems have the ability to transfer information from one cellular location to another. The sensor kinases are mainly integral membrane proteins, perhaps because most are responsive to external signals and the genes that need to be activated are somewhere else that only the cytoplasmic response regulator can access. This information transfer from one location to another requires specific recognition between the interacting components, and it is recognition that represents the potential Achilles heel in the signaling system. Erroneous recognition between an activated sensor kinase and inappropriate response regulators would lead to regulation of the wrong genes.
That signal propagation in two-component pathways requires precise interaction between phosphoryl donors and acceptors to ensure the correct response does not require profound insight. However, it is not a trivial matter either. Bacteria such as Escherichia coli, Bacillus subtilis, and Synchocystis species with sizeable genomes and, therefore, a large repertoire of genes for adaptability possess 30 to 40 different pairs of two-component systems, each dedicated to unique signals and genes (4, 7, 8). Even more amazing is Nostoc punctiformis with 145 sensor kinases and 103 response regulators identified in a partially sequenced genome (www.jgi.doe.gov). Progenitors of these bacteria adopted the two-component system as a useful and precise system of regulation and expanded it by gene duplication and mutation to serve a wide variety of purposes from gene regulation to chemotaxis. Bioinformatic studies comparing the sensor kinases and response regulators clearly show the presence of two major and several minor families of two-component systems within each bacterium that retain high amino acid identity and similarity within the family (4, 7). Yet these highly similar systems must process different signals, interact only with their partner, and activate unique genes. The question arises, how is fidelity achieved in such signal transduction systems? How do newly duplicated two-component systems evolve specificity?
DOMAIN COMPOSITION OF TWO-COMPONENT AND PHOSPHORELAY PROTEINS
Sensor kinases are generally divisible into two domains; an N-terminal stimulus detection domain followed by an autokinase domain consisting of a histidine-containing phosphotransferase subdomain and an ATP-binding subdomain (Fig. 1). Stimulus detection domains are heterogeneous in size and amino acid sequence, reflecting the variety of stimuli detected. In contrast, the autokinase domains with which the response regulator must interact are of similar length and show many conserved amino acid motifs indicative of a common evolutionary origin. Thus, in integral membrane sensor kinases, which constitute the majority of sensor kinases, the cytoplasmic response regulator must find its partner among highly similar autokinase domains protruding internally from the cytoplasmic membrane.
Response regulator transcription factors consist of two domains; the N-terminal regulator domain accepts phosphoryl groups and regulates the activity of the C-terminal DNA-binding domain. The regulatory domain consists of about 120 amino acids and folds into a structure common to all domains of this type. The DNA-binding domains that determine the promoter specificity of the transcription factor are more heterogeneous in sequence and structure. While the vast majority of response regulators are transcription factors, it is important to point out that some response regulators do not have a C-terminal domain and others have an enzyme as the C-terminal domain. Examples of both may be found in chemotaxis signaling pathways (10).
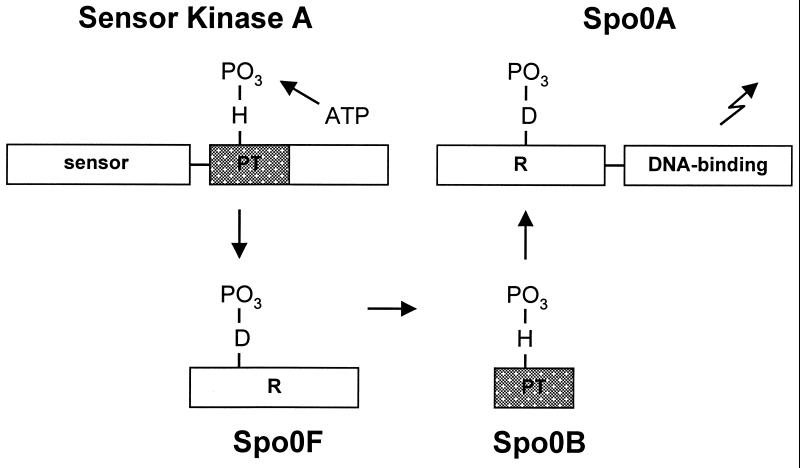
More-complex types of two-component-based systems, termed phosphorelays, are used in bacteria for pathways responding to multiple signal inputs (3). All eukaryotic two-component-based systems are phosphorelays (11). In these systems, the first regulatory domain phosphorylated by the sensor kinase relays its phosphoryl group to a second phosphotransferase domain that serves as the primary phosphoryl donor to the response regulator or transcription factor (Fig. 2). The first three components may be on separate proteins as in the B. subtilis phosphorelay or combined in a multidomain protein, as in the BvgS sensor kinase of Bordetella pertussis (14, 15). All eukaryotic sensor kinases are multidomain, with the sensor kinase and its target regulatory domain on a contiguous polypeptide. The structure of the second phosphotransferase of the B. subtilis phosphorelay Spo0B (16) differs from those of other phosphorelays where an HpT (His-containing phosphotransfer) domain is used. Spo0B and Hpt-type domains are both four-helix bundles on which the active-site histidine is located but differ in the construction of the bundle. Regardless of the domain arrangement, the mechanism of recognition and phosphoryl transfer is most likely conserved. Signal propagation relies on phosphoryl transfer, which requires precise recognition between the highly conserved regions of these proteins, the autokinase domain and the response regulator. In order to understand protein recognition and the mechanism of phosphoryl transfer, it is necessary to understand how these conserved regions of the two proteins fit together.
FIG. 2.
Signal pathway in the sporulation phosphorelay. Phosphorelays have an additional regulatory domain (Spo0F) and phosphotransfer domain (Spo0B) in the signal transduction pathway that directs phosphoryl transfer in a His-Asp-His-Asp sequence. In the sporulation phosphorelay, these domains occur on separate proteins, but in other phosphorelays, one or more domains may be attached to the sensor kinase as a polydomain protein.
STRUCTURES OF THE INTERACTING DOMAINS
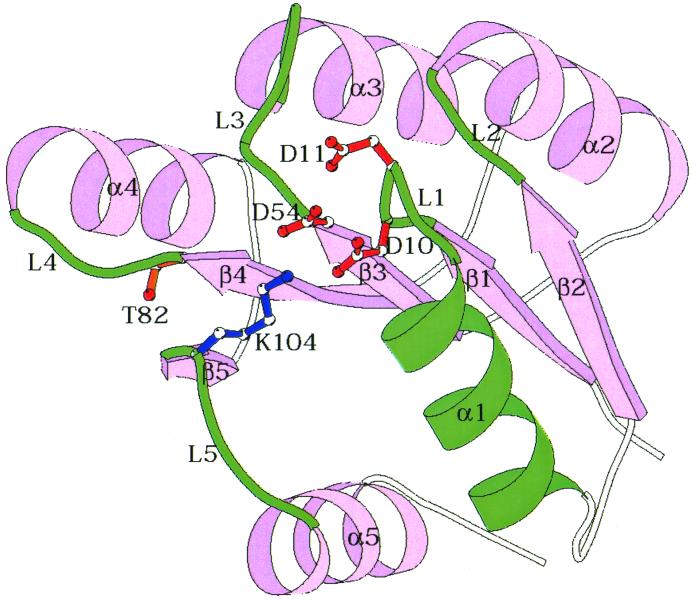
The regulatory domains of response regulators have an α-β structure with five α-helices arranged around a central β-sheet comprised of five parallel β-strands (9). The active-site aspartate is located at one end of the molecule surrounded by the loops connecting the β-strands to the α-helices (Fig. 3). Invariant amino acids in the active site include the phosphorylatable aspartate located at the end of β-strand 3, two acidic residues, usually aspartates, from loop 1, a lysine from loop 5, and a threonine from loop 4. These invariant residues are involved in conserved biochemical functions occurring at the active site, such as divalent metal binding, stabilization of the phosphorylated state, and catalysis. The aspartate that accepts the phosphoryl group is somewhat buried by the side chains of the loop residues. It is this surface of the regulatory domain around the phosphorylated aspartate that must make precise interaction with the histidine-containing phosphotransferase region of the autokinase domain to effect phosphotransfer.
FIG. 3.
Schematic α-carbon structure of Spo0F. Side chains of the five conserved active-site residues, D10, D11, D54, T82, and K104, are shown. Loops connecting the β-strands and α-helices as well as α1-helix are shown in green. Side chains from these residues are involved in binding histidine-containing phosphotransfer domains.
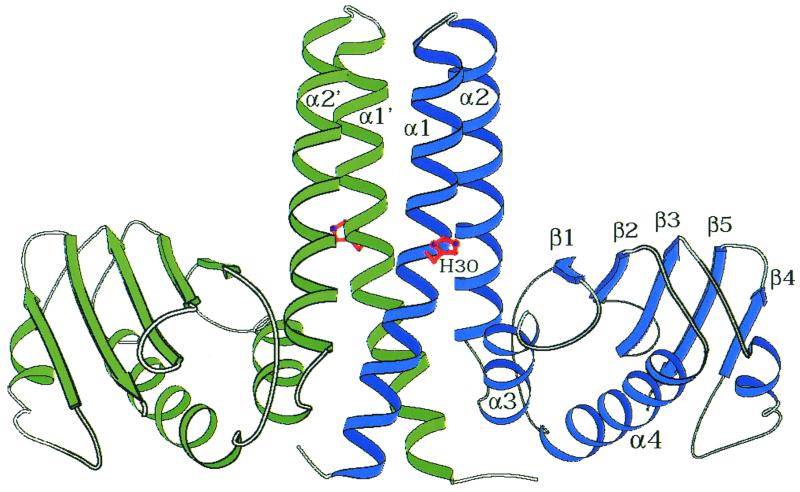
Less is known about the structure and surfaces of the sensor kinase with which the response regulator must interact. However, structural studies of isolated domains revealed that the active-site histidine resides on a four-helix bundle which serves as a dimerization surface for sensor kinases and for phosphotransferases such as Spo0B (Fig. 4) (2, 12, 16). Thus, both Spo0B and the sensor kinases have two histidines and two active sites generated when the two protomers dimerize. In contrast, a single active site is found in a similar four-helix bundle generated from a single polypeptide chain that makes up the active site around the histidine in the chemotaxis sensor kinase CheA (18) and in the Hpt proteins that serve as phosphotransferases in some phosphorelays (5).
FIG. 4.
Structure of the Spo0B dimer. The four-helix bundle on which the active-site histidine resides is formed by the dimerization of two protomers of Spo0B.
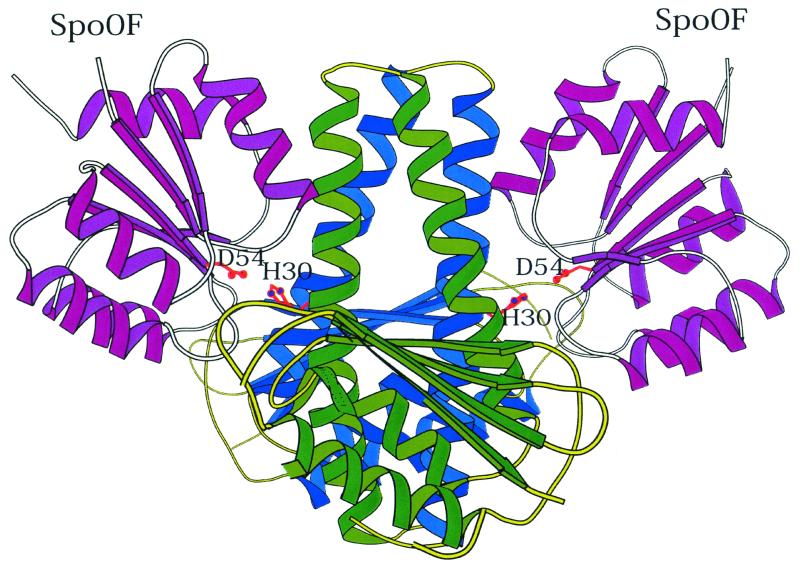
How phosphotransfer domains and regulatory domains fit together was revealed by generation of a cocrystal between the Spo0F response regulator and the Spo0B phosphotransferase (17). Since the four-helix bundle of the Spo0B dimer has two active sites, the cocrystal contained two Spo0F molecules per Spo0B dimer (Fig. 5). Each Spo0F molecule was arranged such that the α1-helix of Spo0F was associated with the α1-helix of one protomer of the four-helix bundle (Fig. 6). This interaction between the two helices aligns the histidine of Spo0B with the aspartate of Spo0F in precisely the correct configuration and distance for phosphotransfer. Spo0F also contacts the four-helix bundle of Spo0B via the residues in loop 4 interacting with the α2-helix of the second protomer (Fig. 6). In view of the close similarity of the four-helix bundles of Spo0B and sensor kinases, the Spo0F-Spo0B structure should be a paradigm for response regulator-sensor kinase interaction. By comparing the residues known to be involved in the surface of interaction of these two proteins in related bacterial species or in evolutionarily close relatives of the proteins in the same organism, it should be possible to determine how interaction surfaces evolve and gain molecular specificity.
FIG. 5.
Orientation of binding by Spo0F and Spo0B as revealed in a cocrystal. The protomers of Spo0B (green and blue) are viewed 90° from the orientation of Spo0B in Fig. 4. The two Spo0F molecules (magenta) are oriented so that their active-site aspartate (D54) is juxtaposed to the histidine (H30) of Spo0B.
FIG. 6.
Orientation of amino acid side chains involved in Spo0F-Spo0B interaction. The four-helix bundle of Spo0B is shown in green, and the SpoOF regions are shown in red. The Spo0B residues arising from helices of the second protein are marked with a prime.
INTERACTION SURFACE RESIDUES RESIST EVOLUTIONARY CHANGE
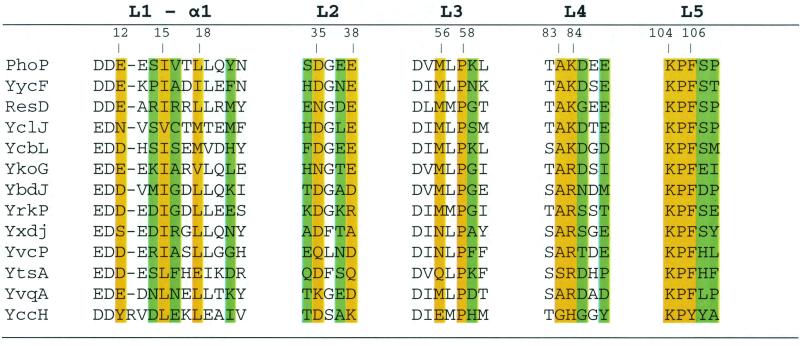
Crystallographic and alanine-scanning studies revealed that those residues of Spo0F making up the loops connecting the β-strands and α-helices surrounding the phosphorylatable aspartate are responsible for interaction with Spo0B and sensor kinase A, the sensor kinase for Spo0F (13, 17). Examination of the evolution of these residues in comparison to the remainder of residues making up Spo0F in the related Bacillus halodurans and Bacillus anthracis species revealed that 20 of 21 (95%) of interaction residues were identical, whereas 50% of the remaining residues were identical in all three organisms (Fig. 7). Thus, residues making up a surface of interaction are resistant to further evolutionary change in true orthologous response regulators, and this conservation may be a general feature of protein-protein interaction.
FIG. 7.
Comparison of Spo0F residues in three Bacillus species. Sequences from three Bacillus species, B.subtilis (Bsu), B. anthracis (Ban), and B. halodurans (Bha), are shown. The positions of the β-strands and α-helices are indicated. Asterisks indicate residues that are identical in all three proteins, and colons indicate functional conservation of residues. Periods indicate residues of the same chemical class, e.g. charged, hydrophobic, etc. Residues that interact with Spo0B (vertical lines) and those that interact with α1-helix or α2′-helix are indicated. Results from alanine-scanning studies (13) are shown as residues affecting the rate of reaction (▵) or equilibrium (○) between Spo0F and Spo0B when changed to alanine.
WHAT THE INTERACTION SURFACE REVEALS ABOUT RECOGNITION
When the analysis of all of the two-component systems of an organism became feasible upon completion of genome sequences, it was noticed that most of the response regulator transcription factors could be placed in families on the basis of homology in the DNA-binding domains (7). Thus, the OmpR or NarL family contains many members with primary structure similarity in the DNA-binding domain and this sequence conservation is explainable now that the tertiary structure of the domain has been elucidated (1, 6). In addition, the sensor kinase members of each family showed considerable sequence conservation around the active-site histidine (4), especially the region immediately C terminal to the phosphorylated histidine that is now known to be the primary site of interaction of the four-helix bundle with the response regulator (17). This suggested that the catalytic autokinase domain of the sensor kinase and both domains of the response regulator evolved as a unit. These observations are most consistent with families arising from expansion by gene duplication. Thus, the question arises, how does the interaction surface of a newly duplicated two-component system evolve away from its immediate progenitor?
The surface by which two-component proteins interact serves not only as the means for recognition of compatible pairs of signaling proteins but also as the catalytic site for phosphoryl transfer between them. Therefore, it should not be surprising that the active-site residues, including those involved in catalysis and those residues closely associated with catalytic residues, are resistant to evolutionary change since the environment of the active site is the primary determinant of directionality and equilibrium in phosphoryl transfer. Relatively minor side chain changes in these residues have profound deleterious effects on both of these properties (13). Because the invariant residues are responsible for catalysis, recognition must be the province of the other residues surrounding the active site that are part of the surface of interaction. Do these residues evolve at random, or is there some pattern to evolution of interaction surfaces?
In order to identify residues involved in recognition, it made sense to examine in detail a closely related family of two-component systems that has relatively recently evolved by gene duplication and whose members must have acquired unique recognition properties to tell each other apart. Furthermore, it seemed intuitive to examine the family within a single bacterial species since it is there that recognition discrimination is important. The OmpR family serves as a convenient measure of the evolution of recognition specificity in sensor kinase-response regulator pairs that multiplied by gene duplication. This is the largest family of two-component signal transduction systems in the bacteria whose genomes have been sequenced. In order to compare these related response regulators from B. subtilis, the residues making up the surface of interaction with the sensor kinase have been superimposed on the Spo0F interaction surface revealed by the crystal structure of the Spo0F-Spo0B complex and by alanine-scanning mutagenesis studies.
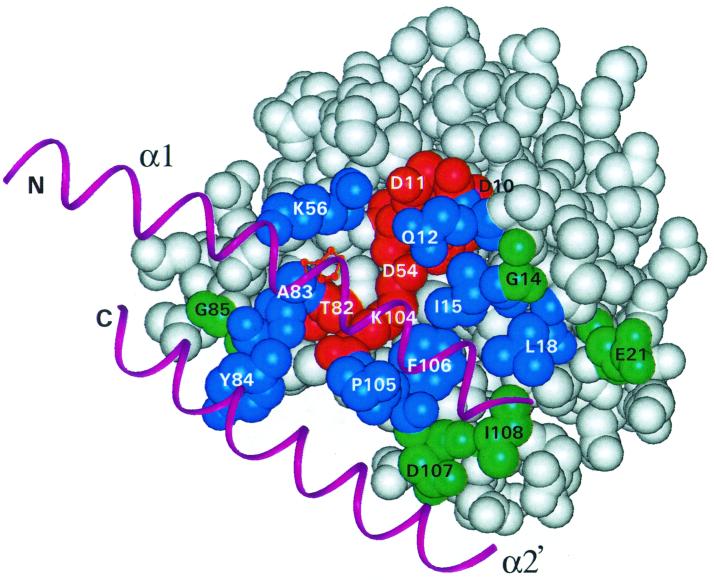
Three groups of response regulator residues defined by their degree of evolutionary conservation make up the interaction surface. The aforementioned catalytic residues, including the phosphorylated aspartate D54, two additional acidic residues, D10 and D11, a threonine, T82, and a lysine, K104, are present in virtually all regulatory domains (Fig. 3). Both variable and conserved residues are found in the amino acids making up the five loops connecting the β-strands to α-helices (L1 to L5 [Fig. 3]) involved in interaction (Fig. 8). Those residues of Spo0F making hydrophobic interaction with the α1- and α2′-helices of the Spo0B phosphotransfer domain, and probably the same helices of this domain of sensor kinase A, are very highly conserved within the OmpR family response regulators. These residues include residues 12, 15, 18, 56, 83, 84, 104, 105, and 106; residues 12, 104, and 105 also make hydrogen bond interactions with the α1-helix. Residue 84, which makes contact with the α2′-helix, is highly conserved within families. We propose that these residues play the role of anchoring the response regulator to the α1- and α2′-helices to ensure that the two proteins fit together in the correct orientation, bringing the phosphorylated histidine of the sensor kinase in accurate juxtaposition and distance to the catalytic aspartate of the response regulator. Since these “anchor” residues interact with the α1-helix of the sensor kinase just C terminal to the histidine, the familial conservation of α1-helix residues previously observed is now explainable (4).
FIG. 8.
Conserved and variable residues at the interaction surface of response regulators of the OmpR family. The surface is composed of side chains from the loops (L1 to L5) and α1-helix. Conserved residues (mustard) variable residues (green) are indicated. The numbers indicate amino acid positions of Spo0F.
The anchor residues may have little, if any, role in determining specificity of interaction within the same family. However, they undoubtedly play a role in discrimination between families. Residues in loop 2 contact the C-terminal domain of Spo0B and may or may not be involved in recognition with sensor kinases. The folding of the ATP domain of sensor kinases is similar to that of the C-terminal domain of Spo0B, but the relative orientation of the ATP-binding domains with the four-helix bundles in sensor kinases is not known. Hence, it is difficult to predict if the interactions seen with the C-terminal domain of Spo0B will be preserved in sensor kinases. Among the residues making contact with the α1- and α2′-helices are those with high variability from response regulator to response regulator that are the likely determinants of recognition specificity. These “recognition” residues may include residues 14, 21, 85, 87, 107, and 108. There appears to be a random assortment of residue types at these positions, and they differ in charge, size, and hydrophobicity, or hydrophilicity. We propose it is the combination of these residues across the interaction surface that erects a barrier to productive interaction of the anchor residues with nonpartner sensor kinases.
The surfaces of these two interacting proteins are best described as mosaics with a fixed pattern of anchor and recognition residues (Fig. 9). Anchor residues determine how the two proteins will fit together, and recognition residues either allow the fit or prevent the two proteins from fitting together by introducing charge and size constraints at several points within the surface. The greater the incompatibilities in recognition residues between the wrong partners of two-component systems, the less chance that signals will go astray. A similar pattern of anchor and recognition residues may be found in all the families of two-component systems.
FIG. 9.
Residues on the interaction surface of OmpR family response regulators. Catalytic residues are colored red. Conserved (anchor) residues are colored blue, and variable (recognition) residues are colored green. The positions of the α1- and α2′-helices of Spo0B are indicated by the magenta Cα trace. The residues and numbers are those of Spo0F.
CONCLUSIONS
The surface of interaction of a response regulator with the phosphoryl donor surface of the phosphotransfer domain of a sensor kinase is a mosaic of amino acid residues arranged in a pattern. The mosaic pattern is very similar in all members of a related family of response regulators. Three types of residues make up the pattern: essentially invariant catalytic residues, anchor residues, and recognition residues. Anchor residues make the interactions required to bind the proteins together in the correct orientation for catalysis. They are highly similar in all members of the family. Recognition residues also make contact with the sensor kinase surface but are extremely variable in individual members of the same family. By virtue of differences in size, charge, and hydrophobicity or hydrophilicity, recognition residues prevent heterologous interaction between nonmated pairs of sensor kinases and response regulators, ensuring fidelity in signal propagation.
Residues making up the surface of interaction between sensor kinases and response regulators do not change in true orthologues in related bacterial species. It would not be surprising if this conservation were true for all associating proteins and domains within a protein that are required to make productive interaction.
ACKNOWLEDGMENTS
This research was supported, in part, by grants GM19416 and GM54246 from the National Institute of General Medical Sciences, National Institutes of Health, USPHS.
Footnotes
Publication 14136-MEM from The Scripps Research Institute.
REFERENCES
- 1.Baikalov I, Schroder I, Kaczor-Grzeskowiak M, Grzeskowiak K, Gunsalus R P, Dickerson R E. Structure of the Escherichia coli response regulator NarL. Biochemistry. 1996;35:11053–11061. doi: 10.1021/bi960919o. [DOI] [PubMed] [Google Scholar]
- 2.Bilwes A M, Alex L A, Crane B R, Simon M I. Structure of CheA, a signal-transducing histidine kinase. Cell. 1999;96:131–141. doi: 10.1016/s0092-8674(00)80966-6. [DOI] [PubMed] [Google Scholar]
- 3.Burbulys D, Trach K A, Hoch J A. The initiation of sporulation in Bacillus subtilis is controlled by a multicomponent phosphorelay. Cell. 1991;64:545–552. doi: 10.1016/0092-8674(91)90238-t. [DOI] [PubMed] [Google Scholar]
- 4.Fabret C, Feher V A, Hoch J A. Two-component signal transduction in Bacillus subtilis: how one organism sees its world. J Bacteriol. 1999;181:1975–1983. doi: 10.1128/jb.181.7.1975-1983.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kato M, Mizuno T, Shimizu T, Hakoshima T. Insights into multistep phosphorelay from the crystal structure of the C-terminal HPt domain of ArcB. Cell. 1997;88:717–723. doi: 10.1016/s0092-8674(00)81914-5. [DOI] [PubMed] [Google Scholar]
- 6.Martinez-Hackert E, Stock A M. The DNA-binding domain of OmpR: crystal structure of a winged helix transcription factor. Structure. 1997;5:109–124. doi: 10.1016/s0969-2126(97)00170-6. [DOI] [PubMed] [Google Scholar]
- 7.Mizuno T. Compilation of all genes encoding two-component phosphotransfer signal transducers in the genome of Escherichia coli. DNA Res. 1997;4:161–168. doi: 10.1093/dnares/4.2.161. [DOI] [PubMed] [Google Scholar]
- 8.Mizuno T, Kaneko T, Tabata S. Compilation of all genes encoding bacterial two-component signal transducers in the genome of the cyanobacterium, Synechocystis sp. strain PCC 6803. DNA Res. 1996;3:407–414. doi: 10.1093/dnares/3.6.407. [DOI] [PubMed] [Google Scholar]
- 9.Stock A M, Mottonen J M, Stock J B, Schutt C E. Three-dimensional structure of CheY, the response regulator of bacterial chemotaxis. Nature. 1989;337:745–749. doi: 10.1038/337745a0. [DOI] [PubMed] [Google Scholar]
- 10.Stock A M, Robinson V L, Goudreau P N. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 11.Thomason P, Kay R. Eukaryotic signal transduction via histidine-aspartate phosphorelay. J Cell Sci. 2000;113:3141–3150. doi: 10.1242/jcs.113.18.3141. [DOI] [PubMed] [Google Scholar]
- 12.Tomomori C, Tanaka T, Dutta R, Park H, Saha S K, Zhu Y, Ishima R, Liu D, Tong K I, Kurokawa H, Qian H, Inouye M, Ikura M. Solution structure of the homodimeric core domain of Escherichia coli histidine kinase EnvZ. Nat Struct Biol. 1999;6:729–734. doi: 10.1038/11495. [DOI] [PubMed] [Google Scholar]
- 13.Tzeng Y-L, Hoch J A. Molecular recognition in signal transduction: the interaction surfaces of the Spo0F response regulator with its cognate phosphorelay proteins revealed by alanine scanning mutagenesis. J Mol Biol. 1997;272:200–212. doi: 10.1006/jmbi.1997.1226. [DOI] [PubMed] [Google Scholar]
- 14.Uhl M A, Miller J F. Central role of the BvgS receiver as a phosphorylated intermediate in a complex two-component phosphorelay. J Biol Chem. 1996;271:33176–33180. doi: 10.1074/jbc.271.52.33176. [DOI] [PubMed] [Google Scholar]
- 15.Uhl M A, Miller J F. Integration of multiple domains in a two-component sensor protein: the Bordetella pertussis BvgAS phosphorelay. EMBO J. 1996;15:1028–1036. [PMC free article] [PubMed] [Google Scholar]
- 16.Varughese K I, Madhusudan, Zhou X Z, Whiteley J M, Hoch J A. Formation of a novel four-helix bundle and molecular recognition sites by dimerization of a response regulator phosphotransferase. Mol Cell. 1998;2:485–493. doi: 10.1016/s1097-2765(00)80148-3. [DOI] [PubMed] [Google Scholar]
- 17.Zapf J W, Sen U, Madhusudan, Hoch J A, Varughese K I. A transient interaction between two phosphorelay proteins trapped in a crystal lattice reveals the mechanism of molecular recognition and phosphotransfer in signal transduction. Struct Fold Des. 2000;8:851–862. doi: 10.1016/s0969-2126(00)00174-x. [DOI] [PubMed] [Google Scholar]
- 18.Zhou H, Lowry D F, Swanson R V, Simon M I, Dahlquist F W. NMR studies of the phosphotransfer domain of the histidine kinase CheA from Escherichia coli: assignments, secondary structure, general fold, and backbone dynamics. Biochemistry. 1995;34:13858–13870. doi: 10.1021/bi00042a018. [DOI] [PubMed] [Google Scholar]